Abstract
Extra-hepatic compartments might contribute to hepatitis C virus (HCV) persistence and extra-hepatic manifestations. Therefore, we investigated HCV infection in colonic tissue in patients with chronic hepatitis C (CHC) and its relationship with HCV pathogenesis. Colonic biopsies were collected from three groups with CHC infection: treatment naïve (TN; n=12), non-responders (NR; n=10) to anti-HCV therapy (pegylated interferon-α and ribavirin) and sustained virologic response (SVR; n=10) and from a fourth healthy control group (n=10). Liver biopsies were examined to assess inflammation and fibrosis. HCV infection and colonic T regulatory (Treg) frequency were detected by immunohistochemistry. HCV core and NS3 proteins were detected in B cells and macrophage/monocytes of 42 % and 25 % of TN and 50 % and 30 % of NR, respectively, but not in SVR or control group. The numbers of cells expressing HCV proteins were positively correlated with both HCV viral load and colonic Treg frequency. A significant negative correlation between HCV-expressing cells with both liver inflammation and fibrosis was identified. Our study provides evidence that HCV can infect B cells and macrophages of the colon. The correlations between HCV infection in colonic tissue and HCV viral load and liver pathology underline the significance of this extra-hepatic infection in HCV pathogenesis and response to therapy.
Keywords: HCV, Extra-hepatic infection, HCV NS3, Liver pathology.
Introduction
In Egypt, it is estimated that 15 % of Egyptians have serologic evidence of hepatitis C virus (HCV) infection (Frank et al., 2000). HCV is also a major cause of chronic liver infection that often leads to chronic hepatitis which may progress to cirrhosis and hepatocellular carcinoma (Kuo et al., 1989; Revie & Salahuddin, 2011).
HCV is an ssRNA virus which produces negative strand RNA as a replicative intermediate. During the HCV replication cycle, one large precursor protein is synthesized from an open reading frame then cleaved to produce structural and non-structural (NS) proteins (Griffin et al., 2003; Hetta, 2014; Revie & Salahuddin, 2011). NS proteins are not found in the virion. Therefore, presence of NS proteins inside cells suggests that HCV infection occurred in those cells (Revie & Salahuddin, 2011).
Although HCV is primarily a hepatotropic virus, a broad spectrum of extra-hepatic manifestations may be associated with HCV infection (Agnello & De Rosa, 2004; Blackard et al., 2006; Gumber & Chopra, 1995; Mayo, 2003; Sène et al., 2004). It is not known if these effects are mediated by direct tissue infection or via soluble proteins.
HCV was believed to infect only hepatocytes (Revie & Salahuddin, 2011). However, recent reports have reported HCV infection in other cell types such as peripheral blood mononuclear cells (PBMC) (Castillo et al., 2005; Chang et al., 1996; Manzin et al., 1994; Saleh et al., 1994; Wang et al., 1992), thyrocytes (Blackard et al., 2013), endothelial cells (Fletcher et al., 2012), B cells, T cells, monocytes, macrophages and macrophage-like cells such as Kupffer cells and dendrocytes (Castillo et al., 2005). In addition, it has been proposed that PBMC could serve as privileged reservoirs for HCV, a possible source of recurrent HCV infection after liver transplantation (Feray et al., 1992).
Immunohistochemistry (IHC) detection of HCV NS proteins was used successfully for analysis of HCV infection in tissues (Gouda et al., 2010; Yan et al., 2000b). The IHC studies were used successfully to identify the extra-hepatic sites for HCV infection in many tissues such as intestinal tissues (Deforges et al., 2004), pancreas (Yan et al., 2000b), kidney (Sansonno et al., 1997), adrenal glands (Yan et al., 2000b), spleen (Yan et al., 2000b), gall bladder (Yan et al., 2000b), heart (Yan et al., 2000a) and lymph nodes (Pal et al., 2006). Gut immune cells are intrinsically linked to the liver responses because there is a shared lymphocyte re- circulation between the gut and liver through the portal circulation (Adams et al., 2008; Gualdi et al., 1996). Additionally, the liver is considered an important toleragenic organ for all foreign proteins consumed. The tolerance effect is probably mediated through the T regulatory (Treg) cells which are shared between the gut and the liver (Adams et al., 2008; Gualdi et al., 1996). Almost all gut mucosal lymphocytes which provide protection against gut pathogens enter the liver (Adams et al., 2008). In this study, we investigated extra-hepatic infection of HCV in human colonic tissue from HCV-infected patients by IHC and identified the relationship between HCV infection in the colonic tissue and HCV pathogenesis, including the frequency of colonic Treg, outcome of anti-HCV therapy, viral persistence and degree of liver inflammation.
Methods
Patients’ recruitment.
A hospital-based cross-sectional study was conducted to detect extra-hepatic infection of HCV in colonic tissue and relationship with HCV pathogenesis in chronic hepatitis C (CHC) patients. All participants were recruited from Assiut Liver Institute for Treatment of Hepatitis C Virus and Assiut University Hospitals outpatient clinics, Assiut, Egypt, between January 2011 and September 2012. Verbal and signed consent forms were obtained from participants according to an institutional review board protocol approved by Assiut University, College of Medicine Institutional Review Board.
Inclusion criteria for the patients were positive for HCV antibodies by ELISA and HCV RNA by real-time PCR without selection by race, age or gender. Exclusion criteria were pregnancy, history of Schistosoma infection, inflammatory bowel diseases or suspected inflammatory bowel diseases, autoimmune diseases including rheumatoid arthritis and any patients on systemic immunomodulators.
Patients with CHC were grouped into four groups: treatment naïve (TN; n=12), non-responders (NR) to therapy (n=10) and patients with sustained virologic response (SVR) to therapy (n=10). The fourth group was healthy control subjects (n=10) who were non-HCV-infected, had a routine colonoscopic examination for colon cancer screening and were negative. All liver biopsies were taken before the start of therapy. In the TN group, colon biopsies and blood samples were taken at the same time of liver biopsy. In NR, colon biopsies and blood samples were taken 6 months after the start of therapy. The time gap between liver biopsies and both blood and colon biopsy in SVR was approximately 1.5 year. Therapy for CHC was performed according to the standard of care in Egypt during the time this study was conducted. It consisted of pegylated interferon-α and ribavirin. A detailed study flow chart for extra-hepatic replication of HCV in the colon tissue and its relationship with HCV pathogenesis is supplied as a supplementary file (available as Supplementary Material with the online version of the paper).
