Abstract
Protein-protein interactions (PPIs) are emerging as a promising new class of drug targets. Here, we present a novel high-throughput approach to screen inhibitors of PPIs in cells. We designed a library of 50,000 human peptide binding motifs and used a pooled lentiviral system to express them intracellularly and screen for their effects on cell proliferation. We thereby identified inhibitors that drastically reduced the viability of a pancreas cancer line (RWP1) while leaving a control line virtually unaffected. We identified their target interactions computationally, and validated a subset in experiments. We also discovered their potential mechanisms of action including apoptosis and cell cycle arrest. Finally, we confirmed that synthetic lipopeptide versions of our inhibitors have similarly specific and dosage dependent effects on cancer cell growth. Our screen reveals new drug targets and peptide drug leads and it provides a rich dataset covering phenotypes for inhibition of thousands of interactions.
Keywords: Cancer, peptide therapeutics, cell viability, lentivirus, next-generation sequencing, protein-protein interaction
Introduction
The productivity of drug discovery research programs has been in a long and steady decline since their heyday in the 1960s 1. One reason for this is a dearth in proven drug targets, with much research focus given to very few protein families proven to be “druggable” 2. Protein kinases comprise an instructive case; long thought to be undruggable, it took the success of imatinib in the 1990s for major kinase-inhibitor programs to be launched, resulting in a multitude of widely used drugs 3. Likewise, protein-protein interactions (PPIs) present a promising class of drug targets 4 that were long thought to be undruggable 5. Recently, inhibitors against a number of PPIs have emerged as candidates for new anti-cancer drugs 6, with notable examples being the inhibitors to the Bcl2-BH3 interaction 7, 8 and the p53-MDM2 interaction 9–12. These examples share two common features: The pro-apoptotic effects of inhibiting these interactions were first confirmed using peptide inhibitors before the costly development of small-molecule inhibitors. Second, the inhibitory peptides were based on short peptide sequences known as linear motifs (i.e., short conserved sequence motifs) 13, which are widespread and mediate a large fraction of all PPIs 14.
Here, we establish a high-throughput method for the screening of peptide inhibitors targeting motif-mediated interactions. We combine computational methods, oligonucleotide microarray synthesis and pooled lentiviral screening methodology; we screen directly for a desired phenotype – specific inhibition of cancer cell proliferation in this case. Briefly, we design a library of roughly 50,000 human peptide motifs and obtain their coding sequences using commercially available oligonucleotide arrays. We then clone this library into a lentiviral vector, such that the short peptide can be produced intracellularly on a GFP protein tag and perform a pooled lentiviral dropout screen analogous to established RNAi systems 15. We subsequently identify and verify the target interactions of peptides showing strong phenotypes. These identified interactions are likely druggable as they can evidently be inhibited by our peptides. Thus, our identified peptides represent valuable lead compounds for further drug development. This screen is highly scalable and a proteome-wide screen (using all ~500,000 potential motifs in the human proteome) can easily be envisioned. We thus present an exciting new venue for drug discovery.
Results
Construction of lentiviral library and pooled screening
The lentivirus system is a flexible and efficient way to deliver expression vectors into mammalian cells 16. It has been successfully applied and highly optimized for delivery of shRNAs in RNAi screens 15. We adopt it here to express GFP-tagged peptides intracellularly in order to explore their potential as PPI inhibitors. We then used an oligonucleotide library we previously developed 17 that contains 50,549 potential human peptide motifs (synthesized using custom oligonucleotide microarrays) and cloned these into lentiviral vectors (Fig. 1 and Online methods). A conceptually related approach was recently published that identifies bioactive extracellular peptides 18.
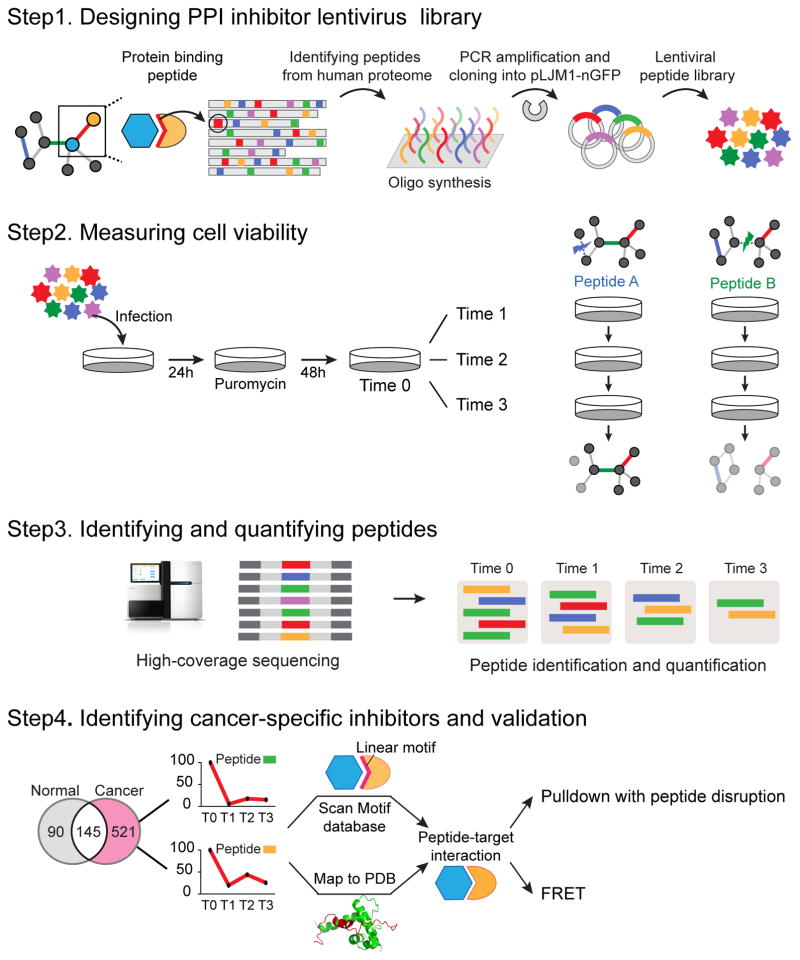
Figure 1. Overview of library design and identification of cancer-specific inhibitors.
(A) A library of 50,549 peptides containing interaction motifs was designed, synthesized on oligonucleotide arrays and cloned into a lentiviral library. (B) A dropout screen was performed with expression of the peptides after puromycin selection and cell collection of four time points in triplicates (at 0, 10, 15 and 24 days for RWP1 cells and 0, 9, 14 and 23 days for HEK283T cells). (C) Effect on cell viability of each peptide was quantified using high-coverage sequencing, with the read count of each peptide being an indication of the viability of cells expressing it. (D) The target of each peptide was identified using structural and bioinformatics methods and subsequently validated using pulldown and Förster resonance energy transfer (FRET) experiments.
We then performed a dropout viability screen to systematically identify PPIs involved in cell proliferation and survival whose inhibition could be therapeutically exploited. To this end, we performed a comparative study between a human pancreatic cancer cell line (RWP1, as a model for pancreatic cancer cells) and a human embryonic kidney cell line (HEK293T, as a control cell line). We will presume here that, to first approximation, effects seen in RWP1 and not in HEK293T are specific to this cell line, and, after further validation, to pancreas cancer. Of course, these cell lines are poor representations of cancerous and normal tissues, respectively, and observed effects will need to be validated in animal models. Cells were infected with lentivirus expressing GFP-tagged peptides for 24 hours, at a multiplicity of infection of 0.3, to ensure an integration of one virus particle per cell. Cells were subjected to puromycin selection for 48 hours in order to remove non-infected cells. The remaining cells were propagated, harvested at different time points in independent triplicates when cells reached confluency, and collected again at a final time point of 30 days post-infection (Fig. 1). For each cell line and each time point, genomic DNA was extracted and the peptide coding sequences amplified using Illumina barcoded primers. Finally, these were sequenced using a HiSeq sequencer. On average, we identified 49,750 peptides (98.42% of original library) at each time point of each cell line (Supplementary Results, Supplementary Table 1).
