Abstract
New approaches in regenerative medicine and vasculogenesis have generated a demand for sufficient numbers of human endothelial cells (ECs). ECs and their progenitors reside on the interior surface of blood and lymphatic vessels or circulate in peripheral blood; however, their numbers are limited, and they are difficult to expand after isolation. Recent advances in human induced pluripotent stem cell (hiPSC) research have opened possible avenues to generate unlimited numbers of ECs from easily accessible cell sources, such as the peripheral blood. In this study, we reprogrammed peripheral blood mononuclear cells, human umbilical vein endothelial cells (HUVECs), and human saphenous vein endothelial cells (HSVECs) into hiPSCs and differentiated them into ECs. The phenotype profiles, functionality, and genome stability of all hiPSC-derived ECs were assessed and compared with HUVECs and HSVECs. hiPSC-derived ECs resembled their natural EC counterparts, as shown by the expression of the endothelial surface markers CD31 and CD144 and the results of the functional analysis. Higher expression of endothelial progenitor markers CD34 and kinase insert domain receptor (KDR) was measured in hiPSC-derived ECs. An analysis of phosphorylated histone H2AX (γH2AX) foci revealed that an increased number of DNA double-strand breaks upon reprogramming into pluripotent cells. However, differentiation into ECs restored a normal number of γH2AX foci. Our hiPSCs retained a normal karyotype, with the exception of the HSVEC-derived hiPSC line, which displayed mosaicism due to a gain of chromosome 1. Peripheral blood from adult donors is a suitable source for the unlimited production of patient-specific ECs through the hiPSC interstage. hiPSC-derived ECs are fully functional and comparable to natural ECs. The protocol is eligible for clinical applications in regenerative medicine, if the genomic stability of the pluripotent cell stage is closely monitored.
Keywords: : induced pluripotent stem cells, endothelial differentiation, peripheral blood mononuclear cells
Introduction
Endothelial cells (ECs) form a thin layer on the interior surface of blood and lymphatic vessels. They regulate various physiological processes, such as blood hemostasis, vascular tone, interaction of the vessel wall with blood elements, and the formation of new blood vessels [1]. On the contrary, ECs are involved in pathological states, such as cancer, atherosclerosis, and other diseases [2, 3]. Therefore, ECs represent an important in vitro model for studies of vascular development and drug screens [4, 5]. In regenerative medicine, ECs have been used to generate the cellular lining of vascular grafts [6, 7]. Currently, several clinical trials are being conducted on endothelial progenitor cells (EPCs), mainly as myocardial infarction and peripheral vascular disease treatments (reviewed in Chong et al. [8]).
The first human ECs were isolated from umbilical cord [human umbilical vein endothelial cells (HUVECs)] [9] and became a popular model for vascular research. Adult ECs are also commonly isolated from saphenous vein [human saphenous vein endothelial cells (HSVECs)] [10], usually from patients undergoing bypass or varicose vein surgery. ECs used for the treatment of ischemic conditions or other diseases are mainly populations of circulating EPCs that are usually positive for CD34 surface marker alone or in combination with kinase insert domain receptor (KDR; also known as vascular endothelial growth factor receptor 2; VEGFR-2) or CD133 [11–13]. However, ECs from blood vessels or peripheral blood can be obtained in limited numbers and are difficult to expand. To overcome these hurdles, pluripotent stem cells (PSCs) can be used. In vitro methods for EC production from PSCs have been recently introduced. This approach ensures a consistent and potentially unlimited source of ECs for in vitro studies and regenerative medicine.
The key question to be addressed is if PSC-derived ECs are comparable to ECs isolated from human tissues and safe for future clinical applications. Both human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs) have the capacity to differentiate into ECs [14–21]. In contrast to hESCs, hiPSCs are easier to obtain and do not generate ethical controversy. The first hiPSCs were created from skin fibroblasts [22, 23], and this cell type is still among the most frequently used source of hiPSC. However, uncomfortable procedures for harvesting skin biopsies and the time requirements for establishing fibroblast cell lines limit the use of fibroblasts for reprogramming.
Peripheral blood overcomes these issues and the quality of hiPSC derived from mononuclear cells [peripheral blood mononuclear cells (PBMCs)] and those derived from fibroblasts is equivalent, indistinguishable from hESCs [24]. Several methods for PBMCs reprogramming have been published [24–30], varying mainly in the type of reprogramming vectors and media used for ex vivo PBMCs expansion. The composition of the expansion media containing the optimal cocktail of cytokines is the crucial factor for successful reprogramming. Preferential induction of the proliferation of hematopoietic progenitor cells, as described in Shah et al. [31], may be the key factor in highly efficient routine PBMCs reprogramming.
The potential of hiPSC-based therapies in regenerative medicine is hindered by genomic instability. The processes of cellular reprogramming and subsequent in vitro culture of hiPSCs have been reported to compromise genomic stability, particularly through introduction of DNA double-strand breaks (DSBs) [32–35]. The genome instability in PSCs may eventually result in karyotypic abnormalities, such as chromosomal and subchromosomal aberrations. Gains of chromosomes 1, 12, 17, 20, and X represent the most common events observed in hESCs and hiPSCs (summarized in Weissbein et al. [36]).
In our study, we reprogrammed PBMCs, HUVECs, and HSVECs with episomal vectors and subsequently created hiPSC-derived ECs. Our goal was to verify that hiPSC-derived ECs are phenotypically and functionally comparable to HUVECs and HSVECs. Genomic stability was a high priority during the reprogramming/differentiation process; therefore, the numbers of DSBs were measured by counting phosphorylated histone H2AX (γH2AX) foci, together with a karyotype analysis. Experiments were conducted in a virus-free and DNA nonintegrating setting without feeder cells, which are the main criteria for future clinical applications.
Materials and Methods
Ethics approval and consent to participate
Studies were performed according to the amended Declaration of Helsinki. Institutional Review Board of the St. Anne's University Hospital Brno and the Faculty of Medicine, Masaryk University Brno approved the protocol used in our study, including the use of human PSCs. All patients gave written informed consent. Protocols for teratoma studies in animals were reviewed and approved by the Institutional Review Board at the Faculty of Medicine, Masaryk University Brno and conformed to the national guidelines of the Czech Republic.
Cell isolation and culture
Peripheral blood samples and pieces of saphenous vein were collected from patients undergoing varicose vein surgery. PBMCs were isolated from 6 mL of peripheral blood using Histopaque-1077 density gradient centrifugation (Sigma-Aldrich, St. Louis, MO). The total count of isolated PBMCs was 7.4 × 106. Before reprogramming, PBMCs were cultured on low-attachment culture dish in complete PBMC medium (cPBMC) consisting of StemPro®-34 serum-free medium, supplemented with 2 mM l-Glutamine (both from Thermo Fisher Scientific, Waltham, MA), ZellShield® (Minerva Biolabs, Berlin, Germany) and the growth factors stem cell factor (SCF), flt-3 Ligand (Flt-3L; both 100 ng/mL), interleukin (IL)-3 (20 ng/mL), and IL-6 (10 ng/mL; all from Peprotech, Rocky Hill, NJ) for 3 days. The initial seeding density was 2 × 105 cells/cm2.
An ∼10 cm-long piece of human saphenous vein was washed with phosphate-buffered saline (PBS) and cut into 1.5–2 cm-long pieces. Vein samples were incubated with 0.3% Collagenase II (Thermo Fisher Scientific) in Hank's medium (Sigma-Aldrich) for 50 min in a humidified 37°C incubator with an atmosphere of 5% (v/v) CO2. The cells from the digested tissue were washed with medium, centrifuged and seeded on T25 EasYFlasks™ (Thermo Fisher Scientific) in Endothelial Growth Medium-2 (EGM-2; Lonza, Basel, Switzerland) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich) and ZellShield (Minerva Biolabs) at a density of 3.5 × 104 cells/cm2. After the first passage, the cells were cultured on tissue-culture plates coated with 0.1% gelatin. Passaging was performed using trypsin/EDTA (0.5 mg/mL trypsin with 0.2 mg/mL EDTA; Sigma-Aldrich) when cells reached ∼90% confluence. HUVECs were purchased from Thermo Fisher Scientific and maintained under the same conditions as HSVECs.
Generation of hiPSCs
hiPSCs were generated from PBMCs (hiPSC-PB; line ID CBIA-26), HUVECs (hiPSC-HU; line ID CBIA-19), and HSVECs (hiPSC-HS; line ID CBIA-25), using genome nonintegrating episomal vectors (Epi5™ Episomal hiPSC Reprogramming Kit; Thermo Fisher Scientific), according to the manufacturer's instructions. In brief, reprogramming factors (Oct3/4, Sox2, Klf4, Lin28, and c-Myc) were delivered in episomal vectors with an oriP/EBNA-1 (Epstein–Barr nuclear antigen-1) backbone [37]. Electroporation was performed at 1,600 V in three pulses for 10 ms for 2 × 105 cells in the Neon electroporator (Thermo Fisher Scientific).
