Abstract
Immune cells of myeloid origin, including microglia, macrophages, and myeloid-derived suppressor cells adopt immunosuppressive phenotypes that support gliomagenesis. Here, we tested an a priori hypothesis that single nucleotide polymorphisms (SNPs) in genes related to glioma-associated myeloid cell regulation and function are also associated with patient survival after glioma diagnosis. Subjects for this study were 992 glioma patients treated at The University of Texas MD Anderson Cancer Center in Houston, Texas between 1992 and 2008. Haplotype-tagging SNPs in 91 myeloid-associated genes were analyzed for association with survival by Cox regression. Individual SNP- and gene-based tests were performed separately in glioblastoma (WHO grade IV, n=511) and lower-grade glioma (WHO grade II-III, n=481) groups. After adjustment for multiple testing, no myeloid-associated gene variants were significantly associated with survival in glioblastoma. Two SNPs, rs147960238 in CD163 (p=2.2 x 10−5) and rs17138945 in MET (p=5.6 x 10−5) were significantly associated with survival of patients with lower-grade glioma. However, these associations were not confirmed in an independent analysis of 563 lower-grade glioma cases from the University of California at San Francisco Adult Glioma Study (p=0.65 and p=0.41, respectively). The results of this study do not support a role for inherited polymorphisms in myeloid-associated genes in affecting survival of patients diagnosed with glioblastoma or lower-grade glioma.
Keywords: Myeloid-derived suppressor cells, macrophages, microglia, immune suppression, glioma, glioblastoma, survival, genetic polymorphism
INTRODUCTION
Gliomas, which account for approximately 75% of primary malignant brain tumors [1], exist in a dynamic microenvironment that influences tumor development and ultimately patient survival. Although gliomas are highly infiltrated by immune cells of the myeloid lineage including microglia, macrophages, and myeloid-derived suppressor cells (MDSCs; collectively, glioma-associated myeloid cells), these cells have been shown to adopt immunosuppressive and tumor-supportive phenotypes in response to tumor-secreted signals [2–5]. Furthermore, the degree of myeloid-derived cell infiltration has been shown to correlate with glioma grade in human specimens and survival in mouse models of glioma [6–8]. Given these observations, we hypothesized that germline polymorphisms in genes central to the function of glioma-associated myeloid cells may be associated with the prognosis of patients with glioblastoma or lower-grade glioma.
MATERIALS AND METHODS
Study subjects and data collection
Participants in the discovery population for this study included 992 adult glioma patients of Caucasian ethnicity (ICD-O-3 codes 9380-9340) diagnosed and treated at The University of Texas MD Anderson Cancer Center (MD Anderson) in Houston, Texas from 1992 to 2008 [9, 10]. Treatment and survival data (dates of death or last contact) were collected retrospectively from medical record review through August 2016, and dates of death were confirmed by querying the Social Security Death Index (a database created from the United States Social Security Administration’s Death Master File). Glioblastoma patients were restricted to those who received both chemotherapy and radiotherapy. The study was approved by the Institutional Review Boards of both MD Anderson and Baylor College of Medicine, and written informed consent was obtained from each participant. An independent population of 563 patients with lower-grade glioma who were included in the University of California at San Francisco Adult Glioma Study [11] served as a validation set for this analysis.
Selection of genes, SNPs, and genotyping assays
We identified genes related to the biology of glioma-associated myeloid cells for analysis in this study. In total, 91 genes were selected based on a systematic review of the literature by our research group and included those encoding transcription factors, cytokines and chemokines, receptors, enzymes, and other genes as previously described for which genotyping data were available [12] (Table 1). Genotyping for the discovery population was conducted using the Illumina Human 610-Quad Bead Chip platform, and genotype imputation was performed to increase genomic resolution, as previously described [9, 11]. A total of 2,040 study population-specific haplotype-tagging SNPs (r2≥0.8) with minor allele frequency (MAF) ≥ 1% were identified using Haploview Tagger software [13] in the 91 gene regions, plus 5 kb upstream of the selected genes.
Table 1.
