Abstract
Rhabdomyosarcoma (RMS), a cancer characterized by skeletal muscle features, is the most common soft tissue sarcoma of childhood. While low and intermediate-risk groups have seen improved outcomes, high-risk patients still face a 5-year survival of <30%, a statistic that has not changed in over 40 years. Understanding the biologic underpinnings of RMS is critical. The developmental pathways of Notch and YAP have been identified as potent but independent oncogenic signals that support the embryonal variant of RMS (eRMS). Here, the cross-talk between these pathways and the impact on eRMS tumorigenesis is reported. Using human eRMS cells grown as 3D rhabdospheres, which enriches in stem cells, it was found that Notch signaling transcriptionally upregulates YAP1 gene expression and YAP activity. Reciprocally, YAP transcriptionally upregulates the Notch ligand genes JAG1 and DLL1 and the core Notch transcription factor RBPJ. This bidirectional circuit boosts expression of key stem cell genes including SOX2, which is functionally required for eRMS spheres. Silencing this circuit for therapeutic purposes may be challenging, since the inhibition of one node (for example pharmacologic Notch blockade) can be rescued by upregulation of another (constitutive YAP expression). Instead, dual inhibition of Notch and YAP is necessary. Finally, supporting the existence of this circuit beyond a model system, nuclear Notch and YAP protein expression are correlated in human eRMS tumors, and YAP suppression in vivo decreases Notch signaling and SOX2 expression.
Implications: This study identifies a novel oncogenic signaling circuit driving eRMS stemness and tumorigenesis, and provides evidence and rationale for combination therapies co-targeting Notch and YAP.
Keywords: Notch, YAP, rhabdomyosarcoma, stemness
Introduction
Rhabdomyosarcoma (RMS) is the most common soft tissue sarcoma of childhood and adolescence. The embryonal histologic variant (eRMS), the most common subtype, is characterized at the protein level by skeletal muscle histogenesis and at the nucleic acid level by an unstable genome with high rates of copy number alterations, structural variations, and sequence mutations including in the RAS oncogene (1). While low-risk patient groups have a favorable prognosis, high-risk groups have a 5-year survival rate of <30% (2). Since there are not yet approved targeted therapies for eRMS, successful treatment relies on combinations of cytotoxic chemotherapy, radiation, and surgery. While these modalities are usually effective, many patients relapse or develop treatment resistance (3). This ability of eRMS cells to persist in the body is due in part to their “stemness” properties, as cancer stem cells are chemoresistant and demonstrate plasticity in the face of toxic insults (reviewed in (4)). Therefore, it is crucial to understand the signaling mechanisms that drive eRMS tumor cell stemness and plasticity, which may identify novel drug targets and combinations.
As with many pediatric malignancies, eRMS demonstrates dysregulation of developmental pathways, including Hedgehog, WNT, Notch and Hippo (1,5–8). Here we focus on two of these pathways, Notch and Hippo. Notch signaling regulates cell fate determination and stem cell proliferation during development and adult tissue maintenance. Signaling is initiated when a Notch receptor binds a Notch ligand on a neighboring cell, resulting in extracellular and intracellular cleavages (through γ-secretase) to generate an active intracellular notch (ICN) moiety. ICN translocates to the nucleus, forms a complex with the RBPJ transcription factor and a MAML transcriptional co-activator, and drives expression of pro-growth genes. In skeletal muscle, high Notch expression maintains the stem cell population, while low Notch expression augments myogenic differentiation. Notch signaling is upregulated in several human cancer types (reviewed in (9)). In eRMS, Notch receptors 1–4, ligands JAG1 and DLL1, the transcription factor RBPJ, and the Notch target genes HEY1 and HES1 have been found upregulated (7,10–14). Functionally, Notch signaling promotes eRMS proliferation and self-renewal (7,15), inhibits myogenic differentiation (7), and increases eRMS cell mobility and invasiveness (11,12). While Notch pathway inhibition via RNAi or γ-secretase inhibitors (GSIs) in pre-clinical tumor models has proven effective (7,15), Notch inhibitors in the clinic have faced dose-limiting toxicities (16).
The Hippo tumor suppressor pathway also has critical roles during development and regeneration, including limiting organ size. Active Hippo signaling suppresses the downstream oncoprotein YAP, a potent transcriptional co-activator. When Hippo signaling is silenced, YAP translocates to the nucleus where it binds and co-activates transcription factors, including the TEA domain (TEAD) family. YAP/TEAD signaling drives proliferation and survival in many cell types (reviewed in (17)), and also has a role in cellular stemness as it is required in the stem cell compartment for tissue regeneration (18,19). Remarkably, ectopic YAP expression can convert differentiated mammalian mammary, pancreatic, and neuronal cells into stem cells of the same lineage (20). In human cancer, YAP1 is found upregulated via genomic amplification or increased expression (21–23). Recently, YAP has also been found upregulated in eRMS tumors (5,6), where it supports tumorigenesis through stimulation of proliferation and inhibition of myogenic differentiation and apoptosis (6,24). YAP1 suppression in vivo using RNAi or the YAP-TEAD inhibitor verteporfin (VP) suppresses tumor growth in vivo (6,24). However, tumor growth is not ablated, suggesting there are additional signals maintaining cell proliferation and survival.
While developmental pathways are often defined as linear conduits that influence cell and organismal fate, in reality they function as networks that cross-talk (reviewed in (25,26)). Unidirectional signaling between Notch and Hippo occurs frequently in metazoan development and occasionally in human malignancy. For example, in neural stem cells Notch transcriptionally regulates YAP (27), while in intestinal stem cells (28,29), adult hepatocytes (30), colon cancer cells (29), and hepatocellular carcinoma (31), YAP activates Notch. This cross-talk is strikingly cell-type and context specific, because even within one organism (e.g. Drosophila melanogaster), Notch activates YAP in the lymph gland, inhibits Hippo signaling in the wing disc, and Notch-Hippo have an inverse relationship in oocytes (32–34). Crosstalk between these two pathways in the pediatric cancer eRMS has not yet been studied.
Here we investigate the signaling crosstalk between Notch and Hippo, and its impact on eRMS stemness and tumorigenesis, using eRMS cells grown as rhabdospheres. We describe a novel bidirectional circuit whereby YAP and Notch directly transcriptionally regulate each other, and increase expression of eRMS stem cell genes including SOX2. This circuit can be targeted genetically and pharmacologically to inhibit eRMS cell growth, stemness, and tumorigenesis. This is the first finding of a bidirectional circuit between Notch and YAP that supports stemness in a pediatric cancer, and this knowledge will be crucial to designing rational combination therapies.
Materials and Methods
Generation of Cell Lines and Constructs
The human eRMS cell line RD (35) was a gift from Tim Triche (Children’s Hospital of Los Angeles, CA, USA) in 2005. SMS-CTR (36) and Rh36 (37) human eRMS cell lines were gifts from Brett Hall (Columbus Children’s Hospital, OH, USA) in 2006. Cell line authentication was performed in July 2014 (Rh36) and September 2016 (RD, SMS-CTR) using STR analysis (Promega GenePrint 10) conducted by the Duke University DNA Analysis Facility (Durham, NC, USA). CCA (38) human eRMS cell line was a gift from Marielle Yohe (National Cancer Institute (NCI), MD, USA) in 2017 with cell line authentication performed in 2013 at the NCI. JR-1 (39) and CT-TC (40) human eRMS cell lines were gifts from Peter Houghton (Greehey Children’s Cancer Research Institute, TX, USA) in 2017. RAS mutational status is described in (41–45), except for JR-1 which harbors an NRAS mutation (via personal correspondence with the Houghton lab). All cell lines were grown in RPMI-1640 + 10% FBS in 5% CO2 except CCA which were grown in DMEM + 20% FBS in 7.5% CO2. YAP shRNA (46,47), SOX2 shRNA (48), RBPJ shRNA (49,50), and ICN constructs (7) were previously described. pCW107-Notch1 ICN and pCW107-Luciferase constructs were gifts from Kris Wood (Duke University) (51). shRNA annealed oligos were ligated into pLKO.1 puro (Addgene 8453), Tet-pLKO-puro (Addgene 21915), and Tet-pLKO-neo (Addgene 21916) lentiviral plasmids. pInducer20 was a gift from Stephen Elledge (Addgene plasmid #44012), and pInducer20-YAP5SA was a gift from Fernando Camargo (Harvard University, Cambridge, MA, USA).
Sphere Assays
RD and Rh36 rhabdospheres were cultured as described (52). A protocol to culture SMS-CTR spheres was developed, in which sphere media was supplemented with 4× bFGF, 4× EGF, and 50µg/ml insulin. This protocol was also used for the JR-1, CCA, and CT-TC cell lines since they had not previously been cultured as rhabdospheres. Sphere experiments were performed in either 96-well, 6-well, 10cm, or 25ml ultra-low attachment plates or flasks (Corning). All conditions were plated at the same cell density and spheres were allowed to form over 48hrs before genetic or pharmacologic manipulation. To induce shRNA or cDNA expression, spheres were manually dissociated and treated with 4µg/ml (RD) or 5µg/ml (SMS-CTR) doxycycline (dox) daily for 4 days. Unless noted, all shRNA experiments utilized the dox-inducible system. At the end of experiments, spheres were photographed and collected. Spheres were measured and quantified using Image J (NIH), 4 photographs per condition. The length and width of each sphere was measured and averaged, then spheres were categorized into four groups (<0.2mm, 0.2–0.5mm, 0.5–1mm, and >1mm in diameter) based on a neurosphere protocol (53).
Limiting Dilution Assays
After expression and selection for shRNA constructs, cells were plated at 1000, 100, 10, or 1 cell/well in sphere conditions in ultra-low attachment 96-well plates, 48 wells per condition. Wells were scored positive (≥ 1 sphere/well) or negative for sphere formation after 1 week in culture. Since dox-inducible shRNAs require daily media change, here we utilized constitutively expressed shRNAs to minimize cell loss and error. Sphere forming frequency and statistics were calculated using ELDA software (54).
Quantitative Real Time PCR (qRT-PCR) and Semi-quantitative PCR (RT-PCR)
PCR was performed as described (5). Primer sets for this work are listed in Supplementary Table 1.
BrdU and Viability Assays
BrdU assays to measure cell proliferation were performed in six replicates as described (5), after cells were grown as spheres for 48 hours. Trypan blue cell counting was used to measure cell viability after SOX2 suppression in spheres. MTT assays were performed to assess the impact of VP and RO492910 (see Drug Studies) on cell viability. RD cells were plated at 2,000 cells/well in a 96-well plate, six replicates per condition. The next day, 3µM VP and varying concentrations of RO492910 were added and cells incubated for 72 hours. Conversion to formazan was performed as described (7).
