Abstract
Objective
This paper introduces the objectives, methods and results of ontology development in the EU co-funded project Advancing Clinico-genomic Trials on Cancer – Open Grid Services for Improving Medical Knowledge Discovery (ACGT). While the available data in the life sciences has recently grown both in amount and quality, the full exploitation of it is being hindered by the use of different underlying technologies, coding systems, category schemes and reporting methods on the part of different research groups. The goal of the ACGT project is to contribute to the resolution of these problems by developing an ontology-driven, semantic grid services infrastructure that will enable efficient execution of discovery-driven scientific workflows in the context of multi-centric, post-genomic clinical trials. The focus of the present paper is the ACGT Master Ontology (MO).
Methods
ACGT project researchers undertook a systematic review of existing domain and upper-level ontologies, as well as of existing ontology design software, implementation methods, and end-user interfaces. This included the careful study of best practices, design principles and evaluation methods for ontology design, maintenance, implementation, and versioning, as well as for use on the part of domain experts and clinicians.
Results
To date, the results of the ACGT project include (i) the development of a master ontology (the ACGT-MO) based on clearly defined principles of ontology development and evaluation; (ii) the development of a technical infra-structure (the ACGT Platform) that implements the ACGT-MO utilizing independent tools, components and resources that have been developed based on open architectural standards, and which includes an application updating and evolving the ontology efficiently in response to end-user needs; and (iii) the development of an Ontology-based Trial Management Application (ObTiMA) that integrates the ACGT-MO into the design process of clinical trials in order to guarantee automatic semantic integration without the need to perform a separate mapping process.
Keywords: Ontology, Cancer research, Translational medicine, Ontological engineering, Clinical trial administration
1. Introduction
Life sciences are currently at the center of an information revolution. The development of new techniques and tools is making possible the collection and organization of biological information at an unprecedented level of detail and in extremely large quantities. With respect to cancer research, the use of high-throughput technologies has resulted in an explosion of information and knowledge about cancers and their treatment. Because it is a complex multifactorial disease group that affects a significant portion of the population worldwide, cancer is a prime target for focused multidisciplinary efforts using these new and powerful technologies [1].
However, the lack of an open and shared information infrastructure is preventing clinical research institutions from being able to mine and analyze disparate data sources. Our inability to share technologies and data that have been developed by different organizations is severely hampering the research process. As a result, very few cross-site studies and multi-center clinical trials are being performed. In addition to this, it has proven to be impossible in most cases to seamlessly integrate data acquired from multiple levels of investigation (e.g., to integrate data from studies focused on the molecular elements of cancer with those focused on what happens at the level of organs, and those that focus on the entire individual).
The vision of the ACGT project (Advancing Clinico-genomic Trials on Cancer – Open Grid Services for Improving Medical Knowledge Discovery) is to contribute to the resolution of these problems by developing an ontology-driven, semantic grid services infrastructure that will enable efficient execution of discovery-driven analytical workflows in the context of multi-centric, postgenomic clinical trials. The ultimate objective of the ACGT project is the development of a secure semantic grid services infrastructure which will (a) facilitate seamless and secure access to heterogeneous, distributed multilevel databases; (b) provide a range of semantically rich re-usable, open tools for the analysis of such integrated, multilevel clinico-genomic data; (c) achieve these results in the context of discovery-driven (eScience) workflows and dynamic VOs; and (d) fulfill these objectives while complying with existing ethical and legal regulations.
In this paper we focus on the ACGT Master Ontology, the principles that guided its development, and the strategies employed for its evaluation and maintenance. We will present in detail the various ways in which the ontology has been utilized to address specific problems, such as semantic data integration by means of a mediator tool and the development of an open-source Ontology-based Trial Management Application.
2. The ACGT Master Ontology
2.1. Technical details
The ACGT Master Ontology (ACGT-MO) is implemented in OWLDL, 1 the description-logics based subtype of the Web Ontology Language (OWL) [2] and can be freely downloaded from http://www.ifomis.org/acgt.
The initial development or beta version of the ACGT-MO was published in June 2007 and it has been further expanded since that time in order to integrate and respond to the needs of users, both clinical and technical. The developers are now working toward version 1.0. At the moment the ontology contains 1667 classes, 288 object properties, 15 data properties and 61 individuals. An ontology of this size is difficult to present in its entirety in a journal paper. Therefore, we have limited ourselves here to providing figures containing selected details of the ontology (Figs. 1–7). For the interested reader, the complete owl-file of the ACGT-MO can be downloaded, accessed and viewed freely from http://www.ifomis.org/acgt/1.0.
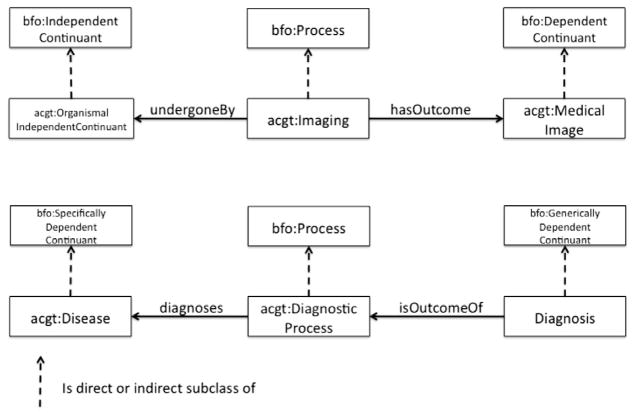
Fig. 1.
Relations between specific information objects (Medical Image, Diagnosis) and processes, independent and other dependent continuants.
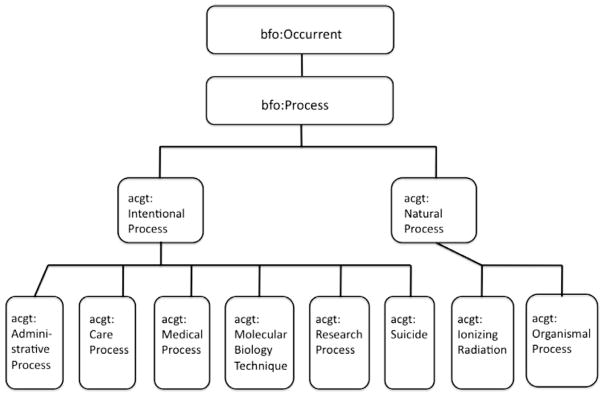
Fig. 7.
ACGT-specific subclasses to Process within the BFO hierarchy.
The ontology has been freely available since it was first published on the Internet in 2007 and comments and criticism of domain and ontology experts has been and is still invited.
There is currently an effort to reduce the number of object properties by around 60%. The reasons for this effort are both practical and principled. Practically speaking, it has become clear that 288 object properties are too many for most end-users to keep track of and utilize efficiently. On the other hand, from the standpoint of the ontology itself there are a number of redundant object properties, for instance undergoes_Process and undergoes_MedicalProcess, which considerations of simplicity and economy recommend eliminating wherever possible.
2.2. Scope
The ACGT-MO developers set out to comprehensively represent the domain of cancer research and management, with special emphasis on mammary carcinoma (“breast cancer”), Wilms’ tumor (nephroblastoma) and rhabdoid tumor. The development of the MO was guided and reviewed by researchers from two pre-existing clinical trials, namely a breast cancer-related trial on Topoisomerase II Alpha Gene Amplification and Protein Overexpression Predicting Efficacy of Epirubicin (TOP) [3] and “Nephroblastoma (Wilms’ Tumor) – Clinical Trial and Study SIOP 2001” by the International Society of Paediatric Oncology [4]. In order to achieve the aim of supporting unified data annotation for these trials, the developers had to shape the MO as a cross-section of a multitude of sub-domains, all of which are vitally important to clinical cancer management and research. In effect, the outcome of this effort is best seen, not as a comprehensive domain ontology, but rather as an application ontology tailored to the needs of the ACGT software system, and as functionally-driven toward the services to be described in Section 6 below. A domain ontology is an ontology that has a clear-cut and distinguishable subject matter, one unified by the kinds of objects that it contains, by the dominance of a particular set of concepts and distinctions pertinent to these objects, and often by certain characteristic methods of inquiry as well. Paradigm examples of domain ontologies include representations of basic scientific subject matters, such as anatomy, cytology, the different areas of genetics, etc. The ACGT-MO, by contrast, tackles a mixed bag of aspects arising from clinical cancer management and cancer research. As a result of this, a single clearly delineated domain to which the ACGT-MO applies cannot be easily identified. The MO, for instance, must represent administrative issues, as well as therapy- and laboratory-related facets of cancer in clinical reality. In designing it to do this we have been cautious to avoid the problem of use-mention mistakes that often occur in medical information systems. The use-mention distinction is violated when discourse that is intended to be about an object or kind of thing is phrased in such a way that it refers to the linguistic term for that thing rather than the thing itself. Consider the following two sentences:
Neoplasm is synonymous with tumor.
A neoplasm can be both, malign or benign.
The first statement is not a statement about neoplasms at all but rather a statement about the term “Neoplasm”, whereas the second is really a statement about actual things, namely neoplasms. Correctly formulated, (1) should be written as follows “Neoplasm” is synonymous with “tumor”. This example might seem relatively obvious, but in complex medical information systems statements about terms are quite often confused with or substituted for statements about the things in reality that the terms are intended to refer to. If an information system does not contain a sharp distinction between sentences of type one and type two, then consider what would happen if the system containing the above two sentences also contained the information: Neoplasm is a word. This would permit inference to the conclusion that there is some word that is either malign or benign, which is either false or, if true, not true in the same sense in which a neoplasm is malign or benign. So, a single use-mention confusion introduces either falsity or ambiguity into the information system, while many such confusions could truly compromise the overall quality of the data the system contains.
Thus, for the development of the ACGT-MO it was crucial to avoid this kind of mistake, especially since we needed to represent both the clinical reality and the various kinds of documentation of clinical reality in the domain of our research. In order to guarantee this, our ontology includes a class called acgt:InformationObject, which includes items such as reports about entities, identifiers of entities and so on. ACGT is an extension of an Upper Ontology, namely Basic Formal Ontology (BFO) and we choose to make acgt:InformationObject a subclass of bfo:GenericallyDependentContinuant. A bfo:GenericallyDependentContinuant is defined as a continuant [snap:Continuant] that is dependent on some other independent continuant [snap:IndependentContinuant] bearer such that every instance of a generically dependent continuant D requires some instance of an independent continuant C, but which particular instance of C serves as the bearer of D can change from time to time [5]. For example, Leo Tolstoy’s novel War and Peace (generically dependent continuant D) requires instantiation in some paper or electronic bearer (e.g., a book or a pdf file) C, but it is not particularly important for the existence of the novel as such which particular bearer instantiates it. We will elaborate in more detail on the use of BFO and its structure in Section 3.
Examples of representations of detailed, real world clinical trial data are given in Section 6.2.1, where the Ontology-based Trial Management Application is described.
Fig. 1 shows a number of examples linking objects and processes from clinical reality to documentation items that are the results of these, as well as the subclass relation that each of these entities (the objects, processes and documentation items) stand into various BFO classes. Fig. 2 shows ACGT-specific relations as sub-relations of relations imported to the ACGT-MO from an external source.
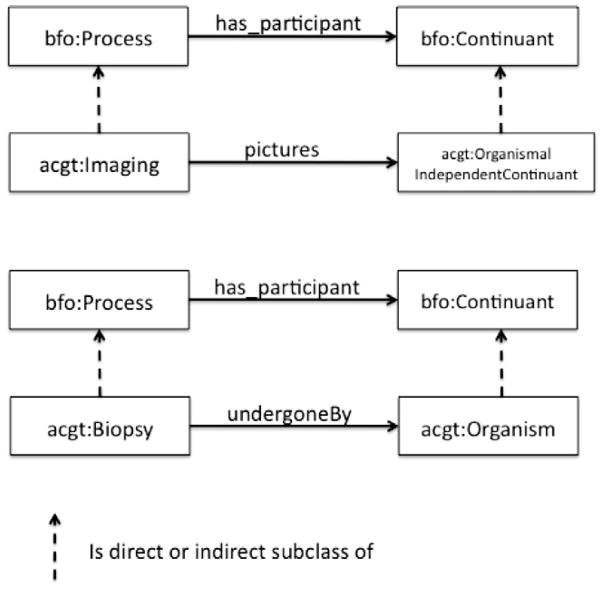
Fig. 2.
