Parahydrogen is used to give efficient NMR detection of array of amines, amides, alcohols, carboxylates, carbonates, and phosphates.
Abstract
Hyperpolarization turns weak nuclear magnetic resonance (NMR) and magnetic resonance imaging (MRI) responses into strong signals, so normally impractical measurements are possible. We use parahydrogen to rapidly hyperpolarize appropriate 1H, 13C, 15N, and 31P responses of analytes (such as NH3) and important amines (such as phenylethylamine), amides (such as acetamide, urea, and methacrylamide), alcohols spanning methanol through octanol and glucose, the sodium salts of carboxylic acids (such as acetic acid and pyruvic acid), sodium phosphate, disodium adenosine 5′-triphosphate, and sodium hydrogen carbonate. The associated signal gains are used to demonstrate that it is possible to collect informative single-shot NMR spectra of these analytes in seconds at the micromole level in a 9.4-T observation field. To achieve these wide-ranging signal gains, we first use the signal amplification by reversible exchange (SABRE) process to hyperpolarize an amine or ammonia and then use their exchangeable NH protons to relay polarization into the analyte without changing its identity. We found that the 1H signal gains reach as high as 650-fold per proton, whereas for 13C, the corresponding signal gains achieved in a 1H-13C refocused insensitive nuclei enhanced by polarization transfer (INEPT) experiment exceed 570-fold and those in a direct-detected 13C measurement exceed 400-fold. Thirty-one examples are described to demonstrate the applicability of this technique.
INTRODUCTION
Nuclear magnetic resonance (NMR) is one of the most powerful methods for the study of materials, and magnetic resonance imaging (MRI) plays a vital role in clinical diagnosis. However, the low sensitivity of these techniques limits their applicability. The hyperpolarization method dynamic nuclear polarization (DNP) improves the detectability of analytes such as pyruvate to the level that the MRI-based diagnosis of disease is now possible (1). Parahydrogen (p-H2), which is cheap to prepare and exists as a pure nuclear spin state, was shown to enhance the strength of an NMR signal in 1987 (2), although these methods have not yet been used clinically. This may reflect the fact that p-H2 was originally used to sensitize chemically modified hydrogenation products (3, 4), and only recently has a method been developed where the original identity of the sensitized analyte is retained (5). This approach, signal amplification by reversible exchange (SABRE), harnesses p-H2 in the form of metal-bound hydride ligands and transfers hyperpolarization into a weakly bound substrate (6–8) via the small J-couplings that connect them (9). Ligand exchange then builds up a pool of hyperpolarized substrate according to Scheme 1A (10). SABRE is successful for analytes with multiple bonds to nitrogen such as nicotinamide (11), isoniazid (12), pyrazole (13), and acetonitrile (14), with 1H polarizations of 50% (11) and 15N values of 20% (15) being achieved. Furthermore, although it works for other nuclei (11, 16–20), it fails to sensitize many classes of analytes.
Scheme 1. (A) Hyperpolarization via SABRE and (B) hyperpolarization via SABRE-RELAY.
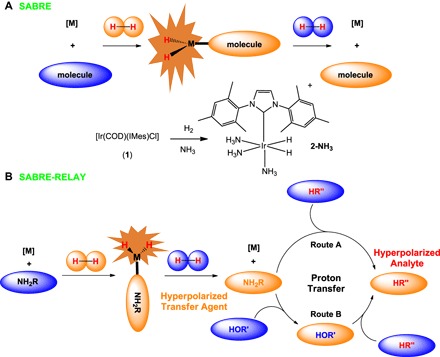
SABRE is used to hyperpolarize the transfer agent NH2R, where R is H or CH2Ph or CH2CH2Ph (etc.), which relays polarization to the analyte (HR″, route A), and R″ is amide, carboxyl, phosphate, or alkoxide (etc.). This process involves both proton exchange and spin-spin interactions and may be mediated by an intermediary HOR′, where R′ is H or suitable scaffold (route B). Center: Reaction scheme shows the formation of SABRE active 2-NH3, which leads to NH3.
Here, we describe a method where p-H2 hyperpolarizes a range of amines, amides, carboxylic acids, alcohols, phosphates, and carbonates without changing their chemical identity. Our method starts with the hyperpolarization of ammonia (the hyperpolarization transfer agent). Subsequently, polarization is relayed into the specified analyte through proton exchange, as outlined in Scheme 1B. Spontaneous low-field transfer then creates the hyperpolarized analyte, which we detect. We called this approach SABRE-RELAY and predict that, when it is fully optimized, it will have a major impact on NMR and MRI in accordance with the fact that we exemplify it for 31 analytes.