Immunoblotting
Immunoblotting was performed as described, using 30–100µg of lysate per sample (7), and the following antibodies: anti-YAP (Cell Signaling #4912, 1:1000), anti-phospho-YAP Ser127 (Cell Signaling #4911, 1:1000), anti-CD133 (DSHB #HB7, 1:50), anti-OCT4 (Cell Signaling #2750, 1:250–1:500), anti-RBPJ (Cell Signaling #5313, 1:1000), anti-JAG1 (Cell Signaling #2620, 1:1000), anti-Nucleolin (Abcam #ab22758, 1:1000) and anti-Actin (Sigma #A2066, 1:1000 or Sigma #A5441, 1:5000). Densitometry was performed using ImageJ (NIH), and all values were normalized to the loading control.
Immunohistochemistry
Paraffin-embedded formalin-fixed xenograft tumor samples were sectioned and stained. YAP (Cell Signaling #4912, 1:40) and SOX2 IHC (Cell Signaling #3579, 1:100) were performed as described (5,55). Ki67 (Dako #M7240) staining was performed per the manufacturer’s protocol. YAP, SOX2, and Ki67 staining was scored on a scale of 0–3 by two blinded scorers (0=negative (no brown staining), 1=weak staining, 2=moderate staining, 3=strong staining), similar to previous work (56). Four images were scored per tumor and averaged. Human RMS tissue microarrays (TMAs) were generated from tissue collected after informed consent through the Children’s Oncology Group (COG) and obtained from the Biopathology Center at Nationwide Children’s Hospital (Columbus, OH, USA). The samples were deidentified and approved for use by the IRB at Duke University. Notch1 and YAP TMA staining was previously described (5,7).
Drug Studies
RO4929097 (GSI) was obtained from Selleck Chemicals and resuspended in DMSO at 10mg/ml. Verteporfin was obtained from Proactive Molecular Research P17–0440, dissolved in DMSO at 100mg/ml and diluted to 0.1µM in cell culture media.
ChIP Assays
5–10×106 RD cells were plated as spheres for 24–48 hours. Spheres were then cross-linked with 1% formaldehyde for 10–15 min, quenched with 0.125M glycine for 10min, and washed with PBS. Cells were resuspended in lysis buffer (50mM Tris pH7.5, 150mM NaCl, 5mM EDTA, 1% Triton-X, 0.1% SDS, 0.5% sodium deoxycholate) and sonicated (Misonix XL-2000) for 14 cycles (12 sec on, 2 min off). Cell debris was pelleted, and chromatin was precleared with Protein G agarose beads (Millipore) for 2 hours at 4°C. RBPJ antibody (Cell Signaling #5313) was added at 1:50 and rotated overnight at 4 °C. Protein G beads were added the next day for 3 hours with rotation at 4 °C. Beads were washed according to the Abcam protocol and DNA was eluted with elution buffer (Santa Cruz) at 67°C for 2 hours with rotation. Crosslinks were reversed overnight followed by a proteinase K digestion for 1hr. DNA was purified using the Qiagen PCR Purification kit. ChIP enrichment was evaluated using semi-quantitative PCR followed by quantitation using ImageJ (NIH) similar to previous published work (57). ChIP primers are listed in Supplementary Table 1.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism (GraphPad). Unless noted, data is presented as the mean and SE. One-way ANOVA, two-way ANOVA, Pearson’s correlation coefficient, and unpaired T-test were used as appropriate. P values were considered significant at *, P< 0.05; **, P<0.01; ***, P<0.001; and ****, P<0.0001.
Results
eRMS rhabdospheres are enriched in Notch and YAP signaling
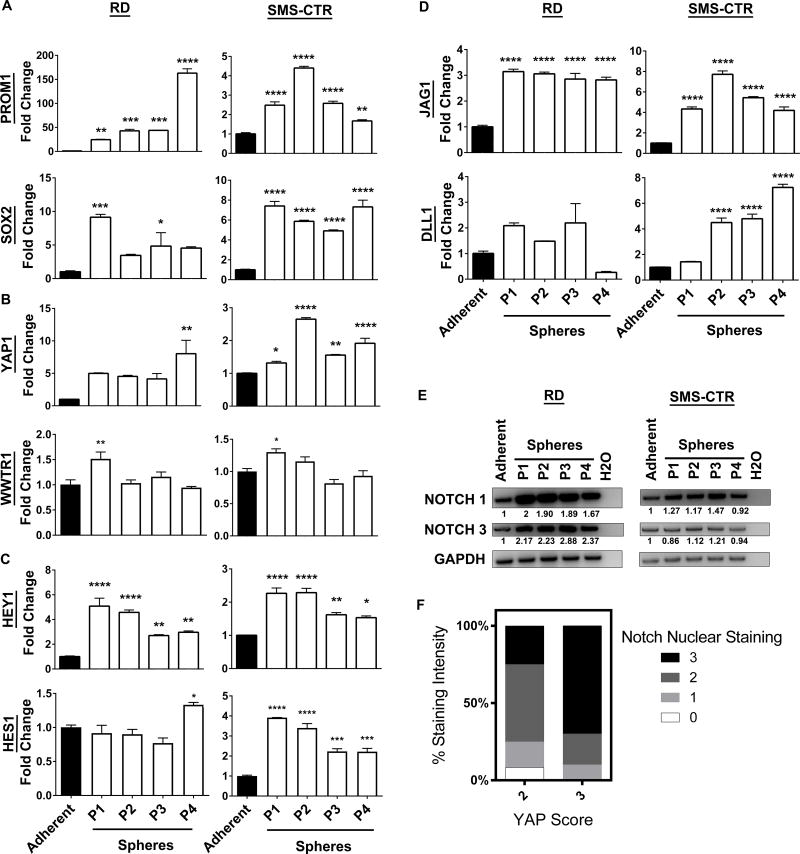
To dissect the roles of Notch-YAP signaling in eRMS, we turned to a model system in which human eRMS cells are cultured in suspension as spheres, termed “rhabdospheres". This microenvironment not only maintains three-dimensional cell-cell contact, which more accurately mimics tumors compared to adherent monolayers, but also permits the study of eRMS cell stemness (52). For the current study, we analyzed gene expression by qRT-PCR and compared rhabdospheres propagated over four passages to eRMS cells grown in monolayers. We relied on two patient-derived eRMS cell lines, RD and SMS-CTR, which harbor oncogenic RAS mutations (41–43), reflecting the high-risk eRMS mutational profile (1,58). As expected (52), expression of the stem cell genes PROM1 (CD133) and SOX2 increased when RD and SMS-CTR cells were grown as spheres (Fig.1A). Intriguingly, the transcriptional co-activator YAP1 (but not its homolog TAZ, encoded by WWTR1) (Fig.1B) and the Notch pathway readout genes HEY1 and HES1 (Fig.1C) were also increased in both cell lines, suggesting YAP and Notch expression may support stemness. To gain insight into the mechanism of increased Notch pathway activation, we investigated the expression of Notch ligands and receptors previously implicated in eRMS tumorigenesis (7,10,13,59), and found increased expression of the Notch ligands JAG1 and DLL1 (Fig.1D) and the Notch receptors NOTCH1 and NOTCH3 (Fig.1E).
Figure 1. Notch and YAP signaling are active in eRMS spheres.
(A) Stem cell markers PROM1 and SOX2 increase in eRMS rhabdospheres over four passages as compared to adherent cells in RD (left) and SMS-CTR (right) cells as assayed by qRT-PCR. (B) YAP1, but not WWTR1 (TAZ), is increased. (C) HEY1 and HES1 are also increased. (D) Notch ligands JAG1 and DLL1 are increased in spheres. (E) Notch receptors NOTCH1 and NOTCH3 are increased as shown by RT-PCR. Densitometry quantitation shown below blots and normalized to loading controls. (F) Human tissue microarrays (TMAs) that were stained and scored (0–3 scale) for nuclear Notch1 and YAP were analyzed. Higher YAP scores correlate with higher nuclear Notch1 scores (n=22 eRMS tumors). P= passage number. *, P<0.05; **, P<0.01; ***, P<0.001; and ****, P<0.0001.
To gain insight into the prevalence of Notch and YAP upregulation in eRMS spheres, we examined four additional eRMS cell lines bearing oncogenic RAS mutations (Rh36, JR-1, CCA, and CT-TC) (Supplementary Fig.1,2). While there was some variability, all cell lines showed an increase in PROM1 and/or SOX2, all cell lines showed an increase in one or more Notch pathway components, and one of four cell lines showed an increase in YAP1 when cultured as spheres. In summary, culturing eRMS cells as rhabdospheres increased cell stemness as measured by PROM1 and SOX2, and Notch and YAP pathway gene profiles, particularly in the RD and SMS-CTR spheres. This system was used to study the role of Notch-YAP signaling in eRMS stemness and tumorigenesis, with all subsequent in vitro studies using RD and SMS-CTR rhabdospheres.
Notch and YAP protein expression correlate in human eRMS tumors
While both Notch and YAP signaling were upregulated in RD and SMS-CTR rhabdospheres, and Notch and YAP were previously shown to be upregulated in human eRMS samples, it was unknown whether they were co-expressed in individual human tumors. Therefore, we re-analyzed human RMS tissue microarrays (TMAs) that had previously been stained using IHC for nuclear Notch1 or YAP protein, respectively (5,7). We correlated the intensity of Notch1 staining with the intensity of YAP staining, and found that those tumor cores with a YAP score of 3 had higher nuclear Notch1 scores (p=0.06) (Fig.1F). There were no eRMS cores with a YAP score of 0 or 1 in this dataset, demonstrating that YAP is highly expressed in eRMS as a whole. This correlation is consistent with a previous observation that YAP and Notch pathway transcriptional signatures are enriched in human RMS tumors compared to skeletal muscle (6). These data suggest there is a subset of eRMS patients whose tumors have concomitant high activity of both pathways, but it is not known if these individuals have a poorer outcome.
