Abstract
Background: Overall survival remains unsatisfactory in stage II/III gastric cancer, even after curative resection and adjuvant chemotherapy. Tensin 4 (TNS4), a cell adhesion factor, is associated with cancer-cell motility and migration. Patients and Methods: We examined the clinical significance of TNS4 gene expression in 134 patients with stage II/III gastric cancer who underwent adjuvant chemotherapy with S-1. TNS4 gene expression in surgical specimens was measured by quantitative reverse-transcription polymerase chain reaction (RT-PCR). Results: TNS4 gene expression levels were significantly higher in cancer tissue than in adjacent normal mucosa. High TNS4 gene expression was associated with significantly poorer 5-year overall survival than was low expression. On multivariate analysis, TNS4 gene expression was an independent prognostic factor. Conclusion: Overexpression of the TNS4 gene is a useful independent predictor of outcomes in patients with stage II/III gastric cancer who undergo surgery and receive adjuvant chemotherapy with S-1.
Keywords: Tensin 4, TNS4, gastric cancer, adjuvant chemotherapy, S-1, prognostic factor
Gastric cancer is the fifth most common cancer in the world and the third leading cause of cancer-related death (1). The outcomes of gastric cancer have been improved by gastrectomy with D2 lymph-node dissection and advances in chemotherapy (2). The Adjuvant Chemotherapy Trial of S-1 for Gastric Cancer (ACTS-GC) demonstrated that 1 year of adjuvant chemotherapy with S-1, an oral fluoropyrimidine, significantly improves outcomes in patients with stage II/III gastric cancer who undergo gastrectomy with D2 dissection (3). However, recurrence rates after curative resection and adjuvant chemotherapy remain high in gastric cancer. In the ACTS-GC study, the 5-year overall survival (OS) rate was 84.2% in those with stage II disease, 67.1% in those with stage IIIA disease, and 50.2% in those with stage IIIB disease, even after adjuvant chemotherapy with S-1 (4). Further improvements in outcomes are likely to require personalized therapy based on biomarker analysis.
We have searched for biomarkers of gastric cancer using DNA microarray analysis in patients with stage II/III gastric cancer who underwent curative resection and received adjuvant chemotherapy with S-1. We found that expression of the human tensin 4 (TNS4) gene in gastric cancer tissue was 23.52-times higher than that in paired adjacent normal mucosa (unpublished data). We, thus, focused on the clinical significance of TNS4 gene expression in gastric cancer.
TNS4 is a member of the tensin gene family (5) and is also known as COOH-terminus tensin-like molecule (CTEN) (6). This gene family comprises four members (TNS1, TNS2, TNS3, and TNS4) and their products are localized in the cytoplasmic tails of integrins at focal adhesions. Tensins play important roles in various biological processes, such as cell adhesion, migration, proliferation, differentiation, apoptosis, and invasion (6-9). TNS4 expression is up-regulated in many cancer types (10-14), suggesting that overexpression of TNS4 may play a critical role in tumorigenesis.
In this study, we evaluated the clinical significance of the relative expression of TNS4 in patients with stage II/III gastric cancer who underwent curative resection followed by adjuvant chemotherapy with S-1.
Patients and Methods
Patients and samples. We studied surgical specimens of cancer tissue from 134 patients with stage II/III gastric cancer who received no preoperative therapy. The patients underwent curative resection and postoperatively received adjuvant chemotherapy with S-1. As a reference group, we concurrently studied TNS4 status and survival in the 103 patients who did not receive adjuvant S-1 chemotherapy. No patient had any other malignancy. All patients were treated at the Department of Surgery, Yokohama City University, Japan and the Gastroenterological Center, Yokohama City University Medical Center and the Gastrointestinal Surgery, Kanagawa Cancer Center between 2002 and 2010. Informed consent was obtained from each patient, while the Ethics Committees of Yokohama City University, Yokohama City University Medical Center (approval number: 18-7A-4), and Kanagawa Cancer Center approved the protocol (approval number: epidemiological study-29) before initiation of the study.
Each tissue sample was embedded in optimum cutting temperature compound (Sakura Finetechnical Co., Ltd., Tokyo) and immediately stored at −80˚C until use. Tissue specimens were stained with hematoxylin and eosin and examined histopathologically. Sections that consisted of >80% carcinoma cells were used to prepare total RNA.
Cell lines. Human gastric cancer cell lines (MKN1, MKN7, MKN45, MKN74, NUGC3, NUGC4, and Kato III) were provided by the Japanese Cancer Research Bank, Tokyo, Japan. Cell lines were maintained in Roswell Park Memorial Institute 1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Equitec-Bio, Ingram, TX, USA), 100 U/ml penicillin G, and streptomycin (Invitrogen). The cells were incubated in 5% CO2 at 37˚C and passaged every 3-4 days, except for MKN7, which was subcultured at 7-day intervals.
