Abstract
Background
Autologous transplantation of mesenchymal stem cells (MSCs) may be a viable option for the treatment of several diseases. Evidence indicates that MSCs release extracellular vesicles (EVs) that shuttle miRNAs to damaged parenchymal cells, activating an endogenous repair program. However, whether comorbidities, such as metabolic syndrome (MetS) interfere with the packaging of cargo of MSC-derived EVs has never been explored. We hypothesized that MetS modulates the miRNA content packed within MSC-derived EVs.
Methods
MSCs were collected from swine abdominal adipose tissue after 16 weeks of Lean or Obese diet (n=7 each). Next-generation miRNA sequencing (miRNA-seq) was performed to identify miRNAs enriched in MSC-derived EVs, and their predicted target genes. Functional pathway analysis of the top 50 target genes of the top 4 miRNAs enriched in each group was performed using FunRich.
Results
Fourteen and 8 miRNAs were enriched in Lean- and MetS- EVs, respectively. Target genes of miRNAs enriched in MetS-EVs were implicated in the development of MetS and its complications, including diabetes-related pathways, validated transcriptional targets of AP1 family members Fra1 and Fra2, Class A/1 (Rhodopsin-like receptors), and Peptide ligand-binding receptors. Contrarily, miRNAs enriched in Lean EVs target primarily EphrinA-EPHA and the Rho family of GTPases.
Conclusions
MetS alters the miRNA content of EVs derived from porcine adipose tissue MSCs. These alterations could impair the efficacy of autologous MSCs and limit its therapeutic use in subjects with MetS. Our findings may assist in developing adequate regenerative strategies to preserve the reparative potency of MSCs in individuals with MetS.
Keywords: Mesenchymal stem cells, extracellular vesicles, microRNA, metabolic syndrome
Introduction
Mesenchymal stem/stromal cells (MSCs) are undifferentiated cells with high proliferative capacity that play fundamental roles in repair and self-renewal of tissues. MSCs can differentiate not only into mesenchymal lineage cells, but also into several other cell lineages, including osteoblasts, chondrocytes, myocytes, and adipocytes [1]. Furthermore, these cells possess important pro-angiogenic, immunomodulatory, and anti-inflammatory properties, and can be isolated from bone marrow, adipose, peripheral blood, skeletal muscle, etc., all of which make MSCs an attractive source for tissue engineering and regenerative medicine.
Accumulating evidence indicates that MSCs release extracellular vesicles (EVs) that shuttle their genetic and protein cargo to damaged parenchymal cells, activating an endogenous repair program[2] [3]. We have previously shown that EVs released from porcine adipose tissue-derived MSCs transport gene regulatory information to modulate angiogenesis, adipogenesis, and other cell pathways in recipient cells [4, 5]. We have also shown that EVs are enriched with proteins linked to a broad range of biological functions, including angiogenesis, inflammation, apoptosis, and extracellular matrix remodeling[5] [6].
For clinical purposes, the use of autologous MSCs, with comes with an implicit need to deliver cells from the same individual, is much preferable compared to allogeneic transplantation. Therefore, it is critical to know whether comorbidities may compromise the integrity and reparative potential of MSCs and their EV progeny.
Metabolic syndrome (MetS) encircles several cardiovascular risk factors, including obesity, hypertension, hyperlipidemia, and insulin resistance, associated with the risk of developing cardiovascular disease and type-2 diabetes [7]. We have previously developed and validated a novel porcine model of MetS achieved by feeding animals a high-carbohydrate and cholesterol diet for 16 weeks, which recapitulates many features of human MetS [8]. Interestingly, we found that adipose tissue-derived MSCs isolated from MetS pigs show increased propensity for differentiation into adipocytes and premature death compared to Lean-MSCs [9]. Yet, whether MetS-induced MSC changes interfere with the packaging of cargo of their daughters EVs has never been explored.
MSC-derived EVs are selectively enriched with micro-RNAs (miRNAs), small non-coding RNAs that mediate post-transcriptional control of gene expression by directing their target mRNAs for degradation or translational repression. We have shown that EVs isolated from porcine adipose tissue MSCs contain miRNAs capable of modulating survival and metabolic activities of recipient cells [4] [5], but whether MetS alters the miRNA content of MSC-derived EVs remains unknown.
The current study tested the hypothesis that MetS modulates the miRNA cargo packed within MSC-derived EVs. For this purpose, we comprehensively characterized the miRNA expression profile of porcine adipose tissue MSC-derived EVs using high-throughput RNA sequencing (RNA-seq) analysis. An important finding of our study is that MetS-EVs show distinctive miRNA cargo compared to their Lean counterparts, targeting different gene transcripts. miRNAs enriched in MetS-EVs target primarily genes involved in diabetes, peptide ligand-binding receptors, and AP1 family members, whereas those upregulated in Lean-EVs target RhoA activity, EphirinA-EPHA pathway, and secretin family receptors. These observations provide a better understanding of the vulnerability of this repair system and contribute towards development of strategies for improving the utility and efficacy of MSC-derived EV therapy.
Methods
Experimental design
Animals and Protocol
Three-month-old female domestic pigs were studied with the approval of the Institutional Animal Care and Use Committee. Animals were randomized into 2 groups: MetS and Lean pigs (n =7 each). MetS pigs were fed a high-cholesterol/carbohydrate diet (5B4L, protein 16.1%, ether extract fat 43.0%, and carbohydrates 40.8%, Purina Test Diet, Richmond, IN) and Lean pigs a standard chow (13% protein, 2% fat, 6% fiber, Purina Animal Nutrition LLC, MN) for a total of 16 weeks. All animals have free access to water.
Systemic measurements
After 16 weeks of diet, body weight, blood pressure, and heart rate were recorded. Total cholesterol, low-density lipoprotein (LDL), high-density lipoprotein (HDL), and triglyceride (TG) levels were measured by enzymatic methods and insulin resistance by the homeostasis model assessment of insulin resistance (HOMA-IR). After completion of all studies, animals were euthanized with a lethal intravenous dose of sodium pentobarbital, and subcutaneous abdominal adipose tissue collected for MSC and EV isolation.
