Abstract
DNA methylation is a stable and heritable epigenetic modification in the mammalian genome and is involved in regulating gene expression to control cellular functions. The reversal of DNA methylation, or DNA demethylation, is mediated by the ten-eleven translocation (TET) protein family of dioxygenases. Although it has been widely reported that aberrant DNA methylation and demethylation are associated with developmental defects and cancer, how these epigenetic changes directly contribute to the subsequent alteration in gene expression or disease progression remains unclear, largely owing to the lack of reliable tools to accurately add or remove DNA modifications in the genome at defined temporal and spatial resolution. To overcome this hurdle, we designed a split-TET2 enzyme to enable temporal control of 5-methylcytosine (5mC) oxidation and subsequent remodeling of epigenetic states in mammalian cells by simply adding chemicals. Here, we describe methods for introducing a chemical-inducible epigenome remodeling tool (CiDER), based on an engineered split-TET2 enzyme, into mammalian cells and quantifying the chemical inducible production of 5-hydroxymethylcytosine (5hmC) with immunostaining, flow cytometry or a dot-blot assay. This chemical-inducible epigenome remodeling tool will find broad use in interrogating cellular systems without altering the genetic code, as well as in probing the epigenotype−phenotype relations in various biological systems.
Keywords: Chemistry, Issue 130, DNA methylation, epigenetics, chromatin accessibility, DNA demethylation, chemical biology, TET2, 5-hydroxymethylcytosine, transcription, gene expression, epigenome editing
Introduction
DNA methylation, mostly refers to the addition of a methyl group to the carbon 5 position of cytosine to form 5-methylcytosine (5mC), is catalyzed by DNA methyl-transferases (DNMTs). 5mC acts as a major epigenetic mark in the mammalian genome that often signals for transcriptional repression, X-chromosome inactivation and transposon silencing1. The reversal of DNA methylation is mediated by the Ten-elven translocation (TET) protein family. TET enzymes belong to the iron (II) and 2-oxoglutarate dependent dioxygenases which catalyze the successive oxidation of 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxycytosine (5caC). TET-mediated 5mC oxidation imposes an additional layer of epigenetic control over the mammalian genome. The discovery of TET has sparked intense interest in the epigenetic field to unveil the biological functions of TET proteins and their major catalytic product 5hmC. 5hmC is not only an intermediate during TET-mediated active DNA demethylation2,3,4, but also acts as a stable epigenetic mark5,6,7,8. Although DNA hydroxymethylation is highly correlated with gene expression and aberrant changes in DNA hydroxymethylation are associated with some human disorders9,10,11, the causal relations between epigenetic modifications on DNA and the phenotypes often remain challenging to be established, which can be partially ascribed to the lack of reliable tool to accurately add or remove DNA modifications in the genome at defined temporal and spatial resolution.
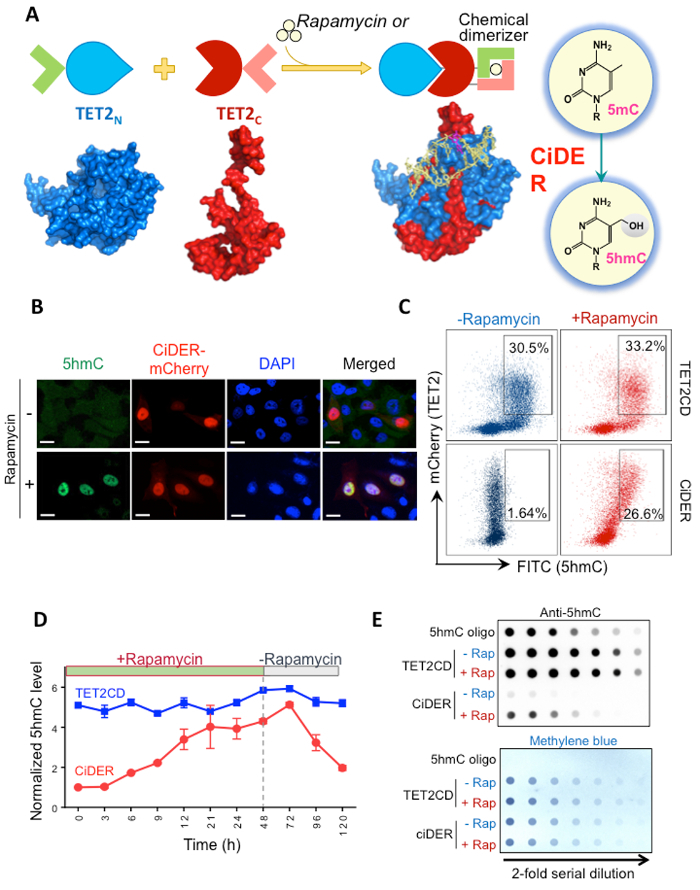
