SUMMARY
STIM1 and STIM2 are ER membrane proteins that sense decreases in ER-luminal free Ca2+ and—through a conformational change in the STIM cytoplasmic domain— control gating of the plasma membrane Ca2+ channel ORAI1. To determine how STIM1 conveys a signal from the ER lumen to the cytoplasm, we have studied the Ca2+-dependent conformational change of engineered STIM1 proteins in isolated ER membranes and, in parallel, physiological activation of these proteins in cells. We find that conserved ‘sentinel’ features of the CC1 region help to prevent activation while Ca2+ is bound to STIM ER-luminal domains. Reduced ER-luminal Ca2+ drives a concerted conformational change, in which STIM luminal domains rearrange and the STIM transmembrane helices and initial parts of the CC1 regions pair in an extended coiled coil. This intradimer rearrangement overcomes the relatively weak CC1-SOAR/CAD interactions that hold STIM in an inactive conformation, releasing the SOAR/CAD domain to activate ORAI channels.
INTRODUCTION
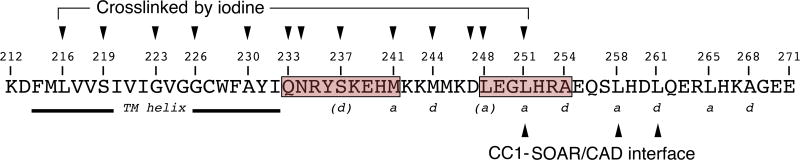
STIM proteins are ER membrane proteins that regulate store-operated Ca2+ entry (Roos et al., 2005; Liou et al., 2005). This pathway is triggered during physiological signalling by the lowering of free Ca2+ concentration in the ER lumen, which causes STIM to relocate within the ER membrane to sites where the ER comes into close apposition to the plasma membrane. Once there, STIM recruits and gates plasma membrane ORAI calcium channels (reviewed in Soboloff et al., 2012; Prakriya and Lewis, 2015; Derler et al., 2016). STIM1 monitors ER Ca2+ levels and gates ORAI channels using separate protein modules— an ER-luminal domain (residues 23–213 in human STIM1) senses Ca2+ (Liou et al., 2005; Zhang et al., 2005; Mercer et al., 2006; Spassova et al., 2006), and the SOAR/CAD domain (‘STIM1 ORAI activating region’, residues 344–442; or ‘CRAC activation domain’, residues 342–448) interacts with and gates ORAI channels (Yuan et al., 2009; Park et al., 2009; Kawasaki et al., 2009). These two functional modules of STIM1 are connected by the conserved STIM1 transmembrane helices (residues 214–232) and the cytoplasmic CC1 region (‘predicted coiled coil 1’, residues 233–343) (Figure 1).
Figure 1. Structural hypotheses on STIM1 activation tested in this work.
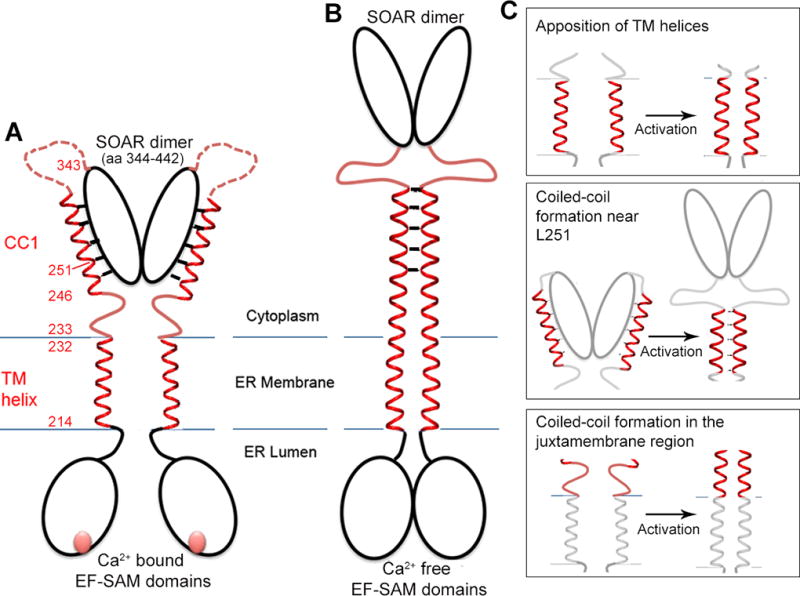
A, Cartoon of the inactive STIM1 dimer. The EF-SAM domains comprise residues 58–201 of each monomer; the transmembrane (TM) helices, residues 214–232; the CC1 regions, residues 233–343; and SOAR/CAD, residues 344–442. The CC1-SOAR/CAD interface of inactive STIM1 is portrayed as spanning CC1 residues L248–L261, based on literature cited in the text. The cartoon depicts one possible configuration of inactive STIM1, in which the Ca2+-bound STIM1 EF-SAM domains do not interact and the transmembrane helices are spatially separated. An alternative configuration in which the luminal domains in cells are stabilized in close proximity, but the transmembrane helices are held apart, would yield similar data in our experimental tests. B, Cartoon of the hypothesized active state of STIM1. The defining feature of active STIM1 is the release of SOAR/CAD from CC1. It has been proposed that SOAR/CAD release is triggered by burial of L251 and surrounding residues of the CC1-SOAR/CAD interface in the core of a CC1-CC1 coiled coil (Zhou et al., 2013). C, The three hypotheses tested— Upper, that the TM helices come together upon activation; Middle, that activation coincides with the formation of a coiled-coil in the region of CC1 containing L251; Lower, that activation entails helical apposition along the entire segment connecting the TM helices and the CC1-SOAR/CAD interface surrounding L251. The STIM1 mutants examined and their phenotypes are summarized in Table S1.
Ca2+ sensing is assigned to the EF-hand in the STIM luminal domain based on targeted mutations in the EF-hand that render STIM insensitive to Ca2+ (Liou et al., 2005; Zhang et al., 2005; Mercer et al., 2006; Spassova et al., 2006). The isolated STIM1 and STIM2 luminal domains with Ca2+ bound are monomeric, and their structures have been determined (Stathopulos et al., 2008; Zheng et al., 2011). When the Ca2+ concentration is lowered below levels typical for the ER lumen of a resting cell, isolated STIM luminal domains undergo a conformational change— which has not yet been defined— and dimerize (Stathopulos et al., 2006; Stathopulos et al., 2008; Furukawa et al., 2014). For simplicity of expression, we will refer to the corresponding change in the luminal domains of full-length STIM as ‘dimerization’, although current evidence is equally consistent with a reorientation of luminal domains that are already in physical contact. Dimerization of luminal domains explains, in part, the STIM-STIM FRET that can be observed in cells upon depletion of ER Ca2+ stores. A further higher-order oligomerization through the SOAR/CAD domains may also contribute to STIM-STIM FRET in cells (discussed critically in Prakriya and Lewis, 2015). Dimerization (or oligomerization) is on the direct path to physiological signalling, since the STIM-STIM FRET change precedes movement of STIM to ER-plasma membrane junctions (Liou et al., 2007; Muik et al., 2008; Covington et al., 2010), and, conversely, artificial dimerization (or oligomerization) of an engineered STIM1 drives its movement to ER-plasma membrane junctions and its gating of ORAI current (Luik et al., 2008). Note, though, that only luminal-domain dimerization, and not higher-order oligomerization, is intrinsic to the initial activation step investigated here (Zhou et al., 2013).
Dimerization restricted to the ER lumen is only a first step in communication between the ER lumen and plasma membrane ORAI channels. It was recognized early that loss of bound Ca2+ from the luminal domain is most likely coupled to a broader conformational change in STIM (Liou et al., 2005). With increased understanding of STIM targeting and interactions, the notion was refined into the proposals that a conformational change in the STIM cytoplasmic domain exposes the polybasic plasma-membrane targeting motif (residues 671–685), or the ORAI-interacting SOAR/CAD domain, or both (Liou et al., 2007; Luik et al., 2008; Park et al., 2009; Korzeniowski et al., 2010).