MSC and EV isolation, characterization, and culture
MSCs were collected from swine subcutaneous abdominal fat tissue (5–10g), digested in collagenase-H, filtered, and cultured for 3 weeks in advanced MEM medium (Gibco/Invitrogen) supplemented with 5% platelet lysate. The third passage was collected and kept in Gibco Cell Culture Freezing Medium. Cellular phenotype was examined by the expression of the MSCs markers CD73 and CD105.
EVs were isolated from MSC supernatants (10×106 cells) by the ultracentrifugation method, as previously described [4]. Samples were centrifuged (2,000g for 20min), and the supernatant collected and subsequently centrifuged (100,000g for 1h) at 4°C. EVs were collected, suspended in wash buffer medium 199, and centrifuged one more time (100,00g for 1hr).
MSCs and their daughters EVs were examined on transmission electron microscopy (JEOL 1200 EXII, Mayo Clinic’s electron microscopy core). In addition, negative staining of MSC supernatant with 2% uranyl acetate was performed to examine EV morphology.
EVs were then characterized based on the expression of both EV (CD9 and CD29) and MSC (CD73 and CD105) surface markers by western blot (AbD Serotec cat#: MCA1189 and AA120-175, and Abcam cat#: ab115289 and ab135528).
miRNA Sequencing and data analysis
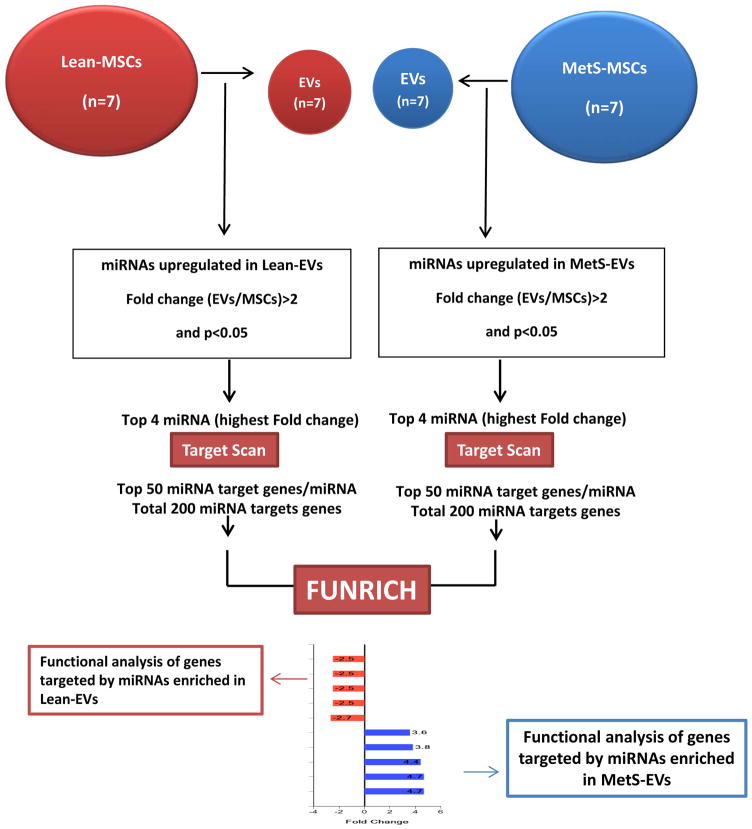
Fig. 1 summarizes our high throughput miRNA sequencing and functional analysis. MSC and EV miRNA sequencing was performed as previously described [4], and the data analyzed using the CAP-miRSeq-v1.1 workflow [10], which starts with unaligned FASTQs, generates aligned BAMs, and then excel files containing raw and normalized known mature miRNA expression counts. EdgeR2.6.2 [11, 12] was used to perform the differential expression analysis to identify miRNAs enriched in Lean- and MetS-EVs compared to their parent MSCs (fold change >2.0 and p≤0.05). TargetScan (Release7.1) was used to predict target genes of the top 4 (highest fold change) miRNAs enriched in Lean- and MetS-EVs. Functional pathway analysis of top 50 target genes of top 4 miRNAs enriched in Lean- and MetS-EVs (total 200 miRNAs each) was performed using FunRich (http://www.funrich.org), an open access standalone tool that allows the generation of graphical representations. Gene targets of miRNAs enriched in Lean- and MetS-EVs were compared on different categories, including cellular component, molecular function, biological process, and biological pathway.
Figure 1. Overview of micro-RNA (miRNA) sequencing and functional analysis.
High throughput miRNA sequencing analysis was performed in both swine adipose tissue-derived mesenchymal stem cells (MSCs) and their daughter extracellular vesicles (EVs) (n=7 each). Differentially expression analysis was performed to identify miRNAs enriched in EVs. miRNA predicted targets of top 4 miRNAs enriched in Lean- and MetS- EVs (highest fold change) were identified using TargetScan. Functional pathway analysis of top 50 target genes of top 4 miRNAs enriched in Lean- and MetS- EVs (total 200 miRNAs each) was performed using FunRich.
Validation of miRNA Sequencing analysis
To validate expression of representative miRNAs, the expression of miR-205, miR-486, and miR-194a in Lean-MSCs and Lean-EVs, and the expression of miR-146a-3p and miR-30c-1-3p in MetS-MSCs and MetS-EVs was measured by quantitative polymerase chain reaction (qPCR). Total RNA was isolated from 5×105 –1×106 MSC samples, as previously described. All primers were from ThermoFisher Scientific (miR-205:000509, miR-486:001278, miR-194a:005656, miR-146a-3p:465157, and miR-30c-1-3p:472491).
Statistical analysis
Statistical analysis was performed using JMP 10.0 (SAS Institute, Cary, NC, USA). Data were expressed as mean ± standard deviation. Student’s t-test was used to evaluate statistically significant differences between Lean and MetS groups. Statistical significance was accepted if p≤0.05.