Notch pathway activation increases YAP signaling
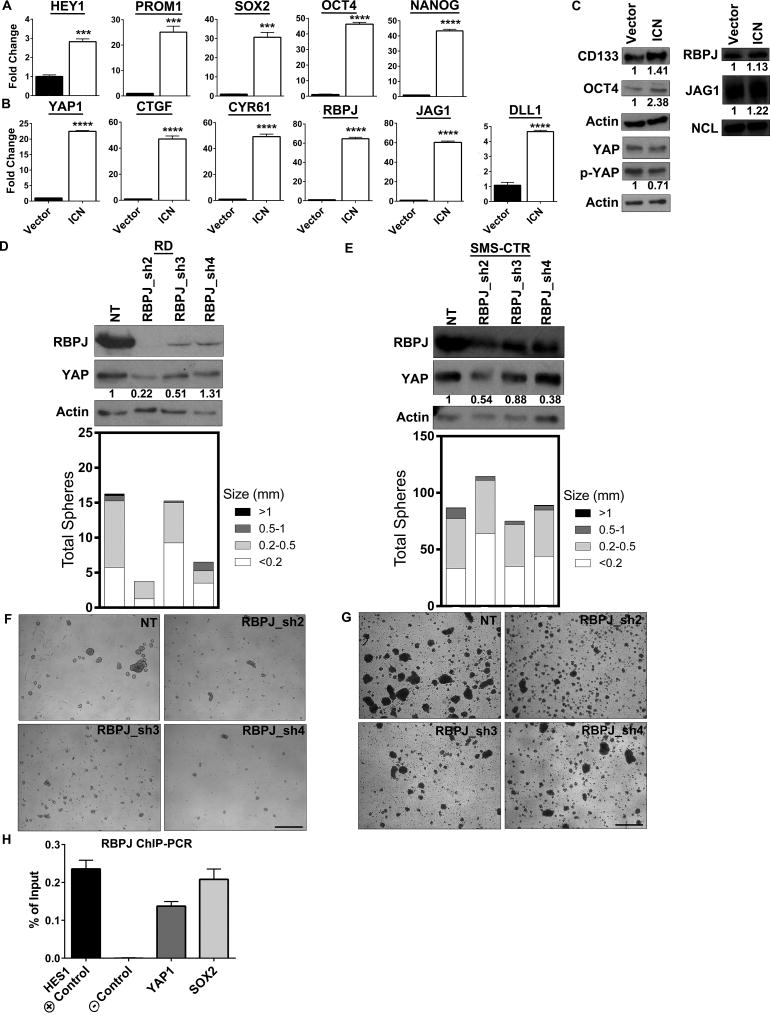
Since Notch signaling regulates YAP expression and activity in several metazoan cell types, we explored whether gain-of-function of Notch signaling could do the same in eRMS (Notch → YAP). To this end, we overexpressed the Notch1 ICN domain in RD spheres. Phenotypically, ICN overexpression increased sphere number and size compared to vector (Supplementary Fig.3A,B). This is in concordance with a recent report that overexpression of Notch1ΔE (a constitutively active Notch1 construct lacking the extracellular domain but still sensitive to GSIs) in RD spheres increases total sphere number (15). As expected for a positive control, constitutive activation of Notch signaling via ICN boosted expression of HEY1 (although interestingly not HES1, data not shown) (Fig.2A). PROM1, SOX2, and additional stem cell genes OCT4 and NANOG were also increased (Fig.2A), consistent with Notch’s role in maintaining stemness (27,60). Last, YAP1 and its target genes were increased, with both CTGF and CYR61 upregulated over 40-fold (Fig.2B). At the protein level, ICN overexpression increased the stem cell genes CD133 and OCT4 and decreased the levels of phosphorylated (inactive) YAP (Fig.2C). Collectively, these data suggest that in eRMS Notch regulation of YAP occurs at both the transcriptional and post-translational levels.
Figure 2. Notch regulates YAP expression and activation.
(A) Expression of the Notch ICN increases levels of HEY1 and stem cell genes PROM1, SOX2, OCT4, and NANOG in RD spheres as assessed by qRT-PCR. (B) YAP1 mRNA levels and YAP target genes CTGF, CYR61, RBPJ, JAG1, and DLL1 are increased. (C) CD133, OCT4, RBPJ, and JAG1 protein levels increase with ICN expression as assessed by immunoblot. Total YAP protein levels do not change, but phospho-YAP levels decrease with ICN expression. Densitometry quantitation shown below blots and normalized to loading controls. Actin blots represent lysates from separate experiments. (D,E) RBPJ suppression by shRNA in RD and SMS-CTR spheres decreases RBPJ and YAP protein levels (top) and decreases sphere number and size (bottom). (F,G) Images of RD and SMS-CTR spheres with RBPJ suppression. (H) RBPJ ChIP-PCR in RD spheres shows RBPJ binding to the HES1 promoter (positive control) but not to a genomic region lacking RBPJ binding sites (negative control). RBPJ also binds the YAP1 promoter and SOX2 promoter. ***, P<0.001; and ****, P<0.0001. NT= non-targeting control. Scale bars: 1.7mm.
Notch pathway suppression decreases YAP levels and sphere formation
Since Notch gain-of-function increased YAP signaling in rhabdospheres, we next evaluated the effect of Notch loss-of-function on YAP through shRNA suppression of the transcription factor RBPJ. Three independent shRNAs to RBPJ decreased RBPJ protein expression as expected, but also decreased YAP protein levels in RD and SMS-CTR spheres (Fig.2D,E, top). Constitutive RBPJ shRNAs were used in the RD spheres due to insufficient knockdown in the dox-inducible system (data not shown). To determine the phenotypic consequences of RBPJ loss, spheres were photographed and compared to the non-targeting (NT) control. Suppression of RBPJ inhibited sphere formation in both RD and SMS-CTR cultures (Fig.2F,G). To quantify the phenotype, we counted the total sphere number and measured the average sphere diameter. Spheres were categorized into four groups based on size (<0.2mm, 0.2–0.5mm, 0.5–1mm, and >1mm), permitting comparison of sphere number and size after genetic manipulation. In RD cells, RBPJ suppression resulted in fewer and smaller spheres, while in SMS-CTR cells, RBPJ loss resulted in smaller spheres only (Fig.2D,E, bottom). In summary, Notch signaling through RBPJ regulates YAP expression and is required for eRMS sphere formation.
RBPJ directly regulates YAP1 and SOX2 genomic loci
To determine whether Notch signaling increases YAP1 mRNA at the genomic level, we performed RBPJ ChIP-PCR in RD rhabdospheres. PCR for the positive (HES1 promoter) and negative (a region on chromosome 14 lacking RBPJ binding sites) controls showed that RBPJ is enriched at the HES1 promoter but not at the negative control, as expected (Fig.2H). PCR for the occupancy of RBPJ on the YAP1 and SOX2 promoters, which are known to contain potential RBPJ binding sites, shows that RBPJ is indeed present (Fig.2H). These data demonstrate that Notch signaling is regulating YAP1 and SOX2 expression at least in part by direct transcriptional activation.
YAP suppression decreases Notch signaling
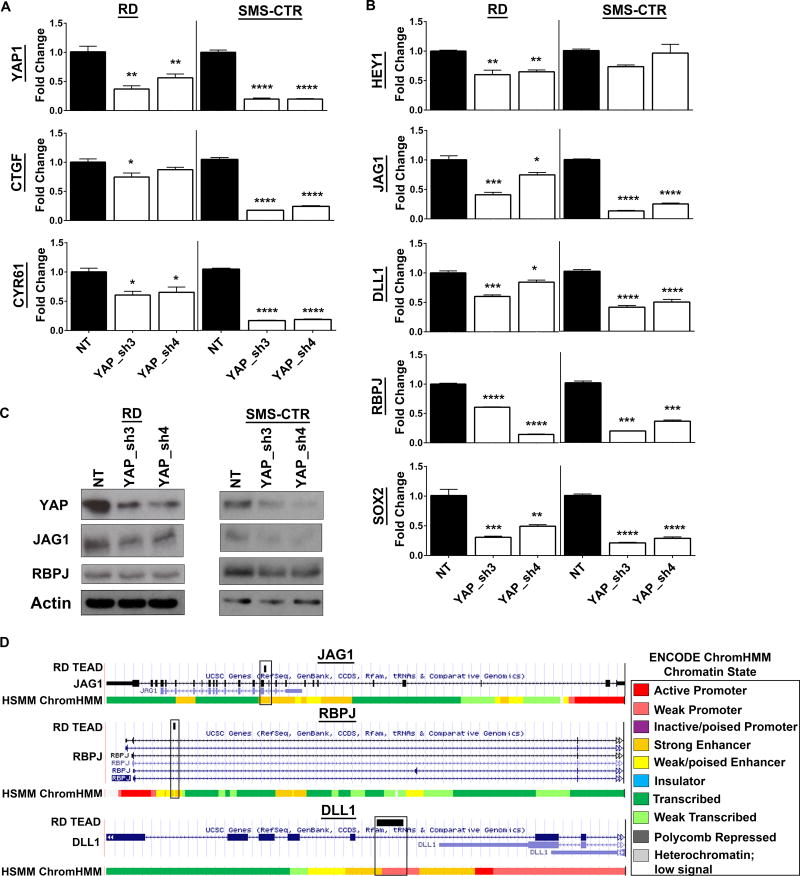
Since Notch signaling can activate YAP and its downstream targets, we next assessed whether the reverse could occur (YAP → Notch). Two independent shRNAs to YAP decreased YAP1 mRNA levels and the expression of YAP target genes (CTGF, CYR61) in RD and SMS-CTR spheres (Fig.3A). YAP suppression also decreased Notch signaling, as shown by a decline in HEY1 (but not HES1, data not shown) expression (Fig.3B). The decline in HEY1 was associated with a decrease in expression of the Notch ligands JAG1 and DLL1, and the transcription factor RBPJ (Fig.3B). The decreases in YAP1, JAG1, and RBPJ mRNA expression were maintained at the protein level (Fig.3C). These experiments demonstrate that YAP can regulate Notch signaling in eRMS spheres.
Figure 3. YAP suppression decreases Notch signaling in vitro.
(A) YAP shRNAs decrease YAP1, and YAP target gene CTGF and CYR61 levels in RD (left) and SMS-CTR (right) spheres as assessed by qRT-PCR. (B) YAP shRNAs also decrease HEY1, JAG1, DLL1, RBPJ, and SOX2 levels. (C) Validation of YAP, JAG1, and RBPJ suppression at the protein level by immunoblot. (D) TEAD ChIP-Seq in RD cells shows TEAD peaks in the JAG1 (top), RBPJ (middle), and DLL1 (bottom) genes. Chromatin state is determined by the ENCODE HMM in human skeletal muscle myoblasts (HSMMs), a cell type that is a possible cell of origin for eRMS and often used as a control tissue type (82–84). NT= non-targeting control. *, P<0.05; **, P<0.01; ***, P<0.001; and ****, P<0.0001.