RNA extraction and complementary DNA (cDNA) synthesis. Total RNA isolated from cancer tissue and adjacent normal mucosa was prepared with the use of Trizol (Gibco, Life Tech, Gaithersburg, MD, USA). Complementary DNA (cDNA) was synthesized from 0.4 mg of total RNA with the use of an iScript cDNA synthesis kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA). After synthesis, the cDNA was diluted to 0.2 mg/ml with water and stored at −20˚C until use.
Oligonucleotide primers for TNS4 cDNA amplification by reverse–transcription polymerase chain reaction (RT-PCR). The oligonucleotide primers for TNS4 were as follows: sense primer 5’-CACCATGAAGTTCGTGATG-3’ and antisense primer 5’-CGGTA TGAAGAGCTGTCC-3’. We used β-actin (ACTB) as the internal control. The oligonucleotide primers for ACTB were as follows: sense primer 5’-AGTTGCGTTACACCCTTTCTTGAC-3’ and antisense primer 5’-GCTCGCTCCAACCGACTGC-3’. Amplifi-cation of TNS4 was performed for 40 cycles of 1 min at 95˚C, 1 min at 60˚C and 1 min at 72˚C. Amplification of ACTB was performed for 40 cycles of 1 min at 95˚C, 1 min at 60˚C, and 1 min at 72˚C. A 10-μl aliquot of each amplified PCR product was electrophoresed on a 3% agarose gel containing ethidium bromide.
Quantitative real-time RT-PCR. Quantitative real-time PCR was performed with iQ SYBR Green Supermix (Bio-Rad Laboratories). PCR reactions were carried out in a total volume of 15 μl, which included 0.2 μg of cDNA, 0.4 μM of each primer, 7.5 μl of iQ SYBR Green Supermix containing dATP, dCTP, dGTP and dTTP at concentrations of 400 μM each, as well as 50 units/ml of iTag DNA polymerase. The PCR consisted of 3 min at 95˚C, followed by 40 cycles of denaturation of the cDNA for 10 sec at 95˚C, annealing for 10 sec at 60˚C (60˚C for ACTB) and a primer extension for 20 sec at 72˚C, followed by 10 min at 72˚C. To distinguish specific from nonspecific products and primer dimmers, melting curve analyses were performed. To evaluate specific mRNA expression in samples, a standard curve was produced for each run, measuring three points of the human control cDNA (Clontech Laboratories, Inc., CA, USA). The concentration of each sample was calculated by relating its crossing point to a standard curve.
Immunohistochemical analysis. Immunohistochemical studies of TNS4 were performed on formalin-fixed, paraffin-embedded surgical specimens obtained from patients with gastric cancer. The tissue sections were deparaffinized and soaked in 10 mM citrate buffer (pH 6.0) at 121˚C for 15 min to retrieve cell antigens. After blocking, the sections were incubated overnight at 4˚C to allow antigen–antibody reactions to occur. Peroxidase-labeled polymer (EnVision+, rabbit; DAKO, Glostrup, Denmark) was used to detect signals of antigen–antibody reactions. All sections were counterstained with hematoxylin. Primary polyclonal antibodies against TNS4 (Atlas Antibodies, Stockholm, Sweden) were used at a dilution of 1:10.
Statistical analysis. Study samples were divided into two groups (low-expression group, n=121; high-expression group, n=13) according to the TNS4 expression level. Gene expression levels of gastric cancer were then compared with those of adjacent normal mucosa with the use of the Wilcoxon test. A univariate Cox proportional-hazards model was used to evaluate the relations of OS to TNS4 and potential prognostic variables, including age, gender, tumor size, histological type, depth of invasion, lymph node metastasis, lymphatic invasion, venous invasion, and tumor node metastasis (TNM) stage. Cutoff points for TNS4 were evaluated with the use of a multivariate Cox proportional-hazards model, including prognostic factors that were significantly related to overall survival in univariate analysis. The optimal cutoff point was selected by the minimum p-value method, whereas the internal validity of the cutoff point was evaluated with a two-fold cross-validation approach (15). Relations between gene expression and potential prognostic variables were evaluated with the chi-square test. The postoperative survival rate was analyzed by the Kaplan–Meier method and differences in survival rates were assessed with the log-rank test. All p-values of less than 0.05 were considered to indicate statistical significance. All statistical analyses were performed using the Dr. SPSS II program, version 11.0.1J for Windows (SPSS, Inc., Chicago, IL, USA) and SAS, version 9.3 (SAS Institute, Cary, NC, USA).