Analysis of the pooled dropout screen
By analyzing the read count of peptide coding sequences, we assess the relative abundance of a particular peptide (and thereby the viability of the cell line that expresses it) along the time course (Fig. 1). Analogous to previously established analysis methods for shRNA, we develop a dropout score that measures the magnitude of a peptide’s effect on cell viability. Briefly, the dropout score is a weighted average of the slope of the cell viability (see Online methods). In our data, dropout scores of less than −2.5 represent a reduction in cell viability by more than 50% (Supplementary Fig. 1A). Dropout scores of peptides are shown in Supplementary Dataset 1. When comparing dropout scores in RWP1 cells with those of HEK293T cells, we found that the distribution of dropout score in RWP1 cells is different from that of HEK293T cells implying that peptides have different effect on cell viability depending on cell type (Supplementary Fig. 1B). Indeed, the correlation between dropout scores in RWP1 cells and dropout scores in HEK293T cells is low (Pearson’s correlation coefficient is 0.047; Supplementary Fig. 1C), which in part underlines the differences in response of the two cell lines to our inhibitors.
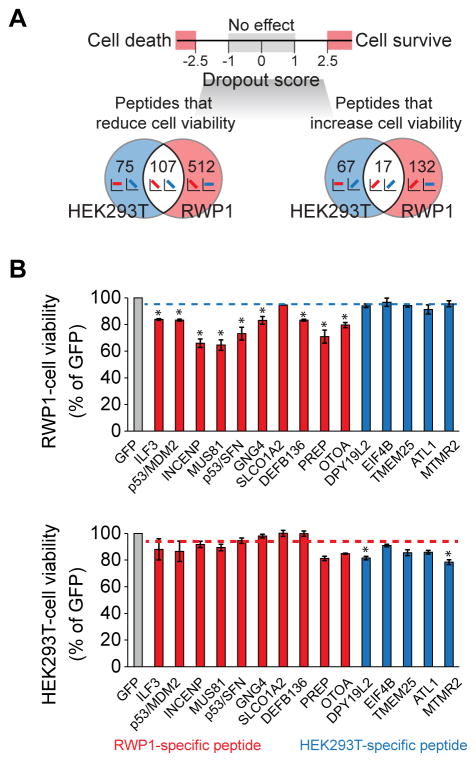
Next, we examined peptides that show distinct effect on cell viability in both cell lines (Fig 2A, see Online methods). We identified 512 peptide inhibitors that significantly reduce cancer cell viability (i.e., have a dropout score of RWP1 cells < −2.5) but do not affect HEK293T cell viability (−1 < dropout score of HEK293T cells < 1, Supplementary Dataset 2).
Figure 2. Identification and experimental validation of cancer-specific inhibitors.
(A) Venn diagram of identified peptides leading to dropout/increase specifically in RWP1 (cancer) or HEK293T (normal) cells, or leading to common dropout/increase. (B) Experimental validation of the effect of peptides on cell viability in single-lentivirus infection experiments. Red bars represent cancer (RWP1)-specific peptides and blue bars represent normal (HEK293T)-specific peptides. Blue dashed line indicates average cell viability of HEK293T-specific peptides in cancer (RWP1) cells. Meanwhile, red dashed line indicates average cell viability of RWP1-specific peptides in normal (HEK293T) cells. Statistical analysis was performed using Student’s t-test (2-tails) by comparing the cell viability of each RWP1-specific peptide against those of HEK293T-specific peptides (upper panel). Comparison of cell viability of each HEK293T-specific peptide against those of RWP1-specific peptides was also performed (lower panel). * P-value < 0.05. Experiments were done in triplicate. Data represent mean values ± s.d.
We then analyzed the cellular functions of proteins containing the peptides that lead to the reduction of cancer cell viability. Among these proteins, there is a significant enrichment of proteins involved in regulation of signaling pathways, cell death, cell cycle and nucleotide metabolism; processes that are critical for cancer pathogenesis (Supplementary Fig. 1D; hypergeometric test, P-value < 0.001). Importantly, the set of cancer-specific inhibitors are also highly enriched in cancer-related proteins (hypergeometric test, P-value: 2.13×10−23), with a strong correlation between the enrichment and the dropout score in RWP1 cancer cells, but not in HEK293T cells (Supplementary Fig. 1E). Interestingly, among our top candidates of cancer-specific inhibitors is a peptide derived from the p53 tumor suppressor, which is known to form part of the p53-MDM2 binding interface 19, 20. This peptide has previously been used to design various candidate compounds for anti-cancer therapy 9, 10, thus supporting the soundness of our approach.
We next validated the effect of 15 peptides shown by our screen to impair the cell viability of RWP1 or HEK293T cells (Supplementary Fig. 2A) in individual infection experiments (see Online methods for details). 9 out 10 of the tested RWP1-specific peptides conferred a decrease in RWP1 cell viability very similar to the effect observed in the pooled screen (average 75.63%, compared to 78.29% estimated effect after 72h in the pooled screen, see Online Methods), but had little or no effect on the viability of HEK293T cells (average 91.37%, compared to 104.45% in the pooled screen; Fig. 2B and Supplementary Table 2). Likewise, we could reproduce the effects of HEK293T-specific peptides through individual validations (84.45% and 86.00% of cell viability in HEK293T cells, in validation and pooled screens, respectively). Meanwhile, they did not affect the viability of RWP1 cells (94.31% and 99.12% of cell viability in validation and pooled screens, respectively; Fig. 2B and Supplementary Table 2). Overall, about 86% of the effects on cell viability observed in the pooled screen were observed in single peptide experiments. The correlation of the quantitative cell viability effects between the pooled and single peptide assays is about 0.4 (Pearson’s correlation coefficient, see Supplementary Fig. 2B). These peptides also showed a similar effect on the cell viabilities of other pancreatic cell lines (Panc 02.03; Supplementary Fig. 2C and Mia Paca-2; Supplementary Fig. 2D). We thus conclude that the results from pooled screens are reproducible in single-peptide infection experiments.
Determination of target interactions
Given a set of peptides with high efficacy we sought to determine their molecular targets. As the inhibitors in our library are peptides containing specific human motifs that mediate protein-protein interactions, it is likely they exert their phenotypic effects by inhibiting the very interactions they mediate. We therefore identified PPIs potentially targeted by the inhibitory peptides using two complementary approaches.
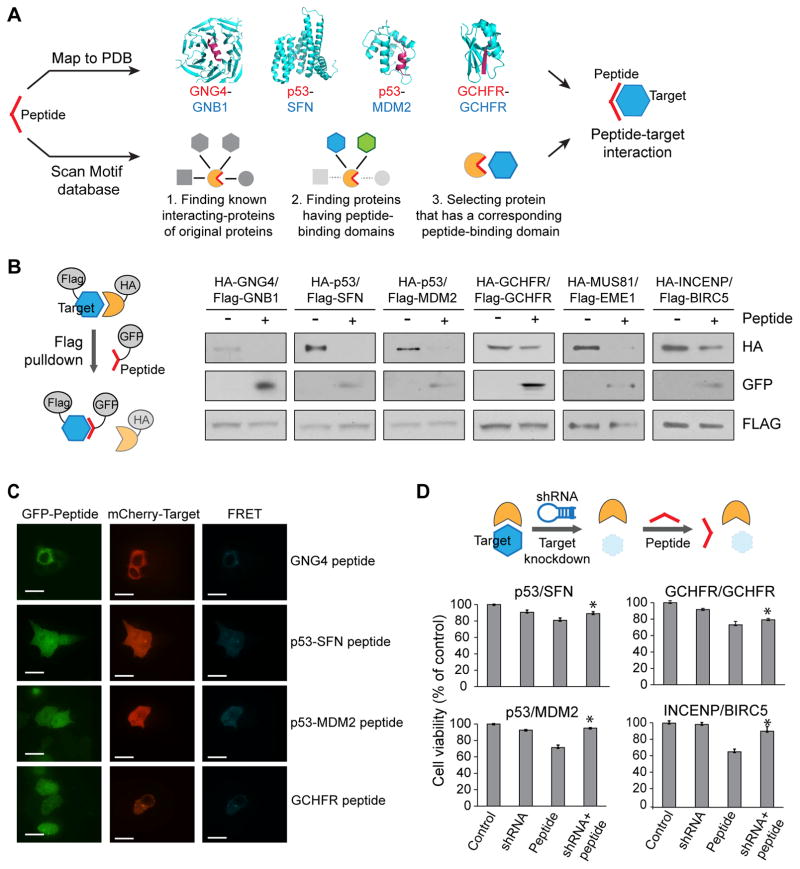
First, we mapped inhibitory peptides to known PDB structures of PPIs using straightforward sequence matching (Fig. 3A top). For example, the peptide KFFCTIL (derived from guanine nucleotide-binding protein subunit gamma-4, GNG4; red in Fig. 3A) maps to a structure of the GNG2/GNB1 complex (guanine nucleotide-binding protein subunit gamma-2/guanine nucleotide-binding protein subunit beta-1 complex; PDB ID: 1OMW), and it is reasonable to assume that it inhibits the homologous GNG4/GNB1 interaction. Using this method, we identified 133 PPIs as putative targets for our inhibitory peptides (Supplementary Dataset 3). Second, we exploited the compiled knowledge of PPIs mediated by short peptide motifs (Fig. 3A bottom). Briefly, we search all known linear motifs contained in our library and identify the interactions mediated by them in known PPI databases 21 by overlapping information about the binding motif 22 and the corresponding protein domain 23. We identified 973 such peptide-target interactions involving linear motifs that can bind to 12 known peptide-binding domain families (Supplementary Dataset 3).
