Abstract
Acid phosphatases play a crucial role in food processing industries to reduce phosphate content of food. Here in acid phosphatase from the seeds of Macrotyloma uniflorum has been purified to homogeneity using UNOsphere-S cation exchange chromatography followed by gel filtration with 81.85 fold purification. Molecular weight of purified enzyme was 55,000 (± 1040) Daltons under physiological conditions. It was a heterodimer of subunits having molecular weights 27,093 and 28,241 Daltons as determined by MALDI-TOF analysis. The optimum pH and temperature for the purified enzyme was 5.0 and 50 °C respectively. The enzyme was stable in the pH range 3.5–5.5 and showed temperature stability up to 60 °C. Substrate specificity of enzyme was checked with different substrates namely, p-nitrophenyl phosphate (p-NPP), ATP, ADP, glucose 6-phosphate, glucose-1-phosphate, fructose 6-phosphate, phenyl phosphate, α-naphthyl-phosphate, pyridoxyl phosphate and β-glycerophosphate. Enzyme showed high substrate specificity towards p-NPP, phenyl phosphate, ATP and α-naphthyl phosphate. Km and Vmax of enzyme were found to be 0.934 mM and 1.333 mM/min respectively with respect to p-NPP as a substrate. Chemical modification studies showed that tryptophan was present at the active site of the enzyme.
Electronic supplementary material
The online version of this article (doi:10.1007/s13197-017-2941-9) contains supplementary material, which is available to authorized users.
Keywords: Acid phosphatase, Macrotyloma uniflorum, Purification, Biochemical characterization
Introduction
Acid phosphatases or orthophosphoric monoester phosphohydrolases (E.C. 3.1.3.2.) are a group of enzymes that catalyze the hydrolysis of a variety of orthophosphate esters as well as transphosphorylation reactions in acidic medium (Berman et al. 1990). They are one of the most ubiquitous enzymes and are widely distributed in nature. They are found in yeasts, fungi, seeds of higher plants, fruits as well as in many animal tissues such as prostate gland of man and monkey, kidney, liver, spleen, erythrocytes, blood plasma etc. (Nicanuzia dos Prazeres et al. 2004; Sambuk et al. 2011; Tabaldi et al. 2007). This wide distribution in lower and higher organisms suggests that they might be involved in fundamental reactions in the organisms. In plants, acid phosphatases act as phosphate (Pi) scavenger. They mobilize Pi during growth and under stressed conditions like drought and Pi deprivation (Plaxton and Carswell 1999; Yenigün and Güvenilir 2003). Acid phosphatases play a crucial role in dephosphorylation of casein to reduce phosphate content of milk and thus reducing the phosphate toxicity (Molina et al. 2007). In many food applications, enzymatic dephosphorylation of casein proteins is preferred over chemical dephosphorylation for modification of caseins without compromising their functional properties (Molina et al. 2007). In this context, currently extensive studies on structure, function, catalytic properties and enzyme kinetics of acid phosphatases from various plant sources are being carried out. These studies may be of significance in approaching all possible perspectives in agricultural and industrial fields.
Although, a number of acid phosphatases from plant source have been reported, detailed studies on the kinetic parameters and chemical modification of active sites of only a few acid phosphatases have been explored. Thus, in the present investigation, isolation and biochemical characterization of acid phosphatase from the seeds of M. uniflorum has been carried.
Macrotyloma uniflorum (Lam.) Verdc. is commonly known as horse gram and is one of the lesser known beans. It is extensively cultivated in dry areas of Australia, Burma, India, and Sri Lanka. It is a native crop to the south-east Asian subcontinent and tropical Africa (Sudha et al. 1995). Macrotyloma uniflorum seeds have been used for the treatment of heart conditions, asthma, bronchitis, leukoderma, urinary discharges, and obesity. It has been used to reduce crystalluria and for lyses of kidney stones. It possesses slowly digestible starch, which is considered to have low postprandial glucose response when consumed by diabetic patients (Gupta et al. 2011). Macrotyloma uniflorum seeds have been used in the formulations of astringents, anthelmintic, expectorants and ophthalmic tonics.
Materials and methods
Materials
Chemicals and biochemicals used were procured from Sisco Research Laboratory, India, Hi-Media, India and Sigma Aldrich. All solutions were prepared in double distilled water unless otherwise specified.
Protein estimation
Protein estimation was carried out by the method of Lowry et al. (1951) using bovine serum albumin (BSA) as a standard.