Results
Systemic characteristics
Table 1 shows the systemic characteristics of all animals at 16 weeks. Body weight and blood pressure were higher in MetS pigs compared to Lean, whereas heart rate was similar between the groups. Fasting glucose levels were comparable, but insulin and HOMA-IR score were higher in MetS versus Lean, as were total cholesterol, HDL, LDL, and triglyceride levels.
Table 1.
Systemic characteristic of Lean and MetS pigs (n=7 each)
| Parameter | Lean | MetS |
|---|---|---|
| Body Weight (Kg) | 77.7±1.6 | 93.3±0.4* |
| Systolic blood pressure (mmHg) | 126.0±2.2 | 146.0±1.5* |
| Diastolic blood pressure (mmHg) | 89.2±1.4 | 110.4±1.3* |
| Mean blood pressure (mmHg) | 101.4±1.4 | 122.3±1.4* |
| Heart rate (beats/min) | 73.5±0.7 | 74.6±0.5 |
| Fasting glucose (mg/dl) | 106.0±2.9 | 108.8±2.2 |
| Fasting insulin (μU/ml) | 0.2±0.1 | 0.6±0.1* |
| HOMA-IR score | 0.6±0.1 | 1.9±0.2* |
| Total cholesterol (mg/dl) | 81.0±7.2 | 372.0±73.1* |
| LDL cholesterol (mg/dl) | 33.5±7.3 | 441.4±118.2* |
| HDL cholesterol (mg/dl) | 45.8±4.1 | 143.8±33.0* |
| Triglycerides (mg/dl) | 8.4±1.0 | 14.5±0.6* |
P≤0.05 vs. Lean
Characterization of Lean- and MetS- MSC-derived EVs
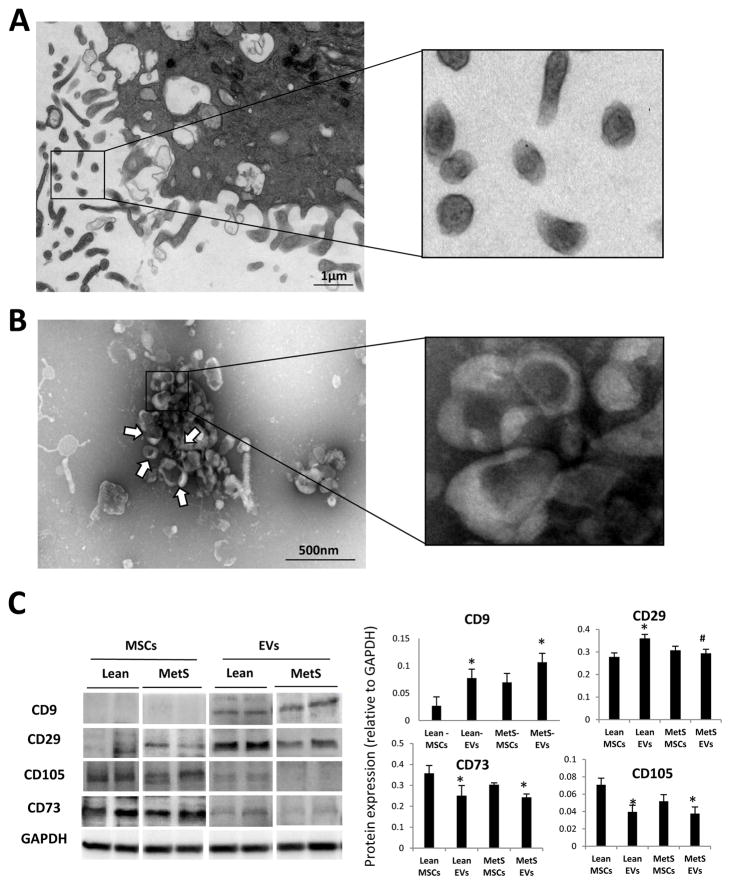
Transmission and scanning electron microscopy demonstrated that cultured MSCs release numerous EVs (Fig. 2A). Negative staining of MSC culture supernatants showed EV clusters with the traditional “cup-like” morphology (Fig. 2B). MSC-derived EVs expressed both MSC (CD73 and CD105) and EV (CD9 and CD29) markers (Fig. 2C). Expression of MSC markers was higher in Lean- and MetS- MSCs compared to their respective EVs, whereas expression of CD9 was higher in both Lean- and MetS- EVs compared to their parent cells. Contrarily, expression of CD29, higher in Lean-EVs versus Lean-MSCs, did not differ between MetS-MSCs and MetS-EVs.
Figure 2. Characterization of MSC-derived EVs.
A: Transmission electron microscopy of culture MSCs releasing EVs. B: Negative staining of MSC culture supernatants showing EV clusters (arrows) with the classic “cup-like” morphology. C: Characterization of Lean- and MetS- EVs by protein expression of common EV (CD9 and CD29) and MSC (CD105 and CD73) markers.