Figure 3. Characterization of peptide-target interactions.
(A) Schematic view of the identification of interactions between peptides and targets. Peptides are either mapped directly to structures of protein interactions in the PDB or are mapped to known domain-motif interactions where both the linear motif and the protein domain involved in the interaction are known. (B) Evaluation of peptide-induced disruption between target and source protein in pulldown experiments. In all four tested cases, the expression of the peptide leads to disruption of the interaction. Also, the peptide is pulled down by its putative binding partner. Experiments were done in triplicate. Full gel images in Supplementary Fig. 11. (C) FRET assay for the determination of binding of peptides to targets. Strong FRET signal is seen, indicating direct in vivo interactions between the peptide and its putative binding partner. White scale bars represent 15 μm. (D) shRNA-induced cell viability rescue experiment. Cell viabilities are measured with cells stably expressing shRNA or transfected with the peptide. For the rescue experiment, cells stably expressing shRNA are transfected with the peptide. * P-value < 0.05. Experiments were done in triplicate. Data represent mean values ± s.d.
Among the 1,106 identified targets interactions, we find 131 that are inhibited by peptides found to have anti-cancer efficacy (cancer-specific inhibitors; dropout score < −2.5 in RWP1 and −1 < dropout score < 1 in HEK293T). Interestingly, we found that most interacting partners of these peptides (79.17%, 76 out of 96 partners) are involved in cancer pathogenesis (hypergeometric test, P-value = 4.08×10−12; Supplementary Dataset 3). For example, GNB1 (GNB1/GNG4 interaction is the putative target of the peptide KFFCTIL) is associated with the apoptosis pathway in breast cancer 24.
Network edgetics
Inspired by the paradigm of network medicine 25 and edgetics 26, we further examined the importance of the identified target interactions in a global network context. We thus mapped the 1,106 peptide-target interactions in a global protein-protein interaction network 21. We found that target interactions of cancer-specific inhibitors have significantly higher edge-betweenness (average edge-betweenness is 9.67) than target interactions of peptides with no viability effect in cancer (average edge-betweenness is 6.92 and Mann-Whitney U test, P-value = 3.74×10−3; Supplementary Fig. 3A). Furthermore, edge-betweenness of cancer-specific inhibitors is higher than those of target interactions of other peptides that are essential to both cell types (average edge-betweenness is 9.35) or only normal cells (average edge-betweenness is 7.51; Supplementary Fig 3A). This indicates that cancer-specific inhibitors-target interactions occupy more central and important interactions. Consistent with this, removal of peptide-target interactions of cancer-specific inhibitors lead to a larger increase in the number of interaction components (Bootstrap test, 1,000 replicates, P-value = 0.01) and decrease in the size of the largest component (Bootstrap test, 1,000 replicates, P-value = 0.02) in the PPI network than random interactions or target interactions of other peptides (Supplementary Fig. 3B).
Confirmation of PPI inhibition
We then sought to validate that the identified interactions are indeed inhibited by our peptide motifs. We thus performed a number of different validation assays aimed at determining whether A) our peptide can indeed mediate the interaction, i.e., bind to one of the interactors B) whether it will break the interaction and C) whether its phenotypic effect is likely mediated by breaking said interaction.
To address points A) and B), we performed pulldown experiments in HEK293T cells (Fig. 3B). We co-transfected cells with the target protein fused to a FLAG tag, the source protein of our peptide sequence fused to HA tag and the peptide fused to GFP (green fluorescent protein). The inhibition of the interaction was monitored by performing FLAG pull-down assays followed by Western blotting. We chose a total of nine to validate out of a total of 131 peptides that had both strong anti-proliferative activity and that we could map to target interactions (see above). We selected the based on highest scores and availability of target interaction partners as clones in Openfreezer, Out of these nice, we found that six (GNB1, SFN, MDM2, GCHFR, INCENP and MUS81) significantly pulled down the GFP-tagged peptides, indicating an interaction between the peptide and the target protein (Fig. 3B). Moreover, addressing the second point, the observed interactions between full-length proteins GNG4/GNB1, p53/SFN, INCENP/BIRC5, MUS81/EME1 and p53/MDM2 were significantly decreased upon co-expression of the peptides, thus supporting an inhibitory function (Fig 3B).
We then sought to obtain additional evidence for direct interactions between the inhibitory peptides and the putative target proteins. To this end, we applied Förster resonance energy transfer (FRET), an effective method to monitor direct protein–protein interactions in live specimen 27. We used FRET to evaluate the interaction between our peptides fused to GFP and their potential interacting protein partners fused to mCherry fluorescent protein. Interaction between four of the peptides and their interacting partners was monitored using FRET microscopy imaging (we tested the ones with positive pulldowns). Positive FRET signals were observed between p53/MDM2, p53/SFN, GCHFR and GNG4 peptides and their respective target proteins (Fig. 3C), supporting binary interactions in the cell based assays. By contrast no FRET signal was detected in control experiments (cells with GFP and/or mCherry; Supplementary Figure 4A). For quantification, we analyzed images of 25 cells for each interacting pair and found that more than 80% of cells have positive FRET signal (Supplementary Fig. 4B).
Although peptides tend to be more selective protein ligands than small molecules, non-selective effects cannot be excluded. Indeed, it is a possibility that even the cancer-specific inhibitors whose target interactions we validated (e.g., the inhibitor of the p53/SFN interaction) have other targets and that inhibition of the p53/SFN interaction alone does not lead to the desired phenotype. We therefore attempted to determine the underlying cause of the reduction in cancer cell viability conferred by our identified inhibitory peptides. To this end, we performed rescue-experiments based on the assumption that if the disruption of a target interaction is causing a phenotype, then removal of the target (and hence removal of the interaction) would eliminate the phenotypic effect of the peptide. A corollary would be that the phenotype is likely mediated by freeing the other interaction partner from the interaction (as is the case for p53/MDM2, where the freed P53 leads to apoptosis 28). We therefore compared the effect of the peptide in cell lines with the target present to their effect in cell lines where we previously knocked down the target gene using shRNA (Fig. 3D, Supplementary Fig. 5A and Supplementary Fig. 5B). We tested the 6 pairs that showed positive pulldown results above. In four cases, knockdown of the target showed rescue of the peptide phenotype, indicating that disruption of the target interaction is indeed the main mediator of the peptides’ phenotypes. These validated peptides include ETFSDLW (inhibiting p53/MDM2; Student’s t-test, P-value = 0.009), TEGPDSD (inhibiting p53/SFN; Student’s t-test, P-value = 0.008) and VWCLHKE (inhibiting GCHFR/GCHFR; Student’s t-test, P-value = 0.01). However, knockdown of GNB1 did not rescue the peptide phenotype, suggesting the possibility that the peptides are mediating their effects through binding to other proteins. Levels of gene inhibition (LNX1, MDM2, GCHFR, GNB1 and SFN) induced by shRNAs ranged from 53% to 94% (Supplementary Fig. 5B).
