Abstract
The KCNH2 or human ether-a go-go-related gene (hERG) encodes the Kv11.1 potassium channel that conducts the rapidly activating delayed rectifier potassium current in the heart. The expression of Kv11.1 C-terminal isoforms is directed by the alternative splicing and polyadenylation of intron 9. Splicing of intron 9 leads to the formation of a functional, full-length Kv11.1a isoform and polyadenylation of intron 9 results in the production of a nonfunctional, C-terminally truncated Kv11.1a-USO isoform. The relative expression of Kv11.1a and Kv11.1a-USO plays an important role in regulating Kv11.1 channel function. In the heart, only one-third of KCNH2 pre-mRNA is processed to Kv11.1a due to the weak 5′ splice site of intron 9. We previously showed that the weak 5′ splice site is caused by sequence deviation from the consensus, and that mutations toward the consensus sequence increased the efficiency of intron 9 splicing. It is well established that 5′ splice sites are recognized by complementary base-paring with U1 small nuclear RNA (U1 snRNA). In this study, we modified the sequence of U1 snRNA to increase its complementarity to the 5′ splice site of KCNH2 intron 9 and observed a significant increase in the efficiency of intron 9 splicing. RNase protection assay and western blot analysis showed that modified U1 snRNA increased the expression of the functional Kv11.1a isoform and concomitantly decreased the expression of the non-functional Kv11.1a-USO isoform. In patch-clamp experiments, modified U1 snRNA significantly increased Kv11.1 current. Our findings suggest that relative expression of Kv11.1 C-terminal isoforms can be regulated by modified U1 snRNA.
Keywords: hERG, U1 snRNA, Splicing, Alternative Polyadenylation, Arrhythmia, Long QT Syndrome
1. Introduction
KCNH2 or human ether-à-go-go-related gene (hERG) encodes the Kv11.1 potassium channel that conducts the rapidly activating delayed rectifier K+ current (IKr) in the heart. (Warmke and Ganetzky, 1994, Sanguinetti et al., 1995, Trudeau et al., 1995, Zhou et al., 1998). Mutations in KCNH2 cause long QT syndrome type 2 (LQT2) (Curran et al., 1995). Several Kv11.1 isoforms, Kv11.1a, Kv11.1b, Kv11.1a-USO and Kv11.1b-USO, have been identified (Larsen, 2010). The Kv11.1a isoform represents the full-length Kv11.1 channel consisting of 1159 amino acids. Kv11.1b lacks the first 376 amino acids of Kv11.1a and has an alternate 36 amino acid N-terminus. The C-terminal isoforms Kv11.1a-USO and Kv11.1b-USO contain the truncated USO C-terminus, in which the last 359 amino acids of Kv11.1a/b are replaced by an alternate 88 residue C-terminal end (Larsen 2010). Functional studies have shown that Kv11.1a and Kv11.1b isoforms generate Kv11.1 currents with distinct gating properties (Sanguinetti et al., 1995, Trudeau et al., 1995, Lees-Miller et al., 1997, London et al., 1997, Zhou et al., 1998), whereas the Kv11.1a-USO and Kv11.1b-USO isoforms fail to form functional channels when expressed in mammalian cells (Kupershmidt et al., 1998, Guasti et al., 2008, Gong et al., 2010, Stump et al., 2012). Coexpression of Kv11.1a and Kv11.1a-USO in equimolar concentrations has no obvious effect on Kv11.1 current (Kupershmidt et al., 1998). When an excess of Kv11.1a-USO or Kv11.1b-USO is coexpressed with Kv11.1a or Kv11.1b, a decrease in Kv11.1 current is observed (Kupershmidt et al., 1998, Guasti et al).
We have previously shown that Kv11.1a and Kv11.1a-USO are produced from a single KCNH2 pre-mRNA precursor by use of alternative splicing and alternative polyadenylation sites (Gong et al., 2010). Kv11.1a is produced by the splicing of intron 9 and polyadenylation at a distal poly(A) site in exon 15, whereas Kv11.1a-USO is generated by the silencing of the 5′ splice site of intron 9 and polyadenylation at an intronic poly(A) site in intron 9. Because formation of Kv11.1a and Kv11.1a-USO is mutually exclusive, the expression of one C-terminal isoform is generated at the expense of the other C-terminal isoform. The importance of alternative splicing and alternative polyadenylation of KCNH2 pre-mRNA in regulation of Kv11.1 isoform expression is underscored by our recent finding that the disease-causing KCNH2 mutation IVS9-2delA disrupted normal splicing and resulted in exclusive polyadenylation of intron 9, leading to the switch from the functional Kv11.1a isoform to the non-functional Kv11.1a-USO isoform (Gong et al., 2014a). Thus, the relative expression of the C-terminal isoforms plays an important role in regulation of Kv11.1 channel function as well as in the pathogenesis of long QT syndrome.