Selective Enrichment of Enriched miRNAs in EVs
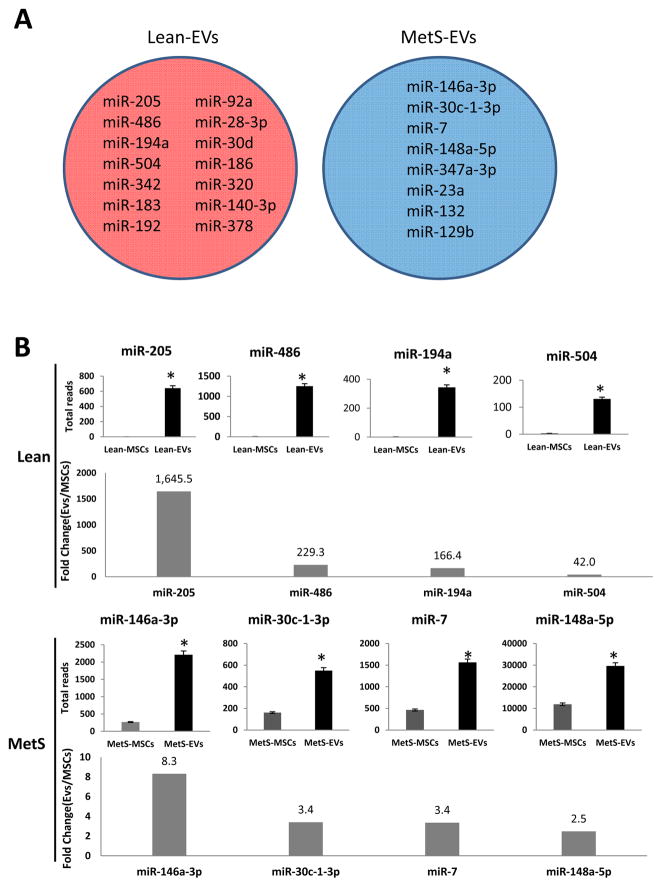
Of all annotated miRNAs (n=326), only 14 (miR-205, miR-486, miR-194a, miR-504, miR-342, miR-183, miR-192, miR-92a, miR-28-3p, miR-30d, miR-186, miR-320, miR-140-3p, miR-378) and 8 (miR-146a-3p, miR-30c-1-3p, miR-7, miR-148a-5p, miR-374a-3p, miR-23a, miR-132, miR-129b) distinct miRNAs were selectively enriched in Lean- and MetS- EVs, respectively (Fig 3A). Total reads and Fold Change (EVs/MSCs) of the top 4 miRNAs enriched (highest fold change) in Lean- and MetS- EVs were presented with (Fig 3B).
Figure 3. miRNAs enriched in Lean- and MetS- EVs.
A: Of all annotated miRNAs (n=326), only 14 (miR-205, miR-486, miR-194a, miR-504, miR-342, miR-183, miR-192, miR-92a, miR-28-3p, miR-30d, miR-186, miR-320, miR-140-3p, miR-378) and 8 (miR-146a-3p, miR-30c-1-3p, miR-7, miR-148a-5p, miR-374a-3p, miR-23a, miR-132, miR-129b) distinct miRNAs were selectively enriched in Lean- and MetS- EVs, respectively. B: Total reads and fold change (EVs/MSCs) of top 4 (highest fold change) miRNAs enriched in Lean-EVs (top) and MetS-EVs (bottom).
Validation of miRNA Sequencing analysis
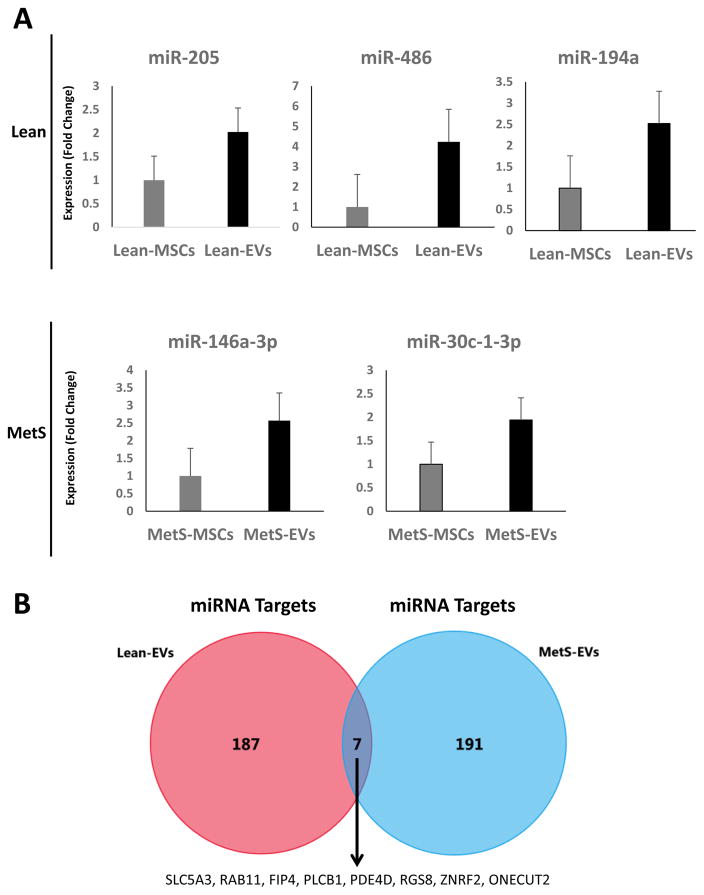
Expression of the candidate miRNAs followed the same patterns as the miRNAseq findings. Specifically, miR-205, miR-486, and miR-194a were higher in Lean-EVs compared to their parent MSCs, whereas expression of miR-146a and miR-30c-1-3p were higher in MetS-EVs versus MetS-MSCs (Fig. 4A).
Figure 4. Target genes of miRNAs enriched in Lean- and MetS- EVs.
Quantitative polymerase chain reaction (qPCR) of candidate miRNAs followed the same patterns as the miRNAseq findings. miR-205, miR-486, and miR-194a were higher in Lean-EVs compared to their parent MSCs, whereas expression of miR-146a and miR-30c-1-3p were higher in MetS-EVs versus MetS-MSCs. B: Venn diagram showing distribution of miRNA target genes of miRNAs enriched in Lean- and MetS- EVs. A total of 194 and 198 target genes were identified for miRNAs enriched in Lean- and MetS- EVs, but only 7 EV-enriched miRNA targets overlapped between these 2 groups. A total of 194 and 198 target genes were identified for miRNAs enriched in Lean- and MetS- EVs, but only 7 EV-enriched miRNA targets overlapped between these 2 groups.
Functional analysis of miRNA targets
Functional analysis to the top 50 target genes of the top 4 miRNAs enriched in Lean- and MetS- EVs was performed using FunRich. There were only 7 common miRNA target genes between the Lean-EVs and the MetS-EVs, which encode proteins mainly related to transport (SLC5A3, RAB11FIP4), signal transduction (PLCB1, PDE4D, RGS8), protein metabolism (ZNRF2), and cell differentiation (ONECUT2) (Fig 4B).
