Abstract
Pseudomonas aeruginosa use small signaling molecules such as acyl homoserine lactones (AHLs), which play an important role in release virulence factors and toxin for further establishment of host infection. Thus, involving with the QS system would provide alternative ways of preventing the pathogenicity. In the present study, totally six medicinal plants (Terminalia bellerica, Celastrus paniculatus, Kingiodendron pinnatum, Schleichera oleosa, Melastoma malabathricum, Garcinia gummi-gutta) were screened for anti-QS activity using biomonitor strain of Chromobacterium violaceum CV12472. The primary screening of antimicrobial activity of all the plant extracts have inhibited the growth of tested bacterial species. Of these at the sub-minimum inhibitory concentration the methanol extract of T. bellerica (0.0625–0.5 mg/ml) has significantly inhibited violacein production (20.07–66.22%) in C. violaceum (CV12472). Consequently, the extract of T. bellerica has reduced the production of pyocyanin, exopolysaccharide and biofilm formation in P. aeruginosa strains. Fluorescence and scanning electron microscopy analysis confirmed the reduction of biofilm formation in P. aeruginosa strains when treated with T. bellerica. GC–MS analysis showed the active compounds inhibited the production of virulence factors of P. aeruginosa. The results suggest the possible use of this T. bellerica as an anti-QS and anti-biofilm agent to control Pseudomonas infection. Interference of QS provides an important means for the inhibition of bacterial virulence and thus aids in treatment strategies.
Keywords: Meningitis, Acyl homoserine lactone, Violacein, Pyocyanin, Biofilm
Abbreviations: AHLs, acyl homoserine lactones; QS, quorum sensing; EPS, exopolymeric substance; 3O-C12-HSL, N-(3-oxododecanoyl)-l-homoserine lactone; C4-HSL, N-butyryl-homoserine lactone
Graphical abstract
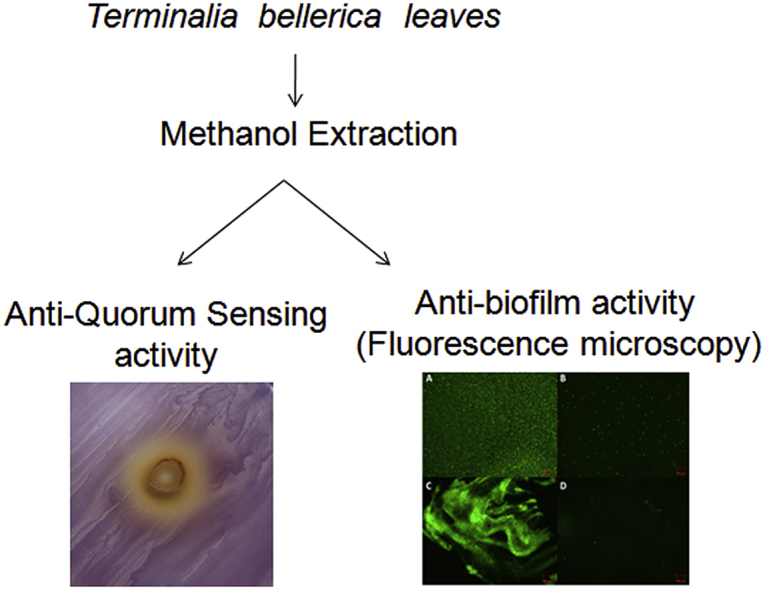
1. Introduction
Pseudomonas aeruginosa is one of the most important opportunistic human pathogens. It is capable of infecting patients suffering from cystic fibrosis, chronic obstructive pulmonary diseases, immunocompromised patients and AIDS patients.1 In addition, P. aeruginosa is able to cause severe burn wound infections, urinary tract infection, bloodstream infections, nosocomial infections2 and meningitis.3 In hospital environment, this organism can easily infect patients who are immunocompromised and it rarely infects healthy people and hospital coworkers.4 P. aeruginosa releases a variety of virulence factors, which are mainly involved in the progression of disease through enforcing the adhesion, modifying the immune response, evading from phagocytosis and destroying the host tissue.5 The ability of P. aeruginosa to communicate and coordinate with other cells in the population using small signaling molecules such as acyl homoserine lactones (AHLs) is called quorum sensing (QS).6 QS is an intercellular communication system, which can efficiently control the gene expression in a cell-density dependent manner.6 Once the threshold concentration is reached, the organisms release virulence factors for further establishment of host infection.6 P. aeruginosa possesses two QS system namely las and rhl which utilize N-acyl homoserine lactones (AHLs) as signaling molecules.7 These signaling molecules, synthesized by bacterial cells, diffuse out the cells and it binds to transcriptional regulators. There are two QS systems in P. aeruginosa (LasR/I and RhlI/R) which have been identified to regulate the expression of virulence factors. The LasI is essential for the production of the AHL molecule, N-(3-oxododecanoyl)-l-homoserine lactone (3O-C12-HSL) and lasR is the transcriptional regulator.8 The second QS system comprises RhlI and RhlR proteins. The RhlI synthase mediates the synthesis of the signal molecule, N-butyryl-homoserine lactone (C4-HSL) and RhlR is the transcriptional regulator.9 The las system controls virulence factors such as LasB elastase, LasA protease, alkaline protease, exotoxin A and biofilm formation. Similarly, rhl system controls the production of pyocyanin pigment, rhamnolipids, LasB elastase and hydrogen cyanide.7, 10, 11 These virulence factors are involved in cellular toxicity and acute infection.12 Treatment strategies of Pseudomonas infections are greatly challenged by the emergence of drug resistant strains and also on account of biofilm formation by this bacterium.