The splicing of KCNH2 intron 9 is inefficient as only one-third of KCNH2 pre-mRNA is processed to Kv11.1a in the heart (Kupershmidt et al., 1998, Gong et al., 2010). We previously showed that the inefficient splicing of intron 9 is due to the presence of a weak 5′ splice site, which deviates from the consensus sequence at four positions (Gong et al., 2010). The initial step of splicing involves the recognition of the 5′ splice site by the U1 small nuclear ribonucleoprotein, which is mediated by base-pair complementarity with its RNA component the U1 small nuclear RNA (U1 snRNA) (Zhuang and Weiner, 1986). It is likely that deviation of the 5′ splice site of KCNH2 intron 9 from the consensus sequence reduces its base-pair complementarity to U1 snRNA, thereby leading to inefficient intron 9 splicing. In the present study, we tested the hypothesis that increasing U1 snRNA complementarity to the 5′ splice site of KCNH2 intron 9 would enhance the efficiency of intron 9 splicing and shift the alternative processing of KCNH2 pre-mRNA toward the production of the Kv11.1a isoform. Our findings indicate that the expression of the functional Kv1 1.1a isoform can be upregulated by modified U1 snRNA.
2. Methods
2.1 Plasmid constructs and transfection
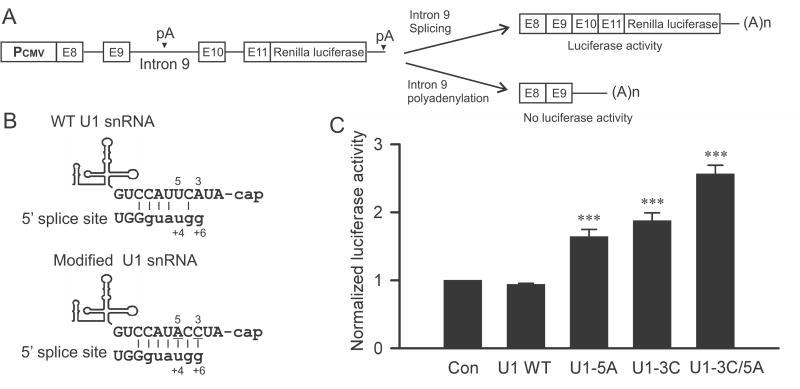
The minigene luciferase reporter construct was generated by subcloning the Renilla luciferase gene downstream of a splicing competent KCNH2 minigene, as previously described (Gong et al., 2014b). The expression of the minigene luciferase reporter is driven by a CMV promoter (Fig. 1 A). The vector also contains the firefly luciferase gene driven by the SV40 promoter, which was used as a control for transfection efficiency. HEK293 cells were transiently transfected with the minigene luciferase reporter construct using the Effectene method (Qiagen, Valencia, CA). After 24 h, cells were harvested and assayed for both firefly and Renilla luciferase activity using the Dual-Luciferase assay kit (Promega, Madison, WI). Data were analyzed by normalizing Renilla luciferase activity to firefly luciferase activity.
Fig. 1.
Effect of modified U1 snRNA on splicing of KCNH2 intron 9 using a luciferase reporter construct. (A) Diagram of the KCNH2 minigene luciferase reporter construct. (B) Diagram of the modified U1 snRNA that increases its complementarity to the 5′ splice site of KCNH2 intron 9 at the +4 and +6 positions. (C) Histogram showing the effect of modified U1 snRNA on luciferase activity (n=6-7, ***P < 0.001).
The generation of the full-length KCNH2 splicing-competent construct composed of Kv11.1a cDNA from exon 1 to exon 6 and the KCNH2 genomic DNA from intron 6 to poly(A) site was described previously (Gong et al., 2010). The full-length KCNH2 splicing-competent construct was subcloned into a modified pcDNA5 vector, in which the BGH poly(A) signal was deleted. Stably transfected Flp-In CV-1 cells were generated by the cotransfection of the full-length KCHN2 splicing-competent construct (0.1 μg) with the Flp recombinase expression vector pOG44 (0.9 μg) using the Effectene method and selected with 100 μg/ml hygromycin.
2.2 Construction of U1 snRNA recombinant adenovirus
The U1 snRNA construct in the pUC13 vector was generously provided by Dr. Alan Weiner from University of Washington. Modification of U1 snRNA at position 3 and 5 was performed by PCR. The AdEasy vector kit was used to generate WT and modified U1 snRNA recombinant adenoviruses (Stratagene, La Jolla, CA). First, the WT and modified U1 snRNA were subcloned into pShuttle vector and recombined with the pAdEasy plasmid in E. coli strain BJ5183. The pAdEasy/U1 snRNA plasmids were transfected into HEK293 cells. After 2 days, the transfected cells were cultured in growth medium containing 1.25% SeaPlaque-agarose (Lonza, Rockland, MD) to promote the formation of recombinant viral plaques. Approximately three weeks following transfection, individual plaques were picked, amplified in HEK293 cells, and purified over a discontinuous CsCl gradient (Gong et al., 2007).
