Summary
Background
Restless legs syndrome is a prevalent chronic neurological disorder with potentially severe mental and physical health consequences. Clearer understanding of the underlying pathophysiology is needed to improve treatment options. We did a meta-analysis of genome-wide association studies (GWASs) to identify potential molecular targets.
Methods
In the discovery stage, we combined three GWAS datasets (EU-RLS GENE, INTERVAL, and 23andMe) with diagnosis data collected from 2003 to 2017, in face-to-face interviews or via questionnaires, and involving 15 126 cases and 95 725 controls of European ancestry. We identified common variants by fixed-effect inverse-variance meta-analysis. Significant genome-wide signals (p≤5 × 10−8) were tested for replication in an independent GWAS of 30 770 cases and 286 913 controls, followed by a joint analysis of the discovery and replication stages. We did gene annotation, pathway, and gene-set-enrichment analyses and studied the genetic correlations between restless legs syndrome and traits of interest.
Findings
We identified and replicated 13 new risk loci for restless legs syndrome and confirmed the previously identified six risk loci. MEIS1 was confirmed as the strongest genetic risk factor for restless legs syndrome (odds ratio 1·92, 95% CI 1·85–1·99). Gene prioritisation, enrichment, and genetic correlation analyses showed that identified pathways were related to neurodevelopment and highlighted genes linked to axon guidance (associated with SEMA6D), synapse formation (NTNG1), and neuronal specification (HOXB cluster family and MYT1).
Interpretation
Identification of new candidate genes and associated pathways will inform future functional research. Advances in understanding of the molecular mechanisms that underlie restless legs syndrome could lead to new treatment options. We focused on common variants; thus, additional studies are needed to dissect the roles of rare and structural variations.
Funding
Deutsche Forschungsgemeinschaft, Helmholtz Zentrum München–Deutsches Forschungszentrum für Gesundheit und Umwelt, National Research Institutions, NHS Blood and Transplant, National Institute for Health Research, British Heart Foundation, European Commission, European Research Council, National Institutes of Health, National Institute of Neurological Disorders and Stroke, NIH Research Cambridge Biomedical Research Centre, and UK Medical Research Council.
Introduction
Despite the prevalence of restless legs syndrome being up to 10% in populations of European ancestry, its genetic basis and underlying pathophysiology remain unclear. The restless legs syndrome phenotype is an unusual composite of sensory and motor symptoms that present with distinct circadian rhythmicity. The symptoms worsen or are only present in the evening or at night and markedly lessen in the early morning. Patients feel an overwhelming urge to move, often in conjunction with unpleasant sensations, usually in the legs. Rest and inactivity provoke the symptoms, whereas movement and other external stimuli lead to temporary relief.1 Due to the chronic progressive nature of the disorder, it has long-lasting effects on patients' mental and physical health. People with restless legs syndrome have substantially impaired sleep, reduced overall quality of life, and increased risk of depression, anxiety disorders, hypertension, and, possibly, cardiovascular disease.2 Around 2–3% of the general population have severe restless legs syndrome, and most need chronic treatment with dopaminergics, α2δ ligands, or even opioids.1 However, long-term use of dopaminergics can lead to severe side-effects, including the worsening of symptoms (augmentation). Hence, there is an urgent need for alternative treatments.
Research in context.
Evidence before this study
We searched PubMed for articles published up to July, 2017, with combinations of the search term “restless legs AND (genomewide OR genome-wide OR GWAS)”, without restrictions on language of publication. This search yielded 42 original articles, including reports on genetic linkage analyses and genome-wide association studies (GWASs). Although linkage results on restless legs syndrome have not been reproducible, GWASs so far have revealed six risk loci, with the strongest signal being in MEIS1, a member of the three aminoacid loop extension homeobox gene class. Subsequent studies have suggested altered embryonic development of the striatum is also important in the aetiology of restless legs syndrome. Nevertheless, the pathogenesis needs further elucidation. Changes in dopaminergic signalling and brain iron deficiency seem to be involved and are targeted by approved drugs. Other suggested causes include peripheral hypoxia and unknown metabolic factors related to uraemia and pregnancy. The current recommended treatment for restless legs syndrome, although effective, can lead to serious adverse effects, including worsening of symptoms, thus alternatives are needed.
Added value of this study
We did a meta-analysis of three GWAS datasets, which yielded a total sample size more than one order of magnitude larger than any previously published restless legs syndrome GWAS. We discovered 13 new risk loci for restless legs syndrome, taking the total from six to 19. Assessment of these candidate genes will enable more granular dissection of the pathogenesis of restless legs syndrome, which could improve determination of shared genetic architecture with other neurological phenotypes and the prospects of developing novel and more effective treatment options. Our results strongly support the link to neurogenesis, changes in neuronal circuit formation, synaptogenesis, and axonal guidance, thereby strengthening the concept of restless legs syndrome as a neurodevelopmental disorder. The novel risk loci include the genes CRBN, which encodes cereblon, and MEIS2, which encodes its physiological substrate homeobox protein Meis2. As thalidomide targets this interaction, this drug could be a candidate for the treatment of restless legs syndrome in patients beyond reproductive age.
Implications of all the available evidence
The genes and associated pathways identified provide a much-needed basis for future investigation of restless legs syndrome, informing which pathophysiological and pharmacological concepts to examine in laboratory and clinical trials. Our findings further suggest investigating the role of neurodevelopmental processes in restless legs syndrome and the mechanism of the interaction between cereblon and homeobox protein Meis2 in the context of assessing the repurposing of thalidomide.
The likelihood of developing symptoms of restless legs syndrome is strongly affected by genetic factors. Family and twin studies have estimated that heritability is 50–60%.3 Individual genetic risk variants and their putative target genes, however, were identified only when genome-wide association studies (GWASs) became feasible. Six risk loci have so far been identified in this way,4, 5, 6, 7 and have notably shaped research by uncovering potential new pathophysiological mechanisms. Genes in these loci provided reliable entry points for functional investigations at the molecular level and for animal studies.8, 9, 10, 11
Given these genetically driven advances, we did a meta-analysis of GWAS on restless legs syndrome in people of European ancestry, followed by replication in an independent dataset. We hypothesised that this approach would enable us to detect novel risk loci and pathways associated with restless legs syndrome that would provide further insights into the molecular mechanisms underlying the disorder, and yield possible novel therapeutic targets or avenues for the repurposing of existing drugs.
Methods
Study populations and phenotype definitions
For the discovery meta-analysis, we combined three GWAS datasets. The EU-RLS-GENE consortium GWAS includes cases and population-matched controls recruited in eight European countries, Canada, and the USA. People with restless legs syndrome were recruited in specialist outpatient clinics for movement disorders and in sleep units. Restless legs syndrome was diagnosed in face-to-face interviews by an expert neurologist, based on the International Restless Legs Syndrome Study Group diagnostic criteria.1, 12 A subset of the samples had been used in previous GWASs for restless legs syndrome (appendix p 10).4, 6, 7
The INTERVAL study13, 14 included whole-blood donors recruited in England, enrolled between 2012 and 2014. The validated Cambridge-Hopkins Restless Legs Questionnaire15 was used to identify patients with restless legs syndrome and to exclude mimicking disorders. Probable and definite cases were combined in one group.
The 23andMe GWAS dataset comprised samples drawn from participants of the customer base of 23andMe (Mountain View, CA, USA), which is a genetic testing company. The restless legs syndrome phenotype was determined with one research question, “Have you ever been diagnosed with restless legs syndrome?”, which had three response options: yes, no, and not sure. We include respondents who answered “yes” as cases and those who answered “no” as controls, and we excluded those who answered “not sure”.
For the replication study, we used the 23andMe customer base to identify a distinct population. We used the same selection methods as for the discovery population.
Participants of all the original studies had provided informed consent, and the study protocols had been approved by the respective ethics committees. Demographics of the study samples are provided in the appendix (p 10).