Relations between ACGT-specific classes and their superclasses from BFO.
All these pre-requisites make the ACGT-MO an application ontology, one unified primarily by the goals or ends that it is designed to achieve or facilitate.
In what follows, we will show how the practical constraints introduced by real-world software development needs have interacted in innovative ways with the design principles that we hold to be necessary for high quality ontology development.
2.3. Aim
The ACGT-MO is an application ontology and its main role, in the context of the translational medicine research framework within which it is developed and applied, is to support data integration across the borders of countries and disciplines, languages and professional terminologies; as well as integration of newly gathered data with data already stored.
As a result, the ACGT-MO is heavily used in the context of the ACGT Semantic Mediation Process – the scientific details of which are elaborated in Section 2.4. In specific, the two key systems exploiting the MO are the ACGT Semantic Mediator (Section 6.1) and the Ontology-based Trial Management Application (ObTiMA).
As for ObTiMA, the current version of the system aims to support clinical trial set up, design and managed. In this context, the MO is utilized as a global schema for data annotation. We foresee that Version 2 of ObTiMA will include decision support with respect to many critical issues for clinical trial set up and management. Such functional requirements are, nevertheless, out of scope for the ACGT project and the development of this functionality will go hand in hand with a process of ontology development towards the needs of such services. As a conclusion, the ACGT-MO does not aim to provide a comprehensive coverage of the complete domain neither in terms of class coverage nor in terms of class definition. Thus the development of new services and the expansion of the ontology itself are processes that will occur gradually and in tandem.
2.4. The ACGT-MO and semantic integration in the ACGT infrastructure
The requirements for the technical infrastructure of the ACGT (the ACGT Platform) are that it be able to support the semantic integration of heterogeneous data sources in cancer research and management. These requirements have been met by designing a federated environment, one that involves independent tools, components and resources that have been developed based on open architectural standards, and which are customizable and capable of dynamic reconfigurations.
In defining the initial architectural blueprint for such an environment a layered approach was selected, one providing different levels of abstraction and classification of functionality into groups of homologous software entities [6]. In specifying this architectural blueprint for the ACGT Platform, similar specifications from other relevant projects were thoroughly studied. Of particular relevance are the Cancer Biomedical Informatics Grid (caBIG) in the US and the CancerGrid project in the UK. One result of this is that the infrastructure being developed for the ACGT Platform uses a set of services and service registrations that are standard for the entire community of cancer clinical trial research. Further, in our approach the required security services and components are in place throughout the ACGT architecture so as to make available the user management, access rights management and enforcement, and trust bindings that are facilitated by Grid and domain-specific security requirements like pseudonymization and anonymization.
As stated previously, one of the key scientific goals of the ACGT is that of achieving semantic integration of heterogeneous, distributed and multilevel clinical and genomic data. Achieving this goal is thus also one of the key scientific and technological challenges of the ACGT. There are a number of different approaches to the achievement of semantically consistent data integration. The main methods fall into the following three categories: model alignment, using semantic tags or metadata, and developing shared conceptual reference models or ontologies [7].
The first approach, model alignment, creates mappings among models to support their semantic interoperability [8–10]. On this approach, alignment is achieved by identifying a relationship directly between synonymous terms in different models, e.g., if ‘biological cell’ appears in one model and ‘cell’ appears in another, where it is clear on investigation that these are intended to refer to the same thing in the two different models, then a mapping is established.
The second method is to use semantic tags or metadata [11], such as those used by the Dublin Core Metadata Initiative [12]. On this sort of approach, mappings are created not directly between data sources, but either between a data source and a meta-data set or between different metadata sets.
The third approach is to develop a core ontology or “shared conceptual reference model” to serve as the common ground for all of the systems to be integrated, and/or for purposes of defining a shared metadata set [13–15]. This third approach is more exact and centralized than the second, insofar as it provides a single frame of reference to which other models are to be mapped or, better, in terms of which entries in other models can be structured and defined.
In responding to the challenging objective of achieving semantically consistent integration of multilevel biomedical data, the ACGT project is pursuing – from among the various alternatives just described – the third: the use of a shared conceptual reference model or ontology. As a result, our semantic integration approach requires the definition and integration of three main components, which together comprise the core of the Semantic Mediation layer. These components are (a) The ACGT Master Ontology on Cancer (ACGT-MO) representing the shared conceptual model of the domain. (b) The mappings between ontology elements and data access services schemas. (c) The Semantic Mediator (SM), a software controlling the translation of queries and the integration of results. Additional components that are used for overcoming several issues in the data integration process are the Mapping Tool, the Data Cleaning module (for retrieved instances), and the Query Preprocessing Module (for literal homogenization in queries).
3. Principles guiding the development of the ACGT-MO
Ontology development is an activity that is constrained from multiple directions and that is subject to multiple, sometimes conflicting, demands: On the one hand, there are practical constraints set by the function or service the ontology-driven system is intended to achieve. On the other hand there are currently a growing number of ontologies, many of which have overlapping or similar contents and/or goals. The only way to ensure that ontologies in the future will be able to keep their promise of unifying the semantics underlying data organization and exchange in computer systems is to be aware of this situation and thus of the need to continually work toward harmonization.
Keeping this in mind, the ACGT-MO has been developed on the assumption that no new ontology should be developed if good pre-existing ontologies already cover its intended domain. Thus, a detailed and thorough review was conducted in order to determine whether developing a new ontology from scratch would indeed be necessary for achieving the goals of the ACGT project. This review covered the Systemized Nomenclature of Medicine – Clinical Terms (SNOMED-CT) [16], the Unified Medical Language System (UMLS) [17], and the National Cancer Institute Thesaurus (NCIT) [18] among others. Existing research literature on the ontology underlying each of these resources was taken into consideration. The conclusion reached was that none of the domain-specific terminologies currently in existence would be used, since none of them fully satisfied the quality criteria that have been adopted by the ACGT developers, criteria that are further discussed below.
In order to provide an idea of the kinds of problems that were discovered, some of the most severe issues identified with the three resources mentioned above are listed here:
-
SNOMED-CT:
Multiple Inheritance (Example: Repair of inguinal hernia (procedure) (ConceptID: 44558001) is_a Inguinal region repair (procedure) (ConceptID: 120205009) & is_a Repair of hernia of abdominal wall (ConceptID: 84744001) [19,20].
UnknownX classes (Example: Unknown living organism (ConceptID: 89088004) [19].
Imprecise usage of the is_a relation (Example: Both testes (ConceptID: 42774007) is_a Structure of bilateral paired structures (ConceptID: 422525002). It is debatable whether both testes of an individual form a structure; it might be safe to say they form a set, though) [19,20].
-
UMLS
While the UMLS uses an Upper Ontology, it is reported to have consistency problems with respect to keeping processes and functions separate, in particular where processes executing functions are involved [21].
-
NCIT
Use of non-formal is_a relations (Example: Other Organism Groupings is_a Organism) [18].
NCIT lacks a coherent Upper Ontology. Biological Process and Biological Function are synonymous in the thesaurus, and thus would refer to the same set of individuals [22]. Furthermore, there is no distinction between physical entities and realizable entities (e.g., roles, functions), which leads to incoherent classifications (Example: Infectious Agent: Virus is not a subclass of Virus, but of Other Organism Groupings. Virus and Other Organism Groupings are both subclasses of Organism [18].
Re-use of one existing ontology, namely the Foundational Model of Anatomy (FMA) [23] was approved, while the decision to reuse the OBO Relation Ontology (RO) [24] was made both because the OBO Relation Ontology is a high quality relation ontology by current standards and because making use of the OBO Relation Ontology is a pre-requisite for becoming a member of the OBO Foundry [25], something which was part of the ACGT evaluation strategy from the beginning (Section 5.3).
A virtue of these latter ontologies is that they stick to specific well-defined and explained methods of ontology development, based on sound theoretical principles. For instance, they seek to develop ontologies with a logical structure that can support algorithmic processing, with a concern for the reality to which the terms in an ontology relate (so that the ontology rests on a clear distinction between entities in reality and the documents or data entries used to represent them), and a concern for the interoperability of the ontology being developed with other representations of related domains of entities [26].
The basic principles and methods that have been selected and employed in the development process of the ACGT-MO are the following, which are first listed here, then subsequently explained in greater detail below:
Adopting a radically restrictive definition of the term “ontology”.
Enforcing a strict subsumption hierarchy, based on a formally specified is_a relation.
Avoiding (non-trivial) multiple inheritance in the hierarchy of universals.
Avoiding “UnknownX” and related classes.
Using an Upper Ontology, namely Basic Formal Ontology.
Using OBO Relation Ontology (RO).
3.1. The adoption of a radically restrictive definition of the term “ontology”, in compliance with the principles of realism
The following definition of ‘ontology’ has recently been proposed [27], and contains most of the crucial elements presupposed by the ACGT-MO understanding of ontology: “an ontology is a representational artifact whose representational units are intended to designate universals in reality and the relations between them”. This definition of an ontology has two parts. The first identifies an ontology as a representational artifact consisting of representational units, while the second has to do with what the representational units in such an artifact are intended to refer to, namely “universals and relations between them” in reality. Here we will first say a few things about universals, then clarify the understanding of “representational artifact” that is being employed.
To begin with universals: when a biologist studies an animal, a particular cat for example, it is normally not because the biologist is interested in the features of this very cat, but rather that she is interested in the cat (and others like it that she may study) as instances of a general kind, as being a potential source of information about cats in general. It is normally this kind of general or abstract information that sciences are interested in capturing. In the history of philosophy and science, universals have been proposed and understood as that which is general or abstract in reality; as the entities or principles that scientists are really seeking knowledge of when they seek truths that apply to and explain all members of a species or all kinds of DNA or all particles in the universe. Universals can thus be seen as a sort of theoretical explanation of the structure, order and regularity that is to be found in nature, and as what all members of a natural kind, grouping or species (such as Oxygen or the cat just mentioned) have in common, at some level of abstraction. Universals are repeatable in the sense that they can be instantiated by more than one object and at more than one time (that they instantiate the universal “Cat” is what all particular cats – cat1, cat2, cat3, etc. – have in common). As opposed to universals, particulars are the individual denizens of reality. Particulars instantiate universals, but cannot themselves be instantiated, and it is in virtue of instantiating the same universal that two particulars will be similar in some respect (e.g., both being cats, both being chromosomes, etc.). Universals can also be related to each other in various ways. For example, the universal “Cat” is related to the universal “Mammal” in the relation of species to genus, since all cats are mammals.
Given all of this, saying that an ontology is a representation of universals and relations between them in reality has a twofold purpose. The primary purpose is to establish that an ontology is a structured collection of information about kinds or types of things, rather than about individuals. The goal of an ontology is first and foremost to codify and articulate relations between general truths that apply to whole classes of things, not just to single individuals or members of those classes in the world. The second purpose of emphasizing the representation of universals in an ontology is to stress the point that the representation of information about a whole group or kind (universal) and the representation of information about specific individuals (particulars) are different and should be represented differently and kept separate in an ontology. This is especially important in an application ontology such as the ACGT, the goals of which will sometimes require representing specific individuals or institutions, as well as general or abstract kinds of things.