Acid phosphatase assay
Acid phosphatase activity was determined essentially according to the procedure of Campbell et al. (1978) by measuring the amount of p-nitrophenol (p-NP) released from the substrate p-nitrophenyl phosphate (p-NPP) disodium salt (Campbell et al. 1978). A test system containing 200 µl acetate buffer (pH 5.0, 200 mM), 200 µl of 2 mM substrate and 50 µl enzyme was incubated at room temperature for 10 min. The reaction was arrested by adding 1.0 ml of 0.5 M NaOH. The p-nitrophenol thus liberated was estimated at 405 nm.
One unit of enzyme (U) is defined as the amount of enzyme that hydrolyses 1 µM of substrate per minute. The molar extinction coefficient of p-nitrophenol is 1.77 × 104 M−1 cm−1. Specific activity of acid phosphatase is defined as the amount of enzyme that liberates 1.0 µM p-nitrophenol/minute/mg of protein.
Acid phosphatase activity was also determined using other substrates like ATP, ADP, glycerophosphate and others. When other substrates were used, the enzyme activity was assayed by measuring the amount of phosphate released. For inorganic phosphate determination, the reaction was terminated by the addition of 1 ml 3% ammonium molybdate (in 200 mM acetate buffer, pH 4.0), followed by addition of 0.1 ml of 1% ascorbic acid (in 200 mM acetate buffer, pH 4.0). The colour was developed for 30 min and the absorbance was recorded at 700 nm. The amount of inorganic phosphate produced was calculated using a molar extinction coefficient of 4000 M−1 cm−1 (Lowry and Lopez 1946).
Isolation of acid phosphatase from M. uniflorum seeds
Macrotyloma uniflorum seed meal (100 g) was extracted with 500 ml of physiological saline (0.145 M NaCl) for 4 h at 4 °C. The extract was centrifuged at 10,000 rpm (12,080 g) and subjected to fractional precipitation with ammonium sulphate. The proteins precipitating between 30 and 80% saturation of ammonium sulphate were collected by centrifugation, dissolved in minimum amount of distilled water, extensively dialysed against distilled water and finally against acetate buffer (pH 5.0, 50 mM). The dialyzed protein solution, clarified by centrifugation (Fraction A) was used to isolate acid phosphatase by ion exchange chromatography on a column of UNOsphere-S a cation exchanger.
Ion exchange chromatography of M. uniflorum acid phosphatase on UNOsphere-S column
The slurry of UNOsphere-S was stirred with 1 M NaCl (3–5 volumes) washed and then equilibrated with acetate buffer (pH 5.0, 50 mM, 3–5 volumes) A glass column (L = 20 cm, Φ = 2 cm) with capacity of 40 ml was packed (under gravity) with above UNOsphere-S in acetate buffer (pH 5.0, 50 mM). Above Fraction ‘A’ was loaded on the column (15 ml, 20 mg/ml) followed by washing of the column with the equilibrating buffer. The adsorbed proteins were eluted with a discontinuous salt gradient (0.05, 0.1, 0.2, 0.5 and 1 M NaCl) in the same buffer. Fractions of 5 ml each were collected at a flow rate of 15 ml/h and monitored for protein content (by measuring the absorbance at 280 nm) and acid phosphatase activity. Acid phosphatase rich fractions were pooled, lyophilized and subjected to further purification by gel filtration on a Sephadex G-100 column. The acid phosphatase obtained after gel filtration was used for further studies.
SDS-PAGE
SDS-PAGE was carried out to check the presence or absence of subunits in the purified protein, according to the method of Laemmli (1970). Protein bands were visualized by the method of silver staining.
Activity staining of acid phosphatase
The native PAGE performed at pH 4.5 was washed 3–4 times with 0.1 M acetate buffer (pH 5.0). The gel was incubated for 2–3 h at R.T. in the reagent containing 0.05 g of α-naphthyl phosphate, 0.05 g of Fast blue RR, 1.0 g of sodium chloride and 0.05 gm magnesium chloride dissolved in 50 ml of acetate buffer (0.1 M, pH 5.0) to visualise reddish brown bands (Sadasivam and Manickam 2008).
Determination of molecular weight of purified acid phosphatase by gel filtration
The molecular weight of the acid phosphatase was determined by gel filtration on a Sephadex G-100 column (L = 17 cm, Φ = 2 cm, capacity 50 ml) using the following protein standards. Alcohol dehydrogenase (Mr = 150,000 Da), Bovine serum albumin (Mr = 66,000 Da), Pepsin (Mr = 35,000 Da), Trypsin (Mr = 23,000 Da) and Lysozyme (Mr = 14,300 Da).
SDS-PAGE
SDS-PAGE was performed by the method of Laemmli (1970). Standard molecular weight markers (Banglore Genei, mid-range markers) were run simultaneously as a reference to compare the molecular weight of the purified acid phosphatase. The molecular weight of the purified acid phosphatase was calculated by plotting a graph of Log of molecular weight × 104 versus Relative mobility of proteins.
