Abstract
Targeted therapies against the human epidermal growth factor receptor 2 (HER2) have radically changed the outcome of patients with HER2-positive breast cancers. However, a minority of cases displays a heterogeneous distribution of HER2-positive cells, which generates major clinical challenges. To date, no reliable and standardized protocols for the characterization and quantification of HER2 heterogeneous gene amplification in large cohorts have been proposed. Here, we present a high-throughput methodology to simultaneously assess the HER2 status across different topographic areas of multiple breast cancers. In particular, we illustrate the laboratory procedure to construct enhanced tissue microarrays (TMAs) incorporating a targeted mapping of the tumors. All TMA parameters have been specifically optimized for the silver in situ hybridization (SISH) of formalin-fixed paraffin-embedded (FFPE) breast tissues. Immunohistochemical analysis of the prognostic and predictive biomarkers (i.e., ER, PR, Ki67, and HER2) should be performed using automated procedures. A customized SISH protocol has been implemented to allow a high-quality molecular analysis across multiple tissues that underwent different fixation, processing, and storage procedures. In this study, we provide a proof-of-principle that specific DNA sequences could be localized simultaneously in distinct topographic areas of multiple and heterogeneously processed breast cancers using an efficient and cost-effective method.
Keywords: Immunology, Issue 130, Breast cancer, human epidermal growth factor receptor 2 (HER2), heterogeneity, tissue microarray, in situ hybridization, high-throughput molecular analysis
Introduction
HER2 is a proto-oncogene that is overexpressed and amplified in 15 - 30% of all invasive breast cancers1,2. HER2 overexpression is inferred by the presence of >10% cells with strong membrane immunohistochemical (IHC) staining (3+), while the gene amplification can be assessed when either the HER2/centromere ratio is ≥2 or the gene copy number is ≥6, on counting at least 20 cells by in situ hybridization (ISH)3.
Intra-tumor genetic heterogeneity has been widely described in breast cancers, being a potentially adverse contributor to biomarkers evaluation and treatments response4. According to the College of American Pathologists (CAP), HER2 heterogeneity exists if HER2 is amplified in >5% and <50% of infiltrating tumor cells5. Regrettably, the actual incidence of HER2 spatial heterogeneity in breast cancers remains a subject of controversy among pathologists, with some authors maintaining that it is an exceedingly rare event, and others suggesting that up to 40% of cases are HER2-heterogeneous1,5,6,7,8,9,10. Despite the biological mechanisms that underpin this condition are not yet fully clarified, the prognostic and clinical impacts of intra-tumor HER2 heterogeneity are crucial for breast cancer patients11.
Recently, bright-field molecular techniques, such as chromogenic ISH (CISH) and silver ISH (SISH), have emerged as reliable methods to detect HER2 heterogeneity in FFPE tissues, with some advantages compared to fluorescent ISH (FISH)12. Regrettably, the bulk analysis of single cases remains impractical in large-cohort research studies. Several groups have suggested that the combination of histochemistry, IHC, and ISH with TMA technologies could represent a valuable strategy in the study of cancer biology13,14,15,16. With this widely adopted method, tissue samples from different patients can be analyzed concurrently, minimizing the tissue and reagents employed and thereby fostering the uniform analysis of a large series of cases14. However, no protocols are available for the simultaneous high-throughput molecular characterization of multiple tissue samples that underwent different processing in terms of reagents, fixation times, and conservation methods employed, such as archival blocks.
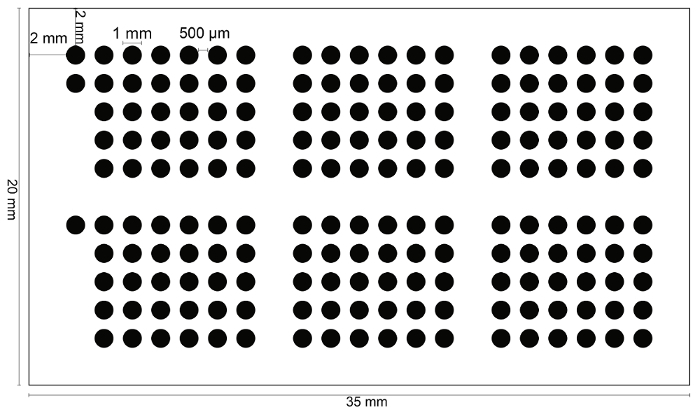
Given the prognostic and clinical implications of HER2 spatial heterogeneity in breast cancers, we developed an integrated molecular platform to assess it in large series of heterogeneously processed cases. Here, we portray the laboratory strategies to generate and analyze the intra-tumor heterogeneity of HER2 amplification in high-yield TMAs of breast cancer by means of SISH. The following protocol has been developed for tumors measuring >5 mm (>pT1b according to the TNM 2017)17. For smaller lesions, we recommend performing the analysis on full-face serial sections. Our procedure allows for the simultaneous IHC and SISH analysis of up to 30 breast cancers, encompassing a mean of 6 distinct areas (range 4 - 8) for each case. Altogether, 180 tissue cores of 1 mm in diameter, with 500 µm between the cores, and 2 mm between the grid and edges will be generated for each TMA block.
