Abstract
Alternative splicing of the oncogene MDM2 is induced in response to genotoxic stress. MDM2-ALT1, the major splice variant generated, is known to activate the p53 pathway and impede full-length MDM2’s negative regulation of p53. Despite this perceptible tumor-suppressive role, MDM2-ALT1 is also associated with several cancers. Furthermore, expression of MDM2-ALT1 has been observed in aggressive metastatic disease in pediatric rhabdomyosarcoma (RMS), irrespective of histological subtype. Therefore, we generated a transgenic MDM2-ALT1 mouse model that would allow us to investigate the effects of this splice variant on the progression of tumorigenesis. Here we show when MDM2-ALT1 is ubiquitously expressed in p53 null mice it leads to increased incidence of spindle cell sarcomas, including RMS. Our data provide evidence that constitutive MDM2-ALT1 expression is itself an oncogenic lesion that aggravates the tumorigenesis induced by p53 loss. On the contrary, when MDM2-ALT1 is expressed solely in B cells in the presence of homozygous wild-type p53 it leads to significantly increased lymphomagenesis (56%) when compared to control mice (27%). However, this phenotype is observable only at later stages in life (≥18 months). Moreover, flow cytometric analyses for B cell markers revealed an MDM2-ALT1-associated decrease in the B cell population of the spleens of these animals. Our data suggest that the B cell loss is p53 dependent and is a response mounted to persistent MDM2-ALT1 expression in a wild-type p53 background. Overall our findings highlight the importance of an MDM2 splice variant as a critical modifier of both p53-dependent and p53-independent tumorigenesis, underscoring the complexity of MDM2 post-transcriptional regulation in cancer. Furthermore, MDM2-ALT1-expressing p53 null mice represent a novel mouse model of fusion-negative RMS.
Keywords: MDM2, splicing, lymphoma, p53 independent, rhabdomyosarcoma
Introduction
Rhabdomyosarcoma (RMS) is a type of soft tissue sarcoma of mesenchymal origin that primarily arises in patients less than 10 years of age. Histologically, RMS is classified into three main subtypes: embryonal (eRMS), alveolar (aRMS), and anaplastic. Of these, embryonal and alveolar tumors constitute the majority of cases and often present as high-grade metastatic disease with poorer prognoses. At the molecular level, aRMS tumors are frequently characterized by chromosomal rearrangements resulting in fusion proteins PAX3-FOXO1 (55%–75% aRMS) or PAX7-FOXO1 (10%–22% aRMS). eRMS and translocation-negative aRMS possess lesions in pathways such as MYCN (amplification), insulin receptor, NFκB, RAS, and Sonic Hedgehog (1–7). Several models of RMS have been developed that underscore its genetic heterogeneity and developmental plasticity (6–15). However, these only account for genetic aberrations observed in a small percentage of patients. For instance, PAX-FOXO1 translocation-based models represent only the aRMS subtype while those bearing perturbations in molecular pathways such as p53, IGF2, MYCN or RAS individually represent < 35% of cases (16). Hence, there remains the need for an in vivo platform to model molecular lesions that are more pervasive across RMS subtypes.
Murine Double Minute 2 (MDM2) is an E3 ubiquitin ligase that binds and targets the tumor suppressor p53 for proteasome-mediated degradation (17–21). Additionally, MDM2 inhibits p53’s transcriptional activity (22–25). Amplification or overexpression of MDM2 is oncogenic and a hallmark of several tumor types, most commonly soft tissue sarcomas (26). Alternative transcripts of MDM2 have been observed in RMS, breast, ovarian, lymphoma, and bladder cancers (27–38). MDM2-ALT1, an isoform typically induced in response to genotoxic stress, is incapable of binding and inactivating p53 and modulates the p53 pathway to sustain a fine-tuned stress response (39–42). However, several in vitro and in vivo studies have described transformative properties for MDM2-ALT1 (28, 29, 33, 40–45). Recently, we demonstrated that MDM2-ALT1 is constitutively expressed in 85% aRMS and 70% eRMS tumors (33). Moreover, MDM2-ALT1 strongly correlated with high-grade metastatic disease in both major subtypes (8, 42), independent of p53 mutations or PAX-FOXO1 fusion status making this to date the most common biomarker characterized, irrespective of RMS histology.
To ascertain whether MDM2-ALT1 contributes to rhabdomyosarcomagenesis, we generated a mouse model that expresses MDM2-ALT1 in a Cre recombinase-dependent (Cre) manner. Using this model, we report that ubiquitous MDM2-ALT1 expression is capable of accelerating tumorigenesis and is sufficient to drive RMS formation in p53 heterozygous and null mice. We attribute these effects to direct emergence of its tumorigenic functions in the absence of p53. However, when expressed in B cells in a wild-type p53 background we observed that MDM2-ALT1-associated tumorigenesis manifests at later stages of life. Together, our model has enabled us to dissect distinct facets of MDM2-ALT1-mediated tumorigenesis and elucidate its dual functionality as an oncogene and a tumor suppressor. Importantly, we demonstrate that MDM2-ALT1 can direct RMS tumor formation recapitulating many of the histological and immunohistochemical features of fusion-negative RMS.
Results
Generation of a transgenic mouse model for Cre-dependent MDM2-ALT1 expression
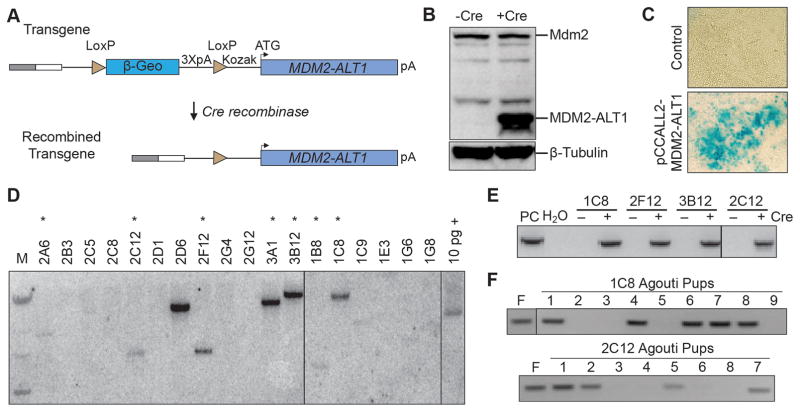
To characterize its role in tumor progression we developed a mouse model of Cre-dependent MDM2-ALT1 expression. We generated a construct that expresses MDM2-ALT1 upon Cre-mediated excision of an upstream β-geo cassette and three polyA signals (Figure 1A and 1B), confirmed by recombination in Escherichia coli (Figure S1) (46). This construct was electroporated into embryonic stem (ES) cells followed by selection for neomycin resistance. β-geo cassette expression was confirmed by β-gal staining (Figure 1C). Individual clones were screened by Southern hybridization for single insertion sites as indicated by a single band following hybridization of EcoRV-digested genomic DNA with a transgene-specific probe (Figure 1D). ES cell clones 2C12 and 1C8 (Figure 1E) were selected and mouse lines were generated using blastocyst injection. Once germline transmission was confirmed (Figure 1F) the chimeric mice were backcrossed into a C57BL/6 background to generate MDM2-ALT1 transgenic animals.
Figure 1. Design and generation of a Cre-inducible MDM2-ALT1 transgenic mouse model.
A. The human MDM2-ALT1 cDNA sequence was cloned into the pCCALL2 vector downstream of a β-geo cassette that is flanked by LoxP sites for recombination. Expression is driven by a CMV enhancer (gray box) and a chicken β-actin promoter (white box). Three downstream polyadenylation signals (3XpA) prevent transcriptional read-through. In the absence of Cre recombinase, only the β-geo cassette is expressed. When Cre is present, the β-geo cassette and polyadenylation signals are excised facilitating expression of the MDM2-ALT1 transgene. B. NIH/3T3 cells were transfected with either the non-recombined pCCALL2-MDM2-ALT1 (−Cre) construct or the recombined (+Cre) plasmids isolated from the BNN132 Escherichia coli strain. Expression of the MDM2-ALT1 protein from the transgene was confirmed by immunoblotting (MDM2 antibody AF1244). C. ES cells were electroporated with the pCCALL2-MDM2-ALT1 construct, selected for G418 resistance, and subjected to β-gal staining. ES cells that were successfully electroporated stained positive confirming LacZ expression from the β-geo cassette. D. Southern blot analysis was performed using a radiolabeled probe against the 3XpA sequence to detect clones with single transgene integrations. We showed that seven ES cell lines (*) are positive for the transgene integration event at a single site. E. To confirm that the inserted transgene maintained recombination competence at the genomic integration sites, the MDM2-ALT1 lines expressing embryonal stem cells were electroporated with a Cre-expressing vector (+) or nonelectroporated (−). PCR for Cre-mediated recombination shows the expected 743 base pair product in BNN pCCALL2 MDM2-ALT1 positive control (PC) and electroporated samples. F. Agouti pups bred from the chimeric founder mouse lines (F) for 1C8 and 2C12 were screened for the germline transmission of the transgene by PCR using primers specific for the MDM2-ALT1 exon 3–12 junction and the polyadenylation sequence. Genomic DNA from a chimeric founder mouse served as a positive control. Mice carrying the transgene produced a 629 base pair amplicon.
MDM2-ALT1 is capable of accelerating tumorigenesis and favors the development of RMS in p53 null mice
Given the conflicting evidence of MDM2-ALT1’s function as both a tumor suppressor and a potential oncogene, we hypothesized that an inactivating mutation in a critical tumor suppressor pathway would be necessary to observe its oncogenic role. Therefore, we generated tumor cohorts that ubiquitously express MDM2-ALT1 on a p53 null background using the early-embryonic driver Sox2-Cre. Our preliminary tumor cohorts comprised both 1C8 and 2C12 animals. Both lines expressed MDM2-ALT1 in tissues analyzed (Figures 2, and S2). Experimental (MDM2-ALT1+/−; Sox2-Cre+/−; p53−/−) mice in both 1C8 and 2C12 cohorts predominantly developed spindle cell sarcomas like rhabdomyosarcomas and osteosarcomas (Figures 2B and S3A). The control (MDM2-ALT1−/−; Sox2-Cre+/−; p53−/−) animals from either line did not develop these tumor types. In these initial tumor cohorts with limited mouse numbers, neither the 1C8 nor the 2C12-derived animals displayed altered life expectancy between control and experimental groups (Figure S3B and data not shown). For this reason, we chose to further characterize one line to investigate MDM2-ALT1 mediated tumorigenesis. We expanded the 2C12 line because of its robust transgene expression and fecundity.
