Abstract
The A3 adenosine receptor (A3AR) subtype is a novel, promising therapeutic target for inflammatory diseases, such as rheumatoid arthritis (RA) and psoriasis, as well as liver cancer. A3AR is coupled to inhibition of adenylyl cyclase and regulation of mitogen-activated protein kinase (MAPK) pathways, leading to modulation of transcription. Furthermore, A3AR affects functions of almost all immune cells and the proliferation of cancer cells. Numerous A3AR agonists, partial agonists, antagonists, and allosteric modulators have been reported, and their structure–activity relationships (SARs) have been studied culminating in the development of potent and selective molecules with drug-like characteristics. The efficacy of nucleoside agonists may be suppressed to produce antagonists, by structural modification of the ribose moiety. Diverse classes of heterocycles have been discovered as selective A3AR blockers, although with large species differences. Thus, as a result of intense basic research efforts, the outlook for development of A3AR modulators for human therapeutics is encouraging. Two prototypical selective agonists, N6-(3-Iodobenzyl)adenosine-5′-N-methyluronamide (IB-MECA; CF101) and 2-chloro-N6-(3-iodobenzyl)-adenosine-5′-N-methyluronamide (Cl-IB-MECA; CF102), have progressed to advanced clinical trials. They were found safe and well tolerated in all preclinical and human clinical studies and showed promising results, particularly in psoriasis and RA, where the A3AR is both a promising therapeutic target and a biologically predictive marker, suggesting a personalized medicine approach. Targeting the A3AR may pave the way for safe and efficacious treatments for patient populations affected by inflammatory diseases, cancer, and other conditions.
Keywords: A3 adenosine receptor, inflammation, cancer, drug development, therapy
1 INTRODUCTION
The relevance of adenosine in the immune system has been established based on mounting scientific evidence showing that the nucleoside represents a paracrine inhibitor of inflammation, regulating the onset, extension, and termination of the inflammatory process and acting through four G protein coupled receptors (GPCRs), designated as A1, A2A, A2B, and A3 adenosine receptors (ARs).1 Following inflammation, metabolic alterations occur leading to an increase of extracellular adenosine that is present in the low nanomolar range under physiological conditions, while in stressful conditions it can rise to micromolar levels.2 Adenosine in the extracellular milieu is largely formed by hydrolysis/dephosphorylation of ATP, ADP, and AMP through specific ectonucleotidases termed ectonucleoside triphosphate diphosphohydrolase (CD39) and ecto-5′-nucleotidase (CD73).3,4 Intracellular levels of adenosine are derived from hydrolysis of AMP and S-adenosylhomocysteine (SAH) through cytosolic 5′-nucleotidase, and SAH hydrolase, respectively. Adenosine activity is extinguished through its phosphorylation to AMP by adenosine kinase (AK) or deamination to inosine by adenosine deaminases (ADA1 and ADA2), with ADA present also extracellularly.2 The existence of concentrative nucleoside transporters (CNTs) and equilibrative nucleoside transporters (ENTs) regulates the extra- and intracellular adenosine concentrations.5
The A3AR, the last of the four subtypes to be discovered, was cloned sequentially in rat, sheep, and human,6–8 but it was not shown to respond as an AR from the outset. One of the first activities of this receptor to be reported was the induction of histamine from rat basophilic cells.9 The discovery and initial characterization of the A3AR, and the exploration of its biological paradoxes, has led to the synthesis and biological characterization of a multitude of receptor probe molecules and clinically relevant candidate molecules, including orthosteric agonists and antagonists as well as allosteric enhancers. This discovery of the A3AR as a fourth AR has spawned current and projected clinical trials of several A3AR agonists and potentially of a selective A3AR allosteric enhancer, as well.10
Using mechanisms triggered by adenosine to inhibit the immune system is a very exciting area of research, and increasing attention is focused on their elucidation in the context of developing new anti-inflammatory strategies. Thus, today the A3ARsubtype is considered a novel, very promising therapeutic target and predictive biological marker, given its overexpression in inflammatory and cancer cells, compared to low levels found in healthy cells.11
The aim of this review is to summarize the state and the progress of the field of A3AR modulators and their clinical development in the context of inflammation and cancer and other conditions, with an emphasis on rheumatoid arthritis (RA), psoriasis, and hepatocellular carcinoma.
2 MOLECULAR BIOLOGY OF A3AR
The A3AR, the only AR subtype cloned before its pharmacological identification, was initially isolated from rat testis and then from a variety of species. The A3AR structure had a sequence homology of only 74% in rat versus sheep and human, versus 85% between sheep and human, suggesting significant interspecies differences in ligand recognition. This is manifested in different pharmacological profiles of the species homologs, especially with respect to antagonist binding, which have made the characterization of this AR subtype difficult.12 There are also species differences in the biological roles of the A3AR, for example, as the main mechanism for adenosine–induced release of inflammatory mediators in rat mast cells, but not in those of human.13
The A3ARis located on human chromosome 1p21-p13 and consists of a single chain of 318 amino acids.14 The A3AR gene presents two exons separated by a single intron of about 2.2 kb.15 Its promoter region has putative binding sites for multiple transcription factors: The upstream sequence has a CCAAT sequence, as well as consensus binding sites for SP1, NF-IL6, GATA1, and GATA3 transcription factors, the latter of which is important for the A3AR-dependent role in immune function. Two species of mRNA code for the hA3AR (sizes 2 and ~5 kb). Variants of the A3AR have been shown to be associated with coronary heart disease, autism spectrum disorder, and aspirin-induced urticaria.16–18 Recently, the A3AR 3′-UTR (untranslated region) of the mRNA was found to be targeted by the proinflammatory microRNA (miR-206) in ulcerative colitis leading to downregulation of A3AR mRNA/protein expression in colon cells.19 The A3AR is expressed in diverse tissues at relatively low levels, compared to A1AR and A2AAR. Genomic analysis of the expression of the A3AR gene in various human tissues (Table 1A) shows highest levels in testes, the spinal cord, and various brain regions, bladder, lung, adipose tissue, and whole blood. The highest expression reached 12.4 RPKM (reads per kilobase of transcript per million mapped reads), while comparable data for A1AR and A2AAR and exceeded 20 RPKM at maximal levels in specific tissues. This suggests that potential use of A3AR ligands in pain and other nervous system disorders is supported by the presence of the receptor in these tissues, although the cell type is not determined in this RNA sequencing data. Various cancer tumors also show major alteration in A3AR expression in comparison to normal tissue. As accessed from a public cancer database (Table 1B), in 393 unique genomic analyses of cancerous tumors, 25 showed a significant increase in A3AR (P < 10−4) and 28 showed a significant decrease compared to normal tissue of the same type. The most prominent increases were in brain cancer (particularly glioblastoma and astrocytoma) and kidney cancer (particularly renal clear cell carcinoma). Thus, the approach of using of A3AR ligands in a wide range of cancers coincides with significant changes in the receptor expression level in tumors.
TABLE 1.
Expression of the A3AR RNA in Normal and Cancerous Tissues from Public Databases
| (A) Exon expression for the A3AR gene in various postmortem human tissues, from RNA sequencing dataa,b | |
|---|---|
| Tissue | RPKMb |
| Testes | 12.401 |
| Brain (spinal cord, cervical C-1) | 5.612 |
| Brain (substantia nigra) | 4.268 |
| Adrenal gland | 3.884 |
| Spleen | 3.495 |
| Small intestine (terminal ileum) | 2.778 |
| Brain (amygdala) | 2.405 |
| Brain (hypothalamus) | 2.201 |
| Nerve (tibial) | 2.102 |
| Brain (hippocampus) | 1.99 |
| Bladder | 1.764 |
| Lung | 1.747 |
| Adipose (subcutaneous) | 1.73 |
| Whole blood | 1.709 |
| Colon (transverse) | 1.604 |
| Artery (coronary) | 1.517 |
| (B) Alteration in the level of A3AR in cancerous tumors compared to normal tissuec | |
| Tumor | Percentile (no. of analyses)c |
| Upregulation | |
| Brain and CNS cancerd | 1% (4/29) |
| Kidney cancer | 1% (6/20) |
| Breast cancer | 5% (11/43) |
| Esophageal cancer | 10% (1/9) |
| Downregulation | |
| Bladder cancer | 5% (3/10) |
| Colorectal cancer | 5% (12/33) |
| Sarcoma | 5% (1/31) |
| Brain and CNS cancer | 10% (1/29) |
| Cervical cancer | 10% (1/10) |
| Myeloma | 10% (1/8) |
Data from The Genotype-Tissue Expression (GTEx) Project. The data were accessed from the GTEx portal (http://www.gtexportal.org/home/) on February 9, 2017, GTEx Analysis Release V6p (dbGaP Accession phs000424.v6.p1).
RPKM stands for reads per kilobase of transcript per million mapped reads for the A3AR gene (ADORA3, gencode ID ENSG00000121933.13). The highest 16 values are shown from a total of 53 tissues assayed.
Percentile refers to the best gene rank percentile for the analyses within the cell. The data were accessed from http://oncomine.org on February 9, 2017, using gene summary visualization for ADORA3. Ratio refers to the number of analyses out of the total number that met the criterion of p < 10−4 for the change in expression in cancer versus normal tissue.
Highly significant upregulation of ADORA3 noted in numerous analyses of glioblastoma and astrocytoma.
As is common to the GPCR superfamily, the A3AR is characterized by seven transmembrane (TM) domains and an intracellular C-terminal region, with Ser and Thr residues serving as potential phosphorylation sites relevant for rapid receptor desensitization. Following agonist stimulation, the A3AR undergoes phosphorylation at the C-terminus by GPCR kinases and subsequent internalization through clathrin-coated pits.20–24 Interestingly, by mutational studies, it has been reported that the highly conserved Trp (W6.48) in TM6 is essential for the active conformation of A3AR necessary to trigger a series of intracellular pathways for signal transmission, to interact with β-arrestin2, and to undergo receptor internalization.25 Furthermore, use of a novel fluorescent A3AR agonist has allowed for the observation of colocalization with internalized receptor–arrestin complexes.26
The energetics of A3AR ligand interactions has been studied using a thermodynamic approach. The thermodynamic parameters of ligand binding at all ARs are similar within either agonist or antagonist classes, which reflects a common ligand receptor interaction mechanism with other ARs This commonality is proposed to explain the difficulty in designing selective adenosine ligands.27–29
3 DISTRIBUTION IN IMMUNE AND CANCER CELLS
The A3AR is highly expressed in several immune cell types, as well as in cancer cells.11 In particular, the native human A3AR was first revealed in human eosinophils and subsequently in neutrophils, monocytes, macrophages, foam cells, dendritic cells, lymphocytes, splenocytes, bone marrow cells, lymphonodes, synoviocytes, chondrocytes, osteoblasts, and mast cells.9,13,30–63 Overall, the presence of the A3ARin almost all the cells involved in inflammatory processes suggests their potential involvement in a number of inflammatory pathologies, spanning from wound healing and remodeling to lung injury, inflammatory bone loss, autoimmune, and eye diseases.2 In addition, high A3AR expression has been observed using biochemical methods in many of types of cancer cells, including astrocytoma, melanoma, lymphoma, sarcoma, glioblastoma, colon, liver, pancreas, prostate, thyroid, lung, breast, and renal carcinomas.64–89 This expression pattern reflects a demonstrated role for this subtype in tumor biology.
4 MEDICINAL CHEMISTRY OF THE A3 ADENOSINE RECEPTOR
4.1 Adenosine derivatives as agonists of the A3 adenosine receptor
The first efforts to develop A3AR selective agonists (Table 2) were performed at the US National Institutes of Health (NIH),90 and culminated in the report of N6-(3-iodobenzyl)adenosine-5′-N-methyluronamide (IB-MECA 1, Fig. 1), which is ~50-fold selective for the A3AR in rat compared to the A1AR and A2AAR.91,92 The first few years of medicinal chemical optimization of the affinity and selectivity of A3AR agonists relied entirely on comparison of binding affinities at the rat ARs,56 because the human homologues were not available initially.7 A successful approach was to combine multiple A3AR enhancing substitutions in adenosine analogues. Thus, IB-MECA contained a substituent that enhanced affinity at the ARs including A3AR, for example, a 5′-N-alkyluronamide, with an N6-benzyl substituent that maintained affinity at this subtype but reduced affinity at the A1AR and A2AAR. Initially, an unsubstituted N6-benzyl group served this purpose, and later a halo atom at the 3-position of the benzyl ring was shown to increase A3AR affinity and selectivity.92 Optimization of the 5′-N-alkyluronamide demonstrated that methyl was more favorable for A3AR binding than larger alkyl groups. A combination with 4-amino-3-iodo substitution of the N6-benzyl group maintained high affinity, but not high selectivity at the A3AR; thus, compound 3 became a widely used high-affinity radioligand in cells and membranes highly expressing this receptor.56 A 3-isothiocyanatobenzyl group was also tolerated at the N6-position, which provided the first selective chemically reactive affinity label of the rat A3AR, termed ICBM(N6-(3-isothiocyanatobenzyl)-5.-N-methylcarboxamidoadenosine) 4.93 In a subsequent study of the structure–activity relationship (SAR), a third position of derivatization was explored: the C2-position.94 It was noted that 2-[p-(2-carboxyethyl)phenyl-ethylamino]-5′-N-ethylcarboxamidoadenosine (CGS21680 1695), originally introduced as an A2AAR-selective agonist, surprisingly displayed affinities (nanomolar, all human homologues) in the order A2AAR (27) > A3AR (67) > A1AR (289).149 Based on this initial observation, it was apparent that the A3AR binding site was flexible in the ability to accommodate a variety of C2 substituents, including sterically bulky groups. Thus, the order of potency of CGS21680 was A2AAR > A3AR > A1AR ≫ A2BAR. Nevertheless, the 2-chloro analogue 2 of IB-MECA was the focus at that time as an A3AR agonist of increased selectivity, since other C2 modifications were not yet systematically explored. Later, an extended C2-alkynyl group, initially in the form of 6-hexynyl, was shown to be tolerated in adenosine derivatives in binding to the A3AR.96,97 This modification was also compatible with a 5′-N-ethyluronamide group, an observation that led to the identification of HE-NECA 7 as a potent, but nonselective A3AR agonist.98,99
TABLE 2.
Affinity of Selected Nucleoside Derivatives in Binding at Human ARs
| Compound | pKi value | |||
|---|---|---|---|---|
|
| ||||
| A1AR | A2AAR | A3AR | ||
| Agonists | ||||
| 1120 | IB-MECA | 7.29 | 5.50 | 8.74 |
| 2120 | Cl-IB-MECA | 6.66 | 5.27 | 8.85 |
| 799,103 | HE-NECA | 7.22 | 8.19 | 8.62 |
| 899,103 | PE-NECA | 6.25 | 6.21 | 8.21 |
| 999,103 | PHP-NECA (R,S) | 8.57 | 8.51 | 9.38 |
| 1099,103 | PEMADO | 4.48 | 4.38 | 8.52 |
| 11104 | 4.27 | 4.98 | 8.60 | |
| 13110 | CP-608,039 | 5.14 | <4.3 | 8.24 |
| 14a126 | <5 | <5 | 7.81 | |
| 14b126 | 6.71 | 5.36 | 9.42 | |
| 15a132 | LC257 | 5.79 | <4 | 8.74 |
| 15b132 | 5.42 | <5.30 | 8.70 | |
| 1695 | CGS21680 | 6.54 | 7.57 | 7.17 |
| 20105 | MRS3558 | 6.59 | 5.64 | 9.54 |
| 21119 | MRS5151 | 4.83 | ~5 | 8.62 |
| 22117 | MRS3630 | 7.74 | 5.49 | 8.43 |
| 25120,121 | MRS5698 | <5 | <5 | 8.52 |
| 26121 | MRS5679 | <5 | <5 | 8.51 |
| 27121 | MRS5980 | <5 | <5 | 9.15 |
| 29122 | MRS5841 | <5 | <5 | 8.72 |
| 32134 | MRS5919 | <5 | <5 | 8.22 |
| Antagonists and partial agonists | ||||
| 33136,142 | <4 | <4 | 6.19 | |
| 35128 | MRS1292 | ND | ND | 7.53a |
| 37109 | 5.80 | 5.32 | 7.91 | |
| 39139 | 5.60 | 6.47 | 8.38 | |
| 41143 | <4 | 8.14 | 7.93 | |
| 42175 | MRS3771 | 5.23 | <5 | 7.54 |
| 45125 | MRS5776 | <5 | <5 | 7.70 |
| 46140 | <5 | 5.13 | 8.31 | |
| 47141 | 9.36 | 7.11 | 8.52 | |
pKi at rat A3AR = 7.31.
ND, not determined.
FIGURE 1.
Structures of adenosine or 1,3-dialkylxanthine riboside derivatives that act as agonists of the A3AR. Compound 16 (CGS21680, not shown) is an A2AAR agonist
Unlike the C2-position, modification of the 2′ and 3′ hydroxyl groups was highly detrimental to A3ARbinding affinity of simple adenosine analogues,96 but a 3′-deoxy analogue of 2 (structure not shown) was later found to be a selective, full agonist at the rat A3AR with a binding Ki value of 33 nM.100 The ribose 5′-position was also amenable to modification beyond 4′-CH2OH and 5′-N-alkyluronamides. For example, 5′-methyl ether analogue NNC53-0055 6 was an agonist at the A3AR,101 and 5′-alkylthioethers were tolerated at the A3AR.102 Acylation of the N6-NH reduced affinity at the A3AR in comparison to the mono-alkylated analogues.97 The flexibility of substitution at the N6-position compatible with A3AR affinity was higher than initially indicated in the report on 1. For example, small alkyl and alkoxy groups, such as N6-methyl in 10 and N6-methyloxy in 6 and 11, could be appended to the nitrogen.99,101–104 However, a small alkyl group at the N6-position often reduced affinity at the rat A3AR compared to the human homologue. The human A3AR tolerated larger N6 substituents, such as the preferred 1S,2R stereoisomer of N6-cyclopropylphenyl in 17.105 The adenine moiety could be replaced with other heterocyclic nucleobases, leading to the retention of A3AR affinity and selectivity, but only in limited cases. For example, xanthine-7-ribosides such as DBXRM 5 were shown to fully activate the A3AR by virtue of an intact 5′-N-methyluronamide.106 Recently, in silico screening using an A2AAR crystal structure identified alternative nucleobases that, when ribosylated, retained receptor affinity and efficacy at the A3AR and other ARs. 107 Various pyridine-3,5-dicarbonitrile derivatives also bind to and activate ARs as atypical agonists, but they are not selective for the A3AR.108
The prototypical A3AR selective agonists 1 (CF101, Piclidenoson) and 2-chloro-N6-(3-iodobenzyl)-adenosine-5′-N-methyluronamide (Cl-IB-MECA 2, CF102, Namodenoson) are both in clinical trials for inflammation and cancer, respectively.10 They have demonstrated safety and clinical efficacy in Phases I and II trials. IB-MECA is now about to enter larger Phase III trials for RA and psoriasis, while Cl-IB-MECA is now about to enter a Phase II trial for primary liver cancer. 1 and 2 have also become widely used pharmacological probes with 2 being more selective for the rat and human A3AR. However, at the mouse A3AR, an exceptionally high affinity with Ki value of 87 pM was noted for 1, leading to 69-fold and >10,000-fold selectivity in comparison to mouse A1AR and A2AAR, respectively.109 Two other agonists of the A3AR, 12 and 13, were considered for clinical application to anti-ischemic cardioprotection.110 Compound 13 was unusually water soluble due to the presence of a 3′-amino group, which is largely protonated in physiological medium. Another highly selective A3AR agonist 15b containing a C2-pyrazolyl group was reported.111
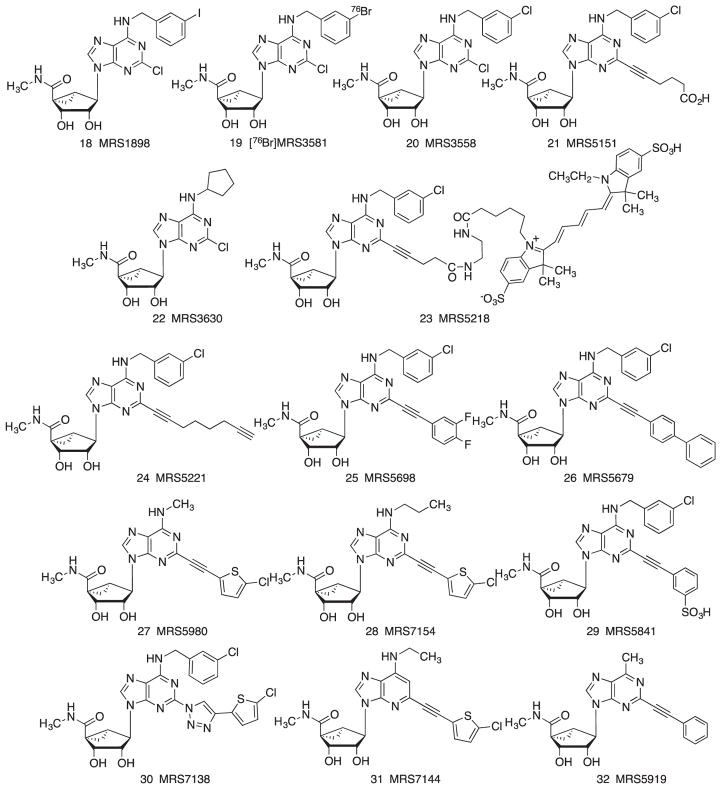
In 2000, Jacobson et al. reported that conformationally constraining a ribose-like moiety in the form of bicyclic ring increased the A3AR selectivity.112 The methanocarba group, that is, a bicyclo[3.1.0]hexane in place of the tetrahydrofuryl group of native ribose, had been applied earlier to antiviral drugs,113 and this was the first report of its incorporation in signaling ligands. Two isomeric forms of the methanocarba ring system, depending on the fusion point of the cyclopropane and cyclopentane rings, enforce either a North (N) (Fig. 2) or South (S) envelope conformation of the pseudoribose. A priori, the conformational preference of the ARs in binding ribose was not known, but the (N) isomer of simple adenosine was found to have >100-times the affinity of the corresponding (S) isomer at the A3AR. Thus, this derivatization approach of using chemically constrained rings can be used to probe the conformational preference of a given receptor. Among the four ARs, the greatest affinity gain with the (N)-methanocarba ring occurred at the A3AR. Numerous A3AR selective agonists were subsequently reported that included this major modification that enhanced affinity and selectivity at the A3AR compared to other ARs. The (N)-methanocarba modification is compatible with many other A3AR affinity-enhancing groups, such as 5′-N-alkyluronamides and N6-benzyl groups.114 Incorporation of the (N)-methanocarba modification in IB-MECA 1 resulted in MRS1898 18, a potent and moderately selective A3AR agonist that was later radiolabeled to provide a selective radioligand for the rodent A3AR.115 Other halogens could be placed at the 3-position, such as bromo 19 and chloro 20; the 3-bromo equivalent 19 provided a selective A3AR agonist as a 76Br-labeled positron emitter for receptor imaging studies.116
FIGURE 2.
