Abstract
Gonadotropin-releasing hormone (GnRH) is required for pubertal onset and reproduction, thus the control of GnRH transcription is tightly regulated during development and adulthood. GnRH neuron development depends on transcription factors of the homeodomain family. For example, Ventral anterior homeobox 1 (Vax1) is necessary to maintain GnRH expression after embryonic day 13 in the mouse. To further our understanding of the mechanisms by which VAX1 regulates GnRH gene expression, we asked whether VAX1 interacts with other transcription factors to modify GnRH expression levels. Using the GnRH cell lines, GN11 and GT1-7, we found that activation of PKC enhances expression of the immediate early gene cFos in both GN11, and GT1-7, and represses expression of Vax1 in GT1-7. Further, VAX1 interacts with cFOS while bound to the GnRH promoter. In immature GN11 cells, VAX1 and cFOS enhance GnRH expression, whereas VAX1 and cFOS have a repressive role in the mature GT1-7 cells.
Keywords: Gonadotropin-releasing hormone, cFOS, Ventral anterior homeobox 1, transcription regulation, development
1. Introduction
Pubertal onset and fertility both depend on proper developmental migration and maturation of gonadotropin-releasing hormone (GnRH) neurons, with improper GnRH neuron development or regulation impairing pubertal onset and fertility (Herbison et al., 2008; Mason et al., 1986). Despite the critical role of GnRH in reproduction, we still have gaps in our understanding of the transcriptional program involved in GnRH neuron function (Balasubramanian et al., 2010; Stamou et al., 2015). The slow advances in the field are in great part due to challenges associated with the in vivo study of GnRH neurons, including a long GnRH neuron migration path, the scattered localization of GnRH neurons throughout the anterior hypothalamic area, the sparsity of this cell population (represented by approximately 800 GnRH neurons in the mouse), and the long projections GnRH neurons send from the preoptic area to the median eminence, where they release GnRH in a pulsatile fashion (Fueshko and Wray, 1994; Messina et al., 2016; Tarozzo et al., 1994).
Most of our current knowledge of GnRH neuron development and control of Gnrh1 expression comes from the use of well-established immortalized mouse GnRH neuron cell lines, represented by the immature, migratory GN11 cells (Radovick et al., 1990), and the mature, non-migratory GT1-7 cells (Mellon et al., 1990). Indeed, these cell lines have helped identify novel transcription factors of the homeodomain binding family that are required for normal GnRH neuron development and GnRH expression, including Orthodenticle homeobox 2 (Otx2) (Diaczok et al., 2011; Kelley et al., 2000; Larder et al., 2013), Ventral anterior homeobox 1 (Vax1) (Hoffmann et al., 2014; Hoffmann et al., 2016), Sine oculis-related homeobox (Six) 3 (unpublished data) and Six6 (Larder et al., 2011). In addition, GN11 and GT1-7 cell lines have repeatedly proven to be a valid in vitro model recapitulating many characteristics of immature and mature GnRH neurons respectively, allowing for advances in our understanding of factors important for GnRH neuron maturation, and Gnrh1 expression (Cariboni et al., 2007; Givens et al., 2005; Glidewell-Kenney et al., 2013; Magni et al., 2007; Tang et al., 2005).
Despite the great advances in the field of GnRH neuron development, it is still unclear what combination of transcription factors and input from the surrounding environment are required to specify and fine-tune Gnrh1 expression. The significance of homeodomain transcription factors in fertility is starting to be appreciated, where knock-out (KO) of Six6 leads to loss of GnRH neurons during late development (Larder et al., 2011), and loss of Six3, a SIX homeodomain transcription factor related to SIX6, is required for the development of the olfactory system and proper GnRH neuron migration (unpublished). In addition, Vax1, a transcription factor not detected in the immature GN11 cells and highly expressed in the mature GT1-7 cells, is necessary to maintain GnRH expression after embryonic day 13 (E13) in the mouse (Hoffmann and Mellon, 2016; Hoffmann et al., 2016). This suggests that the onset of expression of Vax1 in GnRH neurons during development is critical in GnRH transcription, and that the transcriptional program of VAX1 is required for appropriately adjusting GnRH expression during development.
Interestingly, a physical interaction between developmental homeodomain transcription factors and immediate early genes (IEG), such as members of the activating protein-1 (AP1) family, has been found to regulate transcriptional activity by either enhancing or silencing transcriptional activity of these IEG-homeodomain transcription factor complexes (Jeong et al., 2004; Kessel et al., 1988; Schaefer et al., 2001). The idea that IEGs and homeodomain transcription factors interact and form transcriptionally active complexes could be important from a developmental and cellular maturation standpoint, allowing specific expression of key genes to unique cell populations (Jeong et al., 2004). Indeed, during development, the level, duration and timing of transcription factors are imperative for cellular specification and embryogenesis (Briscoe and Small, 2015; Kicheva and Briscoe, 2015; Lewis, 2008). The rapid expression of an IEG, such as FBJ osteosarcoma oncogene (cFOS) or Jun proto-oncogene (cJUN), could thus regulate transcriptional activity of homeodomain transcription factors during a very short developmental time-frame. Further, cFOS is strongly induced in mature GnRH neurons in response to the phorbol esther, phorbol 12-myristate 13-acetate (TPA), which activates protein kinase C (PKC), leading to recruitment of cFOS to the GnRH regulatory region, and repression of Gnrh1 expression (Glidewell-Kenney et al., 2013; Glidewell-Kenney et al., 2014). The neurokinin 3 receptor, a receptor critical in fertility (Balasubramanian et al., 2010; Topaloglu et al., 2009; Yang and Seminara, 2012), also represses Gnrh1 expression through cFOS. The pathway induced by activation of the neurokinin 3 receptor leads to induction of PKC, and recruitment of transcription factors, including serum response factor (SRF), and Elk-1, which in their turn regulate cFos, leading to a repression of Gnrh1 expression. Based on the importance of IEG’s, and particularly cFOS in the regulation of Gnrh1, we asked whether cFOS could modulate the level of GnRH expression during GnRH neuron maturation, through interaction with homeodomain transcription factors.
2. Material and Methods
2.1 Cell culture
GT1-7 (Mellon et al., 1990), GN11 (kindly provided by Sally Radovick), and αT3-1 (Windle et al., 1990) cell lines were cultured in DMEM (Mediatech Inc., Herndon, VA), containing, 10 % fetal bovine serum (Gemini Bio, West Sacramento, CA), and 1× penicillin-streptomycin (Life Technologies, Inc./Invitrogen, Grand Island, NY) in a humidified 5 % CO2 incubator at 37°C. For luciferase assays GN11 and GT1-7 cells were seeded into 24-well plates (Nunc, Roskilde, Denmark) at 75,000 and 200,000 per well, respectively. Cells transfected for quantitative real time PCR (qRT-PCR) were plated into 10 cm dishes (Nunc) at 3 million (GT1-7) and 1 million (GN11, NIH3T3) cells per dish. O/N transfection of cells was done approximately 21 h after plating. At the time of phorbol 12-myristate13-acetate 100 nM (TPA, Tocris Bioscience, dissolved in dimethylsulfoxide) or DMSO (1/2000; Sigma) treatment, media was changed to DMEM containing 0.1 % BSA. To determine the impact of the 21 h TPA 100 nM treatment on cell morphology, live GN11 and GT1-7 cell were visualized using the Keyence BZ-X700 Microscope (Keyence, Laguna Hills, CA). To increase the visibility of the cells, adjustments of brightness, contrast, and color balance were done with Image J (National Institutes of Health, Bethesda) and applied to the entire image.
2.2 Quantitative real-time PCR
Untreated cells, or cells treated with DMSO or TPA 100 nM were harvested in TRIzol® (Invitrogen) and total RNA from GT1-7, GN11 and αT3 cells extracted according to manufacturer’s recommendations. DNA was eliminated by the use of the DNA-free™ kit (Applied Biosystems, Foster City, CA), whereas cDNA was obtained by reverse transcription of RNA using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). cDNA products were detected using iQ SYBR Green Supermix (BioRad) on the CFX Connect real-time detection system (Bio-Rad). Primers to the coding sequences for mouse Otx2 (F: TCGAAGAGCTAAGTGCCGCC, R: GCAATGGTTGGGACTGAGG), cJun (F: GTCCCCTATCGACATGGAGTCT, R: GAGTTTTGCGCTTTCAAGGTTT), cFos (F GGCAAAGTAGAGCAGCTATCTCCT, R: TCAGCTCCCTCCTCCGATTC), Six6 (F: TCGATGTTCCAGCTGCCCAT, R: TGGAAAGCCACGATGGCTCT), Gnrh1 (F: TGCTGACTGTGTGTTTGGAAGGCT, R: TTTGATCCACCTCCTTGCGACTCA), Vax1 (F: CCGGATCCTAGTCCGAGATGCC, R: TCTCCCGGCCCACCACGTAT), Ppia (F: AAGTTCCAAAGACAGCAGAAAAC, R: CTCAAATTTCTCTCCGTAGATGG), H2afz (F: TCACCGCAGAGGTACTTGAG, R: GATGTGTGGGATGACACCA) synthesized by Integrated DNA Technologies (San Diego, CA) and Dlx1 and Dlx5 (Qiagen, Germantown, MD), permitted amplification of the indicated cDNA products. Data was expressed by the 2−Δ ΔCT method by normalizing the gene of interest to the housekeeping genes Ppia and H2afz. Data are expressed as fold change compared to control, or as indicated in the figure legends. Data represent mean fold change ± SEM from a minimum of three independent RNA samples for each data point.