YAP directly regulates Notch signaling
To evaluate whether YAP activates Notch signaling at the genomic level, we interrogated previously published TEAD ChIP-Seq data from RD ChIP experiments (6). Since as a transcriptional co-activator YAP does not bind DNA, but exerts most of its effects by binding to the TEAD transcription factor, TEAD ChIP is considered a reasonable surrogate for YAP (61), represented here as “YAP/TEAD.” Based on published ChIP-Seq studies, YAP/TEAD activates transcription by binding to genomic enhancers in intronic or intergenic regions (61–63). YAP/TEAD can also bind promoters but this is a rarer event, accounting for only ~4% of YAP/TEAD genomic binding sites (61). In RD cells, we found TEAD ChIP peaks within strong enhancer regions of the JAG1 and RBPJ genes, and within a weak promoter of the DLL1 gene (Fig.3D). TEAD peaks were also found in enhancer regions of NOTCH1, NOTCH2, and HEY1 (Supplementary Fig.4), but the significance of these peaks is unknown and should be explored. These ChIP-Seq data show that YAP/TEAD directly regulate JAG1, RBPJ, and DLL1 to activate Notch signaling. This is complementary to the Notch gain-of-function studies where ICN overexpression results in YAP upregulation (Fig.2B), leading to an increase in the YAP target genes including RBPJ, JAG1, and DLL1 at the mRNA and protein level (Fig.2B,C). Together these data reveal a bidirectional Notch-YAP signaling circuit whereby Notch regulates YAP (Notch → YAP), and YAP in turn signals to and activates Notch (YAP → Notch). Since Notch and YAP both have roles in regulating stem cells and antagonizing myogenic differentiation in eRMS, we hypothesized that this circuit may drive stemness during eRMS tumorigenesis.
YAP promotes stemness and proliferation in vitro and in vivo
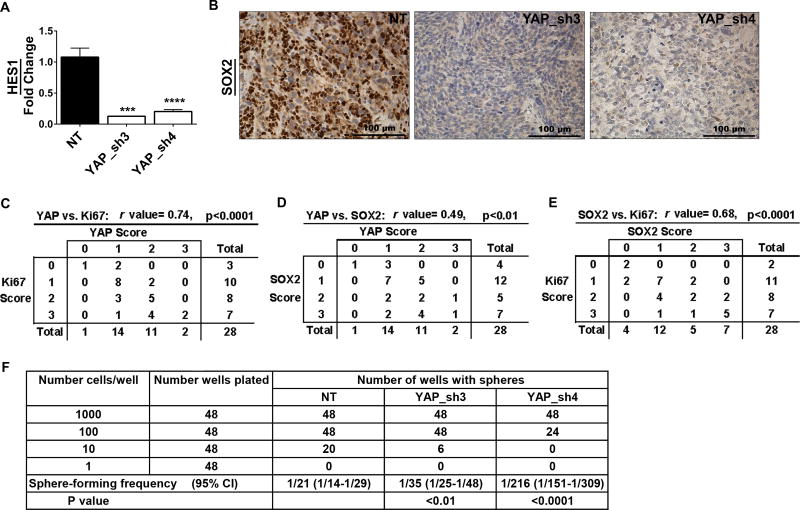
Since Notch signaling supported stem cell gene expression in eRMS spheres, we next investigated the role for downstream YAP in stemness. In addition to dampening Notch signaling, YAP suppression in vitro decreased expression of stem cell genes SOX2 (Fig.3B), OCT4, and NANOG (Supplementary Fig.5). To assess whether YAP impacts stemness in vivo, we analyzed previously generated SMS-CTR xenograft tumors stably expressing YAP shRNAs (24). YAP suppression in vivo decreased Notch signaling, as assessed by a decline in HES1 mRNA (Fig.4A), but also decreased SOX2 protein expression (Fig.4B). While we had previously shown that YAP1 suppression decreased YAP and Ki67 protein expression in vivo (24), to correlate YAP status with proliferation and stemness, we quantified IHC expression of these proteins and calculated a correlation coefficient. As expected, YAP expression positively and significantly correlated with Ki67 expression (Fig.4C). However, YAP expression also positively and significantly correlated with SOX2 expression, suggesting that YAP may regulate SOX2 (Fig.4D). SOX2 expression also positively and significantly correlated with Ki67, consistent with a role in self-renewal (Fig.4E). These data suggest that YAP also regulates Notch signaling and stemness in vivo.
Figure 4. YAP suppression in vivo decreases Notch signaling and SOX2 levels.
(A) YAP shRNAs decrease HES1 expression in SMS-CTR xenograft tumors as assessed by qRT-PCR. (B) Representative images of SOX2 IHC in the tumors. YAP, Ki67, and SOX2 IHC were scored on a scale of 0–3 for all tumors (NT, YAP_sh3, YAP_sh4). The r value represents a positive and significant correlation of expression of (C) YAP and Ki67, (D) YAP and SOX2, and (E) SOX2 and Ki67. (F) Limiting dilution assay in RD spheres after YAP suppression. The average sphere-forming frequency is shown, with the expected range in parentheses. NT= non-targeting control. ***, P<0.001; and ****, P<0.0001.
To functionally analyze the role of YAP in eRMS stemness, we performed limiting dilution assays (LDAs) after YAP suppression in RD cells. While NT control cells readily formed spheres at 1000, 100, and 10 cells/well, cells with YAP suppression showed decreased sphere-forming capacity, dropping from 1/20 in NT control cells to 1/35 (sh3) or 1/216 (sh4) for the spheres expressing the YAP shRNAs (Fig.4F). This identifies a functional role for YAP in promoting eRMS cell stemness.
Last, to determine whether YAP regulated SOX2 directly, we again turned to the RD TEAD ChIP-Seq data. However, there were no peaks associated with the SOX2 gene (data not shown), suggesting that YAP’s control of SOX2 is either indirect through secondary signaling, or TEAD-independent. While future studies are needed to elucidate the mechanism, these data demonstrate a role for YAP in driving eRMS cell stemness, both through upregulation of stem cell genes and functionally.
SOX2 is necessary for stemness in eRMS cells
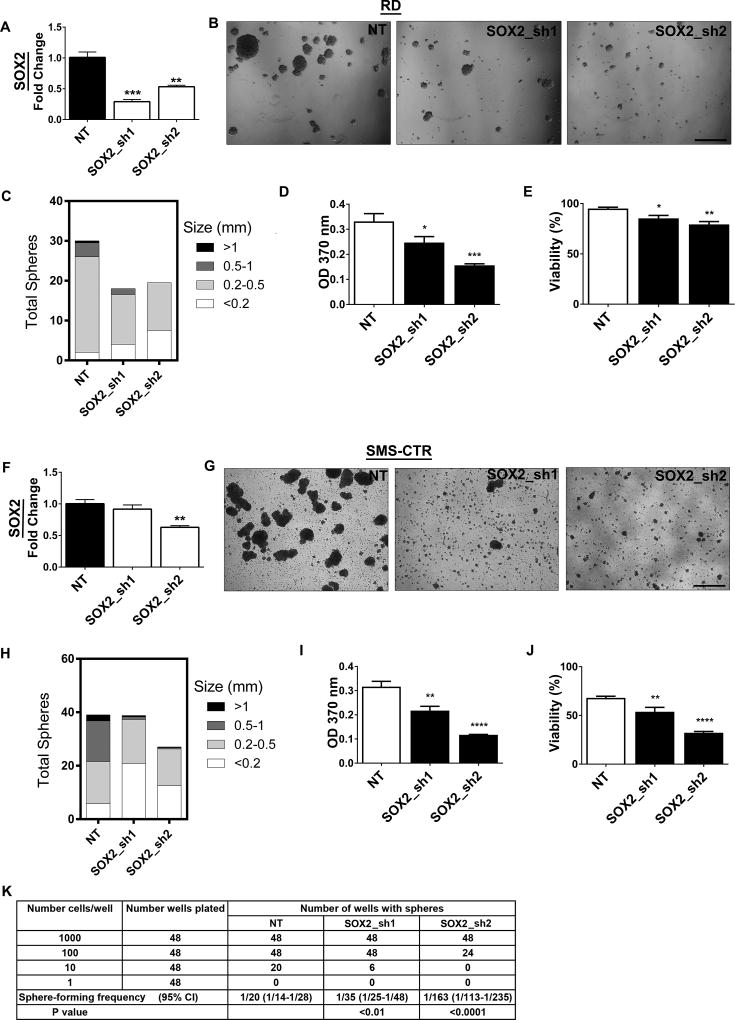
While SOX2 is a cancer stem cell gene in other cancer types, the role of SOX2 in eRMS is largely unknown. SOX2 suppression in RD spheres decreased SOX2 mRNA levels (Fig.5A) and inhibited sphere formation (Fig.5B), decreasing sphere number and size (Fig.5C). Mechanistically, SOX2 suppression inhibited cell proliferation (Fig. 5D) and decreased cell viability (Fig.5E), assessed by BrdU incorporation and trypan blue cell counting, respectively. Similar changes in sphere formation, proliferation, and viability were observed in SMS-CTR spheres after SOX2 suppression (Fig.5F–J). To further define the functional role of SOX2, we performed LDAs. Similar to the LDA with YAP suppression, loss of SOX2 resulted in decreased sphere-forming capacity (Fig.5K). These data suggest that SOX2, a Notch-YAP target gene, is an important regulator of stemness in eRMS cells.
Figure 5. SOX2 is necessary for stemness in eRMS cells in vitro.
(A) SOX2 shRNAs decrease SOX2 levels in RD spheres by qRT-PCR. (B) Representative images of RD spheres after SOX2 knockdown. SOX2 suppression results in decreased (C) sphere number and size, (D) cell proliferation as measured by BrdU assay, and (E) viability as measured by trypan blue cell counting. SOX2 suppression in SMS-CTR spheres similarly decreases (F) SOX2 mRNA, (G,H) sphere formation, (I) proliferation, and (J) viability. (K) Limiting dilution assay in RD spheres after SOX2 suppression. The average sphere-forming frequency is shown, with the expected range in parentheses. NT= non-targeting control. **, P<0.01; ***, P<0.001. Scale bars: 1.7mm.
