Abstract
Parkinson’s disease (PD) is a neurodegenerative disorder characterized by progressive loss of dopaminergic neurons in the substantia nigra of the human brain, leading to depletion of dopamine production. Dopamine replacement therapy remains the mainstay for attenuation of PD symptoms. Nonetheless, the potential benefit of current pharmacotherapies is mostly limited by adverse side effects, such as drug-induced dyskinesia, motor fluctuations and psychosis. Non-dopaminergic receptors, such as human A2A adenosine receptors, have emerged as important therapeutic targets in potentiating therapeutic effects and reducing the unwanted side effects. In this study, new chemical entities targeting both human A2A adenosine receptor and dopamine D2 receptor were designed and evaluated. Two computational methods, namely support vector machine (SVM) models and Tanimoto similarity-based clustering analysis, were integrated for the identification of compounds containing indole-piperazine-pyrimidine (IPP) scaffold. Subsequent synthesis and testing resulted in compounds 5 and 6, which acted as human A2A adenosine receptor binders in the radioligand competition assay (Ki = 8.7–11.2 μM) as well as human dopamine D2 receptor binders in the artificial cell membrane assay (EC50 = 22.5–40.2 μM). Moreover, compound 5 showed improvement in movement and mitigation of the loss of dopaminergic neurons in Drosophila models of PD. Furthermore, in vitro toxicity studies on compounds 5 and 6 did not reveal any mutagenicity (up to 100 μM), hepatotoxicity (up to 30 μM) or cardiotoxicity (up to 30 μM).
Introduction
Parkinson’s disease (PD) is a neurodegenerative disorder characterized by cardinal motor features including tremor, rigidity, bradykinesia and postural instability. It is pathologically associated with loss of dopaminergic neurons in the substantia nigra of the human brain, leading to depletion of dopamine production [1]. Over the years, development of pharmacotherapy for PD has been largely focused on improving motor symptoms caused by dopamine deficiency. Among these pharmacotherapies, dopamine replacement therapy represents the major therapeutic approach to alleviate symptoms by restoring dopamine levels. L-DOPA (Fig 1), a metabolic precursor of dopamine, remains the most effective dopamine replacement therapy for improving motor deficits. It is able to cross the blood brain barrier (BBB) and is efficiently converted into dopamine by enzymatic decarboxylation [2]. Nevertheless, chronic administration of L-DOPA has been associated with side effects such as dyskinesia, end-of-dose deterioration of function and a switch between mobility and immobility (on/off phenomenon) in the treated patients [3,4]. Hence, L-DOPA is often co-administered with other adjuvant drugs to overcome these side effects. For instance, it is co-administered with dopamine agonists to increase the activity of the dopamine system, or monoamine oxidase B (MAO B) inhibitors and catechol-O-methyltransferase (COMT) inhibitors to prevent the metabolism of dopamine by these enzymes, thus increasing dopamine concentration in the brain. However, these adjuvant drugs are still inadequate in reducing the parkinsonian motor disabilities [5].
Fig 1. Structures of L-DOPA, KW-6002 and quinpirole.
In recent years, non-dopaminergic receptors have been identified to play key roles in the pathophysiology of PD. Among these targets, A2A adenosine receptor (A2AAR) has gained much attention as an important pharmacological target in counteracting the motor symptoms of PD [6]. It is co-expressed with dopamine D2 receptors in striato-pallidal neurons where these receptors are known to form heterodimeric complexes [7,8]. The stimulation of A2AAR has been shown to decrease the affinity of D2 receptor agonists. Studies have demonstrated that blockade of A2AAR through the action of antagonists amplifies the therapeutic effect of L-DOPA and reduces the L-DOPA-induced dyskinesia [8–10]. In addition, A2A antagonists were also reported to exert a neuroprotective effect in which they were able to prevent the onset and development of PD [11]. For these reasons, the combination of hA2AAR antagonists and L-DOPA has been investigated for improved efficacy relative to dopamine replacement mono-therapy.
Indeed, tremendous effort has been made towards the development of effective drugs alongside the identification of new therapeutic targets for treatment of PD symptoms. In the past, pharmacotherapies for PD have mostly focused on selective compounds targeting individual proteins (“one compound-one target” approach), particularly the dopamine receptors. Subsequently, discovery of heterodimeric A2A adenosine receptor / dopamine D2 receptor complexes in the striatum has steered the development of combination therapies (“cocktail drug-multiple targets”) containing an adenosine A2A receptor antagonist and either L-DOPA or a dopamine D2 receptor agonist [8,9]. This has been corroborated by marked enhancement of anti-PD activity in 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-treated marmosets administered with combination therapy consisting of KW-6002 (an adenosine A2A receptor antagonist, Ki hA2A = 12 nM, Fig 1) and L-DOPA or of KW-6002 and quinpirole (a dopamine D2 receptor agonist, Ki D2 = 4.8 nM, Fig 1) [12]. Nonetheless, these combination therapies are often associated with side effects arising from drug-drug interactions and varying pharmacokinetic or pharmacodynamic profiles of each component drug. Consequently, “one compound-multiple targets” strategy has emerged as an alternative approach to the management of PD. In such approach, a single drug compound is designed to possess pharmacological activities to multiple targets of interest. The single entity can potentially eliminate side effects derived from interactions amongst drugs in the combination therapies, and improve compliance, especially in elderly patients who are commonly prescribed multiple medications to control the PD.
In our present study, we have successfully employed two virtual screening methods to identify novel scaffolds that simultaneously bind the two receptors—adenosine A2A receptor and dopamine D2 receptor—implicated in the PD pathophysiology [13–21].
Results and discussion
Design rationale
Adenosine A2A receptor antagonists were identified from the existing literature. Each compound with reported binding or antagonistic activity at A2A receptor therein was manually drawn by ChemDraw [22], and the relevant pharmacological data were noted down on ChemFinder. A total of 1969 adenosine A2A receptor antagonists, of which 1595 had reported binding data (inhibition constant Ki, in most cases), were collected from 69 publications, and were classified into 94 major scaffolds. Most scaffolds were composed of either a xanthine or nitrogen-containing heterocyclic nucleus. Of these selected 1595 compounds reported to bind at A2A receptor subtype, 418 compounds (S1 Table) were tested for binding at A2A receptor and further shown to be active in advanced assays relevant to PD, including the ability to block cAMP generation [13], mouse catalepsy model [14], rat catalepsy model [15], as well as Ca2+ mobilisation assessment via a fluorescence imaging plate reader (FLIPR) assay [16]. Similarly, a total of 810 dopamine D2 receptor agonists were collected from 71 publications which focused on identifying agonists for D2 receptor. Of these 810 reported compounds, 569 were described as dopamine D2 receptor agonists with reported binding data, and were classified into 78 major scaffolds. Of these 569 compounds, 332 (S1 Table) were tested for binding at D2 receptor and further shown to be active in various advanced assays relevant to PD [17–19]. Moreover, an additional 465 compounds (295 for adenosine A2A receptor and 170 for dopamine D2 receptor) were collected from the MDDR database.
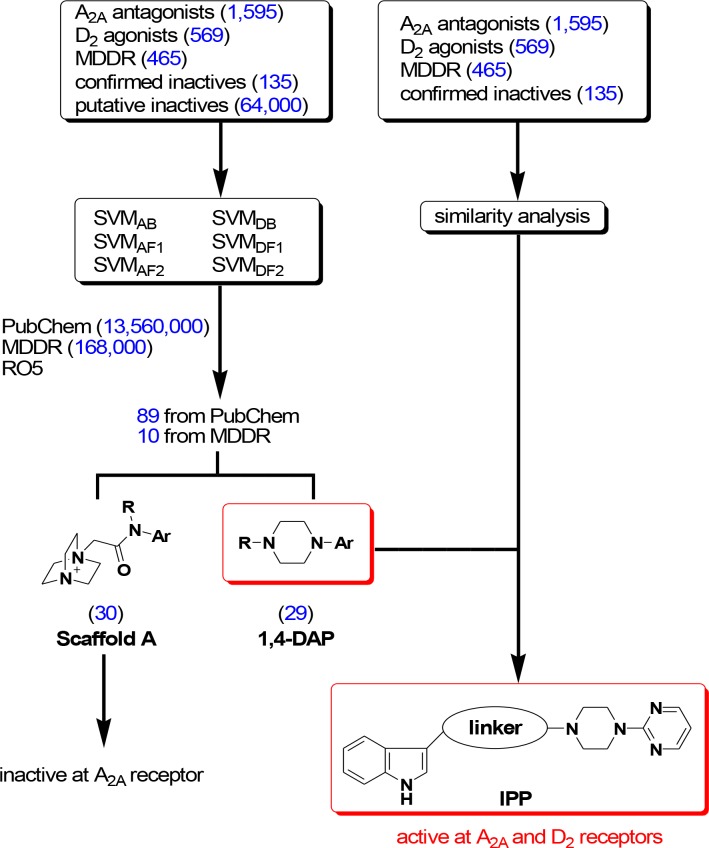
With compound collection from literature and putative inactive families [20] at hand (135 in total, 96 from A2A and 39 from D2 compounds), two ligand-based computational tools—SVM models and Tanimoto similarity-based clustering analysis—were used to analyze these collections. Six SVM models were developed and used for screening 13.56 million compounds in the PubChem database and 168,000 compounds in the MDDR database. A total of 172 hits (162 from PubChem, 10 from MDDR) were identified, and after filtering by Lipinski’s rule of 5, a total of 99 hits (89 from PubChem, 10 from MDDR) were collected (S2 and S3 Tables). Analysis of these 99 hits led to the identification that compounds bearing 1-aza-4-azoniabicyclo[2.2.2]octane moiety (Scaffold A, Fig 2) and compounds bearing 1,4-disubstituted aromatic piperazine (1,4-DAP, Fig 2) constituted the highest percentage in numbers—30.3% for Scaffold A and 29.3% for 1,4-DAP, respectively. Four representative compounds bearing Scaffold A were selected and tested at A2A receptor through the in vitro radioligand displacement assay but did not show binding up to 100 μM. Therefore, the 1,4-DAP scaffold has become the focus of the present study.
Fig 2. Flowchart of virtual screening.
In parallel, a Tanimoto similarity-based clustering analysis using two-dimensional fingerprints [21] was carried out on the collected A2A antagonists and D2 agonists. A total of 1.5 million pairs of compounds were generated. Each pair is composed of an A2A antagonist and a D2 agonist. The degree of structural similarity between an A2A antagonist and a D2 agonist was calculated and expressed by Tanimoto coefficient (Tc). Based on their computed Tc values, the A2A antagonists and D2 agonists were further clustered in a dendrogram to identify regions with high overlapping opportunities. Collectively, it was revealed that compounds having indole and pyrimidine placed at the two terminal ends with a linker of up to 4 carbon atoms (as suggested by the spacers used in both A2A and D2 compounds collected in the original dataset [13–22]) had the potential to bind the two receptors simultaneously. These findings, together with the input of 1,4-DAP identified by SVM models, suggested that pyrimidine was the likely aromatic group in 1,4-DAP and the placement of an indole ring at the other terminal end may be important for binding at the two receptors. Hence, compounds with the substructure indole-piperazine-pyrimidine (IPP, Fig 2) were designed, synthesized, and evaluated in various in vitro and in vivo assays. Of note, small methyl groups were introduced at position 4 and 6 of the pyrimidine, as these substituents were reported to enhance the affinity profile at A2A receptors by ~30 fold, while still displaying a very high structural similarity (Tc > 0.9) with a D2 ligand (S1 Fig).
Chemistry
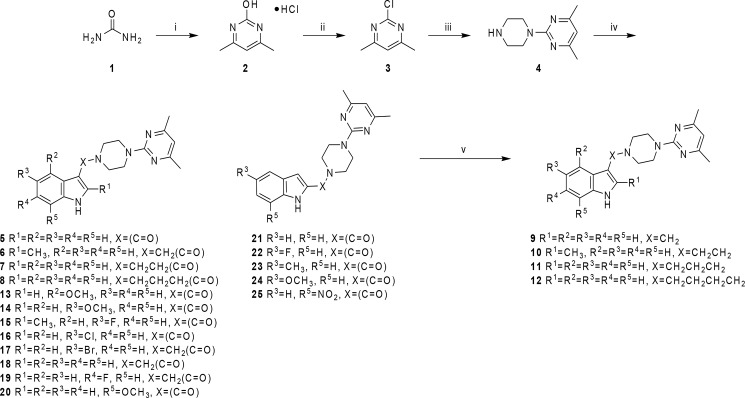
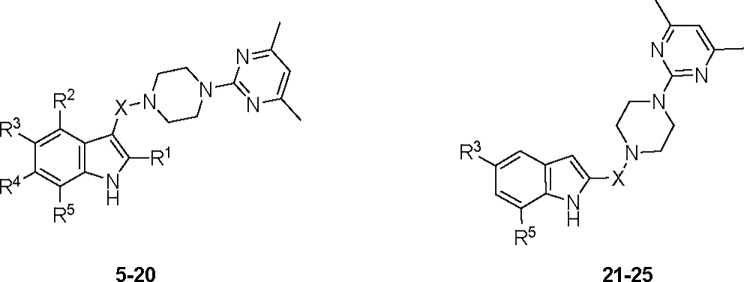
The designed compounds with IPP scaffold were synthesized according to Fig 3. The mixture of urea 1 and 2,4-pentanedione under acidic conditions resulted in the formation of 4,6-dimethylpyrimidin-2-ol hydrochloride (2) with a 78% yield. Following this, treating 2 with phosphorus oxychloride gave 2-chloro-4,6-dimethylpyrimidine (3) with a 91% yield [23]. In order to minimize the formation of 2-[4-(4,6-dimethylpyrimidin-2-yl)piperazin-1-yl]-4,6-dimethylpyrimidine, five equivalents of piperazine were used for coupling with 3 under basic conditions. With 4,6-dimethyl-2-(piperazin-1-yl)pyrimidine (4) on hand, four indole-3-acids (i.e. indole-3-carboxylic acid, 2-methylindole-3-acetic acid, indole-3-propionic acid and indole-3-butyric acid) were selected for 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC)-mediated amide formation to generate compounds 5–8 bearing different lengths of the linker between indole and piperazine rings. To study the effect of the carbonyl group on biological activities, reduction by lithium aluminium hydride (LiAlH4) was performed on 5–8 to give 9–12. Similar procedures were adopted for the preparation of compounds 13–25.
