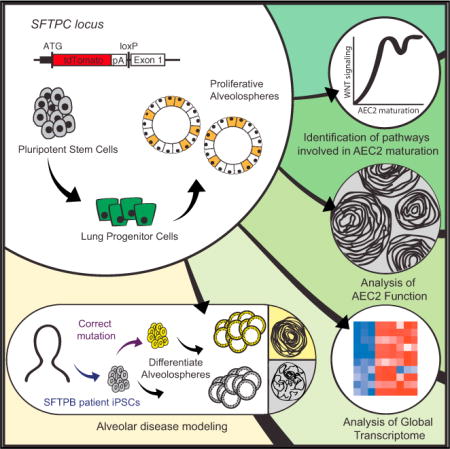
SUMMARY
Lung alveoli, which are unique to air-breathing organisms, have been challenging to generate from pluripotent stem cells (PSCs) in part because there are limited model systems available to provide the necessary developmental roadmaps for in vitro differentiation. Here we report the generation of alveolar epithelial type 2 cells (AEC2s), the facultative progenitors of lung alveoli, from human PSCs. Using multicolored fluorescent reporter lines, we track and purify human SFTPC+ alveolar progenitors as they emerge from endodermal precursors in response to stimulation of Wnt and FGF signaling. Purified PSC-derived SFTPC+ cells form monolayered epithelial “alveolospheres” in 3D cultures without the need for mesenchymal support, exhibit self-renewal capacity, and display additional AEC2 functional capacities. Footprint-free CRISPR-based gene correction of PSCs derived from patients carrying a homozygous surfactant mutation (SFTPB121ins2) restores surfactant processing in AEC2s. Thus, PSC-derived AEC2s provide a platform for disease modeling and future functional regeneration of the distal lung.
Graphical Abstract
INTRODUCTION
Pulmonary alveolar epithelial type 2 cell (AEC2) dysfunction has been implicated as a primary cause of pathogenesis in many poorly understood lung diseases. In particular, studies have shown that mutations affecting genes highly expressed in AEC2s, such as SFTPC, SFTPB, and ABCA3, result in neonatal respiratory distress or early-onset interstitial lung disease (Whitsett et al., 2015). Studying AEC2s from patients with these mutations might provide insight into the mechanisms by which early AEC2 dysfunction can lead to a wide variety of lung diseases.
Despite the broadly acknowledged need for human AEC2s in primary cell culture, a pure source of expandable AEC2s has not been previously achieved. AEC2s proliferate poorly ex vivo and transdifferentiate into type I AECs (AEC1s) when isolated from human lungs (Foster et al., 2007; Borok et al., 1998). Methods that do show maintenance of the AEC2 phenotype in culture require addition of mesenchymal feeders (Barkauskas et al., 2013). Because AEC2s are also relatively inaccessible for study in the developing human embryo, it is difficult to correlate findings in mice with human lung development. These obstacles have limited research of alveolar development and disease and have prevented the engineering of approaches to correct the genetic lesions that cause AEC2-initiated lung diseases.
Using induced pluripotent stem cells (iPSCs) and directed differentiation to generate AEC2s de novo would provide opportunities to study normal human AEC2 development and to understand the pathogenesis of alveolar diseases. Several groups have reported differentiation protocols that result in lung progenitors expressing the transcriptional regulator NKX2-1 as well as more differentiated populations containing cells expressing alveolar marker genes such as SFTPC (Huang et al., 2014; Gotoh et al., 2014; Dye et al., 2015; Hawkins et al., 2017). Next logical steps for the field include comparison of iPSC-derived putative AEC2s (iAEC2s) to primary controls, assessment of their maturation relative to the developing human lung, and evaluation of their ability to model human alveolar disease in vitro.
AEC2s have several critical roles in the distal lung. First, they are the facultative progenitors of the alveolus (Williams and Mason, 1977; Barkauskas et al., 2013; Desai et al., 2014), responding to lung parenchymal injury in mice by self-renewing or differentiating into AEC1s. AEC2s also synthesize surfactant, modulating alveolar surface tension (Kikkawa et al., 1975), and are able to respond to innate immune stimuli, protecting against infection (Williams and Mason, 1977). Several surfactant proteins are expressed in AEC2s, but only one, surfactant protein C (SFTPC), is reported to be highly specific to AEC2s (Kalina et al., 1992). Even then, although SFTPC may be specific to the AEC2s in adults, it is expressed as early as weeks 12–15 in human development (Khoor et al., 1994) and embryonic day 10.5 (E10.5) in mouse development in the distal lung bud (Wert et al., 1993). Mature AEC2s are characterized not only by expression of SFTPC but also by the ability to assemble functional lamellar bodies (LBs; Williams and Mason, 1977), the organelle in which surfactant is processed, a benchmark that has not yet been evaluated in iPSC-derived lung epithelial cells.
In this study, we employ human PSC lines with fluorescent reporters (GFP and/or tdTomato) targeted to the endogenous NKX2-1 and SFTPC loci, respectively, to isolate putative SFTPC+ alveolar cells for transcriptomic analysis compared with primary controls. We find that differentiating NKX2-1+ lung epithelial progenitor cells without mesenchymal co-culture can generate alveolospheres containing SFTPC+ cells. These alveolospheres display canonical AEC2 functional capacities, including innate immune responsiveness and the production of lamellar bodies able to package surfactant. Guided by time series global transcriptomic profiling of PSC derivatives, we find that AEC2 maturation involves modulation of Wnt signaling activity and that the highest differentially expressed transcripts in iPSC-derived AEC2s encode genes associated with surfactant biogenesis. Finally, we correct the mutation in an iPSC line from an infant homozygous for an SFTPB mutation known to cause lethal neonatal respiratory distress syndrome, thus restoring surfactant processing. This human model system is likely to facilitate disease modeling, developmental studies, drug screening, and future regenerative gene or cell therapies for a variety of diseases affecting lung alveoli.
RESULTS
SFTPC Reporter PSC Lines Allow Visualization of Distal Lung Differentiation and Isolation of Putative iAEC2s
During mouse development, AEC2s derive from SFTPC+ distal lung bud progenitors, which, in turn, arise from less differentiated NKX2-1+ SFTPC− foregut endoderm-derived lung epithelial precursors. To observe this putative sequence of AEC2 development in human cells in real time, we first used gene editing to target fluorochrome reporter constructs (GFP and tdTomato) to the endogenous NKX2-1 and SFTPC loci, respectively, of human PSCs representing multiple individual genetic backgrounds (BU3 NKX2-1GFP;SFTPCtdTomato, C17 NKX2-1GFP; SFTPCtdTomato, and RUES2 SFTPCtdTomato; Figure 1A; Figures S1A and S1B). Using a published lung-directed differentiation protocol (Figure S1C) established by Huang et al. (2014), we observed sequential in vitro differentiation of dual-targeted iPSC lines (C17 and BU3) into uncolored foregut endoderm followed by NKX2-1GFP+/SFTPCtdTomato− putative primordial lung progenitors and then NKX2-1GFP+/SFTPCtdTomato+ cells (Figure S1C; Movie S1). In keeping with in vivo observations (Minoo et al., 1999; Boggaram, 2009), all PSC-derived SFTPCtdTomato+ cells co-expressed the NKX2-1GFP reporter (Figure S1D). Flow cytometry sorting of NKX2-1GFP+/SFTPCtdTomato+ cells enriched for expression of both NKX2-1 and SFTPC transcripts (Figure S1D); however, both the efficiency of differentiation and SFTPC expression levels within the SFTPCtdTomato+ population, compared with primary fetal alveolar epithelial control cells, were initially low (Figures S1D and S2).
Figure 1. NKX2-1GFP and SFTPCtdTomato Reporters Allow Visualization of Distal Lung Differentiation and Isolation of Putative iAEC2s.
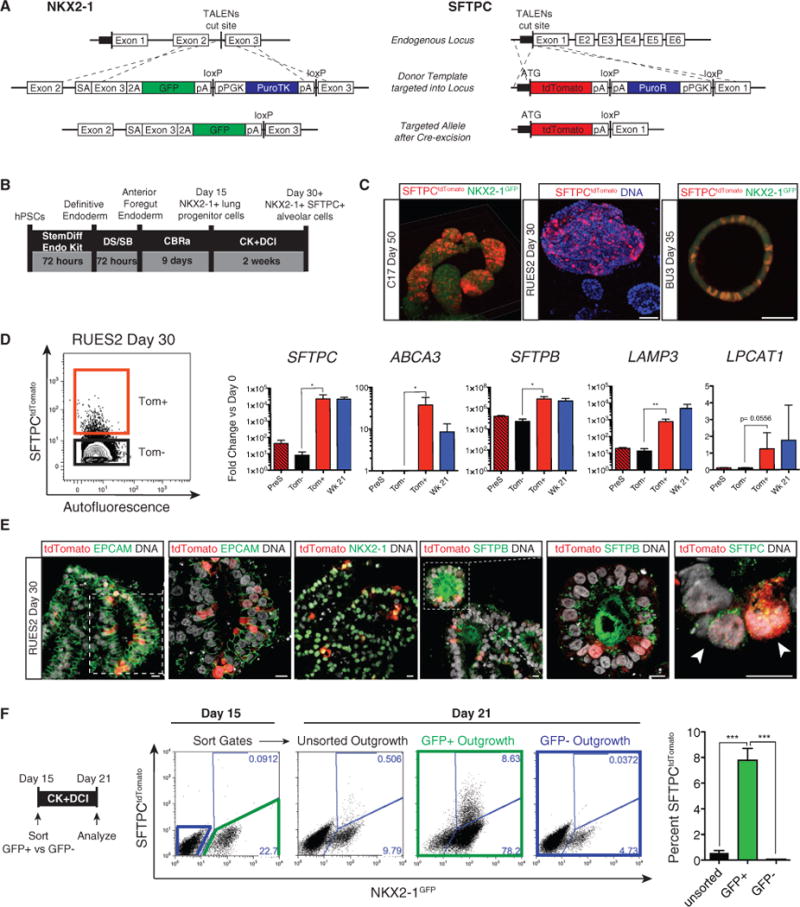
(A) Transcription activator-like effector nucleases (TALEN) targeting strategy and edited NKX2-1GFP and SFTPCtdTomato loci post Cre-mediated antibiotic cassette excision.
(B) Differentiation protocol from PSC to putative iAEC2s used hereafter.
(C) Representative images of “alveolospheres” late in differentiation showing expression of endogenous NKX2-1GFP and SFTPCtdTomato (C17 and RUES2 show 3D whole-mount confocal fluorescence microscopy reconstructions; BU3 shows live imaging on an inverted microscope). Scale bars, 100 μm.
(D) Representative flow cytometry of SFTPCtdTomato expression in day 30 RUES2. qRT-PCR of pre-sorted, sorted tdTomato+ (Tom+), and tdTomato− (Tom−) samples compared with primary week 21 human fetal distal lung control. Bars represent mean fold change (2−ΔΔCt) ± SD in 3 biological replicates separated on day 0; *p % 0.05, **p % 0.01 by unpaired, one-tailed Student’s t test. (E) Representative immunofluorescence microscopy with antibodies against tdTomato (red), EPCAM, NKX2-1, proSFTPB, and proSFTPC (green) in day 30 RUES2 alveolospheres. Nuclei were stained with Hoechst (gray). Scale bars, 10 μm.
(F) Day 21 representative flow cytometry analysis of C17 day 15 unsorted, sorted NKX2-1GFP+, or sorted NKX2-1GFP− outgrowth. Bars represent mean ± SD in 3 biological replicates separated on day 0; ***p % 0.001 by unpaired, two-tailed Student’s t test.
See also Figures S1 and S2.
To optimize SFTPC differentiation efficiency, we sequentially withdrew one factor at a time from each stage of differentiation (NKX2-1+ progenitor induction stage versus SFTPC+ induction stage), observing that only three factors (CHIR99021 [CHIR], BMP4, and retinoic acid [RA]; hereafter called CBRa) were sufficient for the specification of NKX2-1+ progenitors, as published previously (Gotoh et al., 2014; Rankin et al., 2016). For subsequent SFTPC induction within this NKX2-1+ population, only two exogenous factors (CHIR and keratinocyte growth factor [KGF]; hereafter called CK) were sufficient in the presence of known lung maturation additives (dexamethasone, cyclic AMP, and 3-isobutyl-1-methyxanthine [IBMX]; hereafter called DCI; Gonzales et al., 2002; Figures S2A and S2B). We found no further SFTPC induction efficiency with the addition of other reported distalizing factors, such as BMP4, epidermal growth factor (EGF), and fibroblast growth factor 10 (FGF10), or with additional inhibitors of bone morphogenetic protein (BMP), transforming growth factor β (TGF-β), or Notch signaling (Figure S2B and data not shown). Testing our optimized differentiation protocol (Figure 1B) in the RUES2, BU3, and C17 lines, we observed efficient induction of endodermal NKX2-1 with CBRa by day 15 (Figures S2C and S2E). After subsequent exposure to CK+DCI, we noted rapid emergence of SFTPCtdTomato+ cells within 3 to 7 days (Figures 1B and 1C). For all three PSC lines, tdTomato+ cells could be visualized scattered throughout epithelial spheres in 3D cultures (Figure 1C), and the yield by day 30 ranged from 0.2–4.4 SFTPCtdTomato+ cells per day 15 input NKX2-1+ progenitor cell (Figure S2E). By day 30 of differentiation, these tdTomato+ cells were highly enriched in transcripts encoding surfactant proteins as well as transcripts associated with lamellar body biogenesis, SFTPC, SFTPB, ABCA3, LAMP3, and LPCAT1 (Figure 1D). Notably SFTPC, SFTPB, and ABCA3 were expressed in sorted tdTomato+ cells at levels higher than primary fetal lung controls (week 21 gestation fetal alveolar epithelial cells, ~85%–90% SFPTC+). At the protein level, epithelial spheres that included tdTomato+ cells also expressed NKX2-1 nuclear protein, membranous epithelial cell adhesion molecule (EPCAM), punctate cytoplasmic proSFTPC, and intracellular as well as secreted SFTPB protein (Figure 1E).
Human Putative Alveolar Cells Derive from an NKX2-1+ Primordial Progenitor
Next we asked whether the early NKX2-1+ population represented the entire pool of progenitors from which SFTPC+ alveolar cells might arise. We differentiated day 15 unsorted cells and sorted NKX2-1GFP+ versus NKX2-1GFP− cells in parallel (Figure 1F). For both the C17 and BU3 lines, we found that NKX2-1GFP+ sorted cells gave rise to SFTPC+ cells, whereas GFP– sorted cells did not. The mixed (unsorted) population resulted in a lower efficiency of alveolar differentiation, suggesting that the NKX2-1GFP− population does not contain a lineage that promotes alveolar differentiation, such as lung-specific mesenchyme (Figure 1F; Figure S2D). To extend this approach to PSCs that do not contain an NKX2-1 knockin reporter, we next developed a strategy to identify AEC2-competent NKX2-1+ progenitors in the RUES2 cell line, which has a single SFTPCtdTomato reporter. We have previously shown that CD47hi/CD26lo day 15 cells are highly enriched in NKX2-1+ lung progenitors (Hawkins et al., 2017). In keeping with these findings, we observed that sorting for RUES2 CD47hi/CD26lo cells on day 15 identified the entirety of the SFTPC-competent population, enriching the SFTPC+ yield in the day 21 population approximately 4-fold over unsorted cells (Figure S2C).
Having demonstrated that SFTPC+ cells derive via an NKX2-1+ progenitor intermediate, we next sought to test whether Wnt activation was necessary and acting directly on these progenitors. Hence, we purified NKX2-1GFP+ cells on day 15 and repeated our differentiation protocol in the presence or absence of CHIR (Figure 2A; Figure S2A). By day 21, induction of the SFTPCtdTomato reporter was evident in 7.17% ± 0.89% of cells in the presence of CHIR, whereas only rare cells (0.64% ± 0.34%) expressed the tdTomato reporter in the absence of CHIR, even when the cultures were maintained up to day 35. Similar findings were observed in C17, BU3, and RUES2 cell lines (Figure 2A; Figure S2A; data not shown), and these results are consistent with our prior observation (McCauley et al., 2017) that CHIR distalizes NKX2-1+ human lung progenitors while suppressing proximal airway fates. Although KGF was dispensable for initial induction of the tdTomato reporter (Figures S2A and S2B), we found that cells proliferated poorly in the absence of KGF or FGF10 (data not shown). In addition, KGF, when combined with CHIR, was more consistent in promoting the outgrowth of tdTomato+ cells by day 21 compared with FGF10+CHIR (Figure S2B), as predicted by prior mouse lung explant culture experiments (Liu and Hogan 2002).