Determination of molecular weight of purified acid phosphatase by MALDI-TOF MS
The molecular mass was also determined by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight (MALDI-TOF) mass spectrophotometry, using Voyager DE-STR (Applied Biosystems, USA) equipped with a 337 nm nitrogen laser. The protein (0.5 mg/ml) was mixed with the matrix (sinapinic acid) in deionized water (1:1) and 2 μl of the sample was spotted on to a MALDI target plate, dried at room temperature and analysed.
Effect of pH on activity and stability of acid phosphatase
The optimal pH of the enzyme was determined by performing the standard enzyme assay in the appropriate buffers. Buffers used were acetate buffer (100 mM, pH 3.0–5.0), sodium phosphate buffer (100 mM, pH, 6.0–8.0) and Glycine–NaOH buffer (100 mM, pH 8.0–10.0). Corresponding controls were run simultaneously. The pH stability of the enzyme was determined by measuring the residual activity after incubating the enzyme at each desired pH for 12 h at room temperature.
Effect of temperature on activity and stability of acid phosphatase
The optimal temperature of the enzyme was determined by performing the standard enzyme assay at temperatures ranging from 10 to 90 °C. The thermal stability of the enzyme was examined by measuring the residual enzyme activity after incubating the enzyme at each desired temperature for 30 min.
Effect of metal ions on the activity of acid phosphatase
The purified acid phosphatase was demetallised by incubating with 10 mM EDTA (1:1 v/v) for 3 h at 37 °C. The enzyme was then dialysed exhaustively against milli Q water. Effect of metal ions on enzyme activity was checked by preincubating the demetallised enzyme with 5 mM metal ions (Ca2+, Mg2+, Zn2+, K+, Fe3+, Mn2+, Ni2+, Al3+, Na+, Cu2+, Co2+, Hg2+ and Pb2+) for 30 min at room temperature. After incubation, the enzyme activity was checked in all the samples essentially as described earlier. Corresponding controls without metal ions were run simultaneously.
Atomic absorption spectrophotometry
The purified acid phosphatase was analyzed for presence of metal ions (Mg2+, Zn2+, Fe2+, Mn2+, Cu2+and Cr2+) by atomic absorption spectroscopy using a Perkin Elmer model 3100 Atomic Absorption Spectrophotometer.
Substrate specificity
The substrate specificity of acid phosphatase was checked using the following substrates each of 2 mM concentration (in acetate buffer pH 5.0, 50 mM); adenosine triphosphate, adenosine diphosphate, glucose-6-phosphate, glucose-1-phosphate, fructose-6-phosphate, phenyl phosphate, α-naphthyl phosphate, pyridoxyl-6-phosphate, glycerophosphate and p-nitrophenyl phosphate. Enzyme assay was performed essentially as described above.
Kinetic studies
The rate of acid phosphatase activity was determined at different substrate concentrations (0.025–2 mM) (for the substrates p-nitrophenyl-phosphate, ATP, ADP and glycerophosphate) in a total reaction volume of 1.5 ml containing 100 µl of enzyme, 200 µl of buffer (acetate buffer, pH 5.0, 100 mM) and 200 µl of substrate, essentially as described above. A double reciprocal plot was drawn according to the method of Lineweaver and Burk to determine the Km and Vmax of the enzyme.
Chemical modification studies
Chemical modification studies of the purified enzyme were carried out using the reagents N-bromosuccinimide (NBS), p-chloromercuribenzoate (PCMB), phenylmethylsulfonylfluoride (PMSF), 2,2-dithiobisnitrobenzoic acid (DTNB), phenyl glyoxal (PG), N-acetylimidazole (NAI), diethylpyrocarbonate (DEPC), N-ethylmaleimide (NEM) and citraconic anhydride which specifically modify tryptophan, cysteine, serine, arginine, tyrosine, histidine and lysine respectively. 100 µl of pure acid phosphatase was incubated with 100 µl of 10 mM chemical modification reagent at 37 °C for 30 min. After the incubation period, the residual enzyme activity was determined essentially as described above. Corresponding controls without the modifying reagent were run simultaneously.