Protocol
This study was approved by the Institutional Review Boards from IRCCS Ca' Granda Foundation, Policlinico Hospital, Milan, Italy.
1. Selection of Patients and Tissue Specimens
Retrieve the archival slides of all cases to be analyzed, including all the available hematoxylin and eosin (H&E) and IHC slides from the original diagnosis and, if present, one H&E of matched non-neoplastic breast tissue (e.g., surgical margin).
- Perform case reviews. NOTE: For this task, at least two pathologists with experience in breast pathology should discuss the diagnostic slides and resolve disagreements collegially. Use either a conventional microscope or telepathology tools (favor the latter in multicentric studies). If any, exclude all discordant cases.
- Perform the histological re-classification of all cases according to the latest edition of the World Health Organization (WHO) classification of tumors of the breast18.
- Ensure that the normal tissue for each case, if present in the archival clinical samples, comprises at least one non-neoplastic terminal ductal-lobular unit.
- Identify all neoplastic areas that show heterogeneous cytological, architectural, or IHC features using a bright-field microscope (to be jointly performed by a pathologist and the technician(s) who will construct the TMAs).
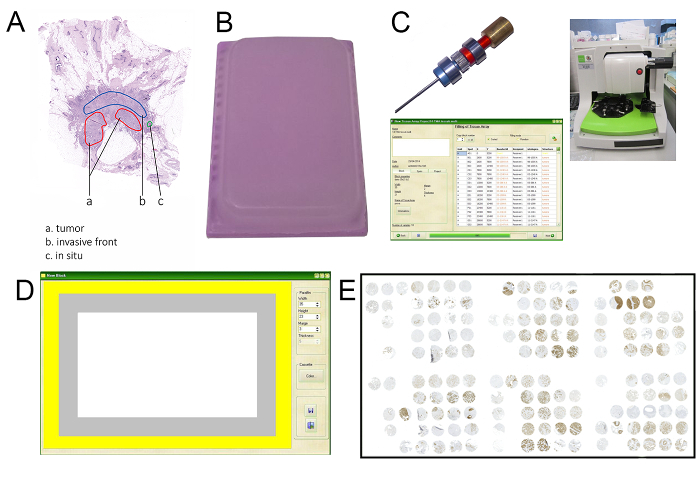
Highlight the discrete topographic areas with a marker on the glass slide or with the proper tool on a digital slide software (Figure 1A).
Based on the selected slides, retrieve the corresponding FFPE blocks from the archives. NOTE: To optimize the TMA construction, to avoid tissue depletion, and to preserve the TMA punch structural integrity, cases with <1 mm-thick residual tissue should be excluded.
2. Design and Construct TMAs Based on the Kononen Technique
- Create and open a new spreadsheet file incorporating the records of the distinct areas of each case. Include the following data on separate columns, as exemplified in Table 1: Case ID, Block ID, Area ID, tissue type (e.g., normal tissue, invasive component histotype, in situ component histotype), topography (e.g., tumor core, invasive front), morphology (e.g., nuclear grade, architecture, mitoses, cytological features), biomarkers status (i.e., estrogen receptor (ER), progesterone receptor (PR), Ki67, and HER2), and all other IHC features available.
- Save the document.
- Create a TMA project.
- Install and open the TMA designer software.
- Define the TMA recipient block(s): access through the menu File > New > Block or by using CTRL+ B.
- Click in the first field to fill or press the Tab key, and type in dimensions of the paraffin block: width 35 (mm), height 20 (mm). Type "2" as the "margin" value (i.e., distance from paraffin edge in mm) (Figure 1B). Save the data by clicking the proper button. The new template of recipient block is now saved and ready to be used in new tissue array templates.
- Create a tissue array project: access to the menu File > New > Tissue Array or by using CTRL+ T.
- Fill in the name of the Tissue array, comments, and author, then click "next." Click the "import" icon, choose the template file of recipient block previously created, and click "next."
- Choose 1 mm as the spot size by clicking the round button. Choose 500 µm as spot spacing which is the space between two centers of neighboring spots. Click on the calculator icon to calculate the total number of spots created, which should be 231. Click "next."
- Define the spot numbering mode by clicking once on it. Click on the button "define" to create grids and sub-grids. Delete the central line (line 6) and the columns 8 and 15. Maintain 3 spots of line 1 for orientation. The final map should encompass a total number of 183 spots, as represented in Figure 2.
- Define the list of tissue types: access through the menu File > New > List of tissue types or by using CTRL+ L.
- Insert the tissue descriptions previously defined. Press down arrow or click on the + button to add a line.
- Define the project: access through the menu File > New > Booster project or by using CTRL+ P.
- Fill in the name of the tissue array, comments, and author, then click "next." Import the spreadsheet file, list of tissue types, and the tissue array model previously created by clicking the proper icons.