Figure 2. Putative iAEC2s Proliferate and Differentiate in Long-Term Culture.
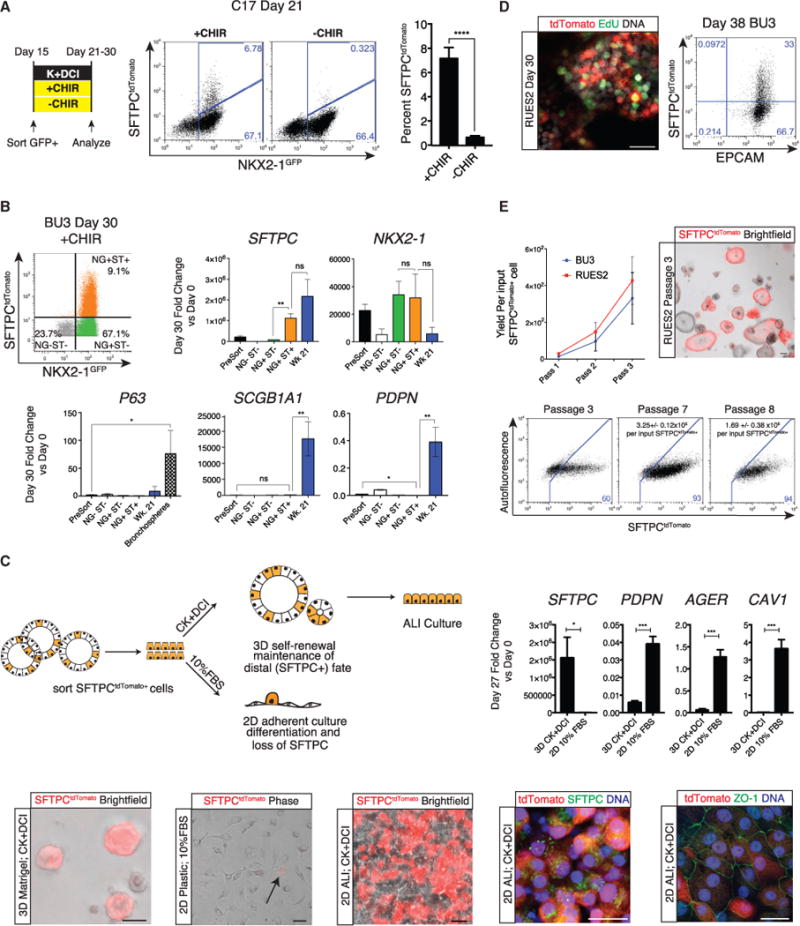
(A) Day 21 representative flow cytometry analysis of C17 day 15 NKX2-1GFP+ outgrowth cultured from days 15–21 with or without addition of CHIR to K+DCI base medium. Bars represent mean ± SD, n = 6 biological replicates separated on day 0; ****p % 0.0001 by unpaired, two-tailed Student’s t test.
(B) Day 30 representative flow cytometry analysis of BU3 iPSCs sorted on day 15 based on NKX2-1GFP+ expression, replated for outgrowth from days 15–30, and then sorted for analysis from each gate: NKX2-1GFP− SFTPCtdTomato− (NG−ST−), NKX2-1GFP+ SFTPCtdTomato− (NG+ST−), and NKX2-1GFP+ SFTPCtdTomato+ (NG+ST+). Shown is qRT-PCR of these populations in addition to unsorted (Pre-Sort) day 30 cells, week 21 primary human fetal distal lung controls, and BU3 iPSC-derived proximal spheres generated as described in McCauley et al. (2017) (Bronchospheres). Bars represent mean fold change (2−ΔΔCt) ± SD, n = 3 biological replicates separated on day 15.
(C) Schematic showing SFTPCtdTomato+ cells from day 22 alveolospheres sorted either into 3D culture in CK+DCI medium or 2D culture (tissue culture-treated plastic) in 10% fetal bovine serum (FBS) medium. Shown is representative phase/bright-field and tdTomato fluorescence microscopy of live cells after 7 days in 2D plastic, 3D Matrigel, or after 2D ALI culture conditions. Scale bars, 50 μm; the arrow indicates tdTomato expression in 2D cultured cells. Also shown is representative immunofluorescence microscopy for tdTomato (red), ZO-1 or proSFTPC (green), and DNA (Hoechst, blue) in RUES2 alveolospheres cultured in ALI. Scale bars, 25 μm. We also show qRT-PCR of day 22 sorted SFTPCtdTomato+ cells cultured under 3D versus 2D conditions for 5 days, with bars representing mean fold change (2−ΔΔCt) ± SD, n = 3 biological replicates; *p % 0.05, ***p % 0.001 by unpaired, two-tailed Student’s t test.
(D) Representative confocal immunofluorescence microscopy of EdU (green), anti-tdTomato (red), and DNA (Hoechst, blue) after 24-hr 5-ethynyl-2′-deoxyuridine (EdU) incubation. Scale bar, 25 μm. Representative flow cytometry analysis of day 38 BU3 iPSC-derived alveolospheres shows co-expression of EPCAM and tdTomato proteins.
(E) Graph showing yield in cell number per input RUES2 or BU3 SFTPCtdTomato+ sorted cell plated on day 22 or 30, respectively. Shown is representative live-cell imaging on passage 3 of SFTPCtdTomato+ sorted RUES2 alveolospheres. Scale bar, 100 μm. Also shown is representative flow cytometry of passage 3, 7, and 8 RUES2-derived alveolospheres from the outgrowth of SFTPCtdTomato+ cells with SFTPCtdTomato+ cell yields indicated (mean ± SD) per input sorted tdTomato+ cell.
See also Figure S3.
We next sought to determine whether other lung lineages were co-developing in these cultures, focusing in particular on the frequent NKX2-1+/SFTPC− cells in our differentiations. Hence, we sorted each population for profiling on day 30 using each combination of the NKX2-1GFP and SFTPCtdTomato dual reporters present in our BU3 iPSC line (Figure 2B). In the presence of CHIR and KGF, we observed significant enrichment of SFTPC in the NKX2-1GFP+/SFTPCtdTomato+ population (NG+ST+; Figure 2B), whereas NKX2-1 was expressed equally in NKX2-1GFP+/SFTPCtdTomato+ cells compared with NKX2-1GFP+/SFTPCtdTomato− (NG+ST−) cells, as expected. In marked contrast, we observed no expression of the airway club cell marker SCGB1A1 in any population and enriched expression of the basal cell marker P63 in only the GFP− non-lung population (Figure 2B), findings in keeping with our observation that there is inefficient proximal airway patterning of iPSC-derived NKX2-1+ progenitors in the presence of high levels of CHIR-stimulated canonical Wnt signaling (McCauley et al., 2017).
In our distalizing medium (CK+DCI; Figures 1B and 2B), we found little evidence of AEC1 differentiation in any population on day 30, as evidenced by low to undetectable levels of podo-planin (PDPN), advanced glycosylation end-product specific receptor (AGER), and AQP5 (Figure 2B; Figure S3A), suggesting that, as has been published for primary adult human AEC2s maintained in 3D sphere cultures (Barkauskas et al., 2013), PSC-derived SFTPC+ cells do not detectably transdifferentiate into AEC1 in 3D cultures. Although NKX2-1+/SFTPC− cells in the day 30 population were not proximal lung cells or AEC1s, we found that they did express ABCA3 and LPCAT1 at high levels, similar to primary fetal AECs (Figures S3A), raising the possibility that the NKX2-1+/SFTPC− population may represent a less mature distal lung population compared with tdTomato+ cells, which express significantly higher levels of SFTPC.
Putative iAEC2s Display Self-Renewal and Differentiation Capacities
Given the absence of AEC1 differentiation under distal 3D culture conditions, we tested the capacity of PSC-derived SFTPCtdTomato sorted cells to differentiate when transferred to conditions that have been published as generating AEC1s from primary AEC2s, such as 2D culture in serum-containing medium without CK+DCI (Dobbs et al., 1988; Borok et al., 1998). In contrast to SFTPCtdTomato+ cells maintained under 3D distal conditions, we observed that sorted PSC-derived SFTPCtdTomato+ cells replated under these AEC1 culture conditions for 5–7 days rapidly flattened into senescent squamous-like cells, significantly downregulated SFTPC, lost visible tdTomato reporter gene expression, and significantly upregulated 3 AEC1 marker genes—PDPN, AGER, and CAV1 (Figure 2C). Cells plated in 2D air-liquid interface (ALI) cultures in the presence of sustained CK+DCI slowly reached confluence and maintained SFTPCtdTomato+ expression in most cells while forming an epithelial barrier with ZO-1-positive tight junctions (Figure 2C).
Proliferation is a well-characterized property of both fetal and adult AEC2s (Barkauskas et al., 2013; Desai et al., 2014), although long-term in vitro AEC2 proliferation has been shown to require mesenchymal feeders. We found that, in the absence of mesenchymal cells, iPSC-derived SFTPCtdTomato+ cells within alveolospheres could be labeled with EdU (Figure 2D), indicating their ability to proliferate. In addition, tdTomato+ cells sorted to purity on day 22 or day 30 continued to increase in number during subsequent serial passaging and to re-form alveolospheres after each passage (Figure 2E). After passaging, the resulting alveolospheres were initially composed of both tdTomato+ and tdTomato− cells, providing direct evidence of a lineage relationship between the two populations. With further passaging, however, the descendants of tdTomato+ sorted cells maintained stable expression in the vast majority (>90%; Figure 2E) of outgrowth cells, resulting in alveolospheres that could be serially passaged at least 8 times, yielding 1.7 ± 0.4 × 106 RUES2-derived tdTomato+ cells on passage 8 per input day 22-derived SFTPCtdTomato+ cell (Figure 2E). Similar results were obtained for BU3 sorted tdTomato+ cells, and, in each case, SFTPCtdTomato reporter gene expression as well as mRNA expression of alveolar transcripts at or above control levels were maintained following multiple passages and freeze-thaw cycles (Figure 2E; Figure S3B; data not shown). When we analyzed colony-forming efficiency in primary adult AEC2s, we found that, in either proprietary small airway growth medium (SAGM) or CK+DCI medium, they did not establish colonies or spheres without the addition of mesenchymal (MRC5) feeders. In contrast, in CK+DCI medium, the addition of mesenchymal feeders did not significantly change PSC-derived alveolosphere colony-forming efficiency (Figure S3C). Taken together, these results indicate a significant self-renewal potential of PSC-derived SFTPC+ cells independent of feeder cells.
Putative iAEC2s Express Lamellar Bodies that Synthesize and Secrete Surfactant
The in vivo ultrastructural and biochemical characteristics of developing fetal and adult alveolar epithelia provide an extensive roadmap with which to compare putative iAEC2s (Williams and Mason 1977). Although alveolar progenitors express SFTPC or other surfactant markers in vivo as early as weeks 12–15 of human gestation (Khoor et al., 1994), they do not express functional lamellar bodies, the classic marker of AEC2 maturity, until after week 24 (Figure 3A). Hence, we assessed whether putative iAEC2s express these highly specialized organelles. Semi-thin plastic sections of PSC-derived alveolo-spheres revealed small inclusions clustering in the apical (luminal) side of the alveolosphere monolayer in a subset of cells (Figure 3B), and ultrastructural analysis by transmission electron microscopy (TEM) showed these to be lamellar body-like inclusions (LBLs; Figure 3C; Figure S4). As has been reported for adult AEC2 in vivo, LBLs in alveolospheres were approximately 1 μm in diameter, with some LBLs expressing central dense cores. Even after multiple passages, day 70 alveolospheres contained LBLs (Figure S4A), further supporting the self-renewing capacity of putative iAEC2s. Within the cytoplasm of most cells, regardless of the presence of LBLs, we observed large areas reminiscent of glycogen-rich regions (Figures 3B and 3C; Figure S4) known to be expressed in fetal AEC2s just prior to birth (Ridsdale and Post, 2004). Mature SFTPB and SFTPC protein forms, detected by immunogold labeling, appeared to localize to LBLs and their precursor organelles, multivesicular bodies (MVBs) (Figure 3E), findings consistent with those reported for mature AEC2s in vivo (Korimilli et al., 2000; Brasch et al., 2004).
Figure 3. Putative iAEC2s Express Lamellar Bodies that Contain Surfactant.
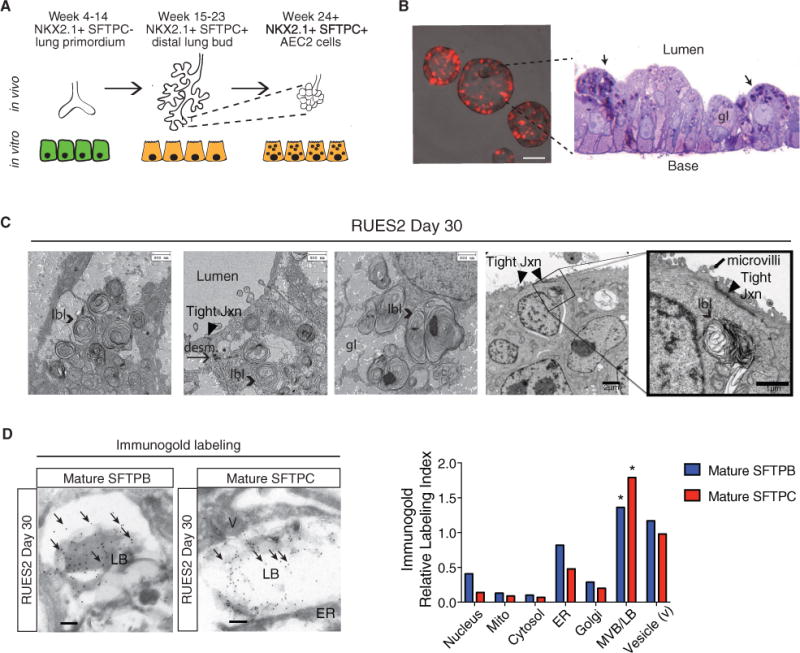
(A) Temporal expression of NKX2-1 and SFTPC during human fetal lung development.
(B) Representative image of live day 30 RUES2-derived alveolospheres (scale bar, 100 μm) and plastic section stained with toluidine blue. Arrows indicate cells with putative lamellar body-like inclusions. gl, glycogen lake.
(C) TEM of day 30 RUES2-derived alveolospheres. LBL, lamellar body-like inclusion; tight jxn, tight junction; desm, desmosome.
(D) Immunogold labeling of mature SFTPB and SFTPC in day 30 RUES2 alveolospheres. Scale bars, 0.2 μm. The significance of the relative labeling index in each cellular compartment (χ2 statistics and contingency table analyses) is indicated. *p < 0.0001.
See also Figure S4.
To further test the functionality of putative iAEC2s, we asked whether the cells were able to process proSFTPB protein to its fully mature 8-kD form (Figure 4A; Guttentag et al., 1998) because the last cleavage step in proSFTPB processing occurs in the functional MVBs and lamellar bodies of post-week 24 gestation human AEC2s (Brasch et al., 2004). We analyzed a time series of protein extracts prepared from RUES2 cells during distal lung differentiation. Western blots immunostained with antibodies able to discern fully processed mature 8-kD SFTPB (PT3 antibody) versus proSFTPB precursor forms (NFLANK antibody; Figures 4A and 4B) revealed increasing production of both the precursor and mature forms of SFTPB beginning on day 22 of differentiation and increasing over time through day 36 (Figure 4B). Compared with primary fetal human AEC2 controls, PSC-derived cells similarly expressed 42-kD, 25-kD, and 10-kD precursor forms as well as mature 8-kD SFTPB protein. Moreover, as with primary controls, SFTPB appeared to be efficiently processed in alveolospheres because the predominant SFTPB form detected was the mature form (Figure 4B). The larger precursor SFTPB forms were only visible when the NFLANK-specific antibody was used to detect the proSFTPB region that is present prior to final cleavage into the mature form.
