SUMMARY
Many networks throughout the nervous system are organized into topographic maps, where the positions of neuron cell bodies in the projecting field correspond with the positions of their axons in the target field. Previous studies of topographic map development show evidence for spatial patterning mechanisms, in which molecular determinants expressed across the projecting and target fields are matched directly in a point-to-point mapping process. Here, we describe a novel temporal mechanism of topographic map formation that depends on spatially regulated differences in the timing of axon outgrowth and functions in parallel with spatial point-to-point mapping mechanisms. We focus on the vagus motor neurons, which are topographically arranged in both mammals and fish. We show that cell position along the anterior-posterior axis of hindbrain rhombomere 8 determines expression of hox5 genes, which are expressed in posterior but not anterior vagus motor neurons. Using live-imaging and transplantation in zebrafish embryos, we additionally reveal that axon initiation is delayed in posterior vagus motor neurons independent of neuron birth time. We show that hox5 expression directs topographic mapping without affecting time of axon outgrowth, and that time of axon outgrowth directs topographic mapping without affecting hox5 expression. The vagus motor neuron topographic map is therefore determined by two mechanisms that act in parallel: a hox5-dependent spatial mechanism akin to classic mechanisms of topographic map formation, and a novel axon outgrowth-dependent temporal mechanism in which time of axon formation is spatially regulated to direct axon targeting.
Keywords: Topographic map development, vagus motor neurons, axogenesis, axon initiation, hoxa5a, hoxb5a, hoxb5b, zebrafish
eTOC BLURB
The development of topographic maps classically relies upon spatial patterning mechanisms. Barsh & Moens show that in zebrafish vagus motor neurons, map formation is determined by a hox5-dependent spatial mechanism that acts in parallel with a novel temporal mechanism dependent upon the time of axon outgrowth.
INTRODUCTION
Topographic maps are a common organizational motif in neurobiology and are crucial for the ability of animals to accurately perceive and respond to their external environment. These maps are characterized by an arrangement of neurons in which neighbor-neighbor relationships are preserved between cell bodies and axon targets, creating representations of the outside world inside the brain [1]. The best-studied topographic maps form through a spatial patterning process, where precise patterns of gene expression in neurons and their target areas direct axons to the proper location in a point-to-point matching system [2,3]. For example, in the retinotopic map, gradients of EphA receptor expression in the retina and EphrinA ligand expression in the tectum are matched such that high and low EphA-expressing neurons innervate opposite regions of the tectum, while neighbor neurons expressing similar EphA levels innervate the same region [4–9]. Similarly, in the spinal musculotopic map, dorsal and ventral limb-innervating neurons occupy spatially distinct locations and reach their respective targets due to the presence of repulsive ligands expressed by the inappropriate targets [10–14]. Thus, spatial patterning mechanisms underlie topographic map development in both sensory and motor systems.
In addition to classic spatial patterning, previous studies have also proposed a temporal mechanism of topographic map development. In the Drosophila photoreceptor map, sequential expression of a temporal identity transcription factor during neurogenesis directs targeting to different layers of the medulla [15]. Additionally, in the mouse olfactory topographic map, cell position correlates with birth order and therefore with time of axon arrival in the target region [16,17]. However, though these studies demonstrate correlations between temporal patterning and topographic connectivity, timing-based mechanisms for topographic map formation have been difficult to explicitly test, as they require live imaging and the ability to manipulate the temporal environment.
Here, to dissect the relative contributions of both spatial and temporal mechanisms of topographic map development, we focus on vagus motor neurons (motor neurons of cranial nerve X, or mX neurons) in the zebrafish embryo as a model of topographic map development. In all vertebrates, mX neurons are located in the most posterior segment of the hindbrain, rhombomere 8 (r8) [18]. In humans, mX neurons innervate pharyngeal arch-derived muscles important for speech and swallowing. In fish, mX neurons innervate homologous muscles to move the pharynx, gills, and pharyngeal teeth. In both fish and humans, the vagus also supplies preganglionic parasympathetic innervation of visceral targets [19,20]. Previous studies in fish and mammals have shown that mX neurons in the adult animal are arranged topographically, with anterior neurons innervating more anterior pharyngeal arches and posterior neurons innervating posterior pharyngeal arches and visceral targets [21,22]. However, it is unclear how the vagus topographic map is established during development.
We describe two parallel and independent mechanisms for vagus topographic map development. First, we show that hox5 genes are expressed in posterior mX neurons and direct mX neuron targeting towards posterior targets. Second, we demonstrate that temporal regulation of axon initiation influences axon targeting. While anterior and posterior mX neurons are born at the same time, axons emerge from posterior mX neurons later than from anterior mX neurons, and late axon arrival in the peripheral target area directs axons to posterior targets. Finally, we demonstrate that these two mechanisms are independent. This work therefore establishes a new temporal mechanism which acts in parallel with hox-dependent spatial patterning mechanisms to govern the robust development of a topographic map.
RESULTS
Characterization of embryonic vagus motor neuron topographic map
Vagus (mX) neurons, detectable with isl1-driven fluorescent proteins [23], are born ventrally in hindbrain r8 and migrate dorsolaterally to form the vagus motor nucleus by 36 hours post fertilization (hpf) [24]. Axons emerge basally, first exiting the hindbrain at 27 hpf, and travel anteriorly in a single fascicle before making a characteristic ventral turn at the otic vesicle towards the pharyngeal arches (PAs) (Figure 1A) [25]. The axons divide peripherally into 5 branches, which are all present by 3 days post fertilization (dpf): 4 branches innervate PA4, 5, 6, and 7, and a 5th posterior branch innervates visceral targets (Figure 1A) [23]. In time lapse movies, the axon branches appear sequentially in anterior-to-posterior order, with the PA4 branch emerging first, and the PA7 and visceral branches emerging last (Figure 1B, Movie S1).
Figure 1. Topographic mapping by vagus motor neurons.
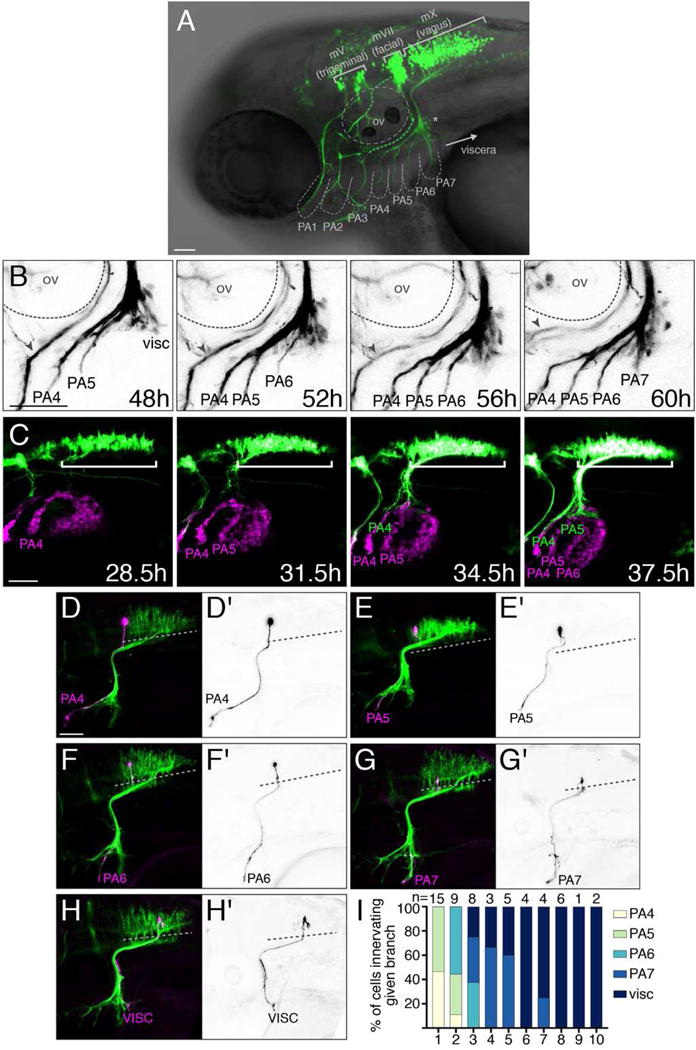
(A) Lateral view of a zebrafish embryo expressing Tg(isl1:Kaede) at 3 dpf. Brackets: cranial motor nuclei; PA: pharyngeal arches; ov: otic vesicle. Asterisk marks a subset of vagus sensory cells labeled by islet1. (B) mX axon branches innervate PAs sequentially. See Movie S1. Gray arrowhead indicates the glossopharyngeal motor nerve (mIX). (C) tcf21:mCherry+ PA muscle precursors (magenta) appear sequentially prior to mX axon entry. White bracket indicates mX nucleus expressing isl1:eGFPCAAX (green). See Movie S2. (D–H) mX neurons are arranged in a topographic map. Single neuron labeled by Tg(isl1:eGFPCAAX) in magenta or black-on-white (D′–H′) on Tg(isl1:mRFP) background (green). Dotted lines indicate the length of the mX territory. (I) Quantification of (D–H). mX territory was divided into 10 equal-length bins along the A-P axis. In all figures, anterior is left and dorsal is up. Scale bars are 50 μm. See also Figure S1.
To simultaneously visualize the appearance of mX axons and their PA targets, we time lapsed embryos expressing Tg(isl1:eGFPCAAX) and Tg(tcf21:mCherry), which marks the progenitors of the PA muscles that are the targets of branchiomotor innervation [26,27]. Tcf21:mCherry+ cells appear in an anterior-to-posterior sequence, with each PA emerging about 6–8 hours before mX axon innervation (Figure 1C, Movie S2). Thus the sequential outgrowth of mX branches correlates with the sequential appearance of their PA targets.
To understand how the mX neuron topographic map is established, we first determined whether it is detectable in the zebrafish embryo. We injected 1-cell stage embryos expressing Tg(isl1:mRFP) with isl1:eGFPCAAX, titrated to a low level such that only a single mX neuron was labeled with eGFPCAAX. We divided the mX territory into 10 equal-length bins, and determined the axon target of each single mX neuron based upon its relative position in the mX territory (Figure 1D–H). We note that single mX neurons only ever project into a single PA (n = 57/57). We found that mX neurons in bin 1 exclusively innervated PA4 and PA5, while mX neurons in bins 4–10 exclusively innervate PA7 and the viscera (Figure 1I). We confirmed this single-cell mapping approach with photoconversion experiments using Tg(isl1:Kaede), photoconverting defined regions of the mX territory and subsequently visualizing the photoconverted Kaede in the peripheral branches (anterograde labeling), and conversely photoconverting specific branches and subsequently visualizing the photoconverted Kaede in cell bodies (retrograde labeling) (Figure S1). While we observed overlap between mX neurons innervating neighboring PAs, our single-neuron and photoconversion mapping approaches consistently describe a vagus topographic map at 3 dpf that is sufficiently resolved to allow us to investigate its developmental basis.
