Abstract
Ocular blast injury is a major medical concern for soldiers and explosion victims due to poor visual outcomes. To define the changes in gene expression following a blast injury to the eye, we examined retinal ribonucleic acid (RNA) expression in 54 mouse strains 5 days after a single 50-psi overpressure air wave blast injury. We observe that almost 40% of genes are differentially expressed with a false discovery rate (FDR) of <0.001, even though the nominal changes in RNA expression are rather small. Moreover, we find through machine learning approaches that genetic networks related to the innate and acquired immune system are activated. Accompanied by lymphocyte invasion into the inner retina, blast injury also results in progressive loss of visual function and retinal ganglion cells (RGCs). Collectively, these data demonstrate how systems genetics can be used to put meaning to the transcriptome changes following ocular blast injury that eventually lead to blindness.
Keywords: : axon injury, ocular blast injury, ocular immune system, systems genetics, transcriptome
Introduction
Since the introduction of improvised explosive devices into modern warfare, the incidence rate of ocular trauma has increased dramatically from 0.65% of all battle injuries to about 13%.1 Based on severity, these can be subdivided into penetrating or open-globe and closed-globe injuries. Whereas open-globe injuries usually require immediate medical attention, closed-globe injuries can go unnoticed. However, the latter can also lead to decreased vision.2 For example, boxers, who frequently sustain blunt trauma to the eye, report decreasing visual function in almost half of cases.3 Similarly, patients suffering from paintball ocular injuries show a visual acuity less than 20/200 in almost 60% of all reported cases.4 The decline in vision is gradual and might not be apparent at initial examination. Recent experimental data suggest subtle axonal damage underlying the deleterious short- and long-term effects of ocular blast trauma in rodents.5–7 The short-term (hours to days) effects include diminished pupillary reflex to red and blue light, increased cell death pathway markers, and reactive gliosis. Visual function declines after a month post-blast, accompanied by gradual thinning of the nerve fiber layer and chronic pattern electroretinogram (ERG) deficits.5 Exposure to one single blast wave is sufficient to lead to decreased axon density and glial scarring in the optic nerve observable as late as 10 months after injury, indicating that the late-onset decline in visual function is partly due to degeneration of optic nerve axons and retinal ganglion cell (RGC) damage.6 These rodent data are consistent with a report on visual dysfunction in veterans, which was strongly associated with blast injury one year after injury.8 Thus, there seems to be a subacute phase during which retinal ganglion cells slowly degenerate, paving the way for gradual vision loss. Despite these findings, no study has systematically investigated the transcriptional changes during this sensitive time.
The present study was designed to define the influence of closed-globe blast injury on the retinal transcriptome. We examined gene expression in 52 BXD recombinant inbred (RI) mouse strains and their parental strains C57BL/6J and DBA/2J 5 days after exposure to a single 50-psi blast wave directed to the eye. Microarray analysis was carried out at both gene and exon level, whereas the use of RI strains allowed for the discovery of gene regulatory networks by linking deoxyribonucleic acid (DNA) sequence variants to corresponding differences in gene expression. The result is a system-wide map of gene interactions that take place 5 days after blast injury. Comparing these data with a naive control group reveals several co-expression modules, among which we find an unexpected cross talk of innate and acquired immune systems. Even though the nominal changes in messenger RNA (mRNA) expression are subtle, the mutual correlation of immune-system–related genes increases, in some cases drastically, suggesting activation of genetic networks.
Methods
Animals: Strains, sex, and age
The DoD Retina Blast (Mar16) dataset contains the data of 213 Affymetrix® Mouse Gene 2.0 ST microarrays. With a total of 52 BXD strains and 2 parental strains (C57BL/6J and DBA/2J), this dataset is genotypically identical to our previously published DoD Normal Retina (May15) dataset and allows for strain-to-strain comparison. Almost all strains are represented by four independent biological samples usually comprising retinas from two male and two female mice between 66 and 114 days of age with a median of 76 days (Supplementary Fig. 1; see online supplementary material at http://www.liebertpub.com). Animals were maintained on a 12-h light–12-h dark cycle in a parasite-free facility with food and water ad libitum. All procedures involving animals were approved by the Animal Care and Use Committee of Emory University, Animal Use and Care Review Office (ACURO) of the U.S. Army Medical Research and Materiel Command (USAMRMC), and were in accordance with the Association for Research in Vision and Ophthalmology (ARVO) Statement for the Use of Animals in Ophthalmic and Vision Research.