- Click "select all" and define the number of spots for each tissue type; click on the "match" button to apply settings. A table listing the total number of cores that will be sampled and transferred to recipient blocks in this project will appear. Save the project and close the program.
- Prepare the acceptor block(s) (Figure 1B).
- Fill cleansed metal molds with elasticized paraffin at 65 °C.
- Leave the paraffin blocks to cool at room temperature overnight inside of the metal molds.
- Extract the paraffin blocks from the metal molds. If cracks occur, discard the block and repeat steps 2.3.1-2.3.3.
- Construct the TMA using an automated or semi-automated arrayer19.
- Retrieve all selected FFPE blocks and the corresponding H&E sections with annotations.
- Turn on the arrayer and insert the acceptor block(s) on the arrayer carousel.
- Open the arrayer control station software, click on "New Tissue Array", and load the project previously created (Figure 1C).
- Click "Continue" and select the position of the donor block(s) from the menu.
- Define the starting position onto each recipient block: use the arrow keys to move the laser light and align it with the top-left edge on the paraffin of the recipient block.
- Click "next" and repeat for the vertical alignment.
- Click "next" to automatically create a hole in the previously defined coordinate of the recipient (acceptor) block.
- Empty the punch.
- Position the donor block for sampling and remove a tissue core from the previously annotated area of interest.
- Insert the donor tissue core into the hole in the recipient (acceptor) block.
- Remove the donor block from the arrayer.
- Repeat steps 2.4.5-2.4.11 until the TMA is completed.
Put the tissue side of the newly generated TMA block on a sterile blank slide and place them in the lab oven at 45 °C for 5 min.
Gently press the block on the slide for 5 s to flatten the tissue cores. Repeat once (for a total of 2 times).
Remove the TMA block together with the slide from the oven, flip them, and gently detach the block from the slide.
Cover the TMA tissue surface with a thin layer of elasticized paraffin at 65 °C.
Leave the TMA block at room temperature for 2 h.
Transfer the TMA block at 4 °C for 24 h. NOTE: The TMA block can be stored at 4 °C until exhaustion.
3. Histochemical and Immunohistochemical Analyses
- Cut TMA sections.
- Place the TMA acceptor block(s) with the paraffin-side down on a -10 °C surface for 10 min to chill the paraffin.
- Fill a floatation bath with ultrapure water and set heat to 38 °C.
- Place a new blade on a rotary microtome and insert the block into the microtome chuck so the wax block faces the blade and is aligned in the vertical plane.
- Approach the block carefully with the blade and cut a few thin sections to ensure the positioning is correct; adjust if necessary.
- Cut 3 µm-thick sections.
- Pick up the sections using tweezers and float them on the water bath surface.
- Pick the sections out the water bath on regular and IHC/ISH glass slides.
- Place the slides on a rack and store them in the oven at 45 °C overnight. NOTE: Once prepared, the TMA sections should be stained within 24 h.
- Perform histochemical staining (H&E) using an autostainer.
- Insert the slides in the autostainer rack and place them at 60 °C for 10 min.
- Open the load drawer of the autostainer and insert the rack with the TMA slides in one of the load stations with a standard H&E clip facing outwards.
- Once the drawer is closed, the clip will be automatically recognized by the system and the following protocol will start: oven station at 37 °C (6 min), xylene (2 min), xylene (2 min), ethanol 100% (2 min), ethanol 100% (2 min), ethanol 95% (2 min), ethanol 95% (2 min), distilled water (4 min), Carazzi's hematoxylin (9 min), low-pressure running water (2 min), eosin 1% (1 min), low-pressure running water (2 min), ethanol 95% (20 s), ethanol 95% (20 s), ethanol 95% (20 s), ethanol 95% (20 s), ethanol 100% (15 s), ethanol 100% (15 s), xylene (30 s), xylene (30 s).
- Unload the racks from the drawer and mount the slide with a cover slip.
- Collect a drop of mount with a pipette and put it on an outer edge of the cover slide.
- Place the wet side of the cover slip on the slide and let the mount dry for 30 min.
- Perform IHC staining for ER, PR, Ki67, and HER2 using an automated immunostainer. NOTE: Before beginning a staining run, start up the system, and check the consumables reagent bottles (Table of Materials) and waste containers.
- Place the TMA slides to analyze at 60 °C for 10 min.
- Start the immunostainer instrument and software.
- At the Home View, click the Protocols button, and then click Create/Edit Protocols.
- Select the standard DAB (3, 3'-diaminobenzidine) IHC procedure to modify the basic template.
- Flag "Deparaffinization" in the box list and set the Medium Temperature at 72 °C.
- Flag the cc1 unmasking solution, set the Medium Temperature at 95 °C, and the incubation time at 36 min.
- Flag the Primary Antibody box, insert the ID of the ER inline dispenser (the instrument software will recognize the dispenser on the reagent carousel), and set the incubation time at 16 min.