Colonic tissue biopsies.
After proper colonic preparation, three rectal biopsies (from the descending colon, 1–3 mm in size using biopsy forceps) were obtained from each patient and controls via flexible sigmoidoscopy or colonoscopy.
Cell lines.
The Huh7.5 cell line – generated by curing an HCV replicon-containing cell line with interferon (Blight et al., 2002) – was provided by Apath LLC. The Huh7.5JFH1 cell line – which produces infectious HCV genotype 2a virions – was provided by Dr J. Blackard (Kong et al., 2012).
Serological and liver function tests.
Liver function tests (serum alanine transaminase [ALT], aspartate transaminase [AST] and albumin) were performed using a chemical analyser Hitachi 911 (Boehringer Mannheim). Serological markers such as HBsAg, anti-HCV and anti-HIV were tested using commercially available microparticle enzyme immunoassay kits (AXSYM; Abbott Laboratories) as specified by the manufacturer.
Determination of HCV viral load by real-time PCR.
HCV viral loads were determined in patients’ plasma using real-time PCR as previously described (Hetta et al., 2015). Briefly, RNA was extracted from 140 µl of patient serum with a viral RNA Mini Kit (catalogue no. 52904; Qiagen), according to the manufacturer’s instructions. Quantitative measurement of HCV RNA in real time was detected using a PCR kit (artus® HCV RG RTPCR supplied by Qiagen, catalogue no. 4518263) as specified by the manufacturer.
Assessment of liver inflammation and fibrosis.
Liver biopsy specimens were obtained from all patients included in the study and examined to assess inflammatory score and fibrosis stage. Hepatic histopathologic findings were interpreted independently of clinical and biochemical data by a single pathologist, according to the criteria described by the METAVIR scoring system.
Demonstration of extra-hepatic infection of HCV in colonic tissue.
Extra-hepatic infection of HCV was examined in colonic tissue using fluorescent IHC as previously described for detection of HCV core and HCV NS3 proteins (Gouda et al., 2010). Briefly, colon biopsy samples were fixed in 10 % buffered formalin, dehydrated in ethanol and embedded in paraffin. Tissue sections (5 µm thickness) were treated with sodium citrate buffer for antigen retrieval, and the non-specific sites were blocked with PBS containing 10 % normal donkey serum and 3 % IgG-free, protease-free bovine serum albumin (BSA). Tissue sections were incubated either with monoclonal mouse anti-Hepatitis C Virus Core 1b (C7-50) antibody (ab2740; Abcam) diluted 1 : 100 or mouse monoclonal anti-HCV NS3 antibody (clone MMM33) (Novocastra) diluted 1 : 50 in PBS containing 0.05 % Tween 20 (PBST) and 1 % BSA. Then the slides were incubated for 1 h at room temperature with Alexa Fluor® 488 Donkey Anti-Mouse (Jackson Immunoresearch) diluted to 1 : 100 in PBST with 1 % BSA.
To identify the cell type where HCV replicates, the sections were incubated with two primary antibodies in a sequential manner. First, the slides were incubated with mouse monoclonal anti-HCV NS3 antibody and then were incubated with either rabbit monoclonal anti-CD3 antibody (clone SP97) (catalogue no. ab99964; Abcam) or rabbit monoclonal anti-CD19 antibody or rabbit monoclonal anti-CD14 antibody (clone SP97) (catalogue no. ab99964; Abcam). The slides were washed extensively, then incubated for 1 h at room temperature in a sequential manner with two secondary antibodies, Alexa Fluor® 594 Donkey Anti-Rabbit (Jackson Immunoresearch) and Alexa Fluor® 488 Donkey Anti-Mouse diluted 1 : 100 in PBST with 1 % BSA. Negative controls were performed by omitting the primary antibodies. 4′,6-Diamidino-2-phenylindole diluted 1 : 1000 was used for nuclear staining. The slides were examined by Zeiss LASER scanning confocal microscopy (LSM710) with high-power fields (HPF) (1 HPF = 0.4 mm2). The presence of HCV antigens in colonic tissue was examined, and the number of cells expressing HCV core protein and NS3 was quantified in HCV-infected patients and matched normal healthy controls. Positive cells were quantified in 10 HPFs of colonic tissues. Data were presented as total number of positive cells/10 HPFs.
Examination of the frequency of Treg cells in colonic tissue.
The frequency of Treg cells in colonic biopsies was evaluated using fluorescent IHC as previously described (Hetta et al., 2015). Briefly, two primary antibodies from Abcam, mouse monoclonal anti-CD3 antibody (clone PS1) (catalogue no. ab699) and rabbit monoclonal anti-Forkhead box protein P3 (FoxP3) antibody (clone SP97) (catalogue no. ab99964), were incubated in a sequential manner with two secondary antibodies from Jackson Immunoresearch, Alexa Fluor® 594 Donkey Anti-Rabbit, and Alexa Fluor® 488 Donkey Anti-Mouse. The number of colonic Treg and CD3+ T cells was recorded using Zeiss LASER scanning confocal microscopy (LSM710) with HPF. Ten HPFs were examined in each slide and averaged. Data were calculated as % frequency of Treg as follows:
Statistical analysis.
Data were given as range (minimum, maximum) or mean±sd. The Student t-test was used to examine the difference between two groups with a significance value at P≤0.05. The ANOVA test was used to examine the difference between groups of the enrolled subjects. Correlations between parameters measured were calculated using Spearman’s correlation coefficient for patients and controls.
Results
Characterization of the enrolled subjects
Demographic and clinical characteristics of the enrolled subjects are summarized in Table 1. The healthy control subjects were significantly older than TN, NR and SVR groups (P=0.002, P=0.005 and P=0.002, respectively), while there was no significant age difference among TN, NR and SVR groups. The healthy controls included more females than the TN group (P=0.02), while there was no significant difference in gender between the control group compared to the NR or SVR group and no significant difference among the TN, NR and SVR groups.