Ocular blast injury procedure
Ocular blast injury was performed using a previously described model.6 Briefly, animals were deeply anesthetized with 67 mg/kg tribromoethanol and secured with tape on a semi-open plastic tube sleigh. The head was safely positioned between Styrofoam nuggets to minimize blast exposure to the brain. The sleigh was then inserted into a hollow plastic cylinder with the right eye of the mouse directly facing a 7-mm-wide hole, which was then placed in front of a custom short airgun barrel. Before every blast procedure, the output pressure was checked at the position of the eye with a pressure sensor (Honeywell; Morris Plains, NJ) and re-calibrated, if necessary, to an output of 49 ± 1 psi (Supplementary Fig. 1). The pressure sensor was fixed in place and placed flush against the tube opening. Because the thickness of the outer and inner tube (∼6 mm), it was not possible to position the eye closer toward the tip of the barrel than 6 mm as this would have resulted in inappropriate pressure on the eye due to squeezing it out of its orbit. Thus, there was an approximately 6-mm difference in distance of the pressure transducer and the eye to the gun barrel tip. Following a single blast, eyes were carefully investigated for signs of macroscopic damage. Eyes were lubricated with GenTeal® and mice were allowed to wake up on a heating pad. There was an overall mortality rate of 5% associated with the blast procedure. Of the 240 mice in the blast experiment, 10 died under anesthesia or during recovery. After recovery from anesthesia, 2 mice died the following day.
Functional assessment and Thy1-CFP flat mounts
Thy1-CFP mice (n = 4–7 per group) bred on a C57BL/6 background were subjected to blast and their eyes were fixed in Z-FIX (Anatech Ltd.; Battlecreek, MI), and washed 3 times in phosphate-buffered saline (PBS). Retinas were dissected, mounted on slides with rails in Vectashield Hardset (Vector Laboratories; Burlingame, CA), and coverslipped.
Thy1-CFP fluorescence, RGC counting, and soma size assessment
Total fluorescence of Thy1-CFP flat mounts was measured by quantifying green channel intensity using Photoshop CS5 without applying any further image enhancements (control n = 3; blast n = 4). Retinal ganglion cell (RGC) soma size was automatically measured in square pixels using a custom script in CellProfiler in two single frames (outer and inner) per retinal quadrant each, resulting in approximately 2000 to 4000 RGCs identified per animal.9 RGCs were counted using flat mounted retinas from Thy1-CFP mice. Briefly, each flat mount was divided into 8 regions, such that regions 1 through 4 were close to the optic nerve head (ONH) and regions 5 through 8 were toward the periphery of each flat mount. Each region consisted of a “cutbox,” which was 636.5 μm × 636.5 μm in dimension and was prepared in Adobe Photoshop. Representative regions for each flat mount were selected and the number of CFP-positive RGC bodies were counted manually using the count tool in Photoshop. Data were averaged per group (blast, control) and determined to be significant if p < 0.05 (Welch's t test).
Optokinetic tracking
Contrast sensitivity and visual acuity thresholds were measured by optokinetic tracking (OptoMotry, CerebralMechanics. Inc.; Lethbridge, Alberta).10 Briefly, the mouse was placed on a central elevated platform in the optometry chamber surrounded by four monitors projecting a virtual rotating cylinder with sinusoidal gratings of vertical light and dark bars. A video camera mounted on the top of the chamber tracked the behavior of the mouse, which followed the moving gratings by turning its head, allowing for determination of spatial frequency (“acuity”) and contrast thresholds. Contrast sensitivity function data are expressed as the inverse of the contrast thresholds.