2.3 RNase protection assay
RNA isolation and the RNase protection assay (RPA) were performed as previously described (Gong et al., 2010). Briefly, antisense RNA riboprobes were transcribed in vitro in the presence of biotin-14-CTP. Yeast RNA was used as a control for the complete digestion of the probes by RNase. The relative intensity of each band was quantified using ImageJ software and adjusted for the number of biotin-labeled cytidines in each protected fragment. The expression level of the hygromycin B resistance gene from the KCNH2 gene constructs was used to normalize relative expression of Kv11.1 isoforms.
2.4 Immunoblot analysis
Immunoblot analysis was performed as previously described (Gong et al., 2010). The cell lysates were subjected to SDS-polyacrylamide gel electrophoresis and then electrophoretically transferred onto nitrocellulose membranes. The membranes were incubated with an anti-Kv11.1 antibody against the N-terminus of Kv11.1a and Kv11.1a-USO proteins (H-175, Santa Cruz, Santa Cruz, CA) at a 1:600 dilution and visualized with the ECL detection kit (Amersham, Piscataway, NJ). The expression level of hygromycin B phosphotransferase (HPT) encoded by the hygromycin B resistant gene was used as loading control (Gong et al., 2010).
2.5 Patch-clamp recordings
Membrane currents were recorded in whole cell configuration using suction pipettes as previously described (Zhou et al., 1998). The bath solution contained (in mM) 137 NaCl, 4 KCl, 1.8 CaCl2, 1 MgCl2, 10 glucose, and 10 HEPES (pH 7.4 with NaOH). The pipette solution contained (in mM) 130 KCl, 1 MgCl2, 5 EGTA, 5 MgATP, and 10 HEPES (pH 7.2 with KOH). All patch-clamp experiments were performed at 22-23 °C. Kv11.1 current was activated by depolarizing steps between –70 and +50 mV from a holding potential of –80 mV and Kv11.1 tail current was recorded following repolarization to –50 mV.
2.6 Statistical analysis
Data are presented as mean ± standard error of mean. Student's t-test was used for comparison between two groups. ANOVA with Bonferroni correction was used for comparisons between more than two groups. P < 0.05 is considered statistically significant.
3. Results
3.1 Splicing of KCNH2 intron 9 can be improved by complementary base changes in U1 snRNA
We have shown previously that the splicing of KCNH2 intron 9 is inefficient due to the presence of non-consensus nucleotides at +4 and +6 positions of the 5′ splice site. When both +4 and +6 nucleotides are changed to the consensus sequence, the Kv11.1a isoform is exclusively produced (Gong et al., 2010). To test whether the inefficient splicing of KCNH2 intron 9 is due to base-pair mismatch between U1 snRNA and the 5′ splice site, we changed the U1 snRNA at position 3 (3A>C) or 5 (5U>A) to make them complementary to the +6g or +4u nt of the 5′ splice site of KCNH2 intron 9 (Fig. 1B), respectively. We cotransfected the minigene luciferase reporter construct with the wild-type (WT) or modified U1 snRNA into HEK293 cells. In the minigene luciferase reporter construct, the splicing of intron 9 would generate active luciferase and polyadenylation of intron 9 would result in no luciferase activity (Gong et al., 2014b). As shown in Fig. 1C, cotransfection with modified U1 snRNA at position 3 (3A>C) or 5 (5U>A) increased the luciferase activity compared to WT U1 snRNA (P < 0.001). When U1 snRNA was modified at both 3 and 5 positions (3A>C/5U>A), the luciferase activity was further increased compared to 3A>C or 5U>A alone (P < 0.001). These results demonstrate that the weak 5′ splice site of KCNH2 intron 9 is due to base-pair mismatches between U1 snRNA and the 5′ splice site, and that the splicing efficiency of KCNH2 intron 9 can be increased by complementary base changes in U1 snRNA.
3.2 Regulation of Kv11.1 isoform expression by modified U1 snRNA
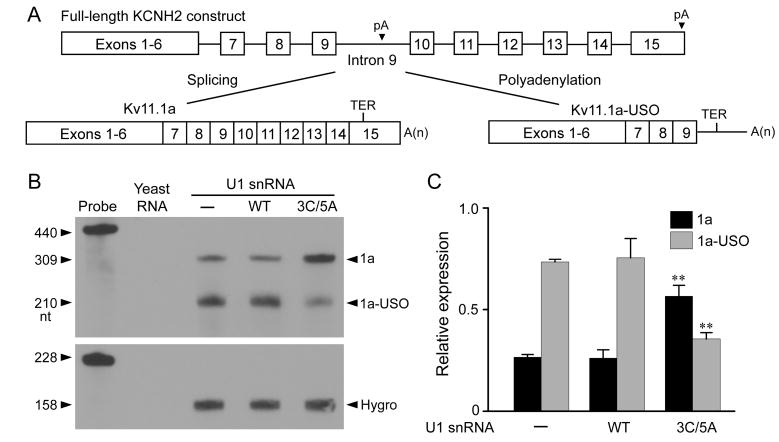
To test whether the modified U1 snRNA can regulate the relative expression of Kv11.1 isoforms, we used the full-length KCNH2 splicing-competent construct composed of the Kv11.1a cDNA from exon 1 to exon 6 and the genomic sequence spanning intron 6 to the poly(A) signal of the KCNH2 gene (Fig. 2A) (Gong et al., 2010). If intron 9 is spliced and the poly(A) site in exon 15 is used, the full-length Kv11.1a transcript will be produced. If the transcript is polyadenylated at the intron 9 poly(A) site, the Kv11.1a-USO isoform will be generated. When expressed in Flp-In CV-1 cells, the full-length KCNH2 splicing-competent construct underwent alternative processing to generate the Kv11.1a and Kv11.1a-USO isoforms. Treatment with modified U1 snRNA (3A>C/5U>A) adenovirus resulted in an increase in the Kv11.1a transcript and a decrease in the Kv11.1a-USO transcript compared to the control WT U1 snRNA adenovirus treatment (Fig. 2B, C, P < 0.01). This result suggests that relative expression of Kv11.1a and Kv11.1a-USO isoforms can be modulated by a modified U1 snRNA with increased complementarity to the 5′ splice site of KCNH2 intron 9.
