Abstract
Circular RNAs (circRNAs) are a class of noncoding RNAs with continuous, covalently closed circular structures. Investigators have shown previously that circRNAs are regulators of gene expression in mammals. These tissue-specific transcripts are produced primarily by exonic or intronic sequences of housekeeping genes. Several biosynthetic models have been proposed for circRNAs, and consensus is lacking. CircRNAs are widely expressed in the cytoplasm and highly conserved, what is more, unlike other noncoding RNAs, circRNAs are relatively stable. These properties suggest special roles of circRNAs, such as microRNA (miRNA) sponges, regulators of selective splicing, or even protein-coding sequences. The expression of circRNAs is associated with many pathologic conditions; therefore, circRNAs may have utility as biomarker for the diagnosis or prediction of diseases. Authors previously have demonstrated that circRNAs can regulate the expression of a variety of disease-related miRNAs. The circRNA-miRNA-target gene interaction network regulates several pathways that inhibit or promote the occurrence of certain diseases. Based on their potential clinical relevance, circRNAs are a crucial topic of disease prevention and treatment research. Herein, we review current research regarding circRNAs and explore their potential clinical applications for thoracic diseases diagnosis and treatment.
Keywords: Circular RNAs (circRNAs), thoracic diseases, diagnosis, treatment
Introduction
In the 1970s, circular RNAs (circRNAs) were identified as byproducts of RNA transcription with high thermal stability (1,2). With the emergence of next-generation sequencing—including RNA-sequencing technology—circRNAs have been further elucidated as a new class of noncoding RNAs with regulatory functions. Unlike linear RNAs, circRNAs are characterized by covalently closed loops with stable ring structure, species conservatism, and tissue specificity; these molecules are not readily cleaved by nucleases (3). Derived from exonic or intronic gene regions, circRNAs are abundant in eukaryotes and have been garnering attention because of their specificity of expression, complexity of regulation, and important role in pathogenesis (4-7). Like microRNAs (miRNAs) and long noncoding RNAs, circRNAs have become a new research hotspot in the RNA field. This review focuses on new advances in the field of circRNAs and summarizes the relationship between circRNAs and thoracic diseases.
Biosynthetic mechanism of circRNAs
With developments in RNA-sequencing and bioinformatics analyses, the total number of known circRNAs has reached more than 30,000 (8-10). The complexity of circRNA expression in certain species is correlated with its evolutionary degree (11). Despite the large number of circRNAs and their diverse distribution among species, the precise biosynthetic mechanism of circRNAs still is unclear. The results of recent investigations have demonstrated some general biosynthetic processes of circRNAs. A rapid transcription event is coordinated by cis-regulatory elements, such as reverse complementary sequences or sequences recognized by RNA-binding proteins (RBPs), and intracellular accumulation results, similar to linear RNA biosynthesis (12). A single gene may encode multiple molecular forms of circRNAs, and once transcribed, circRNAs undergo alternative splicing (13) to yield circRNA-unique cassette exons that are absent of linear RNAs (14). The biosynthetic processes of certain tissue-specific or disease-specific circRNAs appears to be precisely regulated (15).
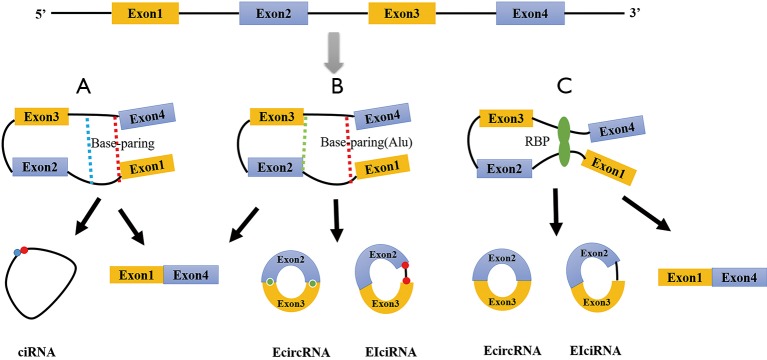
Two biosynthetic models have been proposed for circRNAs: direct back-splicing (6) and lariat-driven circularization (16,17) (Figure 1). In the model of direct back-splicing, circRNA biosynthesis involves noncanonical RNA splicing. Exons of pre-RNA connect the downstream 5' splice site to an upstream 3' splice site to generate a circRNA. Reverse-complementary sequences in introns are needed for this process (e.g., Alu elements with opposite directions) (18). The reverse-complementary sequence of introns in RNA splicing also may directly help the introns form circRNAs (19). This model can explain the biosynthesis of ciRNAs (i.e., circRNAs derived from introns), EIciRNAs (i.e., circRNAs derived from both exons and introns), and ecircRNAs (i.e., circRNAs derived from exons).