Figure 4. Putative iAEC2 Lamellar Bodies Function to Synthesize and Secrete Surfactant.
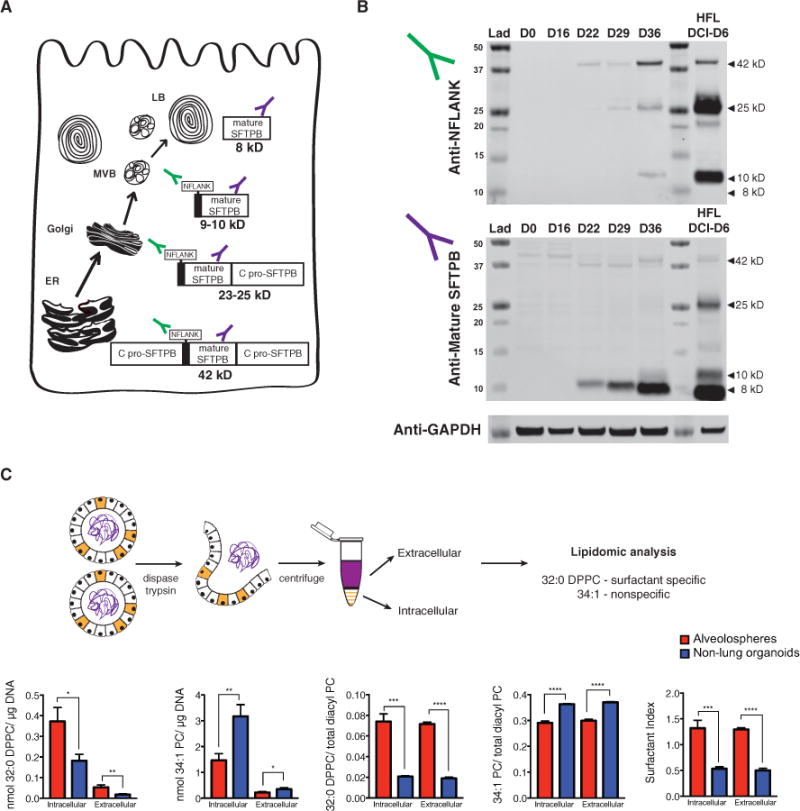
(A) Cellular compartments in which proSFTPB processing into mature SFTPB occurs. MVB, multivesicular body.
(B) Western blot of a time course of alveolosphere differentiation (days 0, 16, 22, 29, and 36) and 6-day DCI-cultured primary week 21 human fetal distal lung (HFL DCI-D6) as a positive control. Top: use of an anti-NFLANK antibody that binds to the N-pro region of pro-SFTPB intermediates. Bottom: use of an anti-mature SFTPB antibody (PT3) that binds all SFTPB forms, including the 8-kD mature form.
(C) Dissociation protocol for isolation of intracellular and extracellular components of alveolospheres. Both absolute (nanomoles of lipid/micrograms of DNA in cell samples) and relative (nanomoles of lipid/nanomoles of total diacyl phosphatidylcholine) are shown for both surfactant-specific 32:0 dipalmitoyl phos-phatidylcholine (DPPC) and nonspecific 34:1 phosphatidylcholine (PC). The surfactant index was calculated as the sum of the surfactant-specific 30 and 32 PCs divided by the sum of non-surfactant-specific 36 PCs. Bars represent mean ± SD for each analysis, n = 3 biological replicates separated on day 15; *p % 0.05, **p % 0.01, ***p % 0.001, ****p % 0.0001 by unpaired, two-tailed Student’s t test.
To determine whether putative iAEC2s synthesize and secrete surfactant-specific 32:0 dipalmitoyl phosphatidylcholine (DPPC), we performed lipidomic analysis on both the intracellular and extracellularmaterial from iPSC-derived alveolospheres.Because these spheres appear to be polarized with the apical surface pointing inward, we first dissociated them to free the secreted products and then performed the analysis on the supernatant fractions and the cells separately (Figure 4C). Both the relative and absolute amounts of DPPC were significantly higher in the intracellular and extracellular material from the alveolospheres generated from PSCs via NKX2-1+ progenitors (isolated by CD47hi/CD26lo sorting) compared with control spheres generated from endodermal progenitors depleted of NKX2-1+ cells. Taken together, these results show that PSC-derived alveolo-spheres contain cells with functional lamellar bodies that synthesize and secrete surfactant, suggesting that they contain phenotypically mature AEC2-like cells, hereafter referred to as iAEC2s.
Global Transcriptomic Profiling of PSC-Derived Lung Progenitors and Their Differentiated iAEC2 Progeny
We next sought to define the global transcriptomes of PSC-derived lung progenitors and their SFTPC+ and SFTPC− progeny by performing a time series analysis using RNA sequencing (RNA-seq). We analyzed 3 different time points in RUES2 differentiation: day 0 undifferentiated cells, day 15 lung progenitors highly enriched in NKX2-1+ cells by CD47hi/CD26lo sorting (hereafter called CD47+), and the outgrowth of these purified progenitors in 3D culture sorted again on day 35 based on SFTPCtdTomato+ (Tom+) and SFTPCtdTomato− (Tom−) gating (Figure 5A). For comparison with primary cells, we simultaneously sequenced RNA from purified primary fetal (21 week gestation) distal alveolar epithelial progenitors and sorted adult human AEC2s. To evaluate the effect of cell culture on primary fetal alveolar cells, parallel samples of the fetal cells were also exposed to 4 days of culture in DCI medium.
Figure 5. Global Transcriptomic Profiling of PSC-Derived Lung Progenitors and Their Differentiated iAEC2 Progeny.

(A) Time points in alveolosphere differentiation from which samples were taken for RNA-seq.
(B) Principal-component analysis (PCA) of gene expression variance across all samples based on 30,000 transcripts.
(C) Top: qRT-PCR of each sample, with mean fold change (2−ΔΔCt) ± SD, n = 3 biological replicates (RUES2 samples), cells isolated for RNA separately (Wk 21), cells cultured for 4 days separately (Wk 21 DCI), and cells from separate lungs (Adult AEC2). *p % 0.05, **p % 0.01 by unpaired, two-tailed Student’s t test. Bottom: Log2 expression of SFTPC across all samples, with the dotted line representing “noise” because these levels of expression are not consistently detected by qRT-PCR.
(D) Heatmap of row-normalized expression of selected lineage markers across PSC-derived and primary samples.
(E) Heatmap of the top 10 genes upregulated in day 35 Tom+ cells versus day 15 progenitors (ranked by fold change, FDR % 0.05). Known AEC2 genes are shown in bold.
(F) Heatmap of the top 50 genes differentially expressed in day 35 Tom+ cells versus day 15 cells (ranked by FDR, FDR % 0.01). Known AEC2 genes are shown in bold.
(G) Heatmap of supervised hierarchical clustering based on the top 300 genes differentially expressed in day 35 Tom+ versus day 15 cells (ranked by absolute fold change, FDR % 0.01).
(H) Left: experimental design. Center: western blot for IκB-alpha, phospho-Stat3 (Tyr705), and pan-actin. Right: qRT-PCR of each sample, with mean fold change (2−ΔΔCt) ± SD, n = 3 biological replicates separated on day 35; *p % 0.05, **p % 0.01 by unpaired, two-tailed Student’s t test.
See also Figure S5.
By principal-component analysis (PCA) of 30,000 transcripts in each sample, we found that PSC-derived cells after 35 days of differentiation clustered closer to primary cells on the PC1 axis (Figure 5B). Fetal primary cells clustered closer to PSC-derived cells after cell culture in DCI medium, whereas sorted adult AEC2s clustered separately from all other samples. In keeping with this finding, profiling of both the canonical marker SFTPC (Figure 5C) as well as a selected set of 22 markers of AEC2s, AEC1s, and early lung endoderm (Figure 5D) showed that expression patterns most closely resembled primary AECs in the sorted PSC-derived cells on day 35. To identify genes differentially expressed in Tom+ cells with differentiation, we compared the sorted day 35 Tom+ cells with their day 15 precursors (Figures 5E–5G; Figure S5C). Notably, we found that the set of the 50 most differentially expressed transcripts in day 35 Tom+ cells was highly enriched in AEC2-specific marker genes (Figure 5F). Strikingly, 7 of the top 10 most differentially upregulated genes in Tom+ cells encoded proteins related to surfactants or lamellar body biogenesis, and these genes were expressed in PSC-derived Tom+ cells at levels similar to adult AEC2s (Figure 5E). Indeed, hierarchical clustering analysis of all samples based on the top 300 differentially expressed genes in the Tom+ population indicated that these cells clustered closest to adult AEC2s (Figure 5G). Consistent with our culture data suggesting a lineage relationship between Tom+ and Tom− cells, we found that these two NKX2-1+ progenitor derived populations displayed similar gene expression profiles by both PCA of 30,000 transcripts and hierarchical clustering analyses of the top 300 genes upregulated from day 15 to 35 in Tom+ cells (Figures 5B and 5G; Figures S5B and S5D). Although both day 35 populations expressed multiple AEC2 marker genes, the level of SFTPC expression was significantly upregulated in the Tom+ versus the Tom− population (Figure 5C; Figure S5D). Upregulation of SFTPC and other markers of AEC2 maturation (SFTPA1, SFTPA2, and napsin A [NAPSA]) in the Tom+ versus Tom− population was validated by qRT-PCR at this same day 30 time point in iPSC-derived cells from an independent genetic background (BU3; Figures S5D and S5F), further suggesting that the Tom+ population represented more mature iAEC2s. In addition to this increase in maturity, we found that, over the course of 3 passages, Tom+ cells gave rise to a significantly higher number of Tom+ cells per input cell than NKX2-1GFP+ Tom− cells (Figure S5G).
We next focused on potential gene expression differences between the various other samples. First, we looked at the tran-scriptomic differences between PSC-derived Tom+ cells and primary cells, expecting to see major differences in global gene expression between iAEC2s and primary adult AEC2s because of the effects of accelerated development of iAEC2s outside of the alveolar niche in submerged sterile cultures versus the effects of life-long maturation of adult AEC2s in an air-breathing, multilineage, non-sterile environment. Not surprisingly, gene set enrichment analysis (GSEA) of Tom+ cells versus adult AEC2s revealed that the gene sets differentially expressed in adult AEC2s involved upregulation of immune pathways and oxidant stress (Figures S5A and S5C).
Focusing next on the gene expression differences between day 15 and day 35 Tom+ cells, GSEA revealed the Janus kinase (JAK)/STAT3/interleukin-6 (IL-6) and tumor necrosis factor alpha (TNF-α)/nuclear factor κB (NF-κB) signaling pathways comprised the top 2 upregulated signaling pathways in the day 35 Tom+ population (Figure S5C). To determine whether PSC-derived day 35 cells are capable of functional signaling through these pathways, we stimulated PSC-derived alveolospheres with cytokines known to activate each pathway in pulmonary epithelial cells (Quinton et al., 2007, 2008): oncostatin M (OSM) for the JAK/STAT3 pathway and TNF-α/IL-1β for the TNF-α/NF-κB pathway. Exposure to OSM ligand resulted in significant and rapid activation of STAT3 signaling, indicated by increased phosphorylated STAT3 protein and downstream SOCS3 mRNA expression, whereas exposure to TNF-α and IL-1β activated NF-κB signaling, as evidenced by a rapid decrease in IκB protein and an increase in the expression of NF-κB-dependent cytokines, IL-8, and granulocyte-macrophage colony-stimulating factor (GM-CSF) (Figure 5H). Taken together, these results indicate that PSC-derived alveolospheres are capable of performing another important function of AEC2s: immune signaling in response to canonical ligands.
Last, we re-examined the few and subtle gene expression differences between PSC-derived day 35 Tom+ and Tom− cells (Figure 6A). Only 203 genes were differentially expressed between Tom+ and Tom− cells, and of these, only 30 were upregulated in the Tom+ population, including SFTPC (false discovery rate [FDR] < 0.05; Figure 6A). The 173 genes downregulated in Tom+ compared with Tom− cells included non-lung endodermal transcripts, implying that Tom+ sorting depleted the iAEC population of these alternative endodermal lineages. Because the global transcriptomic profiles of iPSC-derived Tom+ and Tom− cells were so similar when each population originated from sorted day 15 lung progenitors, we further considered the possibility that each population was largely composed of closely related distal alveolar epithelia.
Figure 6. Temporal Regulation of Wnt Signaling Promotes iAEC2 Maturation and Proliferation.
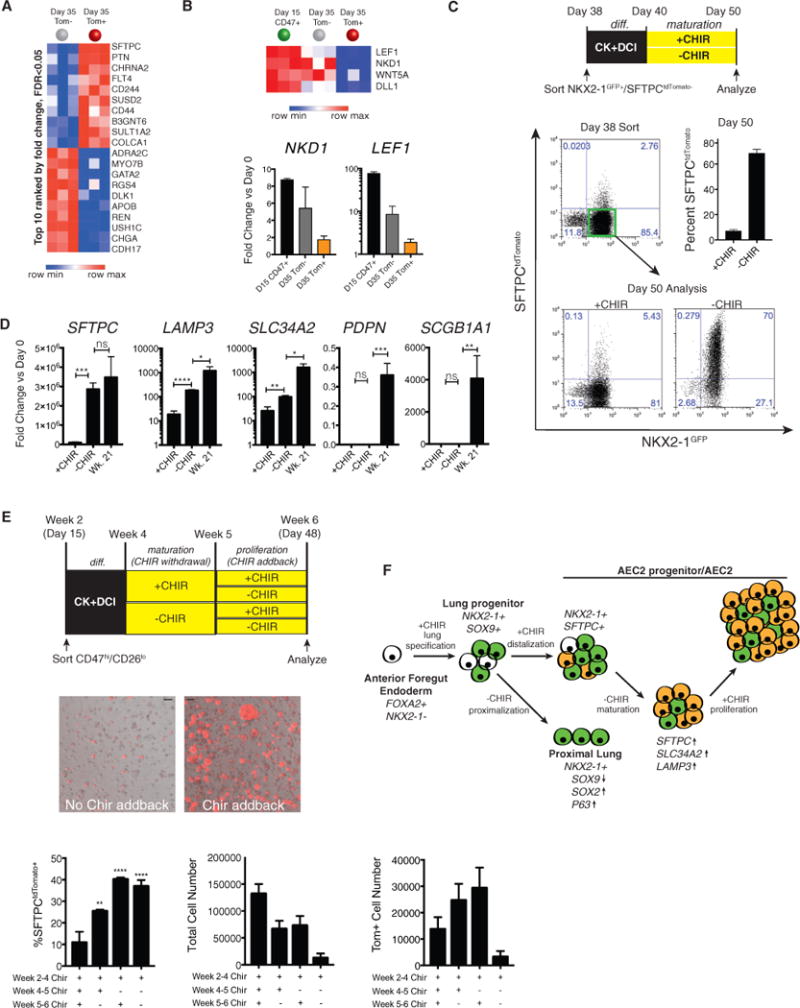
(A) Heatmap of the top 10 upregulated and downregulated genes in day 35 Tom+ versus Tom− populations (ranked by fold change, FDR % 0.05).
(B) Heatmap of selected differentially expressed genes downregulated in Tom+ versus Tom− populations (FDR % 0.05) from the MSigDB v5.1 Hallmark Wnt/β-Catenin Signaling database. Shown is qRT-PCR of day 15 and day 35 Tom− and Day 35 Tom+ samples, with fold change (2−ΔΔCt) ± SD, n = 3 biological replicates.
(C) Schematic showing the late CHIR withdrawal experiment. Shown is a representative flow cytometry analysis of day 38 sort gates and day 50 outgrowth of the BU3 NKX2-1GFP+/SFTPCtdTomato− population cultured with or without CHIR from days 40–50. Bars show mean ± SD, n = 3 biological replicates separated on day 38.