2.3 Transfection
Transient transfections for luciferase assays and qRT-PCR were performed using PolyJet™ (SignaGen Laboratories, Rockville, MD), according to the manufacturer’s recommendations. For luciferase assays, cells were co-transfected as indicated in the figure legends, with 150 ng/well luciferase reporter plasmids and expression plasmids at the indicated concentration. 100 ng/well thymidine kinase-β-galactosidase reporter plasmid was added and served as an internal control. Transfection efficiency was evaluated by staining for the thymidine kinase-β-galactosidase reporter, and showed successful transfection of 15.3 % ± 1.6 GN11 cells, and of 9.8 % ± 1.7 GT1-7 cells. Plasmids were previously described (Ely et al., 2011; Glidewell-Kenney et al., 2013; Glidewell-Kenney et al., 2014; Hoffmann et al., 2016; Iyer et al., 2010; Larder et al., 2011; Nelson et al., 2000). Site-directed mutagenesis of the two homeodomain binding sites were performed using the Quick Change Mutagenesis Kit (Agilent), following manufacturer’s instructions. The −59 bp site in the 1 Kb-cFos-luciferase promoter was mutated from ATT to GGC, and the −313 bp site was mutated from ATT to CGG. To equalize the amount of DNA transfected into cells, we systematically equalized plasmid concentrations by adding varying concentrations of the appropriate empty vector.
2.4 Luciferase assay
Cells were harvested 21 h after transfection in lysis buffer [100 mM potassium phosphate (pH 7.8) and 0.2 % Triton X-100]. Luciferase and β-galactosidase assays were performed as previously described (Givens et al., 2005). Luciferase values are normalized to β-galactosidase values to control for transfection efficiency. Values are normalized to pGL3 or corresponding backbone of expression plasmid as indicated in the figure legends. Data represent the mean ± SEM of at least three independent experiments done in triplicate.
2.5 Statistical analysis
All experiments were repeated a minimum of 3 independent times. Statistical analyses of data were done by Student’s t-test, One or Two Way ANOVA in GraphPad Prism 7 (GraphPad Software, La Jolla, CA) as indicated in figure legends. Statistical significance was set at 95 %. Data are depicted as mean ± SEM.
3. Results
3.1 TPA treatment of GnRH neuronal cell lines causes transcriptional regulation of both cFos and homeodomain transcription factors, enhancing Gnrh1 expression in GN11 cells, and reducing Gnrh1 expression in GT1-7 cells
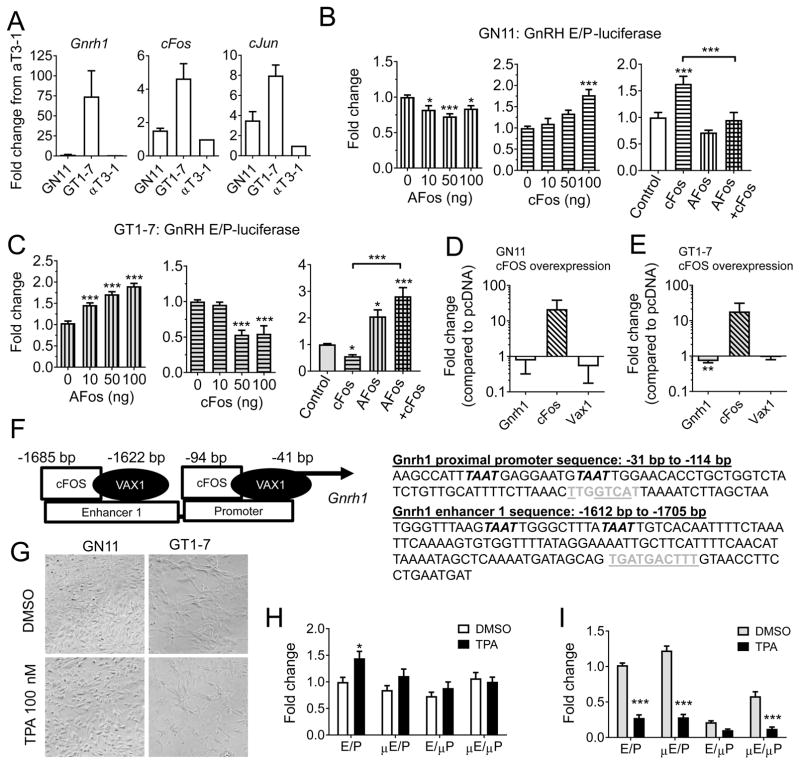
To better understand the molecular events that enhance GnRH expression during GnRH neuron maturation, we asked whether cFOS might have differential roles in immature and mature GnRH neurons, represented by the immortalized GnRH neuronal cells GN11 and GT1-7, respectively. By qRT-PCR, we found that the mature GnRH cell line, GT1-7, expressed relatively high levels of Gnrh1, cFos and cJun, whereas the immature GnRH cells, GN11, and the control cell line, αT3-1, expressed lower levels of these transcripts (Figure 1A). To determine if cFOS plays a role in Gnrh1 expression during GnRH neuron maturation, we transiently transfected GN11 and GT1-7 cells with expression vectors for cFOS, or a dominant-negative cFOS protein, termed AFOS (Olive et al., 1997), and evaluated the level of transcription of the GnRH enhancer and promoter (GnRH E/P) construct driving the expression of luciferase. AFOS overexpression in GN11 reduced GnRH E/P-luciferase levels at all the studied concentrations (One-way ANOVA, F(3, 40) = 7.374, n = 5, p = 0.0005), whereas cFOS overexpression enhanced GnRH E/P-luciferase levels (One-way ANOVA, F(3, 32) = 11.73, p < 0.0001, n = 5, Figure 1B). AFOS was able to reduce cFOS enhanced GnRH E/P-luciferase levels (One-way ANOVA, F = (3, 22) = 20.91, p < 0.0001, n = 3–5, Figure 1B). In contrast in GT1-7 cells, AFOS enhanced (One-way ANOVA, F(3, 42) = 42.12, p < 0.0001, n = 4), and cFOS repressed GnRH E/P-luciferase levels (One-way ANOVA, F(3, 24) = 17.14, p < 0.0001, n = 4–5, Figure 1C). This effect was specific, as AFOS was able to reverse the effect of cFOS repression on the GnRH E/P-luciferase promoter (One-way ANOVA, F(3,29) = 16.02, p < 0.0001, n = 3–5, Figure 1C). To determine if the effect of cFOS would be recapitulated on the endogenous Gnrh1 gene, we overexpressed cFOS in GN11 and GT1-7 cells. We were unable to detect an increase in Gnrh1 expression by qRT-PCR in GN11 cells at the studied time point (Figure 1D), whereas a slight decrease in Gnrh1 expression was seen in GT1-7 cells (Figure 1E). The minor effect of cFOS overexpression on endogenous gene expression was likely due to the small proportion of cells transfected (9–15 %). It has previously been shown that TPA, a phorbol ester strongly activating PKC, induces cFos expression in GT1-7 cells (Bruder et al., 1992; Bruder and Wierman, 1994; Glidewell-Kenney et al., 2013). The enhanced cFos expression leads to repression of Gnrh1 in GT1-7 cells through binding of cFOS to AP1-half sites in the Gnrh1 regulatory region (Figure 1F) (Glidewell-Kenney et al., 2013). To determine the extent to which regulation of Gnrh1 expression by TPA was mediated by cFOS-binding to the identified AP1-half sites (Figure 1F), we treated both GN11 and GT1-7 cells with DMSO or TPA (100 nM, 18–20 h). This treatment was associated with morphological changes in both GN11 and GT1-7 cells, where GN11 cells appeared more round after TPA treatment than cells treated with vehicle, whereas prolongations of GT1-7 cells appeared thinner (Figure 1G). We next evaluated the transcription levels in response to TPA of the GnRH E/P-luciferase plasmid, GnRH μE/P-luciferase (AP1-half site mutated in the E), GnRH E/μP-luciferase (AP1-half site mutated in the promoter) or both (GnRH μE/μP, Figure 1F, sequences underlined). In GN11 cells, TPA slightly increased the expression of GnRH E/P-luciferase, and mutation of either of the AP1-half sites abolished the TPA-dependent increase (Two-way ANOVA, promoter constructs: (F3,73) = 4.882, p = 0.0038; treatment F(1,73) = 7.164, p = 0.0092; interaction F(3,73) = 1.974, n = 5–6, p = 0.1254, Figure 1H), supporting a previous report on enhanced expression of the human GnRH promoter-driving luciferase expression in this same cell line in response to TPA (Zakaria et al., 1996). In GT1-7 cells, TPA-repressed Gnrh1 expression through AP1-halfsites in the promoter (Two-way ANOVA, promoter constructs: F(3,74) = 87.08, p < 0.0001; treatment: F(3,74) = 376, p < 0.001; interaction F(3,74) = 37.81 p<0.0001, n = 4–6; Figure 1I). Specifically, mutating the GnRH E/μP reduced the repressive effect of TPA by ~50 %. However, mutation of both AP1-half sites did not abolish the effects of TPA, suggesting there are either additional sites within the cFOS-binding region, or TPA induces additional signaling pathways, which repress Gnrh1. To identify additional pathways or transcription factors which are important for PKC-regulated Gnrh1 expression, we analyzed the GnRH enhancer and promoter region, and found that the AP1-half sites bound by cFOS were in close proximity to ATTA sites, which are known binding-sites of homeodomain transcription factors (Figure 1F).