Reprogrammed cells were then seeded at a density of 2 × 104 cells/cm2 on plates that had been precoated with the Matrigel™ matrix (Thermo Fisher Scientific) in cPBMC medium for PBMCs or in EGM-2 for HUVECs and HSVECs. At day 7 after reprogramming, the medium was changed to mTeSR™1 (Stemcell Technologies, Vancouver, Canada). hiPSC colonies were mechanically picked from days 17 to 25 and transferred to separate wells of a 12-well plate. Subsequent passaging was performed using 0.5 mM EDTA (Thermo Fisher Scientific). One hour before passaging, the cells were pretreated with 10 μM Y-27632 (ROCK inhibitor; Selleckchem, Houston, TX). hiPSCs were maintained on tissue-culture plates that had been precoated with Matrigel in mTeSR1 medium supplemented with ZellShield. The medium was changed daily.
Immunocytochemistry
Pluripotency markers were detected with primary antibodies against Oct3/4 (Santa Cruz Biotechnology, Dallas, TX), Sox2 (R&D Systems, Minneapolis, MN), and Nanog (Cell Signaling Technology, Danvers, MA) as previously described [38]. In brief, cells on plates were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100 (both from Sigma-Aldrich). Cells were incubated with primary antibodies overnight at 4°C, followed by 2-h incubation with secondary antibody conjugated with Alexa 488 (Cell Signaling Technology). Nuclei were stained with Hoechst dye (bisbenzimide H33258; 1 μg/mL; Sigma-Aldrich). Fluorescent signals were detected under an inverted Olympus IX71 microscope (Olympus, Hamburg, Germany).
Flow cytometry
Flow cytometry was used to determine the expression of cell surface antigens. Specifically, antibodies against the pluripotency markers SSEA-4 (Phycoerythrin-conjugated antibody; PE; clone MC-813-70; R&D Systems) as well as Tra-1-60 and Tra-1-81 (both PE; REA157, resp. REA246; both from Miltenyi Biotec, Bergisch Gladbach, Germany) were used. The expression of CD31 (Allophycocyanin-conjugated antibody; APC; AC128; Miltenyi Biotec), CD34 (PE; AC136; Miltenyi Biotec), CD144 (PE; REA199; Miltenyi Biotec), and KDR (PE; ES8-20E6; Miltenyi Biotec) and the uptake of dil-labeled and acetylated low-density lipoprotein (Dil-Ac-LDL; Alpha Diagnostics, San Antonio, TX) were measured to characterize ECs. For isotype controls were used antibodies Mouse IgG3 (PE; 133316; R&D Systems), REA Control (PE; REA293; Miltenyi Biotec), and Mouse IgG1 (APC; IS5-21F5; Miltenyi Biotec).
Cells were harvested as a single-cell suspension using trypsin/EDTA and resuspended in PBS containing 0.5% bovine serum albumin and 2 mM EDTA. Cells were incubated with fluorochrome-conjugated antibodies for 30 min at 4°C, followed by washes with PBS. Cells were incubated with 10 μg/mL Dil-Ac-LDL for 4 h to assess low-density lipoprotein (LDL) uptake. Samples were measured on a BD FACS Canto II flow cytometer (Becton–Dickinson, Heidelberg, Germany). BD FACSDiva (Becton–Dickinson) and Flowing Software (Cell Imaging Core, Turku Centre for Biotechnology, Turku, Finland) were used to analyze the data.
Detection of immunoglobulin and T-cell receptor gene recombinations
Genomic DNA was isolated from (1) hiPSC-PB, (2) PBMCs from healthy donor, and (3) human dermal neonatal fibroblasts (HDFn; Thermo Fisher Scientific) using DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. DNA concentration was determined spectrophotometrically (NanoDrop ND-1000; NanoDrop Technologies, Wilmington, DE). Rearranged immunoglobulin (Ig) and T-cell receptor (TCR) genes were analyzed by polymerase chain reaction (PCR) assays developed by European BIOMED-2 collaborative study [39]. Sixty-three primers were used in seven multiplex PCR tubes to detect (1) complete VH-JH rearrangement of immunoglobulin heavy chain (IGH) gene (three tubes), (2) complete Vβ-Jβ rearrangement of TRB gene (two tubes), and (3) TRG gene rearrangements (two tubes).
The PCR cocktail, final volume 50 μL, contained 100 ng of genomic DNA, 25 pmol of each primer, 1 × Green GoTaq® Reaction Buffer (Promega, Madison, WI), 0.2 mM dNTP, and 1–2 U of GoTaq G2 DNA Polymerase (Promega). PCR was performed in a DNA Engine (PTC-200) Peltier Thermal Cycler (Bio-Rad Laboratories, Hercules, CA) and the cycling conditions were preactivation at 95°C for 7 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 30 s, and extension at 73°C for 30 s, and a final extension at 74°C for 7 min. The PCR products of Ig/TCR genes were ultraviolet-visualized on 2% ethidium bromide-stained agarose gel. The presence of the expected size product was checked based on a 100 bp DNA Ladder (New England Biolabs, Ipswich, MA).
Endothelial differentiation
Our protocol for the endothelial differentiation of hiPSCs was adapted from a method published by Orlova et al. [40]. In brief, hiPSCs were differentiated on Matrigel in BPEL medium [41] supplemented with the following growth factors: 25 ng/mL Activin A, 30 ng/mL BMP4, 50 ng/mL VEGF165, and 1.5 mM CHIR99021, a small molecule inhibitor. On the 3rd and 7th days, the medium was replaced with BPEL medium supplemented with 50 ng/mL VEGF and 10 mM SB43152. On the 10th day, cells were harvested, analyzed using flow cytometry and immunomagnetically separated using CD31-Microbeads (Miltenyi Biotec). The differentiation procedure generated 10%–45% CD31-positive cells. After separation, hiPSC-derived ECs were cultured on fibronectin-coated dishes in EGM-2 supplemented with 50 ng/mL VEGF.
Tube formation assay
A 96-well μ-plate for angiogenesis (Ibidi, Planegg, Germany) was coated with 50 μL/well of growth factor-reduced Geltrex™ (Thermo Fisher Scientific) and incubated at 37°C for 1 h. Cells were seeded at density of 5,000 cells/well in EGM-2 supplemented with 50 ng/mL VEGF and incubated in a 37°C incubator with a 5% CO2 atmosphere for 24 h to allow tubes to form.
The number of complete rings formed during tube formation assay (TFA) was used to quantify the capability of ECs to form tubes. To count the number of rings in each well, we applied the following procedure. First, we subsampled the original images by a factor of four to reduce the size of images and still have the analyzed structures (rings) in a sufficient detail. The subsampling also reduced the noise in images, and therefore no further noise suppression was needed. Second, we calculated local standard deviation of intensity pixels in windows of size 3 × 3 because the cells as well as their connections had much larger standard deviation from the local mean intensity than the background. It helped us to use a single threshold to segment pixels belonging to cells and their connections. We used a minimum error method to set the appropriate threshold [42]. To remove small background as well as foreground structures in the segmented images, we applied the alternating sequential filter based on area closings and openings [43]. To count the number of rings we calculated the number of salient maxima of the Euclidian distance transform. As the salient maxima, we considered all maxima with a distance smaller than 200 pixels from the segmented structures (cells and their connections). It means that only rings with a radius smaller than ∼400 μm were counted. The centers of the detected rings are visualized by the black cross.
Chemotaxis migration assay
The cells were grown in Millicell® hanging cell culture inserts (Merck Millipore, Billerica, MA) in 24-well plates. hiPSC-derived ECs, HUVECs, and HSVECs were seeded onto the inside of the insert in EGM-2 without VEGF at density of 2.5 × 105 cells per insert. Control HDFn were seeded in Fibroblast medium (DMEM medium, supplemented with 20% FBS, 2 mM l-glutamine, and 100 μM nonessential amino acids; all from Life Technologies). Basolateral side was filled with EGM-2 or fibroblast medium containing 50 ng/mL VEGF. Cells were incubated in a 37°C incubator with a 5% CO2 atmosphere for 18 h to allow the cells to migrate through the membrane. Cells on inserts were fixed with 4% paraformaldehyde (Sigma-Aldrich), permeabilized with 100% methanol (Lach-Ner, Neratovice, Czech Republic), and stained with 20 × diluted Giemsa stain (Sigma-Aldrich). Inner side of the insert was scraped with cotton swab and only cells on the outer side were counted under the microscope. For each cell culture insert, three representative images were counted. Each cell line was grown in three independent inserts.