Figure 1.
Biosynthesis models of circRNAs. (A) CiRNA formation by direct back-splicing; (B) ecircRNA and EIciRNA formation by direct back-splicing; (C) ecircRNA and EIciRNA formation by lariat-driven circularization.
In the model of lariat-driven circularization, the introns “skip” multiple exons to form an O-shaped lariat during the splicing process of pre-RNA. This could isolate exon-containing RNA fragments and yield ecircRNAs or EIciRNAs after exon splicing. Some RBPs (20-22) may be involved in this process, including the RNA splicing protein Quaking (QKI), a regulator of circRNA biosynthesis during human epithelial-mesenchymal transition (20). The role of QKI in the biosynthesis of circRNAs from pre-RNA indicates that the process is not stochastic but rather is a complex system regulated by specific cellular mechanisms (20).
Studies addressing circRNAs often involve large-scale histological screening to identify and evaluate specific circRNA molecules. Many circRNAs remain uncharacterized with regard to specific conditions of biosynthesis, the dynamic process, and the relationship between other linear and circRNAs of the same corresponding gene (23).
Functions of circRNAs and their roles in thoracic diseases
Hallmarks of circRNAs are huge quantities, a high degree of sequence diversity, and tissue and disease specificity (24). As such, circRNAs have no common mode of action and regulate gene expression and protein synthesis in many different ways. The results of current research suggest that circRNAs function via four mechanisms: (I) circRNAs regulate parental gene expression in cis via polymerase II (Pol II) (5,19); (II) circRNAs can act as competitive endogenous RNAs (ceRNAs) to regulate gene expression by miRNA sponge effects (25-27); (III) circRNAs play biological functions through the formation of complexes with proteins (21,28); and (IV) circRNAs can translate protein via base-modification N6-methyladenosine (m6A) (29).
Evidence regarding the structure and functions of circRNAs has indicated that these molecules play a significant role in the pathogenesis of atherosclerosis (30), neurological disorders (31), diabetes (32), cancer (8), and other diseases (33). Herein, we summarize the role of circRNAs in thoracic diseases and their possible mechanisms (Table 1).
Table 1. The roles of circRNAs in thoracic diseases and their possible mechanisms.
| CircRNAs | Diseases | Features and potential function | Ref. |
|---|---|---|---|
| hsa_circ_0067934 | ESCC | Correlate with tumor stage and inhibit cell proliferation | (34) |
| Cir-ITCH | ESCC; lung cancer | Sponge miR-7, miR-17, and miR-214 to increase ITCH expression, then degrade Dvl2 phosphorylation to further inhibit the Wnt signaling pathway | (35,36) |
| CircRNA_001059, CircRNA_000167 | Esophageal cancer | RT resistance | (37) |
| CDR1as | Lung cancer | Sponge miR-7 to inhibit apoptosis of cancer cells | (38) |
| MI | Sponge miR-7a to regulate PARP and SP1 | (39) | |
| Circ-ZEB1 | Lung cancer | Sponge miR-200 to regulate ZEB1 expression | (40) |
| Circ_100876 | NSCLC | Correlate with tumor stage and survival time | (41) |
| CircSLC8A1-1 | Heart disease | Most abundant heart-specific circRNAs | (42) |
| RBM20-dependent TTN circRNAs | DCM | Correlate with pathogenesis of DCM | (43) |
| HRCR | HCM | Sponge miR-233 to regulate ARC | (44) |
| CircRNA_081881 | AMI | Sponge miR-548 to regulate PPARγ | (45) |
| MICRA | MI | Predict LV function | (46) |
| hsa_circ_0124644 | CAD | Diagnostic biomarker | (47) |
| CircRNA_000203 | MF | Sponge miR-26b-5p to regulate Colla2 and CTGF | (48) |
ESCC, esophageal squamous cell carcinoma; RT, radiation therapy; MI, myocardial infarction; LV, left ventricular; NSCLC, non-small cell lung cancer; DCM, dilated cardiomyopathy; HCM, hypertrophic cardiomyopathy; AMI, acute myocardial infarction; CAD, coronary artery disease; MF, myocardial fibrosis; TTN, titin; ARC, apoptosis repressor with CARD domain.
