Abstract
Conditions of the tumor microenvironment, such as hypoxia or nutrient starvation, play critical roles in cancer progression and malignancy. However, the role of acidic extracellular pH in tumor aggressiveness and its underlying mechanism has not been extensively studied compared to hypoxic or nutrient starvation conditions. In addition, a well-defined culture method to mimic the acidic extracellular tumor microenvironment has not been fully reported.
Here we present a simple in vitro culture method to maintain acidic extracellular pH using reduced bicarbonate and increased lactate or HCl concentrations in the culture medium. The medium pH was sustained for at least 24 h and gradually decreased by 72 h following culture of PANC-1 and AsPC-1 pancreatic cancer cells. Three distinct acidic media conditions in this study highly upregulated pH-responsive genes such as MSMO1, INSIG1, and IDI1 compared to hypoxia or nutrient starvation. The upregulation of these genes can be used as a marker of acidic pH. These simple techniques are beneficial to elucidate underlying mechanisms of tumor malignancy under acidic tumor microenvironment. Therefore, our extracellular acidic pH culture system enables discovery of cellular acidic pH responses not only in cancer cells but also in primary cells, such as renal tubular cells, in relation to the other acidic disorders including, diabetic ketoacidosis, lactic acidosis, renal tubular acidosis, and respiratory acidosis.
Keywords: Cancer Research, Issue 129, Extracellular low pH, tumor microenvironment, lactate, hypoxia, nutrient starvation, culture method, medium
Introduction
The tumor microenvironment plays critical roles in tumor progression and cancer cell metabolism1,2,3. Cancer cells are often exposed to conditions such as hypoxia, nutrient deprivation, and acidic extracellular pH (pHe). However, the role of pHe in tumor progression has not been clarified as extensively as in hypoxia or nutrient starvation. The pHe in the tumor tissue can become acidic, reaching approximately pH 6.84,5. The acidic pH arises from aerobic and anaerobic glycolytic excretion of protons (H+) and lactate by proliferating cancer cells5,6.
Recent studies revealed that acidic pHe-induced histone deacetylation, fatty acid oxidation, and exocytosis of acidic lysosomes for adaption to the severe acidic environment7,8,9,10. However, the mechanisms through which extracellular acidification affects cancer behavior and the identity of key regulators in the acidic pH tumor microenvironment have not been fully determined. Furthermore, several reports described various acidic pH media using unclear concentrations of bicarbonate, Tris, PIPES, and HEPES buffer or lactate and HCl, but there are few reports to demonstrate the stability of the adjusted medium nor comprehensive comparison of several distinct acidic culture media7,8,9,10
To elucidate the key regulators and metabolic changes in cancer cells in the context of extracellular acidification, we established a simple in vitro culture model to maintain an acidic pHe and examined the role of pHe in cancer cells11. Using this method, we maintained an acidic culture medium with a pHe of 6.8 at 37 °C under 5% CO2, using reduced bicarbonate concentrations in the culture medium. pH 7.4 was used as normal and control medium. The medium pH was sustained for at least 24 h and gradually decreased by 72 h during culture of PANC-1 and AsPC-1 pancreatic cancer cells. Because enhanced glycolysis accelerates excretion of not only protons but also lactate7,8,9, we also established a culture method mimicking lactate-induced acidosis by adding lactate rather than reducing the bicarbonate concentration. Additionally, HCl-induced acidic pHe in the medium allows us to exclude the possibility that cellular responses to acidic pH culture medium are not due to a reduced concentration of bicarbonate. Moreover, using various media with a pH of 6.4 to 7.4 with different bicarbonate concentration, we can assess the extent of pHe effects on cellular responses.
Protocol
1. Preparation of Acidic Culture Medium
- Preparation of control (pH 7.4) and low-pH (pH 6.8) culture medium
- Dissolve 4.75 g of DMEM powder without L-glutamine in 500 mL of water.
- Prepare 0.33 M NaHCO3 solution in a pressure-tight bottle. Caution: It is important to use a pressure tight bottle since NaHCO3 emits CO2 gas when heated and thermally decomposed.
- Autoclave the medium and solution. Allow these buffers to cool to 25 °C.
- Add 5 mL of 200 mM L-glutamine, 5 mL of penicillin-streptomycin, and 50 mL of fetal bovine serum to 500 mL of DMEM without bicarbonate.