Table 1. Demographic and clinical data characteristics of the enrolled subjects.
| Demographic characteristics | TN, n=12 | NR, n=10 | SVR, n=10 | Controls, n=10 |
|---|---|---|---|---|
| Sex, male : female | 9 : 1 | 9 : 1 | 8 : 2 | 4 : 6 |
| Age, range (Avg±sd) | 21–55 (43±9.7) | 21–54 (44.3±9.66) | 20–51 (38.3±9.97) | 42–64 (56±7.15) |
| HCV viral load (106 IU ml−1), Avg±sd | 1.44±1.43 | 1.34±0.94 | na | na |
| ALT (IU ml−1), Avg ± sd | 69.5±17.40 | 47.5±17.44 | 45.8±29.9 | 21±7.4 |
| AST (IU ml−1), Avg ± sd | 54.9±24 | 34.6±14.2 | 31.4±26.6 | 23±11.6 |
| Distribution of liver inflammatory grade | ||||
| Stage 1–2 | 6 | 7 | 6 | na |
| Stage 3–4 | 6 | 3 | 4 | |
| Distribution of liver fibrosis stage | ||||
| Stage 1–2 | 8 | 8 | 8 | na |
| Stage 3–4 | 4 | 2 | 2 |
TN, Chronic HCV naïve to therapy; NR, non-responder; SVR, sustained virologic response; Avg, average number; na, not applicable.
Upper normal limit of ALT = 45; upper normal limit of AST = 30.
ALT levels were significantly higher in TN (mean±sd, 69.5±17.40 U ml−1) compared to healthy controls (21±7.4 U ml−1), SVR (45.8±29.9 U ml−1) and NR groups (47.5±17.44 U ml−1) (P<0.0001, P=0.006 and P=0.01, respectively). Also, ALT levels were significantly (P=0.007) higher in the NR compared to the control group, while there were no significant differences in ALT levels between the SVR and healthy control groups. AST levels were significantly higher in the TN group (mean±sd, 54.9±24 U ml−1) compared to healthy controls (23±11.6 U ml−1), SVR (31.4±26.6 U ml−1) and NR groups (34.6±14.2 U ml−1) (P=0.0017, P=0.017 and P=0.05, respectively), while there was no significant difference in AST levels among NR, SVR and healthy control groups (Table 1).
We compared the viral load in CHC and NR. There was no significant difference in HCV viral load between the TN group (mean±sd, 1.44×106±1.43×106 IU ml−1) and the NR group (1.34×106±0.94×106 IU ml−1). There was also no significant difference between all the HCV-infected groups in liver METAVIR inflammatory grade and fibrosis stage before treatment. No liver biopsies were available after anti-HCV treatment or for the healthy controls.
Detection of HCV core protein and NS3 protein in colonic tissue of HCV-infected patients
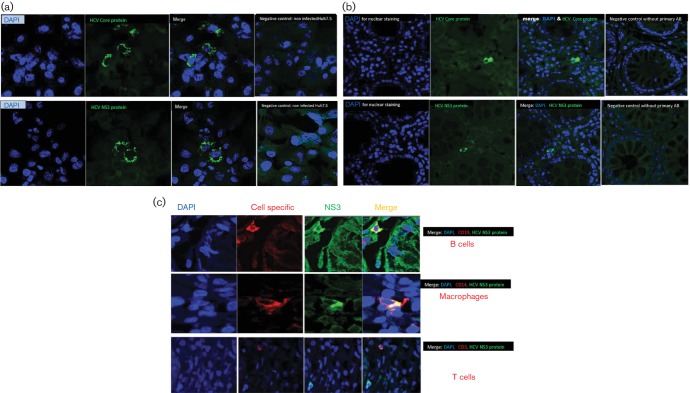
To confirm the feasibility and the specificity of our immunohistochemical staining protocol of HCV core and NS3 proteins, we examined the expression of those proteins in Huh7.5 cell lines infected (Huh7.5JFH1) or uninfected (Huh7.5) with HCV using monoclonal antibodies against HCV core and NS3 proteins. HCV antigens were detected by indirect fluorescent immunohistochemical staining of HCV core and NS3. The cells expressing HCV antigens were identified by the specific green cytoplasmic signals (Alexa Fluor® 488), and no positive staining was observed in HCV-uninfected Huh7.5 cell line (Fig. 1a).
Fig. 1.
(a) Confirmation of the specificity of the immunohistochemistry of HCV core and NS3 antigens using Huh7.5 cell lines. The specificity of the immunohistochemical staining of HCV antigens was confirmed on Huh7.5JFH1 cell line infected with HCV and uninfected Huh7.5 cell line using primary mouse monoclonal antibodies against HCV core and NS3 proteins. The cells expressing HCV antigens were identified by the specific green cytoplasmic signals (Alexa Fluor® 488 Donkey Anti-Mouse secondary antibody). No positive staining was observed in HCV-uninfected Huh7.5 cell line. (b) Immunohistochemical detection of extra-hepatic infection of HCV in colonic tissue of HCV-infected patients: HCV core protein and NS3 as markers of HCV replication were detected by indirect fluorescent immunohistochemical staining and confocal imaging using mouse monoclonal anti-core antibody and mouse monoclonal anti-HCV NS3 antibody. Positive cells were identified by the green cytoplasmic signals (by Alexa Fluor® 488 Donkey Anti-Mouse secondary antibody). No positive signals were detected in the negative control without primary antibodies. (c) Identification of the cell types in colonic tissue with extra-hepatic HCV infection. To examine cell types where HCV infects extra-hepatically, double immunofluorescence staining on the colonic tissue was performed. Colonic tissue sections were stained with mouse monoclonal anti-HCV NS3, as a marker of HCV infection, and antibody against a specific cell marker (for T cells [anti-CD3], for B [anti-CD19] lymphocytes and for macrophage/monocytes [anti-CD14]). The cells in which HCV infects were identified by co-expression of NS3 and the cell-specific marker. HCV NS3 was expressed in B cells and macrophage but not in T cells.
To detect the presence of HCV antigens in colonic tissue, we examined the expression of HCV core (structural protein) and NS3 (NS protein) in colonic biopsies from HCV-infected patients and control groups. HCV antigens were detected by indirect fluorescent immunohistochemical staining of HCV core and NS3 using monoclonal anti-HCV core and mouse anti-HCV NS3 antibodies. The cells expressing HCV antigens were identified by the specific green cytoplasmic signals (Fig. 1b).