Myeloid-associated genes selected for study
| Functional group | n | Genes included |
|---|---|---|
| Transcription factors | 17 | HIF1A, HIF2A/EPAS1, NFKB1, IRF1, IRF3, IRF4, IRF5, IRF7, STAT1, STAT3, STAT5A, STAT6, KLF2, PPARD, PPARG, CEBPB, GATA3 |
| Cytokines & chemokines | 25 | IL1A, IL1B, IL2, IL6, IL10, IL12A, IL15, IL17A, TNF, TGFB1, IFNG, CCL2, CCL5, CCL8, CCL13, CCL15, CCL17, CCL18, CCL19, CCL23, CXCL9, CXCL10, CXCL11, CXCL12, MIP1A/CCL3 |
| Receptors | 26 | IL1RN, IL1RAP, IL2RA, IL4R, IL15RA, HRH1, HRH4, TFRC, EGFR, VEGFR1/FLT1, MET, CCR1, CCR2, CCR3, CCR7, TLR2, TLR3, TLR4, TLR5, ADORA1, ADORA3, P2RY5/LPAR6, P2RY12, CXCR4, CX3CR1, CR3/ITGAM |
| Enzymes | 6 | ARG1, NOS2, PTGS2, SYK, LCK, SRC |
| Others | 17 | SPP1, IKBKB, MT1MMP/MMP14, MMP2, MMP9, CIITA, C3, CD209, CD280/MRC2, CD279/PDCD1, CD276, CD163, CD80, CD86, CCDC26, C14orf139/LINC00341, KCNN2 |
Statistical methods
Survival time was defined as the time between pathological diagnosis and date of death, if known, or date of last contact (patients in the latter group were censored as of the date of last contact). We computed hazard ratios (HR) and 95% confidence intervals (CI) using Cox proportional hazards regression for individual SNPs of interest under an additive allelic model for glioblastoma and lower-grade glioma patient groups with P-values determined by the Wald method. Proportional hazards assumptions were checked for each variable via examination of Schoenfeld residuals and log-log survival plots; assumptions were met for all variables except for surgery among glioblastoma patients, which was remedied by performing Cox regression stratified by surgery type to allow for different baseline hazards. Glioblastoma models were adjusted for age and sex, whereas lower-grade glioma models were adjusted for age, sex, extent of surgery (gross total/partial/biopsy), chemotherapy (yes/no), and radiotherapy (yes/no). Gene-based tests were performed using the CoxKM method [14], and all analyses were performed using R (https://www.r-project.org). Adjustment for multiple comparisons was performed by calculation of False Discovery Rate (FDR)-adjusted p values according to the method of Benjamini and Hochberg [15].
RESULTS
Discovery set patient characteristics
Characteristics for the glioblastoma (WHO grade IV) and lower-grade glioma (WHO grade II–III) discovery set populations are presented in Table 2A and Table 2B, respectively. As of August 2016, a date of death was known for 458 of 511 (89.6%) glioblastoma patients and 299 of 481 (62.2%) lower-grade glioma patients, and the median survival times (MSTs) were 1.6 and 6.8 years, respectively. Approximately two-thirds of patients in both groups were male, and the mean age at diagnosis was 52.6 years for glioblastoma subjects and 40.4 years for lower-grade glioma subjects. At the univariate level, sex, age at diagnosis, and extent of surgery were each statistically significantly-associated with survival of glioblastoma, whereas age at diagnosis, tumor grade, tumor histology, extent of surgery, and treatment with radiotherapy and chemotherapy were each associated with survival of lower-grade glioma.
Table 2A.