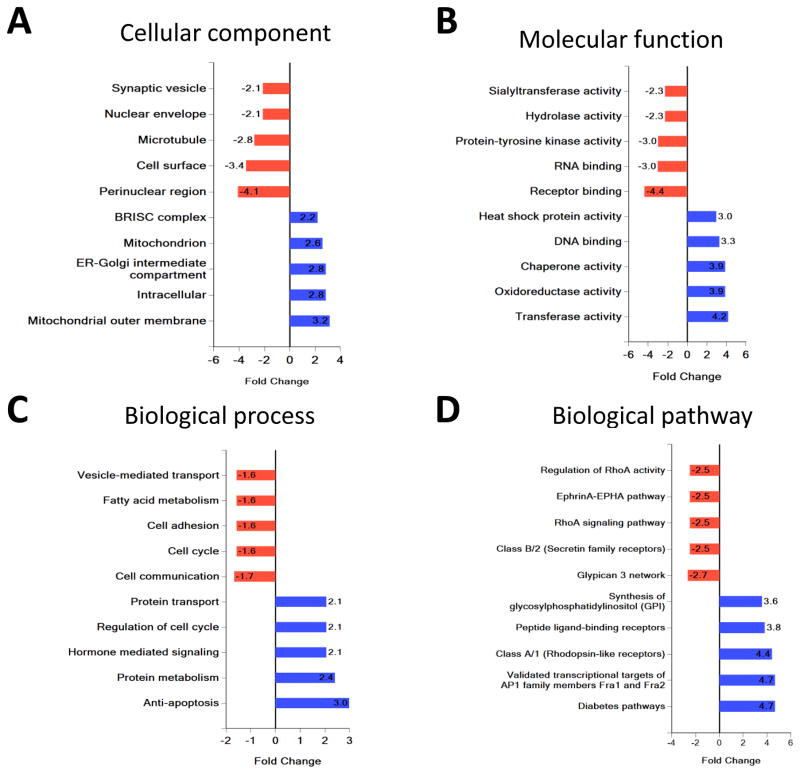
FunRich compared the cellular component, molecular function, biological process, and biological pathway of target genes of miRNAs enriched in Lean-EVs and MetS-EVs.
Analysis of cellular component revealed that target genes of miRNAs enriched in Lean-EVs encode proteins associated with cytoplasmic and cell surface components, whereas target genes of miRNAs enriched in MetS-EVs encode structural and functional organelle proteins (ER-Golgi and mitochondria) (Fig 5A). Classification by molecular function and biological process showed that target genes of miRNAs enriched miRNA in Lean-EVs were associated with receptor binding, RNA binding, the hydrolase and protein tyrosine kinase activity, cell communication, cell adhesion, fatty acid metabolism, and vesicle-mediated transport (Fig 5BC). However, target genes of miRNAs enriched miRNA in MetS-EVs were implicated in DNA binding, transferase activity, oxido-reductase and chaperone, apoptosis, protein metabolism, hormone mediated signaling, and protein transport.
Figure 5. Functional pathway analysis of target genes of miRNAs enriched in Lean- and MetS- EVs.
Classification of gene targets of miRNAs enriched in Lean- and MetS- EVs by cellular component, molecular function, biological process, and biological pathway.
Biological pathway analysis revealed that target genes of miRNAs enriched in Lean-EVs encode proteins involved in the Glypican 3 network, Class B/2 (Secretin family receptors), EphrinA-EPHA pathway, RhoA signaling pathway, and regulation of RhoA activity (Fig 5D). Contrarily, target genes of miRNAs enriched in MetS-EVs encode proteins involved in the Diabetes pathways, validated transcriptional targets of AP1 family members Fra1 and Fra2, Class A/1 (Rhodopsin-like receptors), Peptide ligand-binding receptors pathway, and synthesis of glycosylphosphatidylinositol (GPI).
Discussion
The current study demonstrates that MetS modulates the miRNA cargo packed within porcine adipose tissue MSC-derived EVs. We comprehensively determined the molecular characteristics of EVs using miRNA-seq to interrogate miRNA expression in EVs and their parent MSCs, and conducted a functional analysis of targeted genes of top miRNAs enriched in Lean- and the MetS- EVs using FunRich. Our data demonstrate that the miRNA content of Lean- and MetS- EVs is distinct. MetS-EVs contain miRNAs that modulate pathways involved in the development of MetS and its complications, including obesity, diabetes, and insulin signaling. Contrarily, miRNAs enriched in Lean-EVs are capable of modulating the EphrinA-EPHA and the Rho family of GTPases, modulate several cellular processes including cell migration, axon guidance, synapse formation and angiogenesis. These observations could greatly contribute towards development of regenerative strategies to preserve the reparative potency of MSCs in individuals with MetS.
Although miRNAs enriched in EVs may differ across species, they have the potential to modulate a similar plethora of pathways in recipient cells, including angiogenesis, inflammation, cell proliferation and migration. For example, our study found that miR-486, a miRNA that modulate angiogenesis, is selectively enriched in porcine MSCs derived EVs, which correlates with findings in human bone marrow- and adipose- MSC-derived EVs [13] [14]. However, mouse MSCs modulate angiogenesis partly by packing miR-210 in their daughter EVs [15]. Similarly, miR-194 [16] (enriched in porcine EVs), miR-10 [13] (enriched in human EVs), and miR-133b [17] (enriched in mouse EVs) are capable of modulating cell differentiation. Taken together, these observations indicate that despite distinct miRNA content, MSC-derived EVs from different species share their ability to regulate several cellular pathways in recipient cells.
EVs represent physiologically relevant and powerful components of the MSC secretome. These new players in cell-to-cell communication serve as vehicles for transfer of membrane and cytosolic proteins, lipids, and genetic information. In particular, microRNAs have recently been identified as mediators for many phenotypic changes induced in recipient cells by EVs. However, the determinants of their miRNA composition remain unknown. Interestingly, we found different miRNA cargo profiles between MetS-EVs and their Lean counterparts, targeting different gene transcripts. Merely 7 common miRNAs target-genes, which encode proteins mainly related to transport, signal transduction, protein metabolism, and cell differentiation, were identified. Furthermore, FunRich pathway analysis of non-overlapped target genes regulated by microRNAs showed completely different enrichment of signaling pathways between Lean-EVs and MetS-EVs.