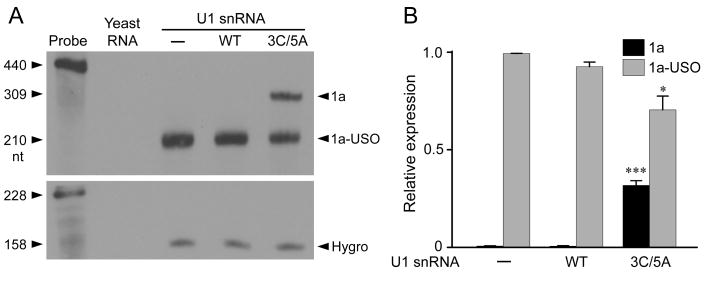
Fig. 2.
Regulation of Kv11.1 isoform expression by modified U1 snRNA in the full-length KCNH2 splicing-competent construct. (A) The structure of the full-length KCNH2 splicing-competent construct. (B) RPA analysis of the effect of modified U1 snRNA on Kv11.1 isoform expression. Flp-In CV-1 cells stably expressing the full-length KCNH2 splicing-competent construct were treated with WT or modified U1 snRNA adenovirus (500 MOI) for 48 hours. (C) RPA signals were quantified, normalized to hygromycin resistance gene (Hygro), and plotted as the relative expression of the total uninfected (1a+1a-USO) Kv11.1 mRNA (n=3, **P < 0.01).
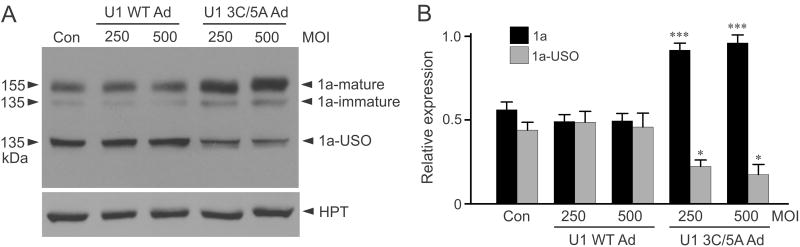
3.3 Upregulation of Kv11.1a channel protein by modified U1 snRNA
To determine whether modified U1 snRNA-induced modulation of Kv11.1 C-terminal isoform expression leads to the isoform switch at the protein level, we analyzed Kv11.1 protein expression by immunoblot. When expressed in Flp-In CV-1 cells, the full-length KCNH2 splicing-competent construct produced three protein bands at about 155 kDa, 135 kDa and 100 kDa. The 155 kDa band represents the fully glycosylated mature form of Kv11.1a, the 135 kDa band represents the core-glycosylated immature form of Kv11.1a, and the 100 kDa band represents the core-glycosylated form of Kv11.1a-USO (Gong et al., 2010). Flp-In CV-1 cells stably expressing the full-length KCNH2 splicing-competent construct were treated with different concentrations of WT or modified U1 snRNA adenovirus. Treatment with modified U1 snRNA (3A>C/5U>A) significantly increased the level of Kv11.1a protein (P < 0.001) and decreased the Kv11.1a-USO protein level compared to WT U1 snRNA control (P < 0.05) (Fig. 3).
Fig. 3.
Regulation of Kv11.1 protein expression by modified U1 snRNA. (A) Flp-In CV-1 cells stably expressing the full-length KCNH2 splicing-competent construct were treated with WT or modified U1 snRNA adenovirus (250 or 500 MOI) for 48 hours. The expression level of hygromycin B phosphotransferase (HPT) encoded by the hygromycin B resistant gene served as a loading control. Cell lysates were subjected to SDS-PAGE and probed with antibodies against the N-terminus of Kv11.1 and against HPT. (B) The level of protein bands was quantified, normalized to HPT, and plotted as the relative expression of the total uninfected (1a+1a-OSU) Kv11.1 protein (n=3, *P < 0.05, ***P < 0.001).