Structures of (N)-methanocarba-adenosine derivatives that act as agonists of the A3AR
The A3AR-favoring (N)-methanocarba-5′-N-methyluronamide scaffold could also be combined with an A1AR-favoring substituent (N6-cyclopentyl) to provide a dual acting A1/A3AR agonist 22 that displayed cardioproprotective properties, as both receptors demonstrate anti-ischemic properties in heart.117
The C2-position could also be derivatized with (aryl)alkylthio groups, which resulted in moderate A3AR affinity but not selectivity.118 The C2-position substitution was expanded to include alkynyl and arylalkynyl groups that were compatible with both the riboside and (N)-methanocarba series.97,103,104 C2-ethynyl and arylethynyl groups were shown to further increase A3AR selectivity of adenosine analogues, that is, ribosides 7–11 and (N)-methanocarba derivatives such as 23–29.119–123 A carboxylic acid congener 21 is an A3AR agonist that displayed greater water solubility and the option of conjugation without losing receptor affinity. A lower homologue of 21 was used as a carboxy-bearing pharmacophore to condense with an amine-functionalized Cy5 fluorophore in the fluorescent A3AR-selective agonist MRS5218 23 of high A3AR affinity. 23 is selective for the A3AR in human and mouse and is suitable for characterization of the receptor in live cells using flow cytometry.124 Compound 24 contains a terminal alkyne group, as well as an alkyne at the C2 fusion position, and is suitable for click coupling to carriers such as gold nanoparticles with the retention of A3AR affinity and selectivity. A 3,4-difluorophenyl ethynyl group in 25 was particularly conducive to attaining high A3AR affinity in multiple species. Thus, the Ki value of 25 was approximately 3 nM at both the human and mouse A3ARs. The tolerance of the receptor for extended C2 substituents was surprising—even a biphenyl substituent in 26 preserved high A3AR affinity, which was explained based on conformational plasticity of TM2.120,125 A3AR-selective agonist 29 contains a sulfonate group that renders it unable to diffuse through biological barriers such as the blood–brain barrier. Thus, it is useful in vivo for distinguishing peripheral and central A3AR effects.122 The preferred placement of a sulfonate group on the scaffold of C2-arylethynyl-methanocarba-adenosine-5′-N-methyluronamides was predicted successfully using computational modeling of the receptor interactions at the human and mouse A3ARs.
In 2003, the group of Lak Shin Jeong in South Korea synthesized thionucleoside analogues that were shown to be highly potent and selective as A3AR agonists.126 The SAR upon modification of thionucleosides at the C2, N6 and 5′-positions was explored in detail. The 4′-thio modification of adenosine analogues was found to be compatible with many other A3AR affinity-enhancing groups, such as 5′-N-alkyluronamides and a range of N6 substitutions. Compounds 14a and 14b are analogues of IB-MECA 1 and Cl-IB-MECA 2, respectively, which were found to be highly potent and selective in A3AR binding. Compound 14b has been shown to suppress angiogenesis, a property that might be beneficial in treating cancer, diabetic retinopathy, and inflammatory diseases.127
To summarize the SAR described, A3AR affinity and selectivity of agonists are based on substitution at the C2, N6, and 5′ positions of adenosine,128 and only limited ribose functional group substitution of nucleosides is tolerated at this receptor. Some potent A3AR agonists such as 13 contain a 3′-amino-3′-deoxy modification of adenosine,110 but this modification does not apply universally. Although highly specific A3AR agonists were obtained in SAR studies, their binding Ki values were not directly predictive of the magnitude of an in vivo protective response, for example, in reducing chronic neuropathic pain. Thus, it became necessary to measure parameters other than simple binding affinities (such as half-life, duration of response, and maximal efficacy in vivo) to select for molecules with translational potential. An in vivo phenotypic screen in real time of the action of A3AR agonists to reduce or prevent chronic neuropathic pain was adopted for a comparison of diverse substitutions at these positions on the adenosine scaffold.121 The data obtained in a mouse model of neuropathic pain, that is, chronic constriction injury (CCI) of Bennett and Xie,129 allowed the chemists to steer the SAR in the direction of compounds that displayed high efficacy in reducing hyperalgesia and a long duration of action in vivo upon oral administration. The CCI model was ideally suited for the comparison of antinociceptive activity of A3AR agonists because of their high potency (greater than the molar potency of other pain medications) and because they lacked activity in tests of acute pain, such as the hot plate test and tail flick assay.130 The efficacy and duration of action of novel A3AR agonists after p.o.per os administration indirectly indicated favorable oral bioavailability and pharmacokinetics, at least with respect to chronic neuropathic pain. Thus, this phenotypic screen proved to be an invaluable guide in the extension of the SAR in this compound series.
This in vivo phenotypic screen confirmed that C2-phenylalkynyl analogues were among the preferred A3AR agonists. The terminal cyclic group in the C2-alkyne series was then varied to include diverse 6-membered rings, 5-membered heterocyclic rings, and cycloalkyl rings.121 With respect to A3AR binding and selectivity, many of these groups, including substituted phenyl rings, maintained high A3AR affinity and selectivity. However, the in vivo phenotypic screen identified 5-membered heterocyclic rings, such as thienyl derivatives, as being particularly potent and efficacious in vivo in the chronic neuropathic pain model. A 5-chlorothienylethynyl group in MRS5980 27 and MRS7154 28 was found to prolong the protective action of A3AR agonists in the CCI model. The substitution of the N1 group of the adenine moiety with CH in 31 was well tolerated in A3AR binding and activation and in the CCI model.
Due to the possibility that the C2-arylethynyl group could serve as a Michael reaction acceptor in nucleophilic attacks, alternative bioisosteric extensions at the C2-position were compared. The aryltriazolyl group in MRS7138 30 was found to mimic the geometry of the corresponding arylethynyl group when the (N)-methanocarba nucleoside was receptor bound,131 and this substituent would not have liability as a potential Michael acceptor. This conformational relationship was predicted using molecular docking to a homology model of the A3AR. C2-triazole substitution (two positional isomers) was previously found to be compatible with A3AR binding in the riboside series, as in 15a and 15b.132 As a postscript to that effort to replace the C2-arylethynyl group, this ethynyl group was found to be relatively unreactive toward thiols such as glutathione, and the risk of such compounds depleting liver glutathione was shown to be very small.133
The surprising finding that substitution of the exocylic NH of adenine with H or CH3 in MRS5919 32 allowed full activation of the A3AR emphasized that the loss of otherwise important recognition elements in a ligand can be compensated by other affinity enhancing moieties on the nucleoside.134 Moreover, the ribose moiety is the main effector of receptor activation, while adenine modifications tend to change the subtype selectivity but usually do not have a major effect on the agonist efficacy. However, there are exceptions to the above generalization, including various N6 substituents and C2 substituents that produce partial agonism or antagonism at the A3AR, as has been summarized,135 and nucleoside derivatives with reduced efficacy are discussed below.
4.2 Nucleoside derivatives as partial agonists and antagonists of the A3 adenosine receptor
Selected nucleoside derivatives that act as antagonists or low efficacy agonists at the A3AR are shown in Figure 3. The truncation of hydroxyl groups of adenosine nucleosides, that were demonstrated to be A3AR-selective agonists, was first explored in 1995.96–99 The goal was to reduce the hydrophilicity of the nucleosides to increase bioavailability without loss of receptor affinity. A secondary goal was to probe the effect on intrinsic efficacy of the truncated nucleosides as A3AR agonists, although it was not immediately achieved.100 The conversion of adenosine agonists into antagonists by complete removal of the ribose ring, that is, in adenine derivatives, was previously demonstrated, but the pharmacological characteristics of intermediate structures, that is, those with partially truncated ribose moieties, were unknown. Among the four ARs, the A3AR appears to be the easiest with respect to conversion of nucleoside agonists into antagonists, and numerous examples have been reported.109,128,132,135–141 However, it should be noted that the degree of efficacy can vary, depending on the functional assay and the receptor expression level. Thus, a modified nucleoside that behaves as an A3AR antagonist in one system, such as binding of a radiolabeled guanine nucleotide, might still activate the receptor under different circumstances, such as measurement of inhibition of adenylate cyclase.115
FIGURE 3.
Structures of (N)-methanocarba and riboside derivatives of adenosine or 1,3-dialkylxanthine that act as partial agonists or antagonists of the A3AR
Cristalli and co-workers took another approach to achieve A3AR antagonism. The presence of 8-alkynyl substituents on adenosine (4′-CH2OH) analogues, such as 33, reduced the ability of the A3AR-selective nucleoside to activate the receptor, as is consistent with antagonism.136,142 The theme of reduced efficacy in truncated nucleosides, rigid nucleosides, and otherwise modified ribosides was developed in pharmacological studies of Gao et al.137 The requirement of an H-bond donating group at the 5′-position of nucleoside analogues for A3AR activation was also demonstrated. A spirolactam 35 related structurally to IB-MECA 1 retained selectivity of binding to the rat and human A3ARs, but completely lacked the ability to activate the receptors and was shown to be a functional antagonist. Thus, a degree of flexibility of the 5′-amide, which is capable of forming multiple H bonds with the receptor, was required for full A3AR activation. Xanthine-7-riboside 34 was a partial agonist at the rat A3AR but an antagonist at the A1AR.106 An N6 substituent that converted A3AR agonist activity into antagonism was the N6-(2,2-diphenyl)ethyl group in 36.105 Curiously, the corresponding rigidified N6-fluorenylmethyl analogue (structure not shown), upon addition to an aryl-aryl bind to 36, became a full agonist. The combination of a N6-benzyl-type substituent with 2-chloro in 38 reduced the efficacy at the A3AR to nearly zero, although its residual efficacy could be expanded to full efficacy in the presence of A3AR PAM LUF6000 95 (where PAM is positive allosteric modulator).108 Modification at the 5′-position as an ester 37 produced a low-efficacy partial agonist, while 5′-N,N-dialkyluronamide in 42 resulted in antagonism at the A3AR.109 Thus, conformational factors at various regions surrounding the adenosine core and H-bonding around the 5′-position are determinants of A3AR efficacy.128
The 4′-truncation of the A3AR nucleosides was explored in great detail for the 4′-thionucleosides 138,139 leading to compounds 39 and 40, which were shown to be antagonists using a functional assay of guanine nucleotide binding. However, the 2-hexynyl group of compound 41 added a second activity to this series of A3AR ligands, that is, 41 was a combined potent A3AR antagonist and A2AAR agonist.143 Truncation of the nucleoside ribose-like moiety in the (N)-methanocarba series also led to A3AR-selective antagonists and partial agonists,109 including a radiolabeled 3-bromo analogue 44 for positron emission tomography (PET).116 Compound 45 is a selective antagonist of both the human and mouse A3ARs.120,125 Compound 46 is a selective A3AR antagonist with renal protective properties.139 Compound 47 is a mixed A1/A3AR antagonist that also displays functional agonism at the A2AAR, which displayed greater potency than predicted from its only moderate affinity in A2AAR binding assays.141
4.3 Non-nucleoside derivatives as antagonists of the A3 adenosine receptor
For more than 20 years, the advance of potent and selective A3AR antagonists as promising therapeutic choices for a range of diseases has been a prime subject of medicinal chemistry research. The pharmaceutical industry and academic communities have focused on the synthesis and screening evaluation of numerous heterocyclic compounds to discover potent and highly selective A3AR antagonists due to their potential therapeutic applications.144
A3AR antagonists belong to different structural groups including monocyclic, bicyclic, and tricyclic aromatic compounds (Table 3). Several xanthine or purine analogues were examined first, but none showed significant affinity or selectivity at rat A3AR.96,100,144,145 Consequently, different classes of compounds, that could be classified as nonxanthine derivatives135,144–149 and the lately nucleoside-derived antagonists, have been discovered as highly potent and selective A3AR antagonists.
TABLE 3.
Affinity of Selected A3AR Antagonists
| Compound | pKi value or % inhibition at 10 μM | |||
|---|---|---|---|---|
|
| ||||
| A1AR | A2AAR | A3AR | ||
| Monocyclic systems | ||||
| 48155 | MRS1334 | 5.54 (r) | <10%(r) | 5.41 (r) |
| 8.57 (h) | ||||
| 49158 | MRS1505 | 4.38 (r) | 4.62 (r) | 6.09 (r) |
| 8.10 (h) | ||||
| 50162 | 17% (h) | 43%(h) | 8.46 (h) | |
| 51164 | ISVY130 | 1% (h) | 10% (h) | 8.44 (h) |
| 52165 | SYJA385 | 7% (h) | 10%(h) | 6.41 (h) |
| 53167 | 24% (h) | 28% (h) | 9.10 (h) | |
| 54146 | <5 (h) | <5 (h) | 9.39 (h) | |
| 55146 | <6.18 (h) | <6.08 (h) | 9.44 (h) | |
| 8.80 (r) | ||||
| Bicyclic systems | ||||
| 56169 | VUF5574 | 52% (r) | 43%(r) | 8.39 (h) |
| 57170 | 4% (h) | 1% (h) | 7.71 (h) | |
| 58171 | 0% (h) | 19%(h) | 9.11 (h) | |
| 59135 | 5.10 (h) | 6.08 (h) | 7.59 (h) | |
| 60173 | 6% (h) | 8%(h) | 7.60 (h) | |
| 61174 | 6.37 (h) | 5.09 (h) | 8.22 (h) | |
| 62175 | MRS3777 | 26% (h) | 16%(h) | 7.33 (h) |
| 63176 | 5.98 (h) | 5.50 (h) | 9.74 (h) | |
| 64176 | <5 (h) | <5 (h) | 8.54 (h) | |
| 65177 | 5% (h) | 1% (h) | 8.92 (h) | |
| 66179 | 1% (h) | 1%(h) | 10.57 (h) | |
| Tricyclic systems | ||||
| 67180 | CGS15943 | 8.46 (h) | 9.40 (h) | 7.02 (h) |
| 68182 | MRS1220 | 7.28 (r) | 8.00 (r) | 9.19 (h) |
| 69183 | <5 (h) | <5 (h) | 8.09 (h) | |
| 70184 | <6 (h) | 6.98 (h) | 8.94 (h) | |
| 71184 | <6 (h) | <6 (h) | 8.16 (h) | |
| 72185 | 42% (b) | 3% (b) | 8.68 (h) | |
| 73186 | <6 (h) | <6 (h) | 8.05 (h) | |
| 74188 | 25% (b) | 14% (b) | 8.33 (h) | |
| 0% (h) | ||||
| 75189 | 6.59 (b) | 0% (b) | 9.10 (h) | |
| 7.96 (h) | 2% (h) | |||
| 76190 | 0% (b) | 8.06 (b) | 8.68 (h) | |
| 77192 | 5.57 (h) | <5 (h) | 8.80 (h) | |
| 78193 | MRE3008-F20 | <5 (r) | 5.70 (r) | 9.54 (h) |
| 79203 | MRE3005-F20 | 6.60 (h) | 7.22 (h) | 10.40 (h) |
| 80149 | 5.47 (h) | <5.3 | 8.01 (h) | |
| 81204 | ND | ND | 79% (h) | |
| 82204 | ND | ND | 16% (h) | |
| 83196 | <6 (h) | <6 (h) | 7.74 (h) | |
| 84143 | 32% (h) | 49% (h) | 9.29 (h) | |
| 85143 | OT-7999 | 4% (h) | 31%(h) | 9.02 (h) |
| 86207 | PSB-10 | 5.77 (h) | 5.56 (h) | 9.36 (h) |
| 87209 | KF-26777 | 5.74 (h) | 6.33 (h) | 9.70 (h) |
| 88211 | 24% (h) | 0% (h) | 8.66 (h) | |
| 89200 | <6 (h) | <6 (h) | 9.10 (h) | |
| 90200 | <6 (h) | <6 (h) | 8.46 (h) | |
| 91195 | 5.60 (h) | <5.3 (h) | 8.84 (h) | |
| 92195 | 5.52 (h) | 5.82 (h) | 8.71 (h) | |
h, human; r, rat; and b, bovin.
ND = not determined.
In this section, we summarize the medicinal chemistry of A3AR antagonists updating our previous reviews on this field.150–153
4.3.1 Monocycles
1,4-Dihydropyridines and pyridines
After the first evidence that 1,4-dihydropyridines (DHPs) exerted antagonistic activity at the A3AR in addition to L-type calcium channel inhibition, Jacobson et al. designed a series of substituted DHPs in the attempt to separate the two different activities.154 In this study, the replacement of the methyl ester at the 5-position of nifedipine with a bulkier 4-nitrobenzyl ester, along with the introduction of phenylethynyl and phenyl moieties at positions 4 and 6, respectively, led to 48 (MRS1334, Fig. 4). This compound showed high affinity and selectivity as an A3AR antagonist without inhibiting L-type calcium channels.155 In addition, a 3,5-diacyl-2,4-dialkylpyridine series was delineated by the oxidation of the corresponding DHP derivatives, and the best profile against A3AR was achieved with 49 (MRS1505, Fig. 4) in which the position 4 of the pyridine ring was substituted with small alkyl groups such as ethyl chain.156,157 General SAR of pyridine derivatives revealed that structural requirements responsible for enhancement of A3AR affinity and selectivity did not completely reflect that of the DHP parent compounds.158 Among this series, were also reported fluorinated and hydroxylated pyridine derivatives159 and an extension of this study performed by Jacobson and co-workers described a series of N-alkylpyridinium salts as water soluble A3AR antagonists although with lower potency than the pyridine analogues.160 A pyridine-based A3AR antagonist PET ligand [18F]FE@SUPPY was introduced.161
FIGURE 4.
Monocycle-based A3AR antagonists
Pyrimidines
Within the classes of bi- and tricyclic ARs antagonists, the pyrimidine nucleus present in the endogenous modulator adenosine, is a frequent substructural scaffold. 4-Amino-6-hydroxy-2 mercaptopyrimidines derived from chain opening of a series of triazolopyrimidinones have been synthesized by Cosimelli et al.162 Introduction of the propylsulfanyl and p-chlorobenzyloxy moieties at 2 and 6 positions, respectively, combined with an acetamide group at 4-position of the pyrimidine ring led to compound 50 (Fig. 4), a potent and selective human A3R antagonist (Ki = 3.5 nM). Similar structures characterized by two regioisomeric series of diaryl 2- or 4-amidopyrimidines such as N-[2,6-bis(4-methoxyphenyl)pyrimidin-4-yl]acetamide 51 (ISVY130, Fig. 4) were reported by Sotelo and co-workers. Some of the ligands in this series exhibited good selectivity and affinity with Ki values of < 10 nM at the A3AR.163,164
Pyrazin-2(1H)-ones
Very recently, Sotelo and co-workers published a novel series of compounds, including a simplified pyrazin-2(1H)one scaffold, as A3AR antagonists and with better pharmacokinetic properties.165 These new derivatives obtained by the Ugi-based multicomponent reaction were less potent than many other A3AR antagonists reported in the literature. The entire library of compounds, including the most potent compound of this series with a Ki value of 386 nM (52, SYJA385, Fig. 4), was subjected to a computational study to determine a rational hypothesis for their binding model.165
Thiadiazole and thiazoles
IJzerman co-workers identified thiadiazole and thiazole analogues as A3AR antagonists by chemical structure simplification of corresponding bicyclic quinazoline and isoquinoline nuclei, respectively.166 Among them, compound N-[3-(4-methoxyphenyl)-[1,2,4]thiadiazol-5-yl]acetamide 53 (Fig. 4) was claimed as the best compound of the series exhibiting a Ki value of 0.79 nM and acting as antagonist in a cyclic AMP (cAMP) functional assay.167 SAR optimization by introduction of a 5-(pyridine)-4-yl moiety on the 2-aminothiazole ring revealed a series of potent and selective compounds such as N-[4-(3,4,5-trimethoxyphenyl)-5-(pyridin-4-yl)thiazol-2-yl]-acetamide 54, with subnanomolar affinity for human A3AR (Ki = 0.4 nM) and 1000-fold selectivity against the other AR subtypes.168
The QSAR analysis of thiazole and thiadiazole A3AR antagonists indicated that their binding affinity increased with decreasing lipophilicity and in the presence of small alkyl moieties such as amide functions (acetamide or propionamide).168 In addition, the introduction of substituents, such as benzoyl, nicotinoyl (e.g., compound 55, Fig. 4) and isonicotinoyl moieties in position 2 of the thiazole ring, led to potent and selective antagonists at both human and rat A3ARs.
4.3.2 Bicycles
Quinazolines, phthalazines, and quinoxalines
A structure–affinity study reported by IJzerman co-workers indicated that introduction of a phenyl or heteroaryl substituent on the 2-position of the quinazoline scaffold or the equivalent 3-position of the isoquinoline improved the A3AR affinity in comparison to the unsubstituted derivatives. Combinations of the best substituents in the two series led to the potent and selective human A3AR antagonist N-(2-methoxyphenyl)-N-(2-(pyridin-3-yl)quinazolin-4-yl)urea 56 (VUF5574; Fig. 5) with a Ki value of 4 nM. 169
FIGURE 5.
Bicycle-based A3AR antagonists
Subsequently, the 2-amino/2-oxoquinazoline-4-carboxamide compounds, resulting from an in silico molecular simplification approach of the 2-aryl-1,2,4-triazolo[4,3-a]quinoxalin-1-one skeleton, were published by Morizzo et al. as
A3AR antagonists. One example of this series is compound 57 (Fig. 5) that showed good affinity and selectivity at the A3AR.170 With a similar approach on the triazoloquinazolinone nucleus, a new series of 2-phenylphthalazin-ones was identified as promising A3AR antagonists. Molecular manipulations by introduction of amide and ureide functional groups at the 4-position of the phthalazinone ring led to compound 58 (Fig. 5) with the best activity profile.171 Additionally, the 2-(4-methyl-1H-benzo[d]imidazol-2-yl)-quinoxaline 59 (Fig. 5) is noteworthy for the novelty of its design strategy utilizing a 3D database searching approach.172
Imidazo[1,2-a]pyrazines
Very recently, the imidazo[1,2-a]pyrazine nucleus was reported as a suitable core for the design of new AR antagonists. Within this series of compounds, a N-(2,6-diphenylimidazo[1,2-a]pyrazin-8-yl)-4-methoxybenzamide 60 (Fig. 5) showed good A3AR affinity with a Ki value of 25 nM. The molecular docking study of these compounds was also carried out to describe the potential binding mode of the new derivatives to their refined target receptor model.173
Adenines and adenine-like derivatives
The first class of selective A3ARantagonists characterized by a bicyclic structure strictly correlated to the adenine core was identified by Biagi et al.174 The adenine-like structure of these new N6-ureidosubstituted-2-phenyl-9-benzyl-8-azaadenines was responsible for their activity as antagonists, while the phenylcarbamoyl group ensured selectivity at the A3AR. The SAR studies based on the systematic substitutions of 2, 6, and 9 positions of the bicyclic system led to improved A1/A3 selectivity with compound 61 (Fig. 5).