Sample processing, RNA isolation, and microarray hybridization
Five days after the blast procedure, mice were given an overdose of tribromoethanol and sacrificed by rapid cervical dislocation. Retinas were then dissected from eyes and directly placed into 160 U/mL Ribolock® (Thermo Scientific; Walton, MA) in Hank's Balanced Salt Solution (Sigma; St. Louis, MO) on ice. Tissue was immediately stored at −80°C. RNA was isolated using a Qiacube® and the RNeasy Mini Kit (Qiagen; Hilden, Germany) according to the manufacturer's instructions. The isolation included on-column DNase1 treatment to remove contaminating genomic DNA. All tissue was harvested between 10 a.m. and noon to minimize circadian differences in gene expression. RNA integrity was assessed on a Bioanalyzer 2100 (Agilent; Santa Clara, CA) and RNA integrity number (RIN) values for all animals ranged from 8.3 to 10 with a median of 9.5 (Supplementary Fig. 1; see online supplementary material at http://www.liebertpub.com). Each retina was hybridized to a separate GeneChip® Mouse Gene 2.0 ST (Affymetrix; Santa Clara, CA) according to the manufacturer's protocol. Microarray hybridization was performed by two different core laboratories: the Molecular Resource Center of Excellence at the University of Tennessee (Dr. William Taylor, Director) and the Emory Integrated Genomics Core (Dr. Michael E. Zwick, Director, and Robert B. Isett, Technical Director). In a separate experiment, we tested a set of arrays from C57BL/6J retinas at each facility to determine if there were batch effects or other confounding differences between core laboratories, but were not able to detect any. Therefore, data from both facilities were included in the analysis.
Quantitative PCR
For validation of microarray expression data, genes were randomly chosen from four BXD strains in both blast and normal situations. Exon-specific primers were designed using NCBI PrimerBlast and verified to be specific to the target by melting curve and gel analysis. Amplification efficiency for all primers was >90%. Primer sequences are given in Supplementary Figure 2 (see online supplementary material at http://www.liebertpub.com). First strand synthesis was carried out at 42°C using Quantitect Reverse Transcription Kit (Qiagen) and a mix of oligo(d)T primer and random hexamers. After incubation in genomic DNA (gDNA) eraser for 5 min, 350 μg of total RNA were retrotranscribed and diluted 10-fold with ultra-pure H2O. Quantitative polymerase chain reaction (PCR) was carried out in 10 μL reactions using QuantiTect SYBR Green Master Mix (Qiagen) according to the manufacturer's instructions on a Mastercycler realplex2 (Eppendorf; Hamburg, Germany) with annealing temperature set to 60°C. Technical triplicates were averaged and normalized against Ppia, which was identified to be a stably expressed housekeeping gene in the retina with the help of all retinal databases found on GeneNetwork. Fold-changes were calculated in log2 using the ddCt method and compared with the microarray results by linear regression models.
Data processing, statistical analysis, and WGCNA
Microarray data were normalized using the Robust Multi-array Average (RMA) method.11 Expression levels were log2-transformed, z-scored, and multiplied by a factor of 2 before a constant of 8 was added to avoid negative expression values and make the data comparable on GeneNetwork (see GeneNetwork extended methods). Data from probes with a mean expression level lower than the fifth percentile and probes whose sequence did not have a unique BLAT hit were filtered out. Differential expression was assessed by pairwise comparison of expression values across all strains.12 P-values were adjusted for multiple comparisons using the false discovery rate (FDR), and a stringent cutoff of 0.001 was used to decide on statistical significance. The following parameters were chosen for weighted gene co-expression network analysis (WGCNA): a thresholding soft power of 7, for which both networks approached approximate scale-free topology. Signed topological overlap matrices were created separately and scaled appropriately to make them comparable. Modules were assigned by applying adaptive branch pruning to hierarchical clustering dendrograms with the deepSplit parameter set to 2, a minimum module size of 100, and the cutHeight set at 0.995. All analyses were performed in the R 3.1.1 statistical programming environment. The ggplot2 package for the R environment was used for most plots.13
Gene enrichment analyses and network graphs
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment were assessed by submitting Affymetrix Probe IDs to WebGestalt.org.14 Reported p-values were adjusted for multiple comparisons using Benjamini-Hochberg's FDR. Network graphs were created with Cytoscape version 3.4.