The HCV core protein and NS3 were detected only in TN and NR groups but neither in SVR nor healthy controls as shown in Table 2. HCV core protein was expressed in 10 patients; of these 10, there were only six patients expressing HCV NS3. The HCV core protein was detected in five of 12 (42 %) in TN and five of ten (50 %) in NR groups but not in SVR or healthy control groups. Additionally, NS3 protein was detected in colonic tissue in three of 12 (25 %) in TN, and three of ten (30 %) in NR groups but not in SVR or healthy control groups.
Table 2. The average number of cells expressing HCV core protein and HCV NS3 in colonic tissue of the study groups.
| TN, n=12 | NR, n=10 | SVR, n=10 | Controls, n=10 | |
|---|---|---|---|---|
| Number of cells containing HCV core protein/10 HPFs (average±sd) |
0.58±0.9 | 0.6±0.7 | 0.0±0.0 | 0.0±0.0 |
| Range | 0–3 | 0–2 | 0 | 0 |
| % of positive samples | 41.67 | 50 | 0 | 0 |
| Number of cells expressing HCV NS3 protein/10 HPF (average±sd) |
1.67±3.37 | 1±1.9 | 0.0±0.0 | 0.0±0.0 |
| Range | 0–10 | 0–5 | 0 | 0 |
| % of positive samples | 25 | 30 | 0 | 0 |
Identification of the colonic cells with HCV protein expression
To identify the cell type in which HCV infects extra-hepatically, we examined the surface markers of the cells in which HCV infects. We used antibodies against a specific marker for T cells (anti-CD3), B cells (anti-CD19) and macrophage/monocytes (anti-CD14). As shown in Fig. 1c, NS3 and core of HCV were expressed mainly in B cells and macrophage/monocytes but not in T cells.
Relationship between the number of cells expressing HCV core and NS3 proteins in colonic tissue and HCV viral load
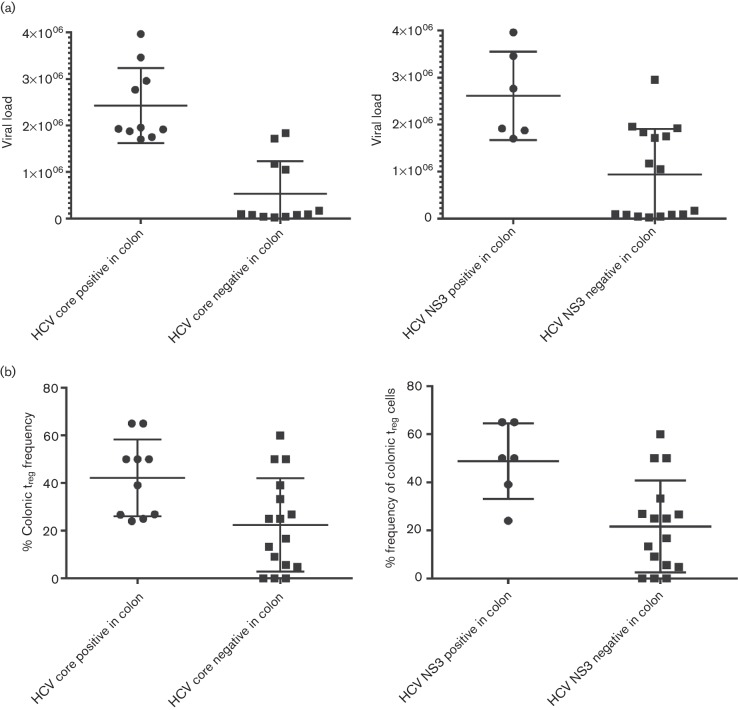
To investigate the impact of HCV infection in colonic tissue on HCV pathogenesis and response to therapy, we examined the relationship between the number of colonic cells expressing HCV core and NS3 proteins with viral load in the TN and NR groups. There was a significant difference (P<0.0001) in the viral load between the groups expressing HCV core and without HCV core expression. Also, there is a significant difference (P=0.007) in the viral load between the groups expressing HCV NS3 and without HCV NS3 expression as shown in Fig. 3a. There was a significant positive correlation between viral load with both the number of cells expressing HCV core protein (P=0.0001 and r=0.663) and the number of cells expressing HCV NS3 (P<0.0001 and r=0.874) in colonic tissue.
Fig. 3.
(a) Relationship between HCV viral load and both the number of cells expressing HCV core and number of cells expressing NS3 proteins in colonic tissue. There was a significant difference (P<0.0001) in the viral load between the groups expressing HCV core and without HCV core expression. Also, there is a significant difference (P=0.007) in the viral load between the groups expressing HCV NS3 and without HCV NS3 expression. (b) Relationship between colonic Treg frequency and the number of cells expressing HCV core and NS3 proteins in colonic tissue. There was a significant difference (P=0.0184) in the colonic Treg frequency between the groups expressing HCV core and without HCV core expression. Also, there is a significant difference (P=0.0121) in the viral load between the groups expressing HCV NS3 and without HCV NS3 expression.
Relationship between colonic Treg and HCV expression in colonic tissues
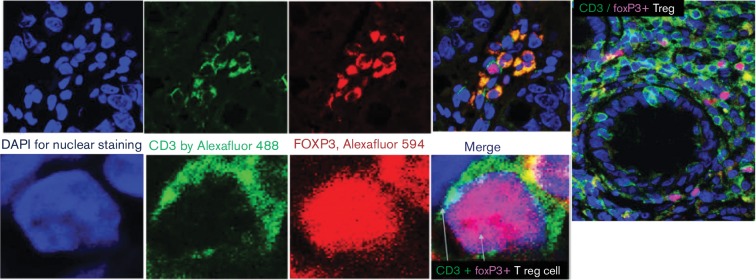
In a previous study by our laboratory (Hetta et al., 2015), colonic Treg cells were detected by indirect fluorescent immunohistochemical staining of CD3 and FoxP3 and correlated with HCV pathogenesis. Treg cells were identified by the green surface CD3 (Alexa Fluor 488) and the red nuclear FoxP3 (Alexa Fluor 594) as demonstrated in Fig. 2. In this study, we examined the relationship between frequency of colonic Treg and HCV infection in colonic tissue. There were a significant difference (P=0.0001) in the absolute number of Treg cells in the group with HCV core expression (average Treg = 7.5 cell/HPF) compared with the group without HCV core expression (average Treg = 1.9 cell/HPF) in the colon and a significant difference (P=0.0002) in the group with NS3 core expression in the colon (average Treg = 9.3 cell/HPF) compared with the group without HCV NS3 expression (average Treg = 1.8 cell/HPF). There was a significant difference (P=0.0184) in the colonic Treg frequency between the groups expressing HCV core and without HCV core expression. Also, there is a significant difference (P=0.0121) in the viral load between the groups expressing HCV NS3 and without HCV NS3 expression as shown in Fig. 3b. A significant positive correlation was identified between the frequency of colonic mucosal Treg and the frequency of cells expressing either HCV core protein (P=0.002 and R=0.61) or HCV NS3 in colonic tissue (P=0.002 and R=0.6).