Results
Patients. The demographic and clinical characteristics of the patients are summarized in Table I. Of the 134 patients, 94 (70.1%) had stage III disease.
Table I. Baseline patient characteristics (n=134).

TNS4: Tensin 4.
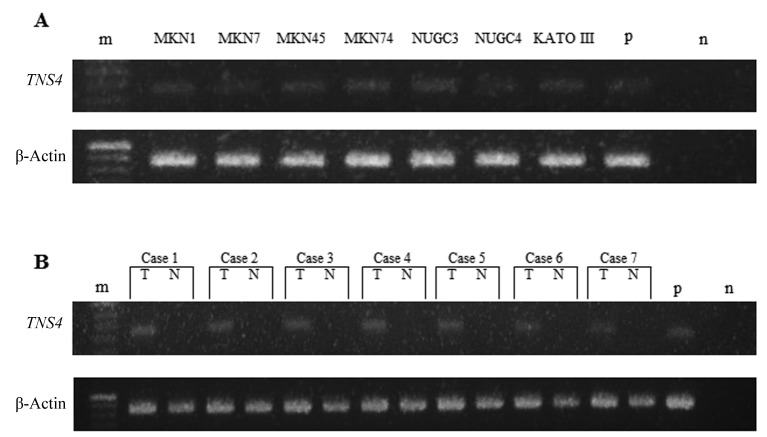
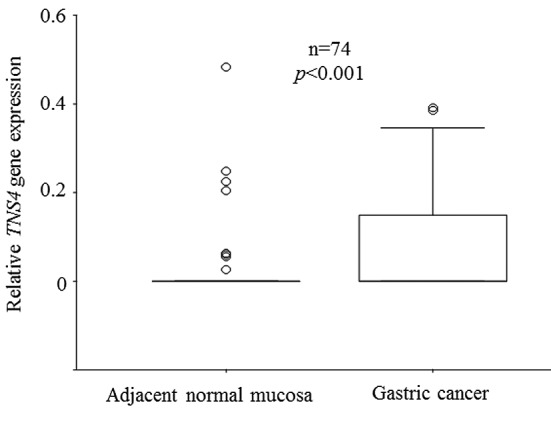
TNS4 mRNA expression in human gastric cancer. Expression of TNS4 mRNA in human gastric cancer cell lines and clinical samples was analyzed by RT-PCR (Figure 1). TNS4 mRNA expression was observed in the human gastric cell lines studied (MKN1, MKN7, MKN45, MKN74, NUGC3, NUGC4, and Kato III) (Figure 1A). We also performed RT-PCR analysis of TNS4 in specimens of gastric cancer and paired normal adjacent mucosa obtained from seven patients. TNS4 mRNA expression in these clinical samples was significantly higher in cancer tissue than in paired normal adjacent mucosa (Figure 1B). We confirmed TNS4 mRNA expression in clinical samples (n=74) by real-time RT-PCR. Expression levels of the TNS4 gene were higher in cancer than in adjacent normal mucosa (p<0.001; Figure 2).
Figure 1. Tensin 4 (TNS4) and β-actin mRNA expression in seven gastric cancer cell lines (a) and clinical samples (b) on reverse-transcription polymerase chain reaction. T: Tumor, N: adjacent normal mucosa, n: negative control, p: positive control, m: ladder. The product sizes of TNS4 and β-actin were 129 bp and 171 bp, respectively.
Figure 2. Comparison of tensin 4 (TNS4) expression between gastric cancer tissue and adjacent normal mucosa. Box boundaries: 25th and 75th percentiles of the observed values; capped bars: 10th and 90th percentiles; solid line: median; circle: outlier. p-Values were calculated by the Wilcoxon signed-rank test. TNS4 gene expression was significantly higher in cancer tissue than in adjacent normal mucosa (p<0.001).
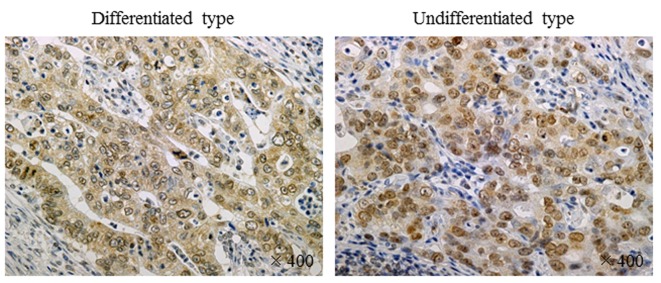
Immunohistochemistry of TNS4 expression. Expression of TNS4 protein was evaluated by immunohistochemical analysis of resected specimens of gastric cancer. Positive staining for TNS4 was mainly observed in the cytoplasm and was markedly more intense in human gastric cancer cells than in stromal cells in both differentiated and undifferentiated types of gastric cancer (Figure 3).
