Abstract
GB virus C (GBV-C) is a non-pathogenic flavivirus that may play a role in modulating HIV disease. Multiple genotypes of GBV-C that have been identified to date that may differentially regulate HIV; however, the number of complete GBV-C sequences published to date is very limited. We sequenced full-length GBV-C genomes from four individuals with HIV/HCV co-infection in the United States. Intergenotypic recombination was evident in two of these individuals. Evaluation of additional full-length GBV-C genomes would facilitate the creation of full-length, replication-competent molecular clones of GBV-C to evaluate the phenotypic diversity of GBV-C genotypes and provide important molecular data on this understudied virus.
Keywords: GB virus C (GBV-C), full-length, complete genome, diversity, genotype, recombination
GB virus C (GBV-C) is a member of the Flaviviridae family of positive-sense, single-stranded RNA viruses. The GBV-C genome consists of a 5′ untranslated region (5′UTR) that contains an internal ribosomal entry site and is followed by a single open reading frame (ORF) that encodes for a polyprotein of approximately 3000 amino acids (reviewed by Mohr & Stapleton, 2009). This polyprotein encodes for at least two structural proteins – envelope protein 1 (E1) and envelope protein 2 (E2) – as well as for multiple non- structural proteins – NS2 to NS5. To date, at least six GBV-C genotypes have been identified (Feng et al., 2011; Muerhoff et al., 2005, 2006). These are typically classified by phylogenetic comparison of the 5′UTR. These genotypes are not evenly distributed worldwide and display significant geographic restriction with genotype 1 in West Africa, genotypes 1 and 2 in Europe and the United States, genotype 3 in parts of Asia, genotype 4 in Southeast Asia and genotype 5 in South Africa (Katayama et al., 1998; Muerhoff et al., 1996, 1997, 2005; Mukaide et al., 1997; Okamoto et al., 1997; Tucker & Smuts, 2000; Tucker et al., 1999).
Multiple studies have demonstrated a beneficial effect of ongoing GBV-C replication on HIV disease resulting in lower HIV viral loads, slower CD4 cell decline and prolonged AIDS-free survival compared to persons without GBV-C infection (Stapleton, 2003; Tillmann et al., 2001; Williams et al., 2004). However, some other studies have not shown such a survival advantage (Birk et al., 2002; Björkman et al., 2004). While it has been suggested that GBV-C genotype may play a role in modulating HIV disease progression (Alcalde et al., 2010; Muerhoff et al., 2003; Schwarze-Zander et al., 2006), which could explain the divergent findings among these studies, additional investigation is required to evaluate this hypothesis adequately. As a necessary first step, we characterized complete ORF sequences from individuals with GBV-C infection in the United States.
Eighteen subjects were considered as part of a prospective cohort of HIV/HCV co-infected patients designed to characterize changes in HCV viral load and alanine aminotransferase (ALT) levels following combination anti-retroviral therapy (ART) initiation as described in detail previously (Sherman et al., 2014). All patients provided informed consent and the study protocols were approved by the Institutional Review Boards at the enrolling sites (University of Cincinnati, Virginia Commonwealth University and New York University).
For the current study, the first 12 individuals from the prospective cohort were screened for GBV-C infection at week 8 post-ART initiation. Viral RNA was extracted from 140 μl of serum using the QIAamp Viral RNA Mini Kit (Qiagen) following the manufacturer’s instructions. GBV-C RNA was detected by nested reverse transcriptase polymerase chain reaction (RT-PCR) using primers corresponding to the 5′UTR as previously described (Schwarze-Zander et al., 2006). PCR products were analysed by agarose gel electrophoresis for the presence of a 256-nucleotide band corresponding to nucleotides 107–362 of GenBank accession number AY196904. Complete ORF sequences were amplified as overlapping fragments using multiple primers as outlined in Table S1a, b (available in the online Supplementary Material).