(D) qRT-PCR of day 50 ± CHIR populations and week 21 human fetal distal lung, with bars representing mean fold change (2−ΔΔCt) ± SD, n = 3 biological replicates from differentiations separated on day 38.
(E) Schematic of CHIR addback experiment and representative images of live RUES2 alveolospheres (bright-field/tdTomato overlay; CHIR weeks 4–5 (days 32–39), ± CHIR addback weeks 5–6 (days 39–48). Scale bar, 100 μm. Bar graphs show percent Tom+, total cell number, and total number of Tom+ cells in day 48 populations ± CHIR withdrawal and ± CHIR addback, with 30,000 cells plated on day 32. Bars represent mean ± SD of 3 differentiations separated on day 32.
(F) Schematic of putative effects of Wnt stimulation on lung epithelial differentiation. *p % 0.05, **p % 0.01, ***p % 0.001 by unpaired, two-tailed Student’s t test for all panels.
See also Figure S6.
Temporal Regulation of Wnt Activity Promotes iAEC2 Maturation
Based on the significantly higher SFTPC, SFTPA1, SFTPA2, and NAPSA expression in the otherwise similar Tom+ versus Tom− cells on day 35 (Figure S5F), we further explored the possibility that Tom+ cells might represent a more mature state of iAEC2s compared with Tom− cells. GSEA analysis to screen for developmental pathways that might distinguish the two populations revealed that Wnt signaling was the top differentially expressed developmental pathway (Figure S5B). Although both cell populations were cultured in the presence of CHIR, Wnt signaling was unexpectedly downregulated in Tom+ cells compared with Tom− cells, with decreased expression of LEF1 and NKD1 (validated in the RUES2 and BU3 cell lines by qRT-PCR; Figure 6B; Figure S6A), two Wnt targets most associated with canonical Wnt signaling levels in our PSC lung model system (McCauley et al., 2017). Hence, to test the hypothesis that decreased Wnt signaling regulates maturation of PSC-derived distal cells, we sorted the distalized NKX2-1GFP+/SFTPCtdTomato− population on day 38 for further 3D culture in the presence or absence of continued Wnt stimulation with CHIR (Figure 6C). We found that withdrawal of CHIR for 10 days (days 40–50) resulted in markedly increased efficiency of differentiation into SFTPC+ progeny (68.7% ± 2.8), with significant upregulation of SFTPC mRNA as well as other markers of AEC2 maturation—LAMP3 and SLC34A2 (Figures 6C and 6D).
Notably, following this period of CHIR withdrawal, we observed decreased proliferation and size of alveolospheres (data not shown), findings that may reflect the low-Wnt pre-al-veologenesis stage of alveolar development evident in mice (Frank et al., 2016). In the same study, an increase in Wnt signaling was shown to correspond to the increase in alveolar epithelial proliferation at the onset of alveologenesis (Frank et al., 2016). Consistent with these findings, when we repeated these experiments, adding back CHIR following a 1-week period of withdrawal (Figure 6E), we found that the population re-exposed to CHIR maintained expression of the SFTPCtdTomato reporter and increased proliferation, evident as increased resulting SFTPCtdTomato+ cell number (Figure 6E). Thus, alveolo-spheres (derived from sorted CD47hi/CD26lo on day 15; week 2) exposed to CK+DCI medium from weeks 2–6 with CHIR withdrawn from weeks 4–5 and added back from weeks 5–6 displayed at least 40% SFTPCtdTomato+ cells and expressed high levels of alveolar transcripts, SFTPC, SFTPB, and ABCA3, and additional AEC2 maturation transcripts, SFTPA1, SFTPA2, NAPSA, pepsinogen C (PGC), SLC34A2, and LAMP3, with only LAMP3 being expressed at a lower level than primary (week 21) human fetal lung alveolar cell controls (Figure 6E; Figure S6). The maturation protocol (CHIR withdrawal followed by addback; Figure 6E) was reproduced in the BU3 line and resulted in a total yield of 77.7 ± 30.8 SFTPCtdTomato+ cells on day 66 per input day 15 sorted NKX2-1+ progenitors, representing a 2-fold increase in the resulting numbers of tdTomato+ cells compared with parallel controls maintained in sustained CHIR. The resulting tdTomato+ cells could be purified and serially passaged as alveolospheres that retained tdTomato expression even after freeze-thaw cycles (Figures S6E and S6F). Taken together, these results suggest that, although Wnt stimulation is required for distal lung epithelial differentiation from primordial NKX2-1+ progenitor cells, long-term sustained Wnt stimulation may prevent these distal cells from committing to a fully mature AEC2 phenotype. Thus, temporal modulation of Wnt signaling promotes iAEC2 maturation and self-renewal.
iPSC-AEC2s Enable In Vitro Modeling of Genetic Alveolar Disease
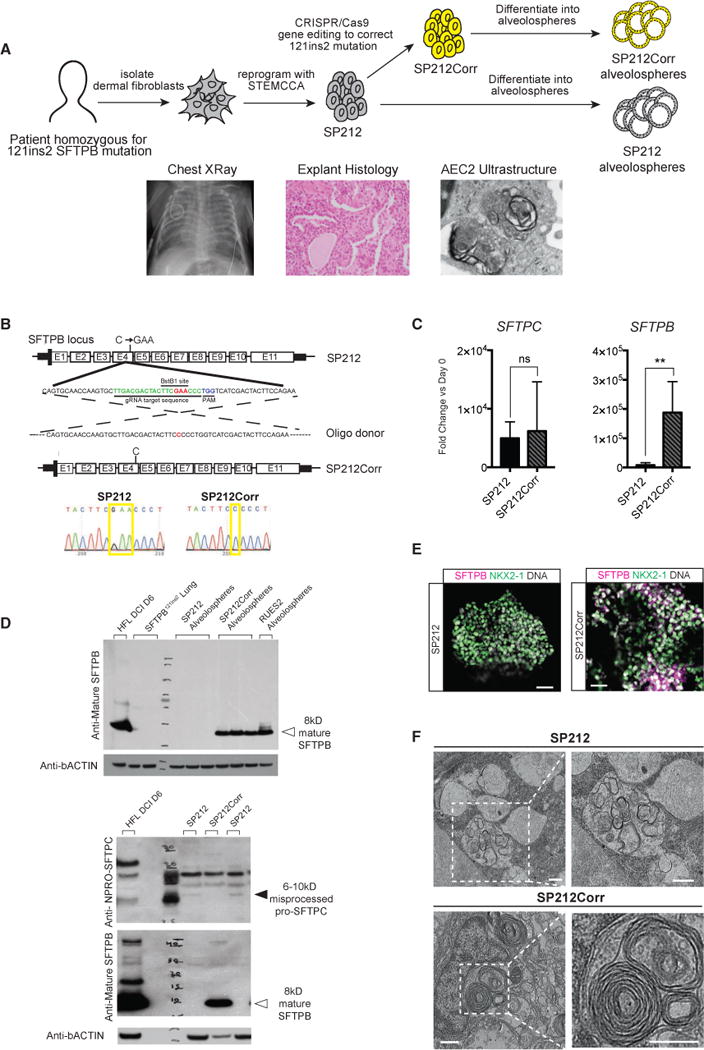
Finally, we sought to derive alveolospheres from disease-specific iPSCs made from an infant homozygous for the 121ins2 mutation in SFTPB, a monogenic cause of neonatal respiratory distress that requires lung transplantation for survival (Figure 7A). Based on prior work, SFTPB mutations do not perturb SFTPB locus transcription, but the unstable mutant mRNA in patients is only detectable at 8% of normal levels and carries a premature stop codon with consequent loss of detectable SFTPB protein, inability to form lamellar bodies, misprocessing of SFTPC protein, and failure to synthesize surfactant (Nogee et al., 1994; Beers et al., 2000). To develop an in vitro model of this disease based on patient-specific cells, we reprogrammed dermal fibroblasts from a patient homozygous for the SFTPB121ins2 mutation (Figure S7A). We gene-edited the resulting iPSC line (SP212) using CRISPR/Cas9 technology to correct both mutant SFTPB alleles in a footprint-free manner, resulting in a corrected line with two normal SFTPB alleles (hereafter called SP212Corr; Figure 7B; Figures S7B–S7E). We differentiated both lines via CD47hi/CD26lo sorted progenitors into alveolospheres expressing similar levels of SFTPC mRNA (Figure 7C) to perform head-to-head comparisons of the pre- versus post-gene-corrected cells (SP212 versus SP212Corr). SP212 alveolospheres expressed lower levels of SFTPB transcript than SP212Corr al-veolospheres (Figure 7C), no detectable SFTPB protein, and no detectable lamellar bodies by TEM (Figures 7D–7F). Notably, in two out of three differentiations, we observed the appearance of an aberrant, misprocessed 6- to 10-kD proSFTPC protein form in SP212 alveolospheres, a form resulting from the residual N-terminal flanking dodecapeptide that cannot be cleaved from proSFTPC in the absence of lamellar bodies (Figure 7F). In SP212Corr alveolospheres, correction of the SFTPB mutation resulted in increased SFTPB mRNA, the appearance of detectable lamellar bodies, reconstitution of the mature 8-kD form of SFTPB protein, and disappearance of the aberrant proSFTPC protein form, verifying that all deficiencies were the direct result of the SFTPB121ins2 mutation (Figures 7C–7F).
Figure 7. iPSC-Derived AEC2s Enable In Vitro Modeling of Genetic Alveolar Disease.
For a Figure360 author presentation of Figure 7, see the figure legend at http://dx.doi.org/10.1016/j.stem.2017.08.014
(A) Radiograph and histopathologic samples of lung of a child from which iPSCs were generated. The schematic shows the process of correcting both alleles carrying the homozygous 121ins2 SFTPB mutations in iPSCs derived from dermal fibroblasts, resulting in pre-correction (SP212) and post-correction (SP212Corr) iPSC lines.
(B) SFTPB exon4 genomic sequence, with the 121ins2 C > GAA mutation shown in red, CRISPR guide RNA target sequence shown in green, the protospacer adjacent motif (PAM) cutting site shown in blue, and the oligo-based donor design with corrected base shown in red. Pre- and post-correction DNA sequencing chromatograms with yellow boxes show the 121ins2 mutation site sequence.
(C) qRT-PCR of day 35 SP212 and SP212Corr alveolospheres, with bars representing mean fold change (2−ΔΔCt) ± SD, n = 3 biological replicates separated on day 0, **p % 0.01 by unpaired, two-tailed Student’s t test.
(D) Top: western blot for mature SFTPB (immunoblotted with PT3 antibody) on SP212 and SP212Corr alveolospheres (n = 3 separated on day 15) as well as RUES2 alveolospheres, 6-day DCI-cultured week 21 human fetal distal lung, and lung samples from a different patient with the same SFTPB121ins2 mutation. Bands showing mature 8-kD SFTPB are present only in normal week 21 controls, RUES2 alveolospheres, and SP212Corr alveolospheres (closed arrowhead). Bottom: western blots for mature SFTPB (PT3) and proSFTPC (NPRO-SFTPC) in 2 different alveolosphere samples of SP212 and 1 sample of SP212Corr, all separated on day 0, with an 6- to 10-kD abnormal/misprocessed proSFTPC band (closed arrowhead) present only in the SP212 samples and 8-kD mature SFTPB (open arrowhead) present only in the cultured week 21 and SP212Corr samples.
(E) Representative immunofluorescence microscopy of proSFTPB (magenta), NKX2-1 (green), and DNA (Hoechst, gray) in SP212 and SP212Corr alveolospheres. Scale bars, 50 μm.
(F) Representative TEM of SP212 and SP212Corr alveolospheres. Scale bars, 500 nm.
See also Figure S7.
DISCUSSION
Our results demonstrate differentiation of phenotypically mature AEC2-like cells, referred to as iAEC2s, from human PSCs of multiple genetic backgrounds. Formed in 3D cultures via an NKX2-1+ endodermal lung progenitor intermediate, the resulting cells express distal lung alveolar epithelial mRNAs and proteins as well as functional lamellar bodies that process surfactant. Contrary to prior reports of the necessity of feeder cells for culturing primary adult AEC2s, we derived and serially passaged “epithelial-only” alveolospheres without using mesenchymal co-culture. We found that the emergence of SFTPC+ cells from NKX2-1+ precursors in culture occurred rapidly in the presence of Wnt stimulation via CHIR within 2–7 days (17–22 total differentiation days) and was augmented by the additional presence of stimulants of FGF signaling together with corticosteroids and cyclic AMP (DCI medium), consistent with studies that have previously shown that Wnt signaling rapidly promotes distal airway patterning and alveologenesis in vivo (Mucenski et al., 2005; Shu et al., 2005; Frank et al., 2016) or in vitro (McCauley et al., 2017), KGF promotes distal epithelial outgrowth ex vivo (Liu and Hogan, 2002), and hormones, such as corticosteroids, promote AEC2 maturation (Gonzales et al., 2002; Ballard et al., 2010).
Although we did observe lamellated inclusions in the cytoplasm of alveolospheres, lamellated inclusions mistakenly referred to as lamellar bodies that are not enriched in surfactant proteins or phospholipid have been known to occur in a variety of cell types that neither display an AEC2 phenotype nor package surfactant in culture, such as A549 cells (Mason and Williams, 1980). Based on their functional capacity to process SFTPB protein to its 8-kD isoform and produce DPPC surfactant phospholipid, we conclude that alveolospheres express true lamellar bodies and represent a maturity level comparable with primary AEC2s post-week 24 of gestation. Despite the presence of a subset of relatively mature cells, the unsorted alveolospheres as well as their sorted SFTPC-high or -low components can be sequentially passaged and maintain proliferative capacity over months without evidence of senescence. Because this in vitro model generates spheres of iAEC2s in close contact without intervening AEC1s, it does not represent all epithelial components of the alveolar microenvironment, but it provides a convenient tool for interrogating diseases postulated to affect only AEC2s, such as the SFTPB mutations we have studied. Although we do not detect AEC1 markers within 3D alveolospheres, when SFTPCtdTomato+ iAEC2s are plated in 2D culture, they downregulate SFTPC and upregulate the AEC1 markers PDPN, CAV1, and AGER, as has been shown in primary human AEC2 culture. These capacities of self-renewal and differentiation are key features that have defined primary AEC2s in vivo as the progenitors of the distal lung and are required for the survival of air-breathing mammals. Further studies are needed, however, to understand both the signaling pathways and the developmental time windows involved in AEC2-to-AEC1 conversion in vivo and in vitro during directed differentiation.
On a whole-transcriptome level, iAEC2s clustered closer to cultured primary fetal AECs than to adult or fetal AEC2s. Not surprisingly, however, there are many ways in which iAEC2s, differentiated in submerged culture over only 30–35 days in vitro, are not identical to primary AEC2s exposed to a lifetime of air breathing in adults. Several of the gene sets enriched in primary adult AEC2s compared with iAEC2s appeared to involve immune responses and oxidant stress pathways, as would be expected. However, we found that alveolospheres treated with immune cytokines do respond by activating the IL-6/JAK/STAT3 and TNF-α/NF-κB pathways. Because AEC2s are known to be important immune modulators in vivo, it is important that iAEC2s allow for studies of immune function.
Although we were surprised to find that SFTPCtdTomato+ and SFTPCtdTomato− cells cluster so closely together by PCA as well as by supervised hierarchical clustering analyses, it is likely that these populations, both deriving from NKX2-1+ lung endo-derm in distalizing conditions, represent very similar cells, some of which have progressed to express high levels of AEC2 maturation markers, and others that may remain “stuck” as less mature cell types expressing significantly lower, albeit easily detected, levels of these markers.
Interestingly, GSEA showed that the Wnt/β-Catenin signaling pathway was downregulated in SFTPCtdTomato+ cells, and late CHIR withdrawal resulted in both a dramatic increase in percent SFTPCtdTomato+ cells in the previously SFTPCtdTomato− population as well as an increase in expression of mature AEC2 transcripts within this population. This finding suggests that overstimulation with CHIR may actually inhibit full alveolar differentiation. We have previously shown that early CHIR withdrawal (day 15) results in proximalization of NKX2-1+ lung progenitors (McCauley et al., 2017), and we now see that late withdrawal of CHIR, following distalization, promotes alveolar differentiation, consistent with the low-Wnt pre-alveologenesis stage of AEC2 development recently reported in mice by Frank et al. (2016). Furthermore, as predicted by these mouse studies, adding back Wnt stimulation following maturation stimulates proliferation of the resulting human iAEC2s.