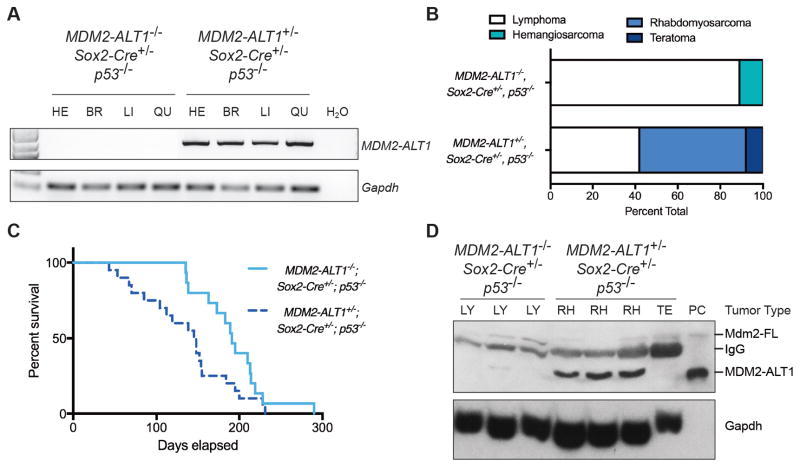
Figure 2. MDM2-ALT1-expressing mice show decreased survival and increased rhabdomyosarcoma compared to control mice.
A. RT-PCR assay showing the expression of MDM2-ALT1 in the heart (HE), brain (BR), liver (LI) and quadriceps muscle (QU) of experimental 2C12-MDM2-ALT1-positive mice, but not in the control MDM2-ALT1-negative mice. Gapdh expression serves as endogenous control. B. Graph depicting the tumor spectrum differences between primary malignancies of the control (MDM2-ALT1−/−; Sox2-Cre+/−; p53−/−, n = 11) and experimental mice (2C12-MDM2-ALT1+/−; Sox2-Cre+/−; p53−/−, n = 14) as determined by a comparative pathologist. 89% of the control tumors assessed were lymphomas. However, the tumor spectrum in experimental mice showed a greater proclivity for spindle cell sarcomas such as RMS (50%). One case of teratoma was also diagnosed. C. Kaplan-Meier curve showing that experimental mice (2C12-MDM2-ALT1+/−; Sox2-Cre+/−; p53−/−, n = 15) mice have an earlier onset of tumorigenesis and statistically significant decrease in viability compared to mice lacking the transgene (MDM2-ALT1−/−; Sox2-Cre+/−; p53−/−, n = 21, p = 0.0091 Gehan-Breslow-Wilcoxon test). D. Immunoblot showing expression of MDM2-ALT1 in three of four experimental tumors (rhabdomyosarcoma, RH; teratoma, TE), but not in the tumors of control mice (lymphoma, LY). The positive control (PC) for MDM2-ALT1 expression is lysate from MCF7 cells transfected with the MDM2-ALT1 expression plasmid. Gapdh serves as loading control.
In a larger cohort of age-matched control and experimental 2C12 mice, we observed that MDM2-ALT1 expression significantly accelerated tumor onset in the p53 null background and decreased median survival from 193 days (control n = 15) to 147 days (experimental n = 21) (Figure 2C; Table S1), but exhibited no differences in weight or muscle size between control and experimental animals (data not shown). Both control and experimental animals developed lymphoblastic T cell lymphomas (Figure 2B), characteristic of p53 null mice (47). However, MDM2-ALT1 induced the tumor spectrum of the p53 null mice to shift toward spindle cell sarcomas in the form of RMS (from 0% of controls to 42% of experimental mice; Figure 2B). We also confirmed expression of MDM2-ALT1 transcripts in five representative tumors from the transgene-positive mice (Figure S2C). At the protein level, three RMS tumors from the experimental mice showed detectable MDM2-ALT1 expression (Figure 2D). We did not detect expression in the teratoma and this tumor is unlikely to be MDM2-ALT1 driven but more likely a result of p53 loss as teratomas have been previoiusly observed in mice with a p53 null genotype (48). We also were not able to detect MDM2-ALT1 protein expression in the normal tissues of the mice that developed RMS (Figure S4). The malignancies were diagnosed by their strong immunoreactivity for CD3 (lymphomas, Figure 3A), or by staining for skeletal muscle lineage markers desmin, myogenin, and alpha-smooth muscle actin (RMS; Figure 3B) (49–51).
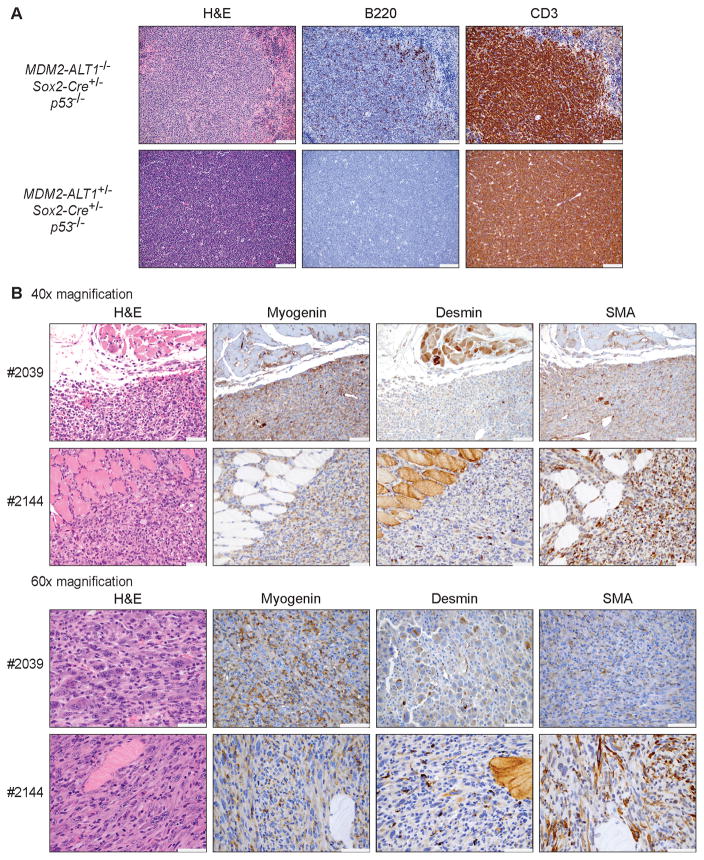
Figure 3. Immunophenotypes of lymphomas and expression of muscle-specific markers in rhabdomyosarcomas of MDM2-ALT1 mice.
A. Immunohistochemical analyses of representative lymphomas from control (MDM2-ALT1−/−; Sox2-Cre+/−; p53−/−, female – six months, top panel) and experimental (2C12-MDM2-ALT1+/−; Sox2-Cre+/−; p53−/−, male – five months, bottom panel) mice. Hematoxylin and eosin staining (H&E) of a control spleen with lymphoma depicts lymphoid sheath and marginal/mantle zones, which are disrupted by neoplastic lymphocytes and surrounded by red pulp with extramedullary hematopoiesis. The majority of neoplastic cells display membrane staining for T cells (CD3) with multifocal lymphocytes in the splenic white pulp at the marginal/mantle zones retaining B cell (B220) immunoreactivity. Experimental thymic lymphoma shows strong cytoplasmic staining for CD3 and relatively few neoplastic lymphocytes staining for B220 (scale = 100 μm). B. Immunohistochemical analyses of representative RMS from two 2C12-MDM2-ALT1+/−; Sox2-Cre+/−; p53−/− mice (#2039, 5-month male and #2144, 2-month male). Myogenin plays an essential role in myogenesis and has been shown to be an extremely sensitive and specific marker for RMS (50). Alpha-smooth muscle actin has also been shown to be a reliable marker for RMS (51). Immunohistochemistry for myogenin and alpha-smooth muscle actin (SMA) shows no staining in normal muscle tissue and moderate to marked cytoplasmic staining in multifocal tumor cells. Desmin is a muscle-specific type III intermediate filament that has been shown to be a highly accurate marker for RMS, distinguishing it from other pediatric sarcomas (49). There is diffuse cytoplasmic immunoreactivity for desmin in normal skeletal muscle, and weak staining in multifocal tumor cells, with rare scattered tumor cells exhibiting marked cytoplasmic staining. Both 40x and 60x magnification is shown (scale = 50 μm).
To confirm that these phenotypes were attributable to MDM2-ALT1, we generated an additional control without Cre alleles (2C12-MDM2-ALT1+/−; Sox2-Cre−/−; p53−/− n = 8). These mice showed no change in survival compared to controls expressing Cre without the MDM2-ALT1 transgene (MDM2-ALT1−/−; Sox2-Cre+/−; p53−/− n = 15) (p = 0.2735, data not shown). This finding argues against a transgene insertion site effect in the phenotypes we observe in our transgenic mice. To further rule out integration site effects we performed Targeted Locus Amplification (TLA) sequencing to precisely map the insertion locus of our mice (Figure S5 and S6) (52). In the 1C8 founder line the transgene integrated into a centromeric region of chromosome 18. For the 2C12 founder line, the transgene integrated into the coding region of Sbno1 on chromosome 5, which resulted in a deletion of ~3 kb of Sbno1. Expression of Sbno1 transcript and protein is reduced by approximately half in 2C12-MDM2-AT1+/− mice compared to control cohorts (Figure S7). Sbno1 has been shown to play a role in brain and central nervous system (CNS) development (53, 54). Interestingly, Sbno1 has also been indicated to have a higher mutational frequency in breast and lung cancers and is suggested to be a potential oncogene (55, 56). However, deletion of a single copy of Sbno1 does not give rise to CNS defects or breast or lung cancer in our 2C12 mice. Importantly, we do not see evidence of novel Sbno1 isoforms being expressed because of the transgene integration event as evidenced by western blot, and expression of genes surrounding Sbno1 was not affected (Figures S7 and S8). These results indicate that MDM2-ALT1 expression not only exacerbates tumorigenesis but also promotes spindle cell sarcoma formation in the absence of a functional p53 pathway, and this effect is not likely related to disruption of the Sbno1 gene.
MDM2-ALT1 expression induces expression of phospho-Ser394 MDM2
To explore the mechanisms driving tumorigenesis in our MDM2-ALT1-expressing p53 null mice we examined a subset of tumors from control (n = 3) and experimental (n = 4) animals for differences in common tumor suppressor and oncogenic pathways often deregulated in cancers. Importantly, we selected pathways that converge with the MDM2 regulatory network regardless of p53 status and queried whether some of these are perturbed in these tumors (Figure 4A). We chose this approach because MDM2-ALT1 is known to modulate full-length MDM2, and direct its function and localization (40, 42, 43). To examine if any observed changes are truly independent of p53 status, we also overexpressed MDM2-ALT1 in cell lines with wild-type p53 (Figure 4B). When we examined expression of the candidate factors including Slug (57), XIAP (58) and signaling pathways such as PI3K (59) (pAkt, Akt and Pten panels) and NFκB (60) (Rela panel), we did not observe MDM2-ALT1-specific differences in tumors from p53 null mice or in the transfected cell lines (Figure 4).