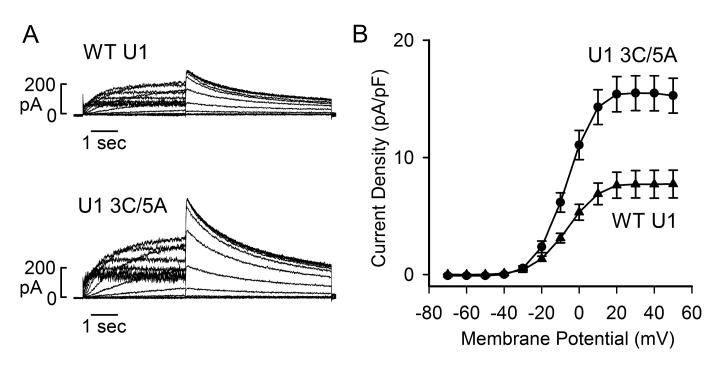
3.4 Modified U1 snRNA increased Kv11.1 channel current
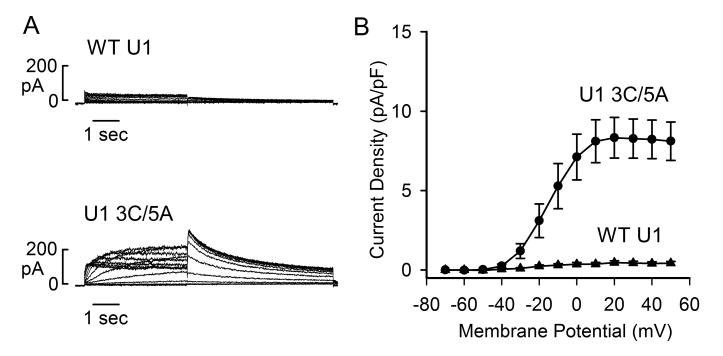
To study functional consequence of modified U1 snRNA, we performed patch-clamp recordings of Kv11.1 channel current. Flp-In CV-1 cells stably expressing the full-length KCNH2 splicing-competent construct were treated with WT or modified U1 snRNA adenovirus for 48 hours. Treatment with modified U1 snRNA (3A>C/5U>A) significantly increased Kv11.1 current compared to the WT U1 snRNA control (Fig. 4). The maximum tail current densities in WT and modified U1 snRNA-treated cells were 7.7 ± 1.2 pA/pF and 15.5 ± 1.5 pA/pF (P < 0.01), respectively. These patch-clamp experiments are consistent with the results of immunoblot and demonstrate that modified U1 snRNA can increase the Kv11.1 channel current.
Fig. 4.
Effect of modified U1snRNA on Kv11.1 channel current. (A) Representative currents recorded from Flp-In CV-1 cells stably expressing following treatment with WT or modified U1 snRNA adenovirus (500 MOI) for 48 hours. (B) I-V plot of tail current density measured at –50 mV following test voltages from –70 to +50 mV for WT (triangle, n=6) and modified U1 snRNA (3A>C/5U>A) (circle, n=8) adenoviruses.
3.5 Effect of modified U1 snRNA on Kv11.1 isoform expression in canonical intron 9 poly(A) signal construct
We have previously shown that competition between the weak 5′ splice site and the weak poly(A) signal in intron 9 plays an important role in regulation of the relative expression of Kv11.1a and Kv11.1a-USO isoforms. When the weak intron 9 poly(A) signal was changed to the strong canonical poly(A) signal, the Kv11.1a-USO isoform was predominantly expressed (Gong et al., 2010). To test whether the modified U1 snRNA can induce the isoform expression from Kv11.1a-USO to Kv11.1a in the presence of a strong intron 9 poly(A) signal, we treated CV-1 cells stably expressing the full-length KCNH2 splicing-competent construct containing the canonical intron 9 poly(A) signal with WT and modified U1 snRNA adenoviruses. RPA analysis showed that in the presence of the WT U1 snRNA, Kv11.1a-USO was predominantly expressed. Treatment with modified U1 snRNA (3A>C/5U>A) significantly increased the expression of the Kv11.1a isoform (P < 0.001) and decreased the expression of the Kv11.1a-USO isoform compared to WT U1 snRNA (P < 0.05) (Fig. 5). In patch clamp studies, treatment with modified U1 snRNA significantly increased Kv11.1 current compared to the WT U1 snRNA control (Fig. 6). The maximum tail current densities in WT and modified U1 snRNA-treated cells were 0.5 ± 0.1 pA/pF and 8.3 ± 1.3 pA/pF (P < 0.001), respectively. These findings suggest that modified U1 snRNA is capable of upregulating the Kv11.1a isoform expression in the presence of a strong intron 9 poly(A) signal. However, the maximum current density upregulated by modified U1 snRNA is lower in the canonical intron 9 poly(A) signal than in the weak, non-canonical intron 9 poly(A) signal (Fig. 4, P < 0.01).
Fig. 5.