- Flag the Counterstaining box, select Hematoxylin I, and set the incubation time at 12 min.
- Click the "Save As" button and name the protocol.
- Repeat the steps 3.3.3-3.3.9 for the remaining antibodies using the following primary antibodies incubation times: PR 16 min, Ki67 16 min, and HER2 12 min. Use pre-diluted ready-to-use (RTU) antibodies.
- At the Home View, click the Create Label button.
- Click the Protocols button and add the ER, PR, Ki67, and HER2 protocols for each of the TMA to analyze.
- Click the Close /Print button.
- As the label template appears, enter the TMA IDs for the label.
- Click the Print button to print the labels and then apply them on the TMA slides.
- Click the instrument icon and set the instrument startup mode as "Ready".
- Open the hood that covers the reagents carousel, place the detection system, and primary antibodies, then close the hood.
- Click the front button on a slide drawer to open it, load the TMA slides, and close the drawer by clicking the same button.
- Set the instrument startup mode as "Running" and follow the instructions.
- At the end of the run, unload the TMA slides by pressing the Open All Drawers with Completed Slides button on the instrument Slide Control and rinse them in warm soapy water to remove any remaining reagents.
- Wash the slides under low-pressure cold running water, dehydrate the TMA slides, and apply a glass coverslip to each slide.
- Perform IHC analysis of ER, PR, and HER2 following the American Society of Clinical Oncology (ASCO)/CAP guidelines, and of Ki67 according to the recommendations of the International Ki67 in Breast Cancer working group (Table 2). NOTE: This analysis should be performed separately in the discrete tissue spots by a pathologist with an experience in breast pathology.
- Assess the HER2 expression as: a) score 3+ if circumferential membrane staining that is complete and intense, b) score 2+ if circumferential membrane staining is incomplete and/or weak/moderate and within > 10% of the invasive tumor cells or complete and circumferential membrane staining is intense and within ≤ 10% of the invasive tumor cells, c) score 1+ if incomplete membrane staining is faint/barely perceptible and within > 10% of the invasive tumor cells, d) negative if no staining is present or membrane staining is incomplete and faint/barely perceptible and within ≤ 10% of the invasive tumor cells3,24.
- Assess the Ki67 index as the percentage of cells with nuclear staining in at least 1,000 neoplastic cells randomly selected over 10 high-power fields (magnification, 400X)25.
4. SISH Analysis of HER2
Cut TMA sections (repeat step 3.1).
- Perform SISH for HER2 using an automated immunostainer.
- Place the TMA slides to analyze at 60 °C for 10 min.
- Start the immunostainer instrument and software.
- At the Home View, click the Protocols button, and then click Create/Edit Protocols.
- Select the "U Dual ISH CKT" procedure to modify the basic template.
- Flag "Drying" in the box list and set the temperature at 63 °C for 20 min.
- Flag "Cell Conditioning" in the box list, then "Cell Conditioning CC2", and set the temperature at 86 °C and incubation time at 4 min.
- Flag CC2 Mild (Cycle 1), Standard (Cycle 2), and Extended (Cycle 3) for 8 min, 12 min, and 8 min, respectively.
- Flag "ISH-Protease 3" in the box list and set the incubation time at 16 min.
- Select the probes HER2DNP and CHR17DIG and set the incubation time at 6 h.
- Set the washing at 76 °C for 8 min.
- Set the SISH multimer (SIL ISH DNP HRP) incubation time at 32 min.
- Set the silver chromogen (SIL ISH DNP CHRC) incubation time at 4 min.
- Set the red multimer (RED ISH DIG AP) incubation time at 24 min.
- Set the red chromogen (RED ISH DIG FR) incubation time at 8 min.
- Flag "Counterstaining" in the box list, select "HEMATOXYLIN II", and set the incubation time at 8 min.
- Flag "Post-Counterstaining" in the box list, select "BLUING REAGENT", and set the incubation time at 4 min.
- Repeat steps from 3.3.11-3.3.15 (select modified U Dual ISH CKT protocol).
- Open the hood that covers the reagents carousel, place the washing and the ISH detection systems, then close the hood.
- Repeat steps from 3.3.18-3.3.21.
Perform SISH analysis for HER2: assess the HER2 gene amplification as positive if the HER2/CEP17 ratio is ≥2.0 with an average HER2 copy number ≥4.0 signals per cell, and negative if the HER2/CEP17 ratio is <2.0 with an average HER2 copy number <4.0 signals/cell3,24. NOTE: Similar to IHC, this analysis should be performed separately in the discrete tissue spots by a pathologist with experience in breast pathology.
