Abstract
It has been hypothesized that a condensed nervous system with a medial ventral nerve cord is an ancestral character of Bilateria. The presence of similar dorsoventral molecular patterns along the nerve cords of vertebrates, flies, and an annelid has been interpreted as support for this scenario. Whether these similarities are generally found across the diversity of bilaterian neuroanatomies is unclear, and thus the evolutionary history of the nervous system is still contentious. To assess the conservation of the dorsoventral nerve cord patterning, we studied representatives of Xenacoelomorpha, Rotifera, Nemertea, Brachiopoda, and Annelida. None of the studied species show a conserved dorsoventral molecular regionalization of their nerve cords, not even the annelid Owenia fusiformis, whose trunk neuroanatomy parallels that of vertebrates and flies. Our findings restrict the use of molecular patterns to explain nervous system evolution, and suggest that the similarities in dorsoventral patterning and trunk neuroanatomies evolved independently in Bilateria.
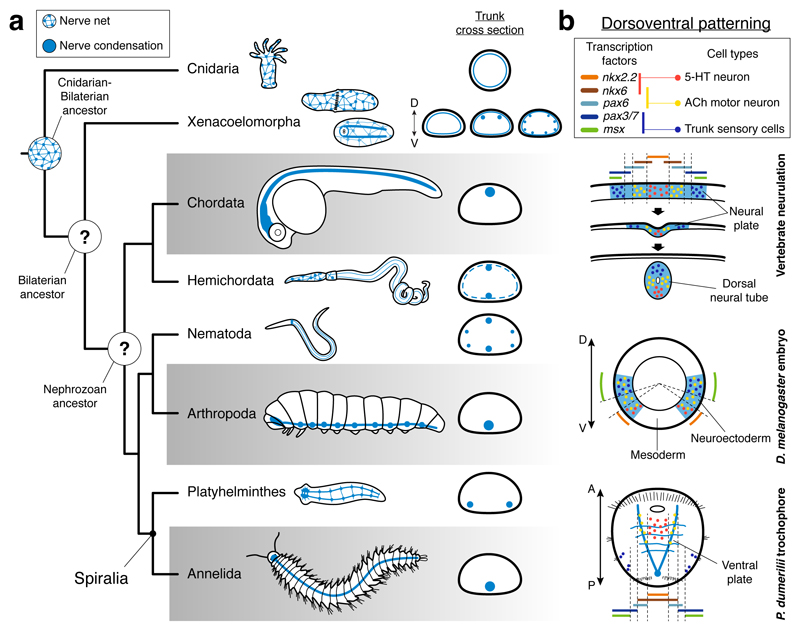
The nervous systems of Bilateria, and in particular their trunk neuroanatomies, are morphologically diverse1 (Fig. 1a). Groups such as arthropods, annelids, and chordates exhibit a medially condensed nerve cord, which is ventral in arthropods and annelids, and dorsal in chordates. In contrast, other lineages have multiple paired longitudinal nerve cords distributed at different dorsoventral (DV) levels. There are even bilaterians with only weakly condensed basiepidermal nerve nets, similar to those in cnidarians (Fig. 1a), which supports that this net-like neural arrangement predates the Cnidaria–Bilateria split2,3 (Fig. 1a). However, the earliest configuration of the bilaterian central nervous system (CNS) is still vividly debated2,4–7 (Fig. 1a), and thus it is unclear when and how often nerve cords evolved in Bilateria.
Figure 1. CNS evolution and dorsoventral patterning.
(a) A nerve net is ancestral for Cnidaria and Bilateria. The neuroanatomical diversity hampers the reconstruction of the CNS evolution in Bilateria. (b) A central argument for an ancestral medially condensed VNC for Bilateria is the similar deployment of DV-TFs in vertebrates, Drosophila, and the P. dumerilii larva. The staggered expression of these genes concurs with specific neuronal populations.
The conserved deployment of signaling molecules and transcription factors (TFs) along the bilaterian anteroposterior and dorsoventral axes grounds most scenarios for the evolution of the CNS2,7–13. In particular, the similar expression of the TFs nkx2.1/nkx2.2, nkx6, pax6, pax3/7, and msx in the ventral neuroectoderm of the fly Drosophila melanogaster and the annelid Platynereis dumerilii, and the dorsal neural plate of vertebrates (Fig. 1b) is a core argument to propose an ancestral CNS comprising a medial ventral nerve cord (VNC) in Bilateria2,4,7,8,13,14. In P. dumerilii and vertebrates, and to some extent in Drosophila, the staggered expression of these genes correlates with the spatial location of neuronal cell types along their trunks8,10,13. Serotonergic neurons form in the ventromedial nkx2.2+/nkx6+ region, cholinergic motoneurons develop in the nkx6+/pax6+ area, and dbx+ interneurons and lateral sensory trunk neurons differentiate in the more dorsolateral pax6+/pax3/7+ and pax3/7+/msx+ domains, respectively (Fig. 1b). The dorsoventral arrangement of these TFs and neuronal cell types is absent in hemichordates11,12,15, nematodes16,17, and planarians18 (Supplemental Data Table 1), consistent with the idea that the most recent ancestor of Bilateria had a dorsoventrally patterned, medially condensed VNC that has been repeatedly lost in these and perhaps other groups13. However, there is an alternative explanation – that a CNS with a single nerve cord and the similar dorsoventral patterning is the trait that repeatedly evolved, and thus was absent in the most recent common bilaterian ancestor5,9,11,12.
Neuroectodermal patterning in Xenacoelomorpha
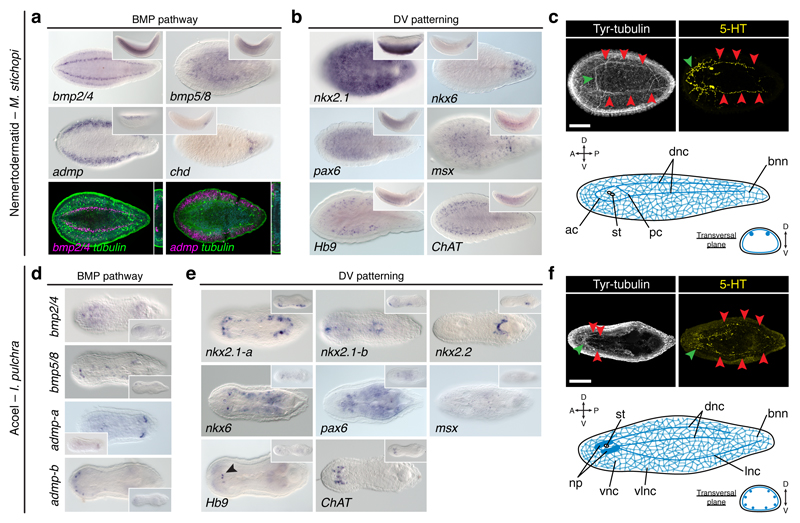
To explore the conservation of neuroectodermal patterning systems in Bilateria, we first studied Xenacoelomorpha (Extended Data Fig. 1), which is the sister group to all remaining bilaterian lineages19,20 (i.e. Nephrozoa). We focused our analyses on Xenoturbella bocki, the nemertodermatids Meara stichopi and Nemertoderma westbladi, and the acoel Isodiametra pulchra. As in the acoel Hofstenia miamia21 and most other bilaterians7,11, these xenacoelomorphs differentially express anteroposterior marker genes along their primary body axis22,23 (Extended Data Fig. 2a, c; Extended Data Fig. 3). The BMP pathway, which has an ancestral DV patterning role21,24 and an anti-neural role in Drosophila and vertebrates10, is also similarly deployed in all studied xenacoelomorphs21, with bmp ligands expressed dorsally and antagonists located more ventrolaterally (Fig. 2a, d; Extended Data Fig. 2d; Extended Data Fig. 4). However, the DV-TFs that we found in our genomic resources (Supplementary Information Table 1) did not show a clear staggered arrangement (Fig. 2b, e). Therefore, Xenacoelomorpha only exhibits the anteroposterior and BMP ectodermal patterning systems, which is reminiscent of the cnidarian condition25.
Figure 2. Dorsoventral patterning in Xenacoelomorpha.
(a) bmp ligands and admp are expressed dorsally; chd is expressed ventroposteriorly. (b) nkx2.1, nkx6, and msx are expressed ventrally; pax6 is expressed broadly; Hb9 and ChAT are in the nerve cords. (c) M. stichopi CNS (green arrowheads indicate the anterior commissures; red arrowheads indicate the nerve cords). (d) bmp ligands are expressed dorsally; admp-a is expressed posteroventrally; admp-b is expressed anterolaterally. (e) nkx2.1 paralogs and nkx2.2 are expressed ventrally; nkx6 is expressed laterally; pax6 throughout the body; msx in isolated cells; Hb9 and ChAT are in the brain. (f) I. pulchra CNS (green arrowheads indicate the brain; red arrowheads indicate the nerve cords). Insets are lateral views. ac, anterior commissure; bnn, basiepidermal nerve net; dnc, dorsal nerve cord; lnc, lateral nerve cord; np, neuropile; pc, posterior commissure; st, statocyst; vlnc, ventrolateral nerve cord; vnc, ventral nerve cord. Scale bars, 100 µm.
Importantly, ectodermal patterning systems are deployed independently of the trunk neuroanatomy in Xenacoelomorpha. Similar to cnidarians, xenacoelomorphs have a uniformly distributed, diffuse basiepidermal nerve net3,26–28. Xenoturbella species have only this network27. However, nemertodermatids have additional longitudinal basiepidermal nerve cords26, located dorsally in M. stichopi29 (Fig. 2c), and ventrally in N. westbladi (Extended Data Fig. 2e). The acoel I. pulchra has also four pairs of subepidermal nerve cords distributed along the DV axis28 (Fig. 2f). Genes commonly involved in neurogenesis (Extended Data Fig. 5a, d) and neural transmission (Extended Data Fig. 2b, f; Extended Data Fig. 5b, c, e) are consistently expressed in the sensory structures and neural condensations in these species. However, the DV-TF nkx6 does not co-localize with the motoneuron marker ChAT in the trunk of M. stichopi and I. pulchra, while the relation of pax6+ cells to this and another motoneuron marker (Hb9) is unclear in both species (Fig. 2b, e). Therefore, the diversity of neuroanatomies of Xenacoelomorpha contrasts with the more conserved deployment of ectodermal anteroposterior and BMP patterning systems. This, together with the observation that disruption of BMP signalling does not affect CNS development (Extended Data Fig. 6) support that the anti-neural role of the BMP pathway evolved after the Xenacoelomorpha-Nephrozoa split. Likewise, the expression of DV-TFs unrelated to the distinct trunk neuroanatomies suggests that the dorsoventral patterning of the nerve cords also evolved after the Xenacoelomorpha–Nephrozoa split.