CircRNAs in esophageal cancer
Esophageal cancer is among the most prevalent of the thoracic and digestive cancers. Esophageal squamous cell carcinoma (ESCC) is the most deadly of these cancers, and the prognosis of ESCC remains poor (34). CircRNA hsa_circ_0067934 was found to be upregulated in ESCC and promote cell proliferation (35). In a retrospective analysis of 51 patients with ESCC, the expression of circRNA hsa_circ_0067934 was significantly higher in the tumor than in adjacent tissues. Moreover, expression of hsa_circ_0067934 was positively correlated with tumor stage. Results of in vitro analyses suggested that hsa_circ_0067934 interference significantly inhibited cell proliferation and led to cell cycle arrest (35). In another study of ESCC in which 684 tumors were compared with paired adjacent nontumor tissues, the expression level of the circRNA cir-ITCH generally was low in ESCC. This molecule competitively sponged miR-7, miR-17, and miR-214, leading to an increase in ITCH expression, the latter of which could degrade Dvl2 phosphorylation and inhibited Wnt signaling pathways to block the growth of ESCC (37).
Radiation therapy (RT) is essential in the treatment of esophageal cancer; however, RT resistance can diminish the therapeutic effects of RT and can lead to local tumor recurrence or treatment failure. Su et al. observed differential expression of circRNAs between the RT-resistant esophageal cancer cell line, KYSE-150R, and its parental cell line, KYSE-150 (37). In the study, these authors identified 57 upregulated and 17 downregulated circRNAs. CircRNA_001059 and circRNA_000167 were considered most likely to be associated with RT resistance of KYSE-150 (49). However, these authors did not determine the mechanism by which these circRNAs influence RT resistance.
CircRNAs in lung cancer
Lung cancer is one of the most common cancers and is the leading cause of cancer-related deaths worldwide (38). CircRNA CDR1as was found to be frequently coexpressed with the miRNA miR-7 in lung cancer. Elevated miR-7 was associated with poor prognosis of lung cancer. Moreover, CDR1as could sponge a large number of miR-7, incrementally inducing miR-7 inhibition and apoptosis in lung cancer cell lines. In combination with CDR1as, miR-671 could activate endonucleolytic cleavage, induce breakage in CDR1as, and release sponged miR-7 (36). Furthermore, cir-ITCH can suppress lung cancer proliferation via the same mechanism as in ESCC (40). The ZEB1 gene-derived circRNA can act as a ceRNA and sponge miR-200, a well-known lung cancer-related miRNA that has been shown to function as a regulator of ZEB1 gene expression in lung cancer. Thus, ZEB1 gene-derived circRNA affects lung cancer by competing with miR-200 (50).
Non-small cell lung cancer (NSCLC) accounts for approximately 85% of all primary cases of lung cancer, and the prognosis is poor (41). A major breakthrough in NSCLC research was the identification of circRNA_100876, a novel prognostic biomarker for NSCLC. The expression of circRNA_100876 was significantly elevated in NSCLC tissue compared with paired adjacent nontumor tissues, and levels of this circRNA were closely correlated with lymph node metastasis and tumor staging in NSCLC. Results of Kaplan-Meier survival analysis indicated that the overall survival time of NSCLC patients with low levels of circRNA_100876 expression was significantly longer than those with high levels of circRNA_100876 (51).
CircRNAs in heart disease
CircRNAs are closely related to cardiac development, physiology, and pathology. Werfel et al. analyzed the expression and localization of circRNAs in tissues from humans, mice, and rats at different developmental stages and in physiologic and pathologic conditions. The authors suggested that circRNAs were closely related to heart function, but the mechanism involved required further exploration. In particular, several highly conserved circRNA molecules arising from the titin (TTN) gene were of great research value (42) in the field of biomarkers or their mechanisms. Another group evaluated the expression profiles of circRNAs in heart specimens of humans and mice by means of RNA-sequencing. These investigators showed that the expression levels of 15,318 circRNAs in human myocardial tissue and 3,017 circRNAs in mouse tissue were generally correlated with the expression levels of their cognate linear RNAs. Moreover, myocardium-specific genes—such as TTN, RYR2, and DMD—had the highest content of circRNAs. The most abundant circRNA was circSLC8A1-1 (52). These findings open a new avenue of research regarding circRNAs in heart disease.