Turning now to the notion of a representational artifact: a representational artifact is an entity that makes pre-existing cognitive representations from the minds of its author or authors publicly available. Representational artifacts include things such as signs, books, pictures and diagrams and have the key feature of including ledgers or rules for their interpretation. Thus, maps do not simply come color coded, they also come with a key or table that makes it possible to interpret their color coding as representing certain kinds of things (countries, oceans, mountain ranges, etc.), and the words in which these tables and keys are written themselves have publicly available rules for their interpretation as referring to things in the world, namely the semantics of natural language itself. According to the above definition then, an ontology is just a highly sophisticated kind of representational artifact. Viewing an ontology in this way leads naturally to two ideas, both of which have functioned as principles in the development of the ACGT-MO:
When constructing a representational artifact for use in science, such as an ontology, based on cognitive representations or concepts in the minds of individual subjects, the goal is not to accurately represent in a publicly accessible way the representations or concepts that exist in those individual’s minds, but rather the things in reality that these representations are representations of. Recognition of this principle is also the point, in the above definition, of saying that an ontology is a representation of universals in reality.
There is a fundamental distinction between using such artifacts to make reference to things in reality, i.e., the entities that they are representations of (e.g., “cats are mammals” or “Cancer is a disease”), on the one hand, and mentioning such artifacts by engaging in discourse about them on the other (e.g., ‘cat’ is a three letter English word or ‘Cancer’ is a term defined in the ACGT-MO). The construction of coherent functional ontologies requires that this use-mention distinction be strictly consistently applied and respected.
The following is an example of a conflation of the use and the mention of a term, taken from an old (and now corrected) definition of ‘mouse’ in BIRNLex:
‘mouse’ is defined as the “name for the species mus musculus”.
The problem with a definition such as this is that it provides information about the word ‘mouse’, rather than information about the biological species “mouse” in reality that is the intended object of scientific study and discourse, thus mentioning the word rather than using it. One goal major goal in the development of the ACGT-MO has been to carefully avoid this sort of potential use-mention confusion.
3.2. Enforcing a strict subsumption hierarchy, based on a formally specified is_a relation, as opposed to a loose “subclass” hierarchy
The great majority of currently existing ontologies incorporate relations that connect their terms (“nodes”). Such relations, however, are sometimes being used in very informal ways, often providing no definitions at all, so that the resulting logical interconnections are far from clear. Even the basic taxonomical relation is_a (as in “Dolphin is_a Mammal”), the foundation of any ontology, is not always used in a consistent or clear fashion. A formal is_a relation should at the very least ensure that an instance of a class is also an instance of its parent class (e.g., that if Tibbles is an instance of the class/universal Cat, and Cat is_a Mammal, then Tibbles is an instance of the class mammal), which is not what always happens in the case of loosely defined taxonomies as encountered in many well-known contemporary ontologies, both formal and domain-specific. Lassila [28] gives the following example of this kind of inaccuracy, taken from Yahoo: “[…] the general category apparel includes a subcategory women (which should more accurately be titled women’s apparel) which then includes subcategories accessories and dresses. While it is the case that every instance of a dress is an instance of apparel (and probably an instance of women’s dress), it is not the case that a dress is a woman and it is also not the case that a fragrance (an instance of a women’s accessory) is an instance of apparel. This mixing of categories such as accessories in web classification schemes is not unique to Yahoo – it appears in many web classification schemes”. While such inaccuracy may be tolerable in the context of shopping for clothes, it seems much less tolerable in the context of serious scientific and medical classification and research, and it has been strictly avoided in the development of the ACGT-MO.
3.3. Avoiding (non-trivial) multiple inheritance in the hierarchy of universals
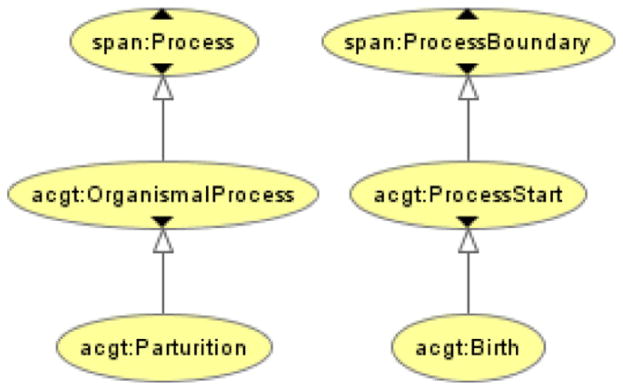
We also embrace the principle that a properly constructed ontology should steer clear of an asserted taxonomical tree that allows multiple parent classes for the same child class (i.e., one child that inherits from multiple parents, so-called “multiple inheritance”). The central aim is to avoid the polysemy, or assignment of multiple meanings to a single term, that often results from multiple inheritances. In the ACGT-MO we chose to deal with polysemy by undertaking a disambiguation of naturally-occurring polysemic terms; e.g., Birth in natural language denotes, among others, both the beginning of Life (a ProcessBoundary), and a Process simpliciter – namely the very process of giving birth. The latter can also be encountered in the specialty literature under the more specific term of Parturition (with proper part Labor), which we chose to adopt, while leaving the term Birth under its former, more common, reading (see Fig. 3).
Fig. 3.
Resolving polysemy.
Related to the multiple inheritance avoidance principle, we subscribe to the principle according to which sibling classes in an ontology should be disjoint. The principle of disjointness says that two sibling classes should not share any members. In terms of “universals”, the principle says that a given particular cannot be an instantiation of two sibling universals. This is one reason why it is important to have the category Role in an ontology. If groups like Physicians and Patients are primitive subclasses of the class Human Being it follows that a particular person cannot be both a physician and a patient. Nevertheless, we know that this occurs in reality. Therefore it is important to represent Physician and Patient as a Role that can be realized by a Human Being, thus avoiding multiple inheritance on the basis of a principled distinction between individuals and the roles that they can, at various times, play or take on.
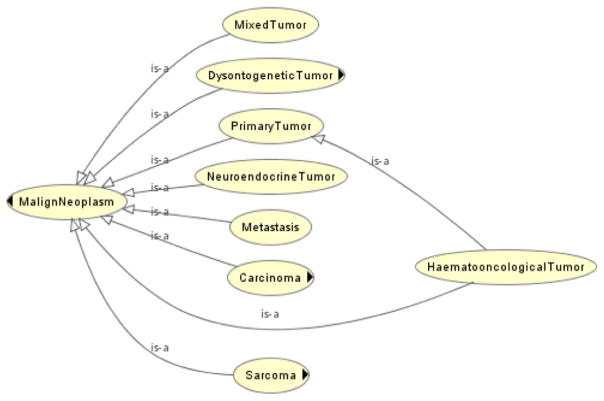
An important exception to the disjointness rule has been tolerated in the ACGT-MO, due to circumstances relating to the mapping process. The architecture of the system built around the ontology comprises, among others, several independently-developed cancer databases (breast cancer, nephroblastoma, Rhabdoid tumor etc.), databases whose terms (fields, cells, records etc.) are supposed to be mapped onto the ACGT-MO as part of the unifying function of the ontology-driven system (the “mapping process”). Querying the ontology (the SPARQL query language [29] has been used for this) can thus be automatically translated/mapped into querying the databases themselves. Unfortunately, the mapping of SPARQL queries would have been considerably hindered by the existence of OPTIONAL and FILTER blocks – blocks normally required by a definition of the PrimaryTumor class in terms of the non-existence of tumors whose metastasis that primary tumor is.2 We have, hence, opted to add both the PrimaryTumor and Metastasis classes to the asserted taxonomy, even though this violates the completeness desideratum often mentioned for clean ontology development: aside from haematooncological tumors, all other tumors (mixed, dysontogenic, neuroendocrine, carcinoma and sarcoma) have both instances that belong in the PrimaryTumor class and the Metastasis class (see Fig. 4). Note that the two classes, which are not built according to the best practice, PrimaryTumor and Metastasis, should ideally be conceived as roles.
Fig. 4.
Disjointness violations in the ACGT-MO.
It is also worth noting that as of this writing, the ACGT-MO includes rather few disjointness stipulations, as there is considerable content-related debate in this respect; we do, however, expect that further versions will make progress towards exhibiting disjoint classes more fully and faithfully. Prompted by similar considerations, we do not exclude further violations of the disjointness rule in the future, even though we would prefer that the amount of such exceptions be kept as low as possible.
3.4. Avoiding UnknownX and related classes
A common procedure among developers of medical databases, terminologies, and ontologies, is the inclusion of classes of type UnknownX, such as “UnspecifiedTumorStage” or “UnknownAffiliation”. “Universals” like these do not, however, have any instances, but merely indicate a lack of data or knowledge. Hence they represent an illegitimate epistemic intrusion into what should otherwise constitute a faithful picture of reality, of what there is. The alleged instances of these “universals” also do not exhibit any shared properties, at least not in most cases, which further speaks against treating them as genuine kinds or classes of things, scientifically speaking. Still, daily clinical care cannot get by without accounting for such lack of knowledge, e.g., to highlight that a certain test still needs to be done for a patient. Therefore information models have to be created indicating the state of epistemic knowledge (and “non-knowledge”) at some point in time, while their actual content can/should, in turn, be modeled based on ontological classes and definitions. Hence if we provide classes only for reality modeling but not for knowledge modeling, then the model would need an additional source for performing the later. In order to avoid this problem, while yet striving to anchor the master ontology in reality as much as possible, we have opted to include some minor epistemic classes via the import of well known and widely used medical classifications like the German version of the TNM [30].
Here (see Fig. 5) TNM’s MX class reads “Presence of distant metastasis cannot be assessed”. It is important to note that “ClinicalClassTCM” is not an object-like entity but a subclass of quality.
Fig. 5.
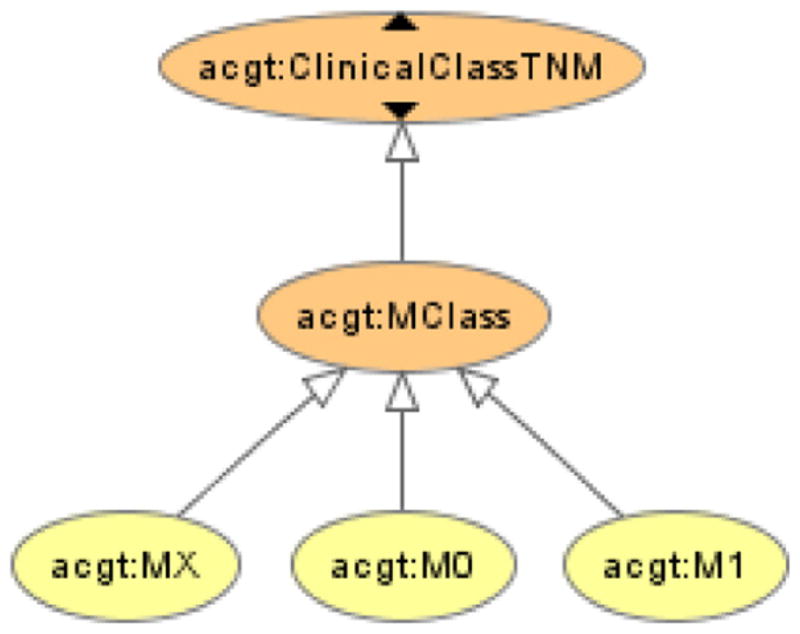
TNM’s MX class.
3.5. Using an Upper Ontology, namely Basic Formal Ontology
The Standard Upper Ontology (SUO) working group of IEEE defines Upper Ontology as follows:
An Upper Ontology is limited to concepts that are meta, generic, abstract and philosophical, and therefore are general enough to address (at a high-level) a broad range of domain areas. Concepts specific to given domains will not be included; however, this standard will provide a structure and a set of general concepts upon which domain ontologies (e.g., medical, financial, engineering, etc.) could be constructed [31].
Smith and Brochhausen [26] identify the use of an Upper Ontology framework for reality representation as a basic harmonizationfostering feature. Upper-level ontologies can provide not merely basic categories and basic structure ensuring good ontology organization, but also a set of tested principles that can be re-used by others in the development of specific domain ontologies.