Characteristics of glioblastoma study subjects
| Characteristic | N (%) | MST (y) | HR (95% CI) | P |
|---|---|---|---|---|
| Total/events | 511/458 | 1.6 | ||
| Sex | 0.041 | |||
| Female | 185 (36.2) | 1.8 | Reference | |
| Male | 326 (63.8) | 1.5 | 1.22 (1.01–1.48) | |
| Age at diagnosis (y) | <0.001 | |||
| Mean ± SD | 52.6 ± 11.7 | |||
| 18–39 | 66 (12.9) | 2.7 | Reference | |
| 40–69 | 408 (79.8) | 1.5 | 2.03 (1.52–2.72) | |
| 70+ | 37 (7.2) | 1.7 | 2.12 (1.38–3.26) | |
| Surgery | <0.001 | |||
| Gross-total resection | 167 (32.7) | 2.0 | Reference | |
| Partial resection | 161 (31.5) | 1.4 | 1.57 (1.24–1.97) | |
| Biopsy only | 82 (16.0) | 1.2 | 1.83 (1.39–2.40) | |
| Unknown | 101 (19.8) |
SD: standard deviation; MST: median survival time; HR: hazard ratio
Table 2B.
Characteristics of lower-grade glioma study subjects
| Characteristic | N (%) | MST (y) | HR (95% CI) | P |
|---|---|---|---|---|
| Total/events | 481/299 | 6.8 | ||
| Sex | 0.20 | |||
| Female | 180 (37.4) | 8.4 | Reference | |
| Male | 301 (62.6) | 6.4 | 1.17 (0.92–1.48) | |
| Age at diagnosis (y) | <0.001 | |||
| Mean ± SD | 40.4 ± 12.2 | |||
| 18–39 | 251 (52.2) | 8.3 | Reference | |
| 40–69 | 223 (46.4) | 4.8 | 1.60 (1.27–2.01) | |
| 70+ | 7 (1.5) | 1.2 | 4.43 (2.07–9.49) | |
| Grade | <0.001 | |||
| II | 145 (30.1) | 10.3 | Reference | |
| III | 267 (55.5) | 4.6 | 1.59 (1.22–2.07) | |
| Unknown | 69 (14.3) | |||
| Histology | <0.001 | |||
| Oligodendroglioma | 148 (30.8) | 11.2 | Reference | |
| Oligoastrocytoma | 43 (8.9) | 7.4 | 1.22 (0.78–1.91) | |
| Astrocytoma | 234 (48.6) | 4.6 | 1.81 (1.38–2.38) | |
| Unknown | 56 (11.6) | |||
| Surgery | <0.001 | |||
| Gross-total resection | 86 (17.9) | 15.6 | Reference | |
| Partial resection | 133 (27.7) | 6.7 | 1.59 (1.09–2.32) | |
| Biopsy only | 203 (42.2) | 4.8 | 2.08 (1.46–2.95) | |
| Unknown | 59 (12.3) | |||
| Radiotherapy | 0.040 | |||
| Yes | 406 (84.4) | 6.6 | Reference | |
| No | 21 (4.4) | 14.4 | 0.50 (0.26–0.97) | |
| Unknown | 54 (11.2) | |||
| Chemotherapy | 0.006 | |||
| Yes | 383 (79.6) | 6.6 | Reference | |
| No | 25 (5.2) | NA | 0.39 (0.20–0.77) | |
| Unknown | 73 (15.2) |
SD: standard deviation; MST: median survival time; HR: hazard ratio
Myeloid-associated gene variants and survival
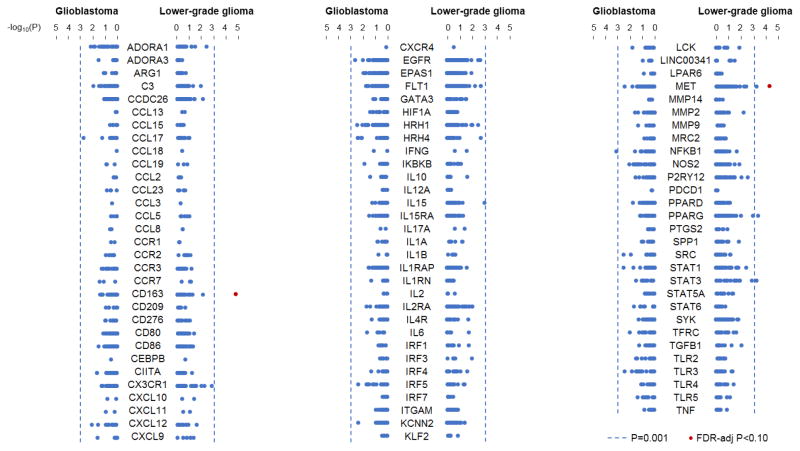
Haplotype-tagging SNPs (n=2,040) in 91 myeloid-associated genes were evaluated for association with glioma survival. All individual SNP associations with survival in patients with glioblastoma and lower-grade glioma are illustrated in Figure 1, and the results for the top-associated SNPs in each glioma subtype are presented in Table 3. The SNP most strongly associated with survival of glioblastoma patients was rs147177288 in NFKB1 (HR=2.53; 95% CI: 1.47–4.37; p=8.7x10−4); however, this association was not significant after adjustment for multiple testing (FDR-adjusted p=0.73). In the lower-grade glioma analysis, five SNPs were associated with survival at p<0.001, two of which remained significant after adjustment for multiple comparisons (FDR-adjusted p<0.10). The results indicated inferior survival for carriers of the C allele at rs147960238 in CD163 (HR=5.47, 95% CI: 2.49–11.99, p=2.2x10−5, FDR-adjusted p=0.046) and for carriers of the G allele at rs17138945 in MET (HR=2.27, 95% CI: 1.52–3.38, p=5.6x10−5, FDR-adjusted p=0.057). In gene-based analyses, the strongest associations with survival in patients with glioblastoma and lower-grade glioma were observed for IL6 (p=0.013) and STAT3 (p=0.034), respectively; however, neither was significant after correction for the 91 genes analyzed.
