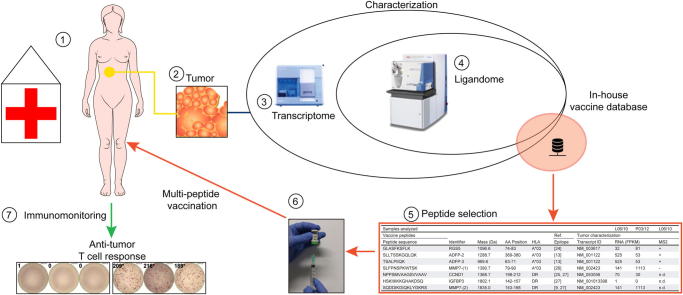
Graphical abstract
Abbreviations: CCA, cholangiocarcinoma; CT, computerized axial tomography; CCND, cyclin D; DMSO, dimethyl sulfoxide; ELISPOT, enzyme linked immune-spot assay; FFPE, formaldehyde fixed-paraffin embedded tissue; FPKM, fragments per kilobase mapped; 18FDG, fluorodeoxyglucose (18F); HLA, human leucocyte antigen; iCCA, intrahepatic cholangiocarcinoma; ICS, intracellular cytokine staining; IDH, isocitrate dehydrogenase; KMT2C, lysine N-methyltransferase 2C; LLoQ, lower limit of quantification; Mbp, megabase pairs; MMP, matrix metallo-proteinase, PBRM, protein polybromo-1; PBMC, peripheral blood mononuclear cells, PET, positron emission tomography; RGS, regulator of G-protein signaling, SIMOA, single molecule array; SSP-PCR, single specific primer polymerase chain reaction; WES, whole exome sequencing; WTS, whole transcriptome sequencing
Keywords: Primary liver cancer, Cholangiocarcinoma, Immunotherapy, Peptides, HLA, Immunopeptidome, Anti-tumor T cell response
Abstract
Background & Aims
We report a novel experimental immunotherapeutic approach in a patient with metastatic intrahepatic cholangiocarcinoma. In the 5 year course of the disease, the initial tumor mass, two local recurrences and a lung metastasis were surgically removed. Lacking alternative treatment options, aiming at the induction of anti-tumor T cells responses, we initiated a personalized multi-peptide vaccination, based on in-depth analysis of tumor antigens (immunopeptidome) and sequencing.
Methods
Tumors were characterized by immunohistochemistry, next-generation sequencing and mass spectrometry of HLA ligands.
Results
Although several tumor-specific neo-epitopes were predicted in silico, none could be validated by mass spectrometry. Instead, a personalized multi-peptide vaccine containing non-mutated tumor-associated epitopes was designed and applied. Immunomonitoring showed vaccine-induced T cell responses to three out of seven peptides administered. The pulmonary metastasis resected after start of vaccination showed strong immune cell infiltration and perforin positivity, in contrast to the previous lesions. The patient remains clinically healthy, without any radiologically detectable tumors since March 2013 and the vaccination is continued.
Conclusions
This remarkable clinical course encourages formal clinical studies on adjuvant personalized peptide vaccination in cholangiocarcinoma.
Lay Summary
Metastatic cholangiocarcinomas, cancers that originate from the liver bile ducts, have very limited treatment options and a fatal prognosis. We describe a novel therapeutic approach in such a patient using a personalized multi-peptide vaccine. This vaccine, developed based on the characterization of the patient’s tumor, evoked detectable anti-tumor immune responses, associating with long-term tumor-free survival.
Introduction
Cholangiocarcinomas (CCA), heterogeneous malignant epithelial tumors originating from hepatic ductal cells, are typically diagnosed in a late stage and have a particularly poor prognosis [1]. CCA are rare in the Western world (0.5–1.5/100,000 persons), while they are the second most frequent primary liver cancer worldwide [2]. Risk factors include liver cirrhosis, viral hepatitis, hepatobiliary flukes and primary sclerosing cholangitis [1]. A recent systematic meta-analysis of 4,756 patients with intrahepatic cholangiocarcinoma (iCCA) reported 5-year survival rates between 5 and 56% and a median survival of 28 months [3]. Surgical resection of tumors is so far the only effective therapeutic strategy and neither adjuvant chemotherapy nor radiotherapy has shown any clear benefit [3]. Prognosis of lymph node metastasis in iCCA is usually fatal and the reported 5-year survival rates are 0–8% [4], [5].
Novel treatment options using targeted agents off-label or loco-regional therapies have only shown limited success in CCA [6]. Intra and inter tumor genetic heterogeneity among iCCA, evidenced by whole exome sequencing (WES), calls for personalized therapies [7]. An encouraging new immunotherapy, using adoptive transfer of ex vivo expanded autologous CD4+ T cells targeting a mutated antigen, yielded spectacular clinical results in a patient with non-resected metastatic CCA [8]. This success, previously unheard of in CCA, points towards the possible effectiveness of immunotherapies. Furthermore, multi-peptide vaccine-induced immune responses correlate positively with increased survival in cancer [9], [10]. In line with such promising immunotherapeutic developments, we report our findings in a patient with metastatic iCCA, treated with a personalized multi-peptide vaccination. Since initial diagnosis and first tumor resection, the patient had tumor recurrences twice in the liver, as well as a lung metastasis, underlining the aggressiveness of the disease. The vaccine was designed based on WES, whole transcriptome sequencing (WTS) and HLA ligandome analysis of the tumors and induced long-term functional vaccine specific T cells. Remarkably, the tumor did not metastasize further and the patient is currently tumor-free, five years after initial diagnosis and 41 months after initiation of vaccination, suggesting therapeutic effectiveness.