Biofilms are a conglomerate of microbes enclosed in a self-secreted exopolymeric substance (EPS) that environs the bacterial population. It is mainly composed of polysaccharides, proteins, DNAs, lipids and other macromolecules.13 The biofilm shields the bacterial population from clearance by the immune system and contributes to the pathogenesis of chronic infections such as cystic fibrosis and other pulmonary illnesses.14 Biofilm formation and maintenance of its architecture are QS-dependent phenomena.15 EPS is important for the development of biofilm matrix and provides an effective barrier that restricts the entry of antibiotics and antimicrobial peptides.16 QS-controlled virulence factors and biofilm formation are vital for the development of acute and chronic infections, particularly in Gram-negative bacteria such as P. aeruginosa. These factors result in bacterial persistence and reduced sensitivity to antimicrobials.16 Hence, there is an increasing need to search potent anti-QS and antibiofilm compounds from the natural sources. Medicinal plants and plants derived essential oils, that interfere the regulation of QS system and biofilm formation could be powerful allies for conventional antibiotics in the struggle against P. aeruginosa infections.17
From time immemorial, plants have been used as traditional medicines and are being considered as safe. Terminalia bellerica belongs to the family of ‘Combretaceae’ commonly known as bastard myrobalan. T. bellerica is a traditional folk medicine which has been used to treat various ailments.18 There are various reports indicative of a wide range of pharmacological activities of T. bellerica such as antimicrobial, antidiarrhoeal, antidiabetics, antioxidant, antianalgesic, anti-inflammatory and antifibrotic.18, 19 The main objective of this study was to evaluate the anti-QS and antibiofilm activity of T. bellerica against the P. aeruginosa. To our best knowledge, this is the first study towards analyzing the anti-QS and antibiofilm potential of T. bellerica against P. aeruginosa.
2. Materials and methods
2.1. Collection of plants and methanolic extraction of medicinal plants leaves
Terminalia bellerica leaves were collected from Kuvempu Nagar, Mysore, Karnataka (between 12° 17′ 2.6376″ N Latitude 76° 37′ 43.7016″ E Longitude). Celastrus paniculatus leaves were collected from Sakleshapura hill, Sakleshapura, Hassan district, Karnataka (between 12° 17′ 2.6376″ N Latitude 76° 37′ 43.7016″ E Longitude). Kingiodendron pinnatum, Schleichera oleosa, Garcinia gummi-gutta and Melastoma malabathricum leaves were collected from Pilikula Nisargadhama, Mangalore, Karnataka (between 12° 17′ 2.6376″ N Latitude 76° 37′ 43.7016″ E Longitude) Plants were identified by taxonomically and the plants herbarium were maintained in the Department of Studies in Microbiology, University of Mysore, Mysore (Table 1). The plant leaves were collected in sterile polyethylene bags and brought to the laboratory. The leaves were washed thrice with sterile distilled water and the leaves were dried under shade. The dried leaves were grinded in a coarse powder by the mechanical grinder. Ten gram of coarse each powder was added with 100 ml of methanol and the extraction was kept for 48 h with agitation at 150 rpm. After 48 h the extract was filtered with a Whatman no. 1 filter. The filtered extract was dried using rotary flash evaporator in a hot condition (50 °C). The collected crude extracts were further dried with vacuum concentrate and stored at 4 °C for further analysis.
Table 1.
Medicinal properties of selected plants tested for anti-QS activity against C. violaceum (CV12472).
| Accession no | Species | Family | Parts | Medicinal value |
|---|---|---|---|---|
| MGMB-002 | Terminalia bellerica | Combretaceae | Bark, fruits, seed and whole plant | Antimicrobial, antidiarrhoeal, antidiabetics, antioxidant, antianalgesic, anti-inflammatory and antifibrotic |
| MGMB-003 | Celastrus paniculatus | Celastraceae | Bark, leaves, seed oil and root | Antibacterial, antiarthritic, anorexia, arthritis, asthma and cough |
| MGMB-004 | Kingiodendron pinnatum | Fabaceae | Leaves | Antimicrobial and antioxidant |
| MGMB-005 | Schleichera oleosa | Sapindaceae | Bark an oil | Antibacterial, analgesic, malaria, ache, lung cancer and burns |
| MGMB-006 | Melastoma malabathricum | Melastomataceae | Leaves, roots, flowers and shoots | Ache, digestive disorder and astringent |
| MGMB-007 | Garcinia gummi-gutta | Clusiaceae | Fruits and leaves | Antibacterial, type 2 diabetes mellitus, antiulcer and diuretic activity |
2.2. Bacterial strains and culture conditions
The clinical isolate of P. aeruginosa CI-01 (GenBank accession no. KU870518) was obtained from a meningitis patient admitted to Mysore Medical College and Hospital, Mysore, Karnataka. C. violaceum (CV12472) was routinely cultured aerobically in Luria-Bertani broth (LB) and the cultures were incubated at 30 °C for 24 h. P. aeruginosa PAO1 and P. aeruginosa CI-01 were cultured in Nutrient Broth (NB). The cultures were incubated at 37 °C for 24 h in a rotary shaker (100 rpm).
2.3. 16S rRNA gene sequencing
The genomic DNA was isolated using the phenol chloroform extraction method.20 The pathogen was identified by 16S rRNA gene sequence analysis using the universal primers, 27F and 1492R as described previously.21
2.4. Antibiotic sensitivity test
A clinical isolate of P. aeruginosa CI-01 and P. aeruginosa PAO1 were sub-cultured in LB broth and cultures were incubated at 37 °C in a shaker incubator. The optical density (OD) was adjusted to 0.5 McFarland (1 × 108 CFU/ml) standard. The clinical isolate P. aeruginosa strains were swabbed on the Muller Hinton Agar (MHA) (Himedia, India) plates. The antibiotics discs (Himedia, India) were gently placed on the center of the agar plates. The plates were incubated at 37 °C for 24 h. After incubation, the zone of inhibition was measured using standard scale (Himedia, India) and the results were analyzed by the standard deviation. The test was performed in triplicates and the results were interpreted according to Clinical and Laboratory Standards Institute (CLSI) guidelines, (2012).22
2.5. Quantification of violacein
In quantitative analysis, the methanol crude extracts were loaded into test tubes (LB broth). 10 μl of an overnight culture of C. violaceum (CV12472) was loaded and the tubes were incubated at 30 °C for 18 h. After 18 h of incubation, all the tubes were centrifuged at 6000 rpm for 10 min. The culture supernatant was discarded and to the pellets, 500 μl of dimethyl sulfoxide (DMSO) was added and the tubes were vortexed vigorously until the violacein was extracted. Then the tubes were centrifuged at 10,000 rpm for 15 min. The extracted violacein (200 μl) was loaded into a microtiter plate (Nest Biotech Co., Ltd, China) and quantified spectrophotometrically (Thermo Scientific, Mumbai, India) at 585 nm. The percentage of growth of the treated strain was compared with that of the untreated control by measuring the optical density (OD) at 600 nm.23
2.6. Quantification of pyocyanin
The pyocyanin quantification assay was performed by using the method as previously described.24 The relative concentration of pyocyanin in the supernatant of P. aeruginosa strains was measured at 520 nm.24 The percentage of growth of the treated strains was compared with that of the untreated control strains by measuring the OD at 600 nm.