Figure 1.
Association of variants in myeloid-associated genes with survival in glioma patients. Dots represent SNPs tested for association in the corresponding myeloid-associated gene with survival of glioblastoma patients or lower-grade glioma patients, and they are plotted according to the −log10(p value) from single-SNP tests. A significance level of p = 0.001 is denoted by the dotted line, and SNPs with False Discovery Rate-adjusted p values < 0.10 are colored in red.
Table 3.
Top SNP associations with glioma patient survival
| Glioma subtype | Gene | rsID | MA | MAF | Info1 | Discovery | Validation | ||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| HR (95% CI) | P | HR (95% CI) | P | ||||||
| Glioblastoma | NFKB1 | rs147177288 | C | 0.016 | 0.98 | 2.53 (1.47–4.37) | 8.69x10−4 | - | - |
| CCL17 | rs62037107 | T | 0.020 | 0.97 | 2.04 (1.30–3.20) | 1.91x10−3 | - | - | |
| EGFR | rs28557040 | G | 0.042 | 0.99 | 1.73 (1.21–2.48) | 2.48x10−3 | - | - | |
| SRC | rs6017962 | G | 0.026 | 0.96 | 1.78 (1.21–2.62) | 3.33x10−3 | - | - | |
| STAT1 | rs113635552 | C | 0.016 | 0.85 | 1.82 (1.22–2.72) | 3.37x10−3 | - | - | |
|
| |||||||||
| Lower-grade glioma | CD163 | rs147960238 | C | 0.014 | 0.88 | 5.47 (2.49–11.99) | 2.23x10−5 * | 0.82 (0.36–1.88) | 0.65 |
| MET | rs17138945 | G | 0.038 | 0.99 | 2.27 (1.52–3.38) | 5.61x10−5 * | 0.84 (0.56–1.27) | 0.41 | |
| PPARG | rs116793915 | G | 0.016 | 0.88 | 3.25 (1.68–6.29) | 4.49x10−4 | - | - | |
| STAT3 | rs139679582 | A | 0.012 | 0.90 | 3.00 (1.60–5.64) | 6.14x10−4 | - | - | |
| MET | rs38841 | G | 0.361 | 1.00 | 0.72 (0.60–0.87) | 6.49x10−4 | - | - | |
MA: minor allele; MAF: minor allele frequency; HR: hazard ratio
Info denotes “information score” reflecting quality of genotype imputation in discovery population
False discovery rate-adjusted p value < 0.10
Replication results
Associations between rs147960238 and rs17138945 and survival of lower-grade glioma were evaluated in an independent cohort of 563 patients with grade II and grade III glioma participating in the University of California at San Francisco Adult Glioma Study [11]. The median survival time for this population was 9.6 years, and dates of death were recorded for 274 (48.7%) of the subjects. The median age at diagnosis was 44 years, and 57% of subjects were male. The distribution of tumor histologies did not differ significantly from those in the discovery set (p=0.20). Minor allele frequencies for rs147960238 (0.014) and rs17138945 (0.042) were consistent with the discovery set population. When tested under the same model as employed in the discovery analysis, these variants were not associated with lower-grade glioma survival (rs147960238: HR=0.82, 95% CI: 0.36–1.88, p=0.65; rs17138945: HR=0.84; 95% CI: 0.56–1.27; p=0.41).