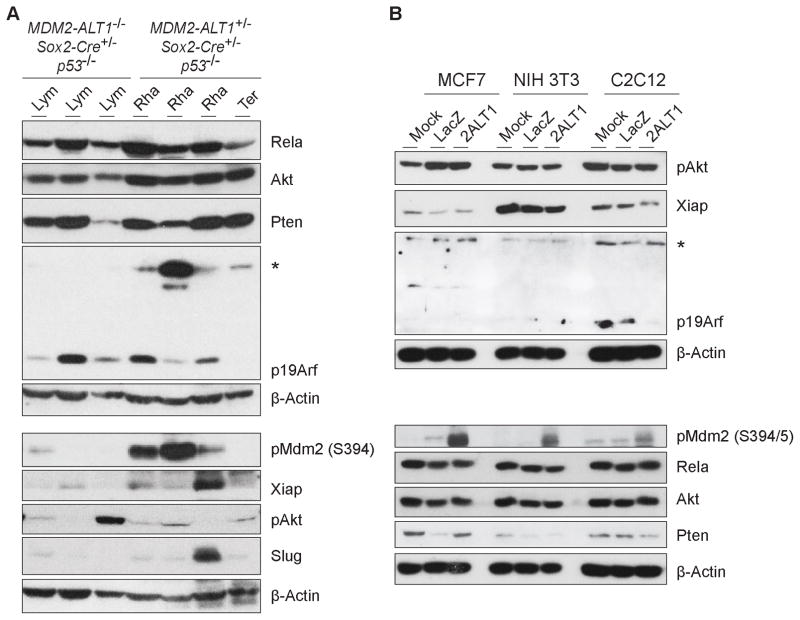
Figure 4. MDM2-ALT1-expressing tumors show upregulation of phospho-MDM2 as compared to control mice lymphomas.
A. Immunoblot showing expression of factors in experimental tumors (2C12-MDM2-ALT1+/−; Sox2-Cre+/−; p53−/− rhabdomyosarcoma, Rha; teratoma, Ter) as compared to control tumors (MDM2-ALT1−/−; Sox2-Cre+/−; p53−/− lymphoma, Lym). Three out of four experimental tumors showed robust expression of phospho-MDM2 Ser394, whereas one out of three control tumors showed low baseline expression. Immunoblotting analysis was performed once inspecting 3 control and 4 experimental tumors. B. Immunoblot showing expression of proteins implicated in the pathogenesis of MDM2-mediated tumorigenesis in transfected cells from both mouse (C2C12, NIH 3T3) and human (MCF7) after 48 hours. Upon transient expression of MDM2-ALT1 (2ALT1), there is an increase in phosphorylation of MDM2 at Ser394/5 in all three cell lines as compared to LacZ (negative control) or mock transfected samples. β-actin was used as a loading control. Higher migrating bands of unknown origin (*) in the p19Arf panel in both A and B are not phosphorylated or ubiquitinated forms of p19Arf.
Full-length Mdm2 was expressed to similar levels in control and experimental tumors (Figure 2D). Interestingly, all three MDM2-ALT1-positive RMS tumors showed increased phosphorylation of Mdm2 at Ser394, an Atm kinase target site critical for regulating p53-mediated stress response (Figure 4A, pMdm2 panel) (61–63). Additionally, all cell lines transiently transfected with MDM2-ALT1 (Figure 4B) showed increased phosphoMDM2 Ser394 levels (S395 in humans), indicating a p53-independent response to MDM2-ALT1 expression. However, when queried for any correlation between Atm activation and MDM2-ALT1 expression we did not see any difference in levels of phospho-Atm (S1981), the kinase active form (data not shown).
MDM2-ALT1 expression with heterozygous p53 exacerbates tumorigenesis
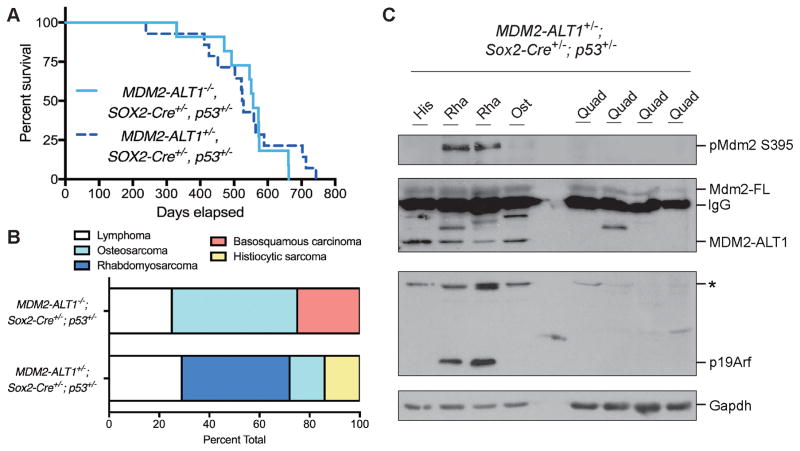
In the absence of functional p53 we observed that MDM2-ALT1 directly contributed to tumorigenesis. However, the consequences of constitutive MDM2-ALT1 expression in the context of wild-type p53 remain unclear. This is because of its conflicting role as an upregulator of p53 and the observation that MDM2-ALT1 does not correlate with p53 mutations in patients (32, 33, 39, 41, 42, 64). To examine whether or not MDM2-ALT1-mediated oncogenesis required loss of p53, we expressed MDM2-ALT1 in the context of mice expressing one functional p53 allele. p53 heterozygous mice are haploinsufficient (65) and are prone to tumorigenesis with median onset around nine months (66). Expression of MDM2-ALT1 did not alter the life expectancy of the experimental (2C12-MDM2-ALT1+/−; Sox2-Cre+/−; p53+/−, n = 14) animals compared to controls (MDM2-ALT1−/−; Sox2-Cre+/−; p53+/−, n = 11) (Figure 5A). However, the experimental mice showed a change in the tumor spectrum with increased incidence of RMS correlating with MDM2-ALT1 expression in a manner similar to the p53 null cohort (Figure 5B). Furthermore, the MDM2-ALT1-positive RMS tumors from the p53 heterozygous cohort exclusively showed upregulation of pMdm2 S394 compared to other tumors and corresponding normal tissue (Figure 5C). When we sequenced the hotspot regions of the wild-type p53 allele (exons 5 to 9) in these tumors (n = 4) (67), we observed no mutations indicating that MDM2-ALT1 induced tumorigenesis did not result from direct inactivation of the remaining p53 allele.
Figure 5. Mice expressing MDM2-ALT1 and heterozygous for p53 show increased rhabdomyosarcoma and upregulation of phospho-MDM2 compared to control p53 heterozygous mice.
A. Kaplan-Meier analysis shows no significant difference in life expectancy between control (MDM2-ALT1−/−; Sox2-Cre+/−; p53+/−, n = 11) and experimental mice (2C12-MDM2-ALT1+/−; Sox2-Cre+/−; p53+/−, n = 14, p = 0.6650 Gehan-Breslow-Wilcoxon test). B. Graph depicting the tumor spectrum differences between primary malignancies of the control (n = 4) and experimental mice (n = 7) as determined by a comparative pathologist. The most common malignancy in p53 heterozygous null animals is osteosarcoma. Our control mice displayed approximately 50% osteosarcoma, however the tumor spectrum in experimental mice showed decreased occurrence of osteosarcoma (14%) and a greater proclivity for RMS (43%). C. Tumors from four mice (histiocytic sarcoma, His; rhabdomyosarcoma, Rha; and osteosarcoma, Ost) were analyzed for protein expression and quadriceps muscles from each corresponding mouse (Quad) were used for comparison. All tumors show higher expression of MDM2-ALT1 compared to the normal muscle tissue, whereas increased expression of pMdm2 (S395) was RMS-specific. Higher migrating bands of unknown origin (*) in the p19Arf panel are not phosphorylated or ubiquitinated forms of p19Arf.
MDM2-ALT1 expression with wild-type p53 exacerbates tumorigenesis in aged mice and demonstrates defects in cell cycle progression prior to tumor onset
To appraise the effects of MDM2-ALT1 in mice with fully functional p53, we expressed MDM2-ALT1 in B cells where any p53-associated cell death or apoptosis will not have overt systemic effects such as embryonic lethality (44) or major organ failure. Importantly, MDM2-ALT1 expression has been observed in lymphomas and Mdm2-b increases the incidence of B cell lymphomas, underscoring the relevance of the system (29, 44, 45, 68). We crossed the MDM2-ALT1 transgenics with mice expressing Cre under the control of B cell-specific CD19 promoter and generated tumor cohorts of experimental (2C12-MDM2-ALT1+/−; CD19-Cre+/−; p53+/+) and control mice (MDM2-ALT1−/−; CD19-Cre+/−; p53+/+). We confirmed B cell-specific MDM2-ALT1 expression in the spleens of experimental animals (Figure 6A). We monitored control and experimental mice until they developed masses that met endpoint criteria for a maximum of 2 years and 21 days. When MDM2-ALT1 was limited to B cells, we did not observe significant differences in life expectancy or changes in tumor spectrum between control (n = 15) and experimental (n = 18) mice (Figure 6B, and data not shown). We determined p53 status by sequencing cDNA from spleens of four experimental and four control animals for p53 mutation hotspot regions (exons 5 to 9) (67). All samples examined were wild-type for p53 at this locus enabling us to estimate that overt somatic p53 mutations do not feature in our model (data not shown).