2.7. Quantification of exopolysaccharide (EPS)
Extraction and quantification of EPS was performed using the method described elsewhere.25 Briefly, the test pathogens were grown in LB broth with T. bellerica extract at the concentration ranging from 0.0625 to 0.5 mg/ml and without T. bellerica extract (control). The tubes were incubated at 37 °C for 18 h. Then the tubes were centrifuged at 10,000 rpm for 15 min. The supernatant was discarded and the bacterial cell pellets were resuspended in 50 ml of high salt buffer for 15 min and tubes were centrifuged at 10,000 rpm for 30 min and the equal volume of ethanol was added to the collected supernatant and centrifuged at 10,000 rpm for 30 min. One ml of the precipitated EPS solution was mixed with one ml of cold 5% phenol and five ml of concentrated sulfuric acid to develop a red color. The intensity of the color (OD) was measured at 490 nm.26
2.8. Biofilm inhibition assay
Static microtiter plate assay was used to evaluate the inhibition of biofilm formation by T. bellerica against the test pathogens. Briefly, different concentration (0.0625–0.5 mg/ml) of T. bellerica extract was loaded to each well containing 170 μl of Tryptic Soy Broth (TSB) and was mixed well. An overnight culture (10 μl) of test pathogens was loaded and the microtiter plate was incubated at 37 °C for 24 h. After 24 h, the planktonic cells were removed without disturbing the biofilm and the planktonic cells were read at 600 nm. The plate was washed with sterilized distilled water for removing excess of planktonic cells. Crystal violet (10 μl, 0.1% (w/v) in water) was added to the wells and the plate was incubated for 15 min at room temperature. After 15 min, the crystal violet was removed from the wells and gently washed with sterile distilled water to remove the unbound crystal violet stain. Finally, the adhered biofilm bounded crystal violet was eluted in ethanol (95%) and the absorbance was measured at 585 nm (Thermo scientific, Mumbai, India). The presence of growth (planktonic cells) of the treated strains was compared with that of the untreated control strains by measuring the OD 600 nm.25
2.9. Fluorescence microscopy analysis
To perform microscopic analysis, the methanol extract of T. bellerica was loaded into freshly prepared 5 ml of TSB with a cover slip (18 mm). To this, 10 μl of an overnight culture of test pathogens were loaded and the control plate without plant extract was also maintained. All the plates were incubated at 37 °C for 24 h. After 24 h, the cover slips were removed from the plates without disturbing biofilm and the cover slips were rinsed with sterilized distilled water. This step was repeated two to three times until the removal of unbound planktonic cells from the cover slips. It was then air dried and stained with 0.1% acridine orange for 2 min. Finally, the cover slips were gently rinsed with distilled water. Both treated and control cover slips were observed under fluorescence microscopy (excitation at 490 nm; emission at 525 nm).25
2.10. Scanning electron microscopy analysis
Scanning electron microscopy analysis was performed based on a previously described method.25 Briefly, the treated and control cover slips were fixed in a glutaraldehyde solution and the cover slips were rinsed with deionized water. Finally, cover slips were serially dehydrated with 70% ethyl alcohol for a few seconds. The nitrogen gas was applied for drying. After critical point drying, they were coated with gold using a sputter-coating system (Time duration 75 s, current 10 mA and gold platinum) and then examined with a scanning electron microscope (Tescan, Czech Republic). The images were taken at 20 kV. Images using VEGATC software are saved for further study.
2.11. Gas chromatography – mass spectrum analysis
GC–MS analysis was performed based on a previously described method.25 Briefly, the methanol extract of T. bellerica was performed by a Shimadzu QP-2010 gas chromatograph coupled with a Shimadzu GCMSQP-2010 plus detector (Shimadu) using a SGE BPX-5 (30.0 m × 0.25 μm i.d., film thickness 0.25 μm) fused silica capillary column. Helium was used as a carrier gas at a constant flow rate of 0.8 ml/min. The methanol extract of T. bellerica was dissolved in methanol and 1 μl of the sample was injected (split ratio 100:1) into GC–MS using AOC5000 auto injector for analysis. Identification of volatile organic constituents was confirmed by published electron impact-mass spectra (EI-MS) in the National Institute for Standard and Technology (NIST).
2.12. Statistical analysis
All the experiments were performed in triplicates. Statistical significance for the quantification of violacein, pyocyanin, EPS and biofilm were determined by one-way ANOVA. All the experiments were statistically analyzed by using software GraphPad Prism 5.03.