Fig. 2.
Characterization of colonic Treg by double immunofluorescence staining with anti-CD3 and anti-FoxP3 antibody. Colonic Treg cells were detected by indirect fluorescent immunohistochemical staining of CD3 and FoxP3 using mouse monoclonal anti-CD3 antibody (PS1) and rabbit monoclonal anti-FoxP3 antibody (SP97). Treg cells were identified by the green surface CD3 (by Alexa Fluor® 488 Donkey Anti-Mouse secondary antibody) and the red nuclear FoxP3 (by Alexa Fluor® 594 Donkey Anti-Rabbit secondary antibody). FoxP3 also was expressed intra-cytoplasmically in some proliferation cells and appeared as an orange colour due to the merge between red FoxP3 and green CD3.
Relationship between HCV infection in colonic tissues and liver pathology in TN group
Significant negative correlation was found between the HCV core expressing cells with METAVIR inflammatory grade (P=0.04; r=−0.459; 95 % confidence interval [CI] = −0.738 to −0.046) and fibrosis stage (P=0.04; r=−0.437 with 95 % CI = −0.725 to −0.019).
Additionally, there was a significant negative correlation between the number of cells expressing HCV core and the serum ALT (P=0.037; r=−0.45 with 95 % CI = −0.731 to −0.032), but not AST (P=0.729 and r=−0.078). In contrast, no significant correlations were identified between the number of cells expressing HCV NS3 protein and the scores of liver inflammation, fibrosis and ALT and AST levels
Discussion
In this study, we investigated the extra-hepatic infection of HCV in colonic tissue of HCV-infected patients using IHC techniques. Additionally, the relationships between HCV infection in colonic tissue and the frequency of colonic Treg, outcome of anti-HCV therapy, viral persistence and liver inflammation were examined.
In our study, IHC was used to detect extra-hepatic replication of HCV in human colonic tissue (Liao et al., 2011). Extra-hepatic infections were examined by detection of the NS3 protein and we identified the type of cells where HCV infects by using antibodies against specific cell surface markers such as CD19 for B cells, CD14 for macrophage/monocytes and CD3 for T cells.
HCV infections in colonic tissue of 42 subjects, TN (n=12), NR (n=10), SVR (n=10) and control (n=10) groups, were examined. HCV core protein was detected in 42 % of the TN group (5/12) and 50 % of the NR group (5/10), but not in the SVR (0 %) or healthy control groups (0 %). HCV core protein was expressed in 10 patients; of these 10, only six patients expressed HCV NS3. Since HCV core is present as a free protein in the circulation (Yao et al., 2001) and could be engulfed by cells (Barth et al., 2005), we confirmed extra-hepatic HCV infection data by investigating NS3 expression as a marker of extra-hepatic infection. NS3 is not present as a free protein in the circulation or in the virion (Revie & Salahuddin, 2011), and therefore, identification of NS3 inside the cells is a strong marker of HCV infection (Revie & Salahuddin, 2011). NS3 protein was detected in colonic tissue of 25 % of TN (3/12) and 33 % of NR groups (3/10), but not in SVR or healthy control groups (0 %). Our findings are supported by a previous study by Yan et al. (2000b), who reported an extra-hepatic infection of HCV in the intestine of a sub-population of HCV-infected patients using IHC detection of HCV NS3 (60 %; 3/5), HCV NS5 (40 %; 2/5) or ISH (20 %; 1/5) or RT-PCR for the minus strand (20 %; 1/5). However, the difference between our study and the previous study by Yan et al. is that they detected extra-hepatic infection of HCV in the intestinal epithelial cells but we detected HCV infection in the B cells and macrophage/monocytes and not in the intestinal epithelial cells. The reasons for this difference are not clear but may be due to differences in viral load or the site of biopsies. Miglioresi et al. (2003) analysed HCV gastric localization in 15 patients and compared viraemia with the status of HCV in gastric biopsy specimens and PBMC. They reported that all HCV-infected patients with positive viraemia were positive for the presence of HCV in tissue and PBMC. The finding of a positive hidden compartment for HCV and simultaneous negative viraemia had also been reported by others (McHutchison et al., 2002).
Extra-hepatic infection of HCV has been reported in B cells (Pham et al., 2008; Sansonno et al., 1996), T cells (Pham et al., 2008; Sansonno et al., 1996), monocytes (Sansonno et al., 1996), macrophages (Coquillard & Patterson, 2009; Sansonno et al., 1996), M cells (Pham et al., 2008) and Kupffer cells (Castillo et al., 2005). In our study, we detected extra-hepatic infection of HCV in colonic tissue B cells and macrophage, but not in the T cells, in three of twelve (25 %) in TN and three of ten (30 %) in NR but not in SVR or healthy control group. The reason for this low detection rate may be due to the limited sensitivity of the IHC technique at the low levels of viremia which is supported by the positive correlation between the viral load and the number of cells expressing HCV NS3 protein. However, we believe that the specificity of the IHC technique is high since we did not detect any extra-hepatic infection of HCV in SVR or healthy control group but only in CHC patients with high viral load.
Based on the previous data, the gastrointestinal tissue could act as an extra-hepatic compartment for HCV and may be involved in virologic relapse after viral clearance, with a possible source of re-infection for hepatocytes through the portal circulation.