Fig 3.
Reagents and conditions: (i) 2,4-pentanedione, 37% HCl, EtOH, reflux, 24 h, 78%; (ii) POCl3, reflux, 10 h, 91%; (iii) piperazine, K2CO3, H2O, 45–50°C, 4.5 h, 93%; (iv) indole-3-acid or indole-2-acid, EDC. HCl, EtOAc, DMF, 23 → 50°C, 4~4.5 h, 50~74%; (v) LiAlH4, THF, 0 → 23°C, 4–21.5 h, 76–97%.
Binding affinity studies at human adenosine receptors
The synthesized IPP compounds were tested in competition binding assays at human (h) A1, A2A, A2B and A3 adenosine receptors expressed in Chinese Hamster Ovary (CHO) cells (Fig 4and Table 1). Based on the results obtained, it was shown that the presence of a carbonyl group between indole and piperazine rings has led to derivatives with higher affinity at the human A2A (hA2A) receptor than the corresponding compounds without a carbonyl group (i.e. compound 5, Ki hA2A = 11.2 μM, hA1/hA2A >9, hA3/hA2A >9 versus compound 9, Ki hA2A > 30 μM; compound 6, Ki hA2A = 8.71 μM, hA1/hA2A >11, hA3/hA2A >11 versus compound 10, Ki hA2A = 34.4 μM, hA1/hA2A >3, hA3/hA2A = 1.16). This observation indicates the importance of the carbonyl group towards the binding affinity at the hA2A receptor. In addition, it was found that extension of the length of the middle linker from two carbon atoms to three or four carbon atoms resulted in complete loss of A2A affinity (compounds 7 and 8: Ki hA2A > 100 μM). Therefore, it was not unexpected that the reduced forms of compounds 7 and 8 (i.e. compounds 11 and 12) did not show binding up to 100 μM.
Fig 4. Structures of compounds 5–25 tested at human adenosine receptor subtypes.
Table 1. Binding affinity (Ki, μM) of compounds 5–25 at human adenosine receptor subtypes.
| Cpd | R1 | R2 | R3 | R4 | R5 | X | hA1a | hA2Ab | hA2Bc | hA3d | hA1/hA2A | hA3/hA2A |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | H | H | H | H | H | C = O | >100 | 11. 20 (9.86–12.70) |
>20 | >100 | >9 | >9 |
| 6 | CH3 | H | H | H | H | CH2C = O | >100 | 8.71 (6.06–12.50) |
>20 | >100 | >11 | >11 |
| 7 | H | H | H | H | H | CH2CH2C = O | N.D. | >100 | >20 | N.D. | N.D. | N.D. |
| 8 | H | H | H | H | H | CH2CH2CH2C = O | N.D. | >100 | >20 | N.D. | N.D. | N.D. |
| 9 | H | H | H | H | H | CH2 | >100 | >30 | >20 | >30 | N.D. | N.D. |
| 10 | CH3 | H | H | H | H | CH2CH2 | >100 | 34.40 (23.80–49.90) |
>20 | 39.80 (32.50–48.80) |
>3 | 1.16 |
| 11 | H | H | H | H | H | CH2CH2CH2 | N.D. | >100 | >20 | N.D. | N.D. | N.D. |
| 12 | H | H | H | H | H | CH2CH2CH2CH2 | N.D. | >100 | >20 | N.D. | N.D. | N.D. |
| 13 | H | OCH3 | H | H | H | C = O | >100 | 18.80 (15.50–22.70) |
>20 | >30 | >5 | >2 |
| 14 | H | H | OCH3 | H | H | C = O | >100 | 12.70 (10.50–15.40) |
>20 | >30 | >8 | >2 |
| 15 | CH3 | H | F | H | H | CH2C = O | >100 | 18.20 (15.50–21.20) |
>20 | >30 | >6 | >2 |
| 16 | H | H | Cl | H | H | C = O | >100 | 15.50 (13.50–17.80) |
>20 | 19.50 (13.70–27.80) |
>7 | 1.26 |
| 17 | H | H | Br | H | H | CH2C = O | >100 | >100 | >20 | >100 | N.D. | N.D. |
| 18 | H | H | H | H | H | CH2C = O | >100 | 27.60 (21.40–35.70) |
>20 | >30 | >4 | >1 |
| 19 | H | H | H | F | H | CH2C = O | >100 | 26.90 (20.30–35.60) |
>20 | >30 | >4 | >1 |
| 20 | H | H | H | H | OCH3 | C = O | >100 | 3.63 (2.01–6.57) |
>20 | >30 | >28 | >8 |
| 21 | — | — | H | — | H | C = O | >100 | >100 | >20 | >100 | N.D. | N.D. |
| 22 | — | — | F | — | H | C = O | >100 | >100 | >20 | >100 | N.D. | N.D. |
| 23 | — | — | CH3 | — | H | C = O | >100 | >100 | >20 | >100 | N.D. | N.D. |
| 24 | — | — | OCH3 | — | H | C = O | >30 | >30 | >20 | >30 | N.D. | N.D. |
| 25 | — | — | H | — | NO2 | C = O | 4.82 (4.37–5.32) |
29.70 (23.50–37.60) |
>20 | >30 | 0.16 | >1 |
aDisplacement of specific [3H]-2-chloro-6-cyclopentyladenosine ([3H]-CCPA) binding at human A1 receptors expressed in CHO cells (n = 3–6).
bDisplacement of specific [3H]-5’-N-ethylcarboxamidoadenosine ([3H]-NECA) binding at human A2A receptors expressed in CHO cells (n = 3–6).
cKi values of the inhibition of NECA-stimulated adenylyl cyclase activity in CHO cells expressing human A2B receptors (n = 3–6).
dDisplacement of specific [3H]-2-hexyn-1-yl-N6-methyladenosine ([3H]-HEMADO) binding at human A3 receptors expressed in CHO cells (n = 3–6).
Data are expressed with 95% confidence limits. N.D., not determined.
Additionally, studies on substitution at the indole C4, C5, C6 and C7 positions were also conducted to investigate their effect on the hA2A binding affinity by replacing the hydrogen atoms with substituent groups, including halogens and methoxy group. It was observed that such replacement did not significantly enhance the binding affinity at the hA2A receptor as compared to that displayed by compounds 5 and 6. For example, at the indole C4 position, introduction of a methoxy group (compound 13, Ki hA2A = 18.8 μM hA1/hA2A >5, hA3/hA2A >2) did not produce appreciable difference in hA2A binding from corresponding derivative without the methoxy substitution (compound 5, Ki hA2A = 11.2 μM, hA1/hA2A >9, hA3/hA2A >9). At the indole C5 position, similar findings were noted with methoxy (compound 14, Ki hA2A = 12.7 μM, hA1/hA2A >8, hA3/hA2A >2 versus compound 5), fluorine (compound 15, Ki hA2A = 18.2 μM, hA1/hA2A >6, hA3/hA2A >2 versus compound 6), and chlorine (compound 16, Ki hA2A = 15.5 μM, hA1/hA2A >7, hA3/hA2A = 1.26 versus compound 5) substitutions. However, the presence of bromine at the C5 position was found to be detrimental to the hA2A binding affinity (compound 17, Ki hA2A > 100 μM versus compound 18, Ki hA2A = 27.6 μM, hA1/hA2A >4, hA3/hA2A >1). It is probable that the large size of bromine at the C5 position causes steric clash with adjacent residues in the binding pocket, thus leading to ineffective binding.
Likewise, at the indole C6 position, introduction of fluorine did not demonstrate significant change in the hA2A affinity (compound 19, Ki hA2A = 26.9 μM, hA1/hA2A >4, hA3/hA2A >1 versus compound 18). Nonetheless, it was found that methoxy group substitution at the indole C7 position had led to a 3-fold improvement in hA2A affinity (compound 20, Ki hA2A = 3.63 μM, hA1/hA2A >28, hA3/hA2A >8 versus compound 5). The enhanced binding observed in compound 20 could be attributed to the oxygen atom of the methoxy substituent participating in hydrogen bonds with neighbouring residues in the binding cavity.
Furthermore, the binding affinity of compound 6 was compared with that of compound 18 (compound 6, Ki hA2A = 8.71 μM hA1/hA2A >11, hA3/hA2A >11 versus compound 18, Ki hA2A = 27.6 μM, hA1/hA2A >4, hA3/hA2A >1). From such comparison, it was noted that the absence of a methyl group at the indole C2 position in compound 18 has led to a 3.2-fold decrease in hA2A affinity. This finding suggests that the C2-methyl group contributes to the binding affinity at the hA2A receptor to a certain extent.
An additional study was also carried out to determine the effect of a linker at the indole C2 position towards the hA2A binding affinity. This provides better understanding of differences in indole C2 and C3 extension towards affinity at the hA2A receptor. From the results obtained, it was revealed that except for compound 25, the incorporation of linker at the indole C2 position rendered derivatives (compounds 21–24) inactive at the hA2A receptor. This observation suggested the piperazine-pyrimidine moiety was not well tolerated when it was extended from the indole C2 position. Notably, 7-nitro indolyl derivative 25 showed modest affinity at the hA2A receptor (Ki hA2A = 29.7 μM, hA1/hA2A = 0.16, hA3/hA2A >1). It is speculated that the oxygen atoms on the nitro group could likely form hydrogen bonding with adjacent water molecules or neighbouring residues in the binding pocket. In fact, such 7-nitroindole-2-susbtituted derivative (compound 25, with the highest hA2A affinity in the indole-2 series) is reminiscent of 7-methoxyindole-3-substituted derivative (compound 20, with the highest hA2A affinity in the indole-3 series). The oxygen atoms of methoxy and nitro group could possibly engage in similar hydrogen bonding interaction with residues in the vicinity.
Binding affinity studies at human dopamine D2 receptor
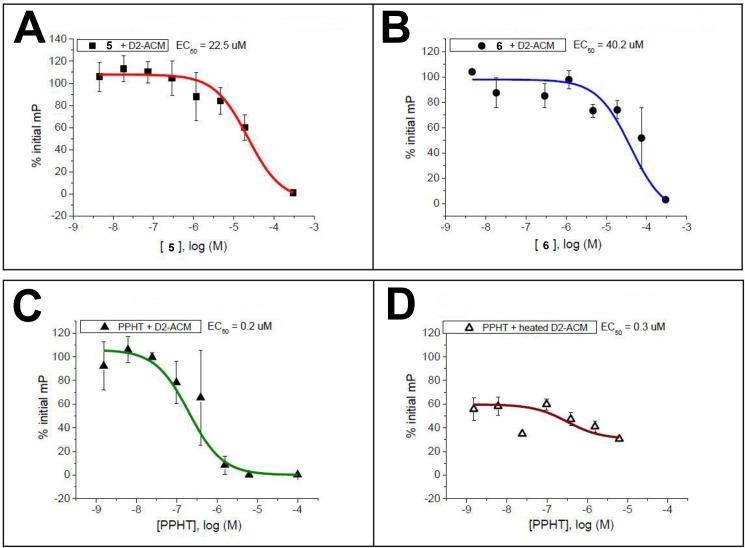
In addition to the human A2A receptor binding affinity assay, the newly synthesized IPP derivatives were further examined for their binding affinity toward human dopamine D2 receptor. Among these, compounds 5 and 6 with hA2A binding affinity in the low micromolar range were selected for the polymersome-based dopamine D2 receptor binding assay. In such assay, boron-dipyrromethene N-(p-aminophenethyl)spiperone (BODIPY-NAPS, a fluorescent ligand) was incubated with D2 receptor-functionalised polymersomes; spiperone is a selective D2-like antagonist with reported Ki of 0.06 nM for D2 receptor [24]. The mixture was then incubated with solutions containing eight different concentrations (ranging from 3 nM to 0.3 mM) of compounds 5 and 6, and the measured fluorescence intensity against ligand concentration was plotted accordingly (Fig 5A and 5B). Notably, dose-dependent reduction in fluorescence was observed for both compounds, indicating that strong binding (Ki = 0.06 nM) of spiperone with D2 receptor can be competitively replaced by compounds 5 and 6.
Fig 5.
Dose-response curves of compound 5 (A), compound 6 (B) and (±)-PPHT.HCl (C) in D2R-proteopolymersomes-based fluorescence polarisation (FP) competition assay with BODIPY-NAPS. The non-binding control (D) was achieved by denaturing the D2R-proteopolymersomes with heat, resulting in a curve that fluorescence intensity did not decrease much when the concentration of (±)-PPHT.HCl increased. For the highest concentrations, experiments were repeated only in duplicate due to solubility issues.
Furthermore, (±)-2-(N-phenethyl-N-propyl)amino-5-hydroxytetralin hydrochloride ((±)-PPHT.HCl), a potent dopamine D2 receptor agonist with a Ki of 13.3 nM determined by competition binding experiments with [3H]spiperone [24], was tested for its ability to displace BODIPY-NAPS. Similarly, a sigmoidal decrease in fluorescence with increasing concentration of (±)-PPHT.HCl was noted (Fig 5C); this observation was characteristic of D2 agonist interactions in the D2R-functionalized polymersomes. As illustrated in Fig 5A–5C, the dose-response curves of compounds 5 and 6 coherently resembled that of the (±)-PPHT.HCl. The EC50 values of compound 5, compound 6 and (±)-PPHT.HCl were determined to be 22.5 μM, 40.2 μM and 0.2 μM, respectively. Besides, treatment of denatured D2R-proteopolymersomes with PPHT.HCl was performed (Fig 5D) for comparison with Fig 5C, showing the necessity of proteopolymersomes’ integrity.
Adenylyl cyclase inhibition studies at human dopamine D2 receptors
D2 receptor activation is mediated by heterotrimeric (i.e. α, β and γ subunits) Gi/o proteins. In the absence of endogenous dopamine or agonists, Gα is bound to guanosine 5’-diphosphate (GDP) and Gβγ. Upon D2 receptor activation, the conformational change of the receptor results in the GDP release, guanosine 5’-triphosphate (GTP) binding, and dissociation of Gα from Gβγ. The released Gα then interacts with and inhibits adenylyl cyclase, leading to a decrease in the adenosine 3’,5’-cyclic monophosphate (cAMP) production. Thus, measuring the extent to which a compound inhibits cAMP accumulation has been one of the functional assays for D2 receptor activation [25].
