Abstract
Reactive oxygen species (ROS) have been implicated in tumorigenesis (tumor initiation, tumor progression, and metastasis). Of the many cellular sources of ROS generation, the mitochondria and the NADPH oxidase family of enzymes are possibly the most prevalent intracellular sources. In this article, we discuss the methodologies to detect mitochondria-derived superoxide and hydrogen peroxide using conventional probes as well as newly developed assays and probes, and the necessity of characterizing the diagnostic marker products with HPLC and LC-MS in order to rigorously identify the oxidizing species. The redox signaling roles of mitochondrial ROS, mitochondrial thiol peroxidases, and transcription factors in response to mitochondria-targeted drugs are highlighted. ROS generation and ROS detoxification in drug-resistant cancer cells and the relationship to metabolic reprogramming are discussed. Understanding the subtle role of ROS in redox signaling and in tumor proliferation, progression, and metastasis as well as the molecular and cellular mechanisms (e.g., autophagy) could help in the development of combination therapies. The paradoxical aspects of antioxidants in cancer treatment are highlighted in relation to the ROS mechanisms in normal and cancer cells. Finally, the potential uses of newly synthesized exomarker probes for in vivo superoxide and hydrogen peroxide detection and the low-temperature electron paramagnetic resonance technique for monitoring oxidant production in tumor tissues are discussed.
Keywords: Reactive oxygen species, Triphenylphosphonium cation, Oxidative phosphorylation, Oxidative stress, Mitochondrial inhibition
1. Introduction
“Nonetheless, from a biological point of view, it is beginning to look as if ROS are neither cellular heroes nor villains—but instead something that occupies that always entertaining, captivating and fertile middle ground.” Holmstrom and Finkel (Nature Reviews)[1]
Holmstrom and Finkel elucidated the dual nature of reactive oxygen species (ROS) that elicits both harmful and beneficial effects in cells and the state of the ROS in diseases including cancer [1]. Also, the authors emphasized the need to appreciate the differing chemistry of various ROS (e.g., superoxide radical anion [O2•–] and hydrogen peroxide [H2O2]) in redox-dependent pathways, highlighting the importance of developing methods to detect oxidants in vivo. In the present article, we address some of the gaps in our knowledge concerning ROS and redox signaling in cancer biology. Further, we discuss state-of-the-art assays and probes for detecting O2•–, H2O2, and other oxidants in tumor cells in response to treatment with OXPHOS-targeting drugs, and their potential applications for the detection of mitochondria-derived ROS during tumorigenesis and metabolic reprogramming. The paradoxical role of ROS in tumor proliferation and tumor suppression [2] is discussed in the context of redox signaling mechanisms. Similarly, the paradoxical effects of antioxidants in tumorigenesis and tumor progression are discussed. Understanding the roles of mitochondrial ROS and redox signaling pathways in cancer biology may help in the discovery of relatively nontoxic and targeted therapies.
2. ROS: The most cited, most popular, yet most ambiguous term
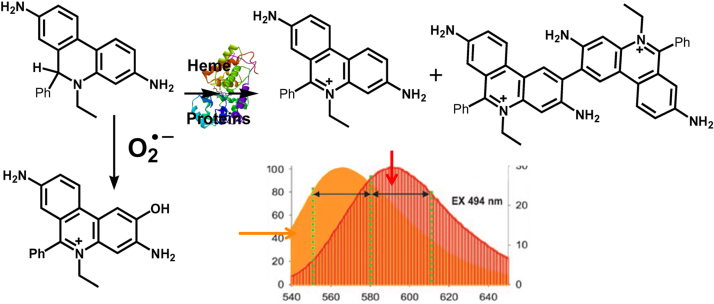
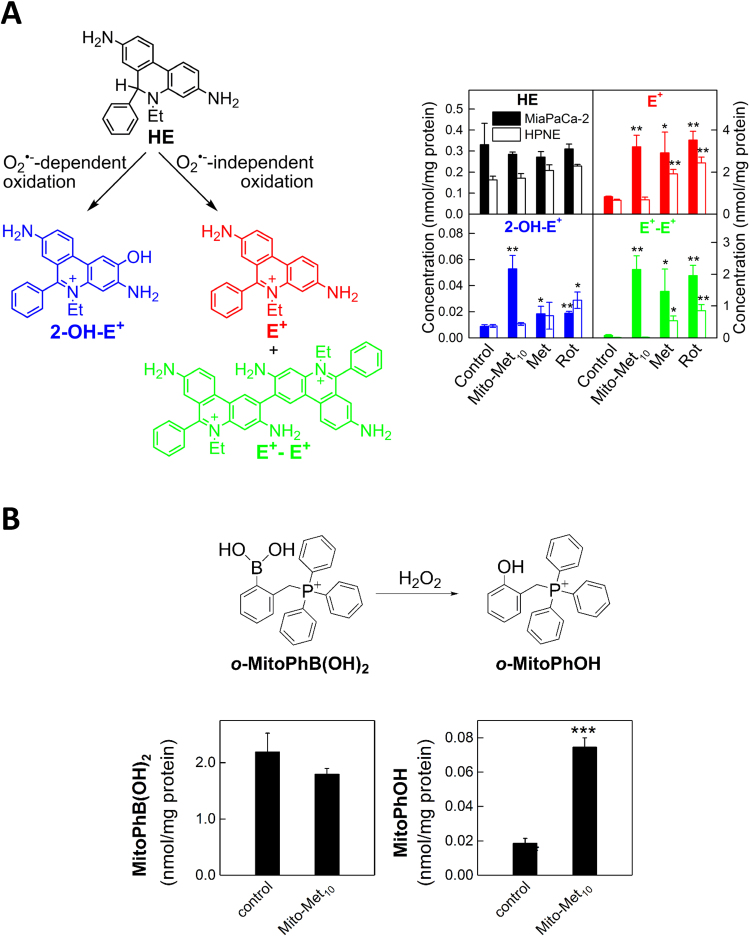
The term “ROS” does not relate to a single species; rather, it covers a range of small molecule oxidizing, nitrosating, nitrating, halogenating, and thiol-reactive species, produced in biological systems. The use of ROS as an umbrella term for oxidants has been previously criticized because of its nonspecificity and ambiguity [3], [4]. One of the authors of this article (BK) was also critical of using ROS as an umbrella term for all oxidants [3]. However, ROS as a term for small-molecule oxidants is now universally embraced and frequently used in novel biological settings by investigators in many areas of research, including cancer biology. Thus, it was decided that the same umbrella term, ROS, would be adapted for oxidants. That said, the lack of proper characterization of the structure of oxidants could seriously hamper our efforts to uncover new and novel oncogenic signaling pathways involved. In order to fully understand the signaling roles of ROS, it is essential to understand more about the nature and identity of the species, whether it is O2•–, H2O2, lipid hydroperoxide, or an electrophile such as 4-hydroxynonenal derived from lipid oxidation. Proper identification of the structure of the ROS will also help us understand the mechanisms of action of drugs and drug resistance in cancer. In some ways, ROS levels and signaling are also modulated by other signaling molecules like nitric oxide (•NO) via a nearly diffusion-controlled reaction between •NO and O2•– [5], generating a potent oxidizing and nitrating molecule, peroxynitrite (ONOO–), also referred to as reactive nitrogen species (RNS). Although there is ample evidence for the occurrence of this type of mechanism and its biological relevance in cardiovascular and neurobiological systems [6], [7], there is very little published data on the •NO and O2•– interaction and its signaling ramifications in cancer biology. Many probes (fluorescent and chemiluminescent) have been previously employed to identify ROS, but there is still a lot of confusion in this field due to a lack of mechanistic rigor and the artifacts generated from reductive/oxidative activation of the probes themselves [8], [9]. Most of these limitations have, however, been previously addressed [10], [11]. Irrespective of the methodology used to detect ROS, it is clear that oxidants are involved, either as a major player or as a bystander, in the underlying biology. On the positive side, there now exist more specific probes and assays for selective identification of various ROS. Published data from independent laboratories are in agreement that identification of specific products formed from ROS interaction with fluorescent probes is crucial for determining the identity of ROS [12], [13]. A reaction between O2•– and hydroethidine (HE) results in the formation of a very specific product, 2-hydroxyethidium (2-OH-E+); this product is not formed from the reaction between HE and other biologically relevant oxidants such as H2O2, singlet oxygen, lipid hydroperoxides, peroxynitrite, HOCl, and •NO2 [14], [15]. This marker product derived from the O2•– and HE reaction (2-OH-E+) can be unambiguously detected by rapid high-performance liquid chromatography (HPLC) and liquid chromatography–mass spectrometry (LC-MS) methods [16]. However, numerous publications using HE-derived fluorescence still posit that ethidium (E+) is the product of the O2•– reaction with HE, whereas it has been clearly established by us and others that E+ is not the product of the reaction between O2•– and HE (Fig. 1) [17], [18]. Evidence also exists that the reaction chemistry between O2•– and other analogs of HE including Mito-SOX is similar to that of HE [19], [20]. The lack of appreciation and the misconception of the chemistry and the mechanism of action of O2•– with HE, Mito-SOX (a mitochondria-targeted HE), and other HE analogs are responsible for the multitude of publications in biomedical research, including cancer, that suggest or conclude the intermediacy of O2•– formation [21], [22].
Fig. 1.
HE-derived nonspecific oxidation and O2•–-specific hydroxylation products. The fluorescence spectra of the nonspecific oxidation product E+ (10 µM, right axis) and O2•–-dependent hydroxylation product 2-OH-E+ (10 µM, left axis) in the presence of DNA show a significant overlap. (Obtained from Ref. [15]. Reprinted from H. Zhao, J. Joseph, H.M. Fales, E.A. Sokoloski, R.L. Levine, J. Vasquez-Vivar, B. Kalyanaraman, Detection and characterization of the product of hydroethidine and intracellular superoxide by HPLC and limitations of fluorescence, Proceedings of the National Academy of Sciences of the United States of America 102(16) (2005) 5727–5732, Copyright 2005 National Academy of Sciences.).
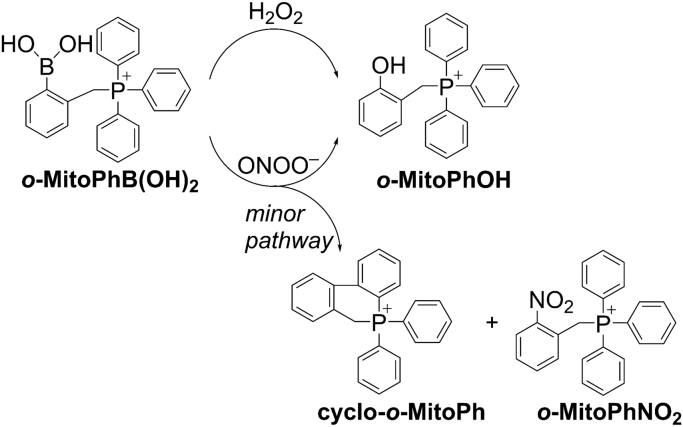
Most assays to detect H2O2 are based on peroxidatic oxidation of probes such as Amplex Red in an extracellular milieu [23]. So far, very little information is available on chemical probes that react directly with H2O2 to form a diagnostic product. However, recently activity in this area has increased [24], [25], [26]. Boronate-based fluorescence probes react with H2O2 stoichiometrically (albeit very slowly, with the rate constant of 1–2 M−1 s−1) to form fluorescent products [27], [28]. Boronates also react with peroxynitrite nearly a million times faster than with H2O2, forming a major product (90%) that is the same as the product derived from the boronate/H2O2 reaction and a very characteristic and diagnostic minor product (5–10%) [27], [29]. If the product that is highly diagnostic for peroxynitrite is not detected, it is likely that the major product is not formed from peroxynitrite (Fig. 2). Mitochondria-targeted boronates (meta-MitoB) were used to detect H2O2 in vivo [30], [31]. We used an isomer, ortho-MitoB, to detect H2O2 because of its ability to distinguish between peroxynitrite and H2O2 [28], [32], [33]. Predicting the cellular response (activation of signaling pathways) to specific ROS requires a thorough understanding of its chemical properties in a biological setting.
Fig. 2.
H2O2and peroxynitrite-induced formation of major and minor products derived from o-MitoPhB(OH)2. Reaction between H2O2 and o-MitoB stoichiometrically generates the only product, o-Mito-PhOH, whereas the reaction between ONOO– and o-MitoB(OH)2 produces a major product, o-Mito-PhOH, and a minor cyclized product that is highly characteristic for reaction with ONOO–. (Obtained from Ref. [55]. Reprinted from Interface Focus, 7, B. Kalyanaraman, G. Cheng, M. Hardy, O. Ouari, A. Sikora, J. Zielonka, M. Dwinell, Mitochondria-targeted metformins: anti-tumor and redox signaling mechanisms, 20160109, Copyright 2017.).
3. Mitochondria, Nox, and ROS
Two major sources of ROS in cancer are mitochondria and nicotinamide adenine dinucleotide phosphate (NADPH) oxidases [34], [35]. Mitochondrial respiratory chain complexes are generally thought to be responsible for generating ROS, O2•–, and H2O2, in particular. Research in the early 1970s by Chance and collaborators provided the first evidence for mitochondrial generation of ROS [36]. Although O2•– formation in mitochondria was not convincingly demonstrated, Chance and coworkers demonstrated mitochondrial generation of H2O2 using a sensitive spectrophotometric method [36]. H2O2 was measured in the cytosolic extracts derived from mitochondria using the absorption changes that occur during the catalytic cycle of cytochrome c peroxidase and H2O2. This is a fundamentally significant discovery revealing an aberrant oxygen metabolism (albeit less than 1%) during mitochondrial respiration [37].
Complex III in the mitochondrial respiratory chain could form O2•– when mitochondria were treated with the inhibitor, antimycin. Mitochondrial complex I is another source of O2•– generation in the presence of rotenone that inhibits complex I [38]. Superoxide from complex I is also formed under conditions of a high proton motive force and reduced coenzyme Q pool (i.e., a situation known as the reverse electron transport mechanism wherein electrons are driven back through complex I) [39].
That mitochondria also generate O2•– and H2O2 under in vivo conditions is supported by the existence of manganese superoxide dismutase (MnSOD) and other antioxidant enzymes (peroxidases and peroxiredoxins) in the mitochondrial matrix, and by the pathological consequences (e.g., mitochondrial oxidative stress including DNA damage) resulting from their deficiency.
Nox enzymes are emerging as a promising target for anticancer drug development due to mounting evidence that suggests that NADPH/Nox-derived ROS inhibit tumor apoptosis and stimulate tumor proliferation [40], [41]. Several Nox isoforms (e.g., Nox2 and Nox4) have been proposed as potential therapeutic targets in the treatment of cancer and other diseases [42]. Unlike other redox enzymes for which ROS generation is an “accidental” byproduct of their primary catalytic function, the only known function of Nox enzymes (Nox1-5, Duox1-2) is generation of ROS (e.g., O2•– and H2O2) [43]. Nox2 forms both O2•– and H2O2 (via dismutation of O2•–); however, published reports suggest that Nox4 primarily generates H2O2 (90%) [44], [45]. A major impediment to advancing Nox research in cancer biology is the lack of availability of selective inhibitors of Nox isoforms [46]. This, in turn, had been due to the lack of assays selective for O2•– and H2O2 using specific probes, but this hurdle has been largely overcome with recent discoveries of new probes and sensitive assays for detection of ROS and RNS [47].
Oncogenic KRAS was reported to promote ROS/RNS generation by increasing the expression and activity of Nox enzymes at the tumor cell membrane [48]. However, it is likely that Nox activity is modulated by changes in mitochondrial bioenergetics. Although there are reports in the vascular biology literature of potential “cross-talk” between mitochondrial ROS and Nox activation [49], there is no information, to our knowledge, on the modulatory role of mitochondrial metabolism on Nox/ROS metabolism and oxidative signaling in cancer biology. Although, this particular aspect is outside the scope of the present review, understanding how modifications of cancer cell bioenergetics and metabolism affect the NADPH/Nox/ROS pathway or phosphorylation of the upstream target in Nox activation could be highly significant in metabolism-based drug therapeutics.