Materials and methods
Next-generation-sequencing
As part of a research project (IndividuaLIVER, approved by the Institutional Review Board at the University Hospital of Tübingen), WES of tumor and normal tissue was performed for L06/10, L04/12 and P03/13 (Fig. 1). In addition, the transcriptome of tumor tissue of L06/12 and P03/13 was sequenced. Further details are provided in Supplementary Tables 6–8; and Supplementary Tables 11 and 14.
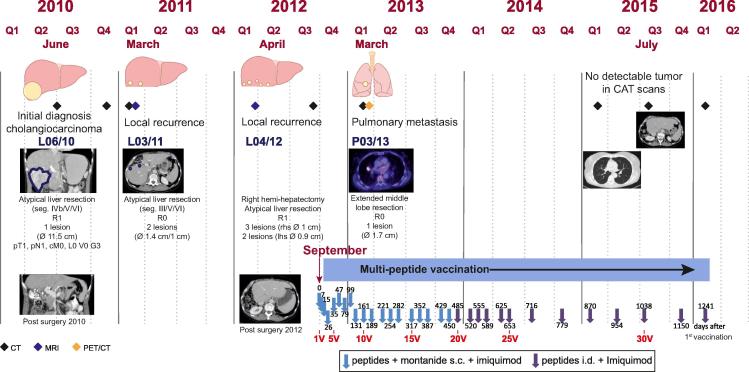
Fig. 1.
Clinical course and therapy. The course of the disease is subdivided in quarters of the year (Q) and the interventions performed (surgery/vaccination) are shown. Clinical imaging results for relevant events are depicted exemplarily in chronological order. Further exemplary imaging results depicting the course of disease can be accessed in Supplementary Figs. 1–15. The multi-peptide vaccination schedule is also indicated chronologically annotated for days (after initiation) and count of vaccinations. Seg, segment; R-classification, R0: no residual tumor; R1: microscopic residual tumor; R2: macroscopic residual tumor; lhs/rhs, left/right hand side; classification of malignant tumours (TNM Classification) (UICC): T = tumor extent (0–4); N = positive lymphnodes (0–1); M = Metastasis (0–1); L = lymphinvasion (0–1); V = venous invasion (0–1); G = grading (1–3); p = histopathological staging; c = clinical staging; Ø diameter; s.c., subcutaneous; i.d., intradermal; nV = nth vaccination.
A total of four tumor samples (Formalin-Fixed, Paraffin Embedded tissue (FFPE) shavings; L06/10; L03/11; L04/12; P03/13) and one reference sample (blood) were sequenced with a custom cancer panel (SureSelect XT; Agilent, Waldbronn, Germany) covering 1.566 Mbp of coding sequence (Supplementary Table 3). On average a two-fold higher read count was generated for the tumor samples compared to the reference sample (39,900,242 reads vs. 17,748,620 reads). This yielded a mean coverage depth of 1,065x for the tumor samples with 98% of regions of interest covered with at least 100x and 90% of the regions covered with at least 500x (For further details see Supplementary Tables 4 and 5). The reference sample was sequenced to a mean coverage depth of 705x and a 98% 100x-coverage of the target region. Bioinformatic analysis for WES, WTS and panel sequencing followed established in-house pipelines. Further details including the bioinformatics analysis pipeline can be found in the Supplementary materials and methods.
HLA typing
Two-digit HLA class I and II typing was performed by SSP-PCR at the Department of Transfusion Medicine, University of Tübingen, following clinical routines. Typing at four-digit resolution using the WES data was performed by OptiType [11] for HLA class I and further manual curation for HLA class II alleles, determining the patient’s HLA allelotype to be: HLA-A∗03:01/A∗29:01, B∗07:05/B∗35:01, C∗04:01/C∗15:05 and HLA-DRB1∗01:01/DRB1∗11:01/DRB3∗02:02, DQB1∗03:01/DQB1∗05:01. The results of OptiType are in accordance with the PCR-based two-digit HLA typing.
Mass spectrometry
HLA ligands were immunoprecipitated from cryopreserved tumor tissues and liver tissue (L03/10) as previously described [12] using the pan-HLA class I antibody, W6/32 (manufactured in-house). HLA ligands were purified using 3 kDa cut-off centrifugal filters (Amicon, Maerck Millipore, Carrightwohill, Ireland), desalted (C18 ZipTip; Merck Millipore, Darmstadt, Germany), concentrated (vacuum centrifuge; Bachofer, München, Germany) and analyzed by LC-MS/MS as previously described [13]. Further details are provided in Supplementary materials and methods and Supplementary Table 17. An individualized protein database containing tumor-specific somatic mutations identified by WES/WTS was generated in FASTA format and used for spectral annotation using the Mascot search engine (v2.2.0.4, Matrix Science, Boston, MA).