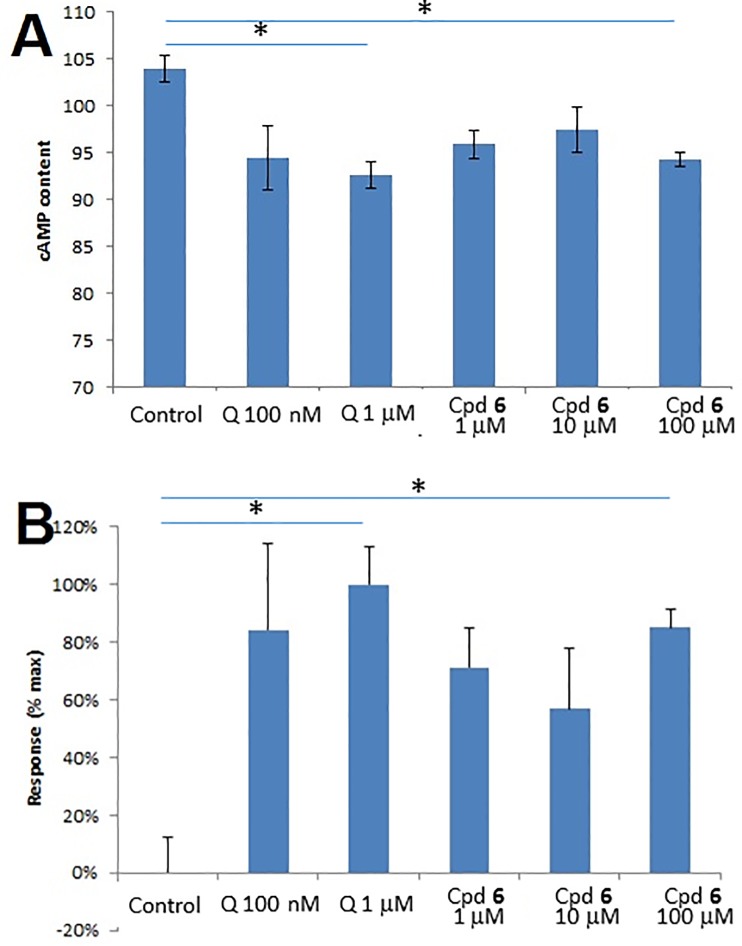
Hence, adenylyl cyclase inhibition studies on compound 6 were carried out to determine whether the compound was able to activate D2 receptors. In this functional assay, Chinese Hamster Ovary (CHO) cells were transfected with D2 receptor, pre-treated with compound 6 or quinpirole, and stimulated with forskolin and 3-isobutyl-1-methylxanthine (IBMX). Quinpirole, a potent D2 receptor agonist with a Ki of 4.8 nM [12], acted as the reference agent. Forskolin (an activator of adenylyl cyclase) and IBMX (an inhibitor of phosphodiesterase) were used to elevate the basal levels of cAMP. As shown in Fig 6A, the control referred to the high cAMP content resulted from the stimulation with forskolin/IBMX in the absence of either quinpirole or compound 6. Subsequently, different concentrations of either quinpirole or compound 6 were added to investigate whether, and if so, to which extent the added ligand could reverse the cAMP effect induced by forskolin/IBMX. The concentration-dependent decrease in cAMP accumulation when the D2 receptor-expressing cells were treated with quinpirole suggested that quinpirole-induced D2 receptor conformation caused the Gi protein to bind and inhibit the adenylyl cyclase [26]. Activation with quinpirole at the concentration of 1 μM resulted in an 11% decrease (calculated by (103.97–92.63)/103.97 = 11%) in intracellular cAMP concentration with p-value of 0.0232, and the result of 1 μM quinpirole was normalised relative to the control (Fig 6B). We evaluated the relative efficacy of our compounds by normalizing the resulting cAMP levels relative to that of forskolin/IBMX-treated cells (0%) and 1 μM quinpirole-mediated inhibitory response (100%) (Fig 6B). Three concentrations of compound 6 (1 μM, 10 μM and 100 μM) were evaluated, and their respective capability to inhibit cAMP accumulation was measured. There was no concentration-dependent response for compound 6 observed in this cAMP assay. The optimal inhibition occurred at 100 μM of compound 6 (p = 0.0232), which showed comparable potency to 100 nM of quinpirole. In Fig 6B, the y-axis values for all three concentrations of compound 6 were above zero, suggesting that compound 6 inhibited cAMP accumulation induced by forskolin/IBMX and therefore acted as a D2 receptor agonist.
Fig 6. Inhibition of cAMP accumulation induced by compound 6 in comparison with the reference compound quinpirole (Q).
The CHO cells were transfected with D2 receptor, pre-treated with the indicated concentrations of quinpirole or compound 6, and activated with forskolin/IBMX. (A) The cAMP concentration (in the unit of picomole) measured by ELISA. (B) Normalisation to control (0%) and 1 μM quinpirole (100%). *p<0.05. Error bars = standard error of the mean (SEM).
Drosophila models of PD
Although most PD cases occur in a sporadic manner, there are increasing studies indicating the association of PD with genetic etiologies [27]. In particular, mutations in both Parkin and LRRK2 genes have been recognized to be the predominant causes for early- [28] and late-onset [29] hereditary PD cases, respectively. Of all the genetic models developed for investigating the molecular events regulated by PD mutant genes, the Drosophila model has gained significant traction due to its highly conserved homologues of many human genes. In fact, both Parkin [30] and LRRK2 [31] mutants in Drosophila models faithfully phenocopy many of the characteristics of PD, including reduction of dopaminergic (DA) neurons in the brain. Moreover, the closest human homologue of the Drosophila adenosine receptor, DmAdoR, is hA2A receptor, and hence using Drosophila models to assess A2A ligands such as compounds 5 and 6 provides better correlation. Furthermore, D2-like receptors [32] have been shown to regulate locomotion in the Drosophila [33]. In view of the relevance of Drosophila to human A2A and D2 receptors, this in vivo system was used to test the efficacy of compounds 5 and 6.
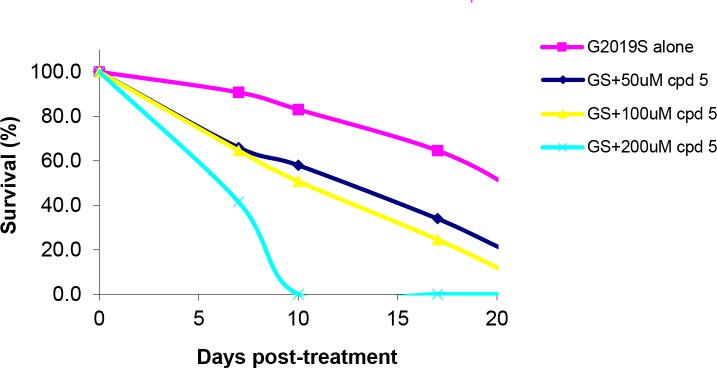
Notably, there is high prevalence of G2019S mutant in the LRRK2-associated PD cases [34,35]. Studies have shown that expression of the LRRK2-G2019S mutantion in Drosophila caused more severe PD phenotypes, including locomotor dysfunction, loss of dopaminergic neurons, and early mortality, relative to that of wild-type LRRK2 [31]. Prior to evaluating compounds 5 and 6 in the Drosophila model of PD, the effect of compound concentrations on fly viability was studied. At the ligand concentration of 500 μM, both compounds were lethal to G2019S flies (data not shown). Lower ligand concentrations were attempted, and at concentrations in the 50–100 μM range, whilst treatment with compound 6 still resulted in toxicity, compound 5 was able to delay the G2019S fly mortality (Fig 7). As there was no remarkable difference in the survival rate between 50 and 100 μM for compound 5, this range was selected for compound 5 for the subsequent climbing assay and quantification of DA neurons.
Fig 7. Survival percentages of mutant LRRK2 G2019S flies after 7 days, 10 days and 17 days of treatment with various concentrations (50 μM, 100 μM, 200 μM) of compound 5.
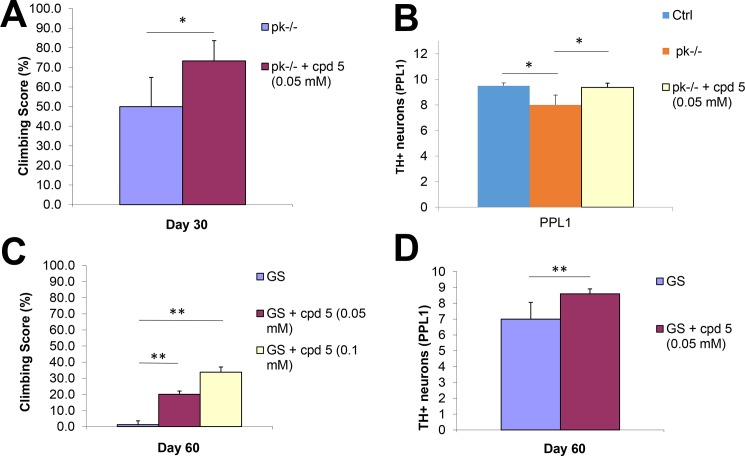
Parkin-null (pk-/-) flies were first used to evaluate compound 5. It was found that compound 5-treated parkin-null flies not only exhibited improvement in climbing scores compared with untreated mutant flies (Fig 8A), but they also showed reduction of DA neuron loss in the PPL1 cluster [36] (Fig 8B), the cluster used widely in the parkin-null flies. Compound 5 was subsequently examined for its ability to ameliorate LRRK2 G2019S-induced PD phenotypes. It was noted that compound 5-treated LRRK2 G2019S flies displayed improvement in climbing (in a concentration-dependent manner) (Fig 8C) as well as mitigation of DA neuron degeneration (Fig 8D). Taken together, these findings indicate that compound 5 acts as a suppressor of DA neuron dysfunction in two Drosophila genetic PD models—one with parkin-null which represents recessive PD, and one with transgene LRRK2 G2019S-mutant, which represents dominant PD.
Fig 8.
Treatment of compounds 5 in both parkin-null (A and B) and mutant LRRK2 G2019S-expressing (C and D) flies. A: Climbing score of untreated and compound 5 (50 μM)-treated parkin-null flies (where 100% is the climbing score of normal flies). B: DA neuronal count (PPL1 cluster) of normal flies (control), parkin-null flies, and compound 5 (50 μM)-treated parkin-null flies. C: As in A, except that parkin-null flies were substituted with LRRK2 G2019S-expressing flies (the control is represented by the mutant flies). Two concentrations, 50 μM and 100 μM, of compound 5 were tested. D: As in B, except that parkin-null flies were substituted with LRRK2 G2019S-expressing flies. (*p < 0.05, **p < 0.01).
Notwithstanding the promising results that we have obtained from the Drosophila system, we recognized that there are inherent limitations of the fly model. For example, by virtue of their vastly different brain architecture from their human counterparts, fly PD models cannot recapitulate fully the phenotypic and pathologic features of the human condition. Moreover, some known disease-associated factors like α-synuclein are not expressed in the fly brain. Nevertheless, they do exhibit salient features of PD such as age-dependent dopaminergic neurodegeneration and associated locomotion deficits. Importantly, like PD patients, fly PD models also respond positively to the L-DOPA treatment, making them suitable model for rapid drug evaluation.
Toxicity studies
In the toxicity studies, in vitro assays for mutagenicity and cytotoxicity were performed on compounds 5 and 6. These compounds were investigated for their mutagenic potential through the Ames assay [37]. Two Salmonella strains TA98 and TA100 with pre-existing mutations (His-) were selected. Both strains are unable to synthesize histidine (one of the growth requirements for these strains) and therefore cannot form colonies on the agar plate. If the incubation of one such strain with a given chemical results in the formation of many colonies, it would suggest that chemical induces back-mutation (His- → His+) and is therefore considered as a mutagen.
Together with the compounds, the Salmonella strains were incubated in the presence (+S9) and absence (–S9) of a metabolizing system “S9 mix” consisting of 9000 g supernatant fraction of rat liver microsomes, nicotinamide adenine dinucleotide phosphate (NADP) and cofactors. Exogenous “S9 mix” was included due to the lack of CYP metabolizing enzymes in Salmonella typhimurium. In the assay, agar plates +S9 and–S9 were used to detect pro-mutagens for cases where the native chemical was not mutagenic but its metabolites were mutagens, and direct-acting mutagens, respectively. 2-aminoanthracene (2-AA) was used as positive control chemical for TA98 +S9 (S9B Fig) and TA100 +S9 (S9D Fig), while 4-nitroquinoline N-oxide (4-NQO) was included as positive control chemical for TA98 –S9 (S9A Fig) and TA100 –S9 (S9C Fig). Methotrexate was used as the negative control for all four incubations (S9A–S9D Fig), and DMSO was used as the solvent control (vehicle). As shown in S9 Fig, the number of His+ revertants induced by compounds 5 and 6 at two concentrations, 10 μM and 100 μM, was even lower than that induced by methotrexate in each incubation, suggesting that these two compounds have no significant mutagenicity and are therefore considered as non-mutagens.
Besides the mutagenicity study, compounds 5 and 6 were also subjected to cell viability assays using transforming growth factor-alpha mouse hepatocyte (TAMH) and HL-1 cardiomyocyte to assess the cytotoxic potential. TAMH lines were treated with compounds 5 and 6 in eight different concentrations (100 μM, 33.3 μM, 11.1 μM, 3.7 μM, 1.23 μM, 0.41 μM, 0.13 μM, 0.045 μM), and the percentage of viable cells was plotted against the logarithm of ligand concentration (S10B and S10C Fig). Both compounds at concentrations less than 30 μM were shown to be non-cytotoxic in the TAMH lines. Acetaminophen, a drug with direct hepatotoxic potential [38], was used as the positive control in this assay (S10A Fig). Similar to the viability study using TAMH lines, there was no toxicity observed at ligand concentrations less than 30 μM in the HL-1 cell (S10E and S10F Fig). Doxorubicin, a chemotherapeutic agent known to cause dose-dependent cardiotoxicity [39], was used as the positive control in this assay (S10D Fig).