Treg plays a fundamental role in maintaining immune homeostasis and the balance between the tissue-damaging and protective effects of the immune response (Cabrera et al., 2004; Keynan et al., 2008; Sturm et al., 2010). In a previous study by our laboratory (Hetta et al., 2015), we reported that the frequency of colonic Treg in HCV-infected patients was higher than those in the controls. In this study, a significant positive correlation between frequency of colonic Treg and the frequency of cells with either HCV core protein (P=0.002 and R=0.61) or HCV NS3 in colonic tissue (P=0.002 and R=0.6) was identified. Additionally, there was a significant difference (P<0.0001) in the viral load between the groups expressing HCV core or NS3 and without HCV core or NS3 expression. The elevated frequency of Treg may lead to weak HCV-specific immune responses in CHC (Cabrera et al., 2004), with subsequent increase in viral load and more chance of extra-hepatic HCV infection.
Significant negative correlations between the HCV core expressing cells, but not in the NS3 expressing cells, with METAVIR inflammatory grade, fibrosis stage and the ALT liver enzyme were identified in this study. HCV core protein can induce Treg cells in CHC (MacDonald et al., 2002). The increase in Treg frequency in NR and TN groups might be due to the effect of HCV core protein, which might lead to suppression of the HCV-specific immune responses and consequently a failure in viral clearance in TN and/or NR in HCV-treated patients. Moreover, HCV core protein has other deleterious effects on the immune system. Lee et al. (2001) demonstrated that HCV core protein, which is either expressed within macrophage cells or exogenously added to them, is capable of suppressing IL-12 production by activated APCs. In addition, HCV core protein was shown to exert an inhibitory effect on the stimulatory capacity of macrophages in mixed lymphocyte reaction (Losikoff et al., 2012). Taken together, these results suggested that HCV core protein could play significant roles in suppressing the effective HCV-specific Th1 immunity through inhibition of IL-12 secretion and inhibition of macrophage functions, which may contribute to persistent HCV infection. Additionally, Treg is shared between the gut and the liver through the portal circulation (Adams et al., 2008; Gualdi et al., 1996), and consequently can migrate through the portal circulation and decrease liver inflammation and fibrosis.
However, the association between HCV NS3 protein expression and severity of liver disease is questionable. Liao et al. (2011) and Kasprzak et al. (2004) reported that tissue expression of NS3 protein does not correlate with histological grade or stage of liver tissues or clinical features in patients with CHC. In our study, we did not find correlation between the number of cells expressing HCV NS3 protein and ALT, AST and METAVIR inflammatory grade and fibrosis stage. These findings are supported by the fact that HCV infection is not cytopathic (Spengler & Nattermann, 2007) and may not cause liver damage (Spengler & Nattermann, 2007). In contrast, HCV NS3 induces both CD4+ and CD8+ effector HCV-specific T cells and stimulates viral clearance (Liao et al., 2011).
One of the limitations of our study was the lack of data on the frequency of intra-hepatic Treg cells because we did not have access to the liver biopsies at the time of the study for all the patients for IHC examination. Therefore, we could not compare the frequency of Treg in colon tissue with that in the liver. Additionally, we did not have serial biopsies for the enrolled subjects to examine the kinetics of HCV extra-hepatic replications during treatment. It is difficult and risky and may be unethical to obtain multiple colon biopsies from the same patient at different time intervals during therapy.
In conclusion, our study provides evidence that HCV can infect B cells and macrophages of the colon tissue The correlations between HCV infection in colonic tissue and HCV viral load and liver pathology highlight the potential significance of this extra-hepatic infection in HCV pathogenesis and offer a possible explanation for differential responses to therapy including late relapse that have been observed in clinical trials.
Acknowledgements
We would like to thank all the participants in this study, particularly the patients. We thank all members of the Department of Medical Microbiology and Immunology, Faculty of Medicine, Assiut University, Assiut, Egypt. We also thank Dr Jason Blackard for supplying us with Huh7.5JFH1 and Huh7.5 cell lines and the Cincinnati Children’s Hospital Medical Center Pathology Core Facility at the Digestive Health Center. This investigation was supported by an Egyptian Government Scholarship for H. F. H. the Grant Office, Faculty of Medicine, Assiut University, Egypt; Merck Investigator Initiated Studies (IISP no. 38879 to M. T. M. S.) and National Institutes of Health Grant (K24DK070528 to K. E. S.) and was supported in part by Public Health Service Grant (P30 DK078392; The Gene Expression Microarray Core Cincinnati Children’s Hospital Medical Center) and National Institutes of Health Grant (NIH P30 DK078392; Core of the Digestive Disease Research Core Center in Cincinnati).