Here we report the use of a chemical-inducible epigenome remodeling tool (CiDER) to overcome the hurdle facing studies of causal relationships between DNA hydroxymethylation and gene transcription. The design is based on the assumption that the catalytic domain of TET2 (TET2CD) can be split into two inactive fragments when expressed in mammalian cells and its enzymatic function can be restored by taking a chemically inducible dimerization approach (Figure 1A). To establish a split-TET2CD system, we selected six sites in TET2CD, composed of a Cys-rich region and a double-stranded β-helix (DSBH) fold, on the basis of a reported crystal structure of TET2-DNA complex that lacks a low complexity region12. A synthetic gene encoding rapamycin-inducible dimerization module FK506 binding protein 12 (FKBP12) and FKBP rapamycin binding domain (FRB) of the mammalian target of rapamycin14,15, along with a self-cleaving peptide T2A polypeptide sequence16,17, was individually inserted into the selected split sites within TET2CD. The selection of TET2 as our engineering target is based on the following considerations. First, somatic mutations in TET2 with concomitant reduction in DNA hydroxymethylation are frequently observed in human disorders including myeloid disorders and cancer10, which provides useful information on sensitive sites to be avoided for insertion. Second, a large fraction of the TET2 catalytic domain, particularly the low complexity region, is dispensable for the enzymatic function12, thus enabling us to craft a minimized split-TET2 for inducible epigenetic modifications. After screening over 15 constructs, a construct that exhibited highest rapamycin-inducible restoration of its enzymatic activity in the mammalian system was chosen and designated as CiDER18. We describe herein the use of mCherry-tagged CiDER to achieve inducible DNA hydroxymethylation and epigenetic remodeling with rapamycin, and present three methods for validating CiDER-mediated 5hmC production in a model cellular system HEK293T.
Protocol
1. Cell Culture, Plasmid Transfection and Chemical Induction
Culture an adherent cell line (e.g., the human embryonic kidney HEK293T cell) in Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% heat-inactivated fetal bovine serum, 100 U/mL penicillin and streptomycin at 37 °C with 5% CO2.
- Transfect cells with the CiDER plasmid.
- At least 18 h before transfection, seed 0.3 x 106 cells in 2.5 mL of appropriate DMEM per well in a 6-well plate and incubate cells at 37 °C overnight to reach 60-80% confluency.
- Pre-warm the transfection reagent to room temperature before use.
- Place 250 µL of serum-free medium in a sterile tube. NOTE: This amount is tailored for a 6-well plate with 80% confluency. If needed, proportionally adjust the medium amounts according to the sizes of plates or cell numbers.
- Add 7.5 µL of transfection reagent to the diluted DNA mixture and gently mix by pipetting.
- Incubate the mixture at room temperature for 20-30 min.
- Change pre-warmed media and add the mixture drop-wise to the wells, and gently shake the cell culture plate to evenly distribute the transfection reagent and DNA mixture.
- Incubate cells and mixture overnight, and then replace the transfection reagent containing media with 2ml of fresh complete DMEM.
- Chemical induction
- Dilute 2 mM of rapamycin stock (dissolved in DMSO) to a concentration of 20 µM in appropriate complete medium.
- Add 20 µL of diluted rapamycin to transfected cells maintained in 2 mL medium to reach a final concentration of 200 nM. NOTE: After incubation for 48 h, cells are ready for quantification of 5hmC generation or any other downstream applications. If needed, the induction efficiency can be monitored by taking out cells every 3 h for further 5hmC quantification as described below and shown in Figure 1.
- For better quantification of 5hmC induction efficiency, seed separate wells to monitor cells every 3 hours.
2. Quantification of Rapamycin-induced 5hmC Production by Immunostaining
- 5hmC immunofluorescence staining NOTE: Add sufficient amounts of the fixation solution, composed of 4% paraformaldehyde, 0.2% surfactant (such as Triton X-100) in PBS with 3 N HCl, to cover all the cells in plate. For example, 500 µL of solution would be enough for a well in a 6-well plate. Perform all steps at room temperature.