- For control (pH 7.4) medium, add 2.4 mL of 0.33 M NaHCO3 solution per 100 mL of DMEM without bicarbonate. For low-pH (pH 6.8) medium, add 0.6 mL of 0.33 M NaHCO3 solution per 100 mL of DMEM without bicarbonate.
- Check the medium pH after 24 h of incubation at 37 °C under 5% CO2. NOTE: The amount of NaHCO3 solution needs to be adjusted depending on the medium pH, which can differ depending on the CO2 content of the incubator and air pressure. Ideally, for the first time, it is better to measure the medium pH with various bicarbonate concentrations to determine the appropriate amount of NaHCO3 solution to add to DMEM.
- Collect culture medium immediately after removing the culture dish from the CO2 incubator and measure the pH using a pH meter. Keep the medium temperature at 37 °C using a water bath if necessary. Measure the medium pH three times independently. NOTE: NaHCO3 solution should be stored at 4 °C and each media should be freshly prepared. Commercially available products which can be used in this study are listed in Table of Materials.
- Preparing lactate-induced or HCl-induced acidic culture medium
- Add 74 µL of lactate solution or 125 µL of 1 M HCl to 100 mL of control medium (prepared in step 1.1.5).
- Check the medium pH after 24 h of incubation at 37 °C under 5% CO2. NOTE: The amount of lactate or HCl solution also needs to be adjusted according to the medium pH, which can differ depending on the CO2 content in the incubator and air pressure. Ideally, it is better to measure the medium pH with serial lactate or HCl concentrations to determine the appropriate amount for adjusting to the desired pH of the medium.
2. Harvesting and Treating Cells
Start cultivating cells at 37 °C under 5% CO2 at least 1 week before performing experiments. NOTE: The cell lines used in this study are listed in Table of Materials.
Passage cells when they reach 70 - 90% confluency. Do not allow the cells to become confluent. Change the medium every 2 - 3 days during daily cultivation.
Wash the cells by adding 5 mL PBS. Aspirate the PBS and add detaching enzyme such as 1 mL Trypsin.
Incubate at 37 °C until the cells are rounded and start to detach.
Add 5 mL of culture medium to the detached cells and deactivate enzyme. Transfer the cell suspension to a 15 mL tube and centrifuge for 3 min at 300 x g. Aspirate the supernatant and re-suspend the cells with culture medium.
Count the viable cells using a hemocytometer and Trypan-Blue.
Seed 5.0 x 105 cells/well of a 10-cm dish in 10 mL of medium. Incubate the cells for 24 h at 37 °C under 5% CO2.
Aspirate the culture medium, wash the cells with 5 mL PBS, and add 10 mL of the acidic culture medium prepared in Section 1. Incubate the cells for 24 h at 37 °C under 5% CO2.
3. RNA Isolation
Aspirate culture medium and wash cells with 5 mL PBS. Disrupt the cells by adding RNA extraction reagent (see Tables of Materials).
Scrape the dish briefly, then remove the RNA extraction reagent with a pipette and deposit the RNA extraction reagent/cell lysate into a 1.5 mL tube. Leave at room temperature for 5 min.
Add 250 µL chloroform and shake the tube vigorously for about 15 s. Leave at room temperature for 5 min and centrifuge at ≥8,000 x g for 5 min.
Carefully remove the aqueous phase using a pipette. Leave behind some of the aqueous phase. Place in another 1.5 mL tube.
Add 550 µL isopropanol to the aqueous phase and mix gently. Leave at room temperature for 5 min. Centrifuge at maximal speed (≥20,000 x g) for 20 min at 4 °C.
Pour off the isopropanol and add 1 mL 75% ethanol in DEPC treated water. Mix gently. Centrifuge at 8,000 x g for 5 min.
Pipette off the ethanol and let the pellets air-dry.
Add approximately 15 - 25 µL (depending on the yield) of DEPC treated water to the RNA pellet. Mix thoroughly.
Measure the concentration of isolated RNA by measuring the OD at 260 nm using a spectrophotometer.
4. cDNA Synthesis
Combine the following components in a PCR reaction tube: up to 5 µg total RNA, 1 µL Oligo dT (50 µM) or Random Primer Mix (60 µM), 1 µL 10 mM dNTP, and nuclease-free water to a total volume of 14 µL.
Mix and briefly centrifuge the components using a small benchtop centrifuge (~5 s). Denature the sample RNA/primer for 5 min at 65 °C. Spin briefly (~5 s) using a small benchtop centrifuge and put the sample promptly on ice for at least 1 min.