Starting from reversine, 2-(4-morpholinoanilino)-N6-cyclohexyladenine, with a moderate activity as A3AR antagonists (Ki = 0.66 μM), Perreira et al. explored the SAR of related derivatives in order to improve A3AR potency and selectivity.175 A series of reversine analogues was synthesized by substitution of 2- and N6-positions of the adenine core. One of the most remarkable compounds in terms of hA3AR affinity and selectivity resulted when the N6-cyclohexyl moiety of reversine was combined with a 2-phenyloxy group (compound 62, MRS3777, Fig. 5).175 Some analogues from this study were shown to be inactive at 10 μM in the rat, reflecting the typical species dependence of binding of most known nonnucleoside A3AR antagonists.
The pyrazolo[3,4-d]pyrimidine scaffold, a bicyclic system structurally connected to the adenine nucleus that resulted from simplification of tricycles pirazolo-triazolo-pyrimidine, was recently described by Taliani et al.176 The SAR profile of this series indicated that the presence of an amide or ureide functionality at the 4-position (compounds 63 and 64, respectively) along with a phenyl ring at the 6-position was essential for promoting A3AR affinity and selectivity. The N2-position was characterized by substantial steric tolerance; in fact, both small methyl group (63, Fig. 5) and bulkier benzyl moiety (64, Fig. 5) were well tolerated. Compound 63, which showed a subnanomolar A3AR affinity and high selectivity versus the other AR subtypes, has been suggested as a promising lead compound for the development of adjuvant agents in glioma chemotherapy. In a related effort, a series of junction isomers of pyrazolo[3,4-d]pyrimidine derivatives were synthesized by Lenzi et al. applying a molecular simplification approach to the tricyclic pyrazolo[3,4-c]quinolin-4-one skeleton.177 The binding results of the junction isomers were successful and the new derivatives maintained high affinity for the hA3AR increasing also the selectivity versus the other AR subtypes. Aryl/arylalkyl substitution at the 5-position of such derivatives was poorly tolerated for A3AR binding affinity, while small groups at the same position were shown to enhance the ligand–receptor interaction. In addition, the substitution of the 2-phenyl ring with a 4-methoxy group led to 2-(4-methoxyphenyl)-5-methyl-2H-pyrazolo[4,3-d]pyrimidin-7(6H)-one 65 (Fig. 5), the most potent compound of this series. Very recently, a large number of 2-arylpyrazolo[4,3-d]pyrimidin-7-amine or 7-acylamine derivatives were synthesized as potent A3AR antagonists.178,179 The pyrazolopyrimidines bearing a 4-methoxyphenyl or a 2-thienyl group at the 5-position showed high hA3AR affinity and selectivity. 4-Methoxy-N-(2-phenyl-5-(thiophen-2-yl)-2H-pyrazolo[4,3-d]pyrimidin-7-yl)benzamide 66 (Ki = 0.027 nM, Fig. 5) is one of the most potent and selective A3AR antagonists in this structural class.179
4.3.3 Tricycles
Triazoloquinazoline
Jacobson and co-workers first described the N5-acylation of the free amino group of well-known 9-chloro-2-(furan-2-yl)-[1,2,4]triazolo[1,5-c]quinazoline-5-amine 67 (CGS15943, Fig. 6).180,181 This structural modification yielded 68 (MRS1220, Fig. 6), which was enhanced in both affinity and A3AR selectivity.181,182 The removal of the chlorine atom at 9-position of the triazoloquinazoline 67 along with the replacement of the 5-phenylacetamido and the 2-furyl moieties with a linear alkyl chain and a 4-Br-phenyl group, respectively, led to 69 (Fig. 6). This compound was found to be a potent and selective A3AR antagonist.183
FIGURE 6.
Tricycle-based A3AR antagonists
Very recently, a new series of triazoloquinazolines was reported as A3AR antagonists. Two examples are the 3,5-diphenyl[1,2,4]triazolo[4,3-c]quinazoline 70 (Ki =1.16 nM, Fig. 6) and the 5′-phenyl-1,2-dihydro-3′H-spiro[indole-3,2′-[1,2,4]triazolo[1,5-c]quinazolin]-2-one 71 (Ki = 6.94 nM, Fig. 6).184
Pyrazolo[3,4-c]/[4,3-c]quinolines
2-Arylpyrazolo[3,4-c]quinolin-4-ones, 4-amines, and 4-amino-substituted derivatives are reported as potent and selective hA3AR antagonists.185 Most of them showed a nanomolar hA3AR affinity and different degrees of selectivity that were strictly dependent on the presence and nature of the substituent on the 4-amino group. The benzoamide derivative 72 (Fig. 6) was the most potent and selective among the three reported series of compounds.
The pyrazole[4,3-c]quinoline-4-one scaffold was adapted to A3AR antagonists as structural isomers of the previous nucleus by Baraldi et al. Among the pyrazole[4,3-c]quinoline-4-ones, compounds that contained a 2-p-substituted phenyl (CH3, OCH3, and Cl) group showed good hA3AR affinity and excellent selectivity in comparison to the other AR subtypes (e.g., compound 73, Fig. 6).186
Triazolo[4,3-a]/[1,5-a]quinoxaline
Colotta et al. identified 1,2,4-triazolo[4,3-a]quinoxalin-1-one derivatives as promising hA3AR antagonists.187 The SAR study recognized the appropriate substitutions at 2, 4, and 6 positions of the tricyclic template. In particular, the introduction of the 4-oxo or 4-N-amide functions afforded selective and potent A3AR antagonists such as compounds 74188 and 75189, respectively, confirming the importance of both nuclear or extranuclear carbonyl functionality for A3AR affinity (Fig. 6). A series of 2-aryl-8-chloro-1,2,4-triazolo[1,5-a]quinoxalines has also been synthesized and evaluated in radioligand binding assay at both bovine and human ARs190,191 showing a similar SAR profile to that of the 2-arylpyrazolo[3,4/4,3-c]quinolines185, 186 and triazolo[4,3-a]quinoxaline.189 One of the representative derivatives was a 2-(4-methoxyphenyl)-[1,2,4]triazolo[1,5-a]quinoxalin-4(5H)-one 76 (Fig. 6).
1,2,3-Triazolo[1,2-a][1,2,4]benzotriazolones
A3AR antagonists based on aminophenyl-triazolobenzotriazinone have been reported by Da Settimo et al. A lead optimization strategy focused on the structural modifications provided that the suitable groups were attached to the 5-amino group and in the 4′-and/or 9-positions. The best result was obtained with compound 77 (Fig. 6), which showed a Ki value of 1.6 nM at the A3AR and no significant affinity at the other ARs.192
Pyrazolo[4,3-e]-1,2,4-triazolo[1,5-c]pyrimidines
The pyrazolo-triazolo-pyrimidine (PTP) scaffold, due to its strong structural correlation with the nonselective antagonists CGS15943 (67, Fig. 6) 180 and to the adenine nucleus present in the endogenous modulator adenosine, has been investigated in depth as a prototypical template for adenosine antagonists.
Rigorous research efforts were made on this scaffold in order to obtain potent A2A- and A3AR antagonists.153 A series of PTP derivatives (MRE series) reported by Baraldi’s group were obtained by the structure–activity optimization based on the introduction of different substituents at the 5, 7, 8, and 9 positions.145,149,193,194,196–200 The N7-substituted derivatives proved to be predominantly hA2AAR antagonists, while the combination of a small alkyl chain at the N8-pyrazole position with a (substituted)phenylcarbamoyl chain at the N5-position led to potent and selective hA3AR antagonists. 198 One of the most active and selective compounds in this series was 78 (MRE3008-F20, Fig. 7) with a Ki value of 0.29 nM.193 The corresponding tritium-labeled analogue as the first potent and selective radiolabeled antagonist for the A3AR was prepared. It bound to the hA3AR expressed in Chinese hamster ovary (CHO) cells with a KD value of 0.82 nM (Bmax = 297 fmol/mg protein).201,202 The isosteric replacement of the phenyl ring with a 4-pyridylmoiety yielded 79 (MRE3005-F20, Fig. 7) that maintained the high affinity at the A3AR with enhanced water solubility.203 Subsequently, replacement of pyridin-4-ylmoiety of MRE3005-F20 with substituted piperidine rings led to the preparation of the hydrochloride salt of 1-(cyclohexylmethyl)piperidin-4-yl (80, Fig. 7).149
FIGURE 7.
Tricycle-based A3AR antagonists
Synthesis of fluorosulfonyl- and bis(β-chloroethyl)amino-phenylamino-pyrazolo[4,3-e]1,2,4-triazolo[1,5-c]pyrimidines as irreversible A3AR antagonists was performed by Baraldi’s group to provide useful tools for structure–activity studies. Electrophilic groups, specifically sulfonyl fluoride and nitrogen mustard (bis-(β-chloroethyl)amino) moieties, have been incorporated at the 4-position of the aryl urea group (compounds 81 and 82, respectively, Fig. 7).204 While compounds containing a fluorosulfonyl moiety proved to be irreversible antagonists at the hA3AR (at 100 nM, 79% of inhibition), the corresponding nitrogen mustard derivatives were unable to covalently bind the target receptor subtype. This difference in the receptor interaction between the 81 and 82 series has been explained on the basis of chemical reactivity of the two different groups.
Very recently a new series of N,N′-disubstituted guanidines of the PTP scaffold, prepared in a one-pot reaction, was described as A3AR antagonists. The best compound of this series was 83 (Fig. 7) bearing a N,N-(4-nitrophenyl)guanidine moiety at 5-position of the tricyclic nuleus.196
Triazolopurines
[1,2,4]Triazolo[5,1-i]purines structurally related to the triazoloquinazolines have been identified as A3AR antagonists.183,205 Within this family, the 5-n-butyl-8-(4-n-propoxyphenyl)-3H-[1,2,4]triazolo[5,1-i]purine 84 (Fig. 7) showed a good potency and selectivity at A3AR. Among the reported structures, compound 85 (OT-7999, Fig. 7) demonstrated a significant reduction of intraocular pressure in cynomolgus monkeys at 2–4 hr following topical application.205
Tricyclic xanthines
Caffeine and theophylline are the classical nonselective xanthine antagonists of ARs that display micromolar affinity at human AR subtypes. At the rat A3AR, caffeine and theophylline are weaker in affinity.
Initial SAR studies at the A3AR were carried out using multiple substituted xanthines, many of which retained selectivity for the A3AR.90 An interesting approach was based on the ring annelation of xanthine derivatives, which permitted several research groups to discover different tricyclic systems that showed dissimilar affinity to AR subtypes.206 A series of imidazo[2,1-i]purinones as tricyclic xanthines (PSB series) was developed by Müller et al. as human A3AR antagonists with improved water solubility.207 For example, 86 (PSB-10, Fig. 7) showed a subnanomolar A3AR affinity and a good selectivity compared to the other AR subtypes, and its tritiated form ([3H]PSB-11) exhibited a KD value of 4.9 nM (Bmax = 3500 fmol/mg of protein).208 Another similar compound is 2-(4-bromophenyl)-4-propyl-7,8-dihydro-1H-imidazo[2,1-i]purin-5(4H)-one 87, also designated KF-26777 (Fig. 7), was endowed with high affinity and selectivity.209
The pyrido[2,1-f]purine-2,4-diones, another series of tricyclic xanthines, have been reported to exert affinity at A3AR in the low nanomolar range. Different substituents at the 1 and 8 positions of the new scaffold were evaluated, and the SAR studies led to 3-(cyclopropylmethyl)-1-(4-methylbenzyl)pyrido[2,1-f]purine-2,4(1H,3H)-dione 88 (Fig. 7) showing the best A3AR binding profile of the series with total selectivity.210,211
Replacement of the pyridine nucleus of the pyrido[2,1-f]purine-2,4-dione scaffold with different 5-membered heterocycles has been extensively examined by Baraldi’s group. The SAR studies led to both series of pyrrolo[2,1-f]purine-2,4-dione and imidazo[2,1-f]purine-2,4-diones as A3AR antagonists.199 Among the examined molecules, the imidazo[2,1-f]purine-ones were 2- to 10-fold more potent than the corresponding pyrrolo[2,1-f]purine-ones (e.g., 89 vs. 90, respectively, Fig. 7) and the most favorable affinity and selectivity at A3AR was obtained by introduction of small alkyl chain at the 7-position of the main scaffold (compound 89).200
More recently, replacement of the trichlorophenyl ring at 2-position of PSB-10 86 and congeners with differently substituted five-membered heterocycles, like 1,3- and 1,5-disubstituted pyrazoles or 3-substituted isoxazoles (R-enantiomer 91 and 92, respectively, Fig. 7) was investigated by Baraldi’s group.195 The 2-heterocyclic substitution induced excellent affinity and selectivity for the hA3AR subtype. Docking of the most potent compound (91) in complex with a hA3ARhomology model furnished a general survey of the hypothetical binding mode of the newly described derivatives.195
4.4 Allosteric modulators of the A3 adenosine receptor
Allosteric modulators of GPCRs bind at a location that is distinct from the binding site for a native ligand, that is, the orthosteric site, and this phenomenon has been reported for the A3AR.212 In theory, the modulation may be positive, that is, with a PAM enhancing the activity of a directly acting (orthosteric) agonist, or negative, in the case of a NAM. A3AR PAMs have been shown to enhance the potency and/or efficacy of agonists. When administered in vivo, they would be expected to be silent, with respect to A3AR activity, unless either an endogenous or exogenous agonist would be present. Therefore, treatment with an A3AR-selective PAM might display greater event- or site-specific action than an exogenous agonist, because it would magnify the effect of locally released adenosine, that is, in response to stress of an organ or tissue.
The SAR of three classes of A3AR PAMs have been explored in detail: 1H-imidazo-[4,5-c]quinolin-4-amines (93 – 97),213–215 2,4-disubstituted quinolines (98), 216 and 3-(2-pyridinyl)isoquinolines (99, 100).217 Representative key members of these structural classes are shown in Figure 8. In addition to these heterocycles, allosteric modulation of this receptor by compounds and ions that are not specific to the A3AR, such as amiloride analogues and sodium, have been studied.212,218
FIGURE 8.
Allosteric modulators of the A3AR
The principal assay used to screen for allosteric modulators of the A3AR has been to examine effects on the dissociation rate of an agonist radioligand, [125I]I-AB-MECA 3.212 Many of the PAMs that have been reported also compete with the radioligand for specific binding. Thus, the objective in early studies was to identify lead molecules that impeded the dissociation rate, with minimal competitive binding potency.213,217 At a concentration of 10 μM, the imidazoquinolinamines and DU124182 93 and DU124183 94 reduced the dissociation rate of the radioligand from the receptor by roughly half, and the phenylamino derivative 94 was more potent as an allosteric enhancer.213 Dichloro substitution and expanding the cycloalkyl group in LUF6000 95, MRS5049 96, and MRS5190 97 were later shown to produce potent allosteric enhancement with less prominent competitive inhibition.214,215 An amide LUF6096 98 was particularly selective for allosteric enhancement of the A3AR compared to 95, but it displayed a short half-life in vivo. The residues of the A3AR that are associated with the allosteric enhancement compared to orthosteric ligand binding were probed using stite-directed mutagenesis.218
When multiple effector mechanisms induced by agonist Cl-IB-MECA were compared, 95 was found to modulate each activity in a different manner, that is, this imidazoquinolinamine appeared to be a functionally biased PAM.108,219 For example, 95 had no effect on phosphorylation of ERK1/2, a small effect on β-arrestin2 translocation, and intense effects on cAMP inhibition and cell hyperpolarization. The agonist-enhancing effect of 95 was probe-dependent, that is, it had different degrees of modulation of different AR agonists, considered receptor “probes.” Notably, 95 greatly increased the efficacy of the naturally occurring nucleoside inosine, which is normally a weak and only partially efficacious agonist at the A3AR. Inosine is a metabolite of adenosine and, like adenosine, is elevated in concentration when stress to an organ is present. Thus, 95 could produce therapeutic benefit in vivo partially from its enhancing action on inosine, as well as endogenous adenosine. 95 also enhanced the efficacy of nucleoside antagonists of the A3AR, such as MRS542 38, to produce full agonism at the A3AR. This indicates that nucleoside antagonists might behave as antagonists in a given functional model, but there is a latent agonism that can be amplified in the presence of a PAM. In contrast, nonnucleoside A3AR antagonists did not display any latent agonism in the presence of an A3AR PAM.
LUF6000 95 has anti-inflammatory effects in rat models of arthritis and a model of liver inflammation in mice,220 suggesting its potential use in the treatment of autoimmune inflammatory diseases. LUF6096 98 was shown to protect the heart in a model of canine cardiac ischemia,218 suggesting the potential use of A3AR PAMs in the treatment of ischemic conditions.
4.5 Structural characterization of the A3 adenosine receptor
Although there is currently no X-ray crystallographic structure of the A3AR available, effective use of modeling techniques has provided a window into its orthosteric binding site (Fig. 9). In particular, docking of agonists in conjunction with data from site-directed mutagenesis has demonstrated the close similarity of the A3AR to the X-ray crystallographic structure of the hA2AAR.221 Complexes of the A2AAR with four different agonists, namely 6-(2,2-diphenylethylamino)-9-((2R,3R,4S,5S)-5-(ethylcarbamoyl)-3,4-dihydroxytetrahydrofuran-2-yl)-N-(2-(3-(1-(pyridin-2-yl)piperidin-4-yl)ureido)ethyl)-9H-purine-2-carboxamide (UK432,097); adenosine; 5′-Nethylcarboxamidoadenosine (NECA); and 2-[p-(2-carboxyethyl)phenylethyl-amino]-5′-N-ethylcarboxamidoadenosine (CGS21680) (PDB IDs: 3QAK, 2YDO, 2YDV and 4UG2, respectively), have been crystallized and their structures determined.222–224 The complexes revealed a common interaction pattern anchoring the adenosine moiety in the orthosteric binding site of the A2AAR involving several conserved amino acid residues that are predicted to serve the same function in the A3AR binding site. For example, the side chain Asn6.55 (using standard notation for numbering of TMs225) coordinates by H-bonding in a bidentate fashion with the adenine N6H and N7 in both ARs. His7.43 is in contact with the 2′-hydroxyl group of ribose. Also, Phe168 (EL2) is predicted to form π–π stacking with the adenine ring, as in the A2AAR. However, there are some differences in the interaction of nucleoside ligands with A3AR compared to A2AAR that account for pharmacological differences of such ligands, with respect to their affinity, selectivity, and efficacy. For example, His6.52, which forms an H-bond to the 5′-carbonyl group of potent A2AAR agonists occurs as Ser6.52 in the A3AR and is not predicted to closely associate with typical nucleoside ligands. This might explain why binding of 4′-truncated nucleosides is maintained at the A3AR relative to other ARs.
FIGURE 9.
(A) Docking pose of 3,4-difluorophenyl agonist analogue MRS5980 (27) at the hA3AR homology model, in which TM2 is based on its position in the active β2 adrenergic receptor. Residues interacting with the ligand (magenta carbon atoms) are labeled. H-bond and π–π interactions are represented as green solid and cyan dashed lines, respectively. Nonconserved ARs residues are in italics. (B) Docking pose of biphenyl agonist analogue MRS5679 (26) at the hA3AR homology model. Residues interacting with the ligand (violet carbon atoms) are labeled and H-bond interactions are represented as green solid lines. The degree of displacement of TM2 with respect to the TM bundle in hA3AR homology models based on the hA2AAR (red ribbon), hybrid hA2AAR-β2 adrenergic receptor (purple ribbon), and hybrid hA2AAR-opsin (violet ribbon) templates is highlighted with an arrow. TM1 is omitted to aid visualization
Conformational plasticity of the A3AR has been proposed to account for the high-affinity binding of rigid C2-arylalkynyl agonists such as MRS5980 27.120 When docking this class of compounds in a homology model of the A3AR derived exclusively from the agonist-bound “active-like” A2AAR structure, there was a steric clash with the extracellular tip of TM2. An outward movement of TM2, as observed in several other active GPCR structures such as the β2-adrenergic receptor and opsin, was therefore hypothesized to occur also for the A3AR. Thus, a hybrid homology model in which TM2 assumed its highly displaced position in opsin was required to dock biphenyl derivative MRS5679 26, and the other TMs followed their orientation in the agonist-bound A2AAR.226 This proposed outward movement of TM2 was also associated with the degree of activational bias of C2-extended A3AR agonists for cell survival, in a comparison of five different functional readouts.131
Molecular dynamics (MD) simulation of the A3AR in complex with agonists have been reported.227 The analysis of the conformational changes of conserved Trp243 (6.48) as a result of agonist (Cl-IB-MECA) binding suggested that the ligand was able to promote and stabilize an expected conformational switch involved in receptor activation.
A supervised MD (SuMD) study simulated the approach of PAM LUF6000 95 toward a putative allosteric binding site on the A3AR that contained an adenosine molecule in the orthosteric site.228 In the modeled ternary complex, 95 occupied the upper region of the orthosteric binding site by directly interacting with the agonist. The study suggested that 95 might exert its PAM activity by acting as “pocket cap.”
GPCRs may form characteristic dimers, either homo- or heterodimers, and the pharmacological implications of such dimerization remain to be fully characterized. Homodimers of the A3AR have been detected using fluorescent methods.229 Indications are that in the future, the discovery of A3AR ligands will rely heavily on computational methods to define the dynamic interaction of the receptor with its ligands and with other proteins, including other GPCR protomers.
5 INTRACELLULAR PATHWAYS IN IMMUNE AND CANCER CELLS
A schematic diagram showing the main signaling pathways triggered by adenosine through A3AR activation in different cellular types is reported in Figure 10. The A3AR preferentially couples to Gi protein to inhibit cAMP accumulation, and it may also couple to Gq protein to mediate stimulation of phospholipase C (PLC), which then increases calcium concentrations.230 The activation of PLC, and even the Ca2+ effects observed at high concentrations of A3AR agonists, could conceivably be triggered by mechanisms other than Gq, such as Gβγ subunits. In addition, a Gq protein kinase C (PKC) dependent mechanism has been related to the apoptosis-inducing factor upregulation mediated by high doses of adenosine and of an A3AR agonist in a human bladder cancer cell line.78 However, PKC was recruited by A3AR stimulation in order to increase tumor necrosis factor alpha (TNF-α) release in activated macrophages.231
FIGURE 10.