T cell in vitro immunomonitoring
The patient received 32 vaccinations in total between September 2012 and February 2016 (ongoing). EDTA-anticoagulated blood (45 ml) and serum (5.5 ml) was drawn at two time points before vaccination and at regular intervals during the course of vaccination. T cell responses to all vaccinated peptides (Table 1) were monitored in peripheral blood mononuclear cells (PBMCs) isolated from blood drawn before vaccination (either during the pre-vaccination screening (scr) or at the first vaccination appointment (1V)) as well as during vaccinations 5V, 6V, 8V, 10V, 12V, 15V, 22V and 25V. PBMCs were pre-stimulated in vitro using the vaccinated peptides and relevant negative control peptides and expanded using IL-2 for 12 days (see details in Supplementary materials and methods) [14], [15]. Responses to HLA class I peptides were monitored by IFN-γ ELISPOT and HLA-peptide multimer staining, while responses to HLA class II peptides and RGS-5 peptide were determined by IFN-γ ELISPOT and ICS (for IFN-γ, TNF-β, IL-2 and IL-10) (all techniques are elaborated in Supplementary materials and methods).
Table 1.
Composition of the multi-peptide vaccine.
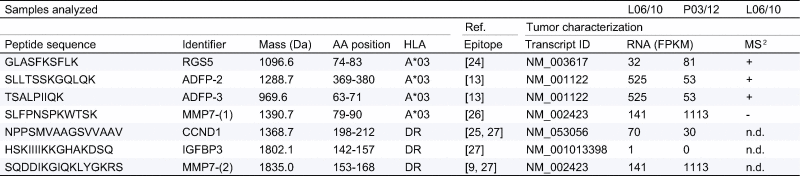 |
Peptides are listed alongside their sequence, identifier of the core protein, molecular mass and position of the peptide in the respective corresponding source protein and HLA-restrictions of the peptides. Characterization of the patient’s tumor, HLA ligandome analysis (LC-MS/MS) as well as whole transcriptome sequencing (WTS) of two tumor manifestations (L06/10 – primary tumor; P03/13 lung metastasis) are shown with respective FPKM values (fragments per kilobase mapped) for the transcripts of interest. AA (amino acid); RGS (regulator of G-protein signaling); ADFP (adipose differentiation-related protein/Perlipin); CCND (cyclin D); IGFBP (Insulin-like growth factor-binding protein); MMP (matrix metalloproteinase); MS2 (tandem mass spectrometry); Ref. (References).
Histopathology
Tissue sections from paraffin embedded diagnostic tumor material obtained after surgical resections (L06/10; L03/11; L04/12; P03/13) were assessed by a consultant pathologist and FFPE shavings (>90% tumor fraction) used to isolate DNA for panel sequencing (above) and IDH1 PCR. Further sections were used for additional immunohistochemical (IHC) analyses to complement CK7, Hep Par 1, Napsin A and TTF1 stainings, which had already been assessed during routine histopathology.
IHC staining was performed on an automated immunostainer, following the manufacturer’s protocol (Benchmark, Ventana Medical Systems, Tucson, AZ). Respective commercially available antibodies used, sources and dilutions are summarized in Supplementary Table 1. Sections were assessed by a consultant pathologist as negative (−), positive with low (+), intermediate (++) and strong (+++) staining in immunohistochemistry or by evaluating positive cells in 10 high power fields counted and cell densities for the epithelial (E) and stromal (S) compartment respectively.
Results
Clinical course
A 56-year-old female patient, previously asymptomatic and healthy without any established risk factors for primary liver cancers, was sonographically diagnosed with a big unilocular mass in the right hepatic lobe (Ø 11.5 cm) in June 2010. Following atypical liver tri-segment resection (segments IVb, V, VI) and radical systemic lymphadenectomy, the tumor was staged as an incompletely resected iCCA (pT1, pN1 (1/20), (pL0, pV0), cM0/R1) (abbreviated as L06/10) (Fig. 1). Histological assessment indicated a poorly differentiated (G3) monomorphic adenocarcinoma with partially solid appearance, displaying moderate desmoplastic stroma. The tumor cells, with extensive eosinophilic cytoplasm and hyperchromatic and moderately polymorphic nuclei, were strongly positive for cytokeratin (CK) 7 and negative for Hep Par 1 (Fig.2B), supporting the diagnosis of iCCA [16]. Adjacent liver tissue showed no steatosis/fibrosis or any indication of liver pathology. The disease recurred rapidly and two new lesions required another atypical liver resection (segments III, V, VI; R0) nine months later (L03/11) and a third one combined with a right hemi-hepatectomy (R1) 22 months after initial diagnosis (L04/12) (Fig. 1). Both recurring liver lesions, L03/11 and L04/12, were also analyzed by immunohistochemistry (Fig.2B and Supplementary Table 2). At this stage, faced with a fatal disease without any standard treatment options, based on expert opinions and a positive individual medical risk-benefit assessment, a multi-peptide vaccine was considered as a last resort (according to principle 35 Declaration of Helsinki (Seoul 2008)). The patient had previously volunteered to participate in a scientific research project (IndividuaLIVER) aimed at the development of personalized tumor vaccines, which enabled extensive molecular characterization of the tumors.