Vagus motor neuron position determines axon target
We first asked whether the position of an mX neuron in r8 determines its axon target. To address this question, we used single-neuron transplantation to manipulate the spatial environment of individual postmitotic neurons [28]. At 27 hpf, when postmitotic mX neurons are present along the anterior-posterior axis of r8 but before they have extended axons, we transplanted postmitotic mX neurons from a Tg(isl1:Kaede) donor to the same (homotopic) or different (heterotopic) position in r8 of a Tg(isl1:GFP) host embryo. To ensure accurate developmental age, we used the precise “prim staging” method to stage all embryos at the time of transplantation ([29] and see STAR Methods for details). We transplanted 1–5 mX neurons (median = 2) into each host embryo, and then assessed targeting of the donor neuron(s) at 3 dpf (Figure 2A). Singly transplanted neurons only projected into a single PA, similar to our earlier single-cell labeling experiments (n = 8/8 embryos; e.g. Figure 2B,E). As expected, in control anterior → anterior homotopic transplants, donor neurons innervated primarily anterior targets (Figure 2B,F), and in control posterior → posterior homotopic transplants, donor neurons innervated primarily posterior targets (Figure 2D,F). In heterotopic anterior → posterior transplants, donor neurons innervated primarily posterior targets (Figure 2C,F). Likewise, in heterotopic posterior → anterior transplants, donor neurons innervated primarily anterior targets (Figure 2E,F). These results show that the anterior-posterior position of an mX neuron in r8 determines its peripheral target.
Figure 2. Vagus motor neuron position determines axon target.
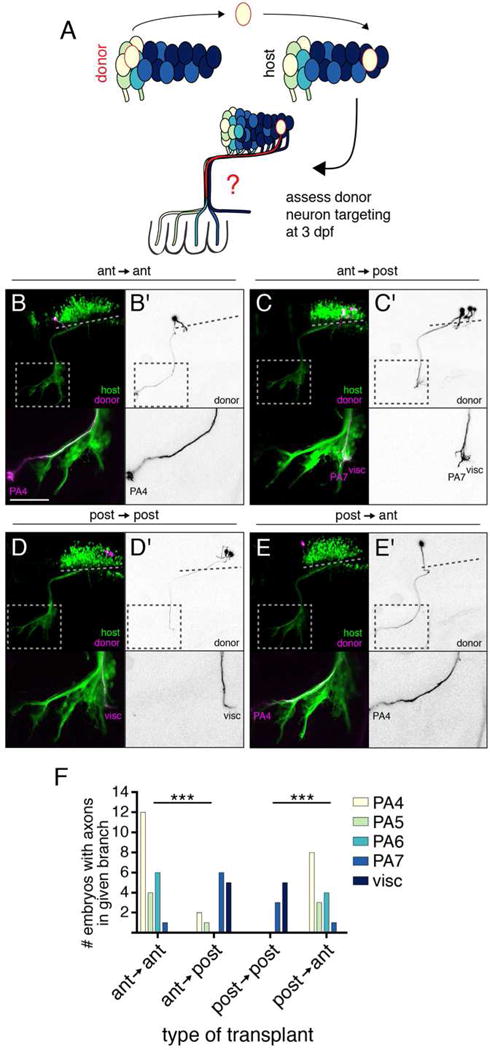
(A) Schematic of postmitotic neuron transplantation approach. mX neurons are transplanted homotopically or heterotopically (pictured) before axon formation and donor axon targeting is assayed at 3 dpf. (B–E) Examples of homotopic (B,D) and heterotopic (C,E) transplants. Donor-derived neurons are marked by Tg(isl1:Kaede) in magenta and black-on-white (B′–E′). Host motor neurons are marked by Tg(isl1:GFP) in green. Dotted lines indicate length of mX territory, dotted boxes indicate region shown in lower panels. (F) Quantification of transplant results showing number of host embryos with a donor axon in a given branch. Statistical analysis done with Fisher’s exact test (see STAR Methods for details). Ant → ant n = 14 host embryos; ant → post n = 12; post → post n = 7; post → ant n = 12.
hox5 expression distinguishes anterior and posterior mX neurons
To understand how cell position determines axon target, we sought to identify differences between anterior and posterior mX neurons. Hox genes are well-known for their role in anterior-posterior patterning of the vertebrate hindbrain including motor neurons; however, a role in vagus-specific topographic mapping has not been described [30]. We therefore sought to determine whether hox4, hox5, and hox6 paralogs, which are known to have anterior expression limits in the posterior hindbrain and anterior spinal cord [31–34], were differentially expressed in anterior versus posterior mX neurons by RNA in situ hybridization at 27 hpf. We found that hox4 genes have anterior limits anterior to the mX territory, and hox6 genes have anterior limits posterior to the mX territory (Figure S2). However, hoxa5a, hoxb5a, and hoxb5b all have diffuse anterior limits of expression within the mX territory (Figure 3A and S2E–G).
Figure 3. hox5 expression distinguishes anterior and posterior mX neurons.
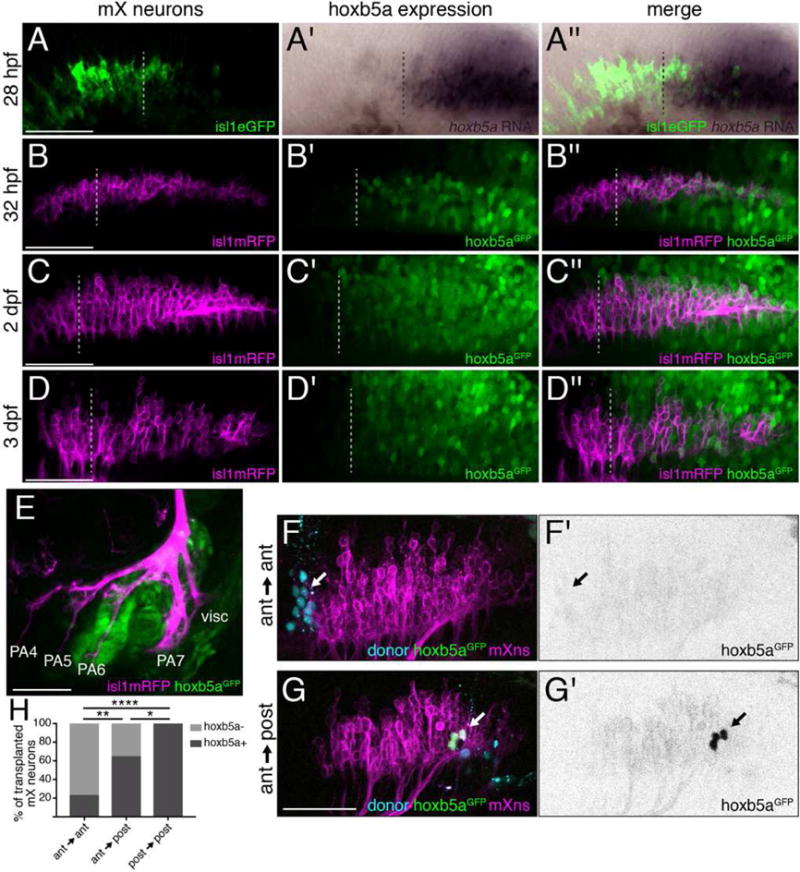
(A) hoxb5a is expressed in the posterior mX territory. RNA in situ hybridization of 28 hpf embryo for hoxb5a expression (purple) followed by immunostaining against Tg(isl1:eGFPCAAX) to label motor neurons (green). Dotted lines in (A–D) indicate anterior limit of hoxb5a expression. See also Figure S2. (B–E) hoxb5aGFP is expressed in posterior mX neurons, PA6, and PA7. hoxb5aGFP (green) and Tg(isl1:mRFP) (magenta) at 32 hpf (B), 2 dpf (C), or 3 dpf (D,E). (F,G) Anterior mX neurons marked by lineage dye (cyan) and isl1:mRFP (magenta) transplanted homotopically (F) or heterotopically (G) are hoxb5aGFP-negative (arrow in F) or hoxb5aGFP-positive, respectively (arrow; green in G, black in G′). Host motor neurons express Tg(isl1:mRFP) (magenta). (H) quantification of (F,G). Analysis done by Fisher’s exact test. Ant → ant n = 22 neurons, 6 embryos; ant → post n = 28 neurons, 9 embryos; post → post n = 13 neurons, 5 embryos.
To visualize hox5 expression and motor neurons together with high resolution in live embryos, we targeted GFP to the endogenous hoxb5a locus ([35] and see STAR Methods for details) and examined expression of hoxb5aGFP at 1, 2, and 3 dpf (Figure 3B–D). Consistent with RNA in situ expression data, hoxb5aGFP is expressed in a graded pattern, with GFP-negative cells at the anterior end of the mX territory (corresponding to regions 1 and 2 from Figure S1), while 100% of cells in the posterior mX territory (regions 3, 4, and 5) are GFP-positive (Figure 3B and data not shown). The anterior limit of hoxb5aGFP expression is more anterior than the corresponding hoxb5a mRNA expression domain (compare Figure 3A to 3B). This difference likely reflects the different sensitivity of the detection methods, as well GFP perdurance: hoxb5aGFP marks every cell that ever expressed hoxb5a, while the mRNA in situ detects the expression domain at the moment of fixation. hoxb5aGFP is also expressed in PA6 and PA7 (Figure 3E). Thus, hox5 genes are expressed in posterior mX neurons and their posterior PA targets, suggesting hox5 expression may direct posterior mX axon targeting.