Aqueous solubility studies
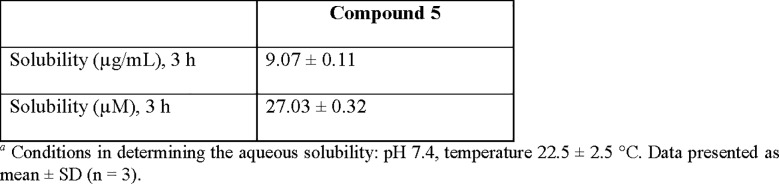
In addition to the pharmacological assays, aqueous solubility of compound 5 was determined and the results were presented in Fig 9. The solubility of compound 5 was found to be 9.07 ± 0.11 μg/mL (27.03 ± 0.32 μM) at 3-hour interval. Based on the solubility guideline for oral absorption by Kerns et al. [40], the following solubility ranges are suggested for lead compound at the drug discovery stage: < 10 μg/mL (low solubility), 10~60 μg/mL (moderate solubility), > 60μg/mL (high solubility). As such, compound 5 is therefore deduced to exhibit marginally low aqueous solubility at ambient temperature in pH 7.4. Nonetheless, the structural modification on compound 5 is underway in our laboratory to enhance its aqueous solubility while improving its binding profiles at both adenosine A2A adenosine receptors and dopamine D2 receptors.
Fig 9. The aqueous solubility of compound 5a.
Conclusion
The significant role of human adenosine A2A receptor and dopamine D2 receptor in the pathogenesis of PD, together with emerging paradigm of drug actions on multiple receptors has spurred the discovery of new compounds to modulate both G protein-coupled receptors. In our study, a new series of indole-piperazine-pyrimidine (IPP) derivatives targeting the two receptors has been successfully synthesized and evaluated through integration of computational tools, synthetic chemistry and pharmacological assays. Compounds 5 and 6 have demonstrated affinity at the human adenosine A2A receptor in the low micromolar range (Ki hA2A = 8.7–11.2 μM). Based on the structure-affinity relationship studies as illustrated above, three important structural features of IPP-containing compounds for the hA2A binding are derived:
At the indole C2 position, the presence of methyl group confers better hA2A binding affinity than that of hydrogen.
The linker bridging indole C3 and piperazine nitrogen should contain a carbonyl group, and the length of the linker is limited to one or two carbon atoms.
At the indole C7 position, methoxy group improves the hA2A binding affinity.
In the proteopolymersome-based D2 receptor binding assay, compounds 5 and 6 have displayed binding to the dopamine D2 receptor with EC50 of 22.5–40.2 μM. In addition, a functional assay for D2 receptor activation was conducted with compound 6 which demonstrated that inhibition of cAMP accumulation in CHO cells by 100 μM was comparable to the activity induced by 100 nM of quinpirole. Such observation has highlighted the D2 receptor agonistic activity of compound 6. Further in vivo testing of compound 5 in the Drosophila model of PD at 50 μM showed improvement in movement as well as mitigation of the loss of dopaminergic neurons. In the in vitro toxicity studies, compounds 5 and 6 did not exhibit mutagenicity up to 100 μM, nor hepatotoxicity or cardiotoxicity up to 30 μM.
In summary, our study has led to the identification of novel IPP scaffolds acting on both human adenosine A2A and dopamine D2 receptors. Structural optimization of this novel scaffold is deemed beneficial in providing insights into structural requirements for future development of new anti-parkinsonian agents.
Experimental section
Chemistry
General
Reactions were constantly monitored by thin layer chromatography (TLC) on silica gel (precoated 60 F254 Merck plates) and carried out under a nitrogen atmosphere. All chemicals are commercial products from Sigma Aldrich or Alfa Aesar. Column chromatography was performed using silica gel 60 (Merck, 70–230 mesh). Compounds were dissolved in HPLC-grade MeOH for accurate mass analysis using ESI time-of-flight (TOF) ionisation mode. 1H and 13C NMR spectra were determined in deuterated chloroform (CDCl3) or deuterated dimethylsulfoxide (DMSO-d6) using Bruker DPX Ultrashield NMR (400 MHz) spectrometer, with chemical shifts given in parts per million (δ) downfield relative to the central peak of the solvents, and J values (coupling constants) were given in Hz. The following abbreviations were used: s = singlet, d = doublet, t = triplet, m = multiplet, br = broad, td = triplet of doublets. High-performance liquid chromatography (HPLC) analysis was carried out for compounds used in biological assays. For HPLC (1): Hewlett-Packard series 1050 HPLC system equipped with a HP-1050 quaternary pump, a degasser, diode array detector, a HP-1100 autosampler, and a LiChrosorb reversed phase C18 (5 μm) column (4.6 × 250 mm) with solvents being CH3CN/H2O. For HPLC (2) and HPLC (3): Agilent HPLC 1200 series instrument on a Zorbax SB-C18, 4.6 mm × 250 mm, 5 μm column with solvents being CH3CN/H2O (0.1% v/v CF3COOH) and MeOH/H2O (0.1% v/v CF3COOH) for HPLC (2) and HPLC (3), respectively. All HPLC samples were prepared by dissolving them in HPLC-grade MeOH. The analysis was performed at 30°C, and the ultraviolet detection was made at wavelength 254 nm. The separations were carried out using gradient elution. The HPLC methods are as follows. HPLC (1): injection volume 5 μL, stop time 20 min, flow rate 1 mL/min, a gradient of 35 → 100% CH3CN for the 0–17 min period and back to CH3CN/H2O (3:7) at 20 min. HPLC (2): injection volume 20 μL, stop time 15 min, flow rate 0.5 mL/min, a gradient of 5 → 95% CH3CN for the 0–12 min period and back to CH3CN/H2O (5:95) at 15 min. HPLC (3): injection volume 20 μL, stop time 15 min, flow rate 0.5 mL/min, a gradient of 5 → 95% MeOH for the 0–12 min period and back to MeOH/H2O (5:95) at 15 min. The purities of all tested compounds were > 95% measured by the peak area of the product divided by that of the total peak areas.
4,6‐Dimethylpyrimidin‐2‐ol hydrochloride (2)
To a suspension of urea 1 (1000 mg, 16.65 mmol) in EtOH (10 mL) were added 2,4-pentanedione (1885 μL, 18.32 mmol) and concentrated hydrochloric acid (2775 μL, 33.3 mmol), and the resulting clear, colourless solution was stirred and refluxed under N2 for 24 h. The mixture was cooled to room temperature and filtered. The filter cake was washed with EtOH and Et2O, and dried in the vacuum oven to afford compound 2 (2096 mg, 78% yield) as an off-white crystalline solid. 1H NMR (400 MHz, DMSO-d6) δ 6.72 (s, 1H), 2.47 (s, 6H).
2‐Chloro‐4,6‐dimethylpyrimidine (3)
The mixture of compound 2 (1998 mg, 12.44 mmol) and phosphorus oxychloride (20 mL) was refluxed for 10 h. The remaining phosphorus oxychloride was evaporated to get a brown oil in the flask. The mixture was cooled in an ice bath, and concentrated aqueous KOH solution was added dropwise cautiously with stirring, until the litmus paper showed pH value that is approximately 8. Diethyl ether (30 mL) was added, and the mixture was stirred for 2.5 h. The water layer was extracted with Et2O (3 × 30 mL) and EtOAc (3 × 30 mL). The organic layers were combined, washed with brine, dried over Na2SO4, filtered and concentrated under reduced pressure. The resulting yellow liquid was put in an ice bath under vacuum to afford compound 3 (1606 mg, 91% yield) as yellow crystals. 1H NMR (400 MHz, DMSO-d6) δ 7.33 (s, 1H), 2.42 (s, 6H).
4,6‐Dimethyl‐2‐(piperazin‐1‐yl)pyrimidine (4)
Into a 250 mL round-bottom flask were charged K2CO3 (3849 mg, 27.85 mmol), piperazine (10903 mg, 126.58 mmol), and H2O (180 mL). The mixture was heated at 45–50°C until it became a clear solution. Compound 3 (3610 mg, 25.32 mmol) was divided into four portions, and each portion was added into the mixture in one-hour interval. After the addition of all amounts of compound 3, the reaction mixture was cooled to room temperature and stirred overnight. The white precipitate was filtered, and the filtrate was collected. The filtrate underwent solid/liquid extraction with EtOAc (3 × 225 mL). Organic layers were collected, washed with brine, dried over Na2SO4, filtered, and concentrated under reduced pressure to give compound 4 (4536 mg, 93% yield) as a white crystalline solid. 1H NMR (400 MHz, DMSO-d6) δ 6.36 (s, 1H), 3.62 (t, J = 5.2 Hz, 4H), 2.69 (t, J = 5.2 Hz, 4H), 2.20 (s, 6H).
General procedure for the coupling reactions to obtain compounds 5–8 and 13–25
To a suspension of indole acid and EDC.HCl were added EtOAc and two drops of DMF, and the mixture was stirred for 1 h at 23°C followed by the addition of compound 4. The reaction was heated at 50°C for 3 h, and solvents were evaporated to dryness under high vacuum. Water was added, sonicated, and filtered. The filter cake was washed with hexane/EtOAc and further purified by crystallization from MeOH/EtOH/Et2O/EtOAc.
3‐[4‐(4,6‐Dimethylpyrimidin‐2‐yl)piperazine‐1‐carbonyl]‐1H‐indole (5)
Yield: 360 mg (50%) as white crystal (re-crystallization from MeOH/Et2O/EtOAc). 1H NMR (400 MHz, DMSO-d6) δ 11.61 (br s, 1H), 7.74–7.72 (m, 2H), 7.45 (d, J = 8 Hz, 1H), 7.16 (td, J = 7.2, 1.2 Hz, 1H), 7.10 (td, J = 7.2, 1.2 Hz, 1H), 6.44 (s, 1H), 3.82–3.80 (m, 4H), 3.71–3.69 (m, 4H), 2.24 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 166.8 (2 × C), 165.7, 161.1, 135.7, 128.1, 126.1, 121.9, 120.3, 120.2, 111.9, 109.6, 109.1, 43.5 (2 × CH2), 40.1–38.9 (2 × CH2), 23.7 (2 × CH3). HRMS-ESI (m/z) [M + H]+ calcd for C19H22N5O, 336.1819; found, 336.1824 (Δ = -1.6 ppm). [M + Na]+ calcd for C19H21N5NaO, 358.1638; found, 358.1639 (Δ = -0.2 ppm). HPLC (1): t = 5.6 min, 96.7% purity. HPLC (3): t = 15.5 min, 98.3% purity.
1‐[4‐(4,6‐Dimethylpyrimidin‐2‐yl)piperazin‐1‐yl]‐2‐(2‐methyl‐1H‐indol‐3‐yl)ethan‐1‐one (6)
Yield: 1130 mg (58%) as light brown crystal (re-crystallization from MeOH/EtOAc (3:1)). 1H NMR (400 MHz, DMSO-d6) δ 10.80 (br s, 1H), 7.45 (d, J = 7.6 Hz, 1H), 7.22 (d, J = 7.6 Hz, 1H), 6.97 (td, J = 7.2, 1.2 Hz, 1H), 6.90 (td, J = 7.2, 1.2 Hz, 1H), 6.41 (s, 1H), 3.74 (s, 2H), 3.62–3.61 (m, 2H), 3.52–3.49 (m, 6H), 2.34 (s, 3H), 2.20 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 169.6, 166.8 (2 × C), 160.8, 135.1, 132.6, 128.2, 120.0, 118.2, 117.8, 110.3, 109.1, 104.0, 45.2, 43.3, 43.2, 41.2, 30.0, 23.6 (2 × CH3), 11.5. HRMS-ESI (m/z) [M + H]+ calcd for C21H26N5O, 364.2132; found, 364.2123 (Δ = 2.4 ppm). [M + Na]+ calcd for C21H25N5NaO, 386.1951; found, 386.1940 (Δ = 3.0 ppm). HPLC (1): t = 9.8 min, 99.9% purity. HPLC (3): t = 15.6 min, 99.6% purity.
1‐[4‐(4,6‐Dimethylpyrimidin‐2‐yl)piperazin‐1‐yl]‐3‐(1H‐indol‐3-yl)propan‐1‐one (7)
Yield: 491 mg (74%) as a transparent crystal (re-crystallization from MeOH/EtOAc (1:1)). 1H NMR (400 MHz, DMSO-d6) δ 10.76 (br s, 1H), 7.53 (d, J = 8.0 Hz, 1H), 7.32 (d, J = 8.0 Hz, 1H), 7.15 (d, J = 2.0 Hz, 1H), 7.06 (td, J = 8.0, 0.8 Hz, 1H), 6.97 (td, J = 8.0, 0.8 Hz, 1H), 6.42 (s, 1H), 3.67–3.64 (m, 2H), 3.62–3.60 (m, 2H), 3.53–3.50 (m, 2H), 3.46–3.44 (m, 2H), 2.95 (t, J = 7.2 Hz, 2H), 2.71 (t, J = 7.2 Hz, 2H), 2.22 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 170.8, 166.8 (2 × C), 160.8, 136.2, 127.1, 122.5, 120.9, 118.3, 118.2, 113.8, 111.3, 109.1, 44.8, 43.4, 43.1, 40.9, 33.4, 23.7 (2 × CH3), 20.6. HRMS-ESI (m/z) [M + H]+ calcd for C21H26N5O, 364.2132; found, 364.2131 (Δ = 0.2 ppm). [M + Na]+ calcd for C21H25N5NaO, 386.1951; found, 386.1949 (Δ = 0.5 ppm). HPLC (1): t = 10.2 min, 98.4% purity. HPLC (3): t = 15.6 min, 98.4% purity.