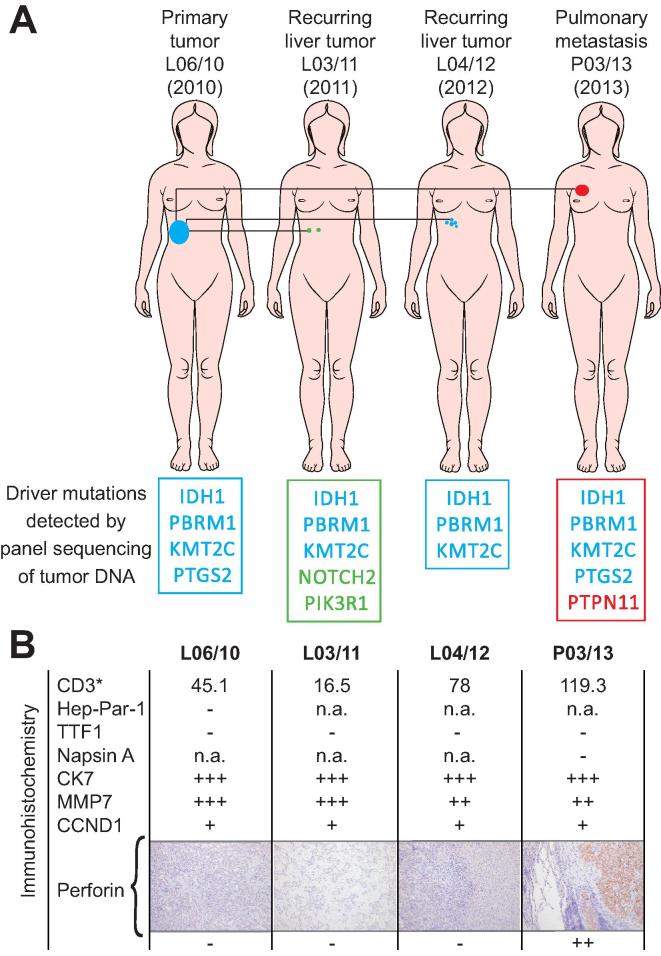
Fig. 2.
Tumor characterization. Characterization of the various surgically resected tumors is shown. (A) Tumor location and putative ancestry based on shared driver mutations are indicated with a color code. (B) Tumors were assessed by immunohistochemistry and qualitative staining patterns (− negative; + slightly positive; ++ moderately positive; +++ strongly positive) are given for different immune and tumor markers. Staining for Hep Par 1 and CK7 supported the initial diagnosis of cholangiocarcinoma (L06/10) and staining of P03/13 for thyroid transcription factor-1 (TTF1) and Napsin A ruled out a primary lung adenocarcinoma. Further, counts for immune cell infiltrates in the epithelial compartment are given for each tumor and microscopic pictures (×100) of perforin staining are shown. More detailed data is provided in Supplementary Table 2.
Next-generation sequencing and HLA ligandome analysis for the identification of vaccine targets
First the tumor was analyzed to design a personalized anti-tumor vaccine based on mutated HLA ligands. We generated comprehensive datasets through sequencing (including WES and WTS) and screened for tumor-specific mutations (Supplementary Tables 8, 11 and 14). Then, using mass spectrometry, we assessed the natural HLA class I ligands on the tumor (HLA ligandome analysis) (Supplementary Table 17). Although we predicted several HLA ligands in silico containing mutations from the sequencing data, we were unable to find direct evidence for their presence in the ligandome (see Supplementary Tables 8–17 for detailed information, and Supplementary materials and methods for experimental procedures). Instead, we found several non-mutated HLA ligands that had been previously identified in other tumor entities (Supplementary Table 17). Based on these observations we selected a multi-peptide vaccine composed of seven tumor-associated epitopes (Table 1) containing four HLA-A∗03 restricted short peptides and three promiscuous HLA-DR long peptides. Three of the short peptides were confirmed as natural HLA ligands on the patient’s tumor tissue by mass spectrometry (L06/10) (Supplementary Figs. 16–21) and source protein transcripts for all but one of the seven vaccine peptides were found to be expressed by WTS (FPKM >25) in the tumor (Table 1 and Supplementary Table 18). The final peptide selection was made by combining the following two aspects, first, known epitopes from our in-house database that had been applied in patients previously, following an individual risk-benefit assessment were considered. Secondly, those peptides were prioritized according to evidence for their presence on the tumor tissue of the patient.
Vaccination, further clinical course and immunomonitoring
After comprehensive and detailed informed consent by multiple attending physicians and a written approval by the patient, immunization using the multi-peptide vaccine was initiated in September 2012. The vaccine contained 300 μg per peptide in 300 μl of 33% DMSO (in H2O) emulsified in montanide ISA 51 VG ad 1 ml. It was applied subcutaneously (s.c.), according to a tapering vaccination schedule, followed by topical application of 250 mg imiquimod twice, with a 12 h interval (Fig. 1).
In March 2013, a suspect solitary pulmonary lesion (Ø 1.7 cm) in the right middle lobe was identified in CT scans (abbreviated as P03/13) and was found to be metabolically active in 18FDG-PET (Fig. 1). At this point, re-inspection of the CT scans performed in September 2012, prior to the start of the vaccination, revealed a small lesion at the same location (Supplementary Figs. 10–13). In an ensuing pleural middle lobe and sleeve resection of the pulmonary artery with lymphadenectomy (N0 (0/9), R0), P03/13 was removed. Histological and immunohistochemical analyses confirmed an iCCA metastasis and ruled out a primary pulmonary malignancy (Fig.2B).
Due to the absence of adverse events apart from expected minor inflammatory reactions at the vaccination site [17] and the lack of therapeutic alternatives, the peptide vaccination was continued as stated above. After 18 s.c. vaccinations, application mode was changed to intradermal (i.d.) (omitting montanide) for further vaccinations (until February 2016). Latest radiological imaging results (CT scan, February 2016), obtained 41 months after initiation of the vaccination showed no evidence of tumor recurrence, neither intra- nor extrahepatic.