Figure 3. Immunohistochemistry of tensin 4 (TNS4) expression. Expression of TNS4 protein was evaluated by immunohistochemical analysis of resected specimens of gastric cancer. Positive staining for TNS4 was observed in the cytoplasm and was markedly more intense in human gastric cancer cells than in stromal cells in both differentiated and undifferentiated types of gastric cancer.
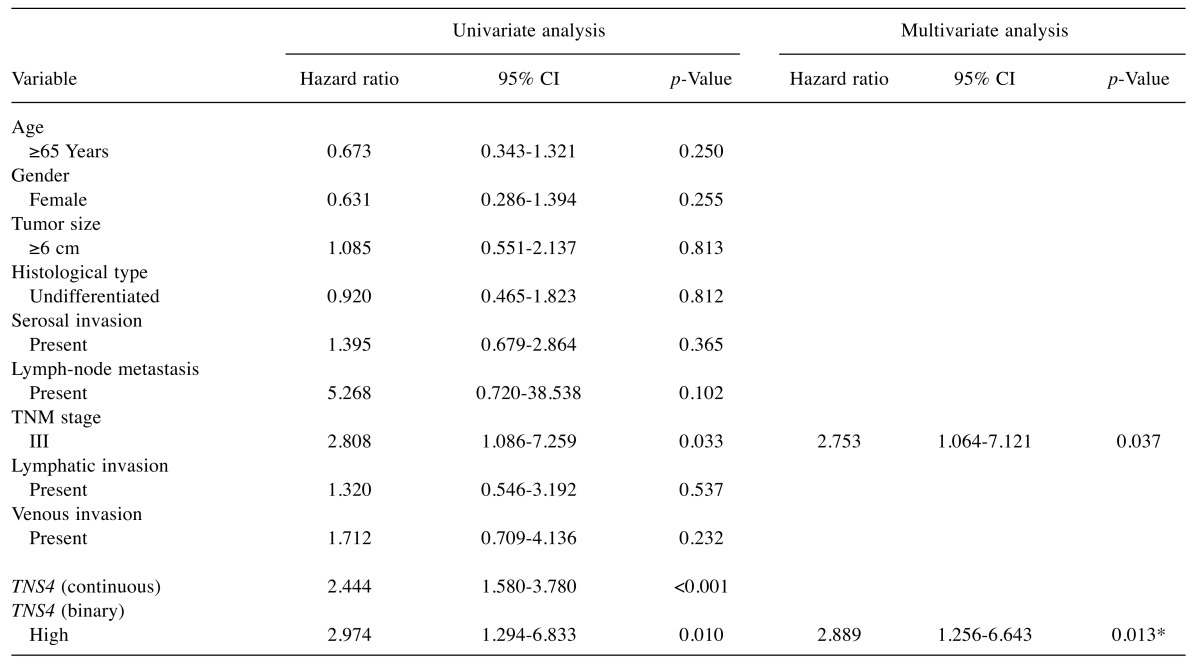
Univariate and multivariate analyses of the relation of clinicopathological factors and TNS4 mRNA expression to outcomes. TNM stage was related to OS on univariate analysis. When 0.625 was used as the cut-off point for TNS4 gene expression level, the p-value was smallest in a multivariate Cox proportional-hazards model including TNM stage. A two-fold cross validation approach showed that categorized TNS4 gene expression (p=0.013) and TNM stage (p=0.037) were significantly related to OS (Table II).
Table II. Univariate and multivariate Cox proportional hazards analysis of clinicopathological factors.
CI: Confidence interval; TNS4: tensin 4. *p-Value was calculated using two-fold validation method.
Relation of TNS4 mRNA expression to clinicopathological features. Study samples were divided into two groups (low-expression group, n=121; high-expression group, n=13) according to the expression of TNS4 mRNA using a cutoff point of 0.625. The relationships between gene expression and clinicopathological features were then examined. TNS4 gene expression levels tended to be positively related to tumor size (Table III).
Table III. Relation between tensin 4 (TNS4) gene expression and clinicopathological features.