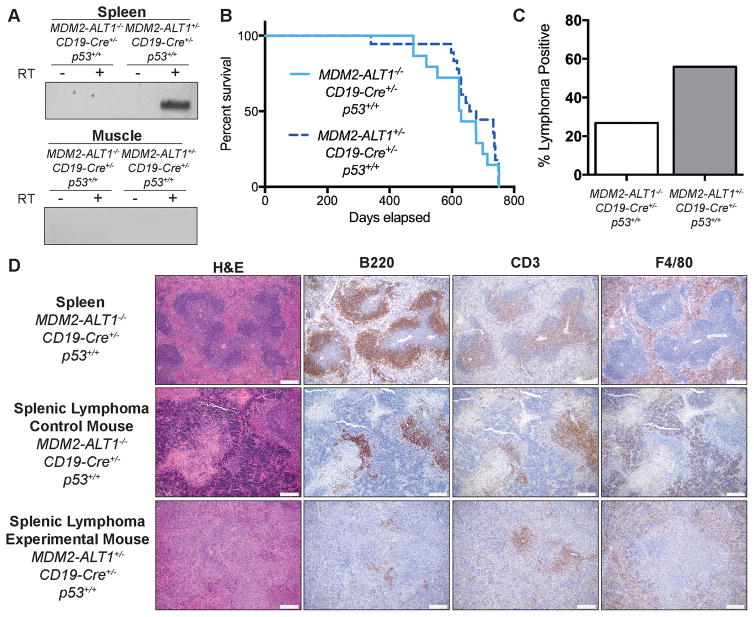
Figure 6. Mice expressing MDM2-ALT1 in B cells exhibit increased incidence of lymphomas compared to controls.
A. Expression of MDM2-ALT1 was confirmed in the spleens (B cells) of experimental mice positive for both the transgene and CD19-Cre alleles (2C12-MDM2-ALT1+/−; CD19-Cre+/−; p53+/+), but not in control mice negative for the MDM2-ALT1 transgene (MDM2-ALT1−/−; CD19-Cre+/−; p53+/+). Importantly, skeletal muscle of the transgene positive mice where the CD19 promoter is not active, did not express MDM2-ALT1. Control reactions without reverse transcription are also shown (-RT) to detect for possible contamination. B. Kaplan-Meier curves show no difference in life expectancy between survival cohorts of experimental (n = 18) and control mice (n = 15, p = 0.2737 Gehan-Breslow-Wilcoxon test). C. Mice expressing MDM2-ALT1 (19/34 lymphoma positive; 56%) in B cells showed significantly higher incidence of lymphomas compared to MDM2-ALT1 negative mice (11/41 lymphoma positive; 27%) at 18 months of age (p = 0.0174 Fisher’s exact test). D. Immunohistochemical analyses of representative splenic lymphomas from control (female – 22 months, middle panel) and experimental (female – 21 months, bottom panel) mice. For comparison, stained sections of a normal mouse spleen (MDM2-ALT1−/−; CD19-Cre+/−; p53+/+) are presented (male – 21 months, top panel). Overall, the architecture of the spleen shows marked disruption by neoplastic lymphocytes, which are more atypical in the experimental lymphoma compared to the lymphoma from the control mouse and the wild-type spleen. B220-positive B cells are significantly lowered in comparison to the normal spleen and the control mouse lymphoma and only a few small aggregates are seen scattered in the red pulp and the white pulp nodules. Additionally, CD3-positive staining is observed throughout the white pulp nodules that have lost normal architecture as compared to staining in the periarterioral lymphoid sheath in the control lymphoma and wild-type spleen. F4/80-positive cells are mainly located in the red pulp surrounding the white pulp nodules with some scattered within the white pulp; a pattern similar to normal macrophage distribution in the spleen (Scale = 200 μm).
It is possible that with the endpoint analyses, phenotypic variations that arose earlier and were less penetrant were not detected. Hence, in subsequent experiments, we harvested cohorts of control and experimental mice before endpoint was reached (18–22 months) and performed complete necropsy and histopathological analyses (control: MDM2-ALT1−/−; CD19-Cre+/−; p53+/+, n = 41 and experimental: 2C12-MDM2-ALT1+/−; CD19-Cre+/−; p53+/+, n = 34). Both experimental and control groups presented with age-associated disorders such as degenerative joint disease, atrophy, and incidental non-neoplastic lesions including fibro-osseous and vascular lesions. Additionally, both experimental and control animals developed non-lymphoid neoplasms common to aging C57BL/6 mice [Table S2 and (69)].
When we evaluated the occurrence of lymphoid neoplasms in these animals we observed that mice from both control and experimental groups developed lymphomas in the spleens and/or lymph nodes. We found at 18 months of age, experimental mice (56% - 19 of 34) showed significantly higher lymphoma incidence compared to controls (27% - 11 of 41) (Figure 6C, Table S2). In our experimental mice, we observed significant disruption of the splenic architecture by a more atypical population of neoplastic lymphocytes in lymphomas when compared to those from the controls in four of five cases examined for hematoxylin and eosin (H&E), B220 (B cell), CD3 (T cell) and F4/80 (macrophage) staining (Figure 6D). The low incidence of tumors in control mice is because lymphomas are commonly associated with aging C57BL/6 mice (69). These results indicate that constitutive MDM2-ALT1 expression in B cells contributed to increased lymphomagenesis in mice of advanced age.
MDM2-ALT1-positive mice show decreased B cell numbers
To examine the origin of the observed lymphoid neoplasms we used flow cytometry for B cell markers (CD19, B220, IgG and IgM) and T cell markers (CD3e and CD5) to immunophenotype the spleens and lymph nodes (axillary, inguinal, and mesenteric) of control (MDM2-ALT1−/−; CD19-Cre+/−; p53+/+, n = 18) and experimental (2C12-MDM2-ALT1+/−; CD19-Cre+/−; p53+/+, n = 15) animals. Concordant with the increased incidence of tumorigenesis, we expected that the spleens and lymph nodes of experimental mice would generally exhibit higher levels of B cell markers than controls. Surprisingly, we found that the experimental mice presented with a significantly lower population of cells expressing B cell markers compared to control animals, but exhibited no change in T cell markers (Figure 7A). This indicates a B cell-specific decrease in cell number potentially due to cell cycle arrest or apoptosis. Hence, we hypothesized that MDM2-ALT1 first led to a widespread lowering of B cell populations in a p53-dependent manner, but eventually overcame such tumor-suppressive constraints and conferred a growth advantage that presented itself as the observed increase in lymphomagenesis.
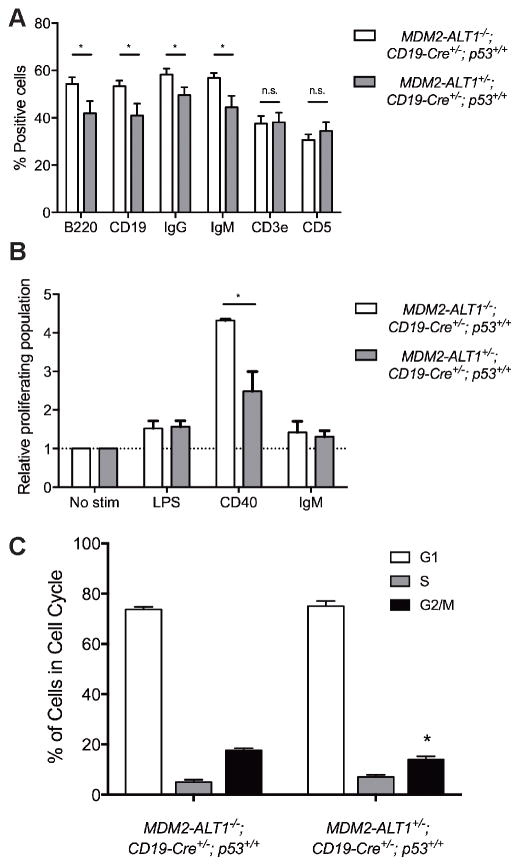
Figure 7. Transgenic mice expressing MDM2-ALT1 in B cells show significantly reduced populations of cells with B cell markers and defects in proliferation in spleens compared to controls.
A. Spleens of age-matched control (MDM2-ALT1−/−; CD19-Cre+/−; p53+/+) and experimental (2C12-MDM2-ALT1+/−; CD19-Cre+/−; p53+/+) mice were harvested at 18 months and the splenocytes were stained for B cell markers CD19, B220, IgG or IgM or T cell markers CD3e or CD5. Compared to MDM2-ALT1-negative control mice (n = 18) the MDM2-ALT1-positive experimental (n = 15) mice showed a statistically significant decrease in the population of cells expressing B cell markers but no changes in T cell population. B. Splenocytes isolated from control (n = 4) and experimental (n = 5) mice were labeled with a fluorescence marker, CFSE, and stimulated for 72 hours with lipopolysaccharides (LPS), CD40 ligand, or anti-IgM molecules. When stimulated with CD40, the MDM2-ALT1 expressing experimental cohort showed a significantly lower proliferative response compared to splenocytes from control mice. The proliferating population was measured by gating for cells displaying low CFSE fluorescence intensity, indicative of dilution of the dye upon cell division. The percent proliferating population from each group was normalized to the respective non-stimulated control set (No stim). * indicates p < 0.05 in all cases as determined by Two-tailed Student’s T test and error bars represent the standard error of the mean. C. Splenocytes from both control (MDM2-ALT1−/−; CD19-Cre+/−; p53+/+, n = 6) and experimental (MDM2-ALT1+/−; CD19-Cre+/−; p53+/+, n = 7) mice were isolated and stained for B cell (B220) and T cell (CD3e) markers, as well as a live/dead discriminator. Cells were then either fixed and stained for propidium iodide and analyzed by flow cytometry. B cells from experimental mice showed a significant reduction of cells in G2/M phase as to controls (p = 0.0435). * indicates p < 0.05 in all cases as determined by Two-tailed Student’s T test and error bars represent the standard error of the mean.
To ascertain whether the decreased numbers were attributed to defective proliferation, we performed ex vivo immunogenic challenge assays on B cells obtained from the experimental (2C12-MDM2-ALT1+/−; CD19-Cre+/−; p53+/+, n = 4) and control (MDM2-ALT1−/−; CD19-Cre+/−; p53+/+, n = 5) animals at 4 months of age. Briefly, we isolated splenocytes from the two groups and cultured and stimulated them for 72 hours with lipopolysaccharides (LPS), CD40 ligand, or anti-IgM molecules. We observed that in comparison with a non-stimulated control, only the CD40 ligand stimulation induced proliferation (Figure 7B). Importantly, cells from the MDM2-ALT1-positive experimental group showed significantly lowered proliferative response compared to those from the control animals. When we examined cell cycle progression of the B cells in four month old cohorts using propidium iodide staining, we observed a modest but statistically significant reduction in the percentage of cells in the G2/M phase in MDM2-ALT1 positive mice (n = 7) compared to controls (n = 6) (Figure 7C). On the other hand, when we assessed apoptosis in B cells of these animals using Annexin V staining, we observed no significant differences between MDM2-ALT1-positive and control mice (Figure S9A). Additionally, we stained for apoptotic genes in splenocytes from mice 7 weeks of age but did not detect differences in expression levels between control (MDM2-ALT1−/−; CD19-Cre+/−; p53+/+, n = 3) and experimental (2C12-MDM2-ALT1+/−; CD19-Cre+/−; p53+/+, n = 4) animals (Figure S9B and C). However, at 20–24 months of age we detected significantly higher Trp53 transcript levels in experimental (2C12-MDM2-ALT1+/−; CD19-Cre+/−; p53+/+ n = 7) mice compared to controls (2C12-MDM2-ALT1+/−; CD19-Cre−/−; p53+/+ n = 2 and 2C12-MDM2-ALT1−/−; CD19-Cre+/−; p53+/+ n = 5) Upon further analysis of p53 target genes that affect cell cycle, we did not detect any significant differences between the cohorts (Figure S10). Taken together, these results suggest that the lowered populations of B cells in the MDM2-ALT1 expressing mice arise from their impaired proliferative capacity along with discernible alterations in p53 levels, detectable at multiple stages during the lifespan of the animals. This aligns with our hypothesis that in the presence of wildtype p53, MDM2-ALT1 fosters incremental changes in p53 expression and possibly its activity that over time, lead toward selection for the observed tumorigenic phenotypes.