For the ACGT-MO the project partners agreed to import Basic Formal Ontology (BFO) [5], an ontology that is also an entry in the OBO Foundry initiative [25]. The latter is a library of ontologies built to meet the same set of quality criteria and to provide ontological reference frameworks for different domains of the life sciences [32].
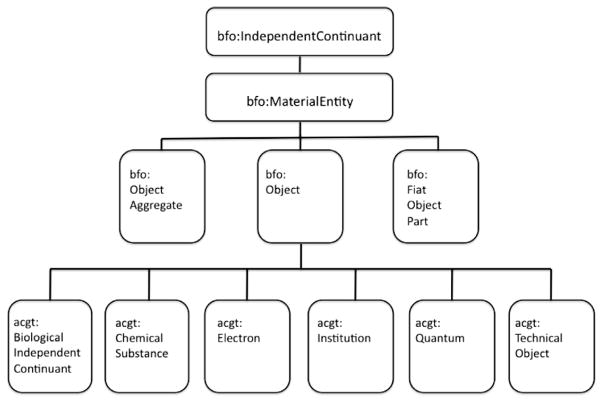
The BFO taxonomy makes use of a basic top-level distinction between two kinds of entities: substantial entities or continuants (entities that endure through time while maintaining their identity) on the one hand, and occurrents or perdurants (entities that happen, unfold, or develop in time) on the other. Corresponding to these two kinds of entities are two basic and distinct perspectives that can be taken on the world, neither of which can fully capture or represent the features of reality represented by the other: these are the SNAP and SPAN perspectives or ontologies, respectively [33]. For our present purposes, it suffices to mention that the SNAP ontology recognizes three major categories of continuants: dependent continuants, independent continuants and spatial regions, while SPAN includes processual entities and spatiotemporal regions. Fig. 6 shows a ACGT-specific subclass structure subsumed under the SNAP branch of BFO, while Fig. 7 gives depicts a detail from the ACGT-specific subclass structure subsumed under the SPAN branch of BFO.
Fig. 6.
ACGT-specific subclasses to Object within the BFO hierarchy.
3.6. Using OBO Relation Ontology (RO) as a source of, and insight for new relations/properties
The ACGT-MO not only represents classes as linked via the basic taxonomical relation (“is_a”), but it also connects them and/or restricts their usage via other semantic relations called “properties” in OWL terminology (e.g., connecting organs and their parts through the parthood relation, and connecting processes and the entities participating in them through the participation relation). Specifically, the OBO Relation Ontology (RO) [24,34] has been used as the basis for representing relations in ACGT because the RO has been specifically developed to account for relations in biomedical ontologies and includes clear and exact definitions specifying the key logical features (transitivity, reflexivity, etc.) of most of the relations it contains. In addition to the benefit of having clearly defined and consistently used relations, using the RO for relation regimentation is also part of the OBO Foundry criteria of ontology excellence. The designers of the ACGT-MO have hence set as one of their goals the inclusion of the ACGT-MO among OBO Foundry ontologies.
As of October 2009, the RO comprises thirteen class-level relations [24]. While the ACGT-MO uses RO, its domain-specific requirements call for more domain-specific relations than the RO currently supplies. For this reason a number of additional relations have had to be defined in the ACGT-MO. In such cases, our goal has been to approximate as closely as possible the clarity and logical explicitness of the RO relations themselves. Table 1 gives examples of non-RO relations in the ACGT-MO.
Table 1.
Non-RO relations in the ACGT-MO.
| addedBy | adds | adverseReactionTo | beginningOf |
| birthOf | causedBy | causes | characterizedBy |
| characterizes | compatibleWith | contralateralTo | denies |
| describedBy | describes | diagnosedBy | diagnoses |
| examinedBy | examines | followUpOf | fulfilledBy |
| hasAdverseReaction | hasBeginning | hasFollowUp | hasHabit |
| hasHistory | hasInfluenceOn | hasInput | hasLegalGuardian |
| hasMetastasis | hasOutput | hasProcessBoundary | hasProtocol |
| hasQuality | hasReason | hasReceptor | hasRelative |
| hasSymptom | hasTherapyAim | implementedBy | implements |
| issuedBy | issues | patientAt | picturedBy |
Due to the fact that the ACGT-MO is an application ontology geared to the specific needs of the ACGT Semantic Mediation Service and the ObTiMA service, no emphasis was laid on providing fully defined classes where representing contingent relations between classes was sufficient to ensure the functionalities aimed at. This is especially important for the way ontological annotations for data are created via ObTiMA. This methodology is explained in detail in Section 6.2.1. In effect the ACGT-MO contains 67 fully defined classes, among them the class “Disease”. The subclasses of “Disease” are represented with relations to other classes, but are not fully defined. How the relations and classes are used to create unified annotations for clinical trial data is shown in detail in Section 6.2.1.
4. Maintenance of the ACGT-MO
The development of medical ontologies, such as the ACGT-MO, reflects the rapid evolution of medical research as a whole. This leads to the known problem of ontology evolution: on the one side, ontologies need to be well-crafted and widely accepted by experts in order to constitute the common agreement on semantics within the operations of our information systems, on the other side there is an urgent need for users to be able to use the latest terminology in their ongoing research.
The ACGT Information Systems use the ACGT-MO as a built-in semantic reference. The challenge is to be able to classify documents (clinical report forms, microbiological processes and findings, etc.) with the latest terminology even before it has been widely approved, and nevertheless to evolve the MO as a stable reference consistent with all documents managed by the system. This implies in particular a need for the ability to represent and retrieve information accurately and precisely at any time.
Two main ontology maintenance processes are described in the literature [35]:
The scientific peer to peer review of concepts and their formal description.
The “democratic” evolution approach, resulting in so-called folksonomies.
The first is a re-active way to keep the ontology up-to-date. Once a concept appears in use, i.e., in literature or databases, decisions about its precise meaning and accepted use are made. These decisions are usually made by a small group of ontology experts whose knowledge of the ontology in question makes their interpretation more or less “authoritative”. They maintain high quality standards, but notoriously lag behind developments in the field. In folksonomies by contrast, anyone can introduce or change a concept as needed. As a result of this, the ontology is always up-to-date and reflects a sort of agreement (of the activists), but the ontology usually lacks the formal consistency necessary for advanced reasoning and runs the risk of having other confusions introduced.
In ACGT an innovative hybrid system was introduced, which combines virtues from both approaches: registered users are allowed to introduce (submit and use) any new class that they wish, on the condition that it is declared as a specialization of some already authorized broader class. This guarantees that it will be possible to locate the newly classified content, which eventually allows an expert team to take over the dialogue about the new class and to make determinations about it, as well as to deal with any content migration associated with it.
4.1. The ACGT Submission System
A major need of the ACGT community was to create a workflow and communication system that would gather all the change requests regarding the content of the ACGT-MO, feed them to the ontology experts in a manageable way, keep the version history of the ACGT-MO, and automate the communication back to the interested parties of any changes taken place. These functional requirements imply that the required information system should have the ability to reclassify content or to rewrite queries involving any authorized new expression that has replaced an old, an obsolete or a previously-used but currently rejected user-provided term. To that end the ACGT Submission System was created. The system is a re-active communication system allowing end-users to criticize and/or submit their own opinion on the existing ACGT-MO to its maintenance team.
The Submission System does not replace ontology development systems such as “Protégé”. Rather, its role is to gather requests for changes, assist the ontology expert by providing access to those requests and by providing a point of reference for the changes in the ontology, and to maintain previous ontology versions on a perclass basis, including the history of related requests. The reason for this is simply that previous classes, versions of or changes to the ontology may well be of relevance in making future decisions about what to include or whether or not to make a change. The ACGT Submission System interfaces with an ontology development system, here Protégé, to implement changes in a particular version of the ACGT-MO and to control the formal consistency of all classes in that version. It (semi-automatically) traces and registers the changes made and relates them to previous versions of the ontology, including changes to individual classes and requests for such changes. The relatively loose coupling with Protégé has the advantage of rendering the ACGT Submission System highly generic and potentially useable with other ontology development systems in the future (Protégé, even though quite popular, is not yet stable enough to encourage a tighter coupling). The system manages the workflow of processing requests, the details of decision-making, and the necessary communications in order to minimize reliance on manual checking and carrying out of these things by human beings. It is inspired by the workflow patterns of well-known international thesaurus development teams such as the Getty Research Institution or English Heritage.
The Submission System can be accessed by authorized users independently through the Web or from within the ObTiMa System described in Section 6.2. Thus, ObTiMa users can add change requests to the ACGT-MO directly from ObTiMa during the process of document definition.
The ACGT Submission System distinguishes three user roles:
The Contributor: A contributor to the system is a person who wishes to comment or suggest changes to the ontology, requesting additions/deletions or modifications of the existing ontology contents.
The Domain Expert: The Domain expert contributes to the system by reviewing the submissions of the Contributors that concern their field of expertise, and informs the Ontology Experts of the necessary changes to the ontology.
The Ontology Expert: The Ontology Expert is trained in logic and formal ontologies and general possesses only minimal domain knowledge. (S)he is responsible for the maintenance of the ontology. (S)he receives all the change requests (submissions), answers them or forwards them to a Domain expert. This communication is automated to the highest degree possible.
The ontology experts can browse through submissions, review the submissions, discuss them with contributors and domain experts, and decide whether they agree or disagree with the proposed changes, leading to either their implementation or their rejection. Any rejection of a proposed change will be accompanied by a declaration of how the correct meaning of a proposed class is to be expressed by the MO (a migration path). In assistance, the system provides the ontology expert with adequate information services about all related class versions and submissions. The system provides automatic feedback in the form of notifications to the Contributors on the status of their submissions, and on the status of the ontology. The system manages the publication of sets of changes to the ontology on a release-by-release basis. A new release can be incorporated into the already running ACGT Information Systems along with migration information.
4.2. The Submission Process
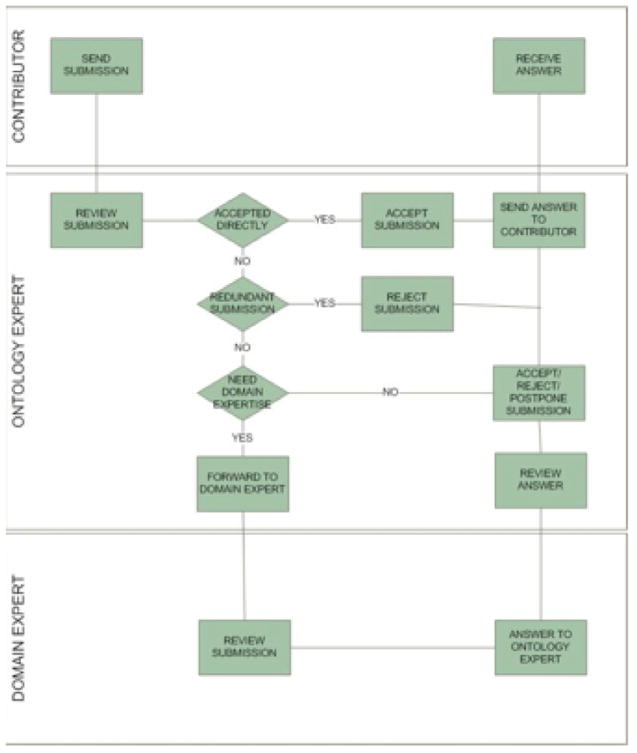
In this subsection the process following a new submission (see Fig. 8) is described in more detail:
Fig. 8.
The Submission Process.
When inserting a new change request (submission) into the System, the end-user automatically receives a notification certifying the submission. Once this is done, the new submission is inserted into the submission pool of the System. These new submissions are sent via mail to the Ontology Expert (a team or an individual), in order to inform her about the new change requests, and the Ontology Expert can see the new submissions to the system by logging into the system.