4. Assays and probes for intracellular detection of ROS (O2•– and H2O2)
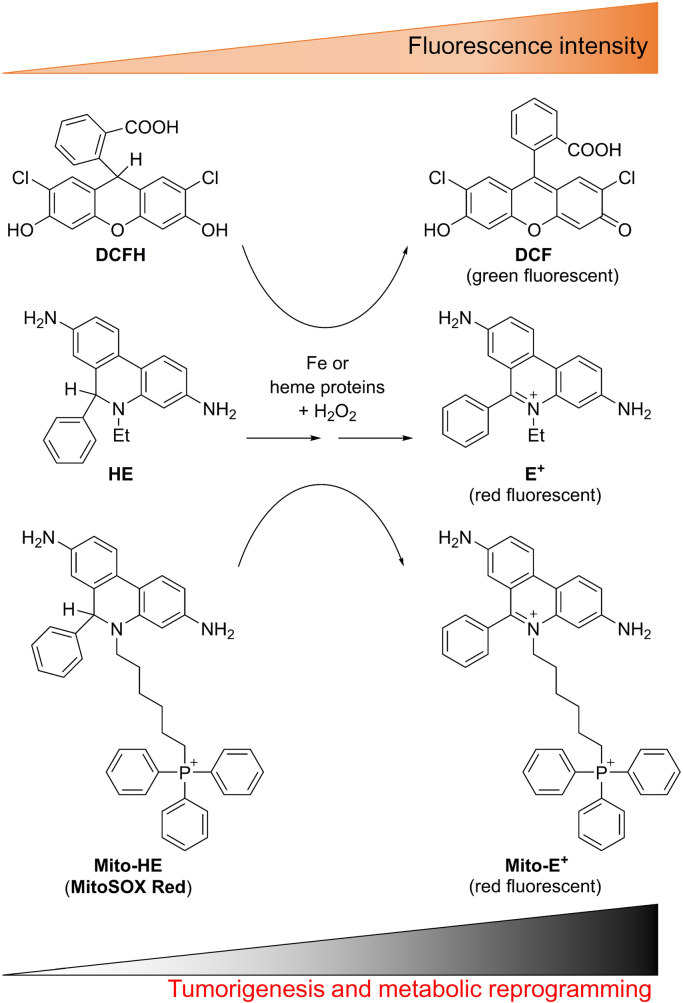
Most publications concerning ROS production in biology, including cancer biology, involve the use of redox dyes—dichlorodihydrofluorescein (DCFH), HE, and Mito-SOX (Fig. 3)—and their oxidation products were detected by fluorescence. Neither O2•– nor H2O2 was reported to react at any appreciable rate with DCFH [50]. Measuring the oxidation of redox dyes by fluorescence is definitely not the same as measuring cellular ROS. This is an important and significant distinction that is validated by a thorough understanding of the chemistry (kinetics, stoichiometry, and analyses of intermediates and products) established for these dyes in the presence of various biologically relevant reactive oxygen and nitrogen species. Investigators have used DCFH-derived green fluorescence for detecting intracellular H2O2 and HE-, and Mito-SOX-derived red fluorescence for detecting intracellular and mitochondrial generation of O2•– [51], [52]. However, most likely the investigators were monitoring the oxidation of the redox dyes (DCFH, HE, and Mito-SOX) through a similar peroxidatic mechanism (iron, heme iron, or cytochrome c-catalyzed) in the presence of O2•– and H2O2 (Fig. 3) [20]. Enhanced fluorescence was related to tumorigenesis and metabolic reprogramming in tumor cells [53], [54]. However, enhanced fluorescence from oxidation of redox dyes cannot be equated to enhanced O2•– formation. As reiterated earlier, it is essential to separate and identify the specific products derived from the O2•– reaction with HE and Mito-SOX under these conditions before implicating the intermediacy of O2•– and/or H2O2 in redox-dependent processes (Fig. 4).
Fig. 3.
Relationship between enhanced oxidation of redox dyes and redox-dependent processes. Enhanced oxidation of redox dyes leading to enhanced fluorescence intensity during tumorigenesis, metabolic reprogramming, and drug resistance.
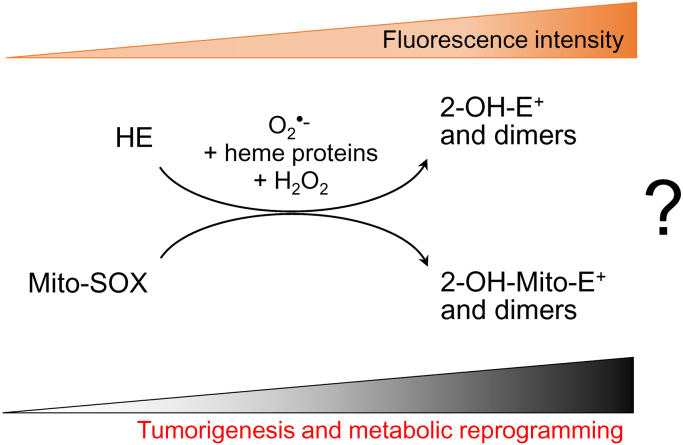
Fig. 4.
Lack of evidence for enhanced O2•–formation during tumorigenesis, metabolic reprogramming, and drug resistance. In order to categorically state that O2•– is formed during redox processes in tumor cells, it is essential to separate and detect the corresponding O2•–-specific hydroxylated products from HE or Mito-SOX.
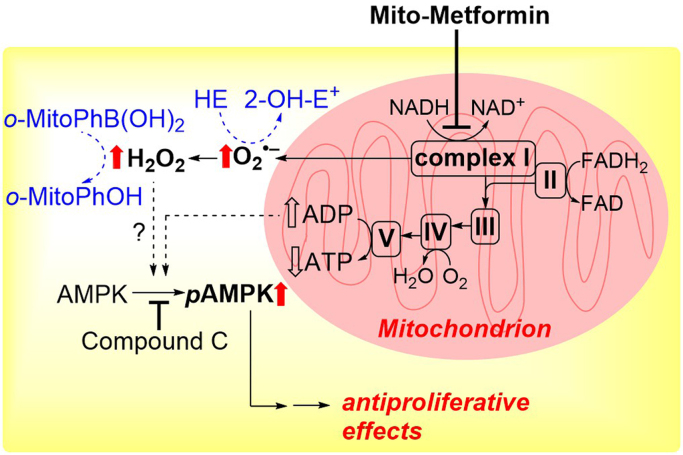
We and others have shown that inhibition of complex I in mitochondria results in increased production of O2•– and other one-electron oxidants [55]. We used HPLC-based analyses to determine products formed from redox dyes [56], [57], [58]. HE is cell-permeable and can be used to detect intracellular O2•–, both cytosolic and mitochondria-derived. Several mitochondria-targeted agents inhibit complex I activity in multiple cancer cell lines (e.g., lung cancer, pancreatic cancer) [59], [60]. Incubation with HE under conditions inhibiting mitochondrial complex I in cancer cells induces formation of O2•– and other oxidants. Fig. 5A shows that Mito-metformin (a mitochondria-targeted derivative of metformin) enhanced the formation of 2-OH-E+, a superoxide-specific product; E+; and diethidium (E+-E+). Because of the interference (spectral overlap) between 2-OH-E+ and E+, it is impossible to use the fluorescence results to make a definite conclusion on the changes in O2•– production. E+ is typically detected at levels at least 10-fold higher than 2-OH-E+ (Fig. 5A).
Fig. 5.
Mito-metformin induces increased O2•–and H2O2formation in pancreatic cancer cells. O2•–-dependent oxidation of the HE probe (A) and H2O2-dependent oxidation of the probe, o-MitoPhB(OH)2 (B) in MiaPaCa-2 cells treated for 24 h with Mito-Met10. (Obtained from Refs. [55], [59]. Reprinted from Cancer Research, 76, G. Cheng, J. Zielonka, O. Ouari, M. Lopez, D. McAllister, K. Boyle, C.S. Barrios, J.J. Weber, B.D. Johnson, M. Hardy, M.B. Dwinell, B. Kalyanaraman, Mitochondria-targeted analogs of metformin exhibit enhanced antiproliferative and radiosensitizing effects in pancreatic cancer cells, 3904-15, Copyright 2016, and from Interface Focus, 7, B. Kalyanaraman, G. Cheng, M. Hardy, O. Ouari, A. Sikora, J. Zielonka, M. Dwinell, Mitochondria-targeted metformins: anti-tumor and redox signaling mechanisms, 20160109, Copyright 2017.).
To detect H2O2 generated in cells, the probe o-MitoPhB(OH)2 is used (Fig. 2). The meta analog, m-MitoPhB(OH)2 (known as the MitoB probe), can also be used [55]. Although both probes react stoichiometrically with H2O2 to form the respective phenolic product, MitoPhOH (also called MitoP), only the ortho-substituted probe forms characteristic cyclized and nitrated minor products upon reaction with peroxynitrite (ONOO–) [28], [32]. Failure to detect the cyclic and nitrated products would definitively indicate that ONOO– is not the oxidant responsible for oxidizing o-MitoPhB(OH)2 to o-MitoPhOH. The meta analog will not form the cyclic minor product upon reaction with ONOO–.
Intracellular H2O2 formation induced by Mito-metformin10 (Mito-Met10) in PDACs was measured using the probe, o-MitoPhB(OH)2. MiaPaCa-2 pancreatic cancer cells were treated for 24 h with Mito-Met10 followed by monitoring of cellular oxidant using o-MitoPhB(OH)2. As shown in Fig. 5B, the intracellular levels of both the probe and the product were quantitated. Mito-Met10 induced an increase in the H2O2-derived product from o-MitoPhB(OH)2. It has been mentioned that H2O2 does not react rapidly with boronates. Peroxiredoxin enzymes react with H2O2 a million times faster than do boronates, and so the extent of the H2O2 reaction with boronate probes is likely to be very small. Protein hydroperoxides react with boronate probes ca. 10 times faster than H2O2, and thus can also be responsible for the oxidation of the probe during increased production of O2•– and H2O2 [61]. As mentioned above, in addition to boronate probes, Amplex Red/horseradish peroxidase may be used to measure extracellularly released H2O2.
In contrast to the frequently used artifact-prone DCFH probe (that self-stimulates H2O2 formation via intermediate radical formation), the MitoB probes do not self-promote H2O2 and, as a result, measurement of the product truly reflects the H2O2 (or other hydroperoxide) induced in cells and not artifactually formed from probe oxidation/reduction.
5. Monitoring intracellular fluorescence derived from redox probes such as HE and Mito-SOX does not measure intracellular or mitochondrial superoxide
HE is a redox active probe that is synthesized from a two-electron reduction of E+ [14], [15]. HE can also be oxidized to E+ through nonspecific mechanisms in cells. In addition, dimeric products (nonfluorescent, e.g. E+-E+) are also formed from HE (Fig. 6) [20]. In cells, E+ is typically present at a much higher concentration than 2-OH-E+ (see for example Fig. 5A). Fig. 1 shows a significant overlap in the fluorescence spectra of E+ and 2-hydroxyethidium. Thus, in an intracellular milieu, where the nonspecific oxidation of HE to E+ is usually much higher, the observed fluorescence intensity is mostly derived from E+ and, therefore, cannot be attributed to the O2•–/HE reaction-derived product, 2-OH-E+. Because the chemistry between O2•– and HE analogs is the same, irrespective of the detection modalities, whether confocal microscopy or flow cytometry, fluorescence-based approaches using the currently available probes are not suitable for selective detection of O2•–. Only the HPLC or LC-MS approach, utilizing authentic standards of the oxidation products, can be used to measure 2-OH-E+ or its analogs. Under the same conditions, the level of the probe and nonspecific E+ and the dimeric products of HE oxidation should also be measured.
Fig. 6.
Dimeric product formed from one-electron oxidation of HE and analogs. (Modified from Ref. [87]. Reprinted from Cell Biochemistry and Biophysics, Modified Metformin as a More Potent Anticancer Drug: Mitochondrial Inhibition, Redox Signaling, Antiproliferative Effects and Future EPR Studies, 75, 2017, 311-7, B. Kalyanaraman, G. Cheng, M. Hardy, O. Ouari, A. Sikora, J. Zielonka, M.B. Dwinell with permission of Springer.).
Mito-SOX is a mitochondria-targeted analog of HE that has a six-carbon aliphatic chain containing a triphenylphosphonium group. Mito-SOX reacts with superoxide, hemes, and one-electron oxidizing agents in exactly the same manner as does HE [20], [62]. Superoxide reacts with Mito-SOX and its radical, forming 2-hydroxylated Mito-SOX (2-OH-Mito-E+). Nonspecific reaction products of Mito-SOX are Mito-E+ and the dimeric products including Mito-E+-Mito-E+. Very similar to 2-OH-E+ and E+ (Fig. 1), both 2-OH-Mito-E+ and Mito-E+ share overlapping fluorescence spectra [63]. Mito-E+ is typically formed in a much higher concentration than 2-OH-Mito-E+. Thus, the fluorescence obtained using Mito-SOX does not measure mitochondrial O2•– but simply indicates the oxidation of Mito-SOX. Unfortunately, a multitude of publications claim to have detected mitochondrial O2•– on the basis of increased fluorescence intensity from cancer cells using Mito-SOX as the ROS detection probe [64], [65]. As with HE, the only way to detect mitochondrial O2•– using Mito-SOX is to measure the marker product, 2-OH-Mito-E+, using HPLC-based techniques. The standard for this product can be synthesized using Mito-SOX and Fremy's salt [17], [66]. Because mitochondria-targeted probes containing the triphenylphosphonium cationic moiety (TPP+) accumulate in tumor cell mitochondria more than in normal cells, it is important to perform a dose-response measurement investigating the effect of probes such as Mito-SOX on mitochondrial function in different cancer cell phenotypes [67].
Mitochondria-derived ROS are often indicated as MROS or mtROS. Mito-SOX is typically used in these studies. An increase in Mito-SOX red fluorescence is equated to mitochondria-derived ROS and by implication to mitochondrial production of O2•–. However, oxidation of the redox probes (DCFH or Mito-SOX) resulting in enhanced fluorescence was observed in cancer cells due to metabolic reprogramming and drug resistance. Unless the hydroxylated product of Mito-SOX oxidation can be isolated and determined to be specific for O2•– (i.e., 2-OH-Mito-E+), attributing increased red fluorescence to mitochondrial O2•– or to mtROS is incorrect!
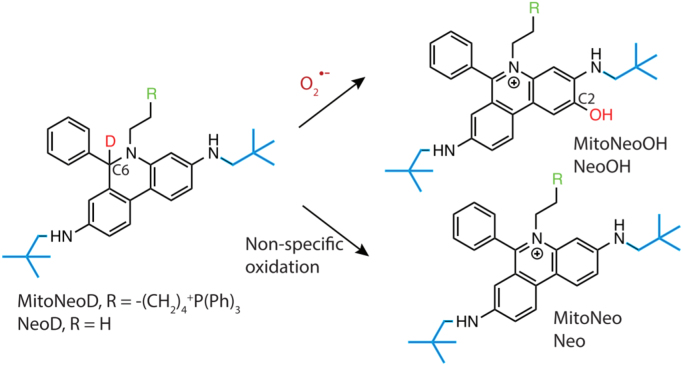
E+ and structurally related analogs intercalate into DNA, and this interaction increases the fluorescence quantum yield. As a result, the fluorescence intensity is enhanced several-fold. The O2•–-specific product, 2-OH-E+, also intercalates into DNA, increasing its fluorescence intensity [14], [15]. Consequently, the fluorescence derived from both E+ and 2-OH-E+ in the intracellular milieu is enhanced, making it difficult to interpret fluorescence changes. Similar limitations apply to Mito-SOX-derived products as well. Hartley, Murphy, and colleagues developed a new mitochondria-targeted O2•– probe, MitoNeoD, which is different from MitoSOX in that oxidation products derived from it do not intercalate into DNA, thereby preventing amplification of the red fluorescence due to the binding of the oxidation product to DNA [68]. MitoNeoD contains a reduced phenanthridinium moiety (modified to prevent DNA intercalation) that reacts with O2•– to form the hydroxylated product, MitoNeoOH (Fig. 7), with the intermediacy of the radical cation of MitoNeoD (formed from one-electron oxidation of the probe). MitoNeoD contains a carbon-deuterium bond that inhibits some pathways of nonspecific oxidation, thereby enhancing selectivity toward O2•–. Finally, this probe possesses a triphenylphosphonium lipophilic cation that facilitates its uptake into mitochondria. It should be emphasized, however, that even the deuterated probe is not selective toward O2•–, and chromatographic analyses are required for specific monitoring of the superoxide product (MitoNeoOH). MitoNeoD and mito-paraquat (a mitochondria-targeted redox cycling agent) were administered to mice, and the hydroxylated product, MitoNeoOH, was detected in the mitochondria isolated from the liver [68]. In the presence of mito-paraquat, the yield of the hydroxylated product was increased, indicating the feasibility of detecting mitochondrial O2•– in in vivo settings, when combined with LC-MS-based analyses of the extracts. Because of the deuterium isotope effect and the bulky substituent attached next to the amino group, it is likely that direct two-electron oxidation of the probe to the E+-type product (MitoNeo cation) is minimized as compared with HE or Mito-SOX [68].