Figure 1.
cFOS differentially regulates GnRH expression in immature and mature GnRH cell lines. A) Endogenous transcript levels of Gnrh1, cFos and cJun were evaluated by qRT-PCR in immature (GN11), and mature (GT1-7) GnRH cells, and the pituitary cell line αT3-1. B) GN11 (white) and C) GT1-7 (grey) cells were transiently transfected with a GnRH enhancer/promoter (GnRH E/P) luciferase construct, in the presence of a dominant negative cFOS plasmid called AFOS (vertical bars), cFOS (horizontal bars), or both (square pattern, AFOS 200 ng and cFOS 100 ng) and the fold change in luciferase levels evaluated. Data is expressed as fold change as compared to control. Statistical analysis by one-way ANOVA, followed by Dunnett’s multiple comparison test to for effects of either AFOS and cFOS, and a Tukey multiple comparison test for the dual treatment with AFOS and cFOS, * p < 0.05, ** p < 0.01, *** p<0.001, or as indicated by bracket, n = 3–5. D) GN11, and E) GT1-7 cells were transiently transfected with an empty vector (pcDNA) or cFOS, and endogenous levels of Gnrh1, Vax1 and cFos evaluated by qRT-PCR. Students t-test; * p > 0.05, n = 4–6. F) Location of ATTA (italic) and AP1-half sites (grey) in the GnRH E/P-luciferase construct. AP1-half sites mutated in G and H are underlined. G) Phase contrast image of GN11 and GT1-7 cells after 20 h DMSO or TPA 100 nM treatment (x20). H) GN11, and I) GT1-7 cells were transiently transfected with GnRH E/P-luciferase with or without AP1-half sites mutated in the enhancer (μE), the proximal promoter (μP) or both (μE/μP) and the capacity of TPA 100 nM to regulate its expression evaluated. Data is expressed as fold change compared to transcript levels in control (DMSO in GnRH E/P-luciferase). Statistical analysis by Two-way ANOVA followed by Sidak’s multiple comparison. * p > 0.05, *** p > 0.001 as compared to DMSO on the same construct, n = 4–6.
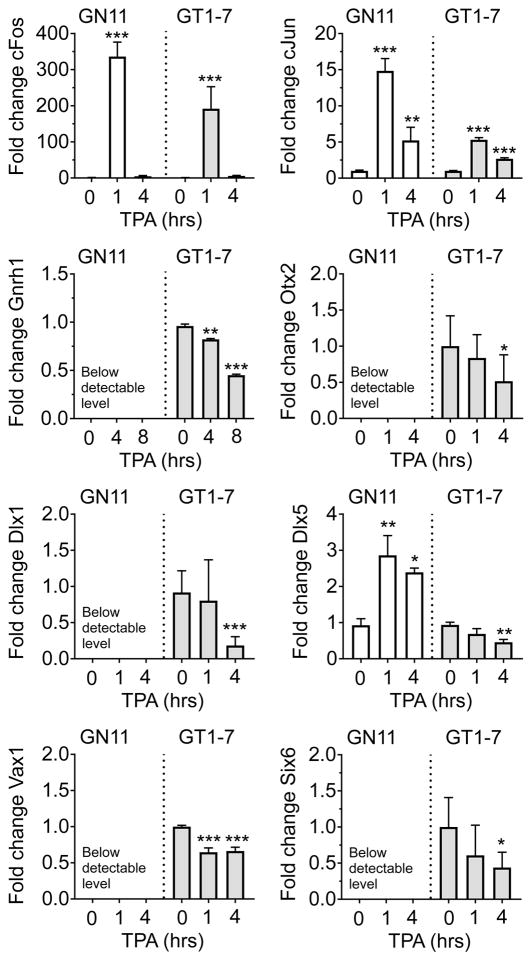
This was of particular interest to us, as homeodomain transcription factors can regulate Gnrh1 expression through ATTA sites, and previously have been shown to be critical in establishing the correct number of GnRH neurons, and maintain GnRH expression (Dateki et al., 2010; Hoffmann et al., 2016; Kelley et al., 2000; Kim et al., 2007; Larder et al., 2011). Based on this, we hypothesized that the effects of TPA are two-fold: involving 1) a PKC-induced increase of cFos, followed by cFOS repression of Gnrh1 expression, and 2) TPA-modulated expression of homeodomain transcription factors. To determine whether TPA regulates expression of homeodomain transcription factors, we treated GN11 and GT1-7 cells with TPA (100 nM) for 1, 4, 8, and 20 h, and evaluated transcript levels of the homeodomain transcription factors Dlx1, Dlx5, Six6, Vax1, and Otx2, as well as Gnrh1, cFos and cJun, by qRT-PCR. We first confirmed the capacity of TPA 100 nM to regulate cFos, cJun, and Gnrh1 in both cell lines. In GN11, TPA enhanced cFos and cJun (One-way ANOVA, cJun, F = 539,4; p < 0.001; cFos = 298.1, p < 0.0001, n = 2–6; Figure 2), whereas TPA was unable to increase Gnrh1 expression to a detectable level at the studied time points in GN11 (1, 4, 8, and 20 h). In GT1-7 cells, TPA enhanced cFos and cJun expression, whereas Gnrh1 was reduced (One-way ANOVA; cJun: F = 135.6, p < 0.0001; cFos: F = 46.38, p < 0.0001; Gnrh1: F = 290, p < 0.0001, n = 2–8) (Bruder et al., 1996; Wetsel et al., 1993). Interestingly, TPA repressed Dlx1, Vax1, Otx2 and Six6 expression in GT1-7 cells, whereas these transcripts were below detectable limit in GN11 (One-way ANOVA; Dlx1: GN11: N/A; GT1-7: F = 11.7, p = 0.0004; Vax1: GN11: N/A; GT1-7: F = 18.38, p < 0.0001; Otx2: GN11: N/A; GT1-7: F = 3.523, p = 0.048; Six6: GN11: N/A; GT1-7: F = 5.017, p = 0.0166, n = 8). TPA enhanced Dlx5 expression in GN11, but repressed this transcript in GT1-7 cells (One-way ANOVA, Dlx5: GN11: F = 8.692, p = 0.0054; GT1-7: F = 6.332, p = 0.0051, n=8). Due to the critical role of VAX1 in maintaining Gnrh1 expression after E13.5 (Hoffmann et al., 2016), and the rapid effect of TPA on Vax1 transcript levels, which were detectable at 1 h of TPA treatment, we decided to focus on this transcription factor for the current study.
Figure 2.
PKC activation rapidly changes transcript levels of genes coding several homeodomain transcription factors in GnRH neurons. The capacity of 100 nM TPA to regulate immediate early genes (cFos, cJun), Gnrh1 and homeodomain transcription factor expression levels was evaluated by qRT-PCR in GN11 (white) and GT1-7 (grey) cells. One-Way ANOVA followed by Dunnett’s multiple comparison test as compared to DMSO (0 h), * p < 0.05 ** p < 0.01, *** p < 0.001. N=2–8.
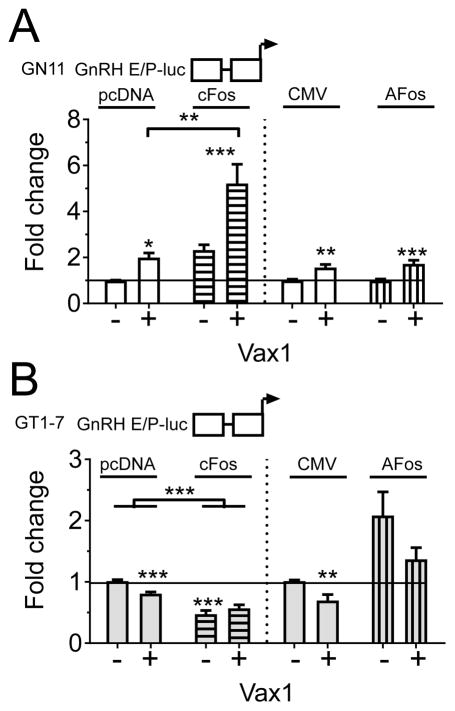
To further our understanding of the molecular mechanisms regulating the developmental expression of Gnrh1, we asked whether the PKC-pathway (and specifically cFOS) is important for determining the level of VAX1-regulated GnRH expression in GT1-7 and/or GN11 cells. We first asked what the consequences of high levels of VAX1, with and without cFOS, would be on Gnrh1 transcription in immature and mature GnRH cell lines. We transiently transfected GN11 and GT1-7 cells with the GnRH E/P-luciferase, VAX1, with or without cFOS (100 ng) or AFOS (100 ng) (Figure 3). We confirmed our prior studies showing that VAX1 increased GnRH E/P-luciferase expression in GN11 (Figure 3A), and repressed this construct in GT1-7 (Figure 3B) (Hoffmann et al., 2016). This differential regulation of GnRH E/P-luciferase expression by VAX1 might be linked with the absence of VAX1 in GN11, and the relatively high expression of VAX1 in GT1-7. It should be noted, that due to the absence of endogenous Vax1 expression in GN11 cells, it is possible this cell line more appropriately reflects the role of VAX1 within GnRH neurons. In GT1-7 cells, the combined overexpression of VAX1, in addition to the high endogenous expression of this gene, might lead to a negative feedback mechanism due to excessive levels of VAX1 in these cells. In agreement with what we found before, cFOS enhanced the GnRH E/P-luciferase promoter in GN11 (Two-way ANOVA, overall effect of cFOS, F(1,49) = 40.94, p < 0.0001) and AFOS repressed this construct, although no overall effect of AFOS was detected by Two-way ANOVA, F(1,38) = 0.5458, p = 0.4646 in GN11. A significant interaction between cFOS and VAX1, but not AFOS and VAX1, was detected by Two-way ANOVA (cFOS-VAX1 interaction: F(1,49) = 7.129, p = 0.013; AFOS-VAX1 interaction: F(1,38) = 0.5458, p = 0.4646). As seen before in GT1-7 cells (Figure 3B), cFOS repressed GnRH E/P-luciferase levels (Two way ANOVA overall effect of cFOS, F(1,54) = 81.31, p < 0.0001), and AFOS enhanced GnRH E/P-luciferase (Two-way ANOVA, overall effect AFOS, F(1,13) = 17.55, p = 0.0011). The overall effect of VAX1 was abolished in the presence of cFOS (Two-way ANOVA, F(1,54) = 1.639, p = 0.206), whereas this was not the case in the presence of AFOS (F(1,13) = 6.191, p = 0.0271).
Figure 3.
The transcription factors VAX1 and cFOS transition from transcriptional activators to repressors of GnRH E/P-luciferase during GnRH neuron maturation. A) GN11 (white), and B) GT1-7 (grey) cells, were co-transfected with GnRH E/P-luciferase, with or without VAX1 (+, −), cFOS (horizontal bars) and AFOS (vertical bars). The fold change in GnRH E/P-luciferase expression was normalized to control (GnRH E/P-luciferase with empty vector). Two-way ANOVA followed by Sidak’s multiple comparison test, * p < 0.05, ** p < 0.01; *** p < 0.001, as compared to empty vector, or as indicated by bracket.
