Abstract
We here report the main characteristics of a new anaerobic bacterial genus and species ‘Lactomassilus timonensis,’ strain Marseille-P4641T (CSUR = P4641), isolated by microbial culturomics from the milk of a 35-year-old healthy lactating mother from Mali.
Keywords: Culturomics, Human breast milk microbiota, Lactomassilus timonensis, Taxonomy
In an assessment study of breast milk microbiota by culturomics [1], we isolated a new anaerobic bacterial species from the milk of a 35-year-old lactating Malian woman. We previously reported Veillonella massiliensis [2] as the first culturomics species isolated from human milk of a French woman. Here we report the first culturomics species isolated from the breast milk of an African woman. This is important because milk is a main vector for the vertical transmission of the human gut microbiota [3]. Moreover, healthy milk microbiota is critical for the later development of healthy mature anaerobic gut microbiota in offspring [3]. Accordingly, this new species could be a potential natural probiotics for children and could more generally benefit human health. Written consent was obtained from each participant before sampling according to the Declaration of Helsinki and council for international organizations of medical sciences (CIOMS) 2016. The study and the consent procedure were approved by the ethics committee of Institut fédératif de recherche (IFR) 48 under consent number 09-022, 2010, and by the FMPOS institutional ethics committee (Mali, comité d'éthique-Faculté de Médecine de Pharmacie et d'OdontoStomatologie (CE-FMPOS)).
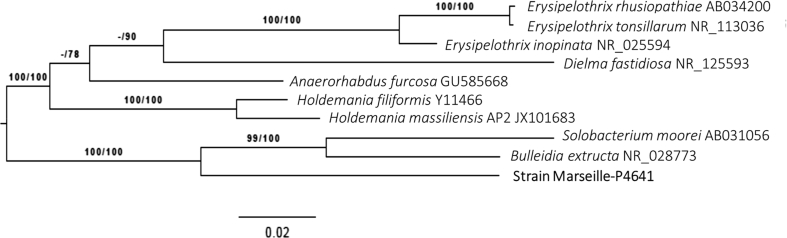
Initial growth was obtained after 21 days of preincubation in a blood culture bottle (BACTEC Lytic/10 Anaerobic/F Culture Vials; Le Pont de Claix, Isère, France) enriched with 4 mL of sheep's blood and 4 mL of rumen under anaerobic atmosphere at 37°C. Light colonies with a mean diameter of 1 to 2 mm and agar-grown (Columbia agar + 5% sheep's blood; bioMérieux, Marcy l'Etoile, France) were observed after 48 hours of incubation in anaerobic conditions for each isolate. Cells were Gram positive and rod shaped. Sequencing of the 16S rRNA gene was performed because colonies were not identified by matrix-assisted desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) screening on a Microflex BioTyper spectrometer (Bruker Daltonics, Bremen, Germany). The 16S rRNA gene was sequenced using fD1–rP2 primers as previously described, using a 3130-XL sequencer (Applied Biosciences, Saint Aubin, France) [4]. Strain Marseille-P4641T exhibited 90% and 85.5% of 16S rRNA sequence similarity with Solobacterium moorei RCA59-74 = CIP 106864 = JCM 10645 and Bulleidia extructa strain W1219, respectively [5], [6], the phylogenetically closest species with standing in nomenclature (Fig. 1). The 16S rRNA sequence divergences of >1.3% and >5% (9.3%) suggest a new species and new genus, respectively [7]. Phylogenies were inferred by the Genome-to-Genome Distance Calculator (GGDC) Web server [8] (http://ggdc.dsmz.de/) using the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) phylogenomics pipeline [9] adapted to single genes. These results suggest the creation of a new genus and species for which the names Lactomassilus and L. timonensis are proposed. In addition, we investigated the presence of 16S rRNA from Strain Marseille-P4641T in the high throughput DNA and RNA sequence read archive (SRA) using an open resource online (https://www.imngs.com). We found metagenomics sequences with a similarity greater than 97% with Strain Marseille-P 4641T in several gut metagenomes (human, bovine and pig gut, food, freshwater and waste).