Discussion
MDM2-ALT1 presents a conundrum because its ability to antagonize MDM2 and MDMX and stabilize p53 is directly at odds with its transformative properties. For instance, MDM2-ALT1 modifies the p53 pathway inducing p53-dependent G1/S phase cell cycle arrest when overexpressed (29, 42). Similarly, in vivo models that ubiquitously expressed Mdm2-b with intact p53 were unable to obtain transgenic animals presumably due to p53-associated embryonic lethality (44). In contrast, Mdm2-b expressed from a tissue-specific promoter increased tumor incidence (44). Likewise, in vitro (28, 33, 44) and adoptive transfer assays (45) demonstrated its tumorigenic properties. These conflicting attributes are only partially explained in the context of mutant p53 where MDM2-ALT1 exacerbates oncogenesis by stabilizing dominant-negative p53 (43). In fact, malignancies associated with MDM2-ALT1 often lack p53 mutations (33, 64, 70) leaving unresolved, the nature of its tumorigenicity.
We developed a mouse model of Cre-dependent MDM2-ALT1 expression that enables tissue-specific assessment of its physiological activity. We show that in the absence of p53, MDM2-ALT1 significantly accelerates tumorigenesis in the 2C12 line. This is a powerful extension of previous in vitro experiments where MDM2-ALT1 has been shown to confer a growth advantage to p53 null cells (28, 44). Importantly, this underscores its distinct oncogenicity because full-length MDM2 and another cancer-associated splice variant Mdm2-a (human MDM2-ALT2) were unable to significantly alter tumor onset or survival in p53 null mice (71, 72), despite exhibiting growth-promoting properties in vitro (28, 71).
Importantly, we show that MDM2-ALT1, upon ubiquitous expression in a p53 null background, is sufficient to increase in spindle cell sarcomas, especially RMS, a malignancy strongly associated with MDM2-ALT1 expression (28, 33, 36). Similarly, when MDM2-ALT1 is expressed in a p53 heterozygous null background we demonstrated a shift in tumor spectrum from osteosarcoma [a common neoplasm for p53+/− mice (73)] to RMS. Patient RMS tumors presenting with MDM2-ALT1 typically retain the wild-type p53 alleles and do not present with overt mutations (33). However, a recent meta-analysis demonstrated that p53 signaling was inactive in 85% of PAX3-FOXO1 (n = 62) and 75% PAX7-FOXO1 (n = 24) aRMS tumors (74). This indicates that despite a dearth of mutations in the p53 gene, RMS tumors possess a defunct p53 pathway uncoupled via alternate means. Indeed, many genetic mouse models of RMS-associated lesions still require a mutation in the CDKN2A or TRP53 locus to observe a fully penetrant tumor phenotype (9, 10, 66, 74, 75).
Since MDM2-ALT1 is sufficient to alter tumorigenesis in p53 null and heterozygous animals, this supports a p53-independent component in MDM2-ALT1-mediated transformation. To identify p53-independent mechanisms underlying MDM2-ALT1 expression we examined tumors from MDM2-ALT1-positive animals for perturbations in tumor suppressive and oncogenic pathways that are subject to MDM2-mediated regulation, irrespective of p53 status (48, 58, 72, 76–87). Strikingly, we observed an elevation in the phosphorylation of Mdm2 at Ser394/395 in association with MDM2-ALT1 expression in both p53-deficient RMS tumors (p53 heterozygous and p53 null) and cell lines with wild-type p53, indicating p53-independent induction. Phospho-Ser394 Mdm2 is stimulated upstream of p53 upon DNA damage and controls its levels during and after stress response by affecting MDM2 oligomerization and E3 ligase activity (61–63, 88, 89). Recent studies have indicated that this is a crucial means of modulating MDM2 function and its deregulation impacts oncogenesis (63, 90). Constitutive phospho-Ser394 Mdm2 expression can have several implications including: a) MDM2-ALT1, whose expression is physiologically induced only under genotoxic stress (33, 39, 40), mimics DNA-damage conditions upon ectopic overexpression triggering phospho-Ser394 Mdm2 (41–43), and/or b) oncogene-induced DNA replication stress (91–93). Because most cancers lack functional DNA repair pathways, their hyper-replicative state leads to widespread genomic instability often selecting for deletions or chromosomal rearrangements favorable for cancer promotion (91–95). This behavior has been reported for several oncogenes (96–99) including full-length MDM2 and MDMX (84, 100, 101). It is possible that similar mechanisms are involved in MDM2-ALT1-mediated transformation exacerbating the tumor sensitivity ordained by p53 loss. Finally, we have shown previously that MDM2-ALT1-positive patient RMS tumors constitutively exhibit several features of stress-response signaling (33). Stress-associated phospho-Ser394 Mdm2 in MDM2-ALT1 mouse tumors suggests that our RMS model recapitulates this aspect of human disease.
When MDM2-ALT1 is expressed in a wild-type p53 background, we show for the first time a concurrence of phenotypes demonstrative of both its tumor suppressive and oncogenic potential. MDM2-ALT1 expression increased lymphomagenesis at an advanced age (≥ 18 months), but also presented a concomitant decrease in splenic B cell populations. Additionally, MDM2-ALT1-expressing B cells did not proliferate as efficiently in response to antigenic stimulants as control cells and displayed cell cycle defects and increased p53 expression concordant with MDM2-ALT1’s ability to induce p53-dependent cell cycle arrest in vitro (Figure 7 and S10). This finding is in accordance with our previous data wherein we demonstrate that exogenous MDM2-ALT1 expression is sufficient to induce cell cycle arrest and upregulate p53 target genes involved in cell cycle control (42). We conclude that deficiencies in B cell proliferation coupled with altered p53 expression contributed to the delayed manifestation of oncogenesis in MDM2-ALT1-positive mice (44) via oncogene-induced senescence, a state in which pre-cancerous cells with intact tumor suppressor pathways set up barriers to tumorigenesis via accumulation of DNA damage signals (92, 93, 102). Altogether, under wild-type p53 the anti-proliferative effects of MDM2-ALT1 will predominate and unlike the p53 null scenario, the tumor-promoting functions will not be immediately apparent. It is likely that a critical level of exposure to MDM2-ALT1 is required, exerting selective pressure to surmount or evade extant tumor-suppressive pathways. Finally, our results reflect previous studies showing late-onset tumorigenesis (80–100 weeks) or senescence in mice expressing mouse homologs of MDM2-ALT1 or MDM2-ALT2 in the presence of an intact p53 pathway (44, 71).
In summary, we show that MDM2-ALT1 promotes tumorigenesis. However, tumor patterns, time of onset, and mechanisms depend upon the nature of its interaction with the p53 tumor suppressor pathway. The preponderance of RMS tumors in our MDM2-ALT1-positive mice when p53-deficient, distinguishes it as a novel model for fusion-negative RMS. Moreover, it represents a critical tool to assess genetic interactions of MDM2-ALT1 with other RMS-associated lesions such as PAX-FOXO1 (alveolar), ΔNp73 over-expression, Arf deletions and Ras mutations (16, 33, 36, 75). Importantly, our system presents an efficient platform for modeling various MDM2-ALT1 associated malignancies to capture the intricacies of its functionality, which eventually will facilitate design and testing of novel therapeutic strategies.
Materials and Methods
Generation of transgenic MDM2-ALT1 lines
All animal experiments were performed according to institute standards approved by the Institute Animal Care and Use Committee (IACUC) at The Research Institute at Nationwide Children’s Hospital. The MDM2-ALT1 transgene was generated by cloning the human cDNA sequence into the mammalian expression vector pCCALL2 (46). The pCCALL2-MDM2-ALT1 construct was linearized with StuI, electroporated into ES cells, and screened for G418-resistant colonies and β-gal staining. Southern analysis of EcoRV-digested genomic DNA was conducted to determine ES cell lines with single insertion events. The 1C8 and 2C12 lines were used for blastocyst injection and chimeric mice were generated using standard transgenesis protocols. Following germline transmission, chimeric mice were crossed to C57BL/6 mice. To generate MDM2-ALT1; CD19-Cre cohorts, MDM2-ALT1 mice were bred to CD19-Cre mice (B6N.129P2-Cd19tm1(cre)Cgn/J). To generate the MDM2-ALT1-p53 tumor cohorts with both male and female mice, the following strains were utilized: F3 MDM2-ALT1, Sox2-Cre (B6.Cg-Tg(Sox2-cre)1Amc/J; strain 008454), and p53 null mice (B6.129S2-Trp53tm1Tyj/J; strain 002101). Animals were genotyped using primers outlined in Table S3. For the ex vivo proliferation assay the F3 and F7 MDM2-ALT1 animals were used. Since there were no treatment groups, animals were not randomized, but we were blinded during the analysis of phenotypes including tumor diagnosis and cell cycle studies. To map the transgene insertion site, TLA was performed by Cergentis (Utrecht, The Netherlands) with two transgene-specific primer pairs.
Cell culture, growth, and transfection
Cell lines were obtained from ATCC and maintained in DMEM (MCF7, verified by STR analysis by Genetica DNA Laboratories) or RPMI (C2C12 and NIH 3T3) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific), L-glutamine, and penicillin/streptomycin (Corning). MCF7 cells were negative for mycoplasma contamination. For transfection of MDM2-ALT1 or LacZ overexpression plasmids, Lipofectamine 3000 (Thermo Fisher Scientific) was used according to manufacturer’s protocol. Cells were harvested after 48 hours in RIPA buffer and proteins were separated by SDS-PAGE.