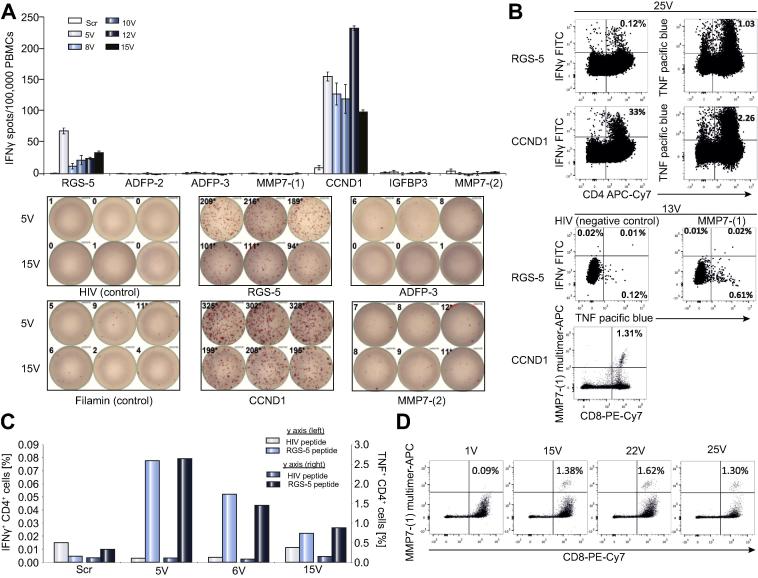
In vitro T cell immunomonitoring showed a rapid (as early as the 5th vaccination i.e., 5V) and persisting (detectable until the last monitored time point- 25V) induction of T cells specific for three vaccine peptides derived from regulator of G-protein signaling 5 (RGS5), cyclin D1 (CCND1) and matrix metalloproteinase-7 (MMP7-(1)) (Fig. 3). T cell cytokine response was detected against CCND1 by IFN-γ ELISPOT (Fig.3A) and was confirmed to be a CD4+ response by intracellular cytokine staining (ICS) (Fig.3B left lower panel). The RGS5 peptide elicited a CD4+ T cell response (Fig.3B, 3C). Indeed, binding of this short peptide to HLA class II molecules was proven by mass spectrometry following immunoprecipitation and its ability to elicit CD4 T cell responses was observed in other patients (unpublished own data). Such observations are not uncommon and have been reported by others [18]. We also detected MMP7-(1) specific CD8+ T cells by HLA-peptide multimer staining (Fig.3D), these cells did not produce any IFN-γ but TNF upon peptide specific stimulation (Fig.3B).
Fig. 3.
Immunomonitoring of patient PBMCs for vaccine induced T cell responses. PBMCs were pre-stimulated using either peptide pool I (RGS-5, ADFP-2, ADFP-3, MMP7-(1) and HIV-A03) or peptide pool II (CCND1, IGFBP3, MMP7-(2) and Filamin-A), and expanded for 12 days using IL-2. (A) 300,000 cells (Class I binding peptides) or 200,000 (for class II binding peptides) were re-stimulated in triplicates (duplicates for pre-vaccination ‘scr’ time point) in an IFN-γ ELISPOT assay using individual peptides. The top panel shows the normalized spot counts for individual peptide stimulations and the bottom panel shows examples of scanned ELISPOT wells. At least 700,000 cells were re-stimulated with the respective peptides, in the presence of Brefeldin A and GolgiStop and 12 h later, intracellular IFN-γ and TNF were stained and analysed for B and C. (B) Exemplary dot plots of IFN-γ and TNF production by CD4(+) cells in response to RGS5 and CCND1 peptides, collected at the 25th vaccination (25V) are shown in the left panel. TNF production by CD8(+) cells in response to MMP7-(1) peptide are shown in the right panel (top). MMP7-(1) multimer staining of the corresponding vaccination time point (13V) is shown in the right panel (bottom). (C) Frequencies of the RGS5 peptide induced cytokine(+) live CD4(+) lymphocytes are shown (left y axis: IFN-γ right y axis: TNF). (D) At least 600,000 cells were analyzed by HLA-peptide multimers (class I). MMP7-(1) multimer(+) cells within live CD4(−) lymphocytes are shown and the % of multimer(+) CD8(+) lymphocytes are indicated.
Since IL-10 induction in serum has been reported to be associated with prolonged imiquimod application in a mouse model [19], we measured serum levels of IL-10 during the course of vaccination. IL-10 levels remained below the lower limit of quantification (LLoQ) using a Luminex based sandwich immunoassay. Nevertheless, we were able to detect baseline levels of IL-10 (1.46 to 4.55 pg/ml) using an in-house developed and technically validated, ultrasensitive single molecule array (SIMOA) immunoassay (unpublished) on the Quanterix platform (Supplementary Table 20; Supplementary materials and methods), coherent with data published for healthy subjects [20], [21].