Sequences were aligned with a database reference using Clustal X 2.1 (Larkin et al., 2007). The GenBank reference sequences used to confirm GBV-C genotype included AB013500, U36380 and KC618399 (genotype 1); U63715, AF121950, AF309966, AF031827, AF031828, AB003289, U44402, AF081782, D87255, AY196904, AF104403, D90600 and U45966 (genotype 2); AB003288, AB003293, D87263, D87262, D87712, D87708, D90601, D87711, D87709, AB008342, AB008335, AB003290, D87710, D87713, AF006500 and U94695 (genotype 3); HQ331233, HQ331234, HQ331235, AB003292, AB21287 and AB18667 (genotype 4); KC618398, KC618400, KC618401 and AY949771 (genotype 5); AB008336 and AB003291 (genotype 6). Putative recombinants included D87715, U75356, AB013501 and AB021287. Outlier sequences included AF070476 – isolated from a chimpanzee – and GB virus A references AF023425, NC001837, U22303 and AF023424. The statistical robustness and reliability of the branching order within the phylogenetic tree was confirmed by bootstrap analysis using 1000 replicates. Additional phylogenetic inference was performed using a Bayesian Markov chain Monte Carlo (MCMC) approach as implemented in the Bayesian Evolutionary Analysis by Sampling Trees (BEAST) v1.8.0 program (Drummond et al., 2012) under an uncorrelated log-normal relaxed molecular clock and the generalized time-reversible (GTR) model with nucleotide site heterogeneity estimated using a gamma distribution. The BEAST MCMC analysis was run for a chain length of 200 000 000 to yield sample size values >500 indicating sufficient sampling. The maximum clade credibility tree was selected from the posterior tree distribution after a 10 % burn-in using TreeAnnotator v1.7.5. Pairwise distances were calculated in MEGA v7.0.
To identify possible recombination events, bootscanning analysis of complete ORF sequences was performed as implemented in SimPlot version 3.5.1 using the Kimura 2-parameter with a 600 base pair (bp) window, a 20 bp step increment and 1000 bootstrap replicates (Lole et al., 1999). Each complete ORF sequence was compared to consensus sequences generated using available GenBank references for genotypes 1 (n=3), 2 (n=13), 3 (n=16), 4 (n=5) and 5 (n=4). Because of the limited number of sequences available for genotype 6, reference AB003291 was included. If >80 % of the permuted trees showed similarity to more than one genotype across the ORF analysed, the ‘parental’ sequences were retained within a second bootscanning analysis along with the consensus genotype 4 sequence as an outlier, since no GBV-C isolates from this cohort were identified as belonging to genotype 4 and the query sequence. Sequences were submitted to GenBank under the accession numbers KU685420–KU685423.
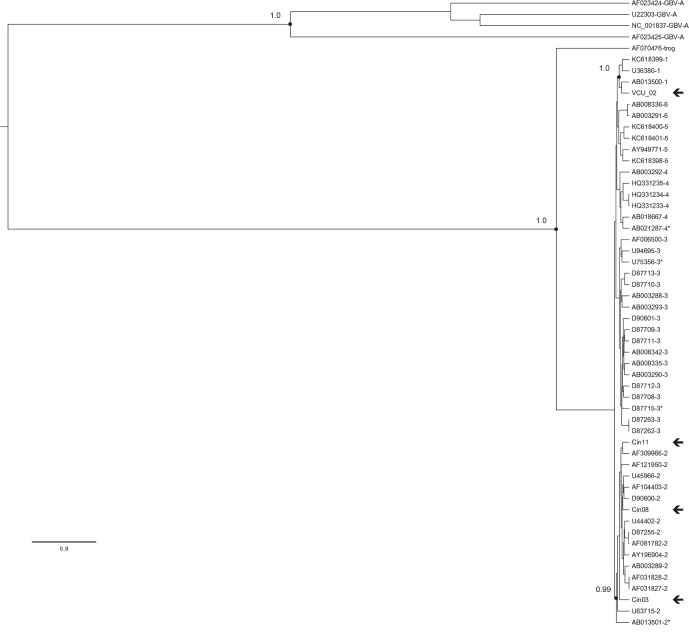
GBV-C RNA was detected in 5 of 12 (41.7 %) HIV/HCV co-infected individuals screened for GBV-C infection. Overlapping genomic fragments of varying length were generated using multiple primer combinations, although fragments were not always positive for every sample (Table S1a, b). Full-length ORF sequences were obtained for four of five individuals; however, a small fragment of Cin06 did not provide high-quality sequence data and was removed from further analysis. Using a Bayesian inference approach, three samples – Cin03, Cin08 and Cin11 – were identified as belonging to GBV-C genotype 2 (Fig. 1). One sample – VCU02 – belonged to GBV-C genotype 1. Pairwise comparisons showed that VCU02 was 10.3 %–11.8 % different from genotype 1 references. Cin03, Cin08 and Cin11 were 7.8 %–11.6 % different from genotype 2 references and did not cluster amongst themselves. The primary purpose of the original cohort was not to evaluate the impact of GBV-C on HIV disease progression; thus, comparison of demographic or clinical variables based on GBV-C RNA status was not possible. As shown in Table S2, GBV-C RNA-positive individuals included two black males, two white females and one black female. At the time of GBV-C RNA amplification, CD4 cell counts for these individuals ranged from 376 to 744, while HIV viral loads ranged from undetectable to 2437.