Genotyping, quality control, imputation, and statistical analysis
Genotyping, quality-control procedures, imputation, and statistical analysis methods of the individual studies are described in detail in the appendix (pp 4–5, pp 11–12). In brief, all studies did association analyses under an additive model by logistic regression on genotype dosage, adjusted for age, sex, and components from either multidimensional scaling or principal components analysis to correct for population stratification.
Meta-analysis procedures
For the discovery stage, summary statistics from each individual GWAS dataset were subjected to further quality control with EasyQC, version 8.5, and we did a fixed-effect inverse-variance meta-analysis with METAL, release 2011-03-25 (appendix p 5). To address heterogeneity between studies in the identified association signals, we did a random-effects meta-analysis with METASOFT, version 2.0.1 (appendix p 5). Genomic control was done in each study separately before meta-analysis by calculating the inflation factor λ and adjusting for it.16
To define independent genome-wide significant signals (p≤5 × 10−9) in the discovery meta-analysis results, we used a two-step procedure (appendix p 5). Briefly, we first assigned variants to clusters with the “clump” command in PLINK software (version 1.90b3.36) based on the association p value and short-range (500 kb) linkage disequilibrium (LD), with each clump defined by one lead single-nucleotide polymorphism (SNP). Second, we tested for statistical independence of the lead SNPs with the stepwise model selection procedure implemented in GCTA, version 1.25.3, taking into account long-range LD (10 Mb) between lead SNPs (appendix p 5). Finally, we did standard conditional analysis in the remaining independent SNP clusters, adjusted for the top-associated lead SNP, to identify secondary independent signals within the cluster.
For the joint analysis of the discovery and replication stages, summary statistics for independent association signals were subjected to fixed-effect inverse-variance and random-effects meta-analysis, as in the discovery stage (appendix p 5).
Heritability, partitioned heritability, and genetic correlation analysis
Heritability and partitioned heritability were estimated by LD-score regression (with LDSC, version 1.0.0) and using the summary statistics of the discovery meta-analysis without genomic control correction. Partitioned heritability analysis used publicly available partitioned LD scores for 52 functional categories based on the phase 3 dataset of the 1000 Genomes Project, as precomputed by the developers of LDSC. For the genetic correlation analysis, we used the LD Hub database (version 1.2.2), which provides access to summary-level GWAS statistics of more than 200 traits. The analytical procedures and settings are detailed in the appendix (pp 5–6). To estimate the variance explained by genome-wide significant association signals, we calculated Nagelkerke's pseudo-R2 in the EU-RLS-GENE dataset with tenfold cross validation.
Genetic risk score analysis
Genetic risk profiles were generated by estimating weighted polygenic risk scores in the EU-RLS-GENE dataset. The weights were based on the effect size estimates of the discovery meta-analysis (fixed-effect model). Polygenic risk scores were calculated with PRSice software, version 1.25 (appendix p 6).
Biological interpretation of association signals
As a first step, we did a literature-based annotation of protein-coding genes located in the genome-wide significant risk loci, defined based on LD, and in their vicinity (appendix p 6). To identify the biological and cellular pathways underlying the association signals, we applied bioinformatic methods for gene prioritisation and did enrichment analyses for pathways and tissues. In restless legs syndrome, the overall number of risk loci is small, restricting the use of standard tools, such as DEPICT.17 Moreover, the enrichment analysis could be hampered by the clinical heterogeneity of restless legs syndrome, its complex phenotype with somatosensory and motor symptoms, and pleiotropic effects of the candidate genes. Therefore, to enable efficient exploration of our data, we developed a new algorithm, called BI-ENRICH, which builds on the concept of biclustering used in gene-expression analysis (appendix pp 6–8). Additionally, we used the DEPICT software (version rel19413) to do gene prioritisation for each locus and to search for gene set and tissue enrichment among these genes (appendix p 8).
Data sharing
BI-ENRICH code is available on GitHub. GWAS summary statistics will be made available for researchers (appendix p 3).
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
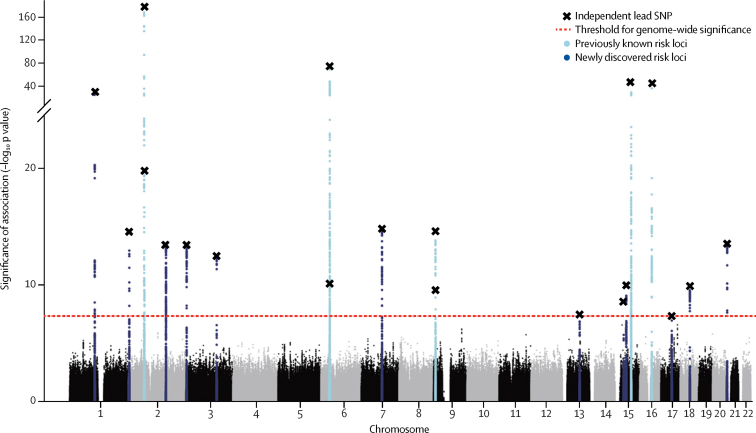
The discovery stage association study was done in 15 126 cases and 95 725 controls: 6228 and 10 992 from EU-RLS-GENE; 3065 and 24 923 from INTERVAL; and 5833 and 59 810 from 23andMe. After quality control, 6 864 281 SNPs and indels with minor allele frequency 1% or greater were available for statistical analysis. We identified 20 independent association signals meeting genome-wide significance (figure 1, table 1, appendix p 13). No secondary independent association signals with genome-wide significance were found in these 20 regions by standard conditional analysis. We assigned the 20 independent association signals to 19 independent genomic risk loci (two mapped to the same gene, PTPRD, and are <500 kb apart; appendix p 65). Lead SNPs rs1836229 and rs62535767 of these two signals were not correlated (r2=0·0002, EU-RLS-GENE dataset). Genomic control showed negligible inflation of the median test statistic (λ1000=1·004, rescaled to adjust for the large sample size), which LD-score regression revealed was mainly due to polygenicity (intercept 1·018), as is expected for a common complex disorder, rather than being caused by population stratification or other confounders. We observed between-study heterogeneity at some loci, but all signals kept genome-wide significance in the random-effects meta-analysis (appendix p 14).
Figure 1.
Manhattan plot showing results of the discovery meta-analysis
Due to their close proximity, two independent risk loci on chromosome 2 (both previously known) and on chromosome 6 (one previously known, one new) appear as single peaks. Thus, 19 loci (six previously known and 13 new) are represented. SNP=single-nucleotide polymorphism.
Table 1.