Immunostaining, microscopy, and lymphocyte quantification
For staining retinal flat mounts, C57BL/6J (n = 4–5 per time-point) mice were deeply anesthetized with tribromoethanol and perfused through the heart with 0.9% saline followed by 4% paraformaldehyde in phosphate buffer (pH 7.4). The retinas were dissected from the globe and washed 3 times in PBS with 1% Triton X-100 (Sigma) added. Tissue was then blocked in 5% BSA (Sigma) with 0.5% Triton X-100 for 1 h at room temperature. The retinas were then transferred into directly labeled primary antibodies: CD3 (HM3420, Life Technologies; 1:1000), CD4 (ab51467, Abcam; 1:1000), and CD8 (MCD0828TR, Life Technologies; 1:1000). After overnight incubation at 4°C, retinas were rinsed, placed on glass slides, and coverslipped. The whole mounts were examined with a NikonTi inverted microscope with C1 confocal scanner (Nikon Instruments; Melville, NY) at 40× to identify labeled cells. Each retina was systematically scanned in the X–Y plain and Z-stacks were taken through the entire thickness of the retina. After merging all 40 × images together to one picture, lymphocytes were manually counted per whole retina.
Results
Experimental design and quality of the data
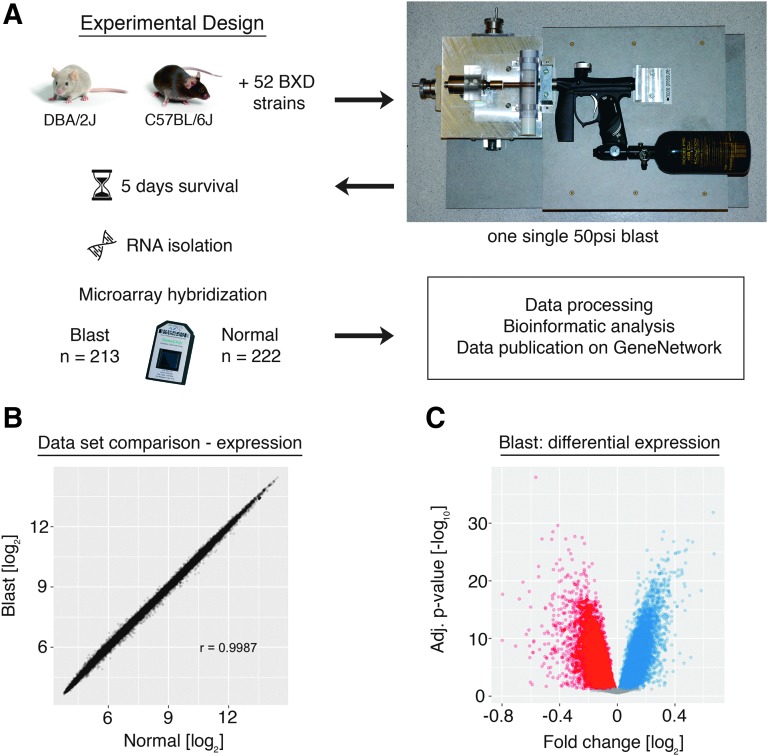
To define the changes in gene expression occurring 5 days after an over-pressure blast to the eye (Fig. 1A), the blast array dataset was compared with a previously published dataset from naive retina.15 The changes in expression were relatively modest and ranged from −0.8 to +0.6-fold on a log2 scale. The magnitude of these changes is below the arbitrary two-fold difference accepted by many microarray studies. We made a conscious decision not to analyze our results using this arbitrary cutoff for biological relevance. Instead, we controlled for statistical outliers by setting a 100-fold more stringent FDR than is usual for these kinds of micorarray studies.16
FIG. 1.