RESULTS
We achieve SABRE-RELAY by reacting ammonia with the most versatile of the current SABRE catalysts, [IrCl(COD)(IMes)] (21, 22) (1) [where IMes is 1,3-bis(2,4,6-trimethylphenyl)imidazol-2-ylidene and COD is cycloocta-1,5-diene] and H2, to form [Ir(H)2(IMes)(NH3)3]Cl (2-NH3) according to Scheme 1. When this reaction is competed in dichloromethane-d2, 2-NH3 exhibits equatorial and axial NH3 ligand signals at δ 2.19 and 2.88 in the corresponding 1H NMR spectrum, alongside a broad NH3 response at δ 0.47, as detailed in Fig. 1A. When this sample is examined after exposure to a 2-bar pressure of p-H2 gas at 60 G, the resulting 1H NMR signal for free NH3 now shows an ~10-fold signal enhancement per proton, with the bound NH3 ligand signal at δ 2.19 showing a 3-fold enhanced response. These observations confirm that 2-NH3 undergoes SABRE to produce hyperpolarized ammonia. When the same process is repeated in methanol-d4, 2-NH3 exhibits a hydride resonance at δ −23.2 that rapidly separates into several components as H-D exchange proceeds to form an array of isotopologues. However, when p-H2 is used, a hyperpolarized NMR signal is readily seen at δ 5.06 for the exchangeable proton of CD3OH, which exhibits a 32-fold intensity gain over its thermally equilibrated signal. Therefore, we added a 5% loading of H2O, relative to iridium, to the CD2Cl2 sample and reexamined it. Under these conditions, the free NH3 signal gain resulting from SABRE proved to increase to 40-fold per proton, whereas the corresponding equatorial ligand signal now showed an 85-fold per proton gain (Fig. 1B). In addition, the free H2O signal was enhanced by 75-fold per proton, a result that compares well with other solvent signal enhancements (23–25).
Fig. 1. Hyperpolarization of NH3 under SABRE.
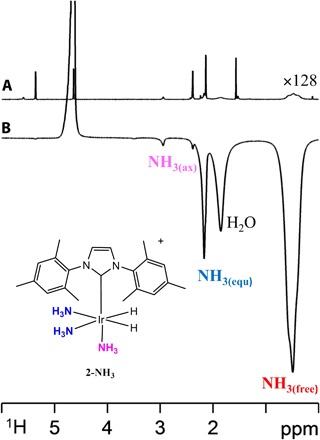
(A) Thermally polarized control 1H NMR spectrum showing peaks for 2-NH3, NH3, and H2 at 298 K in dichloromethane-d2, ×128 vertical expansion relative to (B). (B) Corresponding single-scan 1H NMR spectrum in the presence of p-H2, with the hyperpolarized responses for H2O, NH3(free), Ir-NH3(equatorial), and Ir-NH3(axial) of 2-NH3 indicated.
Exchange spectroscopy measurements were then used to confirm that free NH3 and the equatorially bound NH3 ligand of 2-NH3 are in chemical exchange, with the observation of further exchange peaks between free NH3 and H2O demonstrating the rapid transfer of protons between them. On the basis of this selectivity, we conclude that, when the ammonia is bound, proton exchange between NH3 and H2O is suppressed because the nitrogen lone pair is involved in bonding to the metal center. Consequently, it now becomes hyperpolarized by SABRE. Proton exchange proceeds, though, after NH3 dissociation, and this leads to the observation of hyperpolarization in the chemical exchange–averaged response of H2O (or HOCD3) according to Scheme 1B. Now, we show how it is possible to harness this proton exchange process to hyperpolarize the NMR signals of a series of added analytes.
First, we consider whether the SABRE hyperpolarization of NH3 can be relayed into the 1H and 13C responses of a series of alcohols CH3(CH2)nOH (where n = 0 to 7). To do this, we prepared a range of dichloromethane-d2 solutions that contained [Ir(H)2(IMes)(NH3)3]Cl (2-NH3), NH3, and 1 μl of each alcohol (typical concentration, 20 mM). After hyperpolarization transfer from p-H2, strong signals resulted in the associated single-scan 1H NMR spectra, which reached up to 650-fold intensity gains per alcohol CH proton for 1-propanol, averaging at 265 across the series (see the Supplementary Materials). When the same p-H2 transfer process was undertaken and a fully coupled 13C NMR measurement was made instead of a 1H NMR measurement, molecule-diagnostic 13C and 1H-13C refocused insensitive nuclei enhanced by polarization transfer (INEPT)–based responses could also be recorded in one scan at 9.4 T for all the alcohols, as illustrated in Fig. 2B for 1-pentanol, with the associated signal gains reaching 570-fold for the Cα signal of 1-hexanol. The SABRE-RELAY effect results in the detection of hyperpolarized NMR signals for all the spin-1/2 nuclei in these molecules. In addition, as with SABRE, the hyperpolarized NMR terms reflect a mixture of longitudinal single-spin and higher-order states, whose relative amplitudes depend on the magnetic field that the sample experiences during the polarization transfer step (16, 26). Furthermore, by reducing the concentrations of these analytes below the concentration of NH3, it is possible to improve on SABRE-RELAY efficiency. This is beneficial when studying low-concentration analytes because when propanol was studied, the 1H NMR signal gains seen for its OH resonance increased by 100% on moving from a 15 to 1.5 mM concentration (fig. S4), whereas its CH resonances showed a ca. 50% improvement in enhancement level; a 1H-13C refocused INEPT response was still clearly visible in one scan, where the three signals from the OH end were 639, 538, and 603 times larger, respectively, than those in the corresponding 13C response. This polarization transfer method is also applicable to complex branched alcohols, and when a sample of 13C-labeled glucose was analyzed, a single-scan 13C response could be seen for all the expected α and β form signals, which serves to illustrate the wider significance of this effect (fig. S15E). Furthermore, our studies show that, when SABRE-RELAY is carried out under anhydrous conditions with straight-chain alcohols, superior results are obtained.