Vagus motor neuron position determines hox5 expression
Our expression experiments demonstrated that mX neuron position correlates with hox5 expression. We next asked whether mX neuron position determines hox5 expression by transplanting postmitotic mX neurons at 27 hpf from hoxb5aGFP-positive donor embryos into hoxb5aGFP-negative host embryos, and assessing GFP expression in donor-derived mX neurons at 3 dpf. Host embryos expressed Tg(isl1:mRFP) so that we could correlate hoxb5aGFP expression in the transplanted neuron(s) with their position in the host mX territory. As expected, 100% of donor mX neurons (n = 13 neurons) maintained GFP expression in control homotopic posterior → posterior transplants (Figure 3H). Also as expected, in control anterior → anterior transplants, only 23% of donor mX neurons (n = 22 neurons) were GFP+ (Figure 3F,H). However, when anterior mX neurons were transplanted heterotopically into posterior r8, 64% (n = 28 neurons, p = 0.0046 by Fisher’s exact test compared to anterior → anterior) of the donor mX neurons were GFP+ (Figure 3G,H). Due to GFP perdurance, we were not able to assess hoxb5a downregulation in posterior → anterior transplants. These results suggest that the position of an mX neuron in r8 determines its hox5 expression. The ability of transplanted post-mitotic neurons to turn on expression of hoxb5a (Figure 3) and to change their axon targeting after transplantation (Figure 2) indicates that mX neuron fate remains plastic after birth.
hox5 expression drives posterior axon targeting
Our transplant experiments demonstrated that both hox5 expression and posterior PA targeting are induced in an anterior mX neuron that is placed in the posterior mX territory. We therefore sought to test whether hox5 expression causes posterior PA targeting. Given that there are three hox5 genes with overlapping expression in posterior r8 in zebrafish, and that Hox5 genes are known to act redundantly in other processes [36], we reasoned that a targeted gain-of-function approach would most efficiently allow us to test the ability of hox5 genes to impact mX axon targeting. We constructed plasmids encoding UAS:hox5gene-p2a-eGFPCAAX and injected the DNA into 1-cell stage Tg(isl1:Gal4);Tg(isl1:mRFP) embryos. This strategy allowed us to generate embryos with membrane-GFP-labeled hox5-expressing mX neurons scattered sparsely throughout the vagus territory (Figure 4A–E). If hox5 expression directs axons towards posterior targets, we predicted that hox5-expressing mX neurons would innervate posterior targets with a higher than expected frequency. We found that while hox5-expression had no effect on the overall distribution of mX neurons compared to control eGFPCAAX expression (Figure 4E), hox5-expressing neurons were biased toward innervating more posterior PAs compared to eGFPCAAX-expressing control neurons (Figure 4A–D, F). This was true for neurons expressing any of the three hox5 genes: hoxa5a, hoxb5a, or hoxb5b, though the effect of hoxb5b expression was less dramatic.
Figure 4. hox5 expression directs mX axons to posterior targets.
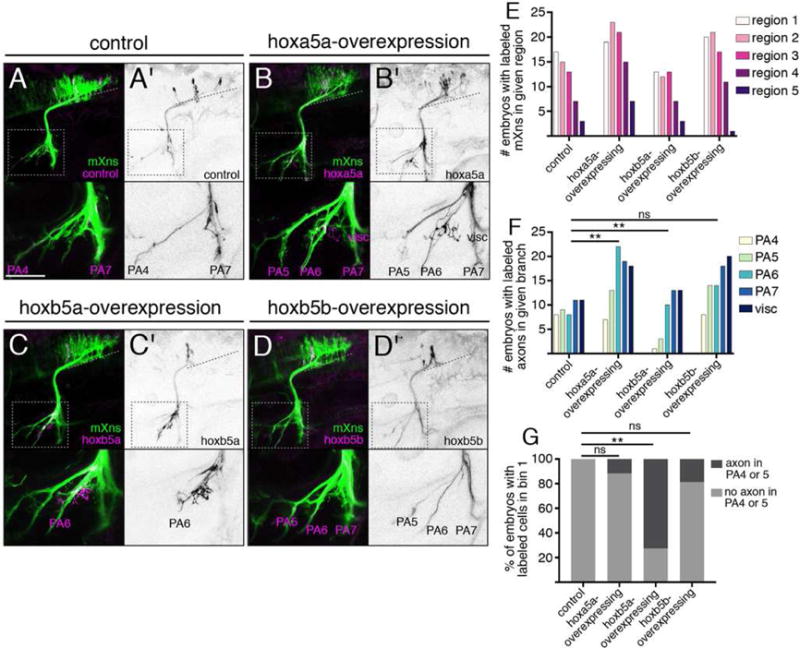
(A–D) Expression of control isl1:eGFPCAAX (A) or isl1:hox5-P2A-eGFPCAAX expression construct (B–D) (magenta and (A′–D′) black) in a Tg(isl1:mRFP) background (green). Dotted lines indicate length of mX territory, dotted boxes indicate region shown in bottom panels. (E) Quantification of distribution of labeled neurons after hox5 expression as in (A–D). Regions are based on morphological boundaries as in Figure S1. (F) Quantification of labeled axon targeting in (A–D). Analysis was done using Fisher’s exact test comparing control to each ectopic expression condition with respect to each axon branch. Conditions are considered different if they differ in at least one branch. In both (E) and (F), control n = 17 embryos, hoxa5a n = 24, hoxb5a n = 20, hoxb5b n = 28. (G) Analysis of axon targeting in cases where a labeled mX neuron was located within the most anterior region of the mX territory (bin 1, see Figure 1I). Ectopic hoxb5a-expressing mX neurons in bin 1 avoid PA4 and PA5. Analysis done by Chi-square followed by Fisher’s exact test comparing each hox5 construct to control. Control n = 9 embryos, hoxa5a n = 17, hoxb5a n = 11 embryos, hoxb5b n = 16.
To further examine how hox5 expression instructs the formation of the mX topographic map, we focused on embryos with hox5-expressing neurons in the anterior-most region of the mX territory that normally exclusively innervates PA4 and PA5 (bin 1 in Figure 1I). In control experiments, 100% of embryos with labeled cells in bin 1 also had labeled axons in the PA4 or PA5 branch, consistent with the single-neuron mapping data (Figure 4G). However, in embryos with hoxb5a-expressing neurons in bin 1, only 27% of the embryos had labeled axons in the PA4 or PA5 branch (p = 0.0014, Figure 4G). The effect of ectopic hoxa5a or hoxb5b expression in bin 1 neurons was less dramatic, with 88% and 81%, respectively, of embryos having labeled axons in the PA4 or PA5 branch (Figure 4G). Together, these results strongly suggest that hox5 expression biases mX axon targeting to posterior targets.
Axon formation and arrival in the periphery is delayed in posterior mX neurons
The anterior-posterior sequence of PA innervation by mX neurons (Figure 1B,C, Movies S1,S2) suggested that there may be earlier developmental differences between anterior and posterior mX neurons that impact axon targeting. We crossed the Tg(isl1:Gal4) line to the variegated Tg(UAS:kaede) line to mosaically label mX neurons, then photoconverted posterior mX neurons and time lapsed starting at 29 hpf when mX neurons are present throughout r8 but axons have only just begun to emerge (Figure 5A, Movie S3). Anterior mX axons exit the brain and turn ventrally to extend into the periphery at 29±1.9 hpf (95% confidence interval (CI) = 26.71–32.6 hpf), consistent with previous literature [24]. However, posterior mX neurons, which were also present at the beginning of the time lapse, show extensive protrusive activity but remain multipolar until axons are finally detected at 37.7±2.7 hpf (95% CI = 35.9–39.5 hpf), when anterior mX axons had already extended into the PAs (Figure 5A,B, Movie S3). Posterior growth cones then extend quickly, following tracts of previously extended anterior mX axons (Movie S3). This suggests that the delayed appearance of posterior mX axons is due to delayed axon initiation, rather than slower extension.
Figure 5. Time of axon formation and arrival in the head periphery is delayed in posterior mX neurons independent of birthdate.
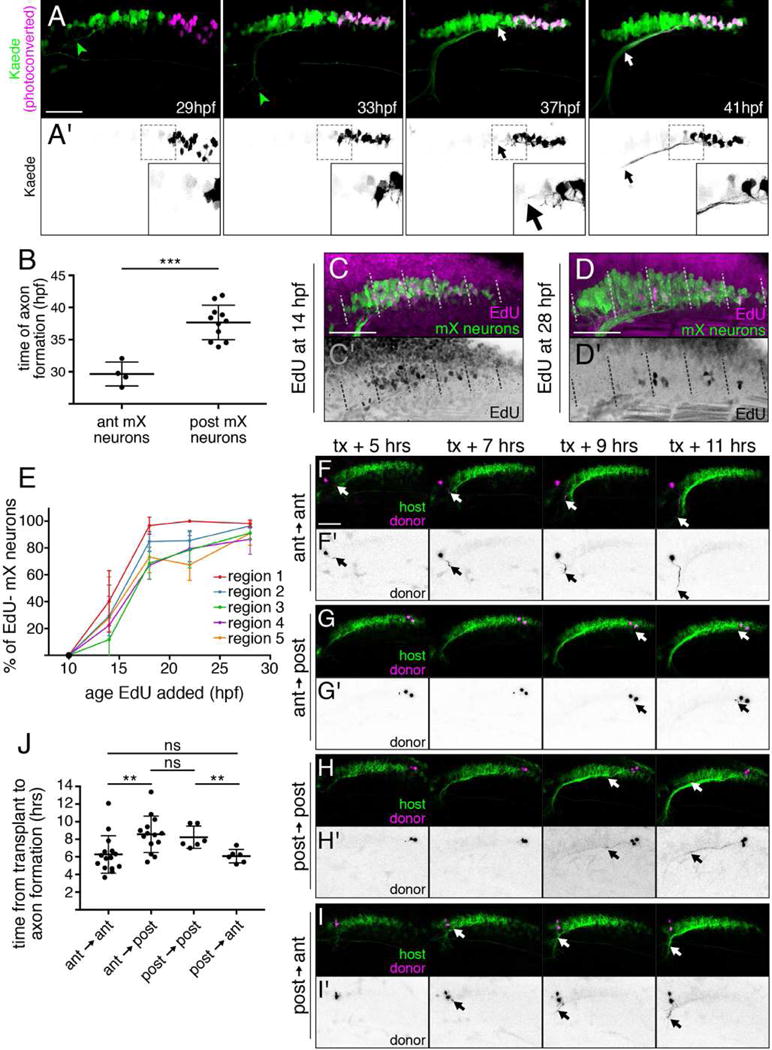
(A) Stills from representative time lapse (see Movie S3) of embryo expressing Tg(isl1:Gal4);Tg(UAS:kaede). Photoconverted posterior mX neurons (magenta (A) or black (A′), white arrow) initiate axons 8 hours after anterior mX neurons (green, green arrowhead). (B) Quantification of (A). Each point represents a single embryo. Analysis done by unpaired t test. (C,D) Embryo expressing Tg(isl1:GFP) (green) was incubated in EdU (magenta and (C′,D′) black) to label cells born after 14 hpf (C) or 28 hpf (D). Embryos were fixed at 48 hpf and the mX territory was divided into 5 equal-length regions from anterior (1) to posterior (5). (E) Quantification of (C,D) showing percentage of postmitotic (EdU−) mX neurons (isl1:GFP+) in each region at each time point ([# EdU−GFP+ cells]/[# GFP+ cells]). Each point represents mean; error bars show standard deviation. n = 5 embryos for all time points except 14 hpf, where n = 4 embryos. (F–I) Transplanting an anterior mX neuron heterotopically into the posterior delays axon formation (F,G) while transplanting a posterior mX neuron into the anterior hastens axon formation (H,I). Donor neurons marked by Tg(isl1:kaede) in magenta and (F′–I′) black, and host neurons marked by Tg(isl1:mRFP) in green. See also Movies S4, S5. (J) Quantification of (F–I). Each point represents a single embryo. Analysis done by unpaired t test.