Karyotype analysis
Karyotype analyses were performed by Cytogenetic Laboratory Brno (Cytogenetická Laboratoř Brno, s.r.o., Brno, Czech Republic). In brief, hiPSCs at passage 16 or higher were grown to ∼90% confluence and exposed to 10 μg/mL colcemid for 1 h. Harvested cells were exposed to a hypotonic solution (culture media diluted with deionized water at a 1:3 ratio) and fixed four times with methanol/acetic acid (3:1). Cells were then dropped onto glass slide and incubated at room temperature overnight. For Giemsa-banding, glass slides were incubated at 95°C for 10 min, washed with Sorensen's phosphate buffer at 50°C, and stained with Wright's stain for 1.5 min. After washing, the karyotype was determined by microscopic examination. Fifty mitosis events per sample were analyzed using “LUCIA Cytogenetics” software (Laboratory Imaging, Prague, Czech Republic).
DSB visualization by quantifying γH2AX foci and analyzing images
Cells were first seeded onto microscope slides coated with Matrigel or gelatin in four-well plates to analyze the number of γH2AX foci in G1 phase of the cell cycle. Four hours before fixation, a nucleoside analog of thymidine, EdU (5-ethynyl-2′-deoxyuridine; Thermo Fisher Scientific), was added at a final concentration of 10 μM to visualize cells in S/G2 phase of the cell cycle.
The cells were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. An overnight incubation with a primary antibody against γH2AX (BioLegend, San Diego, CA) was followed by a 1 h incubation with a secondary antibody conjugated with Alexa Fluor 555 (Cell Signaling Technology). Samples were stained with the Click-iT® EdU Alexa Fluor® 488 Imaging Kit (Thermo Fisher Scientific) to visualize EdU, according to the manufacturer's instructions. Finally, the nuclei were stained with Hoechst dye. Fluorescent signals were detected using the Zeiss Axiovert 200 M system (Carl Zeiss, Oberkochen, Germany). Images were captured using a CoolSNAP HQ2 CCD camera in the wide-field mode (Photometrics, Tucson, AZ) at −30°C. Images of thirty slices at 0.3 μm intervals were acquired in each field at a resolution of 1,392 × 1,040 pixels. The pixel size of the images was 124 × 124 nm.
The open-source software Acquiarium [44] was used to acquire and analyze the images (http://cbia.fi.muni.cz/projects/acquiarium.html), as previously described in detail [35]. In brief, the nucleus of each cell, which was stained with Hoechst dye, was automatically recognized and defined the area, in which we counted γH2AX foci and assessed the intensity of the EdU signal. We used the eMax algorithm developed by Štěpka et al. to identify the γH2AX foci [45]. The EdU signal was quantified based on the total intensity calculated in the nucleus. The threshold for the separation of EdU-negative (G1) and EdU-positive (S/G2) cells was computed in MATLAB (Mathworks, Natick, MA) using the Otsu method.
Teratoma formation
The in vivo differentiation experiments were performed in duplicate for each hiPSC line. Six NOD SCID GAMMA mice were injected (three intramuscularly and three subcutaneously), and all mice developed teratomas after ∼8 weeks. In brief, hiPSCs were grown to near confluency on 6 cm Petri dishes and harvested with 0.5 mM EDTA. Cells were washed with PBS and resuspended in 40 μL of cold PBS. An equal volume (40 μL) of Matrigel was added. The suspension was maintained on ice until it was injected into a mouse. The histological analysis of the teratomas was performed by Dr. Eva Mecova from the Department of Histology and Embryology, Masaryk University Brno.
Statistical analysis
Data sets were compared using Student's t-test or the Mann–Whitney test. P < 0.05 was considered statistically significant.
Results
Generation of hiPSCs
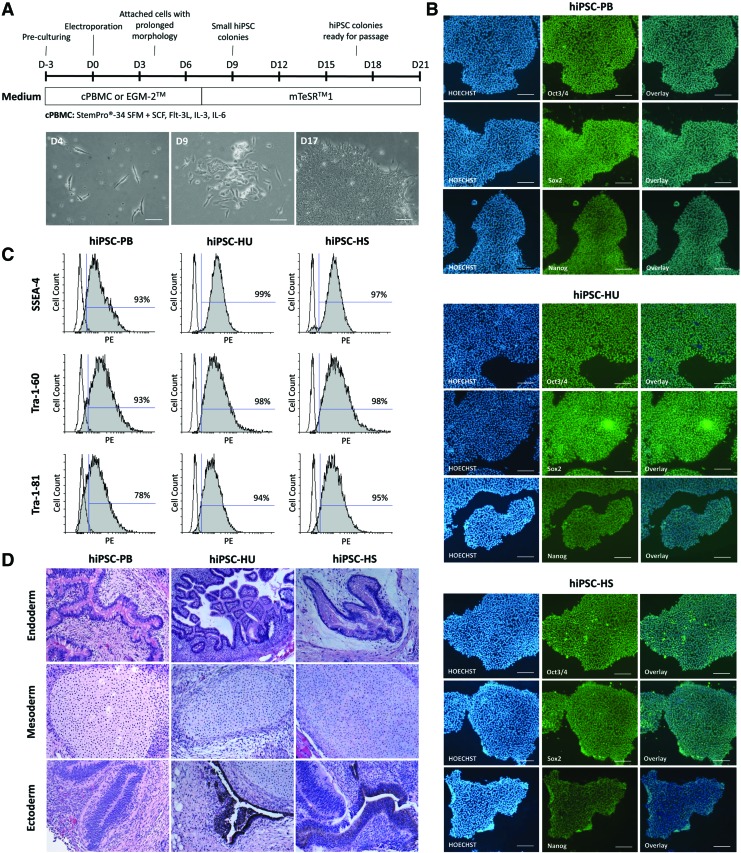
This study was performed on human cells that originated from the peripheral blood and endothelial tissue. PBMCs and HSVECs were isolated from adult tissue donors, whereas HUVECs represents a neonatal cell type. PBMCs, HUVECs, and HSVECs were reprogrammed into hiPSCs using episomal vectors [37]. Figure 1A illustrates the timeline of the reprogramming process, including the appropriate media for each cell type. The first attached cells with an elongated morphology were observed on day 4 after transfection. Small hiPSC colonies developed from these cells at approximately day 9. More than 15 hiPSC colonies emerged in each well on day 17, and these clones were mechanically passaged. At least 10 clones from each cell type were expanded and stored in liquid nitrogen.
FIG. 1.
hiPSC reprogramming and characterization. (A) Experimental timeline for the reprogramming of PBMCs and ECs into hiPSCs. Cell morphology observed during the reprogramming of PBMCs at days 4, 9, and 17 (scale bar = 100 μm). (B) Immunofluorescence staining for the pluripotency markers Oct3/4, Sox2, and Nanog in hiPSCs (scale bar = 200 μm). (C) Flow cytometry-based detection of the pluripotency markers SSEA-4, Tra-1-60, and Tra-1-81 in hiPSCs. (D) Teratoma formation in immunodeficient mice after transplantation of hiPSCs; teratomas contained tissues from all three germ layers. ECs, endothelial cells; hiPSC, human induced pluripotent stem cell; PBMCs, peripheral blood mononuclear cells. Color images are available online at www.libeberpub.com/scd
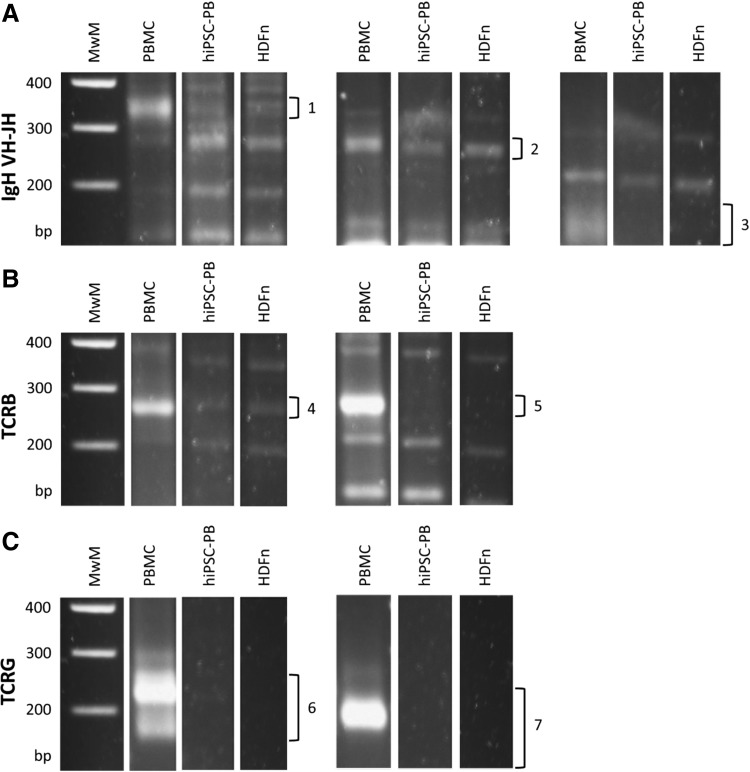
The pluripotency of the generated hiPSC lines—hiPSC-PB, hiPSC-HU, and hiPSC-HS—was characterized. These hiPSCs displayed a typical hESC-like morphology and expressed the pluripotency markers Oct3/4, Sox2 and Nanog (Fig. 1B). Flow cytometry confirmed the expression of the surface pluripotency markers SSEA-4, Tra-1-60, and Tra-1-81 (Fig. 1C). In vivo teratoma formation revealed cell types representative of the three germ layers (Fig. 1D). To reveal which specific subpopulation of PBMCs was reprogrammed, we performed PCR assays with seven multiplex PCR tubes (Fig. 2). No rearrangements in IGH gene (B lymphocytes), TRB gene, or TRG gene (T lymphocytes) were detected, suggesting that nonlymphoid mononuclear cell fraction was reprogrammed. The hiPSC-PB line therefore arose either from hematopoietic progenitor cell or from the monocyte fraction.