In addition, detailed immunohistochemical analyses of tumor tissue indicated that both CCND1 and MMP7 were indeed constantly expressed in the excised tumors during the course of the disease (Fig.2B). Strikingly, stromal T cell infiltration was more pronounced in P03/13, resected six months after initiation of vaccination, than in the earlier liver lesions, evidenced by CD3 staining (Fig.2B). Further, the cytotoxic molecule perforin, previously absent in the liver tumors, was strongly positive in the pulmonary metastasis, confirming the presence of activated cytotoxic T cells within the tumor (Fig.2B). Somatic mutations were detected in seven potential tumor suppressor/oncogenes in a somatic cancer panel (Fig.2A and detailed in Supplementary Table 4). Among these seven mutations, three (in IDH1, PBRM1 and KMT2C) were detected in all the four tumors analyzed verifying the common cellular origin of all different malignant lesions that occurred over time.
Discussion
Cancer immunotherapy has recently gained considerable momentum given the clinical success of checkpoint inhibitors. Particularly in metastatic iCCA, spectacular results have been obtained using the adoptive transfer of mutation-specific CD4+ T cells in a patient [8]. Faced with a malignant disease with recurring tumor, lymphatic metastasis, extremely poor prognosis and no established standard therapies, we decided to explore experimental therapeutic alternatives. Initially we intended to design a personalized peptide vaccine targeting individual tumor mutations presented by HLA class I molecules to induce tumor reactive T cells [22], [23]. However, based on WES (Supplementary Tables 8, 11 and 14) the in silico predicted mutated HLA ligands (Supplementary Tables 9, 10, 12, 13, 15 and 16) could not be confirmed as naturally presented HLA ligands by mass spectrometry (Supplementary Table 17), indicating their low abundance or sub-clonal presentation in the patient’s tumor samples. Our study in leukemia patients identified non-mutated epitopes as valid targets of clinically relevant anti-cancer T cell responses, suggesting that non-mutated ligands may indeed be tumor-specific due to altered antigen presentation on tumor cells [13]. We therefore pursued a personalized vaccination approach based on non-mutated ligands presented by the patient’s tumor. Consequently, we selected a vaccine containing seven tumor-associated peptides (TUMAPs) that had been shown to be immunogenic in cancer patients and/or in vitro T cell assays [9], [13], [24], [25], [26], [27], to secure a positive individual risk-benefit ratio combined with best available evidence for target representation on the patient’s tumor and immunogenicity. Of the four short peptides in the vaccine, transcripts of their source proteins were expressed in the patient’s tumors while three could additionally be confirmed as natural HLA class I ligands by mass spectrometry (Table 1, Supplementary Figs. 16–21, and Supplementary Tables 17 and 18). We also included three promiscuous HLA-DR binding long peptides in the vaccine to simultaneously induce CD4 help (Table 1).
The multi-peptide vaccination induced functional T cell responses against 3 out of the 7 peptides (Fig. 3). Analysis of the pulmonary metastasis (P03/13), which pre-existed in an indiscernible stage prior to vaccination (Supplementary Figs. 10–13) and was resected 6 months after the start of the vaccination, showed a higher infiltration of CD3+ cells than the tumors resected before the vaccination. P03/13 also displayed strong expression of perforin (Fig.2B), which was absent in all other tumor lesions. Although the microenvironment in lung and liver tissue may be fundamentally different and influence immune cell recruitment, our data clearly suggests vaccination induced infiltration of anti-tumor T cells into the lung metastasis but cannot provide absolute certainty.
Given the established genetic heterogeneity of iCCA [7], we identified known driver mutations such as IDH1 [7], KMT2C [7] and PBRM1 [28] and addressed the ancestry of the tumor lesions (Fig.2A). These findings enabled us to safely rule out any de novo malignancy. The inherent individual tumor heterogeneity at the level of malignant driver mutations suggests that metastasis was a rather early event [7]. Although some of the mutations evidenced might activate targetable signaling pathways, no approved targeted therapy was available for our patient much less any evidence for effectiveness in the adjuvant setting.
To our knowledge this is the first report on a personalized adjuvant peptide-based immunotherapy in iCCA. The remarkable tumor-free status and the extended survival of the patient, extremely rare in metastatic iCCA, make this a compelling case worth reporting. The vaccine-induced T cell responses as well as the increased T cell infiltration into the lung metastasis underline the induction of anti-tumor immunosurveillance by the vaccine and can be held responsible for the prevention of any further metastasis and the current status of the patient. Based on our results and previous reports of immunotherapy in CCA, we propose adjuvant personalized peptide-based immunotherapy in iCCA as a highly promising new therapy well worth exploring in formal clinical trials.
Note added in proof
After this manuscript was resubmitted, the patient presented with a new lesion in the liver, which then could be completely resected. A thorough analysis of this tissue is being performed, in particular with respect to characterizing the tumor immune infiltrate and signs of tumor immune escape.
Financial support
The IndividuaLIVER study was supported by a grant (01GU0804 to H-G.R., S.S., P.B., O.R., A.K. and F.F.) and iVacALL (O.K.) of the German Federal Ministry of Education and Research (BMBF). P.A.C., C.G., K.L., (SFB685 Z5), S.S., H-G.R. (SFB685), O.K., M.W., and S. Nahnsen (Core Facilities Initiative QBiC) are supported by the Deutsche Forschungsgemeinschaft (DFG). H-G.R. and S.S. acknowledge support from European Research Council (ERC) (MUTAEDITING) and Deutsches Konsortium für Translationale Krebsforschung (DKTK).