One explanation for the observed delay in posterior mX axon formation could be that posterior mX neurons are born later than anterior ones. To test this possibility, we birthdated mX neurons with (2′S)-2′-deoxy-2′-fluoro-5-ethynyluridine (EdU). We added EdU to Tg(isl1:GFP) embryos at 10, 14, 18, 22, or 28 hpf and fixed embryos at 48 hpf. We then identified mX neurons by GFP expression and determined whether each neuron underwent its last division before (EdU−) or after (EdU+) the time point at which EdU was added. We divided the mX territory into 5 equal-length regions and determined the proportion of EdU− neurons in each region for each treatment time (Figure 5C–E). If time of axon formation correlates with time of birth, we would expect EdU− mX neurons to appear progressively from anterior to posterior. However, this is not what we observed: postmitotic (EdU−) mX neurons appear at all anterior-posterior levels of the motor nucleus between 10 hpf-28 hpf, with no significant difference in the onset of neurogenesis between the five regions assayed (Figure 5E, p = ns by one-way ANOVA of all regions at 14 hpf). Surprisingly therefore, the delayed axon initiation by posterior mX neurons does not reflect a later birthdate. We conclude that timing of axon formation is an independently regulated event along the anterior-posterior axis of the mX territory.
Vagus motor neuron position determines time of axon formation
Given that cell position correlates with time of axon formation, we sought to test whether cell position could determine time of axon formation. We therefore applied time lapse imaging to our postmitotic mX neuron transplant approach, after mX neurons were transplanted homotopically or heterotopically at 27 hpf. In control anterior → anterior transplants, mX neurons initiated axons 6.3±2.1 hours post transplantation (hpt) (95% CI = 5.1–7.5 hpt) (Figure 5F,J Movie S4). However, transplanting mX neurons heterotopically from anterior → posterior delayed time of axon formation, with axons appearing 8.6±2 hpt (95% CI = 7.4–9.8 hpt) (Figure 5G,J, Movie S4, p = 0.0067 by unpaired t-test). Consistent with this result, in control posterior → posterior transplants, mX neurons initiated axons late (8.2±1.3 hpt, 95% CI = 6.9–9.6 hpt), while transplanting neurons heterotopically from posterior → anterior hastened time of axon formation (6.1±0.8hpt, 95% CI = 5.3–6.9 hpt) (Figure 5H-J, Movie S5, p = 0.0051 by unpaired t-test). While the difference in time of axon formation between control and heterotopic transplants was smaller than that observed between endogenous anterior and posterior mX neurons (likely due to the transplantation procedure itself), these results suggest that the position of an mX neuron within r8 determines its time of axon formation.
Time of axon arrival in the periphery determines axon targeting
Our transplant experiments demonstrated that mX neurons positioned in anterior r8 will initiate axon formation early (Figure 5) and will innervate anterior axon targets (Figure 2). Conversely, mX neurons positioned in posterior r8 will initiate axon formation late and will innervate posterior axon targets. We therefore sought to test the hypothesis that in addition to hox5 expression, time of axon formation also influences mX axon targeting. We simulated a delay in axon initiation by transplanting anterior mX neurons from a younger donor (25.9±2 hpf) to the anterior mX territory of an older host (31.8±2.1h hpf) such that the extending axon of the transplanted neuron joins the fascicle with late-arriving posterior mX axons (Figure 6A). If axon targeting is determined by anterior-posterior position alone, then this transplanted mX neuron should innervate anterior targets. Alternatively, if timing of axon outgrowth influences axon targeting independently of position, then it should innervate a posterior targets. We found that compared to stage-matched transplants, where donor-derived anterior mX neurons innervated primarily anterior PAs (Figures 2B, 6B,D), anterior mX neurons transplanted homotopically but heterochronically into older host embryos innervated primarily posterior PAs (Figure 6C,D). This change in axon targeting is not due to induction of ectopic hox5 expression, because when we did the equivalent heterochronic transplant using our hoxb5aGFP line as a donor, we did not see induction of GFP expression in the transplanted neurons (Figure 6E,F). These results indicate that surprisingly, delaying axon growth into the periphery is sufficient to shift axon targeting to posterior targets.
Figure 6. Time of mX axon outgrowth determines peripheral target independently of hox5 expression.
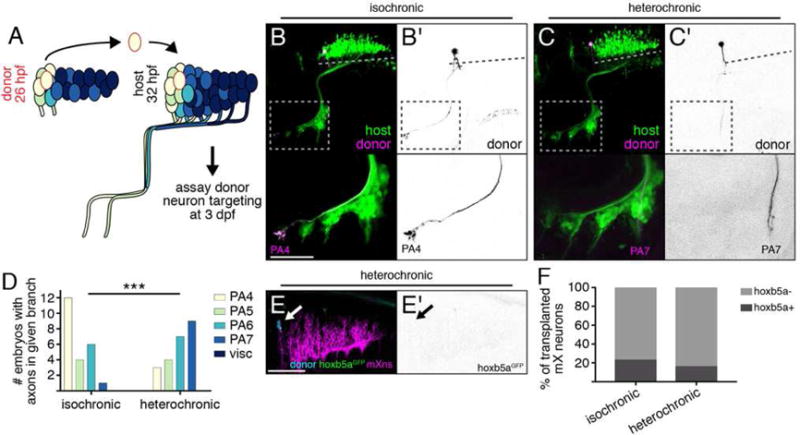
(A) mX neurons are transplanted from anterior to anterior (homotopic) from a 26 hpf donor to a 32 hpf host, and donor axon targeting is assayed at 3 dpf. (B,C) Anterior mX neurons transplanted isochronically (B) innervate anterior targets while anterior mX neurons transplanted heterochronically (C) innervate posterior targets. Donor neurons are marked by Tg(isl1:Kaede) (magenta or (B′,C′) black). Host motor neurons are marked by Tg(isl1:GFP) (green). Dotted lines indicate length of mX territory. Dotted boxes indicate region shown in bottom panels. (D) Quantification of transplant results. Isochronic control transplants are the same data shown in Figure 2F (anterior → anterior). Analysis done using Fisher’s exact test, see STAR Methods for details. Isochronic n = 14 host embryos, heterochronic n = 16. (E) Change in targeting after heterochronic transplantation is not due to induction of hox5 expression. Transgenes are as described in Fig. 3F,G (F) Quantification of (E). Isochronic control transplants are the same data shown in Figure 3F. Heterochronic n = 19 cells, 11 embryos.
hox5 expression and time of axon initiation direct mX axon targeting independently
Our experiments demonstrate that the spatio-temporal environment of mX neurons influences their topographic mapping to the PAs. Position along the anterior-posterior axis of r8 determines hox5 expression and time of axon formation, and both hox5 expression and delayed axon initiation can target axons to more posterior PAs. We asked whether hox5 expression is responsible for delayed axon initiation, which in turn drives posterior PA targeting, or whether the two are independent. If hox5 expression delays axon initiation, we expected that in our hox5 ectopic expression approach, hox5-expressing mX neurons in the anterior mX territory would initiate axon formation late, at a time appropriate for posterior mX neurons. We therefore used our injection strategy to ectopically express hox5 genes sparsely throughout the mX territory, then imaged and quantified the time of axon formation in ectopic hox5-expressing mX neurons within the anterior-most quarter of the mX territory. Control anterior mX neurons expressing isl1:eGFPCAAX formed axons at 29.8±1.6 hpf (95% CI = 28.4–31.05 hpf), consistent with our earlier results (Figures 5B, 7A,E, Movie S6). Surprisingly, we observed no significant difference in the time of axon initiation by anterior mX neurons expressing hoxa5a, hoxb5a, or hoxb5b compared to the control construct (Figure 7B–E, Movies S7–S9). Thus, regardless of hox5 expression, anterior mX neurons initiate axon formation by approximately 30 hpf, an average of approximately 8 hours before posterior mX neurons do. Together, these results indicate that delayed axon initiation guides axons to posterior targets independently of hox5 expression, and hox5 expression guides axons to posterior targets independently of timing of axon initiation.
Figure 7. hox5 acts independently of time of axon formation.
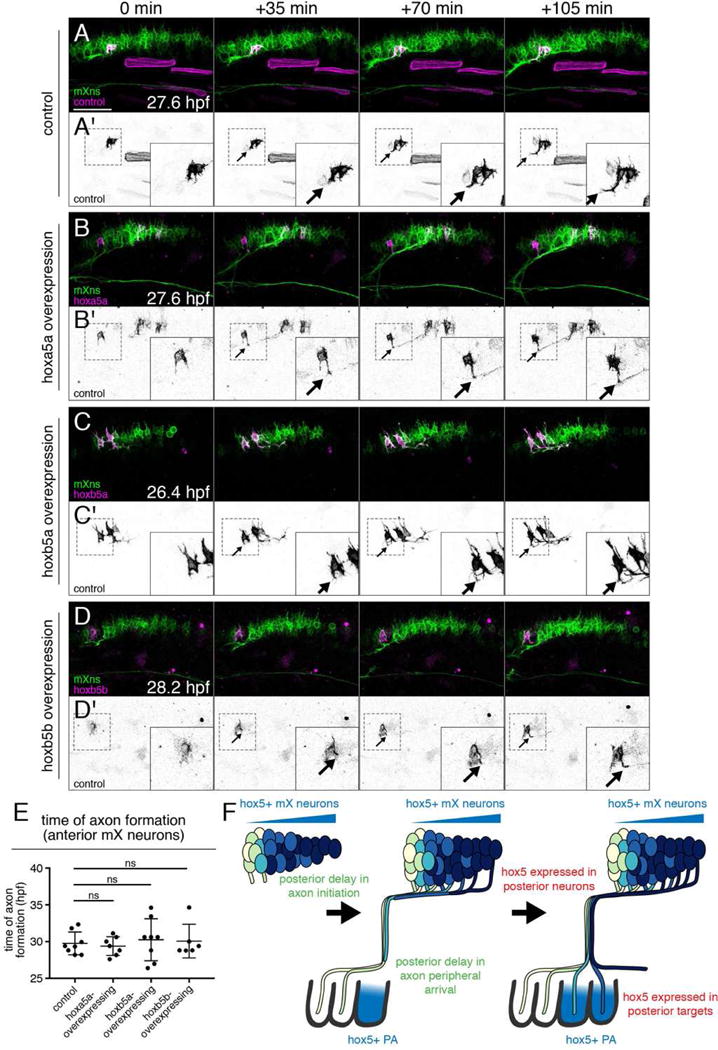
(A–D) hox5 expression does not delay the time of axon formation in anterior mX neurons. Stills from representative time lapses of control (A) or ectopic hox5-expressing (B–D) mX neurons (magenta or black in A′–D′) in a Tg(isl1:mRFP) background (green). Arrows indicate the emerging axon. See Movies S6–S9. (E) Quantification of (A–D). Each point represents a single embryo. Analysis done by unpaired t test. (F) Model of dual mechanisms governing mX neuron topographic map development. Anterior mX neurons arrive in the head periphery when only anterior PAs are available for innervation. Delayed axon initiation in posterior mX neurons results in delayed arrival in the periphery when posterior PAs have formed. Additionally, hox5 genes are expressed in posterior mX neurons and bias their axons towards posterior, hox5-expressing PAs.