Results and discussion
Isolation and purification of acid phosphatase from the seeds of M. uniflorum
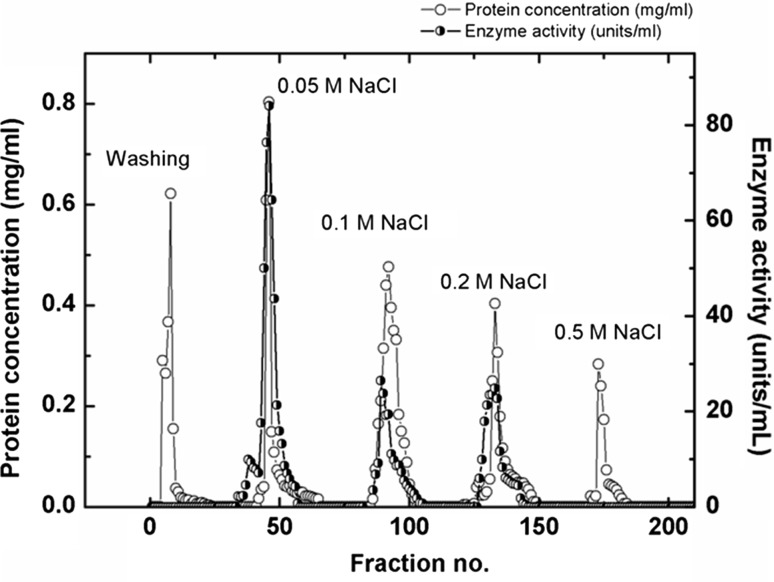
As seen in the elution profile (Fig. 1), three peaks corresponding to acid phosphatase activity were eluted with 0.05, 0.1 and 0.2 M NaCl respectively, suggesting the presence of three isoforms of acid phosphatase. However, the bulk of the acid phosphatase activity (142.91 U/mg) was eluted as a major peak with 0.05 M NaCl (suggesting that it is the major isoform of the enzyme in M. uniflorum seeds) with 46.26 fold of purification. The acid phosphatase rich fractions (of the major isoform) were then pooled, lyophilized and further purified by gel filtration on Sephadex G-100 column to obtain homogenous enzyme with specific activity of 252.5 U/mg and 81.85 fold of purification. All further studies were done using the 0.05 M NaCl eluted major isoform of acid phosphatase.
Fig. 1.
Elution profiles of UNOsphere-S ion exchange chromatography, Proteins were eluted with increase in gradient of NaCl (0.05–0.5 M) in acetate buffer (50 mM, pH 5.0)
Homogeneity of the M. uniflorum acid phosphatase (major isoform) was checked by PAGE under cationic (pH 4.5) as well as anionic (pH 8.3) conditions. A single band was observed under both, cationic as well as anionic conditions.
Activity staining of acid phosphatase
After performing PAGE at pH 4.5 the acid phosphatase bands were also visualized by activity staining using fast blue RR. Fig. S1 (Lane 1) shows the presence of these bands, suggesting the presence of three isoforms of the enzyme. Lane 2 shows the acid phosphatase activity bands corresponding to 0.05 M NaCl eluted fractions. Multiple forms of acid phosphatases have been reported in many plant sources (Ferreira et al. 1998). The different isoforms catalyses the same chemical reactions but differ in their amino acid sequence and kinetic parameters. Presence of multiple isoforms permits the fine adjustment of metabolism to meet the needs of given tissues or developmental stages depending on their physiological makeup and the kind of environment in which they function (Duff et al. 1994). Expression of multiple forms of acid phosphatases is regulated by physiological and environmental factors (Barrett-Lennard and Greenway 1982).