Supplementary Data
Supplementary File 1
Abbreviations:
- ALT
alanine transaminase
- AST
aspartate transaminase
- BSA
bovine serum albumin
- CHC
chronic hepatitis C
- CI
confidence interval
- FoxP3
Forkhead box protein P3
- HCV
hepatitis C virus
- HPF
high-power field
- IHC
immunohistochemistry
- NR
non-responder
- NS
non-structural
- PBMC
peripheral blood mononuclear cell
- PBST
PBS containing 0.05 % Tween 20
- SVR
sustained virologic response
- TN
treatment naïve
- Treg
T regulatory
References
- Adams D. H., Eksteen B., Curbishley S. M.(2008). Immunology of the gut and liver: a love/hate relationship. Gut 57838–848. 10.1136/gut.2007.122168 [DOI] [PubMed] [Google Scholar]
- Agnello V., De Rosa F. G.(2004). Extrahepatic disease manifestations of HCV infection: some current issues. J Hepatol 40341–352. 10.1016/j.jhep.2003.10.009 [DOI] [PubMed] [Google Scholar]
- Barth H., Ulsenheimer A., Pape G. R., Diepolder H. M., Hoffmann M., Neumann-Haefelin C., Thimme R., Henneke P., Klein R., et al. (2005). Uptake and presentation of hepatitis C virus-like particles by human dendritic cells. Blood 1053605–3614. 10.1182/blood-2004-05-1952 [DOI] [PubMed] [Google Scholar]
- Blackard J. T., Kemmer N., Sherman K. E.(2006). Extrahepatic replication of HCV: insights into clinical manifestations and biological consequences. Hepatology 4415–22. 10.1002/hep.21283 [DOI] [PubMed] [Google Scholar]
- Blackard J. T., Kong L., Huber A. K., Tomer Y.(2013). Hepatitis C virus infection of a thyroid cell line: implications for pathogenesis of hepatitis C virus and thyroiditis. Thyroid 23863–870. 10.1089/thy.2012.0507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blight K. J., McKeating J. A., Rice C. M.(2002). Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol 7613001–13014. 10.1128/JVI.76.24.13001-13014.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera R., Tu Z., Xu Y., Firpi R. J., Rosen H. R., Liu C., Nelson D. R.(2004). An immunomodulatory role for CD4(+)CD25(+) regulatory T lymphocytes in hepatitis C virus infection. Hepatology 401062–1071. 10.1002/hep.20454 [DOI] [PubMed] [Google Scholar]
- Castillo I., Rodríguez-Iñigo E., Bartolomé J., de Lucas S., Ortíz-Movilla N., López-Alcorocho J. M., Pardo M., Carreño V.(2005). Hepatitis C virus replicates in peripheral blood mononuclear cells of patients with occult hepatitis C virus infection. Gut 54682–685. 10.1136/gut.2004.057281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T. T., Young K. C., Yang Y. J., Lei H. Y., Wu H. L.(1996). Hepatitis C virus RNA in peripheral blood mononuclear cells: comparing acute and chronic hepatitis C virus infection. Hepatology 23977–981. 10.1002/hep.510230506 [DOI] [PubMed] [Google Scholar]
- Coquillard G., Patterson B. K.(2009). Determination of hepatitis C virus-infected, monocyte lineage reservoirs in individuals with or without HIV coinfection. J Infect Dis 200947–954. 10.1086/605476 [DOI] [PubMed] [Google Scholar]
- Deforges S., Evlashev A., Perret M., Sodoyer M., Pouzol S., Scoazec J. Y., Bonnaud B., Diaz O., Paranhos-Baccalà G., et al. (2004). Expression of hepatitis C virus proteins in epithelial intestinal cells in vivo. J Gen Virol 852515–2523. 10.1099/vir.0.80071-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher N. F., Wilson G. K., Murray J., Hu K., Lewis A., Reynolds G. M., Stamataki Z., Meredith L. W., Rowe I. A., et al. (2012). Hepatitis C virus infects the endothelial cells of the blood-brain barrier. Gastroenterology 142:e636, 634–643. 10.1053/j.gastro.2011.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank C., Mohamed M. K., Strickland G. T., Lavanchy D., Arthur R. R., Magder L. S., El Khoby T., Abdel-Wahab Y., Aly Ohn E. S., et al. (2000). The role of parenteral antischistosomal therapy in the spread of hepatitis C virus in Egypt. Lancet 355887–891. 10.1016/S0140-6736(99)06527-7 [DOI] [PubMed] [Google Scholar]
- Féray C., Samuel D., Thiers V., Gigou M., Pichon F., Bismuth A., Reynes M., Maisonneuve P., Bismuth H., Bréchot C.(1992). Reinfection of liver graft by hepatitis C virus after liver transplantation. J Clin Invest 891361–1365. 10.1172/JCI115723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouda I., Nada O., Ezzat S., Eldaly M., Loffredo C., Taylor C., Abdel-Hamid M.(2010). Immunohistochemical detection of hepatitis C virus (genotype 4) in B-cell NHL in an Egyptian population: correlation with serum HCV-RNA. Appl Immunohistochem Mol Morphol 1829–34. 10.1097/PAI.0b013e3181ae9e82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin S. D., Beales L. P., Clarke D. S., Worsfold O., Evans S. D., Jaeger J., Harris M. P., Rowlands D. J.(2003). The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, Amantadine. FEBS Lett 53534–38. 10.1016/S0014-5793(02)03851-6 [DOI] [PubMed] [Google Scholar]
- Gualdi R., Bossard P., Zheng M., Hamada Y., Coleman J. R., Zaret K. S.(1996). Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev 101670–1682. 10.1101/gad.10.13.1670 [DOI] [PubMed] [Google Scholar]
- Gumber S. C., Chopra S.(1995). Hepatitis C: a multifaceted disease. Review of extrahepatic manifestations. Ann Intern Med 123615–620. 10.7326/0003-4819-123-8-199510150-00008 [DOI] [PubMed] [Google Scholar]
- Hetta H. F.(2014). Gut immune response in the presence of hepatitis C virus infection. World J Immunol 452–62. 10.5411/wji.v4.i2.52 [DOI] [Google Scholar]
- Hetta H. F., Mekky M. A., Khalil N. K., Mohamed W. A., El-Feky M. A., Ahmed S. H., Daef E. A., Nassar M. I., Medhat A., et al. (2015). Association of colonic regulatory T cells with hepatitis C virus pathogenesis and liver pathology. J Gastroenterol Hepatol 301543–1551. 10.1111/jgh.12936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasprzak A., Biczysko W., Adamek A., Wysocki J., Zabel M., Jurczyszyn D., Chmielewski M., Surdyk-Zasada J.(2004). Studies on tissue expression of HCV proteins (NS3 and C) in chronic hepatitis C using the ImmunoMax technique. Scand J Gastroenterol 39387–388. 10.1080/00365520310008818 [DOI] [PubMed] [Google Scholar]
- Keynan Y., Card C. M., McLaren P. J., Dawood M. R., Kasper K., Fowke K. R.(2008). The role of regulatory T cells in chronic and acute viral infections. Clin Infect Dis 461046–1052. 10.1086/529379 [DOI] [PubMed] [Google Scholar]