Regulation of Kv11.1 isoform expression by modified U1 snRNA in canonical poly(A) signal construct. (A) RPA analysis of mRNA from Flp-In CV-1 cells stably expressing the full-length KCNH2 splicing-competent construct containing the canonical poly(A) signal following the treatment with WT or modified U1 snRNA adenovirus. (B) RPA signals were quantified, normalized to hygromycin resistance gene (Hygro), and plotted as the relative expression of the total uninfected (1a+1a-USO) Kv11.1 mRNA (n=3, *P < 0.05, **P < 0.01).
Fig. 6.
Effect of modified U1 snRNA on Kv11.1 channel current in canonical poly(A) signal construct. (A) Representative currents recorded from Flp-In CV-1 cells stably expressing the full-length KCNH2 splicing-competent construct containing the canonical poly(A) signal following treatment with WT or modified U1 snRNA adenovirus (500 MOI) for 48 h. (B) I-V plot of tail current density measured at –50 mV following test voltages from –70 to +50 mV for WT (triangle, n=7) and modified U1 snRNA (3A>C/5U>A) (circle, n=8) adenoviruses.
4. Discussion
In the human heart, the splicing of intron 9 is inefficient with only one-third of KCNH2 pre-mRNA being processed to the functional Kv11.1a isoform and two-thirds of KCNH2 pre-mRNA being processed to the C-terminally truncated, non-functional Kv11.1a-USO isoform. We have previously demonstrated that the 5′ splice site of intron 9 deviates from the consensus sequence, which reduces the efficiency of intron 9 splicing (Gong et al., 2010). Compared to the consensus sequence (CAG/gtaagt), the 5′ splice site of KCNH2 intron 9 (TGG/gtatgg) differs at four positions. Our present findings indicate that deviation from consensus sequence weakens the complimentary base-pairing between the 5′ splice site and the U1 snRNA, thus decreasing the efficiency of intron 9 splicing.
The initial step of splicing process involves recognition of the 5′ splice site by U1 small nuclear ribonucleoprotein. This recognition is mediated by complementary base-pairing between the 5′ splice site and U1 snRNA, the RNA component of the U1 small nuclear ribonucleoprotein. The average number of base-pairs formed by 5′ splice site and U1 snRNA is seven, although the number varies from five to nine (Zhuang and Weiner, 1986). The deviation of the 5′ splice site of intron 9 from the consensus sequence reduces the strength of binding between the 5′ splice site and U1 snRNA, as U1 snRNA can presumably form only five base-pairs with this suboptimal splice site. It has been reported that the degree of complementarity between the 5′ splice site and U1 snRNA is an important determinant of splicing efficiency (Zhuang and Weiner, 1986). Our results provide direct evidence that modified U1 snRNA with increased base-pair complementarity to the 5′ splice sites of intron 9 enhances efficiency of intron 9 splicing, resulting in an increase in the expression of the Kv11.1a isoform and a concomitant decrease in the expression of the Kv11.1a-USO isoform. The complementary base changes at positions 3 and 5 in the modified U1 snRNA (U1-3C/5A) would create seven contiguous base-pairs between U1 snRNA and the 5′ splice site of intron 9 (Fig. 1B). Thus, the degree of complementarity between the 5′ splice site and U1 snRNA plays an important role in determining splicing efficiency of the KCNH2 intron 9.
Modification of U1 snRNA with increased complementarity to mutant 5′ splice sites was originally employed to demonstrate the importance of base-pair binding of U1 snRNA with 5′ splice site in the initial step of the splicing process (Zhuang and Weiner, 1986). Modified U1 snRNA has been used to rescue 5′ splice site disease-causing mutations. Mutation-adapted U1snRNAs with complementary changes to match mutated 5′ splice sites are able to correct splicing defects in various cellular models of human genetic diseases (Baralle et al., 2003, Zhang et al., 2004, Pinotti et al., 2008, Balestra et al., 2014, Glaus et al., 2011, Schmid et al., 2011). The present findings indicate that modified U1 snRNA can be used to increase the splicing efficiency of the intrinsically weak KCNH2 intron 9 splice site and upregulate the expression of the functional Kv11.1a isoform. We recently identified a KCNH2 splice site mutation that causes long QT syndrome by isoform switch from the functional Kv11.1a isoform to the non-functional Kv11.1a-USO isoform (Gong et al., 2014a). Modified U1 snRNA may thus represent an ideal strategy to increase the expression of Kv11.1a from the WT allele and overcome the isoform switch caused by the mutant allele.