DV patterning in Brachiopoda
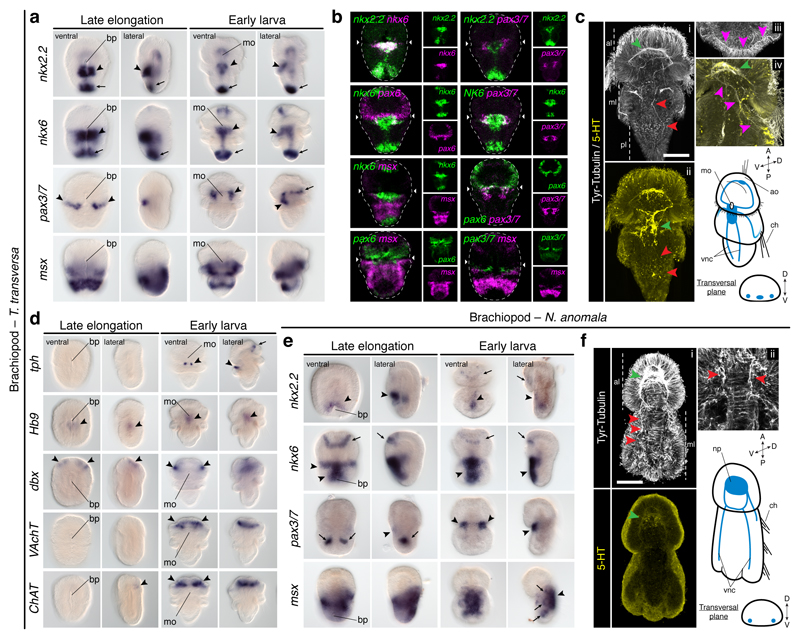
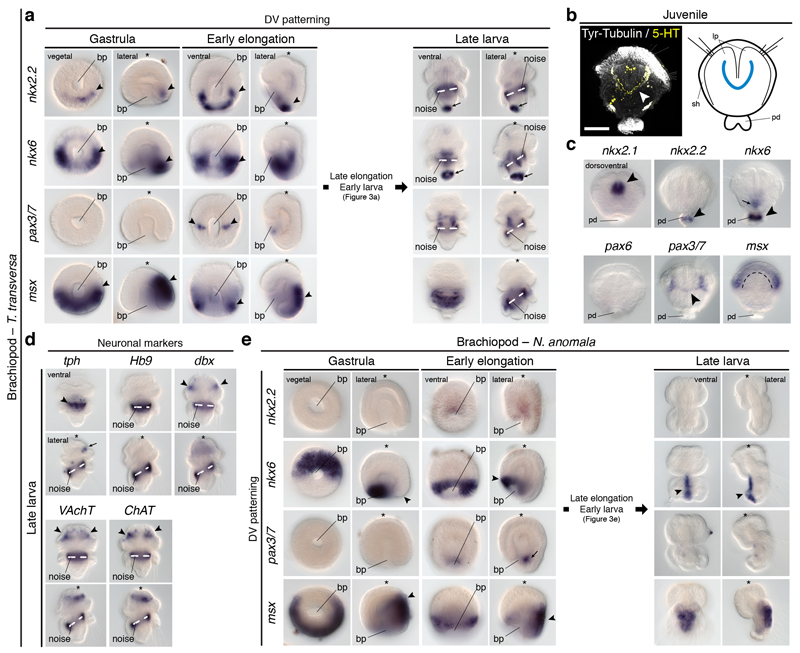
To investigate the conservation of the dorsoventral nerve cord patterning in Nephrozoa, we focused on Spiralia30, one of the three major nephrozoan clades. Although some lineages have a medially condensed VNC (i.e. Annelida), a main pair of VNCs is widespread and probably homologous in Spiralia5. We first studied the brachiopod Terebratalia transversa, where we identified staggered expression of DV-TFs in the anterior ventral midline of the larval trunk. At this stage, nkx2.131 and pax632 are expressed in the apical lobe, albeit pax6 expression projects slightly into the mantle lobe. However, there is a medial nkx2.2+/nkx6+ domain, a more lateral nkx6+/pax6+/pax3/7+ region, and a broad, dorsolateral msx+ area in the anterior ventral ectoderm of the larval ‘trunk’ (i.e. mantle and pedicle lobes) (Fig. 3a, b; Extended Data Fig. 7a). Additionally, a narrow line of cells below the apical-mantle boundary crossing the ventral midline expresses pax3/7 (Fig. 3a, b; Extended Data 7a). These expression domains disappear in the highly modified adult body (Extended Data Fig. 7a–c). The staggered expression of DV-TFs in the ventral anterior ectoderm of the trunk only partially correlates with the larval neuroanatomy, which consists of an anterior condensation and a medial accumulation of serotonergic cells on the ventral side, from where pairs of neurites innervate the chaetae and posterior end (Fig. 3c). The DV-TFs do not co-express with most neuronal markers13, which are mostly expressed in the anterior region (Fig. 3a, d; Extended Data Fig. 7a, d). Only two tph+ clusters in the medial serotonergic condensation of the larval trunk co-localise with the nkx2.2+/nkx6+ medial domain. Therefore, the brachiopod T. transversa resembles vertebrates, arthropods, and P. dumerilii in the presence of a ventral serotonergic nkx2.2+/nkx6+ area8,10,13,33, as well as in the nkx6, pax6, pax3/7 and msx dorsolateral domains, which are however not apparently connected to any neural trunk structure.
Figure 3. Dorsoventral patterning in Brachiopoda.
(a) nkx2.2 and nkx6 are in the trunk midline (arrowheads), posterior tip (arrows), gut, and apical cells (nkx6); pax3/7 is expressed laterally (arrowheads) and in the apical lobe (arrow); msx is in the mantle and ventral pedicle. (b) There is an nkx2.2+/nkx6+ medioventral region, and a more lateral nkx6+/pax6+/pax3/7+ anterior trunk domain. (c) T. transversa larval CNS (green arrowheads indicate the neuropile in i, and the trunk serotonergic condensation in ii; red arrowheads mark the VNCs; pink arrowheads indicate the innervation of the chaetae). (d) Only tph is expressed in the trunk (arrows and arrowheads indicate expression areas). (e) nkx2.2 and nkx6 are in the trunk ventral midline (arrowheads), apical lobe (arrows), and gut; pax3/7 is in the mesoderm (arrows), and in two ventrolateral trunk domains (arrowheads); msx is in the trunk, shell epithelium (arrowhead), and mesoderm (arrows). (f) N. anomala larval CNS (green arrowhead indicates the neuropile; red arrowheads in i mark the VNCs; red arrowheads in ii indicate the innervation of the chaetae). ao, apical organ; bp, blastopore; ch, chaetae; mo, mouth; np, neuropile; vnc, ventral nerve cord. Scale bars, 50 µm.
The staggered ectodermal expression of DV-TFs in the anteroventral trunk of T. transversa is largely conserved in the brachiopod Novocrania anomala. In this brachiopod, nkx2.131 and pax632 are expressed in the apical lobe, and nkx2.2 and nkx6 are expressed medially in the trunk (Fig. 3e). As in T. transversa, nkx6 extends more laterally at the anterior trunk, where it co-localizes with pax3/7 in the early larva, and msx is broadly detected in the trunk (Fig. 3e; Extended Data Fig. 7e). Therefore, N. anomala has also a medial ventral nkx2.2+/nkx6+ domain, but remarkably, this domain does not co-localise with any serotonergic condensation, which is lacking in the larval CNS of this brachiopod (Fig. 3f). Therefore, the conserved staggered expression of the DV-TFs in the anteroventral larval trunk is not necessarily connected to the CNS, suggesting that this patterning system may rather be patterning only the ectoderm in Brachiopoda.
DV patterning in Nemertea
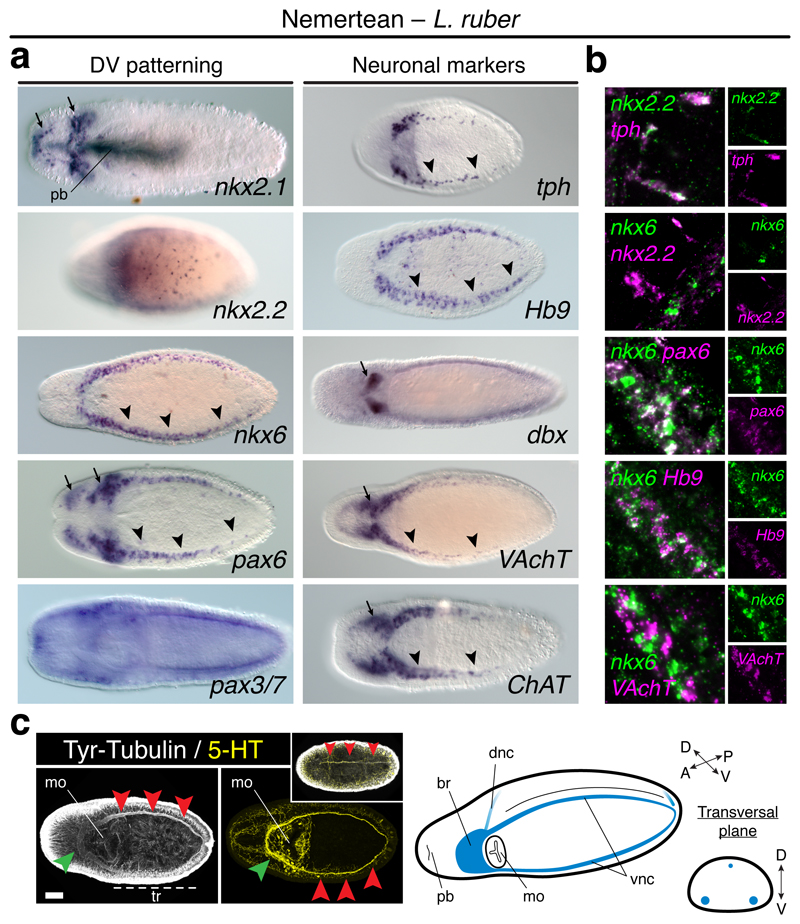
Similar to brachiopods, some DV-TFs show staggered expression along the trunk ventral side of the nemertean Lineus ruber. In this worm, DV-TFs are first detected in the larval imaginal discs (Extended Data Fig. 8a). In metamorphic and definitive juveniles, nkx2.1 is expressed in the head and proboscis, and pax3/7 is broadly expressed (Fig. 4a; Extended Data Fig. 8a). However, nkx2.2, nkx6, and pax6 are detected in isolated ventrolateral cells, as well as in cephalic domains (nkx2.2, nkx6, pax6) and isolated trunk cells (nkx2.2) (Fig. 4a; Extended Data Fig. 8a). Remarkably, nkx2.2 and nkx6 do not co-localise, but nkx6 and pax6 do (Fig 4b). These staggered domains relate to the disposition of the VNCs of L. ruber (Fig. 4c). Furthermore, nkx2.2+ cells co-express the serotonergic marker tph, and nkx6+ cells express the motoneuron marker Hb9, but not VAchT (Fig. 4a, b). Therefore, the staggered expression of the DV-TFs nkx2.2, nkx6, and pax6 are linked to the ventral trunk CNS and some neuronal cell type markers in L. ruber, which is similar to the situation described in vertebrates and P. dumerilii8,10,13,33.