Fig. 1.
Phylogenetic tree showing position of ‘Lactomassilus timonensis’ strain Marseille-P4641T relative to other phylogenetically close species. Maximum likelihood (ML) tree was inferred under general time reversible (GTR) + GAMMA model and rooted by midpoint rooting. Branches are scaled in terms of expected number of substitutions per site. Numbers above branches are support values when >60% from ML left and maximum parsimony (MP) right bootstrapping. Scale bar indicates 2% nucleotide sequence divergence.
Lactomassilus (lacto.massilus, N.L. masc.) is from lacto, ‘milk,’ and massilus, referring to Massilia, the Latin name of Marseille, where the strain was isolated. ‘L. timonensis’ (ti.mon.ensis, N.L. gen. n.) refers to timonensis, the name of La Timone hospital, where the strain was isolated. The species Lactomassilus timonensis and the strain Marseille-P4641T are the type species and the type strain of the new genus and the new species Lactomassilus gen. nov. and L. timonensis sp. nov., respectively.
Microbial culturomics has again succeeded in extending the microbial catalogue of the healthy human microbiome. This could hasten future medical progress because milk microbiota are a critical determinant of human health.
MALDI-TOF MS spectrum
The MALDI-TOF MS spectrum of this strain is available online (http://mediterranee-infection.com/article.php?laref=256&titre=urms-database).
Nucleotide sequence accession number
The 16S r RNA gene sequence was deposited in GenBank under accession number LT934541.
Deposit in a culture collection
Strain Marseille-P4641T (CSUR P 4641T) was deposited in the Collection de Souches de l’Unité des Rickettsies (CSUR = P 4641).
Conflict of interest
None declared.
Acknowledgements
The authors thank M. Lardière for English-language review. This work was funded by the Fondation Méditerranée Infection and by Malaria Research and Training Center-Département d’épidémiologie et des affections parasitaires/Unité mixte de recherche internationale (MRTC-DEAP/UMI3189) Centre national de la recherche scientifique (CNRS) and Fondation Mérieux (FMERIEUX) grants.
References
- 1.Lagier J.C., Khelaifia S., Alou M.T., Ndongo S., Dione N., Hugon P. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Togo A.H., Des Robert C., Bonnet M., Fournier P.E., Raoult D., Million M. ‘Veillonella massiliensis,’ a new anaerobic species isolated from human colostrum. Hum Microbiome J. 2017;4:20–21. [Google Scholar]
- 3.Million M., Diallo A., Raoult D. Gut microbiota and malnutrition. Microb Pathog. 2017;106:127–138. doi: 10.1016/j.micpath.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Drancourt M., Bollet C., Carlioz A., Martelin R., Gayral J.P., Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol. 2000;38:3623–3630. doi: 10.1128/jcm.38.10.3623-3630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kageyama A., Benno Y. Phylogenic and phenotypic characterization of some Eubacterium-like isolates from human feces: description of Solobacterium moorei gen. nov., sp. nov. Microbiol Immunol. 2000;44:223–227. doi: 10.1111/j.1348-0421.2000.tb02487.x. [DOI] [PubMed] [Google Scholar]
- 6.Downes J., Olsvik B., Hiom S.J., Spratt D.A., Cheeseman S.L., Olsen I. Bulleidia extructa gen. nov., sp. nov., isolated from the oral cavity. Int J Syst Evol Microbiol. 2000;50:979–983. doi: 10.1099/00207713-50-3-979. [DOI] [PubMed] [Google Scholar]
- 7.Stackebrandt E., Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152–155. [Google Scholar]
- 8.Meier-Kolthoff J.P., Auch A.F., Klenk H.P., Göker M. Genome sequence–based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meier-Kolthoff J.P., Hahnke R.L., Petersen J., Scheuner C., Michael V., Fiebig A. Complete genome sequence of DSM 30083T, the type strain (U5/41T) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand Genomic Sci. 2014;9:2. doi: 10.1186/1944-3277-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]