Fig. 1.
Phylogenetic inference of complete open reading frame sequences for Cin03, Cin08, Cin11 and VCU02 (denoted by ←) based on a Bayesian MCMC approach as implemented in the BEAST program. GenBank reference sequences are indicated by their accession numbers and genotype. Relevant posterior probabilities are shown. The scale bar indicates 0.9 nucleotide substitutions per site. Previously identified recombinant sequences are denoted by an asterisk.
We and others have reported recombination among GBV-C genotypes (Neibecker et al., 2011; Worobey & Holmes, 2001). Thus, the four full-length ORF sequences were analysed for recombination events. As shown in Fig. 2(a, b), Cin03 and Cin08 were non-recombinant genotype 2 isolates. In contrast, Cin11 was identified as a genotype 2 → 1 → 2 recombinant isolate, while VCU02 was shown to be a genotype 1 → 2 → 1 → 2 → 1 recombinant isolate (Fig. 2c, d).
Fig. 2.
Bootscanning analysis of recombination for complete ORF sequences using a 600 bp window, a 20 bp step increment and 1000 bootstrap replicates. Genotypes 1 and 2 are shown as the red and green lines, respectively, while the grey line represents the outlier genotype 4. The dashed line indicates the 80 % threshold used to denote significance between genotypes. (a) Cin03 is a non-recombinant genotype 2 isolate. (b) Cin08 is a non-recombinant genotype 2 isolate. (c) Cin11 is a genotype 2 → 1 → 2 recombinant isolate. (d) VCU02 is a genotype 1 → 2 → 1 → 2 → 1 recombinant isolate.
Genotypic variation among RNA viruses can have profound biological consequences. The existence of multiple GBV-C genotypes has led several authors to suggest that differences in GBV-C strains circulating within populations might impact HIV disease (Berzsenyi et al., 2005; Kaye et al., 2005; Muerhoff et al., 2003). For instance, Muerhoff et al. reported that CD4 cell counts were lower in subjects infected with GBV-C subtype 2a compared to those with subtype 2b (Muerhoff et al., 2003). We observed a significant difference in CD4 cell counts in HIV-positive persons co-infected with GBV-C genotype 2 compared to GBV-C genotype 1 even after controlling for race, HIV viral load and ART use (Schwarze-Zander et al., 2006). GBV-C genotype 2 was also marginally more sensitive to interferon-based HCV therapy than genotype 1. Similarly, another study reported lower CD4 cell counts associated with genotype 1 compared to genotype 2b (Alcalde et al., 2010). However, in a recent study, no statistical difference in CD4 cell counts was found among HIV/HCV co-infected persons based on GBV-C genotype, although the predominance of a single genotype did not permit a rigorous multi-genotype comparison (Berzsenyi et al., 2009).