Intracellular pathways of A3AR in immune and cancer cells. Schematic diagram showing the main signaling pathways triggered by adenosine through A3AR activation in different cellular types. A3AR preferentially couples to Gi. PLC activation, and even the Ca2+ effects observed at high concentrations of A3AR agonists, could conceivably be triggered by mechanisms other than Gq, such as Gβγ subunits
A huge amount of data supports a link between A3AR and MAPKs in several cellular models.232 A3AR-mediated activation of extracellular signal-regulated kinases (ERK1/2) and mitogenesis modulation was found in human fetal astrocytes, CHO cells expressing the human A3AR (CHO-hA3), microglia, colon carcinoma, glioblastoma, melanoma and in foam cells.43,74,233–240 However, an inhibition of ERK activation has been revealed in A375 melanoma, prostate cancer, and glioma cells, leading to a reduction of cell proliferation as well as a decrease of TNF-α release in RAW 264.7 cells.22,73,241,242 The A3AR also activates p38 MAPKs in different cellular models including CHO-hA3, hypoxic melanoma, glioblastoma, and colon carcinoma cells,235,237,243 while reducing p38 MAPKs in human synoviocytes.53 Concerning c-Jun N-terminal kinase (JNK), its activation by A3AR has been retrieved in microglia and glioblastoma cells, leading to cell migration and matrix metalloproteinase 9 (MMP-9) stimulation, respectively.74,244 MEK/ERK1/2 and PI3K/Akt signaling pathways downstream of the A3AR control multiple resistance-associated protein 1 (MRP1) transporter substrate in glioblastoma cells, demonstrating a chemosensitizing effect by pharmacological blockade of A3AR and consequent reduction in tumor size.245
A pathway involving Akt phosphorylation protects RBL-2H3 and glioblastoma cells from apoptosis, while the same pathway produced an antiproliferative effect and increase in MMP-9 in A375 and glioblastoma cells, respectively.74,236,246,247 On the other hand, A3AR-mediated activation of the phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)/Akt signaling pathway enhances pigmentation in B16melanoma cells and in human skin explants.85
The PI3K/Akt and nuclear factor (NF) κB signaling pathways are the mediators of the anti-inflammatory A3AR-mediated effects, as observed in activated BV2 microglial cells, monocytes, adjuvant-induced arthritis, and in mesothelioma.75,248–253 Akt inhibition is involved in the reduction of A3AR-mediated of hypoxia inducible factor 1 (HIF-1α) accumulation in murine astrocytes.254 In addition, the block of PI3K/Akt/mammalian target of rapamycin (mTOR) signaling through A3AR suppresses angiogenesis in endothelial cells.127
Furthermore, an A3AR-mediated decrease in the protein kinase A (PKA) level was responsible for: (i) an increase in glycogen synthase kinase 3β (GSK-3β), leading to a downregulation of beta-catenin and its transcriptional gene products cyclin D1 and c-Myc, and (ii) a decrease in the level of NF-κB DNA-binding capacity. This effect through A3AR activation has been reported in melanoma, hepatocellular carcinoma, synoviocytes from RA patients, and in adjuvant-induced arthritis rats.89,255–257 Interestingly, a downregulation of A3AR mRNA/protein expression in colon cells after ulcerative colitis by miR-206 led to an increase of NF-κB and the downstream cytokine (TNF-α/IL-8/IL-1β) expression in the mouse colon, producing a proinflammatory effect.19
6. BIOLOGICAL ROLE AND THERAPEUTIC APPLICATIONS IN MODELS OF IMMUNE DISORDERS AND CANCER
6.1 Preclinical studies in immune cells
The A3AR is expressed in almost all cells of the immune system acting as essential mediators of adenosine’s role in inflammation.258–260
A growing body of evidence suggests a key role of the A3AR in neutrophil behavior. In recent years, it has been reported that the A3AR is distributed in a highly polarized fashion on the neutrophils cell membrane and contribute to their chemotaxis and migration. Specifically, A3AR is translocated to the neutrophil leading edge, and adenosine generated by ecto-ATPases/nucleotidases results in autocrine stimulation of A3AR-enhanced polarized migration, following its initiation by ATP activating the P2Y2 receptor.34,261–263 This was confirmed also in a sepsis model of A3ARknockout (KO) mice where a reduction in the recruitment of neutrophils to the lung and peritoneum was observed.264 In contrast, in human breast cancer cells, the A3AR is expressed in multiple leading edges where it promotes cell migration with numerous directional changes. Indeed, exogenous adenosine may simultaneously stimulate A3AR on all leading edges of the cell, inducing it to spread out in opposing directions resulting in arrest of cell motility.263 Furthermore, it has been found that the endogenous A3AR aggregates into plaque-like microdomains that affect cytoskeletal remodeling. In addition, they promote the formation of membrane protrusions, also named cytonemes, which enable neutrophils to capture pathogens.36 In contrast, data in early literature reported that chemotaxis and, in addition, oxidative burst were inhibited by A3AR activation with anti-inflammatory effects.32,33,35
Adenosine modulates monocyte-macrophage functions, being responsible for both inflammatory mediator production and resolution induction. For example, A3AR stimulation inhibits the respiratory burst, interleukin (IL) 1β, TNF-α, chemokine macrophage inflammatory protein (MIP) 1α, interferon regulatory factor 1, iNOS (inducible nitric oxide synthase), and CD36 gene expression.38,40,41,249,265–267 However, adenosine reduced the expression of adhesion molecules on monocytes and decreased cytokine production, effects that were potentiated by an A3AR antagonist.268 In addition, A3AR stimulation increased TNF-α production in activated macrophages.231
A functional A3AR is expressed in dendritic cells, antigen-presenting entities activating naive T lymphocytes and starting primary immune responses.269,270 In particular, the A3AR in the human immature elements has been found to induce elevated Ca2+ levels, actin polymerization, and chemotaxis, while in mature dendritic cells, the A3AR is down-regulated and decreases TNF-α release.44,46
T cells represent the major actors in adaptive immunity and play a crucial role in the battle against infections and tumors. Both cytotoxic (CD8+) and helper (CD4+) T cells express A3AR.48,271 Initial studies attributed to the A3AR an immunosuppressive role toward T cell mediated immune responses, but the mechanisms involved have not been investigated.272,273 Interestingly, oral administration of an A3AR agonist increased the cytotoxic activity of mouse NK cells and serum IL-12, thus reducing in vivo growth of melanoma cells.274 Indeed, ex vivo activation of CD8+ T cells with an A3AR agonist improves adoptive immunotherapy for melanoma.275 A3AR has been found to be upregulated in peripheral blood mononuclear cells (PBMCs) obtained from patients affected by different autoimmune disorders such as RA, Crohn’s disease, and psoriasis. This effect has been ascribed to the increase of TNF-α resulting in an upregulation of NF-κB and the cAMP response element binding protein (CREB), acting as A3AR transcription factors.276 Therefore, A3AR could be indicated as a biological predictive marker in autoimmune inflammatory pathologies. Furthermore, an immunosuppressive effect has been confirmed in lymphocytes derived from patients affected by RA, where the A3AR, upregulated with respect to healthy subjects, inhibited the NF-κB signaling, inflammatory cytokines, and MMPs production. Their density inversely correlated with indexes used to assess RA disease activity, supporting the importance of A3AR in the control of RA joint inflammation.277 Furthermore, A3AR activation in rat models hampered damage of cartilage, formation of osteoclast/osteophyte, bone destruction, and generation of pannus and of lymphocyte formation.278,279 Similarly, an A3AR anti-inflammatory effect, that is, NF-κB-TNF-α-dependent has also been found in synoviocytes of RA patients.257 Interestingly, this mechanism has been suggested also in A3AR-induced hepatoprotection against ischemia/reperfusion (IR) injury.280
The A3ARhas long been recognized as a major contributor of rodent mast cell activation by increasing their degranulation and, more recently, this factor has been observed also in both primary human and LAD2 mast cells.9,13,63,82–84,281 A3AR activation with an agonist significantly increased IL6, IL8, VEGF, amphiregulin, and osteopontin genes in human mast cells, affecting severe asthma.285,286 Indeed, prolonged treatment with an agonist produced A3AR downregulation responsible for the suppression of its basal inhibitory effect on cytokine production. This response was obtained only at a transcriptional level, suggesting that, at variance with rodents, in humans the primary role of the A3AR is to act as a modulator rather than a stimulator of mast cell responses.287
Activation of the A3AR induced hypothermia through the induction of mast cell degranulation, consequent histamine release, and activation of central histamine H1 receptors.288
6.2 Preclinical studies in cancer cells
The role of A3AR in cancer has been extensively studied using selective agonist and antagonist ligands, often with contrasting results. The efficacy of antagonists as anticancer drugs has been supported starting by the concept that hypoxia, characteristic of solid tumors, increases adenosine levels and stabilizes HIF-1α, the most important factor regulating cellular responses to the lack of oxygen.289 In this context, it has been reported that A3AR stimulation induced HIF-1α accumulation in different cancer cell lines.235,243 This has been seen to lead to an increase in angiopoietin-2 and/or VEGF, depending on the cell model investigated.237 Accordingly, it has been found that A3AR stimulation increased microvessel density, expression of proangiogenic factors, macrophage tumor infiltration, and cytokine production in in vivo melanoma tumor models.290 Furthermore, a basal stimulation through A3AR was responsible for an increased MRP1 expression in glioblastoma cells. As a consequence, A3AR antagonist administration provoked a potentiating effect on the reduction in tumor size induced by the chemotherapic vincristine.245 In addition, an increase of glioblastoma cell invasion, through A3AR and MMP-9 stimulation, was found, as already shown in macrophages.74,291 However, a potential therapeutic effect of agonists in cancer has been also reported moving from initial studies on the observation that tumor metastases were infrequent in striated muscles. Interestingly, it has been found that in addition to adenosine, natural agonists of A3AR were secreted from muscle cells, contributing to the systemic anticancer and chemoprotective activity exerted by muscle-conditioned medium. This evidence explained the rarity of tumor metastases in muscle and represented the rationale for the utility of A3AR agonists in cancer.292,293 Further studies reported that A3AR activation inhibited telomerase activity and exerted cytostatic effects in tumor cells.10,255,294,295 Interestingly, cancer tissues and the interstitial fluid of several tumors contained high levels of adenosine able to activate ARs, among which the A3AR subtype was the most expressed.230,296,297 For example, an A3AR upregulation has been found in human colorectal and hepatocellular carcinomas that was reflected in peripheral blood cells, thus making this AR subtype a possible marker for cancer, reflecting receptor status in remote tumor tissue.87–89 As for the role of A3AR in tumors, pro- and antiproliferative effects due to their activation have been documented in several cancer cell types.69,74,75,298–307 A3AR activation decreased prostate cancer cell migration in vitro and in vivo and inhibited cell proliferation, inducing G1 cell cycle arrest and apoptosis.70,73,308
Even though contrasting results about the role of A3AR agonists and antagonists in cancer still are present in literature data, only the therapeutic utility of A3AR agonists has been supported by in vivo studies. These studies encompassed syngeneic xenograft, orthotopic and metastatic experimental animal models of colon, prostate, melanoma, and hepatocellular carcinomas, in which IB-MECA and Cl-IB-MECA were administered orally in view of their stability and bioavailability. These agonists inhibited cell growth in syngeneic and lung metastatic models of murine melanoma, and potentiated cyclophosphamide effect.10 Also in xenograft models, IB-MECA inhibited the development of human colon and prostate tumors in nude mice and increased 5-fluorouracil or taxol antitumor effect. Furthermore, it blocked primary and liver metastases of colon carcinoma cells inoculated in the spleen. Finally, Cl-IB-MECA reduced hepatocellular tumor growth, liver inflammation, and cancer pain in rat bone residing breast cancer.10,76,86,89
6.3 Clinical trials
The scientific evidence obtained through in vitro and in vivo experiments led to the progression of A3AR agonists in clinical studies for the therapy of inflammatory and cancer pathologies. Importantly, they were found safe and well tolerated in all preclinical, Phases I and II human clinical studies. According to clinicaltrials.gov, the agonist IB-MECA (Piclidenoson, CF101) entered in the following clinical trials (NCT numbers):
RA (Phase II, NCT00280917; Phase II, NCT01034306; Phase II, NCT00556894) in which it showed significant antirheumatic effect as a standalone drug. Interestingly, a direct significant correlation was found between receptor expression at baseline and patients’ response to the drug. CF101 treatment resulted in an ACR20 (American College of Rheumatology Criteria for disease improvement) of 48.6%, statistically significantly higher than that of the placebo group (25%) at week 12. CF101 treatment also showed superiority in ACR50 and ACR70 values versus placebo. These data suggest that the A3AR is a promising therapeutic target in RA and can be used also as a biologicalmarker to predict patients’ response to CF101. This is a unique type of a personalized medicine approach, which may pave the way for a safe and efficacious treatment for this patient population.
Plaque psoriasis (Phase II, NCT00428974; Phase II/III, NCT01265667) in which even though the primary endpoint, which was a 75% reduction in the psoriasis area and severity score (PASI75) at week 12, was not reached, however 24.7% of subjects achieved PASI90 at week 32.309,310 In addition, CF101 was more efficacious than apremilast (Otezla), a PDE4 inhibitor, on week 32 while having an excellent safety profile, suggesting its promise as a chronic treatment.
Ocular hypertension (NCT01033422), keratoconjunctivitis sicca (dry eye disease; Phase II, NCT00349466; Phase III, NCT01235234), RA in cotreatment with methotrexate (NCT00280917) failed to reach their endpoints.
Furthermore, additional trials are expected in the near future for the treatment of: RA (Phase III, NCT02647762); osteoarthritis of the knee (Phase II, NCT00837291).
The agonist Cl-IB-MECA (Namodenoson, CF102) has been in the following clinical trials:
Advanced hepatocellular carcinoma (Phase I/II, NCT00790218), finding that it is able to induce a median overall survival (OS) of 7.8 months.311 In patients with advanced HCC and Child Puh B, CF102 induced an OS of 9.3 months (literature data demonstrate OS of 3.5 months). A global Phase II study in this patient population is currently ongoing.
Chronic hepatitis C genotype 1 (Phase I/II, NCT00790673).
Additional trials of CF102 are expected soon for the treatment of: nonalcoholic steatohepatitis (Phase II, NCT02927314) and hepatocellular carcinoma (Phase II, NCT02128958).
An A3AR allosteric enhancer (CF602, LUF6000 95) demonstrated an anti-inflammatory effect in vivo. It is currently under development for treating sexual dysfunction.
7 CONCLUSIONS
The impact of the A3AR on the drug discovery process and development is rapidly expanding, and its significance for human health should not be underestimated. Purine scientists are well advised to remain optimistic for the three following reasons. First, A3AR is overexpressed in cancer and inflammatory cells in comparison to healthy cells, findings that are mirrored in the PBMCs of patients affected by these pathologies. Second, highly selective A3AR agonists are now available as tool compounds and potential clinical molecules. They induce both anti-inflammatory/anticancer effects in pathological cells, as well as protective functions in normal cells, have been synthesized. Third, clinical data that correlate a high expression of A3AR at baseline prior to agonist treatment with a beneficial response in patients suggest that modulation of the A3AR may lead to a personalized medicine approach. Thus, new findings with A3AR ligands appear to open new opportunities to fight inflammatory diseases, cancer, and other conditions.
Abbreviations
- ADA
adenosine deaminase
- AK
adenosine kinase
- AR
adenosine receptor
- cAMP
cyclic AMP
- CCI
chronic constriction injury
- CD73
ecto-5′-nucleotidase
- CHO-hA3
Chinese hamster ovary cells expressing the human A3AR
- Cl-IB-MECA
2-Chloro-N6-(3-iodobenzyl)-adenosine-5′-Nmethyluronamide
- CNT
concentrative nucleoside transporter
- CREB
cAMP response elements binding protein
- ENT
equilibrative nucleoside transporter
- ERK1/2
extracellular signal-regulated kinases
- GPCR
G protein coupled receptor
- GSK-3β
glycogen synthase kinase-3β
- HIF-1α
hypoxia inducible factor 1
- IB-MECA
N6-(3-Iodobenzyl)adenosine-5′-N-methyluronamide
- IL
interleukin
- IR
ischemia/reperfusion
- JNK
c-Jun N-terminal kinase
- KO
knockout
- MIP
macrophage inflammatory protein
- MMP-9
matrix metalloproteinase 9
- MRP1
multiple resistance-associated protein 1
- mTOR
mammalian target of rapamycin
- NF-κB
nuclear factor κB
- PET
positron emission tomography
- PI3K
phosphatidylinositol-4,5-bisphosphate 3-kinase
- PKA
protein kinase A
- PKC
protein kinase C
- PLC
phospholipase C
- SAH
S-adenosylhomocysteine
- TM
transmembrane domain
- TNF-α
tumor necrosis factor α
- PBMC
peripheral blood mononuclear cell
- RA
rheumatoid arthritis.
Biographies
Kenneth A. Jacobson is a Senior Investigator and Chief of the Laboratory of Bioorganic Chemistry and the Molecular Recognition Section at the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health in Bethesda, Maryland, USA. Dr. Jacobson is a medicinal chemist with interests in the structure and pharmacology of G protein coupled receptors, especially receptors for adenosine and for purine and pyrimidine nucleotides. He was inducted into the American Chemical Soc. Medicinal Chemistry Hall of Fame in 2009.
Stefania Merighi received her doctoral degree in Biology Summa cum Laude in 1997 and her Ph.D. degree in Cellular and Molecular Pharmacology in 2003 from the University of Ferrara. Currently, she is Senior Researcher of Pharmacology at the University of Ferrara. Her research activities primarily focus on the pharmacological, biochemical, and molecular study of GPCRs in oncology and neuroinflammation.
Katia Varani received her doctoral degree in Biology in 1988 and her Ph.D. degree in Cellular and Molecular Pharmacology in 1995 from the University of Ferrara. She is currently Associate Professor of Pharmacology in the Medical Sciences Department of the University of Ferrara. Her research activities primarily focus on the pharmacological, biochemical, and molecular study of adenosine receptors.
Pier Andrea Borea received his degree in Chemistry from the University of Ferrara in 1967. He is currently President of the Evaluation Board of the University of Ferrara. He contributed to some 400 publications in international journals and about 20 chapters in international books. His main field of interest is represented by the study, at the molecular level, of drug–receptor interactions.
Stefania Baraldi received her doctoral degree in Pharmaceutical Chemistry and Technology with 110 cum Laude (University of Ferrara, 2005), her Ph.D. in Pharmaceutical Science/medicinal Chemistry (University of Ferrara, 2009) and her second-level master’s degree in Cosmetic Science and Technology with Top Marks (University of Ferrara, 2012). Her scientific interests have focused on the design and synthesis of ligands in the adenosine field and Endocannabinoid System modulators for the treatment of pain and inflammation. Her research activity has been supported by the company King Pharmaceutical (North Carolina, USA) now part of Pfizer.
Mojgan Aghazadeh Tabrizi received her doctoral degree in Pharmacy (University of Ferrara, 1987), Ph.D. in Medicinal Chemistry (University of Ferrara, 1992). She has worked at the Department of Pharmaceutical Sciences of Ferrara University focusing her interests on the adenosine receptors ligands and antitumor compounds in collaboration with the company King Pharmaceuticals (North Carolina, USA), now part of Pfizer. Since 2008, she has been part of a research project on the treatment of pain and inflammation involving the endocannabinoid system. She is author/coauthor of several scientific publications in the field of medicinal chemistry.
Romeo Romagnoli graduated in Pharmaceutical Chemistry and Technology at the University of Ferrara in 1990, and in 1995 he received his Ph.D. in Organic Chemistry. From 1999 to 2014, he has held a position of Assistant Professor in Pharmaceutical Chemistry at the University of Ferrara. Actually he is Associate Professor in Pharmaceutical Chemistry at the same University. His scientific interests have focused on new synthetic methods for the preparation of natural substances or biologically active analogues, the medicinal chemistry of ligands for adenosine receptors subtypes and antitumor agents. He has published more than 230 research papers and 20 international patents.
Pier Giovanni Baraldi received his doctoral degree in Chemistry in 1974 from the University of Ferrara where he is currently Full Professor of Medicinal Chemistry. He has published more than 400 scientific papers including about 50 patents and he participated in more than 90 Medicinal Chemistry meetings as plenary speaker. His research interests have focused on the design and synthesis of minor groove alkylating agents, combretastatin analogs, ligands for ARs, cannabinoid receptors, and TRP channel modulators. He has been promoter of several scientific collaborations with national and international pharmaceutical companies.
Antonella Ciancetta received hermaster’s degree in Pharmaceutical Chemistry and Technology in 2007 and her Ph.D. degree in Drug Sciences in 2010 from the University of Chieti, Italy. She is currently a Visiting Fellow at the National Institutes of Health in Bethesda, Maryland, USA in Dr. Jacobson’s laboratory. Her research focuses on the application and development of structure-based molecular modeling techniques for the design of GPCR ligands and the elucidation of their binding mechanisms.
Dilip K. Tosh is a Staff Scientist in the Molecular Recognition Section, Laboratory of Bioorganic Chemistry, NIDDK, National Institutes of Health in Bethesda, Maryland, USA. He is a medicinal chemist in the field of G protein coupled receptors and transporters, especially interested in adenosine receptors and other targets of nucleosides and nucleotides. He received his Ph.D. in Synthetic Organic Chemistry from the Indian Institute of Technology, Bombay, India.
Zhan-Guo Gao is a Staff Scientist in the Molecular Recognition Section, Laboratory of Bioorganic Chemistry, NIDDK, National Institutes of Health in Bethesda, Maryland, USA. He is a pharmacologist in the field of G protein coupled receptors, especially interested in adenosine receptors and P2Y receptors.
Stefania Gessi received her Doctor degree in Biology Summa cum Laude from University of Ferrara in 1994 and her PhD in Cellular and Molecular Pharmacology in 2000. She is currently Associate Professor of Pharmacology in the Medical Sciences Department of the University of Ferrara. Her research activity focuses on the pharmacological, biochemical and molecular study of adenosine receptors in health and diseases.