Representative Results
Overall, 444 invasive breast cancers were incorporated in 15 TMAs specifically optimized for ISH analyses. Among the 2,664 spots sampled, 2,651 (99.5%) were representative of the previously selected areas and therefore considered amenable for subsequent analyses. Intra-tumor heterogeneity was determined by means of IHC and SIH, with a particular focus on the heterogeneous distributions of HER2-positive clones in the distinct topographic areas of the tumors. Table 3 depicts the biologic characteristics of the cases analyzed, focusing on the ER, PR, Ki67, and HER2 status within different histotypes. In particular, 18% of cases were found to display HER2-positive cells in >10% of tumor cells, and therefore matched the characteristics for HER2 positivity assessment. Interestingly, this value is closer to the lower end of the reported incidence range of HER2-positive breast cancers. On the other hand, the HER2 status was variable in the discrete areas of both HER2-positive and HER2-negative breast cancer (Figure 3, Table 4), providing further credence to the notion that breast cancer is an extremely heterogeneous disease at the genomic level. The failure rate of the SISH analysis was 0.8%, with a total number of 2,629/2,651 spots in which the dinitrophenol-tagged probe bound to the target sequences.
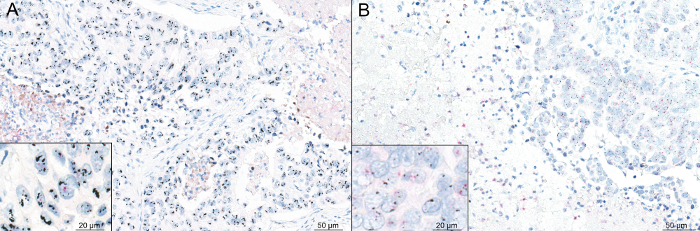
Figure 1: Representation of cornerstone phases in the realization of a high-throughput platform for the analysis of HER2 heterogeneity in large cohorts of breast cancers. (A) Selection of the areas to sample should include the identification and highlight of all areas with cytological, architectural, or IHC heterogeneity. (B) One of the most critical steps of this procedure is the creation of a high-quality acceptor block, given that even a microscopic crack could infer the consistency of the subsequent in situ hybridization analyses. (C) It is important to employ a digitally guided arrayer with a 1 mm-diameter needle to construct high-yield TMA based on the Kononen technique. (D) The creation of the TMA project should start from the definition of the dimensions and borders of the TMA. (E) All IHC and ISH analyses should be performed using digital pathology tools, in order to quickly navigate across the TMA. Please click here to view a larger version of this figure.
Figure 2: Planimetric representation of the TMA realized for this protocol. A total number of 180 spots of 1 mm diameter, 500 µm apart, allow for an optimal in situ hybridization. Denser TMAs provide unproductive results in terms of overall quality and uniformity of the reactions across all spots, with a higher failure rate on a single-spot basis. Note the "orientation" spots in the top- and middle-left of the grid. Please click here to view a larger version of this figure.
Figure 3: Representative micrographs showing the silver in situ hybridization analysis of an HER2-heterogeneous breast cancer. In this paradigmatic example, two distinct spots of a high-grade invasive breast cancer of no special type (BR_121) showed different results in terms of HER2 gene (black) amplification compared to the centromeric enumeration probe 17 (red). In particular, one area belonging to the tumor core and showing cytokeratin 7 irregular staining pattern was HER2 amplified (A), while a single spot from the invasive front (i.e., tumor edge) displayed a wild-type HER2 pattern (B). Scale bars = 50 µm (20 µm in the insets). Please click here to view a larger version of this figure.
| Case ID | Block ID | Area ID | Tissue | Topography | Morphology | ER | PR | Ki67 | HER2 | Other IHC features |
| BR_121 | A5 | a | Normal | Surgical margin | ||||||
| BR_121 | A1 | a | NST | Tumor core | G2 | CK7+ | ||||
| BR_121 | A1 | b | NST | Tumor core | G3 | CK7+/- | ||||
| BR_121 | A3 | a | NST | Tumor core | Mucinous stroma | 90 | 100 | 12 | 2+ | |
| BR_121 | A3 | b | NST | Invasive front | Tubular | 100 | 100 | 35 | 2+ | |
| BR_121 | A4 | a | NST | Invasive front | G3 | |||||
| BR_121 | A3 | c | DCIS (EIC+) | Adjacent to invasive front | High-grade | 100 | 100 | 45 | 2+ |
Table 1: Representative record of one case included in the study. This case (BR_121) was a high-grade invasive carcinoma of no special type with focal myxoid stroma, showing a minor tubular component at the periphery of the lesion, and focal-irregular loss of CK7 immunoexpression. The immunohistochemical features, including biomarkers reporting, refers to the analyses performed at the time of diagnosis. NST, invasive carcinoma of no special type; DCIS, ductal carcinoma in situ; EIC+, extensive intraductal component; CK7, cytokeratin 7.