Fig. 2. Single-scan NMR spectra of 15.3 mM pentanol (CH3CH2CH2CH2CH2OH, color-coded structure shown) in dichloromethane-d2 solution resulting from the action of NH3, 2-NH3, and p-H2.
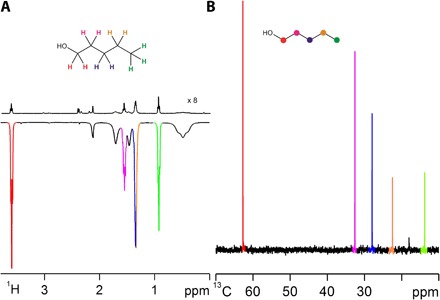
(A) Upper 1H NMR spectrum in the thermally polarized control, ×8 vertical expansion, relative to lower SABRE-RELAY spectrum. (B) Single-scan SABRE-RELAY 1H-13C refocused INEPT NMR spectrum (see fig. S8B for the corresponding thermal control trace).
Our next goal was to expand on the range of materials that can be sensitized by this method. We started with pyruvic acid but found that its addition to a solution of 2-NH3 and NH3 resulted in ammonium salt precipitation, which acted to limit hyperpolarization efficacy. This can be overcome by the addition of a pH modifier such as Cs2CO3, but working with the corresponding sodium salt proved optimal. When 13C-labeled sodium pyruvate, acetate, or propanoic acid samples were studied in the presence of p-H2, strong 1H and 13C signals were seen; the 13C signal gain for propionic acid was 109-fold. Furthermore, sodium dihydrogen phosphate, adenosine 5′-triphosphate disodium, and 13C-labeled sodium hydrogen carbonate provided strong 31P and 13C responses (Fig. 3, A and B), whereas the amides acetamide, urea, and methacrylamide showed substantial 1H, 13C, and 15N signal gains; for urea, a 13C signal gain of 408-fold was observed. These studies could be completed with NH3/H2O or NH3/CH3OH, as detailed in the Supplementary Materials, to promote the necessary proton exchange, and the observations establish that analytes containing the four common functional groups—OH, NH2CO, POH, and COOH—can be used. In some cases, we see evidence for Schiff-base condensation at long reaction times but could suppress this process by adding water.
Fig. 3. Single-scan SABRE-RELAY NMR spectra recorded in dichloromethane-d2 with NH3 and 2-NH3 in the presence of p-H2.
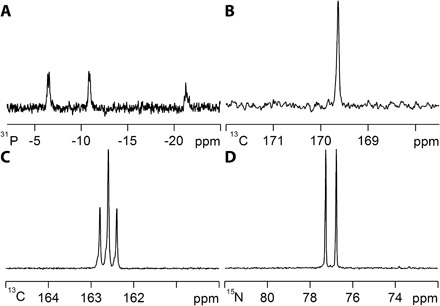
(A) Sodium adenosine 5′-triphosphate, 1H-31P refocused INEPT spectrum (OH transfer) and (B) sodium 13C–labeled pyruvate, 13C NMR spectrum. Single-scan SABRE-RELAY NMR spectra recorded with PEA and 2-PEA in the presence of p-H2 for (C) 15N-13C–labeled urea, 13C NMR spectrum, 25 mM concentration, and (D) 15N-13C–labeled urea, 15N NMR spectrum, 25 mM concentration. The corresponding thermally polarized spectra are detailed in figs. S29A, S18A, S23A, and S23C and yield no signal.