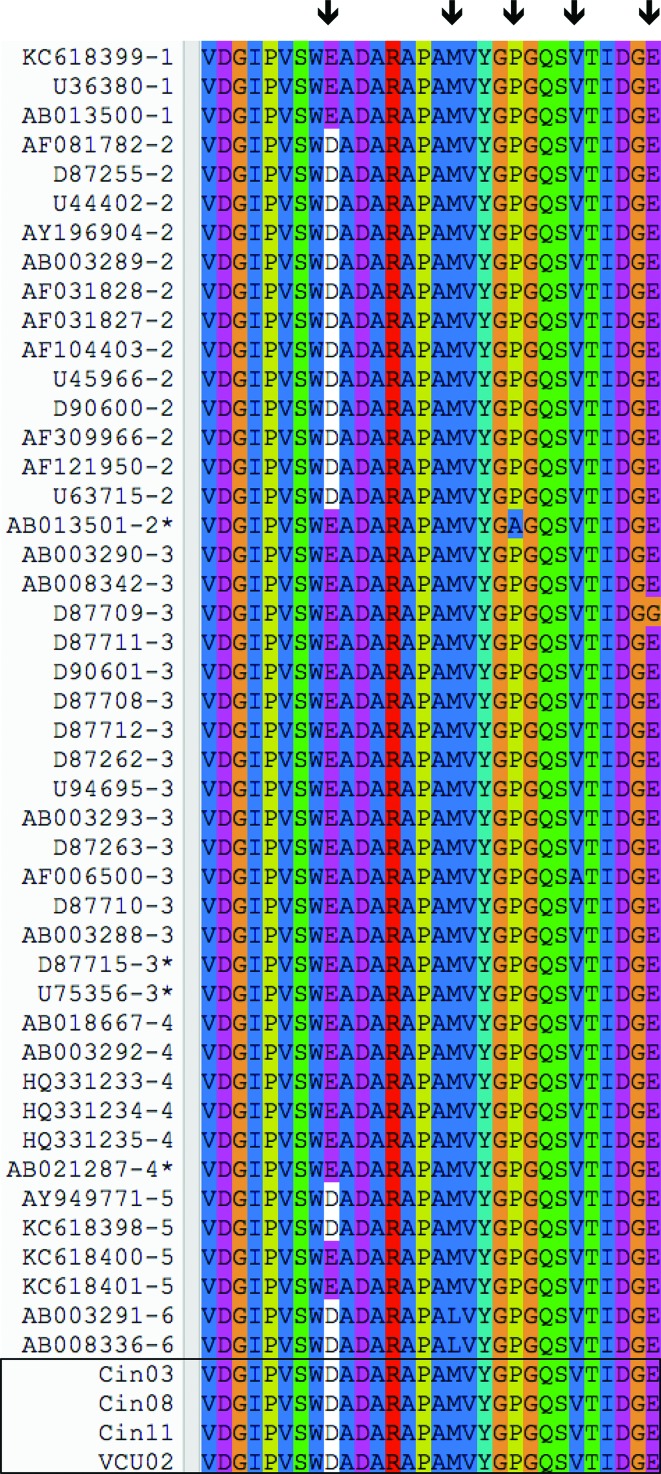
Whether GBV-C genotypes differ in their biological phenotypes is unclear as in vitro comparison of viral genotypes is rare, and only a single GBV-C molecular clone is currently available (Xiang et al., 2000). Nevertheless, several studies have demonstrated that sensitivity to interferon and cell tropism differ among GBV-C variants (Fogeda et al., 2000; Kato et al., 1998; Shimizu et al., 1999; Xiang et al., 2005). For instance, Xiang et al. demonstrated that the NS5A protein from an interferon-resistant individual inhibited RNA-activated protein kinase (PKR) in vitro, while an interferon-sensitive NS5A did not inhibit PKR function (Xiang et al., 2005). Subsequent analysis identified a 30-amino acid segment of NS5A that was sufficient to inhibit HIV replication (Chang et al., 2007; Xiang et al., 2006). This region is relatively well conserved among the existing full-length GBV-C references, as well as among the additional sequences described in the current analysis (Fig. 3). However, several polymorphic sites are present; thus, the impact of viral variability and the roles of genotypically diverse GBV-C proteins in regulating other cellular pathways require additional evaluation. Importantly, clinical GBV-C isolates also vary in their ability to persist in culture, and sequence variability in key regulatory regions may affect growth in PBMC cultures (George et al., 2003).
Fig. 3.
The 30-amino acid segment of NS5A that inhibits HIV replication is relatively well conserved among the existing full-length GBV-C references (shown by their accession number and genotype) published to date and the additional sequences described in the current analysis (boxed sequences). Asterisks denote previously reported recombinant sequences, while arrows denote polymorphic sites.