Association results for lead single-nucleotide polymorphisms reaching genome-wide significance in the discovery meta-analysis
| Chromosome | Position (bp) | Effect allele | Other allele | Effect-allele frequency | Protein-coding gene context |
Discovery stage meta-analysis |
Replication stage |
Joint stage meta-analysis |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p value | Odds ratio (95% CI) | p value | Odds ratio (95% CI) | p value | Odds ratio (95% CI) | |||||||
| rs12046503 | 1 | 107195339 | T | C | 0·59 | PRMT6*†, NTNG1†‡ | 3·32 × 10−31 | 0·85 (0·84–0·87) | 2·03 × 10−29 | 0·90 (0·89–0·92) | 3·25 × 10−63 | 0·88 (0·86–0·89) |
| rs10208712 | 2 | 4034446 | G | A | 0·36 | DCDC2C*† | 3·78 × 10−15 | 0·90 (0·88–0·91) | 7·74 × 10−19 | 0·92 (0·91–0·94) | 1·41 × 10−34 | 0·91 (0·90–0·92) |
| rs113851554 | 2§ | 66750564 | T | G | 0·07 | MEIS1*†‡¶ | 1·1 × 10−180 | 2·16 (2·04–2·29) | 4·80 × 10−236 | 1·82 (1·75–1·89) | 2·00 × 10−280 | 1·92 (1·85–1·99) |
| rs1820989 | 2§ | 68069890 | C | A | 0·53 | MEIS1¶, C1D, *APLF† | 1·23 × 10−20 | 0·88 (0·86–0·90) | 1·98 × 10−39 | 0·89 (0·87–0·90) | 1·39 × 10−58 | 0·88 (0·87–0·90) |
| rs80319144 | 2 | 159199835 | T | C | 0·24 | CCDC148*‡, PKP4†‡, TANC1‡ | 3·18 × 10−14 | 0·89 (0·85–0·92) | 1·40 × 10−22 | 0·90 (0·89–0·92) | 2·55 × 10−26 | 0·90 (0·88–0·92) |
| rs1848460 | 3 | 3448144 | T | A | 0·26 | CNTN4‡, CRBN*‡, LRRN1‡ | 5·38 × 10−14 | 1·13 (1·08–1·17) | 1·93 × 10−9 | 1·06 (1·04–1·08) | 2·01 × 10−13 | 1·07 (1·05–1·10) |
| rs35987657 | 3 | 130535567 | G | A | 0·33 | ATP2C1*‡, ASTE1† | 4·37 × 10−13 | 0·90 (0·88–0·91) | 3·34 × 10−23 | 0·91 (0·90–0·93) | 3·96 × 10−38 | 0·90 (0·89–0·92) |
| rs17636328 | 6 | 37490531 | G | A | 0·20 | RNF8†, CCDC167*, MDGA1†‡ | 6·43 × 10−11 | 0·89 (0·85–0·92) | 7·63 × 10−18 | 0·90 (0·89–0·92) | 2·55 × 10−26 | 0·90 (0·88–0·92) |
| rs61192259 | 6§ | 38453962 | A | C | 0·59 | BTBD9*†‡¶, GLO1† | 1·36 × 10−78 | 1·31 (1·28–1·34) | 1·05 × 10−112 | 1·22 (1·20–1·25) | 3·58 × 10−202 | 1·26 (1·25–1·28) |
| rs10952927 | 7 | 88359060 | G | A | 0·13 | ADAM2‡, STEAP4‡, ZNF804B*‡ | 1·86 × 10−15 | 1·17 (1·13–1·22) | 5·01 × 10−17 | 1·12 (1·09–1·14) | 1·73 × 10−34 | 1·13 (1·11–1·15) |
| rs1836229 | 9§ | 8820573 | G | A | 0·48 | PTPRD*†‡ | 1·94 × 10−15 | 0·90 (0·88–0·91) | 1·57 × 10−29 | 0·90 (0·89–0·92) | 7·36 × 10−42 | 0·90 (0·89–0·91) |
| rs62535767 | 9§ | 9290311 | T | C | 0·32 | PTPRD*†‡ | 3·13 × 10−10 | 0·91 (0·88–0·95) | 8·77 × 10−7 | 0·95 (0·93–0·97) | 3·23 × 10−9 | 0·94 (0·93–0·96) |
| rs340561 | 13 | 72848156 | T | G | 0·20 | DACH1*†‡, DIS3† | 3·93 × 10−8 | 1·09 (1·05–1·14) | 4·91 × 10−7 | 1·05 (1·03–1·07) | 3·23 × 10−9 | 1·06 (1·04–1·08) |
| rs996064 | 15 | 36208998 | T | A | 0·06 | DPH6*, MEIS2‡ | 2·96 × 10−9 | 1·21 (1·14–1·28) | 5·45 × 10−21 | 1·22 (1·17–1·27) | 3·39 × 10−27 | 1·22 (1·17–1·26) |
| rs111652004 | 15 | 47360367 | T | G | 0·10 | SEMA6D*†‡ | 1·05 × 10−10 | 0·84 (0·80–0·89) | 3·83 × 10−17 | 0·87 (0·84–0·90) | 2·69 × 10−16 | 0·86 (0·83–0·89) |
| rs868036 | 15§ | 68055013 | T | A | 0·32 | SMAD3†, MAP2K5*¶, SKOR1¶, CLN6† | 1·09 × 10−48 | 0·80 (0·77–0·83) | 9·23 × 10−70 | 0·85 (0·84–0·87) | 5·48 × 10−69 | 0·84 (0·83–0·86) |
| rs45544231 | 16§ | 52632730 | G | C | 0·42 | TOX3*‡¶ | 4·72 × 10−48 | 0·81 (0·79–0·83) | 4·36 × 10−87 | 0·84 (0·83–0·86) | 7·27 × 10−133 | 0·83 (0·81–0·84) |
| rs12450895 | 17 | 46772776 | A | G | 0·21 | HOXB cluster†‡, PRAC1* | 4·87 × 10−8 | 1·09 (1·05–1·14) | 2·01 × 10−10 | 1·07 (1·05–1·09) | 4·27 × 10−14 | 1·08 (1·06–1·10) |
| rs12962305 | 18 | 41870243 | T | C | 0·25 | SETBP1*‡ | 1·37 × 10−10 | 1·11 (1·06–1·15) | 6·59 × 10−5 | 1·04 (1·02–1·06) | 1·11 × 10−7 | 1·05 (1·03–1·07) |
| rs365032 | 20 | 62795405 | G | A | 0·27 | MYT1*†‡ | 3·36 × 10−14 | 1·13 (1·08–1·17) | 7·83 × 10−36 | 1·13 (1·11–1·15) | 1·73 × 10−34 | 1·13 (1·11–1·15) |
All p values were obtained by fixed-effect inverse-variance meta-analyses. The threshold for genome-wide significance was p≤5 × 10−8. For each locus, only the selected protein-coding genes are listed by genomic position (direction 5′ to 3′ on chromosome). GWAS=genome-wide association study. Position=GRCh37/hg19 coordinates. bp=base pair.
Selected by nearest gene.
Selected by BI-ENRICH prioritisation (nominal p<0·05).
Selected by manual annotation.
Association data for all 20 signals were obtained from the replication dataset, which included 30 770 cases and 286 913 controls (appendix p 10). All 20 association signals of the discovery stage were replicated, three at a Bonferroni-corrected level of p<0·0025, and 17 meeting genome-wide significance (table 1).
In the joint analysis of discovery and replication stages, all loci but one had genome-wide significance in the fixed-effect meta-analysis (table 1). All loci in the joint random-effects meta-analysis had genome-wide significance (appendix p 15). Overall, 13 new risk loci were identified and all six known genomic risk loci for restless legs syndrome were replicated (figure 1, table 1). The MEIS1 locus on chromosome 2 was confirmed as the strongest genetic risk factor for restless legs syndrome (table 1). The lead SNP, rs113851554, located in a putative regulatory element in intron eight of MEIS1, is a low-frequency variant with odds ratio (OR) estimates of 1·82–2·16, which clearly distinguish it from the other risk loci for restless legs syndrome. The amount of heritability of restless legs syndrome attributable to all SNPs available in our dataset in the LD-score regression was 19·6%. Focusing on the 20 independent association signals, these explained 11·7% of the observed variance (60% of the SNP heritability). In the assessment of distribution of genetic risk, calculated from polygenic risk scores based on the 20 association signals (appendix p 6), individuals in the highest risk group (polygenic risk score >4·4, 99·5% quantile), had a significantly increased risk of restless legs syndrome (OR 17·6, 95% CI 8·5–42·3, p=6·9 × 10−26). As this group represents only a very small subset of the population, we also compared the genetic risk profiles of the 25% quantile (polygenic risk score <2·48) and the 75% quantile (polygenic risk score >3·15) of our population, which gave an OR of 5·9 (95% CI 5·3–6·5, p=8·5 × 10−304). The distributions of the polygenic risk scores, however, substantially overlapped between the cases and controls, which precluded the use of these scores for risk prediction (appendix p 43).