FIG. 2.
Detection of Ig and TCR gene recombinations in hiPSC-PB. Genomic DNA from PBMCs is provided as a positive control, while fibroblast line (HDFn) serves as a negative control. Sixty-three primers were used in seven multiplex PCR tubes. (A) Complete VH-JH rearrangement of IGH gene. Three tubes were used with valid sizes of amplicon (1) 310–360 bp, (2) 250–295 bp, and (3) 100–170 bp. (B) Complete Vβ-Jβ rearrangement of TRB gene. Two tubes were used with valid sizes (4) 240–285 bp and (5) 240–285 bp. (C) TRG gene rearrangements. Two tubes were used with valid sizes (6) 145–255 bp and (7) 80–220 bp. MwM, molecular weight marker. HDFn, human dermal neonatal fibroblast; hiPSC-PB, hiPSC derived from PBMCs; Ig, immunoglobulin; PCR, polymerase chain reaction; TCR, T-cell receptor.
All the characterization methods confirmed that PBMCs, HUVECs, and HSVECs were reprogrammed into pluripotent cells that closely resembled hESCs.
Endothelial differentiation of hiPSCs
The main goal of our research was to compare ECs that had differentiated from hiPSCs with somatic ECs isolated from human donors and to prove that these cells are closely related in terms of their phenotype profiles and functionality. Three hiPSC lines, hiPSC-PB, hiPSC-HU, and hiPSC-HS, were differentiated in vitro using a previously published protocol [40], and the derived ECs were purified using CD31+ microbeads.
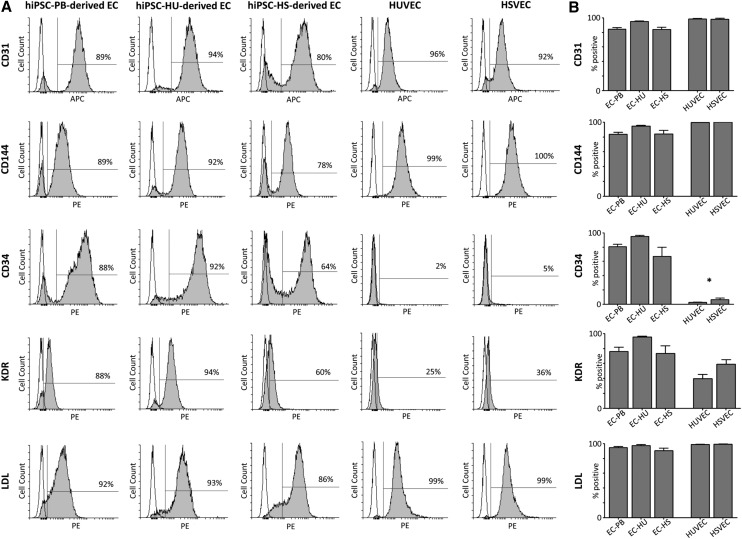
The isolated ECs displayed an endothelial morphology and expressed typical endothelial surface markers within at least four passages following isolation (Fig. 3A). The phenotype profile of hiPSC-derived ECs generally resembled the control EC types, HUVECs and HSVECs, based on the expression of the endothelial surface markers CD31 and CD144. Specifically, as visualized in the graph shown in Fig. 3B, the percentage of the CD31-positive cells among hiPSC-derived ECs ranged from 84% to 95%, whereas their somatic EC counterparts, HUVECs and HSVECs, averaged more than 98% CD31-positive cells. Between 84% and 94% of hiPSC-ECs expressed CD144, whereas ∼100% of HUVECs and HSVECs expressed CD144. For the markers CD34 and KDR, we observed high expression in the EC populations that had differentiated from hiPSCs, in contrast to the HUVEC and HSVEC controls. Approximately 81%, 95%, and 67% of EC-PB, EC-HU, and EC-HS expressed CD34, respectively, but <6% of their somatic EC counterparts were CD34-positive. A similar trend was observed for KDR: 76%, 95%, and 73% of hiPSC-derived ECs were positive compared with 40% and 59% of HUVECs and HSVECs, respectively.
FIG. 3.
Flow cytometry-based analysis of hiPSC-derived ECs and the EC controls. (A) Representative histograms showing the positivity for the cell surface markers CD31, CD144, CD34, KDR, and LDL in ECs derived from hiPSCs-PB, hiPSC-HU, and hiPSC-HS complemented with HUVECs and HSVECs. (B) Percentage of cells positive for each endothelial marker. The mean value was calculated from at least three independent differentiations of each hiPSC line (±SEM). The asterisk indicates a statistically significant (P < 0.05) difference in percentage of positive hiPSCs-derived ECs compared with their EC counterparts, as detected using Student's t-test. hiPSC-HS, HSVEC-derived hiPSCs; hiPSC-HU, HUVEC-derived hiPSCs; HSVECs, human saphenous vein endothelial cells; HUVECs, human umbilical vein endothelial cells; KDR, kinase insert domain receptor; LDL, low-density lipoprotein; SEM, standard error of the mean.
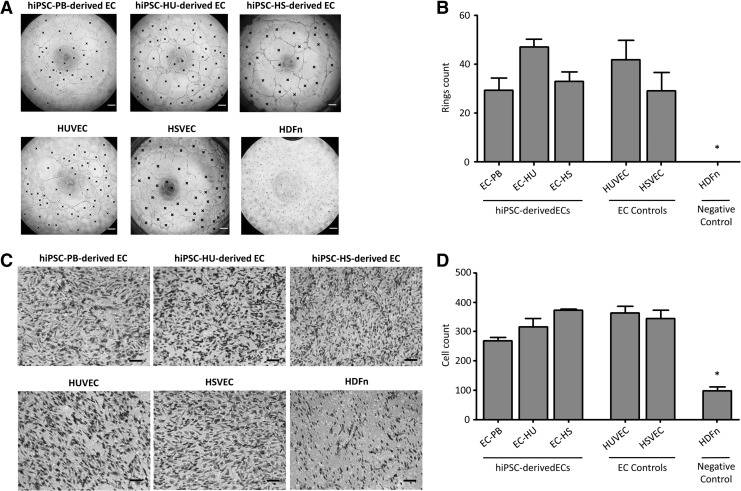
Three elementary tests were performed to verify the functional properties of the ECs—(1) an LDL uptake assay, (2) a TFA, and (3) a chemotaxis migration assay. ECs derived from all three hiPSC lines displayed increased LDL uptake (Fig. 3A, B). More than 90% of cells in all measured EC samples were LDL-positive 4 h after the administration of the substance. Our hiPSC-ECs also formed tubes of the similar quality as the control ECs, HUVECs and HSVECs (Fig. 4A, B). The number of complete rings formed during the TFA averaged between 29 and 47 for hiPSC-derived ECs. Mean number of rings for HUVEC and HSVEC were 42 and 29, respectively. No rings were formed in HDFn sample. Finally, chemotaxis migration assay was performed to show the function of our hiPSC-derived ECs. Figure 4C and D illustrate that hiPSC-derived ECs are attracted by VEGF in the similar manner (between 268 and 373 cells per one field of view) as HUVEC and HSVEC (363 and 344, respectively). Substantially less migrating cells were observed in control fibroblast sample (98 HDFn cells).
FIG. 4.