| Marker | Clone | Company | Dilution | Dewaxing | Antigen retrieval | Antibody incubation | Scoring system |
| ER | SP1 | Ventana | RTU | EZ prep at 72 °C | cc1 at 95 °C for 36 min | 16 min | ASCO/CAP guidelines23 |
| PR | 1E2 | Ventana | RTU | EZ prep at 72 °C | cc1 at 95 °C for 36 min | 17 min | ASCO/CAP guidelines23 |
| HER2 | 4B5 | Ventana | RTU | EZ prep at 72 °C | cc1 at 95 °C for 36 min | 18 min | ASCO/CAP guidelines3 |
| Ki67 | 30-9 | Ventana | RTU | EZ prep at 72 °C | cc1 at 95 °C for 36 min | 12 min | International Ki67 in Breast Cancer working group recommendations25 |
Table 2: List of antibodies, clones, dilutions, antigen retrieval methods, and scoring systems adopted for immunohistochemical analyses. ER, estrogen receptor; PR, progesterone receptor; RTU, ready-to-use.
| Histotype | n (%) | ER+ (%) | PR+ (%) | Ki67-low (%) | Ki-67-high (%) | HER2+ (%) |
| Invasive carcinoma of no special type | 344 (77.5) | 301 (87.5) | 257 (74.7) | 85 (24.7) | 259 (75.3) | 71 (20.6) |
| Invasive lobular carcinoma | 53 (11.9) | 53 (100.0) | 44 (83.0) | 36 (67.9) | 17 (32.0) | 4 (7.5) |
| Invasive carcinoma, mixed-type | 9 (2.0) | 8 (88.9) | 6 (66.7) | 0 (0.0) | 9 (100.0) | 1 (11.1) |
| Tubular carcinoma | 8 (1.8) | 8 (100.0) | 8 (100.0) | 8 (100.0) | 0 (0.0) | 0 (0.0) |
| Mucinous carcinoma | 11 (2.4) | 11 (100.0) | 9 (81.8) | 7 (63.6) | 4 (36.4) | 0 (0.0) |
| Micropapullary carcinoma | 7 (1.6) | 7 (100.0) | 7 (100.0) | 5 (71.4) | 2 (28.6) | 2 (28.6) |
| Invasive carcinoma with apocrine featueres | 5 (1.1) | 3 (60.0) | 1 (20.0) | 1 (20.0) | 4 (80.0) | 3 (60.0) |
| Papillary carcinoma | 1 (0.2) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (100.0) | 0 (0.0) |
| Medullary carcinoma | 4 (0.9) | 0 (0.0) | 1 (25.0) | 0 (0.0) | 4 (100.0) | 0 (0.0) |
| Metaplastic carcinoma | 2 (0.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (100.0) | 0 (0.0) |
| Total | 444 | 391 (88.0) | 333 (75.0) | 142 (32.0) | 302 (68.0) | 81 (18.2) |
Table 3: Breast cancer histotypes, receptor status, and HER2 heterogeneity status. ER, estrogen receptor; PR, progesterone receptor; Ki67-low, Ki67 index < 18%; Ki67-high, Ki67 >18%.
| Interpretable neoplastic spots | HER2-positive spots (%) | Hormone receptors status | HER2-heterogeneous cases |
| 6 | 5 (83) | positive | 10 |
| 6 | 5 (83) | negative | 1 |
| 6 | 4 (67) | positive | 7 |
| 6 | 4 (67) | negative | 1 |
| 6 | 2 (33) | negative | 1 |
| 6 | 1 (17) | positive | 1 |
| 5 | 3 (60) | positive | 10 |
| 5 | 1 (20) | positive | 2 |
| 4 | 3 (75) | positive | 9 |
| 4 | 2 (50) | positive | 8 |
| 4 | 1 (25) | negative | 1 |
| 3 | 2 (67) | positive | 5 |
| 3 | 1 (33) | positive | 2 |
| 46 | 25 (54) | 53 (93) | 57 |
Table 4: HER2 expression, hormone receptor status, and HER2-heterogeneous cases. Among 57 HER2-heterogeneous cases, 4 (7%) cases were ER-negative and PR-negative, confirming the clinical importance of assessing HER2 heterogeneity.
Discussion
Here, we have detailed the laboratory strategies to perform SISH analyses of the HER2 gene and its corresponding centromere in high-yield TMAs of heterogeneously processed breast cancers. This method is cost-effective and can be carried out in most laboratories for the study of HER2 gene amplification heterogeneity in large cohorts of breast cancers retrieved form pathology archives.
Due to the clinical importance of HER2 testing in breast cancer and the challenges generated by its heterogeneous expression, we developed a high-throughput testing protocol to assess intra- and inter-tumor HER2 genetic heterogeneity. The analyzed areas include pre-invasive and invasive components, on the basis of specific cytological, architectural, and IHC features.
There are several critical phases that should be managed carefully in this protocol. One essential step is the selection of the regions of interest. We have determined that the annotation of the different topographic areas through a multidisciplinary approach is pivotal to ensure the quality of the final molecular analysis. In particular, the joint revision of the diagnostic slides performed by a pathologist and a technician increase enormously the number of adequate spots (i.e., spots reflecting the area identified on the slide) and subsequently reduce the failure rate of the single-spots analysis. However, several tumor spots can be lost after serial sections for multiple IHC and ISH analyses. To overcome this inconvenience, we recommend constructing TMAs in duplicate or triplicate, if at all feasible based on tissue availability. Furthermore, we highlight the importance of defining strict laboratory procedures for the creation of acceptor FFPE blocks for the TMAs. Indeed, we have observed that a minimal crack in the TMA block might infer the entire ISH analysis in terms of quality, reproducibility, and clarity of the reaction. If cracks occur, the donor block should be disregarded for TMA construction. It should be acknowledged, however, that IHC could be performed even in suboptimal TMAs, and no evidence of technical problems during such analysis have been observed in our experience.