Fig. 7.
O2•–-specific and nonspecific oxidation products formed from the mitochondrial superoxide probe, MitoNeoD. (Modified from Ref. [68]. Reprinted from Cell Chemical Biology, 24, M.M. Shchepinova, A.G. Cairns, T.A. Prime, A. Logan, A.M. James, A.R. Hall, S. Vidoni, S. Arndt, S.T. Caldwell, H.A. Prag, V.R. Pell, T. Krieg, J.F. Mulvey, P. Yadav, J.N. Cobley, T.P. Bright, H.M. Senn, R.F. Anderson, M.P. Murphy, R.C. Hartley, MitoNeoD: A Mitochondria-Targeted Superoxide Probe, 1285-98.e12, https://doi.org/10.1016/j.chembiol.2017.08.003, https://creativecommons.org/licenses/by/4.0/, Copyright 2017.).
6. Redox signaling, drug resistance, and metabolic reprogramming in cancer cells: The role of mitochondrial ROS
As described previously, to better understand the role of ROS in redox (reduction-oxidation) signaling and vice versa, it is important to determine the identity of ROS (O2•–, H2O2, lipid, or protein oxidation products). Previously, it was thought that ROS are always cytotoxic to both cancer and normal cells. This has been the basis for cytotoxic chemotherapy and radiation therapy, and the therapeutic goal has been to increase the therapeutic window, enhancing tumor cytotoxicity and decreasing toxicity normal cells. So, an effective therapeutic strategy was to inhibit ROS levels in normal cells through enhanced antioxidant mechanisms. However, it is not as simple as it appears. Studies show that ROS (H2O2 in particular) stimulate cancer cell proliferation through modifications in the signaling pathways (e.g., PI3/Akt). On the other hand, some studies report that antioxidant supplementation and upregulation of antioxidant pathways actually enhanced tumor survival by decreasing ROS cytotoxicity [69], [70]. A major limitation of the newly proposed role is that there is very little quantitative information on ROS (modest and high levels) and their effect on cancer cell proliferation/cancer cell apoptosis. As reported previously, ROS formation and metabolic changes are intertwined in cancer cells [1]. The redox status of various cancer cells appears to be dependent on the metabolic reprogramming that occurs during tumorigenesis, progression, and metastasis of cancer cells. Metabolic reprogramming also induces endogenous antioxidant machinery [71]. Increased detoxification of ROS has also been shown to promote tumorigenesis [72]. Furthermore, low levels of ROS activate signaling pathways for cell proliferation and survival [73]. Thus, it is difficult to predict cancer cell response to ROS modulation based on in vitro cell culture studies. With radiation therapy, however, tumors are irradiated using a focused X-ray beam, and the tumor cell killing is enhanced and the damage to collateral tissue is minimized. Radiation therapy induces ROS (e.g., hydroxyl radical)-mediated DNA damage in tumor cells. However, cancer tissues are more hypoxic than normal tissues, so the lack of oxygen is one of the major limitations of radiation therapy.
Increased ROS formation and metabolic reprogramming were shown to occur in cis-platin resistant cells [74], [75]. The resistance to antitumor drugs is a major impediment in chemotherapy [76], [77]. Generally, widely used antitumor drugs like cis-platin elicit positive response in lung cancer patients, but with continued use, the patients develop resistance to cis-platin therapy and this treatment fails. Most chemotherapeutic drugs are subjected to chemoresistance. Studies using cis-platin-resistant cell lines derived from patients who failed cis-platin chemotherapy revealed metabolic reprogramming [78]. These resistant cells relied exclusively on oxidative metabolism and exhibit increased OXPHOS (mitochondrial respiration) as determined by using the Agilent Seahorse Extracellular Flux Analyzer [79]. However, the signaling pathway responsible for enhanced OXPHOS in cis-platin-resistant lung cancer cells was not determined.
As described earlier, increased fluorescence due to using redox dyes simply points to their enhanced oxidation; it does not provide evidence in support of enhanced ROS (O2•–, H2O2, or lipid hydroperoxides) formation. Although ROS detection and identification remain questionable, the investigators report a novel role for the drug riluzole's ability to counteract cis-platin's resistance in lung cancer cells using the oxidation of redox probes [75].
7. Oxidative phosphorylation-inhibiting drugs: Mitochondria-targeted agents and antiproliferative effects
Cancer cells use multiple metabolic pathways to acquire nutrients to support their ever-increasing energetic needs. These include glycolysis, glutaminolysis, and fatty acid oxidation. Treatment with antiglycolytic drugs induces a compensatory increase in OXPHOS. In addition, inhibitors of kinase drugs (which often decrease glycolysis) enhance drug resistance and the OXPHOS mechanism [80]. Cancer-cell-specific OXPHOS inhibitors are needed to counteract this compensatory response, and currently available mitochondrial inhibitors are not very selective.
Cancer cells expressing oncogenic mutations in isocitrate dehydrogenase 1 and 2 (IDH1/2) showed increased dependence on oxidative metabolism and are more sensitive to pharmacological inhibition of OXPHOS. Mutant IDH1 cells exhibited higher sensitivity to electron transport chain inhibition under hypoxia [81]. Results indicate that IDH1 mutant cells are more dependent on complex I of the electron transport chain and, consequently, are more susceptible to complex I inhibition. Anti-OXPHOS drugs (mitochondria-targeted drugs) are likely to exhibit enhanced antiproliferative effects in mutant IDH1 cells under hypoxic conditions.
Oxidative phosphorylation-inhibiting (anti-OXPHOS) drugs refer to mitochondria-targeted or mitochondrial drugs that decrease the rate of mitochondrial respiration through inhibition of electron transport chain proteins such as complex I. Delocalized cations (e.g., rhodamine dye) accumulate into mitochondria and inhibit mitochondrial respiration of cancer cells. Molecules conjugated to the TPP+ moiety (e.g., Mito-Q, Mito-CP, Mito-chromanol or Mito-vitamin-E, Mito-metformin; Fig. 8 and Fig. 9) accumulate preferentially into mitochondria driven by an increased negative membrane potential [82], [83], [84]. Because of the increased mitochondrial membrane potential in cancer cells as compared with normal, non-transformed cells, TPP+-containing molecules accumulate in tumor mitochondria to a higher extent and longer than in normal cell mitochondria. Both Mito-CP and Mito-vitamin E potently and selectively inhibit complex I-mediated mitochondrial respiration or oxygen consumption. These molecules are also retained in cancer cell mitochondria more than in normal cell mitochondria and, therefore, cause decreased mitochondrial respiration in cancer cells for a prolonged period of time. Although Mito-SOX and mito-boronates are used as ROS probes, it should be noted that these probes contain a TPP+ moiety and can inhibit OXPHOS like other TPP+-containing mitochondria-targeted compounds [67], [85].
Fig. 8.
Some examples of TPP+-based positively charged anticancer agents. Note that it is critical to clearly define the complete structure indicating the side chain length of mitochondria-targeted TPP+-based compounds. (Modified from Ref. [84]. Adapted with permission from J. Zielonka, J. Joseph, A. Sikora, M. Hardy, O. Ouari, J. Vasquez-Vivar, G. Cheng, M. Lopez, B. Kalyanaraman, Mitochondria-Targeted Triphenylphosphonium-Based Compounds: Syntheses, Mechanisms of Action, and Therapeutic and Diagnostic Applications, Chemical Reviews 117(15) (2017) 10043-120. Copyright 2017 American Chemical Society.).
Fig. 9.
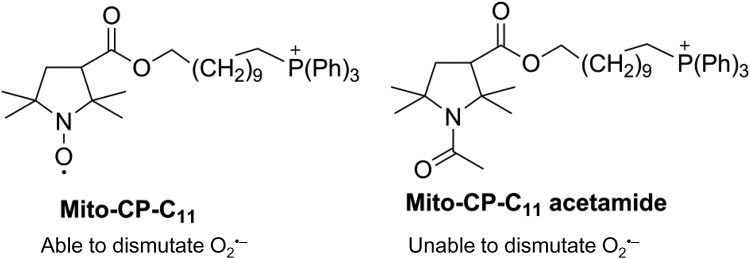
Chemical structures of Mito-CP and its redox inactive analog, Mito-CP-acetamide.
Metformin and Mito-metformin inhibit cancer cell proliferation through the inhibition of complex I, ROS generation, activation of AMP-activated protein kinase (AMPK), and inhibition of mTOR function. Anti-OXPHOS drugs are different from rapamycin, which directly blocks mTOR leading to AMPK activation. Mito-metformin10 is the most potent of all Mito-metformin analogs tested in inhibiting complex I-mediated oxygen consumption [59]. The IC50 values for inhibiting oxygen consumption decrease with increasing carbon–carbon side chain length. Inhibition of pancreatic cancer cell proliferation measured under these conditions follows the same pattern, pointing to the causative role of complex I inhibition in the antiproliferative effects of Mito-metformins.
8. Establishing the mechanism using the right control: An example using a mitochondria-targeted nitroxide
Anti-OXPHOS drugs containing TPP+ typically stimulate ROS formation in cancer cells [86], [87]. At the same time, these drugs inhibit cancer cell proliferation [84]. H2O2 generated by oncogene activation enhanced tumor cell proliferation via stimulation of mitogen-activated protein kinase (MAPK) [88], as discussed in a subsequent section. Mito-CP containing the 10-carbon side chain is paradoxical in that it contains a nitroxide group that can function as a superoxide dismutase (SOD) mimetic while stimulating mitochondrial ROS through inhibition of complex I. We had previously used Mito-CP to mimic mitochondrial SOD activity and protect endothelial cells from oxidant-induced damage [89]. Nitroxides such as carboxy proxyl (CP) and Tempol have previously been used as SOD mimetics (like the SOD enzyme, CP catalytically dismutates O2•– to H2O2 and O2) [90]. Mito-CP is a CP analog that is synthesized by conjugating a TPP+ group to CP via an 11-carbon alkyl chain (Fig. 9). Mito-CP protected endothelial cells from oxidative injury induced by exposure to a steady flux of H2O2 and lipid hydroperoxides [89]. Mito-CP prevented the inactivation of mitochondrial aconitase (an endogenous marker of O2•– production) in endothelial cells, and this effect was attributed to dismutation of O2•– by Mito-CP. The “untargeted” nitroxide, CP (as a control), did not protect against endothelial cell damage or inactivation of aconitase.
In studies using cancer cells, Mito-CP was found to significantly inhibit cancer cell proliferation [91], [92], [93], [94], [95]. This finding supported the view that mitochondria-generated O2•– plays a role in cancer cell proliferation and that Mito-CP inhibits cell proliferation by removing O2•–. In control experiments, an alkyl TPP+ and CP were used. This combination did not selectively inhibit cancer cell growth, suggesting that CP (containing the nitroxide moiety) should be conjugated to TPP+ for efficacy.
More recently, a Mito-CP analog, Mito-CP-Ac, that contains an acetamide group in place of the nitroxide of Mito-CP, was used [94]. The Mito-CP-Ac is nearly the same as Mito-CP except for the replacement of the nitroxide group by an acetamide group (Fig. 9). Mito-CP-Ac does not possess the O2•– scavenging ability so it was an ideal control molecule. Mito-CP-Ac inhibited cancer cell proliferation as effectively as did Mito-CP. Mito-CP-Ac was not metabolized to Mito-CP intracellularly [94]. This finding challenged our original interpretation that Mito-CP inhibited cancer cell proliferation by dismutating O2•–. We subsequently revised our original proposal that the antiproliferative effect of Mito-CP is caused by its ability to dismutate O2•– or scavenge ROS and that a likely mechanism for the antiproliferative effects of Mito-CP and Mito-CP-Ac in cancer cells might be linked to their abilities to induce mitochondrial stress and activate redox signaling mechanisms rather than the SOD mechanism [94].
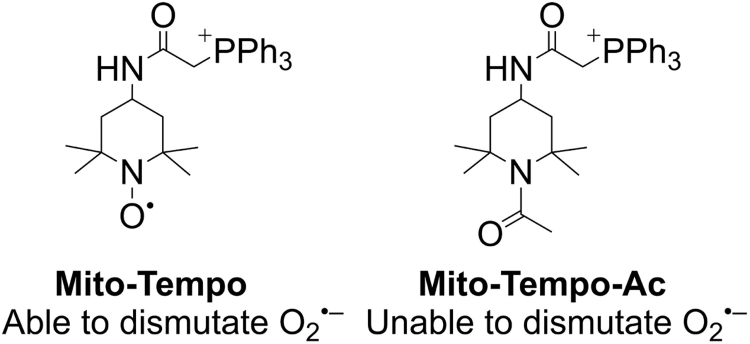
The results obtained from using both Mito-CP and Mito-CP-Ac have implications in understanding the effects of other mitochondria-targeted nitroxides including Mito-Tempol and Mito-Tempo [96]. Tempol, amino Tempol, and other Tempol/Tempo analogs are six-membered nitroxides whereas the CP family consists of a five-membered nitroxide. Mitochondria-targeted Tempo or Tempol analogs synthesized via attachment to the TPP+ moiety using linkers such as varying alkyl chain lengths are typically used to test the involvement of mitochondrial O2•– (also referred to as MROS) and O2•–-induced redox signaling in cancer cells [65]. It is also likely that Mito-Tempo is acting as a chain-breaking antioxidant via its hydroxylamine form. On the basis of Mito-CP and Mito-CP-Ac results in cancer cells, it is clear that one has to use the most appropriate control (i.e., Mito-Tempo-Ac) (Fig. 10) to absolutely verify the role of O2•–. Mito-Tempo-Ac lacks the nitroxide moiety needed to dismutate O2•– but will accumulate in mitochondria stimulating other stress-induced redox signaling mechanisms (such as Mito-Tempol) in cancer cells.
Fig. 10.
Chemical structures of Mito-Tempo and Mito-Tempo acetamide.
Investigators recently showed that lipid electrophiles generated endogenously during the inflammatory response form adducts with mitochondrial proteins [97]. In the presence of Mito-Tempo, a marked decrease in lipid electrophile adduction to proteins was observed. It was concluded that O2•– generated in the mitochondrial electron transport chain is a precursor responsible for lipid electrophile generation. This is indeed a novel mechanism that should be further corroborated using a more appropriate control (e.g., Mito-Tempo-Ac) that is similar to Mito-Tempo but devoid of the nitroxide moiety and its superoxide dismutating ability.
9. Peroxiredoxin enzymes: A family of thiol peroxidases
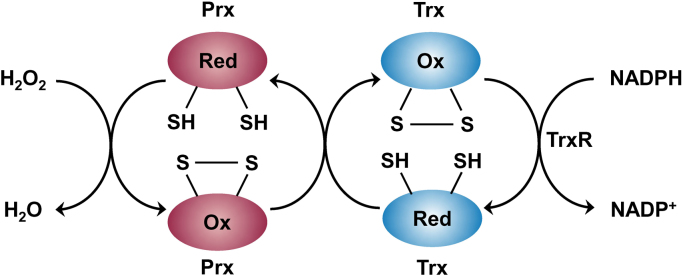
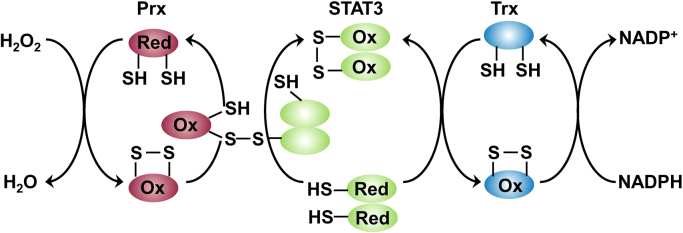
Peroxiredoxins are a family of redox sensing thiol peroxidase enzymes that detoxify H2O2 in combination with thioredoxin (Trx) and thioredoxin reductase (TrxR). Mitochondria typically contain two peroxiredoxin isoenzymes (Prx3 and Prx5). Although low molecular weight thiols such as cysteine and glutathione and redox-regulated proteins with low pKa cysteines react with H2O2 rather slowly (k = 1–10 M−1 s−1), protein thiol peroxidases (e.g., peroxiredoxins and glutathione peroxidases) react rapidly with H2O2 (k = 105–108 M−1 s−1) [98], [99]. The antioxidant potency of peroxiredoxins is vastly enhanced due to redox-coupled processes with the Trx/TrxR system (Fig. 11). Prx3 is present in mitochondria at a fairly high concentration (60 μM) and reacts with H2O2 very rapidly (k = 107 M−1 s−1) compared with catalase. Although the enzyme glutathione peroxidase-1 (GPx1) also reacts with H2O2 rapidly (k = 6 × 107 M−1 s−1), its concentration in mitochondria is relatively low (~ 2 μM). Thus, Prx3 is the major H2O2 detoxifying enzyme (~ 90% of H2O2) in mitochondria [100].