Based on our findings, alterations in the miRNA enriched in MetS-EVs or Lean-EVs, and the target genes that they might modulate, follow specific patterns. Evidently, miRNAs enriched in MetS-EVs inhibit target genes involved in normal function or biological pathways that affect MSCs target cells, induce MetS, or modulate its complications (blue in Fig. 5D). On the other hand, some miRNA contained in Lean MSC-derived EVs are absent from MetS-EVs, which may thus fail to inhibit their target genes, and thereby impair target cell function and pathways, further molding the phenotype of MetS (red in Fig. 5D). For example, miRNAs enriched in MetS-EVs target genes involved in diabetes-related pathways. These genes encode endoplasmic reticulum (ER) membrane associated proteins, participate in insulin-maturation process, and transfer proinsulin from the ER to the Golgi apparatus [18], and might impair insulin secretion and diabetes. In contrast, some of the miRNAs enriched in Lean-EVs, such as miR-205, inhibit target genes like YES Proto-Oncogene 1, Src Family Tyrosine Kinase (YES1), which are involved in both the Glypican-3 network and Ephrin-A (EPHA) pathways, and can interact with IFIH1, a susceptibility gene of type-1 diabetes [19]. However, because the content of these miRNAs is decreased in MetS-EVs, their ability to inhibit YES1 may be blunted, which might facilitate development of diabetes.
The metabolic pathways of diabetes involve intersections of genetics, transcriptomics, and metabolite profiling [20], as well as Synthesis of GPI-anchored proteins pathway, the β-cell secretion plays a key role in these pathways, as the proinsulin newly synthesized in the rough ER is transferred to the cis- and then trans-Golgi network, where the insulin matures and is secreted from the cells [21]. miRNAs enriched in MetS-EVs, such as miR-146-3p, miR-30c-1-3p, miR-7, and miR-148a-5p target genes encoding the ER membrane receptor and protein, including the signal sequence receptor (SSR), Kruppel Like Factor 4 (KLF4), Phosphatidylinositol Glycan Anchor Biosynthesis Class H (PIGH), and Phosphatidylinositol Glycan Anchor Biosynthesis Class C (PIGC), involved transportation of insulin from ER to the Golgi. Importantly, downregulation of SSR causes ER stress [22, 23], and inhibition of the KLF4 decreases insulin sensitivity in the normal cell and tissue [24]. Likewise, changes in the expression of PIGH and PIGC may alter the first step of glycosylphosphatidylinositol biosynthesis [25].
Furthermore, target genes of miRNAs enriched in MetS-EVs included the AP1 family members Fra1 and Fra2, Class A/1 (Rhodopsin-like receptors) and Peptide ligand-binding receptors. The AP1 family members Fra1 and Fra2 modulates cell proliferation, differentiation and oxidative stress in Diabetes Mellitus [26]. Specifically, miR-7, enriched in MetS-EVs, may lead to inhibition of Specificity protein-1 (SP1), a gene associated with the AP1 family members Fra1 and Fra2 pathway, resulting ROS-induced tissue injury [27]. Additionally, miRNAs enriched in MetS-EVs target Peptide ligand-binding receptors, a subset of Class A/1 involved in cell proliferation and differentiation, and hormone transfer. For example, miR-148a-5p, enriched in MetS-EVs may lead to downregulation of Thyroid Stimulating Hormone Receptor (TSHR) and Neuromedin B Receptor (NMBR) that modulate leptin expression, energy balance, and adiposity, and play a role in insulin-induced insulin receptor activation [28].
Unlike miRNAs enriched in MetS-EVs, those enriched in Lean-EVs target primarily EphrinA-EPHA and the Rho family of GTPases, pathways that modulate several cellular processes including cell migration, axon guidance, synapse formation and angiogenesis. For example, EphrinA1 (miR-205 target) and EphA2 (miR-486 target), which are involved the EphrinA-EPHA pathway, can mediate the epithelial-mesenchymal transition [29]. Similarly, Mitogen-Activated Protein Kinase Kinase Kinase 7 (MAP3K7), targeted by miR-486-5p, can induce TGF-β1-activated kinase-1, regulating inflammation and fibrosis [30], as well as ER-stress and leptin resistance [31]. Lastly, target genes of miRNAs enriched in Lean-EVs target Class B/2 (Secretin family receptors), including Calcitonin Receptor Like Receptor (CALCRL), targeted by miR-205, which can modulate tissue inflammation [32]. Taken together, our findings indicate that MetS alters the miRNA content of EVs derived from porcine adipose tissue-derived MSCs.
Limitations
Our study is limited by the small number of samples and the short duration of MetS. Nevertheless, MetS pigs developed several features of human MetS and the content of their EVs differ from their Lean counterparts. Although our functional analysis focused on the top 4 miRNAs and their top 200 target genes, it sufficed to identify differences in the cargo of Lean- and MetS- EVs. Nevertheless, additional studies are needed to explore whether the miRNA content of MSC-derived EVs has distinct potential to alter the phenotype of their recipient cells.
Conclusions
Our study used next-generation sequencing analysis to identify miRNA expression signatures of EVs isolated from Lean- and MetS- porcine adipose tissue-derived MSCs. We found that the EV miRNA cargo differed between Lean- and MetS- MSCs, and functional analysis of miRNA target genes revealed that Lean- and MetS- EVs modulate different signaling pathways. miRNAs enriched in MetS-EVs target transcription factors, as well as genes implicated in the development of MetS and its complications, including validated transcriptional targets of AP1 family members Fra1 and Fra2, Class A/1 (Rhodopsin-like receptors), and Peptide ligand-binding receptors. Contrarily, miRNAs enriched in Lean MSC-derived EVs target primarily EphrinA-EPHA and the Rho family of GTPases. Therefore, our data provide insight into the mechanisms underpinning the protective properties of EVs derived from MSCs, and demonstrate that MetS alter the cargo of EVs. These observations may assist in developing adequate regenerative strategies to treat patients with multiple disorders.