Molecular weight of acid phosphatase
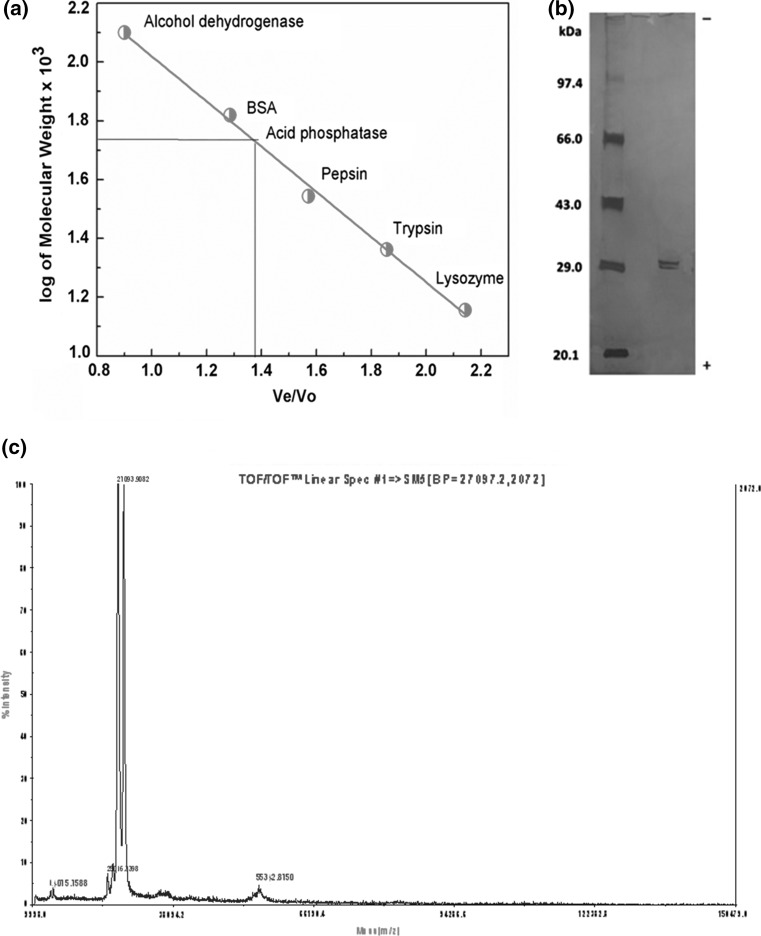
Molecular weight of acid phosphatase was determined by gel filtration chromatography, SDS-PAGE and MALDI-TOF analysis. Molecular weight of acid phosphatase (major isoform) determined using gel filtration chromatography was 55,000 (± 1040) Daltons (Fig. 2a). Whereas, SDS-PAGE of M. uniflorum acid phosphatase (major isoform) showed the presence of two very closely spaced narrow bands of ~ 29 kDa in presence and in absence of β-mercaptoethanol, suggesting that M. uniflorum acid phosphatase is a heterodimer of non-identical subunits (Fig. 2b). MALDI-TOF analysis showed the presence of two peaks of molecular weights of 27,093 and 28,241 Daltons corresponding to the two subunits of acid phosphatase (Fig. 2c). These observations suggest that under physiological condition the M. uniflorum acid phosphatase (major isoform) is a heterodimer of two subunits corresponding to molecular weights of 27,093 and 28,241 Daltons.
Fig. 2.
Molecular weight determination by a gel filtration, b SDS-PAGE, c MALDI-TOF analysis
Effect of pH on activity and stability of acid phosphatase
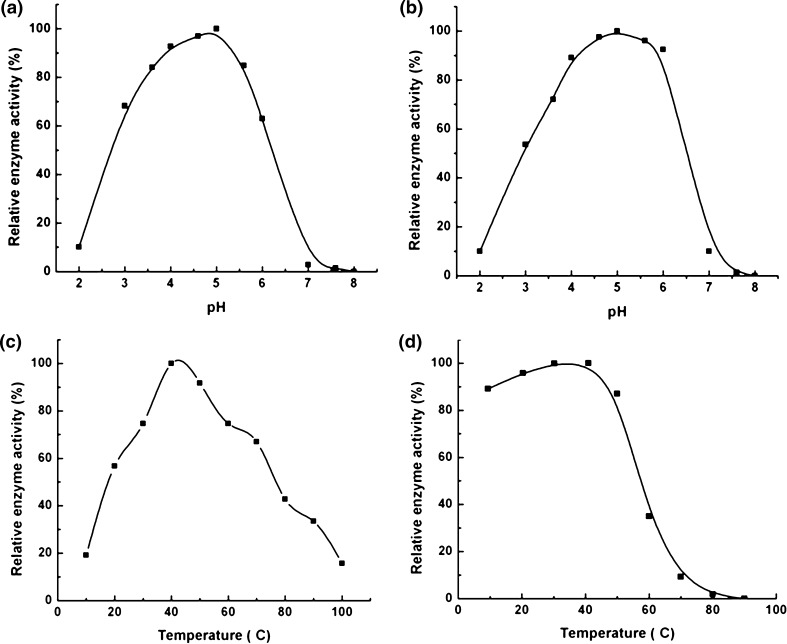
The effect of pH on acid phosphatase activity from M. uniflorum seeds is shown in Fig. 3a. The optimum pH for acid phosphatase activity was 5.0. Similar observations have been reported for the acid phosphatases from different plant sources. Most of the acid phosphatases require acidic pH for their catalytic activity. Chickpea acid phosphatase were found to have maximum enzyme activity at pH 5.0 (Asaduzzaman et al. 2011).
Fig. 3.
a, b Shows temperature and pH optima of acid phosphatase respectively. c, d Shows effect of temperature and pH on the activity of acid phosphatase respectively
As seen in Fig. 3b, acid phosphatase from M. uniflorum seeds was stable in the acidic pH range (3.5–6.5), with virtually no activity above pH 6.5. The maximum stability was observed at pH 5.0.