Fig. 11.
Peroxiredoxin pathway of H2O2metabolism. (Modified from Ref. [103]. Reprinted by permission from Macmillan Publishers Ltd: Nature Chemical Biology, M.C. Sobotta, W. Liou, S. Stöcker, D. Talwar, M. Oehler, T. Ruppert, A.N.D. Scharf, T.P. Dick, Peroxiredoxin-2 and STAT3 form a redox relay for H2O2 signaling, Nature Chemical Biology 11(1) (2015) 64–70, copyright 2015.).
In the first step of the reaction, the cysteine residue of Prx3 reacts with H2O2 to form a protein sulfenic acid (Cys-SOH) that combines with another Prx3 cysteine to form an intermolecular disulfide bond. In the second step, the oxidized Prx3 is converted back to its reduced state by Trx2 and TrxR2. In the presence of excess H2O2, Prx3 cysteine sulfenic acid undergoes hyperoxidation to form an inactive Prx3 cysteine sulfonic acid (Cys-SOOH) [101], [102].
Because of its relative stability and selective reactivity with cellular components, intracellularly generated H2O2 may diffuse over large distances (several microns) and participate in signal transduction. Thus, any involvement of ROS (H2O2 in particular) in tumorigenesis needs to be considered in the context of the presence and reactivity of peroxiredoxins and glutathione peroxidases and other redox partners involved (Fig. 11). Emerging studies in cancer are recognizing the role of transcription factors such as STAT3 in the redox signaling pathway and cell proliferation [103], as discussed in a subsequent section.
10. How does oxidative stress regulate cell signaling and proliferation of tumor cells?
One of the mechanisms by which tumor cells maintain non-cytotoxic levels of ROS is through upregulation of antioxidant enzyme expression (e.g., controlled via the nuclear factor erythroid 2 [Nrf2] pathway) and reductive cofactors (NADPH and GSH). At nontoxic levels, H2O2 can regulate cancer cell signaling through oxidation of cysteine residues. For example, H2O2 inactivates the PTEN molecule, a tumor suppressor, by oxidizing the active site cysteine to a disulfide, which prevents inactivation of the PI3 pathway [104]. Endogenous oncogene-mediated generation of ROS (H2O2 in particular) enhances tumor cell proliferation by stimulating MAPK/extracellular signal regulated kinase (ERK) pathways by inhibiting the action of MAPK phosphatases (via oxidation of the active site cysteines). ROS therefore can modulate/regulate cellular signaling factors. For example, ROS generated from mitochondria are required for KRAS lung cancer growth resulting from MAPK/ERK activation [95]. Increased ROS can also activate transcription factors like the nuclear factor kappa B (NF-κB), which enhances cancer cell proliferation [105].
Alternatively, ROS levels can be increased to a cytotoxic level in breast cancer cells and breast cancer models by using inhibitors of GSH and Trx pathways. Inhibiting the antioxidant enzyme, glutathione peroxidase, through suppression of fumarate in the Krebs cycle, enhanced ROS levels and cancer cell growth [106]. As a result of the paradoxical role of ROS in cancer cells, both pro- and antioxidant approaches have shown tumor enhancing and tumor regressing effects [107]. Similar to autophagy and mitophagy, which have been shown to be both pro- and antitumorigenic, mitochondria-generated ROS can exert tumor promoting and tumor suppressing effects. These are also dependent on tumor stage. The good and bad aspects of ROS have previously been addressed in relation to aging [108]. The signaling aspects of ROS (induction of host defense pathway via a hormetic mechanism) are considered to be good/pro-survival, whereas ROS-induced oxidative damage to DNA, proteins, and lipids is considered bad/pro-death. Because antioxidants could inhibit both the good and bad aspects of ROS activities, the clinical interventions using antioxidants could be problematic in the absence of the right balance [109]. It is likely that we face the same type of ROS conundrum in cancer biology research as well. In addition, reports suggest that a subset of melanoma tumor cells exhibit metabolic reprogramming heterogeneity with different bioenergetic and ROS detoxification capabilities [110]. Clearly, a more detailed understanding of how cancer cells reprogram metabolism to combat oxidative damage is vital prior to clinical intervention with antioxidants.
Recent literature suggests that reprogramming of cellular metabolism plays a crucial role in tumorigenesis [111]. Cancer cells acquire energy to proliferate through metabolic reprogramming as follows: altered glucose utilization, glutamine addiction or glutaminolysis, and lipid metabolism. Modulating the pathway generating cellular reductants (NADPH, GSH) can affect ROS formation/scavenging and tumorigenesis. Elevation of glutamine utilization (increased catabolism of glutamine, glutamine addiction) is one of the hallmarks of metabolic reprogramming in tumors. Glutamine is converted to glutamate by glutaminase (also upregulated by transcription factor cmyc) and glutamate is oxidized to α-ketoglutarate that enters into the TCA cycle. Transaminases use α-ketoglutarate to support redox homeostasis.
Sirtuin 3 (SIRT3) is a major deacetylase in mitochondria that is also considered as a ROS mitigator in cells. SIRT3 targets MnSOD and isocitrate dehydrogenases. Reports suggest that SIRT3 could have a dual role in cancer, both as an oncogene and tumor suppressor [112]. Tumor suppressors, such as p53, regulate glutaminase, an enzyme involved in the formation of glutamate required for synthesis of glutathione [113], [114]. Loss of p53 results in increased levels of ROS and oxidative damage in cancer cells [115].
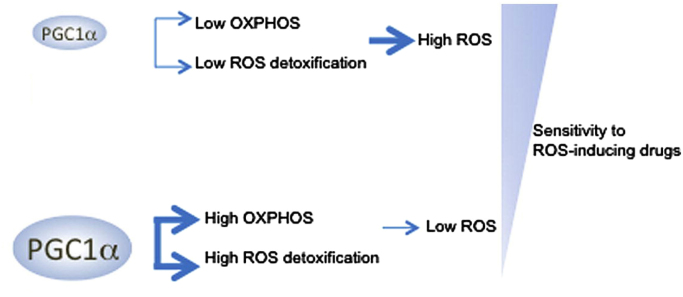
PPARγ coactivator-1α (PGC1α) is a transcription factor that is involved in mitochondrial biogenesis, oxidative metabolism, and metabolic reprogramming [116], [117]. The role of PGC1α in tumorigenesis, progression, and metastasis appears to be dependent on cancer type, reflecting the influence of microenvironment (nutrient status and tissue hypoxia) and genetic perturbations on the metabolic state of cancer cells [118]. PGC1α exhibits both pro- and antitumor function in cancer cells [119], [120]. PGC1α increases mitochondrial respiration and biogenesis and helps cancer cells cope with enhanced metabolic and oxidative stress through enhanced expression of ROS detoxification enzymes and increased resistance to cancer therapy. PGC1α reportedly decreases mitochondria-generated ROS [110]. Inhibiting PGC1α activity or its expression has been shown to enhance ROS levels and increase stabilization of the HIF1α protein, inducing a switch in metabolism from OXPHOS to glycolysis [121]. PGC1α-positive melanoma cells are more sensitive to disruption of mitochondrial respiration as they depend on PGC1α for survival. Conversely, PGC1α-negative melanoma cells with compromised antioxidant enzyme activity are more susceptible to ROS-generating drugs and oxidative stress (Fig. 12).
Fig. 12.
PGC1α regulates mitochondrial biogenesis and oxidative stress in melanoma cells. (Modified from Ref. [110]. Reprinted from Cancer Cell, 23, F. Vazquez, J.H. Lim, H. Chim, K. Bhalla, G. Girnun, K. Pierce, C.B. Clish, S.R. Granter, H.R. Widlund, B.M. Spiegelman, P. Puigserver, PGC1alpha expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress, 287–301, Copyright 2013, with permission from Elsevier.).
The role of PGC1α in tumorigenesis is paradoxical and is dependent on the type of cancer and its phenotype. PGC1α is reported to be pro- and antitumorigenic. In contrast with melanoma cells, PGC1α suppresses prostate cancer metastasis and inhibits renal cell carcinoma [117]. PGC1α overexpression impaired renal cancer cell growth by enhancing ROS and oxidative stress. PGC1α also enhances the susceptibility of renal carcinoma cells, RCC4, to cytotoxic therapy (radiation or doxorubicin) [121]. PGC1α enhanced mitochondrial content, ROS formation, and oxidative damage leading to impaired tumor growth [117], [120]. Thus, measurement of PGC1α levels in tumor subsets could help design appropriate anticancer drug therapy.
11. Do antioxidants enhance tumorigenesis and tumor metastasis?
It is well known that ROS (hydroxyl radical, H2O2, and O2•– and redox metal ions) can cause oncogenic mutations, and that treatment with antioxidants and antioxidant enzymes (ebselen, catalase, SOD, and SOD mimetics) inhibits the initiation and progression of some cancers [122]. However, several studies report the opposite results. The cancer biology field shares the same concerns (as the cardiovascular field) with respect to antioxidant supplementation. Antioxidant supplementation shows both beneficial and deleterious effects [123]. Of significance was the report demonstrating that antioxidant supplementation enhanced lung cancer progression in mouse models of BRAF- and KRAS-induced lung cancer. Two functionally different compounds were used to test the effect of antioxidants on cancer: a lipid-soluble vitamin E (a bona fide lipid peroxidation inhibitor and a chain-breaking antioxidant that terminates lipid peroxidation by scavenging ROS, lipid peroxyl radical) and a water-soluble drug, N-acetylcysteine (NAC), that is not a conventional antioxidant that reacts with ROS (O2•– and H2O2) at an appreciable rate compared with the endogenous antioxidant enzymes. However, NAC supplementation enhances the intracellular glutathione levels that will decrease ROS levels through enhanced activity of antioxidant enzymes. Despite limitations and problems in data interpretation with respect to ROS, this study is clearly provocative and questions the indiscriminate supplementation of antioxidants such as isoflavones, beta-carotenes, vitamin E, and NAC for suppressing tumor progression [124]. Oxidative stress was reported to inhibit melanoma metastasis [125]. Supplementation with antioxidants (NAC) promoted metastasis by inhibiting oxidative stress.
The paradoxical behavior of ROS and antioxidants in cancer is not unique in that similar effects have been observed in other fields of research including aging and cardiovascular diseases [108], [126]. Several years ago, it was shown that beta-carotene may have an adverse effect on the incidence of lung cancer and cardiovascular disease in a clinical trial involving smokers [127]. Although the pro-oxidant mechanism of action is not known, this study pointed out the inadequacy in our understanding of the pro- and antioxidant mechanisms of action in humans.
12. Therapeutic targeting of signaling pathways
ROS-mediated signaling pathways, elevated in many tumors, are involved in cell growth and proliferation, differentiation, survival, and metabolism. ROS, in particular H2O2, may act as second messengers in cell signaling. H2O2 regulates protein activity via direct or indirect reversible oxidation of protein tyrosine phosphatases, protein and receptor tyrosine kinases, and transcription factors [128], [129].
12.1. PI3/Akt signaling pathway
One of the hallmarks of cancer is the hyperactivation of the phosphoinositide 3-kinase (PI3K)-Akt signaling pathway and is an attractive target for antitumor therapy. Akt is a proto-oncogene that is activated in many cancers. In addition to growth factors, ROS (possibly H2O2) are involved in the mechanisms responsible for PI3K signaling [130]. The activated PI3K subsequently phosphorylates phosphatidylinositol-4,5-biphosphate (PIP2) to phosphatidylinositol-3,4,5-triphosphate (PIP3) that acts as a signaling molecule to recruit phosphoinositide-dependent kinase-1 (PDK1) and protein kinase B (PKB, also known as Akt). The tumor suppressor phosphatase, PTEN, downregulates the PI3K/Akt signaling pathway by dephosphorylation of PIP3 back to PIP2 [131]. ROS (importantly H2O2) oxidize the cysteinyl group in PTEN molecules such that PTEN is inactivated, thereby resulting in activation of the PI3K/Akt signaling pathway and tumor progression. Akt is hyperactivated by other mechanisms as well, and is involved in the regulation of multiple cellular activities. Akt inhibits the forkhead box O (FOXO) transcription factor and subsequently regulates apoptosis, leading to enhanced tumor growth [132], [133].
12.2. MAPK/Erk1/2 pathway
ROS such as H2O2 can oxidatively modify the cysteinyl residue of proteins (e.g., protein tyrosine phosphatases [PTPs], protein tyrosine kinases [PTKs], and protein kinase C [PKC]) and subsequently activate downstream kinase cascade (e.g., MAPKs). One of the most extensively studied MAPK pathway is the ERK1/2 signaling pathway. ERK1/2 activation is linked to regulation of cancer cell survival, proliferation, and metastasis [134]. Mutations in genes encoding RAS and RAF modulate cancer development through the RAS-RAF-MEK-ERK kinases axis.
12.3. AMPK/mTOR
Mitochondria-targeted therapeutics (e.g., Mito-metformin) activate AMPK [59]. AMPK is a master energy sensor within the cell. AMPK activation is initiated by the enhanced intracellular ratio of AMP to ATP. AMPK activation results in the upregulation of ATP-generating pathways and decreased ATP-consuming pathways. AMPK regulates the redox state by mitigating the NADPH depletion that occurs via increased fatty acid oxidation and decreased fatty acid synthesis. The relationship between AMPK activation and cancer cell proliferation became evident in light of the protective effects induced by AMPK inhibitors. Inhibiting AMPK signaling by dorsomorphin (compound C) reversed the antiproliferative effect of mitochondria-targeted therapeutics such as Mito-metformin [59]. Mito-metformin stimulated O2•– and H2O2 formation in pancreatic cancer cells (Fig. 5) [55], [59], [87]. Mito-metformin-induced inhibition of complex I was thought to be responsible for formation of O2•– and H2O2. We proposed that H2O2 derived from complex I inhibition was responsible for the AMPK activation and mTOR inhibition that are related to the inhibition of proliferation (Fig. 13). A recent report also suggested that mitochondrial ROS were responsible for the AMPK activation that was linked to oxidation of calmodulin kinase II [135]. Inhibiting calmodulin kinase II suppressed AMPK activation. Additional experiments using AMPK knockout cells and xenografts are needed to gain a complete understanding of the role of ROS in the AMPK signaling mechanism.
Fig. 13.
Mito-metformin and activation of AMPK: Potential signaling role of H2O2. (Obtained from Ref. [59]. Reprinted from Cancer Research, 76, G. Cheng, J. Zielonka, O. Ouari, M. Lopez, D. McAllister, K. Boyle, C.S. Barrios, J.J. Weber, B.D. Johnson, M. Hardy, M.B. Dwinell, B. Kalyanaraman, Mitochondria-targeted analogs of metformin exhibit enhanced antiproliferative and radiosensitizing effects in pancreatic cancer cells, 3904-15, Copyright 2016.).
12.4. Signal transducer and activator of transcription: STAT3/peroxiredoxin and transmission of H2O2 signaling via redox relay
As discussed above, thiol peroxidase, peroxiredoxin, effectively and rapidly scavenges H2O2 in both the cytosolic and mitochondrial compartments in cells. Recently, investigators reported an additional novel mechanism for the redox activity of thiol peroxidase, peroxiredoxin-2 (Prx2), in that Prx2 acts as a signal transductor and a redox regulator (Fig. 14). Rather than directly oxidizing thiols in STAT3, H2O2 rapidly reacts with Prx2 cysteinyl thiol to form an oxidized Prx2 (Prx2 sulfenic acid and disulfide). The oxidized Prx2 forms a disulfide exchange intermediate with STAT3, leading to oxidation of the STAT3 thiols and affecting its transcriptional activity. Oxidized STAT3 can be reduced back to its active form by the Trx/TrxR system, using NADPH as the ultimate electron donor (Fig. 14). This model shows how redox regulating proteins may transmit oxidizing and reducing equivalents from H2O2 and NADPH, respectively, to control the activity of redox-sensitive proteins. STAT-dependent regulation of ROS and oxidative metabolism in normal and cancer cells is an emerging area of research [101], [103].
Fig. 14.