DISCUSSION
Gliomas have been shown to inhibit effective antitumor immune responses via a variety of local and systemic immunosuppressive mechanisms [16, 17]. Myeloid-derived immune cells including microglia, macrophages, and MDSCs are potent mediators of this immunosuppressive activity [18–20]. In this study, we sought to test the specific hypothesis that polymorphisms in genes involved in the immunosuppressive behavior of glioma-associated myeloid cells are associated with survival of patients with glioblastoma or lower-grade glioma.
An analysis of 511 glioblastoma patients did not provide support for our hypothesis at the individual SNP- or gene-level. In an analysis of 481 lower-grade glioma patients, rs147960238, which is located in the 10th intron of CD163, and rs17138945, which is located in the 2nd intron of MET, were significantly associated with survival. CD163 is a hemoglobin/haptoglobin complex receptor that is expressed on macrophages and microglia and may play a role in macrophage-mediated anti-inflammatory responses [21, 22]. MET is a receptor tyrosine kinase and proto-oncogene that is also involved in the expansion of MDSC populations [23]. The associations observed at these genes were not reproduced in an independent analysis of 563 patients with lower-grade glioma who participated in the University of California at San Francisco Adult Glioma Study. It is plausible that detection of these effects in the replication set was hindered by differences between study populations such as in treatment received or in the underlying molecular subtypes of glioma. We were unable to control for treatment variables with greater specificity due to limitations of the data available. However, the associations observed with the low-frequency variants at rs147960238 and rs17138945 in the discovery population may also be attributable to chance, highlighting the importance of independent validation in studies of this nature.
Taken together, although the immunosuppressive effects of glioma-associated myeloid cells are well-documented, the results of this study do not support a significant role for inherited genetic variation in genes governing this behavior in survival of glioma patients. Notably, a similar conclusion was reached in a recent analysis of variants in myeloid-derived suppressor cell pathway genes and survival of ovarian cancer patients [24]. Our study had ≥80% power at α=0.05/2,040=2.45x10−5 to detect hazard ratios of at least 1.42 and 1.78 in the glioblastoma analysis and 1.53 and 1.94 in the lower-grade glioma analysis in variants with minor allele frequencies of 40% and 10%, respectively. Thus, it is plausible that associations may exist with smaller effect sizes and/or with rare variants; it is also possible that associations were missed in genes involved in myeloid cell regulation that were not included for analysis in our study. However, this study did not detect reproducible associations between polymorphisms in known myeloid-associated genes and survival in glioma patients, despite sufficient statistical power to detect moderate and large effects.
Acknowledgments
We are grateful to all of the patients and individuals for their participation, and we would also like to thank the clinicians and other hospital staff members, cancer registries and the study staff members in the respective centers who contributed to the data collection. This publication was supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through UCSF-CTSI Grant Number UL1 RR024131. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. The collection of cancer incidence data used in this study was supported by the California Department of Public Health as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885; the National Cancer Institute’s Surveillance, Epidemiology and End Results Program under contract HHSN261201000140C awarded to the Cancer Prevention Institute of California, contract HHSN261201000035C awarded to the University of Southern California, and contract HHSN261201000034C awarded to the Public Health Institute; and the Centers for Disease Control and Prevention’s National Program of Cancer Registries, under agreement # U58DP003862-01 awarded to the California Department of Public Health. The ideas and opinions expressed herein are those of the author(s) and endorsement by the State of California Department of Public Health, the National Cancer Institute, and the Centers for Disease Control and Prevention or their Contractors and Subcontractors is not intended nor should be inferred. We thank David M. Wildrick, Ph.D., for editorial assistance with the manuscript.