To examine the role of the hyperpolarization transfer agent, we replaced NH3 with benzylamine (BnNH2) or phenethylamine (PEA). Both react with 1 and H2, forming [Ir(H)2(IMes)(NH2Bn)3]Cl (2-BnNH2) and [Ir(H)2(IMes)(PEA)3]Cl (2-PEA), respectively. For the corresponding 2-BnNH2 sample, signal gains for free BnNH2 of 72-fold (NH), 53-fold (CH), and 170-fold (aromatic), respectively, per proton are observed (Fig. 4), and these measurements can be repeated if the same sample is probed with p-H2 several days after the first observation was made. PEA proved to perform better than BnNH2, with the corresponding NH2 signal gain being 108-fold per proton for a 10-fold loading of 1 with signal gains of 50-fold (NCH2), 45-fold (CH2), 92-fold (ortho), 50-fold (meta), and 20-fold (para) resulting for the other groups. These observations show how polarization transfer through the aliphatic carbon chain into the aromatic protons is possible. BnNH2 and PEA also proved suitable for SABRE-RELAY. In the case of PEA, the efficiency of urea hyperpolarization was found to improve (Fig. 3, C and D) over that achieved with NH3, although the measured response of 13C-labeled glucose was found to reduce. Furthermore, replacing BnNH2 with its d7-form, C6D5CD2NH2, led to further improvements in observed analyte response level because the initially created SABRE hyperpolarization was now optimally focused into just the NH2 protons.
Fig. 4. Hyperpolarization of benzylamine (BnNH2) under SABRE.
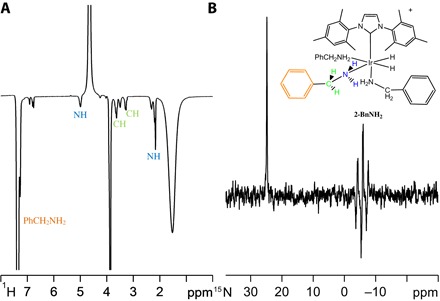
(A) 1H NMR spectrum of hyperpolarized BnNH2 achieved via 2-BnNH2 (top right) under SABRE in dichloromethane-d2 solution after transfer at 60 G (see fig. S32A for the corresponding thermally equilibrated NMR spectrum). The enhanced signals are color-coded; NH2 (blue), CH2 (light green), and Ph (orange) for the bound equatorial BnNH2 ligand of 2-BnNH2, NH2 and CH2 of the free material, and H2O. (B) Corresponding 15N NMR spectrum recorded using 15N-labeled BnNH2 after transfer in a μ-metal shield showing the free (left) and equatorial ligand (right) responses.
Given the wide range of amine pKb values (27), it may be possible to remove the need for an auxiliary base when dealing with acidic analytes through a process of amine variation. Therefore, we conclude that studies on the role of the amine will be important for the optimization of SABRE-RELAY and may even allow the introduction of selectivity into the hyperpolarization process. Furthermore, because improvements in analyte detectability with SABRE can be easily achieved by varying the polarization transfer field, reducing relaxation within the analyte, and optimizing the catalyst lifetime while minimizing its relaxivity, we expect the signal gains reported here to be similarly improved upon in the future (5).
DISCUSSION
In summary, we have shown that SABRE-RELAY can be used to hyperpolarize a wide range of biologically relevant materials. In the initial step, SABRE is used to enhance the NH proton response of the selected hyperpolarized transfer agent (the free amine) by between 10- and 120-fold per proton. When this is achieved in the presence of propanol, proton exchange results in its OH signal being amplified by between 250- and 500-fold. The nonequilibrium magnetic state of the OH proton is then successfully relayed into its aliphatic 1H resonances such that the corresponding signals are amplified by between 650- and 790-fold per proton. We used this 1H signal gain to record a single-scan 1H-13C refocused INEPT NMR spectrum using just 1 × 10−7 moles of material, although direct transfer to 13C means that a weaker fully coupled 13C response can also be seen. On the basis of these signal gains, we hope that this route can be developed to allow the phenotyping of urine via lower-field 13C detection in the future as an alternative to the current high-field 1H detection methods (28). However, because exchangeable protons feature heavily in biochemical NMR, we expect harnessing this effect to be of significant interest to biochemists, especially if it is augmented with high-field transfer via the “low-irradiation generation of high tesla–SABRE” approach (29). In addition, because hyperpolarized urea, glucose, and pyruvate reflect successful MRI probes of disease (30, 31), when SABRE-RELAY is coupled with catalyst removal and biocompatibility, we expect this route to become clinically important because it can theoretically deliver a continuously hyperpolarized bolus (32). Moreover, because studies of catalysis with p-H2 have made significant contributions to process optimization (33–37), we expect this approach to provide insight into important reactions such as transfer hydrogenation (38), hydroamination (39), and N2 fixation in the future (40).
MATERIALS AND METHODS
Experimental design
The measurements undertaken in this work were completed on a 400-MHz Avance III spectrometer and involved 1H, 13C, 15N, and 31P detection, as detailed in the Supplementary Materials. Enhancement values were determined according to the methods defined here, and sample details allowed the repetition of these measurements, which involved the following procedures.