Several threads of evidence have pointed to the conclusion that the initial part of CC1 holds SOAR/CAD in an inactive state in resting cells (Korzeniowski et al., 2010; Muik et al., 2011; Yang et al., 2012; Zhou et al., 2013; Yu, F. et al., 2013; Yu, J. et al., 2013; Cui et al., 2013; Fahrner et al., 2014; Ma et al., 2015; Korzeniowski et al., 2017; Ma et al., 2017). STIM1(L251S) activates ORAI1 channels independently of store depletion, and STIM1(233–474) carrying the L251S substitution is physically extended compared to wildtype STIM1(233–474) (Muik et al., 2011). Moreover, the isolated STIM1 cytoplasmic domain (STIM1(233–685), abbreviated STIM1CT) is folded back on itself by a CC1-SOAR/CAD interaction in a way that would retain SOAR/CAD near the ER in resting cells, and either the L251S mutation or artificially bringing together the N termini of paired CC1 segments in the wildtype STIM1CT dimer releases the interaction (Zhou et al., 2013). More recently, an engineered STIM1 truncated at residue 261 was shown to tether the SOAR/CAD domain at the ER in resting cells, and to release it in response to store depletion (Ma et al., 2015). Although a second region spanning CC1 residues 310–337 might have an auxiliary role in masking SOAR/CAD in resting cells (Korzeniowski et al., 2010; Yang et al., 2012; Yu, F. et al., 2013; Yu, J. et al., 2013; Cui et al., 2013), the CC1 region from residue 233–271 plays a dominant role in retaining SOAR/CAD near the ER, and thereby keeps SOAR/CAD inactive in gating ORAI channels (Muik et al., 2011; Zhou et al., 2013; Fahrner et al., 2014; Ma et al., 2015).
Based on our experiments with the isolated STIM1 cytoplasmic domain, we proposed a specific mechanism that could link dissociation of Ca2+ from the STIM luminal domain to an activating conformational change in the STIM1 cytoplasmic domain (Zhou et al., 2013). However, a direct experimental test of the proposed mechanism has been lacking. The current study addresses four interrelated questions (Figure 1)— Does a reduction in Ca2+ concentration sensed by the STIM1 luminal domain bring together the STIM1 transmembrane segments? If so, does this intradimer rearrangement have the same Ca2+ concentration dependence as physiological activation of STIM1? Does the STIM1 conformational change sequester the region around residue L251 that retains the SOAR/CAD domain near the ER in a resting cell? And, last, is STIM1 activation accounted for by the specific mechanism of coiled coil formation in the initial portion of CC1?
RESULTS
Lowering Ca2+ concentration results in association of STIM1 transmembrane segments
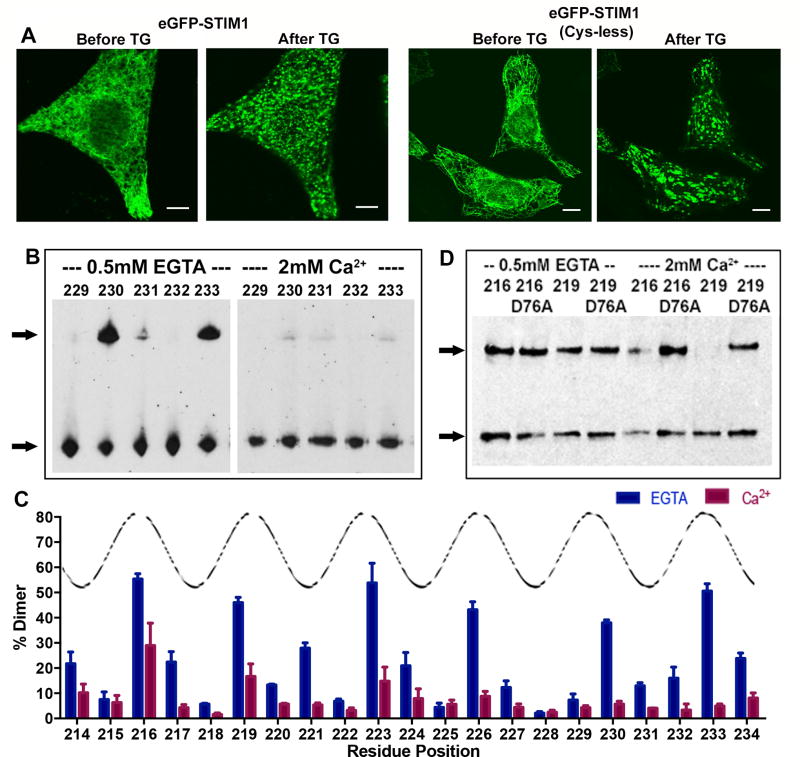
To test whether lowering Ca2+ concentration brings together the transmembrane segments of full-length STIM1, we first verified that engineered cysteineless STIM1 expressed in HeLa cells shows normal ER distribution in store-replete cells and responds normally to ER store depletion (Figure 2A). Then we expressed STIM1 proteins with single engineered cysteine insertions at residues 214–234, isolated HeLa cell membranes, and triggered disulfide crosslinking of STIM1 by addition of iodine to probe the proximity of STIM1 transmembrane helices. Crosslinked dimeric STIM1 was detected by its mobility on a nonreducing SDS-polyacrylamide gel.
Figure 2. Disulfide crosslinking to assess packing of STIM1 transmembrane helices.
A, Confocal micrographs of HeLa cells expressing GFP-tagged wild-type STIM1 or GFP-tagged cysteine-less STIM1, as indicated, at rest (Before TG) and after depleting ER Ca2+ stores by treatment with 1 µM thapsigargin (After TG). Scale bars, 10 µm. B, Western blots showing crosslinking of the specified single-cysteine mutants of STIM1 in isolated cellular membranes incubated in the presence of EGTA or Ca2+. The samples were subjected to nonreducing SDS-polyacrylamide gel electrophoresis, and the blots were probed with anti-STIM1 antibody. Crosslinked STIM1 dimer is marked by the upper arrow, and STIM1 monomer by the lower arrow. C, Crosslinking efficiencies at STIM1 residues 214–234, defined as the percentage of STIM1 in the crosslinked dimer band, in the presence of EGTA (blue) or Ca2+ (magenta). Peaks of the superposed curve define one face of the transmembrane helix. Data from three biological replicates. Error bars report SEM. D, Western blot showing the effect of the activating mutation D76A on crosslinking of STIM1216C and STIM1219C, in the presence of EGTA or Ca2+. See also Figure S1.
STIM1 incubated in the presence of 2 mM Ca2+, to mimic replete stores, shows little crosslinking of most transmembrane residues, with variably some crosslinking at residues 216, 219, and 223 (Figure 2B, 2C, Figure S1). In contrast, STIM1 from membranes incubated in 0.5 mM EGTA, thus essentially in the absence of Ca2+, shows crosslinking in a helical pattern, prominent at residues 216, 219, 223, 226, 230, and 233. Disulfide crosslinking can be affected by the relative orientation of the cysteine sidechains as well as the distance, but the marked increase in crosslinking along the entire length of the transmembrane helices indicates a change in helix-helix proximity. The assay with isolated membranes necessarily exposes both luminal and cytoplasmic domains of STIM1 to Ca2+ or EGTA. To probe whether the low crosslinking in the presence of Ca2+ depends on Ca2+ binding to the STIM1 luminal domain EF-hand, we introduced a D76A replacement in the EF-hand, which renders STIM1 in cells insensitive to physiological changes in Ca2+ (Liou et al., 2005). Correspondingly, the crosslinking of STIM1(D76A) in vitro is insensitive to changes in Ca2+ (Figure 2D). The crosslinking of these D76A proteins in the presence of Ca2+ also rules out the remote possibility that Ca2+ interferes directly with the crosslinking reaction. Additional controls with the cysteineless protein or with the single-cysteine mutants incubated without iodine confirmed that the high crosslinking at residues 216, 219, 223, 226, 230, and 233 in EGTA depends on oxidation of the introduced cysteines (Figure S1).
These results imply that the STIM1 transmembrane segments are not closely associated when luminal Ca2+ is high, and that loss of bound Ca2+ from the STIM luminal domain brings together the transmembrane segments in a way that transmits a signal from the luminal domain to the cytoplasmic end of the transmembrane segments at residues 230 and 233.