DISCUSSION
Topographic maps are of high interest to neurobiologists given their functional importance and prevalence throughout the central nervous system [1,37]. Here, we identify a vagus motor neuron topographic map in the zebrafish embryo and present two mechanisms that function in parallel to govern its initial formation. First, we demonstrate that hox5 gene expression in posterior mX neurons drives posterior axon targeting. Second, we introduce a novel mechanism of topographic map development dependent upon delayed axon formation and outgrowth by posterior mX neurons (Figure 7F). We note that the embryonic vagus map is not highly resolved, in that adjacent neurons can innervate different PAs. It is likely that, as for other topographic maps, later competition- and/or activity-dependent mechanisms further resolve the vagus map [1].
hox5 expression drives posterior axon targeting
A major finding from our work is that hox5 genes are expressed in posterior mX neurons and posterior PAs, and direct posterior mX axons to posterior targets. Hox transcription factors are known to play roles in specifying neuron identity and axon targeting [30] and to function in topographic map development in both motor and sensory systems [38–40]. Hox genes affect axon targeting through both intermediate transcription factors and direct regulation of cell-surface guidance and adhesion molecules [30]. However, hox5 target genes that direct targeting of posterior mX neurons remain unknown. Given the expression of hoxb5a in both the posterior mX neurons and their target region, an intriguing possibility is that hox5 acts through regulation of a homophilic adhesion molecule. Homophilic cadherin interactions have been shown to affect connectivity in the Drosophila visual system [15,41]. In vertebrates, cadherins also affect axon targeting decisions in the visual system, though they are thought to act heterophilically [42]. An important goal of future experiments will be to identify genes that direct posterior mX axon targeting downstream of hox5.
Time of axon initiation is regulated independently of birth order
A second major finding from our work is that axon initiation is delayed in posterior mX neurons despite the fact that anterior and posterior mX neurons have similar birth times, and that this difference in timing plays a critical role in the formation of the topographic map. Differential timing of axon outgrowth has been proposed to direct axon targeting to different regions of the brain [43] or to different targets within a topographic map [15,16]. However, in these cases, temporal mechanisms for topographic map formation have been difficult to test directly without the means of manipulating the temporal environment of projecting neurons. Furthermore in these examples, differences in neuron birthdate underlie differences in timing of axon formation [15,17,44]. Our system is the first we are aware of where topographic mapping depends on the timing of axon outgrowth independently of neuron birthdate.
Our findings raise the question of how timing of axon initiation is controlled in mX neurons. Axon initiation is a complex process that can vary between neuron types. Some neurons inherit polarity information from progenitor cells that determines the subcellular site of axon specification [45,46]. Others, including cortical neurons, adopt a multipolar morphology during migration before transitioning to a bipolar morphology, following which one neurite, typically the trailing process, is specified as an axon [47–49]. Many studies of axon initiation have elucidated intracellular signaling pathways that ultimately result in localized microtubule stabilization to specify one neurite as the future axon [50–57]. These cell-autonomous events can be biased by localized extracellular cues, including diffusible signaling molecules such as BDNF and TGF-beta, as well as contact-mediated cues in the neuronal microenvironment [49,53,58,59].
In our system, we see that both anterior and posterior mX neurons complete their dorsal migration and exhibit extensive multipolar protrusions before axon formation. Our transplant experiments suggest that the difference in axon initiation between anterior and posterior mX neurons is most likely due to a localized non-cell autonomous cue that either promotes axon initiation in anterior mX neurons, similar to BDNF and TGF-beta signaling in mammalian hippocampal and cortical neurons [53,58], or prevents axon initiation in posterior mX neurons, similar to the role of Semaphorin3A in rat cortical neurons [60]. Determining the nature of this cue is an important goal for future work.
Late axon arrival in the periphery drives posterior axon targeting
Posterior mX axons arrive in the periphery later than anterior mX axons due to the delay in axon initiation. Our heterochronic transplant experiments demonstrate that late-arriving axons are guided to posterior targets, but how this guidance occurs is unclear. Our favored hypothesis is that the PAs exhibit short, sequential competence windows for innervation, perhaps mediated by sequential expression of an attractive guidance cue in each PA during their successive anterior-posterior development [20]. The best candidate for a PA-derived mX guidance cue is HGF, which is expressed in the PAs and signals as a chemoattractant through the Met receptor on cranial motor axons [61]. Future research will determine whether HGF-Met signaling is required for PA innervation in zebrafish, and whether HGF is transiently expressed at a critical stage of PA development as our model predicts.
Temporal and spatial mechanisms act in parallel to direct topographic map development
Our findings suggest that hox5 expression and delayed axon outgrowth function in parallel in a “belt-and-suspenders” manner to promote the robust targeting of posterior mX axons to posterior PAs and the viscera, thereby generating the vagus topographic map. Timing of mX axon outgrowth could direct initial targeting of axons to PAs that are accessible at the time axons reach the periphery, and a hox5-dependent mechanism could act to ensure that axons of posterior hox5+ mX neurons do not innervate anterior (hox5−) PAs. The work we present here demonstrates that two mechanisms act in parallel to instruct the development of the vagus topographic map, and future studies will elucidate if and how the spatial hox5-dependent mechanism and temporal axon outgrowth-dependent mechanism cooperate in topographic map development.
STAR METHODS
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Cecilia Moens (cmoens@fredhutch.org).
Experimental model and subject details
Zebrafish care and maintenance
Danio rerio animals were raised at the Fred Hutchinson Cancer Research Center facility in accordance with IACUC-approved protocols. All experiments were carried out in accordance with IACUC standards. Fish were bred and maintained according to standard protocols [62]. For all embryo manipulations done between 23–35hpf, embryos were staged according to the highly accurate prim staging method. For embryo manipulations between 10–23 hpf, embryos were staged by somite staging [29]. Sex is not a relevant biological variable in our experiments, as they are carried out before sex is determined in zebrafish [63]. Transgenic lines used in this study include Tg(isl1-hsp70l:Kaede) (unpublished, Anita Ng and Victoria Prince) gift of the Prince Lab, Tg(isl1:Gal4)fh452 [64], Tg(UAS:Kaede)s1999 [65], gift of the Baier Lab, Tg(isl1:mRFP)fh1 [66], TgBAC(tcf21:mCherry-NTR)pd108 [26], and Tg(isl1:GFP)rw0 [25]. Lines generated for this study include hoxb5aGFPfh468 and Tg(isl1:eGFPCAAX)fh474 (see details below).
Method Details
Generation of hoxb5aGFP
hoxb5aGFP was generated using the CRISPR/Cas9-mediated knock-in strategy outlined in [35]. The donor plasmid was Mbait-hsp70-GFP, gift of the Higashijima lab. The CRISPR gRNA sequence was GTTACAAATGATGACGAGAC. Due to the possibility that plasmid insertion may disrupt endogenous hoxb5a expression, the hoxb5aGFP line was maintained in a heterozygous state.
Plasmid construction and injection
The following plasmids were generated for this study: isl1-hsp70:eGFP-CAAX-polyA, 10XUAS:hoxa5a-p2a-eGFP-CAAX-polyA, 10XUAS:hoxb5a-p2a-eGFP-CAAX-polyA, and10XUAS:hoxb5b-p2a-eGFP-CAAX-polyA. The 10XUAS plasmid and eGFP-CAAX sequence were obtained from the Tol2kit [67]. The islet1 promoter consisted of the zCREST enhancer upstream of hsp70 [64]. The hox gene sequences were amplified from 24hpf cDNA using the following primers:
| hoxa5a: | forward | ATGAGCTCTTATTTCGTCAATTCAT |
| reverse | AGGCCGGTATCCGCTTCCTG | |
| hoxb5a: | forward | ATGAGCTCTTACTTTGTAAACTCG |
| reverse | TGGTTGGAAAGCGCTACCTG | |
| hoxb5b: | forward | ATGAGCTCTTATTTTCTAAACTCG |
| reverse | ATTTTGAAACGCGCTCCCCG |
A pDONR 221 vector containing p2a-NLSeGFP (gift of Jeremy Rabinowitz) was modified to remove the NLS and add a CAAX domain for membrane localization. hox genes were cloned upstream of p2a-eGFPCAAX. Final DNA constructs were assembled in the pBHR4R3 plasmid (gift of the Brockerhoff lab) using the Gateway system (Life Technologies). Embryos were injected at the one-cell stage with 100 pg of plasmid together with 90 pg of Tol2 transposase mRNA. To create the Tg(isl1:eGFPCAAX)fh474 line, injected embryos were raised to adulthood and outcrossed to identify germline-transmitting founders.