3. Results
3.1. Quantification of violacein
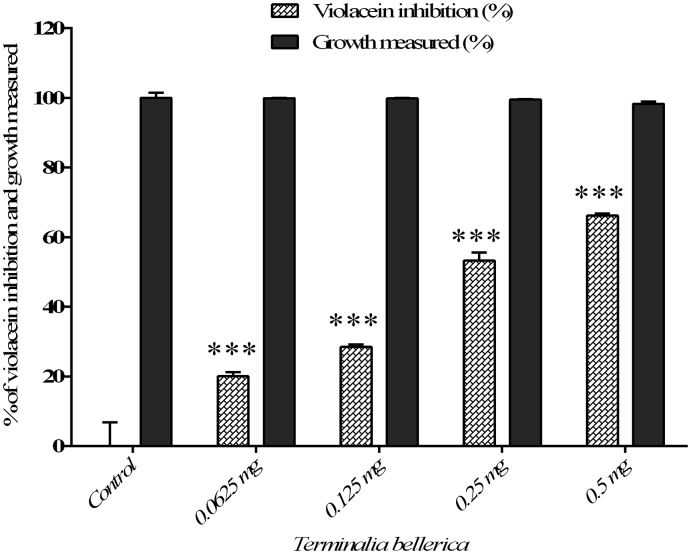
In the qualitative and quantitative analysis, C. paniculatus, K. pinnatum, S. oleosa, M. malabathricum, G. gummi-gutta did not inhibited the violacein production in C. violaceum (CV12472) (Table 2). The methanol extract of T. bellerica has exhibited concentration dependent inhibitory activity in violacein pigment production against C. violaceum (CV12472). A maximum of 66.22% was observed at a concentration of 0.5 mg/ml and the extract of T. bellerica did not inhibit the growth of C. violaceum (CV12472) (Table 2) (Fig. 1).
Table 2.
Antimicrobial and anti-QS activity of medicinal plants against C. violaceum (CV12472).
| Medicinal plants | Antimicrobial activity | Anti-QS activity |
|---|---|---|
| Terminalia bellerica | + | + |
| Celastrus paniculatus | + | – |
| Kingiodendron pinnatum | + | – |
| Schleichera oleosa | + | – |
| Melastoma malabathricum | + | – |
| Garcinia gummi-gutta | + | – |
Note: ‘+’ positive, ‘−’ negative.
Fig. 1.
The quantitative assessment of violacein inhibition and cell growth of C. violaceum CV12472 treated with T. bellerica plant extract. The data represent the mean values of three independent experiments (p < 0.001).
3.2. Molecular identification and determination of antibiotic resistant pattern in P. aeruginosa
The clinical isolate was identified as P. aeruginosa CI-01 based on 16S rRNA gene sequence analysis (GenBank accession no. KU870518). The isolate was found to be resistant to tested antibiotics Similarly, the control strain of PAO1 was observed to be resistant to antibiotics, but remained sensitive to meziocillin, gatifloxacin, ciprofloxacin, tobramycin, amikacin, norfloxacin, levofloxacin, netillin, ofloxacin and gentamicin (Table 3).
Table 3.
Antibiogram of P. aeruginosa PAO1 and clinical isolate of P. aeruginosa CI-01.
| No | Antibiotics | P. aeruginosa PAO1 | P. aeruginosa CI-01 |
|---|---|---|---|
| 1 | Meziocillin | 22.0 ± 1.7 | R |
| 2 | Gatifloxacin | 34.3 ± 1.1 | 37.0 ± 1.7 |
| 3 | Ciprofloxacin | 39.3 ± 1.1 | 37.0 ± 5.1 |
| 4 | Tobramycin | 28.3 ± 2.8 | 26.0 ± 1.7 |
| 5 | Amikacin | 23.3 ± 0.5 | 20 ± 0 |
| 6 | Piperacillin/Tazobactam | R | R |
| 7 | Ceftriaxone | R | R |
| 8 | Azlocillin | R | R |
| 9 | Norfloxacin | 33.0 ± 2.6 | 34.6 ± 0.5 |
| 10 | Levofloxacin | 35 ± 0 | 35.3 ± 2.5 |
| 11 | Netillin | 19.3 ± 1.1 | 17.6 ± 0.5 |
| 12 | Ofloxacin | 33.0 ± 3.4 | 30.0 ± 1.0 |
| 13 | Gentamicin | 23 ± 0 | 20 ± 0 |
| 14 | Carbenicillin | R | R |
| 15 | Ticarcillin/Clavulanic acid | 22.6 ± 1.1 | R |
| 16 | Piperacillin | R | R |
R, resistant.
3.3. Quantification of pyocyanin
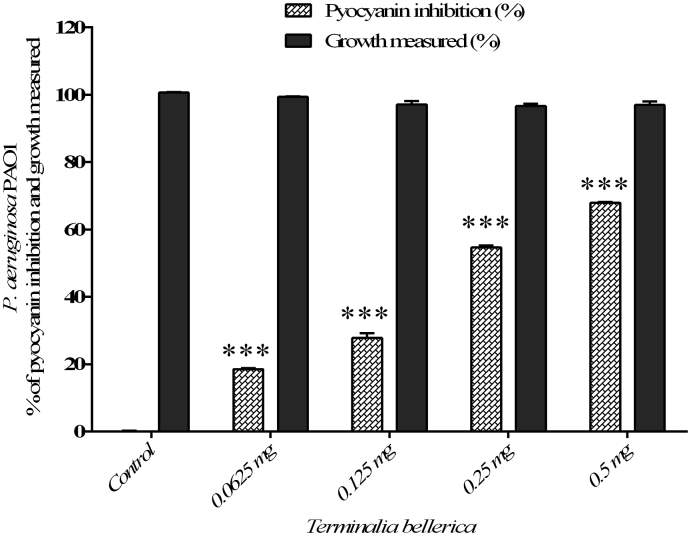
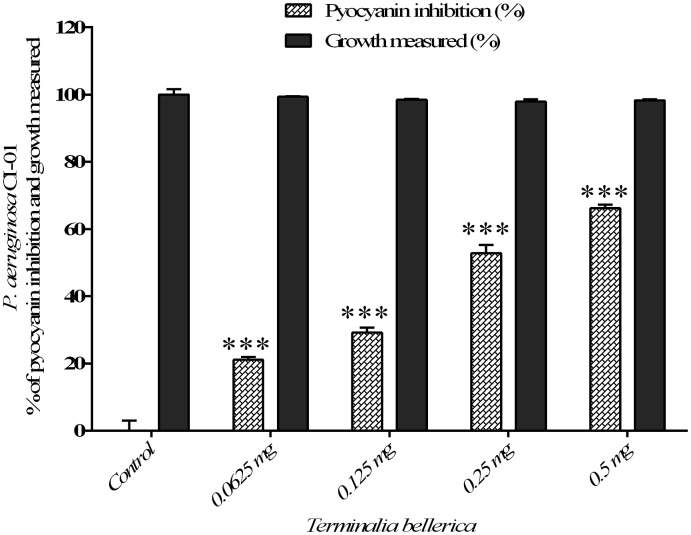
The efficacy of methanol extract of T. bellerica has showed significant reduction in pyocyanin production in P. aeruginosa PAO1 to the level of 18.55%, 27.83%, 54.71% and 67.99% when treated with 0.0625, 0.125, 0.25 and 0.5 mg/ml concentration, respectively (Fig. 2) and did not inhibit the growth of the bacterial cells. Similarly, a maximum of 66.21 % was observed at a concentration of 0.5 mg/ml and a minimum of 21.14% inhibition of pyocyanin production was observed at a concentration of 0.0625 mg/ml and the extract of T. bellerica did not inhibit the growth of P. aeruginosa CI-01 (Fig. 3).