3.2 VAX1 is a transcriptional activator of cFos in immortalized GnRH neurons
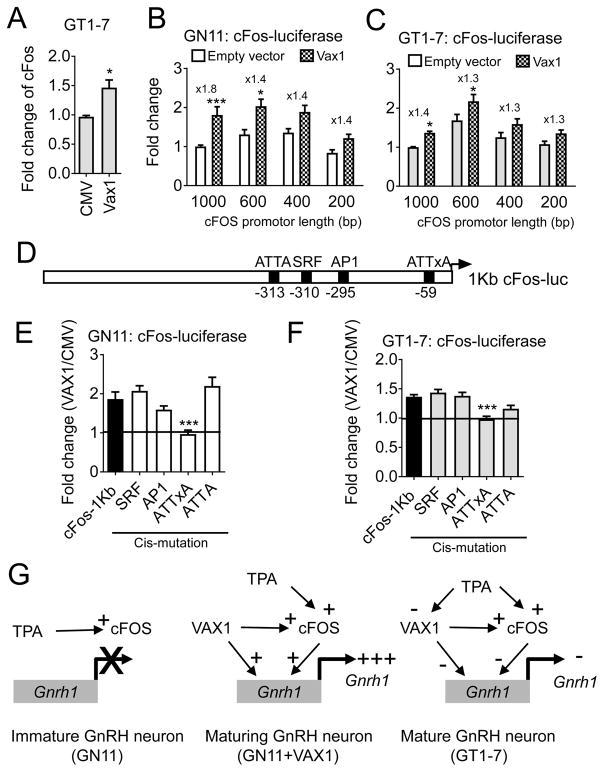
As transcription factors, VAX1 and cFOS target numerous transcripts within a cell, and thus could regulate Gnrh1 expression indirectly. We first asked whether cFOS regulated Vax1 expression, a transcription factor known to have direct effects on GnRH expression (Hoffmann et al., 2016). We performed qRT-PCR in GN11 and GT1-7 cells overexpressing cFOS, and did not detect any changes in endogenous Vax1 expression levels at the studied time point (Figure 1D, E), suggesting the synergistic effect of VAX1 and cFOS was not due to cFOS regulation of Vax1 expression. In contrast, VAX1 overexpression in GT1-7 cells increased cFos expression (Figure 4A) indicating VAX1 can regulate endogenous cFos expression. To determine if part of the effect of VAX1 regulation of the Gnrh1 promoter was due to an indirect effect through VAX1-driven cFOS expression, we asked if VAX1 could regulate the cFos promoter in GN11 and GT1-7 cells. Transient transfections of GN11 and GT1-7 cells with different lengths of the cFos-regulatory region revealed that depending on cFos-luciferase promoter length the transcription level of the construct changed (Two-way ANOVA, cFos-luciferase promoter length, GN11: F(3,74) = 6.94, p = 0.0004; GT1-7: F(3,72) = 22.12, p < 0.0001). In additional VAX1 activated this promoter in both GN11 and GT1-7 cells, and this effect was maintained down to the −200 bp cFos-promoter (Figure 4B, C). Two-way ANOVA statistical analysis determined an overall effect of VAX1 in GN11: F(1,74) = 32.73, p < 0.0001 and GT1-7: F(1,72) = 25.53, p < 0.0001, but no interaction between cFos-promoter length and VAX1 was detected (Two-way ANOVA, GN11: F(3,74) = 0.9166, p = 0.4372; GT1-7: F(3,72) = 0.3465, p = 0.7917). As a homeodomain transcription factor, VAX1 binds ATTA and related sites. We identified a partial ATTA site within −200 bp of the cFos transcriptional start site, and a second ATTA site at −313 bp from the transcriptional start site (Figure 4D). To establish whether VAX1 regulated transcription through the identified ATTA sites, we created cis-mutations of the two ATTA sites in the proximal cFos regulator region (Figure 4D). Mutation of the partial VAX1 binding site at −59 bp, but not at −313 bp, abolished VAX1 regulation of this promoter in both cell lines, whereas mutations in binding sites for other transcription factors, such as cFOS (AP1), and serum response factor (SRF), did not affect VAX1 regulation of this promoter (Figure 4D–F, One-way ANOVA, GN11: F(5, 64) = 6.267, p < 0.0001; GT1-7: F(5, 59) = 15.4, p < 0.0001). Thus, VAX1 can directly regulate the GnRH promoter, but also mediates GnRH expression through cFOS (Figure 4G).
Figure 4.
VAX1 enhances cFos expression in GnRH neurons. A) GT1-7 cells (grey) were transiently transfected with an empty vector (CMV) or the VAX1 expression vector, and endogenous levels of cFos evaluated by qRT-PCR. Students t-test, * p > 0.05, n = 4–6. B) GN11 (white), and C) GT1-7 cells were transiently transfected with different lengths of the cFos promoter driving luciferase expression. Data was normalized to the 1Kb-cFos luciferase promoter. Data were analyzed by Two-way ANOVA followed by Sika’s test, * p < 0.05; *** p < 0.001, as compared to empty vector on the studied promoter length, n = 4–7. D). Schematic of transcription factor binding sites of the 1 Kb-cFos promoter. E–F) The effect of VAX1-regulated transcription in GN11 or GT1-7 cells after cis-mutations in transcription factor binding sites of the 1 Kb-cFos promoter were assessed. Data is expressed as fold change of VAX1/CMV. Statistical analysis by One-way ANOVA, followed by Dunnett’s multiple comparison text as compared to control (cFos-luciferase 1Kb), *** p < 0.001. n = 4–6. G) Schematic recapitulating VAX1 and cFOS control of the GnRH regulatory region in immature and mature GnRH neurons. + : activator, - : repressor.
3.3 cFOS binding to the regulatory region of Gnrh1 controls the capacity of VAX1 to modulate Gnrh1 expression in immortalized GnRH neurons
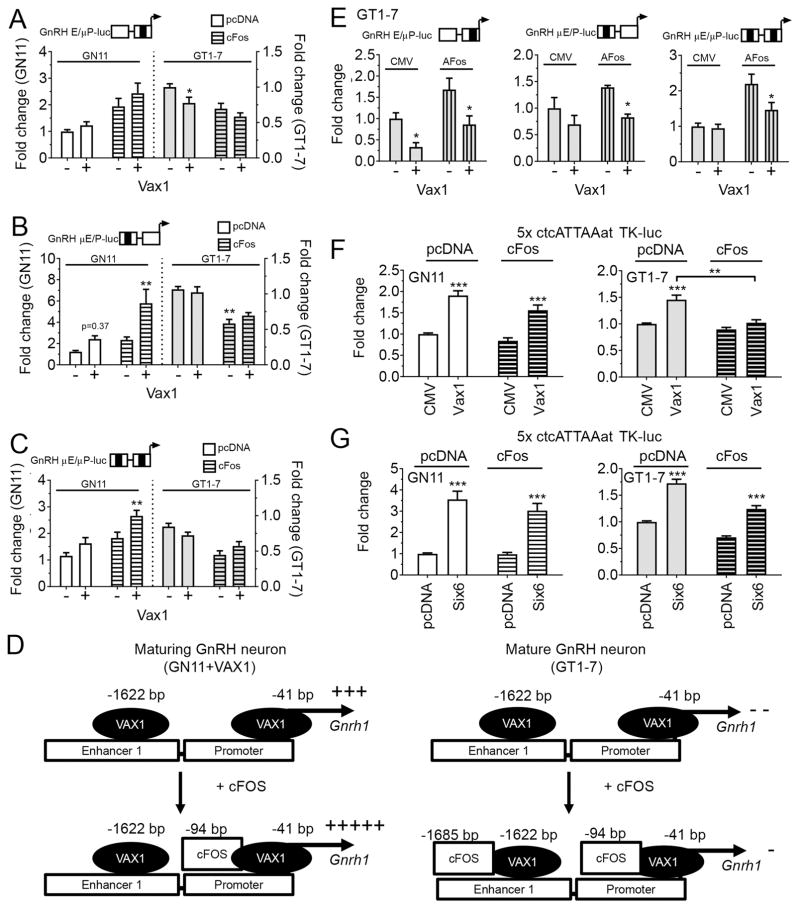
IEGs, such as cFOS and cJUN, regulate transcription by directly interacting with homeodomain transcription factors. To determine if an interaction occurred between cFOS and VAX1 on the Gnrh1 promoter, and if this interaction might change during GnRH neuron development, we asked whether cFOS binding to AP1-half sites in the GnRH E/P-luciferase reporter was required for the synergistic effect of VAX1 and cFOS in GN11 (Figure 3A, Two-way ANOVA, interaction cFOS and VAX1, p = 0.013). Mutation of the AP1-half site in the GnRH promoter (GnRH E/μP) reduced the capacity of VAX1 to regulate this construct in GN11 (fold change in transcriptional induction by VAX1 on GnRH E/P versus GnRH E/μP was reduced by 37 %). The effect of cFOS overexpression on transcription of GnRH E/P and GnRH E/μP (Two-way ANOVA, F(1,48) = 15.75, p = 0.0002) was reduced by 20 % from cFos+GnRH E/P = 2.4 to cFos+GnRH E/μP = 1.9. Mutation of the AP1-half site in the enhancer slightly enhanced VAX1 regulation of this plasmid in GN11 (VAX1+GnRH E/P = 2.0 and VAX1+GnRH μE/P = 2.4, Figure 5B, Two-way ANOVA, F(1,48) = 1.7575, p = 0.189; cFOS-VAX1 interaction on GnRH E/μP F(1.48) = 0.2394, p = 0.6269). Interestingly, the synergistic effect of cFOS and VAX1 observed on GnRH E/P (Figure 3A), was slightly enhanced on the GnRH μE/P construct (Two-way ANOVA, effect of cFOS: F(1,46) = 11,21, p = 0.0016; VAX1: F(1,46) = 12.09, p = 0.0011; interaction: F(1,46) = 2.855, p = 0.096), and changed from 5.3 on GnRH E/P to 5.8 on GnRH μE/P. Further the effect of VAX1-cFOS on GnRH μE/μP (Two-way ANOVA, effect of cFOS: F(1,67) = 19.81, p < 0.0001; VAX1: F(1,67) = 11.54, p = 0.0011; interaction: F(1,67) = 0.912, p = 0.3430) reduced their effect to 2.8, which represents a reduction of 52 %, and a loss of interaction. This suggests cFOS binding to the promoter is involved in the enhanced GnRH expression when cFOS and VAX1 are overexpressed (Figure 5A). In GT1-7 cells, GnRH E/μP abolished VAX1’s capacity to repress GnRH expression in the presence of increased levels of cFOS (GnRH E/μP, Two-way ANOVA, cFOS: (F1,47) = 13.03, p = 0.0007; VAX1: F(1,47) = 5.894, p = 0.0191; interaction F(1,47) = 0.6418, p = 0.427, Figure 5A), whereas mutation of the AP1-half site in the enhancer abolished VAX1 regulation of GnRH μE/P (Figure 5B pcDNA; and C, CMV, GnRH μE/P, Two way ANOVA cFOS: F(1,68) = 50.71, p < 0.0001; VAX1: F(1,68) = 0.357, p = 0.5518; interaction F(1,47) = 1.795, p = 0.1848), and eliminated the capacity of VAX1 to repress transcription (Figure 5B, GT1-7). Further, mutations of both AP1-half sites abolished the capacity of VAX1 to repress this construct (Figure 5C, Two-way ANOVA, VAX1: F(1,85) = 0.0005, p = 0.9823) without impacting cFOS repression (Two way ANOVA cFOS: (F1,85) = 28.42, p < 0.0001). Interestingly, a significant interaction between cFOS and VAX1 was detected (Two-way ANOVA, F(1,85) = 5.742, p = 0.00188), where VAX1 went from slightly repressing the construct in absence of cFOS, to a slight reversal of cFOS repression (Figure 5B, C). To assess the potential role of endogenous cFOS on VAX1 driven GnRH E/P-luciferase expression, basal cFOS was reduced using the dominant negative cFOS plasmid, termed AFOS, a plasmid we had found to be efficient in reverting cFOS regulation of this construct (Figure 1B, C). Surprisingly, the AP1-half site mutations in the GnRH E/P-luciferase reporter only modestly affected the capacity of AFOS to change GnRH E/P luciferase transcription levels (transcription level of AFOS on GnRH E/P = 2.1, GnRH E/μP = 1.7, GnRH μE/P = 1.4, GnRH μE/μP = 2.2). We confirmed the requirement of the partial AP1-half site in the enhancer (E) as required for VAX1 repression (Figure 5D), although the overall effect of VAX1 still reached statistical significance on all constructs (Two-way ANOVA of VAX1 with or without AFOS on GnRH E/μP: F(1,10) = 1.897, p = 0.0014; GnRH μE/P: F(1,11) = 8.891, p = 