Functional characterization of ECs derived from hiPSC-PB, hiPSC-HU, and hiPSC-HS. HUVEC and HSVEC serve as positive controls, while HDFn are provided as negative controls. (A) Tube formation assay. The number of complete rings was assessed by image analysis described in Materials and Methods section and labeled by black cross. Scale bar = 200 μm. (B) Quantification of the tube formation assay. Each column indicates mean number of complete rings counted in three independent wells of 96-well plate (±SEM). Asterisk indicates statistically significant (P < 0.05) difference in the HDFn ring count compared with the ECs ring counts, as detected using Student's t-test. (C) Chemotaxis migration assay. Only the cells on the outer side of the membrane which were attracted by VEGF (50 ng/mL) are shown. Scale bar = 100 μm. (D) Quantification of the chemotaxis migration assay. Each column indicates mean number of cells counted in three independent cell culture inserts (±SEM). For each insert, three images were manually counted. Asterisk indicates statistically significant (P < 0.05) difference in the HDFn cell count compared with the ECs cell counts, as detected using Student's t-test.
Based on these results, the directed in vitro differentiation of hiPSCs produced ECs that resembled HUVECs and HSVECs.
Reprogramming and differentiation affect the cell cycle speed and DSB number
Cellular reprogramming introduces serious changes into the genome and alters the cell fate. If hiPSC-derived cells are used in the clinic, their genomic stability must be monitored. Therefore, we next focused on detecting possible genomic abnormalities in our cells.
We analyzed the cell cycle speed, DSB numbers, and karyotypes. The number of γH2AX foci, a measure of DSB, was counted in (1) hiPSCs derived from PBMC, HUVEC, and HSVEC somatic founders, (2) ECs differentiated from all three hiPSCs lines, and finally (3) control somatic ECs (HUVECs and HSVECs). The numbers of γH2AX foci depend on the cell cycle phase, as substantially more foci are observed in the nuclei of cells in S/G2 phases than in cells in G1 phase due to the presence of replication-related DSBs. In this study, we compared somatic ECs with PSCs, cell types that differ in the lengths of their cell cycle phases. We used a highly sensitive method that was previously published by our team to obtain the most precise results [35].
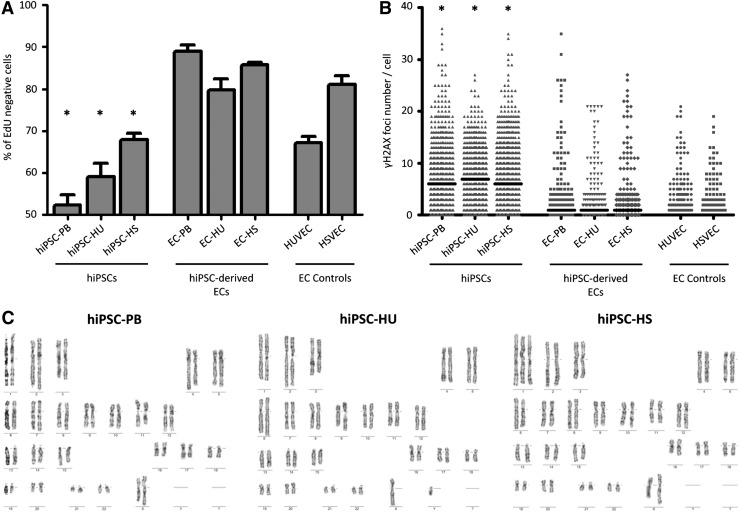
Individual cells were first separated according to their actual cell cycle phase by labeling newly synthesized DNA with EdU. The EdU signal strength in each cell was plotted on a histogram, which allowed us to calculate the threshold for the number of cells in G1 phase as described in the Materials and Methods section. Figure 5A illustrates the distribution of EdU-negative (G1 phase) cells among samples. Reprogramming into pluripotent cells speeds up the cell cycle, as manifested by the decrease in the number of cells in G1 phase in all hiPSC lines. In somatic EC controls, the percentages of HUVECs and HSVECs in G1 phase were 67% and 81%, respectively, and decreased to 52%, 59%, and 68% for hiPSC-PB, hiPSC-HU, and hiPSC HS, respectively. As expected, the differentiation of hiPSCs into ECs had the opposite effect and slowed the cell cycle again. Approximately 89% of ECs derived from hiPSC-PB, 80% of ECs derived from hiPSCs-HU, and 86% of ECs derived from hiPSC-HS were in G1 phase. Only the EdU-negative groups (G1 phase) were used for the subsequent analysis to exclude replication-related DSBs (S/G2 phase).
FIG. 5.
Cell cycle speed and genome stability during the reprogramming and differentiation processes. (A) Percentage of EdU-negative (G1 phase) cells among hiPSCs, hiPSC-derived ECs, and EC controls. Mean value (±SEM; n = 3). Asterisks indicate statistically significant (P < 0.05) decreases in the percentages of hiPSCs in G1 phase compared with their EC counterparts, as detected using Student's t-test. (B) Number of γH2AX foci per cell in hiPSCs, hiPSC-derived ECs and EC controls. The bar represents the median. Asterisks indicate statistically significant (P < 0.05) differences between hiPSCs and ECs, as confirmed by the Mann–Whitney test. (C) Cytogenetic data from hiPSC lines. Approximately 100% of cells possess a normal karyotype in hiPSC-PB and hiPSC-HU lines (passages 27 and 16, respectively). Representative aneuploid karyotype detected in 80% of cells in the hiPSC-HS line, in which a gain of chromosome 1 was observed (passage 17). EdU, 5-ethynyl-2′-deoxyuridine; γH2AX, phosphorylated histone H2AX.
The numbers of γH2AX foci were counted in cells in G1 phase to determine whether the process of reprogramming to pluripotent cells and subsequent endothelial differentiation influenced the numbers of DSBs. As shown in Fig. 5B, substantially larger numbers of γH2AX foci were observed in the EdU-negative groups of hiPSCs lines than in all ECs, regardless of whether original somatic ECs or ECs derived from hiPSCs were analyzed. Specifically, in hiPSCs, the median numbers of foci per cell were 6, 7, and 6 for hiPSC-PB, hiPSC-HU, and hiPSC-HS, respectively. The median number of γH2AX foci per cell in ECs differentiated from these hiPSCs decreased to 1 for all samples. The numbers of foci in hiPSC-derived ECs more closely resembled control ECs, in which no foci were detected in each cell.
Finally, we performed a karyotype analysis of all three hiPSC lines to determine whether a faster cell cycle and larger number of DSBs in hiPSCs led to chromosomal abnormalities (Fig. 5C). A normal karyotype was observed in the hiPSC-PB (46, xx) and hiPSC-HU (46, xy) cell lines. A heterogeneous cell population was detected in the hiPSC-HS line, as 80% of the cells gained chromosome 1 (47, xx). Thus, the genome stability of hiPSCs is challenged during in vitro culture and should be closely monitored.
Discussion
ECs are valuable tools in regenerative medicine. Their use in the de novo regeneration of injured veins and the lining of vascular grafts is promising. However, the sources of ECs are limited, and therefore, new methods for ECs production are being developed. In our study, we produced ECs from hiPSCs and compared them with ECs isolated from donors (HUVECs and HSVECs) to confirm that the derived ECs resembled natural ECs. The hiPSCs used in this project were generated from three somatic cell types. We focused on the most easily accessible tissue—peripheral blood—as well as hiPSCs derived from HUVECs and HSVECs.
PBMCs offer several advantages over cell types that are traditionally used for hiPSC generation, such as dermal fibroblasts or, less often, ECs. Surgical removal of the skin tissue is painful and leaves a scar, which discourages potential donors. Fibroblasts or ECs are usually collected from donors during a planned surgery, such as plastic surgery or varicose vein surgery, which limits the opportunities to obtain tissue sample from patients with specific diseases, such as rare genetic disorders. In contrast, the routine collection of a few milliliters of blood is a minimally invasive procedure. The existence of blood banks is another argument favoring blood cells as a source for hiPSC production. The total amount of time needed for the derivation of the primary cell line is an important factor. A few weeks are needed to expand cells from skin tissue in vitro, whereas only 3 days of preculture are sufficient for PBMCs before reprogramming (Fig. 1A). The establishment of our HSVEC lines from vein samples usually requires between 2 and 3 weeks.