SABRE-RELAY polarization transfer method with NH3
The polarization transfer experiments that were reported in this study were conducted in 5-mm NMR tubes that were equipped with a J. Young’s tap. Samples for these polarization transfer experiments were based on a 5 mM solution of [IrCl(COD)(IMes)] and the indicated substrate and NH3 loadings in methanol-d4 or dichloromethane-d2 (0.6 ml). The samples were degassed before the introduction of NH3. Subsequently, p-H2 at a pressure of ca. 3 bar was added. Then, samples were shaken for 10 s in the specified fringe field of an NMR spectrometer before they were rapidly transported into the magnet for subsequent interrogation by NMR spectroscopy. This whole process takes ca. 15 s to achieve.
SABRE-RELAY polarization transfer method with BnNH2 or PEA
The polarization transfer experiments that were reported were conducted in 5-mm NMR tubes that were equipped with a J. Young’s tap. Samples for these polarization transfer experiments were based on a 5 mM solution of [IrCl(COD)(IMes)], the indicated BnNH2 or PEA loading, and the indicated additional substrate at the specified loading in dichloromethane-d2 (0.6 ml). The samples were degassed before the introduction of p-H2 at a pressure of ca. 3 bar. Samples were then shaken for 10 s in the specified fringe field of an NMR spectrometer before they were rapidly transported into the magnet for subsequent interrogation by NMR spectroscopy.
Supplementary Material
Acknowledgments
We thank A. J. Holmes, M. Fekete, and A. Ruddlesden for their help. Funding: We thank the Wellcome Trust (092506 and 098335), the Engineering and Physical Sciences Research Council (EP/R51181X/1), and the University of York for supporting this work. Author contributions: W.I., P.J.R., and S.B.D. all contributed equally to the preparation of this manuscript. W.I. and P.J.R. collected the raw data, and all authors were involved in the interpretation of results and design of experiments. Competing interests: W.I., P.J.R., and S.B.D. are inventors on a patent application filed by the University of York based on this work (patent no. GB 1711967.8, filed 25 July 2017). The authors declare that they have no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All relevant plasmids and experimental data may be requested from the authors. Raw data can be found via DOI: 10.15124/d93f83bf-9fc7-4cf8-8267-504a0304a516.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/1/eaao6250/DC1
section S1. SABRE-RELAY polarization transfer method with NH3
section S2. SABRE-RELAY polarization transfer method with BnNH2 or PEA
section S3. Polarization enhancement quantification procedures
section S4. NMR spectrometer details
section S5. Pulse sequence details
section S6. SABRE-RELAY spectra
fig. S1. INEPT pulse sequence.
fig. S2. DEPT pulse sequence.
fig. S3. SABRE-RELAY NMR spectra methanol.
fig. S4. SABRE-RELAY NMR spectra ethanol.
fig. S5. SABRE-RELAY NMR spectra propanol.
fig. S6. SABRE-RELAY NMR spectra propanol, low concentration.
fig. S7. SABRE-RELAY NMR spectra butanol.
fig. S8. SABRE-RELAY NMR spectra pentanol.
fig. S9. SABRE-RELAY NMR spectra hexanol.
fig. S10. SABRE-RELAY NMR spectra heptanol.
fig. S11. SABRE-RELAY NMR spectra octanol.
fig. S12. SABRE-RELAY NMR spectra isopropanol.
fig. S13. SABRE-RELAY NMR spectra tert-butanol.
fig. S14. SABRE-RELAY NMR spectra d-glucose.
fig. S15. SABRE-RELAY NMR spectra d-glucose-13C.
fig. S16. SABRE-RELAY NMR spectra glycerol.
fig. S17. SABRE-RELAY NMR spectra sodium acetate-13C.
fig. S18. SABRE-RELAY NMR spectra sodium pyruvate-13C.
fig. S19. SABRE-RELAY NMR spectra sodium acetate-1,2 13C2.
fig. S20. SABRE-RELAY NMR spectra propionic acid-13C.
fig. S21. SABRE-RELAY NMR spectra sodium hydrogen carbonate-13C.
fig. S22. SABRE-RELAY NMR spectra urea-13C.
fig. S23. SABRE-RELAY NMR spectra urea-13C-15N2.
fig. S24. SABRE-RELAY NMR spectra urea-13C-15N2.
fig. S25. SABRE-RELAY NMR spectra acetamide.
fig. S26. SABRE-RELAY NMR spectra methacrylamide.
fig. S27. SABRE-RELAY NMR spectra cyclohexyl methacrylamide.
fig. S28. SABRE-RELAY NMR spectra mono sodium dihydrogen orthophosphate.
fig. S29. SABRE-RELAY NMR spectra adenosine 5′-triphosphate disodium salt.
fig. S30. SABRE-RELAY NMR spectra ammonia in methanol.
fig. S31. SABRE-RELAY NMR spectra ammonia in dichloromethane.
fig. S32. SABRE-RELAY NMR spectra benzylamine.
fig. S33. SABRE-RELAY NMR spectra benzylamine-15N.
fig. S34. SABRE-RELAY NMR spectra, mixture of urea, propanol, and PEA.
table S1. Alcohol 1H SABRE-RELAY signal enhancement values.
table S2. Alcohol 13C SABRE-RELAY signal enhancement values.
table S3. NMR data for 2-NH3.
table S4. NMR data for 2-BnNH2.