Despite that this protocol is specifically optimized for HER2 SISH analyses of high-yield breast TMAs, it is of note that other analyses, such as FISH and IHC, can be reliably performed in several tissue types, as previously described14,19,26. However, we have herein defined the ideal number of spots and the topographic characteristic of the TMA to ensure high-quality and reproducible SISH analyses of the HER2 gene in breast cancer specimens. Another significant point is represented by the reshaping of standard automatized SISH protocols to match the peculiarity of the multiple samples to be analyzed. Indeed, the manufacturers' guidelines are generally made for the molecular analyses of conventional full-face sections, and therefore are not able to reach high standards on heterogeneously processed tissues samples, such as in TMAs from archival FFPE blocks. To this end, we have provided a step-by-step description of a reliable customized protocol for SISH analyses in these problematic samples.
It would be extremely beneficial to explore the heterogeneity of HER2 expression and gene amplification in respect to the heterogeneous distribution of other clinically actionable molecular biomarkers in breast cancer. To this end, further translational research studies are needed to explore the validity of our method for other clinically relevant molecular analyses, such as matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI-MSI)27. Recently, the management of FFPE tissues for MALDI-MSI analysis has become feasible, albeit challenging. This in situ proteomic technique allows for the visualization of the spatial distribution of proteins and peptides in pathological tissue sections, including breast cancer.
This protocol has several critical steps that can potentially interfere with the final analysis. First, particular attention should be paid towards the different mechanisms underlying HER2 amplification. For example, chromosome 17 polysomy is a genetic aberration that may occur in breast cancer11, representing a well-known alternative mechanism for the increased HER2 copy number. This condition, albeit not frequent, may affect not only the cancer biological features and patient management but also the ISH analyses. Second, it has been described that intra-tumor genetic heterogeneity occur also at a single-cell level and not only in different neoplastic areas. This protocol is not able to increase the detection power of this particular condition compared to full-face sections. Furthermore, it is important to highlight that the TMA-based study of spatial heterogeneity has some intrinsic limitations, including the lack of a comprehensive analysis of the whole section. This study, however, should be considered a proof-of-principle that the heterogeneity of HER2 gene amplification can be assessed by means of a high-throughput and cost-effective platform, using standard laboratory equipment. Further studies analyzing serial full-face sections of HER2-heterogeneous cases, coupled with cutting-edge molecular studies and patients' clinical data will be required to validate this protocol.
In conclusion, the comprehensive mapping of HER2 status in heterogeneous HER2 breast cancer specimens and the investigation of potential driver genetic mutations in HER2-negative components should rely on the analysis of multiple neoplastic regions. This analysis would require unbearable costs and efforts using standard diagnostic facilities. This method represents a reliable tool for the simultaneous characterization of HER2 genetic heterogeneity in large sets of multiple and heterogeneously processed breast cancers.
Disclosures
The authors have no conflicts of interests.
Acknowledgments
None.
References
- Allison KH, Dintzis SM, Schmidt RA. Frequency of HER2 heterogeneity by fluorescence in situ hybridization according to CAP expert panel recommendations: time for a new look at how to report heterogeneity. Am J Clin Pathol. 2011;136(6):864–871. doi: 10.1309/AJCPXTZSKBRIP07W. [DOI] [PubMed] [Google Scholar]
- Montemurro F, Scaltriti M. Biomarkers of drugs targeting HER-family signalling in cancer. J Pathol. 2014;232(2):219–229. doi: 10.1002/path.4269. [DOI] [PubMed] [Google Scholar]
- Wolff AC, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31(31):3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- Ng CK, Pemberton HN, Reis-Filho JS. Breast cancer intratumor genetic heterogeneity: causes and implications. Expert Rev Anticancer Ther. 2012;12(8):1021–1032. doi: 10.1586/era.12.85. [DOI] [PubMed] [Google Scholar]
- Vance GH, et al. Genetic heterogeneity in HER2 testing in breast cancer: panel summary and guidelines. Arch Pathol Lab Med. 2009;133(4):611–612. doi: 10.5858/133.4.611. [DOI] [PubMed] [Google Scholar]