Effect of temperature on activity and stability of acid phosphatase
Acid phosphatase from M. uniflorum seeds showed maximum catalytic activity at 50 °C (Fig. 3c). Similar observations have been reported for the acid phosphatases from cotyledons of Psoralea corylifolia L., from garlic seedling which also shows optimum enzyme activity at 50 °C (Yenigün and Güvenilir 2003). Acid phosphatase activity from Vigna mungo L. seedlings showed highest activity at 55 °C (Asaduzzaman et al. 2011).
Temperature stability studies of purified acid phosphatase from M. uniflorum seeds, carried out, at various temperatures showed that the enzyme was stable up to 55 °C. Further heating of the enzyme resulted in about 45% loss in enzyme activity at 60 °C with a total loss in activity at 80 °C (Fig. 3d). This suggests that the acid phosphatase from the M. uniflorum seeds is mildly thermostable. These observations are similar to those reported for the acid phosphatase from Vigna mungo seeds which was found to be stable up to 55 °C and showed no activity at 80 °C (Asaduzzaman et al. 2011). The acid phosphatase from garlic seedlings was showed stability up to 60 °C (Yenigün and Güvenilir 2003).
Many tropical plants have shown the presence of thermostable enzymes in order to withstand and acclimatize to the environment temperature variations, which are essential for plant survival (Vogt et al. 1997). The thermal stability of an enzyme is most important for its biotechnological applications (Vogt et al. 1997). Acid phosphatases are being used in food industries for processing of casein to reduce phosphate content, thereby reducing phosphate toxicity (Molina et al. 2007). Phosphorylated compounds synthesized by using acid phosphatases are being used as nutritional supplements or taste enhancer in the food industries (Babich et al. 2012). Owing to these widespread applications, pH and thermal stability of enzyme is of great significance (Leisola et al. 2001).
Substrate specificity
Phosphorus plays a vital role in energy transfer, metabolic regulations and is an important constituent of biomolecules like proteins, phospholipids and nucleic acids (Ferreira et al. 1998). Acid phosphatases catalyse the hydrolysis of a wide range of orthophosphate monoesters. Plant acid phosphatases thus exhibit broad substrate specificity at optimum pH below 6.0 (Nicanuzia dos Prazeres et al. 2004).
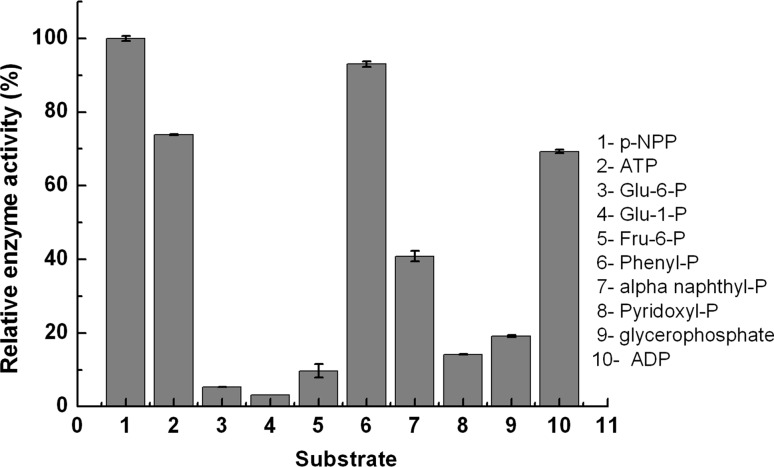
Substrate specificity of acid phosphatase from M. uniflorum seeds was studied using different substrates namely, p-nitrophenyl phosphate (p-NPP), ATP, ADP, glucose- 6-phosphate, glucose-1-phosphate, fructose-6-phosphate, phenyl phosphate, α-naphthyl-phosphate, pyridoxyl phosphate and β-glycerophosphate (Fig. 4). The enzyme showed high catalytic activity towards p-NPP, phenyl phosphate, ATP and ADP whereas it was observed to be less active towards glucose-6-phosphate, glucose-1-phosphate, fructose-6-phosphate and pyridoxyl-6-phosphate.
Fig. 4.
Effect of different substrates on Macrotyloma uniflorum seed acid phosphatase activity
Effect of substrate concentration on acid phosphatase activity
The effect of substrate concentrations on acid phosphatase activity was studied by using p-NPP, ATP and ADP as substrates. A plot of 1/[V] verses 1/[S] was drawn according to the method of Lineweaver and Burk to determine the value of Km and Vmax in each case (Supplementary Fig. S2). Km and Vmax of acid phosphatase for p-NPP were found to be 0.934 mM and 1.333 mM/min respectively (Fig. S2a). Km and Vmax of acid phosphatase with ATP as a substrate was 5 mM and 0.988 mM/min respectively (Fig. S2b) whereas, Km and Vmax of acid phosphatase for ADP was 8.7 mM and 0.738 mM/min respectively (Fig. S2c).