In the sequence, the Ontology Expert reviews the new submission. The submission may be directly accepted, being seen as redundant, or the Ontology Expert may need domain expert advice. If it is accepted, the contributor receives a notification. It is redundant if it refers to something already covered by the MO. In such a case it is rejected along with an explanation. If more domain expertise is needed, the Ontology Expert sends the submission to the Domain Expert (a group or individual). The Domain Expert will be informed via mail about the submission. After the Domain Expert has checked the submission, he can either reformulate it and send it back to the Ontology Expert or introduce an Implementation Proposal for the request. Either way, the Domain expert sends the submission back and the Ontology Expert accepts, reject, or postpones the submission and sends an answer, i.e., the way it will be implemented or not implemented, to the Contributor.
At release time, all contributors are once again notified that their accepted submissions have been released in an authorized version.
5. Evaluation of the ACGT-MO
5.1. Criteria of ontology evaluation
Within ontology-driven computing there is a clear need to begin focusing more heavily on ontology evaluation, particularly since the spread of ontological engineering over the last years has fostered the development of a multitude of ontologies, often representing the same or similar domains. On the one hand it is good to see that ontologies are becoming more and more a common solution for interoperability problems. On the other hand, the vast number of ontology artifacts that are available leaves engineers who are potentially interested in utilizing ontologies with the problem of evaluating the different ontologies that are available, and of identifying the ontology that will be most appropriate for their concerns. Yet, the development of shared standards for evaluating ontologies seems to be moving rather slowly. Furthermore, the development of multiple domain ontologies that are not interoperable with one another is a threat to the promise of semantic interoperability held out by ontology-driven systems.
Ref. [36] provides a description of four different methodologies for ontology evaluation. Yet, when it comes to evaluating the usability of the methods themselves, the authors are merely checking whether or not a methodology is actually in use, regardless of the outcome it produces.
It is widely accepted that there is a central distinction to be drawn between two different evaluation strategies, namely “glass box” or “component” evaluation and “black box” or “task based” evaluation. This distinction applies to evaluation processes for ontologies and ontology-driven systems as well [36,37]. The two strategies must be seen as complementary, each providing testing for different kinds of significant qualities.
Glass box evaluation is used for the evaluation of the ontology as such, on its adequacy as a logically structured representation of some domain of reality. It evaluates aspects such as domain coverage, the fitness of the ontology for a given task, and everything that has to do with logical and structural virtues of the artifact at hand, plus an assessment of the modules out of which the ontology is built [37].
Hartmann et al. stress that glass box evaluating should start in the design phase of an ontology (type 1), should accompany the entire development process (type 2), and should continue after the release of the ontology (type 3). Typically, type 1 and type 2 evaluations are done by the ontology engineer developing the artifact in question, whereas type 3 evaluation is usually carried out by ontology experts outside of the project [36]. Notably, there are no domain experts or end-users involved in these activities.
For the ACGT project four main aspects of glass box evaluation were identified:
Logical soundness;
Domain coverage;
Task orientation;
Re-use of existing ontologies.
By contrast with glass box evaluation, black box evaluation focuses on the adequacy of the ontology as a functional computational system. It measures the performance of an ontology-driven application and is typically carried out using the same interfaces that the end-users are going to employ [36]. Gangemi et al. [37] identify user-friendliness and agreement of domain experts as quintessential measurements to be considered in black box evaluations. Naturally, black box evaluation can be carried out by end-users. In this paper we concentrate on reporting a glass box evaluation technique used on the ACGT-MO and its results. Black box evaluation ought to be carried out once the entirety of the ACGT system is available.
5.1.1. Logical soundness
With respect to ontology development it is crucial to check that the ontology at hand does not contain any contradictory statements. A contradiction free artifact is called consistent. The consistency of the ACGT-MO is constantly and automatically checked using the Pellet reasoner application [38]. Constant consistency checks during the development process are highly important in order to facilitate troubleshooting, once inconsistencies occur, and to facilitate the tracking down of erroneous logical definitions.
5.1.2. Domain coverage
Validating the domain coverage is crucial to ensuring the usability of an ontology. There are a multitude of strategies for this task. For the ACGT-MO we decided to automatically extract term lists from domain-specific publications, namely journal articles on clinical aspects of mamma carcinoma, Wilm’s tumor and rhabdoid tumor. The text corpus used for NLP-based term extraction consisted of slightly more than 3000 abstracts. The resulting term list was then filtered to eliminate non-domain-specific terms, and the number of direct mappings was evaluated. Bearing in mind the fundamental difference between terminology and ontology, it is obvious that the direct mappings give only a hint regarding the actual completeness of domain coverage. Moreover, the study will check to determine to what extent the ACGT-MO provides reference to the things designated by the terms extracted. This effort is still ongoing and results will be published in parallel with this paper.
5.1.3. Task orientation
The task orientation of the ACGT-MO was secured by a joint development of the ontology with all applications of the ACGT project exploiting the ontology. The description of the ACGT ontology development principles and their specifications in Section 3 above indicates that compromises in favor of task orientation were made when necessary. The way the MO deals with PrimaryTumor and Metastasis in relation to the different pathological types of tumors is a good example of this kind of task orientation (see Fig. 4 and the associated discussion in Section 3 above).
Another aspect is that the ACGT-MO needs to give relatively detailed information about constraints, especially for some of the leaf nodes. In order to supply the knowledge basis for creating, for instance, Case Report Forms (CRFs) (as described below) it was unavoidable to represent constraints with cardinality restriction set to 0 (zero), e.g., Chemotherapy has Role min 0 AdjuvantChemotherapy. These kinds of constraints exhibit the high-level of task orientation that has guided the development of the ACGT-MO.
5.1.4. Re-use of existing ontologies
The ACGT-MO re-uses three ontologies of the OBO Foundry [25], which is a library of ontologies built to meet the same quality criteria and to provide ontological reference for different domains of the life sciences. The three ontologies are:
Basic Formal Ontology (BFO) [5];
Relation Ontology (RO) [27];
Foundational Model of Anatomy (FMA) [23].
While the OWL implementations of BFO and RO are directly imported into the OWL file of the ACGT-MO, this was not possible with respect to the FMA. The reason for this was first, that no official version of the FMA in OWL existed when we started the development (there were only two experimental conversions), and second, that the sheer size of the FMA in its entirety was by far too large (it was, e.g., impossible to apply the reasoner to the FMA). The developers of the MO thus decided to include anatomical entities as they occurred in the documentation serving as a blueprint for the targeted studies, and then to represent these in a formal is_a hierarchy using the FMA as a model. The whole upper structure of the ontological representation of anatomical entities in ACGT-MO is thus effectively taken from the FMA.
5.2. The role of the OBO Foundry in the evaluation of the ACGT-MO
From the beginning the ACGT consortium planned to submit the MO to the OBO Foundry in order to secure high quality in ontology development and generate feedback from ontology experts. Most of the criteria of the foundry are already fulfilled by the ACGT-MO, while some others are the subject of ongoing work.
5.3. The use of the ACGT-MO outside the ACGT project
From the beginning the ACGT-MO developers followed strategies of ontology development which state that it is vitally important to treat ontology development as a scientific enterprise, inviting critical discussion among experts to optimize the results and stay clear of idiosyncratic solutions. In April 2009 the developers uploaded the ACGT-MO to the National Center for Biomedical Ontology’s (NCBO) BioPortal [39]. Making it available for interested domain experts and ontologists.
Even though the ACGT project is still ongoing, the ACGT-MO has already experienced interest among other experts in the field of ontology-driven clinical data integration. In specific the ACGT-MO is currently used by the Theseus medico project [40]. Publications on its use within that framework are under preparation. In [41] the ACGT-MO is used as a possible bridging tool between pre-existing health communication standards. Also, the ACGT-MO is used to provide a middle layer for clinical disease management as a basis for disease specific sub-ontologies [42], within the EU project CHRONIUS (FP7-ICT-2007-1-216461 – CHRONIOUS) which focuses on chronic disease management.
6. Exploitation of the MO in the ACGT project
The ACGT project is devoted to the development of a technological infrastructure – namely, the ACGT Platform – aimed at assisting clinicians, bioinformaticians and medical researchers involved in cancer-related clinical trials in their data integration and analysis tasks. The ACGT Platform is comprised of several services designed to facilitate interaction between these groups. Two of these services (the ACGT Semantic Mediator and the ObTiMA system) require a semantic framework describing the domain of cancer for proper functioning. This semantic framework is in both cases provided by the ACGT-MO. The next subsections describe these components in some detail.
6.1. Semantic data integration in ACGT
6.1.1. Ontologies in database integration systems – background
Ontologies have been widely used in recent years to overcome some of the difficulties encountered when integrating heterogeneous databases. In [43], Jakoniene and Lambrix describe specific tasks in database integration that can benefit from the use of ontologies, namely: (i) query formulation, (ii) query rewriting, and (iii) data integration. In query formulation, ontologies can support the process of query composition by providing human-understandable interfaces, alleviating end-users from having to learn complex query languages. Examples of systems employing ontologies for such purposes can be found in [44] and [45]. Regarding the query rewriting process, ontologies are employed to implement schema mappings that allow overcoming the schema heterogeneities present in distributed sources. Queries in terms of a schema can be effectively translated into queries for different schemas using this approach. This is the case of systems such as ONTOFUSION [46] or SEMEDA [47]. Finally, ontologies can be used to solve syntactic heterogeneities in order to correctly join data from heterogeneous sources. Synonymy, granularity differences, or even scale disparities are tackled prior to actual integration with the help of ad hoc ontologies. This is the case in the CREAM framework [48], COIN [49], or OntoDataClean [50].
6.1.2. Semantic Mediation
The ACGT Semantic Mediation (SM) Layer has the goal of providing clients with a seamless interface for integrated querying of a number of heterogeneous data sources. This requires addressing the following challenges: (1) Post-genomic clinical trials comprise a dynamic data environment – i.e., new databases can arrive, or existing ones can change; (2) databases present heterogeneities at different levels – i.e., schema and instances; and (3) results are presented in heterogeneous ways, without any type of annotation. In order to overcome these problems, several approaches were adopted. These approaches are described in the following sections.
6.1.3. Query processing
The query transformation approach adopted in ACGT is a difficult task that can be subdivided in a set of sub-problems to be addressed separately. Among the most important of these, we have identified the following: (i) schema level heterogeneity, (ii) instance level heterogeneity, (iii) performance in query translation and results retrieval, (iv) complexity of the mapping process, and (v) complex query constraints satisfaction [51]. The Semantic Mediator tackles this process as follows: SPARQL was chosen as the query language for the ACGT-SM. When a query is launched, the ACGT-SM splits it into sub-queries for the corresponding ACGT Data Access Service (DAS). Each DAS returns the results in XML, and the ACGT-SM integrates and annotates them to present a result set consistent with the original query. The ACGT-SM follows a Local-as-view (LAV) based approach to solve the data integration problem. The MO acts as the global schema in the mediation process, so local views of the databases are defined using its terminology and relations. These local views maintain semantic and syntactic homogeneity. However, LAV based approaches have problems of scalability when translating the queries. Another problem when dealing with queries against integrated repositories is the issue of identifiers heterogeneity. Queries can be formulated using several literal identifiers expressing the same instance. Hence, for a given query to be transformed, it must pass the filter of the mapping – i.e., a set of correspondences between elements from the databases to elements from the global schema. This filter contains the information needed to translate the semantic information present in the query – i.e., concepts and relations – into the appropriate format.
The LAV semantic query translation process is a difficult task because of the possible incompleteness of the pre-defined global schema – i.e., the global schema is intended to describe the domain, but databases are not taken into account in its production procedure – nor are the views defining the underlying databases. The process of finding the best query rewriting using local views can be an NP-hard problem. This issue has been approached in several projects [52,53], but the problem of scalability is still difficult to overcome. To this end, we propose to constrain the queries that can be formulated by a single user, creating a personalized profile based on requirements gathered using examples.