Acknowledgments
This study was partly supported by the NIH grant numbers: DK73608, DK104273, HL123160, and DK102325, DK106427, the Central Society of Clinical and Translational Research, and Mayo Clinic: Mary Kathryn and Michael B. Panitch Career Development Award.
Footnotes
Conflict of interests
The authors declare no conflict of interest.
References
- 1.Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop D, Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8(4):315–7. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 2.Lai RC, Chen TS, Lim SK. Mesenchymal stem cell exosome: a novel stem cell-based therapy for cardiovascular disease. Regen Med. 2011;6(4):481–92. doi: 10.2217/rme.11.35. [DOI] [PubMed] [Google Scholar]
- 3.Yeo RW, Lai RC, Zhang B, Tan SS, Yin Y, Teh BJ, Lim SK. Mesenchymal stem cell: an efficient mass producer of exosomes for drug delivery. Adv Drug Deliv Rev. 2013;65(3):336–41. doi: 10.1016/j.addr.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Eirin A, Riester SM, Zhu XY, Tang H, Evans JM, O’Brien D, van Wijnen AJ, Lerman LO. MicroRNA and mRNA cargo of extracellular vesicles from porcine adipose tissue-derived mesenchymal stem cells. Gene. 2014;551(1):55–64. doi: 10.1016/j.gene.2014.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eirin A, Zhu XY, Puranik AS, Woollard JR, Tang H, Dasari S, Lerman A, van Wijnen AJ, Lerman LO. Integrated transcriptomic and proteomic analysis of the molecular cargo of extracellular vesicles derived from porcine adipose tissue-derived mesenchymal stem cells. PLoS One. 2017;12(3):e0174303. doi: 10.1371/journal.pone.0174303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eirin A, Zhu XY, Puranik AS, Woollard JR, Tang H, Dasari S, Lerman A, van Wijnen AJ, Lerman LO. Comparative proteomic analysis of extracellular vesicles isolated from porcine adipose tissue-derived mesenchymal stem/stromal cells. Sci Rep. 2016;6:36120. doi: 10.1038/srep36120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cameron AJ, Shaw JE, Zimmet PZ. The metabolic syndrome: prevalence in worldwide populations. Endocrinol Metab Clin North Am. 2004;33(2):351–75. doi: 10.1016/j.ecl.2004.03.005. table of contents. [DOI] [PubMed] [Google Scholar]
- 8.Pawar AS, Zhu XY, Eirin A, Tang H, Jordan KL, Woollard JR, Lerman A, Lerman LO. Adipose tissue remodeling in a novel domestic porcine model of diet-induced obesity. Obesity (Silver Spring) 2015;23(2):399–407. doi: 10.1002/oby.20971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu XY, Ma S, Eirin A, Woollard JR, Hickson LJ, Sun D, Lerman A, Lerman LO. Functional Plasticity of Adipose-Derived Stromal Cells During Development of Obesity. Stem Cells Transl Med. 2016;5(7):893–900. doi: 10.5966/sctm.2015-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Z, Evans J, Bhagwate A, Middha S, Bockol M, Yan H, Kocher JP. CAP-miRSeq: a comprehensive analysis pipeline for microRNA sequencing data. BMC Genomics. 2014;15:423. doi: 10.1186/1471-2164-15-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dudakovic A, Camilleri E, Riester SM, Lewallen EA, Kvasha S, Chen X, Radel DJ, Anderson JM, Nair AA, Evans JM, Krych AJ, Smith J, Deyle DR, Stein JL, Stein GS, Im HJ, Cool SM, Westendorf JJ, Kakar S, Dietz AB, van Wijnen AJ. High-resolution molecular validation of self-renewal and spontaneous differentiation in clinical-grade adipose-tissue derived human mesenchymal stem cells. J Cell Biochem. 2014;115(10):1816–28. doi: 10.1002/jcb.24852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baglio SR, Rooijers K, Koppers-Lalic D, Verweij FJ, Perez Lanzon M, Zini N, Naaijkens B, Perut F, Niessen HW, Baldini N, Pegtel DM. Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res Ther. 2015;6:127. doi: 10.1186/s13287-015-0116-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng S, Gao D, Gao C, Wei P, Niu M, Shuai C. MicroRNAs regulate signaling pathways in osteogenic differentiation of mesenchymal stem cells (Review) Mol Med Rep. 2016;14(1):623–9. doi: 10.3892/mmr.2016.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang N, Chen C, Yang D, Liao Q, Luo H, Wang X, Zhou F, Yang X, Yang J, Zeng C, Wang WE. Mesenchymal stem cells-derived extracellular vesicles, via miR-210, improve infarcted cardiac function by promotion of angiogenesis. Biochim Biophys Acta. 2017 doi: 10.1016/j.bbadis.2017.02.023. [DOI] [PubMed] [Google Scholar]
- 16.Jeong BC, I, Kang H, Hwang YC, Kim SH, Koh JT. MicroRNA-194 reciprocally stimulates osteogenesis and inhibits adipogenesis via regulating COUP-TFII expression. Cell Death Dis. 2014;5:e1532. doi: 10.1038/cddis.2014.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xin H, Li Y, Buller B, Katakowski M, Zhang Y, Wang X, Shang X, Zhang ZG, Chopp M. Exosome-mediated transfer of miR-133b from multipotent mesenchymal stromal cells to neural cells contributes to neurite outgrowth. Stem Cells. 2012;30(7):1556–64. doi: 10.1002/stem.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu M, Wright J, Guo H, Xiong Y, Arvan P. Proinsulin entry and transit through the endoplasmic reticulum in pancreatic beta cells. Vitam Horm. 2014;95:35–62. doi: 10.1016/B978-0-12-800174-5.00002-8. [DOI] [PubMed] [Google Scholar]