Funding: D.I.J. is supported by the CPRIT Post-Graduate Training Program in Integrative Cancer Epidemiology (Award ID RP160097). Work at MD Anderson Cancer Center and Baylor College of Medicine was supported by National Institutes of Health grants R01 CA120813 to A.B.H, P50 CA127001 to A.B.H, R01 CA119215 to M.L.B, R01 CA070917 to M.L.B, R01 CA139020 to M.L.B, and K07 CA181480 to Y.L; American Brain Tumor Association to M.L.B, The National Brain Tumor Society to M.L.B, and The Dr. Marnie Rose Foundation to A.B.H. Work at University of California, San Francisco was supported by the National Institutes of Health (grant numbers R01CA52689, P50CA097257, and R01CA139020), as well as the loglio Collective, the National Brain Tumor Foundation, the Stanley D. Lewis and Virginia S. Lewis Endowed Chair in Brain Tumor Research, the Robert Magnin Newman Endowed Chair in Neuro-oncology, and by donations from families and friends of John Berardi, Helen Glaser, Elvera Olsen, Raymond E. Cooper, and William Martinusen.
Footnotes
COMPLIANCE WITH ETHICAL STANDARDS
Conflict of Interest: All of the authors have declared no conflicts of interest.
Ethical Approval and Informed Consent: This study was approved by the Institutional Review Boards of MD Anderson, Baylor College of Medicine, and University of California, San Francisco, and written informed consent was obtained from each participant. All procedures performed were in accordance with the ethical standards of the institutional research committees and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
References
- 1.Ostrom QT, Gittleman H, Fulop J, Liu M, Blanda R, Kromer C, Wolinsky Y, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2008–2012. Neuro Oncol. 2015;17:iv1–iv62. doi: 10.1093/neuonc/nov189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gabrusiewicz K, Ellert-Miklaszewska A, Lipko M, Sielska M, Frankowska M, Kaminska B. Characteristics of the alternative phenotype of microglia/macrophages and its modulation in experimental gliomas. PLoS One. 2011;6:e23902. doi: 10.1371/journal.pone.0023902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prosniak M, Harshyne LA, Andrews DW, Kenyon LC, Bedelbaeva K, Apanasovich TV, Heber-Katz E, Curtis MT, Cotzia P, Hooper DC. Glioma grade is associated with the accumulation and activity of cells bearing M2 monocyte markers. Clin Cancer Res. 2013;19:3776–3786. doi: 10.1158/1078-0432.CCR-12-1940. [DOI] [PubMed] [Google Scholar]
- 4.Raychaudhuri B, Rayman P, Ireland J, Ko J, Rini B, Borden EC, Garcia J, Vogelbaum MA, Finke J. Myeloid-derived suppressor cell accumulation and function in patients with newly diagnosed glioblastoma. Neuro Oncol. 2011;13:591–599. doi: 10.1093/neuonc/nor042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Umemura N, Saio M, Suwa T, Kitoh Y, Bai J, Nonaka K, Ouyang GF, Okada M, Balazs M, Adany R, Shibata T, Takami T. Tumor-infiltrating myeloid-derived suppressor cells are pleiotropic-inflamed monocytes/macrophages that bear M1- and M2-type characteristics. J Leukoc Biol. 2008;83:1136–1144. doi: 10.1189/jlb.0907611. [DOI] [PubMed] [Google Scholar]
- 6.Morimura T, Neuchrist C, Kitz K, Budka H, Scheiner O, Kraft D, Lassmann H. Monocyte subpopulations in human gliomas: expression of Fc and complement receptors and correlation with tumor proliferation. Acta Neuropathol (Berl) 1990;80:287–294. doi: 10.1007/BF00294647. [DOI] [PubMed] [Google Scholar]