1‐[4‐(4,6‐Dimethylpyrimidin‐2‐yl)piperazin‐1‐yl]‐4‐(1H‐indol‐3‐yl)butan‐1‐one (8)
Yield: 411 mg (66%) as an off-white crystal (re-crystallization from MeOH/EtOAc (2:1)). 1H NMR (400 MHz, DMSO-d6) δ 10.75 (br s, 1H), 7.52 (d, J = 8.0 Hz, 1H), 7.32 (d, J = 8.0 Hz, 1H), 7.11 (d, J = 2.4 Hz, 1H), 7.05 (td, J = 8.0, 0.8 Hz, 1H), 6.96 (td, J = 8.0, 0.8 Hz, 1H), 6.43 (s, 1H), 3.70–3.67 (m, 4H), 3.53–3.50 (m, 2H), 3.46–3.44 (m, 2H), 2.72 (t, J = 7.2 Hz, 2H), 2.41 (t, J = 7.2 Hz, 2H), 2.23 (s, 6H), 1.89 (quintet, J = 7.2 Hz, 2H). 13C NMR (100 MHz, DMSO-d6) δ 170.8, 166.8 (2 × C), 160.1, 136.3, 127.2, 122.3, 120.8, 118.3, 118.1, 114.2, 111.3, 109.1, 44.7, 43.5, 43.1, 40.9, 32.1, 25.6, 24.3, 23.7 (2 × CH3). HRMS-ESI (m/z) [M + H]+ calcd for C22H28N5O, 378.2288; found, 378.2295 (Δ = -1.8 ppm). [M + Na]+ calcd for C22H27N5NaO, 400.2108; found, 400.2110 (Δ = -0.7 ppm). HPLC (1): t = 11.3 min, 99.7% purity. HPLC (3): t = 16.3 min, 99.1% purity.
3‐[4‐(4,6‐Dimethylpyrimidin‐2‐yl)piperazine‐1‐carbonyl]‐4‐methoxy‐1H‐indole (13)
Yield: 219 mg, (69%) as a brown solid. 1H NMR (400 MHz, DMSO-d6) δ 11.40 (br s, 1H), 7.37 (d, J = 1.2 Hz, 1H), 7.09–7.02 (m, 2H), 6.55 (d, J = 7.2 Hz, 1H), 6.42 (s, 1H), 3.80–3.68 (2 × br s, 11H), 2.22 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 166.8 (2 × C), 166.3, 161.1, 152.8, 136.9, 124.1, 122.8, 115.0, 110.4, 109.1, 105.1, 100.1, 55.1, 43.1 (2 × CH2), 40.1–38.9 (2 × CH2), 23.7 (2 × CH3). HRMS-ESI (m/z) [M + H]+ calcd for C20H24N5O2, 366.1925; found, 366.1925 (Δ = -0.2 ppm). [M + Na]+ calcd for C20H23N5NaO2, 388.1744; found, 388.1739 (Δ = 1.3 ppm). HPLC (1): t = 8.7 min, 99.2% purity. HPLC (3): t = 15.4 min, 99.3% purity.
3‐[4‐(4,6‐Dimethylpyrimidin‐2‐yl)piperazine‐1‐carbonyl]‐5‐methoxy‐1H‐indole (14)
Yield: 206 mg, (47%) as light yellow shiny transparent crystals (re-crystallization from MeOH/EtOAc (1:1)). 1H NMR (400 MHz, DMSO-d6) δ 11.50 (br s, 1H), 7.68 (s, 1H), 7.33 (d, J = 8.8 Hz, 1H), 7.21 (d, J = 2.4 Hz, 1H), 6.80 (dd, J = 8.8, 2.4 Hz, 1H), 6.44 (s, 1H), 3.83–3.80 (m, 4H), 3.76 (s, 3H), 3.71–3.69 (m, 4H), 2.24 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 166.8 (2 × C), 165.9, 161.1, 154.3, 130.7, 128.5, 126.8, 112.7, 112.3, 109.3, 109.1, 101.8, 55.3, 43.6 (2 × CH2), 40.1–38.9 (2 × CH2), 23.7 (2 × CH3). HRMS-ESI (m/z) [M + H]+ calcd for C20H24N5O2, 366.1925; found, 366.1930 (Δ = -1.4 ppm). [M + Na]+ calcd for C20H23N5NaO2, 388.1744; found, 388.1742 (Δ = 0.4 ppm). HPLC (1): t = 8.4 min, 99.6% purity. HPLC (3): t = 15.2 min, 99.0% purity.
1‐[4‐(4,6‐Dimethylpyrimidin‐2‐yl)piperazin‐1‐yl]‐2‐(5‐fluoro‐2‐methyl‐1H‐indol‐3‐yl)ethan‐1‐one (15)
Yield: 133 mg, (44%) as transparent colourless crystals (re-crystallization from MeOH/EtOAc (6:5)). 1H NMR (400 MHz, DMSO-d6) δ 10.92 (br s, 1H), 7.22–7.19 (m, 2H), 6.79 (td, J = 9.2, 2.4 Hz, 1H), 6.41 (s, 1H), 3.73 (s, 2H), 3.63–3.62 (m, 2H), 3.52 (s, 6H), 2.33 (s, 3H), 2.21 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 169.4, 166.8 (2 × C), 161.0, 156.7 (d, J = 228.9 Hz, C-F), 135.0, 131.7, 128.6 (d, J = 10.2 Hz), 111.1 (d, J = 9.5 Hz), 109.1, 107.7 (d, J = 25.6 Hz, F-C-C-H), 104.5 (d, J = 4.3 Hz), 102.7 (d, J = 23.3 Hz, F-C-C-H), 45.2, 43.3, 43.2, 41.2, 29.7, 23.7 (2 × CH3), 11.6. HRMS-ESI (m/z) [M + H]+ calcd for C21H25FN5O, 382.2038; found, 382.2042 (Δ = -1.2 ppm). [M + Na]+ calcd for C21H24FN5NaO, 404.1857; found, 404.1852 (Δ = 1.2 ppm). HPLC (1): t = 10.1 min, 99.7% purity. HPLC (3): t = 15.8 min, 98.7% purity.
5‐Chloro‐3‐[4‐(4,6‐dimethylpyrimidin‐2‐yl)piperazine‐1‐carbonyl]‐1H‐indole (16)
Yield: 137 mg, (43%) as pink crystals (re-crystallization from MeOH/EtOAc 1:1). 1H NMR (400 MHz, DMSO-d6) δ 11.83 (br s, 1H), 7.83 (s, 1H), 7.76 (dd, J = 2.0, 0.4 Hz, 1H), 7.47 (dd, J = 8.8, 0.4 Hz, 1H), 7.17 (dd, J = 8.4, 2.0 Hz, 1H), 6.44 (s, 1H), 3.82–3.80 (m, 4H), 3.73–3.70 (m, 4H), 2.24 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 167.3 (2 × C), 165.5, 161.6, 134.7, 130.0, 128.1, 125.4, 122.5, 120.1, 114.0, 109.61, 109.57, 43.9 (2 × CH2), 40.6–39.3 (2 × CH2), 24.2 (2 × CH3). HRMS-ESI (m/z) [M + H]+ calcd for C19H21ClN5O, 370.1429; found, 370.1433 (Δ = -0.9 ppm). [M + Na]+ calcd for C19H20ClN5NaO, 392.1249; found, 392.1245 (Δ = 0.9 ppm). HPLC (1): t = 10.9 min, 99.1% purity. HPLC (3): t = 16.6 min, 98.1% purity.
2‐(5‐Bromo‐7‐fluoro‐2‐methyl‐1H‐indol‐3‐yl)‐1‐[4‐(4,6-dimethylpyrimidin‐2‐yl)piperazin‐1‐yl]ethan‐1‐one (17)
Yield: 130 mg, (50%) as dark brown crystals (re-crystallization from MeOH/EtOAc 9:1). 1H NMR (400 MHz, DMSO-d6) δ 11.53 (br s, 1H), 7.48 (d, J = 1.2 Hz, 1H), 7.04 (dd, J = 10.4, 1.2 Hz, 1H), 6.43 (s, 1H), 3.75 (s, 2H), 3.66–3.64 (m, 2H), 3.61–3.59 (m, 2H), 3.55–3.53 (m, 4H), 2.34 (s, 3H), 2.25 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 169.5, 167.3 (2 × C), 161.4, 148.5 (d, J = 245.7 Hz, C-F), 136.5, 133.8 (d, J = 7.3 Hz), 122.1 (d, J = 13.2 Hz, F-C-C-NH), 117.3 (d, J = 2.9 Hz), 109.8 (d, J = 8.0 Hz), 109.6, 108.7 (d, J = 20.4 Hz, F-C-C-H), 105.9 (d, J = 2.2 Hz), 45.5, 43.8, 43.6, 41.7, 29.7, 24.1 (2 × CH3), 11.9. HRMS-ESI (m/z) [M + H]+ calcd for C21H24BrFN5O, 460.1143; found, 460.1132 (Δ = 2.3 ppm). [M + Na]+ calcd for C21H23BrFN5NaO, 482.0962; found, 482.0950 (Δ = 2.5 ppm). HPLC (1): t = 13.3 min, 95.9% purity. HPLC (3): t = 17.2 min, 96.5% purity.
1‐[4‐(4,6‐Dimethylpyrimidin‐2‐yl)piperazin‐1‐yl]‐2‐(1H‐indol‐3‐yl)ethan‐1‐one (18)
Yield: 414 mg, (46%) as white crystals (re-crystallization from EtOH/EtOAc 1:1). 1H NMR (400 MHz, DMSO-d6) δ 10.90 (br s, 1H), 7.58 (d, J = 7.6 Hz, 1H), 7.34 (d, J = 8.4 Hz, 1H), 7.24 (d, J = 2.0 Hz, 1H), 7.07 (td, J = 7.4, 0.8 Hz, 1H), 6.97 (td, J = 7.4, 0.8 Hz, 1H), 6.41 (s, 1H), 3.81 (s, 2H), 3.66–3.63 (m, 2H), 3.57 (s, 4H), 3.53–3.51 (m, 2H), 2.21 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 169.4, 166.8 (2 × C), 161.0, 136.1, 127.1, 123.5, 121.1, 118.8, 118.4, 111.3, 109.1, 108.1, 45.4, 43.4, 43.1, 41.1, 30.9, 23.7 (2 × CH3). HRMS-ESI (m/z) [M + H]+ calcd for C20H24N5O, 350.1975; found, 350.1980 (Δ = -1.4 ppm). [M + Na]+ calcd for C20H23N5NaO, 372.1795; found, 372.1794 (Δ = 0.2 ppm). HPLC (1): t = 9.2 min, 97.5% purity. HPLC (3): t = 15.3 min, 99.2% purity.
1‐[4‐(4,6‐Dimethylpyrimidin‐2‐yl)piperazin‐1‐yl]‐2‐(6‐fluoro‐1H‐indol‐3‐yl)ethan‐1‐one (19)
Yield: 179 mg, (57%) as light brown crystals (re-crystallization from MeOH/EtOAc 8:5). 1H NMR (400 MHz, DMSO-d6) δ 10.97 (br s, 1H), 7.56 (dd, J = 8.8, 5.6 Hz, 1H), 7.24 (d, J = 2.0 Hz, 1H), 7.11 (dd, J = 10.0, 2.4 Hz, 1H), 6.86–6.81 (m, 1H), 6.42 (s, 1H), 3.81 (s, 2H), 3.66–3.63 (m, 2H), 3.58 (s, 4H), 3.53–3.50 (m, 2H), 2.21 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 169.8, 167.2 (2 × C), 161.5, 159.3 (d, J = 232.6 Hz, C-F), 136.4 (d, J = 12.4 Hz), 124.6 (d, J = 3.7 Hz), 124.5, 120.3 (d, J = 10.2 Hz), 109.6, 108.9, 107.3 (d, J = 24.1 Hz, F-C-C-NH), 97.7 (d, J = 24.8 Hz, F-C-C-NH), 45.8, 43.9, 43.6, 41.6, 31.2, 24.1 (2 × CH3). HRMS-ESI (m/z) [M + H]+ calcd for C20H23FN5O, 368.1881; found, 368.1879 (Δ = 0.6 ppm). [M + Na]+ calcd for C20H22FN5NaO, 390.1701; found, 390.1689 (Δ = 2.9 ppm). HPLC (1): t = 9.6 min, 99.8% purity. HPLC (3): t = 15.6 min, 99.0% purity.
3‐[4‐(4,6‐Dimethylpyrimidin‐2‐yl)piperazine‐1‐carbonyl]‐7‐methoxy‐1H‐indole (20)
Yield: 215 mg, (57%) as an off-white solid.1H NMR (400 MHz, DMSO-d6) δ 11.75 (br s, 1H), 7.59 (d, J = 2.4 Hz, 1H), 7.29 (d, J = 7.2 Hz, 1H), 7.02 (t, J = 8.0 Hz, 1H), 6.73 (d, J = 7.6 Hz, 1H), 6.44 (s, 1H), 3.93 (s, 3H), 3.81–3.78 (m, 4H), 3.69–3.66 (m, 4H), 2.24 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 166.8 (2 × C), 165.8, 161.1, 146.3, 127.6, 127.4, 125.9, 120.9, 112.9, 110.3, 109.1, 102.4, 55.3, 43.5 (2 × CH2), 40.1–38.9 (2 × CH2), 23.7 (2 × CH3). HRMS-ESI (m/z) [M + H]+ calcd for C20H24N5O2, 366.1925; found, 366.1920 (Δ = 1.2 ppm). [M + Na]+ calcd for C20H23N5NaO2, 388.1744; found, 388.1733 (Δ = 2.9 ppm). HPLC (1): t = 9.5 min, 99.8% purity. HPLC (3): t = 15.9 min, 99.3% purity.