Finally, the ultimate test of iAEC2s as a clinically relevant surrogate for primary AEC2s is whether they can recapitulate human alveolar disease in vitro. Indeed, in our studies, iAEC2s generated from a child with severe lung disease because of homozygous 121ins2 SFTPB mutations recapitulated known aspects of this disease, which were rescued in gene-corrected iAEC2s from the same patient. Thus, our work shows generation of phenotypically mature iPSC-derived alveolar organoids that represent a robust in vitro model of both human alveolar development and disease, providing a platform by which new insights can be obtained into the effects of genetic and environmental insults on AEC2 biology.
STAR+METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal to beta-Actin | Sigma | Cat#A1978 |
| Rabbit monoclonal to pan-Actin | Cell Signaling | Cat#8456 |
| Mouse monoclonal to CD26, PE conjugated (clone BA5b) | Biolegend | Cat#302705 |
| Mouse monoclonal to CD47, PerCP-Cy5.5 conjugated (clone CC2C6) | Biolgend | Cat#323110 |
| Mouse monoclonal to EPCAM (clone AUA1) | Abcam | Cat#ab181853 |
| Mouse monoclonal to GAPDH | Chemicon | Cat#MAB374 |
| Mouse IgM monoclonal to HT2-280 | Terrance Biotech | Cat#TB-27AHT2-280 |
| Rabbit polyclonal to IκB | Santa Cruz | Cat#sc-371 |
| Rabbit monoclonal to NKX2-1 (clone EP15847) | Abcam | Cat#ab76013 |
| Mouse monoclonal to NKX2-1 (clone 8G7G3/1) | Abcam | Cat#ab72876 |
| Rabbit polyclonal to phosphoStat3 | Cell Signaling | Cat#9131 |
| Mouse monoclonal to red fluorescent protein (RFP) | Abcam | Cat#65856 |
| Rabbit monoclonal to red fluorescent protein (RFP) | Rockland | Cat#600-401-379 |
| Rabbit polyclonal to Mature SFTPB | Seven Hills | Cat#r28031 |
| Anti-mature SFTPB (PT3) | Beers et al., 1992 | N/A |
| Anti-NFLANK SFTPB | Korimilli et al., 2000 | N/A |
| Rabbit polyclonal to Pro-SFTPB | Seven Hills | Cat#WRAB-55522 |
| Rabbit polyclonal to Mature SFTPC | Seven Hills | Cat#r76694 |
| Anti-NPRO SFTPC | Beers et al., 1994 | N/A |
| Rabbit polyclonal to Pro-SFTPC | Seven Hills | Cat#WRAB-9337 |
| Rabbit polyclonal to ZO-1 | Thermo Fisher | Cat# 61-7300 |
| AffiniPure Donkey Anti-Rabbit IgG (H+L), 488 conjugated | Jackson Immunoresearch | Cat#711-225-152 |
| AffiniPure Donkey Anti-Rabbit IgG (H+L), Cy3 conjugated | Jackson Immunoresearch | Cat#711-165-152 |
| AffiniPure Donkey Anti-Rabbit IgG (H+L), AlexaFluor 647 conjugated | Jackson Immunoresearch | Cat#711-605-152 |
| AffiniPure Donkey Anti-Mouse IgG (H+L), AlexaFluor 647 conjugated | Jackson Immunoresearch | Cat#715-605-150 |
| AffiniPure Donkey Anti-Mouse IgG (H+L), AlexaFluor Cy3 conjugated | Jackson Immunoresearch | Cat#715-165-150 |
| AffiniPure Donkey Anti-Goat IgG (H+L), AlexaFluor 647 conjugated | Jackson Immunoresearch | Cat#305-605-003 |
| Biological Samples | ||
| Primary human lung samples | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Growth Factor Reduced Matrigel | Corning | Cat#356230 |
| SB431542 | Tocris | Cat#1614 |
| Dorsomorphin | Stemgent | Cat#04-0024 |
| CHIR99021 | Tocris | Cat#4423 |
| Recombinant human FGF10 | R&D Systems | Cat#345-FG-025 |
| Recombinant human KGF | R&D Systems | Cat#251-KG-010 |
| Recombinant human BMP4 | R&D Systems | Cat#314-BP |
| Retinoic acid | Sigma | Cat#R2625 |
| Y-27632 dihydrochloride | Tocris | Cat#1254 |
| Recombinant human FGF2 | R&D Systems | Cat#233-FB |
| Recombinant human TGFβ | R&D Systems | Cat#240-B |
| DAPT | Sigma | Cat#D5942 |
| Dexamethasone | Sigma | Cat#D4902 |
| 8-bromoadenosine 3′,5′-cyclic monophosphate sodium salt (cAMP) | Sigma | Cat#B7880 |
| 3-Isobutyl-1-methylxanthine (IBMX) | Sigma | Cat#I5879 |
| Recombinant mouse Noggin | R&D Systems | Cat#1967-NG |
| Recombinant mouse EGF | R&D Systems | Cat#2028-EG-200 |
| Hoechst | Thermo Fisher | Cat#H3570 |
| Puromycin Dihydrochloride | Thermo Fisher | Cat#A1113802 |
| Geneticin Sulfate | Life Technologies | Cat#11811-023 |
| 0.05% trypsin-EDTA | Invitrogen | Cat#25300-120 |
| Defined Fetal Bovine Serum | Thermo Fisher | Cat#NC0652331 |
| Calcein blue | Life Technologies | Cat#C1429 |
| Recombinant human TNFα | R&D Systems | Cat#210-TA-005 |
| Recombinant human IL1β | R&D Systems | Cat#201-LB-005 |
| Recombinant human OSM | R&D Systems | Cat#295-OM-010/CF |
| Dispase | Thermo Fisher | Cat#354235 |
| Glutaraldehyde | Ladd Research | Cat#20100 |
| Osmium Tetroxide | Polysciences | Cat#0223D |
| Uranyl Acetate | Electron Microscopy Sciences | Cat#22400 |
| EMbed 812 | Electron Microscopy Sciences | Cat#14120 |
| 14:0 PC (DMPC) | Avanti Polar Lipids | Cat#850345 |
| Paraformaldehyde | Electron Microscopy Sciences | Cat#19208 |
| Critical Commercial Assays | ||
| RNeasy Mini Kit | QIAGEN | Cat#74104 |
| QIAzol Lysis Reagent | QIAGEN | Cat#79306 |
| TaqMan Fast Universal PCR Master Mix (2X), no AmpErase UNG | Thermo Fisher | Cat#4364103 |
| High-Capacity cDNA Reverse Transcription Kit | Applied Biosystems | Cat#4368814 |
| Click-iT® EdU Alexa Fluor® 488 Imaging Kit | Thermo Fisher | Cat#C10337 |
| EZ-TAL TALE Assembly Kit | System Bioscience | Cat#GE120A-1 |
| Lipfectamine LTX Kit | Thermo Fisher | Cat#15338100 |
| P3 Primary Cell 4D-Nucleofector® X Kit S | Lonza | Cat#V4XP-3032 |
| PAS Kit | Sigma | Cat#395b |
| HeLa Monster transfection reagent | Mirus | Cat#MIR 2904 |
| Deposited Data | ||
| RNA Sequencing Data: “Generation of mature lung alveolar epithelial cells from human pluripotent stem cells” | This paper | GEO: GSE96642 |
| Experimental Models: Cell Lines | ||
| Human: Normal donor induced pluripotent stem cell (iPSC) line (BU3) | Kotton Lab (Kurmann et al., 2015) | http://www.bumc.bu.edu/stemcells |
| Human: Normal donor iPSC line targeted with NKX2-1GFP (BU3GFP) | Kotton Lab (Hawkins et al., 2017) | http://www.bumc.bu.edu/stemcells |
| Human: Cystic fibrosis donor iPSC line targeted with NKX2-1GFP (C17) | Brian Davis Lab (Hawkins et al., 2017) | http://www.bumc.bu.edu/stemcells |
| Human: SFTPB deficiency donor iPSC line (SP212) | This paper | http://www.bumc.bu.edu/stemcells |
| Human: Corrected SFTPB deficiency donor iPSC line (SP212) | This paper | http://www.bumc.bu.edu/stemcells |
| Human: RUES2 embryonic stem cell line | Gift from Dr. Ali H. Brivanlou, Rockefeller University | N/A |
| Human: MRC5 fibroblast cell line | ATCC | CCL-171 |
| Recombinant DNA | ||
| EF1a-TALEN_HD; SFTPC recognition sequence of right TALEN 5′ -TCA CCG GCG GGC TCT CCA TC-3′. | This paper | http://www.kottonlab.com |
| EF1a-TALEN_NN; SFTPC recognition sequence of left TALEN: 5′-TAG CAC CTG CAG CAA GAT GG-3′ | This paper | http://www.kottonlab.com |
| p1303-DV-SFTPC-tdTomato | This paper | http://www.kottonlab.com |
| pHAGE2 EF1a-Cre-IRES-NeoR-W | This paper | http://www.kottonlab.com |
| pHAGE2-Cre-IRES-PuroR | Kotton Lab (Somers et al., 2010) | Addgene #30205 |
| pD1321-AD- SFTPB121ins2 | This paper | |
| Sequence-Based Reagents | ||
| TaqMan Gene Expression Assay Primer/Probe Sets | Thermo Fisher | |
| ABCA3 | Thermo Fisher | Hs00975530 m1 |
| AGER | Thermo Fisher | Hs00542584 g1 |
| AQP5 | Thermo Fisher | Hs00387048 m1 |
| CAV1 | Thermo Fisher | Hs00971716_m1 |
| NKD1 | Thermo Fisher | Hs00263894 m1 |
| LEF1 | Thermo Fisher | Hs01547250 m1 |
| GMCSF | Thermo Fisher | Hs00531296 g1 |
| IL-8 | Thermo Fisher | Hs00174103_m1 |
| LAMP3 | Thermo Fisher | Hs00180880 m1 |
| LPCAT1 | Thermo Fisher | Hs00227357 m1 |
| NAPSA | Thermo Fisher | Hs00362192 m1 |
| NKX2-1 | Thermo Fisher | Hs00968940 m1 |
| P63 | Thermo Fisher | |
| PDPN | Thermo Fisher | Hs00366766 m1 |
| PGC | Thermo Fisher | Hs00160052 m1 |
| SCGB1A1 | Thermo Fisher | Hs00171092_m1 |
| SFTPA1 | Thermo Fisher | Hs00831305 s1 |
| SFTPA2 | Thermo Fisher | Hs04195463 g1 |
| SFTPC | Thermo Fisher | Hs00161628 m1 |
| SFTPB | Thermo Fisher | Hs01090667 m1 |
| SLC34A2 | Thermo Fisher | Hs00197519 m1 |
| SOCS3 | Thermo Fisher | Hs02330328 s1 |
| Oligonucleotide primers | Integrated DNA Technologies | |
| STEMCCA Excision Forward | Somers et al., 2010 | 5′ GGAACTCTTGT GCGTAAGTCGATAG 3′ |
| STEMCCA Excision Reverse | Somers et al., 2010 | 5′ GGAGGCGGCCC AAAGGGAGATCCG3′ |
| tdTomato Targeting Forward | This paper | 5′ GGGTGAGTGAG CTGATTCGAG 3′ |
| tdTomato Targeting Reverse | This paper | 5′ TGACCTCCTCG CCCTTGCTCACCATG 3′ |
| Intact SFTPC Locus Forward | This paper | 5′ CTACGGACACA TATAAGACCCTGGTC 3′ |
| Intact SFTPC Locus Reverse | This paper | 5′ GCTGTGCATCC CACACCT 3′ |
| SFTPCtdTomato Sequencing Primer | This paper | 5′ GGGTGAGTGAG CTGATTCGAG 3′ |
| Puromycin Resistance Cassette Forward | This paper | 5′ ATGACCGAGTA CAAGCCCACG 3′ |
| Puromycin Resistance Cassette Reverse | This paper | 5′ TCAGGCACCGG GCCTGC 3′ |
| SFTPB 121ins2 CRISPR gRNA | This paper | 5′ TTGACGACTACT TCGAACCCTGG 3′ |
| SFTPB 121ins2 correction single-stranded oligonucleotide | This paper | 5′ GAAGCTGCTCA TGCCCCAGTGC AACCAAGTGCTTGACGACTACTTC CCCCTGGTCATCGACTACTTCCA GAACCAGATTGTGAGGCTG 3′ |
| SFTPB 121ins2 Correction Forward | This paper | 5′ ACTCCTTGGCA CTCGTGAAC 3′ |
| SFTPB 121ins2 Correction Reverse | This paper | 5′ GGGTGCTGTGT GTTTGTGTC 3′ |
| Software and Algorithms | ||
| ImageJ | National Institutes of Health | https://imagej.nih.gov/ij/ |
| Other | ||
| StemDiff Definitive Endoderm Kit | StemCell Technologies | Cat#05110 |
| mTeSR1 | StemCell Technologies | Cat#05850 |
| Glutamax | Life Technologies | Cat#35050-061 |
| Gentle Cell Dissociation Reagent | StemCell Technologies | Cat#07174 |
| 0.4mm Pore Polyester Membrane Transwell Insert | Corning | Cat#3470 |
| Microscope Slides with Double Concavity | Eisco | Cat# BI0086B |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by, the Lead Contact, Darrell Kotton (dkotton@bu.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
ESC/iPSC Line Generation and Maintenance
All experiments involving the differentiation of human iPSC lines were performed with the approval of the Institutional Review Board of Boston University (protocol H33122). BU3 and C17 iPSC lines carrying the NKX2-1GFP reporter were obtained from our prior studies (Hawkins et al., 2017). These lines were derived from a normal donor (BU3) (Kurmann et al., 2015) and an individual with cystic fibrosis (C17) carrying a published compound heterozygous CFTR genotype (Crane et al., 2015), respectively. The human embryonic stem cell line RUES2 was a generous gift from Dr. Ali H. Brivanlou of The Rockefeller University. The iPSC line SP212 was derived by reprogramming dermal fibroblasts (see below) of a patient with neonatal respiratory distress syndrome resulting from documented homozygous 121ins2 mutations (c.397delinsGAA (p.P133Efs*95), hg19) in the surfactant protein B (SFTPB) locus. The Institutional Review Board of Washington University, St. Louis, MO, approved procurement of fibroblasts with documented informed consent.
All PSC lines used in this study (BU3, C17, RUES2, SP212 and SP212Corr) displayed a normal karyotype (BU3 and C17, 46XY; RUES2, SP212, and SP212Corr 46XX) when analyzed by G-banding both before and after gene-editing (Cell Line Genetics). Culture conditions used for maintenance and editing of undifferentiated PSCs were as follows: for TALENs targeting, PSC lines were maintained on mitomycin C-inactivated MEFs in human iPSC media (WiCell feeder dependent protocol). For CRISPR targeting and prior to directed differentiation, all PSC lines were maintained in feeder-free conditions, on growth factor reduced matrigel (Corning) in 6-well tissue culture dishes (Corning), in mTeSR1 medium (StemCell Technologies) using gentle cell dissociation reagent for passaging. Further details of iPSC derivation, characterization, and culture are available for free download at http://www.bu.edu/dbin/stemcells/protocols.php.