CircRNAs in dilated cardiomyopathy (DCM)
DCM is a chief cause of heart failure (43). In a recent study, authors suggested that the RNA-binding motif protein 20 (RBM20), an important pathogenic gene of DCM, regulated the biosynthesis of circRNAs from the TTN gene. Results of this study may lend insight into the pathogenesis of DCM. The authors performed circRNA profiling with hypertrophic cardiomyopathy (HCM) and unaffected heart tissues. Eighty circRNAs were found to be expressed from the TTN gene. Some of these were specifically regulated in DCM rather than HCM and corresponded to a region inside TTN gene called the I-band, an important site related to sarcomere strength. Investigators previously had shown that RBM20 was involved in RNA splicing associated with the I-band region. A mutation in RBM20 could lead to abnormal splicing in the I-band region, which could in turn affect functioning of the TTN protein. RBM20 null mice had no detectable expression of these TTN circRNAs. The investigators concluded that these RBM20-dependent TTN circRNAs are highly correlated with pathogenesis of DCM (53).
CircRNAs in HCM
HCM is another major cause of heart failure (44). Wang et al. determined that a novel circRNA termed HRCR could act as an endogenous miR-223 sponge to inhibit and sequester miR-223 activity, resulting in elevated expression of the apoptosis repressor with CARD domain (ARC) (45) MiR-233 and ARC then were found to influence HCM and heart failure in mice. The regulatory pathway comprising HRCR, miR-233, and ARC may have utility as therapeutic targets for the treatment of HCM and heart failure (45).
CircRNAs in myocardial infarction (MI)
Acute myocardial infarction (AMI) poses sizeable morbidity and mortality risks (39). In a study of circRNA expression in human AMI, Deng et al. found that 160 circRNAs were differentially expressed in plasma samples between AMI patients and healthy volunteers. Of these, circRNA_081881 was substantially downregulated in AMI and contained seven competitive binding sites for miR-548. Results of additional bioinformatics analysis showed that miR-548 could regulate the PPARγ gene, a heart-protective factor, in AMI. The pathway constituting circRNA_081881–miR-548–PPARγ was found to influence foam-cell formation (46). In addition to its role in lung cancer, CDR1as promotes MI by sponging miR-7a on expression of its target genes, including PARP and SP1 (47).
After MI, identification of left ventricular (LV) remodeling and dysfunction would be beneficial for personalized medicine. A novel biomarker predicting LV function and outcomes of MI was found: the circRNA MICRA was specific to patients with MI, and detection of MICRA in peripheral blood was predictive of LV function (48).
CircRNAs in coronary artery disease (CAD)
Zhao et al. determined that 22 peripheral blood circRNAs had substantially different levels of expression between patients with CAD and controls. Further study revealed that hsa_circ_0124644 obtained the best AUC (i.e., area under the curve). In a large independent cohort study comprising 137 patients in the CAD group and 115 patients in the control group, it was shown that hsa_circ_0124644 could be used as a diagnostic biomarker for CAD (54).
CircRNAs in myocardial fibrosis (MF)
MF is closely related to hypertension, chronic heart failure, HCM, DCM, diabetes, and other diseases and serves as a potential risk factor for sudden cardiac death (55). Tang et al. found that circRNA_000203 was upregulated in the myocardium in a mouse model of diabetes. This suggested a signaling pathway in the regulation of MF: miR-26b-5p binds to the 3'-UTR (i.e., untranslated region) of Col1a2 and CTGF and prevents their expression. CircRNA_000203 acted as a ceRNA, and its combination with miR-26b-5p could promote the expression of genes related to MF (56).
Conclusions and perspectives
Current research regarding circRNAs is in the early stages, but many investigators have demonstrated that circRNAs likely are formed by direct back-splicing or lariat-driven circularizing pre-RNA and function in regulating transcription or modifying protein. Mechanisms of circRNA biogenesis, regulation, and associated biological functions are not understood fully.