Figure 4. Dorsoventral patterning in Nemertea.
(a) nkx2.1 and nkx2.2 are in the head (arrows), proboscis (nkx2.1), and trunk cells (nkx2.2); nkx6 and pax6 are in the head (arrows) and VNCs (arrowheads); pax3/7 is broadly expressed. Neuronal markers are in the brain (arrows) and VNCs (arrowheads). (b) In the VNCs, nkx2.2+ cells express tph, but not nkx6; nkx6+ cells express pax3/7 and Hb9, but not VAchT. (c) L. ruber CNS (green arrowheads indicate the brain; red arrowheads mark the VNCs and the dorsal neurite in the upper inset). br, brain; dnc, dorsal nerve cord; mo, mouth; tr, trunk; pb, proboscis; vnc, ventral nerve cord. Scale bar, 100 µm.
DV patterning in Rotifera
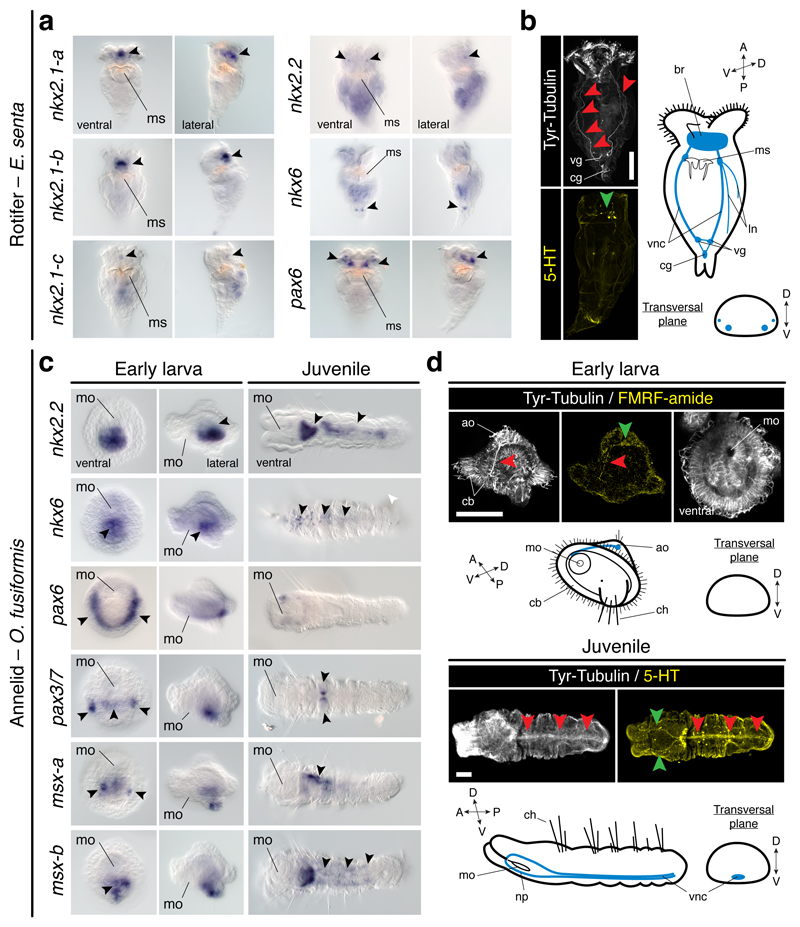
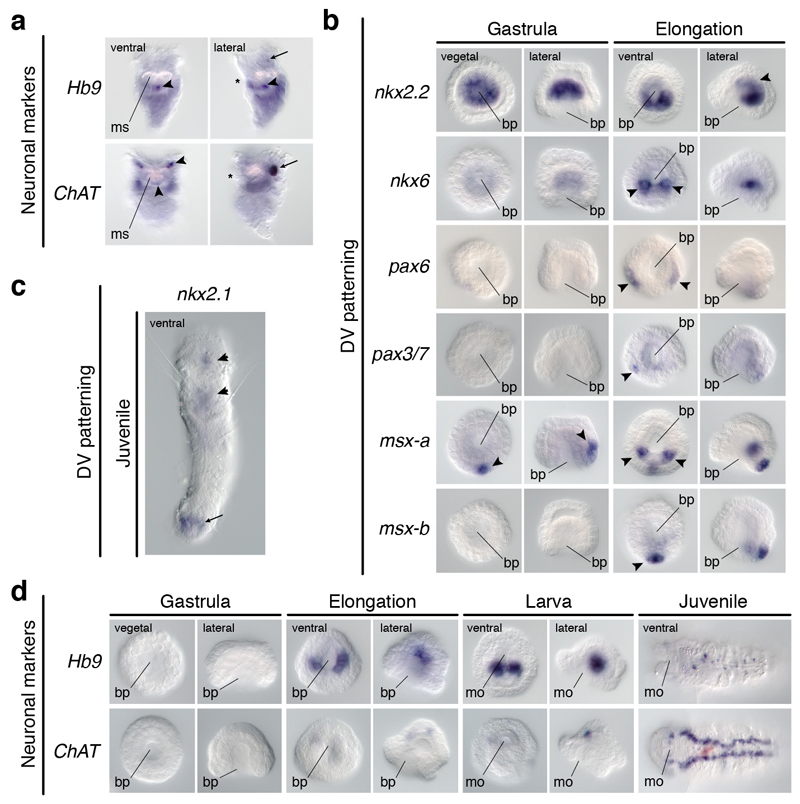
To explore the conservation of the dorsoventral patterning in Spiralia, we studied the rotifer Epiphanes senta, a member of the sister lineage to all remaining Spiralia30. Different from the brachiopod larvae and the nemertean juvenile, E. senta juveniles lack a staggered expression of DV-TFs along their trunks. The three nkx2.1 paralogs, nkx2.2, and pax6 are all in distinct brain domains of the juvenile rotifer (Fig. 5a). Only the gene nkx6 is detected in two posterior trunk cells (Fig. 5a). As in brachiopods and nemerteans, the trunk CNS comprises two VNCs, and additional paired dorsolateral nerves (Fig. 5b). The trunk expression of nkx6 probably corresponds to the vesicle ganglia1, but it is not related to motoneurons, as inferred by the expression of Hb9 and ChAT (Extended Data Fig. 9a). Therefore, spiralians with paired VNCs deploy the DV-TFs without a consistent association with their trunk neuroanatomies.
Figure 5. Dorsoventral patterning in Rotifera and Annelida.
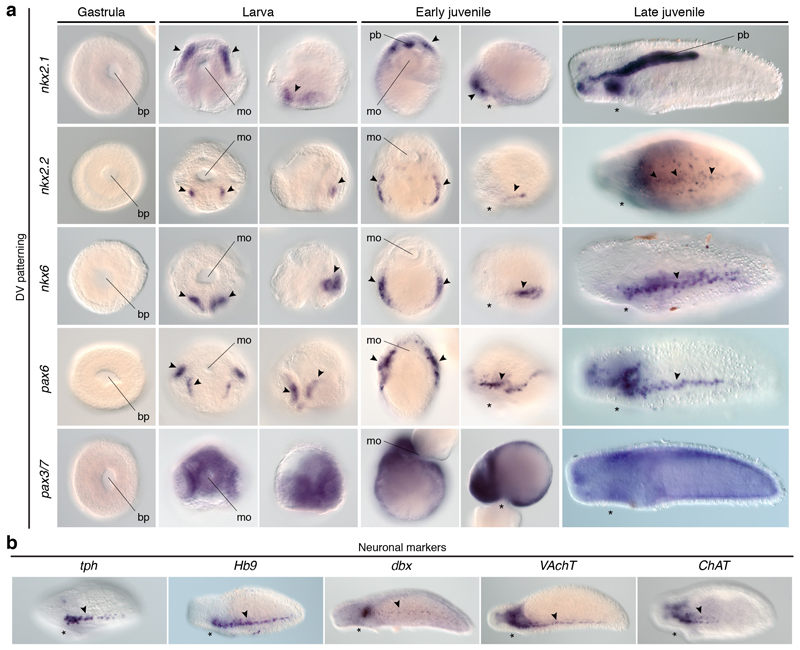
(a) nkx2.1 paralogs, nkx2.2, and pax6 show brain domains (arrowheads); nkx6 is detected posteriorly (arrowheads). (b) E. senta CNS (green arrowhead indicates the brain; red arrowheads indicate the VNCs, and additional neurites). (c) nkx2.2 shows gut expression (arrowhead); nkx6 is in the ventral midline (arrowheads); pax6 is in two lateral larval bands (arrowheads), and juvenile head; pax3/7 is in two ventrolateral larval clusters and midline (arrowheads), but in two trunk clusters in juveniles (arrowheads); msx paralogs are in ventral larval domains and the juvenile VNC (arrowheads). (d) O. fusiformis CNS (green arrowheads indicate the apical larval FMRF-amide cell and juvenile brain; red arrowheads indicate the larval anterior axon and juvenile medial VNC). ao, apical organ; br, brain; cb, ciliary band; cg, caudal ganglion; ch, chaetae; ln, lateral neurites; mo, mouth; ms, mastax; np, neuropile; vg, vesicle ganglia; vnc, ventral nerve cord. Scale bars, 50 µm.