To identify biological pathways related to restless legs syndrome, we annotated protein-coding genes in or close to the genomic risk loci we identified. Six loci contained only one gene and had no genes in the immediate vicinity: three of the newly identified loci, SEMA6D (15q21·1), SETBP1 (18q12·3), MYT1 (20q13·33), and three of the previously described genes, MEIS1 (2p14), PTPRD (9p24·1–p23), and TOX3 (16q12·1). These genes function in axonal pathfinding and signalling, synaptogenesis, neuronal differentiation, and neurogenesis (table 2). Most of the remaining loci also contained genes linked to neurodevelopment, among others (table 2). A basic annotation for additional genes linked to the risk loci and regional association plots for all loci are provided in the appendix (pp 16–19, pp 44–86).
Table 2.
Candidate genes linked to functions in neurodevelopment
| Genes | Functions related to neurodevelopment | |
|---|---|---|
| rs12046503 | NTNG1 | Presynaptic cell-adhesion molecule involved in synapse formation18 |
| rs10208712 | DCDC2C | Encodes neuronal migration protein doublecortin, a member of the DCX protein family of cell-adhesion molecules; unknown function, but other members of the DCX family act in neuronal migration and axonal growth and have been linked to neurological and developmental disorders19 |
| rs113851554 rs1820989 |
MEIS1 | Implicated in neurogenesis, specification of neuronal cell type, and establishing connectivity between neurons and their target field; binds HOX proteins of all paralogue groups, participates in controlling HOX gene expression20 |
| rs80319144 | PKP4 | Encodes the cell-adhesion molecule plakophilin-4, which serves as a scaffold for signalling complexes and plays a part in cell adhesion and neurite outgrowth21 |
| rs1848460 | CRBN, CNTN4 | Cereblon, encoded by CRBN, is the substrate receptor of a Cullin4a RING E3 ubiquitin ligase and regulates assembly and expression of calcium-activated potassium channels in the brain;22, 23 contactin-4, encoded by CNTN4, is a cell-adhesion molecule with an important role in axon guidance, synapse formation, and neuronal network plasticity24 |
| rs17636328 | MDGA1 | Encodes MAM domain-containing glycosylphosphatidylinositol anchor protein 1, which is a trans-synaptic cell-adhesion molecule implicated in synapse development25, 26 |
| rs10952927 | ZNF804B, ADAM22 | ZNF804B, which is highly homologous to ZNF804A, has been associated with schizophrenia and bipolar disorder;27 ADAM22 is a synaptic receptor involved in synaptic transmission and synaptic disorders28 |
| rs1836229 rs62535767 |
PTPRD | Related to functions in axon guidance and synaptogenesis, especially in the formation of excitatory synapses29, 30 |
| rs340561 | DACH1 | Dach1 is a transcription factor acting as a neurogenic cell-fate determining factor31 |
| rs996064 | MEIS2 | Involved in neurogenesis and contributes to determination of dopaminergic-cell fate; binds HOX proteins of all paralogue groups and participates in controlling expression of HOX genes32 |
| rs111652004 | SEMA6D | Involved in axonal pathfinding and signalling; exerts repulsive or attractive effects on axons, depending on the specific combinations of its main receptor with co-receptors; SEMA6D knockout mice show misdirection of proprioceptive axons and their associated oligodendrocytes in the dorsal horn, affecting proper synapse formation33, 34 |
| rs45544231 | TOX3 | Implicated in neurogenesis, specification of neuronal cell type, and establishing connectivity between neurons and their target fields35 |
| rs12450895 | HOXB cluster family | Assign positional identities to neurons along the rostrocaudal axis in hindbrain and spinal cord, which is crucial in the specification of neural subpopulations and their target cells; mouse models show the necessity of Hoxb genes for correct neuronal specification, migration, and circuit formation36, 37 |
| rs365032 | MYT1 | Myt1 kinase is a transcription factor expressed in neural progenitor cells in the central and peripheral nervous systems; involved in neuronal differentiation by suppressing neural progenitor fate and promoting neurogenesis38 |
Only risk loci with candidate genes linked to functions in neurodevelopment are listed. Annotation of the genes was done as described in the appendix (p 6).
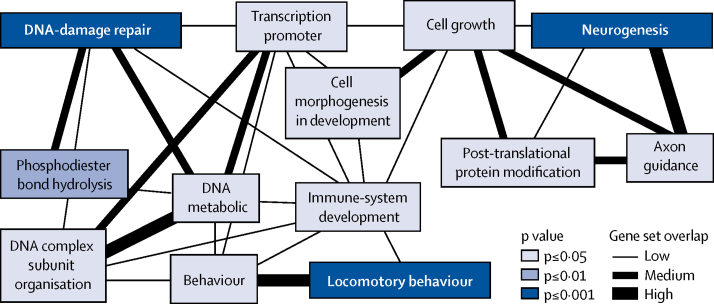
These observations were substantiated by bioinformatic enrichment and gene prioritisation analyses. The BI-ENRICH algorithm showed that most of the top pathways identified for restless legs syndrome were related to neurodevelopment, including neurogenesis (genes MDGA1, MYT1, NTNG1, and SEMA6D), cell-junction organisation (PKP4 and SMAD3), and axon guidance (NTNG1 and SEMA6D). Moreover, locomotor behaviour (BTBD9, CLN6, HOXB8, and MEIS1) was highlighted (figure 2, appendix pp 20–22). Pathways related to DNA repair and maintenance (APLF, ASTE1, DIS3, PRMT6, and RNF8) were also detected by the enrichment analysis. Consistent with these results, BI-ENRICH prioritised genes related to neurodevelopment and DNA-damage repair (appendix pp 23–30). Use of the standard version of DEPICT returned no significant results (appendix pp 31–32). However, including the UniProtKB biological process annotations in the gene-set definitions, genes prioritised by DEPICT were enriched for neurogenesis (false discovery rate <0·05, appendix p 33). Finally, we did stratified LD-score regression to partition the heritability carried by all SNPs to specific functional categories. 14·8% of the SNPs that reside in regions associated with histone marks in the CNS explain 44·7% of the variance. This CNS enrichment is significant (enrichment factor 44·7/14·8=3·0, p=0·0043). Consistent with this observation, tissue enrichment analyses with BI-ENRICH and DEPICT ranked brain and spinal cord tissues at the top, but false discovery rates were greater than 0·2, and only spinal-cord tissue (BI-ENRICH) and mesencephalon (DEPICT) were significant (appendix pp 34–37).
Figure 2.
Representation of significantly enriched functional gene sets found by the BI-ENRICH analysis
Similarities between gene sets are measured with the Jaccard index, with low being J<0·3, medium being J≥0·3 to <0·5, and high being J≥0·5. Empirical p values are shown.