We thus possess evidence that a number of the validated peptides, which show strong phenotypic effects, A) directly interact with their putative target proteins B) disrupt their identified target interactions C) likely lead to the reduction of cancer cell viability by inhibiting the interaction in question.
Downstream effects
Having identified and validated a set of inhibitory peptides, we aimed to characterize their mechanism of action by investigating downstream effects. We specifically focused on the peptides targeting the p53/MDM2 (ETFSDLW), p53/SFN (TEGPDSD), GNG4/GNB1 (KFFCTIL), INCENP/BIRC5 (PSSLAYSLKKH) and MUS81/EME1 (RTLSQLYCSYGPLT) interactions. In addition to the RWP1 and HEK293T cell lines, we analyzed the effect of the peptides in two other pancreatic cell lines, namely Panc 02.03 and Mia paca-2.
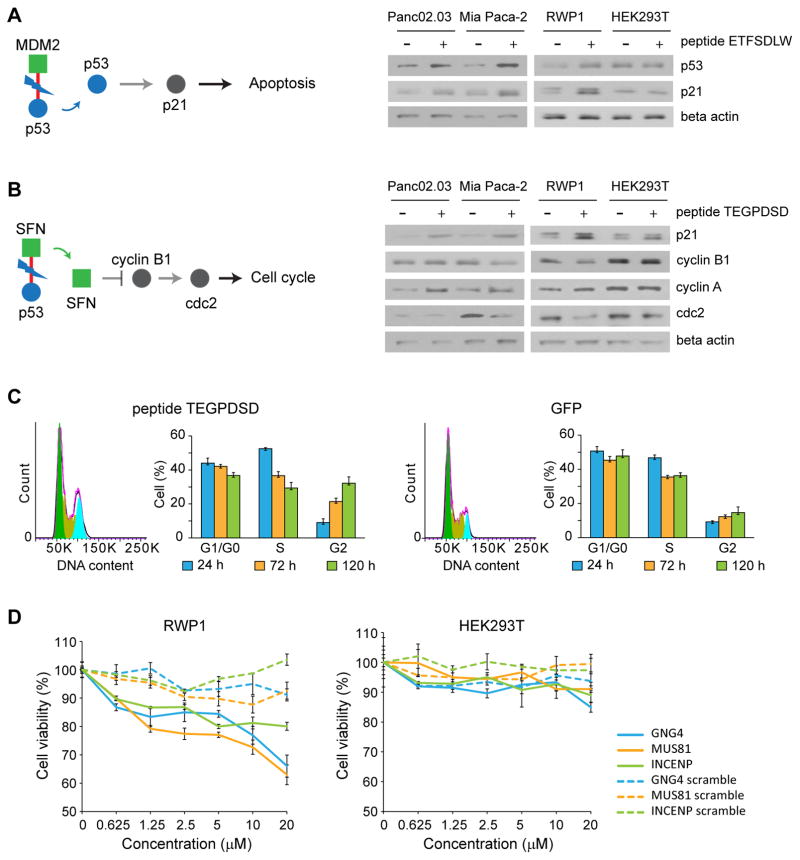
The peptide mediating the interaction between p53 and MDM2 has previously been shown to lead to apoptosis by inducing the accumulation of p53 20. In line with these results, our results indicate that expression of the p53/MDM2 inhibitory peptide ETFSDLW conferred a decrease in degradation of p53, leading to increase in p21 levels in RWP1, Panc 02.03 and Mia Paca-2 but not in HEK293T cells (Fig. 4A). These results suggest that the ETFSDLW peptide inhibits cell proliferation in RWP1, Panc 02.03 and Mia Paca-2 lines by increasing apoptosis.
Figure 4. Mechanism of action of cancer-specific inhibitors.
Downstream effect of peptide (A) ETFSDLW (inhibiting P53/MDM2). As is documented, disruption of this interaction leads to a decrease in degradation of P53, thereby increasing P53 levels, triggering apoptosis.. Experiments were done in triplicate. Full gel images in Supplementary Fig. 11. (B) TEGPDSD (inhibiting p53/SFN). Inhibition of this interaction appears to lead to decrease in cyclin B1 and cdc2 levels (and an increase in cyclin A), suggesting it may trigger cell cycle arrest. Blue circle represents source protein of peptide, and green square represents targets of peptide. Red line represents interaction between cancer-specific inhibitor and target. Experiments were done in triplicate. Full gel images in Supplementary Fig. 11. (C) Effect of peptide TEGPDSD on cell cycle. Fluorescence-activated cell sorting (FACS) analysis confirms that it leads to cell cycle arrest in G2. Experiments were done in triplicate. Data represent mean values ± s.d. (D) Effect of synthetic lipidated peptides on cell viability. Synthetic GNG4, MUS81 and INCENP derived lipopeptides show strong dosage dependent effects on cell viability in RWP1 cells, while scrambled versions do not affect cell viability. Experiments were done in duplicate. Data represent mean values ± s.d.
SFN is linked to cell cycle regulation 29. We therefore hypothesized that it could affect cell proliferation by regulating the expression of different cyclins and possibly lead to cell cycle arrest. Supporting this hypothesis, we found that expression of the p53/SFN inhibitory peptide (TEGPDSD) led to a reduced expression of both cyclin B1 and cdc2 in RWP1 and Mia Paca-2 cells (Fig. 4B). In contrast, the expression of this peptide led to an increase of cyclin A2 expression in RWP1, Panc 02.03 and Mia Paca-2 cells. To confirm the effect of the decrease in cyclin expression, we performed fluorescence-activated cell sorting (FACS) experiments to assess the cell cycle state and found that cells transfected with the SFN peptide indeed appear to arrest in G2 (Fig. 4C and Supplementary Fig. 6A). The percentage of GFP positive cells is similar between GFP alone and SFN peptide through different time points (Supplementary Fig. 6B). These data suggest that the peptide TEGPDSD inhibits cell viability by affecting cell cycle, most probably through a cell cycle arrest in G2. While the link of SFN to the cell cycle was previously been reported 29, we provide the first experimental data supporting the notion that inhibition of the p53/SFN interaction may lead to cell cycle arrest. An alternative interpretation is that 14-3-3 family proteins (SFN belongs to the 14-3-3 protein family) have been shown to stabilize the interaction of p53 with DNA 30 and general 14-3-3 inhibiting peptides have previously been shown to lead to apoptosis 31. As such, our TEGPDSD peptide could also lead to apoptosis via a general 14-3-3 mediated action, though the sequence similarity with previous, phage-derived peptides is low 32.
GNB1 has been linked to the phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K) and thus the AKT pathway 33. Indeed, it has been shown that the C-terminal half of GNB1 binds to Raf1 34, which is the downstream component of Ras signaling pathway. Ras is known regulator for AKT activity. Furthermore, it has been shown that GNB1 (K89E) activated RAS/MAPK and PI3K/AKT pathways 35. We hence hypothesized that the inhibition of the GNB1/GNG4 interaction would affect the activity of AKT by affecting its phosphorylation state. Indeed, expression of the peptide KFFCTIL in RWP1 and Panc 02.03 cells resulted in a decrease of the level of phospho-AKT (S473), and thus presumably AKT activity (Supplementary Fig. 7A). By contrast, it did not affect the level of phospho-AKT in HEK293T cells. Consistent with this, the peptide KFFCTIL also increased the expression of p21 in RWP1 and Panc 02.03 cells suggesting that the peptide may lead to an increase in apoptosis by inhibition of the AKT pathway. While GNB1 has previously been implicated in regulating apoptosis, we provide, to our knowledge, the first indication that inhibition of the GNB1/GNG4 interaction can be used to trigger apoptosis in cancer cell lines.
Knockdown of BIRC5 (survivin, the putative target of our INCENP peptide) causes apoptosis in neuroblastoma via p53 and CASP2 36. Furthermore, it has been shown that DNA damage induces a p53-survivin signaling pathway that results in down-regulation of survivin, cell cycle arrest, and apoptosis 37. In line with these evidences, we demonstrated that expression of the peptide PSSLAYSLKKH leads to an accumulation of p53 and p21 in RWP1, Panc 02.03 and Mia Paca-2 cells (Supplementary Fig. 7B). By contrast, it did not affect the level of p53 and p21 in HEK293T cells.