0.0125; GnRH μE/μP: F(1,16) = 4.59, p = 0.0478). Mutating AP1-half sites in the GnRH E/P did not reveal any interaction between VAX1 and AFOS in GT1-7 cells, (Two-way ANOVA, GnRH E/P: F(1,13) = 0.9098, p = 0.3576; GnRH E/μP: F(1,47) = 0.6418, p = 0.4271; GnRH μE/P: F(1,68) = 1.795, p = 0.1848), although this interaction trended towards significance when both AP1-half sites were mutated (GnRH μE/μP: F(1,16) = 3.418, p = 0.0830), supporting the findings in Figure 5C. Overall, the minor effect of mutating the partial AP1-sites suggests that cFOS might act in part by interacting with VAX1. These results are summarized in Figure 5D. To determine if cFOS can regulate transcription through VAX1, we used a promoter construct containing 5xATTA repeats driving expression of luciferase, the DNA sequence recognized by VAX1. As we have demonstrated previously, VAX1 enhanced expression of the 5xATTA multimer in both GN11 and GT1-7 cells (Figure 5F, Two-way ANOVA, GN11: F(1,43) = 43.1, p < 0.0001; GT1-7: F(1,29) = 30.05, p < 0.0001) (Hoffmann et al., 2016). In GN11, cFOS did not impact VAX1 driven expression of the ATTA-multimer (Two-way ANOVA, F(1,43) = 0.5568, p = 0.4557), whereas cFOS impaired VAX1-driven ATTA-luciferase expression in GT1-7 (Figure 5F, Two-way ANOVA, F(1,29) = 9.819, p = 0.0039). To determine if this effect was specific to VAX1, we repeated the study with SIX6, a homeodomain transcription factor required for GnRH neuron survival and a strong activator of Gnrh1 acting through ATTA sites in the Gnrh1 regulatory region (Larder et al., 2011). As expected, SIX6 strongly increased transcription of the ATTA multimer in both GN11 and GT1-7 cells (Figure 5G, Two-way ANOVA, GN11: F(1,39) = 80.08, p < 0.0001; GT1-7: F(1,43) = 159.8, p < 0.0001), and cFOS overexpression did not significantly affect SIX6 regulation of this construct in either cell line (Figure 5G, Two-way ANOVA, GN11: F(1,39) = 0.9594; GT1-7: F(1,43) = 3.694, p = 0.0613).
Figure 5.
Complex transcriptional interaction between VAX1 and cFOS on the GnRH E/P-luciferase promoter. A–C, E) GN11 (white) and GT1-7 (grey) cells were co-transfected with VAX1, cFOS (horizontal bars), and/or AFOS (vertical bars) and fold change in luciferase expression evaluated on the indicated luciferase reporters. Mutation sites (black bar on schematic over histogram), correspond to the sequences underlined in Figure 1F. Two-way ANOVA followed by Sidak’s post-hoc test * p < 0.05; ** p < 0.01; as compared to control (empty vector). D) Hypothetical model of the role of increased levels of cFOS on VAX1 regulation of the GnRH E/P-luciferase construct. F, G) GN11 and GT1-7 cells were co-transfected with VAX1, SIX6 (200 ng), cFOS (100 ng) or their corresponding empty vectors and fold induction of an ATTA-multimer TK-promoter luciferase reporter evaluated. Two-way ANOVA followed by Sidak’s multiple comparison test, ** < 0.01, *** < 0.001 as compared to empty vector (pcDNA, CMV) or as indicated by bracket, n=3–5.
4. Discussion
4.1 PKC activation in GN11 and GT1-7 cells leads to rapid transcriptional regulation of both cFos and homeodomain transcription factors
GnRH neurons receive a myriad of signals coordinating correct GnRH neuron development and subsequent control of fertility. The requirement of appropriately localized GnRH neurons, in combination with pulsatile GnRH release controls reproductive function and places this neuronal population at the apex of the reproductive axis. Because GnRH release patterns determine reproductive status, and a reduction in GnRH release shuts down the reproductive axis during periods of stress, malnutrition or seasonal changes in day length in seasonal breeders (Lehman et al., 1997; Luo et al., 2016; Mintz et al., 2007), it is unsurprising that the signaling pathways and transcription factors regulating GnRH expression and release are multifold. It is well established that TPA, a PKC activator, enhances cFos, and represses Gnrh1 in the model GnRH neurons GT1-7 through well-defined binding sites on the Gnrh1 regulatory region (Bruder et al., 1992; Bruder et al., 1996; Bruder and Wierman, 1994; Eraly and Mellon, 1995; Glidewell-Kenney et al., 2013; Tang et al., 2005; Wetsel et al., 1993), and chromatin closure (Iyer et al., 2011). Surprisingly, when we investigated to what extent the AP1-half sites identified to bind cFOS were important in TPA-regulation of GnRH expression, we found that mutating these sites only modestly impacted TPA-repression of these reporters in GT1-7 cells, whereas mutation of the AP1-half site in the promoter, abolished the moderate increase in GnRH E/P-luciferase expression by TPA in GN11. This clearly shows that TPA regulates other pathways in order to regulate GnRH expression in GT1-7 cells, and involves TPA-induced signal-transduction in the regulation of Gnrh1 expression in GnRH cells. It should be noted that, although we were able to recapitulate the capacity of TPA to repress Gnrh1 expression in GT1-7 cells, at no point did we detected Gnrh1 in GN11. We were also unable to induce endogenous Gnrh1 expression in GN11 cells by TPA, although TPA enhanced GnRH E/P expression in this cell line. The capacity of TPA to induce GnRH E/P-luciferase in GN11 suggests that, in more mature GnRH neurons, where the GnRH regulatory region is in a more relaxed conformation, TPA might be able to increase Gnrh1 expression. However, due to the few, relatively quick, time points after TPA treatment at which Gnrh1 expression was studied (up to 20 hrs), TPA was unable to allow enhanced Gnrh1 expression, or expression of transcription factors required for GnRH expression, such as VAX1 and SIX6. However, the maintained capacity of TPA to modulate the GnRH E/P-luciferase construct to a great extent when AP1-half sites were mutated, clearly indicated TPA allowed recruitment of other transcription factors than cFOS to the GnRH regulatory region to regulate its expression. Due to the importance of homeodomain transcription factors in GnRH neuron development and function, we speculated that TPA could regulate this group of genes. Thus, we analyzed expression of transcription factors known to be involved in GnRH transcription regulation such as DLX1/5, SIX3/6, VAX1, and OTX2 (Eraly et al., 1998; Hoffmann et al., 2016; Iyer et al., 2010; Kim et al., 2007; Novaira et al., 2012; Tang et al., 2005). In agreement with previous reports, we found that TPA strongly induces cFos and only modestly induces cJun in GT1-7 cells, and represses Gnrh1 in GT1-7 cells (Bruder et al., 1992). Interestingly, TPA rapidly (within 4 h) repressed Dlx1, Dlx5, Vax1, Otx2 and Six6 in GT1-7 cells, and enhanced Dlx5 in GN11, the only homeodomain transcription factor we were able to detect in GN11 of the six studied. We also evaluated Six3 expression, but transcript levels were below the detectable limit in all the studied conditions and therefore Six3 was not included. It is interesting to note that all transcriptional effects observed, aside from the induction of cFos and cJun by TPA, were regulated oppositely in GN11 and GT1-7 cells, supporting the major differences in transcription complexes and chromatin status in these cell lines (Huang et al., 2016; Iyer et al., 2011). Although we only detected endogenous Vax1 in GT1-7 cells, the rapid effect by TPA on Vax1 expression, which we were able to detect at 1h of TPA treatment, made this transcription factor of particular interest to us. As we had observed in our previous study, Vax1 was not expressed in GN11 cells, and highly expressed in GT1-7 cells (Hoffmann and Mellon, 2016; Hoffmann et al., 2016). This differential expression of Vax1, and the loss of Gnrh1 after E13.5 in mice with Vax1 deleted within GnRH neurons (Hoffmann et al., 2016), suggest that the transcriptional program regulated by VAX1 allows for maintenance of GnRH expression after E13.5, and thus VAX1 is turned on in maturing GnRH neurons on or around E13.5. Based on this, we decided to use the GN11 and GT1-7 cells to further our understanding of VAX1 regulation of the GnRH enhancer and promoter (GnRH E/P).