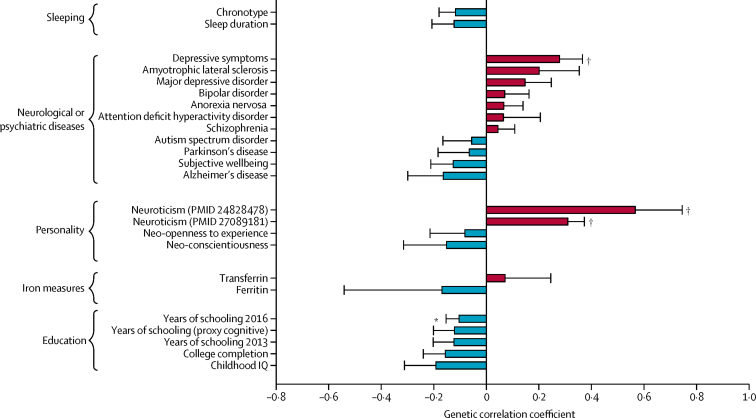
Finally, we used LD-score regression to assess the genetic correlation between restless legs syndrome and diseases and traits of interest (figure 3, appendix p 38). Restless legs syndrome showed some positive correlations with neuropsychiatric diseases, whereas correlations were mainly negative with neurodegenerative disorders (figure 3), although none reached nominal significance. Few supporting data are available from corresponding epidemiological studies. Two small studies showed increased prevalence of restless legs syndrome among people with amyotrophic lateral sclerosis.39, 40 One study showed prevalence of 4% among people with Alzheimer's disease, which is lower than the average 6–12% in elderly people of European descent,41, 42 but making a diagnosis of restless legs syndrome in cognitively impaired individuals is challenging and the findings are difficult to interpret. Data on restless legs syndrome and Parkinson's disease were inconclusive, showing wide-ranging prevalence (0–50%), with some studies proposing restless legs syndrome as a risk factor for Parkinson's disease and others showing protective effects.43 Moreover, dopaminergic treatment of Parkinson's disease has been suggested to precipitate restless legs syndrome in some patients.44 The risk locus on chromosome 15q23 (encompassing MAP2K5 and SKOR1) overlaps with GWAS signals of four different traits, including the posterior cortical atrophy variant of Alzheimer's disease (appendix pp 39–41). The ORs of the shared SNP rs11637445 were 1·48 for posterior cortical atrophy and 0·89 for restless legs syndrome, which is consistent with the negative correlation between these two disorders. We found a significant positive correlation between restless legs syndrome and depressive symptom phenotypes and with neuroticism (figure 3, appendix p 38). These genetic correlations are in line with epidemiological associations, which show increased prevalence of these phenotypes in patients with restless legs syndrome.42, 45 Overall, the correlations between restless legs syndrome and neurological or psychiatric phenotypes were low to moderate. We found a positive genetic correlation with number of children (r=0·22, p=0·0079). Pregnancy is a validated risk factor for restless legs syndrome, and parity increases the risk of developing restless legs syndrome in later life.46 Sleep-related phenotypes and iron-related traits showed correlations in line with published data,1, 47 but none reached nominal significance. Finally, negative genetic correlations were generally seen between restless legs syndrome and measures of educational attainment (figure 3), although this association does not seem to be widely reported.
Figure 3.
Genetic correlation between restless legs syndrome and other traits
Data are mean (SE) correlations, based on linkage disequilibrium score regression in LD-Hub (appendix pp 5–6). PMID=PubMed article unique identifier. *p<0·05. †p<0·005.
Discussion
This association study identified 20 independent association signals that reached genome-wide significance in 19 risk loci, of which 13 had not been previously reported. A subset of these signals showed between-study heterogeneity in our meta-analyses; in a random-effects meta-analysis that accounted for heterogeneity, all reached genome-wide significance. Effect-size estimates and strength of the association signal in these loci varied between studies, probably because of differences in phenotyping methods (clinical face-to-face interview, self-reporting by participants via a validated questionnaire, and self-reporting by use of one question). These differences can lead to variation in the proportions of misclassified cases and controls and might have modified some of the association signals. Moreover, the specific features of the restless legs syndrome phenotype, such as disease severity or preponderance of motor or sensory symptoms, might have varied between the studies. Detailed clinical data were scarce, and the restless legs syndrome phenotypes in the INTERVAL and 23andMe datasets were not clinically validated and, therefore, we cannot further address the underlying causes of heterogeneity. Studies including detailed assessments of restless legs syndrome symptoms are needed to dissect the specific roles of and possible interactions between genetic variants in the risk loci and the resulting phenotypes.
Together, the 20 association signals accounted for 60% of the SNP-based heritability of 19·6% estimated in this study. Our dataset was limited to SNPs and small indels with minor allele frequency of 1% or greater. Therefore, we could not assess the contribution of rarer or structural variants, which are likely to have larger effect sizes and might explain another part of the heritability of restless legs syndrome. Future large-scale whole-genome sequencing efforts might have the power to detect such variants.
The main aim of our study was to provide new clues to understanding the biology of restless legs syndrome. Candidate genes and pathway analyses across the 19 risk loci converge on functions important in the development of the CNS, such as neurogenesis and neural-circuit formation, including axon guidance and synaptogenesis. Additionally, the BI-ENRICH analysis highlighted DNA-damage repair, which is important for development and maintenance of the nervous system, as having a relevant role.48, 49 The involvement of perturbations in neurodevelopmental processes is in line with previous functional studies of MEIS1 that identified the embryonic ganglionic eminences as relevant structures of restless legs syndrome biology.50 Our pathway analysis was designed to have high sensitivity to avoid downward bias, such as false-negative results due to incomplete information provided by the annotation databases. To avoid upward bias (ie, false-positive enrichment results), we corrected by sampling and null-GWAS permutation (phenotype label permutation). Significance of results in the tissue enrichment analyses might have been hindered by a lack of appropriate samples from relevant anatomical regions or developmental stages in the input datasets. Nonetheless, neuronal tissues were consistently prioritised, and nominally significant regions, such as midbrain structures and spinal cord, have previously been implicated in restless legs syndrome.51
At present, major concepts in the pathophysiology of restless legs syndrome address alterations in the dopaminergic neurotransmitter system and in brain iron metabolism, and are supported by evidence from animal, imaging, and human post-mortem studies.47 Even though our enrichment analyses did not specifically highlight corresponding biological processes, the new risk loci offer potential bases for functional studies that might shed light on mechanisms underlying the suspected changes of the dopaminergic neurotransmitter system or brain iron metabolism. Moderate, albeit non-significant, positive genetic correlations are being reported with neuropsychiatric disorders, such as schizophrenia or bipolar disorder, for which evidence of contributing pathological events that affect early neurodevelopment and the correct setup of neuronal circuitry is growing.52, 53
Finally, annotation of the risk loci identified a gene with a product targeted by an existing drug that is readily available for repurposing. The ubiquitin ligase substrate receptor, cereblon, which is encoded by CRBN (locus on chromosome 3p26) is a target of the drug thalidomide, and homeobox protein Meis2, which is encoded by MEIS2 (locus on chromosome 15q14), has been identified as an endogenous substrate of cereblon.33 Thalidomide was initially licensed as a hypnotic, but was withdrawn from the market because of teratogenicity. Thalidomide and its analogues are now used as immunomodulatory drugs in cancer. They block binding of homeobox protein Meis2 to the ubiquitin ligase, thereby modulating its activity.20 Thalidomide and thalidomide-like substances, therefore, could be potential candidates for therapeutic use in restless legs syndrome, including in women beyond reproductive age. Moreover, this finding might also provide clues to the sleep-promoting mechanisms of this drug. Carefully designed clinical trials are needed to further investigate these drugs.
With the 13 new risk loci identified for restless legs syndrome, the total becomes 19. Our results suggest that altered embryonic neurodevelopment, impaired neurogenesis at later age, or both, could underlie restless legs syndrome pathogenesis. The exact molecular mechanisms, the relevant times in life, and the connection to brain iron metabolism and the dopaminergic system are unknown, but the implicated loci, candidate genes, and candidate pathways will provide impetus for further functional research in restless legs syndrome. Finally, the possibility of repurposing thalidomide could lead to translation of these research findings into the care of patients.