Redox signaling of H2O2via peroxiredoxin/Trx redox relay including STAT3 transcription factor. (Obtained from Ref. [103]. Reprinted by permission from Macmillan Publishers Ltd: Nature Chemical Biology, M.C. Sobotta, W. Liou, S. Stöcker, D. Talwar, M. Oehler, T. Ruppert, A.N.D. Scharf, T.P. Dick, Peroxiredoxin-2 and STAT3 form a redox relay for H2O2 signaling, Nature Chemical Biology 11(1) (2015) 64–70, copyright 2015.).
12.5. Keap1 mutations, Nrf2-targeted gene activation, ROS, and glutamine uptake in tumor cells
Tumor cells are metabolically hyperactive during tumorigenesis in order to meet and sustain their energy and building-blocks requirements for enhanced growth. Enhanced metabolism results in ROS generation during tumorigenesis [136]. Tumor cells activate the Nrf2 pathway via metabolic reprogramming to stimulate the expression of antioxidant enzymes, leading to decreased ROS levels [137]. Glutathione synthesis was elevated in Keap1 or Nrf2-mutant lung cancers through enhanced glutaminolysis.
The Keap1 (Kelch-like ECH-associated protein 1) gene is one of the most frequently mutated genes in KRAS-mutant lung adenocarcinoma. It is estimated that approximately 20% of KRAS-mutant lung cancer cells carry the mutated Keap1 gene. Keap1 is a negative regulator of Nrf2, the master transcriptional regulator of the endogenous antioxidant response, and thus is critical for maintaining the redox homeostasis in cells.
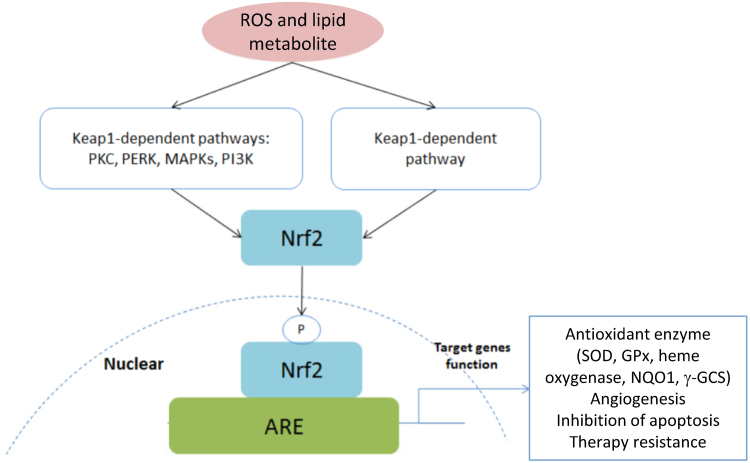
Keap1-Nrf2 is a redox regulating system that plays a critical role in oxidative stress and cytoprotection. Under basal conditions or nonoxidative stress conditions, the Keap1/Nrf2 complex is constantly degraded by the ubiquitin-proteasome pathway. Increased levels of ROS or electrophiles (4-HNE formed from oxidized lipids) result in modification of the cysteinyl residues of Keap1 and stop the negative regulation of Nrf2 by Keap1. Consequently, Nrf2 is stabilized and migrates to the nucleus and activates the expression of target genes that include antioxidative stress genes (heme oxygenase 1, peroxiredoxin 1, MnSOD), detoxifying enzyme genes (NADPH quinone oxidoreductase, glutathione S-transferases), and ABC transporter genes (Fig. 15).
Fig. 15.
Schematic of the Keap1–Nrf2–ARE pathway. (Modified from Ref. [128]. Reprinted from Oncotarget, 8, X. Gao, B. Schottker, Reduction-oxidation pathways involved in cancer development: a systematic review of literature reviews, 51888-906, copyright 2017.).
Nrf2 has a dual mechanism of action in cancer therapies, exhibiting chemotherapy and radiotherapy resistance and proliferative capacity, and promoting host defense against cancer cells. Nrf2 protects against oxidative stress in normal cells, and Nrf2 activation in cancer cells induces drug and radiation resistance. Patients bearing Nrf2-activated tumors exhibit poor prognoses. Activation of Nrf2 is associated with tumor resistance to anticancer drugs via detoxification of ROS and electrophiles induced by the drugs. Inhibitors of the Keap1-Nrf2 pathway enhanced chemosensitivity in lung cancer cells [138].
12.6. ROS and autophagy
Autophagy or “self-eating” is a highly regulated process of removing modified or damaged intracellular proteins that are presumably required to maintain redox homeostasis [139]. Its functional role (cytoprotective or cytotoxic) in normal and cancer cells is different and context-dependent [140]. Enhancement or mitigation of autophagy in tumorigenesis depends on the tissue, the stage, and the type of tumor [141]. It is becoming more evident that ROS generated from mitochondria (and probably from Nox activation) and the resulting oxidative stress is responsible for increasing autophagic flux, which, in turn, decreases ROS levels. Thus, autophagy can play an antitumoral and protumoral role during cancer development [142]. ROS were proposed to serve as intracellular messengers to regulate autophagosome formation. During the initiation phase, where ROS-induced DNA damage is the key event, autophagy plays an antitumoral role. However, during the propagation, progression, and metastatic phases of cancer development, autophagy may play a protumoral role because of decreased ROS-mediated oxidative stress and increased production of nutrients provided by autophagic activation.
Starvation or nutrient deprivation triggers autophagy. Pharmacologically, mitochondria-targeted drugs can promote autophagy through stimulation of bioenergetic stress, increased ROS formation, AMPK activation, and mTOR inhibition [143]. Alternatively, enhanced PI3K-1/Akt and MAPK/ERK1/2 signaling activate mTOR and regulate autophagy [144]. It is clear that the balance between ROS levels and ROS-induced autophagy is critical for tumor progression or regression. Antioxidants and their paradoxical effects in cancer may be linked to the cytoprotective or cytotoxic mechanisms of autophagy.
Antimalarial drugs, chloroquine and hydroxychloroquine, inhibit autophagy and are undergoing clinical trials as antitumor drugs [145]. However, reports also indicate the potentially deleterious adverse effects of inhibiting autophagy in kidney and other organs [146]. Combining chloroquine with the standard-of-care chemotherapies (cis-platin and doxorubicin) was reported to exacerbate nephrotoxicity and possibly cardiotoxicity [147].
12.7. ROS and the tumor microenvironment
T cells in the tumor microenvironment (TME) are effective in recognizing and killing malignant cells. The TME, especially in pancreatic tumors, has been shown to be immunosuppressive, and granulocytic myeloid-derived suppressor cells (MDSC) have been identified as suppressors of the immunoresponse to tumor cells [148]. Previously, Corzo et al. showed that ROS generated from Nox2 were responsible for T cell inactivation or suppression by granulocytic MDSC [149]. The defective T cell reactivity against the tumor is a prominent feature of tumor suppressive microenvironment in PDAC. Because MDSC deficient in Nox2 do not suppress T cell reactivity, it was rationalized that Nox2-generated ROS caused the inactivation of T cells. Inhibiting Nox signaling using small molecular weight compounds could potentially enhance tumor-specific T cell responses.
The role of Nrf2 in the tumor microenvironment is intriguing. MDSC consisting of macrophages, dendritic cells, and neutrophils support tumor development and metastasis by inhibiting innate and adaptive immunity. ROS generated by MDSC in the tumor microenvironment facilitate immunosuppression by MDSC. Nrf2 activation regulates the immunosuppressive action of MDSC by modulating ROS levels [150], [151].
13. In vivo measurements of ROS in tumor tissues
Typically, either protein carbonyls or protein tyrosyl nitrated products are used as in vivo markers of oxidant (ROS/RNS) formation [152]. DMPO has been used to trap protein radicals, and an antibody to DMPO has been used to detect this adduct using an immunospin-trapping method [153]. Some other newly developed probes and techniques for assessing in vivo oxidants are described below.
13.1. MitoB
MitoB (a boronate probe attached to a TPP+ moiety) was used to detect mitochondria-generated H2O2 in a living organism [30], [31]. This probe reacts stoichiometrically with H2O2 to form a diagnostic exomarker, MitoP. One of the advantages of the TPP+ cation is that the positive charge enhances the sensitivity of detection (pmol/g levels) by LC-MS. The MitoP/MitoB ratio was used as a measure of H2O2 generated within mitochondria. The investigators used a meta-substituted boronate probe (meta-MitoB). Because peroxynitrite reacts with this probe a million-fold faster than H2O2 (yielding the same product [MitoP]), it is essential to use a probe that will distinguish between the two species. As indicated previously, in the presence of peroxynitrite, ortho-MitoB will form a cyclized product (not possible with meta-MitoB) that can be used as a specific exomarker for ONOO– (Fig. 2) [28], [32], [33]. Absence of formation of this minor product would indicate that MitoP is formed in peroxynitrite-independent process, most likely by H2O2.
13.2. Mito-NeoD
Recently, Hartley, Murphy, and colleagues synthesized a phenanthridinium-based mitochondria-targeted superoxide probe, MitoNeoD, that is more robust than the existing probes for detecting O2•– formed in animals [68]. As discussed, the chemistry between MitoNeoD and O2•– and other oxidants is similar to that described for HE and Mito-SOX (Fig. 7) [59]. Reaction with O2•– results in the formation of MitoNeoOH from MitoNeoD as a diagnostic marker product. In contrast to HE and Mito-SOX, MitoNeoD was shown to be more robust because of the presence of bulky neopentyl groups and incorporation of deuterium at carbon-6 position [68]. These modifications decrease the extent of nonspecific oxidation to the E+ analog product (MitoNeo) and its DNA intercalation. MitoNeoOH, the superoxide-specific product, is typically detected by LC-MS/MS (after extraction from the tissue) relative to deuterated internal standards [68].
13.3. Low-temperature EPR
Electron paramagnetic resonance (EPR) spectroscopy detects unpaired electrons, including those in free radicals and in many electronic states of transition ions (e.g., FeIII, CuII MnII/III/IV) and in metal clusters (e.g., [2Fe2S]+, [3Fe4S]0/+, [4Fe4S]+). EPR is often diagnostic for individual paramagnetic species, and 12–16 of the approximately 20 signals expected from mitochondria can be assigned depending on the tissue and redox status [154]. EPR is also quantitative and amounts of species in the paramagnetic state can be estimated by computer fitting. EPR of mitochondrial redox centers requires cryogenic temperatures (5–40 K). At such low temperatures, unpaired electrons remain free to migrate between redox centers with appreciable exchange interactions and adopt statistical distributions according to the centers’ midpoint potentials. However, diffusion and active transport of molecules ceases. Therefore, low-temperature EPR of flash-frozen intact tissue or cell samples provides a snapshot of the redox status of the various mitochondrial respiratory chain complexes at the time of freezing, and reports on the midpoint potentials of the individual redox centers and the integrity of their intramolecular electron transfer pathways. EPR also reports on oxidative stress history, most conveniently through interrogation of the oxidative partial disassembly of the mitochondrial aconitase EPR-silent [4Fe4S]2+ to the characteristically EPR-active [3Fe4S]+ center [155], [156]. The use of low-temperature EPR in mitochondrial diseases [154], neurodegeneration and neuroprotection [6], [157], and chromium toxicity [155] has been reported, and the potential for use in tumor investigation appears high.
13.4. Complexes I and II EPR signals as redox status markers
Complexes I and II together generally exhibit up to seven signals due to reduced FeS clusters (complex I: N1b, N2, N3, and N4; complex II: S1, S2, and Rieske cluster) and an additional signal due to oxidized complex II S3. The “g2” resonance positions of each of the reduced [2Fe2S] and [4Fe4S] clusters overlap considerably, giving rise to an intense and characteristic EPR line at g = 1.94 in most tissues under normal physiological conditions. Reduced intensity of this signal indicates a global increase in the redox potential. Two centers, N3 and N4, have very low midpoint potentials, close to the NADH/NAD+ couple, and exhibit weak but well-separated respective g3 EPR resonances with g < 1.9, i.e., well upfield of the other signals. The relative intensities of the N3 and N4 signals and the g = 1.94 signal are, therefore, very sensitive markers for subtle changes in redox status. Computer fitting can reliably separate the complex II signals S1 and S2 from the complex I signals, though not from each other; this provides a way to distinguish global from local phenomena. The signal intensities of the reduced clusters may also be compared with the intensity of the oxidized S3 signal. The S3 signal overlaps substantially with the aconitase [3Fe4S]+ signal but is readily differentiated by a markedly different temperature dependence. Thus, a wealth of information on redox status is available from which various inferences can be made, though complementary techniques may well be required to elucidate the underlying mechanisms of dysfunction [154].
13.5. Aconitase inactivation: EPR detectable biomarker of mitochondrial oxidation
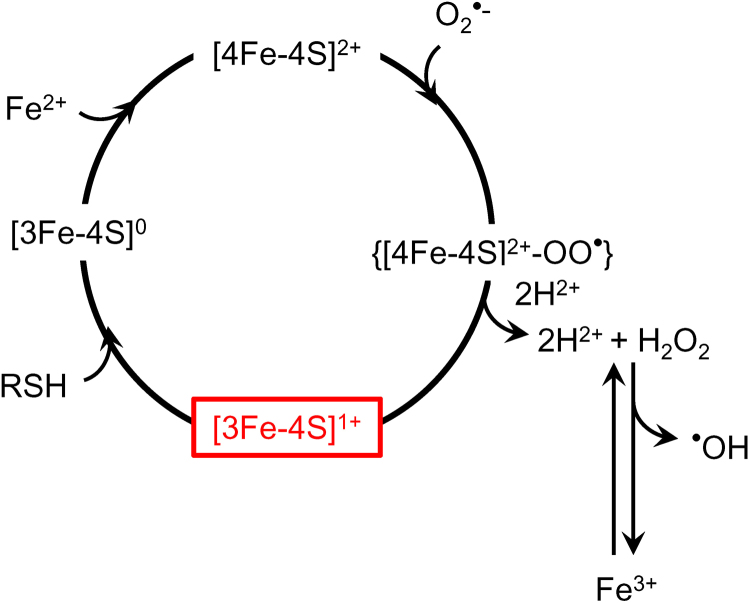
Aconitase inactivation is used as an EPR marker of oxidative stress as O2•– produced from mitochondrial electron transport chain dysfunction reacts with the iron-sulfur centers. Aconitase is converted from the enzymatically active, EPR-silent, [4Fe4S]2+-containing form to the enzymatically inactive, but EPR-active, [3Fe4S]+ form [156], [158] (Fig. 16). The EPR signals from the distinct mitochondrial and cytosolic forms of aconitase are easily distinguishable with the isolated enzymes [159] but may be difficult to differentiate in the complex spectra of tissue and cells unless present at very high levels. Nevertheless, the aconitase signal, with a maximum at around g = 2.02, has been successfully used as a marker for oxidative stress in biological materials [6], [154], [155], [157].
Fig. 16.
O2•–-mediated inactivation of aconitase: A possible source of hydroxyl radical generation in mitochondria. (Modified from Ref. [160]. This research was originally published in Journal of Biological Chemistry. J. Vasquez-Vivar, B. Kalyanaraman, M.C. Kennedy. Mitochondrial aconitase is a source of hydroxyl radical. An electron spin resonance investigation. 2000; 275:14064-9. © the American Society for Biochemistry and Molecular Biology.).
14. Conclusions and future perspectives
In this review, we discussed the use of specific chemical probes and analytical methods in the detection of ROS formed in tumor cells in vitro and in vivo as well as the use of the low-temperature EPR technique to monitor oxidants formed from the endogenous redox-sensing protein, aconitase, in tumor cells. This technique can be translated to the clinic in that the surgically removed tumor tissues from patients can be monitored by EPR at low temperatures. This may be the only technique that will enable detection of the endomarkers of mitochondrial oxidants in isolated human tumors. Monitoring changes in ROS formation in an in vivo setting following drug resistance and metabolic reprogramming in tumors can aid in the development of a precise and effective antitumor drug regimen. The newly discovered in vivo mitochondrial O2•– and H2O2 probes can be used in patient-derived mice xenograft models. The metabolic reprogramming that occurs in hypoxia and drug resistance likely favors mitochondrial OXPHOS and possibly ROS stimulation. An increased understanding of ROS in cancer biology will help us better interpret the redox signaling and therapeutics, and it will help reveal the paradoxical role of ROS and autophagy in tumor growth and antitumor treatments.
Acknowledgements
This research was supported by NIH NCI U01 CA178960 to Michael Dwinell and BK (MPIs), NIH NCI R01 CA208648 to Ming You and BK (MPIs), and the Quadracci Endowment. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This teaching review is the third in a series of publications in this journal and is based on the lectures on cancer biology that BK presented as part of Anna University's Global Initiative Academic Networks (GIAN) program, Chennai, India.