Finally, MUS81 and EME1 play key roles in DNA repair and the maintenance of genome integrity in mammalian cells 38, 39. It has been demonstrated that p53 acts as a critical checkpoint for the MUS81 repair pathway in response to ICL-induced DNA damage 39. In our study, we found that peptide RTLSQLYCSYGPLT increases the expression of p53 and p21 in RWP1 and Panc 02.03 cells but not in Mia Paca-2 and HEK293T cells (Supplementary Fig. 7C). These results suggest that the RTLSQLYCSYGPLT peptide can induce apoptosis through the p53 pathway.
Effects of synthetic inhibitory peptides
Delivery of peptides by transfection or infection (as we have in most experiments described above) is currently not a viable therapeutic option. We hence sought to investigate effects of synthetic analogs of our identified sequences. To make them membrane-permeable we turned to derivatization by a fatty acid 40 and thus synthesized N-terminally palmitoylated versions of the peptides (see Online methods). The choice of peptide was based on a combination of physicochemical properties and effect size. We first established that synthetic peptides indeed bind their intended targets in ITC measurements and find that the INCENP/BIRC5 inhibitory peptide binds its target with low micromolar affinity (Supplementary Fig. 8). We then performed cell viability assays to test whether the lipopeptide analogs would have similar effects as the intracellularly expressed versions. We used scrambled versions of the same peptides (i.e., same amino acid content with the sequence randomized. See Supplementary Table 3 for exact sequences) as controls. We find that the synthetic peptides have similar effects as the transfected ones with micromolar concentrations leading to a roughly 30% decrease of cell viability (see Fig. 4D), suggesting good cellular uptake. Furthermore, the scrambled versions having little to no effects on cell viability indicating that the synthesized peptides have specific effects and indeed inhibit their target protein-protein interaction. These lipopeptide compounds could be developed further as anti-cancer agents.
Discussion
Considering the vast number of protein-protein interactions and their involvement in virtually every biological process, development of potential new inhibitors to them is of course of great clinical importance. If intracellular PPI inhibitors can be established as a new class of drugs, it would enormously expand the number of proteins that can be targeted; thus there is huge potential here for future drugs. While a number of complications may have lessened initial enthusiasm towards intracellular PPI inhibitors, there have been encouraging recent developments, including the success of ABT-199.
We present a new platform to identify peptidic PPI inhibitors based on their phenotypic effect in cell lines and we show that we can successfully identify such inhibitors that have relatively strong effects as synthetic compounds when delivered into cells. As such, our technology holds great promise for the identification of new inhibitors that could potentially be developed into drugs.
We were able to identify a relatively large number of inhibitors that showed strong phenotype. This does partly reflect the fact that our initial library is clearly enriched in motifs that mediate protein-protein interactions. Also, it is likely indicative of the huge potential of PPI inhibitors as potential drugs; as our initial library is far from comprehensive, there are likely many fold more PPIs whose inhibition will lead to strong and specific phenotypic effects that could be exploited for therapeutic intervention.
Of course, it is a long development process from identification of efficacy in cell lines to an actual therapeutic and peptides in particular face a number or hurdles including lack of stability, relatively low specificity and, importantly, difficulty of cellular delivery. Because of the huge potential, this is a highly active area of research with many proposed solutions. For instance, many approaches to deliver proteins or peptides into the cell exist, including lipidation (which we used), liposomes, cell-penetrating peptides, customized bacterial toxins or nanoparticles.
We believe that our screen opens an exciting new venue for drug development, identifies a number of novel inhibitors that can be developed further and provides a rich dataset of the effects of thousands of motif-based peptide inhibitors in cells.
Online Methods
Cloning of the lentiviral library
To identify peptide inhibitors of protein-protein interaction, we used a previously designed human peptide library containing 50,549 heptamer C-terminal sequences, corresponding to 75,797 proteins, including isoforms and cleaved sequences 17. These C-terminal sequences are enriched in domain-motif interaction motifs and we thus use it as a proof-of-concept for our screening methodology. The oligonucleotide library was amplified by PCR with primers containing EcoRI and XmaI restriction sites, purified after restriction digest and gel electrophoresis, then ligated into the recipient lentiviral vector for functional characterization of the peptides. The lentiviral vector used in this process is pLJM1 (Addgene) with the human CMV promoter for highly efficient gene expression, a green fluorescent protein (GFP) as N-terminal tag and puromycin resistance marker to select infected cells. The ligated libraries were then amplified by transformation into Stbl4 E. coli (Life Technologies) and plasmid DNA was prepared from the E. coli cell mixture. Complexity and fidelity of the libraries were assessed by performing Illumina deep sequencing on both the lentiviral plasmid DNA preparation, and on test batches of human embryonic kidney HEK293T and human cancer RWP1 cells infected with lentivirus made with the peptide libraries.
Cell lines and reagents
HEK293T cell line was obtained from the American Type Culture Collection (ATCC; Rockville, MD). RWP1, Mia Paca-2 and Panc02.03 cell lines were kindly provided by Dr. Jason Moffat. HEK293T cells and the pancreatic cancer RWP1, Mia Paca-2 cell lines were maintained in DMEM (ATCC) supplemented with 10% FBS and 1% pen/strep/glutamine, and the appropriate selection antibiotics when required. Panc02.03 cells were maintained in RPMI (ATCC) supplemented with 20% FBS and 1% pen/strep/glutamine, and the appropriate selection antibiotics when required. Antibodies were obtained from the following commercial sources: HA (7392) antibodies (1:2000) were purchased from Santa Cruz. Flag antibodies (A2220) (1:1000) were purchased from Sigma. GFP (33-2600) (1:2000), p21 (33-7000) (1:1000), p53 (13-4100) (1:1000), AKT (AHO1112) (1:1000), phospho-AKT (S473) (44-609G) (1:1000) antibodies were purchased from Invitrogen. Cyclin B1 (4138) (1:1000), cyclin A2 (BF683) (1:2000), cdc2 (9112) (1:1000), beta-actin (8457) (1:1000), anti-mouse (7076) (1:2500) and anti-rabbit (1:2500) HRP-linked (7074) antibodies were purchased from Cell Signaling.
Library Construction, Amplification and Lentiviral Plasmid Construction
The PCR amplification of the oligonucleotides was conducted as previously described 17. The PCR product was further amplified using primer for insertion of restriction sites (EcoRI and XmaI): Primer forward 5′-CTCAAGCTTGCCTCTTCATCTGGC-3′, primer reverse 5′-GCTCTTAAGTTAGCCTCCTCCGGATCC-3′. The PCR product was digested and cloned into pLJM1 lentiviral vector as C-terminal fusion proteins with the green fluorescent protein GFP. The pLJM1 vector contains a CMV promoter and puromycin for the selection marker. The ligated library was amplified by transformation into E. coli (Stbl4) and plasmid DNA was extracted using DNA Midiprep Kit (Qiagen).
Lentiviral Delivery and Dropout Screen
Lentiviruses were made in 6-well format by transfecting packaging cells (293T) with a three-plasmid system as previously described 41, 42. Viral transduction was performed into HEK293T and RWP1 pancreatic cancer cells with a multiplicity of infection of 0.3 (MOI = 0.3). Infected cells were selected in puromycin-containing medium to eliminate uninfected cells and three aliquots of cells were collected for sampling of the initial (T0) cell population. Cells were harvested at confluency at different time-point in a 30 days period.
Genomic DNA preparation and Illumina sample preparation
Genomic DNA from peptide expressing cells at different time-points was extracted using QIAamp DNA Blood Mini Kit. PCR amplifications of peptides from gDNA in parallel with the lentiviral plasmid library (naïve library) were performed using indexed Illumina PCR primers to incorporate both the Illumina adapter sequences and indexing sequences. Each 50 μl reaction contained 3.2 μg of template, 2x PCR buffer, 2x enhancer solution, 300μM each dNTP, 900nM each for Adapter A (5′-AATGATACGGCGACCACCGAAATG-GACTATCATATGCTTACCGTAACTTGAA-3′) and Adapter B (5′-CAAGCAGAA-GACGGCATACGATGTGGATGAATACTGCCATTTGTCTCGAGGTC-3′), 1mMMgSO4, 3.75 units of Platinum Pfx polymerase, and water to 50μl. The PCR reaction was performed by denaturing at 94°C for 5 minutes, followed by (94°C for 30 seconds, 65°C for 30 seconds, 68°C for 30 seconds) x 28, 68°C for 5 minutes, then cooling to 4°C. The resulting 244bp product was purified by electrophoresis in 2% agarose followed by gel extraction. Peptide libraries were quantified using Quant-It assay (Invitrogen) and pooled. The insert size of the pooled library was confirmed on an Agilent Bioanalyzer High Sensitivity DNA chip (Agilent Technologies), and the size corrected concentration determined with RT-qPCR (KAPA biosystems Illumina standards). 11.4 pM of peptide library and 0.6 pM of PhiX control library (Illumina) were denatured and loaded on a HiSeq 2000 V3 150 cycle sequencing kit, with a read length of 150bp.