DV patterning in Annelida
To investigate the conservation of the dorsoventral patterning in Annelida, the only spiralian lineage with a medially condensed VNC1,5, we studied the annelid Owenia fusiformis, which belongs to the sister lineage to all remaining annelids34. Remarkably, this annelid deploys the DV-TFs differently from P. dumerilii13,35. Besides the gut-related expression of nkx2.131, nkx2.2, and nkx6 in embryos and larvae, the ventral ectodermal midline expresses nkx6, pax3/7, and two msx paralogs (Fig. 5c; Extended Data Fig. 9b). Additionally, pax6 and pax3/7 show more lateral larval expression domains (Fig. 5c). However, the ventral ectoderm of the juvenile only expresses nkx6 and msx-b (Fig. 5c; Extended Data Fig. 9c). As in most other annelids1, the adult CNS includes a VNC in O. fusiformis, which is not yet present in the early larva36 (Fig. 5d). In the juvenile, only the expression of nkx6 and msx-b relate to the location of serotonin (Fig. 5d) and motoneuronal markers (Extended Data Fig. 9d). Therefore, the dorsoventral patterning system varies also among annelids with a homologous condensed VNC, and between larval13 and adult stages35 (Extended Data Fig. 10a).
Discussion
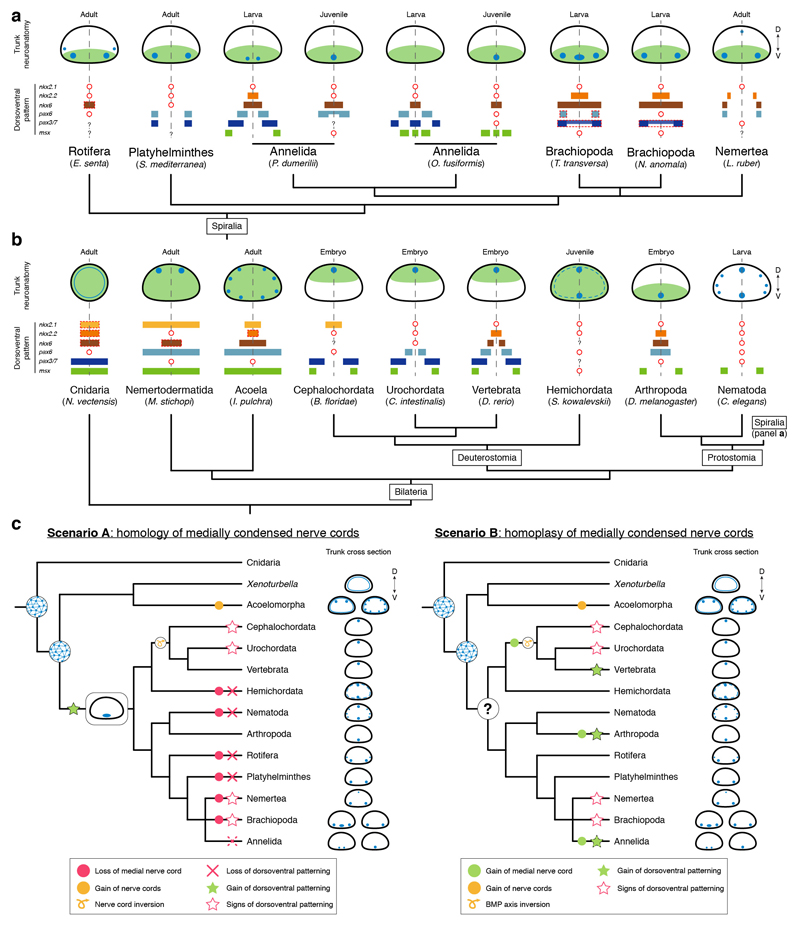
Our study provides compelling evidence that the genes involved in the dorsoventral patterning of vertebrate, Drosophila, and P. dumerilii nerve cords do not show a similar staggered expression in the nerve cords of xenacoelomorphs and many spiralian lineages (Fig. 6a; Extended Data Fig. 10a, b). Although DV-TFs define ectodermal domains in the larval brachiopod trunks and the nemertean juvenile (Fig. 6a), these do not necessarily correlate with the trunk CNS and the location of neuronal markers (Fig. 6a). Indeed, the cell lineage relationships between the early ectodermal expression domains and specific neuronal cell types8,10,13 are unclear, even in Drosophila10,33, and still need to be broadly and functionally tested. Our findings demonstrate that the expression of DV-TFs not only differs between species with multiple nerve cords but also between spiralians that share a medially condensed homologous VNC. A similar case is observed among chordates, where the cephalochordate37 and tunicate38 neural plates only partially show the vertebrate molecular arrangement (Extended Data Fig. 10b; Supplementary Information Table 2), which is likely not a secondary loss given the absence of the dorsoventral patterning in Hemichordata11,12. Therefore, the expression of DV-TFs evolved independently from the trunk neuroanatomy at least in certain bilaterian lineages, which restricts the use of this patterning system to homologize CNS anatomies7,8,14 and neuronal cell types2,8.
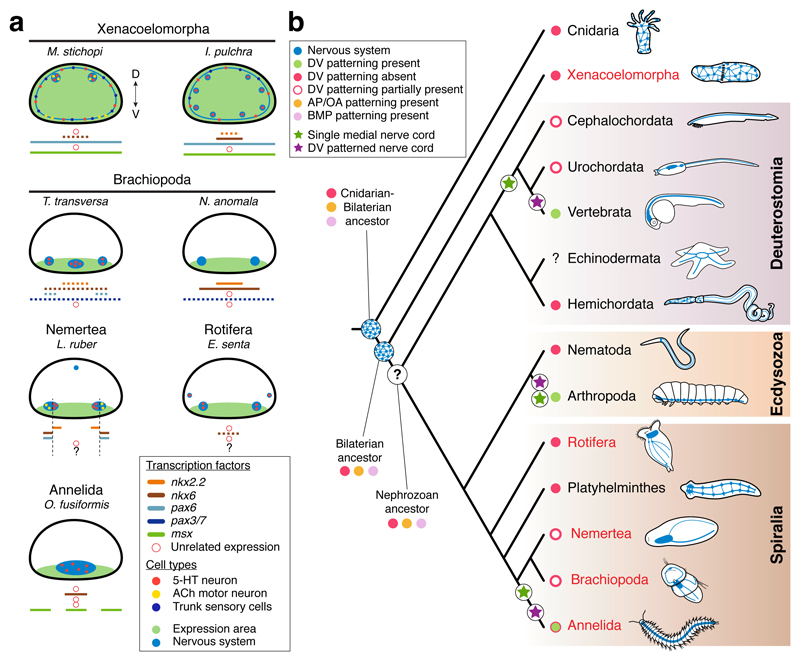
Figure 6. Dorsoventral patterning and CNS evolution.
(a) Gene expression summary. Dotted lines indicate that the expression does not extend along the entire trunk. (b) Proposed scenario for the evolution of neuroectodermal patterning systems in Bilateria. A nerve net, and the anteroposterior (AP) and BMP axial patterning predate the Cnidaria-Bilateria split, and were present in the Bilateria ancestor. The ancestral nephrozoan neuroanatomy remains unclear (question mark). The dorsoventral (DV) patterning system is not tied to the CNS arrangement in Bilateria (as in Chordata and Annelida). In red, lineages analysed in this study. The green dot with red border indicates that there are annelids with and without the DV patterning.
The similarities in the expression of anteroposterior and BMP patterning systems in Cnidaria and Bilateria7,21,25 suggest that these patterning mechanisms predate the Cnidaria–Bilateria split (Fig. 6b). However, these systems are deployed in organisms within these clades with diffuse nerve nets and/or centralized nervous systems, which indicates that their ancient role was probably general body plan regionalization9, and not CNS patterning and neurogenesis2,7. This also limits their use to homologize CNS anatomies. However, the evolution of the dorsoventral patterning of the nerve cords is more complicated (Extended Data Fig. 10c). If the similarities in dorsoventral CNS patterning between vertebrates, flies, and P. dumerilii are homologous and thus reflect the ancestral bilaterian (or nephrozoan) state7,8,13,14, then this patterning system was independently lost/modified a great number of times. The differences between vertebrates and Drosophila in the upstream modulators of DV-TFs and in their functional integration33,39 should thus be regarded as a case of developmental system drift40 over large phylogenetic distances. Alternatively, and more parsimoniously, these differences may indicate that the commonalities in dorsoventral nerve cord organization between vertebrates, arthropods and some annelids evolved convergently (Fig. 6b; Extended Data Fig. 10c). The similar staggered expression domains of DV-TFs in these three lineages, together with the ones uncovered by our study (Figs. 3, 4) may reflect the existence of ancient ectodermal gene regulatory sub-modules17,38,41,42 that got repeatedly assembled for the patterning of bilaterian nerve cords and neuronal cell type specification. Therefore, advancing our understanding of CNS evolution largely relies on functionally identifying the developmental implications of the anteroposterior and dorsoventral patterning systems in diverse bilaterians, before they can be used to homologize particular morphological structures and cell types5,43.