Reverse transcription and polymerase chain reaction (RT-PCR)
RNA isolation was performed using the RNeasy Mini Protocol (Qiagen). RNA with random hexamers was used to synthesize cDNA in reverse transcription reactions using the Transcriptor RT enzyme (Roche Diagnostics). MDM2-ALT1 expression was validated using MDM2-ALT1 genotyping primers (Table S3). Controls lacking RT were used to rule out genomic DNA contamination. Quantitative RT-PCR (qRT-PCR) on transgenic tissues was performed with standard PCR conditions for 40 cycles using an Applied Biosystems 7900HT Fast Real Time PCR system (Life Technologies). To measure Sbno1 expression and surrounding genes, transcript levels were quantified by qRT-PCR using primers outlined in Table S3. To investigate expression of cell cycle target genes RNA from mouse spleens was used in qRT-PCT experiments with the corresponding primers (Table S3).
Sequencing
Transgene sequences were verified from mouse gDNA using standard PCR protocol. Spleens from eight (2C12-MDM2ALT1+/−; CD19-Cre+/−; p53+/+) or four (2C12-MDM2ALT1+/−; Sox2-Cre+/−; p53+/−) mice were analyzed for p53 mutations in exons 5–9 (see Table S3 for sequencing primer details).
Pathological analysis of transgenic mice
Mice were monitored for tumor development biweekly. Mice showing signs of pain or distress or with masses, abscesses and tumors in excess of 2.0 centimeters were humanely euthanized according to endpoint criteria. At 18–22 months, the mice from the 2C12-MDM2-ALT1; CD19-Cre; p53 cohort mice were euthanized, necropsied, and examined for lesions and tissue abnormalities. Mice from the MDM2-ALT1; Sox2-Cre; p53 null cohort were euthanized and necropsied when tumor burden endpoints were reached. Tissue specimens were fixed in 10% neutral buffered formalin, paraffin embedded, and sectioned, followed by hematoxylin and eosin staining, and other histochemical and immunohistochemical stains. Kaplan-Meier statistical analysis and graphic representation of results were performed using GraphPad Prism Software.
Immunoblotting and Immunohistochemistry
Tissue was homogenized using a Tissumizer from Tekmar in RIPA buffer and equal amounts of protein were separated by SDS-PAGE. The following antibodies were used for protein detection: MDM2 clone 2A10 (kind gift from Dr. Arthur Levine); MDM2 (AF1244) from R&D Systems; Gapdh (14C10), Pten (138G6), pAkt S473 (D9E) from Cell Signaling; Rela (C-20), Xiap (E-2), Slug (A-7) from Santa Cruz Biotechnology; p19Arf from Abcam; pMDM2 S395 from Thermo Fisher; and β-Actin (AC-15) from Sigma. Tissue specimens were stained with the following antibodies: B220 from BD Pharmingen; CD3, Myogenin (F5D), and Smooth Muscle Actin (1A4) from Dako; and Desmin from Abcam.
Flow cytometry analyses and ex vivo proliferation assays
Freshly excised spleens and lymph nodes of mice were mashed, filtered, and resuspended in FACS buffer (1X phosphate buffered saline plus 2% fetal calf serum and 0.01% sodium azide). Red blood cells were lysed using ACK lysis buffer (Life Technologies). Cells were stained for 30 minutes at 4°C with a combination of either anti-B220, anti-CD5, anti-IgM and anti-CD19 or anti-IgG, anti-CD3e, anti-IgM and anti-CD19 antibodies, then stained with PE-p53, FITC-Bcl-2, FITC-Annexin V (Life Technologies) or fixed and stained with propidium iodide. IgG FITC and IgM PE Cy7 antibodies were obtained from Southern Biotech while all others were purchased from BD Biosciences unless noted otherwise. Only live cell populations were analyzed on the basis of a live-dead cell discriminator (7AAD, BD Pharmingen). Stained cells were then analyzed by flow cytometry using a BD Biosciences Accuri Cytometer. Data were collected for 10,000 events. For the ex vivo antigen challenge assays, splenocytes were harvested as above and were either untreated or stimulated with LPS (100 ng/mL, Sigma) or CD40 ligand (1 μg/mL, BD Biosciences) or anti-IgM antibodies (50 μg/mL, Southern Biotech) for 72h. All splenocytes were additionally treated with CFSE (Life Technologies) to trace cell division. Flow cytometry was performed and analyzed as mentioned above for B220, CD19, IgG, CD5, IgM markers.
Supplementary Material
Acknowledgments
The authors would like to acknowledge Matthew Rouhier for optimizing conditions for cell cycle analysis of B cells; Juliann Rectenwald and The Ohio State University Comparative Pathology & Mouse Phenotyping Shared Resource, supported in part through NCI Cancer Center Support Grant P30 CA016058; and funding sources Alex’s Lemonade Stand, Sarcoma Foundation of America, NIH (CA133571) awarded to DSC and Pelotonia (awarded to DFC and AGJ). We also thank Dr. Andras Nagy for kindly providing the plasmid used in generating the mouse models.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
References
- 1.Davis RJ, Barr FG. Fusion genes resulting from alternative chromosomal translocations are overexpressed by gene-specific mechanisms in alveolar rhabdomyosarcoma. Proc Natl Acad Sci U S A. 1997;94(15):8047–51. doi: 10.1073/pnas.94.15.8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hachitanda Y, Toyoshima S, Akazawa K, Tsuneyoshi M. N-myc gene amplification in rhabdomyosarcoma detected by fluorescence in situ hybridization: its correlation with histologic features. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 1998;11(12):1222–7. [PubMed] [Google Scholar]
- 3.Minniti CP, Tsokos M, Newton WA, Helman LJ. Specific expression of insulin-like growth factor-II in rhabdomyosarcoma tumor cells. American journal of clinical pathology. 1994;101(2):198–203. doi: 10.1093/ajcp/101.2.198. [DOI] [PubMed] [Google Scholar]
- 4.Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J, et al. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14(5):369–81. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xia SJ, Pressey JG, Barr FG. Molecular pathogenesis of rhabdomyosarcoma. Cancer biology & therapy. 2002;1(2):97–104. doi: 10.4161/cbt.51. [DOI] [PubMed] [Google Scholar]
- 6.Tostar U, Malm CJ, Meis-Kindblom JM, Kindblom LG, Toftgard R, Unden AB. Deregulation of the hedgehog signalling pathway: a possible role for the PTCH and SUFU genes in human rhabdomyoma and rhabdomyosarcoma development. J Pathol. 2006;208(1):17–25. doi: 10.1002/path.1882. [DOI] [PubMed] [Google Scholar]
- 7.Hatley ME, Tang W, Garcia MR, Finkelstein D, Millay DP, Liu N, et al. A mouse model of rhabdomyosarcoma originating from the adipocyte lineage. Cancer Cell. 2012;22(4):536–46. doi: 10.1016/j.ccr.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fleischmann A, Jochum W, Eferl R, Witowsky J, Wagner EF. Rhabdomyosarcoma development in mice lacking Trp53 and Fos: tumor suppression by the Fos protooncogene. Cancer Cell. 2003;4(6):477–82. doi: 10.1016/s1535-6108(03)00280-0. [DOI] [PubMed] [Google Scholar]
- 9.Nanni P, Nicoletti G, De Giovanni C, Croci S, Astolfi A, Landuzzi L, et al. Development of rhabdomyosarcoma in HER-2/neu transgenic p53 mutant mice. Cancer Res. 2003;63(11):2728–32. [PubMed] [Google Scholar]
- 10.Sharp R, Recio JA, Jhappan C, Otsuka T, Liu S, Yu Y, et al. Synergism between INK4a/ARF inactivation and aberrant HGF/SF signaling in rhabdomyosarcomagenesis. Nat Med. 2002;8(11):1276–80. doi: 10.1038/nm787. [DOI] [PubMed] [Google Scholar]
- 11.Chamberlain JS, Metzger J, Reyes M, Townsend D, Faulkner JA. Dystrophin-deficient mdx mice display a reduced life span and are susceptible to spontaneous rhabdomyosarcoma. FASEB J. 2007;21(9):2195–204. doi: 10.1096/fj.06-7353com. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez K, Serinagaoglu Y, Hammond S, Martin LT, Martin PT. Mice lacking dystrophin or alpha sarcoglycan spontaneously develop embryonal rhabdomyosarcoma with cancer-associated p53 mutations and alternatively spliced or mutant Mdm2 transcripts. Am J Pathol. 2010;176(1):416–34. doi: 10.2353/ajpath.2010.090405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosur V, Kavirayani A, Riefler J, Carney LMB, Lyons B, Gott B, et al. Dystrophin and dysferlin double mutant mice: a novel model for rhabdomyosarcoma. Cancer genetics. 2012;205(5):232–41. doi: 10.1016/j.cancergen.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hosur V, Kavirayani A, Riefler J, Carney LM, Lyons B, Gott B, et al. Dystrophin and dysferlin double mutant mice: a novel model for rhabdomyosarcoma. Cancer genetics. 2012;205(5):232–41. doi: 10.1016/j.cancergen.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zanola A, Rossi S, Faggi F, Monti E, Fanzani A. Rhabdomyosarcomas: an overview on the experimental animal models. Journal of Cellular and Molecular Medicine. 2012;16(7):1377–91. doi: 10.1111/j.1582-4934.2011.01518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Brien D, Jacob AG, Qualman SJ, Chandler DS. Advances in pediatric rhabdomyosarcoma characterization and disease model development. Histology and histopathology. 2012;27(1):13–22. doi: 10.14670/hh-27.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387(6630):299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 18.Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21(3):307–15. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou BP, Liao Y, Xia W, Zou Y, Spohn B, Hung M-C. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nature cell biology. 2001;3(11):973–82. doi: 10.1038/ncb1101-973. [DOI] [PubMed] [Google Scholar]