(A) Experimental design. Mice were subjected to a single ocular 50-psi blast wave in the apparatus depicted on the right. Ribonucleic acid (RNA) was isolated 5 days after injury and hybridized to the Affymetrix GeneChip®Mouse Gene ST 2.0 microarray. (B) Correlation of microarray probes between the blast and the normal dataset. In this scatterplot, each dot represents one unique microarray probe. The total correlation (Pearson's r) is 0.999. (C) Volcano plot showing fold-change after blast and its associated logarithmic p-value after adjustment for multiple comparisons using a false discovery rate (FDR) of 0.001. Each dot represents one microarray probe. Red: downregulated probes. Blue: upregulated probes. Gray: no significant change at FDR <0.001. Color image is available online at www.liebertpub.com/neu
We found that 13,971 genes were differentially expressed (Fig. 1B,C) with FDR <0.001. A subset of randomly chosen genes was used to validate the microarray results by quantitative PCR (Pearson r with microarray data = 0.90, Supplementary Fig. 2; see online supplementary material at htpp://www.liebertpub.com). We were able to detect these moderate changes due to the size of both datasets and the quality of RNA samples. The blast injury dataset (DoD Retina After BlastAffyMoGene 2.0 ST (Mar 16), “Blast”) contained 213 independent biological samples from 52 BXD strains plus the two parental strains. For these microarrays, care was taken to produce high-quality RNA. The average RIN score was 9.5 (± 0.03, standard error of the mean [SEM]; Supplementary Fig. 1). The normal retina dataset (DoD Retina Normal AffyMoGene 2.0 ST (May 15), “Normal”) contained a total of 222 microarrays from 55 strains and had an average RIN score of 9.4 (± 0.03, SEM). An optimized RNA isolation protocol combined with the repeatability of tissue dissection results in consistency between each of the biological samples. Tissue surrounding the retina was easily excluded from the sample including the optic nerve, minimizing between-sample variation and contamination by extraneous tissues. Thus, the large number of microarrays in each dataset, the quality of the RNA used to generate the data, and the consistency of tissue isolation allow our group to identify changes in gene expression with a high degree of statistical confidence. These changes may not have been seen in a smaller-sized traditional microarray or RNA-sequencing study. Finally, both datasets are hosted on GeneNetwork.17
Blast injury affects the expression of distinct molecular pathways
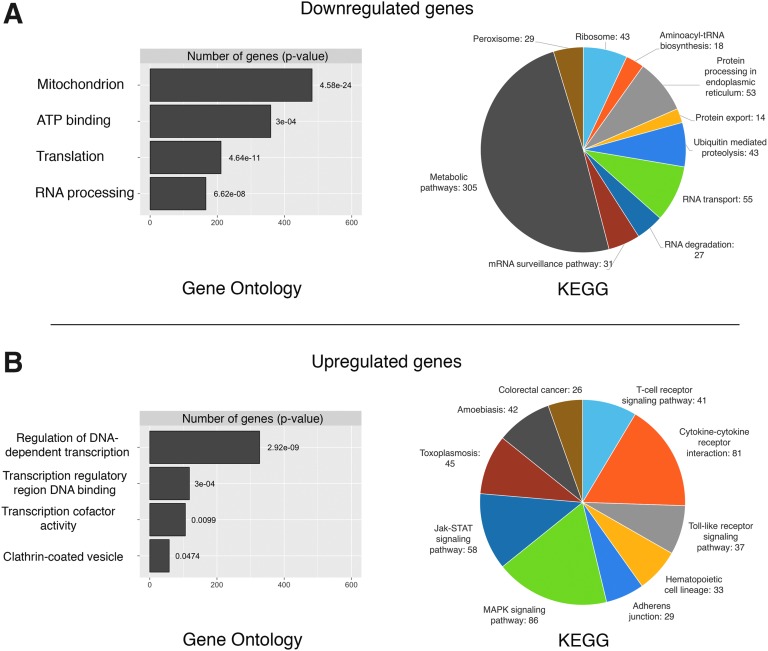
The initial approach to the data was designed to identify differentially expressed genes and then perform functional analysis using gene enrichment profiling. GO and KEGG analysis were used to identify pathways associated with the changes observed following blast injury.18,19 For the downregulated transcripts,we found significant enrichment of genes related to protein turnover and metabolic function. The largest number of significantly downregulated genes was associated with the GO term “Mitochondrion” (Fig. 2A, left panel), whereas for KEGG pathways, the biggest change fell in the “metabolic pathways” category (Fig 2A, right panel). Many genes encoding mitochondrial ribosomes (Mrp*) were found within this cluster. The second largest change in GO enrichment was for the term “ATP binding” (see Supplementary Files 1 and 2 for the full list; see online supplementary material at htpp://www.liebertpub.com). The extent of the downregulation of genes associated with metabolic activity reflects a clear depression of metabolic capacity within the retina as a result of blast injury. The remaining categories for downregulated genes were primarily related to post-transcriptional molecular processes. For example, enrichment in GO terms such as “Translation” or KEGG pathways such as “Ribosome,” “RNA transport,” or “tRNA biosynthesis” collectively indicate protein synthesis dysregulation. Thus, at 5 days after blast injury, there is an overall decrease in genes regulating metabolic processes and genes associated with the production of finished protein products.
FIG. 2.