Much progress has been made to elucidate the relationship between circRNAs and thoracic disease. The characteristic O-shaped closure structure of circRNAs has no free terminus. As such, circRNAs are different from linear RNAs and proteins that are easy to degrade by exonucleases, RNases, or proteases. Therefore, it is expected that clinical applications of circRNAs will be found in tissues, blood (57), saliva (58), and other bodily fluids (59). CircRNAs show promise as a new biomarker that can replace previous indicators (60), or they are possible to build novel models for prognosis together with various other biomarkers and thus improve the diagnosis accuracy and specificity of certain diseases.
Characteristics of circRNAs—including the unique role of ceRNA and protein translation via m6A—suggest that these molecules will provide new targets to interfere with disease pathogenesis and enhance treatment effects, or even as potential targets for developing new drugs (61). As mentioned before, circRNAs could mediate, or sponge miRNAs to further influence the expression of target genes, the circRNA-miRNA-target gene interaction network can also provide us some new treatment target for diseases, such as cancer, atherosclerosis (30), cardiomyopathy, and diabetes mellitus (62). On the other hand, the various peptides encoded by circRNAs can also act as potential treatment target. Moreover, fusion-circRNAs (63), first found in leukemia, and circRNAs in exosomes (64) are a lot worthy of study about their therapeutic potency.
Investigators have not implicated circRNAs in asthma, intractable cough, or several other thoracic diseases. What’s more, the role of circRNAs in exosomes (64) and fusion-circRNAs in thoracic diseases remains unknown. Although multiple features of circRNAs have been determined, there remain many circRNAs that appear extraneous in thoracic disease.
The study of circRNAs broadens our understanding of the eukaryotic genome and provides new directions for comprehensive understanding of common diseases.. Further research regarding circRNAs may uncover biogenesis-induced factors, functions of different types of these molecules, whether circRNAs can interact directly with the genome (akin to long noncoding RNAs), and the kinetic mechanisms of circRNA degradation. Assessments of regulatory networks of noncoding RNAs may explain or overturn the theories of previous studies. We anticipate that the role of circRNAs will attract even more attention as research progresses.
Acknowledgements
Funding: This study was supported by the National Natural Science Foundation of China (Grant No. 81470908).
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Sanger HL, Klotz G, Riesner D, et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A 1976;73:3852-6. 10.1073/pnas.73.11.3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 1979;280:339-40. 10.1038/280339a0 [DOI] [PubMed] [Google Scholar]
- 3.Qu S, Yang X, Li X, et al. Circular RNA: A new star of noncoding RNAs. Cancer Lett 2015;365:141-8. 10.1016/j.canlet.2015.06.003 [DOI] [PubMed] [Google Scholar]
- 4.Lu Z, Filonov GS, Noto JJ, et al. Metazoan tRNA introns generate stable circular RNAs in vivo. RNA 2015;21:1554-65. 10.1261/rna.052944.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015;22:256-64. 10.1038/nsmb.2959 [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA 2015;21:172-9. 10.1261/rna.048272.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.AbouHaidar MG, Venkataraman S, Golshani A, et al. Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt. Proc Natl Acad Sci U S A 2014;111:14542-7. 10.1073/pnas.1402814111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Mo Y, Gong Z, et al. Circular RNAs in human cancer. Mol Cancer 2017;16:25. 10.1186/s12943-017-0598-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012;7:e30733. 10.1371/journal.pone.0030733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grouse L. Galileo in Bethesda: The future of gene editing. J Transl Int Med 2016;4:143-6. 10.1515/jtim-2016-0014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong R, Ma XK, Chen LL, et al. Increased complexity of circRNA expression during species evolution. RNA Biol 2017;14:1064-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Xue W, Li X, et al. The Biogenesis of Nascent Circular RNAs. Cell Rep 2016;15:611-24. 10.1016/j.celrep.2016.03.058 [DOI] [PubMed] [Google Scholar]