Add the following components: 4 µL 5x Reverse Transcription buffer, 1 µL Reverse Transcriptase (200 U/µL), 1 µL RNase Inhibitor (40 U/µL). NOTE: Examples of commercially available kits are shown in Table of Materials
Cap the tube, mix, and then briefly centrifuge the contents using a small benchtop centrifuge (~5 s). Incubate the combined reaction mixture at 50 - 55 °C for 10 min.
Inactivate the reaction by incubating at 80 °C for 10 min.
5. Measurement of Acid-responsive Gene Expression Using Real-time PCR
Combine the following components in a 384-well reaction plate: 1 µL Template cDNA, 0.75 µL 10X N',N'-dimethyl-N-[4-[(E)-(3-methyl-1,3-benzothiazol-2-ylidene)methyl]-1-phenylquinolin-1-ium-2-yl]-N-propylpropane-1,3-diamine, 0.3 µL 50 µM forward primer, 0.3 µL 50 µM reverse primer, 1 µL Taq polymerase, 2.5 µL 10x Taq polymerase buffer, 2.5 µL 20 mM dNTP, 16.65 µL water. NOTE: Example of commercially available kits are shown in Table of Materials. A list of primers is shown in Table 1. A 96-well reaction plate can be also used in this step. Other probes (e.g., Taqman probe) can also be used.
Set up the experiment and the following PCR program on a Real-time PCR System: 50 °C for 2 min, 1 cycle; 95 °C for 10 min, 1 cycle; 95 °C for 15 s and 60 °C for 1 min, 40 cycles; 95 °C for 15 s, 1 cycle; 60 °C for 15 s, 1 cycle; 95 °C for 15 s, 1 cycle.
Place the plate in the qPCR instrument. Start the program. NOTE: Upregulation of the pH responsive genes MSMO1, INSIG1, and IDI1 can be measured by protein levels.
Representative Results
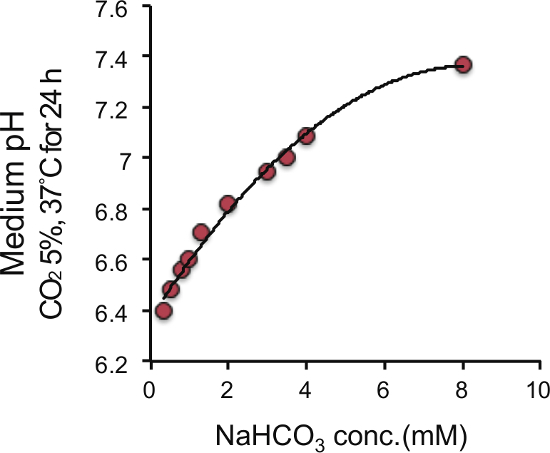
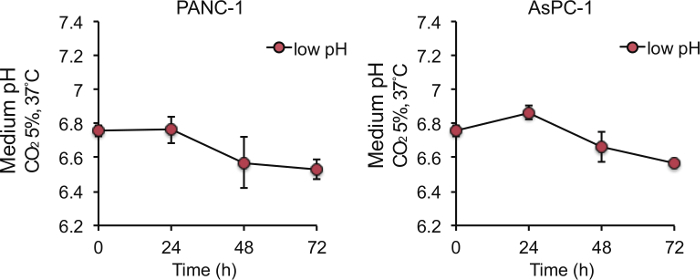
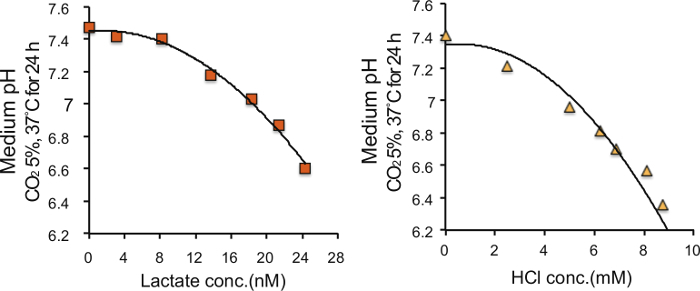
To determine the appropriate bicarbonate concentration, we prepared DMEM at range of 0 - 8 mM NaHCO3 (final concentration in the culture medium) and succeeded in preparing culture media with pH ranging from 6.4 - 7.4 (Figure 1). We prepared DMEM with 8 mM NaHCO3 (pH 7.4) as a control medium, and 2 mM NaHCO3 (pH 6.8) as a medium with acidic pH according to a previous report stating that the extracellular pH reaches pH 6.8 for solid tumors4,5. The pH of the acidic medium was sustained for 24 h, and gradually decreased to approximately pH 6.6 for 72 h during culture of PANC-1 and AsPC-1 cells (Figure 2). Next, to determine the amount of lactate or HCl required to adjust the control medium (pH 7.4) to pH 6.8, we added various amounts of lactate or HCl to the control medium and measured the pH of the medium, and found that the pH of the control medium with a final concentration of 22.5 µM lactate and 6.25 mM HCl reached a value of 6.8 (Figure 3).