References
- 1.Antonioli L, Csóka B, Fornai M, et al. Adenosine and inflammation: What’s new on the horizon? Drug Discov Today. 2014;19:1051–1068. doi: 10.1016/j.drudis.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 2.Borea PA, Gessi S, Merighi S, Varani K. Adenosine as a multi-signalling guardian angel in human diseases: When, where and how does it exert its protective effects? Trends Pharmacol Sci. 2016;37:419–434. doi: 10.1016/j.tips.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Fredholm BB, Jzerman IAP, Jacobson KA, Linden J, Müller CE International union of basic and clinical pharmacology. LXXXI Nomenclature and classification of adenosine receptors—An update. Pharmacol Rev. 2011;63:1–34. doi: 10.1124/pr.110.003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zimmermann H. Extracellular metabolism of ATP and other nucleotides. Naunyn Schmiedebergs Arch Pharmacol. 2000;362:299–309. doi: 10.1007/s002100000309. [DOI] [PubMed] [Google Scholar]
- 5.Molina-Arcas M, Casado FJ, Pastor-Anglada M. Nucleoside transporter proteins. Curr Vasc Pharmacol. 2009;7:426–434. doi: 10.2174/157016109789043892. [DOI] [PubMed] [Google Scholar]
- 6.Meyerhof W, Müller-Brechlin R, Richter D. Molecular cloning of a novel putative G-protein coupled receptor expressed during rat spermiogenesis. FEBS Lett. 1991;284:155–160. doi: 10.1016/0014-5793(91)80674-r. [DOI] [PubMed] [Google Scholar]
- 7.Salvatore CA, Jacobson MA, Taylor HE, Linden J, Johnson RG. Molecular cloning and characterization of the human A3 adenosine receptor. Proc Natl Acad Sci. 1993;90:10365–10369. doi: 10.1073/pnas.90.21.10365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou QY, Li C, Olah ME, Johnson RA, Stiles GL, Civelli O. Molecular cloning and characterization of an adenosine receptor: The A3 adenosine receptor. Proc Natl Acad Sci. 1992;89:7432–7436. doi: 10.1073/pnas.89.16.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramkumar V, Stiles GL, Beaven MA, Ali H. The A3 adenosine receptor is the unique adenosine receptor which facilitates release of allergic mediators in mast cells. J Biol Chem. 1993;268:16887–16890. [PubMed] [Google Scholar]
- 10.Fishman P, Bar-Yehuda S, Liang BT, Jacobson KA. Pharmacological and therapeutic effects of A3 adenosine receptor agonists. Drug Discov Today. 2012;17:359–366. doi: 10.1016/j.drudis.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gessi S, Merighi S, Varani K, Leung E, Mac Lennan S, Borea PA. The A3 adenosine receptor: An enigmatic player in cell biology. Pharmacol Ther. 2008;117:123–140. doi: 10.1016/j.pharmthera.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Alnouri MW, Jepards S, Casari A, Schiedel AC, Hinz S, Müller CE. Selectivity is species-dependent: Characterization of standard agonists and antagonists at human, rat, and mouse adenosine receptors. Purinergic Signal. 2015;11:389–407. doi: 10.1007/s11302-015-9460-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leung CT, Li A, Banerjee J, et al. The role of activated adenosine receptors in degranulation of human LAD2 mast cells. Purinergic Signal. 2014;10:465–475. doi: 10.1007/s11302-014-9409-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atkinson MR, Townsend-Nicholson A, Nicholl JK, Sutherland GR, Schofield PR. Cloning, characterisation and chromosomal assignment of the human adenosine A3 receptor (ADORA3) gene. Neurosci Res. 1997;29:73–79. doi: 10.1016/s0168-0102(97)00073-4. [DOI] [PubMed] [Google Scholar]
- 15.Murrison EM, Goodson SJ, Edbrooke MR, Harris CA. Cloning and characterisation of the human adenosine A3 receptor gene. FEBS Lett. 1996;384:243–246. doi: 10.1016/0014-5793(96)00324-9. [DOI] [PubMed] [Google Scholar]
- 16.Peculis R, Latkovskis G, Tarasova L, Pirags V, Erglis A, Klovins J. A nonsynonymous variant I248L of the adenosine A3 receptor is associated with coronary heart disease in a Latvian population. DNA Cell Biol. 2011;30:907–911. doi: 10.1089/dna.2011.1230. [DOI] [PubMed] [Google Scholar]
- 17.Campbell NG, Zhu C-B, Lindler KM, et al. Rare coding variants of the adenosine A3 receptor are increased in autism: On the trail of the serotonin transporter regulome. Mol Autism. 2013;4:28. doi: 10.1186/2040-2392-4-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim S-H, Nam E-J, Kim Y-K, Ye Y-M, Park H-S. Functional variability of the adenosine A3 receptor (ADORA3) gene polymorphism in aspirin-induced urticaria. Br J Dermatol. 2010;163:977–985. doi: 10.1111/j.1365-2133.2010.09983.x. [DOI] [PubMed] [Google Scholar]
- 19.Wu W, He Y, Feng X, et al. Micro RNA-206 is involved in the pathogenesis of ulcerative colitis via regulation of adenosine A3 receptor. Oncotarget. 2017;8:705–721. doi: 10.18632/oncotarget.13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trincavelli ML, Tuscano D, Marroni M, et al. A3 adenosine receptors in human astrocytoma cells: Agonist-mediated desensitization, internalization, and down-regulation. Mol Pharmacol. 2002;62:1373–1384. doi: 10.1124/mol.62.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trincavelli ML, Tuscano D, Marroni M, Klotz K-N, Lucacchini A, Martini C. Involvement of mitogen protein kinase cascade in agonist-mediated human A3 adenosine receptor regulation. Biochim Biophys ActaMol Cell Res. 2002;1591:55–62. doi: 10.1016/s0167-4889(02)00248-3. [DOI] [PubMed] [Google Scholar]
- 22.Madi L, Bar-Yehuda S, Barer F, Ardon E, Ochaion A, Fishman P. A3 adenosine receptor activation in melanoma cells: Association between receptor fate and tumor growth inhibition. J Biol Chem. 2003;278:42121–42130. doi: 10.1074/jbc.M301243200. [DOI] [PubMed] [Google Scholar]
- 23.Palmer TM, Stiles GL. Identification of threonine residues controlling the agonist-dependent phosphorylation and desensitization of the rat A(3) adenosine receptor. Mol Pharmacol. 2000;57:539–545. [PubMed] [Google Scholar]
- 24.Pugliese AM, Coppi E, Volpini R, et al. Role of adenosine A3 receptors on CA1 hippocampal neurotransmission during oxygen-glucose deprivation episodes of different duration. Biochem Pharmacol. 2007;74:768–779. doi: 10.1016/j.bcp.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stoddart LA, Kellam B, Briddon SJ, Hill SJ. Effect of a toggle switch mutation in TM6 of the human adenosine A3 receptor on Gi protein-dependent signalling and Gi-independent receptor internalization. Br J Pharmacol. 2014;171:3827–3844. doi: 10.1111/bph.12739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stoddart LA, Vernall AJ, Briddon SJ, Kellam B, Hill SJ. Direct visualisation of internalization of the adenosine A3 receptor and localization with arrestin3 using a fluorescent agonist. Neuropharmacology. 2015;98:68–77. doi: 10.1016/j.neuropharm.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 27.Merighi S, Varani K, Gessi S, et al. Binding thermodynamics at the human A(3) adenosine receptor. Biochem Pharmacol. 2002;63:157–161. doi: 10.1016/s0006-2952(01)00825-5. [DOI] [PubMed] [Google Scholar]
- 28.Borea PA, Dalpiaz A, Varani K, Gilli P, Gilli G. Can thermodynamic measurements of receptor binding yield information on drug affinity and efficacy? Biochem Pharmacol. 2000;60:1549–1556. doi: 10.1016/s0006-2952(00)00368-3. [DOI] [PubMed] [Google Scholar]
- 29.Gessi S, Fogli E, Sacchetto V, et al. Thermodynamics of A2B adenosine receptor binding discriminates agonistic from antagonistic behaviour. Biochem Pharmacol. 2008;75:562–569. doi: 10.1016/j.bcp.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 30.Kohno Y, Ji X, Mawhorter SD, Koshiba M, Jacobson KA. Activation of A3 adenosine receptors on human eosinophils elevates intracellular calcium. Blood. 1996;88:3569–3574. [PMC free article] [PubMed] [Google Scholar]
- 31.Morschl E, Molina JG, Volmer JB, et al. A3 adenosine receptor signaling influences pulmonary inflammation and fibrosis. Am J Respir Cell Mol Biol. 2008;39:697–705. doi: 10.1165/rcmb.2007-0419OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bouma MG, Jeunhomme TM, Boyle DL, et al. Adenosine inhibits neutrophil degranulation in activated human whole blood: Involvement of adenosine A2 and A3 receptors. J Immunol. 1997;158:5400–5408. [PubMed] [Google Scholar]
- 33.Gessi S, Varani K, Merighi S, et al. A(3) adenosine receptors in human neutrophils and promyelocytic HL60 cells: A pharmacological and biochemical study. Mol Pharmacol. 2002;61:415–424. doi: 10.1124/mol.61.2.415. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y, Corriden R, Inoue Y, et al. ATP release guides neutrophil chemotaxis via P2Y2 and A3 receptors. Science. 2006;314:1792–1795. doi: 10.1126/science.1132559. [DOI] [PubMed] [Google Scholar]
- 35.van der Hoeven D, Wan TC, Auchampach JA. Activation of the A(3) adenosine receptor suppresses superoxide production and chemotaxis of mouse bone marrow neutrophils. Mol Pharmacol. 2008;74:685–696. doi: 10.1124/mol.108.048066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corriden R, Self T, Akong-Moore K, et al. Adenosine-A3 receptors in neutrophil microdomains promote the formation of bacteria-tethering cytonemes. EMBO Rep. 2013;14:726–732. doi: 10.1038/embor.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mulloy DP, Sharma AK, Fernandez LG, et al. Adenosine A3 receptor activation attenuates lung ischemia-reperfusion injury. Ann Thorac Surg. 2013;95:1762–1767. doi: 10.1016/j.athoracsur.2013.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Broussas M, Cornillet-Lefèbvre P, Potron G, Nguyen P. Inhibition of fMLP-triggered respiratory burst of human monocytes by adenosine: Involvement of A3 adenosine receptor. J Leukoc Biol. 1999;66:495–501. [PubMed] [Google Scholar]
- 39.Broussas M, Cornillet-Lefèbvre P, Potron G, Nguyên P. Adenosine inhibits tissue factor expression by LPS-stimulated human monocytes: Involvement of the A3 adenosine receptor. Thromb Haemost. 2002;88:123–130. [PubMed] [Google Scholar]
- 40.Thiele A, Kronstein R, Wetzel A, Gerth A, Nieber K, Hauschildt S. Regulation of adenosine receptor subtypes during cultivation of human monocytes: Role of receptors in preventing lipopolysaccharide-triggered respiratory burst. Infect Immun. 2004;72:1349–1357. doi: 10.1128/IAI.72.3.1349-1357.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McWhinney CD, Dudley MW, Bowlin TL, et al. Activation of adenosine A3 receptors on macrophages inhibits tumor necrosis factor-alpha. Eur J Pharmacol. 1996;310:209–216. doi: 10.1016/0014-2999(96)00272-5. [DOI] [PubMed] [Google Scholar]
- 42.Szabó C, Scott GS, Virág L, et al. Suppression of macrophage inflammatory protein (MIP)-1alpha production and collagen-induced arthritis by adenosine receptor agonists. Br J Pharmacol. 1998;125:379–387. doi: 10.1038/sj.bjp.0702040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gessi S, Fogli E, Sacchetto V, et al. Adenosine modulates HIF-1 alpha, VEGF, IL-8, and foam cell formation in a human model of hypoxic foam cells. Arterioscler Thromb Vasc Biol. 2010;30:90–97. doi: 10.1161/ATVBAHA.109.194902. [DOI] [PubMed] [Google Scholar]
- 44.Panther E, Idzko M, Herouy Y, Rheinen H, Gebicke-Haerter PJ, Mrowietz U, Dichmann S, Norgauer J. Expression and function of adenosine receptors in human dendritic cells. FASEB J. 2001;15:1963–1970. doi: 10.1096/fj.01-0169com. [DOI] [PubMed] [Google Scholar]
- 45.Fossetta J, Jackson J, Deno G, et al. Pharmacological analysis of calcium responses mediated by the human A3 adenosine receptor in monocyte-derived dendritic cells and recombinant cells. Mol Pharmacol. 2003;63:342–350. doi: 10.1124/mol.63.2.342. [DOI] [PubMed] [Google Scholar]
- 46.Dickenson JM, Reeder S, Rees B, Alexander S, Kendall D. Functional expression of adenosine A2A and A3 receptors in the mouse dendritic cell line XS-106. Eur J Pharmacol. 2003;474:43–51. doi: 10.1016/s0014-2999(03)02041-7. [DOI] [PubMed] [Google Scholar]
- 47.Hofer S, Ivarsson L, Stoitzner P, et al. Adenosine slows migration of dendritic cells but does not affect other aspects of dendritic cell maturation. J Invest Dermatol. 2003;121:300–307. doi: 10.1046/j.1523-1747.2003.12369.x. [DOI] [PubMed] [Google Scholar]
- 48.Gessi S, Varani K, Merighi S, et al. Expression of A3 adenosine receptors in human lymphocytes: Up-regulation in T cell activation. Mol Pharmacol. 2004;65:711–719. doi: 10.1124/mol.65.3.711. [DOI] [PubMed] [Google Scholar]
- 49.Varani K, Massara A, Vincenzi F, et al. Normalization of A2A and A3 adenosine receptor up-regulation in rheumatoid arthritis patients by treatment with anti-tumor necrosis factor alpha but not methotrexate. Arthritis Rheum. 2009;60:2880–2891. doi: 10.1002/art.24794. [DOI] [PubMed] [Google Scholar]
- 50.Varani K, Vincenzi F, Tosi A, et al. A2A adenosine receptor overexpression and functionality, as well as TNF-alpha levels, correlate with motor symptoms in Parkinson’s disease. FASEB J. 2010;24:587–598. doi: 10.1096/fj.09-141044. [DOI] [PubMed] [Google Scholar]
- 51.Bar-Yehuda S, Luger D, Ochaion A, et al. Inhibition of experimental auto-immune uveitis by the A3 adenosine receptor agonist CF101. Int J MolMed. 2011;28:727–731. doi: 10.3892/ijmm.2011.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Varani K, De Mattei M, Vincenzi F, et al. Characterization of adenosine receptors in bovine chondrocytes and fibroblast-like synoviocytes exposed to low frequency low energy pulsed electromagnetic fields. Osteoarthr Cartil. 2008;16:292–304. doi: 10.1016/j.joca.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 53.Varani K, Vincenzi F, Tosi A, et al. Expression and functional role of adenosine receptors in regulating inflammatory responses in human synoviocytes. Br J Pharmacol. 2010;160:101–115. doi: 10.1111/j.1476-5381.2010.00667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stamp LK, Hazlett J, Roberts RL, Frampton C, Highton J, Hessian PA. Adenosine receptor expression in rheumatoid synovium: A basis for methotrexate action. Arthritis Res Ther. 2012;14:R138. doi: 10.1186/ar3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vincenzi F, Targa M, Corciulo C, et al. Pulsed electromagnetic fields increased the anti-inflammatory effect of A2A and A3 adenosine receptors in human T/C-28a2 chondrocytes and hFOB 1. 19 osteoblasts. PLoS One. 2013;8:e65561. doi: 10.1371/journal.pone.0065561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olah ME, Gallo-Rodriguez C, Jacobson KA, Stiles GL. 125I-4-aminobenzyl-5′-N-methylcarboxamidoadenosine, a high affinity radioligand for the rat A3 adenosine receptor. Mol Pharmacol. 1994;45:978–982. [PMC free article] [PubMed] [Google Scholar]
- 57.Carruthers AM, Fozard JR. Adenosine A3 receptors: Two into one won’t go. Trends Pharmacol Sci. 1993;14:290–291. doi: 10.1016/0165-6147(93)90042-I. [DOI] [PubMed] [Google Scholar]
- 58.Fozard JR, Carruthers AM. Adenosine A3 receptors mediate hypotension in the angiotensin II-supported circulation of the pithed rat. Br J Pharmacol. 1993;109:3–5. doi: 10.1111/j.1476-5381.1993.tb13522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hannon JP, Pfannkuche HJ, Fozard JR. A role for mast cells in adenosine A3 receptor-mediated hypotension in the rat. Br J Pharmacol. 1995;115:945–952. doi: 10.1111/j.1476-5381.1995.tb15902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.el-Hashim A, D’Agostino B, Matera MG, Page C. Characterization of adenosine receptors involved in adenosine-induced bronchoconstriction in allergic rabbits. Br J Pharmacol. 1996;119:1262–1268. doi: 10.1111/j.1476-5381.1996.tb16031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fozard JR, Pfannkuche HJ, Schuurman HJ. Mast cell degranulation following adenosine A3 receptor activation in rats. Eur J Pharmacol. 1996;298:293–297. doi: 10.1016/0014-2999(95)00822-5. [DOI] [PubMed] [Google Scholar]
- 62.Hua X, Chason KD, Fredholm BB, Deshpande DA, Penn RB, Tilley SL. Adenosine induces airway hyperresponsiveness through activation of A3 receptors on mast cells. J Allergy Clin Immunol. 2008;122:107–113. 113–117. doi: 10.1016/j.jaci.2008.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gomez G, Zhao W, Schwartz LB. Disparity in FcεRI-induced degranulation of primary human lung and skin mast cells exposed to adenosine. J Clin Immunol. 2011;31:479–487. doi: 10.1007/s10875-011-9517-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kohno Y, Sei Y, Koshiba M, Kim HO, Jacobson KA. Induction of apoptosis in HL-60 human promyelocytic leukemia cells by adenosine A3 receptor agonists. Biochem Biophys Res Commun. 1996;219:904–910. doi: 10.1006/bbrc.1996.0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mousavi S, Panjehpour M, Izadpanahi MH, Aghaei M. Expression of adenosine receptor subclasses in malignant and adjacent normal human prostate tissues. Prostate. 2015;75:735–747. doi: 10.1002/pros.22955. [DOI] [PubMed] [Google Scholar]
- 66.Gessi S, Varani K, Merighi S, et al. Pharmacological and biochemical characterization of A3 adenosine receptors in Jurkat T cells. Br J Pharmacol. 2001;134:116–126. doi: 10.1038/sj.bjp.0704254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Merighi S, Varani K, Gessi S, et al. Pharmacological and biochemical characterization of adenosine receptors in the human malignant melanoma A375 cell line. Br J Pharmacol. 2001;134:1215–1226. doi: 10.1038/sj.bjp.0704352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suh BC, Kim TD, Lee JU, Seong JK, Kim KT. Pharmacological characterization of adenosine receptors in PGT-beta mouse pineal gland tumour cells. Br J Pharmacol. 2001;134:132–142. doi: 10.1038/sj.bjp.0704218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gessi S, Merighi S, Varani K, et al. Adenosine receptors in colon carcinoma tissues and colon tumoral cell lines: Focus on the A(3) adenosine subtype. J Cell Physiol. 2007;211:826–836. doi: 10.1002/jcp.20994. [DOI] [PubMed] [Google Scholar]
- 70.Morello S, Sorrentino R, Porta A, et al. Cl-IB-MECA enhances TRAIL-induced apoptosis via the modulation of NF-kappaB signalling pathway in thyroid cancer cells. J Cell Physiol. 2009;221:378–386. doi: 10.1002/jcp.21863. [DOI] [PubMed] [Google Scholar]
- 71.Merighi S, Mirandola P, Milani D, et al. Adenosine receptors as mediators of both cell proliferation and cell death of cultured human melanoma cells. J Invest Dermatol. 2002;119:923–933. doi: 10.1046/j.1523-1747.2002.00111.x. [DOI] [PubMed] [Google Scholar]
- 72.Merighi S, Simioni C, Gessi S, et al. A(2B) and A(3) adenosine receptors modulate vascular endothelial growth factor and interleukin-8 expression in human melanoma cells treated with etoposide and doxorubicin. Neoplasia. 2009;11:1064–1073. doi: 10.1593/neo.09768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jajoo S, Mukherjea D, Watabe K, Ramkumar V. Adenosine A(3) receptor suppresses prostate cancer metastasis by inhibiting NADPH oxidase activity. Neoplasia. 2009;11:1132–1145. doi: 10.1593/neo.09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gessi S, Sacchetto V, Fogli E, et al. Modulation of metalloproteinase-9 in U87MG glioblastoma cells by A3 adenosine receptors. Biochem Pharmacol. 2010;79:1483–1495. doi: 10.1016/j.bcp.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 75.Varani K, Maniero S, Vincenzi F, et al. A3 receptors are overexpressed in pleura from patients with mesothelioma and reduce cell growth via Akt/nuclear factor-κB pathway. Am J Respir Crit CareMed. 2011;183:522–530. doi: 10.1164/rccm.201006-0980OC. [DOI] [PubMed] [Google Scholar]
- 76.Cohen S, Stemmer SM, Zozulya G, et al. CF102 an A3 adenosine receptor agonist mediates anti-tumor and anti-inflammatory effects in the liver. J Cell Physiol. 2011;226:2438–2447. doi: 10.1002/jcp.22593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hofer M, Dušek L, Hoferová Z, Stixová L, Pospíšil M. Expression of mRNA for adenosine A(1), A(2a), A(2b), and A(3) receptors in HL-60 cells: Dependence on cell cycle phases. Physiol Res. 2011;60:913–920. doi: 10.33549/physiolres.932233. [DOI] [PubMed] [Google Scholar]
- 78.Kanno T, Gotoh A, Fujita Y, Nakano T, Nishizaki T. A(3) adenosine receptor mediates apoptosis in 5637 human bladder cancer cells by G(q) protein/PKC-dependent AIF upregulation. Cell Physiol Biochem. 2012;30:1159–1168. doi: 10.1159/000343306. [DOI] [PubMed] [Google Scholar]
- 79.Nogi Y, Kanno T, Nakano T, et al. AMP converted from intracellularly transported adenosine upregulates p53 expression to induce malignant pleural mesothelioma cell apoptosis. Cell Physiol Biochem. 2012;30:61–74. doi: 10.1159/000339048. [DOI] [PubMed] [Google Scholar]
- 80.Kamiya H, Kanno T, Fujita Y, Gotoh A, Nakano T, Nishizaki T. Apoptosis-related gene transcription in human A549 lung cancer cells via A(3) adenosine receptor. Cell Physiol Biochem. 2012;29:687–696. doi: 10.1159/000312589. [DOI] [PubMed] [Google Scholar]
- 81.Otsuki T, Kanno T, Fujita Y, et al. A3 adenosine receptor-mediated p53-dependent apoptosis in Lu-65 human lung cancer cells. Cell Physiol Biochem. 2012;30:210–220. doi: 10.1159/000339058. [DOI] [PubMed] [Google Scholar]