2‐[4‐(4,6‐Dimethylpyrimidin‐2‐yl)piperazine‐1‐carbonyl]‐1H‐indole (21)
Yield: 241 mg, (23%) as clear crystals (re-crystallization from MeOH/EtOAc 1:1). 1H NMR (400 MHz, DMSO-d6) δ 11.60 (br s, 1H), 7.62 (dd, J = 8.0, 0.4 Hz, 1H), 7.43 (dd, J = 8.0, 0.4 Hz, 1H), 7.21–7.17 (m, 1H), 7.07–7.03 (m, 1H), 6.86 (dd, J = 2.0, 0.8 Hz, 1H), 6.46 (s, 1H), 3.84 (br s, 8H), 2.25 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 166.9 (2 × C), 162.2, 161.0, 136.0, 129.8, 126.9, 123.3, 121.4, 119.8, 112.1, 109.2, 104.3, 43.4 (2 × CH2), 40.1–38.9 (2 × CH2), 23.7 (2 × CH3). HRMS-ESI (m/z) [M + H]+ calcd for C19H22N5O, 336.1819; found, 336.1821 (Δ = -0.8 ppm). [M + Na]+ calcd for C19H21N5NaO, 358.1638; found, 358.1634 (Δ = 1.1 ppm). HPLC (1): t = 11.3 min, 95.0% purity. HPLC (3): t = 16.5 min, 99.3% purity.
2‐[4‐(4,6‐Dimethylpyrimidin‐2‐yl)piperazine‐1‐carbonyl]‐5‐fluoro‐1H‐indole (22)
Yield: 31 mg, (16%) as white silky crystals (re-crystallization from EtOH/EtOAc 1:1). 1H NMR (400 MHz, DMSO-d6) δ 11.72 (br s, 1H), 7.43 (dd, J = 8.8, 4.4 Hz, 1H), 7.37 (dd, J = 9.6, 2.4 Hz, 1H), 7.06 (td, J = 9.6, 2.4 Hz, 1H), 6.84 (s, 1H), 6.46 (s, 1H), 3.84 (br s, 8H), 2.25 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 166.8 (2 × C), 161.9, 161.0, 157.1 (d, J = 231.1 Hz, C-F), 132.7, 131.6, 126.9 (d, J = 10.2 Hz), 113.3 (d, J = 9.4 Hz), 111.9 (d, J = 26.2 Hz, F-C-C-H), 109.2, 105.5 (d, J = 22.6 Hz, F-C-C-H), 104.2 (d, J = 5.1 Hz), 43.3 (2 × CH2), 40.1–38.9 (2 × CH2), 23.7 (2 × CH3). HRMS-ESI (m/z) [M + H]+ calcd for C19H21FN5O, 354.1725; found, 354.1720 (Δ = 0.4 ppm). [M + Na]+ calcd for C19H20FN5NaO, 376.1544; found, 376.1537 (Δ = 0.7 ppm). HPLC (1): t = 11.7 min, 98.0% purity. HPLC (3): t = 16.6 min, 99.7% purity.
2‐[4‐(4,6‐Dimethylpyrimidin‐2‐yl)piperazine‐1‐carbonyl]‐5‐methyl‐1H‐indole (23)
Yield: 70 mg, (35%) as brown crystals (re-crystallization from MeOH/EtOAc 8:5). 1H NMR (400 MHz, DMSO-d6) δ 11.46 (br s, 1H), 7.38 (d, J = 0.8 Hz, 1H), 7.32 (d, J = 8.4 Hz, 1H), 7.02 (dd, J = 8.4, 1.6 Hz, 1H), 6.76 (d, J = 1.6 Hz, 1H), 6.46 (s, 1H), 3.84 (br s, 8H), 2.37 (s, 3H), 2.25 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 167.3 (2 × C), 162.7, 161.5, 134.8, 130.2, 128.8, 127.6, 125.6, 121.1, 112.3, 109.7, 104.3, 43.8 (2 × CH2), 40.6–9.3 (2 × CH2), 24.2 (2 × CH3), 21.6. HRMS-ESI (m/z) [M + H]+ calcd for C20H24N5O, 350.1975; found, 350.1970 (Δ = 1.5 ppm). [M + Na]+ calcd for C20H23N5NaO, 372.1795; found, 372.1785 (Δ = 2.7 ppm). HPLC (1): t = 13.1 min, 98.9% purity. HPLC (3): t = 17.1 min, 98.3% purity.
2‐[4‐(4,6‐Dimethylpyrimidin‐2‐yl)piperazine‐1‐carbonyl]‐5‐methoxy‐1H‐indole (24)
Yield: 139 mg, (36%) as shiny, colourless crystals (re-crystallization from MeOH/EtOAc 8:5). 1H NMR (400 MHz, DMSO-d6) δ 11.45 (br s, 1H), 7.32 (d, J = 8.8 Hz, 1H), 7.07 (d, J = 2.4 Hz, 1H), 6.85 (dd, J = 8.8, 2.4 Hz, 1H), 6.76 (d, J = 1.2 Hz, 1H), 6.46 (s, 1H), 3.83 (br s, 8H), 3.76 (s, 3H), 2.25 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 166.9 (2 × C), 162.2, 161.1, 153.8, 131.3, 130.2, 127.2, 114.4, 113.0, 109.2, 104.1, 102.0, 55.3, 43.3 (2 × CH2), 40.1–38.9 (2 × CH2), 23.7 (2 × CH3). HRMS-ESI (m/z) [M + H]+ calcd for C20H24N5O2, 366.1925; found, 366.1925 (Δ = -0.1 ppm). [M + Na]+ calcd for C20H23N5NaO2, 388.1744; found, 388.1733 (Δ = 2.7 ppm). HPLC (1): t = 10.6 min, 97.2% purity. HPLC (3): t = 16.2 min, 99.2% purity.
{2‐[4‐(4,6‐Dimethylpyrimidin‐2‐yl)piperazine‐1‐carbonyl]‐1H‐indol‐7‐yl}azinic acid (25)
Yield: 137 mg, (36%) as a light yellow solid. 1H NMR (400 MHz, DMSO-d6) δ 11.77 (br s, 1H), 8.20 (d, J = 8.0 Hz, 1H), 8.15 (d, J = 8.0 Hz, 1H), 7.32 (t, J = 8.0 Hz, 1H), 7.04 (d, J = 2.4 Hz, 1H), 6.46 (s, 1H), 3.84 (br s, 4H), 3.71 (br s, 4H), 2.24 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 166.8 (2 × C), 161.5, 161.0, 133.9, 133.0, 131.1, 130.0, 128.1, 120.2, 119.7, 109.3, 104.5, 40.1–38.9 (4 × CH2), 23.7 (2 × CH3). HRMS-ESI (m/z) [M + H]+ calcd for C19H21N6O3, 381.1670; found, 381.1662 (Δ = 1.9 ppm). [M + Na]+ calcd for C19H20N6NaO3, 403.1489; found, 403.1480 (Δ = 2.2 ppm). HPLC (x): t = 11.7 min, 97.3% purity. HPLC (3): t = 16.8 min, 99.0% purity.
General procedure for the reduction reactions to obtain compounds 9–12
To a slurry of LiAlH4 in anhydrous THF at 0°C was added dropwise by syringe the solution of the starting carbonyl compound in anhydrous THF, and the mixture was warmed to 23°C with stirring for 4~21.5 h. At 0°C, the following sequence of dropwise addition was performed with continuous stirring: H2O, 15% aqueous NaOH. The white sticky part was removed by filtration, and the filtrate was concentrated under reduced pressure. The crude product was purified by flash chromatography (EtOAc/hexane).
3‐{[4‐(4,6‐Dimethylpyrimidin‐2‐yl)piperazin‐1‐yl]methyl}‐1H‐indole (9)
Yield: 391 mg, (76%) as a yellow solid (EtOAc/hexane 1:1). 1H NMR (400 MHz, DMSO-d6) δ 10.93 (br s, 1H), 7.64 (d, J = 7.6 Hz, 1H), 7.35 (d, J = 7.6 Hz, 1H), 7.23 (d, J = 2.4 Hz, 1H), 7.07 (td, J = 6.8, 1.2 Hz, 1H), 6.98 (td, J = 6.8, 1.2 Hz, 1H), 6.36 (s, 1H), 3.71–3.68 (m, 4H), 3.65 (s, 2H), 2.43–2.40 (m, 4H), 2.20 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 166.6 (2 × C), 161.1, 136.3, 127.7, 124.7, 121.0, 119.0, 118.5, 111.4, 110.6, 108.6, 53.2, 52.5 (2 × CH2), 43.4 (2 × CH2), 23.7 (2 × CH3). HRMS-ESI (m/z) [M + H]+ calcd for C19H24N5, 322.2026; found, 322.2029 (Δ = -1.0 ppm). [M + Na]+ calcd for C19H23N5Na, 344.1846; found, 344.1845 (Δ = 0.3 ppm). HPLC (1): t = 8.5 min, 100% purity. HPLC (3): t = 14.6 min, 97% purity.
3‐{2‐[4‐(4,6‐Dimethylpyrimidin‐2‐yl)piperazin‐1‐yl]ethyl}‐2‐methyl‐1H‐indole (10)
Yield: 546 mg, (95%) as a yellow solid (EtOAc/hexane 6:1). 1H NMR (400 MHz, DMSO-d6) δ 10.67 (br s, 1H), 7.39 (d, J = 7.6 Hz, 1H), 7.21 (d, J = 7.6 Hz, 1H), 6.96 (td, J = 7.2, 1.2 Hz, 1H), 6.91 (td, J = 7.2, 1.2 Hz, 1H), 6.39 (s, 1H), 3.74–3.72 (m, 4H), 2.83–2.79 (m, 2H), 2.46–2.44 (m, 6H), 2.32 (s, 3H), 2.22 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 166.6 (2 × C), 161.2, 135.1, 131.6, 128.2, 119.8, 118.0, 117.3, 110.3, 108.7, 108.2, 59.1, 52.7 (2 × CH2), 43.4 (2 × CH2), 23.7 (2 × CH3), 21.5, 11.2. HRMS-ESI (m/z) [M + H]+ calcd for C21H28N5, 350.2339; found, 350.2345 (Δ = -1.6 ppm). [M + Na]+ calcd for C21H27N5Na, 372.2159; found, 372.2166 (Δ = -2.0 ppm). HPLC (2): t = 9.0 min, 96.7% purity. HPLC (3): t = 14.5 min, 96.3% purity.
3‐{3‐[4‐(4,6‐Dimethylpyrimidin‐2‐yl)piperazin‐1‐yl]propyl}‐1H‐indole (11)
Yield: 700 mg, (97%) as a white solid (EtOAc/hexane 1:1). 1H NMR (400 MHz, DMSO-d6) δ 10.74 (br s, 1H), 7.51 (d, J = 7.6 Hz, 1H), 7.32 (d, J = 7.6 Hz, 1H), 7.11 (d, J = 2 Hz, 1H), 7.05 (td, J = 7.2, 1.2 Hz, 1H), 6.96 (td, J = 7.2, 1.2 Hz, 1H), 6.38 (s, 1H), 3.71–3.69 (m, 4H), 2.73–2.69 (m, 2H), 2.40–2.34 (m, 6H), 2.21 (s, 6H), 1.83 (quintet, J = 7.6 Hz, 2H). 13C NMR (100 MHz, DMSO-d6) δ 166.6 (2 × C), 161.2, 136.3, 127.2, 122.2, 120.8, 118.3, 118.1, 114.4, 111.3, 108.7, 57.8, 52.8 (2 × CH2), 43.3 (2 × CH2), 27.2, 23.7 (2 × CH3), 22.5. HRMS-ESI (m/z) [M + H]+ calcd for C21H28N5, 350.2339; found, 350.2346 (Δ = -1.9 ppm). [M + Na]+ calcd for C21H27N5Na, 372.2159; found, 372.2166 (Δ = -2.0 ppm). HPLC (2): t = 9.0 min, 100% purity. HPLC (3): t = 14.4 min, 96.2% purity.
3‐{4‐[4‐(4,6‐Dimethylpyrimidin‐2‐yl)piperazin‐1‐yl]butyl}‐1H‐indole (12)
Yield: 510 mg, (97%) as a white solid (EtOAc/hexane 4:6). 1H NMR (400 MHz, DMSO-d6) δ 10.72 (br s, 1H), 7.50 (d, J = 7.6 Hz, 1H), 7.32 (d, J = 7.6 Hz, 1H), 7.10 (d, J = 2.4 Hz, 1H), 7.05 (td, J = 6.8, 0.8 Hz, 1H), 6.95 (td, J = 6.8, 0.8 Hz, 1H), 6.38 (s, 1H), 3.69–3.66 (m, 4H), 2.70 (t, J = 7.2 Hz, 2H), 2.37–2.31 (m, 6H), 2.21 (s, 6H), 1.67 (quintet, J = 7.6 Hz, 2H), 1.52 (quintet, J = 7.6 Hz, 2H). 13C NMR (100 MHz, DMSO-d6) δ 166.6 (2 × C), 161.2, 136.3, 127.2, 122.1, 120.7, 118.3, 118.0, 114.6, 111.3, 108.7, 57.8, 52.7 (2 × CH2), 43.3 (2 × CH2), 27.7, 26.2, 24.5, 23.7 (2 × CH3). HRMS-ESI (m/z) [M + H]+ calcd for C22H30N5, 364.2496; found, 364.2497 (Δ = -0.4 ppm). [M + Na]+ calcd for C22H29N5Na, 386.2315; found, 386.2318 (Δ = -0.8 ppm). HPLC (2): t = 9.3 min, 100% purity. HPLC (3): t = 14.7 min, 98.5% purity.
Biological evaluations
Membrane preparation
A two-step procedure was adopted to prepare membrane for radioligand binding from cells stably transfected with human adenosine receptor subtypes (hA1, hA2A and hA3 expressed on CHO cells) [41]. Firstly, cell fragments and nuclei were removed by using low-speed centrifugation (1,000 g for 10 min). After that, crude membrane fraction from supernatant was then sedimented at 100,000 g for 30 min. The membrane pellet was subsequently re-suspended in the specific binding buffer, frozen in liquid nitrogen and stored at -80°C. For measurement of adenylyl cyclase activity in the hA2B receptors, one-step centrifugation procedure was used; the homogenate was sedimented for 30 min at 54,000 g. The so-obtained crude membrane pellet was re-suspended in 50 mM Tris/HCl, pH 7.4 and used for the adenylyl cyclase assay immediately.