Identifiers heterogeneity in queries is tackled using an ontology- based solution. It makes use of an ontology that describes a data-cleaning domain to let the user define the transformations that must be applied on data. An additional module is responsible for parsing and extracting the identifiers from SPARQL queries, communicating them to the query cleaning system, and recomposing the query with the new identifiers. In order to facilitate interoperability, the module is made accessible via a Web Service interface. The ACGT-SM invokes this service before sending the queries to the ACGT-DAS. Proper ontology instances need to be defined for each of the databases included in the integration schema. This task can be performed along with the mapping process, and a domain expert should be able to carry it out without the assistance of an IT professional – using a dedicated tool, such as Protégé.
6.1.4. The mapping process
The goal of the mapping process is the production of a “mapping file” – i.e., a set of correspondences between the global schema and a given database schema. A correspondence is a pair of semantically equivalent elements in both schemas. In the ACGT approach, the queries are built-in terms of the information contained in the mapping files. In this case, the element used as global schema is the ACGT Master Ontology. Ontologies have been used for semantic homogenization in mediation processes in several previous works [44,45,54,55].
The mapping process usually requires the involvement of a team of experts in different domains. In a real case scenario, at least the following profiles are needed: (i) a Master Ontology authority, (ii) an expert in the database system to be mapped, and (iii) a specialist in the mapping format and mediation process. These three types of professionals collaborate in the definition of semantic correspondences between the database schema and the ontology. This can be a very complex task in the absence of dedicated tools that leverage the processes of navigating the ontology, identifying class level correspondences and creating entries in the mapping language.
The mapping process is a necessary step for adapting legacy data sources, but the ultimate goal of ontology-based information management is to enable the direct and transparent integration of semantically heterogeneous data created in different environments (e.g., clinical research, laboratory data, etc.). ACGT aims to provide solutions that demonstrate the possibility of collecting data in an ontology-governed way. To explore this approach an ontology- based Trial Management Application (ObTiMA) has been developed, one that integrates the ACGT-MO already at the beginning, in the design process of a clinical trial, in order to guarantee that the data collected during the trial has comprehensive metadata in terms of the ACGT-MO without the need to perform a separate mapping process. We will describe ObTiMA in the following section in more detail.
6.2. ObTiMA – an Ontology-based Trial Management Application for ACGT
ObTiMA [56] is an Ontology-based Trial Management Application intended to help design and conduct clinical trials in an end-user-friendly way. To support the whole life cycle of a clinical trial, it utilizes the features provided by the ACGT-MO and the ACGT-SM.
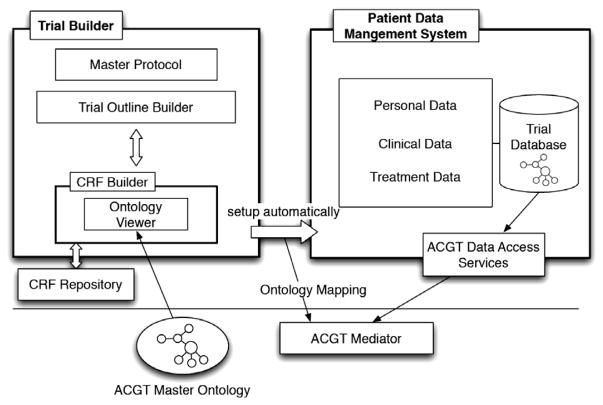
In Fig. 9 the main components of ObTiMA, which are the Trial Builder and the Patient Data Management System, and their interaction with the ACGT-SM are shown. The Trial Builder allows the trial chairman to define the master protocol, the Case Report Forms (CRFs) as well as the treatment plan for the trial, in a way that is both semantically compliant with the ACGT-MO and user-friendly. From these definitions, the Patient Data Management System can be set up automatically in such a way that a medical clinician can collect the patient data during the trial. The data collected in the trial is stored in trial databases whose comprehensive metadata has been rendered in terms of the ACGT-MO. The data can thus be seamlessly integrated into the mediator architecture. It is important to understand that in the first version of ObTiMA the ontology is not used for the purpose of decision support for clinical trial development.
Fig. 9.
Overview of ObTiMA.
To provide a deeper understanding how we have achieved that goal, in the following sections we will describe the various aspects of ontology integration into ObTiMA in more detail. We will first describe how a trial can be set up in an ontology compliant way, and then we will show how seamless data integration of the data collected in the clinical trial can be performed and how the system can handle the evolution of the ontology. We will then discuss the advantages that ObTiMA gains from ontology integration when compared with traditional trial management systems.
6.2.1. Ontology-based trial set up
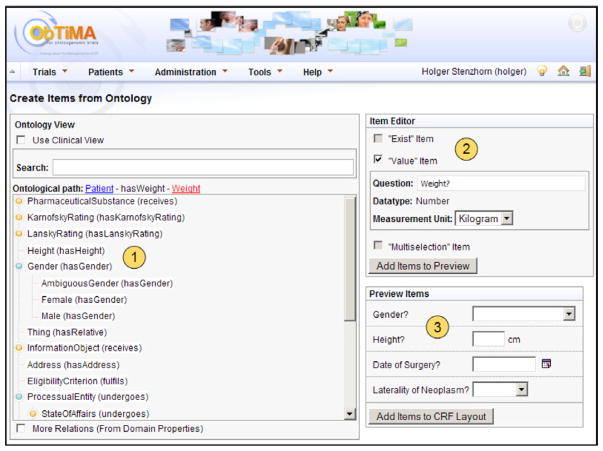
In the design phase of a trial ObTiMA allows a clinical trial chairman to design both treatment plans which guide clinicians through the treatment of a patient and CRFs to collect patient data for full patient documentation. In this phase it is necessary that the trial chairman defines all information to make data integration possible i.e., an ontology description for each question on the CRF and some metadata such as e.g., data type and measurement unit to set up the trial databases. However, clinicians want to focus on the user interfaces of the CRFs and try to integrate and adapt them into the specific workflow of the clinical trial planned. They should not be concerned with theoretical aspects and design principles of databases or ontological metadata. In ObTiMA the trial chairman can adapt the trial database for his trial and define ontological metadata by creating the CRFs for his trials. Therefore, the trial chairman can create the questions on the CRFs, which are also called items in the following, from the ACGT-MO with the help of the “Ontology Viewer”, a graphical user interface that depicts the ontology especially adapted to the task of creating items, which consists of the following sections (see Fig. 10):
Fig. 10.
Ontology Viewer during creation of item “Weight”: (1) Ontology View Section: Ontology description (described as ontological path) for patient’s weight is selected. (2) Item Creation Section: Type Value Item is selected and the attributes for question “patient’s weight” are depicted. (3) Preview Section: Previews of different items are depicted.
The Ontology View Section allows the selection of classes from the ACGT-MO to describe an item in the clinical trial with a path from the ontology. We have designed the Ontology View to overcome the gap between clinical practice and biomedical reality representation. Even if an ontology provides natural language definitions for its entities and relationships (is, in other words, ‘human understandable’) they are still defined in a way that is not based on practical or clinical perceptions of reality. In order to overcome this challenge, we provide an application specific view of the ontology, a view that is meant to assist clinicians in clinical practice. The starting point of each ontology description is the class “Patient”, which is the focal point of each CRF. To this end, when opening the Ontology View, the only classes shown are those that can be related to the class patient, such as e.g., “Weight” (indicating the patient’s weight) or “Tumor” (indicating the patient’s tumor). When selecting e.g., tumor, only classes that can be related with “Tumor”, such as the “Laterality” (indicating the laterality of the patient’s tumor) are shown.
In the Item Creation Section an item can be created from the selected ontology description by selecting a question type, which can be “Value Item”, “Multiple Choice Item” or “Exist Item” (for descriptions see examples below). Only these question types are enabled, which are sensible to create for the selected ontology description. When an item type has been selected, the attributes required in order to create the question on the CRF are shown, e.g., the label, data type or answer possibilities and a preview for the question is shown, where the automatically created attributes can be manually adopted and the item can be added to the Preview Section.
In the Preview Section all created items are shown and the order in which they shall appear on the CRF can be selected.
In our example, the clinician wants to create a query about the patient’s weight (see Fig. 10). In the Ontology View Section he finds a relation between the classes “Patient” and “Weight”. To create the question he simply chooses the class “Weight” and an item type. The user in our example creates a Value Item for his selected ontology descriptions, which will query a float value for the patient’s weight. The attributes required in order to create the question on the CRF are then determined automatically, e.g., the label and data type, and shown in the Item Selection Section.
Beside Value Items, which query number or string values for the last class in the selected ontology description, it is possible to create Multiple Choice and Exist Items in ObTiMA. Multiple Choice Items are questions created from an ontology description of, for example, a superclass, for which answer possibilities can be selected from the ontology, for example, from amongst the subclasses of the superclass. An example is the question “laterality of nephroblastoma” with the answer possibilities “left”, “right” and “bilateral”. To create this question the user has to select the classes “Nephroblastoma” and “Laterality” in the Ontology View. When creating a question of type Multiple Choice Item, the possible answers are automatically determined from the ontology as “left”, “right”, “midline”, “bilateral”, “systemic” and “unknown” from which the user can choose the desired ones.
Exist Items query whether an instance of a class in the ontology description exists for the patient, an example is “Does patient have a nephroblastoma” with answer possibilities “yes” and “no”.
Table 2 provides a real world examples of ontological paths selected from the MO to represent the data collected for a specified question in the CRF. The individual value for a patient is one instance of the class specified. The examples are taken from the SIOP Trial CRFs [4]. It is important to understand that the ontology is used in ObTiMA to help the chair person develop the CRFs and to ensure semantic interoperability of the data gathered at different study sites with each other and with external resources.
Table 2.
Examples from the SIOP Trial for CRF questions and ontological paths. Note that for multiselection items the chairperson can select from the subclasses of the specified class all classes she wants to provide as multiple-choice items.
| Question on CRF | Ontological path | Question type |
|---|---|---|
| “Is the patient undergoing treatment as part of a clinical trial protocol he/she is enrolled in?” | Patient undergoes TherapeuticProcess implements ClinicalTrialProtocol | Exist |
| “What is the tumor structure of the tumor in the patient’s kidney?” | Patient hasPart Kidney has Part Neoplasm hasQuality TumorHomogeneity | Multiselection with answer possibilities x, y and z Value |
| “At what date does radiotherapy start for the patient?” | Patient undergoes Radiotherapy hasDate Date |
6.2.2. Ontology-based data integration for cross-trial analysis
When the trial chairman decides the trial is ready to be conducted, the form-based trial database and the data access services are set up automatically from the definitions done in the design phase. The mapping file (see Section 6.1), which contains the translation for the mediator to query the form-based databases in terms of the ontology, is created automatically and is sent to the mediator. While the trial is being conducted a clinician fills in the CRFs for the patient, without being bothered about annotations from the ontology. ObTiMA stores this filled in data in the trial database. The mediator can, with the help of the mapping file, seamlessly query the data of different clinical trials set up with ObTiMA and other data sources in the ACGT mediator environment. Thus cross-trial meta-analysis in terms of the shared ontology becomes possible.
6.2.3. Ontology evolution in ObTiMA
In clinical trials new therapies or medicines are often introduced, thus it is likely that ontology classes or relations necessary to assemble queries for the CRFs are not yet represented in the shared ontology. In Section 4.1 we have already described how new classes and relations can be requested with the ontology submission system. It would, however, be tedious for the trial chairman to request a change in the submission system manually and wait until the change has been accepted from an ontology expert to be able to create his required question. Therefore, we have implemented a direct interface between ObTiMA and the ACGT Submission System, which allows the chairman of a clinical trial to extend the ontology by creating the questions on the CRF, without being interrupted in the design of the trial.
When the user observes that a class for creating a required question is missing in the ontology, he can create a new class while creating the question in the Ontology View Section. The newly created class is stored as a temporary class in a local copy of the ontology and can be used directly with the ontology description of the question. ObTiMA automatically sends a request to the ontology submission system to have the new class added to the shared ontology. When the ontology expert accepts the request without changes and releases a new version, ObTiMA automatically replaces the local class in the ontology descriptions of the questions. With the same mechanism temporary relations can be added during the creation of a question.