The effect of extracellular acidification using a medium with low pH can be easily evaluated based on upregulation of acidic pH-responsive genes such as MSMO1, IDI1, and INSIG1. Expression of these genes was highly increased under low pH compared to their expression under hypoxia or nutrient starvation (Figure 4). In addition, the upregulation of these genes was not specific to a medium in which the acidic pH was caused by reduced bicarbonate levels, and they were also upregulated by a medium in which acidic pH was caused by the addition of lactate and/or HCl (Figure 5). Comparison of cellular responses to media in which the acidic pH is derived from reduced bicarbonate levels or the addition of lactate or HCl enables us to determine if cellular responses are specific to certain types of acidification. Upregulation of acidic pH responsible genes such as IDI1, MSMO1, and INSIG1 under extracellular low pH also occurs in normal cells (Figure 6), so our method can be applied to not only tumor cells but also normal cells. Under high pH condition, the expression level of pH-responsible genes was not upregulated in PANC-1 and AsPC-1 cells (Figure 7), which can be used as negative control.
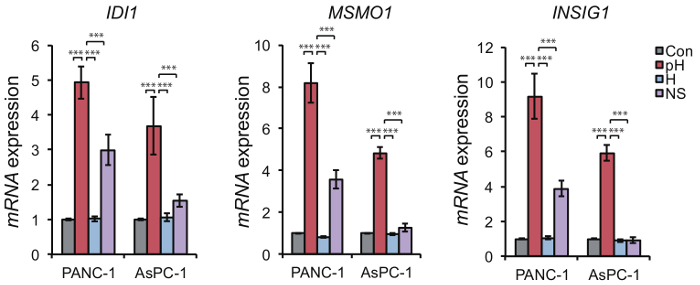
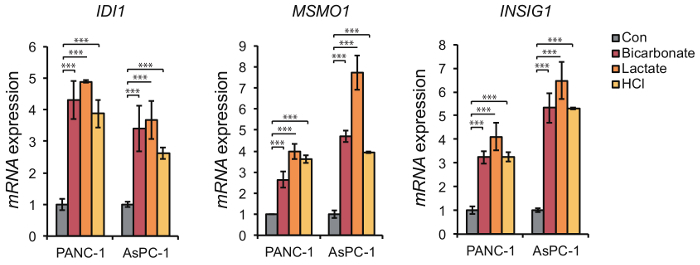
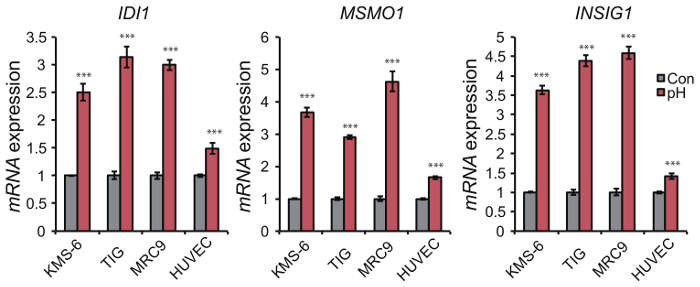
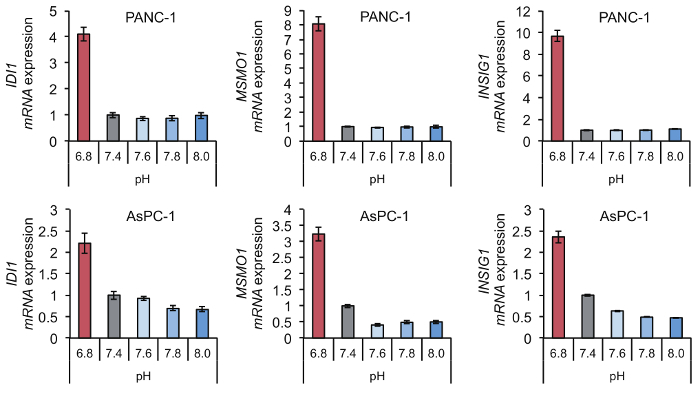
Figure 1. Correlation between NaHCO3 concentration and medium pH. Horizontal axis shows concentration of NaHCO3 and vertical axis shows medium pH at 37 °C under 5% CO2. Please click here to view a larger version of this figure.