- Murthy SS, et al. Assessment of HER2/Neu status by fluorescence in situ hybridization in immunohistochemistry-equivocal cases of invasive ductal carcinoma and aberrant signal patterns: a study at a tertiary cancer center. Indian J Pathol Microbiol. 2011;54(3):532–538. doi: 10.4103/0377-4929.85087. [DOI] [PubMed] [Google Scholar]
- Ohlschlegel C, Zahel K, Kradolfer D, Hell M, Jochum W. HER2 genetic heterogeneity in breast carcinoma. J Clin Pathol. 2011;64(12):1112–1116. doi: 10.1136/jclinpath-2011-200265. [DOI] [PubMed] [Google Scholar]
- Chang MC, Malowany JI, Mazurkiewicz J, Wood M. 'Genetic heterogeneity' in HER2/neu testing by fluorescence in situ hybridization: a study of 2,522 cases. Mod Pathol. 2012;25(5):683–688. doi: 10.1038/modpathol.2011.206. [DOI] [PubMed] [Google Scholar]
- Bartlett AI, et al. Heterogeneous HER2 gene amplification: impact on patient outcome and a clinically relevant definition. Am J Clin Pathol. 2011;136(2):266–274. doi: 10.1309/AJCP0EN6AQMWETZZ. [DOI] [PubMed] [Google Scholar]
- Seol H, et al. Intratumoral heterogeneity of HER2 gene amplification in breast cancer: its clinicopathological significance. Mod Pathol. 2012;25(7):938–948. doi: 10.1038/modpathol.2012.36. [DOI] [PubMed] [Google Scholar]
- Hanna WM, et al. HER2 in situ hybridization in breast cancer: clinical implications of polysomy 17 and genetic heterogeneity. Mod Pathol. 2014;27(1):4–18. doi: 10.1038/modpathol.2013.103. [DOI] [PubMed] [Google Scholar]
- Sanguedolce F, Bufo P. HER2 assessment by silver in situ hybridization: where are we now? Expert Rev Mol Diagn. 2015;15(3):385–398. doi: 10.1586/14737159.2015.992416. [DOI] [PubMed] [Google Scholar]
- Fusco N, et al. The Contrasting Role of p16Ink4A Patterns of Expression in Neuroendocrine and Non-Neuroendocrine Lung Tumors: A Comprehensive Analysis with Clinicopathologic and Molecular Correlations. PLoS One. 2015;10(12):e0144923. doi: 10.1371/journal.pone.0144923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanghali M, et al. Construction of tissue microarrays from core needle biopsies - a systematic literature review. Histopathology. 2016;68(3):323–332. doi: 10.1111/his.12802. [DOI] [PubMed] [Google Scholar]
- Navani S. Manual evaluation of tissue microarrays in a high-throughput research project: The contribution of Indian surgical pathology to the Human Protein Atlas (HPA) project. Proteomics. 2016;16(8):1266–1270. doi: 10.1002/pmic.201500409. [DOI] [PubMed] [Google Scholar]
- Fusco N, et al. HER2 in gastric cancer: a digital image analysis in pre-neoplastic, primary and metastatic lesions. Mod Pathol. 2013;26(6):816–824. doi: 10.1038/modpathol.2012.228. [DOI] [PubMed] [Google Scholar]
- Amin MB, et al. AJCC Cancer Staging Manual. Eight. Springer International Publishing; 2017. [Google Scholar]
- Lakhani SR, Ellis IO, Schnitt SJ, Tan PH, van de Vijver MJ. WHO Classification of Tumours of the Breast. 4th. IARC press; 2012. [Google Scholar]
- Kononen J, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4(7):844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- Fusco N, et al. Resolving quandaries: basaloid adenoid cystic carcinoma or breast cylindroma? The role of massively parallel sequencing. Histopathology. 2016;68(2):262–271. doi: 10.1111/his.12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco N, et al. Genetic events in the progression of adenoid cystic carcinoma of the breast to high-grade triple-negative breast cancer. Mod Pathol. 2016;29(11):1292–1305. doi: 10.1038/modpathol.2016.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco N, et al. Recurrent NAB2-STAT6 gene fusions and oestrogen receptor-α expression in pulmonary adenofibromas. Histopathology. 2017;70(6):906–917. doi: 10.1111/his.13165. [DOI] [PubMed] [Google Scholar]
- Hammond ME, et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28(16):2784–2795. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco N, Bosari S. HER2 aberrations and heterogeneity in cancers of the digestive system: Implications for pathologists and gastroenterologists. World J Gastroenterol. 2016;22(35):7926–7937. doi: 10.3748/wjg.v22.i35.7926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowsett M, et al. Assessment of Ki67 in breast cancer: recommendations from the International Ki67 in Breast Cancer working group. J Natl Cancer Inst. 2011;103(22):1656–1664. doi: 10.1093/jnci/djr393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker TJ, et al. Determining sensitivity and specificity of HER2 testing in breast cancer using a tissue micro-array approach. Breast Cancer Res. 2012;14(3):R93. doi: 10.1186/bcr3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sio G, et al. A MALDI-Mass Spectrometry Imaging method applicable to different formalin-fixed paraffin-embedded human tissues. Mol Biosyst. 2015;11(6):1507–1514. doi: 10.1039/c4mb00716f. [DOI] [PubMed] [Google Scholar]