Inter-genotypic recombination was evident in two of the four full-length ORF sequences analysed in the current study. Similarly, a previous study conducted by Worobey et al. demonstrated that recombination occurs within and between GBV-C genotypes, thus highlighting the important role played by recombination in shaping GBV-C diversity (Worobey & Holmes, 2001). While the contribution of recombination to GBV-C pathogenesis itself has not been examined, studies of other highly recombinogenic viruses such as HIV (reviewed by Blackard et al., 2002) suggest that GBV-C recombination may have important implications for cell tropism, virulence and drug resistance/sensitivity, and may influence the impact of GBV-C on HIV disease progression. However, the primary purpose of the cohort from which these sequences were derived was not to evaluate the impact of GBV-C on HIV disease progression; thus, comparison of demographic or clinical variables based on GBV-C RNA status is not possible, although this has been investigated in other studies as noted above.
Additional analysis of full-length genomes would facilitate the creation of full-length, replication-competent molecular clones of GBV-C to evaluate the phenotypic diversity of GBV-C genotypes and explore the consequences of viral recombination in more depth.
Acknowledgements
This work was supported by the National Institute of Allergy and Infectious Diseases (award AI081564 to JTB and award AI065256 to KES).
Supplementary Data
Supplementary File 1
References
- Alcalde R., Nishiya A., Casseb J., Inocêncio L., Fonseca L. A., Duarte A. J.(2010). Prevalence and distribution of the GBV-C/HGV among HIV-1-infected patients under anti-retroviral therapy. Virus Res 151148–152. 10.1016/j.virusres.2010.04.008 [DOI] [PubMed] [Google Scholar]
- Berzsenyi M. D., Bowden D. S., Roberts S. K.(2005). GB virus C: insights into co-infection. J Clin Virol 33257–266. 10.1016/j.jcv.2005.04.002 [DOI] [PubMed] [Google Scholar]
- Berzsenyi M. D., Bowden D. S., Roberts S. K., Revill P. A.(2009). GB virus C genotype 2 predominance in a hepatitis C virus/HIV infected population associated with reduced liver disease. J Gastroenterol Hepatol 241407–1410. 10.1111/j.1440-1746.2009.05920.x [DOI] [PubMed] [Google Scholar]
- Birk M., Lindbäck S., Lidman C.(2002). No influence of GB virus C replication on the prognosis in a cohort of HIV-1-infected patients. AIDS 162482–2485. 10.1097/00002030-200212060-00017 [DOI] [PubMed] [Google Scholar]
- Björkman P., Flamholc L., Nauclér A., Molnegren V., Wallmark E., Widell A.(2004). GB virus C during the natural course of HIV-1 infection: viremia at diagnosis does not predict mortality. AIDS 18877–886. 10.1097/00002030-200404090-00005 [DOI] [PubMed] [Google Scholar]
- Blackard J. T., Cohen D. E., Mayer K. H.(2002). Human immunodeficiency virus superinfection and recombination: current state of knowledge and potential clinical consequences. Clin Infect Dis 341108–1114. 10.1086/339547 [DOI] [PubMed] [Google Scholar]
- Chang Q., McLinden J. H., Stapleton J. T., Sathar M. A., Xiang J.(2007). Expression of GB virus C NS5A protein from genotypes 1, 2, 3 and 5 and a 30 aa NS5A fragment inhibit human immunodeficiency virus type 1 replication in a CD4+ T-lymphocyte cell line. J Gen Virol 883341–3346. 10.1099/vir.0.83198-0 [DOI] [PubMed] [Google Scholar]