Fig. 2.
The quantitative assessment of pyocyanin inhibition and cell growth: pyocyanin inhibition and cell growth of P. aeruginosa PAO1 treated with T. bellerica plant extract and the data represent the mean values of three independent experiments (p < 0.001).
Fig. 3.
The quantitative assessment of pyocyanin inhibition and cell growth: T. bellerica plant extract inhibited the pyocyanin production and cell growth of P. aeruginosa CI-01 and the data represent the mean values of three independent experiments (p < 0.001).
3.4. Quantification of EPS
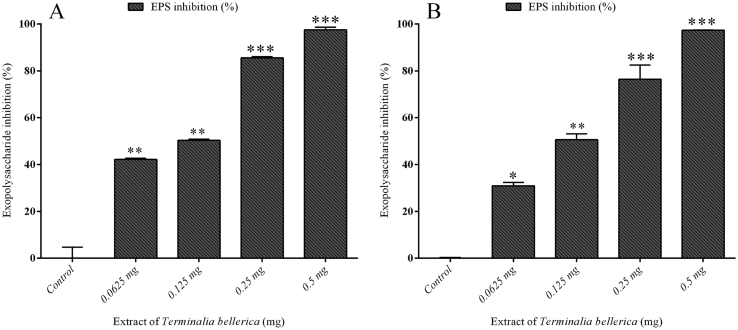
The methanol extract of T. bellerica has reduced the EPS production of test pathogens. In P. aeruginosa PAO1 a maximum reduction was found to be 97.47% at the concentration of 0.5 mg/ml. At a minimum concentration (0.1 mg/ml), the extract of T. bellerica has reduced the EPS production to the level of 42% with compared to that of untreated control (Fig. 4A). Similarly, in a clinical isolate of P. aeruginosa (CI-01), the extract of T. bellerica has potentially inhibited the EPS production at the concentration of 0.0625, 0.125, 0.25 and 0.5 mg/ml to the level of 30%, 50%, 76% and 97% respectively (Fig. 4B).
Fig. 4.
Effect of T. bellerica in EPS production by P. aeruginosa PAO1 and P. aeruginosa CI-01: The T. bellerica inhibited the production of EPS at concentrations ranging from 0.0625 to 0.5 mg/ml (p < 0.05, p < 0.01 and p < 0.001) and the error bar represents standard deviations in three repeated experiments.
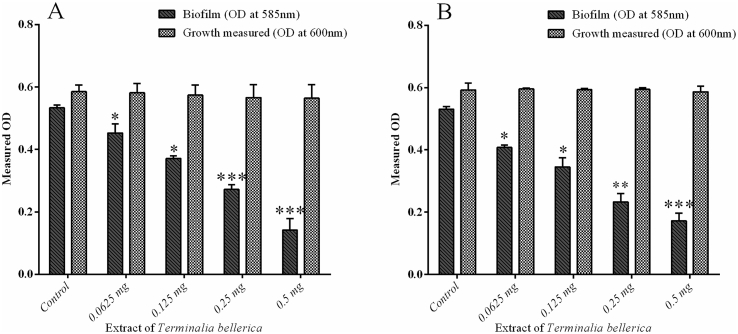
3.5. Biofilm inhibition assay
The present investigation was carried out to study the antibiofilm activity of methanol extract of T. bellerica against test pathogens and C. violaceum CV12472. Treatment with 0.0625, 0.125, 0.25 and 0.5 mg/ml of extract has shown significant reduction in biofilm formation in P. aeruginosa PAO1 to the level of 30.71%, 43.22%, 58.3% and 78.32% respectively (Fig. 5A). The extract of T. bellerica also inhibited the biofilm formation In P. aeruginosa CI-01 to the level of 23.19%, 35%, 56.12% and 67.54% respectively (Fig. 5B).
Fig. 5.
Inhibition of biofilms formation in bacterial pathogens: P. aeruginosa PAO1 treated with T. bellerica extract and untreated control (p < 0.05 and p < 0.001) (A), P. aeruginosa CI-01 treated with T. bellerica extract and untreated control (p < 0.05, p < 0.01 and p < 0.001) (B).