Acknowledgments
Acknowledgments
We thank all colleagues and staff at the participating centres of the EU-RLS-GENE consortium for their help with recruitment of patients. We thank Daniel Lam, Institute of Neurogenomics, Helmholtz Zentrum München, German Research Centre for Environmental Health, Neuherberg, Germany, for critically proofreading the draft paper. We thank the German Restless Legs Syndrome Foundation for continuously supporting our study and NIH Research Cambridge Biomedical Research Centre for funding (RG64219). JW was supported by the Deutsche Forschungsgemeinschaft (grant WI 1820/5-1). WHOe is a Hertie Senior Research Professor, supported by the Charitable Hertie Foundation. MT-L and AM were supported by the Estonian Research Council (grant IUT20-60), European Union Framework for Research and Innovation Horizon 2020 (grant 692145), European Regional Development Fund (project 2014-2020.4.01.15-0012), and the Centre of Excellence for Genomics and Translational Medicine. GMH was supported by the University of Thessaly (2845). OAR and ZW are supported by the National Institutes of Health National Institute of Neurological Disorders and Stroke (P50 NS072187) and Mayo Clinic Neuroscience Focused Research Team (Cecilia and Dan Carmichael Family Foundation, and the James C and Sarah K Kennedy Fund for Neurodegenerative Disease Research). The DESIR study provided control genotype data for the EU-RLS-GENE genome-wide association study. We thank the INTERVAL study coordination teams, University of Cambridge, University of Oxford, and NHS Blood and Transplant (NHSBT), and the blood donation staff at the 25 static centres for help with participant recruitment and study fieldwork. We thank staff at Cambridge BioResource and NHSBT staff for their help with volunteer recruitment and members of the Cambridge BioResource Scientific Advisory Board and Management Committee for supporting this study. We also thank Brendan Burchell, University of Cambridge, for advice on restless leg syndrome in the INTERVAL study. WHOu is supported by grants to his laboratory from the National Institute for Health Research (NIHR), the European Commission (HEALTH-F2-2012-279233), the British Heart Foundation ([BHF] RP-PG-0310-1002 and RG/09/12/28096), and NHSBT, and he is a Senior Investigator for NIHR. NS is supported by the Wellcome Trust (WT098051 and WT091310) and the European Commission Framework Programme 7 (EPIGENESYS 257082 and BLUEPRINT HEALTH-F5-2011-282510). DJR is supported by NIHR (NIHR-RP-PG-0310-1004). JD is supported by BHF (SP/09/002), European Research Council (268834UK), Medical Research Council (G0800270), NIHR (BTRU-2014-10024 and Cambridge Biomedical Research Centre), and NHS Blood and Transplant (11-01-GEN). PV was supported by Progres (Q28/LF1). We thank the research participants and employees of 23andMe for making this work possible. The 23andMe research team provided infrastructure for generating 23andMe data.
Contributors
BS and JW designed and oversaw the study. BS, CZ, AVS, KO, BM-M, and JW did primary interpretation of the data. BS, CZ, KO, and JW wrote the paper, and SB and DAH contributed substantial edits. BS did the EU-RLS-GENE GWAS. CZ did the meta-analyses. ET and BP participated in quality control and statistical analysis of the EU-RLS-GENE GWAS. BM-M supervised statistical analyses for EU-RLS-GENE GWAS and meta-analyses. YD, AS, BH, WPo, DK, KS, CGB, WPa, CT, WHOe, MH, MT-L, AM, GMH, OP, IF, OAR, ZW, WGO, KB, and JW recruited patients for the EU-RLS-GENE GWAS. RPA, CJE, LX, JM, and GAR recruited patients and controls for the EU-RLS-GENE GWAS. MP, PV, CD, AF, LT, AFRS, SHS, CG, and AP provided control samples or genotype data for the EU-RLS-GENE GWAS. JW acquired funding for the EU-RLS-GENE GWAS and all meta-analyses. SB did statistical analysis of the INTERVAL GWAS for restless legs syndrome. ASB and NS were responsible for data curation of the INTERVAL study. ASB, NS, WHOu DJR, JD, and EDA acquired funding for and supervised the INTERVAL study. EDA conceived and oversaw the INTERVAL GWAS for restless legs syndrome. DAH performed data analysis of the 23andMe restless legs syndrome GWAS. All authors contributed and critically reviewed the final version of the manuscript.
DESIR study group
CESP, INSERM U1018, Villejuif: B Balkau, P Ducimetière, E Eschwège; Université Paris Descartes, Paris: F Rancière; INSERM U367, Paris: F Alhenc-Gelas; CHU D'Angers, Angers: Y Gallois, A Girault; Centre de Recherche des Cordeliers, INSERM U1138, Bichat Hospital, Paris: F Fumeron, M Marre, R Roussel; CHU de Rennes, Rennes: F Bonnet; CNRS UMR8090, Lille: A Bonnefond, S Cauchi, P Froguel; Centres d'Examens de Santé: Alençon, Angers, Blois, Caen, Chateauroux, Chartres, Cholet, Le Mans, Orléans, Tours; Institute de Recherche Médecine Générale, Paris: J Cogneau; General practitioners of the Region; Institute Inter-Regional pour la Santé, La Riche: C Born, E Caces, M Cailleau, O Lantieri, JG Moreau, F Rakotozafy, J Tichet, S Vol.
23andMe research team
Michelle Agee, Babak Alipanahi, Adam Auton, Robert K Bell, Katarzyna Bryc, Sarah L Elson, Pierre Fontanillas, Nicholas A Furlotte, David A Hinds, Bethann S Hromatka, Karen E Huber, Aaron Kleinman, Nadia K Litterman, Matthew H McIntyre, Joanna L Mountain, Carrie AM Northover, Steven J Pitts, J Fah Sathirapongsasuti, Olga V Sazonova, Janie F Shelton, Suyash Shringarpure, Chao Tian, Joyce Y Tung, Vladimir Vacic, and Catherine H Wilson.
Declaration of interests
BS has received grants from Deutsche Forschungsgemeinschaft ([DFG] German Research Foundation) and grants from the German restless legs syndrome foundation. YD has received grants and personal fees from UCB and personal and other fees from Bioprojet and Jazz. AS has received travel grants from Habel Medizintechnik, Inspire Medical Systems, OSG, and UCB. BH has received grants and personal fees from UCB, personal fees from Abbvie, Axovant, Benevolent Bio, Janssen Cilag, Lundbeck, Mundipharma, and Ostuka, and other fees from Air Liquide Austria, Habel Medizintechnik Viena, and Vivisol Austria. WPo has received personal fees from AbbVie, AstraZeneca, BIAL, Biogen, Britannia, Cynapsus, Grünenthal, Intec, Ipsen, Lundbeck, Merz Pharmaceuticals, Novartis, Neuroderm, Orion Pharma, Prexton, Teva, UCB, and Zambon. DK has received grants from UNCE 204011/12 (Charles University Prague). WPa has received fees from GlaxoSmithKline and UCB Pharma, personal and other fees from Desitin Arzneimittel, and personal fees from Aesculap, CIN Werner Reichardt, Danish Agency for Science, Deutsches Stiftungszentrum, EBS Technologies, Kenes Israel, Klinikum Bremen-Mitte, Mundipharma, Schaaf Verlagsgesellschaft mbH, Thiel Foundation, and Thieme Verlag KG, and grants and other awards from BMBF and DFG. CT has received fees from Abbvie, Benevolent, Britannia Pharmaceuticals, Gruenenthal, Mundipharma, Servier, UCB, and Vifor Pharma. IF has received grants and personal fees from Philips and ResMed, grants from Weinmann, and personal fees from UCB. ZW has received grants from NIH/NINDS P50 NS072187, from Mayo Clinic Neuroscience Focused Research Team (Cecilia and Dan Carmichael Family Foundation, and the James C and Sarah K Kennedy Fund for Neurodegenerative Disease Research at Mayo Clinic in Florida), personal fees from European Journal of Neurology and Parkinsonism and Related Disorders. ASB has received grants from British Heart Foundation, Medical Research Council, and National Institute for Health Research (NIHR), grants from Biogen, European Research Council, European Union Framework Programme 7, Merck, Novartis, and Pfizer. JD has received grants from British Heart Foundation, European Commission, European Research Council, Medical Research Council, Merck, NHS Blood and Transplant, NIHR, Novartis, Pfizer, and Wellcome Trust, and personal fees and non-financial support from Merck Sharpe and Dohme UK Atherosclerosis, Metabolic Advisory Board, Novartis Cardiovascular, and Pfizer Population Research Advisory Panel. RPA has received personal fees from Luitpold Pharma and grants from AMAG pharmaceuticals. WGO has received grants from Cybapsus, Luitpold, and Lundbeck, and speaker activities for ACADIA, Lundbeck, Neurocrine, TEVA, and UCB Pharma. JM has received personal fees from Merck Pharma, Novartis, and Takeda Pharma. KB has received unrestricted grants to the University of Muenster from Boehringer Ingelheim, German Restless Legs Society, Mundipharma Research, Neurobiotec, Roche, UCB Germany, UCB Switzerland, and Vifor Pharma. DAH is an employee of and has stock options in 23andMe. JW received grants from DFG, Else Kröner-Fresenius-Stiftung, Era Net Neuron, and German RLS Foundation, and personal fees from UCB, and has a patent pending (17 15 6438.8). Members of the 23andMe research team are employees of and have stock, stock options, or both, in 23andMe. The other authors declare no competing interests.