Ca2+ concentration dependence of crosslinking at 230C
Next we compared the Ca2+ concentration dependence of the conformational change in vitro with the known Ca2+ concentration dependence of STIM1 activation in cells (Brandman et al., 2007, Luik et al., 2008). Cysteine inserted at residue 230 near the cytosolic side of the ER membrane both gives a reliable increase in crosslinking in EGTA relative to Ca2+ and serves as an indicator of a conformational change that has propagated across the ER membrane. Further, STIM1(A230C) localization in cells follows wildtype STIM1 localization at rest and after store depletion (Figure 3A). Therefore we used the A230C protein to examine the Ca2+ concentration dependence of crosslinking. Membranes were prepared in rigorously Ca2+-free buffer, known concentrations of Ca2+ were added, and iodine crosslinking and analysis were performed as above.
Figure 3. Crosslinking at A230C as a function of Ca2+ concentration.
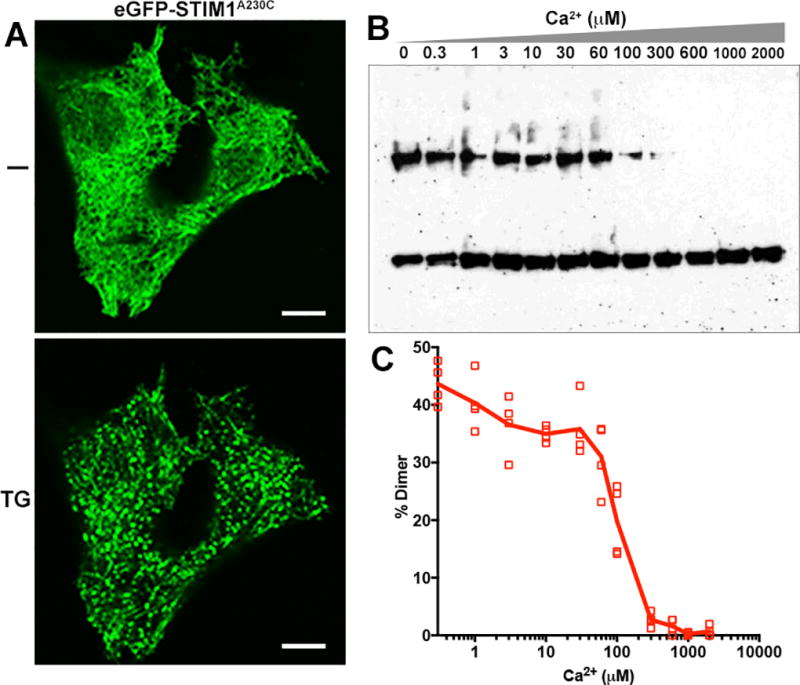
A, Confocal micrographs of HeLa cells expressing GFP-STIM1(A230C), at rest (−) and after store depletion with 1 µM thapsigargin (TG), documenting the response of the A230C protein in cells. Scale bar, 10 µm. B, Crosslinking of STIM1(A230C) in isolated membranes incubated at the specified Ca2+ concentrations, assessed as in Figure 2. C, Efficiency of STIM-STIM dimer formation at each Ca2+ concentration is plotted for four biological replicates.
For these experiments, a modest dilution of the membranes resulted in a higher signal-to-noise ratio than in the earlier series of experiments, with ~40% crosslinking in the presence of EGTA and negligible crosslinking at millimolar concentrations of Ca2+ (Figure 3B, 3C). Inclusion of Ca2+ at concentrations from 1 to 60 µM had at most a marginal effect on the fraction of STIM1 in the dimer band. The crosslinking changed sharply, however, at higher Ca2+ concentrations— 100 µM Ca2+ resulted in a decrease in crosslinking, and 300 µM Ca2+ in almost no crosslinking.
The range over which Ca2+ affects crosslinking in vitro— between 60 µM and 300 µM— agrees well with the range of ER-luminal Ca2+ that controls STIM1 in cells (Brandman et al., 2007; Luik et al., 2008), strengthening the conclusion that the apposition of STIM1 transmembrane helices is physiologically relevant, and supporting use of the crosslinking assay in combination with studies in cells to parse the mechanism that controls STIM1 activation by Ca2+ depletion.
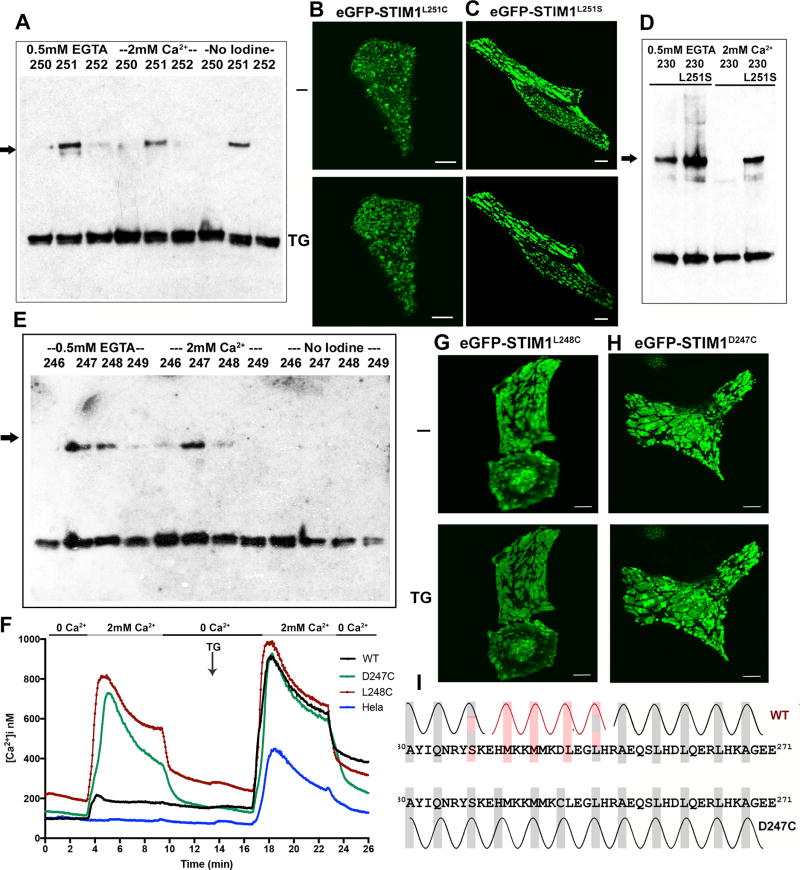
Conformational change extends to residue L251
Residue L251 is implicated in binding and releasing SOAR/CAD (Muik et al., 2011; Zhou et al., 2013), and a specific proposed mechanism for physiological release of SOAR/CAD is that activation of STIM1 buries L251 and other residues of a CC1-SOAR/CAD interface in the core of a partial CC1-CC1 coiled coil (Zhou et al., 2013). We therefore examined STIM1-STIM1 crosslinking at L251C. Consistent with the proposed mechanism, there is prominent crosslinking at 251C in the absence of Ca2+, contrasting with only low levels of crosslinking at positions 250C and 252C (Figure 4A, Figure S2A). There is also considerable crosslinking at 251C in the presence of Ca2+, and there is even substantial background crosslinking before addition of iodine (Figure 4A). The latter can be reversed with more aggressive DTT reduction (not shown). The high background crosslinking could be explained if the L251C mutation favors release of SOAR/CAD from its interaction with CC1, as does the somewhat more polar replacement L251S, and thereby causes activation of STIM even at resting ER Ca2+ levels. This explanation gains independent support from fluorescence microscopy showing that GFP-STIM1(L251C) is partially localized to puncta in resting cells (Figure 4B, Figure S3A). Thus, the conformational change in STIM1 triggered by Ca2+ depletion extends to residue 251, and, extrapolating from STIM1(L251C) to wildtype STIM1, it buries pairs of L251 residues so that they are inaccessible for interaction with SOAR/CAD.
Figure 4. Involvement of CC1 residues 246–252 in a Ca2+-dependent conformational switch.