YAP expression rescues pharmacologic Notch inhibition
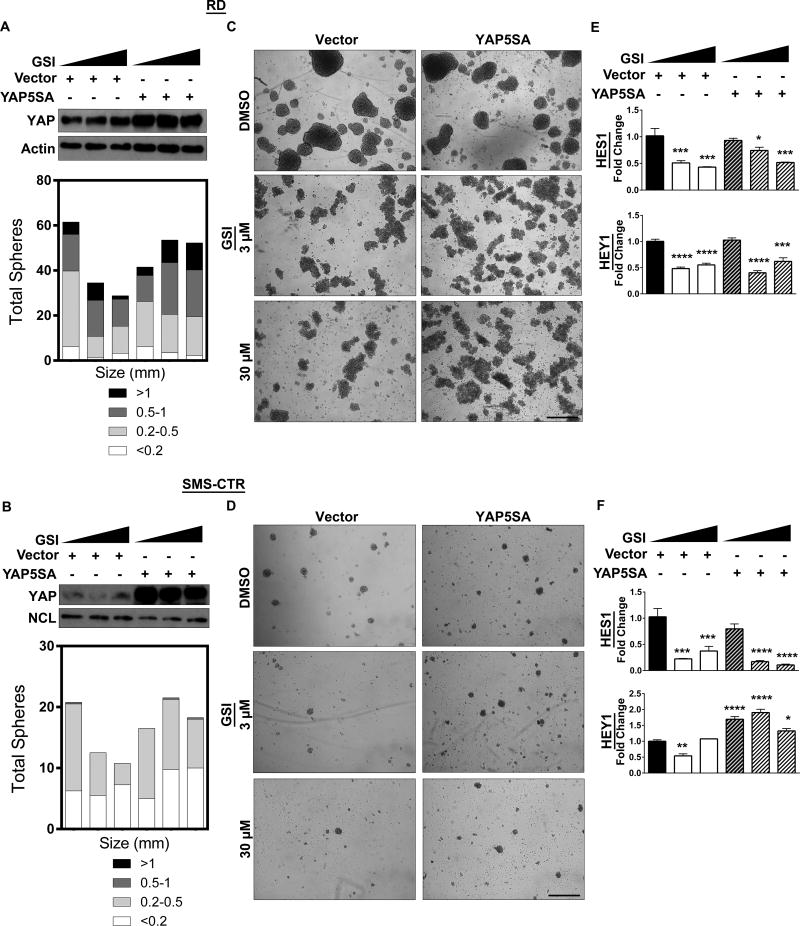
Since Notch signaling supports eRMS tumorigenesis, pharmacologic inhibition of the Notch pathway could be a reasonable eRMS treatment strategy. Indeed, Notch inhibition with GSIs decreases tumor growth in eRMS xenografts (7). However, our finding that YAP can upregulate several Notch pathway components (Fig.3) predicts that YAP could rescue Notch inhibition, leading to treatment resistance. To examine this hypothesis, we expressed a dox-inducible constitutively active YAP (YAP5SA, which is resistant to phosphorylation-mediated cytoplasmic retention and degradation) in the presence of the GSI RO4929097 (Fig.6A,B, top). GSI treatment alone inhibited sphere formation in a dose-responsive fashion (Fig.6C,D, left), and had on-target effects as shown by decreased HES1 and HEY1 expression (Fig.6E,F). (We had already shown that ICN overexpression can rescue Notch signaling after GSI treatment in eRMS cells (7).) While GSI treatment still altered sphere morphology, YAP5SA expression rescued sphere formation (Fig. 6C,D, right) as shown by a restoration of total sphere number and sphere size similar to that of the control (Fig.6A,B, bottom). YAP5SA rescued HEY1 levels in the SMS-CTRs, however, HES1 and HEY1 levels were not fully restored in the RDs (Fig.6E,F). These data show that YAP upregulation can partially rescue Notch inhibition in vitro, and raises the question of whether dual inhibition of these pathways might be more effective in thwarting this circuit and eRMS stem cell plasticity.
Figure 6. YAP expression rescues pharmacologic Notch inhibition.
(A, B, top) Expression of the dox-inducible YAP5SA construct increases total YAP protein levels in RD and SMS-CTR spheres as assessed by immunoblot. (A, B, bottom) Spheres quantified. (C, D, left) Increasing doses (0µM, 3µM, 30µm) of GSI RO4929097 inhibits sphere formation in RD and SMS-CTR spheres. (C, D right) Expression of YAP5SA in the presence of GSI restores sphere formation. (E, F) HES1 and HEY1 levels are decreased after GSI treatment in RD and SMS-CTR spheres, shown by qRT-PCR. *, P<0.05; **, P<0.01; ***, P<0.001; and ****, P<0.0001. Scale bars: 1.7mm.
Dual inhibition silences the Notch-YAP circuit
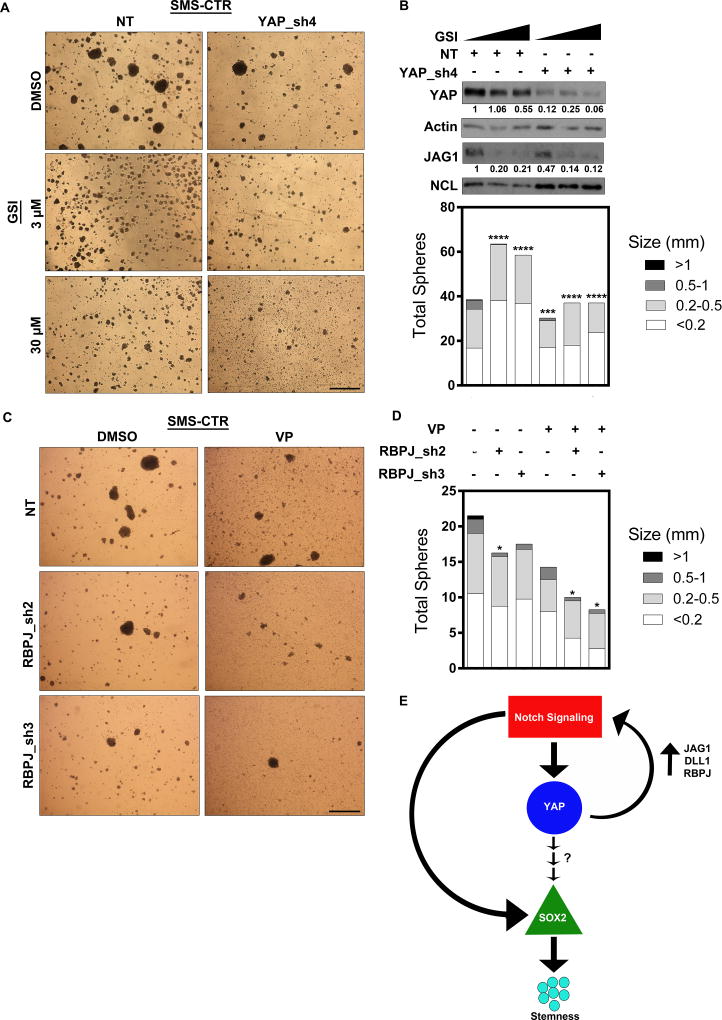
To simultaneously inhibit Notch and YAP pathways in vitro, we combined expression of a YAP1-directed shRNA with the GSI RO4929097. While GSI treatment again inhibited sphere formation (Fig.7A, left), combining GSI treatment with YAP_sh4 in SMS-CTR spheres was most effective at inhibiting sphere formation (Fig7.A, right), causing a smaller sphere size (Fig.7B, bottom) and decreasing total YAP and JAG1 protein levels (Fig.7B, top). Supplementary Fig.6A,B show similar results of dual inhibition in RD spheres. These data suggest that both pathways impact YAP and JAG1 levels, and that dual inhibition is effective in silencing this circuit. As a complementary dual inhibition approach, we combined expression of the RBPJ-directed shRNAs with the YAP-TEAD inhibitor verteporfin (VP). Again, while Notch pathway inhibition and VP treatment each decreased sphere number and size, dual inhibition had the greatest effect (Fig.7C,D). Last, in a third approach to evaluate dual inhibition we combined GSI with VP in RD adherent cells to assess cell viability using an MTT assay. Even in the adherent cells, the drug combination had a greater effect on cell viability than either drug alone (Supplementary Fig.6C), although a more profound effect might be seen in spheres or tumors expressing higher YAP-Notch signaling. In total, these data demonstrate that dual inhibition is more effective at silencing this Notch-YAP circuit, eRMS cell growth, and eRMS stemness than mono-inhibition of either pathway.
Figure 7. Dual inhibition of Notch and YAP inhibits sphere formation and silences the Notch-YAP circuit.
(A, left) Treatment of SMS-CTR spheres with 3µM or 30µm GSI inhibits sphere formation. (A, right) Addition of YAP_sh4 further inhibits SMS-CTR sphere formation. (B, top) GSI treatment alone decreases YAP and JAG1 protein levels. The combination of GSI with YAP_sh4 (last two lanes) almost completely eliminates YAP and JAG1 protein expression in SMS-CTR spheres. Densitometry quantitation shown below blots and normalized to loading controls. (B, bottom) Sphere number and size quantified. Statistical analysis was performed on the total number of spheres greater than 0.5 mm as compared to NT + DMSO. (C) Combination treatment with RBPJ shRNAs and 0.1µM VP in SMS-CTR spheres inhibits sphere formation. (D) Sphere size and number quantified. Statistical analysis was performed on the total number of spheres greater than 0.5 mm as compared to NT + DMSO. (E) Model shows that active Notch signaling in eRMS cells directly increases YAP levels, thus increasing YAP signaling. YAP signaling indirectly increases SOX2, which promotes a stemness phenotype. YAP provides a positive feedback loop by activating Notch signaling through direct upregulation of the ligands JAG1 and DLL1 and the transcription factor RBPJ. Notch signaling also directly upregulates SOX2 levels to contribute to the stemness phenotype. NT= non-targeting control. *, P<0.05; **, P<0.01; ***, P<0.001; and ****, P<0.0001. Scale bars: 1.7mm.
In conclusion, Notch and YAP signaling function in a bidirectional circuit that drives stemness and tumorigenesis in eRMS (Fig.7E). In TMAs, nuclear Notch and YAP are highly upregulated and positively correlate, suggesting this circuit is intact in eRMS human tumors. Mechanistically, Notch signaling directly regulates YAP1 and SOX2 to promote a stem-like phenotype. YAP in turn directly regulates JAG1, DLL1, and RBPJ to activate Notch signaling, and indirectly activates SOX2. Collectively, this circuit sustains stemness in eRMS cells and tumors.
Discussion
Developmental pathways including Notch and Hippo are important regulators of cell growth and fate in normal tissue development and homeostasis, including the myogenic lineage (reviewed in (64,65)), and are therefore tightly regulated. However, in many human cancers developmental pathways have been co-opted by gain-of-function mutations, epigenetic changes, or dysregulated signaling that supports tumor growth. eRMS is no exception. Activating mutations in Notch receptors and genomic amplification of YAP1 have recently been discovered in eRMS patient samples (1,6). Both pathways are also aberrantly activated in eRMS, as demonstrated by >90% and 100% of eRMS tumors staining positive for nuclear Notch1 and YAP, respectively (5,7). Mechanistic studies have elucidated roles for Notch and YAP in promoting eRMS tumor cell proliferation and antagonizing myogenic differentiation. Genetic or pharmacologic inhibition of Notch or YAP delays tumor growth in vivo, drawing attention to them as potential therapeutic targets (6,7,24). However, while mono-inhibition of these pathways may initially be effective, tumors often become resistant. Resistance to GSIs has been observed in T-ALL (66), and YAP activation is a mechanism of resistance to several therapies (67,68). In eRMS patients, relapsed disease is the most common reason for treatment failure (3), suggesting that combination therapies to target collaborating or compensatory pathways may be more effective. While dysregulation of each of the major developmental pathways (Hedgehog, Hippo, Notch, TGF-β, and Wnt) has been analyzed for contribution to eRMS, crosstalk between them is only beginning to be tackled (69,70). Therefore, in this study we analyzed crosstalk between Notch and Hippo signaling in eRMS to understand the potential implications for treatment resistance.