Effect of metal ions on acid phosphatase activity
The pure enzyme was initially incubated with EDTA which resulted in demetallisation of acid phosphatase. The sample was divided into aliquots. In each aliquot a different monovalent, divalent or trivalent metal ions was added (5 mM) and incubated. After the end of incubation time, acid phosphatase activity was measured.
Re-metallization of M. uniflorum acid phosphatase with Mg2+ resulted in restoration of 94% of the enzyme activity (Supplementary Fig. S3). K+, Ca2+ and Na2+ had no effect on the enzyme activity. Acid phosphatase was found to be strongly inhibited by Hg2+ whereas moderate inhibition was observed with Ag2+ (Supplementary Fig. S3). Activity of acid phosphatase was found to be increased slightly in presence of Mg2+ ions suggesting its important role in catalysis. Most of the plant acid phosphatases are reported to be metalloproteins containing Mg2+. EDTA treatment or electro-dialysis of the enzyme solution removes Mg2+ from the enzyme resulting in loss of activity which can be reactivated by adding Mg2+. Acid phosphatase from other plant sources like from A. thaliana seedlings, banana fruits and L. Esculentum were also found be activated in presence of Mg2+ thus, suggesting the importance of Mg2+ in the catalytic action of plant acid phosphatases (Turner and Plaxton 2001; Veljanovski et al. 2006). Atomic absorption spectroscopic studies also confirmed the presence of Mg2+ in native acid phosphatase from M. uniflorum seeds.
The tolerance to various heavy metals is also one of the criteria for selecting enzyme for industrial applications (Leisola et al. 2001). Acid phosphatase, in the present study (isolated from M. uniflorum) was tolerant to most of the metal ions (Supplementary Fig. S3) except Hg2+. Thermostability and selective inhibition of acid phosphatase in presence of Hg2+ is a potential tool for the fabrication of enzyme inhibition based portable biosensor for the detection of Hg2+ (Tagad et al. 2016).
Chemical modification studies
For chemical modification studies, aliquots of enzyme were treated with different concentrations of amino acid modifying reagents and incubated for 30 min. Subsequently, substrate was added and the residual enzyme activity was checked. Chemical modifying reagents that are specific for amino acids were used to study the presence of amino acids at the active site of purified acid phosphatase.
A limited number of reagents were used that were specific for some of the common amino acids that are present at the active site of enzymes. Chemical modification study in presence of NBS showed only about 8% residual enzyme activity suggesting that tryptophan is present at the active site (Supplementary Table S1). In presence of PCMF and phenyl glyoxal the enzyme showed only 27 and 32% residual enzyme activity which suggests the possibility of presence of Ser and Arg at or close to the active site of acid phosphatase. Inhibition of acid phosphatase activity was not observed in presence of PCMB, NAI, NEM, DEPC and citraconic anhydride suggesting that Cys, Tyr, His and Lys may not have a role in catalysis.
Conclusion
Acid phosphatase from the seeds of M. uniflorum has been purified to homogeneity. Molecular weight of purified enzyme was 55,000 (± 1040) Daltons. The optimum pH and temperature for the purified enzyme was 5.0 and 50 °C respectively. The enzyme was found to be fairly thermostable suggesting its suitability for applications in food and pharmaceutical industries. Enzyme showed high substrate specificity towards p-NPP, phenyl phosphate, ATP and α-naphthyl phosphate.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s13197-017-2941-9) contains supplementary material, which is available to authorized users.