A, Crosslinking of STIM1 with single cysteine replacements at residues 250–252, as indicated, assessed as in Figure 2. Representative of three biological replicates. Control samples incubated without addition of iodine were included to determine background crosslinking. The STIM1 dimer band is marked with an arrow. B,C, Confocal micrographs of HeLa cells expressing GFP-STIM1(L251C) (B) or GFP-STIM1(L251S) (C) at rest (−) and after store depletion (TG). Scale bars, 10 µm. D, Effect of the activating mutation L251S on crosslinking of STIM1(A230C), in the presence of EGTA or Ca2+. E, Crosslinking of STIM1 with single cysteine replacements at residues 246–249, as indicated, assessed as in Figure 2. The STIM1 dimer band is marked with an arrow. F, Single-cell [Ca2+]i measurements in HeLa cells expressing wildtype GFP-STIM1 (WT; n = 55), GFP-STIM1(D247C) (D247C; n = 48), GFP-STIM1(L248C) (L248C; n = 50), and non-transfected HeLa cells (HeLa; n = 65). Cells were exposed to solutions containing varied concentrations of CaCl2 or 1 µM thapsigargin (TG) as indicated. G,H, Confocal micrographs of HeLa cells expressing GFP-STIM1(L248C) (G) or GFP-STIM1(D247C) (H) at rest (−) and after store depletion (TG). Scale bars, 10 µm. I, Upper, Provisional model for helix-helix packing of the transmembrane helices and initial parts of CC1 in wildtype STIM1. Closely packed transmembrane helix residues (from Figure 2) are highlighted with grey boxes, predicted coiled-coil core residues in the juxtamembrane segment (from Figure S5) in red, and predicted CC1 core residues beyond L251 (from Figure S5) in grey. Grey and red highlights correspond to different heptad registers of the core residues. Hatched shading of S237 and L251 indicates that the spacing of these residues is compatible with either adjacent heptad assignment. Lower, An alternative model for helix-helix packing of the D247C mutant. Core residues are highlighted with grey boxes. The D>C replacement introduces a favorable core residue at position 247, and could permit a local adjustment in the region from residues 240–247 that creates a coiled coil lacking heptad discontinuities. See also Figures S2, S3, and S4.
To determine whether coupling between the STIM1 transmembrane helices and the CC1-SOAR/CAD interface is bidirectional, we compared the crosslinking of STIM1(A230C) and STIM(A230C/L251S). The L251S mutation activates STIM1 by releasing SOAR/CAD from CC1 (Muik et al., 2011; Zhou et al., 2013) (Figure 4C, Figure S3B). Correspondingly, the presence of the L251S mutation results in efficient crosslinking at 230C both in the absence and in the presence of Ca2+ (Figure 4D, Figure S2B). A corollary of these observations is that the interaction of CC1 with SOAR/CAD is one factor restraining the transmembrane helices and CC1 regions from associating in resting cells.
Residues 246–252
We extended the crosslinking analysis to a broader region of the CC1-SOAR/CAD interface spanning residues 246–252 (Figure 4E, Figure S2A). In line with the prediction that L248 is buried in active STIM1, STIM1(L248C) shows relatively efficient crosslinking in the EGTA condition. The 248C protein also crosslinks appreciably in the presence of Ca2+, indicating that the mutation leads to constitutive activation. The localization of GFP-STIM1(L248C) to puncta in resting cells and its support of constitutive Ca2+ influx confirm that the L248C mutant is preactivated to a considerable extent (Figure 4F, 4G, Figure S2C, Figure S4A). These results dovetail with the previous finding that an L248S replacement is highly effective in causing an activating conformational change in STIM1(233–474) (Muik et al., 2011). Residues 246C, 249C, 250C, and 252C show only low levels of crosslinking, as expected for the proposed coiled coil.
The STIM1(D247C) mutant is constitutively active, exhibiting a high degree of crosslinking that is insensitive to Ca2+ (Figure 4E, Figure S2A), localizing to puncta in unstimulated cells (Figure 4H, Figure S4B), and triggering constitutive Ca2+ influx (Figure 4F, Figure S2C). Crosslinking at both positions 247 and 248 would not be expected in a perfect CC1-CC1 coiled coil. However, as explained in more detail in the Discussion, wildtype STIM1 forms an imperfect coiled coil, and arguably the D247C replacement allows an alternative active conformation that differs minimally from the proposed coiled coil of wildtype STIM1 (Figure 4I).
Juxtamembrane residues
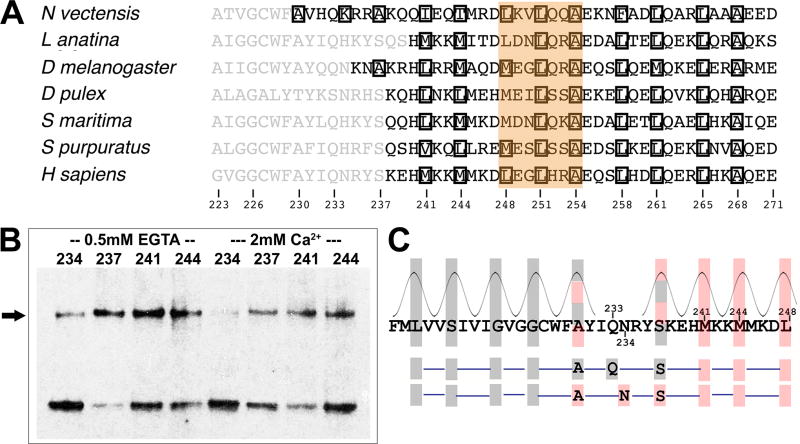
Can the initial parts of two paired CC1 regions also assemble as a coiled coil? A remarkable congruence in the predicted coiled-coil core residues in the juxtamembrane region of STIM proteins across many species, despite wide divergence in the sequences (Figure 5A, Figure S5), led us to focus on positions 237, 241, and 244 of human STIM1. Mutants 237C, 241C, and 244C all crosslink in the EGTA condition, consistent with packing in the core of a coiled coil in active STIM1 (Figure 5B, Figure S6A). The 237C protein shows elevated crosslinking in Ca2+, but still higher crosslinking in EGTA. In fact, the proportion of STIM1 dimer in the S237C samples is the highest seen in our experiments. The 241C and 244C proteins resemble the 248C and 251C mutants in exhibiting substantial crosslinking even in the presence of Ca2+. This crosslinking might again reflect constitutive activation— the point was not further examined— but the salient conclusion is that residue pairs 237-237, 241-241, and 244-244 can pack together. Our data indicate that a continuous coiled coil can form between the transmembrane helices and STIM1 residue L251, and define a tangible basis for concerted transmission of the store depletion signal into the STIM cytoplasmic domain.
Figure 5. Crosslinking of STIM1 juxtamembrane residues.
A, Abbreviated alignment of STIM sequences from varied species. Further examples are shown in Figure S5. Residues are shown in black if COILS (input matrix MTIDK, using weights, window length 14) calculates a coiled-coil probability greater than 0.5. Other residues are shown in grey. Boxed residues are assigned to core a or d positions by COILS. The brown highlight indicates a break in the predicted heptad repeat that is conserved across these species. The species represented are Nematostella vectensis, starlet sea anemone; Lingula anatina, a lingulid brachiopod; Drosophila melanogaster, common fruit fly; Daphnia pulex, a water flea; Strigamia maritima, a centipede; Strongylocentrotus purpuratus, purple sea urchin; Homo sapiens, human. B, Crosslinking of STIM1 with single cysteine replacements at residues 237, 241, or 244. The STIM1 dimer band is marked with an arrow. Crosslinking at these specific positions was tested because residues 241 and 244 align with predicted coiled-coil core residues of STIM1 orthologues across a broad range of species (Figure S5), and the spacing of residue 237 from these predicted core positions is compatible with its placement in a core position. The residue corresponding to S237 is, in fact, predicted to be a core residue in a few species, but the conservation of a polar serine in most species and its proximity to the transmembrane segment render coiled-coil prediction uncertain. Data from three biological replicates. C, Upper, STIM1 sequence from residues 214–248. Closely packed transmembrane helix residues are highlighted with grey boxes, and juxtamembrane residues that crosslink are highlighted in red. There is a shift in the heptad repeat of helix-helix packing centered on positions 233 and 234. Hatched shading indicates that the spacing of the highlighted residue is compatible with either adjacent heptad assignment. Lower, Either residue 233 or residue 234 alone, as a core residue, could support continuation of an imperfect coiled coil across the region of ambiguity. See also Figures S5, S6, and S7.