Motor neuron transplants
To track donor-derived cells, donor embryos were injected at the one-cell stage with 1% cascade-blue dextran (10,000 mw, Invitrogen). Before transplantation, all donor and hosts were screened to confirm they expressed the proper combination of transgenes. Embryos were then anesthetized with 0.4% ethyl 3-aminobenzoate methanesulfonate (ms-222) (Sigma), and embedded in 1.4% low-melting point agarose (Gibco). Embedded embryos were immersed in normal Ringer’s solution containing ms-222 [62]. A small wedge of agar was removed to expose the head. A 10 um diameter transplant pipette was inserted through the 4th ventricle into the hindbrain. Motor neurons were visualized using Tg(isl1:kaede) or Tg(isl1:mRFP) on a Zeiss AxioSkop fixed-stage microscope fitted with a 40X long working distance water-immersion lens. Motor neurons were removed from either the anterior-most or posterior-most end of the vagus territory using an oil-controlled syringe mounted on a hydraulic micromanipulator [68] and transplanted to the anterior-most or posterior-most end of the host vagus territory, visualized by Tg(isl1:GFP) or Tg(isl1:mRFP). In the case of transplanting hoxb5aGFP− mX neurons, we relied upon positional information to isolate hoxb5aGFP− cells, as the GFP signal was too weak to be visualized on the AxioSkop. However, we took donor cells only from the most anterior end of the mX territory, which we knew to be reliably hoxb5aGFP-negative on the confocal, and our control anterior → anterior transplants demonstrated that we successfully isolated a hoxb5aGFP− mX neuron at least 77% of the time (Figure 3F). After transplantation, embryos were staged using the prim method (see above), then unmounted from the agar and allowed to recover in normal Ringer’s solution. For time lapse experiments, embryos were given at least 1 hour of recovery time before remounting for confocal imaging. For axon targeting experiments, embryos were imaged at 3 dpf.
Live imaging
All images were collected on a Zeiss LSM 700 confocal microscope. For Kaede photoconversion, a region of interest was defined using morphological boundaries, and protein was photoconverted used the 405 laser at 10% power. Since initially axons are difficult to distinguish from other dynamic processes, we determined the time of axon initiation in timelapse movies, we tracked the axon backward over time until the earliest timepoint it could be detected.
RNA in situ hybridization
Anesthetized embryos were fixed in 4% paraformaldehyde with 1X PBS (phosphate-buffered saline) and 4% sucrose at 4°C overnight. The RNA in situ hybridization protocol was followed from [69]. For dual in situ and antibody staining, primary antibody staining (chicken anti-GFP, abcam 13970, 1:500) was performed in conjunction with anti-Digoxigenin staining. Upon completion of in situ protocol, samples were re-blocked for 1 hour, stained with secondary antibody (goat anti-chicken 488, Molecular Probes, 1:500), and washed 3× with PBS+0.1% Tween 20. Following staining brain tissue was dissected, cleared step-wise into 75% glycerol, mounted, and imaged on a Zeiss Axioplan2 microscope (color imaging) or Zeiss LSM700 microscope (fluorescent imaging). For dual in situ and antibody imaging, color and fluorescent images were collected with the same pixel dimensions and the fluorescent image was aligned to the color image using the LSM DIC image. Probes for all hox genes were previously published [31,32].
EdU labeling
Embryos were incubated in 0.5 mM (2′S)-2′-deoxy-2′-fluoro-5-ethynyluridine (EdU, Sigma T511293)) diluted in fish water. Embryos were anesthetized and fixed at 48 hpf in 4% paraformaldehyde with 1X PBS (phosphate-buffered saline) and 4% sucrose for 30 minutes at room temperature. After fixation, embryos were permeabilized in PBS + 0.5% TritonX100 for 30 minutes at room temperature, and brain tissue was dissected. To visualized EdU labeling, brains were incubated in a solution containing 10uM Cy5-azide (Lumiprobe A2020), 2 mM copper(II) sulfate (Sigma 45167), and 20 mM sodium ascorbate (Sigma A7631) for 1 hour at room temperature. Following incubation, brains were washed in PBS + 0.5% TritonX100 and processed for immunofluorescence using standard blocking and antibody incubations. The antibody used was chicken anti-GFP (1:250, Abcam) followed by goat anti-chicken 488 (Molecular Probes, 1:250).
Quantification and Statistical Analysis
To determine time of axon initiation, we started at the end of the time lapse when the axon was easily identified and then backtracked through the time lapse until the first frame when that particular protrusion was visible. Statistical analysis was done using GraphPad Prism for all tests except for transplant experiments that used 2×3 Fisher’s exact test (Figure 2), which was done using Vassar Stats. Statistical details are noted in figure legends. For transplant experiments, some embryos had multiple donor mX axons innervating multiple branches. Therefore, to ensure each host embryo was counted only once in the analysis, we grouped embryos by whether they innervated an anterior branch (PA4, PA5, PA6), a posterior branch (PA7, visceral), or both. For all figures, *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
Supplementary Material
Movie S1. Peripheral outgrowth of posterior mX axon branches is delayed compared to anterior mX axon branches. Related to Figure 1. Time lapse of embryo expressing Tg(isl1:kaede) beginning at 42 hpf. mX axon branches appear in the head periphery sequentially from anterior to posterior. Anterior left, dorsal up.
Movie S2. Sequential anterior-to-posterior PA development and mX axon growth. Related to Figure 1. Three concatenated timelapses of embryos expressing Tg(isl1:eGFPCAAX) (green, marking cranial motor neurons) and Tg(tcf21:mCherry) (magenta, marking PA muscle progenitors). The start time for each of the three parts of the movie was determined by prim staging; subsequent time stamps are estimated based on elapsed time. Vagus axons enter each PA 6–8 hours after the onset of tcf21:mCherry expression. Anterior left, dorsal up.
Movie S3. Axon formation is delayed in posterior mX neurons. Related to Figure 5. Time lapse of embryo expressing Tg(isl1:kaede) beginning at 29 hpf. Posterior mX neurons are photoconverted, shown in magenta (left) and black (right). Axons from anterior mX neurons form at 29 hpf and immediately turn ventrally to extend into the periphery, while axons from posterior mX neurons are not detectable until 37 hpf, though they show extensive multipolar protrusive activity. Anterior left, dorsal up.
Movie S4. Vagus motor neuron position determines time of axon formation. Related to Figure 5. Time lapse after transplantation of donor anterior mX neuron, marked by Tg(isl1:kaede) in magenta (left) and black (right), into anterior mX territory (top) or posterior mX territory (bottom). Host mX neurons, marked by Tg(isl1:mRFP), are shown in green. Time stamp indicates hours post transplantation (hpt). In this example, an anterior mX neuron transplanted homotopically initiates axon formation at 4.3 hpt, while an anterior mX neuron transplanted heterotopically initiates axon formation at 9 hpt. Anterior left, dorsal up.
Movie S5. Vagus motor neuron position determines time of axon formation. Related to Figure 5. Time lapse after transplantation of donor posterior mX neuron, marked by Tg(isl1:kaede) in magenta (left) and black (right), into posterior mX territory (top) or anterior mX territory (bottom). Host mX neurons, marked by Tg(isl1:mRFP), are shown in green. Time stamp indicates hours post transplantation (hpt). In this example, a posterior mX neuron transplanted homotopically initiates axon formation at 7.8 hpt, while a posterior mX neuron transplanted heterotopically initiates axon formation at 7 hpt. Anterior left, dorsal up.
Movie S6. hox5 gene expression does not affect time of axon formation. Related to Figure 7 and Movies S7–S9. Time lapse showing mX neuron labeled by control isl1:eGFPCAAX construct in magenta (left) and black (right). All mX neurons express Tg(isl1:mRFP) (green). Time stamp indicates hours post fertilization (hpf). In this example, a control mX neuron in the anterior region forms an axon at 28.2 hpf.
Movie S7. hox5 gene expression does not affect time of axon formation. Related to Figure 7 and Movies S6, S8, S9. Time lapse showing mX neuron labeled by UAS:hoxa5a-p2a-eGFPCAAX construct in magenta (left) and black (right) in embryo carrying Tg(isl1:Gal4). All mX neurons are labeled by Tg(isl1:mRFP) (green). Time stamp indicates hours post fertilization (hpf). In this example, an ectopic hoxa5a-expressing mX neuron in the anterior region forms an axon at 28.2 hpf.
Movie S8. hox5 gene expression does not affect time of axon formation. Related to Figure 7 and Movies S6, S7, S9. Time lapse showing mX neuron labeled by UAS:hoxb5a-p2a-eGFPCAAX construct in magenta (left) and black (right) in embryo carrying Tg(isl1:Gal4). All mX neurons are labeled by Tg(isl1:mRFP) (green). Time stamp indicates hours post fertilization (hpf). In this example, an ectopic hoxb5a-expressing mX neuron in the anterior region forms an axon at 27 hpf.
Movie S9. hox5 gene expression does not affect time of axon formation. Related to Figure 7 and Movies S6–S8. Time lapse showing mX neuron labeled by UAS:hoxb5b-p2a-eGFPCAAX construct in magenta (left) and black (right) in embryo carrying Tg(isl1:Gal4). All mX neurons are labeled by Tg(isl1:mRFP) (green). Time stamp indicates hours post fertilization (hpf). In this example, an ectopic hoxb5b-expressing mX neuron in the anterior region forms an axon at 28.8 hpf.
HIGHLIGHTS.
-
-
Time of axon formation is differentially regulated independent of neuron birthdate
-
-
hox5 genes are expressed in posterior but not anterior vagus motor neurons
-
-
hox5 directs vagus topographic map formation independent of time of axon outgrowth
-
-
Time of axon outgrowth directs vagus topographic map formation independent of hox5
Acknowledgments
We thank Rachel Garcia for excellent animal care and Arish Shah and Jason Stonick for generating constructs for hox5 gene expression and targeting. Victoria Prince, Holger Knaut, Anita Ng, Jeremy Rabinowitz, Herwig Baier, and Lisa Maves generously provided transgenic lines, constructs, and reagents. We thank Jeremy Rabinowitz, Adam Miller, and members of the Moens Lab for discussions and editing. Funding was provided by National Institutes of Health grants R01 NS082567 to C.B.M. and F30 NS093703 to G.R.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
G.R.B. and C.B.M. designed and performed experiments, analyzed data, and wrote the manuscript. G.R.B. and A.J.I. acquired and quantified data and generated images for publication. All authors edited the manuscript.