- 82.Vincenzi F, Targa M, Corciulo C, et al. The anti-tumor effect of A3 adenosine receptors is potentiated by pulsed electromagnetic fields in cultured neural cancer cells. PLoS One. 2012;7:e39317. doi: 10.1371/journal.pone.0039317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nagaya H, Gotoh A, Kanno T, Nishizaki T. A3 adenosine receptor mediates apoptosis in in vitro RCC4-VHL human renal cancer cells by up-regulating AMID expression. J Urol. 2013;189:321–328. doi: 10.1016/j.juro.2012.08.193. [DOI] [PubMed] [Google Scholar]
- 84.Sakowicz-Burkiewicz M, Kitowska A, Grden M, Maciejewska I, Szutowicz A, Pawelczyk T. Differential effect of adenosine receptors on growth of human colon cancer HCT 116 and HT-29 cell lines. Arch Biochem Biophys. 2013;533:47–54. doi: 10.1016/j.abb.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 85.Madi L, Rosenberg-Haggen B, Nyska A, Korenstein R. Enhancing pigmentation via activation of A3 adenosine receptors in B16melanoma cells and in human skin explants. Exp Dermatol. 2013;22:74–77. doi: 10.1111/exd.12028. [DOI] [PubMed] [Google Scholar]
- 86.Varani K, Vincenzi F, Targa M, et al. The stimulation of A(3) adenosine receptors reduces bone-residing breast cancer in a rat preclinical model. Eur J Cancer. 2013;49:482–491. doi: 10.1016/j.ejca.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 87.Gessi S, Cattabriga E, Avitabile A, et al. Elevated expression of A3 adenosine receptors in human colorectal cancer is reflected in peripheral blood cells. Clin Cancer Res. 2004;10:5895–5901. doi: 10.1158/1078-0432.CCR-1134-03. [DOI] [PubMed] [Google Scholar]
- 88.Madi L, Ochaion A, Rath-Wolfson L, et al. The A3 adenosine receptor is highly expressed in tumor versus normal cells: Potential target for tumor growth inhibition. Clin Cancer Res. 2004;10:4472–4479. doi: 10.1158/1078-0432.CCR-03-0651. [DOI] [PubMed] [Google Scholar]
- 89.Bar-Yehuda S, Stemmer SM, Madi L, et al. The A3 adenosine receptor agonist CF102 induces apoptosis of hepatocellular carcinoma via de-regulation of the Wnt and NF-kappaB signal transduction pathways. Int J Oncol. 2008;33:287–295. [PubMed] [Google Scholar]
- 90.van Galen PJ, van Bergen AH, Gallo-Rodriguez C, et al. A binding site model and structure-activity relationships for the rat A3 adenosine receptor. Mol Pharmacol. 1994;45:1101–1111. [PMC free article] [PubMed] [Google Scholar]
- 91.Jacobson KA, Nikodijevic O, Shi D, et al. A role for central A 3-adenosine receptors. FEBS Lett. 1993;336:57–60. doi: 10.1016/0014-5793(93)81608-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gallo-Rodriguez C, Ji X, Melman N, et al. Structure-Activity Relationships of N6-Benzyladenosine-5′-uronamides as A3-Selective Adenosine Agonists. JMed Chem. 1994;37:636–646. doi: 10.1021/jm00031a014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ji XD, Gallo-Rodriguez C, Jacobson KA. A selective agonist affinity label for A3 adenosine receptors. Biochem Biophys Res Commun. 1994;203:570–576. doi: 10.1006/bbrc.1994.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim HO, Ji X, Siddiqi SM, Olah ME, Stiles GL, Jacobson KA. 2-Substitution of N6-benzyladenosine-5′-uronamides enhances selectivity for A3 adenosine receptors. JMed Chem. 1994;37:3614–3621. doi: 10.1021/jm00047a018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gao Z-G, Jeong LS, Moon HR, et al. Structural determinants of efficacy at A3 adenosine receptors: Modification of the ribose moiety. Biochem Pharmacol. 2004;67:893–901. doi: 10.1016/j.bcp.2003.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Siddiqi SM, Jacobson KA, Esker JL, et al. Search for New purine- and ribose-modified adenosine analogs as selective agonists and antagonists at adenosine receptors. JMed Chem. 1995;38:1174–1188. doi: 10.1021/jm00007a014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Baraldi PG, Cacciari B, Pineda de Las Infantas MJ, et al. Synthesis and biological activity of a new series of N6-arylcarbamoyl, 2-(Ar)alkynyl-N6-arylcarbamoyl, and N6-carboxamido derivatives of adenosine-5′-N-ethyluronamide as A1 and A3 adenosine receptor agonists. JMed Chem. 1998;41:3174–3185. doi: 10.1021/jm980147p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Varani K, Cacciari B, Baraldi PG, Dionisotti S, Ongini E, Borea PA. Binding affinity of adenosine receptor agonists and antagonists at human cloned A3 adenosine receptors. Life Sci. 1998;63:PL81–PL87. doi: 10.1016/s0024-3205(98)00289-6. [DOI] [PubMed] [Google Scholar]
- 99.Volpini R, Costanzi S, Lambertucci C, Taffi S, Vittori S, Klotz K-N, Cristalli G. N(6)-alkyl-2-alkynyl derivatives of adenosine as potent and selective agonists at the human adenosine A(3) receptor and a starting point for searching A(2B) ligands. J Med Chem. 2002;45:3271–3279. doi: 10.1021/jm0109762. [DOI] [PubMed] [Google Scholar]
- 100.Jacobson KA, Siddiqi SM, Olah ME, et al. Structure-activity relationships of 9-alkyladenine and ribose-modified adenosine derivatives at rat A3 adenosine receptors. JMed Chem. 1995;38:1720–1735. doi: 10.1021/jm00010a017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mogensen JP, Roberts SM, Bowler AN, Thomsen C, Knutsen LJ. The synthesis of new adenosine A3 selective ligands containing bioisosteric isoxazoles. Bioorg Med Chem Lett. 1998;8:1767–1770. doi: 10.1016/s0960-894x(98)00302-3. [DOI] [PubMed] [Google Scholar]
- 102.van Tilburg EW, von Frijtag Drabbe Kunzel J, de Groote M, IJzerman AP. 2,5′-Disubstituted adenosine derivatives: Evaluation of selectivity and efficacy for the adenosine A(1), A(2A), and A(3) receptor. JMed Chem. 2002;45:420–429. doi: 10.1021/jm010952v. [DOI] [PubMed] [Google Scholar]
- 103.Klotz KN, Camaioni E, Volpini R, Kachler S, Vittori S, Cristalli G. 2-Substituted N-ethylcarboxamidoadenosine derivatives as high-affinity agonists at human A3 adenosine receptors. Naunyn Schmiedebergs Arch Pharmacol. 1999;360:103–108. doi: 10.1007/s002109900044. [DOI] [PubMed] [Google Scholar]
- 104.Volpini R, Dal Ben D, Lambertucci C, et al. N6-methoxy-2-alkynyladenosine derivatives as highly potent and selective ligands at the human A3 adenosine receptor. JMed Chem. 2007;50:1222–1230. doi: 10.1021/jm060963u. [DOI] [PubMed] [Google Scholar]
- 105.Tchilibon S, Kim S-K, Gao Z-G, et al. Exploring distal regions of the A3 adenosine receptor binding site: Sterically constrained N6-(2-phenylethyl)adenosine derivatives as potent ligands. Bioorg Med Chem. 2004;12:2021–2034. doi: 10.1016/j.bmc.2004.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Park K, Hoffmann C, Kim HO, et al. Activation and desensitization of rat A3-adenosine receptors by selective adenosine derivatives and xanthine-7-ribosides. Drug Dev Res. 1998;44:97–105. doi: 10.1002/(SICI)1098-2299(199806/07)44:2/3<97::AID-DDR7>3.0.CO;2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rodríguez D, Chakraborty S, Warnick E, et al. Structure-based screening of uncharted chemical space for atypical adenosine receptor agonists. ACS Chem Biol. 2016;11:2763–2772. doi: 10.1021/acschembio.6b00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gao Z-G, Verzijl D, Zweemer A, et al. Functionally biased modulation of A(3) adenosine receptor agonist efficacy and potency by imidazoquinolinamine allosteric enhancers. Biochem Pharmacol. 2011;82:658–668. doi: 10.1016/j.bcp.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Melman A, Wang B, Joshi BV, Gao Z-G, Castro S, de Heller CL, Kim S-K, Jeong LS, Jacobson KA. Selective A3 adenosine receptor antagonists derived from nucleosides containing a bicyclo[3.1. 0]hexane ring system. Bioorg Med Chem. 2008;16:8546–8556. doi: 10.1016/j.bmc.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.DeNinno MP, Masamune H, Chenard LK, et al. 3′-Aminoadenosine-5′-uronamides: Discovery of the first highly selective agonist at the human adenosine A3 receptor. JMed Chem. 2003;46:353–355. doi: 10.1021/jm0255724. [DOI] [PubMed] [Google Scholar]
- 111.Elzein E, Palle V, Wu Y, Maa T, Zeng D, Zablocki J. 2-Pyrazolyl-N6-substituted adenosine derivatives as high affinity and selective adenosine A3 receptor agonists. JMed Chem. 2004;47:4766–4773. doi: 10.1021/jm049682h. [DOI] [PubMed] [Google Scholar]
- 112.Jacobson KA, Ji X, Li A-H, et al. Methanocarba analogues of purine nucleosides as potent and selective adenosine receptor agonists. JMed Chem. 2000;43:2196–2203. doi: 10.1021/jm9905965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Marquez VE, Siddiqui MA, Ezzitouni A, et al. Nucleosides with a twist. Can fixed forms of sugar ring pucker influence biological activity in nucleosides and oligonucleotides? JMed Chem. 1996;39:3739–3747. doi: 10.1021/jm960306+. [DOI] [PubMed] [Google Scholar]
- 114.Tchilibon S, Joshi BV, Kim S-K, Duong HT, Gao Z-G, Jacobson KA. (N)-Methanocarba 2, N6-disubstituted adenine nucleosides as highly potent and selective A3 adenosine receptor agonists. JMed Chem. 2005;48:1745–1758. doi: 10.1021/jm049580r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gao Z-G, Teng B, Wu H, Joshi BV, Griffiths GL, Jacobson KA. Synthesis and pharmacological characterization of [125I]MRS1898, a high-affinity, selective radioligand for the rat A3 adenosine receptor. Purinergic Signal. 2009;5:31–37. doi: 10.1007/s11302-008-9107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kiesewetter DO, Lang L, Ma Y, et al. Synthesis and characterization of [76Br]-labeled high-affinity A3 adenosine receptor ligands for positron emission tomography. Nucl Med Biol. 2009;36:3–10. doi: 10.1016/j.nucmedbio.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jacobson KA, Gao Z-G, Tchilibon S, et al. Semi-rational design of (north)-methanocarba nucleosides as dual acting A(1) and A(3) adenosine receptor agonists: Novel prototypes for cardioprotection. JMed Chem. 2005;48:8103–8107. doi: 10.1021/jm050726b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Volpini R, Costanzi S, Lambertucci C, et al. Adenosine receptor agonists: Synthesis and binding affinity of 2-(aryl)alkylthioadenosine derivatives. ARKIVOC. 2004;5:301–311. [Google Scholar]
- 119.Tosh DK, Chinn M, Ivanov AA, Klutz AM, Gao Z-G, Jacobson KA. Functionalized congeners of A3 adenosine receptor-selective nucleosides containing a bicyclo[3.1. 0]hexane ring system. JMed Chem. 2009;52:7580–7592. doi: 10.1021/jm900426g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tosh DK, Deflorian F, Phan K, et al. Structure-guided design of A(3) adenosine receptor-selective nucleosides: Combination of 2-arylethynyl and bicyclo[3.1. 0]hexane substitutions. JMed Chem. 2012;55:4847–4860. doi: 10.1021/jm300396n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tosh DK, Finley A, Paoletta S, et al. In vivo phenotypic screening for treating chronic neuropathic pain: Modification of C 2-arylethynyl group of conformationally constrained A3 adenosine receptor agonists. JMed Chem. 2014;57:9901–9914. doi: 10.1021/jm501021n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Paoletta S, Tosh DK, Finley A, et al. Rational design of sulfonated A3 adenosine receptor-selective nucleosides as pharmacological tools to study chronic neuropathic pain. JMed Chem. 2013;56:5949–5963. doi: 10.1021/jm4007966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Volpini R, Buccioni M, Dal Ben D, et al. Synthesis and biological evaluation of 2-alkynyl-N6-methyl-5′-N - methylcarboxamidoadenosine derivatives as potent and highly selective agonists for the human adenosine A3 receptor. JMed Chem. 2009;52:7897–7900. doi: 10.1021/jm900754g. [DOI] [PubMed] [Google Scholar]
- 124.Kozma E, Gizewski ET, Tosh DK, Squarcialupi L, Auchampach JA, Jacobson KA. Characterization by flow cytometry of fluorescent, selective agonist probes of the A(3) adenosine receptor. Biochem Pharmacol. 2013;85:1171–1181. doi: 10.1016/j.bcp.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tosh DK, Paoletta S, Phan K, Gao Z-G, Jacobson KA. Truncated nucleosides as A3 adenosine receptor ligands: Combined 2-arylethynyl and bicyclohexane substitutions. ACSMed Chem Lett. 2012;3:596–601. doi: 10.1021/ml300107e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jeong LS, Lee HW, Jacobson KA, et al. Structure–activity relationships of 2-chloro-N6-substituted-4′-thioadenosine-5′-uronamides as highly potent and selective agonists at the human A3 adenosine receptor. JMed Chem. 2006;49:273–281. doi: 10.1021/jm050595e. [DOI] [PubMed] [Google Scholar]
- 127.Kim GD, Oh J, Jeong LS, Lee SK. Thio-Cl-IB-MECA, a novel A3 adenosine receptor agonist, suppresses angiogenesis by regulating PI3K/AKT/mTOR and ERK signaling in endothelial cells. Biochem Biophys Res Commun. 2013;437:79–86. doi: 10.1016/j.bbrc.2013.06.040. [DOI] [PubMed] [Google Scholar]
- 128.Gao Z-G, Kim S-K, Biadatti T, et al. Structural determinants of A(3) adenosine receptor activation: Nucleoside ligands at the agonist/antagonist boundary. JMed Chem. 2002;45:4471–4484. doi: 10.1021/jm020211+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bennett GJ, Xie YK. A peripheral mononeuropathy in rat that produces disorders of pain sensation like those seen in man. Pain. 1988;33:87–107. doi: 10.1016/0304-3959(88)90209-6. [DOI] [PubMed] [Google Scholar]
- 130.Chen Z, Janes K, Chen C, et al. Controlling murine and rat chronic pain through A3 adenosine receptor activation. FASEB J. 2012;26:1855–1865. doi: 10.1096/fj.11-201541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tosh DK, Paoletta S, Chen Z, et al. Structure-based design, synthesis by click chemistry and in vivo activity of highly selective A3 adenosine receptor agonists. Med Chem Commun. 2015;6:555–563. doi: 10.1039/C4MD00571F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cosyn L, Palaniappan KK, Kim S-K, et al. 2-Triazole-substituted adenosines: A new class of selective A3 adenosine receptor agonists, partial agonists, and antagonists. JMed Chem. 2006;49:7373–7383. doi: 10.1021/jm0608208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fang Z-Z, Tosh DK, Tanaka N, et al. Metabolic mapping of A3 adenosine receptor agonist MRS5980. Biochem Pharmacol. 2015;97:215–223. doi: 10.1016/j.bcp.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tosh DK, Ciancetta A, Warnick E, et al. Purine (N)-methanocarba nucleoside derivatives lacking an exocyclic amine as selective A3 adenosine receptor agonists. JMed Chem. 2016;59:3249–3263. doi: 10.1021/acs.jmedchem.5b01998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jacobson KA, Klutz AM, Tosh DK, Ivanov AA, Preti D, Baraldi PG. Medicinal chemistry of the A3 adenosine receptor: Agonists, antagonists, and receptor engineering. In: Wilson CN, Mustafa SJ, editors. Adenosine Receptors in Health and Disease. Berlin and Heidelberg: Springer-Verlog; 2009. pp. 123–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Volpini R, Costanzi S, Lambertucci C, et al. Introduction of alkynyl chains on C-8 of adenosine led to very selective antagonists of the A(3) adenosine receptor. Bioorg Med Chem Lett. 2001;11:1931–1934. doi: 10.1016/s0960-894x(01)00347-x. [DOI] [PubMed] [Google Scholar]
- 137.Gao Z-G, Joshi BV, Klutz AM, et al. Conversion of A3 adenosine receptor agonists into selective antagonists by modification of the 5′-ribofuran-uronamide moiety. Bioorg Med Chem Lett. 2006;16:596–601. doi: 10.1016/j.bmcl.2005.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jeong LS, Lee HW, Kim HO, et al. Structure–activity relationships of 2-chloro-N6-substituted-4′-thioadenosine-5′-N,N-dialkyluronamides as human A3 adenosine receptor antagonists. Bioorg Med Chem Lett. 2008;18:1612–1616. doi: 10.1016/j.bmcl.2008.01.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jeong LS, Choe SA, Gunaga P, et al. Discovery of a new nucleoside template for human A3 adenosine receptor ligands: D -4′-thioadenosine derivatives without 4′-hydroxymethyl group as highly potent and selective antagonists. JMed Chem. 2007;50:3159–3162. doi: 10.1021/jm070259t. [DOI] [PubMed] [Google Scholar]
- 140.Nayak A, Chandra G, Hwang I, et al. Synthesis and anti-renal fibrosis activity of conformationally locked truncated 2-hexynyl-N(6)-substituted-(N)-methanocarba-nucleosides as A3 adenosine receptor antagonists and partial agonists. J Med Chem. 2014;57:1344–1354. doi: 10.1021/jm4015313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Petrelli R, Torquati I, Kachler S, et al. 5′-C-ethyl-tetrazolyl-N(6)-substituted adenosine and 2-chloro-adenosine derivatives as highly potent dual acting A1 adenosine receptor agonists and A3 adenosine receptor antagonists. J Med Chem. 2015;58:2560–2566. doi: 10.1021/acs.jmedchem.5b00074. [DOI] [PubMed] [Google Scholar]
- 142.Costanzi S, Lambertucci C, Vittori S, Volpini R, Cristalli G. 2- and 8-alkynyladenosines: Conformational studies and docking to human adenosine A3 receptor can explain their different biological behavior. JMol GraphModel. 2003;21:253–262. doi: 10.1016/s1093-3263(02)00161-4. [DOI] [PubMed] [Google Scholar]
- 143.Hou X, Majik MS, Kim K, et al. Structure-activity relationships of truncated C2- or C8-substituted adenosine derivatives as dual acting A2 A and A3 adenosine receptor ligands. JMed Chem. 2012;55:342–356. doi: 10.1021/jm201229j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Borea PA, Varani K, Vincenzi F, et al. The A3 adenosine receptor: History and perspectives. Pharmacol Rev. 2015;67:74–102. doi: 10.1124/pr.113.008540. [DOI] [PubMed] [Google Scholar]
- 145.Kim HO, Ji X, Melman N, Olah ME, Stiles GL, Jacobson KA. Structure-activity relationships of 1,3-dialkylxanthine derivatives at rat A3 adenosine receptors. JMed Chem. 1994;37:3373–3382. doi: 10.1021/jm00046a022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Baraldi PG, Romagnoli R, Saponaro G, Baraldi S, Tabrizi MA, Preti D. A3 Adenosine Receptor Antagonists: History and Future Perspectives. In: Borea PA, editor. A3 Adenosine Receptors from Cell Biology to Pharmacology and Therapeutics. Dordrecht: Springer Netherlands; 2010. pp. 121–147. [Google Scholar]
- 147.Taliani S, Pugliesi I, Bellandi M, La Motta C, Da Settimo F. A3 receptor ligands: Past, present and future trends. Curr Top Med Chem. 2010;10:942–975. doi: 10.2174/156802610791293109. [DOI] [PubMed] [Google Scholar]
- 148.Müller CE, Jacobson KA. Recent developments in adenosine receptor ligands and their potential as novel drugs. Biochim Biophys Acta Biomembr. 2011;1808:1290–1308. doi: 10.1016/j.bbamem.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Baraldi PG, Preti D, Zaid AN, et al. Water-soluble pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidines as human A3 adenosine receptor antagonists. JMed Chem. 2012;55:5380–5390. doi: 10.1021/jm300323t. [DOI] [PubMed] [Google Scholar]
- 150.Baraldi PG, Cacciari B, Romagnoli R, et al. A3 adenosine receptor ligands: History and perspectives. Med Res Rev. 2000;20:103–128. doi: 10.1002/(sici)1098-1128(200003)20:2<103::aid-med1>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 151.Baraldi PG, Tabrizi MA, Fruttarolo F, et al. Recent developments in the field of A3 adenosine receptor antagonists. Drug Dev Res. 2003;58:315–329. doi: 10.1016/s0223-5234(03)00042-4. [DOI] [PubMed] [Google Scholar]
- 152.Baraldi PG, Preti D, Borea PA, Varani K. Medicinal chemistry of A3 adenosine receptor modulators: Pharmacological activities and therapeutic implications. JMed Chem. 2012;55:5676–5703. doi: 10.1021/jm300087j. [DOI] [PubMed] [Google Scholar]
- 153.Baraldi PG, Tabrizi MA, Gessi S, Borea PA. Adenosine receptor antagonists: Translating medicinal chemistry and pharmacology into clinical utility. Chem Rev. 2008;108:238–263. doi: 10.1021/cr0682195. [DOI] [PubMed] [Google Scholar]