As most homeodomain transcription factors VAX1 regulates GnRH expression through ATTA sites, which are abundant and bound by many other homeodomain transcription factors in these cell lines (Hoffmann et al., 2016). One unique feature of VAX1 is its dosage effect on GnRH neuron numbers (Hoffmann et al., 2014; Hoffmann et al., 2016), which places it in a unique position in GnRH neuron development. We therefore hypothesized that, in addition to being able to compete for binding with SIX6 on the GnRH regulatory region (Hoffmann et al., 2016), VAX1 would interact with other transcription factors in these cells. Indeed, Vax1 overexpression in GT1-7 cells enhanced both endogenous cFos expression in GT1-7 cells, and cFos-luciferase expression in GN11 and GT1-7 cells, an effect we found to be mediated by VAX1 action on the ATTA site at −59 bp from the cFos transcriptional start site. Thus, VAX1 is able to regulate GnRH expression through both a direct action on the GnRH regulatory region and indirectly by enhancing cFos expression (Figure 4G).
4.2 Identification of a GnRH neuron maturation-stage dependent interaction between VAX1 and cFOS, a novel mechanism allowing fine-tuning of homeodomain transcription factor regulation of GnRH expression
The timing and dosage of numerous factors within GnRH neurons, as well as their environment, are required to obtain mature GnRH neurons that are localized in the anterior hypothalamic area and express GnRH. Despite the well-established key roles of homeodomain transcription factors in GnRH neuron development, we were surprised by the dramatic effect of Vax1 KO on the number of neurons expressing GnRH in the brain (Hoffmann et al., 2014; Hoffmann et al., 2016). To further our understanding of the role of VAX1 in GnRH gene regulation, we next asked if VAX1 could regulate GnRH expression by interacting with other transcription factors. As a homeodomain transcription factor, VAX1 controls a transcriptional network regulating eye and ventral forebrain development (Bertuzzi et al., 1999; Bharti et al., 2011; Hallonet et al., 1999). There are different strategies utilized by transcription factors to specifically regulate their respective target genes, such as binding to unique sequences (ATTA), and interactions with other transcription factors or co-factors (Cheatle Jarvela and Hinman, 2015; Kicheva and Briscoe, 2015). Prior work found that homeodomain transcription factors can directly interact with members of the AP1 family, probably fine-tuning expression of their common gene targets (Andreucci et al., 2002; Dony and Gruss, 1988; Jeong et al., 2004; Schaefer et al., 2001). Due to the rapid decline in GnRH-expressing neurons in the Vax1 KO, we focused on the IEG cFOS as its expression is regulated on a rapid time scale and is important in GnRH expression, but only plays a minor role in GnRH neuron development (Xie et al., 2015). As VAX1 expression turns on during GnRH neuron maturation, we hypothesized that transfecting the immature GN11 cells with VAX1 would reflect a maturing GnRH neuron (Figure 4G and 5D). It should be noted that one major limitation to the strategy used is the presence of endogenous Vax1 in GT1-7 cells, levels of which we increased further in the transient transfection studies. However, due to the difficulty in transfecting GT1-7 cells, which only reach about 10 % efficiency, we decided not to attempt to use Vax1 siRNA in the current study to understand how VAX1-cFOS dosage impacts Gnrh1 expression levels. However, we have previously shown that reducing endogenous Vax1 by approximately 70 % allows an increase in Gnrh1 expression in GT1-7 cells (Hoffmann et al., 2016), confirming its role as a repressor on the GnRH promoter. In the current study, we found a GnRH neuron maturation-stage dependent transcriptional interaction between VAX1 and cFOS on the GnRH regulatory region (Figure 5D). In GN11 cells, endogenous cFOS (as evaluated by the use of AFOS) had no role on VAX1 regulation of GnRH E/P, but elimination of cFOS was able to reverse the synergistic effect between cFOS and VAX1, showing that cFOS is required for the synergy in this cell line. Indeed, this synergy depended upon cFOS binding principally to the AP1-half site in the GnRH promoter, and did not require direct interaction between cFOS and VAX1, as the synergy was lost on the ATTA-multimer. We next asked if there would also be synergy between cFOS and VAX1 in the mature GnRH cells, GT1-7. First, we confirmed that VAX1 and cFOS reduced GnRH E/P-luciferase levels (Bruder et al., 1996; Bruder and Wierman, 1994; Ely et al., 2011; Hoffmann and Mellon, 2016; Hoffmann et al., 2016; Wetsel et al., 1993). Interestingly, simultaneous overexpression of VAX1 and cFOS leads to GnRH E/P-luciferase transcription levels approximately the same as those observed by cFOS alone (0.56 ± 0.06 versus 0.60 ± 0.06, respectively), showing that cFOS and VAX1 transcriptional regulation of the GnRH E/P-luciferase construct in GT1-7 cells is non-additive, and might involve competition for occupancy of the promoter, due to the close proximity of VAX1 and cFOS binding sites (Figure 1F). To determine whether VAX1 and cFOS were competing for occupancy, we asked if cFOS would still impact VAX1-driven transcription in the absence of AP1-half sites in the GnRH E (GnRH μE/P), the GnRH P (GnRH E/μP), or both (GnRH μE/μP). Interestingly, in contrast to what we expected, cFOS binding to the GnRH E was required for VAX1 repression of GnRH transcription (see Figure 5B and C, GT1-7). To determine if cFOS and VAX1 can participate in the same transcriptional complex, we asked if cFOS overexpression would impact VAX1-driven expression of the ATTA-multimer. Indeed, cFOS was able to abolish VAX1-enhanced expression of the ATTA-multimer in GT1-7 cells, whereas this was not the case in GN11. The fact that cFOS was unable to alter SIX6-enhanced transcription of the ATTA-multimer in either of the two cell lines, suggests that the VAX1-cFOS interaction on the GnRH regulatory region is specific. This indicates that in GT1-7 cells, VAX1 and cFOS possibly form part of the same transcriptional complex on the GnRH promoter. Such large transcriptional complexes have been described and are suggested to be important in specific targeting of particular genes to discrete cell populations (Andreucci et al., 2002; Jeong et al., 2004; Kicheva and Briscoe, 2015; Schaefer et al., 2001). Thus, in GT1-7 cells, there was no interaction between endogenous cFOS and VAX1 on the GnRH E/P-luciferase construct (Figure 5D). AFOS did not impact VAX1 regulation of GnRH E/P-luciferase (no interaction), whereas cFOS binding to the GnRH enhancer or promoter reduced VAX1 repression (Figure 5D). Overall, these data support the previously suggested functional importance of the interaction of cFOS with homeodomain transcription factors as a mechanism to target specific promoters and fine-tune transcriptional regulation (Dony and Gruss, 1988; Schaefer et al., 2001).
In adulthood, the interaction between cFOS and VAX1 might also be physiologically relevant at the time of the preovulatory LH surge or during sexual stimulation, both of which are associated with increased expression of cFos in GnRH neurons (Clarkson et al., 2008; Hoffman et al., 1990; Lee et al., 1992; Pfaus et al., 1994). During conditions with increased cFos expression, the interaction between VAX1 and cFOS would reduce the repressive effect of cFOS on the GnRH promoter, and thus maintain a specific level of GnRH transcription, which would be advantageous in conditions when fertility maintenance is required.
5. Conclusion
We provide exciting new evidence of a transcriptional interaction of cFOS and VAX1 on the Gnrh1 regulatory region, and these interactions dynamically change depending on GnRH neuron maturation stage. We show that cFOS is involved in enhancing GnRH expression during GnRH neuron maturation, an effect potentiated in the presence of VAX1. In contrast, both cFOS and VAX1 repress GnRH expression in the mature GnRH neurons, where increased levels of cFOS seem to counteract VAX1 regulation of this promoter. Thus, our data suggest that concomitant expression of cFOS and VAX1 in GnRH neurons allows fine-tuning of homeodomain transcription factor-regulated GnRH expression in both immature and mature GnRH cell lines.
Highlights.
VAX1 induces cFos expression in the model GnRH cell lines, GN11 and GT1-7
cFOS and VAX1 enhance GnRH expression in the immature mouse GnRH cell line, GN11
TPA has opposite effects on GnRH-luciferase expression in GN11 and GT1-7 cells
TPA regulates expression of homeodomain protein transcripts in GnRH neurons
VAX1 interaction with cFOS on GnRH E/P-luciferase sets the level of GnRH expression
Acknowledgments
This work was supported by National Institutes of Health (NIH) Grants R01 HD072754, R01 HD082567 (to P.L.M.). It was also supported by NICHD/NIH P50 HD012303 as part of the National Centers for Translational Research in Reproduction and Infertility (P.L.M.). P.L.M. was also partially supported by P30 DK063491, P30 CA023100, and P42 ES101337. H.M.H. was partially supported by K99 HD084759. We thank Genevieve E. Ryan, Erica L. Schoeller, and Shanna Newton for critical reading of the manuscript Dr. Dorota Skowronska-Krawczyk for image acquisition.