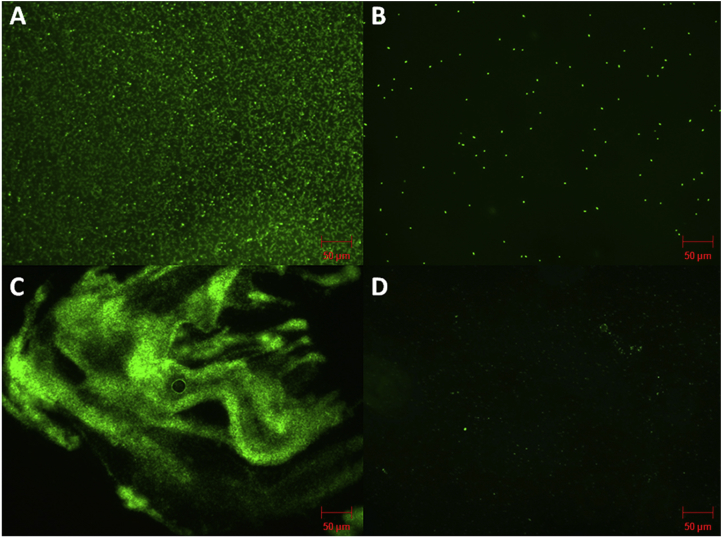
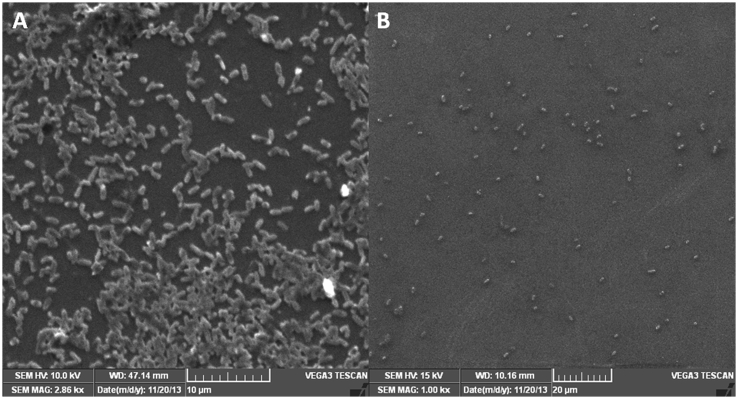
3.6. Fluorescence microscopy and scanning electron microscopy analysis
The antibiofilm activity of the methanol extract of T. bellerica was confirmed by fluorescence and scanning electron microscopy. Test pathogens of P. aeruginosa PAO1 and P. aeruginosa CI-01 were grown on the surface of a glass slide in a static condition to establish biofilm. The treated cultures showed a drastic reduction in biofilm thickness and a thick mass of the biofilm was found adhered to the cover slip in the untreated sample (Fig. 6, Fig. 7A–B). This result indicates the efficacy of extract has potentially prevented the biofilm formation of P. aeruginosa.
Fig. 6.
Fluorescence microscopic images of P. aeruginosa PAO1 and P. Aeruginosa CI-01 biofilms. The cells were stained with acridine orange dye. A) Thick biofilm formation in the non-treated sample of P. aeruginosa PAO1. B) Decreased biofilm formation in P. aeruginosa PAO1 treated with T. bellerica at the concentration of 0.5 mg/ml. C) Thick biofilm formation in the non-treated sample of P. aeruginosa CI-01. D) Decreased biofilm formation in P. aeruginosa CI-01 treated with T. bellerica at the concentration of 0.5 mg/ml compared to the control after 24 h of incubation.
Fig. 7.
Scanning electron microscopy images of P. aeruginosa PAO1 biofilm. A) Control shows biofilm formation after 24 h of incubation. B) The methanol extract of T. bellerica inhibited the biofilm formation at a concentration of 0.5 mg/ml.
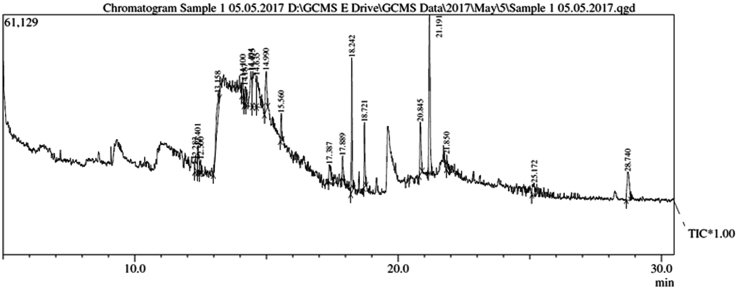
3.7. GC–MS analysis
A total of 20 chemical compounds were identified from the methanol extract of T. bellerica by GC–MS analysis (Table 4). 4-(ethynyloxy)-1-butyne, (E)-1-(1-propenyl)-1-(tosyloxy) cyclopropane, 2-butyn-1-ol, 1,2,3-benzenetriol, N-(Benzyloxycarbonyl)-3,3,3-trifluoroalanine ethyl ester, 2-[2-(benzyloxy)-1-(1-methoxy-1-methylethoxy)ethyl]oxirane, pentanoic acid, 2,5-furandione, dioxolano[b]tricycle[4.1.0.0(1.3)]heptan-2-thione, 3,5-Hexadien-2-ol, 2-methyl, Phthalic acid, nonyl pentyl ester, butyric acid, pentane, 1-octadecyne, cyclopentane, bis-(3,5,5-trimethylhexyl) ether, phytol, 1,2-dibenzyloxybenzene, 4-oxiranylmethoxy, benzylthio thioacyl(piperidinyl) and 2,2′-methylene-bis(4-methyl-6-tert-octylphenol) were present in the methanol extract of T. bellerica. The composition determined for the methanol extract of T. bellerica corresponds to 100% of the entire GC–MS chromatogram (Fig. 8). The present investigation helps to predict the structure of 20 active compounds. Further study may lead to isolation of anti-QS and antibiofilm compounds and their structural elucidation of active compounds will be helpful for further drug development.
Table 4.
Bioactive molecules of methanol extract of T. bellerica as determined by gas-chromatography–mass spectrometer.
| No | Retention time (min) | Compounds | Percentage (%) |
|---|---|---|---|
| 1 | 12.283 | 4-(ethynyloxy)-1-butyne | 2.69 |
| 2 | 12.401 | (E)-1-(1-propenyl)-1-(tosyloxy) cyclopropane | 2.55 |
| 3 | 12.500 | 2-butyn-1-ol, 1,2,3-benzenetriol | 1.05 |
| 4 | 13.158 | N-(Benzyloxycarbonyl)-3,3,3-trifluoroalanine ethyl ester | 4.94 |
| 5 | 14.100 | N-(Benzyloxycarbonyl)-3,3,3-trifluoroalanine ethyl ester | 2.21 |
| 6 | 14.187 | 2-[2-(benzyloxy)-1-(1-methoxy-1-methylethoxy)ethyl]oxirane | 2.00 |
| 7 | 14.404 | pentanoic acid 2,5-furandione | 7.50 |
| 8 | 14.475 | dioxolano[b]tricycle[4.1.0.0(1.3)]heptan-2-thione | 9.86 |
| 9 | 14.635 | 3,5-Hexadien-2-ol, 2-methyl | 6.91 |
| 10 | 14.990 | Phthalic acid | 5.51 |
| 11 | 15.560 | nonyl pentyl ester | 2.17 |
| 12 | 17.387 | butyric acid, pentane | 3.78 |
| 13 | 17.889 | 11-octadecyne | 2.53 |
| 14 | 18.242 | cyclopentane | 9.14 |
| 15 | 18.721 | bis-(3,5,5-trimethylhexyl) ether | 4.51 |
| 16 | 20.845 | phytol | 5.63 |
| 17 | 21.191 | 1,2-dibenzyloxybenzene, | 17.38 |
| 18 | 21.850 | 4-oxiranylmethoxy | 1.61 |
| 19 | 25.172 | benzylthio thioacyl(piperidinyl) | 2.33 |
| 20 | 28.740 | 2,2′-methylene-bis(4-methyl-6-tert-octylphenol) | 5.71 |
Fig. 8.