References
- 1.Holmstrom K.M., Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 2014;15(6):411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 2.Galadari S., Rahman A., Pallichankandy S., Thayyullathil F. Reactive oxygen species and cancer paradox: to promote or to suppress? Free Radic. Biol. Med. 2017;104:144–164. doi: 10.1016/j.freeradbiomed.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Murphy M.P., Holmgren A., Larsson N.G., Halliwell B., Chang C.J., Kalyanaraman B., Rhee S.G., Thornalley P.J., Partridge L., Gems D., Nystrom T., Belousov V., Schumacker P.T., Winterbourn C.C. Unraveling the biological roles of reactive oxygen species. Cell Metab. 2011;13(4):361–366. doi: 10.1016/j.cmet.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winterbourn C.C. Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol. 2008;4(5):278–286. doi: 10.1038/nchembio.85. [DOI] [PubMed] [Google Scholar]
- 5.Beckman J.S., Beckman T.W., Chen J., Marshall P.A., Freeman B.A. Apparent hydroxyl radical production by peroxynitrite: implications for endothelial injury from nitric oxide and superoxide. Proc. Natl. Acad. Sci. USA. 1990;87(4):1620–1624. doi: 10.1073/pnas.87.4.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghosh A., Kanthasamy A., Joseph J., Anantharam V., Srivastava P., Dranka B.P., Kalyanaraman B., Kanthasamy A.G. Anti-inflammatory and neuroprotective effects of an orally active apocynin derivative in pre-clinical models of Parkinson's disease. J. Neuroinflamm. 2012;9:241. doi: 10.1186/1742-2094-9-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pacher P., Beckman J.S., Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol. Rev. 2007;87(1):315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonini M.G., Rota C., Tomasi A., Mason R.P. The oxidation of 2′,7′-dichlorofluorescin to reactive oxygen species: a self-fulfilling prophesy? Free Radic. Biol. Med. 2006;40(6):968–975. doi: 10.1016/j.freeradbiomed.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 9.Zielonka J., Lambeth J.D., Kalyanaraman B. On the use of L-012, a luminol-based chemiluminescent probe, for detecting superoxide and identifying inhibitors of NADPH oxidase: a reevaluation. Free Radic. Biol. Med. 2013;65:1310–1314. doi: 10.1016/j.freeradbiomed.2013.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalyanaraman B., Darley-Usmar V., Davies K.J., Dennery P.A., Forman H.J., Grisham M.B., Mann G.E., Moore K., Roberts L.J., 2nd, Ischiropoulos H. Measuring reactive oxygen and nitrogen species with fluorescent probes: challenges and limitations. Free Radic. Biol. Med. 2012;52(1):1–6. doi: 10.1016/j.freeradbiomed.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalyanaraman B., Hardy M., Zielonka J. A critical review of methodologies to detect reactive oxygen and nitrogen species stimulated by NADPH oxidase enzymes: implications in pesticide toxicity. Curr. Pharmacol. Rep. 2016;2(4):193–201. doi: 10.1007/s40495-016-0063-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardy M., Zielonka J., Karoui H., Sikora A., Michalski R., Podsiadly R., Lopez M., Vasquez-Vivar J., Kalyanaraman B., Ouari O. Detection and characterization of reactive oxygen and nitrogen species in biological systems by monitoring species-specific products. Antioxid. Redox Signal. 2017 doi: 10.1089/ars.2017.7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maghzal G.J., Stocker R. Improved analysis of hydroethidine and 2-hydroxyethidium by HPLC and electrochemical detection. Free Radic. Biol. Med. 2007;43(7):1095–1096. doi: 10.1016/j.freeradbiomed.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 14.Zhao H., Kalivendi S., Zhang H., Joseph J., Nithipatikom K., Vasquez-Vivar J., Kalyanaraman B. Superoxide reacts with hydroethidine but forms a fluorescent product that is distinctly different from ethidium: potential implications in intracellular fluorescence detection of superoxide. Free Radic. Biol. Med. 2003;34(11):1359–1368. doi: 10.1016/s0891-5849(03)00142-4. [DOI] [PubMed] [Google Scholar]
- 15.Zhao H., Joseph J., Fales H.M., Sokoloski E.A., Levine R.L., Vasquez-Vivar J., Kalyanaraman B. Detection and characterization of the product of hydroethidine and intracellular superoxide by HPLC and limitations of fluorescence. Proc. Natl. Acad. Sci. USA. 2005;102(16):5727–5732. doi: 10.1073/pnas.0501719102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zielonka J., Hardy M., Michalski R., Sikora A., Zielonka M., Cheng G., Ouari O., Podsiadly R., Kalyanaraman B. Recent developments in the probes and assays for measurement of the activity of NADPH oxidases. Cell Biochem. Biophys. 2017;75(3–4):335–349. doi: 10.1007/s12013-017-0813-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zielonka J., Hardy M., Kalyanaraman B. HPLC study of oxidation products of hydroethidine in chemical and biological systems: ramifications in superoxide measurements. Free Radic. Biol. Med. 2009;46(3):329–338. doi: 10.1016/j.freeradbiomed.2008.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michalski R., Michalowski B., Sikora A., Zielonka J., Kalyanaraman B. On the use of fluorescence lifetime imaging and dihydroethidium to detect superoxide in intact animals and ex vivo tissues: a reassessment. Free Radic. Biol. Med. 2014;67:278–284. doi: 10.1016/j.freeradbiomed.2013.10.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zielonka J., Kalyanaraman B. Hydroethidine- and MitoSOX-derived red fluorescence is not a reliable indicator of intracellular superoxide formation: another inconvenient truth. Free Radic. Biol. Med. 2010;48(8):983–1001. doi: 10.1016/j.freeradbiomed.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zielonka J., Srinivasan S., Hardy M., Ouari O., Lopez M., Vasquez-Vivar J., Avadhani N.G., Kalyanaraman B. Cytochrome c-mediated oxidation of hydroethidine and mito-hydroethidine in mitochondria: identification of homo- and heterodimers. Free Radic. Biol. Med. 2008;44(5):835–846. doi: 10.1016/j.freeradbiomed.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aird K.M., Allensworth J.L., Batinic-Haberle I., Lyerly H.K., Dewhirst M.W., Devi G.R. ErbB1/2 tyrosine kinase inhibitor mediates oxidative stress-induced apoptosis in inflammatory breast cancer cells. Breast Cancer Res. Treat. 2012;132(1):109–119. doi: 10.1007/s10549-011-1568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Q., Raje V., Yakovlev V.A., Yacoub A., Szczepanek K., Meier J., Derecka M., Chen Q., Hu Y., Sisler J., Hamed H., Lesnefsky E.J., Valerie K., Dent P., Larner A.C. Mitochondrial localized Stat3 promotes breast cancer growth via phosphorylation of serine 727. J. Biol. Chem. 2013;288(43):31280–31288. doi: 10.1074/jbc.M113.505057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dikalov S.I., Harrison D.G. Methods for detection of mitochondrial and cellular reactive oxygen species. Antioxid. Redox Signal. 2014;20(2):372–382. doi: 10.1089/ars.2012.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller E.W., Tulyathan O., Isacoff E.Y., Chang C.J. Molecular imaging of hydrogen peroxide produced for cell signaling. Nat. Chem. Biol. 2007;3(5):263–267. doi: 10.1038/nchembio871. [DOI] [PubMed] [Google Scholar]
- 25.Lippert A.R., Van de Bittner G.C., Chang C.J. Boronate oxidation as a bioorthogonal reaction approach for studying the chemistry of hydrogen peroxide in living systems. Acc. Chem. Res. 2011;44(9):793–804. doi: 10.1021/ar200126t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zielonka J., Sikora A., Hardy M., Joseph J., Dranka B.P., Kalyanaraman B. Boronate probes as diagnostic tools for real time monitoring of peroxynitrite and hydroperoxides. Chem. Res. Toxicol. 2012;25(9):1793–1799. doi: 10.1021/tx300164j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sikora A., Zielonka J., Lopez M., Joseph J., Kalyanaraman B. Direct oxidation of boronates by peroxynitrite: mechanism and implications in fluorescence imaging of peroxynitrite. Free Radic. Biol. Med. 2009;47(10):1401–1407. doi: 10.1016/j.freeradbiomed.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zielonka J., Zielonka M., VerPlank L., Cheng G., Hardy M., Ouari O., Ayhan M.M., Podsiadly R., Sikora A., Lambeth J.D., Kalyanaraman B. Mitigation of NADPH oxidase 2 activity as a strategy to inhibit peroxynitrite formation. J. Biol. Chem. 2016;291(13):7029–7044. doi: 10.1074/jbc.M115.702787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sikora A., Zielonka J., Lopez M., Dybala-Defratyka A., Joseph J., Marcinek A., Kalyanaraman B. Reaction between peroxynitrite and boronates: EPR spin-trapping, HPLC Analyses, and quantum mechanical study of the free radical pathway. Chem. Res. Toxicol. 2011;24(5):687–697. doi: 10.1021/tx100439a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cochemé H.M., Logan A., Prime T.A., Abakumova I., Quin C., McQuaker S.J., Patel J.V., Fearnley I.M., James A.M., Porteous C.M., Smith R.A.J., Hartley R.C., Partridge L., Murphy M.P. Using the mitochondria-targeted ratiometric mass spectrometry probe MitoB to measure H2O2 in living Drosophila. Nat. Protoc. 2012;7(5):946–958. doi: 10.1038/nprot.2012.035. [DOI] [PubMed] [Google Scholar]
- 31.Cochemé H.M., Quin C., McQuaker S.J., Cabreiro F., Logan A., Prime T.A., Abakumova I., Patel J.V., Fearnley I.M., James A.M., Porteous C.M., Smith R.A.J., Saeed S., Carré J.E., Singer M., Gems D., Hartley R.C., Partridge L., Murphy M.P. Measurement of H2O2 within living drosophila during aging using a ratiometric mass spectrometry probe targeted to the mitochondrial matrix. Cell Metab. 2011;13(3):340–350. doi: 10.1016/j.cmet.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sikora A., Zielonka J., Adamus J., Debski D., Dybala-Defratyka A., Michalowski B., Joseph J., Hartley R.C., Murphy M.P., Kalyanaraman B. Reaction between peroxynitrite and triphenylphosphonium-substituted arylboronic acid isomers: identification of diagnostic marker products and biological implications. Chem. Res. Toxicol. 2013;26(6):856–867. doi: 10.1021/tx300499c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zielonka J., Sikora A., Adamus J., Kalyanaraman B. Detection and differentiation between peroxynitrite and hydroperoxides using mitochondria-targeted arylboronic acid. Methods Mol. Biol. 2015;1264:171–181. doi: 10.1007/978-1-4939-2257-4_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chance B., Sies H., Boveris A. Hydroperoxide metabolism in mammalian organs. Physiol. Rev. 1979;59(3):527–605. doi: 10.1152/physrev.1979.59.3.527. [DOI] [PubMed] [Google Scholar]
- 35.Bedard K., Krause K.H. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol. Rev. 2007;87(1):245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 36.Boveris A., Chance B. The mitochondrial generation of hydrogen peroxide. general properties and effect of hyperbaric oxygen. Biochem. J. 1973;134(3):707–716. doi: 10.1042/bj1340707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boveris A. Mitochondrial production of superoxide radical and hydrogen peroxide. Adv. Exp. Med. Biol. 1977;78:67–82. doi: 10.1007/978-1-4615-9035-4_5. [DOI] [PubMed] [Google Scholar]
- 38.Turrens J.F. Mitochondrial formation of reactive oxygen species. J. Physiol. 2003;552(Pt 2):335–344. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murphy M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009;417(1):1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meitzler J.L., Antony S., Wu Y., Juhasz A., Liu H., Jiang G., Lu J., Roy K., Doroshow J.H. NADPH oxidases: a perspective on reactive oxygen species production in tumor biology. Antioxid. Redox Signal. 2014;20(17):2873–2889. doi: 10.1089/ars.2013.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu W., Hu Y., Chen G., Chen Z., Zhang H., Wang F., Feng L., Pelicano H., Wang H., Keating M.J., Liu J., McKeehan W., Wang H., Luo Y., Huang P. Novel role of NOX in supporting aerobic glycolysis in cancer cells with mitochondrial dysfunction and as a potential target for cancer therapy. PLoS Biol. 2012;10(5):e1001326. doi: 10.1371/journal.pbio.1001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lambeth J.D., Krause K.H., Clark R.A. NOX enzymes as novel targets for drug development. Semin. Immunopathol. 2008;30(3):339–363. doi: 10.1007/s00281-008-0123-6. [DOI] [PubMed] [Google Scholar]
- 43.Streeter J., Thiel W., Brieger K., Miller F.J., Jr. Opportunity nox: the future of NADPH oxidases as therapeutic targets in cardiovascular disease. Cardiovasc. Ther. 2013;31(3):125–137. doi: 10.1111/j.1755-5922.2011.00310.x. [DOI] [PubMed] [Google Scholar]
- 44.Nisimoto Y., Diebold B.A., Cosentino-Gomes D., Lambeth J.D. Nox4: a hydrogen peroxide-generating oxygen sensor. Biochemistry. 2014;53(31):5111–5120. doi: 10.1021/bi500331y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Serrander L., Cartier L., Bedard K., Banfi B., Lardy B., Plastre O., Sienkiewicz A., Forro L., Schlegel W., Krause K.H. NOX4 activity is determined by mRNA levels and reveals a unique pattern of ROS generation. Biochem. J. 2007;406(1):105–114. doi: 10.1042/BJ20061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cifuentes-Pagano E., Meijles D.N., Pagano P.J. The quest for selective nox inhibitors and therapeutics: challenges, triumphs and pitfalls. Antioxid. Redox Signal. 2014;20(17):2741–2754. doi: 10.1089/ars.2013.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zielonka J., Cheng G., Zielonka M., Ganesh T., Sun A., Joseph J., Michalski R., O'Brien W.J., Lambeth J.D., Kalyanaraman B. High-throughput assays for superoxide and hydrogen peroxide: design of a screening workflow to identify inhibitors of NADPH oxidases. J. Biol. Chem. 2014;289(23):16176–16189. doi: 10.1074/jbc.M114.548693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suh Y., Lee S.-J. KRAS-driven ROS promote malignant transformation. Mol. Cell. Oncol. 2015;2(1):e968059. doi: 10.4161/23723548.2014.968059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dikalov S. Cross talk between mitochondria and NADPH oxidases. Free Radic. Biol. Med. 2011;51(7):1289–1301. doi: 10.1016/j.freeradbiomed.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tampo Y., Kotamraju S., Chitambar C.R., Kalivendi S.V., Keszler A., Joseph J., Kalyanaraman B. Oxidative stress-induced iron signaling is responsible for peroxide-dependent oxidation of dichlorodihydrofluorescein in endothelial cells: role of transferrin receptor-dependent iron uptake in apoptosis. Circ. Res. 2003;92(1):56–63. doi: 10.1161/01.res.0000048195.15637.ac. [DOI] [PubMed] [Google Scholar]
- 51.Diehn M., Cho R.W., Lobo N.A., Kalisky T., Dorie M.J., Kulp A.N., Qian D., Lam J.S., Ailles L.E., Wong M., Joshua B., Kaplan M.J., Wapnir I., Dirbas F.M., Somlo G., Garberoglio C., Paz B., Shen J., Lau S.K., Quake S.R., Brown J.M., Weissman I.L., Clarke M.F. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458(7239):780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saretzki G., Walter T., Atkinson S., Passos J.F., Bareth B., Keith W.N., Stewart R., Hoare S., Stojkovic M., Armstrong L., von Zglinicki T., Lako M. Downregulation of multiple stress defense mechanisms during differentiation of human embryonic stem cells. Stem Cells. 2008;26(2):455–464. doi: 10.1634/stemcells.2007-0628. [DOI] [PubMed] [Google Scholar]