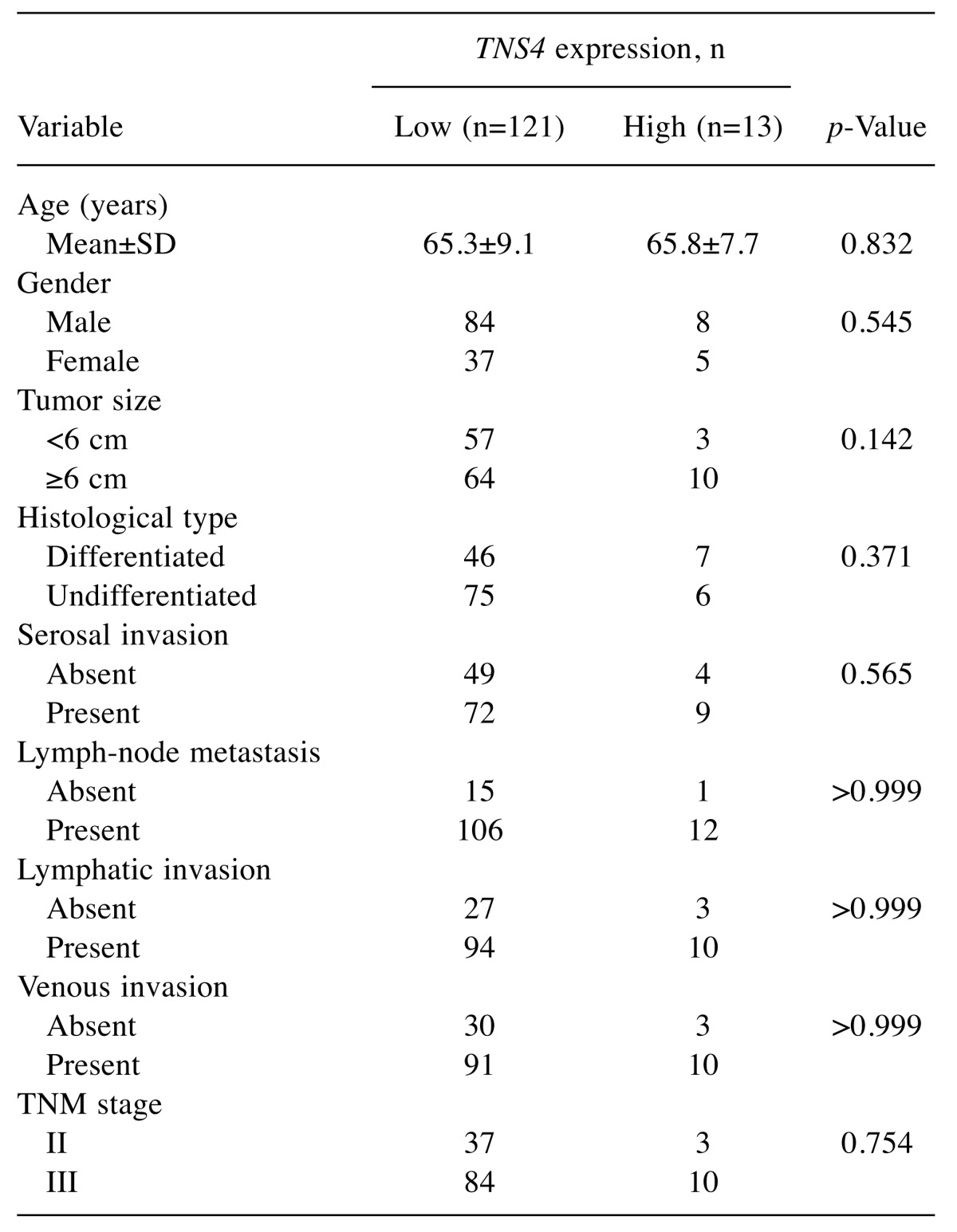
Survival curves according to TNS4 mRNA expression. In the study group as a whole, 5-year OS was poorer in patients with high TNS4 expression than in those with low TNS4 expression (log-rank p-value=0.007; Figure 4). Figure 5 shows the survival curves for the reference group of patients with stage II/III gastric cancer who underwent curative resection but did not receive adjuvant chemotherapy with S-1. There was no significant difference in survival between the patients with low TNS4 expression and those with high TNS4 expression (p=0.440).
Figure 4. Postoperative survival of patients with gastric cancer. Comparison of postoperative survival according to tensin 4 (TNS4) gene expression. In the study group as a whole, the 5-year overall survival rate was poorer in patients with high TNS4 expression (39.5%) than in those with low TNS4 expression (71.0%; log-rank p-value=0.007).