- 19.Nayak RR, Kearns M, Spielman RS, Cheung VG. Coexpression network based on natural variation in human gene expression reveals gene interactions and functions. Genome Res. 2009;19(11):1953–62. doi: 10.1101/gr.097600.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strawbridge RJ, Dupuis J, Prokopenko I, Barker A, Ahlqvist E, Rybin D, Petrie JR, Travers ME, Bouatia-Naji N, Dimas AS, Nica A, Wheeler E, Chen H, Voight BF, Taneera J, Kanoni S, Peden JF, Turrini F, Gustafsson S, Zabena C, Almgren P, Barker DJ, Barnes D, Dennison EM, Eriksson JG, Eriksson P, Eury E, Folkersen L, Fox CS, Frayling TM, Goel A, Gu HF, Horikoshi M, Isomaa B, Jackson AU, Jameson KA, Kajantie E, Kerr-Conte J, Kuulasmaa T, Kuusisto J, Loos RJ, Luan J, Makrilakis K, Manning AK, Martinez-Larrad MT, Narisu N, Nastase Mannila M, Ohrvik J, Osmond C, Pascoe L, Payne F, Sayer AA, Sennblad B, Silveira A, Stancakova A, Stirrups K, Swift AJ, Syvanen AC, Tuomi T, van ‘t Hooft FM, Walker M, Weedon MN, Xie W, Zethelius B, Mu TC, Ongen H, Malarstig A, Hopewell JC, Saleheen D, Chambers J, Parish S, Danesh J, Kooner J, Ostenson CG, Lind L, Cooper CC, Serrano-Rios M, Ferrannini E, Forsen TJ, Clarke R, Franzosi MG, Seedorf U, Watkins H, Froguel P, Johnson P, Deloukas P, Collins FS, Laakso M, Dermitzakis ET, Boehnke M, McCarthy MI, Wareham NJ, Groop L, Pattou F, Gloyn AL, Dedoussis GV, Lyssenko V, Meigs JB, Barroso I, Watanabe RM, Ingelsson E, Langenberg C, Hamsten A, Florez JC Consortium D, Consortium G, Consortium CA, Consortium CD. Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes. 2011;60(10):2624–34. doi: 10.2337/db11-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang SY. The acute effects of glucose on the insulin biosynthetic-secretory pathway in a simian virus 40-transformed hamster pancreatic islet beta-cell line. Endocrinology. 1989;124(4):1980–7. doi: 10.1210/endo-124-4-1980. [DOI] [PubMed] [Google Scholar]
- 22.Chinen K, Sudo K, Takahashi E, Nakamura Y. Isolation and mapping of the human beta-signal sequence receptor gene (SSR2) Cytogenet Cell Genet. 1995;70(3–4):215–7. doi: 10.1159/000134036. [DOI] [PubMed] [Google Scholar]
- 23.Garg B, Pathria G, Wagner C, Maurer M, Wagner SN. Signal Sequence Receptor 2 is required for survival of human melanoma cells as part of an unfolded protein response to endoplasmic reticulum stress. Mutagenesis. 2016;31(5):573–82. doi: 10.1093/mutage/gew023. [DOI] [PubMed] [Google Scholar]
- 24.Saitoh I, Sato M, Soda M, Inada E, Iwase Y, Murakami T, Ohshima H, Hayasaki H, Noguchi H. Tissue-Specific Stem Cells Obtained by Reprogramming of Non-Obese Diabetic (NOD) Mouse-Derived Pancreatic Cells Confer Insulin Production in Response to Glucose. PLoS One. 2016;11(9):e0163580. doi: 10.1371/journal.pone.0163580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe R, Inoue N, Westfall B, Taron CH, Orlean P, Takeda J, Kinoshita T. The first step of glycosylphosphatidylinositol biosynthesis is mediated by a complex of PIG-A, PIG-H, PIG-C and GPI1. EMBO J. 1998;17(4):877–85. doi: 10.1093/emboj/17.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roman-Pintos LM, Villegas-Rivera G, Rodriguez-Carrizalez AD, Miranda-Diaz AG, Cardona-Munoz EG. Diabetic Polyneuropathy in Type 2 Diabetes Mellitus: Inflammation, Oxidative Stress, and Mitochondrial Function. J Diabetes Res. 2016;2016:3425617. doi: 10.1155/2016/3425617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chuang JY, Kao TJ, Lin SH, Wu AC, Lee PT, Su TP, Yeh SH, Lee YC, Wu CC, Chang WC. Specificity protein 1-zinc finger protein 179 pathway is involved in the attenuation of oxidative stress following brain injury. Redox Biol. 2017;11:135–143. doi: 10.1016/j.redox.2016.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fox CS, Heard-Costa NL, Wilson PW, Levy D, D’Agostino RB, Sr, Atwood LD. Genome-wide linkage to chromosome 6 for waist circumference in the Framingham Heart Study. Diabetes. 2004;53(5):1399–402. doi: 10.2337/diabetes.53.5.1399. [DOI] [PubMed] [Google Scholar]
- 29.Wakayama Y, Miura K, Sabe H, Mochizuki N. EphrinA1-EphA2 signal induces compaction and polarization of Madin-Darby canine kidney cells by inactivating Ezrin through negative regulation of RhoA. J Biol Chem. 2011;286(51):44243–53. doi: 10.1074/jbc.M111.267047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma FY, Tesch GH, Ozols E, Xie M, Schneider MD, Nikolic-Paterson DJ. TGF-beta1-activated kinase-1 regulates inflammation and fibrosis in the obstructed kidney. Am J Physiol Renal Physiol. 2011;300(6):F1410–21. doi: 10.1152/ajprenal.00018.2011. [DOI] [PubMed] [Google Scholar]
- 31.Sai K, Morioka S, Takaesu G, Muthusamy N, Ghashghaei HT, Hanafusa H, Matsumoto K, Ninomiya-Tsuji J. TAK1 determines susceptibility to endoplasmic reticulum stress and leptin resistance in the hypothalamus. J Cell Sci. 2016;129(9):1855–65. doi: 10.1242/jcs.180505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma W, Dumont Y, Vercauteren F, Quirion R. Lipopolysaccharide induces calcitonin gene-related peptide in the RAW264.7 macrophage cell line. Immunology. 2010;130(3):399–409. doi: 10.1111/j.1365-2567.2009.03239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]