References
- 1.Cang J, Feldheim DA. Developmental Mechanisms of Topographic Map Formation and Alignment. Annu Rev Neurosci. 2013;36:51–77. doi: 10.1146/annurev-neuro-062012-170341. [DOI] [PubMed] [Google Scholar]
- 2.Triplett JW, Feldheim DA. Eph and ephrin signaling in the formation of topographic maps. Semin Cell Dev Biol. 2012;23:7–15. doi: 10.1016/j.semcdb.2011.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sperry RW. Chemoaffinity in the orderly growth of nerve fiber patterns and connections. Proc Natl Acad Sci U S A. 1963;50:703–10. doi: 10.1073/pnas.50.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng HJ, Nakamoto M, Bergemann AD, Flanagan JG. Complementary gradients in expression and binding of ELF-1 and Mek4 in development of the topographic retinotectal projection map. Cell. 1995;82:371–381. doi: 10.1016/0092-8674(95)90426-3. [DOI] [PubMed] [Google Scholar]
- 5.Drescher U, Kremoser C, Handwerker C, Löschinger J, Noda M, Bonhoeffer F. In vitro guidance of retinal ganglion cell axons by RAGS, a 25 kDa tectal protein related to ligands for Eph receptor tyrosine kinases. Cell. 1995;82:359–370. doi: 10.1016/0092-8674(95)90425-5. [DOI] [PubMed] [Google Scholar]
- 6.Nakamoto M, Cheng HJ, Friedman GC, McLaughlin T, Hansen MJ, Yoon CH, O’Leary DDM, Flanagan JG. Topographically specific effects of ELF-1 on retinal axon guidance in vitro and retinal axon mapping in vivo. Cell. 1996;86:755–766. doi: 10.1016/s0092-8674(00)80150-6. [DOI] [PubMed] [Google Scholar]
- 7.Frisen J, Yates PA, McLaughlin T, Friedman GC, O’Leary DDM, Barbacid M. Ephrin-A5 (AL-1/RAGS) is essential for proper retinal axon guidance and topographic mapping in the mammalian visual system. Neuron. 1998;20:235–243. doi: 10.1016/s0896-6273(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 8.Feldheim DA, Kim YI, Bergemann AD, Frisé J, Barbacid M, Flanagan JG. Genetic Analysis of Ephrin-A2 and Ephrin-A5 Shows Their Requirement in Multiple Aspects of Retinocollicular Mapping. Neuron. 2000;25:563–574. doi: 10.1016/s0896-6273(00)81060-0. [DOI] [PubMed] [Google Scholar]
- 9.Pfeiffenberger C, Yamada J, Feldheim DA. Ephrin-As and patterned retinal activity act together in the development of topographic maps in the primary visual system. J Neurosci. 2006;26:12873–84. doi: 10.1523/JNEUROSCI.3595-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landmesser L. The development of motor projection patterns in the chick hind limb. J Physiol. 1978;284:391–414. doi: 10.1113/jphysiol.1978.sp012546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsuchida T, Ensini M, Morton SB, Baldassare M, Edlund T, Jessell TM, Pfaff SL. Topographic organization of embryonic motor neurons defined by expression of {LIM} homeobox genes. Cell. 1994;79:957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- 12.Kania A, Jessell TM. Topographic motor projections in the limb imposed by {LIM} homeodomain protein regulation of {ephrin-A:EphA} interactions. 2003;38:581–596. doi: 10.1016/s0896-6273(03)00292-7. [DOI] [PubMed] [Google Scholar]
- 13.Luria V, Krawchuk D, Jessell TM, Laufer E, Kania A. Specification of Motor Axon Trajectory by Ephrin-B:EphB Signaling: Symmetrical Control of Axonal Patterning in the Developing Limb. Neuron. 2008;60:1039–1053. doi: 10.1016/j.neuron.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 14.Poliak S, Morales D, Croteau LP, Krawchuk D, Palmesino E, Morton S, Jean-François C, Charron F, Dalva MB, Ackerman SL, et al. Synergistic integration of Netrin and ephrin axon guidance signals by spinal motor neurons Sebastian. Elife. 2015;4:1–26. doi: 10.7554/eLife.10841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petrovic M, Hummel T. Temporal identity in axonal target layer recognition. Nature. 2008;456:800–803. doi: 10.1038/nature07407. [DOI] [PubMed] [Google Scholar]
- 16.Takeuchi H, Inokuchi K, Aoki M, Suto F, Tsuboi A, Matsuda I, Suzuki M, Aiba A, Serizawa S, Yoshihara Y, et al. Sequential arrival and graded secretion of Sema3F by olfactory neuron axons specify map topography at the bulb. Cell. 2010;141:1056–1067. doi: 10.1016/j.cell.2010.04.041. [DOI] [PubMed] [Google Scholar]
- 17.Eerdunfu, Ihara N, Ligao B, Ikegaya Y, Takeuchi H. Differential timing of neurogenesis underlies dorsal-ventral topographic projection of olfactory sensory neurons. Neural Dev. 2017;12:2. doi: 10.1186/s13064-017-0079-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilland E, Baker R. Evolutionary patterns of cranial nerve efferent nuclei in vertebrates. Brain Behav Evol. 2005;66:234–254. doi: 10.1159/000088128. [DOI] [PubMed] [Google Scholar]
- 19.Erman AB, Kejner AE, Hogikyan ND, Feldman EL. Disorders of cranial nerves IX and X. Semin. Neurol. 2009;29:85–92. doi: 10.1055/s-0028-1124027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schilling T, Kimmel C. Musculoskeletal patterning in the pharyngeal segments of the zebrafish embryo. Development. 1997;124:2945–2960. doi: 10.1242/dev.124.15.2945. [DOI] [PubMed] [Google Scholar]
- 21.Bieger D, Hopkins DA. Viscerotopic representation of the upper alimentary tract in the medulla oblongata in the rat: The nucleus ambiguus. J Comp Neurol. 1987;262:546–562. doi: 10.1002/cne.902620408. [DOI] [PubMed] [Google Scholar]
- 22.Morita Y, Finger TE. Topographic and laminar organization of the vagal gustatory system in the goldfish, carassius auratus. J Comp Neurol. 1985;238:187–201. doi: 10.1002/cne.902380206. [DOI] [PubMed] [Google Scholar]
- 23.Higashijima S, Hotta Y, Okamoto H. Visualization of cranial motor neurons in live transgenic zebrafish expressing green fluorescent protein under the control of the islet-1 promoter/enhancer. J Neurosci. 2000;20:206–218. doi: 10.1523/JNEUROSCI.20-01-00206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohata S, Kinoshita S, Aoki R, Tanaka H, Wada H, S T-K, Tsuboi T, Watabe S, Okamoto H. Neuroepithelial cells require fucosylated glycans to guide the migration of vagus motor neuron progenitors in the developing zebrafish hindbrain. Development. 2009;136:1653–1663. doi: 10.1242/dev.033290. [DOI] [PubMed] [Google Scholar]
- 25.Cox J, Lamora A, Johnson S, Voigt M. Diverse mechanisms for assembly of branchiomeric nerves. Dev Biol. 2011;357:305–317. doi: 10.1016/j.ydbio.2011.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Cao J, Dickson AL, Poss KD. Epicardial regeneration is guided by cardiac outflow tract and Hedgehog signalling. Nature. 2015;522:226–30. doi: 10.1038/nature14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagelberg D, Wang J, Su R, Torres-Vázquez J, Targoff KL, Poss KD, Knaut H. Origin, specification, and plasticity of the great vessels of the heart. Curr Biol. 2015;25:2099–2110. doi: 10.1016/j.cub.2015.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eisen J. Determination of primary motoneuron identity in developing zebrafish embryos. Science (80-) 1991;252:569–572. doi: 10.1126/science.1708527. [DOI] [PubMed] [Google Scholar]
- 29.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 30.Philippidou P, Dasen J. Hox genes: choreographers in neural development, architects of circuit organization. Neuron. 2013;80:12–34. doi: 10.1016/j.neuron.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prince VE, Joly L, Ekker M, Ho RK. Zebrafish hox genes: genomic organization and modified colinear expression patterns in the trunk. Development. 1998;125:407–420. doi: 10.1242/dev.125.3.407. [DOI] [PubMed] [Google Scholar]
- 32.Prince VE, Moens CB, Kimmel CB, Ho RK. Zebrafish hox genes: expression in the hindbrain region of wild-type and mutants of the segmentation gene, valentino. Development. 1998;125:393–406. doi: 10.1242/dev.125.3.393. [DOI] [PubMed] [Google Scholar]
- 33.Bruce AEE, Oates AC, Prince VE, Ho RK. Additional hox clusters in the zebrafish: Divergent expression patterns belie equivalent activities of duplicate hoxB5 genes. Evol Dev. 2001;3:127–144. doi: 10.1046/j.1525-142x.2001.003003127.x. [DOI] [PubMed] [Google Scholar]
- 34.Hortopan GA, Baraban SC. Aberrant expression of genes necessary for neuronal development and notch signaling in an epileptic mind bomb zebrafish. Dev Dyn. 2011;240:1964–1976. doi: 10.1002/dvdy.22680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimura Y, Hisano Y, Kawahara A, Higashijima S. Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci Rep. 2014;4:6545. doi: 10.1038/srep06545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hrycaj SM, Dye BR, Baker NC, Larsen BM, Burke AC, Spence JR, Wellik DM. Hox5 genes regulate the Wnt2/2b-Bmp4-signaling axis during lung development. Cell Rep. 2015;12:903–912. doi: 10.1016/j.celrep.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karmakar K, Narita Y, Fadok J, Ducret S, Loche A, Kitazawa T, Genoud C, Di Meglio T, Thierry R, Bacelo J, et al. Hox2 Genes Are Required for Tonotopic Map Precision and Sound Discrimination in the Mouse Auditory Brainstem. Cell Rep. 2017;18:185–197. doi: 10.1016/j.celrep.2016.12.021. [DOI] [PubMed] [Google Scholar]
- 38.Dasen JS, Liu JP, Jessell TM. Motor neuron columnar fate imposed by sequential phases of Hox-c activity. Nature. 2003;425:926–933. doi: 10.1038/nature02051. [DOI] [PubMed] [Google Scholar]
- 39.Dasen JS, Tice BC, Brenner-Morton S, Jessell TM. A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell. 2005;123:477–491. doi: 10.1016/j.cell.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 40.Oury F, Murakami Y, Renaud J, Pasqualetti M, Charnay P, Ren S, Rijli FM. Hoxa2- and Rhombomere-Dependent Development of the Mouse Facial Somatosensory Map. Science (80-) 2006:1408–1413. doi: 10.1126/science.1130042. [DOI] [PubMed] [Google Scholar]
- 41.Nern A, Zhu Y, Zipursky SL. Local N-Cadherin Interactions Mediate Distinct Steps in the Targeting of Lamina Neurons. Neuron. 2008;58:34–41. doi: 10.1016/j.neuron.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duan X, Krishnaswamy A, De la Huerta I, Sanes JR. Type II Cadherins Guide Assembly of a Direction-Selective Retinal Circuit. Cell. 2014;158:793–807. doi: 10.1016/j.cell.2014.06.047. [DOI] [PubMed] [Google Scholar]