Conflict of interest
HG.R. and S.S. are the inventors of patents owned by Immatics biotechnologies GmbH, which develops targets for cancer immunotherapies. HGR is shareholder of Immatics and CureVac. The other authors declare no conflict of interest. The described patient has explicitly consented to publishing her case in this present form.
Authors’ contributions
Drafting of the article: MWL, PAC, CG; study concept and design MWL, HGR, FF, OK, AK, S. Nadalin; data collection: MWL, PAC, KL, CS, IB, FJH, DJK, NT, HS, MG, VY, SC, HB; obtained funding: FF, HGR, SS, OK, OR, AK, CG, PB, MW, S. Nahnsen; data analysis: MWL, PAC, KL, CG, IB, HB, CS, FJH, MS, DJK, NT, HS, MG, MW, CM, S. Nahnsen, NSM, SC, CG, SS, HGR; patient’s clinical characterization and management: DZ, KT, JG, S. Nadalin, HPN, AK, SC; immunological characterization of patient samples: PAC, KL, VY, NSM, CG, HB, IB; FF; in vitro T cell immunomonitoring and data analysis: PAC, KL, NSM, CG; immunopeptidomic characterization of patient samples and data analysis: MWL, DJK, HS, MG, NT, HPN, SS; next-generation sequencing and panel sequencing of patient samples and data analysis: CS, FJH, PB, OR; immuno-histochemical characterization of patient samples and analysis: IB, HB, FF; bioinformatics analysis of patient data and data integration: OK, MW, CM, MS, S. Nahnsen; peptide selection and manufacturing: SS, HGR, MWL, CS, NT, HS, DJK, MW, CM, OR, S. Nahnsen, DZ, FF, S. Nadalin, OK; critical revision of the data (including supplements) and manuscript preparation: MWL, PAC, KL, CS, IB, FJH, NT, HS, MG, MW, CM, NSM, SC, HB; critical revision of the article and supplementary materials: MWL, PAC, KL, CS, IB, FJH, MS, DJK, NT, HS, MG, VY, MW, CM, HPN, OR, PB, S. Nahnsen, AK, S. Nadalin, DZ, JG, KT, NSM, SC, HB, FF, OK, CG, SS, HGR; study supervision: OR, PB, AK, DZ, FF, CG, SS, AK, HGR.
Acknowledgments
We would like to acknowledge the expert technical assistance of Patricia Hrstić, Nicole Bauer and Elisa Rusch. This data has been presented previously at the 15th International Conference on Progress in Vaccination against cancer (PIVAC-15) of the European Association for Cancer Research (EACR) (Poster #5) and 14th Annual Meeting of the Association for cancer Immunotherapy (CIMT 2016) (Poster/short talk #43).
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jhep.2016.06.027.
Supplementary data
References
Author names in bold designate shared co-first authorship
- 1.Razumilava N., Gores G.J. Cholangiocarcinoma. Lancet. 2014;383:2168–2179. doi: 10.1016/S0140-6736(13)61903-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergquist A., von Seth E. Epidemiology of cholangiocarcinoma. Best Pract Res Clin Gastroenterol. 2015;29:221–232. doi: 10.1016/j.bpg.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Mavros M.N., Economopoulos K.P., Alexiou V.G., Pawlik T.M. Treatment and prognosis for patients with intrahepatic cholangiocarcinoma: systematic review and meta-analysis. JAMA Surg. 2014;149:565–574. doi: 10.1001/jamasurg.2013.5137. [DOI] [PubMed] [Google Scholar]
- 4.Nakagohri T., Asano T., Kinoshita H., Kenmochi T., Urashima T., Miura F. Aggressive surgical resection for hilar-invasive and peripheral intrahepatic cholangiocarcinoma. World J Surg. 2003;27:289–293. doi: 10.1007/s00268-002-6696-7. [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto M., Takasaki K., Yoshikawa T. Extended resection for intrahepatic cholangiocarcinoma in Japan. J Hepatobiliary Pancreat Surg. 1999;6:117–121. doi: 10.1007/s005340050093. [DOI] [PubMed] [Google Scholar]
- 6.Subbiah I.M., Subbiah V., Tsimberidou A.M., Naing A., Kaseb A.O., Javle M. Targeted therapy of advanced gallbladder cancer and cholangiocarcinoma with aggressive biology: eliciting early response signals from phase 1 trials. Oncotarget. 2013;4:153–162. doi: 10.18632/oncotarget.832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zou S., Li J., Zhou H., Frech C., Jiang X., Chu J.S. Mutational landscape of intrahepatic cholangiocarcinoma. Nat Commun. 2014;5:5696. doi: 10.1038/ncomms6696. [DOI] [PubMed] [Google Scholar]
- 8.Tran E., Turcotte S., Gros A., Robbins P.F., Lu Y.C., Dudley M.E. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344:641–645. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walter S., Weinschenk T., Stenzl A., Zdrojowy R., Pluzanska A., Szczylik C. Multipeptide immune response to cancer vaccine IMA901 after single-dose cyclophosphamide associates with longer patient survival. Nat Med. 2012;18:1254–1261. doi: 10.1038/nm.2883. [DOI] [PubMed] [Google Scholar]