- To fix cells, add 4% paraformaldehyde in PBS to rapamycin-treated cells for 15 min at ambient temperature. For the control group, use cells expressing mCherry-CiDER without rapamycin treatment. Do not agitate. Caution: Paraformaldehyde is toxic. Wear appropriate protection.
- Discard fixative solution. Incubate fixed cells with 0.2% surfactant in PBS for 30 min to permeabilize cell membrane.
- Discard permeabilization solution. Add 3 N HCl to the permeabilized cells and incubate for 15 min to denature the DNA.
- Discard HCl. Add 100 mM Tris-HCl buffer (pH 8.0) to the cells for 10 min for neutralization of pH.
- Discard all the solution. Wash cells thoroughly with PBS for 3-5 times. Remove PBS thoroughly. NOTE: Add sufficient amounts of PBS for every wash step. For example, 1 mL of PBS is enough for a 6-well plate.
- Block cells by adding the blocking buffer comprising 1% BSA and 0.05% of surfactant (such as Tween 20) in PBS for 1 h with gentle shaking.
- Discard the blocking solution. Add the diluted 5hmC antibody (1:200) to the cells and incubate for 2 h at room temperature or overnight at 4 °C.
- Wash the cells with PBS three times for 15 min.
- Add a diluted fluorophore (FITC)-conjugated secondary antibody (1:200) to the cells and incubate for 1 h.
- Wash the cells with PBS three times extensively to remove residual antibodies.
- Add 250 ng/mL of 4',6-diamidino-2- phenylindole (DAPI) to stain the nucleus for 5 min. Remove the staining solution.
- Mount the slide or glass-bottom culture dish with immunostained cells for confocal imaging. A representative result was shown in Figure 1B.
- Flow cytometry NOTE: Use a 96-well round bottom plate for the following staining procedure. Usually, 150 µL of reagents is sufficient for each well.
- Resuspend transfected and rapamycin-induced cells in FACS buffer (PBS with 1% BSA, 2 mM EDTA). For the control group, use CiDER-expressing cells without rapamycin treatment.
- Fix and permeabilize the cells as described in Step 2.1.
- Add 2 N HCl to the cells to denature the DNA for 10 min.
- Discard HCl. Neutralize and block the cells as described in Step 2.1.
- Add a diluted anti-5hmC antibody (1:200) to the cells and incubate for 1 h at room temperature or overnight at 4 °C.
- Wash the cells with FACS buffer three times. Remove FACS buffer thoroughly.
- Add a diluted fluorophore (FITC)-conjugated secondary antibody (1:400) for fluorescence-activated cell sorting (FACS) analysis, and incubate for 1 h at room temperature.
- Wash the cells with FACS buffer three times.
- Samples are ready for FACS analysis. A representative result was shown in Figure 1C-D.
3. Dot-blot Assay to Quantify 5hmC and 5-methylcytosine (5mC) Amounts
Isolate genomic DNA from transfected and rapamycin-induced cells using a commercial DNA extraction kit. For the control group, use CiDER-transfected cells without rapamycin treatment.
Heat the vacuum oven to 80 °C and pre-soak nitrocellulose membrane in double-distilled H2O and then in 6x saline-sodium citrate (SSC) buffer for 10 min.
Load 60 µL of equal amounts of DNA (total 2 µg in TE buffer) to a 96-well PCR plate. Add 30 µL of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) to the rest wells.
Make two-fold serial dilutions vertically on the 96-well plate by transferring 30 µL of solution on row 1 to the same column position on row 2. Mix again and transfer 30 µL of solution to the wells in the subsequent row till reaching the boundary. Remove the last 30 µL.
Add 20 µL of 1 M NaOH and 10 mM EDTA to each well, seal the plate and heat the mixture for 10 min at 95 °C to completely denature the DNA duplex.
Immediately cool down the samples on ice and add 50 µL of ice-cold 2 M ammonium acetate (pH 7.0) and incubate on ice for 10 min. NOTE: Steps 3.7-3.10 involves the use of vacuum.
Assemble the dot blot apparatus with pre-soaked membrane and remove residual buffer using full vacuum.
Wash the membrane by adding 200 µL of TE buffer to each well of the dot blot apparatus. Turn on vacuum to remove TE buffer.
Load the denatured DNA (prepared in previous steps 3.1-3.6) to each well of the dot blot apparatus. Turn on vacuum to remove solution.