- Kong L., Cardona Maya W., Moreno-Fernandez M. E., Ma G., Shata M. T., Sherman K. E., Chougnet C., Blackard J. T.(2012). Low-level HIV infection of hepatocytes. Virol J 9157. 10.1186/1743-422X-9-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo G., Choo Q. L., Alter H. J., Gitnick G. L., Redeker A. G., Purcell R. H., Miyamura T., Dienstag J. L., Alter M. J., Stevens C. E.(1989). An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science 244362–364. 10.1126/science.2496467 [DOI] [PubMed] [Google Scholar]
- Lee C. H., Choi Y. H., Yang S. H., Lee C. W., Ha S. J., Sung Y. C.(2001). Hepatitis C virus core protein inhibits interleukin 12 and nitric oxide production from activated macrophages. Virology 279271–279. 10.1006/viro.2000.0694 [DOI] [PubMed] [Google Scholar]
- Liao W. H., Tung S. Y., Shen C. H., Lee K. F., Wu C. S.(2011). Tissue expression of the hepatitis C virus NS3 protein does not correlate with histological or clinical features in patients with chronic hepatitis C. Chang Gung Med J 34260–267. [PubMed] [Google Scholar]
- Losikoff P. T., Self A. A., Gregory S. H.(2012). Dendritic cells, regulatory T cells and the pathogenesis of chronic hepatitis C. Virulence 3610–620. 10.4161/viru.21823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald A. J., Duffy M., Brady M. T., McKiernan S., Hall W., Hegarty J., Curry M., Mills K. H.(2002). CD4 T helper type 1 and regulatory T cells induced against the same epitopes on the core protein in hepatitis C virus-infected persons. J Infect Dis 185720–727. 10.1086/339340 [DOI] [PubMed] [Google Scholar]
- Manzin A., Candela M., Paolucci S., Caniglia M. L., Gabrielli A., Clementi M.(1994). Presence of hepatitis C virus (HCV) genomic RNA and viral replicative intermediates in bone marrow and peripheral blood mononuclear cells from HCV-infected patients. Clin Diagn Lab Immunol 1160–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo M. J.(2003). Extrahepatic manifestations of hepatitis C infection. Am J Med Sci 325135–148. 10.1097/00000441-200303000-00006 [DOI] [PubMed] [Google Scholar]
- McHutchison J. G., Poynard T., Esteban-Mur R., Davis G. L., Goodman Z. D., Harvey J., Ling M. H., Garaud J. J., Albrecht J. K., et al. (2002). Hepatic HCV RNA before and after treatment with interferon alone or combined with ribavirin. Hepatology 35688–693. 10.1053/jhep.2002.31870 [DOI] [PubMed] [Google Scholar]
- Miglioresi L., Riva E., Antonelli G., Russo F., Ricci G. L.(2003). Localization of hepatitis C virus in gastrointestinal mucosa: a possible reservoir for relapse. Hepatology 38775. 10.1053/jhep.2003.50322 [DOI] [PubMed] [Google Scholar]
- Pal S., Sullivan D. G., Kim S., Lai K. K., Kae J., Cotler S. J., Carithers R. L., Wood B. L., Perkins J. D., Gretch D. R.(2006). Productive replication of hepatitis C virus in perihepatic lymph nodes in vivo: implications of HCV lymphotropism. Gastroenterology 1301107–1116. 10.1053/j.gastro.2005.12.039 [DOI] [PubMed] [Google Scholar]
- Pham T. N., King D., Macparland S. A., McGrath J. S., Reddy S. B., Bursey F. R., Michalak T.(2008). Hepatitis C virus replicates in the same immune cell subsets in chronic hepatitis C and occult infection. Gastroenterology 134812–822. 10.1053/j.gastro.2007.12.011 [DOI] [PubMed] [Google Scholar]
- Revie D., Salahuddin S. Z.(2011). Human cell types important for hepatitis C virus replication in vivo and in vitro: old assertions and current evidence. Virol J 8346. 10.1186/1743-422X-8-346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh M. G., Tibbs C. J., Koskinas J., Pereira L. M., Bomford A. B., Portmann B. C., McFarlane I. G., Williams R.(1994). Hepatic and extrahepatic hepatitis C virus replication in relation to response to interferon therapy. Hepatology 201399–1404. 10.1002/hep.1840200604 [DOI] [PubMed] [Google Scholar]
- Sansonno D., Iacobelli A. R., Cornacchiulo V., Iodice G., Dammacco F.(1996). Detection of hepatitis C virus (HCV) proteins by immunofluorescence and HCV RNA genomic sequences by non-isotopic in situ hybridization in bone marrow and peripheral blood mononuclear cells of chronically HCV-infected patients. Clin Exp Immunol 103414–421. 10.1111/j.1365-2249.1996.tb08296.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansonno D., Gesualdo L., Manno C., Schena F. P., Dammacco F.(1997). Hepatitis C virus-related proteins in kidney tissue from hepatitis C virus-infected patients with cryoglobulinemic membranoproliferative glomerulonephritis. Hepatology 251237–1244. 10.1002/hep.510250529 [DOI] [PubMed] [Google Scholar]
- Sène D., Limal N., Cacoub P.(2004). Hepatitis C virus-associated extrahepatic manifestations: a review. Metab Brain Dis 19357–381. 10.1023/B:MEBR.0000043982.17294.9b [DOI] [PubMed] [Google Scholar]
- Spengler U., Nattermann J.(2007). Immunopathogenesis in hepatitis C virus cirrhosis. Clin Sci 112141–155. 10.1042/CS20060171 [DOI] [PubMed] [Google Scholar]
- Sturm N., Thélu M. A., Camous X., Dimitrov G., Ramzan M., Dufeu-Duchesne T., Bonorino P., Guillermet C., Brambilla E., et al. (2010). Characterization and role of intra-hepatic regulatory T cells in chronic hepatitis C pathogenesis. J Hepatol 5325–35. 10.1016/j.jhep.2010.02.024 [DOI] [PubMed] [Google Scholar]
- Wang J. T., Sheu J. C., Lin J. T., Wang T. H., Chen D. S.(1992). Detection of replicative form of hepatitis C virus RNA in peripheral blood mononuclear cells. J Infect Dis 1661167–1169. 10.1093/infdis/166.5.1167 [DOI] [PubMed] [Google Scholar]
- Yan F., Hao F., Zhao L.(2000a). Study of expression of hepatitis C virus antigens and viral replication in extrahepatic tissues. Zhonghua Gan Zang Bing Za Zhi 840–42. [PubMed] [Google Scholar]
- Yan F. M., Chen A. S., Hao F., Zhao X. P., Gu C. H., Zhao L. B., Yang D. L., Hao L. J.(2000b). Hepatitis C virus may infect extrahepatic tissues in patients with hepatitis C. World J Gastroenterol 6805–811. 10.3748/wjg.v6.i6.805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z. Q., Nguyen D. T., Hiotellis A., Hahn Y. S.(2001). Hepatitis C virus core protein inhibits human T lymphocyte responses by a complement-dependent regulatory pathway. J Immunol 1675264–5272. 10.4049/jimmunol.167.9.5264 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1