Sequence Mapping for Dropout Screens
Quantification of (RNAi|peptide) dropout screens by sequencing was performed as follows. All sequencing is performed using Illumina HiSeq 2000 with paired end (2×150bp) and filtered for average Phred quality score of 30 to ensure high read quality in the peptide region (21 nucleotide variable sequence). In total, 78,446,361 reads that represent peptides identified at four different time points in two cell lines (RWP1 and HEK293T cells) with three replicates are identified (average 73.33 reads per peptide; Supplementary Table 1). To separate reads that were sequenced on one lane to different samples, demultiplexing is performed by allowing 1 mismatch in barcode and 1 mismatch in constant region. Each read is trimmed at both the 5′- and 3′-end to remove uninformative sequence (adapter, barcode and vector sequence). The 21 nucleotide variable sequence, unique to each peptide, is then mapped against a library of the 50,549 peptide-encoding sequences using Bowtie with 0 mismatches between the reference sequence and read 43. On average, 49,750 peptides (98.42% of naïve lentiviral library) are identified at each time points in each cell line (Supplementary Table 1).
To incorporate measurements from multiple time points in a screen, dropout score is introduced 44. Dropout score assigns a value to each peptide by calculating the slope between the measured read counts at each time point and the number of reads at time zero (T0), producing a weighted average of the fold-changes across time and accounting for the growth rate of the cells.
where, n is the number of time points, Δy is he change in expression intensity at Ti relative to T0, and Δx is the number of doublings for the cell line at Ti relative to T0. To calculate dropout score, we only considered peptides with > 55 reads at time zero (T0). All dropout scores of peptides are shown in Supplementary Dataset 1.
To examine cancer-associations of peptides, we investigated if proteins hosting the peptides that lead to specific regulation of cell viability are known cancer-related proteins. Cancer-related proteins that are human proteins whose functional alterations are casually implicated in oncogenesis (e.g. oncogene product, tumor-suppressor gene products and proteins in core cancer pathway) are selected using CancerGenes database 45.
Identification of cancer-specific inhibitors
For the identification of peptides that show distinct cell viabilities in both cell lines, we selected peptides that are identified in all three replicates of two cell lines (RWP1 and HEK293T) and four time points (standard deviation of three replicates < 0.003). Peptides should reduce at least 50% of cell viability at the last time point. For cancer-specific peptide inhibitors, we selected peptides that significantly reduce RWP1 cell viability (dropout score < −2.5) and show no significant change of HEK293T cell viability (−1 < dropout score < 1). In total, 512 anti-proliferative peptides are identified (Supplementary Dataset 2).
Validation of individual peptides
Oligonucleotides encoding the specific peptides were synthesized and individually cloned into the pLJM1 nGFP lentiviral vector. Cells were infected with individual constructs, and cell viability was assessed using Alamar blue assay after 72 hours post infection. For direct comparison with viabilities from the pooled screens, we interpolated cell viabilities at day 7 using polynomial regression (i.e., while the average viability reduction for the 6 RWP1-specific peptides was to 38.88% over 1 week in the pooled screen, the single cell screen could only be carried out for 72h. Hence, we estimated the effect after 72h from the pooled screen using linear interpolation). Most of the peptides that we experimentally followed up have strong evidences to mediate specific interactions with other proteins. Of 15 peptides, 11 peptides have linear motifs, which recruit selective protein targets and 7 peptides are located within disordered regions, which mediate protein-peptide interactions (Supplementary Table 2).
Alamar Blue Cell Viability Assay
Cells were trypsinized from subconfluent cultures as described earlier, were suspended in culture then seeded into triplicate wells of a 96-well plate (100 μl well–1) at a density of 1.5 × 104 per well at standard culture conditions of 5% CO2 in air at 37°C. Cells were infected with lentivirus expressing peptide at an MOI of 5 for 72h or transfected using plasmid for 72h. Alamar Blue reagent was added to each well (10 uL) and optical density of the plate was measured at 540 and 630 nm with a standard spectrophotometer at 1, 2, and 4 hours after adding Alamar Blue.
Determination of peptide-target interactions
To identify potential interacting targets of peptides, two complementary approaches are used. First, peptide sequences are mapped onto known protein complex structures 46. If peptide sequence has structural information (at least four out of seven amino acids are aligned with known structures with 0 ~ 1 mismatch) and is located at the interface, the other protein in a complex structure is considered as an interacting target of peptide. Second, peptide sequences are scanned against the database of linear motifs that mediate protein-protein interactions. If interacting partner of source protein has a peptide-binding domain that can bind to linear motif of peptide, this interacting partner is considered as a target of peptide. Interacting partners of source proteins are identified from the integrated study of human protein-protein interactions 21. The Eukaryotic Linear Motif database 22 is used to identify linear motifs of peptides, and Pfam database is used to identify peptide-binding domains of interacting partners of source proteins 23.
Enriched signaling pathways in peptide-target interactions
To understand biological roles of interactions between cancer-specific inhibitors and targets, we examined enriched signaling pathways in peptides and their targets using KEGG pathway database 47. The significance of pathway enrichments is evaluated by Benjamini-Hochberg-corrected p-values. Peptides and targets are enriched in several signaling pathways that would likely be related to cancer pathogenesis. Signaling pathways that are involved in junction assembly and regulation are enriched in all types of peptides that reduced cell viability in both cell-types (commonly essential peptides and targets) or single cell type (Normal essential peptides and targets). PI3K-Akt, Jak-Stat signaling pathway, cell cycle and apoptosis pathways are enriched in cancer-specific inhibitor-target interactions whereas there are no enriched pathways in target interactions with peptides that reduced cell viability of both cell types and only normal cells (Supplementary Fig. 9).
Flag pull-down
HEK293T cells were cotransfected with Flag-tagged target protein, HA-tagged source protein and GFP-tagged peptide or GFP. Cells were lysed 48 h after transfections with radioimmune precipitation assay buffer (50 mM Tris-HCl pH7.4, 1% Nonidet P-40, 150 mM NaCl, 1 mM EDTA, 10 mM Na3VO4, 10 mM sodium pyrophosphate, 25 mM NaF, 1× protease inhibitor mixture (Sigma) for 30 min at 4 °C and coimmunoprecipitated with Flag beads (Clontech). The resulting immunocomplexes were analyzed by Western blot using the antibodies indicated in Figure 3. Protein samples were separated on a NuPage Bis·Tris 10% SDS/PAGE gel (Invitrogen) and transferred to PVDF membranes. Transferred samples were immunoblotted with primary anti-HA antibodies (1:2000), followed by incubation with horseradish peroxidase-conjugated anti-rabbit secondary antibodies (Cell Signaling) (1:2500) and detected using enhanced chemiluminescence (GE Healthcare).
FRET microscopy imaging
HEK293T cells were trypsinized from subconfluent cultures as described earlier, were suspended in culture then seeded into a 24-well plate (500 μl/well) containing poly-L-lysine treated glass coverslips at concentrations of 5 × 104 per well at standard culture conditions of 5% CO2 in air at 37°C. Cells were cotransfected with the peptide fused to GFP and the target protein fused to mCherry. Forty-eight hours after transfection, cells were then fixed with 4% paraformaldehyde, washed in PBS, and imaged at room temperature using confocal FRET microscopy 48. For data collection, we used a Leica DMI6000B confocal microscope equipped with a Leica 63×/1.4 NA objective lens and a Hamamatsu EM-CCD digital camera (C9100-13), Argon (Donor excitation: 488nm) and Green HeNe (Acceptor excitation: 543nm) lasers, and band-pass emission filters: 515/30 nm (range¼ 500–545 nm) and 590/70 nm (range¼ 555–625 nm); and imported into Volocity software. Both laser power and photomultiplier tube (PMT) gain levels were kept constant throughout image collection. FRET, GFP fluorescence bleed-through and mCherry emission resulting from the Argon-UV excitation were detected at the FRET channel with the detection window at 530–570 nm. The net FRET image was obtained after subtracting the GFP fluorescence bleed-through and the emission of mCherry from Argon-UV laser excitation.