Wash the membrane by adding 200 µL of 2x SSC and remove residual solutions completely using vacuum.
Disassemble the dot blot apparatus, take out the membrane and rinse the whole membrane with 20 mL of 2x SSC.
Air-dry the membrane for 20 min.
Bake the membrane at 80 °C under vacuum for 2 h.
Add the blocking solution (5% skim milk in TBST) to the membrane and incubate for 1 h at room temperature.
Discard the blocking solution. Add a diluted 5hmC (1:5,000) or 5mC (1:1,000) antibodies to the membrane and incubate overnight at 4 °C.
Wash the membrane with TBST three times for 10 min to remove residual antibodies. Remove TBST thoroughly.
Add an HRP-conjugated secondary antibody (1:10,000 for anti-rabbit HRP(5hmC) or 1:3,000 for anti-mouse HRP(5mC)) to the membrane and incubate for 1 h at room temperature.
Wash the membrane with TBST three times for 10 min.
Add ECL western blotting substrate to the membrane for visualization of 5hmC signals using a chemiluminescence detector. A representative result was shown in Figure 1E.
Representative Results
Chemical inducible 5mC-to-5hmC conversion can be validated by immunostaining at single-cell level, flow cytometry analysis on transfected (mCherry-positive) cell populations, or a more quantitative dot-blot assay as illustrated in Figure 1. The catalytic domain of TET2, or TET2CD, were used throughout the study as a positive control. See references 18-20 for further details regarding the genome-wide mapping of rapamycin-induced changes in 5hmC and chromatin accessibility.
Figure 1: CiDER-mediated inducible DNA hydroxymethylation in HEK293T cells. (A) Design of a chemical-inducible epigenome remodeling (CiDER) tool based on a split TET2 enzyme. (B) Representative immunostaining results in HEK293T cells expressing CiDER-mCherry before (upper panel) and after (lower panel) rapamycin treatment (200 nM). Green, 5hmC, red, CiDER-mCherry, Blue, DAPI staining of nuclei. Scale bar = 10 µm. (C) Quantification of CiDER-mediated 5hmC production by flow cytometry analysis. HEK293T cells transfected with CiDER-mCherry of TET2CD-mCherry (as positive control) were stained with an anti-5hmc primary antibody and an FITC-labeled secondary antibody. Only TET2 (mCherry) and 5hmC (FITC) double-positive cells were gated and indicated. (D) Time course of chemical inducible production of 5hmC in HEK293T cells expressing CiDER following administration or withdrawal of rapamycin (200 nM). n = 5, data shown as mean ± S.D. (E) Dot-blot assay to quantify rapamycin-inducible changes in 5hmC levels in HEK293T cells. HEK293T cells were transfected with either a CiDER or TET2CD construct. A synthetic oligonucleotide with a known amount of 5hmC was used as a positive control (top row). The loading control visualized by ethylene blue staining of total amounts of input DNA was shown in the bottom panel. Please click here to view a larger version of this figure.
Discussion
Here we have illustrated the use of an engineered split-TET2 enzyme to achieve temporal control of DNA hydroxymethylation. Following the discovery of TET family of 5-methycytosine dioxygenase, many studies have been performed to decipher the biological functions of TET proteins and their major catalytic product 5hmC1,2,3,4,5,6,7,8,9,10,11. However, temporal and precise control over DNA modifications in the genome continues to be a demanding challenge for the field. Our CiDER system is one of the few systems utilizing an engineered DNA modifying enzyme to fill this technical gap. The protocol described herein is mostly tailored for model cellular systems, such as the human embryonic kidney (HEK) 293T and the HeLa cell lines. Rapamycin concentrations and treatment time may need to be adjusted to achieve optimal performance in different cell types. To find out the best 5hmC induction time point, a time course experiment, as we described in section 1.3.3, is strongly recommended. Immunofluorescence staining of 5hmC in transfected cells can be used as the most convenient readout. For a more quantitative analysis, a dot-blot assay is recommended but it may take more laborious procedures. For dot-blot assays, the amounts of input DNA must be optimized for different cell types. A careful titration experiment using 2-fold serial dilution with varying DNA loading amounts is strongly recommended.
The CiDER system can be broadly used to dissect the correlation between DNA hydroxymethylation and phenotypic changes in different biological systems. Listed below are exemplary downstream applications or questions that can be addressed with the CiDER system.