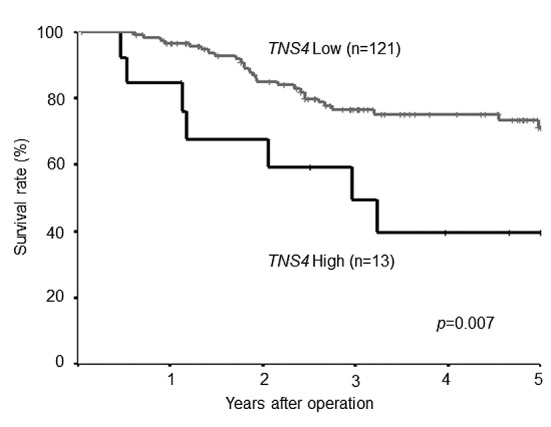
Figure 5. Postoperative survival of patients with gastric cancer not treated with S-1 adjuvant chemotherapy. Comparison of postoperative survival according to tensin 4 (TNS4) gene expression. There was no difference in survival between the patients with low TNS4 expression (62.5%) and those with high TNS4 expression (64.3%) (log-rank p-value=0.44).
Discussion
In this study, we measured TNS4 gene expression in cancer tissue and adjacent normal mucosa in patients with stage II/III gastric cancer who received curative resection followed by adjuvant chemotherapy with S-1 and examined the relationships of relative TNS4 gene expression to clinicopathological factors and treatment outcomes.
We first compared relative mRNA expression levels of the TNS4 gene between gastric cancer tissue and adjacent normal mucosa. In a previous study, expression levels of TNS4 mRNA were higher in colorectal cancer than in normal mucosa (13,14). Another study found that TNS4 mRNA levels in gastric cancer were significantly higher than those in the matched mucosa (16). Our results are consistent with these previous findings: expression levels of TNS4 mRNA were significantly higher in 74 specimens of gastric cancer tissue than in paired adjacent normal mucosa.
We next assessed the relation between TNS4 gene expression levels and outcomes in gastric cancer. Breast cancer-specific survival was reported to be significantly shorter in patients with invasive breast cancer with high TNS4 expression than in those with low TNS4 expression (17). In colon cancer, a previous study showed that patients with high levels of TNS4 transcripts had significantly poorer outcomes than those with low levels (18). Overall survival has been reported to be shorter in patients with gastric cancer with high TNS4 expression than in those with low TNS4 expression (16). Our results confirm and extend the findings of these previous studies. Overall survival at 5 years was significantly poorer in patients with high TNS4 gene expression than in those with low TNS4 gene expression. On univariate and multivariate Cox proportional-hazards regression analysis, a higher level of TNS4 gene expression was a significant independent predictor of 5-year survival in patients with stage II/III gastric cancer after curative surgery and adjuvant chemotherapy with S-1.
We finally examined the relation between TNS4 mRNA expression levels and clinicopathological features in gastric cancer. A previous study reported that overexpression of TNS4 correlated with tumor size, histological grade, and axillary nodal involvement in invasive breast carcinoma (17). In colorectal cancer, high expression of TNS4 was reported to be associated with advanced Duke’s stage and distant metastasis (18). Increased TNS4 mRNA expression was found to significantly correlate with histological grade, serosal invasion, lymph-node metastasis, and peritoneal dissemination in gastric cancer (16). In our study, TNS4 mRNA expression levels tended to be related to tumor size.
The reason why TNS4 gene expression is associated with prognostic factors for stage II/III gastric cancer after curative resection and adjuvant chemotherapy with S-1 remains to be fully elucidated. Available evidence suggests that two main factors are involved. One is the effect of TNS4 on cancer cell motility, and the other is the relation between TNS4 and epidermal growth factor receptor (EGFR), which was a prognostic factor in the biomarker analysis of the ACTS-GC study (19).
Recent in vitro investigations reported that genes of the tensin family positively regulate cell motility and migration (20-23). Previous studies have reported that TNS4 correlates with altered cell morphology and increased cell motility attributed to repression of E-cadherin protein (13). Albasri et al. reported that TNS4 expression was a prognostic factor in colorectal cancer and that a TNS4–integrin-linked kinase (ILK) pathway controlled cell motility and possibly promoted metastasis by knocking down TNS4 and ILK in vitro (18). Moreover, they demonstrated that TNS4 can influence metastatic processes in vivo by intrasplenic injection into nude mice of colorectal cancer cells stably transfected with a TNS4 expression vector, which resulted in shorter OS than in the control.