Here we uncover a novel Notch-YAP circuit that supports stemness in eRMS. While unidirectional signaling between these two pathways has been reported (27–34), and recently a positive feedback loop between Notch and YAP was described in hepatocellular carcinoma (71), this is the first identification of an intact bidirectional circuit supporting stemness in a pediatric cancer. Active Notch signaling directly stimulates YAP1 and the stem cell gene SOX2, while YAP positively feeds back to Notch through direct regulation of Notch pathway components JAG1, DLL1, and RBPJ. YAP also regulates SOX2, either through an indirect mechanism or TEAD-independent signaling. Studies in other cell types show that YAP can regulate SOX2 expression through binding to β-catenin (72) or OCT4 (73), however future studies are needed to determine the mechanism in eRMS. Together, the Notch-YAP-SOX2 signaling circuit maintains stem cell plasticity in eRMS, which may be a source of resistance to traditional chemotherapy (52). We provide evidence that simultaneous dual inhibition of these pathways is more effective at inhibiting cell growth and stemness than inhibiting either pathway alone.
An unexpected but fascinating finding from this study was that while all eRMS cells showed upregulation of SOX2 and Notch pathway components, not all eRMS showed upregulation of YAP1. This variability suggests that other developmental pathways might cooperate with Notch to support stemness in eRMS. For example, the Hedgehog pathway also supports stem cell properties (74) and has been found critical for Rh36, CCA, and CT-TC cell growth (75). Why there might be different developmental pathways underlying stemness in different eRMS tumors is unknown, however we speculate it may be due to factors including but not limited to patient age, genetic background, primary tumor mutational status, anatomic location, cell of origin, and even culturing in vitro, and emphasizes the need for a large repository of eRMS cell lines and tumors for systematic preclinical studies. Perhaps in the future, eRMS tumors will be classified based on expression of developmental pathways, similar to medulloblastoma (76).
Since Notch and Hippo signaling are dysregulated in many adult and pediatric cancers, there are continuing efforts to target these pathways pharmacologically (reviewed in (26,77)). A benefit of combination treatment might be the ability to lower the dose of each drug, minimizing toxicity. However, while dual inhibition is effective in pre-clinical studies, new pharmacologic agents are needed. For Notch pathway inhibition, next-generation GSIs and anti-Notch receptor or ligand monoclonal antibodies are being evaluated in current clinical trials (78–80).For YAP/TEAD inhibition, while VP is FDA-approved as a photosensitizer for the treatment of macular degeneration, it has significant solubility issues and is often not YAP/TEAD-specific (24,81). Novel YAP-directed agents will be crucial. In summary, we describe a Notch-YAP circuit that supports eRMS stemness and tumorigenesis, which can be thwarted by dual inhibition of these pathways. Other Notch/YAP driven human cancers should be assessed for a similar circuit and for responsiveness to combined Notch and YAP inhibition.
Supplementary Material
(A) Stem cell markers PROM1 and SOX2 increase in eRMS rhabdospheres over four passages as compared to adherent cells in Rh36 (left) and JR-1 (right) cells as assessed by qRT-PCR. (B) YAP1 and WWTR1 are not increased in either cell line. (C) HEY1 and HES1 are not increased in Rh36 spheres, but are increased in JR-1 spheres. (D) Notch ligand JAG1 is increased in Rh36 spheres while the ligand DLL1 is increased in JR-1 spheres. (E) Notch receptors NOTCH1 and NOTCH3 are not changed as shown by RT-PCR. GAPDH loading control is shown below. P= passage number. *, P<0.05; **, P<0.01; ***, P<0.001; and ****, P<0.0001.
(A) Expression of stem cell markers PROM1 and SOX2 as assessed by qRT-PCR in eRMS rhabdospheres over four passages as compared to adherent cells in CCA (left) and CT-TC (right) cells. SOX2 is increased while PROM1 is not. (B) YAP1 is increased in CT-TC spheres, while WWTR1 is increased in both lines. (C) HEY1 but not HES1 is increased in both cell lines. (D) Notch ligands JAG1 and DLL1 are increased in CCA spheres but not CT-TC spheres. (E) Notch receptors NOTCH1 and NOTCH3 are minimally changed as shown by RT-PCR. GAPDH loading control is shown below. Lanes are from the same RT-PCR but have been rearranged into this order. P= passage number. *, P<0.05; **, P<0.01; ***, P<0.001; and ****, P<0.0001.
(A) Representative images of Vector and ICN-expressing RD spheres. Scale bars: 100µm. (B) Sphere size and number quantified.
TEAD ChIP-Seq in RD cells shows additional peaks within NOTCH1, NOTCH2, and HEY1 enhancers. Chromatin state is determined by the ENCODE HMM in HSMMs as described in the legend to Fig.3.
YAP suppression decreases OCT4 and NANOG levels as assessed by qRT-PCR in (left) RD and (right) SMS-CTR spheres. NT= non-targeting control. ***, P<0.001; and ****, P<0.0001.
(A) Combination treatment of RD spheres with YAP_sh4 and GSI inhibits sphere formation. (B, top) GSI treatment alone inhibits YAP and JAG1 expression as assessed by immunoblot. The combination of GSI with YAP_sh4 (last two lanes) almost completely eliminates YAP and JAG1 protein expression in RD spheres. Densitometry quantitation shown below blots and normalized to loading controls. (B, bottom) Sphere number and size quantified. Statistical analysis was performed on the total number of spheres greater than 0.5 mm as compared to NT + DMSO but none were significant due to variability. (C) MTT assay combining GSI with 3µM VP in RD adherent cells. ****, P<0.0001. NT= non-targeting control. VP=verteporfin. Scale bars: 1.7mm.
Acknowledgments
We thank Dr. Michael Deel, Kristi Oristian, and Elaine Justice (Duke University) for technical support. We thank the laboratories of Dr. Daniel Wechsler, Dr. Michael Armstrong, Dr. Oren Becher, and Dr. David Kirsch (Duke University) for helpful discussions.
Financial support
This research was supported by NIH grants R01 CA122706 (CML) and T32 CA059365 (KKS), a Hartwell Foundation grant (LC), and the V Foundation for Cancer Research, Hyundai Hope on Wheels, Duke School of Medicine Bridge Grants, and the Glenn and Stacey Schiffman Pediatric Cancer Research Fund (CML).
Footnotes
Conflicts of interest
The authors declare no potential conflicts of interest.
References
- 1.Chen X, Stewart E, Shelat AA, Qu C, Bahrami A, Hatley M, et al. Targeting oxidative stress in embryonal rhabdomyosarcoma. Cancer cell. 2013;24(6):710–24. doi: 10.1016/j.ccr.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perkins SM, Shinohara ET, DeWees T, Frangoul H. Outcome for children with metastatic solid tumors over the last four decades. PloS one. 2014;9(7):e100396. doi: 10.1371/journal.pone.0100396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pappo AS, Anderson JR, Crist WM, Wharam MD, Breitfeld PP, Hawkins D, et al. Survival after relapse in children and adolescents with rhabdomyosarcoma: A report from the Intergroup Rhabdomyosarcoma Study Group. J Clin Oncol. 1999;17(11):3487–93. doi: 10.1200/JCO.1999.17.11.3487. [DOI] [PubMed] [Google Scholar]
- 4.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14(3):275–91. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Crose LE, Galindo KA, Kephart JG, Chen C, Fitamant J, Bardeesy N, et al. Alveolar rhabdomyosarcoma-associated PAX3-FOXO1 promotes tumorigenesis via Hippo pathway suppression. The Journal of clinical investigation. 2014;124(1):285–96. doi: 10.1172/JCI67087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tremblay AM, Missiaglia E, Galli GG, Hettmer S, Urcia R, Carrara M, et al. The Hippo transducer YAP1 transforms activated satellite cells and is a potent effector of embryonal rhabdomyosarcoma formation. Cancer cell. 2014;26(2):273–87. doi: 10.1016/j.ccr.2014.05.029. [DOI] [PubMed] [Google Scholar]
- 7.Belyea BC, Naini S, Bentley RC, Linardic CM. Inhibition of the Notch-Hey1 axis blocks embryonal rhabdomyosarcoma tumorigenesis. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17(23):7324–36. doi: 10.1158/1078-0432.CCR-11-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tostar U, Malm CJ, Meis-Kindblom JM, Kindblom LG, Toftgard R, Unden AB. Deregulation of the hedgehog signalling pathway: a possible role for the PTCH and SUFU genes in human rhabdomyoma and rhabdomyosarcoma development. J Pathol. 2006;208(1):17–25. doi: 10.1002/path.1882. [DOI] [PubMed] [Google Scholar]
- 9.Ranganathan P, Weaver KL, Capobianco AJ. Notch signalling in solid tumours: a little bit of everything but not all the time. Nature reviews Cancer. 2011;11(5):338–51. doi: 10.1038/nrc3035. [DOI] [PubMed] [Google Scholar]
- 10.De Salvo M, Raimondi L, Vella S, Adesso L, Ciarapica R, Verginelli F, et al. Hyper-activation of Notch3 amplifies the proliferative potential of rhabdomyosarcoma cells. PloS one. 2014;9(5):e96238. doi: 10.1371/journal.pone.0096238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masia A, Almazan-Moga A, Velasco P, Reventos J, Toran N, Sanchez de Toledo J, et al. Notch-mediated induction of N-cadherin and alpha9-integrin confers higher invasive phenotype on rhabdomyosarcoma cells. British journal of cancer. 2012;107(8):1374–83. doi: 10.1038/bjc.2012.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roma J, Masia A, Reventos J, Sanchez de Toledo J, Gallego S. Notch pathway inhibition significantly reduces rhabdomyosarcoma invasiveness and mobility in vitro. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17(3):505–13. doi: 10.1158/1078-0432.CCR-10-0166. [DOI] [PubMed] [Google Scholar]