How will TET-mediated 5mC oxidation alter the DNA methylation landscape in various model cellular system? This can now be easily addressed by comparing the DNA methylome and hydroxymethylome before and after rapamycin treatment. How will DNA hydroxymethylation alter chromatin accessibility and histone modifications? A comparison of ATAC-seq and ChIP-Seq results in the same cell before and after administration of rapamycin will tell the answer. Since TET proteins play a suppressive role in certain types of cancer, it will be interesting to test how chemical inducible restoration of TET protein and 5hmC production will impinge on tumor growth. Given the role of TET/5hmC in stem cell differentiation, it remains to be determined how temporally controlled 5hmC production at a defined transitionary stage will impact on lineage specification of stem cells during development.
In its current configuration, the CiDER system can only be used to globally induce 5mc-to-5hmC conversion. Ideally, CiDER can be fused with a catalytically-inactive Cas9 or its orthologues, which will enable loci-specific targeting and inducible DNA methylation editing in the genome. Such precise chemical-inducible epigenome remodeling tool can be widely applied to unambiguously probe the epigenotype-phenotype relations in mammals without altering the genetic code.
Disclosures
The authors declare no competing financial interests.
Acknowledgments
We thank the financial supports from the National Institutes of Health grant (R01GM112003 to YZ), the Welch Foundation (BE-1913 to YZ), the American Cancer Society (RSG-16-215-01 TBE to YZ), the Cancer Prevention and Research Institute of Texas (RR140053 to YH, RP170660 to YZ), the American Heart Association (16IRG27250155 to YH), and the John S. Dunn Foundation Collaborative Research Award Program.
References
- Li E, Zhang Y. DNA methylation in mammals. Cold Spring Harbor Perspectv Biol. 2014;6(5):a019133. doi: 10.1101/cshperspect.a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruijt CG, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152(5):1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- Lio CW, et al. Tet2 and Tet3 cooperate with B-lineage transcription factors to regulate DNA modification and chromatin accessibility. eLife. 2016;5:e18290. doi: 10.7554/eLife.18290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellinger MW, et al. 5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription. Nat Struct Mol Biol. 2012;19(8):831–833. doi: 10.1038/nsmb.2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M, et al. 5-Formylcytosine Could Be a Semipermanent Base in Specific Genome Sites. Angew Chem Int Ed Engl. 2016;55(39):11797–11800. doi: 10.1002/anie.201605994. [DOI] [PubMed] [Google Scholar]
- Ko M, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468(7325):839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Rao A. Connections between TET proteins and aberrant DNA modification in cancer. Trends Genet. 2014;30(10):464–474. doi: 10.1016/j.tig.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Zhao BS, He C. TET family proteins: oxidation activity, interacting molecules, and functions in diseases. Chem Rev. 2015;115(6):2225–2239. doi: 10.1021/cr500470n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, et al. Crystal structure of TET2-DNA complex: insight into TET-mediated 5mC oxidation. Cell. 2013;155(7):1545–1555. doi: 10.1016/j.cell.2013.11.020. [DOI] [PubMed] [Google Scholar]
- Hashimoto H, et al. Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA. Nature. 2014;506(7488):391–395. doi: 10.1038/nature12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszynski LA, Liu CW, Wandless TJJ. Characterization of the FKBP.rapamycin.FRB ternary complex. J Am Chem Soc. 2005;127(13):4715–4721. doi: 10.1021/ja043277y. [DOI] [PubMed] [Google Scholar]
- Clackson T, et al. Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity. Proc Natl Acad Sci U S A. 1998;95(18):10437–10442. doi: 10.1073/pnas.95.18.10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahimi A, et al. Highly efficient multicistronic lentiviral vectors with peptide 2A sequences. Hum Gene Ther. 2009;20(8):845–860. doi: 10.1089/hum.2008.188. [DOI] [PubMed] [Google Scholar]
- Kim JH, et al. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One. 2011;6(4):e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, et al. Engineered Split-TET2 Enzyme for Inducible Epigenetic Remodeling. J Am Chem Soc. 2017;139(13):4659–4662. doi: 10.1021/jacs.7b01459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Pastor WA, Zepeda-Martínez JA, Rao A. The anti-CMS technique for genome-wide mapping of 5-hydroxymethylcytosine. Nat Protoc. 2012;7(10):1897–1908. doi: 10.1038/nprot.2012.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol. 2015;109(21.29):1–9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]