REFERENCES AND NOTES
- 1.Ardenkjaer-Larsen J. H., On the present and future of dissolution-DNP. J. Magn. Reson. 264, 3–12 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Bowers C. R., Weitekamp D. P., Parahydrogen and synthesis allow dramatically enhanced nuclear alignment. J. Am. Chem. Soc. 109, 5541–5542 (1987). [Google Scholar]
- 3.Natterer J., Bargon J., Parahydrogen induced polarization. Prog. Nucl. Magn. Reson. Spectrosc. 31, 293–315 (1997). [Google Scholar]
- 4.Green R. A., Adams R. W., Duckett S. B., Mewis R. E., Williamson D. C., Green G. G. R., The theory and practice of hyperpolarization in magnetic resonance using parahydrogen. Prog. Nucl. Magn. Reson. Spectrosc. 67, 1–48 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Adams R. W., Aguilar J. A., Atkinson K. D., Cowley M. J., Elliott P. I. P., Duckett S. B., Green G. G. R., Khazal I. G., López-Serrano J., Williamson D. C., Reversible interactions with para-hydrogen enhance NMR sensitivity by polarization transfer. Science 323, 1708–1711 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Adams R. W., Duckett S. B., Green R. A., Williamson D. C., Green G. G. R., A theoretical basis for spontaneous polarization transfer in non-hydrogenative parahydrogen-induced polarization. J. Chem. Phys. 131, 194505 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Pravdivtsev A. N., Yurkovskaya A. V., Vieth H. M., Ivanov K. L., Kaptein R., Level anti-crossings are a key factor for understanding para-hydrogen-induced hyperpolarization in SABRE experiments. ChemPhysChem 14, 3327–3331 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Pravdivtsev A. N., Yurkovskaya A. V., Ivanov K. L., Vieth H.-M., Importance of polarization transfer in reaction products for interpreting and analyzing CIDNP at low magnetic fields. J. Magn. Reson. 254, 35–47 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Eshuis N., Aspers R. L. E. G., van Weerdenburg B. J. A., Feiters M. C., Rutjes F. P. J. T., Wijmenga S. S., Tessari M., Determination of long-range scalar 1H–1H coupling constants responsible for polarization transfer in SABRE. J. Magn. Reson. 265, 59–66 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Atkinson K. D., Cowley M. J., Elliott P. I. P., Duckett S. B., Green G. G. R., López-Serrano J., Whitwood A. C., Spontaneous transfer of parahydrogen derived spin order to pyridine at low magnetic field. J. Am. Chem. Soc. 131, 13362–13368 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Rayner P. J., Burns M. J., Olaru A. M., Norcott P., Fekete M., Green G. G. R., Highton L. A. R., Mewis R. E., Duckett S. B., Delivering strong 1H nuclear hyperpolarization levels and long magnetic lifetimes through signal amplification by reversible exchange. Proc. Natl. Acad. Sci. U.S.A. 114, E3188–E3194 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng H., Xu J., Gillen J., McMahon M. T., Artemov D., Tyburn J.-M., Lohman J. A. B., Mewis R. E., Atkinson K. D., Green G. G. R., Duckett S. B., van Zijl P. C. M., Optimization of SABRE for polarization of the tuberculosis drugs pyrazinamide and isoniazid. J. Magn. Reson. 237, 73–78 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ducker E. B., Kuhn L. T., Münnemann K., Griesinger C., Similarity of SABRE field dependence in chemically different substrates. J. Magn. Reson. 214, 159–165 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Mewis R. E., Green R. A., Cockett M. C. R., Cowley M. J., Duckett S. B., Green G. G. R., John R. O., Rayner P. J., Williamson D. C., Strategies for the hyperpolarization of acetonitrile and related ligands by SABRE. J. Phys. Chem. B 119, 1416–1424 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barskiy D. A., Shchepin R. V., Coffey A. M., Theis T., Warren W. S., Goodson B. M., Chekmenev E. Y., Over 20% 15N hyperpolarization in under one minute for metronidazole, an antibiotic and hypoxia probe. J. Am. Chem. Soc. 138, 8080–8083 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mewis R. E., Atkinson K. D., Cowley M. J., Duckett S. B., Green G. G. R., Green R. A., Highton L. A. R., Kilgour D., Lloyd L. S., Lohman J. A. B., Williamson D. C., Probing signal amplification by reversible exchange using an NMR flow system. Magn. Reson. Chem. 52, 358–369 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colell