Figure 2. Change of medium pH of low-pH (pH 6.8) culture medium during 72 h-culture. Medium pH was sustained for 24 h and gradually decreased for 72 h during culture of PANC-1 and AsPC-1 cells. 5.0 x 105 cells were seeded to a 10-cm dish in 10 mL of medium, incubated for 24 h, and changed to low pH culture medium. Data are presented as the mean ± SEM of at least three independent experiments. Please click here to view a larger version of this figure.
Figure 3. Correlation between lactate or HCl concentrations and pH of the medium. The control medium (pH 7.4) was buffered with various concentrations of lactate or HCl. Please click here to view a larger version of this figure.
Figure 4. Transcriptional upregulation of acidic pH-responsive genes in response to low pH. Expression level of MSMO1, IDI1, and INSIG1 was higher in PANC-1 and AsPC-1 cells in the context of low pH (pH) than under hypoxia (H) or nutrient starvation (NS) conditions after 24 h. Data are presented as the mean ± SEM of at least three independent experiments. Student's t-tests were performed for the indicated comparisons. ***p <0.005. Please click here to view a larger version of this figure.
Figure 5. Transcriptional upregulation of acidic pH-responsive genes in response to lactic acidosis and HCl-mediated acidosis. Expression of IDI1, MSMO1, and INSIG1 mRNAs in PANC-1 cells and AsPC-1 cells was determined by quantitative real-time polymerase chain reaction analysis under control (Con; pH 7.4), low pH (pH; pH 6.8, reduced amount of NaHCO3), lactic acidosis (lactate; pH 6.8, increased amount of lactate), and HCl-mediated acidosis (HCl; pH 6.8, increased amount of HCl) conditions for 24 h. Data are presented as the mean ± SEM of at least three independent experiments. Student's t-tests were performed for the indicated comparisons. ***p <0.005. Please click here to view a larger version of this figure.
Figure 6. Transcriptional upregulation of acidic pH-responsive genes in response to low pH in normal cells. Expression level of MSMO1, IDI1, and INSIG1 was upregulated in fibroblastic KMS-6, TIG, and MRC9 cells and endothelial HUVEC cells after 24 h. Data are presented as the mean ± SEM of at least three independent experiments. Student's t-tests were performed for the indicated comparisons. ***p <0.005. Please click here to view a larger version of this figure.
Figure 7. Expression level of acidic pH-responsible genes under high pH (pH 7.6 to 8.0) condition in comparison under low pH (pH 6.8). The expression level of pH-responsible genes such as IDI1, MSMO1, and INSIG1 remains unchanged under high pH (pH 7.6 to 8.0) conditions in PANC-1 and AsPC-1 cells after 24 h. Data are presented as the mean ± SEM of at least three independent experiments. Please click here to view a larger version of this figure.
| Primer | Forward primer sequence | Primer | Reverse pimer sequence |
| ACTB | 5'-AGAAGGAGATCACTGCCCTGGCACC-3' | ACTB | 5'-CCTGCTTGCTGATCCACATCTGCTG-3' |
| MSMO1 | 5'-ATCATGAGTTTCAGGCTCCATT-3' | MSMO1 | 5'-AAGCACGATTCCAATGAAAAAT-3' |
| INSIG1 | 5'-TGGCAGCTTCCCAAGTATTC-3' | INSIG1 | 5'-ACTGCGGGTTGGTAATTGAG -3' |
| IDI1 | 5'-TGGATAAAACCCCTGTGGTG-3' | IDI1 | 5'-CAACATCCGGCATAACTGTG-3' |
Table 1. List of primers used in quantitative real-time PCR analysis.
Discussion
Here, we described a simple acidic pH culture system and its evaluation process. The combination of three methods of medium acidification, i.e., reduced bicarbonate concentration, lactate addition, and HCl addition, enabled us to investigate the pH-response mechanism thoroughly and to compare the cellular response to other tumor microenvironmental conditions such as hypoxia or nutrient starvation.