The local copy of the ontology is always backwards compatible to the current and to all previous versions of the shared ontology. This approach assures that a trial containing temporary classes or relations can already be queried with the current version of the shared ontology by the mediator. Such mediator queries can even include the data filled into items for which the ontology description contains temporary classes or relations, since they can be queried with their super entities.
6.2.4. Advantages of ontology integration
Compared with traditional data management systems that lack ontology support, ObTiMA has the following advantages:
Built-in semantic interoperability between different trials
The procedure of ontology-based trial set up makes possible the direct integration of the data collected in the clinical trial into the semantics of the ontology. This means that data sharing between clinical trials and other data sources in the ACGT mediator architecture becomes possible. This is necessary to leverage the collected data for further research, such as cross-trial meta-analysis. ObTiMA promises to put an end to error-prone coding techniques recently used to map clinical data onto biomedical terminologies. Recent studies show that the accuracy of SNOMED coding is only slightly over 50% given three different scenarios [57,58].
Increased quality of collected data
By using a shared ontology to create a data model, the collected data becomes consistent with the knowledge of the underlying domain, which is coded in the ontology, and data quality increases. Currently ObTiMA ensures, during creation of items, that only classes and relations from the ontology are chosen and that certain restrictions from the ontology such as domain and range restrictions are satisfied. However, currently not all restrictions from the domain ontology, as e.g., number restrictions, can be guaranteed automatically. Therefore, we are currently developing algorithms to further improve data quality [59].
Nevertheless, ObTiMA has been designed to hide the details of the ontology and the ACGT-SM from the user, enabling him or her to concentrate on the workflow of the clinical trial, thus making the system as user-friendly as possible. Furthermore, the assembled ontology descriptions can be used to determine attributes necessary for setting up the database, such as e.g., the data types for items to be entered, and as a consequence enables the user to set up the trial database in a way that is simultaneously user-friendly and semantically compliant.
7. Discussion
7.1. Semantic Mediation in ACGT
The selection of the LAV approach was motivated mainly by the nature of the domain, where the number of available databases grows continuously [60]. This choice implies a relatively small effort when changes in the environment occur – i.e., new databases need to be included, or existing databases change. However, defining new views describing databases remains the bottleneck of the data integration process.
From our point of view, LAV is the most appropriate choice given the domain, but it leads to several issues that must be overcome. One of these is the possible incompleteness of the global schema, which is built without taking into consideration the underlying databases. The ACGT-MO is built using CRFs belonging to the initially selected clinical trials. In the case study, we encountered certain difficulties integrating a DICOM database – most of the terms were present, but some of them not. The ACGT-SM allows the utilization of several ontologies in defining the view. This feature can be used to solve this kind of added semantic heterogeneity. However, it is advisable to use only the ACGT-MO, in order to avoid high complexity – mainly regarding query translation and formulation.
In order to overcome the difficulties of query rewriting associated with the LAV approach, a novel method was proposed for creating user profile-guided domain restrictions. This method makes available only a subset of the global schema to the user, a subset whose construction is based on pre-defined user requirements. The observed benefit of this approach is twofold: (1) The query translation process becomes simpler and (2) query formulation is easier for end-users. However, this method presents one main drawback: its high sensitivity to changes in the structure of the integration – i.e., if new databases are added, or modifications occur in databases already integrated, then existing user profiles may become invalid.
7.2. Comparison of the ACGT strategy with the caBIG approach
Having presented the scientific and technical details of our approach, we feel that it is important to critically review current efforts aimed at addressing similar problems which have adopted a different approach to ours. The most prominent of such efforts is the work of the cancer Biomedical Informatics Grid (caBIG), which is being developed under the leadership of the National Cancer Institute’s Center for Bioinformatics.
7.2.1. Overview of the caBIG data integration platform
caBIG [61] is a grid connecting individuals and institutions to enable the sharing of data and tools. The goal is to speed the delivery of innovative approaches for the prevention and treatment of cancer. caGrid [62] provides the core enabling infrastructure. It is a service-oriented architecture and provides the implementation of the required core services, toolkits and wizards for the development and deployment of community provided services, APIs for building client applications, and some sample client applications for interacting with the current test bed installation. A particular framework and set of tools provided by caGrid is the Cancer Common Ontologic Representation Environment (caCORE), which aims to facilitate the creation of syntactically and semantically interoperable biomedical information services [63].caCORE defines a data model specified using industry standard techniques to define common biological concepts. The main components of caCORE include:
Cancer Bioinformatics Infrastructure Objects (caBIO): Platform independent APIs that reflect an object-oriented view of bio-medical information.
Cancer Data Standards Repository (caDSR): A metadata registry based upon the ISO/IEC11179 standard that is used to register the descriptive information needed to render cancer research data re-usable and interoperable.
Enterprise Vocabulary Services (EVS): Controlled vocabulary resources that support the life sciences domain, implemented in a description-logics framework. EVS vocabularies provide the semantic ‘raw material’ from which data elements, classes and objects are constructed.
It is important to note that the EVS contains, among others, the NCI Thesaurus, whose semantical vices and virtues have been thoroughly discussed in [22].
In caBIG a Model Driven Architecture (MDA) [64] is followed. Following this approach, the designer uses the Unified Modeling Language (UML) to create a graphical model of the functions, components, and behavior of the system.
7.2.2. caBIG vs ACGT – the problem of metadata
In caBig the consistent use of metadata is secured by providing a common meta-model built around the notion of (Common) Data Elements. A data element consists of two parts, a Data Element Concept (DEC) and a Value Domain (VD). The DEC is a formal description of the thing about which we are recording a data value, which is drawn from the Value Domain. Data Element Concepts are further refined into two subcomponents, Object Classes and properties. An Object Class is the entity that is being described by the data element, while the property is a specific attribute of the entity whose value is being recorded. Data Elements also have other associated components, including a Representation which describes the nature of the data that is being recorded (code, text, number) and a Conceptual Domain, which is a means of classifying CDE components (such as Data Element Concepts and Value Domains) for easier search and identification.
A first main difference between the caBIG and the ACGT approaches is that in the case of ACGT no MDA and UML modeling precedes the implementation and the publication of new data sources. In ACGT there is no single registry for models e.g., to preserve all the mappings and local schemas. Therefore the definition of the local database schemas and the accompanied metadata information are not “publicly” available in the same sense that ca-BIG fosters reusability through the central metadata registry. In ACGT the metadata definitions exist inside the “mapping files” but the case of reusing these is irrelevant because the goal is to integrate existing databases rather than designing and implementing new ones based on what is already available.
Furthermore, in ACGT there is a single component (Semantic Mediator) that is responsible for performing the data integration in a transparent way through the appropriate query translations based on the mappings of the local database schemas to the global ACGT Master Ontology. This single authoritative query service allows not only accessing the individual data services using a common terminology and query language, but also permits, unknown to the user, the “fusion” of records coming from different databases and a filtering of the results based on the high-level user criteria. The role of the ACGT Master Ontology is, of course, critical to achieve this level of integration and although caBIG uses the NCI Thesaurus and Metathesaurus more or less for the same purposes, we argue that starting with a formal ontology as a sound theoretical foundation is superior [22]. Furthermore, once a stable and dependable semantic resource is created, the project of providing meta-models linking and defining the data elements can always be undertaken, whereas high-level meta-modeling with an inconsistent semantic resource remains likely to result in further inconsistencies and errors. We hold that the value of the semantic integration in ACGT lies in the fact that an ontology for cancer management is provided that satisfies strict criteria for ontology development. In essence the ACGT approach is a more top down, unified, and ontology-based solution to the semantic data integration problem.
8. Conclusion
The development of the ACGT-MO is a clear-cut example of the parallel development of an ontology and specific applications within a major domain framework. We have shown that this strategy leads to specific design decisions facilitating the use of the ontology and assuring that the necessary amount of knowledge is present in the ontology.
With respect to the relation between a knowledge management system (in this special case a clinical information system) and an ontology, the result is that reality-based ontology development is no opposite to the development of a highly pragmatic information system.
9. Summary table
9.1. What was already known on the topic
Principles of ontology development.
Ontology maintenance and evaluation.
Ontology-based clinical systems.
9.2. What the study added to our knowledge
The study yielded a huge amount of experience in basing a sophisticated knowledge sharing system on reality-based ontology development.
Interdependencies between ontology principles and needs of the knowledge management system have been studied. Solutions to reconcile user needs, technical requirements and theoretical coherence were achieved.
Acknowledgments
The authors thank all members of the ACGT consortium who are actively contributing to addressing the R&D challenges faced. The ACGT project (FP6-2005-IST-026996) is partly funded by the EC and the authors are grateful for this support. The authors thank two anonymous reviewers for valuable comments.
Footnotes
Current level of DL expressivity is SROIQ(D).
A FILTER directive, for example, is a SPARQL construct that specifies that certain classes are to be ignored/filtered out from the results of the query.
References
- 1.Sotiriou C, Pickard MJ. Taking gene-expression profiling to the clinic: when will molecular signatures become relevant to patient care? Nat Rev. 2007;7:545–53. doi: 10.1038/nrc2173. [DOI] [PubMed] [Google Scholar]
- 2.OWL Web Ontology Language Semantics and Abstract Syntax. Available from: http://www.w3.org/TR/owl-semantics/; last visited: 10-28-2009.
- 3.Available from: http://clinicaltrials.gov/ct2/show/NCT00162812; last visited: 04-20-2010.
- 4.International Society of Paediatric Oncology: Nephroblastoma (Wilms Tumour) – clinical trial and study SIOP 2001. Final version January 2002, amended 2004 and 2007, EUDRACT No. 2007-004591-39.
- 5.Basic Formal Ontology (BFO) Available from: http://www.ifomis.org/bfo; last visited: 10-28-2009.
- 6.Tsiknakis M, Brochhausen M, Nabrzyski J, Pucaski J, Sfakianakis S, Potamias G, et al. A semantic grid infrastructure enabling integrated access and analysis of multilevel biomedical data in support of post-genomic clinical trials on Cancer. IEEE Trans Inform Technol Biomed, Special issue on Bio-Grids. 2008;12(2):191–204. doi: 10.1109/TITB.2007.903519. [DOI] [PubMed] [Google Scholar]
- 7.Weng C, Gennari JH, Fridsma DB. User-centered semantic harmonization: a case study. J Biomed Inform. 2007;40:353–64. doi: 10.1016/j.jbi.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Campbell KE, Oliver DE, Shortliffe EH. The unified medical language system: toward a collaborative approach for solving terminologic problems. J Am Med Inform Assoc. 1998;5(1):12–6. doi: 10.1136/jamia.1998.0050012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noy N, Musen M. PROMPT: algorithm and tool for automated ontology merging and alignment; Proceedings of the seventeenth national conference on artificial intelligence (AAAI-2000); 2000 July 31–August 02; Austin, TX. Menlo Park, CA: The AAAI Press; 2000. pp. 550–5. [Google Scholar]
- 10.Klein M. Combining and relating ontologies: an analysis of problems and solutions. In: Gomez-Perez A, Gruninger M, Stuckenshmidt H, Uschold M, editors. Workshop on ontologies and information sharing, IJCAI’01; Seattle, WA. 2001. [Google Scholar]
- 11.Sciore E, Siegel M, Rosenthal A. Using semantic values to facilitate interoperability among heterogeneous information systems. ACM Trans Database Syst. 1994;19(2):254–90. [Google Scholar]
- 12.Dublin Core Metadata Standards. Available from: http://dublincore.org/; last visited: 10-28-2009.