Generation of SP212 iPSCs
Reprogramming of patient-specific dermal fibroblasts (Figure 7 and Figure S7) was performed with a single-integrated excisable copy of the floxed hSTEMCCA lentiviral reprogramming vector (Somers, et.al., 2010) followed by excision with transient Cre-recom-binase exposure. Ten iPSC colonies were mechanically isolated 30 days after lentiviral transduction and expanded on MEF feeders in human “iPSC media” (Somers et al., 2010), composed of DMEM/F12 (Sigma-Aldrich) with 20% KnockOut Serum Replacement (Invitrogen), 1mM nonanimal L-glutamine (Sigma-Aldrich), 0.1mM B-mercaptoethanol, and 10 ng/ml FGF2 (R&D Systems) on 0.1% gelatin (Sigma-Aldrich) coated plates preseeded with mitomycin C-inactivated or irradiated mouse embryonic fibroblast (MEF) feeder cells. Integrated hSTEMCCA copy number was assessed by Southern blot of BamHI-digested gDNA extracts probed for the lentiviral WPRE cassette (Somers et al. 2010), and only iPSC clones with single copy hSTEMCCA integrations were selected for vector excision and further study. The single copy hSTEMCCA lentiviral cassette was removed from two iPSC clones (SP212-1 and SP212-5) via transient transfection of pHAGE2-Cre-IRES-PuroR plasmid DNA (Somers et al., 2010; Addgene #30205) using HeLa Monster transfection reagent (Mirus) according to the manufacturer’s instructions (Somers et al., 2010). Approximately 11-14 days later, colonies were picked and gDNA from each subclone was screened for vector excision by PCR using the following primers and conditions: 5′-GGA ACT CTT GTG CGT AAG TCG ATA G-3′; 5′-GGA GGC GGC CCA AAG GGA GAT CCG-3′; 95 °C for 3 min; followed by 33 cycles of 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 1 min; followed by a single cycle of 72 °C for 5 min. Vector excision was then confirmed by Southern blot using BamHI digested gDNA probed for the WPRE element (Somers et al., 2010) to identify two iPSC lines (each generated from the two separately picked clones, renamed SP212-1-Cr3 and SP212-5-Cr3 to reflect successful vector excision; Figure S7).
SFTPCtdTomato Reporter PSC Line Generation
To generate SFTPCtdTomato knock-in reporter PSCs, TALENs were designed to target the sequences close to the translation initiation (ATG) site of the human SFTPC gene. All TALENs and donor template plasmid maps and sequences are available at http://www.kottonlab.com. The SFTPC TALEN recognition sequences are: left TALEN 5′-TAG CAC CTG CAG CAA GAT GG-3′ and right TALEN 5′-TCA CCG GCG GGC TCT CCA TC-3′. Between the two binding sites is a 22 bp spacer (ATG TGG GCA GCA AAG AGG TCC T). TALENs were constructed using EZ-TAL TALE Assembly Kit (System Bioscience), according to manufacturer’s instructions, and the resulting SFTPC TALENS encoding plasmids were named EF1a-TALEN_NN (SPC left) and EF1a-TALEN_HD (SPC right), respectively.
To deliver the donor template to the SFTPC locus, we generated a donor vector (p1303 DV-SFTPC-tdTomato) containing the tdTomato coding sequence and a floxed PGK promoter-driven puromycin resistance cassette, flanked by left and right arms of homology to the human endogenous SFTPC locus, as follows: we first modified the CReM’s targeting vector, TVGIP-eGFP-puro (generous gift of Gustavo Mostoslavsky and Cesar Sommer, CReM of Boston University and Boston Medical Center; http://www.bumc.bu.edu/stemcells). The GIP-eGFP sequence was replaced with the tdTomato coding sequence. 5′ and 3′ arms of homology to the SFTPC locus were generated by PCR cloning using gDNA extracts of human ES cells (RUES2) as templates. The 5′ arm of homology extends 750 base pairs upstream of the SFTPC ATG start site, and the 3′ arm of homology extends 750 base pairs downstream of the ATG start site.
The TALENs and donor vector plasmids were co-transfected into the following PSC lines: RUES2, C17 NKX2-1GFP, and BU3 NKX2-1GFP (Hawkins et al., 2017) using a lipofectamine based transfection protocol. Each line was plated onto a mitomycin C-inactivated DR4 mouse embryonic fibroblast (MEF) feeder layer and cultured in human iPSC media (WiCell) in a 6 well plate. After the cells reached 50% confluence, they were transfected with the two TALENs and tdTomato donor vector as follows: 3 μg of donor vector and 1.2 μg of each TALEN were added to 275 μL of IMDM and 4 μL of Plus reagent from the Lipofectamine LTX kit (Thermo Fisher), and this mixture was incubated at room temperature for 5 min. 16 μL of lipofectmine LTX from the same kit was added to another 275 μL of IMDM. 275 μL of the DNA mixture was added to 275 μL of the LTX mixture and incubated at room temperature for 30 min. 550 μL of the total mixture was added drop by drop to 1 well of a 6 well plate. 5 hr later, the media was changed, and 48 hr later, 0.7 μg/ml puromycin (Fisher Scientific) was added to the media for 4 days to select antibiotic resistant colonies. After 10 days, individual colonies from each line were picked and screened for targeting using the following primer pairs (Figure S1): 5′ GGG TGA GTG AGC TGA TTC GAG 3′, 5′ TGA CCT CCT CGC CCT TGC TCA CCA TG 3′. To confirm heterozygous targeting, colonies were screened for a remaining intact SFTPC gene using the following primers: 5′ CTA CGG ACA CAT ATA AGA CCC TGG TC 3′, 5′ GCT GTG CAT CCC ACA CCT 3′. DNA sequencing using a primer binding in the genome outside any regions included in targeting plasmids confirmed targeting into the endogenous SFTPC locus (5′ GGG TGA GTG AGC TGA TTC GAG 3′).
Cre-mediated excision of the floxed puromycin resistance cassette was performed using a plasmid containing Cre-recombinase and neomycin resistance (PHAGE2 EF1a-Cre-IRES-NeoR-W; http://www.kottonlab.com) using the same lipofectamine-based protocol described above, with 4 days of 200ng/μl geneticin-based (Life Tech) selection for clones that were transfected with Cre-containing plasmid. Excision of the puromycin cassette was confirmed by PCR using the following primers: 5′ ATG ACC GAG TAC AAG CCC ACG 3′, 5′ TCA GGC ACC GGG CCT GC 3′.
CRISPR-based gene correction of SFTPB121ins2
We used CRISPR/Cas9 technology to target the region adjacent to the 121ins2 mutation (also known as c.397delinsGAA (p.P133Efs*95), hg19) in the human SFTPB gene locus (Figure 7) with a guide RNA (specifically recognizing the 121ins2 mutation but not wild-type) that had the following sequence: 5′ TTG ACG ACT ACT TCG AAC CCT GG 3′. gRNA sequences were commercially cloned into the pD1321-AD plasmid backbone (DNA 2.0 ATUM) that contains a M-dasher-GFP sequence fused to Cas9. Delivery of this plasmid to iPSCs enabled co-expression of Cas9, gRNA, and a GFP reporter. To accomplish footprint-free correction of the SFTPB mutation, a short single stranded DNA oligo (sequence 5′ GAA GCT GCT CAT GCC CCA GTG CAA CCA AGT GCT TGA CGA CTA CTT CCC CCT GGT CAT CGA CTA CTT CCA GAA CCA GAT TGT GAG GCT G 3′) was used as a donor template containing the wild-type SFTPB sequence. SP212 cells maintained in feeder-free conditions in mTeSR1 media were treated with 10 μm Y-27632 (Tocris) for 3 hr, dissociated in Gentle Cell Dissociation Reagent (GCDR, StemCell Technologies) for 10 min at 37 °C and counted using a Luna-II Automated Cell counter (Logos Biosystems). Approximately 5×106 cells were centrifuged at 200 g × 5 min, resus-pended in a mixture containing P3 solution and supplement (P3 Primary Cell 4D-Nucleofector X Kit L, Lonza) as well as 5 μg CRISPR/Cas9 plasmid and 5 μg oligo donor, and nucleofected using program code CB-150 in the 4D nucleofector system (Lonza). Nucleofected cells were replated on 2 wells of a matrigel-coated 6 well plate in mTeSR1 media, and 10 μm Y-27632 was added for 24 hr. After 48 hr, cells were prepared for sorting. They were treated with 10 μm Y-27632 for 3 hr, dissociated into single cell suspension with GCDR for 10-15 min at 37 °C, centrifuged at 200 g × 5 min, resuspended in mTeSR1 media + 10 μm Y-27632, and filtered through a 30 μm filter (Falcon). GFP+ cells were sorted into recovery media (1 part mTeSR1 and 1 part conditioned mTeSR1 supplemented with 0.7ng/ml FGF2, plus 10 μm Y-27632) using a high-speed cell sorter (MoFlo Legacy, Beckman Coulter). 1×104 cells GFP+ sorted cells were plated into a 10cm tissue culture treated dish pre-coated with growth factor-reduced matrigel. Recovery media was changed every other day for the first 5 days, with 10 μm Y-27632 added for the first 24 hr. After 5 days, mTeSR1 was used to feed the cells, and after 10 days, colonies that emerged were of sufficient size for picking for clonal expansion and screening. Individual colonies were picked and screened for correction using the following primers: 5′ ACT CCT TGG CAC TCG TGA AC 3′, 5′ GGG TGC TGT GTG TTT GTG TC 3′. In pre-correction SP212, there is a BstB1 restriction site created by the 121ins2 mutation, and after enzyme digestion, 2 bands at 230bp and 191bp appear. Post-correction, the BstB1 site disappears, and only the uncut 430bp band is seen on a gel. After PCR screening, colonies with the uncut band were further analyzed for correction, which was confirmed by DNA sequencing, resulting in the SP212Corr line.
METHOD DETAILS
Directed Differentiation of PSCs into NKX2-1+ lung progenitors
PSC directed differentiation into NKX2-1 lung progenitors was performed as described previously (Hawkins et al., 2017; Rankin et al., 2016). Briefly, cells maintained on mTESR1 media were differentiated into definitive endoderm using the STEMdiff Definitive Endo-derm Kit (StemCell Technologies), with 1 day addition of supplement A and B, and 2 days addition of supplements B only (Day 4 in the STEMdiff kit protocol). After the endoderm-induction stage, cells were dissociated for 1-2 min at room temperature with GCDR and passaged at a ratio between 1:2 to 1:6 into 6 well plates pre-coated with growth factor reduced matrigel in “DS/SB” anteriorization media, consisting of complete serum-free differentiation medium (cSFDM) base, including IMDM (ThermoFisher) and Ham’s F12 (ThermoFisher) with B27 Supplement with retinoic acid (Invitrogen, Waltham, MA), N2 Supplement (Invitrogen), 0.1% bovine serum albumin Fraction V (Invitrogen), monothioglycerol (Sigma), Glutamax (ThermoFisher), ascorbic acid (Sigma), and primocin with supplements of 10 μm SB431542 (“SB”; Tocris) and 2 μm Dorsomorphin (“DS”; Stemgent). For the first 24 hr after passaging, 10 μm Y-27632 was added to the media. After anteriorization in DS/SB media for 3 days (72 hr), cells were cultured in “CBRa” lung progenitor-induction media for 9-11 days. “CBRa” media consists of cSFDM containing 3 μm CHIR99021 (Tocris), 10 ng/mL recombinant human BMP4 (rhBMP4, R&D Systems), and 100 nM retinoic acid (RA, Sigma), as previously described (Rankin et al., 2016). On Day 15 of differentiation, efficiency of specification of NKX2-1+ lung progenitors was evaluated either by flow cytometry for intracellular NKX2-1 protein, NKX2-1GFP reporter expression, or by expression of surrogate cell surface markers CD47hi/CD26lo based on the method of Hawkins and Kotton (Hawkins et al., 2017).
Cell sorting of NKX2-1+ Lung Progenitors
On day 15 of differentiation, cells were incubated at 37 °C in 0.05% trypsin-EDTA (Invitrogen) for 7-15 min, until they reached single cell suspension. Cells were then washed in media containing 10% fetal bovine serum (FBS, ThermoFisher), centrifuged at 300 g × 5 min, and resuspended in sort buffer containing Hank’s Balanced Salt Solution (ThermoFisher), 2% FBS, 10 μm Y-27632, and 10 μm calcein blue AM (Life Technologies) for dead cell exclusion. Cells not containing the NKX2-1GFP reporter were subsequently stained with CD47-PerCPCy5.5 and CD26-PE antibodies (mouse monoclonal; Biolegend 1:200; 1 × 106 cells in 100 μl) for 30 min at 4 °C, washed with PBS, and resuspended in sort buffer. Cells were passed through a 40 μm strainer prior to sorting (Falcon). Various live cell populations indicated in the text (i.e., GFP+, GFP−, CD47hi/CD26-,CD47lo) were sorted on a high-speed cell sorter (MoFlo Legacy).
NKX2-1+ Lung Progenitor Outgrowth into iAEC2s
Day 15 cells, either sorted (as described above) or unsorted (dissociated as described above without sorting), were resuspended in undiluted growth factor-reduced matrigel (Corning) at a dilution of 25-100 cells/μl, with droplets ranging in size from 20 μL in 96 well plates to 1ml in 10cm tissue culture-treated dishes (Corning). Cells in 3D matrigel suspension were incubated at 37 °C for 20-30 min, then warm media was added to the plates.
Where indicated in the text, outgrowth and distal/alveolar differentiation of cells after day 15 was performed in “CK+DCI” medium, consisting of cSFDM base, with 3 μm CHIR99021, 10 ng/mL rhKGF, and 50 nM dexamethasone (Sigma), 0.1 mM 8-Bromoadenosine 3′, 5′-cyclic monophosphate sodium salt (Sigma) and 0.1 mM 3-Isobutyl-1-methylxanthine (IBMX; Sigma) (DCI). Immediately after re-plating cells on Day 15 10 μm Y-27632 was added to the medium for 24 hr. Additional growth factors or cytokines were added or withdrawn as indicated in the text, including FGF10, TGFb, EGF, OSM (20ng/ml), TNFα (10ng/ml), and IL-1β (10ng/ml) with other concentrations listed in figure legends (Figures S2A and S2B).
Long Term Culture of Alveolospheres
Alveolospheres developed in 3D matrigel culture outgrowth within 3-7 days after day 15 replating, and were maintained in CK+DCI media for weeks to months, as indicated in the text. These spheres were analyzed as follows: Z stack images of live alveolospheres were taken and processed on a Keyence BZ-X700 fluorescence microscope. For some analyses (RT-qPCR, western blot, lipidomic analysis) alveolospheres were released from matrigel droplets, and for other techniques (flow cytometry, cell sorting), they were dissociated into single cell suspension. To release alveolospheres from matrigel, droplets were incubated in dispase (2mg/ml, Fisher) at 37°C for 1 hr, centrifuged at 300 g × 1 min, washed in 1x PBS, then centrifuged again at 300 g x 1 min. To generate single cell suspensions, cell pellets were incubated in 0.05% trypsin and continued through the trypsin-based dissociation protocol described above, after which they could be passaged into fresh matrigel, analyzed by flow cytometry, or sorted as described above. Sorted cells from alveolospheres were replated into matrigel droplets for serial passaging where indicated in the text. For AEC1 differentiation experiments approximately 20,000 sorted SFTPCtdTomato+ cells were plated in 96 well tissue culture plates in Dulbecco’s Modified Eagle Medium (DMEM), 10% FBS, glutamax, and primocin for 4-7 days, then harvested for analysis. For ALI experiments, alveolospheres were dissociated into single cell suspension as described above and cells were transferred into matrigel pre-coated TransWell inserts (Corning) at a confluency of 500,000 cells/cm2. Media was removed from the apical side of the TransWell after 1 week and cells were analyzed 2-3 weeks later. For EdU labeling, alveolospheres in 3D culture were incubated in CK+DCI media with 5uM EdU for 24 hr, then dissociated as described above, fixed, and processed according to the manufacturer’s instructions (Click-iT® EdU Alexa Fluor® 488 Imaging Kit, Thermo Fisher).