Binding assays for hA1, hA2A, hA3 adenosine receptors
In accordance with procedures described previously [41–43], competition binding experiment for human A1 adenosine receptors was carried out for 3 h at 25°C in 200 μL of buffer containing 1 nM [3H]CCPA (KD = 0.61 nM), 0.2 U/mL adenosine deaminase, 20 μL of diluted membranes (50 μg of protein/assay) in 50 mM Tris/HCl, pH 7.4 and test compounds in different concentrations. Non-specific binding was determined using theophylline 1 mM. In a similar manner, binding of [3H]NECA to CHO cells transfected with hA2A adenosine receptors was performed. A mixture of protein with a concentration of 50 μg/assay in buffer, 10 nM [3H]NECA (KD = 20 nM) and test compound in different concentrations were incubated for 3 h at 25°C. Non-specific binding was determined using N6-R-phenylisopropyl adenosine (R-PIA) 100 μM. 41 The binding of [3H]HEMADO to CHO cells transfected with hA3 adenosine receptors was carried out as described earlier [44]. The binding experiment was carried out for 3 h at 25°C in the buffer solution containing 1 nM [3H]HEMADO (KD = 1.1 nM), 50 μg membrane protein in 50 mM Tris-HCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 10 mM MgCl2, pH 8.25 and test compound in different concentrations. Non-specific binding was determined using R-PIA 100 μM.
The assay mixture was then filtered through the built-in filter at the bottom of the 96-well microplate filtration system (Millipore Multiscreen MAFC) and washed three times with 200 μL of cold buffer. After addition of 20 μL of scintillator to the dried filter plates, the filter bound radioactivity was counted on a Wallac Micro-Beta counter. All Ki values were calculated by non-linear curve fitting with the program SCTFIT [45].
Adenylyl cyclase activity assay for hA2B adenosine receptors
Due to the lack of a useful high-affinity radioligand for A2B receptors, the potency of test compounds at hA2B receptor was determined in adenylyl cyclase experiments as described previously [41,42]. Concentration-dependent inhibition of NECA-stimulated adenylyl cyclase (stimulation with 5 μM of NECA, EC50 = 2.4 μM) caused by test compounds was measured in membranes from CHO cells stably transfected with the hA2B adenosine receptors. The membranes were incubated with about 150,000 cpm of [α-32P]adenosine triphosphate ([α-32P]ATP) and test compounds in different concentrations for 20 min. A non-linear regression analysis was applied to calculate the IC50 of the adenylyl cyclase activity assay. From the IC50 values, Ki values were then calculated using the Cheng and Prusoff equation [46].
Dopamine D2 receptor proteopolymersomes
Polymersomes (ABA triblock-copolymer and BD21 diblock-copolymer: ABA stands for PMOXA-PDMS-PMOXA; A = PMOXA = poly(2-methyloxazoline); B = PDMS = poly(dimethylsiloxane) and BD21 stands for [PBd]22-b-[PEO]13; PBd/PEO = polybutadiene/polyethylene oxide) preparation, cloning and in vitro synthesis of dopamine D2 receptor, and purification of proteopolymersomes were performed following previously described procedures [47]. For the replacement assay, ABA-polymersomes were covalently attached to an amino-functionalized glass slide, which was then treated with isopropanol, ultrapure water and a N2-stream. The slide was cut into small chips and each chip was treated with the ethanolic solution of the mixture containing tetrazol(4-(2-phenyl-2H-tetrazol-5-yl)benzoic acid, N-hydroxysuccinimide and N-(3-dimethylaminopropyl)-N’-ethylcarbodiimidehydrochloride, and then incubated for one hour. ABA with 10% methacrylate was dispensed onto the polydimethylsiloxane (PDMS) stamps and incubated for one hour. The photoinducible 1,3-dipolar cycloaddition between the tetrazole and the methacrylate functional group on the polymersomes was induced by 15 min incubation under UV light (254 nm). The chips were incubated with a 30 μM BODIPY-NAPS solution in Tris-MgCl2-NaCl (TMN) buffer for 30 min in the dark at room temperature. Program ImageJ was used for determination of fluorescence intensities.
Adenylyl cyclase inhibition assay for dopamine D2 receptors
CHO cells transfected with D2 receptors were cultured and then serum starved overnight. Cells were treated with various concentrations of ligands (25 μL of 10X solution), stimulated with forskolin (0.5 μM) and 3-isobutyl-1-methylxanthine (IBMX) (0.5 mM), and incubated for 15 minutes. After lysis, cAMP content was determined by ELISA.
Drosophila models of PD
Fly lines for 24B-Gal4 (muscle specific), ddc-Gal4 (dopaminergic neuron-specific), elav-Gal4 (pan-neuronal), UAS-mito-GFP, and UAS-dAMPK-KA were purchased from Bloomington Drosophila Stock Center. The parkin-null mutant flies were kind gifts from Chung J. and Cho K. S. (Korea Advanced Institute of Science and Technology, Daejeon, Korea). To generate transgenic mutant LRRK2 G2019S, cDNA containing a myc-tag at the C terminus was inserted into pUAST plasmid and microinjected into Drosophila embryos (BestGene). Sequencing of cloned products was performed before they were microinjected into the embryos. Climbing assays were performed according to a previously described method [48]. Briefly, 20 female adult flies from each group were randomly selected after being anaesthetised and placed in a vertical plastic column (length 25 cm; diameter 1.5 cm). Age-matched normal flies were used as controls. After a 2-hour recovery period from CO2 exposure, flies were gently tapped to the bottom of the column, and the number of flies that reached the top of column at 1 min was counted. Results are presented as mean ± SEM of the scores obtained from three independent experiments. To study the effect of compounds, flies were fed with cornmeal-agar medium supplemented with the DMSO solution of the compound immediately at post-eclosion (for parkin-null flies) or at day 35 onwards (for LRRK2 mutant flies) for a period of 25 days. Immunohistochemical analysis of whole-mount adult fly brains were prepared according to published protocols [36] and stained with rabbit anti-TH (1:300, Pel-Freez Biologicals) as primary antibody. The stained samples were viewed using an Olympus Fluoview Upright Confocal Microscope. DA neurons were quantified according to previously published method [36]. The size of mito-GFP puncta was measured using the ImageJ program and expressed as mean ± SD (n ≥ 10 DA neurons per experimental group). Statistical significance for all the quantitative data obtained were analyzed using one-way ANOVA with Tukey’s test HSD post hoc test (*p < 0.05, **p < 0.01).
Mutagenicity test
In vitro mutagenicity was performed using a modified Ames Test protocol according to manufacturer’s instructions (MolTox). S. typhimurium strains (TA98 and TA100) were grown from bacterial discs in Oxoid #2 nutrient broth at 37°C in a shaking incubator (~150 rpm) for about 10 h. The cultures were then measured for absorbance with a UV spectrophotometer at 660 nm and to be used at a density of approximately 1.0 to 1.2 absorbance units. For compound treatment, the top agar was melted in a hot water bath or microwave oven and 2 mL volumes were aliquoted into culture tubes. The tubes of agar were then maintained at 45°C for at least 30 to 45 min for temperature equilibration. 100 μL of test compounds at concentration of 1 mM were added to separate tubes containing top agar in duplicates. Additional tubes were set aside as negative (DMSO, methotrexate) and positive (4-NQO and 2-AA) controls. 4-NQO was reported to be a DNA damaging agent in the cell-based assay with no added exogenous metabolic enzymes [49], while 2-AA was reported to be metabolically activated into mutagens by various CYP isozymes [50]. Subsequently, 500 μL of S9 mix was introduced to each tube containing either controls or test compounds. The contents were immediately mixed and decanted onto Minimal Glucose (MG) Agar Plate and swirled to obtain an even distribution of plating mixture over the agar surface. After the agar was set, the plates were then incubated at 37°C for 48 h. Images of colonies were captured and counted with the aid of ImageJ software.
Cytotoxicity test
Immortalised hepatocyte and cardiomyocyte cell lines, TAMH (TGF-α overexpressing mouse hepatocytes) and HL-1 were used respectively as models for in vitro toxicity studies. Both cell lines were cultured in accordance with previously described methods [51,52]. Briefly, TAMH lines between passages 21–35 were grown in serum-free DMEM/Ham’s F12 (Invitrogen, Carlsbad, CA) supplemented 5 μg/mL insulin, 5 μg/mL transferrin, 5 ng/mL selenium (Collaborative Biomedical Products, 354351 Boston, MA), 100 nM dexamethasone, 10 mM nicotinamide and 0.1% v/v gentamicin (Invitrogen) and 0.12% sodium bicarbonate (Sigma 5671). For HL-1 cells, cell culture flasks were first coated with fibronectin/gelatin (25 μg of fibronectin in 2 mL of 0.02% gelatin in water per T25 flask (Sigma G1393 & F1141)) overnight at 37°C and the excess fluid was aspirated thereafter. Cells were then maintained in Claycomb medium (Sigma 51800C) supplemented with FBS (Sigma F2442), 2 mM L-glutamine (Sigma), 10 μM norepinephrine, 100 U/ml penicillin, 100 μg/ml streptomycin and 1X non-essential amino acids. Medium was changed every 24 h. All cultures were maintained in a humidified incubator with 5% carbon dioxide/95% air at 37°C and passaged at 70–90% confluence. TAMH cells were plated into 96-well plates at 12,000 cells per well whilst HL-1 at 15,000 cells per well and incubated overnight. The following day, test compounds were prepared from DMSO stock solutions and diluted to concentrations of 0.03 μM to 100 μM. Medium was aspirated and replaced with respective compound concentrations and incubated for another 24 h at 37°C (n = 6). Subsequently, Cell-Titer-Glo (Promega G7571) assay was performed according to manufacturer’s instructions. The cell-reagent mixture was transferred to a solid white flat-bottom 96-well plate (Greiner 655207) for luminescence reading. Luminescence was recorded with an integration time of 0.25 second with a Tecam Infinite® M200 Microplate reader. Data are expressed as percentage of viable cells compared to DMSO-treated controls. Semi-log graphs were plotted using GraphPad Prism (La Jolla, CA, USA).
Aqueous solubility studies
The 10 μL stock solution of the ligand (10 mM in DMSO) was added to the universal aqueous buffer (pH 7.4), and the mixture was sonicated. 300 μL of the turbid mixture was transferred to the 3 wells of MultiScreen HTS- PCF filter plate (Millipore Corp., Ireland), and the plate was covered and incubated with gentle shaking (250 rpm, 3 h) at room temperature (22.5 ± 2.5°C). After the period of incubation (3 h), the filter plate was placed on a vacuum manifold and the contents were filtered into a 96-well UV plate. After filtration, 200 μL of filtrate was transferred from each well to the PP vial. Absorbances of the solutions were quantified by HPLC-UV and read at 250 nm.
Supporting information
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
Number of His+-revertant colonies grown on the agar plates containing various chemicals and (A) TA98 strain without “S9 mix”, (B) TA98 strain with “S9 mix”, (C) TA100 strain without “S9 mix”, and (D) TA100 strain with “S9 mix”. Blank indicates spontaneously induced revertants without treatment with any drug or solvent.
(DOC)
Cell viability after treatment of TAMH (A-C) and HL-1 (D-F) cells with positive controls and compounds.
(DOC)
Acknowledgments
The authors would like to thank Ms Sonja Kachler for the binding data at A2A adenosine receptors; Dr. Sourabh Banerjee and Dr. Madhavan Nallani for the artificial cell membrane assays; NUS Drug Development Unit for the assays on drug-like properties; Mr. Yang Xuan, Dr. Sam Ramanujulu and Mr. See Cheng Shang for the assistance in high-performance liquid chromatography.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Support provided by ARC FRC (R-148-000-213-112 to GP), A-STAR-SERC (R-148-000-222-305 to GP), Leung Kai Fook (R-148-000-227-720 to GP),and an NGS scholarship to YMS and AT. The funding agencies provided the resources for the synthesis, characterization and the pharmacological testing of the compounds.