Effect of GNG4 peptide on phosphor-AKT induced by serum stimulation
In order to analyze the dynamic of AKT (S473) phosphorylation, we starved RWP1 and HEK293T cells for 24 hours. We induced AKT phosphorylation by incubating cells with 10% serum (FBS) for 20 minutes in the presence or absence of peptide expression. Protein samples were separated on a NuPage Bis·Tris 10% SDS/PAGE gel (Invitrogen) and transferred to PVDF membranes. Transferred samples were immunoblotted with primary anti-AKT (1:1000) or anti-phospho-AKT (S473) (1:1000) antibodies, followed by incubation with horseradish peroxidase-conjugated secondary antibodies (1:2500) (Cell Signaling) and detected using enhanced chemiluminescence (GE Healthcare). Stimulation of cells with serum led to an increase in AKT phosphorylation in both cell lines. Expression of KFFCTIL peptide inhibited the increase in AKT phosphorylation in RWP1 cells but not in HEK293T cells (Supplementary Fig. 10).
Peptide synthesis
The peptides were synthesized on a Liberty Blue Microwave peptide synthesizer (CEM Corporation) using Fmoc chemistry (See Supplementary Table 3). Peptides were N-terminally extended based on their source protein (See Supplementary Table 3). To avoid oxidation, Met residues in the sequence of HVR have been substituted by isosteric norleucine. The following modifications have been introduced to the published protocol of high efficiency peptide synthesis49. The coupling with N,N′-Diisopropylcarbodiimide/ethyl 2-cyano-2-(hydroxyimino) acetate (OXYMA) was performed for 4 min at 90°C for all residue except for Cys and His, for which the reaction was carried out for 10 min at 50°C. Removal of Fmoc group was conducted at 90°C for 2 min for sequences containing no Cys or Asp. All deprotection cycles after Asp and Cys were conducted at room temperature to avoid racemization and aspartimide formation. Low loading Rink Amide MBHA resin (Merck) was used for the synthesis of amidated peptides and Wang resins were used for the synthesis of peptides with free carboxy-termini. The peptides were cleaved from the resin and deprotected with a mixture of 90.0% (v/v) trifluoroacetic acid (TFA) with 2.5% water, 2.5% triisopropyl-silane, 2.5% 2,2′-(ethylenedioxy)diethanethiol and 5% thioanisol. Peptides were purified on a preparative (25 mm × 250 mm) Atlantis C3 reverse phase column (Agilent Technologies) in a 90 min gradient of 0.1% (v/v) trifluoroacetic acid in water and 0.1% trifluoroacetic acid in acetonitrile, with a 10 mL/min flow rate. The fractions containing peptides were analyzed on Agilent 6100 LC/MS spectrometer with the use of a Zorbax 300SB-C3 PoroShell column and a gradient of 5% acetic acid in water and acetonitrile. Fractions that were more than 95% pure were combined and freeze dried.
Purification of protein (BIRC5)
BIRC5 cDNA clone was obtained from Openfreezer in Gateway Entry vector. BIRC5 clone was then transferred into a pET-53-DEST by Gateway LR clonase (Invitrogen). Destination vectors were transformed into E. coli BL21 (DE3) and cultivated to express proteins. Protein expression was induced by 0.5 mM of Isopropyl β-D-1-thiogalactopyranoside at mid-log phase. After growing the culture overnight at 16°C, cells were harvested by centrifugation at 14,000 × g for 10 min. Cells were lysed with a sonicator and proteins were purified using Ni-NTA agarose (Qiagen) according to the product manual. Concentration of the purified proteins was determined by measuring the absorption at 280 nm using extinction coefficients of 16875·M−1·cm−1 and 1490·M−1·cm−1 for BIRC5 and INCENP peptide respectively.
Isothermal titration calorimetry
After the Ni-NTA agarose purification step, protein samples intended for ITC were purified on a Superdex-75 column equilibrated and eluted with the 20mM Tris, 300 mM NaCl and 5 mM BME buffer. Protein purity was analyzed by SDS-PAGE, concentrated using Amicon Ultra-15 centrifugal units. All protein and peptide samples were dialysed overnight at 4 °C against the same buffer, 25 mM Tris, 150 mM NaCl, 5 mM BME, and 5% DMSO at pH 7. Calorimetric titrations were carried out using a MicroCal ITC200 microcalorimeter (Malvern), with an operating cell volume of 300 μL. The ITC measurements were performed at 25°C and stirred speed was set at 700 rpm to ensure rapid mixing in the cell. Each titration was initiated by a 0.4 μL injection, followed by 20 injections spaced 150s, of 2 μL. The titrations were performed using the same protein batch with a concentration of 15 μM BIRC5 in the cell and 100 μM for INCENP peptide. The same concentrations were used for titrations with the scrambled peptides. The binding parameters were obtained by non-linear regression analysis using a one-independent-type-of-sites binding model implemented in the Origin 7.0. Software. A summary of thermodynamics and curve fitting of INCENP peptide to BIRC5 at 25 °C and pH 7.0 is shown in Supplementary Fig. 8.
Statistical analyses
Data are presented as the means ± s.d. Significance of functional enrichment of peptides was evaluated using hypergeometric test. To examine the statistical difference between two groups, two-tailed independent Student’s t-test and Mann-Whitney U test were used. We calculate edge-betweenness of peptide-target network using Python package “NetworkX 1.8” (https://networkx.github.io) and compared the properties of our peptide-target network with 1,000 randomly generated peptide-target networks using bootstrap test. P-value < 0.05 was considered as statistically significant. All statistical analyses are performed using Python package “Numpy 1.7” and “Scipy 0.13.2” (http://numpy.scipy.org).
Supplementary Material
Acknowledgments
We thank the members of the Moffat laboratory for valuable technical assistance with lentiviral screening technology, usage of reagents and equipment. We thank Dr. Andrew Emili, Dr. Tim Hughes and Dr. Michael Garton for helpful comments on the manuscript. We thank Dr. Andrea Musacchio for providing us the INCENP cDNA clone. PMK acknowledges an Operating Grant from the Canadian Institute of Health Research (CIHR MOP-123526) and an Innovation Grant from the Canadian Cancer Society Research Institute (CCSRI# 702884). JM is a Tier 2 Canada Research Chair in Functional Genomics of Cancer.