Methods
Animal collections and sample fixations
Gravid adults were collected from the coasts near Friday Harbor Laboratories, U.S.A. (T. transversa), Espeland Marine Biological Station, Norway (M. stichopi and N. anomala), Fanafjorden, Norway (L. ruber), Station Biologique de Roscoff, France (O. fusiformis), and Gullmarsfjord, Sweden (N. westbladi and X. bocki). Peter Ladurner (University of Innsbruck) kindly provided a stable culture of I. pulchra, which was maintained as previously described44. A stable laboratory culture of E. senta was maintained in glass bowls with 25 mL Jaworski’s medium in controlled environment of 20 °C and a 14:10 h light-dark cycle. They were fed ad libitum with the algae Rhodomonas sp., Cryptomonas sp. and Chlamydomonas reinhardtii. Brachiopod, nemertean and annelid adults were spawned as described elsewhere45–48. Acoelomorph eggs were collected year round (I. pulchra) and in September–October (M. stichopi)29. All samples were fixed in 4% paraformaldehyde in culture medium for 1 h at room temperature. After fixation, samples were washed in 0.1% Tween-20 phosphate buffer saline (PTw), dehydrated through a graded series of methanol, and stored at -20 ºC in pure methanol. Samples used for immunohistochemistry were store in PTw at 4 ºC. Before fixation, larval and juvenile stages were relaxed in 7.4% magnesium chloride, E. senta were relaxed in 10% EtOH and 1% bupivacaine. The eggshells of M. stichopi and I. pulchra eggs were permeabilised with 1% sodium thioglycolate and 0.2 mg/ml protease for 20 min before fixation.
DMH1 treatments
M. stichopi and I. pulchra embryos were collected at the 1–2 cell stage and cultured with regular water changes in cell culture dishes until the desired developmental stage. Control embryos were treated with 0.1% DMSO and experimental embryos were treated with DMH1 (Sigma) up to 10 µM. Seawater containing the DMH1 was changed every day until fixation. Embryos and hatchlings were fixed as described above, and stored in PTw at 4 ºC.
Gene identification and expression analyses
RNAseq data obtained from mixed developmental stages and juveniles/adults was used for gene identification. Gene orthology was based on reciprocal best BLAST hit. For particular gene families, maximum likelihood phylogenetic analyses were conducted with RAxML v8.2.649, after building multiple protein alignments with MAFFT v750 and trimming poorly aligned regions with gblocks v0.91b51 (Supplementary Fig. 1). Whole-mount colorimetric in situ hybridization on brachiopod embryos, L. ruber, O. fusiformis and juvenile E. senta was performed following an already established protocol31,45. Probe concentrations ranged 0.1–1 ng/µl and permeabilisation time was 15 min for M. stichopi and post-metamorphic brachiopod juveniles, 5 min for I. pulchra, and 10 min for the other species. Double fluorescent whole-mount in situ hybridization was performed as described elsewhere31.
Immunohistochemistry
Samples were permeabilised in 0.1–0.5% Triton X-100 phosphate buffer saline (PTx), and blocked in 0.1–1% bovine serum albumin (BSA) in PTx. The antibodies anti-tyrosinated tubulin (Sigma), anti-serotonin (Sigma), and anti-FMRFamide (Immunostar) were diluted in 5% normal goat serum (NGS) in PTx at a concentration of 1:500, 1:200, and 1:200, respectively. Samples were incubated with the primary antibody solutions for 24-72 h at 4 ºC. Followed by several washes in 1% BSA in PTx, samples were incubated overnight with Alexa-conjugated secondary antibodies at a 1:250 dilution in 5% NGS in PTx. Before mounting and imaging, samples were washed several times in 1% BSA in PTx. Nuclei and actin filaments were counterstained with DAPI (Molecular Probes) and BODIPY FL Phallacidin (Molecular Probes).
Imaging
Representative embryos from colorimetric in situ hybridization experiments were cleared in 70% glycerol and imaged with a Zeiss Axiocam HRc connected to a Zeiss Axioscope Ax10 using bright field Nomarsky optics. Fluorescently labelled samples were cleared and mounted in benzyl benzoate/benzyl alcohol (2:1) and scanned in a Leica SP5 confocal laser-scanning microscope. Images were analysed with Fiji and Photoshop CS6 (Adobe) and figure plates were assembled with Illustrator CS6 (Adobe). Brightness/contrast and colour balance adjustments were applied to the whole image, not parts.
Data availability
All newly determined sequences have been deposited in GenBank (accession numbers KY809717–KY809754, KY709718–KY709823, and MF988103–MF988108). Multiple protein alignments used for orthology assignment are available upon request. Extended Data Figure 6c has associated source data.
Extended Data
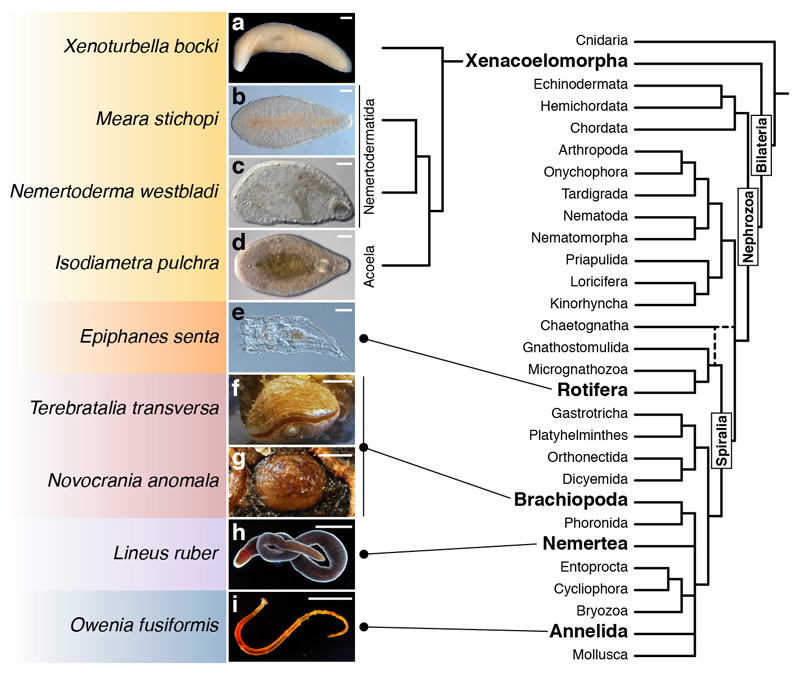
Extended Data Figure 1. Studied species.
(a–i) Images of the adult forms of the studied species within a consensus bilaterian phylogeny19. Color boxes highlight major taxonomical clades. Scale bars, 100 µm in a–e, 0.5 cm in h and g, 1 cm in f, h, and i.
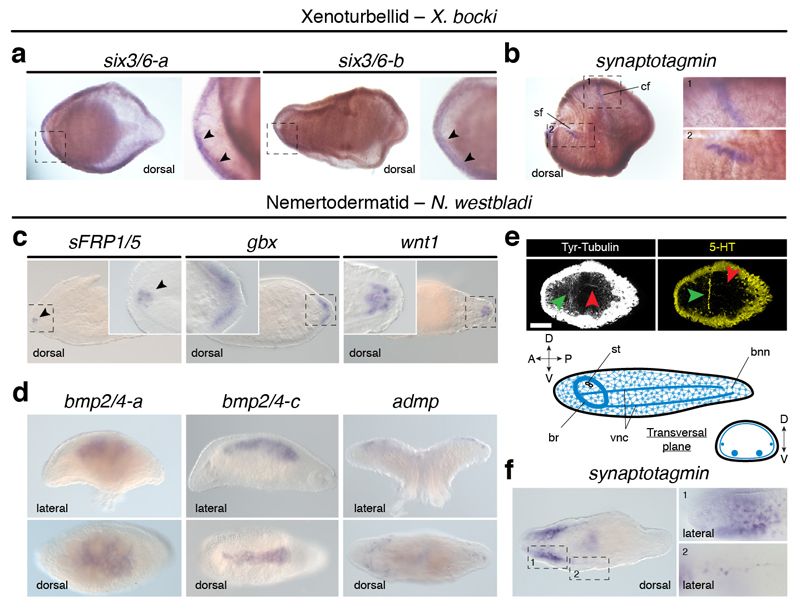
Extended Data Figure 2. Gene expression in Xenoturbella bocki and Nemertoderma westbladi.
(a) Two six3/6 paralogs are expressed in the anterior head margin in X. bocki (arrowheads). (b) The neural marker synaptotagmin (syt) is detected in the circumferential (cf; inset 1) and side (sf; inset 2) sensory furrows in X. bocki. (c) In N. westbladi, the anterior marker sFRP1/5 (arrowhead) and the posterior genes gbx and wnt1 are asymmetrically expressed along the anteroposterior axis of N. westbladi. (d) The BMP ligands bmp2/4-a and bmp2/4-c are expressed dorsally, while the BMP antagonist admp is expressed dorsolaterally. (e) The CNS of N. westbladi comprises an anterior ring-like commissure (green arrowheads) and a main pair of ventral condensations (red arrowhead). (f) The neuronal marker syt is highly expressed in the anterior part (inset 1), and in the nerve cords (inset 2). In the different panels, dotted rectangles indicate magnified areas. In all panels, the anterior pole is to the left. The schematic drawing in c is not to scale. Scale bar, 100 µm in c.
Extended Data Figure 3. Anteroposterior patterning in Xenacoelomorpha.
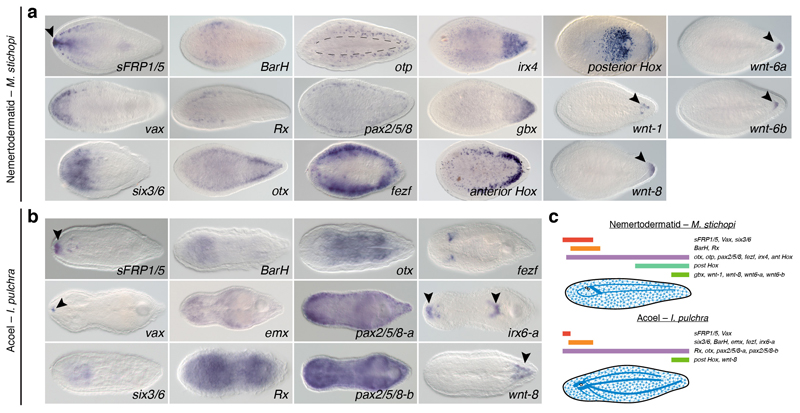
(a, b) Expression of anteroposterior markers in adult specimens of M. stichopi and I. pulchra. In both species, sFRP1/5, vax, six3/6, and BarH are expressed in anterior territories (black arrowheads). In M. stichopi, Rx is also expressed anteriorly, but broadly along the animal body in I. pulchra. In this acoel, emx is detected in the anterior part of the animal (background staining close to the gonads). In the nemertodermatid, the anterior neural markers otx, otp, pax2/5/8, and fezf are expressed along the entire AP axis, in association with the dorsal nerve cords (black dotted lines in otp). In I. pulchra, otx, pax2/5/8-a, and pax2/5/8-b are broadly expressed. In M. stichopi, an irx ortholog is detected in the posterior tip, while it is detected in the anterior tip and around the mouth and copulatory apparatus in the acoel (arrowheads). The gbx ortholog of M. stichopi is expressed posteriorly, and the trunk-related Hox genes are expressed in two lateral rows (anterior Hox) and anteriorly to the mouth and in the posterior tip (posterior Hox). In the nemertodermatid and the acoel, Wnt ligand genes are expressed posteriorly (arrowheads). All images are dorsoventral views with anterior to the left. (c) Schematic summary of anteroposterior expression in the nemertodermatid M. stichopi and the acoel I. pulchra. Drawings are not to scale and the extent of the expression domains are only approximate. The expression of posterior Hox in I. pulchra is based on23.