References
- 1.Obeso JA, Rodriguez-Oroz MC, Rodriguez M, Lanciego JL, Artieda J, Gonzalo N, et al. Pathophysiology of the basal ganglia in Parkinson's disease. Trends Neurosci. 2000;23(10 Suppl):S8–19. . [DOI] [PubMed] [Google Scholar]
- 2.Djaldetti R, Melamed E. New therapies for Parkinson's disease. J Neurol. 2001;248(5):357–62. . [DOI] [PubMed] [Google Scholar]
- 3.Huot P, Johnston TH, Koprich JB, Fox SH, Brotchie JM. The pharmacology of L-DOPA-induced dyskinesia in Parkinson's disease. Pharmacol Rev. 2013;65(1):171–222. doi: 10.1124/pr.111.005678 . [DOI] [PubMed] [Google Scholar]
- 4.Obeso JA, Olanow CW, Nutt JG. Levodopa motor complications in Parkinson's disease. Trends Neurosci. 2000;23(10 Suppl):S2–7. . [DOI] [PubMed] [Google Scholar]
- 5.Yuan H, Zhang ZW, Liang LW, Shen Q, Wang XD, Ren SM, et al. Treatment strategies for Parkinson's disease. Neurosci Bull. 2010;26(1):66–76. doi: 10.1007/s12264-010-0302-z . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwarzschild MA, Agnati L, Fuxe K, Chen JF, Morelli M. Targeting adenosine A2A receptors in Parkinson's disease. Trends Neurosci. 2006;29(11):647–54. doi: 10.1016/j.tins.2006.09.004 . [DOI] [PubMed] [Google Scholar]
- 7.Ferre S, Ciruela F, Woods AS, Lluis C, Franco R. Functional relevance of neurotransmitter receptor heteromers in the central nervous system. Trends Neurosci. 2007;30(9):440–6. doi: 10.1016/j.tins.2007.07.001 . [DOI] [PubMed] [Google Scholar]
- 8.Fuxe K, Ferre S, Genedani S, Franco R, Agnati LF. Adenosine receptor-dopamine receptor interactions in the basal ganglia and their relevance for brain function. Physiol Behav. 2007;92(1–2):210–7. doi: 10.1016/j.physbeh.2007.05.034 . [DOI] [PubMed] [Google Scholar]
- 9.Simola N, Morelli M, Pinna A. Adenosine A2A receptor antagonists and Parkinson's disease: state of the art and future directions. Curr Pharm Des. 2008;14(15):1475–89. . [DOI] [PubMed] [Google Scholar]
- 10.Antonelli T, Fuxe K, Agnati L, Mazzoni E, Tanganelli S, Tomasini MC, et al. Experimental studies and theoretical aspects on A2A/D2 receptor interactions in a model of Parkinson's disease. Relevance for L-dopa induced dyskinesias. J Neurol Sci. 2006;248(1–2):16–22. doi: 10.1016/j.jns.2006.05.019 . [DOI] [PubMed] [Google Scholar]
- 11.Morelli M, Wardas J. Adenosine A2A receptor antagonists: Potential therapeutic and neuroprotective effects in parkinson’s disease. Neurotoxicity Research. 2001;3(6):545–56. doi: 10.1007/BF03033210 [DOI] [PubMed] [Google Scholar]
- 12.Le Witt PA, Guttman M, Tetrud JW, Tuite PJ, Mori A, Chaikin P, Sussman NM. Adenosine A2A receptor antagonist istradefylline (KW6002) reduces off time in Parkinson’s disease: a double-blind, randomized, multicenter clinical trial (6002-US-005). Ann Neurol 2008:63(3):295−302. doi: 10.1002/ana.21315 . [DOI] [PubMed] [Google Scholar]
- 13.Katritch V, Jaakola VP, Lane JR, Lin J, Ijzerman AP, Yeager M, et al. Structure-based discovery of novel chemotypes for adenosine A(2A) receptor antagonists. J Med Chem. 2010;53(4):1799–809. doi: 10.1021/jm901647p ; PubMed Central PMCID: PMCPMC2826142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vu CB, Peng B, Kumaravel G, Smits G, Jin X, Phadke D, et al. Piperazine derivatives of [1,2,4]triazolo[1,5-a][1,3,5]triazine as potent and selective adenosine A2A receptor antagonists. J Med Chem. 2004;47(17):4291–9. doi: 10.1021/jm0498405 . [DOI] [PubMed] [Google Scholar]
- 15.Neustadt BR, Hao J, Lindo N, Greenlee WJ, Stamford AW, Tulshian D, et al. Potent, selective, and orally active adenosine A2A receptor antagonists: arylpiperazine derivatives of pyrazolo[4,3-e]-1,2,4-triazolo[1,5-c]pyrimidines. Bioorg Med Chem Lett. 2007;17(5):1376–80. doi: 10.1016/j.bmcl.2006.11.083 . [DOI] [PubMed] [Google Scholar]
- 16.Gillespie RJ, Bamford SJ, Gaur S, Jordan AM, Lerpiniere J, Mansell HL, et al. Antagonists of the human A(2A) receptor. Part 5: Highly bio-available pyrimidine-4-carboxamides. Bioorg Med Chem Lett. 2009;19(10):2664–7. doi: 10.1016/j.bmcl.2009.03.142 . [DOI] [PubMed] [Google Scholar]
- 17.Giorgioni G, Ambrosini D, Vesprini C, Hudson A, Nasuti C, Di Stefano A, et al. Novel imidazoline compounds as partial or full agonists of D2-like dopamine receptors inspired by I2-imidazoline binding sites ligand 2-BFI. Bioorg Med Chem. 2010;18(19):7085–91. doi: 10.1016/j.bmc.2010.08.005 . [DOI] [PubMed] [Google Scholar]
- 18.Pettersson F, Ponten H, Waters N, Waters S, Sonesson C. Synthesis and evaluation of a set of 4-phenylpiperidines and 4-phenylpiperazines as D2 receptor ligands and the discovery of the dopaminergic stabilizer 4-[3-(methylsulfonyl)phenyl]-1-propylpiperidine (huntexil, pridopidine, ACR16). J Med Chem. 2010;53(6):2510–20. doi: 10.1021/jm901689v . [DOI] [PubMed] [Google Scholar]
- 19.Johnson DS, Choi C, Fay LK, Favor DA, Repine JT, White AD, et al. Discovery of PF-00217830: aryl piperazine napthyridinones as D2 partial agonists for schizophrenia and bipolar disorder. Bioorg Med Chem Lett. 2011;21(9):2621–5. doi: 10.1016/j.bmcl.2011.01.059 . [DOI] [PubMed] [Google Scholar]
- 20.Han LY, Ma XH, Lin HH, Jia J, Zhu F, Xue Y, et al. A support vector machines approach for virtual screening of active compounds of single and multiple mechanisms from large libraries at an improved hit-rate and enrichment factor. J Mol Graph Model. 2008;26(8):1276–86. doi: 10.1016/j.jmgm.2007.12.002 . [DOI] [PubMed] [Google Scholar]
- 21.Willett P. Similarity-based virtual screening using 2D fingerprints. Drug Discov Today. 2006;11(23–24):1046–53. doi: 10.1016/j.drudis.2006.10.005 . [DOI] [PubMed] [Google Scholar]
- 22.Evans DA. History of the Harvard ChemDraw project. Angew Chem Int Ed Engl. 2014;53(42):11140–5. doi: 10.1002/anie.201405820 . [DOI] [PubMed] [Google Scholar]
- 23.Vlad G, Horvath IT. Improved synthesis of 2,2'-bipyrimidine. J Org Chem. 2002;67(18):6550–2. . [DOI] [PubMed] [Google Scholar]
- 24.Bakthavachalam V, Baindur N, Madras BK, Neumeyer JL. Fluorescent probes for dopamine receptors: synthesis and characterization of fluorescein and 7-nitrobenz-2-oxa-1,3-diazol-4-yl conjugates of D-1 and D-2 receptor ligands. J Med Chem. 1991;34(11):3235–41. . [DOI] [PubMed] [Google Scholar]
- 25.Kuhhorn J, Gotz A, Hubner H, Thompson D, Whistler J, Gmeiner P. Development of a bivalent dopamine D(2) receptor agonist. J Med Chem. 2011;54(22), 7911–7919.) doi: 10.1021/jm2009919 [DOI] [PubMed] [Google Scholar]
- 26.Taussig R, Iniguez-Lluhi JA, Gilman AG. Inhibition of adenylyl cyclase by Gi alpha. Science 1993;261(5118), 218–221. [DOI] [PubMed] [Google Scholar]
- 27.Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276(5321):2045–7. . [DOI] [PubMed] [Google Scholar]
- 28.Lucking CB, Durr A, Bonifati V, Vaughan J, De Michele G, Gasser T, et al. Association between early-onset Parkinson's disease and mutations in the parkin gene. N Engl J Med. 2000;342(21):1560–7. doi: 10.1056/NEJM200005253422103 . [DOI] [PubMed] [Google Scholar]
- 29.Kumari U, Tan EK. LRRK2 in Parkinson's disease: genetic and clinical studies from patients. FEBS J. 2009;276(22):6455–63. doi: 10.1111/j.1742-4658.2009.07344.x . [DOI] [PubMed] [Google Scholar]
- 30.Cha GH, Kim S, Park J, Lee E, Kim M, Lee SB, et al. Parkin negatively regulates JNK pathway in the dopaminergic neurons of Drosophila. Proc Natl Acad Sci U S A. 2005;102(29):10345–50. doi: 10.1073/pnas.0500346102 ; PubMed Central PMCID: PMCPMC1177361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Z, Wang X, Yu Y, Li X, Wang T, Jiang H, et al. A Drosophila model for LRRK2-linked parkinsonism. Proc Natl Acad Sci U S A. 2008;105(7):2693–8. doi: 10.1073/pnas.0708452105 ; PubMed Central PMCID: PMCPMC2268198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hearn MG, Ren Y, McBride EW, Reveillaud I, Beinborn M, Kopin AS. A Drosophila dopamine 2-like receptor: Molecular characterization and identification of multiple alternatively spliced variants. Proc Natl Acad Sci U S A. 2002;99(22):14554–9. doi: 10.1073/pnas.202498299 ; PubMed Central PMCID: PMCPMC137921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Draper I, Kurshan PT, McBride E, Jackson FR, Kopin AS. Locomotor activity is regulated by D2-like receptors in Drosophila: an anatomic and functional analysis. Dev Neurobiol. 2007;67(3):378–93. doi: 10.1002/dneu.20355 . [DOI] [PubMed] [Google Scholar]
- 34.Lesage S, Durr A, Tazir M, Lohmann E, Leutenegger AL, Janin S, et al. LRRK2 G2019S as a cause of Parkinson's disease in North African Arabs. N Engl J Med. 2006;354(4):422–3. doi: 10.1056/NEJMc055540 . [DOI] [PubMed] [Google Scholar]
- 35.Ozelius LJ, Senthil G, Saunders-Pullman R, Ohmann E, Deligtisch A, Tagliati M, et al. LRRK2 G2019S as a cause of Parkinson's disease in Ashkenazi Jews. N Engl J Med. 2006;354(4):424–5. doi: 10.1056/NEJMc055509 . [DOI] [PubMed] [Google Scholar]
- 36.Whitworth AJ, Theodore DA, Greene JC, Benes H, Wes PD, Pallanck LJ. Increased glutathione S-transferase activity rescues dopaminergic neuron loss in a Drosophila model of Parkinson's disease. Proc Natl Acad Sci U S A. 2005;102(22):8024–9. doi: 10.1073/pnas.0501078102 ; PubMed Central PMCID: PMCPMC1142368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mortelmans K, Zeiger E. The Ames Salmonella/microsome mutagenicity assay. Mutat Res. 2000;455(1–2):29–60. . [DOI] [PubMed] [Google Scholar]
- 38.James LP, Mayeux PR, Hinson JA. Acetaminophen-induced hepatotoxicity. Drug Metab Dispos. 2003;31(12):1499–506. doi: 10.1124/dmd.31.12.1499 . [DOI] [PubMed] [Google Scholar]
- 39.Aryal B, Jeong J, Rao VA. Doxorubicin-induced carbonylation and degradation of cardiac myosin binding protein C promote cardiotoxicity. Proc Natl Acad Sci U S A. 2014;111(5):2011–6. doi: 10.1073/pnas.1321783111 ; PubMed Central PMCID: PMCPMC3918758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kerns EH, Di L. Chapter 2—Advantages of Good Drug-like Properties In Drug-like Properties: Concepts, Structure Design and Methods, Kerns E.H., and Di L., eds. (San Diego: Academic Press; ), 2008; pp. 6–16. [Google Scholar]
- 41.Klotz KN, Hessling J, Hegler J, Owman C, Kull B, Fredholm BB, et al. Comparative pharmacology of human adenosine receptor subtypes—characterization of stably transfected receptors in CHO cells. Naunyn Schmiedebergs Arch Pharmacol. 1998;357(1):1–9. . [DOI] [PubMed] [Google Scholar]
- 42.Klotz KN, Cristalli G, Grifantini M, Vittori S, Lohse MJ. Photoaffinity labeling of A1-adenosine receptors. J Biol Chem. 1985;260(27):14659–64. . [PubMed] [Google Scholar]
- 43.Lohse MJ, Klotz KN, Lindenborn-Fotinos J, Reddington M, Schwabe U, Olsson RA. 8-Cyclopentyl-1,3-dipropylxanthine (DPCPX)—a selective high affinity antagonist radioligand for A1 adenosine receptors. Naunyn Schmiedebergs Arch Pharmacol. 1987;336(2):204–10. . [DOI] [PubMed] [Google Scholar]
- 44.Klotz KN, Falgner N, Kachler S, Lambertucci C, Vittori S, Volpini R, et al. [3H]HEMADO—a novel tritiated agonist selective for the human adenosine A3 receptor. Eur J Pharmacol. 2007;556(1–3):14–8. doi: 10.1016/j.ejphar.2006.10.048 . [DOI] [PubMed] [Google Scholar]
- 45.De Lean A, Hancock AA, Lefkowitz RJ. Validation and statistical analysis of a computer modeling method for quantitative analysis of radioligand binding data for mixtures of pharmacological receptor subtypes. Mol Pharmacol. 1982;21(1):5–16. . [PubMed] [Google Scholar]
- 46.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22(23):3099–108. . [DOI] [PubMed] [Google Scholar]
- 47.May S, Andreasson-Ochsner M, Fu Z, Low YX, Tan D, de Hoog HP, et al. In vitro expressed GPCR inserted in polymersome membranes for ligand-binding studies. Angew Chem Int Ed Engl. 2013;52(2):749–53. doi: 10.1002/anie.201204645 . [DOI] [PubMed] [Google Scholar]
- 48.Ng CH, Guan MS, Koh C, Ouyang X, Yu F, Tan EK, et al. AMP kinase activation mitigates dopaminergic dysfunction and mitochondrial abnormalities in Drosophila models of Parkinson's disease. J Neurosci. 2012;32(41):14311–7. doi: 10.1523/JNEUROSCI.0499-12.2012 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dahm K, Hubscher U. Colocalization of human Rad17 and PCNA in late S phase of the cell cycle upon replication block. Oncogene. 2002;21(50):7710–9. doi: 10.1038/sj.onc.1205872 . [DOI] [PubMed] [Google Scholar]
- 50.Carriere V, de Waziers I, Courtois YA, Leroux JP, Beaune PH. Cytochrome P450 induction and mutagenicity of 2-aminoanthracene (2AA) in rat liver and gut. Mutat Res. 1992;268(1):11–20. . [DOI] [PubMed] [Google Scholar]
- 51.Claycomb WC, Lanson NA Jr., Stallworth BS, Egeland DB, Delcarpio JB, Bahinski A, et al. HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc Natl Acad Sci U S A. 1998;95(6):2979–84. ; PubMed Central PMCID: PMCPMC19680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu JC, Merlino G, Cveklova K, Mosinger B Jr., Fausto N. Autonomous growth in serum-free medium and production of hepatocellular carcinomas by differentiated hepatocyte lines that overexpress transforming growth factor alpha 1. Cancer Res. 1994;54(22):5964–73. . [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
Number of His+-revertant colonies grown on the agar plates containing various chemicals and (A) TA98 strain without “S9 mix”, (B) TA98 strain with “S9 mix”, (C) TA100 strain without “S9 mix”, and (D) TA100 strain with “S9 mix”. Blank indicates spontaneously induced revertants without treatment with any drug or solvent.
(DOC)
Cell viability after treatment of TAMH (A-C) and HL-1 (D-F) cells with positive controls and compounds.
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.