- Drummond A. J., Suchard M. A., Xie D., Rambaut A.(2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 291969–1973. 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Zhao W., Feng Y., Dai J., Li Z., Zhang X., Liu L., Bai J., Zhang H., et al. (2011). A novel genotype of GB virus C: its identification and predominance among injecting drug users in Yunnan, China. PLoS One 6e21151. 10.1371/journal.pone.0021151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogeda M., López-Alcorocho J. M., Bartolomé J., Arocena C., Martín M. A., Carreño V.(2000). Existence of distinct GB virus C/hepatitis G virus variants with different tropism. J Virol 747936–7942. 10.1128/JVI.74.17.7936-7942.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George S. L., Xiang J., Stapleton J. T.(2003). Clinical isolates of GB virus type C vary in their ability to persist and replicate in peripheral blood mononuclear cell cultures. Virology 316191–201. 10.1016/S0042-6822(03)00585-3 [DOI] [PubMed] [Google Scholar]
- Katayama K., Kageyama T., Fukushi S., Hoshino F. B., Kurihara C., Ishiyama N., Okamura H., Oya A.(1998). Full-length GBV-C/HGV genomes from nine Japanese isolates: characterization by comparative analyses. Arch Virol 1431063–1075. 10.1007/s007050050356 [DOI] [PubMed] [Google Scholar]
- Kato T., Mizokami M., Nakano T., Orito E., Ohba K., Kondo Y., Tanaka Y., Ueda R., Mukaide M., et al. (1998). Heterogeneity in E2 region of GBV-C/hepatitis G virus and hepatitis C virus. J Med Virol 55109–117. [DOI] [PubMed] [Google Scholar]
- Kaye S., Howard M., Alabi A., Hansmann A., Whittle H., Schim van der Loeff M.(2005). No observed effect of GB virus C coinfection on disease progression in a cohort of African woman infected with HIV-1 or HIV-2. Clin Infect Dis 40876–878. 10.1086/428123 [DOI] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 232947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Lole K. S., Bollinger R. C., Paranjape R. S., Gadkari D., Kulkarni S. S., Novak N. G., Ingersoll R., Sheppard H. W., Ray S. C.(1999). Full-length human immunodeficiency virus type 1 genomes from subtype c-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr E. L., Stapleton J. T.(2009). GB virus type C interactions with HIV: the role of envelope glycoproteins. J Viral Hepat 16757–768. 10.1111/j.1365-2893.2009.01194.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muerhoff A. S., Simons J. N., Erker J. C., Desai S. M., Mushahwar I. K.(1996). Identification of conserved nucleotide sequences within the GB virus C 5′-untranslated region: design of PCR primers for detection of viral RNA. J Virol Methods 6255–62. 10.1016/0166-0934(96)02088-5 [DOI] [PubMed] [Google Scholar]
- Muerhoff A. S., Smith D. B., Leary T. P., Erker J. C., Desai S. M., Mushahwar I. K.(1997). Identification of GB virus C variants by phylogenetic analysis of 5′-untranslated and coding region sequences. J Virol 716501–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muerhoff A. S., Tillmann H. L., Manns M. P., Dawson G. J., Desai S. M.(2003). GB virus C genotype determination in GB virus-C/HIV co-infected individuals. J Med Virol 70141–149. 10.1002/jmv.10375 [DOI] [PubMed] [Google Scholar]
- Muerhoff A. S., Leary T. P., Sathar M. A., Dawson G. J., Desai S. M.(2005). African origin of GB virus C determined by phylogenetic analysis of a complete genotype 5 genome from South Africa. J Gen Virol 861729–1735. 10.1099/vir.0.80854-0 [DOI] [PubMed] [Google Scholar]
- Muerhoff A. S., Dawson G. J., Desai S. M.(2006). A previously unrecognized sixth genotype of GB virus C revealed by analysis of 5′-untranslated region sequences. J Med Virol 78105–111. 10.1002/jmv.20510 [DOI] [PubMed] [Google Scholar]
- Mukaide M., Mizokami M., Orito E., Ohba K., Nakano T., Ueda R., Hikiji K., Iino S., Shapiro S., et al. (1997). Three different GB virus C/hepatitis G virus genotypes. Phylogenetic analysis and a genotyping assay based on restriction fragment length polymorphism. FEBS Lett 40751–58. 10.1016/S0014-5793(97)00136-1 [DOI] [PubMed] [Google Scholar]