Abbreviations
- AP1
Activator protein-1
- BSA
Bovine serum albumin
- cFos
FBJ osteosarcoma oncogene
- cJun
jun proto-oncogene
- Dlx
Distal-less homeobox
- DMSO
Dimethylsulfoxide
- E
Embryonic day
- Fgf
Fibroblast growth factor
- FGFR
Fgf receptor
- GnRH
Gonadotropin-releasing hormone
- H2afz
H2A histone family, member Z
- IEG
Immediate early gene
- KO
knock-out
- LH
Luteinizing hormone
- Otx2
Orthodenticle homeobox 2
- PKC
protein kinase C
- Ppia
peptidylprolyl isomerase A
- qRT-PCR
Quantitative real-time
- PCR SRF
serum response factor
- Six
Sine oculis-related homeobox
- TAC
Tachykinin receptor
- TPA
Phorbol 12-myristate 13-acetate
- Vax1
Ventral anterior homeobox 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andreucci JJ, Grant D, Cox DM, Tomc LK, Prywes R, Goldhamer DJ, Rodrigues N, Bedard PA, McDermott JC. Composition and function of AP-1 transcription complexes during muscle cell differentiation. J Biol Chem. 2002;277:16426–16432. doi: 10.1074/jbc.M110891200. [DOI] [PubMed] [Google Scholar]
- Balasubramanian R, Dwyer A, Seminara SB, Pitteloud N, Kaiser UB, Crowley WF., Jr Human GnRH deficiency: a unique disease model to unravel the ontogeny of GnRH neurons. Neuroendocrinology. 2010;92:81–99. doi: 10.1159/000314193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertuzzi S, Hindges R, Mui SH, O’Leary DD, Lemke G. The homeodomain protein vax1 is required for axon guidance and major tract formation in the developing forebrain. Genes Dev. 1999;13:3092–3105. doi: 10.1101/gad.13.23.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti K, Gasper M, Bertuzzi S, Arnheiter H. Lack of the ventral anterior homeodomain transcription factor VAX1 leads to induction of a second pituitary. Development. 2011;138:873–878. doi: 10.1242/dev.056465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J, Small S. Morphogen rules: design principles of gradient-mediated embryo patterning. Development. 2015;142:3996–4009. doi: 10.1242/dev.129452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruder JM, Drebs WD, Nett TM, Wierman ME. Phorbol ester activation of the protein kinase C pathway inhibits gonadotropin-releasing hormone gene expression. Endocrinology. 1992;131:2552–2558. doi: 10.1210/endo.131.6.1446598. [DOI] [PubMed] [Google Scholar]
- Bruder JM, Spaulding AJ, Wierman ME. Phorbol ester inhibition of rat gonadotropin-releasing hormone promoter activity: role of fos and jun in the repression of transcription. Mol Endocrinol. 1996;10:35–44. doi: 10.1210/mend.10.1.8838143. [DOI] [PubMed] [Google Scholar]
- Bruder JM, Wierman ME. Evidence for transcriptional inhibition of GnRH gene expression by phorbol ester at a proximal promoter region. Mol Cell Endocrinol. 1994;99:177–182. doi: 10.1016/0303-7207(94)90006-x. [DOI] [PubMed] [Google Scholar]
- Cariboni A, Hickok J, Rakic S, Andrews W, Maggi R, Tischkau S, Parnavelas JG. Neuropilins and their ligands are important in the migration of gonadotropin-releasing hormone neurons. J Neurosci. 2007;27:2387–2395. doi: 10.1523/JNEUROSCI.5075-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheatle Jarvela AM, Hinman VF. Evolution of transcription factor function as a mechanism for changing metazoan developmental gene regulatory networks. Evodevo. 2015;6:3. doi: 10.1186/2041-9139-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson J, d’Anglemont de Tassigny X, Moreno AS, Colledge WH, Herbison AE. Kisspeptin-GPR54 signaling is essential for preovulatory gonadotropin-releasing hormone neuron activation and the luteinizing hormone surge. J Neurosci. 2008;28:8691–8697. doi: 10.1523/JNEUROSCI.1775-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dateki S, Kosaka K, Hasegawa K, Tanaka H, Azuma N, Yokoya S, Muroya K, Adachi M, Tajima T, Motomura K, Kinoshita E, Moriuchi H, Sato N, Fukami M, Ogata T. Heterozygous orthodenticle homeobox 2 mutations are associated with variable pituitary phenotype. J Clin Endocrinol Metab. 2010;95:756–764. doi: 10.1210/jc.2009-1334. [DOI] [PubMed] [Google Scholar]
- Diaczok D, DiVall S, Matsuo I, Wondisford FE, Wolfe AM, Radovick S. Deletion of Otx2 in GnRH neurons results in a mouse model of hypogonadotropic hypogonadism. Mol Endocrinol. 2011;25:833–846. doi: 10.1210/me.2010-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dony C, Gruss P. Expression of a murine homeobox gene precedes the induction of c-fos during mesodermal differentiation of P19 teratocarcinoma cells. Differentiation. 1988;37:115–122. doi: 10.1111/j.1432-0436.1988.tb00803.x. [DOI] [PubMed] [Google Scholar]
- Ely HA, Mellon PL, Coss D. GnRH Induces the c-Fos gene via phosphorylation of SRF by the calcium/calmodulin kinase II pathway. Mol Endocrinol. 2011;25:669–680. doi: 10.1210/me.2010-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eraly SA, Mellon PL. Regulation of GnRH transcription by protein kinase C is mediated by evolutionarily conserved, promoter-proximal elements. Mol Endocrinol. 1995;9:848–859. doi: 10.1210/mend.9.7.7476968. [DOI] [PubMed] [Google Scholar]
- Eraly SA, Nelson SB, Huang KM, Mellon PL. Oct-1 binds promoter elements required for transcription of the gonadotropin-releasing hormone gene. Mol Endocrinol. 1998;12:469–481. doi: 10.1210/mend.12.4.0092. [DOI] [PubMed] [Google Scholar]
- Fueshko S, Wray S. LHRH cells migrate on peripherin fibers in embryonic olfactory explant cultures: an in vitro model for neurophilic neuronal migration. Dev Biol. 1994;166:331–348. doi: 10.1006/dbio.1994.1319. [DOI] [PubMed] [Google Scholar]
- Givens ML, Rave-Harel N, Goonewardena VD, Kurotani R, Berdy SE, Swan CH, Rubenstein JL, Robert B, Mellon PL. Developmental regulation of gonadotropin-releasing hormone gene expression by the MSX and DLX homeodomain protein families. J Biol Chem. 2005;280:19156–19165. doi: 10.1074/jbc.M502004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glidewell-Kenney CA, Shao PP, Iyer AK, Grove AM, Meadows JD, Mellon PL. Neurokinin B Causes Acute GnRH Secretion and Repression of GnRH Transcription in GT1-7 GnRH Neurons. Mol Endocrinol. 2013;27:437–454. doi: 10.1210/me.2012-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glidewell-Kenney CA, Trang C, Shao PP, Gutierrez-Reed N, Uzo-Okereke AM, Coss D, Mellon PL. Neurokinin B induces c-fos transcription via protein kinase C and activation of serum response factor and Elk-1 in immortalized GnRH neurons. Endocrinology. 2014:en20141263. doi: 10.1210/en.2014-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallonet M, Hollemann T, Pieler T, Gruss P. Vax1, a novel homeobox-containing gene, directs development of the basal forebrain and visual system. Genes Dev. 1999;13:3106–3114. doi: 10.1101/gad.13.23.3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbison AE, Porteous R, Pape JR, Mora JM, Hurst PR. Gonadotropin-releasing hormone (GnRH) neuron requirements for puberty, ovulation and fertility. Endocrinology. 2008;149:597–604. doi: 10.1210/en.2007-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman GE, Lee WS, Attardi B, Yann V, Fitzsimmons MD. Luteinizing hormone-releasing hormone neurons express c-fos antigen after steroid activation. Endocrinology. 1990;126:1736–1741. doi: 10.1210/endo-126-3-1736. [DOI] [PubMed] [Google Scholar]
- Hoffmann HM, Mellon PL. A small population of hypothalamic neurons govern fertility: the critical role of VAX1 in GnRH neuron development and fertility maintenance. Neurosci Commun (Houst) 2016:2. [PMC free article] [PubMed] [Google Scholar]
- Hoffmann HM, Tamrazian A, Xie H, Perez-Millan MI, Kauffman AS, Mellon PL. Heterozygous deletion of ventral anterior homeobox (Vax1) causes subfertility in mice. Endocrinology. 2014;155:4043–4053. doi: 10.1210/en.2014-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann HM, Trang C, Gong P, Kimura I, Pandolfi EC, Mellon PL. Deletion of Vax1 from GnRH neurons abolishes GnRH expression and leads to hypogonadism and infertility. J Neurosci. 2016;36:3506–3518. doi: 10.1523/JNEUROSCI.2723-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang PP, Brusman LE, Iyer AK, Webster NJ, Mellon PL. A Novel Gonadotropin-Releasing Hormone 1 (Gnrh1) Enhancer-Derived Noncoding RNA Regulates Gnrh1 Gene Expression in GnRH Neuronal Cell Models. PLoS One. 2016;11:e0158597. doi: 10.1371/journal.pone.0158597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer AK, Brayman MJ, Mellon PL. Dynamic chromatin modifications control GnRH gene expression during neuronal differentiation and protein kinase C signal transduction. Mol Endocrinol. 2011;25:460–473. doi: 10.1210/me.2010-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer AK, Miller NL, Yip K, Tran BH, Mellon PL. Enhancers of GnRH transcription embedded in an upstream gene use homeodomain proteins to specify hypothalamic expression. Mol Endocrinol. 2010;24:1949–1964. doi: 10.1210/me.2010-0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong KH, Chin WW, Kaiser UB. Essential role of the homeodomain for pituitary homeobox 1 activation of mouse gonadotropin-releasing hormone receptor gene expression through interactions with c-Jun and DNA. Mol Cell Biol. 2004;24:6127–6139. doi: 10.1128/MCB.24.14.6127-6139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley CG, Lavorgna G, Clark ME, Boncinelli E, Mellon PL. The Otx2 homeoprotein regulates expression from the gonadotropin-releasing hormone proximal promoter. Mol Endocrinol. 2000;14:1246–1256. doi: 10.1210/mend.14.8.0509. [DOI] [PubMed] [Google Scholar]