Viewing the juxtamembrane coiled coil as a continuation of the paired transmembrane helices, the crosslinking at 237C, 241C, and 244C requires a shift in the heptad repeat of helix-helix packing (Figure 5C). Shifts of this sort are a common feature of coiled coils, and are accommodated by local adjustments in the helix geometry and the helix-helix contacts (Brown, 1996; Strelkov et al., 2002). The discontinuities in the STIM1 heptad repeat centered on residues 237 and 251 seem likely to have a critical role in STIM1 activation. This point is addressed in the Discussion.
A juxtamembrane ‘sentinel region’
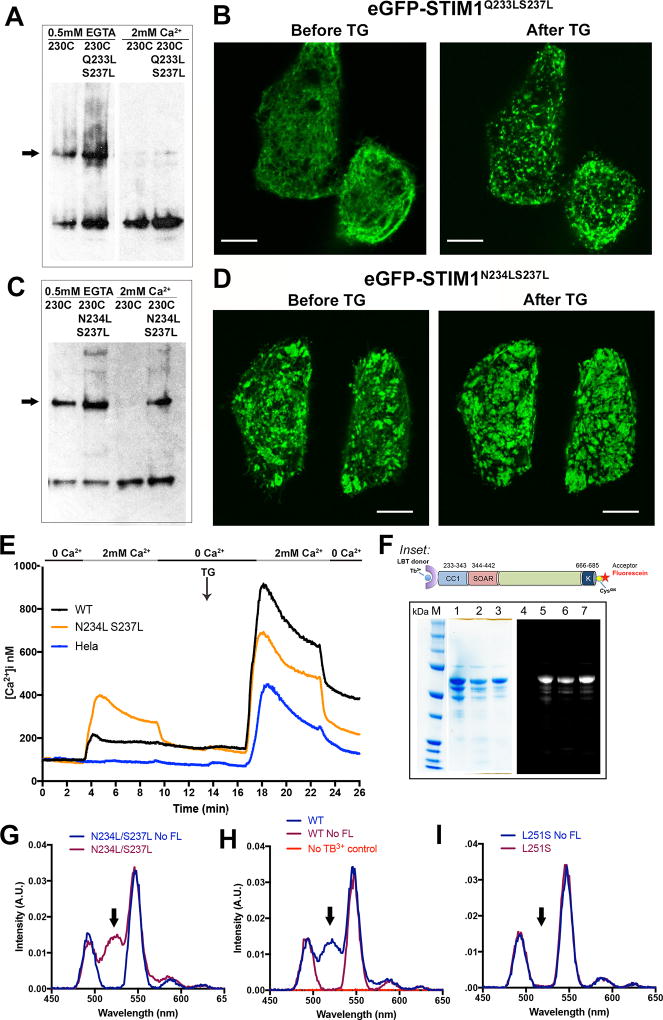
The native STIM1 residues Q233, N234, and S237 are suboptimal core residues for a canonical coiled coil (Lupas and Gruber, 2005), and might act as a brake on the STIM1 conformational change. To explore this possibility, we introduced the pairwise substitutions Q233L/S237L or N234L/S237L, and examined the effect on STIM1 activation in vitro and in cells. STIM1(Q233L/S237L) behaved like wildtype STIM1 in these assays, showing appreciable crosslinking at 230C only in the absence of Ca2+, and forming puncta in cells only upon store depletion (Figure 6A, 6B, Figure S6B). In contrast, STIM1(N234L/S237L) exhibited crosslinking at 230C in the presence of Ca2+, and very pronounced crosslinking in the absence of Ca2+ (Figure 6C, Figure S6B). GFP-STIM1(N234L/S237L) was partially active already in resting cells, with STIM1 puncta visible in most cells, and showed a further consolidation of puncta upon store depletion (Figure 6D). The partial constitutive activity was also evident in Ca2+ influx (Figure 6E, Figure S6C).
Figure 6. Analysis of STIM1 residues Q233, N234, and S237.
A, The effect of additional Q233L/S237L mutations on crosslinking of STIM1(A230C), assessed as in Figure 2. The STIM dimer band is marked with an arrow. Data representative of three biological replicates. B, Confocal micrographs of HeLa cells expressing GFP-STIM1(Q233L/S237L) under the conditions indicated. Scale bars, 10 µm. C, The effect of additional N234L/S237L mutations on crosslinking of STIM1(A230C), assessed as in Figure 2. The STIM dimer band is marked with an arrow. D, Confocal micrographs of HeLa cells expressing GFP-STIM1(N234L/S237L) under the conditions indicated. Scale bar, 10 µm. E, Single-cell [Ca2+]i measurements in HeLa cells expressing wildtype GFP-STIM1 (WT; n = 55), GFP-STIM1(N234L/S237L) (N234L S237L; n = 73), and non-transfected HeLa cells (HeLa; n = 65). Cells were exposed to solutions containing varied concentrations of CaCl2 or 1 µM thapsigargin (TG) as indicated. These experiments were performed together with those of Figure 4F, and the data plotted in Figure 4F for wildtype GFP-STIM1 and non-transfected HeLa cells are repeated here for reference. F, SDS-polyacrylamide gel analysis of the proteins used for energy transfer experiments. In the gel on the left, the samples in lanes 1–3 are the wildtype, L251S, and N234L/S237L proteins, respectively, stained with Coomassie Brilliant Blue. In the separate gel on the right, the sample in lane 4 is unlabeled wildtype protein as a negative control for fluorescence, and the samples in lanes 5–7 are the same fluorescein-labeled proteins as in lanes 1–3 of the Coomassie Brilliant Blue-stained gel. The gel was illuminated with UV light and the resulting fluorescence imaged to detect dye-labeled proteins. Inset: STIM1 proteins designed for Tb3+-acceptor energy transfer measurements comprise the STIM1 cytoplasmic domain, STIM1(233–685), with an engineered lanthanide-binding tag (LBT) at the N terminus to bind Tb3+ and a single engineered cysteine at the C terminus for covalent labelling with an acceptor dye. Fluorescein-5-maleimide was the labelling reagent in these experiments. G–I, Gated luminescence emission spectra of N234L/S237L (G), wildtype (H), and L251S (I) proteins labeled with donor Tb3+ and acceptor fluorescein. The spectrum of each protein labeled with donor Tb3+, but without acceptor, is shown for comparison (No FL). In (H), the specificity control with acceptor-labeled protein, but omitting Tb3+, is also shown. The arrows in G–I mark the expected position of the fluorescein emission peak. Data are representative of two biological replicates. See also Figure S6.
The partial activation of STIM1(N234L/S237L) in resting cells might have reflected a weakening of the CC1-SOAR/CAD interaction, as occurs with STIM1(L251S). In order to test this possibility, we took advantage of the previous finding that mutants at the CC1-SOAR/CAD interface cause a physical extension of the isolated STIM1 luminal domain that can be detected as a loss of luminescence resonance energy transfer (LRET) between a Tb3+ label at the N terminus of STIM1CT and a fluorescent acceptor at the C terminus (Muik et al., 2011; Zhou et al., 2013). In fact, labeled STIM1CT(N234L/S237L) exhibits an LRET signal in this assay comparable to that of wildtype STIM1CT (Figure 6F–6I), demonstrating that the N234L/S237L replacements do not compromise binding at the CC1-SOAR/CAD interface. This implies that they have their primary effect elsewhere in the full-length STIM1, arguably by perfecting the juxtamembrane coiled coil, and release SOAR/CAD only secondarily, by favoring the active coiled-coil configuration over the resting configuration. Minimally, these experiments indicate that there is a fine balance between the resting and activated conformations of STIM1, with N234 and S237 acting as part of a sentinel region, and setting an energetic penalty that minimizes spontaneous activation.