- 53.Finley L.W., Carracedo A., Lee J., Souza A., Egia A., Zhang J., Teruya-Feldstein J., Moreira P.I., Cardoso S.M., Clish C.B., Pandolfi P.P., Haigis M.C. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell. 2011;19(3):416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Costa A., Scholer-Dahirel A., Mechta-Grigoriou F. The role of reactive oxygen species and metabolism on cancer cells and their microenvironment. Semin. Cancer Biol. 2014;25:23–32. doi: 10.1016/j.semcancer.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 55.Kalyanaraman B., Cheng G., Hardy M., Ouari O., Sikora A., Zielonka J., Dwinell M. Mitochondria-targeted metformins: anti-tumour and redox signalling mechanisms. Interface Focus. 2017;7(2):20160109. doi: 10.1098/rsfs.2016.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zielonka J., Joseph J., Sikora A., Kalyanaraman B. Real-time monitoring of reactive oxygen and nitrogen species in a multiwell plate using the diagnostic marker products of specific probes. Methods Enzymol. 2013:145–157. doi: 10.1016/B978-0-12-405883-5.00009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zielonka J., Zielonka M., Sikora A., Adamus J., Joseph J., Hardy M., Ouari O., Dranka B.P., Kalyanaraman B. Global profiling of reactive oxygen and nitrogen species in biological systems: high-throughput real-time analyses. J. Biol. Chem. 2012;287(5):2984–2995. doi: 10.1074/jbc.M111.309062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dranka B.P., Zielonka J., Kanthasamy A.G., Kalyanaraman B. Alterations in bioenergetic function induced by Parkinson's disease mimetic compounds: lack of correlation with superoxide generation. J. Neurochem. 2012;122(5):941–951. doi: 10.1111/j.1471-4159.2012.07836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng G., Zielonka J., Ouari O., Lopez M., McAllister D., Boyle K., Barrios C.S., Weber J.J., Johnson B.D., Hardy M., Dwinell M.B., Kalyanaraman B. Mitochondria-targeted analogues of metformin exhibit enhanced antiproliferative and radiosensitizing effects in pancreatic cancer cells. Cancer Res. 2016;76(13):3904–3915. doi: 10.1158/0008-5472.CAN-15-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng G., Zielonka J., McAllister D., Tsai S., Dwinell M.B., Kalyanaraman B. Profiling and targeting of cellular bioenergetics: inhibition of pancreatic cancer cell proliferation. Br. J. Cancer. 2014;111(1):85–93. doi: 10.1038/bjc.2014.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Michalski R., Zielonka J., Gapys E., Marcinek A., Joseph J., Kalyanaraman B. Real-time measurements of amino acid and protein hydroperoxides using coumarin boronic acid. J. Biol. Chem. 2014;289(32):22536–22553. doi: 10.1074/jbc.M114.553727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kalyanaraman B., Hardy M., Podsiadly R., Cheng G., Zielonka J. Recent developments in detection of superoxide radical anion and hydrogen peroxide: opportunities, challenges, and implications in redox signaling. Arch. Biochem. Biophys. 2017;617:38–47. doi: 10.1016/j.abb.2016.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kalyanaraman B., Dranka B.P., Hardy M., Michalski R., Zielonka J. HPLC-based monitoring of products formed from hydroethidine-based fluorogenic probes--the ultimate approach for intra- and extracellular superoxide detection. Biochim. Biophys. Acta. 2014;1840(2):739–744. doi: 10.1016/j.bbagen.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tan A.S., Baty J.W., Dong L.F., Bezawork-Geleta A., Endaya B., Goodwin J., Bajzikova M., Kovarova J., Peterka M., Yan B., Pesdar E.A., Sobol M., Filimonenko A., Stuart S., Vondrusova M., Kluckova K., Sachaphibulkij K., Rohlena J., Hozak P., Truksa J., Eccles D., Haupt L.M., Griffiths L.R., Neuzil J., Berridge M.V. Mitochondrial genome acquisition restores respiratory function and tumorigenic potential of cancer cells without mitochondrial DNA. Cell Metab. 2015;21(1):81–94. doi: 10.1016/j.cmet.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 65.Porporato P.E., Payen V.L., Perez-Escuredo J., De Saedeleer C.J., Danhier P., Copetti T., Dhup S., Tardy M., Vazeille T., Bouzin C., Feron O., Michiels C., Gallez B., Sonveaux P. A mitochondrial switch promotes tumor metastasis. Cell Rep. 2014;8(3):754–766. doi: 10.1016/j.celrep.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 66.Zielonka J., Vasquez-Vivar J., Kalyanaraman B. Detection of 2-hydroxyethidium in cellular systems: a unique marker product of superoxide and hydroethidine. Nat. Protoc. 2008;3(1):8–21. doi: 10.1038/nprot.2007.473. [DOI] [PubMed] [Google Scholar]
- 67.Cheng G., Zielonka J., Kalyanaraman B. Modulatory effects of MitoSOX On cellular bioenergetics: a cautionary note. Free Radic. Biol. Med. 2011;51(Suppl.):S37–S38. [Google Scholar]
- 68.Shchepinova M.M., Cairns A.G., Prime T.A., Logan A., James A.M., Hall A.R., Vidoni S., Arndt S., Caldwell S.T., Prag H.A., Pell V.R., Krieg T., Mulvey J.F., Yadav P., Cobley J.N., Bright T.P., Senn H.M., Anderson R.F., Murphy M.P., Hartley R.C. MitoNeoD: a mitochondria-targeted superoxide probe. Cell Chem. Biol. 2017;24(10):1285–1298. doi: 10.1016/j.chembiol.2017.08.003. (e12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang J., Yi J. Cancer cell killing via ROS: to increase or decrease, that is the question. Cancer Biol. Ther. 2008;7(12):1875–1884. doi: 10.4161/cbt.7.12.7067. [DOI] [PubMed] [Google Scholar]
- 70.Seifried H.E., McDonald S.S., Anderson D.E., Greenwald P., Milner J.A. The antioxidant conundrum in cancer. Cancer Res. 2003;63(15):4295–4298. [PubMed] [Google Scholar]
- 71.Xiang Y., Stine Z.E., Xia J., Lu Y., O'Connor R.S., Altman B.J., Hsieh A.L., Gouw A.M., Thomas A.G., Gao P., Sun L., Song L., Yan B., Slusher B.S., Zhuo J., Ooi L.L., Lee C.G., Mancuso A., McCallion A.S., Le A., Milone M.C., Rayport S., Felsher D.W., Dang C.V. Targeted inhibition of tumor-specific glutaminase diminishes cell-autonomous tumorigenesis. J. Clin. Investig. 2015;125(6):2293–2306. doi: 10.1172/JCI75836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Storz P. KRas, ROS and the initiation of pancreatic cancer. Small GTPases. 2017;8(1):38–42. doi: 10.1080/21541248.2016.1192714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liou G.Y., Storz P. Reactive oxygen species in cancer. Free Radic. Res. 2010;44(5):479–496. doi: 10.3109/10715761003667554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wangpaichitr M., Kandemir H., Li Y.Y., Wu C., Nguyen D., Feun L.G., Kuo M.T., Savaraj N. Relationship of metabolic alterations and PD-L1 expression in cisplatin resistant lung cancer. Cell Dev. Biol. 2017;6(2) doi: 10.4172/2168-9296.1000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wangpaichitr M., Wu C., Li Y.Y., Nguyen D.J.M., Kandemir H., Shah S., Chen S., Feun L.G., Prince J.S., Kuo M.T., Savaraj N. Exploiting ROS and metabolic differences to kill cisplatin resistant lung cancer. Oncotarget. 2017;8(30):49275–49292. doi: 10.18632/oncotarget.17568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hientz K., Mohr A., Bhakta-Guha D., Efferth T. The role of p53 in cancer drug resistance and targeted chemotherapy. Oncotarget. 2017;8(5):8921–8946. doi: 10.18632/oncotarget.13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Amoedo N.D., Obre E., Rossignol R. Drug discovery strategies in the field of tumor energy metabolism: limitations by metabolic flexibility and metabolic resistance to chemotherapy. Biochim. Biophys. Acta. 2017;1858(8):674–685. doi: 10.1016/j.bbabio.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 78.Hudson C.D., Savadelis A., Nagaraj A.B., Joseph P., Avril S., DiFeo A., Avril N. Altered glutamine metabolism in platinum resistant ovarian cancer. Oncotarget. 2016;7(27):41637–41649. doi: 10.18632/oncotarget.9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nicholls D.G., Darley-Usmar V.M., Wu M., Jensen P.B., Rogers G.W., Ferrick D.A. Bioenergetic profile experiment using C2C12 myoblast cells. J. Vis. Exp.: JoVE. 2010;46:2511. doi: 10.3791/2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pollak M. Targeting oxidative phosphorylation: why, when, and how. Cancer Cell. 2013;23(3):263–264. doi: 10.1016/j.ccr.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 81.Grassian A.R., Parker S.J., Davidson S.M., Divakaruni A.S., Green C.R., Zhang X., Slocum K.L., Pu M., Lin F., Vickers C., Joud-Caldwell C., Chung F., Yin H., Handly E.D., Straub C., Growney J.D., Vander Heiden M.G., Murphy A.N., Pagliarini R., Metallo C.M. IDH1 mutations alter citric acid cycle metabolism and increase dependence on oxidative mitochondrial metabolism. Cancer Res. 2014;74(12):3317–3331. doi: 10.1158/0008-5472.CAN-14-0772-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cheng G., Zielonka J., McAllister D.M., Mackinnon A.C., Jr., Joseph J., Dwinell M.B., Kalyanaraman B. Mitochondria-targeted vitamin E analogs inhibit breast cancer cell energy metabolism and promote cell death. BMC Cancer. 2013;13:285. doi: 10.1186/1471-2407-13-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kalyanaraman B., Cheng G., Hardy M., Ouari O., Lopez M., Joseph J., Zielonka J., Dwinell M.B. A review of the basics of mitochondrial bioenergetics, metabolism, and related signaling pathways in cancer cells: therapeutic targeting of tumor mitochondria with lipophilic cationic compounds. Redox Biol. 2017;14:316–327. doi: 10.1016/j.redox.2017.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zielonka J., Joseph J., Sikora A., Hardy M., Ouari O., Vasquez-Vivar J., Cheng G., Lopez M., Kalyanaraman B. Mitochondria-targeted triphenylphosphonium-based compounds: syntheses, mechanisms of action, and therapeutic and diagnostic applications. Chem. Rev. 2017;117(15):10043–10120. doi: 10.1021/acs.chemrev.7b00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Roelofs B.A., Ge S.X., Studlack P.E., Polster B.M. Low micromolar concentrations of the superoxide probe MitoSOX uncouple neural mitochondria and inhibit complex IV. Free Radic. Biol. Med. 2015;86:250–258. doi: 10.1016/j.freeradbiomed.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pokrzywinski K.L., Biel T.G., Kryndushkin D., Rao V.A. Therapeutic targeting of the mitochondria initiates excessive superoxide production and mitochondrial depolarization causing decreased mtDNA integrity. PLoS One. 2016;11(12):e0168283. doi: 10.1371/journal.pone.0168283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kalyanaraman B., Cheng G., Hardy M., Ouari O., Sikora A., Zielonka J., Dwinell M.B. Modified metformin as a more potent anticancer drug: mitochondrial inhibition, redox signaling, antiproliferative effects and future EPR studies. Cell Biochem. Biophys. 2017 doi: 10.1007/s12013-017-0796-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu K., Shang H., Kong X., Ren M., Wang J.Y., Liu Y., Lin W. A novel near-infrared fluorescent probe for H2O2 in alkaline environment and the application for H2O2 imaging in vitro and in vivo. Biomaterials. 2016;100:162–171. doi: 10.1016/j.biomaterials.2016.05.029. [DOI] [PubMed] [Google Scholar]
- 89.Dhanasekaran A., Kotamraju S., Karunakaran C., Kalivendi S.V., Thomas S., Joseph J., Kalyanaraman B. Mitochondria superoxide dismutase mimetic inhibits peroxide-induced oxidative damage and apoptosis: role of mitochondrial superoxide. Free Radic. Biol. Med. 2005;39(5):567–583. doi: 10.1016/j.freeradbiomed.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 90.Krishna M.C., Russo A., Mitchell J.B., Goldstein S., Dafni H., Samuni A. Do nitroxide antioxidants act as scavengers of O2-. or as SOD mimics? J. Biol. Chem. 1996;271(42):26026–26031. doi: 10.1074/jbc.271.42.26026. [DOI] [PubMed] [Google Scholar]
- 91.Cunniff B., Benson K., Stumpff J., Newick K., Held P., Taatjes D., Joseph J., Kalyanaraman B., Heintz N.H. Mitochondrial-targeted nitroxides disrupt mitochondrial architecture and inhibit expression of peroxiredoxin 3 and FOXM1 in malignant mesothelioma cells. J. Cell. Physiol. 2013;228(4):835–845. doi: 10.1002/jcp.24232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Starenki D., Park J.I. Mitochondria-targeted nitroxide, Mito-CP, suppresses medullary thyroid carcinoma cell survival in vitro and in vivo. J. Clin. Endocrinol. Metab. 2013;98(4):1529–1540. doi: 10.1210/jc.2012-3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hong S.K., Starenki D., Wu P.K., Park J.I. Suppression of B-Raf(V600E) melanoma cell survival by targeting mitochondria using triphenyl-phosphonium-conjugated nitroxide or ubiquinone. Cancer Biol. Ther. 2017;18(2):106–114. doi: 10.1080/15384047.2016.1250987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cheng G., Zielonka J., McAllister D., Hardy M., Ouari O., Joseph J., Dwinell M.B., Kalyanaraman B. Antiproliferative effects of mitochondria-targeted cationic antioxidants and analogs: role of mitochondrial bioenergetics and energy-sensing mechanism. Cancer Lett. 2015;365(1):96–106. doi: 10.1016/j.canlet.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Weinberg F., Hamanaka R., Wheaton W.W., Weinberg S., Joseph J., Lopez M., Kalyanaraman B., Mutlu G.M., Budinger G.R., Chandel N.S. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc. Natl. Acad. Sci. USA. 2010;107(19):8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nazarewicz R.R., Dikalova A., Bikineyeva A., Ivanov S., Kirilyuk I.A., Grigor'ev I.A., Dikalov S.I. Does scavenging of mitochondrial superoxide attenuate cancer prosurvival signaling pathways? Antioxid. Redox Signal. 2013;19(4):344–349. doi: 10.1089/ars.2013.5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Beavers W.N., Rose K.L., Galligan J.J., Mitchener M.M., Rouzer C.A., Tallman K.A., Lamberson C.R., Wang X., Hill S., Ivanova P.T., Brown H.A., Zhang B., Porter N.A., Marnett L.J. Protein modification by endogenously generated lipid electrophiles: mitochondria as the source and target. ACS Chem. Biol. 2017;12(8):2062–2069. doi: 10.1021/acschembio.7b00480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Peskin A.V., Low F.M., Paton L.N., Maghzal G.J., Hampton M.B., Winterbourn C.C. The high reactivity of peroxiredoxin 2 with H(2)O(2) is not reflected in its reaction with other oxidants and thiol reagents. J. Biol. Chem. 2007;282(16):11885–11892. doi: 10.1074/jbc.M700339200. [DOI] [PubMed] [Google Scholar]
- 99.Perkins A., Nelson K.J., Parsonage D., Poole L.B., Karplus P.A. Peroxiredoxins: guardians against oxidative stress and modulators of peroxide signaling. Trends Biochem. Sci. 2015;40(8):435–445. doi: 10.1016/j.tibs.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Andreyev A.Y., Kushnareva Y.E., Murphy A.N., Starkov A.A. Mitochondrial ROS metabolism: 10 years later, biochemistry. Biokhimiia. 2015;80(5):517–531. doi: 10.1134/S0006297915050028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gutscher M., Sobotta M.C., Wabnitz G.H., Ballikaya S., Meyer A.J., Samstag Y., Dick T.P. Proximity-based protein thiol oxidation by H2O2-scavenging peroxidases. J. Biol. Chem. 2009;284(46):31532–31540. doi: 10.1074/jbc.M109.059246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jarvis R.M., Hughes S.M., Ledgerwood E.C. Peroxiredoxin 1 functions as a signal peroxidase to receive, transduce, and transmit peroxide signals in mammalian cells. Free Radic. Biol. Med. 2012;53(7):1522–1530. doi: 10.1016/j.freeradbiomed.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 103.Sobotta M.C., Liou W., Stöcker S., Talwar D., Oehler M., Ruppert T., Scharf A.N.D., Dick T.P. Peroxiredoxin-2 and STAT3 form a redox relay for H2O2 signaling. Nat. Chem. Biol. 2015;11(1):64–70. doi: 10.1038/nchembio.1695. [DOI] [PubMed] [Google Scholar]