References
- Asaduzzaman A, Rahman HM, Yeasmin T. Purification and characterization of acid phosphatase from a germinating black gram (Vigna mungo L.) seedling. Arch Biol Sci. 2011;63:747–756. doi: 10.2298/ABS1103747A. [DOI] [Google Scholar]
- Babich L, Hartog AF, van der Horst MA, Wever R. Continuous-flow reactor-based enzymatic synthesis of phosphorylated compounds on a large scale. Chem Eur J. 2012;18(21):6604–6609. doi: 10.1002/chem.201200101. [DOI] [PubMed] [Google Scholar]
- Barrett-Lennard EG, Greenway H. Partial separation and characterization of soluble phosphatases from leaves of wheat grown under phosphorus deficiency and water deficit. J Exp Bot. 1982;33(4):694–704. doi: 10.1093/jxb/33.4.694. [DOI] [Google Scholar]
- Berman T, Wynne D, Kaplan B. Phosphatases revisited: analysis of particle-associated enzyme activities in aquatic systems. Hydrobiologia. 1990;207(1):287–294. doi: 10.1007/BF00041467. [DOI] [Google Scholar]
- Campbell HD, Dionysius DA, Keough DT, Wilson BE, Jersey Jd, Zerner B. Iron-containing acid phosphatases: comparison of the enzymes from beef spleen and pig allantoic fluid. Biochem Biophys Res Commun. 1978;82:615–620. doi: 10.1016/0006-291X(78)90919-1. [DOI] [PubMed] [Google Scholar]
- Duff SMG, Sarath G, Plaxton WC. The role of acid phosphatases in plant phosphorus metabolism. Physiol Plant. 1994;90:791–800. doi: 10.1111/j.1399-3054.1994.tb02539.x. [DOI] [Google Scholar]
- Ferreira CV, Granjeiro JM, Taga EM, Aoyama H. Purification and characterization of multiple forms of soybean seed acid phosphatases. Plant Physiol Biochem. 1998;36(7):487–494. doi: 10.1016/S0981-9428(98)80173-3. [DOI] [Google Scholar]
- Gupta LH, Badole SL, Bodhankar SL, Sabharwal SG. Antidiabetic potential of α-amylase inhibitor from the seeds of Macrotyloma uniflorum in streptozotocin-nicotinamide-induced diabetic mice. Pharm Biol. 2011;49:182–189. doi: 10.3109/13880209.2010.507633. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leisola M, Jokela J, Pastinen O, Turunen O, Schoemaker H. Industrial use of enzymes. Oxford: Eolss Publishers; 2001. [Google Scholar]
- Lowry O, Lopez J. The determination of inorganic phosphate in the presence of labile phosphate esters. J Biol Chem. 1946;162:421. [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Molina AC, Alli I, Konishi Y, Kermasha S. Effect of dephosphorylation on bovine casein. Food Chem. 2007;101(3):1263–1271. doi: 10.1016/j.foodchem.2006.03.033. [DOI] [Google Scholar]
- Nicanuzia dos Prazeres J, Veríssima Ferreira C, Aoyama H. Acid phosphatase activities during the germination of Glycine max seeds. Plant Physiol Biochem. 2004;42:15–20. doi: 10.1016/j.plaphy.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Plaxton WC, Carswell MC. Metabolic aspects of the phosphate starvation response in plants. In: Lerner HR, editor. Plant responses to environmental stresses: from phytohormones to genome reorganization. New York: Marcel Dekker; 1999. pp. 349–372. [Google Scholar]
- Sadasivam S, Manickam A. Biochemical methods. New Delhi: New Age International Publishers; 2008. [Google Scholar]
- Sambuk EV, Fizikova AY, Savinov VA, Padkina MV. Acid phosphatases of budding yeast as a model of choice for transcription regulation research. Enzym Res. 2011;2011:16. doi: 10.4061/2011/356093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudha N, Mushtari Begum J, Shambulingappa KG, Babu C. Nutrients and some anti-nutrients in horsegram (Macrotyloma uniflorum (Lam.) Verdc.) Food Nutr Bull. 1995;16(1):81–83. [Google Scholar]
- Tabaldi LA, Ruppenthal R, Cargnelutti D, Morsch VM, Pereira LB, Schetinger MRC. Effects of metal elements on acid phosphatase activity in cucumber (Cucumis sativus L.) seedlings. Environ Exp Bot. 2007;59:43–48. doi: 10.1016/j.envexpbot.2005.10.009. [DOI] [Google Scholar]
- Tagad CK, Kulkarni A, Aiyer RC, Patil D, Sabharwal SG. A miniaturized optical biosensor for the detection of Hg2+ based on acid phosphatase inhibition. Optik. 2016;127(20):8807–8811. doi: 10.1016/j.ijleo.2016.06.123. [DOI] [Google Scholar]
- Turner WL, Plaxton WC. Purification and characterization of banana fruit acid phosphatase. Planta. 2001;214(2):243–249. doi: 10.1007/s004250100607. [DOI] [PubMed] [Google Scholar]
- Veljanovski V, Vanderbeld B, Knowles VL, Snedden WA, Plaxton WC. Biochemical and molecular characterization of AtPAP26, a vacuolar purple acid phosphatase up-regulated in phosphate-deprived Arabidopsis suspension cells and seedlings. Plant Physiol. 2006;142:1282–1293. doi: 10.1104/pp.106.087171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt G, Woell S, Argos P. Protein thermal stability, hydrogen bonds, and ion pairs. J Mol Biol. 1997;269(4):631–643. doi: 10.1006/jmbi.1997.1042. [DOI] [PubMed] [Google Scholar]
- Yenigün B, Güvenilir Y. Partial purification and kinetic characterization of acid phosphatase from garlic seedling. Appl Biochem Biotechnol. 2003;107(1–3):677–687. doi: 10.1385/ABAB:107:1-3:677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.