DISCUSSION
Our findings define the structural change that links dissociation of Ca2+ and dimerization of STIM1 luminal domains to an activating conformational change in the STIM1 cytoplasmic domain (Figure 7). Intramolecular dimerization or rearrangement of STIM1 luminal domains causes intramolecular association of the paired transmembrane helices and a concerted replacement of CC1-SOAR/CAD interactions by CC1-CC1 interactions, thereby releasing SOAR/CAD. The transmembrane and CC1 residues that form the helix-helix interface in activated human STIM1 are conserved in STIM1 across vertebrates— and between human STIM1 and STIM2, except for an M>V replacement corresponding to M241— and hence the basic mechanism leading to activation is almost certainly the same.
Figure 7. Summary diagram placing evidence from this study into context.
The STIM1 transmembrane helix and the initial part of CC1, extending to the SOAR/CAD-interacting region, are shown. Residues found to be crosslinked in EGTA in this study are marked with arrowheads, demonstrating apposition of the transmembrane helices and initial parts of CC1 from residue 216 to 251. The crosslinking of 234C and 247C is discussed in the text. Assigned core positions (a or d) in the conserved coiled-coil organization of STIM1 orthologues (Figure 5A; Figure S5) are indicated. L251, L258, and L261 have been shown to be involved in CC1-SOAR/CAD interaction, implying that close apposition of the CC1 helices of active STIM1 might extend to residue 261. The highlighted shifts in the heptad repeat (beige boxes) may have evolved to facilitate switching between the inactive and active conformations of STIM1 and to facilitate the reversible binding and release of SOAR/CAD.
Whereas the physiological signal transmitted by STIM1 is directional, originating in the ER lumen and propagating to the cytoplasm, the protein conformational change is inherently cooperative and therefore nondirectional. The point is neatly illustrated by our finding that the activating conformational change in the L251S mutant propagates ‘backward’ to the transmembrane helices. Rather than focus on the process of physiological activation, then, it may be useful to think of the multiple restraints preventing activation: the CC1-SOAR/CAD interaction, as shown here; the presence of STIM1 luminal domains with bound Ca2+ (Li et al., 2007; Muik et al., 2011; Ma et al., 2017; Korzeniowski et al., 2017); and presumably aspects of the STIM1 sequence itself, given that STIM1 is activated by several point mutations in CC1.
In this context, two shifts in the heptad repeat of STIM1 coiled-coil core residues, centered on residues 237 and 251, deserve attention (Figure 7). Breaks in the heptad repeat in other proteins afford flexibility to the protein backbone and favor protein-protein interactions, often at a cost in stability of the coiled coil (Brown, 1996). The heptad discontinuities in STIM1 can be interpreted in these terms. The shift within the juxtamembrane segment could allow the transmembrane helices the flexibility to link to different configurations of the STIM1 cytoplasmic domain, and could perhaps support protein-protein or protein-lipid interactions that stabilize the inactive state. The shift within the SOAR/CAD-interacting segment, by reducing the favorable energy of CC1-CC1 coiled-coil formation, could lower the energetic barrier to adopting the inactive CC1-SOAR/CAD configuration. Stated succinctly, we believe that the ambivalent coiled-coil sequences of these two segments in CC1 evolved to allow facile switching between the coiled-coil and inactive conformations.
Breaks in the STIM1 heptad repeat are not sufficient in themselves to prevent inappropriate activation. The variant STIM1(N234L/S237L), for example, retains the heptad discontinuities, yet it is partially activated under resting conditions. Thus additional restraints are needed to set the correct balance between physiological responsiveness and unwanted constitutive activation. We have shown here that a ‘sentinel’ feature is built into the juxtamembrane sequence of wildtype STIM1 in the form of polar residues that must pack into the coiled-coil core of active STIM1. The presence of the juxtamembrane sentinel region is compatible with activation, but imposes an energetic restraint on activation that is overcome only when coiled-coil assembly is coupled to the concerted change in STIM1 conformation triggered by dissociation of Ca2+ from the STIM1 luminal domain.
Turning to the SOAR/CAD-interacting segment of CC1, it seems likely that D247 is another sentinel feature. First, as discussed in detail in the next paragraph, the imperfect coiled coil in the L248–L254 region has been strongly conserved in evolution, suggesting that a more stable coiled coil would compete effectively with the CC1-SOAR/CAD interaction even in resting cells. Second, the nonpolar D247C replacement allows coiled coil formation in an alternative register that is minimally different from the proposed active conformation and that has a fully regular heptad repeat (Figure 4I). Third, experimentally, the D247C replacement activates STIM1. And, fourth, STIM sequence alignments show the near invariance of an acidic residue at the position of D247 in those species where the L248–L254 heptad discontinuity is present (Figure 5A; Figure S5). We suggest that D247 serves as a sentinel residue to enforce the heptad discontinuity at L248–L254 by raising the energetic cost of assembling the more regular alternative coiled coil.
The coiled-coil pattern identified in human STIM1 and conserved in vertebrate STIM1 proteins— with the two signature discontinuities in the heptad repeat, polar sentinel residues in the juxtamembrane region, and an acidic residue corresponding to human STIM1 residue 247— is recognizable in many, though not in all, invertebrate STIM proteins (Figure 5A; Figure S5). STIM is reminiscent, in this respect, of other coiled-coil proteins that conserve a biologically successful coiled-coil design, including characteristic discontinuities, across evolution (Offer, 1990; Taylor et al., 2015; Chernyatina et al., 2015). In an informative counterpoint, nematode STIM proteins have evolved a modification of the ancestral pattern that has a regular heptad repeat from the ER membrane to the residue corresponding to position 251 in human STIM1, loss of the acidic residue at position 247, and typically lower coiled-coil propensity in the twenty residues following position 251 (Figure S7). The modified pattern is compatible with store-dependent activation of STIM (Lorin-Nebel et al., 2007; Gao et al., 2009), although the details of the activating conformational change in this case remain to be worked out. Most relevant here, the preservation of the modified coiled coil pattern across a diverse panel of nematodes, again in the face of substantial variations in protein sequence, underscores our conclusion that evolution has matched specific coiled coil patterns in STIM to their specific biological and cellular contexts.
A previous report examined the constitutively activated C227W mutant of STIM1, and concluded that reorganization of the STIM1 transmembrane helices can switch the STIM1 cytoplasmic domain to an active conformation (Ma et al., 2015). Importantly, the same report directly demonstrated in cells that physiologically activated STIM1 releases SOAR/CAD from its intramolecular interaction with CC1. The detailed proposals in that paper regarding the packing of transmembrane helices will need revision in light of our present data. First, we do not detect helix-helix contacts in the region of residues 221–230 in inactive STIM1. Second, the helix-helix contacts of active full-length STIM1 differ to some extent from the observed interactions of the TM-CC1(C227W) fragment. These differences can likely be traced to differences in methodology— the use of much higher protein concentrations in the earlier study, the crosslinking reaction in detergent rather than in an ER membrane environment, and the focus on a TM-CC1 fragment that may lack physical constraints on the transmembrane helices that are present in full-length STIM1. Nevertheless, despite the differences in detail, the two studies together constitute compelling evidence that a rearrangement of the STIM1 transmembrane helices is part of the concerted change that switches STIM1 into an active conformation and that releases SOAR/CAD to engage the ORAI channel.
We note in closing that this analysis and our previous work (Zhou et al., 2013) have focused only on the initial step that releases SOAR/CAD and the STIM polybasic tail. The partial CC1-CC1 coiled coil need not persist in cells during sustained STIM activation, if the active conformation is subsequently stabilized by further STIM oligomerization, by accessory proteins, or by STIM interaction with the plasma membrane and ORAI. Similarly, the analysis has no specific implications for STIM1 secondary structure in the region of CC1 between residues 252 and 343, since a continuous α helix, a broken α helix, or a random coil would all allow CC1 to span the ER-plasma membrane distance.
EXPERIMENTAL PROCEDURES
Additional information can be found in Supplemental Experimental Procedures.