Contributor Information
Juliane Winkelmann, Email: juliane.winkelmann@tum.de.
DESIR study group:
B Balkau, P Ducimetière, E Eschwège, F Rancière, F Alhenc-Gelas, Y Gallois, A Girault, F Fumeron, M Marre, R Roussel, F Bonnet, A Bonnefond, S Cauchi, P Froguel, J Cogneau, C Born, E Caces, M Cailleau, O Lantieri, JG Moreau, F Rakotozafy, J Tichet, S Vol, Michelle Agee, Babak Alipanahi, Adam Auton, Robert K Bell, Katarzyna Bryc, Sarah L Elson, Pierre Fontanillas, Nicholas A Furlotte, David A Hinds, Bethann S Hromatka, Karen E Huber, Aaron Kleinman, Nadia K Litterman, Matthew H McIntyre, Joanna L Mountain, Carrie AM Northover, Steven J Pitts, J Fah Sathirapongsasuti, Olga V Sazonova, Janie F Shelton, Suyash Shringarpure, Chao Tian, Joyce Y Tung, Vladimir Vacic, and Catherine H Wilson
Supplementary Material
References
- 1.Allen RP, Picchietti DL, Garcia-Borreguero D. Restless legs syndrome/Willis-Ekbom disease diagnostic criteria: updated International Restless Legs Syndrome Study Group (IRLSSG) consensus criteria—history, rationale, description, and significance. Sleep Med. 2014;15:860–873. doi: 10.1016/j.sleep.2014.03.025. [DOI] [PubMed] [Google Scholar]
- 2.Allen RP, Bharmal M, Calloway M. Prevalence and disease burden of primary restless legs syndrome: results of a general population survey in the United States. Mov Disord. 2011;26:114–120. doi: 10.1002/mds.23430. [DOI] [PubMed] [Google Scholar]
- 3.Schormair B, Winkelmann J. Genetics of restless legs syndrome: mendelian, complex, and everything in between. Sleep Med Clin. 2011;6:203–215. [Google Scholar]
- 4.Winkelmann J, Schormair B, Lichtner P. Genome-wide association study of restless legs syndrome identifies common variants in three genomic regions. Nat Genet. 2007;39:1000–1006. doi: 10.1038/ng2099. [DOI] [PubMed] [Google Scholar]
- 5.Stefansson H, Rye DB, Hicks A. A genetic risk factor for periodic limb movements in sleep. N Engl J Med. 2007;357:639–647. doi: 10.1056/NEJMoa072743. [DOI] [PubMed] [Google Scholar]
- 6.Schormair B, Kemlink D, Roeske D. PTPRD (protein tyrosine phosphatase receptor type delta) is associated with restless legs syndrome. Nat Genet. 2008;40:946–948. doi: 10.1038/ng.190. [DOI] [PubMed] [Google Scholar]
- 7.Winkelmann J, Czamara D, Schormair B. Genome-wide association study identifies novel restless legs syndrome susceptibility loci on 2p14 and 16q12.1. PLoS Genet. 2011;7:e1002171. doi: 10.1371/journal.pgen.1002171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spieler D, Kaffe M, Knauf F. Restless legs syndrome-associated intronic common variant in Meis1 alters enhancer function in the developing telencephalon. Genome Res. 2014;24:592–603. doi: 10.1101/gr.166751.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Catoire H, Dion PA, Xiong L. Restless legs syndrome-associated MEIS1 risk variant influences iron homeostasis. Ann Neurol. 2011;70:170–175. doi: 10.1002/ana.22435. [DOI] [PubMed] [Google Scholar]
- 10.Freeman A, Pranski E, Miller RD. Sleep fragmentation and motor restlessness in a drosophila model of restless legs syndrome. Curr Biol. 2012;22:1142–1148. doi: 10.1016/j.cub.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drgonova J, Walther D, Wang KJ. Mouse model for PTPRD associations with WED/RLS and addiction: reduced expression alters locomotion, sleep behaviors and cocaine-conditioned place preference. Mol Med. 2015;21:717. doi: 10.2119/molmed.2015.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allen RP, Picchietti D, Hening WA. Restless legs syndrome: diagnostic criteria, special considerations, and epidemiology. A report from the restless legs syndrome diagnosis and epidemiology workshop at the National Institutes of Health. Sleep Med. 2003;4:101–119. doi: 10.1016/s1389-9457(03)00010-8. [DOI] [PubMed] [Google Scholar]
- 13.Moore C, Bolton T, Walker M. Recruitment and representativeness of blood donors in the INTERVAL randomised trial assessing varying inter-donation intervals. Trials. 2016;17:458. doi: 10.1186/s13063-016-1579-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Angelantonio E, Thompson SG, Kaptoge S. Efficiency and safety of varying the frequency of whole blood donation (INTERVAL): a randomised trial of 45 000 donors. Lancet. 2017 doi: 10.1016/S0140-6736(17)31928-1. http://dx.doi.org/10.1016/S0140-6736(17)31928-1 published online Sept 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allen RP, Burchell BJ, MacDonald B, Hening WA, Earley CJ. Validation of the self-completed Cambridge-Hopkins questionnaire (CH-RLSq) for ascertainment of restless legs syndrome (RLS) in a population survey. Sleep Med. 2009;10:1097–1100. doi: 10.1016/j.sleep.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 16.Devlin B, Bacanu S-A, Roeder K. Genomic control to the extreme. Nat Genet. 2004;36:1129–1130. doi: 10.1038/ng1104-1129. [DOI] [PubMed] [Google Scholar]
- 17.Pers TH, Karjalainen JM, Chan Y. Biological interpretation of genome-wide association studies using predicted gene functions. Nat Commun. 2015;6:5890. doi: 10.1038/ncomms6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Q, Goto H, Akiyoshi-Nishimura S. Diversification of behavior and postsynaptic properties by netrin-G presynaptic adhesion family proteins. Mol Brain. 2016;9:6. doi: 10.1186/s13041-016-0187-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dijkmans TF, van Hooijdonk LWA, Fitzsimons CP, Vreugdenhil E. The doublecortin gene family and disorders of neuronal structure. Cent Nerv Syst Agents Med Chem. 2010;10:32–46. doi: 10.2174/187152410790780118. [DOI] [PubMed] [Google Scholar]
- 20.Rataj-Baniowska M, Niewiadomska-Cimicka A, Paschaki M. Retinoic acid receptor β controls development of striatonigral projection neurons through FGF-dependent and Meis1-dependent mechanisms. J Neurosci. 2015;35:14467–14475. doi: 10.1523/JNEUROSCI.1278-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keil R, Schulz J, Hatzfeld M. p0071/PKP4, a multifunctional protein coordinating cell adhesion with cytoskeletal organization. Biol Chem. 2013;394:1005–1017. doi: 10.1515/hsz-2013-0114. [DOI] [PubMed] [Google Scholar]
- 22.Fischer ES, Böhm K, Lydeard JR. Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature. 2014;512:49–53. doi: 10.1038/nature13527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jo S, Lee K-H, Song S, Jung Y-K, Park C-S. Identification and functional characterization of cereblon as a binding protein for large-conductance calcium-activated potassium channel in rat brain. J Neurochem. 2005;94:1212–1224. doi: 10.1111/j.1471-4159.2005.03344.x. [DOI] [PubMed] [Google Scholar]