Extended Data Figure 4. Expression of BMP components in Nemertodermatida and Acoela.
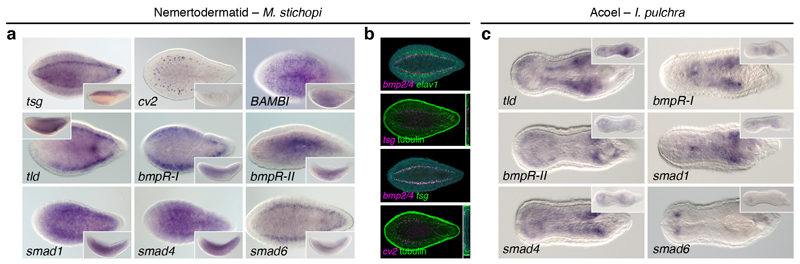
(a) In the nemertodermatid Meara stichopi, the BMP pathway antagonists twisted gastrulation (tsg), and crossveinless 2 (cv2) are expressed dorsally, while the antagonist BAMBI is broadly detected in the ventral side. The gene tolloid (tld) is expressed both dorsally and ventrally. The BMP receptors bmpR-I is expressed dorsolaterally and the bmpR-II is detected more broadly. The genes smad1 and smad4 are expressed broadly and smad6 is expressed along the dorsal nerve cords. (b) bmp2/4 is not expressed in neuronal cells (elav1+ cells), but in cells located medially to the nerve cords (tubulin positive). The cells expressing bmp2/4 also express tsg, and cv2 is expressed dorsally along the nerve cords. (c) In the acoel Isodiametra pulchra, the BMP antagonist tld is expressed ventrally, the bmpR-I is detected in the inner body, and bmpR-II is expressed anteriorly and posteriorly around the copulatory organ. The genes smad1 and smad4 are expressed generally, while smad6 is expressed in two bilaterally symmetrical anterior clusters. All main panels are dorsoventral views, and the insets are lateral views.
Extended Data Figure 5. Expression of neuronal markers in Nemertodermatida and Acoela.
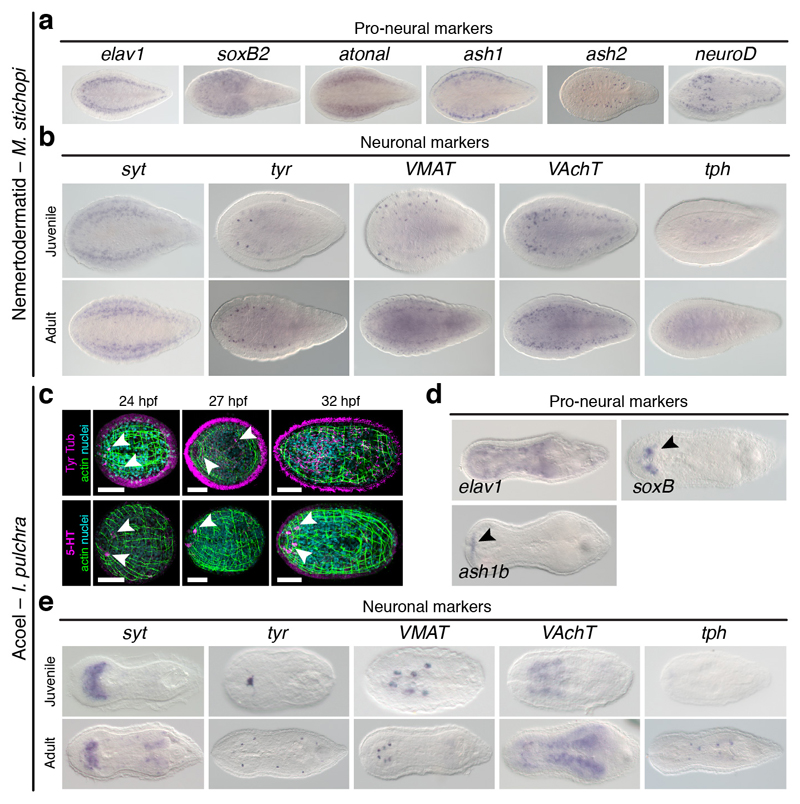
(a) In the nemertodermatid Meara stichopi, the genes associated with neuronal fate commitment elav1, soxB2, ash1, ash2, atonal, and neuroD are detected along the dorsal nerve cords. (b) Similarly, the neuronal markers synaptotagmin (syt), tyrosine hydroxylase (tyr), vesicular monoamine transporter (VMAT), choline acetyltransferase (ChAT), vesicular acetylcholine transporter (VAchT), and tryptophan hydroxylase (tph) are mostly expressed dorsally, along the dorsal nerve cords. (c) Morphology of I. pulchra embryos stained against tyrosinated tubulin (Tyr tub) and serotonin (5-HT), and counterstained with phallacidin (actin bundles) and DAPI (nuclei). The first tubulin-positive cells that resemble neurons appear anteriorly (arrowheads) at 24 hours post-fertilization (hpf). By 32 hpf, the anterior and posterior lobes of the brain, as well as some neurite bundles are visible. Similarly, the first serotonergic cells are detected at 24 hpf in the anterior end (arrowheads). (d) In I. pulchra, the pro-neural marker elav1 is broadly expressed, soxB is detected in the head region (arrowhead), and ash1b in the anterior tip (arrowhead). (e) In I. pulchra, the neuronal marker syt is highly expressed in the anterior neuropile. The marker tyr is detected in the statocyst and isolated cells. VMAT is detected in isolated dorsal cell clusters in the juvenile that concentrate along the adult brain. ChAT and VAchT are expressed in the brain in juveniles and adults (gonadal staining in the adult is background). The gene tph is expressed in isolated ventral cells of the adult. All panels are dorsoventral views with anterior to the left. Scale bars, 50 µm in c.
Extended Data Figure 6. DMH1 treatments in Meara stichopi and Isodiametra pulchra.
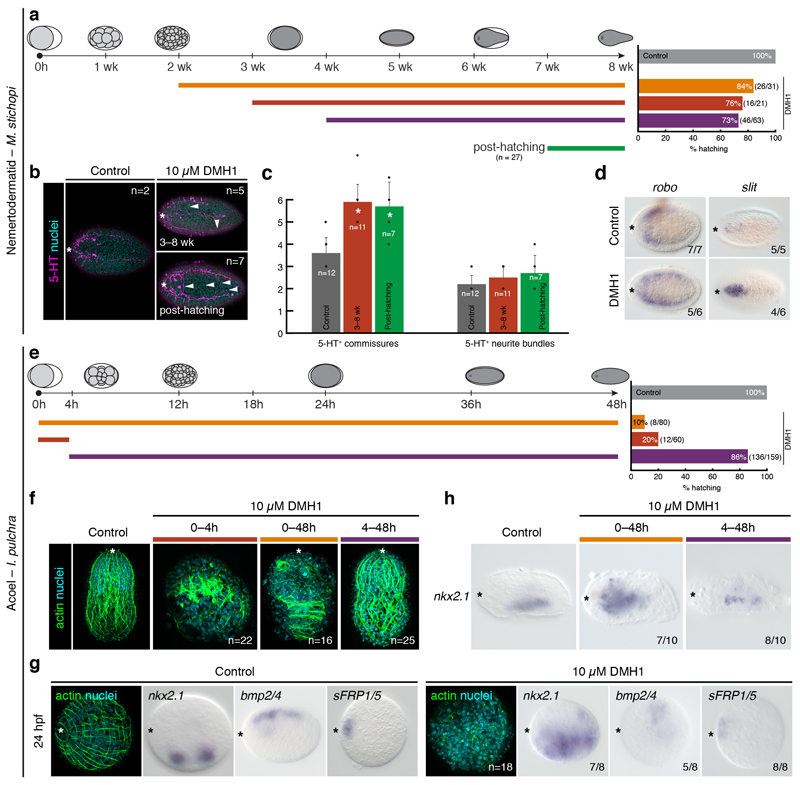
(a) Schematic overview of dorsomorphin homologue 1 (DMH1) treatments in M. stichopi and % of hatching embryos for each experimental condition. (b) M. stichopi embryos incubated with DMH1 from 3 to 8 weeks and after hatching show more serotonergic commissures that control animals. (c) The differences in the number of commissures are significant in both pre-hatching (asterisk; two-tail t-test; p-value < 0.0001) and post-hatching (asterisk; two-tail t-test; p-value < 0.0014) treated embryos. In contrast, the number of serotonin positive neurite bundles is not significantly increased in any of the treatments. (d) Despite the abnormal development of serotonergic axonal tracts, slit and robo genes are expressed similarly. The differences in signal intensity are due to technical variability. (e) Schematic overview of DMH1 treatments in I. pulchra and % of hatching embryos for each experimental condition. (f) Morphological analyses of DMH1 treated embryos. Treatment in early stages affects normal development, while treatments from 4 hours onwards does not significantly compromises embryogenesis. (g) Embryos treated between 0–4 hours post fertilization and fixed at 24 hours of development show expanded expression of the ventral marker nkx2.1, reduced expression of the dorsal gene bmp2/4, and unaffected expression of the anterior marker sFRP1/5. The embryo shows a disorganized morphology, as revealed by actin staining. (h) The expression of the ventral marker nkx2.1 is expanded in early treated embryos (0–48 h), but unaffected in embryos treated after 4 hours of development. In (b, d, f–h), the asterisk marks the anterior pole. In (b, d, f), panels are dorsoventral views, and in (g, h) the panels are lateral views.