Plasmids
The wild-type eGFP-STIM1 construct was made by cloning full-length human STIM1 into pCMV-XL5 with the STIM1 signal peptide preceding the eGFP tag. The HA-tagged STIM1 insert for crosslinking experiments was PCR-amplified from pIRES2-EGFP-HA-STIM1 (Wu et al., 2006) and cloned into pcDNA3.1. The sequence encoding STIM1 residues between the 3×HA tag and residue 58 was deleted, and cysteine codons 227 and 437 were replaced by serine codons. Wild-type STIM1CT for Tb3+-acceptor energy transfer experiments has been described (Zhou et al., 2013). Mutations specified in the text were introduced into these expression plasmids using the Quikchange site-directed mutagenesis kit (Agilent).
Confocal microscopy
HeLa cells expressing wild-type eGFP-STIM1 or specified variants were imaged at room temperature, first in modified Ringer’s soution for the resting-state images, then after at least 5 min in modified Ringer’s solution lacking CaCl2 and containing 1µM thapsigargin for the activated-state images.
STIM1 crosslinking assay
HeLa cells were suspended in dilution buffer containing either 0.5 mM EGTA or 2 mM Ca2+, lysed at 4°C by passage through a 25G syringe needle, and centrifuged to collect cellular membranes. Membranes in resuspension buffer containing either 0.5 mM EGTA or 2 mM Ca2+ were incubated with iodine for 10 min at 4°C. The reactions were quenched with excess iodoacetamide, subjected to SDSPAGE, and STIM1 monomer and crosslinked dimer bands detected by immunoblotting. In experiments on the Ca2+ concentration dependence of crosslinking, the dilution and resuspension buffers were Chelex-treated (Chelex-100 resin, Bio-Rad) with no added EGTA or Ca2+, and oxidation reactions were supplemented to give either a final EGTA concentration of 0.5 mM or final Ca2+ concentrations ranging from 0.3 µM to 2 mM.
Single-cell Ca2+ influx assay
HeLa cells expressing specified eGFP-STIM1 variants and mCherry-ORAI1 were loaded with Fura-2, then alternately illuminated at 340 nm and 380 nm while imaging fluorescence emission at λ > 400 nm (Sharma, Quintana et al., 2013). Ratio images were recorded at intervals of 4 s. Ca2+ concentration was estimated as [Ca2+]i = K*(R−Rmin)/(Rmax−R), where values of K, Rmin, and Rmax were determined from an in situ calibration of Fura-2 in HeLa cells.
LRET assay
Plasmids encoding LBT-STIM1CT-C437S/C686 or its L251S or N234L/S237L variants were expressed and purified as previously described (Zhou et al., 2013), and labeled with fluorescein-5-maleimide. Luminescence resonance energy transfer (LRET) assays were performed according to the protocol in Zhou et al., 2013.
Coiled coil prediction
Sequences of STIM1 orthologues were retrieved from the Ensembl, Ensembl Metazoa, Wellcome Trust Sanger Institute, or KEGG (Kyoto Encyclopedia of Genes and Genomes) databases. Individual sequences were trimmed to the segments corresponding to Homo sapiens STIM1(201–300) or, in cases of less extensive alignment, an equivalent length of sequence flanking the transmembrane region and submitted to the COILS server (Max Planck Institute for Developmental Biology). COILS predictions (input matrix MTIDK, and using weights) are quoted for the narrowest window (window length 14) in order to maximize sensitivity to local sequence features.
Statistical methods
STIM1 crosslinking was quantitated as the fraction of the summed monomer and dimer signal in the dimer band. Cytoplasmic Ca2+ was quantitated as peak Ca2+ concentrations recorded before store depletion and after store depletion. Data are plotted as mean ± SEM. Statistical significance was tested using an unpaired, one-tailed Student’s t-test, with Bonferroni adjustment for multiple testing in Figure S2C.
Supplementary Material
Acknowledgments
We thank Dr Anjana Rao for discussions and comments on the manuscript. This work was funded by US National Institutes of Health grants AI084167 (to PGH) and AI040127 (to Anjana Rao and PGH).
Footnotes
AUTHOR CONTRIBUTIONS
AG and PGH designed the experiments, and AG supervised their implementation in the laboratory. VR developed the STIM1 crosslinking assay. NH, VR, and AG carried out the protein engineering, disulfide crosslinking assays, and recombinant protein purifications. AG performed the confocal microscopy, Ca2+ imaging, and LRET assays. AG and PH analyzed the data and wrote the manuscript with input from the other authors.
DECLARATION OF INTERESTS
PGH is a founder and scientific advisor of CalciMedica, Inc. The other authors declare that they have no competing financial interests.
References
- Brandman O, Liou J, Park WS, Meyer T. STIM2 is a feedback regulator that stabilizes basal cytosolic and endoplasmic reticulum Ca2+ levels. Cell. 2007;131:1327–1339. doi: 10.1016/j.cell.2007.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JH. Heptad breaks in alpha-helical coiled coils: stutters and stammers. Proteins. 1996;26:134–145. doi: 10.1002/(SICI)1097-0134(199610)26:2<134::AID-PROT3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Chernyatina AA, Guzenko D, Strelkov SV. Intermediate filament structure: the bottom-up approach. Curr. Opin. Cell Biol. 2015;32:65–72. doi: 10.1016/j.ceb.2014.12.007. [DOI] [PubMed] [Google Scholar]
- Covington ED, Wu MM, Lewis RS. Essential role for the CRAC activation domain in store-dependent oligomerization of STIM1. Mol. Biol. Cell. 2010;21:1897–1907. doi: 10.1091/mbc.E10-02-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui B, Yang X, Li S, Lin Z, Wang Z, Dong C, Shen Y. The inhibitory helix controls the intramolecular conformational switching of the C-terminus of STIM1. PLoS One. 2013;8:e74735. doi: 10.1371/journal.pone.0074735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derler I, Jardin I, Romanin C. Molecular mechanisms of STIM/Orai communication. Am. J. Physiol. Cell. Physiol. 2016;310:C643–C662. doi: 10.1152/ajpcell.00007.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrner M, Muik M, Schindl R, Butorac C, Stathopulos P, Zheng L, Jardin I, Ikura M, Romanin C. A coiled-coil clamp controls both conformation and clustering of stromal interaction molecule 1 (STIM1) J. Biol. Chem. 2014;289:33231–33244. doi: 10.1074/jbc.M114.610022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa Y, Teraguchi S, Ikegami T, Dagliyan O, Jin L, Hall D, Dokholyan NV, Namba K, Akira S, Kurosaki T, Baba Y, Standley DM. Intrinsic disorder mediates cooperative signal transduction in STIM1. J. Mol. Biol. 2014;426:2082–2097. doi: 10.1016/j.jmb.2014.03.006. [DOI] [PubMed] [Google Scholar]
- Gao S, Fan Y, Chen L, Lu J, Xu T, Xu P. Mechanism of different spatial distributions of Caenorhabditis elegans and human STIM1 at resting state. Cell Calcium. 2009;45:77–88. doi: 10.1016/j.ceca.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Lange I, Feske S. A minimal regulatory domain in the C terminus of STIM1 binds to and activates ORAI1 CRAC channels. Biochem. Biophys. Res. Commun. 2009;385:49–54. doi: 10.1016/j.bbrc.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzeniowski MK, Wisniewski E, Baird B, Holowka D, Balla T. Molecular anatomy of the early events in STIM1 activation; oligomerization or conformational change? J. Cell Sci. 2017;130:2821–2832. doi: 10.1242/jcs.205583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzeniowski MK, Manjarres IM, Varnai P, Balla T. Activation of STIM1-Orai1 involves an intramolecular switching mechanism. Sci. Signal. 2010;3:ra82. doi: 10.1126/scisignal.2001122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Lu J, Xu P, Xie X, Chen L, Xu T. Mapping the interacting domains of STIM1 and Orai1 in Ca2+ release-activated Ca2+ channel activation. J. Biol. Chem. 2007;282:29448–29456. doi: 10.1074/jbc.M703573200. [DOI] [PubMed] [Google Scholar]