- 154.Jacobson KA, Park KS, Jiang JL, et al. Pharmacological characterization of novel A3 adenosine receptor-selective antagonists. Neuropharmacology. 1997;36:1157–1165. doi: 10.1016/s0028-3908(97)00104-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jiang J, van Rhee AM, Chang L, et al. Structure–activity relationships of 4-(phenylethynyl)-6-phenyl-1,4-dihydropyridines as highly selective A3 adenosine receptor antagonists. JMed Chem. 1997;40:2596–2608. doi: 10.1021/jm970091j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jiang J, van Rhee AM, Melman N, Ji X, Jacobson KA. 6-Phenyl-1,4-dihydropyridine derivatives as potent and selective A3 adenosine receptor antagonists. JMed Chem. 1996;39:4667–4675. doi: 10.1021/jm960457c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Van Rhee AM, Jiang JL, Melman N, Olah ME, Stiles GL, Jacobson KA. Interaction of 1,4-dihydropyridine and pyridine derivatives with adenosine receptors: Selectivity for A3 receptors. JMed Chem. 1996;39:2980–2989. doi: 10.1021/jm9600205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Li A-H, Moro S, Melman N, Ji X, Jacobson KA. Structure–activity relationships and molecular modeling of 3,5-diacyl-2,4-dialkylpyridine derivatives as selective A3 adenosine receptor antagonists. JMed Chem. 1998;41:3186–3201. doi: 10.1021/jm980093j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Li AH, Moro S, Forsyth N, Melman N, Ji XD, Jacobson KA. Synthesis, CoMFA analysis, and receptor docking of 3,5-diacyl-2,4-dialkylpyridine derivatives as selective A3 adenosine receptor antagonists. JMed Chem. 1999;42:706–721. doi: 10.1021/jm980550w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Xie R, Li A, Ji X, Melman N, Olah ME, Stiles GL. Selective A 3 adenosine receptor antagonists: Water-soluble 3, 5-diacyl-1, 2, 4-trialkylpyridinium salts and their oxidative generation from dihydropyridine precursors. J Med Chemitry. 1999;42:4232–4238. doi: 10.1021/jm990234x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Haeusler D, Grassinger L, Fuchshuber F, et al. Hide and seek: A comparative autoradiographic in vitro investigation of the adenosine A3 receptor. Eur J Nucl MedMol Imaging. 2015;42:928–939. doi: 10.1007/s00259-014-2985-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cosimelli B, Greco G, Ehlardo M, et al. Derivatives of 4-amino-6-hydroxy-2-mercaptopyrimidine as novel, potent, and selective A3 adenosine receptor antagonists. JMed Chem. 2008;51:1764–1770. doi: 10.1021/jm701159t. [DOI] [PubMed] [Google Scholar]
- 163.Yaziji V, Coelho A, El Maatougui A, et al. Divergent solution-phase synthesis of diarylpyrimidine libraries as selective A3 adenosine receptor antagonists. J Comb Chem. 2009;11:519–522. doi: 10.1021/cc900044k. [DOI] [PubMed] [Google Scholar]
- 164.Yaziji V, Rodríguez D, Gutiérrez-de-Terán H, et al. Pyrimidine derivatives as potent and selective A3 adenosine receptor antagonists. JMed Chem. 2011;54:457–471. doi: 10.1021/jm100843z. [DOI] [PubMed] [Google Scholar]
- 165.Azuaje J, Carbajales C, González-Gómez M, et al. Pyrazin-2(1H)-ones as a novel class of selective A3 adenosine receptor antagonists. FutureMed Chem. 2015;7:1373–1380. doi: 10.4155/fmc.15.69. [DOI] [PubMed] [Google Scholar]
- 166.van Muijlwijk-Koezen JE, Timmerman H, Vollinga RC, Frijtag von Drabbe Künzel J, de Groote M, Visser S, IJzerman AP. Thiazole and thiadiazole analogues as a novel class of adenosine receptor antagonists. JMed Chem. 2001;44:749–762. doi: 10.1021/jm0003945. [DOI] [PubMed] [Google Scholar]
- 167.Jung K-Y, Kim S-K, Gao Z-G, et al. Structure–activity relationships of thiazole and thiadiazole derivatives as potent and selective human adenosine A3 receptor antagonists. Bioorg Med Chem. 2004;12:613–623. doi: 10.1016/j.bmc.2003.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Bhattacharya P, Leonard JT, Roy K. Exploring 3D-QSAR of thiazole and thiadiazole derivatives as potent and selective human adenosine A3 receptor antagonists. JMol Model. 2005;11:516–524. doi: 10.1007/s00894-005-0273-6. [DOI] [PubMed] [Google Scholar]
- 169.VanMuijlwijk-Koezen JE, Timmerman H, van der Goot H, et al. Isoquinoline and quinazoline urea analogues as antagonists for the human-adenosine A3 receptor. JMed Chem. 2000;43:2227–2238. doi: 10.1021/jm000002u. [DOI] [PubMed] [Google Scholar]
- 170.Morizzo E, Capelli F, Lenzi O, et al. Scouting human A3 adenosine receptor antagonist binding mode using a molecular simplification approach: From triazoloquinoxaline to a pyrimidine skeleton as a key study. J Med Chem. 2007;50:6596–6606. doi: 10.1021/jm070852a. [DOI] [PubMed] [Google Scholar]
- 171.Poli D, Catarzi D, Colotta V, et al. The identification of the 2-phenylphthalazin-1(2 H)-one scaffold as a new decorable core skeleton for the design of potent and selective human A3 adenosine receptor antagonists. J Med Chem. 2011;54:2102–2113. doi: 10.1021/jm101328n. [DOI] [PubMed] [Google Scholar]
- 172.Novellino E, Cosimelli B, Ehlardo M, et al. 2-(Benzimidazol-2-yl)quinoxalines: A novel class of selective antagonists at human A 1 and A 3 adenosine receptors designed by 3D database searching. JMed Chem. 2005;48:8253–8260. doi: 10.1021/jm050792d. [DOI] [PubMed] [Google Scholar]
- 173.Poli D, Falsini M, Varano F, Betti M, et al. Imidazo[1,2-a]pyrazin-8-amine core for the design of new adenosine receptor antagonists: Structural exploration to target the A3 and A2A subtypes. Eur J Med Chem. 2017;125:611–628. doi: 10.1016/j.ejmech.2016.09.076. [DOI] [PubMed] [Google Scholar]
- 174.Biagi G, Bianucci AM, Coi A, et al. 2,9-Disubstituted-N6-(arylcarbamoyl)-8-azaadenines as new selective A3 adenosine receptor antagonists: Synthesis, biochemical and molecular modelling studies. Bioorg Med Chem. 2005;13:4679–4693. doi: 10.1016/j.bmc.2005.04.063. [DOI] [PubMed] [Google Scholar]
- 175.Perreira M, Jiang J, Klutz AM, et al. “Reversine” and its 2-substituted adenine derivatives as potent and selective A3 adenosine receptor antagonists. 2005;48:4910–4918. doi: 10.1021/jm050221l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Taliani S, La Motta C, Mugnaini L, et al. Novel N2-substituted pyrazolo[3,4-d]pyrimidine adenosine A3 receptor antagonists: Inhibition of A3-mediated human glioblastoma cell proliferation. JMed Chem. 2010;53:3954–3963. doi: 10.1021/jm901785w. [DOI] [PubMed] [Google Scholar]
- 177.Lenzi O, Colotta V, Catarzi D, et al. 2-Phenylpyrazolo[4,3-d]pyrimidin-7-one as a new scaffold to obtain potent and selective human A3 adenosine receptor antagonists: New insights into the receptor–antagonist recognition. JMed Chem. 2009;52:7640–7652. doi: 10.1021/jm900718w. [DOI] [PubMed] [Google Scholar]
- 178.Squarcialupi L, Colotta V, Catarzi D, et al. 2-Arylpyrazolo[4,3-d]pyrimidin-7-amino derivatives as new potent and selective human A3 Adenosine Receptor Antagonists. Molecular Modeling Studies and Pharmacological Evaluation. J Med Chem. 2013;56:2256–2260. doi: 10.1021/jm400068e. [DOI] [PubMed] [Google Scholar]
- 179.Squarcialupi L, Catarzi D, Varano F, et al. Structural refinement of pyrazolo[4,3-d]pyrimidine derivatives to obtain highly potent and selective antagonists for the human A3 adenosine receptor. Eur J Med Chem. 2016;108:117–133. doi: 10.1016/j.ejmech.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 180.Gatta F, Del Giudice M, Borioni A, Borea PA, Dionisotti S, Ongini E. Synthesis of imidazo[1,2-c]pyrazolo[4,3-e]pyrimidines, pyrazolo[4,3-e]1,2,4-triazolo[1,5-c]pyrimidines and 1,2,4-triazolo[5,1-i]purines: New potent adenosine A2 receptor antagonists. Eur J Med Chem. 1993;28:569–576. [Google Scholar]
- 181.Kim Y, Ji X, Jacobson KA. Derivatives of the triazoloquinazoline adenosine antagonist (CGS15943) are selective for the human A3 receptor subtype. JMed Chem. 1996;2623:4142–4148. doi: 10.1021/jm960482i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kim YC, De Zwart M, Chang L, et al. Derivatives of the triazoloquinazoline adenosine antagonist (CGS 15943) having high potency at the human A(2B) and A3 receptor subtypes. JMed Chem. 1998;41:2835–2845. doi: 10.1021/jm980094b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Okamura T, Kurogi Y, Hashimoto K, et al. Structure–activity relationships of adenosine A3 receptor ligands: New potential therapy for the treatment of glaucoma. Bioorg Med Chem Lett. 2004;14:3775–3779. doi: 10.1016/j.bmcl.2004.04.099. [DOI] [PubMed] [Google Scholar]
- 184.Burbiel JC, Ghattas W, Küppers P, et al. 2-Amino[1,2,4]triazolo[1,5-c]quinazolines and derived novel heterocycles: Syntheses and structure-activity relationships of potent adenosine receptor antagonists. ChemMedChem. 2016;1523:1–16. doi: 10.1002/cmdc.201600255. [DOI] [PubMed] [Google Scholar]
- 185.Colotta V, Catarzi D, Varano F, et al. Synthesis and structure–activity relationships of a new set of 2-arylpyrazolo[3,4-c]quinoline derivatives as adenosine receptor antagonists. JMed Chem. 2000;43:3118–3124. doi: 10.1021/jm000936i. [DOI] [PubMed] [Google Scholar]
- 186.Baraldi PG, Tabrizi MA, Preti D, et al. New 2-Arylpyrazolo[4,3-c]quinoline derivatives as potent and selective human A3 adenosine receptor antagonists. JMed Chem. 2005;48:5001–5008. doi: 10.1021/jm050125k. [DOI] [PubMed] [Google Scholar]
- 187.Colotta V, Catarzi D, Varano F, et al. 1,2,4-Triazolo[4,3-a]quinoxalin-1-one: A versatile tool for the synthesis of potent and selective adenosine receptor antagonists. JMed Chem. 2000;43:1158–1164. doi: 10.1021/jm991096e. [DOI] [PubMed] [Google Scholar]
- 188.Colotta V, Catarzi D, Varano F, et al. 1,2,4-Triazolo[4,3-a]quinoxalin-1-one moiety as an attractive scaffold to develop new potent and selective human A3 adenosine receptor antagonists: Synthesis, pharmacological, and ligand-receptor modeling studies. JMed Chem. 2004;47:3580–3590. doi: 10.1021/jm031136l. [DOI] [PubMed] [Google Scholar]
- 189.Lenzi O, Colotta V, Catarzi D, et al. 4-Amido-2-aryl-1,2,4-triazolo[4,3-a]quinoxalin-1-ones as new potent and selective human A3 adenosine receptor antagonists. Synthesis, pharmacological evaluation, and ligand-receptor modeling studies. JMed Chem. 2006;49:3916–3925. doi: 10.1021/jm060373w. [DOI] [PubMed] [Google Scholar]
- 190.Catarzi D, Colotta V, Varano F, et al. 1,2,4-Triazolo[1,5-a]quinoxaline as a versatile tool for the design of selective human A3 adenosine receptor antagonists: Synthesis, biological evaluation, and molecular modeling studies of 2-(Hetero)aryland 2-carboxy-substitued derivatives. JMed Chem. 2005;48:7932–7945. doi: 10.1021/jm0504149. [DOI] [PubMed] [Google Scholar]
- 191.Catarzi D, Colotta V, Varano F, et al. 2-Aryl-8-chloro-1,2,4-triazolo[1,5-a]quinoxalin-4-amines as highly potent A1 and A3 adenosine receptor antagonists. BioorgMed Chem. 2005;13:705–715. doi: 10.1016/j.bmc.2004.10.050. [DOI] [PubMed] [Google Scholar]
- 192.Da Settimo F, Primofiore G, Taliani S, et al. 5-Amino-2-phenyl[1,2,3]triazolo[1,2-a][1,2,4]benzotriazin-1-one: A versatile scaffold to obtain potent and selective A3 adenosine receptor antagonists. JMed Chem. 2007;50:5676–5684. doi: 10.1021/jm0708376. [DOI] [PubMed] [Google Scholar]
- 193.Baraldi PG, Cacciari B, Romagnoli R, et al. Pyrazolo[4,3-e]-1,2,4-triazolo[1,5-c]-pyrimidine derivatives as highly potent and selective human A3 adenosine receptor antagonists. JMed Chem. 1999;42:1–6. doi: 10.1021/jm991114s. [DOI] [PubMed] [Google Scholar]
- 194.Baraldi P, Tabrizi M, Romagnoli R, et al. Pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidine template: Organic and medicinal chemistry approach. Curr Org Chem. 2006;10:259–275. [Google Scholar]
- 195.Baraldi PG, Preti D, Zaid AN, et al. New 2-heterocyclyl-imidazo[2,1-i]purin-5-one derivatives as potent and selective human A3 adenosine receptor antagonists. JMed Chem. 2011;54:5205–5220. doi: 10.1021/jm2004738. [DOI] [PubMed] [Google Scholar]
- 196.Baraldi PG, Baraldi S, Saponaro G, et al. One-pot reaction to obtain N,N′-disubstituted guanidines of pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidine scaffold as human A3 adenosine receptor antagonists. J Med Chem. 2015;58:5355–5360. doi: 10.1021/acs.jmedchem.5b00551. [DOI] [PubMed] [Google Scholar]
- 197.Baraldi PG, Cacciari B, Romagnoli R, et al. Pyrazolo[4,3-e]1,2,4-triazolo[1,5-c]pyrimidine derivatives as highly potent and selective human A3 adenosine receptor antagonists: Influence of the chain at the N8 pyrazole nitrogen. JMed Chem. 2000;43:4768–4780. doi: 10.1021/jm001047y. [DOI] [PubMed] [Google Scholar]
- 198.Baraldi PG, Cacciari B, Moro S, et al. Synthesis, biological activity, and molecular modeling investigation of new pyrazolo[4,3-e]-1,2,4-triazolo[1,5-c]pyrimidine derivatives as human A(3) adenosine receptor antagonists. J Med Chem. 2002;45:770–780. doi: 10.1021/jm0109614. [DOI] [PubMed] [Google Scholar]
- 199.Baraldi PG, Preti D, Tabrizi MA, et al. New Pyrrolo[2,1-f]purine-2,4-dione and Imidazo[2,1-f]purine-2,4-dione derivatives as potent and selective human A3 adenosine receptor antagonists. JMed Chem. 2005;48:4697–4701. doi: 10.1021/jm058008c. [DOI] [PubMed] [Google Scholar]
- 200.Baraldi PG, Preti D, Tabrizi MA, et al. Structure–activity relationship studies of a new series of imidazo[2,1-f]purinones as potent and selective A3 adenosine receptor antagonists. BioorgMed Chem. 2008;16:10281–10294. doi: 10.1016/j.bmc.2008.10.049. [DOI] [PubMed] [Google Scholar]
- 201.Baraldi PG, Cacciari B, Romagnoli R, et al. Synthesis and preliminary biological evaluation of [3 H]-MRE 3008-F20: The first high affinity radioligand antagonist for the human A3 adenosine receptors. Bioorg Med Chem Lett. 2000;10:209–211. doi: 10.1016/s0960-894x(99)00674-5. [DOI] [PubMed] [Google Scholar]
- 202.Varani K, Merighi S, Gessi S, et al. [(3)H]MRE 3008F20: A novel antagonist radioligand for the pharmacological and biochemical characterization of human A(3) adenosine receptors. Mol Pharmacol. 2000;57:968–975. [PubMed] [Google Scholar]
- 203.Maconi A, Moro S, Pastorin G, et al. Synthesis, biological properties, and molecular modeling investigation of the first potent, selective, and water-soluble human A3 adenosine receptor antagonist. JMed Chem. 2002;45:3579–3582. doi: 10.1021/jm020974x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Baraldi PG, Cacciari B, Moro S, et al. Fluorosulfonyl- and bis-(β-chloroethyl)amino-phenylamino functionalized pyrazolo[4,3-e]1,2,4-triazolo[1,5-c]pyrimidine derivatives: Irreversible antagonists at the human a3 adenosine receptor and molecular modeling studies. JMed Chem. 2001;44:2735–2742. doi: 10.1021/jm010818a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Okamura T, Kurogi Y, Nishikawa H, Hashimoto K, Fujiwara H, Nagao Y. 1,2,4-Triazolo[5,1-i]purine derivatives as highly potent and selective human adenosine A3 receptor ligands. JMed Chem. 2002;45:3703–3708. doi: 10.1021/jm010570p. [DOI] [PubMed] [Google Scholar]
- 206.Drabczyńska A, Schumacher B, Müller CE, et al. Impact of the aryl substituent kind and distance from pyrimido[2,1-f]purindiones on the adenosine receptor selectivity and antagonistic properties. Eur J Med Chem. 2003;38:397–402. doi: 10.1016/s0223-5234(03)00051-5. [DOI] [PubMed] [Google Scholar]
- 207.Müller CE, Thorand M, Qurishi R, et al. Imidazo[2,1-i]purin-5-ones and related tricyclic water-soluble purine derivatives: Potent A2A- and A3-adenosine receptor antagonists. JMed Chem. 2002;45:3440–3450. doi: 10.1021/jm011093d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Müller CE, Diekmann M, Thorand M, Ozola V. [3H]8-ethyl-4-methyl-2-phenyl-(8R)-4,5,7,8-tetrahydro-1H-imidazo[2,1-i]-purin-5-one ([3H]PSB-11), a novel high-affinity antagonist radioligand for human A3 adenosine receptors. Bioorg Med Chem Lett. 2002;12:501–503. doi: 10.1016/s0960-894x(01)00785-5. [DOI] [PubMed] [Google Scholar]
- 209.Saki M, Tsumuki H, Nonaka H, Shimada J, Ichimura M. KF26777 (2-(4-bromophenyl)-7,8-dihydro-4-propyl-1Himidazo[ 2,1-i]purin-5(4H)-one dihydrochloride), a new potent and selective adenosine A3 receptor antagonist. Eur J Pharmacol. 2002;444:133–141. doi: 10.1016/s0014-2999(02)01662-x. [DOI] [PubMed] [Google Scholar]
- 210.Priego EM, von Frijtag Drabbe Kuenzel J, IJzerman AP, Camarasa MJ, Pérez-Pérez MJ. Pyrido[2,1-f]purine-2,4-dione derivatives as a novel class of highly potent human A3 adenosine receptor antagonists. JMed Chem. 2002;45:3337–3344. doi: 10.1021/jm0208469. [DOI] [PubMed] [Google Scholar]
- 211.Priego E-M, Pérez-Pérez M-J, von Frijtag Drabbe Kuenzel JK, et al. Selective human adenosine A3 antagonists based on pyrido[2,1-f]purine-2,4-diones: Novel features of hA3 antagonist binding. ChemMedChem. 2008;3:111–119. doi: 10.1002/cmdc.200700173. [DOI] [PubMed] [Google Scholar]
- 212.Jacobson KA, Gao Z-G, Göblyös A, IJzerman AP. Allosteric modulation of purine and pyrimidine receptors. In: Jacobson KA, Indlen J, editors. Advances in pharmacology. San Diego: Elsevier; 2011. pp. 187–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Gao Z-G, Kim SG, Soltysiak KA, Melman N, IJzerman AP, Jacobson KA. Selective allosteric enhancement of agonist binding and function at human A3 adenosine receptors by a series of imidazoquinoline derivatives. Mol Pharmacol. 2002;62:81–89. doi: 10.1124/mol.62.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Göblyös A, Gao Z-G, Brussee J, et al. Structure-activity relationships of new 1H-imidazo[4,5-c]quinolin-4-amine derivatives as allosteric enhancers of the A3 adenosine receptor. JMed Chem. 2006;49:3354–3361. doi: 10.1021/jm060086s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Kim Y, de Castro S, Gao Z-G, Ijzerman AP, Jacobson KA. Novel 2- and 4-substituted 1H-imidazo[4,5-c]quinolin-4-amine derivatives as allosteric modulators of the A3 adenosine receptor. JMed Chem. 2009;52:2098–2108. doi: 10.1021/jm801659w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Heitman LH, Göblyös A, Zweemer AM, et al. Aseries of 2,4-disubstituted quinolines as a new class of allosteric enhancers of the adenosine A3 receptor. JMed Chem. 2009;52:926–931. doi: 10.1021/jm8014052. [DOI] [PubMed] [Google Scholar]
- 217.Gao Z-G, Van Muijlwijk-Koezen JE, Chen A, Müller CE, Ijzerman AP, Jacobson KA. Allosteric modulation of A(3) adenosine receptors by a series of 3-(2-pyridinyl)isoquinoline derivatives. Mol Pharmacol. 2001;60:1057–1063. [PMC free article] [PubMed] [Google Scholar]
- 218.Gao Z-G, Kim S-K, Gross AS, Chen A, Blaustein JB, Jacobson KA. Identification of essential residues involved in the allosteric modulation of the human A(3) adenosine receptor. Mol Pharmacol. 2003;63:1021–1031. doi: 10.1124/mol.63.5.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Gao Z-G, Ye K, Göblyös A, IJzerman AP, Jacobson KA. Flexible modulation of agonist efficacy at the human A3 adenosine receptor by the imidazoquinoline allosteric enhancer LUF6000. BMC Pharmacol. 2008;8:20. doi: 10.1186/1471-2210-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Cohen S, Barer F, Bar-Yehuda S, IJzerman AP, Jacobson KA, Fishman P. A3 adenosine receptor allosteric modulator induces an anti-inflammatory effect: In vivo studies and molecular mechanism of action. Mediators Inflamm. 2014;2014:1–8. doi: 10.1155/2014/708746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Gao Z-G, Chen A, Barak D, Kim S-K, Müller CE, Jacobson KA. Identification by site-directed mutagenesis of residues involved in ligand recognition and activation of the human A3 adenosine receptor. J Biol Chem. 2002;277:19056–19063. doi: 10.1074/jbc.M110960200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Xu F, Wu H, Katritch V, et al. Structure of an agonist-bound human A2A adenosine receptor. Science. 2011;332:322–327. doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Lebon G, Warne T, Edwards PC, et al. Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature. 2011;474:521–525. doi: 10.1038/nature10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Lebon G, Edwards PC, Leslie AGW, Tate CG. Molecular determinants of CGS21680 binding to the human adenosine A2A receptor. Mol Pharmacol. 2015;87:907–915. doi: 10.1124/mol.114.097360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Ballesteros JA, Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. In: Sealfon SC, editor. Methods in Neurosciences. Vol. 25. Ballesteros JA: Weinstein H publisher Academic Press, Inc; 1995. pp. 366–428. [Google Scholar]
- 226.Baltos J-A, Paoletta S, Nguyen ATN, et al. Structure-activity analysis of biased agonism at the human adenosine A3 receptor. Mol Pharmacol. 2016;90:12–22. doi: 10.1124/mol.116.103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Hallmen C, Wiese M. Molecular dynamics simulation of the human adenosine A3 receptor: Agonist induced conformational changes of Trp243. J Comput Aided Mol Des. 2006;20:673–684. doi: 10.1007/s10822-006-9088-5. [DOI] [PubMed] [Google Scholar]
- 228.Deganutti G, Cuzzolin A, Ciancetta A, Moro S. Understanding allosteric interactions in G protein-coupled receptors using supervised molecular dynamics: A prototype study analysing the human A3 adenosine receptor positive allosteric modulator LUF6000. Bioorg Med Chem. 2015;23:4065–4071. doi: 10.1016/j.bmc.2015.03.039. [DOI] [PubMed] [Google Scholar]
- 229.May LT, Bridge LJ, Stoddart LA, Briddon SJ, Hill SJ. Allosteric interactions across native adenosine-A3 receptor homodimers: Quantification using single-cell ligand-binding kinetics. FASEB J. 2011;25:3465–3476. doi: 10.1096/fj.11-186296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Borea PA, Varani K, Vincenzi F, et al. The A3 adenosine receptor: History and perspectives. Pharmacol Rev. 2014;67:74–102. doi: 10.1124/pr.113.008540. [DOI] [PubMed] [Google Scholar]
- 231.Forte G, Sorrentino R, Montinaro A, Pinto A, Morello S. Cl-IB-MECA enhances TNF-α release in peritoneal macrophages stimulated with LPS. Cytokine. 2011;54:161–166. doi: 10.1016/j.cyto.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 232.Merighi S, Simioni C, Lane R, Ijzerman AP. Regulation of second messenger systems and intracellular pathways. In: Borea PA, editor. A3 Adenosine Receptors from Cell Biology to Pharmacology and Therapeutics. Dordrecht, The Netherlands: Springer; 2010. pp. 61–73. [Google Scholar]