Extended Data Figure 7. Gene expression in Brachiopoda.
(a) Gene expression during early gastrulation and elongation, and in late larvae of T. transversa. The gene nkx2.2 is expressed ventroposteriorly (black arrowhead) and in the pedicle lobe of the larva (arrow). nkx6 is detected in two bilateral symmetrical ectodermal posterior clusters (arrowheads) and in the archenteron wall. In the larva, nkx6 is expressed in the pedicle lobe (arrow) and midgut. pax3/7 is first detected in two ventrolateral domains at the prospective apical-trunk boundary (arrowheads), and in the ventral anterior region of the larva. The gene msx is first expressed dorsally, in the future mantle ectoderm (arrowheads), and in the mantle of the larva. (b) In 2-days old post-metamorphic juveniles, the CNS comprises a main serotonergic anterior commissure (white arrowhead; dorsoventral view) that innervates the developing lophophore. The schematic drawing is not to scale, and the blue line represent the commissure. (c) The gene nkx2.1 is expressed in the anterior region (arrowhead), between the lophophores in 2-day old juveniles. The genes nkx2.2 and nkx6 are expressed in the pedicle (arrowheads), and nkx6 is also detected in the gut (arrow). The gene pax6 shows no expression, pax3/7 is detected in the neural commissure (arrowhead), and msx is expressed in the cells at the edge of the mantle (dotted line). (d) Neuronal markers in late larvae of T. transversa. The serotonergic marker tph is expressed in the anteroventral condensation of the mantle lobe (arrowhead) and in dorsal ectodermal cells of the apical lobe (arrow). No expression is detected for Hb9, and the genes dbx, VAchT and ChAT are all detected in the anterior apical neuroectoderm (arrowheads). (e) Gene expression during early gastrulation and elongation, and in late larvae of N. anomala. The gene nkx2.2 is expressed in the anterior blastoporal lip at the onset of axial elongation, and it is not detected in the late larva. nkx6 is asymmetrically expressed around the blastopore, in the putative anteroventral ectoderm (arrowhead). As the blastopore closes, the expression extends posteriorly and concentrates along the midline of the larva (arrowhead). The gene pax3/7 is detected in the posterior mesoderm at the onset of axial elongation (arrow). The gene msx is expressed in the prospective mantle lobe ectoderm (arrowheads) and in the dorsal shell-forming epithelium of the late larva. The asterisks indicate the animal/anterior pole and white dotted lines in (a, d) mark the region of background noise caused by probe trapping in the shell forming ectoderm. Panel orientations are indicated in the first row/column and apply to the rest of the panels in the same column/row. Scale bar, 100 µm in b.
Extended Data Figure 8. Gene expression in the nemertean Lineus ruber.
(a) None of the nerve cord patterning genes is expressed during gastrulation in L. ruber. In the intracapsular larva, nkx2.1 is expressed in the cephalic imaginal discs (arrowheads), nkx2.2 and nkx6 in an anterior and a posterior domain of the trunk imaginal discs (arrowheads) respectively, and pax6 is detected both in the cephalic and the anterior trunk imaginal discs (arrowheads). pax3/7 is broadly expressed. With metamorphosis, nkx2.1 is detected in the head and proboscis, nkx2.2 is detected in the nerve cords and isolated trunk cells (arrowheads), nkx6 is expressed in the nerve cords (arrowheads), pax6 is observed in the head and nerve cords (arrowheads), and pax3/7 remains broadly expressed. All gastrulae are vegetal views. For larvae and early juveniles, the left column is a dorsoventral view and the right column is a lateral view (anterior to the left). All late juvenile pictures are lateral views, with anterior to the left. (b) Lateral views (anterior to the left) of neuronal markers in juveniles. They are all expressed in the ventral nerve cords (arrowheads), and not in the dorsal neurite bundle. In all panels, the asterisk indicates the position of the mouth opening. bp, blastopore; mo, mouth; pb, proboscis.
Extended Data Figure 9. Molecular patterning and motoneuron markers in Rotifera and Annelida.
(a) Expression of the motoneuron markers Hb9 and ChAT in juveniles of the rotifer Epiphanes senta. The gene Hb9 is detected in neurons of the mastax (arrowheads) and weakly in isolated cells of the brain (arrow). The gene ChAT is detected in the brain (arrow), cells of the corona and mastax (arrowheads). (b) Expression of dorsoventral patterning genes in gastrulae and elongating embryos of Owenia fusiformis. nkx2.2 and nkx6 are expressed in the internalized endomesoderm (arrowheads). The gene pax6 is expressed in two lateral rows during elongation (arrowhead) and pax3/7 in two lateral cells (arrowhead). Of the two paralogs, msx-a is first detected in a posterior ectodermal domain (arrowhead) and in two additional bilaterally symmetrical posterior cells (arrowheads) during elongation. The gene msx-b is only detected during elongation in a posterior domain (arrowhead). (c) Ventral view of the expression of nkx2.1 in the juvenile of the annelid O. fusiformis. This gene is detected in the foregut (arrowheads) and hindgut (arrow). (d) Expression of the motoneuron markers Hb9 and ChAT in O. fusiformis. Hb9 is first detected in lateral domains of the archenteron/gut during embryogenesis and larva, and in isolated cells of the ventral trunk of the juvenile. The gene ChAT is detected in three cells of the apical region of the embryo and larva, and in the neuropile and two lateral ventral cords of the juvenile. bp, blastopore; mo, mouth; ms, mastax. The asterisk in (a) marks the position of the mouth.
Extended Data Figure 10. Dorsoventral patterning and the evolution of bilaterian trunk neuroanatomy.
(a, b) Schematic drawings of trunk neuroanatomy (nerve cords in blue) and expression of patterning genes in spiralian (a) and bilaterian (b) lineages. The overall location of patterning genes expression domains with respect to the DV axis and nerve cords is indicated by light green. In (a), the red dashed squared expression of pax6 and pax3/7 in brachiopods indicates that these expression domains are only in the anterior region of the mantle lobe, not all along the trunk. Similarly, the red dashed squared expression of nkx6 in rotifers highlights that this gene is only expressed posteriorly in the trunk. In (b), the red dashed squared expression of nkx2.1, nkx2.2 and nkx6 in Cnidaria indicates that these genes are expressed in the pharynx ectoderm. The red dashed squared expression of nkx6 in M. stichopi shows that this gene is only expressed posteriorly. In the acoel I. pulchra, the red dashed squared expression of nkx2.2 specifies that this gene is only expressed between mouth and copulatory organ. Red circles imply that a gene is not expressed in the trunk or is missing. Question marks indicate that there is no available data about the expression of that particular gene. See Extended Data Table 1 and main text for references. Schematic drawings are not to scale and only represent approximate relative expression domains. (c) Alternative scenarios for the evolution of the dorsoventral patterning and bilaterian nerve cords. In scenario A, the medially condensed nerve cords of vertebrates, arthropods, and annelids are homologous. Therefore, the dorsoventral patterning was lost multiple times both in lineages with medially condensed nerve cords (e.g. the annelid Owenia fusiformis, cephalochordates, and tunicates) and in lineages with multiple nerve cords and diffuse nerve nets. In scenario B, which is supported by this study and is more parsimonious, the similarities in dorsoventral patterning and trunk neuroanatomies of vertebrates, arthropods, and some annelids evolved convergently. The diversity of nerve cord arrangements in nephrozoan lineages hampers reconstructing the ancestral neuroanatomy for this group (question mark). Animal phylogeny based on19
Supplementary Information
Acknowledgements
We thank the staff at the marine stations, current and former members of the Hejnol lab, and Casey Dunn. The Sars Core Budget, the FP7-PEOPLE-2009-RG, and the ERC Community’s Framework Program Horizon 2020 to AH funded this work. The NSF IRFP Postdoctoral Fellowship funded KP. The Carlsberg Foundation funded HSL. The Swedish Research Council funded UJ and JTC.
Footnotes
Author Contributions
JMMD, KP, HSL, AH designed the study. JMMD, KP, AB, AF, AH, UJ, JTC collected the animals. JMMD, KP, AB, HS, AF, AH performed the experiments. JMMD, KP, AH wrote the manuscript. All authors read and approved the final manuscript.