Several protocols for the expansion and reprogramming of PBMCs have been published [25–29]. In these studies, hiPSCs were reprogrammed using different vectors under different culture conditions. One of the most important issues is the composition of medium used for preculturing PBMCs before reprogramming and during the first days after reprogramming. StemPro-34 medium influences the ratio of particular blood cell types in the sample and induces the proliferation of hematopoietic progenitor cells to a greater extent than terminally differentiated lymphocytes. Various combinations of cytokine cocktails used to support the growth of hematopoietic progenitors have been proposed, but all include one or more of the following nine growth factors: SCF, Flt-3L, IL-6, IL-3, IL-1, erythropoietin (EPO), thrombopoietin (TPO), granulocyte colony-stimulating factor (G-CSF), and granulocyte-macrophage CSF (GM-CSF) (reviewed in Heike and Nakahata [46]).
In our study, we enriched the StemPro-34 medium with a combination of the growth factors SCF, Flt-3L, IL-3, and IL-6, which was reported to significantly increase the percentage of human hematopoietic progenitor cells expressing the CD34 surface marker [31] and should facilitate the reprogramming process of PBMCs. Similar medium, with addition of TPO, was used for pre-reprogramming expansion of CD34+ cells by Mack et al. [30].
Our PCR results excluded DNA rearrangements associated with T- or B-lymphocytes in hiPSC-PB cell line, which implies that hematopoietic progenitors were reprogrammed without the need for time-consuming isolation process of CD34+ cells from peripheral blood or mobilizing the blood as reported by Loh et al. [24]. Although we cannot exclude the possibility that monocytes were reprogrammed, it is unlikely because of difficult ex vivo expansion of this cell type [47, 48]. It is of note that our hiPSC-PB contains unmodified DNA without gene recombinations introduced during lymphoid maturation.
Two EC types, HUVECs and HSVECs, were used in the study as EC controls and to complement PBMCs for hiPSC derivation. HUVECs were purchased from Thermo Fisher Scientific and HSVECs were isolated from the saphenous vein of an adult donor. The protocol used to isolate HSVECs in this study is simple and effective. Enzymatic digestion with collagenase II did not result in contamination with other cell types, and a pure population of primary HSVECs was successfully characterized using both surface markers and a functional TFA. Primary ECs have a relatively short life-span in vitro, displaying signs of dedifferentiation into fibroblasts and senescence after a few passages [49]. We were able to expand HUVECs and HSVECs for up to ∼10 passages. Although our hiPSC-ECs did not exceed 10 passages in the majority of differentiations, hiPSC-derived ECs offer a constant and theoretically unlimited source of uniform cells for vascular grafts and in vitro studies. Robust protocols for the differentiation of hiPSCs into ECs have been recently published [20,21,40].
In the present study, we did not detect significant differences in the quality of hiPSCs or hiPSC-derived ECs between the three tested hiPSC lines derived from PBMCs, HUVECs, and HSVECs. All hiPSC lines produced hiPSC-derived ECs that were almost indistinguishable from each other and from the original ECs, based on their expression of the endothelial markers CD31 and CD144, LDL uptake, ability to form tubes, and chemotaxis migration.
Unlike the original ECs, hiPSC-derived ECs expressed higher levels of the endothelial progenitor markers CD34 and KDR. A substantial effort has attempted by other groups to identify and isolate EPCs that are capable of producing functional ECs (reviewed in Pelosi et al. [50]). Physiologically, EPCs are present in the human body as circulating cells that share common endothelial markers with mature ECs, such as CD31, but differ in the expression of certain cell surface antigens, such as CD34 and KDR [51–53]. EPCs are often divided into two subgroups, termed early- and late-outgrowth EPCs [54,55]. Late-outgrowth EPCs, unlike early-outgrowth EPCs, contribute to blood vessels formation and reparation by direct incorporation into their endothelial lining [56, 57]. Based on phenotypical profile (CD31+CD144+CD34+KDR+) and ability to form tubes [56–58], our hiPSC-derived cells resemble late outgrowth EPCs.
Genomic stability is a key issue in the use of hiPSC-derived cells in the clinic. For this reason, we studied the effect of in vitro cellular reprogramming and endothelial differentiation on the cell cycle speed and number of DSBs. In addition, a karyotype analysis of three tested hiPSC lines was performed.
The analysis of the cell cycle speed revealed differences between samples. The lowest percentage of cells in G1 phase was observed in hiPSCs, suggesting that these cells have a higher proliferation rate than their differentiated counterparts (hiPSC-derived ECs and ECs). This result is consistent with previously published data [59–62]. The number of DSBs observed in G1 phase is substantially increased in S/G2 phases as a consequence of replication stress [63–67]. Therefore, we used methodology that had been previously published by our team to reliably compare DSB counts visualized by γH2AX foci between samples with different cell cycle speeds [35]. Fluorescence microscopy enables us to analyze the number of γH2AX foci in each single cell, assign the cell cycle phase to that particular cell and only include G1 phase cells in the DSB analysis. Greater numbers of γH2AX foci were recorded for the hiPSC group compared to hiPSC-derived ECs and primary ECs, suggesting that the reprogramming process increases the number of DSBs, but differentiation restores DSBs to a number typically observed in somatic cells.
High DSB counts in pluripotent cells may increase the risk of acquiring chromosomal or subchromosomal abnormalities and challenge the overall genome stability of the cells. According to Laurent et al., hESC and hiPSC cells contain a greater frequency of subchromosomal copy number variations than somatic cells or tissues [33]. In our study, hiPSC lines hiPSC-PB and hiPSC-HU displayed a normal karyotype. However, the hiPSC-HS culture was mosaic and contained two populations of cells, one with a normal karyotype and one with an abnormal karyotype, characterized by a gain of chromosome 1. Trisomy of chromosome 1 is a common abnormality detected in PSCs. Mayshar et al. identified an extra copy of chromosome 1 in several tested hiPSC lines, but no corresponding aneuploidy was detected in the somatic cells [68]. Amps et al. observed mosaic hESC lines at an early passage, with extra copies of chromosomes 1, 12, 17, 20, or X [69–72].
The chromosomal abnormality we observed in the subset of hiPSCs derived from HSVEC probably results from the clonal selection during passaging and adaptation to culture conditions. Prolonged in vitro culturing of PSCs is associated with karyotypic abnormalities [32, 33] and karyotype analysis is a crucial test for selection of hiPSC clones [73]. Structural alteration, loss, or gain of a particular chromosome must lead to immediate exclusion of the clone or its derivative from any possible clinical application. Our results confirm that prolonged in vitro culturing of hiPSCs should be avoided and the time in culture before differentiation should be kept on necessary minimum.
Based on our data, PBMCs may serve as a safe and reliable source for artificial EC production for clinical purposes. hiPSC-derived ECs are fully functional and comparable with EC controls. In this protocol, cells pass through a pluripotent state, during which the number of DSBs increases and genome stability is challenged. Even if the number of DSBs is reversed upon endothelial differentiation, close monitoring of a normal karyotype is crucial for the possible clinical application of hiPSC-derived ECs.