- 13.Nagao H, Setoguchi T, Kitamoto S, Ishidou Y, Nagano S, Yokouchi M, et al. RBPJ is a novel target for rhabdomyosarcoma therapy. PloS one. 2012;7(7):e39268. doi: 10.1371/journal.pone.0039268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sang L, Coller HA, Roberts JM. Control of the reversibility of cellular quiescence by the transcriptional repressor HES1. Science. 2008;321(5892):1095–100. doi: 10.1126/science.1155998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ignatius MS, Hayes MN, Lobbardi R, Chen EY, McCarthy KM, Sreenivas P, et al. The NOTCH1/SNAIL1/MEF2C Pathway Regulates Growth and Self-Renewal in Embryonal Rhabdomyosarcoma. Cell reports. 2017;19(11):2304–18. doi: 10.1016/j.celrep.2017.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krop I, Demuth T, Guthrie T, Wen PY, Mason WP, Chinnaiyan P, et al. Phase I pharmacologic and pharmacodynamic study of the gamma secretase (Notch) inhibitor MK-0752 in adult patients with advanced solid tumors. J Clin Oncol. 2012;30(19):2307–13. doi: 10.1200/JCO.2011.39.1540. [DOI] [PubMed] [Google Scholar]
- 17.Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the Roots of Cancer. Cancer cell. 2016;29(6):783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, et al. Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell. 2011;144(5):782–95. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai J, Zhang N, Zheng Y, de Wilde RF, Maitra A, Pan D. The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes & development. 2010;24(21):2383–8. doi: 10.1101/gad.1978810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panciera T, Azzolin L, Fujimura A, Di Biagio D, Frasson C, Bresolin S, et al. Induction of Expandable Tissue-Specific Stem/Progenitor Cells through Transient Expression of YAP/TAZ. Cell Stem Cell. 2016 doi: 10.1016/j.stem.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer cell. 2009;16(5):425–38. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lau AN, Curtis SJ, Fillmore CM, Rowbotham SP, Mohseni M, Wagner DE, et al. Tumor-propagating cells and Yap/Taz activity contribute to lung tumor progression and metastasis. The EMBO journal. 2014;33(5):468–81. doi: 10.1002/embj.201386082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu D, Lv X, Hua G, He C, Dong J, Lele SM, et al. YAP regulates cell proliferation, migration, and steroidogenesis in adult granulosa cell tumors. Endocrine-related cancer. 2014;21(2):297–310. doi: 10.1530/ERC-13-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slemmons KK, Crose LE, Rudzinski E, Bentley RC, Linardic CM. Role of the YAP Oncoprotein in Priming Ras-Driven Rhabdomyosarcoma. PloS one. 2015;10(10):e0140781. doi: 10.1371/journal.pone.0140781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barry ER, Camargo FD. The Hippo superhighway: signaling crossroads converging on the Hippo/Yap pathway in stem cells and development. Current opinion in cell biology. 2013;25(2):247–53. doi: 10.1016/j.ceb.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 26.Conti B, Slemmons KK, Rota R, Linardic CM. Recent Insights into Notch Signaling in Embryonal Rhabdomyosarcoma. Curr Drug Targets. 2016;17(11):1235–44. doi: 10.2174/1389450116666150907105756. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Hibbs MA, Gard AL, Shylo NA, Yun K. Genome-wide analysis of N1ICD/RBPJ targets in vivo reveals direct transcriptional regulation of Wnt, SHH, and hippo pathway effectors by Notch1. Stem cells. 2012;30(4):741–52. doi: 10.1002/stem.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17(23):2054–60. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 29.Zhou D, Zhang Y, Wu H, Barry E, Yin Y, Lawrence E, et al. Mst1 and Mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis by inhibition of Yes-associated protein (Yap) overabundance. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(49):E1312–20. doi: 10.1073/pnas.1110428108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yimlamai D, Christodoulou C, Galli GG, Yanger K, Pepe-Mooney B, Gurung B, et al. Hippo pathway activity influences liver cell fate. Cell. 2014;157(6):1324–38. doi: 10.1016/j.cell.2014.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tschaharganeh DF, Chen X, Latzko P, Malz M, Gaida MM, Felix K, et al. Yes-associated protein up-regulates Jagged-1 and activates the Notch pathway in human hepatocellular carcinoma. Gastroenterology. 2013;144(7):1530–42. e12. doi: 10.1053/j.gastro.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferguson GB, Martinez-Agosto JA. Yorkie and Scalloped signaling regulates Notch-dependent lineage specification during Drosophila hematopoiesis. Curr Biol. 2014;24(22):2665–72. doi: 10.1016/j.cub.2014.09.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen HJ, Wang CM, Wang TW, Liaw GJ, Hsu TH, Lin TH, et al. The Hippo pathway controls polar cell fate through Notch signaling during Drosophila oogenesis. Developmental biology. 2011;357(2):370–9. doi: 10.1016/j.ydbio.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 34.Djiane A, Zaessinger S, Babaoglan AB, Bray SJ. Notch inhibits Yorkie activity in Drosophila wing discs. PloS one. 2014;9(8):e106211. doi: 10.1371/journal.pone.0106211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McAllister RM, Melnyk J, Finkelstein JZ, Adams EC, Jr, Gardner MB. Cultivation in vitro of cells derived from a human rhabdomyosarcoma. Cancer. 1969;24(3):520–6. doi: 10.1002/1097-0142(196909)24:3<520::aid-cncr2820240313>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 36.Whang-Peng J, Knutsen T, Theil K, Horowitz ME, Triche T. Cytogenetic studies in subgroups of rhabdomyosarcoma. Genes, chromosomes & cancer. 1992;5(4):299–310. doi: 10.1002/gcc.2870050405. [DOI] [PubMed] [Google Scholar]
- 37.Houghton PJ, Morton CL, Tucker C, Payne D, Favours E, Cole C, et al. The pediatric preclinical testing program: description of models and early testing results. Pediatr Blood Cancer. 2007;49(7):928–40. doi: 10.1002/pbc.21078. [DOI] [PubMed] [Google Scholar]
- 38.De Giovanni C, Nanni P, Nicoletti G, Ceccarelli C, Scotlandi K, Landuzzi L, et al. Metastatic ability and differentiative properties of a new cell line of human embryonal rhabdomyosarcoma (CCA) Anticancer Res. 1989;9(6):1943–9. [PubMed] [Google Scholar]
- 39.Clayton J, Pincott JR, van den Berghe JA, Kemshead JT. Comparative studies between a new human rhabdomyosarcoma cell line, JR-1 and its tumour of origin. British journal of cancer. 1986;54(1):83–90. doi: 10.1038/bjc.1986.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morton CL, Potter PM. Rhabdomyosarcoma-specific expression of the herpes simplex virus thymidine kinase gene confers sensitivity to ganciclovir. J Pharmacol Exp Ther. 1998;286(2):1066–73. [PubMed] [Google Scholar]
- 41.Hinson AR, Jones R, Crose LE, Belyea BC, Barr FG, Linardic CM. Human rhabdomyosarcoma cell lines for rhabdomyosarcoma research: utility and pitfalls. Frontiers in oncology. 2013;3:183. doi: 10.3389/fonc.2013.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schaaf G, Hamdi M, Zwijnenburg D, Lakeman A, Geerts D, Versteeg R, et al. Silencing of SPRY1 triggers complete regression of rhabdomyosarcoma tumors carrying a mutated RAS gene. Cancer research. 2010;70(2):762–71. doi: 10.1158/0008-5472.CAN-09-2532. [DOI] [PubMed] [Google Scholar]
- 43.Shukla N, Ameur N, Yilmaz I, Nafa K, Lau CY, Marchetti A, et al. Oncogene mutation profiling of pediatric solid tumors reveals significant subsets of embryonal rhabdomyosarcoma and neuroblastoma with mutated genes in growth signaling pathways. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18(3):748–57. doi: 10.1158/1078-0432.CCR-11-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 45.Martinelli S, McDowell HP, Vigne SD, Kokai G, Uccini S, Tartaglia M, et al. RAS signaling dysregulation in human embryonal Rhabdomyosarcoma. Genes, chromosomes & cancer. 2009;48(11):975–82. doi: 10.1002/gcc.20702. [DOI] [PubMed] [Google Scholar]
- 46.Zhang J, Smolen GA, Haber DA. Negative regulation of YAP by LATS1 underscores evolutionary conservation of the Drosophila Hippo pathway. Cancer research. 2008;68(8):2789–94. doi: 10.1158/0008-5472.CAN-07-6205. [DOI] [PubMed] [Google Scholar]
- 47.Xia Y, Zhang YL, Yu C, Chang T, Fan HY. YAP/TEAD co-activator regulated pluripotency and chemoresistance in ovarian cancer initiated cells. PloS one. 2014;9(11):e109575. doi: 10.1371/journal.pone.0109575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Justilien V, Walsh MP, Ali SA, Thompson EA, Murray NR, Fields AP. The PRKCI and SOX2 oncogenes are coamplified and cooperate to activate Hedgehog signaling in lung squamous cell carcinoma. Cancer cell. 2014;25(2):139–51. doi: 10.1016/j.ccr.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Asnaghi L, Lin MH, Lim KS, Lim KJ, Tripathy A, Wendeborn M, et al. Hypoxia promotes uveal melanoma invasion through enhanced Notch and MAPK activation. PloS one. 2014;9(8):e105372. doi: 10.1371/journal.pone.0105372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takam Kamga P, Bassi G, Cassaro A, Midolo M, Di Trapani M, Gatti A, et al. Notch signalling drives bone marrow stromal cell-mediated chemoresistance in acute myeloid leukemia. Oncotarget. 2016;7(16):21713–27. doi: 10.18632/oncotarget.7964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martz CA, Ottina KA, Singleton KR, Jasper JS, Wardell SE, Peraza-Penton A, et al. Systematic identification of signaling pathways with potential to confer anticancer drug resistance. Sci Signal. 2014;7(357):ra121. doi: 10.1126/scisignal.aaa18777/357/ra121. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walter D, Satheesha S, Albrecht P, Bornhauser BC, D'Alessandro V, Oesch SM, et al. CD133 positive embryonal rhabdomyosarcoma stem-like cell population is enriched in rhabdospheres. PloS one. 2011;6(5):e19506. doi: 10.1371/journal.pone.0019506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Louis Sharon A, Reynolds BA. Neurosphere and Neural Colony-Forming Cell Assays. Protocols for Neural Cell Culture. Fourth. Springer Protocols Handbooks: Humana Press; 2009. pp. 1–28. [Google Scholar]