Author information
All sequences have been deposited in GenBank (accession numbers KY809717–KY809754, KY709718–KY709823, MF988103–MF988108). Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Schmidt-Rhaesa A, Harzsch S, Purschke G. Structure & Evolution of Invertebrate Nervous Systems. Oxford University Press; 2016. [Google Scholar]
- 2.Arendt D, Tosches MA, Marlow H. From nerve net to nerve ring, nerve cord and brain--evolution of the nervous system. Nat Rev Neurosci. 2016;17:61–72. doi: 10.1038/nrn.2015.15. [DOI] [PubMed] [Google Scholar]
- 3.Hejnol A, Pang K. Xenacoelomorpha's significance for understanding bilaterian evolution. Curr Opin Genet Dev. 2016;39:48–54. doi: 10.1016/j.gde.2016.05.019. [DOI] [PubMed] [Google Scholar]
- 4.Arendt D, Denes AS, Jékely G, Tessmar-Raible K. The evolution of nervous system centralization. Philos Trans R Soc Lond B Biol Sci. 2008;363:1523–1528. doi: 10.1098/rstb.2007.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hejnol A, Lowe CJ. Embracing the comparative approach: how robust phylogenies and broader developmental sampling impacts the understanding of nervous system evolution. Phil Trans R Soc B. 2015;370 doi: 10.1098/rstb.2015.0045. 20150045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holland ND. Early central nervous system evolution: an era of skin brains? Nat Rev Neurosci. 2003;4:617–627. doi: 10.1038/nrn1175. [DOI] [PubMed] [Google Scholar]
- 7.Holland LZ, et al. Evolution of bilaterian central nervous systems: a single origin? Evodevo. 2013;4:27. doi: 10.1186/2041-9139-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arendt D, Denes AS, Jekely G, Tessmar-Raible K. The evolution of nervous system centralization. Philos Trans R Soc Lond B Biol Sci. 2008;363:1523–1528. doi: 10.1098/rstb.2007.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pani AM, et al. Ancient deuterostome origins of vertebrate brain signalling centres. Nature. 2012;483:289–294. doi: 10.1038/nature10838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizutani CM, Bier E. EvoD/Vo: the origins of BMP signalling in the neuroectoderm. Nat Rev Genet. 2008;9:663–677. doi: 10.1038/nrg2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lowe CJ, et al. Anteroposterior patterning in hemichordates and the origins of the chordate nervous system. Cell. 2003;113:853–865. doi: 10.1016/s0092-8674(03)00469-0. [DOI] [PubMed] [Google Scholar]
- 12.Lowe CJ, et al. Dorsoventral patterning in hemichordates: insights into early chordate evolution. PLoS Biol. 2006;4 doi: 10.1371/journal.pbio.0040291. e291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denes AS, et al. Molecular architecture of annelid nerve cord supports common origin of nervous system centralization in bilateria. Cell. 2007;129:277–288. doi: 10.1016/j.cell.2007.02.040. [DOI] [PubMed] [Google Scholar]
- 14.Arendt D, Nübler-Jung K. Comparison of early nerve cord development in insects and vertebrates. Development. 1999;126:2309–2325. doi: 10.1242/dev.126.11.2309. [DOI] [PubMed] [Google Scholar]
- 15.Kaul-Strehlow S, Urata M, Praher D, Wanninger A. Neuronal patterning of the tubular collar cord is highly conserved among enteropneusts but dissimilar to the chordate neural tube. Sci Rep. 2017;7:7003. doi: 10.1038/s41598-017-07052-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okkema PG, Ha E, Haun C, Chen W, Fire A. The Caenorhabditis elegans NK-2 homeobox gene ceh-22 activates pharyngeal muscle gene expression in combination with pha-1 and is required for normal pharyngeal development. Development. 1997;124:3965–3973. doi: 10.1242/dev.124.20.3965. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, et al. Conserved gene regulatory module specifies lateral neural borders across bilaterians. Proc Natl Acad Sci USA. 2017 doi: 10.1073/pnas.1704194114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scimone ML, Kravarik KM, Lapan SW, Reddien PW. Neoblast specialization in regeneration of the planarian Schmidtea mediterranea. Stem Cell Reports. 2014;3:339–352. doi: 10.1016/j.stemcr.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cannon JT, et al. Xenacoelomorpha is the sister group to Nephrozoa. Nature. 2016;530:89–93. doi: 10.1038/nature16520. [DOI] [PubMed] [Google Scholar]
- 20.Ruiz-Trillo I, Riutort M, Littlewood DTJ, Herniou EA, Baguña J. Acoel flatworms: earliest extant bilaterian Metazoans, not members of Platyhelminthes. Science. 1999;283:1919–1923. doi: 10.1126/science.283.5409.1919. [DOI] [PubMed] [Google Scholar]
- 21.Srivastava M, Mazza-Curll KL, van Wolfswinkel JC, Reddien PW. Whole-Body acoel regeneration is controlled by Wnt and Bmp-Admp signaling. Curr Biol. 2014;24:1107–1113. doi: 10.1016/j.cub.2014.03.042. [DOI] [PubMed] [Google Scholar]
- 22.Hejnol A, Martindale MQ. Coordinated spatial and temporal expression of Hox genes during embryogenesis in the acoel Convolutriloba longifissura. BMC Biol. 2009;7:65. doi: 10.1186/1741-7007-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreno E, De Mulder K, Salvenmoser W, Ladurner P, Martinez P. Inferring the ancestral function of the posterior Hox gene within the bilateria: controlling the maintenance of reproductive structures, the musculature and the nervous system in the acoel flatworm Isodiametra pulchra. Evol Dev. 2010;12:258–266. doi: 10.1111/j.1525-142X.2010.00411.x. [DOI] [PubMed] [Google Scholar]
- 24.De Robertis EM, Sasai Y. A common plan for dorsoventral patterning in Bilateria. Nature. 1996;380:37–40. doi: 10.1038/380037a0. [DOI] [PubMed] [Google Scholar]
- 25.Layden MJ, Rentzsch F, Röttinger E. The rise of the starlet sea anemone Nematostella vectensis as a model system to investigate development and regeneration. Wiley Interdiscip Rev Dev Biol. 2016;5:408–428. doi: 10.1002/wdev.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raikova OI, Meyer-Wachsmuth I, Jondelius U. The plastic nervous system of Nemertodermatida. Org Divers Evol. 2016;16:85–104. [Google Scholar]
- 27.Raikova OI, Reuter M, Jondelius U, Gustafsson MKS. An immunocytochemical and ultrastructural study of the nervous and muscular systems of Xenoturbella westbladi (Bilateria inc. sed.) Zoomorphology. 2000;120:107–118. [Google Scholar]
- 28.Achatz JG, Martinez P. The nervous system of Isodiametra pulchra (Acoela) with a discussion on the neuroanatomy of the Xenacoelomorpha and its evolutionary implications. Front Zool. 2012;9:27. doi: 10.1186/1742-9994-9-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Børve A, Hejnol A. Development and juvenile anatomy of the nemertodermatid Meara stichopi (Bock) Westblad 1949 (Acoelomorpha) Front Zool. 2014;11:50. doi: 10.1186/1742-9994-11-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Struck TH, et al. Platyzoan paraphyly based on phylogenomic data supports a noncoelomate ancestry of spiralia. Molecular biology and evolution. 2014;31:1833–1849. doi: 10.1093/molbev/msu143. [DOI] [PubMed] [Google Scholar]
- 31.Martín-Durán JM, Passamaneck YJ, Martindale MQ, Hejnol A. The developmental basis for the recurrent evolution of deuterostomy and protostomy. Nature Ecology & Evolution. 2016;1:0005. doi: 10.1038/s41559-016-0005. [DOI] [PubMed] [Google Scholar]
- 32.Vellutini BC, Hejnol A. Expression of segment polarity genes in brachiopods supports a non-segmental ancestral role of engrailed for bilaterians. Sci Rep. 2016;6:32387. doi: 10.1038/srep32387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cornell RA, Ohlen TV. Vnd/nkx, ind/gsh, and msh/msx: conserved regulators of dorsoventral neural patterning? Curr Opin Neurobiol. 2000;10:63–71. doi: 10.1016/s0959-4388(99)00049-5. [DOI] [PubMed] [Google Scholar]
- 34.Weigert A, et al. Illuminating the base of the annelid tree using transcriptomics. Molecular biology and evolution. 2014;31:1391–1401. doi: 10.1093/molbev/msu080. [DOI] [PubMed] [Google Scholar]
- 35.Vergara HM, et al. Whole-organism cellular gene-expression atlas reveals conserved cell types in the ventral nerve cord of Platynereis dumerilii. Proc Natl Acad Sci USA. 2017;114:5878–5885. doi: 10.1073/pnas.1610602114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Helm C, Vöcking O, Kourtesis I, Hausen H. Owenia fusiformis - a basally branching annelid suitable for studying ancestral features of annelid neural development. BMC Evol Biol. 2016;16:129. doi: 10.1186/s12862-016-0690-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Albuixech-Crespo B, et al. Molecular regionalization of the developing amphioxus neural tube challenges major partitions of the vertebrate brain. PLoS Biol. 2017;15 doi: 10.1371/journal.pbio.2001573. e2001573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stolfi A, Ryan K, Meinertzhagen IA, Christiaen L. Migratory neuronal progenitors arise from the neural plate borders in tunicates. Nature. 2015;527:371–374. doi: 10.1038/nature15758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winterbottom EF, Illes JC, Faas L, Isaacs HV. Conserved and novel roles for the Gsh2 transcription factor in primary neurogenesis. Development. 2010;137:2623–2631. doi: 10.1242/dev.047159. [DOI] [PubMed] [Google Scholar]
- 40.True JR, Haag ES. Developmental system drift and flexibility in evolutionary trajectories. Evol Dev. 2001;3:109–119. doi: 10.1046/j.1525-142x.2001.003002109.x. [DOI] [PubMed] [Google Scholar]
- 41.Cheesman SE, Layden MJ, Von Ohlen T, Doe CQ, Eisen JS. Zebrafish and fly Nkx6 proteins have similar CNS expression patterns and regulate motoneuron formation. Development. 2004;131:5221–5232. doi: 10.1242/dev.01397. [DOI] [PubMed] [Google Scholar]
- 42.Goridis C, Rohrer H. Specification of catecholaminergic and serotonergic neurons. Nat Rev Neurosci. 2002;3:531–541. doi: 10.1038/nrn871. [DOI] [PubMed] [Google Scholar]
- 43.Peter IS, Davidson EH. Evolution of gene regulatory networks controlling body plan development. Cell. 2011;144:970–985. doi: 10.1016/j.cell.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Mulder K, et al. Characterization of the stem cell system of the acoel Isodiametra pulchra. BMC Dev Biol. 2009;9:69. doi: 10.1186/1471-213X-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martín-Durán JM, Vellutini BC, Hejnol A. Evolution and development of the adelphophagic, intracapsular Schmidt's larva of the nemertean Lineus ruber. Evodevo. 2015;6:28. doi: 10.1186/s13227-015-0023-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Freeman G. Regional specification during embryogenesis in the articulate brachiopod Terebratalia. Developmental biology. 1993;160:196–213. doi: 10.1006/dbio.1993.1298. [DOI] [PubMed] [Google Scholar]
- 47.Freeman G. Regional specification during embryogenesis in the craniiform brachiopod Crania anomala. Developmental biology. 2000;227:219–238. doi: 10.1006/dbio.2000.9857. [DOI] [PubMed] [Google Scholar]
- 48.Smart TI, Von Dassow G. Unusual development of the mitraria larva in the polychaete Owenia collaris. Biol Bull. 2009;217:253–268. doi: 10.1086/BBLv217n3p253. [DOI] [PubMed] [Google Scholar]
- 49.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular biology and evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All newly determined sequences have been deposited in GenBank (accession numbers KY809717–KY809754, KY709718–KY709823, and MF988103–MF988108). Multiple protein alignments used for orthology assignment are available upon request. Extended Data Figure 6c has associated source data.