The expression of the full-length Kv11.1a isoform and the truncated Kv11.1a-USO isoform is determined by competition between intron 9 splicing and polyadenylation at the intronic poly(A) site. The increase in splicing efficiency by modified U1 snRNA can shift the balance toward the splicing pathway, thereby leading to the upregulation of the full-length Kv11.1a isoform. Our results also show that the modified U1 snRNA is capable of upregulating Kv11.1a isoform expression in the presence of a strong intron 9 poly(A) signal, although the effect of modified U1 snRNA on upregulation of the Kv11.1a isoform expression is less than that in the presence of the native weak intron 9 poly(A) signal. This finding suggests that relative efficiencies of intron 9 splicing and polyadenylation play an important role in regulation of the relative expression of the Kv11.1a and Kv11.1a-USO isoforms. Regulation of isoform expression by the competition between splicing and intronic polyadenylation has been reported in many genes. A well-studied example is the regulation of the IgM heavy-chain gene, where a membrane-bound form of IgM heavy-chain is produced through splicing of intron 4 and a secreted form of IgM heavy-chain is generated by the utilization of an intronic poly(A) site within intron 4 (Peterson and Perry, 1989). Similarly, many receptor tyrosine kinase genes undergo alternative processing of pre-mRNA through competing splicing and intronic polyadenylation. Membrane-bound receptor tyrosine kinases are generated by intron splicing and secreted isoforms are produced by intronic polyadenylation. Inhibition of U1 snRNA binding to 5′ splice sites by antisense oligonucleotides can activate intronic poly(A) sites, leading to the switch from the full-length, membrane-bound isoforms to the truncated, soluble isoforms of receptor tyrosine kinases (Vorlova et al., 2011). Rather than increasing intronic polyadenylation by the inhibition of splicing, our present results demonstrate that modified U1 snRNA with increased complementary binding to the weak 5′ splice sites of KCNH2 intron 9 increases splicing efficiency and decreases intronic polyadenylation. Thus, modified U1 snRNA with increased complementarity to a weak, nonconsensus 5′ splice site may represent a useful strategy to upregulate the expression of full-length isoforms of other genes that are regulated by competition between splicing and intronic polyadenylation.
In summary, our present findings show that increasing U1 snRNA complementarity to the 5′ splice site of KCNH2 intron 9 results in an increase in the expression of the functional Kv11.1a isoform and a concomitant decrease in the expression of the non-functional Kv11.1a-USO isoform. Modified U1 snRNA with increased complementarity to the 5′ splice site of KCNH2 intron 9 may represent a novel approach to increase Kv11.1 channel function.
Research Highlights.
Kv11.1 C-terminal isoform expression is regulated by KCNH2 intron 9 splicing
Splicing is inefficient due to mismatches between 5′ splice site and U1 snRNA
Increasing U1 snRNA complementarity to 5′ splice site improved splicing efficiency
Modified U1 snRNA increased Kv11.1a expression and Kv11.1 current
Modified U1 snRNA represents a novel approach to increase Kv11.1 channel function
Acknowledgments
This work was supported in part by grants from the NIH HL068854 (ZZ) and American Heart Association 15GRNT23020018 (ZZ).
Abbreviations
- hERG
human ether-a-go-go-related gene
- HPT
hygromycin B phosphotransferase
- LQT2
long QT syndrome type 2
- MOI
multiplicity of infection
- nt
nucleotide
- RPA
RNase protection assay
- U1 snRNA
U1 small nuclear RNA
- WT
wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Balestra D, Faella A, Margaritis P, Cavallari N, Pagani F, Bernardi F, Arruda VR, Pinotti M. An engineered U1 small nuclear RNA rescues splicing defective coagulation F7 gene expression in mice. J Thromb Haemost. 2014;12:177–185. doi: 10.1111/jth.12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baralle M, Baralle D, De Conti L, Mattocks C, Whittaker J, Knezevich A, Ffrench-Constant C, Baralle FE. Identification of a mutation that perturbs NF1 gene splicing using genomic DNA samples and a minigene assay. J Med Genet. 2003;40:220–222. doi: 10.1136/jmg.40.3.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran ME, Splawski I, Timothy KW, Vincent GM, Green ED, Keating MT. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell. 1995;80:795–803. doi: 10.1016/0092-8674(95)90358-5. [DOI] [PubMed] [Google Scholar]