- 7.Roggendorf W, Strupp S, Paulus W. Distribution and characterization of microglia/macrophages in human brain tumors. Acta Neuropathol (Berl) 1996;92:288–293. doi: 10.1007/s004010050520. [DOI] [PubMed] [Google Scholar]
- 8.Kong L-Y, Wu AS, Doucette T, Wei J, Priebe W, Fuller GN, Qiao W, Sawaya R, Rao G, Heimberger AB. Intratumoral mediated immunosuppression is prognostic in genetically engineered murine models of glioma and correlates to immunotherapeutic responses. Clin Cancer Res. 2010;16:5722–5733. doi: 10.1158/1078-0432.CCR-10-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, Simon M, Marie Y, Boisselier B, Delattre JY, Hoang-Xuan K, El Hallani S, Idbaih A, Zelenika D, Andersson U, Henriksson R, Bergenheim AT, Feychting M, Lonn S, Ahlbom A, Schramm J, Linnebank M, Hemminki K, Kumar R, Hepworth SJ, Price A, Armstrong G, Liu Y, Gu X, Yu R, Lau C, Schoemaker M, Muir K, Swerdlow A, Lathrop M, Bondy M, Houlston RS. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, Scheurer ME, El-Zein R, Cao Y, Do KA, Gilbert M, Aldape KD, Wei Q, Etzel C, Bondy ML. Association and interactions between DNA repair gene polymorphisms and adult glioma. Cancer Epidemiol Biomarkers Prev. 2009;18:204–214. doi: 10.1158/1055-9965.EPI-08-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melin BS, Barnholtz-Sloan JS, Wrensch MR, Johansen C, Il’yasova D, Kinnersley B, Ostrom QT, Labreche K, Chen Y, Armstrong G, Liu Y, Eckel-Passow JE, Decker PA, Labussiere M, Idbaih A, Hoang-Xuan K, Di Stefano A-L, Mokhtari K, Delattre J-Y, Broderick P, Galan P, Gousias K, Schramm J, Schoemaker MJ, Fleming SJ, Herms S, Heilmann S, Nothen MM, Wichmann H-E, Schreiber S, Swerdlow A, Lathrop M, Simon M, Sanson M, Andersson U, Rajaraman P, Chanock S, Linet M, Wang Z, Yeager M, GliomaScan C, Wiencke JK, Hansen H, McCoy L, Rice T, Kosel ML, Sicotte H, Amos CI, Bernstein JL, Davis F, Lachance D, Lau C, Merrell RT, Shildkraut J, Ali-Osman F, Sadetzki S, Scheurer M, Shete S, Lai RK, Claus EB, Olson SH, Jenkins RB, Houlston RS, Bondy ML. Genome-wide association study of glioma subtypes identifies specific differences in genetic susceptibility to glioblastoma and non-glioblastoma tumors. Nat Genet. 2017;49:789–794. doi: 10.1038/ng.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gabrusiewicz K, Rodriguez B, Wei J, Hashimoto Y, Healy LM, Maiti SN, Thomas G, Zhou S, Wang Q, Elakkad A. Glioblastoma-infiltrated innate immune cells resemble M0 macrophage phenotype. JCI Insight. 2016;1:e85841. doi: 10.1172/jci.insight.85841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 14.Lin X, Cai T, Wu MC, Zhou Q, Liu G, Christiani DC, Lin X. Kernel machine SNP-set analysis for censored survival outcomes in genome-wide association studies. Genet Epidemiol. 2011;35:620–631. doi: 10.1002/gepi.20610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 16.Vega EA, Graner MW, Sampson JH. Combating immunosuppression in glioma. Future Oncol. 2008;4:433–442. doi: 10.2217/14796694.4.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dix AR, Brooks WH, Roszman TL, Morford LA. Immune defects observed in patients with primary malignant brain tumors. J Neuroimmunol. 1999;100:216–232. doi: 10.1016/s0165-5728(99)00203-9. [DOI] [PubMed] [Google Scholar]