- Neibecker M., Schwarze-Zander C., Rockstroh J. K., Spengler U., Blackard J. T.(2011). Evidence for extensive genotypic diversity and Recombination of GB virus C (GBV-C) in Germany. J Med Virol 83685–694. 10.1002/jmv.22029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H., Nakao H., Inoue T., Fukuda M., Kishimoto J., Iizuka H., Tsuda F., Miyakawa Y., Mayumi M.(1997). The entire nucleotide sequences of two GB virus C/hepatitis G virus isolates of distinct genotypes from Japan. J Gen Virol 78737–745. 10.1099/0022-1317-78-4-737 [DOI] [PubMed] [Google Scholar]
- Schwarze-Zander C., Blackard J. T., Zheng H., Addo M. M., Lin W., Robbins G. K., Sherman K. E., Zdunek D., Hess G., Chung R., AIDS Clinical Trial Group A5071 Study Team (2006). GB virus C (GBV-C) infection in hepatitis C virus (HCV)/HIV-coinfected patients receiving HCV treatment: importance of the GBV-C genotype. J Infect Dis 194410–419. 10.1086/505713 [DOI] [PubMed] [Google Scholar]
- Sherman K. E., Guedj J., Shata M. T., Blackard J. T., Rouster S. D., Castro M., Feinberg J., Sterling R. K., Goodman Z., et al. (2014). Modulation of HCV replication after combination antiretroviral therapy in HCV/HIV co-infected patients. Sci Transl Med 6246ra98. 10.1126/scitranslmed.3008195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y. K., Hijikata M., Kiyohara T., Kitamura Y., Yoshikura H.(1999). Replication of GB virus C (hepatitis G virus) in interferon-resistant Daudi cells. J Virol 738411–8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton J. T.(2003). GB virus type C/Hepatitis G virus. Semin Liver Dis 23137–148. 10.1055/s-2003-39943 [DOI] [PubMed] [Google Scholar]
- Tillmann H. L., Heiken H., Knapik-Botor A., Heringlake S., Ockenga J., Wilber J. C., Goergen B., Detmer J., McMorrow M., et al. (2001). Infection with GB virus C and reduced mortality among HIV-infected patients. N Engl J Med 345715–724. 10.1056/NEJMoa010398 [DOI] [PubMed] [Google Scholar]
- Tucker T. J., Smuts H., Eickhaus P., Robson S. C., Kirsch R. E.(1999). Molecular characterization of the 5′ non-coding region of South African GBV-C/HGV isolates: major deletion and evidence for a fourth genotype. J Med Virol 5952–59. [DOI] [PubMed] [Google Scholar]
- Tucker T. J., Smuts H. E.(2000). GBV-C/HGV genotypes: proposed nomenclature for genotypes 1–5. J Med Virol 6282–83. [DOI] [PubMed] [Google Scholar]
- Williams C. F., Klinzman D., Yamashita T. E., Xiang J., Polgreen P. M., Rinaldo C., Liu C., Phair J., Margolick J. B., et al. (2004). Persistent GB virus C infection and survival in HIV-infected men. N Engl J Med 350981–990. 10.1056/NEJMoa030107 [DOI] [PubMed] [Google Scholar]
- Worobey M., Holmes E. C.(2001). Homologous recombination in GB virus C/hepatitis G virus. Mol Biol Evol 18254–261. 10.1093/oxfordjournals.molbev.a003799 [DOI] [PubMed] [Google Scholar]
- Xiang J., Wünschmann S., Schmidt W., Shao J., Stapleton J. T.(2000). Full-length GB virus C (Hepatitis G virus) RNA transcripts are infectious in primary CD4-positive T cells. J Virol 749125–9133. 10.1128/JVI.74.19.9125-9133.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang J., Martinez-Smith C., Gale M., Chang Q., Labrecque D. R., Schmidt W. N., Stapleton J. T.(2005). GB virus type C NS5A sequence polymorphisms: association with interferon susceptibility and inhibition of PKR-mediated eIF2alpha phosphorylation. J Interferon Cytokine Res 25261–270. 10.1089/jir.2005.25.261 [DOI] [PubMed] [Google Scholar]
- Xiang J., McLinden J. H., Chang Q., Kaufman T. M., Stapleton J. T.(2006). An 85-aa segment of the GB virus type C NS5A phosphoprotein inhibits HIV-1 replication in CD4+ Jurkat T cells. Proc Natl Acad Sci U S A 10315570–15575. 10.1073/pnas.0604728103 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1