- 13.Gao Y, Wang J, Zheng Y, et al. Comprehensive identification of internal structure and alternative splicing events in circular RNAs. Nat Commun 2016;7:12060. 10.1038/ncomms12060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang XO, Dong R, Zhang Y, et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res 2016;26:1277-87. 10.1101/gr.202895.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xin Z, Ma Q, Ren S, et al. The understanding of circular RNAs as special triggers in carcinogenesis. Brief Funct Genomics 2017;16:80-6. [DOI] [PubMed] [Google Scholar]
- 16.Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141-57. 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilusz J. Circular RNA and Splicing: Skip Happens. J Mol Biol 2015;427:2411-3. 10.1016/j.jmb.2015.05.019 [DOI] [PubMed] [Google Scholar]
- 18.Lev-Maor G, Ram O, Kim E, et al. Intronic Alus influence alternative splicing. PLoS Genet 2008;4:e1000204. 10.1371/journal.pgen.1000204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell 2013;51:792-806. 10.1016/j.molcel.2013.08.017 [DOI] [PubMed] [Google Scholar]
- 20.Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015;160:1125-34. 10.1016/j.cell.2015.02.014 [DOI] [PubMed] [Google Scholar]
- 21.Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014;56:55-66. 10.1016/j.molcel.2014.08.019 [DOI] [PubMed] [Google Scholar]
- 22.Ivanov A, Memczak S, Wyler E, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 2015;10:170-7. 10.1016/j.celrep.2014.12.019 [DOI] [PubMed] [Google Scholar]
- 23.Salzman J, Chen RE, Olsen MN, et al. Cell-type specific features of circular RNA expression. PLoS Genet 2013;9:e1003777. 10.1371/journal.pgen.1003777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vidal AF, Sandoval GT, Magalhaes L, et al. Circular RNAs as a new field in gene regulation and their implications in translational research. Epigenomics 2016;8:551-62. 10.2217/epi.16.3 [DOI] [PubMed] [Google Scholar]
- 25.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 26.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 27.Taulli R, Loretelli C, Pandolfi PP. From pseudo-ceRNAs to circ-ceRNAs: a tale of cross-talk and competition. Nat Struct Mol Biol 2013;20:541-3. 10.1038/nsmb.2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chao CW, Chan DC, Kuo A, et al. The mouse formin (Fmn) gene: abundant circular RNA transcripts and gene-targeted deletion analysis. Mol Med 1998;4:614-28. [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res 2017;27:626-41. 10.1038/cr.2017.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holdt LM, Stahringer A, Sass K, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun 2016;7:12429. 10.1038/ncomms12429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Floris G, Zhang L, Follesa P, et al. Regulatory Role of Circular RNAs and Neurological Disorders. Mol Neurobiol 2017;54:5156-65. 10.1007/s12035-016-0055-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu H, Guo S, Li W, et al. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep 2015;5:12453. 10.1038/srep12453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sullenger BA, Nair S. From the RNA world to the clinic. Science 2016;352:1417-20. 10.1126/science.aad8709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ozgul G, Cetinkaya E, Ozgul MA, et al. Efficacy and safety of electromagnetic navigation bronchoscopy with or without radial endobronchial ultrasound for peripheral lung lesions. Endosc Ultrasound 2016;5:189-95. 10.4103/2303-9027.183979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xia W, Qiu M, Chen R, et al. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep 2016;6:35576. 10.1038/srep35576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res 2013;73:5609-12. 10.1158/0008-5472.CAN-13-1568 [DOI] [PubMed] [Google Scholar]
- 37.Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget 2015;6:6001-13. 10.18632/oncotarget.3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dietrich CF, Jenssen C, Herth FJ. Endobronchial ultrasound elastography. Endosc Ultrasound 2016;5:233-8. 10.4103/2303-9027.187866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gong XH, Yu JM, Mao Y, et al. Anticoagulant therapy for non-ST-segment elevation acute coronary syndrome in China: A multi-center observational study. J Transl Int Med 2016;4:25-8. 10.1515/jtim-2016-0006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wan L, Zhang L, Fan K, et al. Circular RNA-ITCH Suppresses Lung Cancer Proliferation via Inhibiting the Wnt/beta-Catenin Pathway. Biomed Res Int 2016;2016:1579490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arya N, Sahai AV, Paquin SC. Credentialing for endoscopic ultrasound: A proposal for Canadian guidelines. Endosc Ultrasound 2016;5:4-7. 10.4103/2303-9027.175875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Werfel S, Nothjunge S, Schwarzmayr T, et al. Characterization of circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol 2016;98:103-7. 10.1016/j.yjmcc.2016.07.007 [DOI] [PubMed] [Google Scholar]