- 54.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347(1–2):70–8. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 55.Ren C, Ren T, Yang K, Wang S, Bao X, Zhang F, et al. Inhibition of SOX2 induces cell apoptosis and G1/S arrest in Ewing's sarcoma through the PI3K/Akt pathway. J Exp Clin Cancer Res. 2016;35:44. doi: 10.1186/s13046-016-0321-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou X, Zhu H, Lu J. PTEN and hTERT gene expression and the correlation with human hepatocellular carcinoma. Pathol Res Pract. 2015;211(4):316–9. doi: 10.1016/j.prp.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 57.Jeziorska DM, Koentges G, Vance KW. Novel cis-regulatory modules control expression of the Hairy and Enhancer of Split-1 (HES1) transcription factor in myoblasts. J Biol Chem. 2012;287(8):5687–97. doi: 10.1074/jbc.M111.286484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paulson V, Chandler G, Rakheja D, Galindo RL, Wilson K, Amatruda JF, et al. High-resolution array CGH identifies common mechanisms that drive embryonal rhabdomyosarcoma pathogenesis. Genes, chromosomes & cancer. 2011;50(6):397–408. doi: 10.1002/gcc.20864. [DOI] [PubMed] [Google Scholar]
- 59.Raimondi L, Ciarapica R, De Salvo M, Verginelli F, Gueguen M, Martini C, et al. Inhibition of Notch3 signalling induces rhabdomyosarcoma cell differentiation promoting p38 phosphorylation and p21(Cip1) expression and hampers tumour cell growth in vitro and in vivo. Cell death and differentiation. 2012;19(5):871–81. doi: 10.1038/cdd.2011.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Konishi H, Asano N, Imatani A, Kimura O, Kondo Y, Jin X, et al. Notch1 directly induced CD133 expression in human diffuse type gastric cancers. Oncotarget. 2016 doi: 10.18632/oncotarget.10967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stein C, Bardet AF, Roma G, Bergling S, Clay I, Ruchti A, et al. YAP1 Exerts Its Transcriptional Control via TEAD-Mediated Activation of Enhancers. PLoS Genet. 2015;11(8):e1005465. doi: 10.1371/journal.pgen.1005465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Galli GG, Carrara M, Yuan WC, Valdes-Quezada C, Gurung B, Pepe-Mooney B, et al. YAP Drives Growth by Controlling Transcriptional Pause Release from Dynamic Enhancers. Mol Cell. 2015;60(2):328–37. doi: 10.1016/j.molcel.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B, et al. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nature cell biology. 2015;17(9):1218–27. doi: 10.1038/ncb3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mourikis P, Tajbakhsh S. Distinct contextual roles for Notch signalling in skeletal muscle stem cells. BMC developmental biology. 2014;14:2. doi: 10.1186/1471-213X-14-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wackerhage H, Del Re DP, Judson RN, Sudol M, Sadoshima J. The Hippo signal transduction network in skeletal and cardiac muscle. Sci Signal. 2014;7(337):re4. doi: 10.1126/scisignal.2005096. [DOI] [PubMed] [Google Scholar]
- 66.Knoechel B, Roderick JE, Williamson KE, Zhu J, Lohr JG, Cotton MJ, et al. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nature genetics. 2014;46(4):364–70. doi: 10.1038/ng.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin L, Sabnis AJ, Chan E, Olivas V, Cade L, Pazarentzos E, et al. The Hippo effector YAP promotes resistance to RAF- and MEK-targeted cancer therapies. Nature genetics. 2015;47(3):250–6. doi: 10.1038/ng.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Muranen T, Selfors LM, Hwang J, Gallegos LL, Coloff JL, Thoreen CC, et al. ERK and p38 MAPK Activities Determine Sensitivity to PI3K/mTOR Inhibition via Regulation of MYC and YAP. Cancer research. 2016 doi: 10.1158/0008-5472.CAN-16-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eichenmuller M, Bauer R, Von Schweinitz D, Hahn H, Kappler R. Hedgehog-independent overexpression of transforming growth factor-beta1 in rhabdomyosarcoma of Patched1 mutant mice. International journal of oncology. 2007;31(2):405–12. [PubMed] [Google Scholar]
- 70.Nitzki F, Cuvelier N, Drager J, Schneider A, Braun T, Hahn H. Hedgehog/Patched-associated rhabdomyosarcoma formation from delta1-expressing mesodermal cells. Oncogene. 2016;35(22):2923–31. doi: 10.1038/onc.2015.346. [DOI] [PubMed] [Google Scholar]
- 71.Kim W, Khan SK, Gvozdenovic-Jeremic J, Kim Y, Dahlman J, Kim H, et al. Hippo signaling interactions with Wnt/beta-catenin and Notch signaling repress liver tumorigenesis. The Journal of clinical investigation. 2016 doi: 10.1172/JCI88486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL, et al. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332(6028):458–61. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bora-Singhal N, Nguyen J, Schaal C, Perumal D, Singh S, Coppola D, et al. YAP1 Regulates OCT4 Activity and SOX2 Expression to Facilitate Self-Renewal and Vascular Mimicry of Stem-Like Cells. Stem cells. 2015;33(6):1705–18. doi: 10.1002/stem.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Satheesha S, Manzella G, Bovay A, Casanova EA, Bode PK, Belle R, et al. Targeting hedgehog signaling reduces self-renewal in embryonal rhabdomyosarcoma. Oncogene. 2016;35(16):2020–30. doi: 10.1038/onc.2015.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tostar U, Toftgard R, Zaphiropoulos PG, Shimokawa T. Reduction of human embryonal rhabdomyosarcoma tumor growth by inhibition of the hedgehog signaling pathway. Genes Cancer. 2010;1(9):941–51. doi: 10.1177/1947601910385449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123(4):465–72. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zanconato F, Battilana G, Cordenonsi M, Piccolo S. YAP/TAZ as therapeutic targets in cancer. Curr Opin Pharmacol. 2016;29:26–33. doi: 10.1016/j.coph.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Diaz-Padilla I, Wilson MK, Clarke BA, Hirte HW, Welch SA, Mackay HJ, et al. A phase II study of single-agent RO4929097, a gamma-secretase inhibitor of Notch signaling, in patients with recurrent platinum-resistant epithelial ovarian cancer: A study of the Princess Margaret, Chicago and California phase II consortia. Gynecol Oncol. 2015;137(2):216–22. doi: 10.1016/j.ygyno.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 79.Chiorean EG, LoRusso P, Strother RM, Diamond JR, Younger A, Messersmith WA, et al. A Phase I First-in-Human Study of Enoticumab (REGN421), a Fully Human Delta-like Ligand 4 (Dll4) Monoclonal Antibody in Patients with Advanced Solid Tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21(12):2695–703. doi: 10.1158/1078-0432.CCR-14-2797. [DOI] [PubMed] [Google Scholar]
- 80.Yen WC, Fischer MM, Axelrod F, Bond C, Cain J, Cancilla B, et al. Targeting Notch signaling with a Notch2/Notch3 antagonist (tarextumab) inhibits tumor growth and decreases tumor-initiating cell frequency. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21(9):2084–95. doi: 10.1158/1078-0432.CCR-14-2808. [DOI] [PubMed] [Google Scholar]
- 81.Yu FX, Zhang K, Guan KL. YAP as oncotarget in uveal melanoma. Oncoscience. 2014;1(7):480–1. doi: 10.18632/oncoscience.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Linardic CM, Downie DL, Qualman S, Bentley RC, Counter CM. Genetic modeling of human rhabdomyosarcoma. Cancer research. 2005;65(11):4490–5. doi: 10.1158/0008-5472.CAN-04-3194. [DOI] [PubMed] [Google Scholar]
- 83.Cam H, Griesmann H, Beitzinger M, Hofmann L, Beinoraviciute-Kellner R, Sauer M, et al. p53 family members in myogenic differentiation and rhabdomyosarcoma development. Cancer cell. 2006;10(4):281–93. doi: 10.1016/j.ccr.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 84.Ciarapica R, Russo G, Verginelli F, Raimondi L, Donfrancesco A, Rota R, et al. Deregulated expression of miR-26a and Ezh2 in rhabdomyosarcoma. Cell Cycle. 2009;8(1):172–5. doi: 10.4161/cc.8.1.7292. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Stem cell markers PROM1 and SOX2 increase in eRMS rhabdospheres over four passages as compared to adherent cells in Rh36 (left) and JR-1 (right) cells as assessed by qRT-PCR. (B) YAP1 and WWTR1 are not increased in either cell line. (C) HEY1 and HES1 are not increased in Rh36 spheres, but are increased in JR-1 spheres. (D) Notch ligand JAG1 is increased in Rh36 spheres while the ligand DLL1 is increased in JR-1 spheres. (E) Notch receptors NOTCH1 and NOTCH3 are not changed as shown by RT-PCR. GAPDH loading control is shown below. P= passage number. *, P<0.05; **, P<0.01; ***, P<0.001; and ****, P<0.0001.
(A) Expression of stem cell markers PROM1 and SOX2 as assessed by qRT-PCR in eRMS rhabdospheres over four passages as compared to adherent cells in CCA (left) and CT-TC (right) cells. SOX2 is increased while PROM1 is not. (B) YAP1 is increased in CT-TC spheres, while WWTR1 is increased in both lines. (C) HEY1 but not HES1 is increased in both cell lines. (D) Notch ligands JAG1 and DLL1 are increased in CCA spheres but not CT-TC spheres. (E) Notch receptors NOTCH1 and NOTCH3 are minimally changed as shown by RT-PCR. GAPDH loading control is shown below. Lanes are from the same RT-PCR but have been rearranged into this order. P= passage number. *, P<0.05; **, P<0.01; ***, P<0.001; and ****, P<0.0001.
(A) Representative images of Vector and ICN-expressing RD spheres. Scale bars: 100µm. (B) Sphere size and number quantified.
TEAD ChIP-Seq in RD cells shows additional peaks within NOTCH1, NOTCH2, and HEY1 enhancers. Chromatin state is determined by the ENCODE HMM in HSMMs as described in the legend to Fig.3.
YAP suppression decreases OCT4 and NANOG levels as assessed by qRT-PCR in (left) RD and (right) SMS-CTR spheres. NT= non-targeting control. ***, P<0.001; and ****, P<0.0001.
(A) Combination treatment of RD spheres with YAP_sh4 and GSI inhibits sphere formation. (B, top) GSI treatment alone inhibits YAP and JAG1 expression as assessed by immunoblot. The combination of GSI with YAP_sh4 (last two lanes) almost completely eliminates YAP and JAG1 protein expression in RD spheres. Densitometry quantitation shown below blots and normalized to loading controls. (B, bottom) Sphere number and size quantified. Statistical analysis was performed on the total number of spheres greater than 0.5 mm as compared to NT + DMSO but none were significant due to variability. (C) MTT assay combining GSI with 3µM VP in RD adherent cells. ****, P<0.0001. NT= non-targeting control. VP=verteporfin. Scale bars: 1.7mm.