- 20.Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Mono- Versus Polyubiquitination: Differential Control of p53 Fate by Mdm2. Science. 2003;302(5652):1972–5. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- 21.Pant V, Lozano G. Limiting the power of p53 through the ubiquitin proteasome pathway. Genes & Development. 2014;28(16):1739–51. doi: 10.1101/gad.247452.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69(7):1237–45. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 23.Ito A, Lai CH, Zhao X, Saito S, Hamilton MH, Appella E, et al. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. Embo j. 2001;20(6):1331–40. doi: 10.1093/emboj/20.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274(5289):948–53. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 25.Brooks CL, Gu W. p53 Regulation by Ubiquitin. FEBS letters. 2011;585(18):2803–9. doi: 10.1016/j.febslet.2011.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fakharzadeh SS, Rosenblum-Vos L, Murphy M, Hoffman EK, George DL. Structure and organization of amplified DNA on double minutes containing the mdm2 oncogene. Genomics. 1993;15(2):283–90. doi: 10.1006/geno.1993.1058. [DOI] [PubMed] [Google Scholar]
- 27.Tamborini E, Della Torre G, Lavarino C, Azzarelli A, Carpinelli P, Pierotti MA, et al. Analysis of the molecular species generated by MDM2 gene amplification in liposarcomas. Int J Cancer. 2001;92(6):790–6. doi: 10.1002/ijc.1271. [DOI] [PubMed] [Google Scholar]
- 28.Sigalas I, Calvert AH, Anderson JJ, Neal DE, Lunec J. Alternatively spliced mdm2 transcripts with loss of p53 binding domain sequences: transforming ability and frequent detection in human cancer. Nat Med. 1996;2(8):912–7. doi: 10.1038/nm0896-912. [DOI] [PubMed] [Google Scholar]
- 29.Sanchez-Aguilera A, Garcia JF, Sanchez-Beato M, Piris MA. Hodgkin’s lymphoma cells express alternatively spliced forms of HDM2 with multiple effects on cell cycle control. Oncogene. 2006;25(18):2565–74. doi: 10.1038/sj.onc.1209282. [DOI] [PubMed] [Google Scholar]
- 30.Matsumoto R, Tada M, Nozaki M, Zhang CL, Sawamura Y, Abe H. Short alternative splice transcripts of the mdm2 oncogene correlate to malignancy in human astrocytic neoplasms. Cancer Res. 1998;58(4):609–13. [PubMed] [Google Scholar]
- 31.Lukas J, Gao DQ, Keshmeshian M, Wen WH, Tsao-Wei D, Rosenberg S, et al. Alternative and aberrant messenger RNA splicing of the mdm2 oncogene in invasive breast cancer. Cancer Res. 2001;61(7):3212–9. [PubMed] [Google Scholar]
- 32.Kraus A, Neff F, Behn M, Schuermann M, Muenkel K, Schlegel J. Expression of alternatively spliced mdm2 transcripts correlates with stabilized wild-type p53 protein in human glioblastoma cells. Int J Cancer. 1999;80(6):930–4. doi: 10.1002/(sici)1097-0215(19990315)80:6<930::aid-ijc20>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 33.Jacob AG, O’Brien D, Singh RK, Comiskey DF, Littleton RM, Mohammad F, et al. Stress-induced isoforms of MDM2 and MDM4 correlate with high-grade disease and an altered splicing network in pediatric rhabdomyosarcoma. Neoplasia. 2013;15(9):1049–63. doi: 10.1593/neo.13286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hori M, Shimazaki J, Inagawa S, Itabashi M, Hori M. Alternatively spliced MDM2 transcripts in human breast cancer in relation to tumor necrosis and lymph node involvement. Pathology international. 2000;50(10):786–92. doi: 10.1046/j.1440-1827.2000.01119.x. [DOI] [PubMed] [Google Scholar]
- 35.Evdokiou A, Atkins GJ, Bouralexis S, Hay S, Raggatt LJ, Cowled PA, et al. Expression of alternatively-spliced MDM2 transcripts in giant cell tumours of bone. Int J Oncol. 2001;19(3):625–32. doi: 10.3892/ijo.19.3.625. [DOI] [PubMed] [Google Scholar]
- 36.Bartel F, Taylor AC, Taubert H, Harris LC. Novel mdm2 splice variants identified in pediatric rhabdomyosarcoma tumors and cell lines. Oncol Res. 2001;12(11–12):451–7. doi: 10.3727/096504001108747459. [DOI] [PubMed] [Google Scholar]
- 37.Bartel F, Meye A, Wurl P, Kappler M, Bache M, Lautenschlager C, et al. Amplification of the MDM2 gene, but not expression of splice variants of MDM2 MRNA, is associated with prognosis in soft tissue sarcoma. Int J Cancer. 2001;95(3):168–75. doi: 10.1002/1097-0215(20010520)95:3<168::aid-ijc1029>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 38.Okoro DR, Arva N, Gao C, Polotskaia A, Puente C, Rosso M, et al. Endogenous human MDM2-C is highly expressed in human cancers and functions as a p53-independent growth activator. PLoS One. 2013;8(10):e77643. doi: 10.1371/journal.pone.0077643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chandler DS, Singh RK, Caldwell LC, Bitler JL, Lozano G. Genotoxic stress induces coordinately regulated alternative splicing of the p53 modulators MDM2 and MDM4. Cancer Res. 2006;66(19):9502–8. doi: 10.1158/0008-5472.CAN-05-4271. [DOI] [PubMed] [Google Scholar]
- 40.Dias CS, Liu Y, Yau A, Westrick L, Evans SC. Regulation of hdm2 by stress-induced hdm2alt1 in tumor and nontumorigenic cell lines correlating with p53 stability. Cancer Res. 2006;66(19):9467–73. doi: 10.1158/0008-5472.CAN-05-3013. [DOI] [PubMed] [Google Scholar]
- 41.Evans SC, Viswanathan M, Grier JD, Narayana M, El-Naggar AK, Lozano G. An alternatively spliced HDM2 product increases p53 activity by inhibiting HDM2. Oncogene. 2001;20(30):4041–9. doi: 10.1038/sj.onc.1204533. [DOI] [PubMed] [Google Scholar]
- 42.Jacob AG, Singh RK, Comiskey DF, Jr, Rouhier MF, Mohammad F, Bebee TW, et al. Stress-Induced Alternative Splice Forms of MDM2 and MDMX Modulate the p53-Pathway in Distinct Ways. PLoS One. 2014;9(8):e104444. doi: 10.1371/journal.pone.0104444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng T, Wang J, Zhao Y, Zhang C, Lin M, Wang X, et al. Spliced MDM2 isoforms promote mutant p53 accumulation and gain-of-function in tumorigenesis. Nature communications. 2013;4:2996. doi: 10.1038/ncomms3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steinman HA, Burstein E, Lengner C, Gosselin J, Pihan G, Duckett CS, et al. An alternative splice form of Mdm2 induces p53-independent cell growth and tumorigenesis. J Biol Chem. 2004;279(6):4877–86. doi: 10.1074/jbc.M305966200. [DOI] [PubMed] [Google Scholar]
- 45.Fridman JS, Hernando E, Hemann MT, de Stanchina E, Cordon-Cardo C, Lowe SW. Tumor promotion by Mdm2 splice variants unable to bind p53. Cancer Res. 2003;63(18):5703–6. [PubMed] [Google Scholar]
- 46.Lobe CG, Koop KE, Kreppner W, Lomeli H, Gertsenstein M, Nagy A. Z/AP, a double reporter for cre-mediated recombination. Dev Biol. 1999;208(2):281–92. doi: 10.1006/dbio.1999.9209. [DOI] [PubMed] [Google Scholar]
- 47.Donehower LA, Harvey M, Vogel H, McArthur MJ, Montgomery CA, Jr, Park SH, et al. Effects of genetic background on tumorigenesis in p53-deficient mice. Mol Carcinog. 1995;14(1):16–22. doi: 10.1002/mc.2940140105. [DOI] [PubMed] [Google Scholar]
- 48.McDonnell TJ, Montes de Oca Luna R, Cho S, Amelse LL, Chavez-Reyes A, Lozano G. Loss of one but not two mdm2 null alleles alters the tumour spectrum in p53 null mice. The Journal of pathology. 1999;188(3):322–8. doi: 10.1002/(SICI)1096-9896(199907)188:3<322::AID-PATH372>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 49.Altmannsberger M, Weber K, Droste R, Osborn M. Desmin is a specific marker for rhabdomyosarcomas of human and rat origin. The American Journal of Pathology. 1985;118(1):85–95. [PMC free article] [PubMed] [Google Scholar]
- 50.Kumar S, Perlman E, Harris CA, Raffeld M, Tsokos M. Myogenin is a Specific Marker for Rhabdomyosarcoma: An Immunohistochemical Study in Paraffin-Embedded Tissues. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2000;13(9):988–93. doi: 10.1038/modpathol.3880179. [DOI] [PubMed] [Google Scholar]
- 51.Rangdaeng S, Truong LD. Comparative Immunohistochemical Staining for Desmin and Muscle-Specific Actin: A Study of 576 Cases. American journal of clinical pathology. 1991;96(1):32–45. doi: 10.1093/ajcp/96.1.32. [DOI] [PubMed] [Google Scholar]
- 52.de Vree PJP, de Wit E, Yilmaz M, van de Heijning M, Klous P, Verstegen MJAM, et al. Targeted sequencing by proximity ligation for comprehensive variant detection and local haplotyping. Nat Biotech. 2014;32(10):1019–25. doi: 10.1038/nbt.2959. [DOI] [PubMed] [Google Scholar]
- 53.Takano A, Zochi R, Hibi M, Terashima T, Katsuyama Y. Function of strawberry notch family genes in the zebrafish brain development. The Kobe journal of medical sciences. 2011;56(5):E220–30. [PubMed] [Google Scholar]
- 54.Takano A, Zochi R, Hibi M, Terashima T, Katsuyama Y. Expression of strawberry notch family genes during zebrafish embryogenesis. Developmental dynamics: an official publication of the American Association of Anatomists. 2010;239(6):1789–96. doi: 10.1002/dvdy.22287. [DOI] [PubMed] [Google Scholar]
- 55.Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, et al. The Consensus Coding Sequences of Human Breast and Colorectal Cancers. Science. 2006;314(5797):268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 56.Suzuki C, Takahashi K, Hayama S, Ishikawa N, Kato T, Ito T, et al. Identification of Myc-associated protein with JmjC domain as a novel therapeutic target oncogene for lung cancer. Molecular cancer therapeutics. 2007;6(2):542–51. doi: 10.1158/1535-7163.MCT-06-0659. [DOI] [PubMed] [Google Scholar]
- 57.Jung CH, Kim J, Park JK, Hwang SG, Moon SK, Kim WJ, et al. Mdm2 increases cellular invasiveness by binding to and stabilizing the Slug mRNA. Cancer letters. 2013;335(2):270–7. doi: 10.1016/j.canlet.2013.02.035. [DOI] [PubMed] [Google Scholar]
- 58.Gu L, Zhu N, Zhang H, Durden DL, Feng Y, Zhou M. Regulation of XIAP translation and induction by MDM2 following irradiation. Cancer Cell. 2009;15(5):363–75. doi: 10.1016/j.ccr.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singh S, Ramamoorthy M, Vaughan C, Yeudall WA, Deb S, Palit Deb S. Human oncoprotein MDM2 activates the Akt signaling pathway through an interaction with the repressor element-1 silencing transcription factor conferring a survival advantage to cancer cells. Cell Death Differ. 2013;20(4):558–66. doi: 10.1038/cdd.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gu L, Findley HW, Zhou M. MDM2 induces NF-kappaB/p65 expression transcriptionally through Sp1-binding sites: a novel, p53-independent role of MDM2 in doxorubicin resistance in acute lymphoblastic leukemia. Blood. 2002;99(9):3367–75. doi: 10.1182/blood.v99.9.3367. [DOI] [PubMed] [Google Scholar]