Acknowledgments
This study was generously supported by the Grant Agency of the Czech Republic (302/12/G157), by the Czech Health Research Council (16-31501A), and by the European Regional Development Fund (FNUSA-ICRC, no. CZ.1.05/1.1.00/02.0123).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Carmeliet P. (2000). Mechanisms of angiogenesis and arteriogenesis. Nat Med 6:389–395 [DOI] [PubMed] [Google Scholar]
- 2.Folkman J. (1995). Angiogenesis in cancer, vascular, rheumatoid and other disease. Nat Med 1:27–31 [DOI] [PubMed] [Google Scholar]
- 3.Cooke JP. (2003). Flow, NO, and atherogenesis. Proc Natl Acad Sci U S A 100:768–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang L, Perrault C, Coelho-Martins J, Hu C, Dulong C, Varna M, Liu J, Jin J, Soria C, et al. (2013). Induction of acquired drug resistance in endothelial cells and its involvement in anticancer therapy. J Hematol Oncol 6:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Folkman J. (2007). Angiogenesis: an organizing principle for drug discovery? Nat Rev Drug Discov 6:273–286 [DOI] [PubMed] [Google Scholar]
- 6.Herring M, Baughman S. and Glover J. (1985). Endothelium develops on seeded human arterial prosthesis: a brief clinical note. J Vasc Surg 2:727–730 [PubMed] [Google Scholar]
- 7.Herring M, Gardner A. and Glover J. (1978). A single-staged technique for seeding vascular grafts with autogenous endothelium. Surgery 84:498–504 [PubMed] [Google Scholar]
- 8.Chong MS, Ng WK. and Chan JK. (2016). Concise review: endothelial progenitor cells in regenerative medicine: applications and challenges. Stem Cells Transl Med 5:530–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaffe EA, Nachman RL, Becker CG. and Minick CR. (1973). Culture of human endothelial cells derived from umbilical veins. Identification by morphologic and immunologic criteria. J Clin Invest 52:2745–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watkins MT, Sharefkin JB, Zajtchuk R, Maciag TM, D'Amore PA, Ryan US, Van Wart H. and Rich NM. (1984). Adult human saphenous vein endothelial cells: assessment of their reproductive capacity for use in endothelial seeding of vascular prostheses. J Surg Res 36:588–596 [DOI] [PubMed] [Google Scholar]
- 11.Kinoshita M, Fujita Y, Katayama M, Baba R, Shibakawa M, Yoshikawa K, Katakami N, Furukawa Y, Tsukie T, et al. (2012). Long-term clinical outcome after intramuscular transplantation of granulocyte colony stimulating factor-mobilized CD34 positive cells in patients with critical limb ischemia. Atherosclerosis 224:440–445 [DOI] [PubMed] [Google Scholar]
- 12.Barsheshet A, Hod H, Shechter M, Sharabani-Yosef O, Rosenthal E, Barbash IM, Matetzky S, Tal R, Bentancur AG, et al. (2008). The effects of external counter pulsation therapy on circulating endothelial progenitor cells in patients with angina pectoris. Cardiology 110:160–166 [DOI] [PubMed] [Google Scholar]
- 13.Baran Ç, Durdu S, Dalva K, Zaim Ç, Dogan A, Ocakoglu G, Gürman G, Arslan Ö. and Akar AR. (2012). Effects of preoperative short term use of atorvastatin on endothelial progenitor cells after coronary surgery: a randomized, controlled trial. Stem Cell Rev 8:963–971 [DOI] [PubMed] [Google Scholar]
- 14.Wang ZZ, Au P, Chen T, Shao Y, Daheron LM, Bai H, Arzigian M, Fukumura D, Jain RK. and Scadden DT. (2007). Endothelial cells derived from human embryonic stem cells form durable blood vessels in vivo. Nat Biotechnol 25:317–318 [DOI] [PubMed] [Google Scholar]
- 15.Kane NM, Meloni M, Spencer HL, Craig MA, Strehl R, Milligan G, Houslay MD, Mountford JC, Emanueli C. and Baker AH. (2010). Derivation of endothelial cells from human embryonic stem cells by directed differentiation: analysis of microRNA and angiogenesis in vitro and in vivo. Arterioscler Thromb Vasc Biol 30:1389–1397 [DOI] [PubMed] [Google Scholar]
- 16.Nourse MB, Halpin DE, Scatena M, Mortisen DJ, Tulloch NL, Hauch KD, Torok-Storb B, Ratner BD, Pabon L. and Murry CE. (2010). VEGF induces differentiation of functional endothelium from human embryonic stem cells: implications for tissue engineering. Arterioscler Thromb Vasc Biol 30:80–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rufaihah AJ, Huang NF, Jamé S, Lee JC, Nguyen HN, Byers B, De A, Okogbaa J, Rollins M, et al. (2011). Endothelial cells derived from human iPSCS increase capillary density and improve perfusion in a mouse model of peripheral arterial disease. Arterioscler Thromb Vasc Biol 31:e72–e79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White MP, Rufaihah AJ, Liu L, Ghebremariam YT, Ivey KN, Cooke JP. and Srivastava D. (2013). Limited gene expression variation in human embryonic stem cell and induced pluripotent stem cell-derived endothelial cells. Stem Cells 31:92–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orlova VV, Drabsch Y, Freund C, Petrus-Reurer S, van den Hil FE, Muenthaisong S, Dijke PT. and Mummery CL. (2014). Functionality of endothelial cells and pericytes from human pluripotent stem cells demonstrated in cultured vascular plexus and zebrafish xenografts. Arterioscler Thromb Vasc Biol 34:177–186 [DOI] [PubMed] [Google Scholar]
- 20.Patsch C, Challet-Meylan L, Thoma EC, Urich E, Heckel T, O'Sullivan JF, Grainger SJ, Kapp FG, Sun L, et al. (2015). Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells. Nat Cell Biol 17:994–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sahara M, Hansson EM, Wernet O, Lui KO, Später D. and Chien KR. (2014). Manipulation of a VEGF-Notch signaling circuit drives formation of functional vascular endothelial progenitors from human pluripotent stem cells. Cell Res 24:820–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K. and Yamanaka S. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872 [DOI] [PubMed] [Google Scholar]
- 23.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318:1917–1920 [DOI] [PubMed] [Google Scholar]
- 24.Loh YH, Agarwal S, Park IH, Urbach A, Huo H, Heffner GC, Kim K, Miller JD, Ng K. and Daley GQ. (2009). Generation of induced pluripotent stem cells from human blood. Blood 113:5476–5479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Staerk J, Dawlaty MM, Gao Q, Maetzel D, Hanna J, Sommer CA, Mostoslavsky G. and Jaenisch R. (2010). Reprogramming of human peripheral blood cells to induced pluripotent stem cells. Cell Stem Cell 7:20–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loh YH, Hartung O, Li H, Guo C, Sahalie JM, Manos PD, Urbach A, Heffner GC, Grskovic M, et al. (2010). Reprogramming of T cells from human peripheral blood. Cell Stem Cell 7:15–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okita K, Yamakawa T, Matsumura Y, Sato Y, Amano N, Watanabe A, Goshima N. and Yamanaka S. (2013). An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells 31:458–466 [DOI] [PubMed] [Google Scholar]
- 28.Chou BK, Mali P, Huang X, Ye Z, Dowey SN, Resar LM, Zou C, Zhang YA, Tong J. and Cheng L. (2011). Efficient human iPS cell derivation by a non-integrating plasmid from blood cells with unique epigenetic and gene expression signatures. Cell Res 21:518–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Liu T, Van Halm-Lutterodt N, Chen J, Su Q. and Hai Y. (2016). Reprogramming of blood cells into induced pluripotent stem cells as a new cell source for cartilage repair. Stem Cell Res Ther 7:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mack AA, Kroboth S, Rajesh D. and Wang WB. (2011). Generation of induced pluripotent stem cells from CD34+ cells across blood drawn from multiple donors with non-integrating episomal vectors. PLoS One 6:e27956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shah AJ, Smogorzewska EM, Hannum C. and Crooks GM. (1996). Flt3 ligand induces proliferation of quiescent human bone marrow CD34+CD38− cells and maintains progenitor cells in vitro. Blood 87:3563–3570 [PubMed] [Google Scholar]
- 32.Taapken SM, Nisler BS, Newton MA, Sampsell-Barron TL, Leonhard KA, McIntire EM. and Montgomery KD. (2011). Karotypic abnormalities in human induced pluripotent stem cells and embryonic stem cells. Nat Biotechnol 29:313–314 [DOI] [PubMed] [Google Scholar]
- 33.Laurent LC, Ulitsky I, Slavin I, Tran H, Schork A, Morey R, Lynch C, Harness JV, Lee S, et al. (2011). Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell 8:106–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shrivastav M, De Haro LP. and Nickoloff JA. (2008). Regulation of DNA double-strand break repair pathway choice. Cell Res 18:134–147 [DOI] [PubMed] [Google Scholar]