The key of this method is to determine the appropriate concentrations of bicarbonate, lactate, and HCl in the culture medium. Measuring the medium pH with various bicarbonate concentrations when performing this method for the first time is important to determine the appropriate amount of NaHCO3 in the medium. During this process, it is necessary to measure the medium pH after 24 h of incubation at 37 °C under 5% CO2, as medium temperature and equilibrium status of CO2 greatly affect the medium pH. Autoclaving of bicarbonate is frequently recommended, as bicarbonate easily becomes flat. Cell density and cell confluency are crucial factors because large numbers of growing cancer cells easily acidify the culture medium even in control samples. In addition, responses to the extracellular acidic pH depend on the condition of the cells and may fade as cells are passaged, so cells with less than 10 passages were used for the experiments.
Our method has significantly demonstrated the stability of medium pH for 24 h and compared several acidic pH media. In this protocol, medium pH of 6.8 was used as a low pH condition, but serial acidic pH between pH 6.4 and 6.9, in addition to serial basic pH media between pH 7.4 and 8.0, were used to investigate cellular response against extracellular acidification.
We mainly focused on cancer cells, but this method can be used for other types of cells within the tumor microenvironment, including stromal cells, immune cells, and vascular endothelial cells. We and others reported that at least some pH sensitive mechanisms in cancer cells are common in normal cells7. It is also possible to apply this method for the analysis of other primary cells as well as embryonic stem cells and induced pluripotent stem cells. One limitation of this culture system could be the difficulty of maintaining the acidic pH condition in long-term experiments.
Acidosis is associated with various types of diseases. This method can be applicable not only in cancer research but also in studying acidic disorders such as diabetic ketoacidosis, renal tubular acidosis, and respiratory acidosis.
Disclosures
The author Ayano Kondo is an employee of Kyowa Hakko Kirin Co., Ltd.
Acknowledgments
We thank the members of the Division of Genome Science and Laboratory for Systems Biology and Medicine, RCAST, The University of Tokyo. We especially thank Dr. H. Aburatani, Dr. T. Kodama, (LSBM, RCAST, The University of Tokyo), Dr. K. Tomizuka, Dr. T. Yoshida, and Dr. A. Kunisato (Kyowa Hakko Kirin Co., Ltd.) for helpful discussions and support. This work was partly supported by Grant-in-Aid for Young Scientist (A) (26710005, T.O.), Grant-in-Aid for Scientific Research on Innovative Areas (26116711 and 16H01567, T.O.), and Grant-in-Aid for Challenging Exploratory Research (16K14605, T.O.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, the Takeda Science Foundation (T.O.), the Kobayashi Foundation of Cancer Research (T.O.), and the Project for Cancer Research and Therapeutic Evolution (P-CREATE) and the Practical Research for Innovative Cancer Control from Japan Agency for Medical Research and Development, AMED (T.O.).
References
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nature reviews. Cancer. 2011;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- Chang CH, et al. Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell. 2015;162(6):1229–1241. doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134(5):703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- Gerweck LE, Seetharaman K. Cellular pH gradient in tumor versus normal tissue: potential exploitation for the treatment of cancer. Cancer research. 1996;56(6):1194–1198. [PubMed] [Google Scholar]
- Webb BA, Chimenti M, Jacobson MP, Barber DL. Dysregulated pH: a perfect storm for cancer progression. Nature Rev. Cancer. 2011;11(9):671–677. doi: 10.1038/nrc3110. [DOI] [PubMed] [Google Scholar]
- Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nature reviews. Cancer. 2004;4(11):891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- McBrian MA, et al. Histone acetylation regulates intracellular pH. Mol Cell. 2013;49(2):310–321. doi: 10.1016/j.molcel.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damaghi M, et al. Chronic acidosis in the tumour microenvironment selects for overexpression of LAMP2 in the plasma membrane. Nat Commun. 2015;6:8752. doi: 10.1038/ncomms9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbet C, et al. Acidosis Drives the Reprogramming of Fatty Acid Metabolism in Cancer Cells through Changes in Mitochondrial and Histone Acetylation. Cell Metab. 2016;24(2):311–323. doi: 10.1016/j.cmet.2016.07.003. [DOI] [PubMed] [Google Scholar]
- Chen JL, et al. The genomic analysis of lactic acidosis and acidosis response in human cancers. PLoS genetics. 2008;4(12):e1000293. doi: 10.1371/journal.pgen.1000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo A, et al. Extracellular Acidic pH Activates the Sterol Regulatory Element-Binding Protein 2 to Promote Tumor Progression. Cell Rep. 2017;18(9):2228–2242. doi: 10.1016/j.celrep.2017.02.006. [DOI] [PubMed] [Google Scholar]