J. F. P., Logan A. W. J., Zhou Z., Shchepin R. V., Barskiy D. A., Ortiz G. X. Jr, Wang Q., Malcolmson S. J., Chekmenev E. Y., Warren W. S., Theis T., Generalizing, extending, and maximizing nitrogen-15 hyperpolarization induced by parahydrogen in reversible exchange. J. Phys. Chem. C 121, 6626–6634 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barskiy D. A., Shchepin R. V., Tanner C. P. N., Colell J. F. P., Goodson B. M., Theis T., Warren W. S., Chekmenev E. Y., The absence of quadrupolar nuclei facilitates efficient 13C hyperpolarization via reversible exchange with parahydrogen. ChemPhysChem 18, 1493–1498 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Zhivonitko V. V., Skovpin I. V., Koptyug I. V., Strong 31P nuclear spin hyperpolarization produced via reversible chemical interaction with parahydrogen. Chem. Commun. 51, 2506–2509 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Burns M. J., Rayner P. J., Green G. G. R., Highton L. A. R., Mewis R. E., Duckett S. B., Improving the hyperpolarization of 31P nuclei by synthetic design. J. Phys. Chem. B 119, 5020–5027 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Weerdenburg B. J. A., Glöggler S., Eshuis N., Engwerda A. H. J. T., Smits J. M. M., de Gelder R., Appelt S., Wymenga S. S., Tessari M., Feiters M. C., Blumich B., Rutjes F. P. J. T., Ligand effects of NHC–iridium catalysts for signal amplification by reversible exchange (SABRE). Chem. Commun. 49, 7388–7390 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Lloyd L. S., Asghar A., Burns M. J., Charlton A., Coombes S., Cowley M. J., Dear G. J., Duckett S. B., Genov G. R., Green G. G. R., Highton L. A. R., Hooper A. J. J., Khan M., Khazal I. G., Lewis R. J., Mewis R. E., Roberts A. D., Ruddlesden A. J., Hyperpolarisation through reversible interactions with parahydrogen. Cat. Sci. Technol. 4, 3544–3554 (2014). [Google Scholar]
- 23.Lehmkuhl S., Emondts M., Schubert L., Spannring P., Klankermayer J., Blümich B., Schleker P. P. M., Hyperpolarizing water with parahydrogen. ChemPhysChem 18, 2426–2429 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Moreno K. X., Nasr K., Milne M., Sherry A. D., Goux W. J., Nuclear spin hyperpolarization of the solvent using signal amplification by reversible exchange (SABRE). J. Magn. Reson. 257, 15–23 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fekete M., Rayner P. J., Green G. G. R., Duckett S. B., Harnessing polarisation transfer to indazole and imidazole through signal amplification by reversible exchange to improve their NMR detectability. Magn. Reson. Chem. 55, 944–957 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Truong M. L., Theis T., Coffey A. M., Shchepin R. V., Waddell K. W., Shi F., Goodson B. M., Warren W. S., Chekmenev E. Y., 15N hyperpolarization by reversible exchange using SABRE-SHEATH. J. Phys. Chem. C 119, 8786–8797 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall H. K., Jr, Correlation of the base strengths of amines1. J. Am. Chem. Soc. 79, 5441–5444 (1957). [Google Scholar]
- 28.Beckonert O., Keun H. C., Ebbels T. M. D., Bundy J. G., Holmes E., Lindon J. C., Nicholson J. K., Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2, 2692–2703 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Theis T., Truong M., Coffey A. M., Chekmenev E. Y., Warren W. S., LIGHT-SABRE enables efficient in-magnet catalytic hyperpolarization. J. Magn. Reson. 248, 23–26 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Månsson S., Johansson E., Magnusson P., Chai C.-M., Hansson G., Petersson J. S., Ståhlberg F., Golman K., 13C imaging—A new diagnostic platform. Eur. Radiol. 16, 57–67 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Kurhanewicz J., Vigneron D. B., Brindle K., Chekmenev E. Y., Comment A., Cunningham C. H., DeBerardinis R. J., Green G. G., Leach M. O., Rajan S. S., Rizi R. R., Ross B. D., Warren W. S., Malloy C. R., Analysis of cancer metabolism by imaging hyperpolarized nuclei: Prospects for translation to clinical research. Neoplasia 13, 81–97 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hövener J.-B., Schwaderlapp N., Lickert T., Duckett S. B., Mewis R. E., Highton L. A. R., Kenny S. M., Green G. G. R., Leibfritz D., Korvink J. G., Hennig J., von Elverfeldt D., A hyperpolarized equilibrium for magnetic resonance. Nat. Commun. 4, 2946 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salnikov O. G., Kovtunov K. V., Barskiy D. A., Khudorozhkov A. K., Inozemtseva E. A., Prosvirin I. P., Bukhtiyarov V. I., Koptyug I. V., Evaluation of the mechanism of heterogeneous hydrogenation of α,β-unsaturated carbonyl compounds via pairwise hydrogen addition. ACS Catal. 4, 2022–2028 (2014). [Google Scholar]