- 233.Hammarberg C, Schulte G, Fredholm BB. Evidence for functional adenosine A3 receptors in microglia cells. J Neurochem. 2003;86:1051–1054. doi: 10.1046/j.1471-4159.2003.01919.x. [DOI] [PubMed] [Google Scholar]
- 234.Soares AS, Costa VM, Diniz C, Fresco P. The combination of Cl-IB-MECA with paclitaxel: A new anti-metastatic therapeutic strategy formelanoma. Cancer Chemother Pharmacol. 2014;74:847–860. doi: 10.1007/s00280-014-2557-y. [DOI] [PubMed] [Google Scholar]
- 235.Merighi S, Benini A, Mirandola P, et al. Adenosine modulates vascular endothelial growth factor expression via hypoxia-inducible factor-1 in human glioblastoma cells. Biochem Pharmacol. 2006;72:19–31. doi: 10.1016/j.bcp.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 236.Merighi S, Benini A, Mirandola P, et al. Hypoxia inhibits paclitaxel-induced apoptosis through adenosine-mediated phosphorylation of bad in glioblastoma cells. Mol Pharmacol. 2007;72:162–172. doi: 10.1124/mol.106.031849. [DOI] [PubMed] [Google Scholar]
- 237.Merighi S, Benini A, Mirandola P, et al. Caffeine inhibits adenosine-induced accumulation of hypoxia-inducible factor-1alpha, vascular endothelial growth factor, and interleukin-8 expression in hypoxic human colon cancer cells. Mol Pharmacol. 2007;72:395–406. doi: 10.1124/mol.106.032920. [DOI] [PubMed] [Google Scholar]
- 238.Neary JT, McCarthy M, Kang Y, Zuniga S. Mitogenic signaling from P1 and P2 purinergic receptors to mitogen-activated protein kinase in human fetal astrocyte cultures. Neurosci Lett. 1998;242:159–162. doi: 10.1016/s0304-3940(98)00067-6. [DOI] [PubMed] [Google Scholar]
- 239.Schulte G, Fredholm BB. Human adenosine A(1), A(2A), A(2B), and A(3) receptors expressed in Chinese hamster ovary cells all mediate the phosphorylation of extracellular-regulated kinase 1/2. Mol Pharmacol. 2000;58:477–482. [PubMed] [Google Scholar]
- 240.Schulte G, Fredholm BB. Signaling pathway from the human adenosine A(3) receptor expressed in Chinese hamster ovary cells to the extracellular signal-regulated kinase 1/2. Mol Pharmacol. 2002;62:1137–1146. doi: 10.1124/mol.62.5.1137. [DOI] [PubMed] [Google Scholar]
- 241.Kim TH, Kim YK, Woo JS. The Adenosine A3 Receptor agonist Cl-IB-MECA induces cell death through Ca2+/ROS-dependent down regulation of ERK and Akt in A172 human glioma cells. Neurochem Res. 2012;37:2667–2677. doi: 10.1007/s11064-012-0855-5. [DOI] [PubMed] [Google Scholar]
- 242.Martin L, Pingle SC, Hallam DM, Rybak LP, Ramkumar V. Activation of the adenosine A3 receptor in RAW 264. 7 cells inhibits lipopolysaccharide-stimulated tumor necrosis factor-alpha release by reducing calcium-dependent activation of nuclear factor-kappaB and extracellular signal-regulated kinase 1/2. J Pharmacol Exp Ther. 2006;316:71–78. doi: 10.1124/jpet.105.091868. [DOI] [PubMed] [Google Scholar]
- 243.Merighi S, Benini A, Mirandola P, et al. A3 adenosine receptors modulate hypoxia-inducible factor-1a expression in human A375 melanoma cells. Neoplasia. 2005;7:894–903. doi: 10.1593/neo.05334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Ohsawa K, Sanagi T, Nakamura Y, Suzuki E, Inoue K, Kohsaka S. Adenosine A3 receptor is involved in ADP-induced microglial process extension and migration. J Neurochem. 2012;121:217–227. doi: 10.1111/j.1471-4159.2012.07693.x. [DOI] [PubMed] [Google Scholar]
- 245.Torres A, Vargas Y, Uribe D, et al. Adenosine A3 receptor elicits chemoresistance mediated by multiple resistance-associated protein-1 in human glioblastoma stem-like cells. Oncotarget. 2016;7:67373–67386. doi: 10.18632/oncotarget.12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Merighi S, Benini A, Mirandola P, et al. A3 adenosine receptor activation inhibits cell proliferation via phosphatidylinositol 3-kinase/Akt-dependent inhibition of the extracellular signal-regulated kinase 1/2 phosphorylation in a375 human melanoma cells. J Biol Chem. 2005;280:19516–19526. doi: 10.1074/jbc.M413772200. [DOI] [PubMed] [Google Scholar]
- 247.Gao Z, Li BS, Day YJ, Linden J. A3 adenosine receptor activation triggers phosphorylation of protein kinase B and protects rat basophilic leukemia 2H3 mast cells from apoptosis. Mol Pharmacol. 2001;59:76–82. doi: 10.1124/mol.59.1.76. [DOI] [PubMed] [Google Scholar]
- 248.La Sala A, Gadina M, Kelsall BL. G(i)-protein-dependent inhibition of IL-12 production is mediated by activation of the phosphatidylinositol 3-kinase-protein 3 kinase B/Akt pathway and JNK. J Immunol. 2005;175:2994–2999. doi: 10.4049/jimmunol.175.5.2994. [DOI] [PubMed] [Google Scholar]
- 249.Lee JY, Jhun BS, Oh YT, et al. Activation of adenosine A3 receptor suppresses lipopolysaccharide-induced TNF-α production through inhibition of PI 3-kinase/Akt and NF-κB activation in murine BV2microglial cells. Neurosci Lett. 2006;396:1–6. doi: 10.1016/j.neulet.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 250.Lee H-S, Chung H-J, Lee HW, Jeong LS, Lee SK. Suppression of inflammation response by a novel A3 adenosine receptor agonist thio-Cl-IB-MECA through inhibition of Akt and NF-κB signaling. Immunobiology. 2011;216:997–1003. doi: 10.1016/j.imbio.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 251.Fishman P, Bar-Yehuda S, Madi L, et al. The PI3K-NF-kappaB signal transduction pathway is involved in mediating the anti-inflammatory effect of IB-MECA in adjuvant-induced arthritis. Arthritis Res Ther. 2006;8:R33. doi: 10.1186/ar1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Haskó G, Németh ZH, Vizi ES, Salzman AL, Szabó C. An agonist of adenosine A3 receptors decreases interleukin-12 and interferon-gamma production and prevents lethality in endotoxemic mice. Eur J Pharmacol. 1998;358:261–268. doi: 10.1016/s0014-2999(98)00619-0. [DOI] [PubMed] [Google Scholar]
- 253.Madi L, Cohen S, Ochayin A, Bar-Yehuda S, Barer F, Fishman P. Overexpression of A3 adenosine receptor in peripheral blood mononuclear cells in rheumatoid arthritis: Involvement of nuclear factor-kappaB in mediating receptor level. J Rheumatol. 2007;34:20–26. [PubMed] [Google Scholar]
- 254.Gessi S, Merighi S, Stefanelli A, Fazzi D, Varani K, Borea PA. A1 and A3 adenosine receptors inhibit LPS-induced hypoxia-inducible factor-1 accumulation in murine astrocytes. Pharmacol Res. 2013;76:157–170. doi: 10.1016/j.phrs.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 255.Fishman P, Bar-Yehuda S, Ohana G, et al. An agonist to the A3 adenosine receptor inhibits colon carcinoma growth in mice via modulation of GSK-3 beta and NF-kappa B. Oncogene. 2004;23:2465–2471. doi: 10.1038/sj.onc.1207355. [DOI] [PubMed] [Google Scholar]
- 256.Fishman P, Bar-Yehuda S, Madi L, Cohn I. A3 adenosine receptor as a target for cancer therapy. Anticancer Drugs. 2002;13:437–443. doi: 10.1097/00001813-200206000-00001. [DOI] [PubMed] [Google Scholar]
- 257.Ochaion A, Bar-Yehuda S, Cohen S, et al. The A3 adenosine receptor agonist CF502 inhibits the PI3K, PKB/Akt and NF-kappaB signaling pathway in synoviocytes from rheumatoid arthritis patients and in adjuvant-induced arthritis rats. Biochem Pharmacol. 2008;76:482–494. doi: 10.1016/j.bcp.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Gessi S, Merighi S, Fazzi D, Stefanelli A, Varani K, Borea PA. Adenosine receptor targeting in health and disease. Expert Opin Investig Drugs. 2011;20:1591–1609. doi: 10.1517/13543784.2011.627853. [DOI] [PubMed] [Google Scholar]
- 259.Antonioli L, Fornai M, Colucci R, et al. Control of enteric neuromuscular functions by purinergic A(3) receptors in normal rat distal colon and experimental bowel inflammation. Br J Pharmacol. 2010;161:856–871. doi: 10.1111/j.1476-5381.2010.00917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Haskó G, Cronstein B. Regulation of inflammation by adenosine. Front Immunol. 2013;4:85. doi: 10.3389/fimmu.2013.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Butler M, Sanmugalingam D, Burton VJ, et al. Impairment of adenosine A3 receptor activity disrupts neutrophil migratory capacity and impacts innate immune function in vivo. Eur J Immunol. 2012;42:3358–3368. doi: 10.1002/eji.201242655. [DOI] [PubMed] [Google Scholar]
- 262.Corriden R, Chen Y, Inoue Y, et al. Ecto-nucleoside triphosphate diphosphohydrolase 1 (E-NTPDase1/CD39) regulates neutrophil chemotaxis by hydrolyzing released ATP to adenosine. J Biol Chem. 2008;283:28480–28486. doi: 10.1074/jbc.M800039200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Ledderose C, Hefti MM, Chen Y, Bao Y, Seier T, Li L, Woehrle T, Zhang J, Junger WG. Adenosine arrests breast cancer cell motility by A3 receptor stimulation. Purinergic Signal. 2016;12:673–685. doi: 10.1007/s11302-016-9531-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Inoue Y, Chen Y, Hirsh MI, Yip L, Junger WG. A3 and P2Y2 receptors control the recruitment of neutrophils to the lungs in a mouse model of sepsis. Shock. 2008;30:173–177. doi: 10.1097/shk.0b013e318160dad4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Sajjadi FG, Takabayashi K, Foster AC, Domingo RC, Firestein GS. Inhibition of TNF-alpha expression by adenosine: Role of A3 adenosine receptors. J Immunol. 1996;156:3435–3442. [PubMed] [Google Scholar]
- 266.Sipka S, Kovács I, Szántó S, et al. Adenosine inhibits the release of interleukin-1β in activated human peripheral mononuclear cells. Cytokine. 2005;31:258–263. doi: 10.1016/j.cyto.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 267.Barnholt KE, Kota RS, Aung HH, Rutledge JC. Adenosine blocks IFN-gamma-induced phosphorylation of STAT1 on serine 727 to reduce macrophage activation. J Immunol. 2009;183:6767–6777. doi: 10.4049/jimmunol.0900331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Takahashi HK, Iwagaki H, Hamano R, et al. Effects of adenosine on adhesion molecule expression and cytokine production in human PBMC depend on the receptor subtype activated. Br J Pharmacol. 2007;150:816–822. doi: 10.1038/sj.bjp.0707126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Gessi S, Sacchetto V, Fogli E, Fozard J. A3 adenosine receptor regulation of cells of the immune system and modulation of inflammation. In: Borea PA, editor. A3 Adenosine Receptors from Cell Biology to Pharmacology and Therapeutics. Dordrecht, The Netherlands: Springer Netherlands; 2010. pp. 235–256. [Google Scholar]
- 270.Koscsó B, Csóka B, Pacher P, Haskó G. Investigational A3 adenosine receptor targeting agents. ExpertOpin Investig Drugs. 2011;20:757–768. doi: 10.1517/13543784.2011.573785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Kumar V. Adenosine as an endogenous immunoregulator in cancer pathogenesis: Where to go? Purinergic Signal. 2013;9:145–165. doi: 10.1007/s11302-012-9349-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Hoskin DW, Butler JJ, Drapeau D, Haeryfar SMM, Blay J. Adenosine acts through an A3 receptor to prevent the induction of murine anti-CD3-activated killer T cells. Int J cancer. 2002;99:386–395. doi: 10.1002/ijc.10325. [DOI] [PubMed] [Google Scholar]
- 273.Hoskin DW, Mader JS, Furlong SJ, Conrad DM, Blay J. Inhibition of T cell and natural killer cell function by adenosine and its contribution to immune evasion by tumor cells (review) Int J Oncol. 2008;32:527–535. [PubMed] [Google Scholar]
- 274.Harish A, Hohana G, Fishman P, Arnon O, Bar-Yehuda S. A3 adenosine receptor agonist potentiates natural killer cell activity. Int J Oncol. 2003;23:1245–1249. [PubMed] [Google Scholar]
- 275.Montinaro A, Forte G, Sorrentino R, et al. Adoptive immunotherapy with Cl-IB-MECA-treated CD8+ T cells reduces melanoma growth in mice. PLoS One. 2012;7:e45401. doi: 10.1371/journal.pone.0045401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Ochaion A, Bar-Yehuda S, Cohen S, et al. The anti-inflammatory target A(3) adenosine receptor is over-expressed in rheumatoid arthritis, psoriasis and Crohn’s disease. Cell Immunol. 2009;258:115–122. doi: 10.1016/j.cellimm.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 277.Varani K, Padovan M, Vincenzi F, et al. A2A and A3 adenosine receptor expression in rheumatoid arthritis: Upregulation, inverse correlation with disease activity score and suppression of inflammatory cytokine and metalloproteinase release. Arthritis Res Ther. 2011;13:R197. doi: 10.1186/ar3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Rath-Wolfson L, Bar-Yehuda S, Madi L, et al. IB-MECA, an A3 adenosine receptor agonist prevents bone resorption in rats with adjuvant induced arthritis. Clin Exp Rheumatol. 2006;24:400–406. [PubMed] [Google Scholar]
- 279.Bar-Yehuda S, Rath-Wolfson L, Del Valle L, et al. Induction of an antiinflammatory effect and prevention of cartilage damage in rat knee osteoarthritis by CF101 treatment. Arthritis Rheum. 2009;60:3061–3071. doi: 10.1002/art.24817. [DOI] [PubMed] [Google Scholar]
- 280.Ohana G, Cohen S, Rath-Wolfson L, Fishman P. A3 adenosine receptor agonist, CF102, protects against hepatic ischemia/reperfusion injury following partial hepatectomy. MolMed Rep. 2016;14:4335–4341. doi: 10.3892/mmr.2016.5746. [DOI] [PubMed] [Google Scholar]
- 281.Zhong H, Shlykov SG, Molina JG, et al. Activation of murine lung mast cells by the adenosine A3 receptor. J Immunol. 2003;171:338–345. doi: 10.4049/jimmunol.171.1.338. [DOI] [PubMed] [Google Scholar]
- 282.Smith SR, Denhardt G, Terminelli C. A role for histamine in cytokine modulation by the adenosine A(3) receptor agonist, 2-Cl-IB-MECA. Eur J Pharmacol. 2002;457:57–69. doi: 10.1016/s0014-2999(02)02645-6. [DOI] [PubMed] [Google Scholar]
- 283.Reeves JJ, Jones CA, Sheehan MJ, Vardey CJ, Whelan CJ. Adenosine A3 receptors promote degranulation of rat mast cells both in vitro and in vivo. Inflamm Res. 1997;46:180–184. doi: 10.1007/s000110050169. [DOI] [PubMed] [Google Scholar]
- 284.Salvatore CA, Tilley SL, Latour AM, Fletcher DS, Koller BH, Jacobson MA. Disruption of the A(3) adenosine receptor gene in mice and its effect on stimulated inflammatory cells. J Biol Chem. 2000;275:4429–4434. doi: 10.1074/jbc.275.6.4429. [DOI] [PubMed] [Google Scholar]
- 285.Pardo A, Gibson K, Cisneros J, et al. Up-regulation and profibrotic role of osteopontin in human idiopathic pulmonary fibrosis. PLoS Med. 2005;2:e251. doi: 10.1371/journal.pmed.0020251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Zhou Y, Lee J-Y, Lee C-M, et al. Amphiregulin, an epidermal growth factor receptor ligand, plays an essential role in the pathogenesis of transforming growth factor-β-induced pulmonary fibrosis. J Biol Chem. 2012;287:41991–42000. doi: 10.1074/jbc.M112.356824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Rudich N, Dekel O, Sagi-Eisenberg R. Down-regulation of the A3 adenosine receptor in human mast cells upregulates mediators of angiogenesis and remodeling. Mol Immunol. 2015;65:25–33. doi: 10.1016/j.molimm.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 288.Carlin JL, Jain S, Gizewski E, et al. Hypothermia in mouse is caused by adenosine A1 and A3 receptor agonists and AMP via three distinct mechanisms. Neuropharmacology. 2017;114:101–113. doi: 10.1016/j.neuropharm.2016.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Melillo G. HIF-1: A target for cancer, ischemia and inflammation—Too good to be true? Cell Cycle. 2004;3:154–155. [PubMed] [Google Scholar]
- 290.Koszałka P, Gołuńska M, Urban A, et al. Specific Activation of A3, A2A and A1 adenosine receptors in CD73-knockout mice affects B16F10 melanoma growth, neovascularization, angiogenesis and macrophage infiltration. PLoS One. 2016;11:e0151420. doi: 10.1371/journal.pone.0151420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Velot E, Haas B, Léonard F, et al. Activation of the adenosine-A3 receptor stimulates matrix metalloproteinase-9 secretion by macrophages. Cardiovasc Res. 2008;80:246–254. doi: 10.1093/cvr/cvn201. [DOI] [PubMed] [Google Scholar]
- 292.Bar-Yehuda S, Barer F, Volfsson L, Fishman P. Resistance of muscle to tumor metastases: A role for a3 adenosine receptor agonists. Neoplasia. 2001;3:125–131. doi: 10.1038/sj.neo.7900138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Fishman P, Bar-Yehuda S, Vagman L. Adenosine and other low molecular weight factors released by muscle cells inhibit tumor cell growth. Cancer Res. 1998;58:3181–3187. [PubMed] [Google Scholar]
- 294.Fishman P, Bar-Yehuda S, Ohana G, Pathak S, Wasserman L, Barer F, Multani AS. Adenosine acts as an inhibitor of lymphoma cell growth: A major role for the A3 adenosine receptor. Eur J Cancer. 2000;36:1452–1458. doi: 10.1016/s0959-8049(00)00130-1. [DOI] [PubMed] [Google Scholar]
- 295.Fishman P, Bar-Yehuda S, Barer F, Madi L, Multani AS, Pathak S. The A3 adenosine receptor as a new target for cancer therapy and chemoprotection. Exp Cell Res. 2001;269:230–236. doi: 10.1006/excr.2001.5327. [DOI] [PubMed] [Google Scholar]
- 296.Blay J, White TD, Hoskin DW. The extracellular fluid of solid carcinomas contains immunosuppressive concentrations of adenosine. Cancer Res. 1997;57:2602–2605. [PubMed] [Google Scholar]
- 297.Antonioli L, Blandizzi C, Pacher P, Haskó G. Immunity, inflammation and cancer: A leading role for adenosine. Nat Rev Cancer. 2013;13:842–857. doi: 10.1038/nrc3613. [DOI] [PubMed] [Google Scholar]
- 298.Lu J, Pierron A, Ravid K. An adenosine analogue, IB-MECA, down-regulates estrogen receptor alpha and suppresses human breast cancer cell proliferation. Cancer Res. 2003;63:6413–6423. [PubMed] [Google Scholar]
- 299.Merighi S, Mirandola P, Varani K, Gessi S, Leung E, Baraldi PG, Tabrizi MA, Borea PA. A glance at adenosine receptors: Novel target for antitumor therapy. Pharmacol Ther. 2003;100:31–48. doi: 10.1016/s0163-7258(03)00084-6. [DOI] [PubMed] [Google Scholar]
- 300.Nakamura K, Yoshikawa N, Yamaguchi Y, Kagota S, Shinozuka K, Kunitomo M. Antitumor effect of cordycepin (3′-deoxyadenosine) on mouse melanoma and lung carcinoma cells involves adenosine A3 receptor stimulation. Anticancer Res. 26:43–47. [PubMed] [Google Scholar]
- 301.Kim H, Kang JW, Lee S, et al. A3 adenosine receptor antagonist, truncated Thio-Cl-IB-MECA, induces apoptosis in T24 human bladder cancer cells. Anticancer Res. 2010;30:2823–2830. [PubMed] [Google Scholar]
- 302.Gessi S, Merighi S, Sacchetto V, Simioni C, Borea PA. Adenosine receptors and cancer. Biochim Biophys Acta Biomembr. 2011;1808:1400–1412. doi: 10.1016/j.bbamem.2010.09.020. [DOI] [PubMed] [Google Scholar]
- 303.Tsuchiya A, Nishizaki T. Anticancer effect of adenosine on gastric cancer via diverse signaling pathways. World J Gastroenterol. 2015;21:10931–10935. doi: 10.3748/wjg.v21.i39.10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Taliani S, Pugliesi I, Bellandi M, La Motta C, Da Settimo F. A3 receptor ligands: Past, present and future trends. Curr Top Med Chem. 2010;10:942–975. doi: 10.2174/156802610791293109. [DOI] [PubMed] [Google Scholar]
- 305.Merighi S, Benini A, Mirandola P, Gessi S, Varani K, Leung E, Maclennan S, Borea PA. A3 adenosine receptor activation inhibits cell proliferation via phosphatidylinositol 3-kinase/Akt-dependent inhibition of the extracellular signal-regulated kinase 1/2 phosphorylation in A375 human melanoma cells. J Biol Chem. 2005;280:19516–19526. doi: 10.1074/jbc.M413772200. [DOI] [PubMed] [Google Scholar]
- 306.Jacobson KA. Adenosine A3 receptors: Novel ligands and paradoxical effects. Trends Pharmacol Sci. 1998;19:184–191. doi: 10.1016/s0165-6147(98)01203-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.D’Alimonte I, Nargi E, Zuccarini M, et al. Potentiation of temozolomide antitumor effect by purine receptor ligands able to restrain the in vitro growth of human glioblastoma stem cells. Purinergic Signal. 2015;11:331–346. doi: 10.1007/s11302-015-9454-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Aghaei M, Panjehpour M, Karami-Tehrani F, Salami S. Molecular mechanisms of A3 adenosine receptor-induced G1 cell cycle arrest and apoptosis in androgen-dependent and independent prostate cancer cell lines: Involvement of intrinsic pathway. J Cancer Res Clin Oncol. 2011;137:1511–1523. doi: 10.1007/s00432-011-1031-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.David M, Akerman L, Ziv M, et al. Treatment of plaque-type psoriasis with oral CF101: Data from an exploratory randomized phase 2 clinical trial. J Eur Acad Dermatol Venereol. 2012;26:361–367. doi: 10.1111/j.1468-3083.2011.04078.x. [DOI] [PubMed] [Google Scholar]
- 310.David M, Gospodinov DK, Gheorghe N, et al. Treatment of plaque-type psoriasis with oral CF101: Data from a phase II/III multicenter, randomized, controlled trial. J Drugs Dermatol. 2016;15:931–938. [PubMed] [Google Scholar]
- 311.Stemmer SM, Benjaminov O, Medalia G, et al. CF102 for the treatment of hepatocellular carcinoma: A phase I/II, openlabel, dose-escalation study. Oncologist. 2013;18:25–26. doi: 10.1634/theoncologist.2012-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]