- Kessel M, Fibi M, Gruss P. Organization of homeodomain proteins. Prog Clin Biol Res. 1988;284:93–104. [PubMed] [Google Scholar]
- Kicheva A, Briscoe J. Developmental Pattern Formation in Phases. Trends Cell Biol. 2015;25:579–591. doi: 10.1016/j.tcb.2015.07.006. [DOI] [PubMed] [Google Scholar]
- Kim HH, Wolfe A, Cohen RN, Eames SC, Johnson AL, Wieland CN, Radovick S. In vivo identification of a 107 bp promoter element mediating neuron-specific expression of mouse GnRH. Mol Endocrinol. 2007;21:457–471. doi: 10.1210/me.2005-0216. [DOI] [PubMed] [Google Scholar]
- Larder R, Clark DD, Miller NL, Mellon PL. Hypothalamic dysregulation and infertility in mice lacking the homeodomain protein Six6. J Neurosci. 2011;31:426–438. doi: 10.1523/JNEUROSCI.1688-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larder R, Kimura I, Meadows J, Clark DD, Mayo S, Mellon PL. Gene dosage of Otx2 is important for fertility in male mice. Mol Cell Endocrinol. 2013;377:16–22. doi: 10.1016/j.mce.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WS, Smith MS, Hoffman GE. cFos Activity Identifies Recruitment of Luteinizing Hormone-Releasing Hormone Neurons During the Ascending Phase of the Proestrous Luteinizing Hormone Surge. J Neuroendocrinol. 1992;4:161–166. doi: 10.1111/j.1365-2826.1992.tb00154.x. [DOI] [PubMed] [Google Scholar]
- Lehman MN, Goodman RL, Karsch FJ, Jackson GL, Berriman SJ, Jansen HT. The GnRH system of seasonal breeders: anatomy and plasticity. Brain Res Bull. 1997;44:445–457. doi: 10.1016/s0361-9230(97)00225-6. [DOI] [PubMed] [Google Scholar]
- Lewis J. From signals to patterns: space, time, and mathematics in developmental biology. Science. 2008;322:399–403. doi: 10.1126/science.1166154. [DOI] [PubMed] [Google Scholar]
- Luo E, Stephens SB, Chaing S, Munaganuru N, Kauffman AS, Breen KM. Corticosterone Blocks Ovarian Cyclicity and the LH Surge via Decreased Kisspeptin Neuron Activation in Female Mice. Endocrinology. 2016;157:1187–1199. doi: 10.1210/en.2015-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magni P, Dozio E, Ruscica M, Watanobe H, Cariboni A, Zaninetti R, Motta M, Maggi R. Leukemia inhibitory factor induces the chemomigration of immortalized gonadotropin-releasing hormone neurons through the independent activation of the Janus kinase/signal transducer and activator of transcription 3, mitogen-activated protein kinase/extracellularly regulated kinase 1/2, and phosphatidylinositol 3-kinase/Akt signaling pathways. Mol Endocrinol. 2007;21:1163–1174. doi: 10.1210/me.2006-0270. [DOI] [PubMed] [Google Scholar]
- Mason AJ, Hayflick JS, Zoeller RT, Young WS, Phillips HS, Nikolics K, Seeburg PH. A deletion truncating the gonadotropin-releasing hormone gene is responsible for hypogonadism in the hpg mouse. Science. 1986;234:1366–1371. doi: 10.1126/science.3024317. [DOI] [PubMed] [Google Scholar]
- Mellon PL, Windle JJ, Goldsmith P, Padula C, Roberts J, Weiner RI. Immortalization of hypothalamic GnRH neurons by genetically targeted tumorigenesis. Neuron. 1990;5:1–10. doi: 10.1016/0896-6273(90)90028-e. [DOI] [PubMed] [Google Scholar]
- Messina A, Langlet F, Chachlaki K, Roa J, Rasika S, Jouy N, Gallet S, Gaytan F, Parkash J, Tena-Sempere M, Giacobini P, Prevot V. A microRNA switch regulates the rise in hypothalamic GnRH production before puberty. Nat Neurosci. 2016 doi: 10.1038/nn.4298. [DOI] [PubMed] [Google Scholar]
- Mintz EM, Lavenburg KR, Blank JL. Short photoperiod and testosterone-induced modification of GnRH release from the hypothalamus of Peromyscus maniculatus. Brain Res. 2007;1180:20–28. doi: 10.1016/j.brainres.2007.08.083. [DOI] [PubMed] [Google Scholar]
- Nelson SB, Lawson MA, Kelley CG, Mellon PL. Neuron-specific expression of the rat gonadotropin-releasing hormone gene is conferred by interactions of a defined promoter element with the enhancer in GT1-7 cells. Mol Endocrinol. 2000;14:1509–1522. doi: 10.1210/mend.14.9.0521. [DOI] [PubMed] [Google Scholar]
- Novaira HJ, Fadoju D, Diaczok D, Radovick S. Genetic mechanisms mediating kisspeptin regulation of GnRH gene expression. J Neurosci. 2012;32:17391–17400. doi: 10.1523/JNEUROSCI.2438-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive M, Krylov D, Echlin DR, Gardner K, Taparowsky E, Vinson C. A dominant negative to activation protein-1 (AP1) that abolishes DNA binding and inhibits oncogenesis. J Biol Chem. 1997;272:18586–18594. doi: 10.1074/jbc.272.30.18586. [DOI] [PubMed] [Google Scholar]
- Pfaus JG, Jakob A, Kleopoulos SP, Gibbs RB, Pfaff DW. Sexual stimulation induces Fos immunoreactivity within GnRH neurons of the female rat preoptic area: interaction with steroid hormones. Neuroendocrinology. 1994;60:283–290. doi: 10.1159/000126760. [DOI] [PubMed] [Google Scholar]
- Radovick S, Wondisford FE, Nakayama Y, Yamada M, Cutler GBJ, Weintraub BD. Isolation and characterization of the human gonadotropin-releasing hormone gene in the hypothalamus and placenta. Mol Endocrinol. 1990;4:476–480. doi: 10.1210/mend-4-3-476. [DOI] [PubMed] [Google Scholar]
- Schaefer LK, Wang S, Schaefer TS. Functional interaction of Jun and homeodomain proteins. J Biol Chem. 2001;276:43074–43082. doi: 10.1074/jbc.M102552200. [DOI] [PubMed] [Google Scholar]
- Stamou MI, Cox KH, Crowley WF. Discovering Genes Essential to the Hypothalamic Regulation of Human Reproduction Using a Human Disease Model: Adjusting to Life in the “-Omics” Era. Endocr Rev. 2015:er20151045. doi: 10.1210/er.2015-1045. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tang Q, Mazur M, Mellon PL. The protein kinase C pathway acts through multiple transcription factors to repress gonadotropin-releasing hormone gene expression in hypothalamic GT1-7 neuronal cells. Mol Endocrinol. 2005;19:2769–2779. doi: 10.1210/me.2004-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarozzo G, Peretto P, Perroteau I, Andreone C, Varga Z, Nicholls J, Fasolo A. GnRH neurons and other cell populations migrating from the olfactory neuroepithelium. Ann Endocrinol (Paris) 1994;55:249–254. [PubMed] [Google Scholar]
- Topaloglu AK, Reimann F, Guclu M, Yalin AS, Kotan LD, Porter KM, Serin A, Mungan NO, Cook JR, Ozbek MN, Imamoglu S, Akalin NS, Yuksel B, O’Rahilly S, Semple RK. TAC3 and TACR3 mutations in familial hypogonadotropic hypogonadism reveal a key role for Neurokinin B in the central control of reproduction. Nat Genet. 2009;41:354–358. doi: 10.1038/ng.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetsel WC, Eraly SA, Whyte DB, Mellon PL. Regulation of gonadotropin-releasing hormone by protein kinases A and C in immortalized hypothalamic neurons. Endocrinology. 1993;132:2360–2370. doi: 10.1210/endo.132.6.8504741. [DOI] [PubMed] [Google Scholar]
- Windle JJ, Weiner RI, Mellon PL. Cell lines of the pituitary gonadotrope lineage derived by targeted oncogenesis in transgenic mice. Mol Endocrinol. 1990;4:597–603. doi: 10.1210/mend-4-4-597. [DOI] [PubMed] [Google Scholar]
- Xie C, Jonak CR, Kauffman AS, Coss D. Gonadotropin and kisspeptin gene expression, but not GnRH, are impaired in cFOS deficient mice. Mol Cell Endocrinol. 2015;411:223–231. doi: 10.1016/j.mce.2015.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Seminara S. Uncovering novel reproductive defects in neurokinin B receptor null mice: Closing the gap between mice and men. Endocrinology. 2012;153:1498–1508. doi: 10.1210/en.2011-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakaria M, Dunn IC, Zhen S, Su E, Smith E, Patriquin E, Radovick S. Phorbol ester regulation of the gonadotropin-releasing hormone (GnRH) gene in GnRH-secreting cell lines: a molecular basis for species differences. Molecular Endocrinology. 1996;10:1282–1291. doi: 10.1210/mend.10.10.9121495. [DOI] [PubMed] [Google Scholar]