- 10.Sasada T., Kibe S., Akagi Y., Itoh K. Personalized peptide vaccination for advanced colorectal cancer. Oncoimmunology. 2015;4 doi: 10.1080/2162402X.2015.1005512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szolek A., Schubert B., Mohr C., Sturm M., Feldhahn M., Kohlbacher O. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics. 2014;30:3310–3316. doi: 10.1093/bioinformatics/btu548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kowalewski D.J., Stevanovic S. Biochemical large-scale identification of MHC class I ligands. Methods Mol Biol. 2013;960:145–157. doi: 10.1007/978-1-62703-218-6_12. [DOI] [PubMed] [Google Scholar]
- 13.Kowalewski D.J., Schuster H., Backert L., Berlin C., Kahn S., Kanz L. HLA ligandome analysis identifies the underlying specificities of spontaneous antileukemia immune responses in chronic lymphocytic leukemia (CLL) Proc Natl Acad Sci U S A. 2015;112:E166–E175. doi: 10.1073/pnas.1416389112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hadrup S.R., Maurer D., Laske K., Frosig T.M., Andersen S.R., Britten C.M. Cryopreservation of MHC multimers: Recommendations for quality assurance in detection of antigen specific T cells. Cytometry Part A. 2015;87:37–48. doi: 10.1002/cyto.a.22575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Widenmeyer M., Griesemann H., Stevanovic S., Feyerabend S., Klein R., Attig S. Promiscuous survivin peptide induces robust CD4+ T-cell responses in the majority of vaccinated cancer patients. Int J Cancer. 2012;131:140–149. doi: 10.1002/ijc.26365. [DOI] [PubMed] [Google Scholar]
- 16.Fan Z., van de Rijn M., Montgomery K., Rouse R.V. Hep par 1 antibody stain for the differential diagnosis of hepatocellular carcinoma: 676 tumors tested using tissue microarrays and conventional tissue sections. Mod Pathol. 2003;16:137–144. doi: 10.1097/01.MP.0000052103.13730.20. [DOI] [PubMed] [Google Scholar]
- 17.Hu Y., Smolkin M.E., White E.J., Petroni G.R., Neese P.Y., Slingluff C.L., Jr. Inflammatory adverse events are associated with disease-free survival after vaccine therapy among patients with melanoma. Ann Surg Oncol. 2014;21:3978–3984. doi: 10.1245/s10434-014-3794-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gross S., Lennerz V., Gallerani E., Mach N., Bohm S., Hess D. Short peptide vaccine induces CD4+ T helper cells in patients with different solid cancers. Cancer Immunol Res. 2016;4:18–25. doi: 10.1158/2326-6066.CIR-15-0105. [DOI] [PubMed] [Google Scholar]
- 19.Lu H., Wagner W.M., Gad E., Yang Y., Duan H., Amon L.M. Treatment failure of a TLR-7 agonist occurs due to self-regulation of acute inflammation and can be overcome by IL-10 blockade. J Immunol. 2010;184:5360–5367. doi: 10.4049/jimmunol.0902997. [DOI] [PubMed] [Google Scholar]
- 20.Kleiner G., Marcuzzi A., Zanin V., Monasta L., Zauli G. Cytokine levels in the serum of healthy subjects. Mediators Inflamm. 2013;2013 doi: 10.1155/2013/434010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong H.L., Pfeiffer R.M., Fears T.R., Vermeulen R., Ji S., Rabkin C.S. Reproducibility and correlations of multiplex cytokine levels in asymptomatic persons. Cancer Epidemiol Biomark Prev. 2008;17:3450–3456. doi: 10.1158/1055-9965.EPI-08-0311. [DOI] [PubMed] [Google Scholar]
- 22.Schumacher T.N., Schreiber R.D. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 23.Vonderheide R.H., Nathanson K.L. Immunotherapy at large: the road to personalized cancer vaccines. Nat Med. 2013;19:1098–1100. doi: 10.1038/nm.3317. [DOI] [PubMed] [Google Scholar]
- 24.Boss C.N., Grunebach F., Brauer K., Hantschel M., Mirakaj V., Weinschenk T. Identification and characterization of T-cell epitopes deduced from RGS5, a novel broadly expressed tumor antigen. Clin Cancer Res. 2007;13:3347–3355. doi: 10.1158/1078-0432.CCR-06-2156. [DOI] [PubMed] [Google Scholar]
- 25.Dengjel J., Decker P., Schoor O., Altenberend F., Weinschenk T., Rammensee H.G. Identification of a naturally processed cyclin D1 T-helper epitope by a novel combination of HLA class II targeting and differential mass spectrometry. Eur J Immunol. 2004;34:3644–3651. doi: 10.1002/eji.200425510. [DOI] [PubMed] [Google Scholar]
- 26.Yokoyama Y., Grunebach F., Schmidt S.M., Heine A., Hantschel M., Stevanovic S. Matrilysin (MMP-7) is a novel broadly expressed tumor antigen recognized by antigen-specific T cells. Clin Cancer Res. 2008;14:5503–5511. doi: 10.1158/1078-0432.CCR-07-4041. [DOI] [PubMed] [Google Scholar]
- 27.Dengjel J., Nastke M.D., Gouttefangeas C., Gitsioudis G., Schoor O., Altenberend F. Unexpected abundance of HLA class II presented peptides in primary renal cell carcinomas. Clin Cancer Res. 2006;12:4163–4170. doi: 10.1158/1078-0432.CCR-05-2470. [DOI] [PubMed] [Google Scholar]
- 28.Jiao Y., Pawlik T.M., Anders R.A., Selaru F.M., Streppel M.M., Lucas D.J. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet. 2013;45:1470–1473. doi: 10.1038/ng.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.