GC–MS chromatogram of methanol extract of T. bellerica.
4. Discussion
From time immemorial plants have been used as a traditional medicine for controlling and curing many of the bacterial infections. Since ancient time, medicinal plants have been considered as one of the indispensable perennial medicinal plants to cure several diseases.18 In the preliminary study, six medicinal plants were tested for anti-QS properties. C. paniculatus, K. pinnatum, S. oleosa, M. malabathricum, G. gummi-gutta plants inhibited the growth of bacterial pathogens and did not inhibited the violacein production in biomonitor strain of C. violaceum (CV12472) (Table 2). T. bellerica possess antimicrobial and anti-QS activity (Table 2). At the highest concentration, the T. bellerica extract has inhibited the growth of the bacterial cells. Similarly, at the sub-MIC level T. bellerica extract has inhibited the production of virulence factors in P. aeruginosa. This possesses question of whether reduced concentration of antimicrobial compounds can create an anti-QS activity by reducing the cell count to a number below the quorum sensing level.28 The methanol extract of T. bellerica has effectively inhibited violacein production by C. violaceum (CV12472) (Fig. 1). The obtained results are comparable with previous reports.27, 28 The efficacy of T. bellerica against QS-controlled virulence factors of test pathogens such as P. aeruginosa PAO1 and a clinical isolate of P. aeruginosa CI-01 was analyzed. Many Gram-negative bacteria release the extracellular virulence factors which are under the control of QS system. In this study, the methanol extract of T. bellerica has significantly inhibited the pyocyanin pigment production in both P. aeruginosa PAO1 and clinical strain of P. aeruginosa CI-01 (Fig. 2, Fig. 3). Similar reports showed a reduction of pyocyanin in P. aeruginosa PAO1 when treated with plant extracts and essential oils.29 EPS is one of the major virulence factors which are mainly involved in biofilm formation and establishing the infection in host tissue. It also prevents the penetration of antibiotics substance into the bacterial cell.30 In the present study, T. bellerica significantly inhibited the EPS production of P. aeruginosa PAO1 and P. aeruginosa CI-01 to a significant level (Fig. 4A and B). The decreased production of EPS may eventually allow proper penetration of antibiotics to bacterial cells. Similarly, previous studies on the antibiofilm activities of Murraya koenigii essential oil showed a comparable inhibition of exopolysaccharide production in P. aeruginosa PAO1.25 Therefore; our results also envisage that treatment with the plant extract of T. bellerica can deteriorate biofilm formation by P. aeruginosa at the site of infection.
Biofilm is one of the virulence factors controlled by the QS molecules of P. aeruginosa which adhere into the living surface of the host cells. Most of the biofilm forming bacteria are resistant to antibiotics because of exopolysaccharide (EPS).31 In the present study, the methanol extract of T. bellerica has potentially inhibited the biofilm formation both in the control strain of P. aeruginosa PAO1 and in a clinical isolate of P. aeruginosa CI-01 (Fig. 5A and B). The fluorescence microscopic analysis revealed that T. bellerica potentially reduced the biofilm formation in both the strains (P. aeruginosa PAO1 and P. aeruginosa CI-01) when compared to the untreated control (Fig. 6A–D). Similarly, scanning electron microscopy analysis revealed that the methanol extract of T. bellerica reduced the biofilm mate on the surface of cover slip. It indicates that the extract potentially inhibited the production of virulence factors and biofilm formation in P. aeruginosa PAO1 (Fig. 7A and B). The GC–MS analysis confirmed the presence of various bioactive compounds with different retention time as illustrated in Table 4. The mass spectrometer analysis the bioactive compounds eluted at different time interval to identify the structure of the bioactive compounds (Fig. 8). These active compounds may lead to inhibit the AHL molecule at a concentration dependent manner.
5. Conclusion
In the current situation, the pathogenic bacterium of P. aeruginosa causes various infections and also it is emerging as a multidrug resistant bacteria making treatment difficult. To overcome this problem the plant's extracts have anti-pathogenic and anti-infective effect against Gram-negative bacteria. In our study, T. bellerica has significantly inhibited the QS-controlled virulence factors and reduced biofilm formation in P. aeruginosa. Based on the microscopy images the extract has drastically reduced the biofilm formation in P. aeruginosa. Therefore, the potent QS inhibition and antibiofilm properties of T. bellerica may be useful either alone or in combination with conventional antibiotic to treat the bacterial infections. Further studies are needed to check the pharmacokinetic properties of this T. bellerica to make use of their clinical value.
Conflict of interest
None declared.
Acknowledgement
The work was supported by the Indian Council of Medical Research, Delhi, India [Grant number 59/42/2011/BMS/TRM]. P. aeruginosa PAO1 was kindly gifted by Prof. Kalai Mathee (Florida International University).
Footnotes
Peer review under responsibility of The Center for Food and Biomolecules, National Taiwan University.