- 18.Wu A, Wei J, Kong L-Y, Wang Y, Priebe W, Qiao W, Sawaya R, Heimberger AB. Glioma cancer stem cells induce immunosuppressive macrophages/microglia. Neuro Oncol. 2010;12:1113–1125. doi: 10.1093/neuonc/noq082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hussain SF, Yang D, Suki D, Aldape K, Grimm E, Heimberger AB. The role of human glioma-infiltrating microglia/macrophages in mediating antitumor immune responses. Neuro Oncol. 2006;8:261–279. doi: 10.1215/15228517-2006-008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohanbash G, McKaveney K, Sakaki M, Ueda R, Mintz AH, Amankulor N, Fujita M, Ohlfest JR, Okada H. GM-CSF promotes the immunosuppressive activity of glioma-infiltrating myeloid cells through interleukin-4 receptor-α. Cancer Res. 2013;73:6413–6423. doi: 10.1158/0008-5472.CAN-12-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaer DJ, Alayash AI, Buehler PW. Gating the radical hemoglobin to macrophages: the anti-inflammatory role of CD163, a scavenger receptor. Antioxid Redox Signal. 2007;9:991–999. doi: 10.1089/ars.2007.1576. [DOI] [PubMed] [Google Scholar]
- 22.Borda JT, Alvarez X, Mohan M, Hasegawa A, Bernardino A, Jean S, Aye P, Lackner AA. CD163, a marker of perivascular macrophages, is up-regulated by microglia in simian immunodeficiency virus encephalitis after haptoglobin-hemoglobin complex stimulation and is suggestive of breakdown of the blood-brain barrier. Am J Pathol. 2008;172:725–737. doi: 10.2353/ajpath.2008.070848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yen BL, Yen M-L, Hsu P-J, Liu K-J, Wang C-J, Bai C-H, Sytwu H-K. Multipotent human mesenchymal stromal cells mediate expansion of myeloid-derived suppressor cells via hepatocyte growth factor/c-met and STAT3. Stem Cell Reports. 2013;1:139–151. doi: 10.1016/j.stemcr.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sucheston-Campbell LE, Cannioto R, Clay AI, Etter JL, Eng KH, Liu S, Battaglia S, Hu Q, Szender JB, Minlikeeva A, Joseph J, Mayor P, Abrams SI, Segal B, Wallace PK, Soh KT, Zsiros EZ, Anton-Culver H, Bandera EV, Beckmann MW, Berchuck A, Bjørge L, Bruegl A, Campbell IG, Campbell SP, Chenevix-Trench G, Cramer D, Dansonka-Mieszkowska A, Dao F, Diergaarde B, Doerk T, Doherty JA, du Bois A, Eccles D, Engelholm SA, Fasching PA, Gayther SA, Gentry-Maharaj A, Glasspool RM, Goodman MT, Gronwald J, Harter P, Hein A, Heitz F, Hillemmanns P, Hogdall C, Høgdall EVS, Huzarski T, Jensen A, Johnatty SE, Jung A, Karlan B, Klapdor R, Kluz T, Konopka B, Krüger Kjær S, Kupryjanczyk J, Lambrechts D, Lester J, Lubiński J, Levine DA, Lundvall L, McGuire V, McNeish IA, Menon U, Modugno F, Ness RB, Orsulic S, Paul J, Pearce CL, Pejovic T, Pharoah P, Ramus SJ, Rothstein J, Rossing MA, Rübner M, Schildkraut JM, Schmalfeldt B, Schwaab I, Siddiqui N, Sieh W, Sobiczewski P, Song H, Terry KL, Van Nieuwenhuysen E, Vanderstichele A, Vergote I, Walsh CS, Webb PM, Wentzensen N, Whittemore AS, Wu AH, Ziogas A, Odunsi K, Chang-Claude J, Goode EL, Moysich KB. No evidence that genetic variation in the myeloid-derived suppressor cell pathway influences ovarian cancer survival. Cancer Epidemiol Biomarkers Prev. 2016;26:420–424. doi: 10.1158/1055-9965.EPI-16-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]