Footnotes
Author Contribution
P.M.K. designed the project provided study guidance and wrote the bulk of the manuscript. S.N. performed most experiments and contributed to writing of the manuscript. J.J. performed all bioinformatics analysis and interpreted results as well as assisted in manuscript preparation. C.C. and M.S. performed affinity measurements and helped with other biochemical experiments. Y.I. provided oligonucleotide library and provided study guidance. N.T. provided synthetic peptides and provided guidance on their use. J.M. helped design the project and provided guidance on lentiviral screening.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Booth B, Zemmel R. Prospects for productivity. Nature reviews. Drug discovery. 2004;3:451–456. doi: 10.1038/nrd1384. [DOI] [PubMed] [Google Scholar]
- 2.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nature reviews. Drug discovery. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 3.Grant SK. Therapeutic protein kinase inhibitors. Cellular and molecular life sciences: CMLS. 2009;66:1163–1177. doi: 10.1007/s00018-008-8539-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mullard A. Protein-protein interaction inhibitors get into the groove. Nature reviews. Drug discovery. 2012;11:173–175. doi: 10.1038/nrd3680. [DOI] [PubMed] [Google Scholar]
- 5.Fuller JC, Burgoyne NJ, Jackson RM. Predicting druggable binding sites at the protein-protein interface. Drug discovery today. 2009;14:155–161. doi: 10.1016/j.drudis.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 6.Nero TL, Morton CJ, Holien JK, Wielens J, Parker MW. Oncogenic protein interfaces: small molecules, big challenges. Nature reviews. Cancer. 2014;14:248–262. doi: 10.1038/nrc3690. [DOI] [PubMed] [Google Scholar]
- 7.van Delft MF, et al. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer cell. 2006;10:389–399. doi: 10.1016/j.ccr.2006.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Souers AJ, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nature medicine. 2013;19:202–208. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- 9.Liu M, et al. D-peptide inhibitors of the p53-MDM2 interaction for targeted molecular therapy of malignant neoplasms. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14321–14326. doi: 10.1073/pnas.1008930107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shangary S, Wang S. Small-molecule inhibitors of the MDM2-p53 protein-protein interaction to reactivate p53 function: a novel approach for cancer therapy. Annual review of pharmacology and toxicology. 2009;49:223–241. doi: 10.1146/annurev.pharmtox.48.113006.094723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang YS, et al. Stapled alpha-helical peptide drug development: a potent dual inhibitor of MDM2 and MDMX for p53-dependent cancer therapy. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E3445–3454. doi: 10.1073/pnas.1303002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khoo KH, Verma CS, Lane DP. Drugging the p53 pathway: understanding the route to clinical efficacy. Nature reviews. Drug discovery. 2014;13:217–236. doi: 10.1038/nrd4236. [DOI] [PubMed] [Google Scholar]
- 13.Neduva V, et al. Systematic discovery of new recognition peptides mediating protein interaction networks. PLoS biology. 2005;3:e405. doi: 10.1371/journal.pbio.0030405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neduva V, Russell RB. Linear motifs: evolutionary interaction switches. FEBS letters. 2005;579:3342–3345. doi: 10.1016/j.febslet.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 15.Sims D, et al. High-throughput RNA interference screening using pooled shRNA libraries and next generation sequencing. Genome biology. 2011;12:R104. doi: 10.1186/gb-2011-12-10-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amado RG, Chen IS. Lentiviral vectors--the promise of gene therapy within reach? Science. 1999;285:674–676. doi: 10.1126/science.285.5428.674. [DOI] [PubMed] [Google Scholar]
- 17.Ivarsson Y, et al. Large-scale interaction profiling of PDZ domains through proteomic peptide-phage display using human and viral phage peptidomes. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:2542–2547. doi: 10.1073/pnas.1312296111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Natarajan V, et al. Peptides genetically selected for NF-kappaB activation cooperate with oncogene Ras and model carcinogenic role of inflammation. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E474–483. doi: 10.1073/pnas.1311945111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poyurovsky MV, et al. The C terminus of p53 binds the N-terminal domain of MDM2. Nature structural & molecular biology. 2010;17:982–989. doi: 10.1038/nsmb.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chene P, et al. A small synthetic peptide, which inhibits the p53-hdm2 interaction, stimulates the p53 pathway in tumour cell lines. Journal of molecular biology. 2000;299:245–253. doi: 10.1006/jmbi.2000.3738. [DOI] [PubMed] [Google Scholar]
- 21.Bossi A, Lehner B. Tissue specificity and the human protein interaction network. Molecular systems biology. 2009;5:260. doi: 10.1038/msb.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dinkel H, et al. The eukaryotic linear motif resource ELM: 10 years and counting. Nucleic acids research. 2014;42:D259–266. doi: 10.1093/nar/gkt1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finn RD, et al. Pfam: the protein families database. Nucleic acids research. 2014;42:D222–230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wazir U, Jiang WG, Sharma AK, Mokbel K. Guanine nucleotide binding protein beta 1: a novel transduction protein with a possible role in human breast cancer. Cancer genomics & proteomics. 2013;10:69–73. [PubMed] [Google Scholar]
- 25.Barabasi AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nature reviews. Genetics. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong Q, et al. Edgetic perturbation models of human inherited disorders. Molecular systems biology. 2009;5:321. doi: 10.1038/msb.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sekar RB, Periasamy A. Fluorescence resonance energy transfer (FRET) microscopy imaging of live cell protein localizations. The Journal of cell biology. 2003;160:629–633. doi: 10.1083/jcb.200210140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J, Wu X, Lin J, Levine AJ. mdm-2 inhibits the G1 arrest and apoptosis functions of the p53 tumor suppressor protein. Molecular and cellular biology. 1996;16:2445–2452. doi: 10.1128/mcb.16.5.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hermeking H, et al. 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Molecular cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 30.Rajagopalan S, Jaulent AM, Wells M, Veprintsev DB, Fersht AR. 14-3-3 activation of DNA binding of p53 by enhancing its association into tetramers. Nucleic acids research. 2008;36:5983–5991. doi: 10.1093/nar/gkn598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masters SC, Fu H. 14-3-3 proteins mediate an essential anti-apoptotic signal. The Journal of biological chemistry. 2001;276:45193–45200. doi: 10.1074/jbc.M105971200. [DOI] [PubMed] [Google Scholar]
- 32.Wang B, et al. Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display. Biochemistry. 1999;38:12499–12504. doi: 10.1021/bi991353h. [DOI] [PubMed] [Google Scholar]
- 33.Ito K, Caramori G, Adcock IM. Therapeutic potential of phosphatidylinositol 3-kinase inhibitors in inflammatory respiratory disease. The Journal of pharmacology and experimental therapeutics. 2007;321:1–8. doi: 10.1124/jpet.106.111674. [DOI] [PubMed] [Google Scholar]
- 34.Pumiglia KM, et al. A direct interaction between G-protein beta gamma subunits and the Raf-1 protein kinase. The Journal of biological chemistry. 1995;270:14251–14254. doi: 10.1074/jbc.270.24.14251. [DOI] [PubMed] [Google Scholar]
- 35.Yoda A, et al. Novel oncogenic mutations in the beta subunit of heteromeric G-proteins identified by functional cDNA library screening. Molecular Cancer Therapeutics. 2013;12(11 Suppl):PR07. [Google Scholar]
- 36.Lamers F, et al. Knockdown of survivin (BIRC5) causes apoptosis in neuroblastoma via mitotic catastrophe. Endocrine-related cancer. 2011;18:657–668. doi: 10.1530/ERC-11-0207. [DOI] [PubMed] [Google Scholar]
- 37.Zhou M, et al. DNA damage induces a novel p53-survivin signaling pathway regulating cell cycle and apoptosis in acute lymphoblastic leukemia cells. The Journal of pharmacology and experimental therapeutics. 2002;303:124–131. doi: 10.1124/jpet.102.037192. [DOI] [PubMed] [Google Scholar]
- 38.Abraham J, et al. Eme1 is involved in DNA damage processing and maintenance of genomic stability in mammalian cells. The EMBO journal. 2003;22:6137–6147. doi: 10.1093/emboj/cdg580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pamidi A, et al. Functional interplay of p53 and Mus81 in DNA damage responses and cancer. Cancer research. 2007;67:8527–8535. doi: 10.1158/0008-5472.CAN-07-1161. [DOI] [PubMed] [Google Scholar]
- 40.Zhang L, Bulaj G. Converting peptides into drug leads by lipidation. Current medicinal chemistry. 2012;19:1602–1618. doi: 10.2174/092986712799945003. [DOI] [PubMed] [Google Scholar]
- 41.Naldini L, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 42.Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nature biotechnology. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- 43.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marcotte R, et al. Essential gene profiles in breast, pancreatic, and ovarian cancer cells. Cancer discovery. 2012;2:172–189. doi: 10.1158/2159-8290.CD-11-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Higgins ME, Claremont M, Major JE, Sander C, Lash AE. CancerGenes: a gene selection resource for cancer genome projects. Nucleic acids research. 2007;35:D721–726. doi: 10.1093/nar/gkl811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berman HM, et al. The Protein Data Bank. Nucleic acids research. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kanehisa M, et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wallrabe H, Elangovan M, Burchard A, Periasamy A, Barroso M. Confocal FRET microscopy to measure clustering of ligand-receptor complexes in endocytic membranes. Biophysical journal. 2003;85:559–571. doi: 10.1016/S0006-3495(03)74500-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collins JM, Porter KA, Singh SK, Vanier GS. High-efficiency solid phase peptide synthesis (HE-SPPS) Organic letters. 2014;16:940–943. doi: 10.1021/ol4036825. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.