Electron Microscopy of Alveolospheres
Alveolospheres were fixed for 3 hr total in 2.5% glutaraldehyde (Ladd Research) in 0.1% cacodylate buffer pH 7.4 at room temperature. An equal volume of 5% glutaraldehyde/.1M cacodylate was added to the Eppendorf tube with alveolospheres in known volume of media, fixed for 1.5 hr, and spun down gently (300 g × 1 min). Fresh 2.5% glutaraldehyde/0.1M cacodylate was added, and the sample was fixed for an additional 1.5 hr at room temperature. The sample was then washed in 0.1M cacodylate three times, post-fixed in 1% Tannic Acid in cacodylate buffer for 5 min at room temperature, washed again 3 times in cacodylate buffer, and post fixed overnight in 1.5% osmium tetroxide (Polysciences) in 0.1M cacodylate buffer in dark at 4 °C. The sample was washed 3-4 times in 0.05M Na Maleate buffer pH 5.2 and block stained in 1.5% Uranyl acetate (Electron Microscopy Sciences, EMS) in 0.025M Na Maleate buffer pH 6.0. Next, the sample was dehydrated quickly through acetone on ice, from 70% to 80% to 90%. Then, it was incubated 2 times in 100% acetone at room temperature for 10 min each, and in propylene oxide at room temperature for 15 min each. Finally, the sample was changed into EMbed 812 (EMS), left for 2 hr at RT, changed into fresh Embed 812 and left overnight at room temperature, after which it was embedded in fresh EMbed 812 and polymerized overnight at 60 °C. Plastic embedded samples were thin sectioned at 70nm and grids were stained in 4% aqueous Uranyl Acetate for 5 min at 60 °C followed by Lead Citrate for 10 min at room temperature.
Sections on grids were imaged on a CM12 Transmission Electron Microscope (Philips), using a TEMCAM F216 camera (TVIPS) at an original magnification of 7875× and and a Morgagni 268 (FEI), using a Veleta camera (Olympus SIS) at original magnifications of 7200× and 14000×.
Immunogold Staining of Alveolospheres
Alveolospheres were processed for immunogold labeling described previously (Ridsdale et al., 2011). Alveolospheres were first fixed in situ with 4% paraformaldehyde (EMS), 0.1% glularaldehyde (EMS), 75 mM L-lysine (Sigma), 10 mM INaO4 (Sigma), and 0.1% CaCl2 in 0.2M HEPES (Sigma), pH 7.2 at room temperature for 10 min, followed by postfixation with fresh fixative at 4 °C overnight. They were embedded with 10% gelatin, cryoprotected with 2.3M Polyvinylpyrrolidone (PVP; M.W. 10,000; Sigma)/sucrose (Sigma) in 0.2M HEPES, pH 7.2, and frozen in liquid nitrogen for cryoultramicrotomy. 70 to 80 nm frozen sections were picked up with mixture of 1.15M PVP/sucrose, 1% methyl cellulose (Sigma), 0.2% uranyl acetate (EMS), and 0.1% glutatraldehyde, transferred to 200 mesh Butvar® coated nickel grids (EMS), and stored at −20 °C until they were ready for immunogold labeling. To localize SFTPB or SFTPC proteins, thawed frozen sections were stained with rabbit polyclonal Ab directed against mature SFTPB (Seven Hills; Lin et al., 1996) or mature SFTPC (Seven Hills; Ross et al., 1999), and 10 nm protein A gold (CMC). Electron micrographs of labeled cells were acquired using a Hitachi TEM 7650 (Hitachi High Technologies America) with an AMT CCD camera (Advanced Microscopy Techniques).
Other Alveolosphere Staining
Toluidine blue staining was performed as follows: 0.5 μm sections (from EMbed plastic blocks) were collected, dried for 30 min on a hot plate, and stained 30-60 s in 0.5% Toluidine blue + 0.5% Borax in dH20, rinsed in dH20, dried and coverslipped. PAS staining was performed according to manufacturer’s instructions using the Sigma PAS kit.
Western Blot Analysis
Cultured cells were treated with lysis buffer (RIPA buffer and 1× Roche Complete Protease Inhibitor cocktail). Buffer-treated cells were removed from the well, incubated on ice for 30 min, and cleared by centrifugation at 15,000 g for 20min. Supernatants were collected and stored at −80 °C until analysis. Protein was measured using the Bio-Rad DC Protein Assay. A total of 35 μg of alveolo-sphere lysate and 25 μg of lysate from AECs isolated from lung explants of 21 wk human lung cultured for 6 d in DCI were resolved on pre-cast 10% NUPAGE gels (Thermo Fisher) and transferred to PVDF membrane (Bio-Rad). Blots were incubated with the following primary antisera: surfactant protein B (PT3, a rabbit polyclonal antibody against bovine mature SP-B; Beers et al., 1992; 1:3000 dilution); NFLANK (rabbit polyclonal antibody against a synthetic peptide of Gln186-Gln200 of the human Pro-SPB amino acid sequence; dilution 1:5000; Korimilli et al., 2000; 1:2000); GAPDH (1:5000, Chemicon), NPRO-SFTPC (dilution 1:3000, Beers et al., 1994), b-actin (dilution 1:10000, Sigma). Species-specific secondary antisera were all conjugated to IR dyes of either 680 or 800 nm wavelengths (Rockland) at a dilution of 1:10000. Visualization was accomplished using the Odyssey Imaging System (LiCOR Biosciences).
Lipidomic Analysis
Alveolospheres were dissociated from matrigel as described above, and incubated in 1ml of trypsin for 5 min to break apart the alveolospheres but leave cells intact. After centrifugation, the trypsin “extracellular sample” was separated from the cell pellet “intracellular sample,” the cell pellet was washed in PBS, and both samples were stored at −80C until ready to process.
For lipid extraction, a modified Bligh & Dyer protocol was used with an internal standard of 14:0 PC (DMPC) from Avanti Polar Lipids). For extracellular samples, we used 500ngPC (0.738nmol) per sample, and for intracellular samples, we used 2000ng PC (2.95nmol) PC per sample. All reagents (water, methanol, chloroform) used were HPLC grade.
Internal standard was added to each disposable glass culture tube prior to addition of a sample. Each cell pellet and extracellular supernatant was resuspended in 1ml water, transferred to a tube, and 3ml methanol:chloroform (2:1) was added, intracellular samples were sonicated for 30 s, and extracelullar samples were vortexed for 30 s. 1ml chloroform and 1ml water were added to each sample, followed by 30 s of vortexing. Samples were centrifuged for 5-10 min at 1500 g and the bottom layer was collected with a glass pasteur pipet and dried under nitrogen gas. Intracellular samples were resuspended in 300 μL methanol, and extracellular samples were resuspended in 100 μL methanol.
Electrospray ionization (ESI)/tandem mass spectrometry and gas chromatography/mass spectrometry (GC/MS) were used, respectively, to measure phosphatidylcholine (PC) composition in the samples. Results are described as “Absolute Quantification” based on alveolosphere DNA amount in ng or as “Relative Quantification” (Absolute Quant of each individual PC species, such as PC 32:0 divided by total acyl PC).
Isolation of Primary AECs
Week 21 human lung tissues were obtained in the Guttentag laboratory under protocols originally reviewed by the Institutional Review Board at the Children’s Hospital of Philadelphia and subsequently reviewed by Vanderbilt University and in the Beers laboratory under a University of Pennsylvania Institutional Review Board exemption and isolated as previously published. The cell stocks used in the present studies were donated to the Kotton laboratory for the purpose of providing reference data. “Week 21” samples were isolated by the overnight culture of lung explants in Waymouth media, a technique that generally yields 86 ± 2% epithelial cells with the remaining cells consisting of fibroblasts with < 1% endothelial cells. “Week 21 DCI” samples were prepared in a similar manner except that the lung explants were also cultured for 4 days in Waymouth’s media supplemented with DCI (10 nM Dexameth-asone, 0.1mM 8-Br cAMP, and 0.1mM 3-isobutyl-1-1methylxanthine), and “HFL DCI-D6” samples were cultured in this media for 6 days. Week 21 epithelial cells do not exhibit features of alveolar type 2 cells including lamellar bodies, whereas Week 21 DCI epithelial cells do exhibit lamellar bodies (Wade et al., 2006; Gonzales et al., 2002).
Isolation of Adult AEC2s
For human primary lung epithelial isolation, 1×1cm pieces of distal human lung obtained from healthy regions of the upper lobe of non-utilized human lungs donated for transplantation were dissected and all airway tissue and pleura was removed. The tissue was then digested using dispase, collagenase I, and DNase using the gentleMACS dissociator (Miltenyi) for 30 min at 37 °C. The resulting cell suspension was passed over 70 μm and 40 μm filters to generate a single cell suspension. Purified human AEC2 cells were obtained by magnetic bead sorting using MACS LS columns (Miltenyi) and the following antibodies: HT2-280 (anti-human AEC2 antibody, IgM, Terrace Biotechnologies) and anti-IgM magnetic beads (Miltenyi). MACS-sorted cells were collected into trizol.
Mesenchymal Co-culture Experiments
For 3D experiments with and without supportive fibroblasts, primary AEC2 cells were obtained via bead sorting for HT2-280 expression as described above. Cells were cultured in a mix of 50% matrigel and 50% SAGM media (Lonza) CK+DCI media in 24 well trans-well inserts, with additional media provided in the lower chamber and changed every 48h. For experiments including supportive fibroblasts, low passage (< passage 10) human lung fibroblasts (MRC5 cells, ATCC, line CCL-171) were added at a 10 fibroblasts to 1 epithelial cell ratio. Cultures were maintained for 2 weeks, and colony forming efficiency was calculated by counting colonies of > 8 visible cells using a 60x objective, and dividing by the number of input epithelial cells. Statistical comparisons were done by ANOVA with follow-up pairwise comparisons done using the method of Bonferroni to control for multiple comparison.
RNA Sequencing
The following samples were harvested from RUES2 PSCs in Qiazol (QIAGEN) for RNA Sequencing analysis. (1) Day 0 samples representing undifferentiated PSCs cultured feeder-free in mTeSR1 media, as described above. (2) Day 15 samples representing CD47hi/CD26lo sorted lung progenitor. Flow cytometry analysis confirmed that this population consisted of between 85%–90% NKX2-1+ cells. (3) Day 35 SFTPCtdTomato+ (Tom+) and SFTPCtdTomato− (Tom−) samples resulting from the outgrowth of the day 15 sorted progenitors. Other samples were harvested from primary cells: (1) Week 21 human fetal distal lung cells (as described above), (2) Week 21 human fetal distal lung cells, cultured in “DCI” media for 4 days (as described above), and (3) Adult AEC2s purified by HT2-280-based sorting (as described above). Sequencing libraries were prepared from total RNA samples using Illumina TruSeq RNA Sample Preparation Kit v2. The mRNA was isolated using magnetic beads-based poly(A) selection, fragmented, and randomly primed for reverse transcription, followed by second-strand synthesis to create double-stranded cDNA fragments. These cDNA fragments were then end-repaired, added with a single ‘A’ base, and ligated to Illumina® Paired-End sequencing adapters. The products were purified and PCR-amplified to create the final cDNA library. The libraries from individual samples were pooled in groups of four for cluster generation on the Illumina cBot using Illumina TruSeq Paired-End Cluster Kit. Each sample was sequenced four per lane on the Illumina HiSeq 2500 to generate more than 30 million single end 100-bp reads.
Reverse Transcriptase Quantitative PCR
RT-qPCR was performed as previously described (Hawkins et al., 2017). Briefly, RNA was isolated according to manufacturer’s instructions using the QIAGEN miRNeasy mini kit (QIAGEN). cDNA was generated by reverse transcription of up to 150ng RNA from each sample using the Applied Biosystems High-Capacity cDNA Reverse Transcription Kit. For qPCR, technical triplicates of either 20 μL reactions (for use in Applied Biosystems StepOne 96-well System) or 12 μL reactions (for use in Applied Biosystems QuantStudio7 384-well System) were prepared with 2 μL of diluted or undiluted cDNA and run for 40 cycles. All primers were TaqMan probes from Applied Biosystems (see all in Key Resources Table). Relative gene expression was calculated based on the average cycle (Ct) value of the technical triplicates, normalized to 18S control, and reported as fold change (2(−ΔΔCT)), with a fold change of 1 being assigned to undifferentiated (day 0) iPSCs or ESCs. Undetected probes were assigned a Ct value of 40 to allow for fold change calculations.
Immunofluorescence Microscopy of Cultured Cells
Alveolospheres were dissociated as described above, washed in PBS, and fixed with 4% paraformaldehyde. Alveolospheres were subsequently either processed for cryosectioning or whole-mount staining. For cryosectioning, fixed alveolospheres were first embedded in low melting temperature agarose (SeaPrep). After incubation in 7.5% and 30% sucrose solution, they were further embedded in OCT and flash frozen. 6 μm sections were cut on a cryotome. Both whole mount alveolospheres and frozen sections were washed in PBS, blocked in 4% normal donkey serum (NDS) with 0.5% Triton X-100 (Sigma) for 30 min, and incubated overnight in primary antibody (see Key Resources Table) in 0.5% Triton X-100 and 4% NDS. Samples were then washed in 4% NDS and incubated with secondary antibody from Jackson Immunoresearch (1:300 anti rabbit IgG (H+L) or anti mouse IgG (H+L)) for 2 hr at room temperature. Nuclei were stained with Hoechst dye (Thermo Fisher, 1:500) and sections were mounted with Prolong Diamond Anti-Fade Mounting Reagent (ThermoFisher) and coverslipped, while whole mount alveolospheres were mounted on cavity slides (Eisco). Both stained whole mount and cryosectioned alveolospheres were visualized with a Zeiss LSM 880 laser scanning confocal microscope.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical Methods
In figures containing RT-qPCR, flow cytometry, or lipidomics data, data was presented as the mean with error bars representing the standard deviation from the mean. Unpaired, two-tailed Student’s t tests were performed on 2 groups of n R 3 replicates each, and the p value threshold to determine significance was set at p = 0.05. Replicates generally represent samples differentiated separately from Day 0, though in some cases they represent separate sorted populations from the same differentiation.
Immunogold Quantification
To determine if immunogold stained mature SFTPB and mature SFTPC localized to specific cellular compartments in PSC-derived cells, gold counts registered on biosynthetic and non-biosynthetic compartments were tabulated and analyzed by relative labeling index (RLI) described by Mayhew (Mayhew, 2011). Briefly, non-destructive counting grids generated by the grid plugin under FIJI was randomly superimposed over the acquired electron micrograph at magnification of 15,000×. Gold counts and sampled grid points registered on the compartments of interest, i.e., multivesicular bodies/lamellar bodies, were collected for estimation of the expected gold counts for selected compartments after normalization to surface areas of selected compartment to total cell surface areas. To determine if gold labeling was specific to the compartments of interests, observed counts (n = 1160 and 2510 total gold particles for SFTPB and SFTPC, respectively) were compared with expected counts by χ2 statistics and contingency table analyses. Any cellular compartment that had RLI > 1 (p < 0.05) and significantly higher partial χ2 value compared to other compartments was considered labeled preferentially by SFTPB or SFTPC antibodies (25% of total χ2 value was arbitrarily chosen for this study).
RNA Sequencing
Fastq files were assessed for quality control using the FastQC program. Fastq files were aligned against the human reference genome (hg19/hGRC37) using the STAR aligner (Dobin et al., 2013). Duplicate reads were flagged using the MarkDuplicates program from Picard tools. Gene counts represented as counts per million (CPM) were computed for Ensembl (v67) gene annotations using the Rsubread R package with duplicate reads removed. Genes with 10% of samples having a CPM < 1 were removed and deemed low expressed. The resultant data was transformed using the VOOM method implemented in limma R package (Law et al., 2014). Voom transformed data was then tested for differential gene expression using standard linear models using the limma package. Multiple hypothesis test correction was performed using the Benjamini–Hochberg procedure (FDR). Heatmaps and PCA plots were generated in R. Gene Set Enrichment Analysis (GSEA) was performed using the camera method implemented on the limma package using gene sets from the Molecular Signatures database (MSigDB).
DATA AND SOFTWARE AVAILABILITY
The accession number for the raw fastq files and VOOM transformed gene expression files reported in this paper is GEO: GSE96642 and is available as well on the Kotton Lab’s Bioinformatics Portal at http://www.kottonlab.com.
Supplementary Material
In Brief.
Jacob et al. differentiate human pluripotent stem cells (PSCs) into type II alveolar cells (iAEC2s). They find that iAEC2s display many of the functions, transcriptomic features, and surfactantprocessing capacities that characterize primary cells. Finally, they derive AEC2s from gene-edited, patient-specific PSCs to model surfactant protein B deficiency in vitro.
Highlights.