The biomarker analysis in the ACTS-GC study showed that EGFR was positively associated with poor outcomes in patients with stage II/III gastric cancer after curative resection and adjuvant chemotherapy with S-1 (19). Previous studies have reported that the Src homology 2 (SH2) domain of TNS4 binds to tyrosine phosphorylated c-Casitas B-lineage lymphoma (c-CBl), and this interaction reduces the level of EGFR ubiquitination and degradation, thereby prolonging EGFR signaling, which enhances the tumorigenicity of cancer cells (24,25). This mechanism is thought to explain why TNS4 would be a prognostic factor in patients with stage II/III gastric cancer after curative resection and adjuvant chemotherapy with S-1.
In conclusion, TNS4 gene expression levels were higher in gastric cancer tissue than in adjacent normal mucosa, and high expression of this gene was significantly related to poor outcomes in patients with stage II/III gastric cancer after curative resection and adjuvant chemotherapy with S-1. Our findings suggest that overexpression of the TNS4 gene is a useful independent predictor of outcomes in patients with stage II/III gastric cancer who undergo curative resection and receive adjuvant chemotherapy with S-1.
Acknowledgements
The Authors thank Kazue Yoshihara for her technical support.
References
- 1.World Health Organization International Agency for Research on Cancer Stomach Cancer Estimated Incidence, Mortality and Prevalence Worldwide in 2012. 2014 http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx. Accessed 13 April 2014. [Google Scholar]
- 2.Bang YJ, Kim YW, Yang HK, Chung HC, Park YK, Lee KH, Lee KW, Kim YH, Noh SI, Cho JY, Mok YJ, Ji J, Yeh TS, Button P, Sirzen F, Noh SH. Adjuvant capecitabine and oxaliplatin for gastric cancer after D2 gastrectomy (CLASSIC): a phase 3 open-label, randomised controlled trial. Lancet. 2012;379:315–321. doi: 10.1016/S0140-6736(11)61873-4. [DOI] [PubMed] [Google Scholar]
- 3.Sakuramoto S, Sasako M, Yamaguchi T, Kinoshita T, Fujii M, Nashimoto A, Furukawa H, Nakajima T, Ohashi Y, Imamura H, Higashino M, Yamamura Y, Kurita A, Arai K. Adjuvant chemotherapy for gastric cancer with S-1, an oral fluoropyrimidine. N Engl J Med. 2007;357:1810–1820. doi: 10.1056/NEJMoa072252. [DOI] [PubMed] [Google Scholar]
- 4.Sasako M, Sakuramoto S, Katai H, Kinoshita T, Furukawa H, Yamaguchi T, Nashimoto A, Fujii M, Nakajima T, Ohashi Y. Five-year outcomes of a randomized phase III trial comparing adjuvant chemotherapy with S-1 versus surgery alone in stage II or III gastric cancer. J Clin Oncol. 2011;29:4387–4393. doi: 10.1200/JCO.2011.36.5908. [DOI] [PubMed] [Google Scholar]
- 5.Al-Ghamdi S, Albasri A, Cachat J, Ibrahem S, Muhammad BA, Jackson D, Nateri AS, Kindle KB, Ilyas M. Cten is targeted by Kras signalling to regulate cell motility in the colon and pancreas. PLoS One. 2011;6:e20919. doi: 10.1371/journal.pone.0020919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lo SH, Lo TB. CTEN, a COOH-terminal tensin-like protein with prostate restricted expression, is down-regulated in prostate cancer. Cancer Res. 2002;62:4217–4221. [PubMed] [Google Scholar]
- 7.Chen H, Ishii A, Wong WK, Chen LB, Lo SH. Molecular characterization of human tensin. Biochem J. 2000;351:403–411. [PMC free article] [PubMed] [Google Scholar]
- 8.Lo SH. Tensin. Int J Biochem Cell Biol. 2004;36:31–34. doi: 10.1016/s1357-2725(03)00171-7. [DOI] [PubMed] [Google Scholar]
- 9.Lo SH, Janmey PA, Hartwig JH, Chen LB. Interactions of tensin with actin and identification of its three distinct actin-binding domains. J Cell Biol. 1994;125:1067–1075. doi: 10.1083/jcb.125.5.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katz M, Amit I, Citri A, Shay T, Carvalho S, Lavi S, Milanezi F, Lyass L, Amariglio N, Jacob-Hirsch J, Ben-Chetrit N, Tarcic G, Lindzen M, Avraham R, Liao YC, Trusk P, Lyass A, Rechavi G, Spector NL, Lo SH, Schmitt F, Bacus SS, Yarden Y. A reciprocal tensin-3-cten switch mediates EGF-driven mammary cell migration. Nat Cell Biol. 2007;9:961–969. doi: 10.1038/ncb1622. [DOI] [PubMed] [Google Scholar]