- 104.Lee C.U., Hahne G., Hanske J., Bange T., Bier D., Rademacher C., Hennig S., Grossmann T.N. Redox modulation of PTEN phosphatase activity by hydrogen peroxide and bisperoxidovanadium complexes. Angew. Chem. (Int. Ed. Engl.) 2015;54(46):13796–13800. doi: 10.1002/anie.201506338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Morgan M.J., Liu Z.G. Crosstalk of reactive oxygen species and NF-kappaB signaling. Cell Res. 2011;21(1):103–115. doi: 10.1038/cr.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jin L., Li D., Alesi G.N., Fan J., Kang H.B., Lu Z., Boggon T.J., Jin P., Yi H., Wright E.R., Duong D., Seyfried N.T., Egnatchik R., DeBerardinis R.J., Magliocca K.R., He C., Arellano M.L., Khoury H.J., Shin D.M., Khuri F.R., Kang S. Glutamate dehydrogenase 1 signals through antioxidant glutathione peroxidase 1 to regulate redox homeostasis and tumor growth. Cancer Cell. 2015;27(2):257–270. doi: 10.1016/j.ccell.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang J., Lin D., Peng H., Huang Y., Huang J., Gu J. Cancer-derived immunoglobulin G promotes tumor cell growth and proliferation through inducing production of reactive oxygen species. Cell Death Dis. 2013;4(12):e945. doi: 10.1038/cddis.2013.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sanz A. Mitochondrial reactive oxygen species: do they extend or shorten animal lifespan? Biochim. Biophys. Acta. 2016;1857(8):1116–1126. doi: 10.1016/j.bbabio.2016.03.018. [DOI] [PubMed] [Google Scholar]
- 109.Gorrini C., Harris I.S., Mak T.W. Modulation of oxidative stress as an anticancer strategy. Nat. Rev. Drug Discov. 2013;12(12):931–947. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- 110.Vazquez F., Lim J.H., Chim H., Bhalla K., Girnun G., Pierce K., Clish C.B., Granter S.R., Widlund H.R., Spiegelman B.M., Puigserver P. PGC1alpha expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell. 2013;23(3):287–301. doi: 10.1016/j.ccr.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Haq R., Fisher D.E., Widlund H.R. Molecular pathways: BRAF induces bioenergetic adaptation by attenuating oxidative phosphorylation, clinical cancer research: an official journal of the American Association for. Cancer Res. 2014;20(9):2257–2263. doi: 10.1158/1078-0432.CCR-13-0898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Torrens-Mas M., Oliver J., Roca P., Sastre-Serra J. SIRT3: oncogene and tumor suppressor in cancer. Cancers. 2017;9(7):90. doi: 10.3390/cancers9070090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kim M.H., Kim H. Oncogenes and tumor suppressors regulate glutamine metabolism in cancer cells. J. Cancer Prev. 2013;18(3):221–226. doi: 10.15430/JCP.2013.18.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lu W., Pelicano H., Huang P. Cancer metabolism: is glutamine sweeter than glucose? Cancer Cell. 2010;18(3):199–200. doi: 10.1016/j.ccr.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Montero J., Dutta C., van Bodegom D., Weinstock D., Letai A. p53 regulates a non-apoptotic death induced by ROS. Cell Death Differ. 2013;20(11):1465–1474. doi: 10.1038/cdd.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tan Z., Luo X., Xiao L., Tang M., Bode A.M., Dong Z., Cao Y. The role of PGC1alpha in cancer metabolism and its therapeutic implications. Mol. Cancer Ther. 2016;15(5):774–782. doi: 10.1158/1535-7163.MCT-15-0621. [DOI] [PubMed] [Google Scholar]
- 117.Vyas S., Zaganjor E., Haigis M.C. Mitochondria and Cancer. Cell. 2016;166(3):555–566. doi: 10.1016/j.cell.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.D'Errico I., Salvatore L., Murzilli S., Lo Sasso G., Latorre D., Martelli N., Egorova A.V., Polishuck R., Madeyski-Bengtson K., Lelliott C., Vidal-Puig A.J., Seibel P., Villani G., Moschetta A. Peroxisome proliferator-activated receptor-gamma coactivator 1-alpha (PGC1alpha) is a metabolic regulator of intestinal epithelial cell fate. Proc. Natl. Acad. Sci. USA. 2011;108(16):6603–6608. doi: 10.1073/pnas.1016354108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lim J.H., Luo C., Vazquez F., Puigserver P. Targeting mitochondrial oxidative metabolism in melanoma causes metabolic compensation through glucose and glutamine utilization. Cancer Res. 2014;74(13):3535–3545. doi: 10.1158/0008-5472.CAN-13-2893-T. [DOI] [PubMed] [Google Scholar]
- 120.D'Errico I., Lo Sasso G., Salvatore L., Murzilli S., Martelli N., Cristofaro M., Latorre D., Villani G., Moschetta A. Bax is necessary for PGC1alpha pro-apoptotic effect in colorectal cancer cells. Cell Cycle. 2011;10(17):2937–2945. doi: 10.4161/cc.10.17.16791. [DOI] [PubMed] [Google Scholar]
- 121.LaGory E.L., Wu C., Taniguchi C.M., Ding C.C., Chi J.T., von Eyben R., Scott D.A., Richardson A.D., Giaccia A.J. Suppression of PGC-1alpha Is critical for reprogramming oxidative metabolism in renal cell carcinoma. Cell Rep. 2015;12(1):116–127. doi: 10.1016/j.celrep.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Durand N., Storz P. Targeting reactive oxygen species in development and progression of pancreatic cancer. Expert Rev. Anticancer Ther. 2017;17(1):19–31. doi: 10.1080/14737140.2017.1261017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chandel N.S., Tuveson D.A. The promise and perils of antioxidants for cancer patients. N. Engl. J. Med. 2014;371(2):177–178. doi: 10.1056/NEJMcibr1405701. [DOI] [PubMed] [Google Scholar]
- 124.Sayin V.I., Ibrahim M.X., Larsson E., Nilsson J.A., Lindahl P., Bergo M.O. Antioxidants accelerate lung cancer progression in mice. Sci. Transl. Med. 2014;6(221):221ra15. doi: 10.1126/scitranslmed.3007653. [DOI] [PubMed] [Google Scholar]
- 125.Piskounova E., Agathocleous M., Murphy M.M., Hu Z., Huddlestun S.E., Zhao Z., Leitch A.M., Johnson T.M., DeBerardinis R.J., Morrison S.J. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature. 2015;527(7577):186–191. doi: 10.1038/nature15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schaar C.E., Dues D.J., Spielbauer K.K., Machiela E., Cooper J.F., Senchuk M., Hekimi S., Van Raamsdonk J.M. Mitochondrial and cytoplasmic ROS have opposing effects on lifespan. PLoS Genet. 2015;11(2):e1004972. doi: 10.1371/journal.pgen.1004972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Omenn G.S., Goodman G.E., Thornquist M.D., Balmes J., Cullen M.R., Glass A., Keogh J.P., Meyskens F.L., Valanis B., Williams J.H., Barnhart S., Hammar S. Effects of a combination of beta carotene and vitamin A on lung cancer and cardiovascular disease. N. Engl. J. Med. 1996;334(18):1150–1155. doi: 10.1056/NEJM199605023341802. [DOI] [PubMed] [Google Scholar]
- 128.Gao X., Schottker B. Reduction-oxidation pathways involved in cancer development: a systematic review of literature reviews. Oncotarget. 2017;8(31):51888–51906. doi: 10.18632/oncotarget.17128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Denu J.M., Tanner K.G. Specific and reversible inactivation of protein tyrosine phosphatases by hydrogen peroxide: evidence for a sulfenic acid intermediate and implications for redox regulation. Biochemistry. 1998;37(16):5633–5642. doi: 10.1021/bi973035t. [DOI] [PubMed] [Google Scholar]
- 130.Sullivan L.B., Chandel N.S. Mitochondrial reactive oxygen species and cancer. Cancer Metab. 2014;2:17. doi: 10.1186/2049-3002-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Keniry M., Parsons R. The role of PTEN signaling perturbations in cancer and in targeted therapy. Oncogene. 2008;27(41):5477–5485. doi: 10.1038/onc.2008.248. [DOI] [PubMed] [Google Scholar]
- 132.Greer E.L., Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24(50):7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- 133.Zhang X., Tang N., Hadden T.J., Rishi A.K. Akt, FoxO and regulation of apoptosis. Biochim. Biophys. Acta. 2011;1813(11):1978–1986. doi: 10.1016/j.bbamcr.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 134.Roberts P.J., Der C.J. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26(22):3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 135.Hart P.C., Mao M., de Abreu A.L., Ansenberger-Fricano K., Ekoue D.N., Ganini D., Kajdacsy-Balla A., Diamond A.M., Minshall R.D., Consolaro M.E., Santos J.H., Bonini M.G. MnSOD upregulation sustains the Warburg effect via mitochondrial ROS and AMPK-dependent signalling in cancer. Nat. Commun. 2015;6:6053. doi: 10.1038/ncomms7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Benfeitas R., Uhlen M., Nielsen J., Mardinoglu A. New challenges to study heterogeneity in cancer redox metabolism. Front. Cell Dev. Biol. 2017;5:65. doi: 10.3389/fcell.2017.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kong B., Qia C., Erkan M., Kleeff J., Michalski C.W. Overview on how oncogenic Kras promotes pancreatic carcinogenesis by inducing low intracellular ROS levels. Front. Physiol. 2013;4:246. doi: 10.3389/fphys.2013.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tong Y.H., Zhang B., Fan Y., Lin N.M. Keap1-Nrf2 pathway: a promising target towards lung cancer prevention and therapeutics. Chronic Dis. Transl. Med. 2015;1(3):175–186. doi: 10.1016/j.cdtm.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yan Y., Finkel T. Autophagy as a regulator of cardiovascular redox homeostasis. Free Radic. Biol. Med. 2017;109:108–113. doi: 10.1016/j.freeradbiomed.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yang S., Wang X., Contino G., Liesa M., Sahin E., Ying H., Bause A., Li Y., Stommel J.M., Dell'antonio G., Mautner J., Tonon G., Haigis M., Shirihai O.S., Doglioni C., Bardeesy N., Kimmelman A.C. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25(7):717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Poillet-Perez L., Despouy G., Delage-Mourroux R., Boyer-Guittaut M. Interplay between ROS and autophagy in cancer cells, from tumor initiation to cancer therapy. Redox Biol. 2015;4:184–192. doi: 10.1016/j.redox.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lorin S., Codogno P., Djavaheri-Mergny M. Autophagy: a new concept in cancer research. Bull. du Cancer. 2008;95(1):43–50. doi: 10.1684/bdc.2008.0565. [DOI] [PubMed] [Google Scholar]
- 143.Hu D., Cao S., Zhang G., Xiao Y., Liu S., Shang Y. Florfenicol-induced mitochondrial dysfunction suppresses cell proliferation and autophagy in fibroblasts. Sci. Rep. 2017;7(1):13554. doi: 10.1038/s41598-017-13860-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kim K.Y., Park K.I., Kim S.H., Yu S.N., Park S.G., Kim Y.W., Seo Y.K., Ma J.Y., Ahn S.C. Inhibition of autophagy promotes salinomycin-induced apoptosis via reactive oxygen species-mediated PI3K/AKT/mTOR and ERK/p38 MAPK-dependent signaling in human prostate cancer cells. Int. J. Mol. Sci. 2017;18(5) doi: 10.3390/ijms18051088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mainz L., Rosenfeldt M.T. Autophagy and cancer - insights from mouse models. FEBS J. 2017 doi: 10.1111/febs.14274. [DOI] [PubMed] [Google Scholar]
- 146.Li T., Liu Y., Zhao J., Miao S., Xu Y., Liu K., Liu M., Wang G., Xiao X. Aggravation of acute kidney injury by mPGES-2 down regulation is associated with autophagy inhibition and enhanced apoptosis. Sci. Rep. 2017;7(1):10247. doi: 10.1038/s41598-017-10271-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kimura T., Takabatake Y., Takahashi A., Isaka Y. Chloroquine in cancer therapy: a double-edged sword of autophagy. Cancer Res. 2013;73(1):3–7. doi: 10.1158/0008-5472.CAN-12-2464. [DOI] [PubMed] [Google Scholar]
- 148.Biswas S.K. Metabolic reprogramming of immune cells in cancer progression. Immunity. 2015;43(3):435–449. doi: 10.1016/j.immuni.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 149.Corzo C.A., Cotter M.J., Cheng P., Cheng F., Kusmartsev S., Sotomayor E., Padhya T., McCaffrey T.V., McCaffrey J.C., Gabrilovich D.I. Mechanism regulating reactive oxygen species in tumor-induced myeloid-derived suppressor cells. J. Immunol. 2009;182(9):5693–5701. doi: 10.4049/jimmunol.0900092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hiramoto K., Satoh H., Suzuki T., Moriguchi T., Pi J., Shimosegawa T., Yamamoto M. Myeloid lineage-specific deletion of antioxidant system enhances tumor metastasis. Cancer Prev. Res. 2014;7(8):835–844. doi: 10.1158/1940-6207.CAPR-14-0094. [DOI] [PubMed] [Google Scholar]
- 151.Beury D.W., Carter K.A., Nelson C., Sinha P., Hanson E., Nyandjo M., Fitzgerald P.J., Majeed A., Wali N., Ostrand-Rosenberg S. Myeloid-derived suppressor cell survival and function are regulated by the transcription factor Nrf2. J. Immunol. 2016;196(8):3470–3478. doi: 10.4049/jimmunol.1501785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gomez-Mejiba S.E., Zhai Z., Della-Vedova M.C., Munoz M.D., Chatterjee S., Towner R.A., Hensley K., Floyd R.A., Mason R.P., Ramirez D.C. Immuno-spin trapping from biochemistry to medicine: advances, challenges, and pitfalls. Focus on protein-centered radicals. Biochim. Biophys. Acta. 2014;1840(2):722–729. doi: 10.1016/j.bbagen.2013.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Baynes J.W., Thorpe S.R. Role of oxidative stress in diabetic complications: a new perspective on an old paradigm. Diabetes. 1999;48(1):1–9. doi: 10.2337/diabetes.48.1.1. [DOI] [PubMed] [Google Scholar]
- 154.Bennett B., Helbling D., Meng H., Jarzembowski J., Geurts A.M., Friederich M.W., Van Hove J.L., Lawlor M.W., Dimmock D.P. Potentially diagnostic electron paramagnetic resonance spectra elucidate the underlying mechanism of mitochondrial dysfunction in the deoxyguanosine kinase deficient rat model of a genetic mitochondrial DNA depletion syndrome. Free Radic. Biol. Med. 2016;92:141–151. doi: 10.1016/j.freeradbiomed.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Myers C.R., Antholine W.E., Myers J.M. The pro-oxidant chromium(VI) inhibits mitochondrial complex I, complex II, and aconitase in the bronchial epithelium: EPR markers for Fe-S proteins. Free Radic. Biol. Med. 2010;49(12):1903–1915. doi: 10.1016/j.freeradbiomed.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kennedy M.C., Emptage M.H., Dreyer J.L., Beinert H. The role of iron in the activation-inactivation of aconitase. J. Biol. Chem. 1983;258(18):11098–11105. [PubMed] [Google Scholar]
- 157.Langley M., Ghosh A., Charli A., Sarkar S., Ay M., Luo J., Zielonka J., Brenza T., Bennett B., Jin H., Ghaisas S., Schlichtmann B., Kim D., Anantharam V., Kanthasamy A., Narasimhan B., Kalyanaraman B., Kanthasamy A.G. Mito-apocynin prevents mitochondrial dysfunction, microglial activation, oxidative damage, and progressive neurodegeneration in mitopark transgenic mice. Antioxid. Redox Signal. 2017;27(14):1048–1066. doi: 10.1089/ars.2016.6905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Johnson M.K., Thomson A.J., Richards A.J., Peterson J., Robinson A.E., Ramsay R.R., Singer T.P. Characterization of the Fe-S cluster in aconitase using low temperature magnetic circular dichroism spectroscopy. J. Biol. Chem. 1984;259(4):2274–2282. [PubMed] [Google Scholar]
- 159.Kennedy M.C., Antholine W.E., Beinert H. An EPR investigation of the products of the reaction of cytosolic and mitochondrial aconitases with nitric oxide. J. Biol. Chem. 1997;272(33):20340–20347. doi: 10.1074/jbc.272.33.20340. [DOI] [PubMed] [Google Scholar]
- 160.Vasquez-Vivar J., Kalyanaraman B., Kennedy M.C. Mitochondrial aconitase is a source of hydroxyl radical. An electron spin resonance investigation. J. Biol. Chem. 2000;275(19):14064–14069. doi: 10.1074/jbc.275.19.14064. [DOI] [PubMed] [Google Scholar]