- Liou J, Fivaz M, Inoue T, Meyer T. Live-cell imaging reveals sequential oligomerization and local plasma membrane targeting of stromal interaction molecule 1 after Ca2+ store depletion. Proc. Natl. Acad. Sci. USA. 2007;104:9301–9306. doi: 10.1073/pnas.0702866104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou J, Kim ML, Heo WD, Jones JT, Myers JW, Ferrell JE, Jr, Meyer T. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr. Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorin-Nebel C, Xing J, Yan X, Strange K. CRAC channel activity in C. elegans is mediated by Orai1 and STIM1 homologues and is essential for ovulation and fertility. J. Physiol. 2007;580:67–85. doi: 10.1113/jphysiol.2006.124883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luik RM, Wang B, Prakriya M, Wu MM, Lewis RS. Oligomerization of STIM1 couples ER calcium depletion to CRAC channel activation. Nature. 2008;454:538–542. doi: 10.1038/nature07065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas AN, Gruber M. The structure of alpha-helical coiled coils. Adv. Protein Chem. 2005;70:37–78. doi: 10.1016/S0065-3233(05)70003-6. [DOI] [PubMed] [Google Scholar]
- Ma G, Zheng S, Ke Y, Zhou L, He L, Huang Y, Wang Y, Zhou Y. Molecular determinants for STIM1 activation during store-operated Ca2+ entry. Curr. Mol. Med. 2017;17:60–69. doi: 10.2174/1566524017666170220103731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma G, Wei M, He L, Liu C, Wu B, Zhang SL, Jing J, Liang X, Senes A, Tan P, Li S, Sun A, Bi Y, Zhong L, Si H, Shen Y, Li M, Lee MS, Zhou W, Wang J, Wang Y, Zhou Y. Inside-out Ca2+ signalling prompted by STIM1 conformational switch. Nat. Commun. 2015;6:7826. doi: 10.1038/ncomms8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer JC, Dehaven WI, Smyth JT, Wedel B, Boyles RR, Bird GS, Putney JW., Jr Large store-operated calcium selective currents due to coexpression of Orai1 or Orai2 with the intracellular calcium sensor, Stim1. J. Biol. Chem. 2006;281:24979–24990. doi: 10.1074/jbc.M604589200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muik M, Frischauf I, Derler I, Fahrner M, Bergsmann J, Eder P, Schindl R, Hesch C, Polzinger B, Fritsch R, Kahr H, Madl J, Gruber H, Groschner K, Romanin C. Dynamic coupling of the putative coiled-coil domain of ORAI1 with STIM1 mediates ORAI1 channel activation. J. Biol. Chem. 2008;283:8014–8022. doi: 10.1074/jbc.M708898200. [DOI] [PubMed] [Google Scholar]
- Muik M, Fahrner M, Schindl R, Stathopulos P, Frischauf I, Derler I, Plenk P, Lackner B, Groschner K, Ikura M, Romanin C. STIM1 couples to ORAI1 via an intramolecular transition into an extended conformation. EMBO J. 2011;30:1678–1689. doi: 10.1038/emboj.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offer G. Skip residues correlate with bends in the myosin tail. J. Mol. Biol. 1990;216:213–218. doi: 10.1016/S0022-2836(05)80309-2. [DOI] [PubMed] [Google Scholar]
- Park CY, Hoover PJ, Mullins FM, Bachhawat P, Covington ED, Raunser S, Walz T, Garcia KC, Dolmetsch RE, Lewis RS. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell. 2009;136:876–890. doi: 10.1016/j.cell.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakriya M, Lewis RS. Store-operated calcium channels. Physiol. Rev. 2015;95:1383–1436. doi: 10.1152/physrev.00020.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos J, DiGregorio PJ, Yeromin AV, Ohlsen K, Lioudyno M, Zhang S, Safrina O, Kozak JA, Wagner SL, Cahalan MD, Veliçelebi G, Stauderman KA. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J. Cell Biol. 2005;169:435–445. doi: 10.1083/jcb.200502019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Quintana A, Findlay GM, Mettlen M, Baust B, Jain M, Nilsson R, Rao A, Hogan PG. An siRNA screen for NFAT activation identifies septins as coordinators of store-operated Ca2+ entry. Nature. 2013;499:238–242. doi: 10.1038/nature12229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soboloff J, Rothberg BS, Madesh M, Gill DL. STIM proteins: dynamic calcium signal transducers. Nat. Rev. Mol. Cell Biol. 2012;13:549–565. doi: 10.1038/nrm3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassova MA, Soboloff J, He LP, Xu W, Dziadek MA, Gill DL. STIM1 has a plasma membrane role in the activation of store-operated Ca2+ channels. Proc. Natl. Acad. Sci. USA. 2006;103:4040–4045. doi: 10.1073/pnas.0510050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopulos PB, Li GY, Plevin MJ, Ames JB, Ikura M. Stored Ca2+ depletion-induced oligomerization of stromal interaction molecule 1 (STIM1) via the EF-SAM region. An initiation mechanism for capacitive Ca2+ entry. J. Biol. Chem. 2006;281:35855–35862. doi: 10.1074/jbc.M608247200. [DOI] [PubMed] [Google Scholar]
- Stathopulos PB, Zheng L, Li GY, Plevin MJ, Ikura M. Structural and mechanistic insights into STIM1-mediated initiation of store-operated calcium entry. Cell. 2008;135:110–122. doi: 10.1016/j.cell.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Strelkov SV, Burkhard P. Analysis of alpha-helical coiled coils with the program TWISTER reveals a structural mechanism for stutter compensation. J. Struct. Biol. 2002;137:54–64. doi: 10.1006/jsbi.2002.4454. [DOI] [PubMed] [Google Scholar]
- Taylor KC, Buvoli M, Korkmaz EN, Buvoli A, Zheng Y, Heinze NT, Cui Q, Leinwand LA, Rayment I. Skip residues modulate the structural properties of the myosin rod and guide thick filament assembly. Proc. Natl. Acad. Sci. USA. 2015;112:E3806–E3815. doi: 10.1073/pnas.1505813112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MM, Buchanan J, Luik RM, Lewis RS. Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J. Cell. Biol. 2006;174:803–813. doi: 10.1083/jcb.200604014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Jin H, Cai X, Li S, Shen Y. Structural and mechanistic insights into the activation of Stromal interaction molecule 1 (STIM1) Proc. Natl. Acad. Sci. USA. 2012;109:5657–5662. doi: 10.1073/pnas.1118947109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Sun L, Hubrack S, Selvaraj S, Machaca K. Intramolecular shielding maintains the ER Ca2+ sensor STIM1 in an inactive conformation. J. Cell. Sci. 2013;126:2401–2410. doi: 10.1242/jcs.117200. [DOI] [PubMed] [Google Scholar]
- Yu J, Zhang H, Zhang M, Deng Y, Wang H, Lu J, Xu T, Xu P. An aromatic amino acid in the coiled-coil 1 domain plays a crucial role in the auto-inhibitory mechanism of STIM1. Biochem. J. 2013;454:401–409. doi: 10.1042/BJ20130292. [DOI] [PubMed] [Google Scholar]
- Yuan JP, Zeng W, Dorwart MR, Choi YJ, Worley PF, Muallem S. SOAR and the polybasic STIM1 domains gate and regulate Orai channels. Nat. Cell Biol. 2009;11:337–343. doi: 10.1038/ncb1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SL, Yu Y, Roos J, Kozak JA, Deerinck TJ, Ellisman MH, Stauderman KA, Cahalan MD. STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature. 2005;437:902–905. doi: 10.1038/nature04147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Stathopulos PB, Schindl R, Li GY, Romanin C, Ikura M. Auto-inhibitory role of the EF-SAM domain of STIM proteins in store-operated calcium entry. Proc. Natl. Acad. Sci. USA. 2011;108:1337–1342. doi: 10.1073/pnas.1015125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Srinivasan P, Razavi S, Seymour S, Meraner P, Gudlur A, Stathopulos PB, Ikura M, Rao A, Hogan PG. Initial activation of STIM1, the regulator of store-operated calcium entry. Nat. Struct. Mol. Biol. 2013;20:973–981. doi: 10.1038/nsmb.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.