References
- 1.Gomez M.I., Prince A. Opportunistic infections in lung disease: Pseudomonas infections in cystic fibrosis. Curr Opin Pharmacol. 2012;7:244–251. doi: 10.1016/j.coph.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 2.Rossolini G.M., Mantengoli E. Treatment and control of severe infections caused by multiresistant Pseudomonas aeruginosa. Clin Microbiol Infect. 2005;11:17–32. doi: 10.1111/j.1469-0691.2005.01161.x. [DOI] [PubMed] [Google Scholar]
- 3.Pai S., Bedford L., Ruramayi R. Pseudomonas aeruginosa meningitis/ventriculitis in a UK tertiary referral hospital. Qjm. 2016;109:85–89. doi: 10.1093/qjmed/hcv094. [DOI] [PubMed] [Google Scholar]
- 4.Sadikot R.T., Blackwell T.S., Christman J.W., Prince A.S. Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. Am J Respir Crit Care Med. 2005;171:1209–1223. doi: 10.1164/rccm.200408-1044SO. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moghaddam M.M. Quorum sensing in bacteria and a glance on Pseudomonas aeruginosa. Clin Microbiol. 2014;3:1–10. [Google Scholar]
- 6.Fuqua W.C., Winans S.C., Greenberg E.P. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pesci E.C., Pearson J.P., Seed P.C., Iglewski B.H. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pearson J.P., Gray K.M., Passador L. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pearson J.P., Passador L., Iglewski B.H., Greenberg E.P. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearson J.P., Pesci E.C., Iglewski B.H. Role of Pseudomonas aeruginosa las and rhl quorum sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams P., Winzer K., Chan W.C., Camara M. Look who's talking: communication and quorum sensing in the bacterial world. Phil Trans R Soc B. 2007;362:1119–1134. doi: 10.1098/rstb.2007.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sawa T., Ohara M., Kurahashi K. In vitro cellular toxicity predicts Pseudomonas aeruginosa virulence in lung infections. Infect Immun. 1998;66:3242–3249. doi: 10.1128/iai.66.7.3242-3249.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vu B., Chen M., Crawford R.J., Ivanova E.P. Bacterial extracellular polysaccharides involved in biofilm formation. Molecules. 2009;14:2535–2554. doi: 10.3390/molecules14072535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen P.O., Givskov M., Bjarnsholt T., Moser C. The immune system vs. Pseudomonas aeruginosa biofilms. FEMS Immunol Med Microbiol. 2010;59:292–305. doi: 10.1111/j.1574-695X.2010.00706.x. [DOI] [PubMed] [Google Scholar]
- 15.Wang S., Yu S., Zhang Z. Coordination of swarming motility, biosurfactant synthesis, and biofilm matrix exopolysaccharide production in Pseudomonas aeruginosa. Appl Environ Microbiol. 2014;80:6724–6732. doi: 10.1128/AEM.01237-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grant S.S., Hung D.T. Persistent bacterial infections, antibiotic tolerance, and the oxidative stress response. Virulence. 2014;4:273–283. doi: 10.4161/viru.23987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koh C.L., Sam C.K., Yin W.F. Plant-derived natural products as sources of anti-quorum sensing compounds. Sensors. 2013;13:6217–6228. doi: 10.3390/s130506217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elizabeth K.M. Antimicrobial activity of Terminalia bellerica. Indian J Clin Biochem. 2005;20:150–153. doi: 10.1007/BF02867416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabu M.C., Kuttan R. Antidiabetic and antioxidant activity of Terminalia belerica. Roxb. Indian J Exp Biol. 2009;47:270–275. [PubMed] [Google Scholar]
- 20.Sambrook J., Russel D.W. Cold Spring Harbor Laboratory Press; New York: 2001. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 21.Lane D.J. 16S/23S rRNA sequencing. In: Stackebrandt E., Goodfellow M., editors. Nucleic Acids Techniques in Bacterial Systematics. John Wiley; New York: 1991. pp. 115–175. [Google Scholar]
- 22.Clinical and Laboratory Standards Institute (CLSI) CLSI; Wayne, PA: 2012. Performance Standards for Antimicrobial Susceptibility Testing; Twenty Second Informational Supplement. M100–S22. [Google Scholar]
- 23.Blosser R.S., Gray K.M. Extraction of violacein from Chromobacterium violaceum provides a new quantitative bioassay for N-acyl homoserine lactone autoinducers. J Microbiol Methods. 2000;40:47–55. doi: 10.1016/s0167-7012(99)00136-0. [DOI] [PubMed] [Google Scholar]
- 24.Essar D.W., Eberly L., Hadero A., Crawford I.P. Identification and characterization of genes for a 2nd anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the second anthranilate synthases and evolutionary implications. J Bacteriol. 1990;172:884–900. doi: 10.1128/jb.172.2.884-900.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sankar Ganesh P., Rai Vittal R. In vitro antibiofilm activity of Murraya koenigii essential oil extracted using supercritical fluid CO2 method against Pseudomonas aeruginosa PAO1. Nat Prod Res. 2015;29:2295–2298. doi: 10.1080/14786419.2015.1004673. [DOI] [PubMed] [Google Scholar]
- 26.Dubois M., Gilles K.A., Hamilton J.K., Rebers P.A., Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–356. [Google Scholar]
- 27.Adonizio A.L., Downum K., Bennett B.C., Mathee K. Anti-quorum sensing activity of medicinal plants in southern Florida. J Ethanopharmacol. 2006;105:427–435. doi: 10.1016/j.jep.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 28.Choo J.H., Rukayadi Y., Hwang J.K. Inhibition of bacterial quorum sensing by vanilla extract. Lett Appl Microbiol. 2006;42:637–641. doi: 10.1111/j.1472-765X.2006.01928.x. [DOI] [PubMed] [Google Scholar]
- 29.Khan M.S.A., Zahin M., Hasan S., Husain F.M., Ahmad I. Inhibition of quorum sensing regulated bacterial functions by plant essential oils with special reference to clove oil. Lett Appl Microbiol. 2009;49:354–360. doi: 10.1111/j.1472-765X.2009.02666.x. [DOI] [PubMed] [Google Scholar]
- 30.Stewart P.S., Costerton J.W. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358:135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 31.Chen L., Wen Y. The role of bacterial biofilm in persistent infections and control strategies. Int J Oral Sci. 2011;3:66–73. doi: 10.4248/IJOS11022. [DOI] [PMC free article] [PubMed] [Google Scholar]