- 13.Doerr M. The CIDOC conceptual reference module: an ontological approach to semantic interoperability of metadata. AI Mag. 2003;24(3):75–92. [Google Scholar]
- 14.Gennari JH, Silberfein A, Wiley JC. Integrating genomic knowledge sources through an anatomy ontology. Proceedings of the Pacific symposium on biocomputing; 2005 January 04–05; Fairmont Orchid (HI). Available from: http://psb.stanford.edu/psb-online/proceedings/psb05/gennari.pdf; last visited: 10-28-2009. [PubMed] [Google Scholar]
- 15.Rosse C, Mejino JLV. A reference ontology for bioinformatics: the foundational model of anatomy. J Biomed Inform. 2003;36(6):478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 16.About SNOMED CT. Available from: http://www.ihtsdo.org/snomed-ct/snomed-ct0/; last visited: 10-28-2009.
- 17.UMLS. Available from: http://www.nlm.nih.gov/research/umls/; last visited: 10-28-2009.
- 18.NCI thesaurus. Available from: http://nciterms.nci.nih.gov/; last visited: 02-02-2010.
- 19.Available from: http://terminology.vetmed.vt.edu/SCT/menu.cfm; last visited: 02-02-2010.
- 20.Bodenreider O, Smith B, Kumar A, Burgun A. Investigating subsumption in SNOMED CT: an exploration into large description logic-based biomedical terminologies. Artif Intell Med. 2007;39(3):183–95. doi: 10.1016/j.artmed.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar A, Smith B. The unified medical language system and the gene ontology: some critical reflections. In: Günter A, Kruse R, Neumann B, editors. KI 2003: Advances in artificial intelligence. Berlin: Springer; 2003. pp. 135–48. Lecture notes in artificial intelligence 2821. [Google Scholar]
- 22.Ceusters W, Smith B, Goldberg L. A terminological and ontological analysis of the NCI thesaurus. Methods Info Med. 2005;44:498–507. [PubMed] [Google Scholar]
- 23.Foundational Model of Anatomy. Available from: http://sig.biostr.washington.edu/projects/fm/; last visited: 10-28-2009.
- 24.OBO Relation Ontology. Available from: http://www.obofoundry.org/ro/; last visited: 10-28-2009.
- 25.The Open Biomedical Ontologies: OBO Foundry. Available from: http://www.obofoundry.org/; last visited: 10-28-2009.
- 26.Smith B, Brochhausen M. Establishing and harmonizing ontologies in an interdisciplinary health care and clinical research environment. In: Blobel B, Pharow P, Nerlich M, editors. EHealth: combining health telematics, telemedicine, biomedical engineering and bioinformatics to the edge. Amsterdam: IOS Press; 2008. pp. 219–34. [Google Scholar]
- 27.Smith B, Kusnierczyk W, Schober D, Ceusters W. Towards a reference terminology for ontology research and development in the biomedical domain. In: Bodenreider O, editor. Proceedings of KR-MED; 2006 January 8; Baltimore, MD. pp. 57–66. [Google Scholar]
- 28.Lassila O, McGuinness D. Technical report KSL-01-02. Knowledge System Laboratory, Stanford University; Stanford: 2001. The role of frame-based representation on the semantic web. [Google Scholar]
- 29.SPARQL Query Language for RDF. Available from: http://www.w3.org/TR/rdf-sparql-query/; last visited: 10-28-2009.
- 30.Wittekind C, Meyer HJ, Bootz F. TNM Klassifikation maligner Tumoren. Berlin, Heidelberg: Springer; 2005. [Google Scholar]
- 31.IEEE P1600.1 Standard Upper Ontology Working Group (SUO WG) Home Page. Available from: http://suo.ieee.org; last visited: 10-28-2009.
- 32.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. 2007;25(11):1251–5. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grenon P, Smith B. SNAP and SPAN: towards dynamic spatial ontology. Spat Cogn Comput. 2004;4:69–103. [Google Scholar]
- 34.Smith B, Ceusters W, Klagges B, Köhler J, Kumar A, Lomax J, et al. Relations in biomedical ontologies. Genome Biol. 2005;6(5):R46. doi: 10.1186/gb-2005-6-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith B. The evaluation of ontologies: editorial review vs. democratic ranking. Proceedings of InterOntology; 26–27 February 2008; Tokyo, Japan. pp. 127–38. [Google Scholar]
- 36.Hartmann J, Spyns P, Giboin A, Maynard D, Cuel R, Suárez-Figueroa MC, et al. Methods for ontology evaluation, Knowledge Web Deliverable D1.2.3. 2004. Available from: http://knowledgeweb.semanticweb.org/semanticportal/deliverables/D1.2.3.pdf; last visited: 10-28-2009.
- 37.Gangemi A, Catenacci C, Ciaramita M, Lehmann J. Modelling ontology evaluation. Proceedings of the third European semantic web conference; 2006 June 11–14; Budva, Montenegro. Berlin: Springer; 2006. pp. 140–54. [Google Scholar]
- 38.Pellet: The Open Source OWL Reasoner. Available from: http://clarkparsia.com/pellet; last visited: 10-28-2009.
- 39.The National Center for Biomedical Ontologies BioPortal. Available from: http://bioportal.bioontology.org/; last visited: 10-28-2009.
- 40.THESEUS: MEDICO – towards scalable semantic image search in medicine. Available from: http://www.theseus-programm.de/en-us/theseus-application-scenarios/medico/default.aspx; last visited: 10-28-2009.
- 41.Oemig F, Blobel B. Semantic interoperability between health communication standards through formal ontologies medical informatics in a united and healthy Europe. In: Adlassnig KP, Blobel B, Mantas J, Masic I, editors. Proceedings of the MIE 2009 – European federation for medical informatics; 2009 August 30–September 03; Sarajevo, Bosnia&Herzegovina. Amsterdam: IOS Press; 2009. pp. 200–4. [PubMed] [Google Scholar]
- 42.CHRONIUS Project Homepage. Available from: http://www.chronious.eu/; last visited: 10-28-2009.
- 43.Jakoniene V, Lambrix P. Ontology-based integration for bioinformatics. In: Collard M, editor. Ontologies-based databases and information systems, first and second VLDB workshops, ODBIS 2005/2006; Trondheim, Norway. September 2–3, 2005; Seoul, Korea. September 11, 2006; Berlin: Springer; 2007. pp. 55–8. Revised Papers. [Google Scholar]
- 44.Mahalingam K, Huhns MN. An ontology tool for query formulation in an agent-based context. Proceedings of the second IFCIS international conference on cooperative information systems; 1997 June 24–27; Kiawah Island, SC. Los Alamitos, CA: The IEEE Computer Society; 1997. pp. 170–8. [Google Scholar]
- 45.Catarci T, Mascio TD, Franconi E, Santucci G, Tessaris S. An ontology based visual tool for query formulation support. In: López de Mántaras R, Saitta L, editors. Proceedings of the 16th European conference on artificial intelligence (ECAI 2004); 2004 August 22–27; Valencia, Spain. Amsterdam: IOS Press; 2004. pp. 208–12. [Google Scholar]
- 46.Pèrez-Rey D, Maojo V, Garcìa-Remesal M, Alonso-Calvo R, Billhardt, Martìn-Sanchez F, et al. ONTOFUSION: ontology-based integration of genomic and clinical databases. Comput Biol Med. 2006;36:712–30. doi: 10.1016/j.compbiomed.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 47.Köhler J, Philippi S, Lange M. SEMEDA: ontology based semantic integration of biological databases. Bioinformatics. 2003;19(18):2420–7. doi: 10.1093/bioinformatics/btg340. [DOI] [PubMed] [Google Scholar]
- 48.Park J, Ram S. Information systems interoperability: what lies beneath? ACM Trans Inform Syst. 2004;22(4):595–632. [Google Scholar]
- 49.Madnick S, Zhu H. Improving data quality through effective use of data semantics. Data Knowledge Eng. 2006;59(2):460–75. [Google Scholar]
- 50.Perez-Rey D, Anguita A, Crespo J. OntoDataClean: ontology-based integration and preprocessing of distributed data. Lect Notes Comput Sci. 2006;4345:262–72. [Google Scholar]
- 51.Martín L, Anguita A, Jiménez A, Crespo J. Enabling cross constraint satisfaction in RDF-based heterogeneous database integration. Proceedings of the 2008 20th IEEE international conference on tools with artificial intelligence; 2008 November 3–5; Dayton, OH. Los Alamitos, CA: The IEEE Computer Society; 2008. pp. 341–8. [Google Scholar]
- 52.Martín L, Bonsma E, Anguita A, Vrijnsen J, García-Remesal M, Crespo J, et al. Data access and management in ACGT: tools to solve syntactic and semantic heterogeneities between clinical and image databases. Advances in conceptual modeling – foundations and applications. 2007;4802:24–33. [Google Scholar]
- 53.Duschka OM, Genesereth MR. Query planning in infomaster. Proceedings of the ACM symposium on applied computing; 1997 February 28–March 02; San Jose, CA. New York: ACM; 1997. pp. 109–11. [Google Scholar]
- 54.Librelotto GR, Souza W, Armalo JC, Henriques PR. Using the ontology paradigm to integrate information systems. Proceedings of the international conference on knowledge engineering and decision support; 2004 July 21–23; Porto, Portugal. pp. 497–504. [Google Scholar]
- 55.Bizer C. D2R MAP – a database to RDF mapping language. Proceedings of the international world wide web conference; 2003 May 20–24; Budapest, Hungary. Available from: http://www2003.org/cdrom/; last visited: 10-28-2009. [Google Scholar]
- 56.Weiler G, Brochhausen M, Graf N, Schera F, Hoppe A, Kiefer S. Ontology based data management systems for post-genomic clinical trials within an European grid infrastructure for cancer research. Proceedings of the 29th annual international conference of the IEEE EMBS; 2007 August 23–26; Lyon, France. pp. 6434–7. [DOI] [PubMed] [Google Scholar]
- 57.Andrews JE, Richesson RL, Krischer J. Variation of SNOMED CT coding of clinical research concepts among coding experts. J Am Med Inform Assoc. 2007;14(4):497–506. doi: 10.1197/jamia.M2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chiang MF, Hwang JC, Yu AC, Casper DS, Cimino JJ, Starren J. Reliability of SNOMED-CT coding by three physicians using two terminology browsers. Proceedings of the AMIA 2006 symposium; 2006 November 11–15; Washington. pp. 131–5. Available from: http://www.ncbi.nlm.nih.gov/pmc/journals/362/; last visited: 10-28-2009. [PMC free article] [PubMed] [Google Scholar]
- 59.Weiler G, Poetzsch-Heffter A, Kiefer S. Consistency checking for workflows with an ontology-based data perspective. Proceedings of 20th international conference for database and expert systems applications; 2009 August 31–September 04; Linz, Austria. Berlin: Springer; 2009. pp. 98–113. Lecture notes in computer science 5690. [Google Scholar]
- 60.Galperin GY. Molecular biology database collection: 2008 update. Nucleic Acids Res. 2008;36:D2–4. doi: 10.1093/nar/gkm1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kakazu KK, Cheung LW, Lynne W. The cancer biomedical informatics grid (caBIG): pioneering an expansive network of information and tools for collaborative cancer research. Hawaii Med J. 2004;63:273–5. [PubMed] [Google Scholar]
- 62.Oster S, Langella S, Hastings S, Ervin D, Madduri R, Phillips J, et al. caGrid 1. 0: an enterprise grid infrastructure for biomedical research. J Am Med Inform Assoc. 2008;15(2):138–49. doi: 10.1197/jamia.M2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Komatsoulis GA, Warzel DB, Hartel FW, Shanbhag K, Chilukuri R, Fragoso G, et al. CaCORE version 3: implementation of a model driven, service-oriented architecture for semantic interoperability. J Biomed Inform. 2008;41(1):106–23. doi: 10.1016/j.jbi.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mellor SJ, Scott K, Uhl A, Weise D. Model-driven architecture. Proceedings of advances in object-oriented information systems: OOIS 2002 workshops; 2002 September 02; Montpellier, France. Berlin: Springer; 2002. pp. 290–7. [Google Scholar]