- 43.Dietrich CF, Fusaroli P, Jenssen C. European Federation of Societies for Ultrasound in Medicine and Biology guidelines 2015 on interventional endoscopic ultrasound. Endosc Ultrasound 2016;5:143-8. 10.4103/2303-9027.183968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dhillon SS, Harris K. Endobronchial ultrasound for the detection of chronic pulmonary artery thrombus. Endosc Ultrasound 2016;5:272-3. 10.4103/2303-9027.187893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang K, Long B, Liu F, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J 2016;37:2602-11. 10.1093/eurheartj/ehv713 [DOI] [PubMed] [Google Scholar]
- 46.Deng YY, Zhang W, She J, et al. GW27-e1167 Circular RNA Related to PPARγ Function as ceRNA of microRNA in Human Acute Myocardial Infarction. J Am Coll Cardiol 2016;68:C51-2. 10.1016/j.jacc.2016.07.189 [DOI] [Google Scholar]
- 47.Geng HH, Li R, Su YM, et al. The Circular RNA Cdr1as Promotes Myocardial Infarction by Mediating the Regulation of miR-7a on Its Target Genes Expression. PLoS One 2016;11:e0151753. 10.1371/journal.pone.0151753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vausort M, Salgado-Somoza A, Zhang L, et al. Myocardial Infarction-Associated Circular RNA Predicting Left Ventricular Dysfunction. J Am Coll Cardiol 2016;68:1247-8. 10.1016/j.jacc.2016.06.040 [DOI] [PubMed] [Google Scholar]
- 49.Su H, Lin F, Deng X, et al. Profiling and bioinformatics analyses reveal differential circular RNA expression in radioresistant esophageal cancer cells. J Transl Med 2016;14:225. 10.1186/s12967-016-0977-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xie W, Yuan S, Sun Z, et al. Long noncoding and circular RNAs in lung cancer: advances and perspectives. Epigenomics 2016;8:1275-87. 10.2217/epi-2016-0036 [DOI] [PubMed] [Google Scholar]
- 51.Yao JT, Zhao SH, Liu QP, et al. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract 2017;213:453-6. 10.1016/j.prp.2017.02.011 [DOI] [PubMed] [Google Scholar]
- 52.Tan WL, Lim BT, Anene-Nzelu CG, et al. A landscape of circular RNA expression in the human heart. Cardiovasc Res 2017;113:298-309. [DOI] [PubMed] [Google Scholar]
- 53.Khan MA, Reckman YJ, Aufiero S, et al. RBM20 Regulates Circular RNA Production From the Titin Gene. Circ Res 2016;119:996-1003. [DOI] [PubMed] [Google Scholar]
- 54.Zhao Z, Li X, Gao C, et al. Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci Rep 2017;7:39918. 10.1038/srep39918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chazal RA. China may hold answers to addressing cardiovascular disease epidemic. J Transl Int Med 2016;4:11-3. 10.1515/jtim-2016-0003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang CM, Zhang M, Huang L, et al. CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci Rep 2017;7:40342. 10.1038/srep40342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang YG, Yang HL, Long Y, et al. Circular RNA in blood corpuscles combined with plasma protein factor for early prediction of pre-eclampsia. BJOG 2016;123:2113-8. 10.1111/1471-0528.13897 [DOI] [PubMed] [Google Scholar]
- 58.Bahn JH, Zhang Q, Li F, et al. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem 2015;61:221-30. 10.1373/clinchem.2014.230433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qu S, Zhong Y, Shang R, et al. The emerging landscape of circular RNA in life processes. RNA Biol 2017;14:992-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abu N, Jamal R. Circular RNAs as Promising Biomarkers: A Mini-Review. Front Physiol 2016;7:355. 10.3389/fphys.2016.00355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen Y, Li C, Tan C, et al. Circular RNAs: a new frontier in the study of human diseases. J Med Genet 2016;53:359-65 10.1136/jmedgenet-2016-103758 [DOI] [PubMed] [Google Scholar]
- 62.Zhao Z, Li X, Jian D, et al. Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetol 2017;54:237-45. 10.1007/s00592-016-0943-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guarnerio J, Bezzi M, Jeong JC, et al. Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell 2016;165:289-302. 10.1016/j.cell.2016.03.020 [DOI] [PubMed] [Google Scholar]
- 64.Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 2015;25:981-4. 10.1038/cr.2015.82 [DOI] [PMC free article] [PubMed] [Google Scholar]