- 35.Simara P, Tesarova L, Rehakova D, Matula P, Stejskal S, Hampl A. and Koutna I. (2017). DNA double-strand breaks in hiPSC reprogramming and long-term in vitro culturing. Stem Cell Res Ther 8:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weissbein U, Benvenisty N. and Ben-David U. (2014). Quality control: genome maintenance in pluripotent stem cells. J Cell Biol 204:153–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II. and Thomson JA. (2009). Human induced pluripotent stem cells free of vector and transgene sequences. Science 324:797–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Šimara P, Tesařová L, Padourová S. and Koutná I. (2014). Generation of human induced pluripotent stem cells using genome integrating or non-integrating methods. Folia Biol (Praha) 60 (Suppl. 1):85–89 [DOI] [PubMed] [Google Scholar]
- 39.van Dongen JJ, Langerak AW, Brüggemann M, Evans PA, Hummel M, Lavender FL, Delabesse E, Davi F, Schuuring E, et al. (2003). Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98–3936. Leukemia 17:2257–2317 [DOI] [PubMed] [Google Scholar]
- 40.Orlova VV, van den Hil FE, Petrus-Reurer S, Drabsch Y, Ten Dijke P. and Mummery CL. (2014). Generation, expansion and functional analysis of endothelial cells and pericytes derived from human pluripotent stem cells. Nat Protoc 9:1514–1531 [DOI] [PubMed] [Google Scholar]
- 41.Ng ES, Davis R, Stanley EG. and Elefanty AG. (2008). A protocol describing the use of a recombinant protein-based, animal product-free medium (APEL) for human embryonic stem cell differentiation as spin embryoid bodies. Nat Protoc 3:768–776 [DOI] [PubMed] [Google Scholar]
- 42.Kittler J. and Illingworth J. (1986). Minimum error thresholding. Pattern Recognit 19:41–47 [Google Scholar]
- 43.Soille P. (2004). Morphological Image Analysis. New York, NY: Springer-Verlag [Google Scholar]
- 44.Matula P, Maška M, Daněk O, Matula M. and Kozubek P. (2009). Acquiarium: free software for acquisition and analysis of 3 images of cells in fluorescence microscopy D. In: 6th International Symposium on Biomedical Imaging IEEE. Boston, pp. 1138–1141 [Google Scholar]
- 45.Štěpka K, Matula P, Wörz S, Rohr K. and Kozubek M. (2015). Performance and sensitivity evaluation of 3D spot detection methods in confocal microscopy. Cytometry A 87:759–772 [DOI] [PubMed] [Google Scholar]
- 46.Heike T. and Nakahata T. (2002). Ex vivo expansion of hematopoietic stem cells by cytokines. Biochim Biophys Acta 1592:313–321 [DOI] [PubMed] [Google Scholar]
- 47.Kim Y, Rim YA, Yi H, Park N, Park SH. and Ju JH. (2016). The generation of human induced pluripotent stem cells from blood cells: an efficient protocol using serial plating of reprogrammed cells by centrifugation. Stem Cells Int 2016:1329459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang XB. (2013). Cellular reprogramming of human peripheral blood cells. Genomics Proteomics Bioinformatics 11:264–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scoumanne A, Kalamati T, Moss J, Powell JT, Gosling M. and Carey N. (2002). Generation and characterisation of human saphenous vein endothelial cell lines. Atherosclerosis 160:59–67 [DOI] [PubMed] [Google Scholar]
- 50.Pelosi E, Castelli G. and Testa U. (2014). Endothelial progenitors. Blood Cells Mol Dis 52:186–194 [DOI] [PubMed] [Google Scholar]
- 51.Asahara T, Murohara T, Sullivan A, Silver M, van der Zee R, Li T, Witzenbichler B, Schatteman G. and Isner JM. (1997). Isolation of putative progenitor endothelial cells for angiogenesis. Science 275:964–967 [DOI] [PubMed] [Google Scholar]
- 52.Peichev M, Naiyer AJ, Pereira D, Zhu Z, Lane WJ, Williams M, Oz MC, Hicklin DJ, Witte L, Moore MA. and Rafii S. (2000). Expression of VEGFR-2 and AC133 by circulating human CD34(+) cells identifies a population of functional endothelial precursors. Blood 95:952–958 [PubMed] [Google Scholar]
- 53.de la Puente P, Muz B, Azab F. and Azab AK. (2013). Cell trafficking of endothelial progenitor cells in tumor progression. Clin Cancer Res 19:3360–3368 [DOI] [PubMed] [Google Scholar]
- 54.Hur J, Yoon CH, Kim HS, Choi JH, Kang HJ, Hwang KK, Oh BH, Lee MM. and Park YB. (2004). Characterization of two types of endothelial progenitor cells and their different contributions to neovasculogenesis. Arterioscler Thromb Vasc Biol 24:288–293 [DOI] [PubMed] [Google Scholar]
- 55.Medina RJ, Barber CL, Sabatier F, Dignat-George F, Melero-Martin JM, Khosrotehrani K, Ohneda O, Randi AM, Chan JKY, et al. (2017). Endothelial progenitors: a consensus statement on nomenclature. Stem Cells Transl Med 6:1316–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Minami Y, Nakajima T, Ikutomi M, Morita T, Komuro I, Sata M. and Sahara M. (2015). Angiogenic potential of early and late outgrowth endothelial progenitor cells is dependent on the time of emergence. Int J Cardiol 186:305–314 [DOI] [PubMed] [Google Scholar]
- 57.Mukai N, Akahori T, Komaki M, Li Q, Kanayasu-Toyoda T, Ishii-Watabe A, Kobayashi A, Yamaguchi T, Abe M, Amagasa T. and Morita I. (2008). A comparison of the tube forming potentials of early and late endothelial progenitor cells. Exp Cell Res 314:430–440 [DOI] [PubMed] [Google Scholar]
- 58.Cheng CC, Chang SJ, Chueh YN, Huang TS, Huang PH, Cheng SM, Tsai TN, Chen JW. and Wang HW. (2013). Distinct angiogenesis roles and surface markers of early and late endothelial progenitor cells revealed by functional group analyses. BMC Genomics 14:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stead E, White J, Faast R, Conn S, Goldstone S, Rathjen J, Dhingra U, Rathjen P, Walker D. and Dalton S. (2002). Pluripotent cell division cycles are driven by ectopic Cdk2, cyclin A/E and E2F activities. Oncogene 21:8320–8333 [DOI] [PubMed] [Google Scholar]
- 60.Becker KA, Ghule PN, Therrien JA, Lian JB, Stein JL, van Wijnen AJ. and Stein GS. (2006). Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J Cell Physiol 209:883–893 [DOI] [PubMed] [Google Scholar]
- 61.Calder A, Roth-Albin I, Bhatia S, Pilquil C, Lee JH, Bhatia M, Levadoux-Martin M, McNicol J, Russell J, Collins T. and Draper JS. (2013). Lengthened G1 phase indicates differentiation status in human embryonic stem cells. Stem Cells Dev 22:279–295 [DOI] [PubMed] [Google Scholar]
- 62.Barta T, Dolezalova D, Holubcova Z. and Hampl A. (2013). Cell cycle regulation in human embryonic stem cells: links to adaptation to cell culture. Exp Biol Med (Maywood) 238:271–275 [DOI] [PubMed] [Google Scholar]
- 63.Adams BR, Golding SE, Rao RR. and Valerie K. (2010). Dynamic dependence on ATR and ATM for double-strand break repair in human embryonic stem cells and neural descendants. PLoS One 5:e10001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang X, Tanaka T, Kurose A, Traganos F. and Darzynkiewicz Z. (2006). Constitutive histone H2AX phosphorylation on Ser-139 in cells untreated by genotoxic agents is cell-cycle phase specific and attenuated by scavenging reactive oxygen species. Int J Oncol 29:495–501 [PubMed] [Google Scholar]
- 65.MacPhail SH, Banáth JP, Yu Y, Chu E. and Olive PL. (2003). Cell cycle-dependent expression of phosphorylated histone H2AX: reduced expression in unirradiated but not X-irradiated G1-phase cells. Radiat Res 159:759–767 [DOI] [PubMed] [Google Scholar]
- 66.Suchánková J, Kozubek S, Legartová S, Sehnalová P, Küntziger T. and Bártová E. (2015). Distinct kinetics of DNA repair protein accumulation at DNA lesions and cell cycle-dependent formation of γH2AX- and NBS1-positive repair foci. Biol Cell 107:440–454 [DOI] [PubMed] [Google Scholar]
- 67.Suzuki K, Okada H, Yamauchi M, Oka Y, Kodama S. and Watanabe M. (2006). Qualitative and quantitative analysis of phosphorylated ATM foci induced by low-dose ionizing radiation. Radiat Res 165:499–504 [DOI] [PubMed] [Google Scholar]
- 68.Mayshar Y, Ben-David U, Lavon N, Biancotti JC, Yakir B, Clark AT, Plath K, Lowry WE. and Benvenisty N. (2010). Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell 7:521–531 [DOI] [PubMed] [Google Scholar]
- 69.Amps K, Andrews PW, Anyfantis G, Armstrong L, Avery S, Baharv H, Baker J, Baker D, Munoz MB, et al. (2011). Screening ethnically diverse human embryonic stem cells identifies a chromosome 20 minimal amplicon conferring growth advantage. Nat Biotechnol 29:1132–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baker DE, Harrison NJ, Maltby E, Smith K, Moore HD, Shaw PJ, Heath PR, Holden H. and Andrews PW. (2007). Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechnol 25:207–215 [DOI] [PubMed] [Google Scholar]
- 71.Draper JS, Smith K, Gokhale P, Moore HD, Maltby E, Johnson J, Meisner L, Zwaka TP, Thomson JA. and Andrews PW. (2004). Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nat Biotechnol 22:53–54 [DOI] [PubMed] [Google Scholar]
- 72.Ben-David U, Mayshar Y. and Benvenisty N. (2011). Large-scale analysis reveals acquisition of lineage-specific chromosomal aberrations in human adult stem cells. Cell Stem Cell 9:97–102 [DOI] [PubMed] [Google Scholar]
- 73.Mandai M, Watanabe A, Kurimoto Y, Hirami Y, Morinaga C, Daimon T, Fujihara M, Akimaru H, Sakai N, et al. (2017). Autologous induced stem-cell-derived retinal cells for macular degeneration. N Engl J Med 376:1038–1046 [DOI] [PubMed] [Google Scholar]