- 34.Godard C., Duckett S. B., Polas S., Tooze R., Whitwood A. C., An NMR study of cobalt-catalyzed hydroformylation using para-hydrogen induced polarisation. Dalton Trans. 2496–2509 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Fox D. J., Duckett S. B., Flaschenriem C., Brennessel W. W., Schneider J., Gunay A., Eisenberg R., A model iridium hydroformylation system with the large bite angle ligand xantphos: Reactivity with parahydrogen and implications for hydroformylation catalysis. Inorg. Chem. 45, 7197–7209 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Blazina D., Duckett S. B., Dyson P. J., Lohman J. A. B., Direct comparison of hydrogenation catalysis by intact versus fragmented triruthenium clusters. Angew. Chem. Int. Ed. 40, 3874–3877 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Colebrooke S. A., Duckett S. B., Lohman J. A. B., Eisenberg R., Hydrogenation studies involving halobis(phosphine)-rhodium(I) dimers: Use of parahydrogen induced polarisation to detect species present at low concentration. Chemistry 10, 2459–2474 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Samec J. S. M., Bäckvall J.-E., Andersson P. G., Brandt P., Mechanistic aspects of transition metal-catalyzed hydrogen transfer reactions. Chem. Soc. Rev. 35, 237–248 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Patel M., Saunthwal R. K., Verma A. K., Base-mediated hydroamination of alkynes. Acc. Chem. Res. 50, 240–254 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Anderson J. S., Rittle J., Peters J. C., Catalytic conversion of nitrogen to ammonia by an iron model complex. Nature 501, 84–87 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/1/eaao6250/DC1
section S1. SABRE-RELAY polarization transfer method with NH3
section S2. SABRE-RELAY polarization transfer method with BnNH2 or PEA
section S3. Polarization enhancement quantification procedures
section S4. NMR spectrometer details
section S5. Pulse sequence details
section S6. SABRE-RELAY spectra
fig. S1. INEPT pulse sequence.
fig. S2. DEPT pulse sequence.
fig. S3. SABRE-RELAY NMR spectra methanol.
fig. S4. SABRE-RELAY NMR spectra ethanol.
fig. S5. SABRE-RELAY NMR spectra propanol.
fig. S6. SABRE-RELAY NMR spectra propanol, low concentration.
fig. S7. SABRE-RELAY NMR spectra butanol.
fig. S8. SABRE-RELAY NMR spectra pentanol.
fig. S9. SABRE-RELAY NMR spectra hexanol.
fig. S10. SABRE-RELAY NMR spectra heptanol.
fig. S11. SABRE-RELAY NMR spectra octanol.
fig. S12. SABRE-RELAY NMR spectra isopropanol.
fig. S13. SABRE-RELAY NMR spectra tert-butanol.
fig. S14. SABRE-RELAY NMR spectra d-glucose.
fig. S15. SABRE-RELAY NMR spectra d-glucose-13C.
fig. S16. SABRE-RELAY NMR spectra glycerol.
fig. S17. SABRE-RELAY NMR spectra sodium acetate-13C.
fig. S18. SABRE-RELAY NMR spectra sodium pyruvate-13C.
fig. S19. SABRE-RELAY NMR spectra sodium acetate-1,2 13C2.
fig. S20. SABRE-RELAY NMR spectra propionic acid-13C.
fig. S21. SABRE-RELAY NMR spectra sodium hydrogen carbonate-13C.
fig. S22. SABRE-RELAY NMR spectra urea-13C.
fig. S23. SABRE-RELAY NMR spectra urea-13C-15N2.
fig. S24. SABRE-RELAY NMR spectra urea-13C-15N2.
fig. S25. SABRE-RELAY NMR spectra acetamide.
fig. S26. SABRE-RELAY NMR spectra methacrylamide.
fig. S27. SABRE-RELAY NMR spectra cyclohexyl methacrylamide.
fig. S28. SABRE-RELAY NMR spectra mono sodium dihydrogen orthophosphate.
fig. S29. SABRE-RELAY NMR spectra adenosine 5′-triphosphate disodium salt.
fig. S30. SABRE-RELAY NMR spectra ammonia in methanol.
fig. S31. SABRE-RELAY NMR spectra ammonia in dichloromethane.
fig. S32. SABRE-RELAY NMR spectra benzylamine.
fig. S33. SABRE-RELAY NMR spectra benzylamine-15N.
fig. S34. SABRE-RELAY NMR spectra, mixture of urea, propanol, and PEA.
table S1. Alcohol 1H SABRE-RELAY signal enhancement values.
table S2. Alcohol 13C SABRE-RELAY signal enhancement values.
table S3. NMR data for 2-NH3.
table S4. NMR data for 2-BnNH2.