- 24.Asami O-A, Zuko A, Kleijer KTE, Burbach JPH. A current view on contactin-4, -5, and -6: implications in neurodevelopmental disorders. Mol Cell Neurosci. 2017;81:72–83. doi: 10.1016/j.mcn.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 25.Loh KH, Stawski PS, Draycott AS. Proteomic analysis of unbounded cellular compartments: synaptic clefts. Cell. 2016;166:1295–1307. doi: 10.1016/j.cell.2016.07.041. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee K, Kim Y, Lee S-J. MDGAs interact selectively with neuroligin-2 but not other neuroligins to regulate inhibitory synapse development. Proc Natl Acad Sci. 2013;110:336–341. doi: 10.1073/pnas.1219987110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun Y, Hu D, Liang J. Association between variants of zinc finger genes and psychiatric disorders: systematic review and meta-analysis. Schizophr Res. 2015;162:124–137. doi: 10.1016/j.schres.2015.01.036. [DOI] [PubMed] [Google Scholar]
- 28.Fukata Y, Yokoi N, Miyazaki Y, Fukata M. The LGI1–ADAM22 protein complex in synaptic transmission and synaptic disorders. Neurosci Res. 2016;116:39–45. doi: 10.1016/j.neures.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi H, Craig AM. Protein tyrosine phosphatases PTPδ, PTPσ, and LAR: presynaptic hubs for synapse organization. Trends Neurosci. 2013;36:522–534. doi: 10.1016/j.tins.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Zhang P, Choi T-Y. Splicing-dependent trans-synaptic SALM3-LAR-RPTP interactions regulate excitatory synapse development and locomotion. Cell Rep. 2015;12:1618–1630. doi: 10.1016/j.celrep.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Honsa P, Pivonkova H, Anderova M. Focal cerebral ischemia induces the neurogenic potential of mouse Dach1-expressing cells in the dorsal part of the lateral ventricles. Neuroscience. 2013;240:39–53. doi: 10.1016/j.neuroscience.2013.02.048. [DOI] [PubMed] [Google Scholar]
- 32.Agoston Z, Heine P, Brill MS. Meis2 is a Pax6 co-factor in neurogenesis and dopaminergic periglomerular fate specification in the adult olfactory bulb. Development. 2014;141:28–38. doi: 10.1242/dev.097295. [DOI] [PubMed] [Google Scholar]
- 33.Yoshida Y, Han B, Mendelsohn M, Jessell TM. PlexinA1 signaling directs the segregation of proprioceptive sensory axons in the developing spinal cord. Neuron. 2006;52:775–788. doi: 10.1016/j.neuron.2006.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leslie JR, Imai F, Fukuhara K. Ectopic myelinating oligodendrocytes in the dorsal spinal cord as a consequence of altered semaphorin 6D signaling inhibit synapse formation. Development. 2011;138:4085–4095. doi: 10.1242/dev.066076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sahu SK, Fritz A, Tiwari N. TOX3 regulates neural progenitor identity. Biochim Biophys Acta. 2016;1859:833–840. doi: 10.1016/j.bbagrm.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 36.Philippidou P, Dasen JS. Hox genes: choreographers in neural development, architects of circuit organization. Neuron. 2013;80:12–34. doi: 10.1016/j.neuron.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Bonito M, Glover JC, Studer M. Hox genes and region-specific sensorimotor circuit formation in the hindbrain and spinal cord. Dev Dyn. 2013;242:1348–1368. doi: 10.1002/dvdy.24055. [DOI] [PubMed] [Google Scholar]
- 38.Vasconcelos FF, Sessa A, Laranjeira C. MyT1 counteracts the neural progenitor program to promote vertebrate neurogenesis. Cell Rep. 2016;17:469–483. doi: 10.1016/j.celrep.2016.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lo Coco D, Piccoli F, La Bella V. Restless legs syndrome in patients with amyotrophic lateral sclerosis. Mov Disord. 2010;25:2658–2661. doi: 10.1002/mds.23261. [DOI] [PubMed] [Google Scholar]
- 40.Limousin N, Blasco H, Corcia P, Arnulf I, Praline J. The high frequency of restless legs syndrome in patients with amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2011;12:303–306. doi: 10.3109/17482968.2011.557736. [DOI] [PubMed] [Google Scholar]
- 41.Talarico G, Canevelli M, Tosto G. Restless legs syndrome in a group of patients with Alzheimer's disease. Am J Alzheimer's Dis Other Demen. 2013;28:165–170. doi: 10.1177/1533317512470208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohayon MM, O'Hara R, Vitiello MV. Epidemiology of restless legs syndrome: a synthesis of the literature. Sleep Med Rev. 2012;16:283–295. doi: 10.1016/j.smrv.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rijsman RM, Schoolderman LF, Rundervoort RS, Louter M. Restless legs syndrome in Parkinson's disease. Parkinsonism Relat Disord. 2014;20:S5–S9. doi: 10.1016/S1353-8020(13)70004-X. [DOI] [PubMed] [Google Scholar]
- 44.Marchesi E, Negrotti A, Angelini M, Goldoni M, Abrignani G, Calzetti S. A prospective study of the cumulative incidence and course of restless legs syndrome in de novo patients with Parkinson's disease during chronic dopaminergic therapy. J Neurol. 2016;263:441–447. doi: 10.1007/s00415-015-7937-7. [DOI] [PubMed] [Google Scholar]
- 45.Winkelmann J, Prager M, Lieb R. ‘Anxietas tibiarum’: depression and anxiety disorders in patients with restless legs syndrome. J Neurol. 2005;252:67–71. doi: 10.1007/s00415-005-0604-7. [DOI] [PubMed] [Google Scholar]
- 46.Berger K, Luedemann J, Trenkwalder C. Sex and the risk of restless legs syndrome in the general population. Arch Intern Med. 2004;164:196–202. doi: 10.1001/archinte.164.2.196. [DOI] [PubMed] [Google Scholar]
- 47.Earley CJ, Connor J, Garcia-Borreguero D. Altered brain iron homeostasis and dopaminergic function in restless legs syndrome (Willis–Ekbom disease) Sleep Med. 2014;15:1288–1301. doi: 10.1016/j.sleep.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 48.Madabhushi R, Gao F, Pfenning AR. Activity-induced DNA breaks govern the expression of neuronal early-response genes. Cell. 2015;161:1592–1605. doi: 10.1016/j.cell.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suberbielle E, Sanchez PE, Kravitz AV. Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-β. Nat Neurosci. 2013;16:613–621. doi: 10.1038/nn.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spieler D, Kaffe M, Knauf F. Restless legs syndrome-associated intronic common variant in Meis1 alters enhancer function in the developing telencephalon. Genome Res. 2014;24:592–603. doi: 10.1101/gr.166751.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rizzo G, Li X, Galantucci S, Filippi M, Cho YW. Brain imaging and networks in restless legs syndrome. Sleep Med. 2017;31:39–48. doi: 10.1016/j.sleep.2016.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pidsley R, Viana J, Hannon E. Methylomic profiling of human brain tissue supports a neurodevelopmental origin for schizophrenia. Genome Biol. 2014;15:483. doi: 10.1186/s13059-014-0483-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O'Shea KS, McInnis MG. Neurodevelopmental origins of bipolar disorder: iPSC models. Mol Cell Neurosci. 2016;73:63–83. doi: 10.1016/j.mcn.2015.11.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.