- 43.Osterhout JA, El-Danaf RN, Nguyen PL, Huberman AD. Birthdate and outgrowth timing predict cellular mechanisms of axon target matching in the developing visual pathway. Cell Rep. 2014;8:1006–17. doi: 10.1016/j.celrep.2014.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kulkarni A, Ertekin D, Lee CH, Hummel T. Birth order dependent growth cone segregation determines synaptic layer identity in the Drosophila visual system. Elife. 2016;5 doi: 10.7554/eLife.13715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zolessi FR, Poggi L, Wilkinson CJ, Chien CB, Harris WA. Polarization and orientation of retinal ganglion cells in vivo. Neural Dev. 2006;1:2. doi: 10.1186/1749-8104-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morgan JL, Dhingra A, Vardi N, Wong RO. Axons and dendrites originate from neuroepithelial-like processes of retinal bipolar cells. Nat Neurosci. 2006;9:85–92. doi: 10.1038/nn1615. [DOI] [PubMed] [Google Scholar]
- 47.Noctor SC, Martínez-Cerdeño V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- 48.Hatanaka Y, Yamauchi K. Excitatory cortical neurons with multipolar shape establish neuronal polarity by forming a tangentially oriented axon in the intermediate zone. Cereb Cortex. 2013;23:105–113. doi: 10.1093/cercor/bhr383. [DOI] [PubMed] [Google Scholar]
- 49.Namba T, Kibe Y, Funahashi Y, Nakamuta S, Takano T, Ueno T, Shimada A, Kozawa S, Okamoto M, Shimoda Y, et al. Pioneering axons regulate neuronal polarization in the developing cerebral cortex. Neuron. 2014;81:814–829. doi: 10.1016/j.neuron.2013.12.015. [DOI] [PubMed] [Google Scholar]
- 50.Shi SH, Jan LY, Jan YN. Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell. 2003;112:63–75. doi: 10.1016/s0092-8674(02)01249-7. [DOI] [PubMed] [Google Scholar]
- 51.Kishi M, Pan YA, Crump JG, Sanes JR. Mammalian SAD kinases are required for neuronal polarization. Science (80-) 2005;307:929–932. doi: 10.1126/science.1107403. [DOI] [PubMed] [Google Scholar]
- 52.Barnes AP, Lilley BN, Pan YA, Plummer LJ, Powell AW, Raines AN, Sanes JR, Polleux F. LKB1 and SAD Kinases Define a Pathway Required for the Polarization of Cortical Neurons. Cell. 2007;129:549–563. doi: 10.1016/j.cell.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 53.Shelly M, Cancedda L, Heilshorn S, Sumbre G, Poo M ming. LKB1/STRAD Promotes Axon Initiation During Neuronal Polarization. Cell. 2007;129:565–577. doi: 10.1016/j.cell.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 54.Chen S, Chen J, Shi H, Wei M, Castaneda-Castellanos DR, Bultje RS, Pei X, Kriegstein AR, Zhang M, Shi SH. Regulation of Microtubule Stability and Organization by Mammalian Par3 in Specifying Neuronal Polarity. Dev Cell. 2013;24:26–40. doi: 10.1016/j.devcel.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Witte H, Neukirchen D, Bradke F. Microtubule stabilization specifies initial neuronal polarization. J Cell Biol. 2008;180:619–632. doi: 10.1083/jcb.200707042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yau KW, vanBeuningen SFB, Cunha-Ferreira I, Cloin BMC, vanBattum EY, Will L, Schatzle P, Tas RP, vanKrugten J, Katrukha EA, et al. Microtubule minus-end binding protein CAMSAP2 controls axon specification and dendrite development. Neuron. 2014;82:1058–1073. doi: 10.1016/j.neuron.2014.04.019. [DOI] [PubMed] [Google Scholar]
- 57.Van Beuningen SFB, Will L, Harterink M, Chazeau A, Van Battum EY, Frias CP, Franker MAM, Katrukha EA, Stucchi R, Vocking K, et al. TRIM46 Controls Neuronal Polarity and Axon Specification by Driving the Formation of Parallel Microtubule Arrays. Neuron. 2015;88:1208–1226. doi: 10.1016/j.neuron.2015.11.012. [DOI] [PubMed] [Google Scholar]
- 58.Yi JJ, Barnes AP, Hand R, Polleux F, Ehlers MD. TGF-beta Signaling Specifies Axons during Brain Development. Cell. 2010;142:144–157. doi: 10.1016/j.cell.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Randlett O, Poggi L, Zolessi FR, Harris WA. The Oriented Emergence of Axons from Retinal Ganglion Cells Is Directed by Laminin Contact In Vivo. Neuron. 2011;70:266–280. doi: 10.1016/j.neuron.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shelly M, Cancedda L, Lim BK, Popescu AT, Cheng P Lin, Gao H, Poo M Ming. Semaphorin3A regulates neuronal polarization by suppressing axon formation and promoting dendrite growth. Neuron. 2011;71:433–446. doi: 10.1016/j.neuron.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Caton A, Hacker A, Naeem A, Livet J, Maina F, Bladt F, Klein R, Birchmeier C, Guthrie S. The branchial arches and HGF are growth-promoting and chemoattractant for cranial motor axons. Development. 2000;127:1751–1766. doi: 10.1242/dev.127.8.1751. [DOI] [PubMed] [Google Scholar]
- 62.Westerfield M. The Zebrafish Book Eugene. University of Oregon Press; 2000. [Google Scholar]
- 63.Siegfried KR. In search of determinants: gene expression during gonadal sex differentiation. J Fish Biol. 2010;76:1879–902. doi: 10.1111/j.1095-8649.2010.02594.x. [DOI] [PubMed] [Google Scholar]
- 64.Davey CF, Mathewson AW, Moens CB. PCP Signaling between Migrating Neurons and their Planar-Polarized Neuroepithelial Environment Controls Filopodial Dynamics and Directional Migration. PLoS Genet. 2016;12 doi: 10.1371/journal.pgen.1005934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Davison JM, Akitake CM, Goll MG, Rhee JM, Gosse N, Baier H, Halpern ME, Leach SD, Parsons MJ. Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev Biol. 2007;304:811–24. doi: 10.1016/j.ydbio.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grant P, Moens C. The neuroepithelial basement membrane serves as a boundary and a substrate for neuron migration in the zebrafish hindbrain. Neural Dev. 2010;5:9. doi: 10.1186/1749-8104-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–99. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- 68.Kemp HA, Carmany-Rampey A, Moens C. Generating chimeric zebrafish embryos by transplantation. J Vis Exp. 2009 doi: 10.3791/1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Peripheral outgrowth of posterior mX axon branches is delayed compared to anterior mX axon branches. Related to Figure 1. Time lapse of embryo expressing Tg(isl1:kaede) beginning at 42 hpf. mX axon branches appear in the head periphery sequentially from anterior to posterior. Anterior left, dorsal up.
Movie S2. Sequential anterior-to-posterior PA development and mX axon growth. Related to Figure 1. Three concatenated timelapses of embryos expressing Tg(isl1:eGFPCAAX) (green, marking cranial motor neurons) and Tg(tcf21:mCherry) (magenta, marking PA muscle progenitors). The start time for each of the three parts of the movie was determined by prim staging; subsequent time stamps are estimated based on elapsed time. Vagus axons enter each PA 6–8 hours after the onset of tcf21:mCherry expression. Anterior left, dorsal up.
Movie S3. Axon formation is delayed in posterior mX neurons. Related to Figure 5. Time lapse of embryo expressing Tg(isl1:kaede) beginning at 29 hpf. Posterior mX neurons are photoconverted, shown in magenta (left) and black (right). Axons from anterior mX neurons form at 29 hpf and immediately turn ventrally to extend into the periphery, while axons from posterior mX neurons are not detectable until 37 hpf, though they show extensive multipolar protrusive activity. Anterior left, dorsal up.
Movie S4. Vagus motor neuron position determines time of axon formation. Related to Figure 5. Time lapse after transplantation of donor anterior mX neuron, marked by Tg(isl1:kaede) in magenta (left) and black (right), into anterior mX territory (top) or posterior mX territory (bottom). Host mX neurons, marked by Tg(isl1:mRFP), are shown in green. Time stamp indicates hours post transplantation (hpt). In this example, an anterior mX neuron transplanted homotopically initiates axon formation at 4.3 hpt, while an anterior mX neuron transplanted heterotopically initiates axon formation at 9 hpt. Anterior left, dorsal up.
Movie S5. Vagus motor neuron position determines time of axon formation. Related to Figure 5. Time lapse after transplantation of donor posterior mX neuron, marked by Tg(isl1:kaede) in magenta (left) and black (right), into posterior mX territory (top) or anterior mX territory (bottom). Host mX neurons, marked by Tg(isl1:mRFP), are shown in green. Time stamp indicates hours post transplantation (hpt). In this example, a posterior mX neuron transplanted homotopically initiates axon formation at 7.8 hpt, while a posterior mX neuron transplanted heterotopically initiates axon formation at 7 hpt. Anterior left, dorsal up.
Movie S6. hox5 gene expression does not affect time of axon formation. Related to Figure 7 and Movies S7–S9. Time lapse showing mX neuron labeled by control isl1:eGFPCAAX construct in magenta (left) and black (right). All mX neurons express Tg(isl1:mRFP) (green). Time stamp indicates hours post fertilization (hpf). In this example, a control mX neuron in the anterior region forms an axon at 28.2 hpf.
Movie S7. hox5 gene expression does not affect time of axon formation. Related to Figure 7 and Movies S6, S8, S9. Time lapse showing mX neuron labeled by UAS:hoxa5a-p2a-eGFPCAAX construct in magenta (left) and black (right) in embryo carrying Tg(isl1:Gal4). All mX neurons are labeled by Tg(isl1:mRFP) (green). Time stamp indicates hours post fertilization (hpf). In this example, an ectopic hoxa5a-expressing mX neuron in the anterior region forms an axon at 28.2 hpf.
Movie S8. hox5 gene expression does not affect time of axon formation. Related to Figure 7 and Movies S6, S7, S9. Time lapse showing mX neuron labeled by UAS:hoxb5a-p2a-eGFPCAAX construct in magenta (left) and black (right) in embryo carrying Tg(isl1:Gal4). All mX neurons are labeled by Tg(isl1:mRFP) (green). Time stamp indicates hours post fertilization (hpf). In this example, an ectopic hoxb5a-expressing mX neuron in the anterior region forms an axon at 27 hpf.
Movie S9. hox5 gene expression does not affect time of axon formation. Related to Figure 7 and Movies S6–S8. Time lapse showing mX neuron labeled by UAS:hoxb5b-p2a-eGFPCAAX construct in magenta (left) and black (right) in embryo carrying Tg(isl1:Gal4). All mX neurons are labeled by Tg(isl1:mRFP) (green). Time stamp indicates hours post fertilization (hpf). In this example, an ectopic hoxb5b-expressing mX neuron in the anterior region forms an axon at 28.8 hpf.