- Gong Q, Stump MR, Deng V, Zhang L, Zhou Z. Identification of Kv11.1 isoform switch as a novel pathogenic mechanism of long QT syndrome. Circ Cardiovasc Genet. 2014a;7:482–90. doi: 10.1161/CIRCGENETICS.114.000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Q, Stump MR, Dunn AR, Deng V, Zhou Z. Alternative splicing and polyadenylation contribute to the generation of hERG1 C-terminal isoforms. J Biol Chem. 2010;285:32233–32241. doi: 10.1074/jbc.M109.095695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Q, Stump MR, Zhou Z. Upregulation of functional Kv11.1 isoform expression by inhibition of intronic polyadenylation with antisense morpholino oligonucleotides. J Mol Cell Cardiol. 2014b;76:26–32. doi: 10.1016/j.yjmcc.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Q, Zhang L, Vincent GM, Horne BD, Zhou Z. Nonsense mutations in hERG cause a decrease in mutant mRNA transcripts by nonsense-mediated mRNA decay in human long-QT syndrome. Circulation. 2007;116:17–24. doi: 10.1161/CIRCULATIONAHA.107.708818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaus E, Schmid F, Da Costa R, Berger W, Neidhardt J. Gene therapeutic approach using mutation-adapted U1 snRNA to correct a RPGR splice defect in patient-derived cells. Mol Ther. 2011;19:936–941. doi: 10.1038/mt.2011.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guasti L, Crociani O, Redaelli E, Pillozzi S, Polvani S, Masselli M, Mello T, Galli A, Amedei A, Wymore RS, Wanke E, Arcangeli A. Identification of a posttranslational mechanism for the regulation of hERG1 K+ channel expression and hERG1 current density in tumor cells. Mol Cell Biol. 2008;28:5043–5060. doi: 10.1128/MCB.00304-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupershmidt S, Snyders DJ, Raes A, Roden DM. A K+ channel splice variant common in human heart lacks a C-terminal domain required for expression of rapidly activating delayed rectifier current. J Biol Chem. 1998;273:27231–27235. doi: 10.1074/jbc.273.42.27231. [DOI] [PubMed] [Google Scholar]
- Larsen AP. Role of ERG1 isoforms in modulation of ERG1 channel trafficking and function. Pflugers Arch. 2010;460:803–812. doi: 10.1007/s00424-010-0855-8. [DOI] [PubMed] [Google Scholar]
- Lees-Miller JP, Kondo C, Wang L, Duff HJ. Electrophysiological characterization of an alternatively processed ERG K+ channel in mouse and human hearts. Circ Res. 1997;81:719–726. doi: 10.1161/01.res.81.5.719. [DOI] [PubMed] [Google Scholar]
- London B, Trudeau MC, Newton KP, Beyer AK, Copeland NG, Gilbert DJ, Jenkins NA, Satler CA, Robertson GA. Two isoforms of the mouse ether-a-go-go-related gene coassemble to form channels with properties similar to the rapidly activating component of the cardiac delayed rectifier K+ current. Circ Res. 1997;81:870–878. doi: 10.1161/01.res.81.5.870. [DOI] [PubMed] [Google Scholar]
- Peterson ML, Perry RP. The regulated production of mu m and mu s mRNA is dependent on the relative efficiencies of mu s poly(A) site usage and the c mu 4-to-M1 splice. Mol Cell Biol. 1989;9:726–738. doi: 10.1128/mcb.9.2.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinotti M, Rizzotto L, Balestra D, Lewandowska MA, Cavallari N, Marchetti G, Bernardi F, Pagani F. U1-snRNA-mediated rescue of mRNA processing in severe factor VII deficiency. Blood. 2008;111:2681–2684. doi: 10.1182/blood-2007-10-117440. [DOI] [PubMed] [Google Scholar]
- Sanguinetti MC, Jiang C, Curran ME, Keating MT. A mechanistic link between an inherited and an acquired cardiac arrhythmia: HERG encodes the IKr potassium channel. Cell. 1995;81:299–307. doi: 10.1016/0092-8674(95)90340-2. [DOI] [PubMed] [Google Scholar]
- Schmid F, Glaus E, Barthelmes D, Fliegauf M, Gaspar H, Nürnberg G, Nürnberg P, Omran H, Berger W, Neidhardt J. U1 snRNA-mediated gene therapeutic correction of splice defects caused by an exceptionally mild BBS mutation. Hum Mutat. 2011;32:815–824. doi: 10.1002/humu.21509. [DOI] [PubMed] [Google Scholar]
- Stump MR, Gong Q, Zhou Z. Isoform-specific dominant-negative effects associated with hERG1 G628S mutation in long QT syndrome. PLoS ONE. 2012;7:e42552. doi: 10.1371/journal.pone.0042552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudeau MC, Warmke JW, Ganetzky B, Robertson GA. HERG, a human inward rectifier in the voltage-gated potassium channel family. Science. 1995;269:92–95. doi: 10.1126/science.7604285. [DOI] [PubMed] [Google Scholar]
- Vorlová S, Rocco G, Lefave CV, Jodelka FM, Hess K, Hastings ML, Henke E, Cartegni L. Induction of antagonistic soluble decoy receptor tyrosine kinases by intronic polyA activation. Mol Cell. 2011;43:927–939. doi: 10.1016/j.molcel.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmke JW, Ganetzky B. A family of potassium channel genes related to eag in Drosophila and mammals. Proc Natl Acad Sci USA. 1994;91:3438–3442. doi: 10.1073/pnas.91.8.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Vincent GM, Baralle M, Baralle FE, Anson BD, Benson DW, Whiting B, Timothy KW, Carlquist J, January CT, Keating MT, Splawski I. An intronic mutation causes long QT syndrome. J Am Coll Cardiol. 2004;44:1283–1291. doi: 10.1016/j.jacc.2004.06.045. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Gong Q, Ye B, Fan Z, Makielski JC, Robertson GA, January CT. Properties of HERG channels stably expressed in HEK 293 cells studied at physiological temperature. Biophys J. 1998;74:230–241. doi: 10.1016/S0006-3495(98)77782-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y, Weiner AM. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell. 1986;46:827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]