SFTPC reporter PSC lines allow isolation of putative PSC-derived AEC2s (iAEC2s)
iAEC2s extensively self-renew, growing as proliferative alveolospheres
Lamellar bodies present in iAEC2s are able to functionally process surfactant
Temporal regulation of Wnt activity promotes PSC-derived AEC2 maturation
Acknowledgments
We dedicate this manuscript to our mentor, teacher, and friend, Dr. Mary C. Williams of Boston University School of Medicine (BUSM), whose work deciphering alveolar epithelial cell biology paved the way for the current project. We wish to thank all members of the Kotton lab for insightful discussions and Drs. Robin Deterding, Larry Nogee, and Aaron Hamvas for guidance and encouragement to pursue our shared mission of advancing research related to pediatric lung disease. We are grateful to Brian R. Tilton of the BUSM Flow Cytometry Core for technical assistance, supported by NIH Grant 1UL1TR001430, and Drs. Greg Miller, Amulya Iyer, and Marianne James of the CReM, supported by grants R24HL123828 and U01TR001810. We thank Adam Gower and Avrum Spira for RNA sequencing. We thank Don Gantz and Lars Knudsen for assistance with electron microscopy. This work was supported by Cystic Fibrosis Foundation grants HAWKIN15XX0 (to F.H.) and DAVISGO (to B.R.D.) and NIH grants TL1TR001410 and F31HL134274 (to A.J.); F31HL129777 (to K.B.M.); HL059959 (to S.H.G.); DAVIS15XX1 (to B.R.D.); R33HL120760 (to F.S.C., J.A.W., and H.H.); R01 HL119436 (to M.F.B.); U01HL099997 and U01HL134745 (to E.E.M., J.A.W., and D.N.K.); and R01HL095993, R01HL122442, R01HL128172, and U01HL134766 (to D.N.K.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and one movie and can be found with this article online at http://dx.doi.org/10.1016/j.stem.2017.08.014.
AUTHOR CONTRIBUTIONS
A.J. and D.N.K. designed the project, performed experiments, and wrote the manuscript. A.J., F.H., K.B.M., K.H., and M.V. performed differentiation experiments. J.C.J. performed the reprogramming and SFTPB gene-editing experiments. J.C.J., A.J., A.C., and B.R.D. designed and targeted the fluoro-chrome-reporter iPSC lines. A.H. and M.O. performed TEM. M.M., W.Z., and E.E.M. provided adult primary samples and performed the RNA-seq analysis. S.J.R., S.K., S.H.G., and M.F.B. performed protein immunoblots and provided primary fetal lung samples. K.T. and L.J.Q. performed the cytokine analysis. H.H., J.W., and F.S.C. performed the lipidomic analysis. C.-L.N., T.E.W., and J.A.W. performed immunogold labeling. F.V.W. and J.W. provided patient samples.
ADDITIONAL RESOURCES
Further protocol information for reprogramming, iPSC/ESC cultures, directed differentiation and the production of lentiviral particles can be found at http://www.bu.edu/dbin/stemcells/protocols.php.
References
- Ballard PL, Lee JW, Fang X, Chapin C, Allen L, Segal MR, Fischer H, Illek B, Gonzales LW, Kolla V, Matthay MA. Regulated gene expression in cultured type II cells of adult human lung. Am J Physiol Lung Cell Mol Physiol. 2010;299:L36–L50. doi: 10.1152/ajplung.00427.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkauskas CE, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, Randell SH, Noble PW, Hogan BLM. Type 2 alveolar cells are stem cells in adult lung. J Clin Invest. 2013;123:3025–3036. doi: 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beers MF, Bates SR, Fisher AB. Differential extraction for the rapid purification of bovine surfactant protein B. Am J Physiol. 1992;262:L773–L778. doi: 10.1152/ajplung.1992.262.6.L773. [DOI] [PubMed] [Google Scholar]
- Beers MF, Kim CY, Dodia C, Fisher AB. Localization, synthesis, and processing of surfactant protein SP-C in rat lung analyzed by epitope-specific antipeptide antibodies. J Biol Chem. 1994;269:20318–20328. [PubMed] [Google Scholar]
- Beers MF, Hamvas A, Moxley MA, Gonzales LW, Guttentag SH, Solarin KO, Longmore WJ, Nogee LM, Ballard PL. Pulmonary surfactant metabolism in infants lacking surfactant protein B. Am. J Respir Cell Mol Biol. 2000;22:380–391. doi: 10.1165/ajrcmb.22.3.3645. [DOI] [PubMed] [Google Scholar]
- Boggaram V. Thyroid transcription factor-1 (TTF-1/Nkx2.1/TITF1) gene regulation in the lung. Clin Sci. 2009;116:27–35. doi: 10.1042/CS20080068. [DOI] [PubMed] [Google Scholar]
- Borok Z, Danto SI, Lubman RL, Cao Y, Williams MC, Crandall ED. Modulation of t1alpha expression with alveolar epithelial cell phenotype in vitro. Am J Physiol. 1998;275:L155–L164. doi: 10.1152/ajplung.1998.275.1.L155. [DOI] [PubMed] [Google Scholar]
- Brasch F, Johnen G, Winn-Brasch A, Guttentag SH, Schmiedl A, Kapp N, Suzuki Y, Müller KM, Richter J, Hawgood S, Ochs M. Surfactant protein B in type II pneumocytes and intra-alveolar surfactant forms of human lungs. Am J Respir Cell Mol Biol. 2004;30:449–458. doi: 10.1165/rcmb.2003-0262OC. [DOI] [PubMed] [Google Scholar]
- Crane AM, Kramer P, Bui JH, Chung WJ, Li XS, Gonzalez-Garay ML, Hawkins F, Liao W, Mora D, Choi S, et al. Targeted correction and restored function of the CFTR gene in cystic fibrosis induced pluripotent stem cells. Stem Cell Reports. 2015;4:569–577. doi: 10.1016/j.stemcr.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai TJ, Brownfield DG, Krasnow MA. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature. 2014;507:190–194. doi: 10.1038/nature12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbs LG, Williams MC, Gonzalez R. Monoclonal antibodies specific to apical surfaces of rat alveolar type I cells bind to surfaces of cultured, but not freshly isolated, type II cells. Biochim Biophys Acta. 1988;970:146–156. doi: 10.1016/0167-4889(88)90173-5. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye BR, Hill DR, Ferguson MAH, Tsai YH, Nagy MS, Dyal R, Wells JM, Mayhew CN, Nattiv R, Klein OD, et al. In vitro generation of human pluripotent stem cell derived lung organoids. eLife. 2015;4:e05098. doi: 10.7554/eLife.05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster CD, Varghese LS, Skalina RB, Gonzales LW, Guttentag SH. In vitro transdifferentiation of human fetal type II cells toward a type I-like cell. Pediatr Res. 2007;61:404–409. doi: 10.1203/pdr.0b013e3180332c6d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DB, Peng T, Zepp JA, Snitow M, Vincent TL, Penkala IJ, Cui Z, Herriges MJ, Morley MP, Zhou S, et al. Emergence of a Wave of Wnt Signaling that Regulates Lung Alveologenesis by Controlling Epithelial Self-Renewal and Differentiation. Cell Rep. 2016;17:2312–2325. doi: 10.1016/j.celrep.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales LW, Guttentag SH, Wade KC, Postle AD, Ballard PL. Differentiation of human pulmonary type II cells in vitro by glucocorticoid plus cAMP. Am J Physiol Lung Cell Mol Physiol. 2002;283:L940–L951. doi: 10.1152/ajplung.00127.2002. [DOI] [PubMed] [Google Scholar]
- Gotoh S, Ito I, Nagasaki T, Yamamoto Y, Konishi S, Korogi Y, Matsumoto H, Muro S, Hirai T, Funato M, et al. Generation of alveolar epithelial spheroids via isolated progenitor cells from human pluripotent stem cells. Stem Cell Reports. 2014;3:394–403. doi: 10.1016/j.stemcr.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttentag SH, Beers MF, Bieler BM, Ballard PL. Surfactant protein B processing in human fetal lung. Am J Physiol. 1998;275:L559–L566. doi: 10.1152/ajplung.1998.275.3.L559. [DOI] [PubMed] [Google Scholar]
- Hawkins F, Kramer P, Jacob A, Driver I, Thomas DC, McCauley KB, Skvir N, Crane AM, Kurmann AA, Hollenberg AN, et al. Prospective isolation of NKX2-1-expressing human lung progenitors derived from pluripotent stem cells. J Clin Invest. 2017;127:2277–2294. doi: 10.1172/JCI89950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SXL, Islam MN, O’Neill J, Hu Z, Yang YG, Chen YW, Mumau M, Green MD, Vunjak-Novakovic G, Bhattacharya J, Snoeck HW. Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat Biotechnol. 2014;32:84–91. doi: 10.1038/nbt.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalina M, Mason RJ, Shannon JM. Surfactant protein C is expressed in alveolar type II cells but not in Clara cells of rat lung. Am J Respir Cell Mol Biol. 1992;6:594–600. doi: 10.1165/ajrcmb/6.6.594. [DOI] [PubMed] [Google Scholar]
- Khoor A, Stahlman MT, Gray ME, Whitsett JA. Temporalspatial distribution of SP-B and SP-C proteins and mRNAs in developing respiratory epithelium of human lung. J Histochem Cytochem. 1994;42:1187–1199. doi: 10.1177/42.9.8064126. [DOI] [PubMed] [Google Scholar]
- Kikkawa Y, Yoneda K, Smith F, Packard B, Suzuki K. The type II epithelial cells of the lung. II. Chemical composition and phospholipid synthesis. Lab Invest. 1975;32:295–302. [PubMed] [Google Scholar]
- Korimilli A, Gonzales LW, Guttentag SH. Intracellular localization of processing events in human surfactant protein B biosynthesis. J Biol Chem. 2000;275:8672–8679. doi: 10.1074/jbc.275.12.8672. [DOI] [PubMed] [Google Scholar]
- Kurmann AA, Serra M, Hawkins F, Rankin SA, Mori M, Astapova I, Ullas S, Lin S, Bilodeau M, Rossant J, et al. Regeneration of Thyroid Function by Transplantation of Differentiated Pluripotent Stem Cells. Cell Stem Cell. 2015;17:527–542. doi: 10.1016/j.stem.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Akinbi HT, Breslin JS, Weaver TE. Structural requirements for targeting of surfactant protein B (SP-B) to secretory granules in vitro and in vivo. J Biol Chem. 1996;271:19689–19695. doi: 10.1074/jbc.271.33.19689. [DOI] [PubMed] [Google Scholar]
- Liu Y, Hogan BLM. Differential gene expression in the distal tip endoderm of the embryonic mouse lung. Gene Expr Patterns. 2002;2:229–233. doi: 10.1016/s1567-133x(02)00057-1. [DOI] [PubMed] [Google Scholar]
- Mason RJ, Williams MC. Phospholipid composition and ultra-structure of A549 cells and other cultured pulmonary epithelial cells of presumed type II cell origin. Biochim Biophys Acta. 1980;617:36–50. doi: 10.1016/0005-2760(80)90222-2. [DOI] [PubMed] [Google Scholar]
- Mayhew TM. Quantifying immunogold localization on electron microscopic thin sections: a compendium of new approaches for plant cell biologists. J Exp Bot. 2011;62:4101–4113. doi: 10.1093/jxb/err176. [DOI] [PubMed] [Google Scholar]
- McCauley KB, Hawkins F, Serra M, Thomas DC, Jacob A, Kotton DN. Efficient Derivation of Functional Human Airway Epithelium from Pluripotent Stem Cells via Temporal Regulation of Wnt Signaling. Cell Stem Cell. 2017;20:844–857.e6. doi: 10.1016/j.stem.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoo P, Su G, Drum H, Bringas P, Kimura S. Defects in tracheoesophageal and lung morphogenesis in Nkx2.1(−/−) mouse embryos. Dev Biol. 1999;209:60–71. doi: 10.1006/dbio.1999.9234. [DOI] [PubMed] [Google Scholar]
- Mucenski ML, Nation JM, Thitoff AR, Besnard V, Xu Y, Wert SE, Harada N, Taketo MM, Stahlman MT, Whitsett JA. Beta-catenin regulates differentiation of respiratory epithelial cells in vivo. Am J Physiol Lung Cell Mol Physiol. 2005;289:L971–L979. doi: 10.1152/ajplung.00172.2005. [DOI] [PubMed] [Google Scholar]
- Nogee LM, Garnier G, Dietz HC, Singer L, Murphy AM, deMello DE, Colten HR. A mutation in the surfactant protein B gene responsible for fatal neonatal respiratory disease in multiple kindreds. J Clin Invest. 1994;93:1860–1863. doi: 10.1172/JCI117173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinton LJ, Jones MR, Simms BT, Kogan MS, Robson BE, Skerrett SJ, Mizgerd JP. Functions and regulation of NF-kappaB RelA during pneumococcal pneumonia. J Immunol. 2007;178:1896–1903. doi: 10.4049/jimmunol.178.3.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinton LJ, Jones MR, Robson BE, Simms BT, Whitsett JA, Mizgerd JP. Alveolar epithelial STAT3, IL-6 family cytokines, and host defense during Escherichia coli pneumonia. Am J Respir Cell Mol Biol. 2008;38:699–706. doi: 10.1165/rcmb.2007-0365OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin SA, Han L, McCracken KW, Kenny AP, Anglin CT, Grigg EA, Crawford CM, Wells JM, Shannon JM, Zorn AM. A Retinoic Acid-Hedgehog Cascade Coordinates Mesoderm-Inducing Signals and Endoderm Competence during Lung Specification. Cell Rep. 2016;16:66–78. doi: 10.1016/j.celrep.2016.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridsdale R, Post M. Surfactant lipid synthesis and lamellar body formation in glycogen-laden type II cells. Am J Physiol Lung Cell Mol Physiol. 2004;287:L743–L751. doi: 10.1152/ajplung.00146.2004. [DOI] [PubMed] [Google Scholar]
- Ridsdale R, Na CL, Xu Y, Greis KD, Weaver T. Comparative proteomic analysis of lung lamellar bodies and lysosome-related organelles. PLoS ONE. 2011;6:e16482. doi: 10.1371/journal.pone.0016482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross GF, Ikegami M, Steinhilber W, Jobe AH. Surfactant Protein C in Fetal and Ventilated Preterm Rabbit Lungs. Am J Physiol. 1999;277:L1104–L1108. doi: 10.1152/ajplung.1999.277.6.L1104. [DOI] [PubMed] [Google Scholar]
- Shu W, Guttentag S, Wang Z, Andl T, Ballard P, Lu MM, Piccolo S, Birchmeier W, Whitsett JA, Millar SE, Morrisey EE. Wnt/beta-catenin signaling acts upstream of N-myc, BMP4, and FGF signaling to regulate proximal-distal patterning in the lung. Dev Biol. 2005;283:226–239. doi: 10.1016/j.ydbio.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Somers A, Jean JC, Sommer CA, Omari A, Ford CC, Mills JA, Ying L, Sommer AG, Jean JM, Smith BW, et al. Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells. 2010;28:1728–1740. doi: 10.1002/stem.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade KC, Guttentag SH, Gonzales LW, Maschhoff KL, Gonzales J, Kolla V, Singhal S, Ballard PL. Gene induction during differentiation of human pulmonary type II cells in vitro. Am J Respir Cell Mol Biol. 2006;34:727–737. doi: 10.1165/rcmb.2004-0389OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wert SE, Glasser SW, Korfhagen TR, Whitsett JA. Transcriptional elements from the human SP-C gene direct expression in the primordial respiratory epithelium of transgenic mice. Dev Biol. 1993;156:426–443. doi: 10.1006/dbio.1993.1090. [DOI] [PubMed] [Google Scholar]
- Whitsett JA, Wert SE, Weaver TE. Diseases of pulmonary surfactant homeostasis. Annu Rev Pathol. 2015;10:371–393. doi: 10.1146/annurev-pathol-012513-104644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MC, Mason RJ. Development of the type II cell in the fetal rat lung. Am Rev Respir Dis. 1977;115:37–47. doi: 10.1164/arrd.1977.115.S.37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