- 11.Sasaki H, Yukiue H, Kobayashi Y, Fukai I, Fujii Y. CTEN mRNA expression is correlated with tumor progression in thymoma. Tumour Biol. 2003;24:271–274. doi: 10.1159/000076141. [DOI] [PubMed] [Google Scholar]
- 12.Sasaki H, Moriyama S, Mizuno K, Yukiue H, Konishi A, Yano M, Kaji M, Fukai I, Kiriyama M, Yamakawa Y, Fujii Y. CTEN mRNA expression was correlated with tumor progression in lung cancers. Lung Cancer. 2003;40:151–155. doi: 10.1016/s0169-5002(03)00037-0. [DOI] [PubMed] [Google Scholar]
- 13.Albasri A, Seth R, Jackson D, Benhasouna A, Crook S, Nateri AS, Chapman R, Ilyas M. C-Terminal Tensin-like (CTEN) is an oncogene which alters cell motility possibly through repression of E-cadherin in colorectal cancer. J Pathol. 2009;218:57–65. doi: 10.1002/path.2508. [DOI] [PubMed] [Google Scholar]
- 14.Liao YC, Chen NT, Shih YP, Dong Y, Lo SH. Up-regulation of C-terminal tensin-like molecule promotes the tumorigenicity of colon cancer through beta-catenin. Cancer Res. 2009;69:4563–4566. doi: 10.1158/0008-5472.CAN-09-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mazumdar M, Smith A, Bacik J. Methods for categorizing a prognostic variable in a multivariable setting. Stat Med. 2003;22:559–571. doi: 10.1002/sim.1333. [DOI] [PubMed] [Google Scholar]
- 16.Sakashita K, Mimori K, Tanaka F, Kamohara Y, Inoue H, Sawada T, Hirakawa K, Mori M. Prognostic relevance of tensin 4 expression in human gastric cancer. Ann Surg Oncol. 2008;15:2606–2613. doi: 10.1245/s10434-008-9989-8. [DOI] [PubMed] [Google Scholar]
- 17.Albasri A, Aleskandarany M, Benhasouna A, Powe DG, Ellis IO, Ilyas M, Green AR. CTEN (C-terminal tensin-like), a novel oncogene overexpressed in invasive breast carcinoma of poor prognosis. Breast Cancer Res Treat. 2011;126:47–54. doi: 10.1007/s10549-010-0890-3. [DOI] [PubMed] [Google Scholar]
- 18.Albasri A, Al-Ghamdi S, Fadhil W, Aleskandarany M, Liao YC, Jackson D, Lobo DN, Lo SH, Kumari R, Durrant L, Watson S, Kindle KB, Ilyas M. Cten signals through integrin-linked kinase (ILK) and may promote metastasis in colorectal cancer. Oncogene. 2011;30:2997–3002. doi: 10.1038/onc.2011.26. [DOI] [PubMed] [Google Scholar]
- 19.Terashima M, Kitada K, Ochiai A, Ichikawa W, Kurahashi I, Sakuramoto S, Katai H, Sano T, Imamura H, Sasako M. Impact of expression of human Epidermal growth factor receptors EGFR and ERBB2 on survival in stage II/III gastric cancer. Clin Cancer Res. 2012;18:5992–6000. doi: 10.1158/1078-0432.CCR-12-1318. [DOI] [PubMed] [Google Scholar]
- 20.Ishii A, Lo SH. A role of tensin in skeletal-muscle regeneration. Biochem J. 2001;356:737–745. doi: 10.1042/0264-6021:3560737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H, Duncan IC, Bozorgchami H, Lo SH. Tensin 1 and a previously undocumented family member, tensin 2, positively regulate cell migration. Proc Natl Acad Sci USA. 2002;99:733–738. doi: 10.1073/pnas.022518699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen H, Lo SH. Regulation of tensin-promoted cell migration by its focal adhesion binding and Src homology domain 2. Biochem J. 2003;370:1039–1045. doi: 10.1042/BJ20021308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamashita M, Horikoshi S, Asanuma K, Takahara H, Shirato I, Tomino Y. Tensin is potentially involved in extracellular matrix production in mesangial cells. Histochem Cell Biol. 2004;121:245–254. doi: 10.1007/s00418-004-0626-8. [DOI] [PubMed] [Google Scholar]
- 24.Hong SY, Shih YP, Li T, Carraway KL 3rd, Lo SH. CTEN prolongs signaling by EGFR through reducing its ligand-induced degradation. Cancer Res. 2013;73:5266–5276. doi: 10.1158/0008-5472.CAN-12-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lo SH. C-Terminal tensin-like (CTEN): A promising biomarker and target for cancer. Int J Biochem Cell Biol. 2014;51:150–154. doi: 10.1016/j.biocel.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]