- 61.Cheng Q, Chen L, Li Z, Lane WS, Chen J. ATM activates p53 by regulating MDM2 oligomerization and E3 processivity. EMBO J. 2009;28(24):3857–67. doi: 10.1038/emboj.2009.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheng Q, Cross B, Li B, Chen L, Li Z, Chen J. Regulation of MDM2 E3 ligase activity by phosphorylation after DNA damage. Mol Cell Biol. 2011;31(24):4951–63. doi: 10.1128/MCB.05553-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gannon HS, Woda BA, Jones SN. ATM phosphorylation of Mdm2 Ser394 regulates the amplitude and duration of the DNA damage response in mice. Cancer Cell. 2012;21(5):668–79. doi: 10.1016/j.ccr.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ognjanovic S, Martel G, Manivel C, Olivier M, Langer E, Hainaut P. Low Prevalence of TP53 Mutations and MDM2 Amplifications in Pediatric Rhabdomyosarcoma. Sarcoma. 2012;2012:492086. doi: 10.1155/2012/492086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lynch CJ, Milner J. Loss of one p53 allele results in four-fold reduction of p53 mRNA and protein: a basis for p53 haplo-insufficiency. Oncogene. 2006;25(24):3463–70. doi: 10.1038/sj.onc.1209387. [DOI] [PubMed] [Google Scholar]
- 66.Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, et al. Tumor spectrum analysis in p53-mutant mice. Current Biology. 1994;4(1):1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 67.Walker DR, Bond JP, Tarone RE, Harris CC, Makalowski W, Boguski MS, et al. Evolutionary conservation and somatic mutation hotspot maps of p53: correlation with p53 protein structural and functional features. Oncogene. 1999;18(1):211–8. doi: 10.1038/sj.onc.1202298. [DOI] [PubMed] [Google Scholar]
- 68.Dang J, Kuo ML, Eischen CM, Stepanova L, Sherr CJ, Roussel MF. The RING domain of Mdm2 can inhibit cell proliferation. Cancer Res. 2002;62(4):1222–30. [PubMed] [Google Scholar]
- 69.Haines DC, Chattopadhyay S, Ward JM. Pathology of aging B6;129 mice. Toxicologic pathology. 2001;29(6):653–61. doi: 10.1080/019262301753385988. [DOI] [PubMed] [Google Scholar]
- 70.Taylor AC, Shu L, Danks MK, Poquette CA, Shetty S, Thayer MJ, et al. P53 mutation and MDM2 amplification frequency in pediatric rhabdomyosarcoma tumors and cell lines. Medical and pediatric oncology. 2000;35(2):96–103. doi: 10.1002/1096-911x(200008)35:2<96::aid-mpo2>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 71.Volk EL, Fan L, Schuster K, Rehg JE, Harris LC. The MDM2-A Splice Variant of MDM2 Alters Transformation In vitro and the Tumor Spectrum in Both Arf- and p53-Null Models of Tumorigenesis. Molecular Cancer Research. 2009;7(6):863–9. doi: 10.1158/1541-7786.MCR-08-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jones SN, Hancock AR, Vogel H, Donehower LA, Bradley A. Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc Natl Acad Sci U S A. 1998;95(26):15608–12. doi: 10.1073/pnas.95.26.15608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415(6867):45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 74.Abraham J, Nuñez-Álvarez Y, Hettmer S, Carrió E, Chen H-IH, Nishijo K, et al. Lineage of origin in rhabdomyosarcoma informs pharmacological response. Genes & Development. 2014;28(14):1578–91. doi: 10.1101/gad.238733.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Keller C, Arenkiel BR, Coffin CM, El-Bardeesy N, DePinho RA, Capecchi MR. Alveolar rhabdomyosarcomas in conditional Pax3:Fkhr mice: cooperativity of Ink4a/ARF and Trp53 loss of function. Genes Dev. 2004;18(21):2614–26. doi: 10.1101/gad.1244004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xiao ZX, Chen J, Levine AJ, Modjtahedi N, Xing J, Sellers WR, et al. Interaction between the retinoblastoma protein and the oncoprotein MDM2. Nature. 1995;375(6533):694–8. doi: 10.1038/375694a0. [DOI] [PubMed] [Google Scholar]
- 77.Yang JY, Zong CS, Xia W, Yamaguchi H, Ding Q, Xie X, et al. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nature cell biology. 2008;10(2):138–48. doi: 10.1038/ncb1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fu W, Ma Q, Chen L, Li P, Zhang M, Ramamoorthy S, et al. MDM2 acts downstream of p53 as an E3 ligase to promote FOXO ubiquitination and degradation. J Biol Chem. 2009;284(21):13987–4000. doi: 10.1074/jbc.M901758200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou S, Gu L, He J, Zhang H, Zhou M. MDM2 regulates vascular endothelial growth factor mRNA stabilization in hypoxia. Mol Cell Biol. 2011;31(24):4928–37. doi: 10.1128/MCB.06085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gu L, Zhang H, He J, Li J, Huang M, Zhou M. MDM2 regulates MYCN mRNA stabilization and translation in human neuroblastoma cells. Oncogene. 2012;31(11):1342–53. doi: 10.1038/onc.2011.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang JY, Zong CS, Xia W, Wei Y, Ali-Seyed M, Li Z, et al. MDM2 promotes cell motility and invasiveness by regulating E-cadherin degradation. Mol Cell Biol. 2006;26(19):7269–82. doi: 10.1128/MCB.00172-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tang YA, Lin RK, Tsai YT, Hsu HS, Yang YC, Chen CY, et al. MDM2 overexpression deregulates the transcriptional control of RB/E2F leading to DNA methyltransferase 3A overexpression in lung cancer. Clin Cancer Res. 2012;18(16):4325–33. doi: 10.1158/1078-0432.CCR-11-2617. [DOI] [PubMed] [Google Scholar]
- 83.Lin HK, Wang L, Hu YC, Altuwaijri S, Chang C. Phosphorylation-dependent ubiquitylation and degradation of androgen receptor by Akt require Mdm2 E3 ligase. EMBO J. 2002;21(15):4037–48. doi: 10.1093/emboj/cdf406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bouska A, Lushnikova T, Plaza S, Eischen CM. Mdm2 promotes genetic instability and transformation independent of p53. Mol Cell Biol. 2008;28(15):4862–74. doi: 10.1128/MCB.01584-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wade M, Wang YV, Wahl GM. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends in cell biology. 2010;20(5):299–309. doi: 10.1016/j.tcb.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brown DR, Thomas CA, Deb SP. The human oncoprotein MDM2 arrests the cell cycle: elimination of its cell-cycle-inhibitory function induces tumorigenesis. EMBO J. 1998;17(9):2513–25. doi: 10.1093/emboj/17.9.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lundgren K, Montes de Oca Luna R, McNeill YB, Emerick EP, Spencer B, Barfield CR, et al. Targeted expression of MDM2 uncouples S phase from mitosis and inhibits mammary gland development independent of p53. Genes Dev. 1997;11(6):714–25. doi: 10.1101/gad.11.6.714. [DOI] [PubMed] [Google Scholar]
- 88.Maya R, Balass M, Kim ST, Shkedy D, Leal JF, Shifman O, et al. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 2001;15(9):1067–77. doi: 10.1101/gad.886901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stiff T, Walker SA, Cerosaletti K, Goodarzi AA, Petermann E, Concannon P, et al. ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J. 2006;25(24):5775–82. doi: 10.1038/sj.emboj.7601446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Carr MI, Roderick JE, Gannon HS, Kelliher MA, Jones SN. Mdm2 Phosphorylation Regulates Its Stability and Has Contrasting Effects on Oncogene and Radiation-Induced Tumorigenesis. Cell reports. 2016;16(10):2618–29. doi: 10.1016/j.celrep.2016.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434(7035):907–13. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 92.Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444(7119):638–42. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 93.Bartek J, Bartkova J, Lukas J. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene. 2007;26(56):7773–9. doi: 10.1038/sj.onc.1210881. [DOI] [PubMed] [Google Scholar]
- 94.Miron K, Golan-Lev T, Dvir R, Ben-David E, Kerem B. Oncogenes create a unique landscape of fragile sites. Nature communications. 2015;6:7094. doi: 10.1038/ncomms8094. [DOI] [PubMed] [Google Scholar]
- 95.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nature reviews Molecular cell biology. 2010;11(3):220–8. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 96.Felsher DW, Bishop JM. Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proc Natl Acad Sci U S A. 1999;96(7):3940–4. doi: 10.1073/pnas.96.7.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mai S, Fluri M, Siwarski D, Huppi K. Genomic instability in MycER-activated Rat1A-MycER cells. Chromosome Res. 1996;4(5):365–71. doi: 10.1007/BF02257272. [DOI] [PubMed] [Google Scholar]
- 98.Denko NC, Giaccia AJ, Stringer JR, Stambrook PJ. The human Ha-ras oncogene induces genomic instability in murine fibroblasts within one cell cycle. Proc Natl Acad Sci U S A. 1994;91(11):5124–8. doi: 10.1073/pnas.91.11.5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ellsworth RE, Ellsworth DL, Patney HL, Deyarmin B, Love B, Hooke JA, et al. Amplification of HER2 is a marker for global genomic instability. BMC cancer. 2008;8:297. doi: 10.1186/1471-2407-8-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Carrillo AM, Bouska A, Arrate MP, Eischen CM. Mdmx promotes genomic instability independent of p53 and Mdm2. Oncogene. 2014:0. doi: 10.1038/onc.2014.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Alt JR, Bouska A, Fernandez MR, Cerny RL, Xiao H, Eischen CM. Mdm2 binds to Nbs1 at sites of DNA damage and regulates double strand break repair. J Biol Chem. 2005;280(19):18771–81. doi: 10.1074/jbc.M413387200. [DOI] [PubMed] [Google Scholar]
- 102.Hong S, Pusapati RV, Powers JT, Johnson DG. Oncogenes and the DNA damage response: Myc and E2F1 engage the ATM signaling pathway to activate p53 and induce apoptosis. Cell Cycle. 2006;5(8):801–3. doi: 10.4161/cc.5.8.2638. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.