(A) The left-hand plot shows the number and associated adjusted p-value of significantly downregulated genes and their top four Gene Ontology (GO) terms. The right-hand pie chart indicates significant Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment for the same genes. (B) Identical to (A), but for significantly upregulated genes after blast injury. Color image is available online at www.liebertpub.com/neu
When we examined the genes that were upregulated following blast injury, a very different picture emerged. Most importantly and in contrast to downregulated genes, GO analysis was enriched in genes related to pre-transcriptional processes, such as transcriptional regulation (Fig. 2B, left panel). Additionally, upregulated genes were specific for diverse KEGG pathways, a good half of which were related to immune system processes (Fig. 2B, right panel). For example, the pathway “Cytokine-cytokine interaction” contained many cytokines from the CC and CXL subfamily as well as the transforming growth factor (TGF)-beta family. Additionally, Interferons (IFNs) alpha, beta, epsilon, and gamma were found upregulated within this category. These data point to a difference in transcriptional regulation as well as an increase in expression of immune response genes following blast injury, suggesting activation of the immune system similar to what our group had previously described following optic nerve crush (ONC).20
Network analysis of expression changes after blast injury
Gene enrichment profiling of significantly differentially expressed genes detected changes in several metabolic processes and revealed a role for the immune system following blast injury. Although it is known that isolated traumatic brain injury (TBI) in rodents and humans results in activation of an inflammatory cascade, we wondered if these transcriptional changes would be recapitulated in the mouse retina as well.21 Because simply looking at gene expression changes between two conditions does not reveal any information about the inherently dynamic nature of gene networks, we expanded our analysis by using unbiased machine learning algorithms that compared gene co-expression patterns across the BXD strain set.
First, we performed hierarchical clustering of expression data using weighted gene co-expression network analysis, which partitioned our data into 25 modules. These modules can be thought of as functionally different compartments of the retinal transcriptome, forming groups of highly interconnected transcripts that may shape a pathway.22,23 In general, there was very good preservation of modules between conditions (aggregate eigengene correlation = 0.86, see also Supplementary Fig. 3; see online supplementary material at htpp://www.liebertpub.com), suggesting that the general gene network architecture in the retina is well conserved after blast injury and the changes seen are due to dysregulation of a small number of genes only. Genes in each module were collected and subjected to GO profiling in order to reveal the module's closest functional annotation. Among the modules with the largest drop in preservation (or the biggest changes between conditions) were three modules whose top GO terms were significantly enriched in immune system and metabolic processes, mirroring the gene enrichment profiling results (the black, blue, and dark green module, Supplementary Fig. 3D). We then performed GO analysis separately for up- and downregulated genes in these three modules. This revealed a strong over-representation for the terms “T-cell activation” (adj. p = 0.003), “Cytokine signaling” (adj. p = 0.02), and “regulation of gene expression” (adj. p < 1e-8) for upregulated genes, whereas downregulated ones were enriched for “primary metabolic process” (adj. p < 1e-4) and “cellular protein metabolic process” (adj. p = 0.004).
Another way to examine gene network differences between conditions would be to look at changes of gene connectivity. This measure assigns an arbitrary number to a gene that represents how well its expression correlates to other genes.24 Changes in connectivity mirror the dynamic nature of gene regulatory networks; an increase can be thought of as activation of a gene network and vice versa.25 We performed GO analysis for the top percentile of genes with changes in connectivity. GO trees were very detailed, with genes having the highest increase in connectivity enriched in the terms “cell adhesion” (adj. p = 0.025), “macrophage apoptotic process” (adj. p = 0.019), “extracellular region” (adj. p = 0.014), “regulation of immunoglobulin-mediated immune response” (adj. p < 0.001), as well as “isotype switching to IgG subtypes” (adj. p < 0.001). On the other hand, genes whose connectivity dropped were associated with the GO terms “axonogenesis,” “synaptic transmission,” “dendrite” (all adj. p < 1e-8), “synapse” (adj. p < 1e-12), “protein kinase activity,” and “ATP binding” (both adj. p < 0.001), indicating that normal transcriptional regulation of these molecular processes or entities was impaired after blast injury.
In summary, these results suggest three dominating biologically relevant processes as a result of ocular blast injury: loss of synaptic transmission, impaired cell metabolism, as well as activation of the immune system.
Activation of innate and acquired immune system
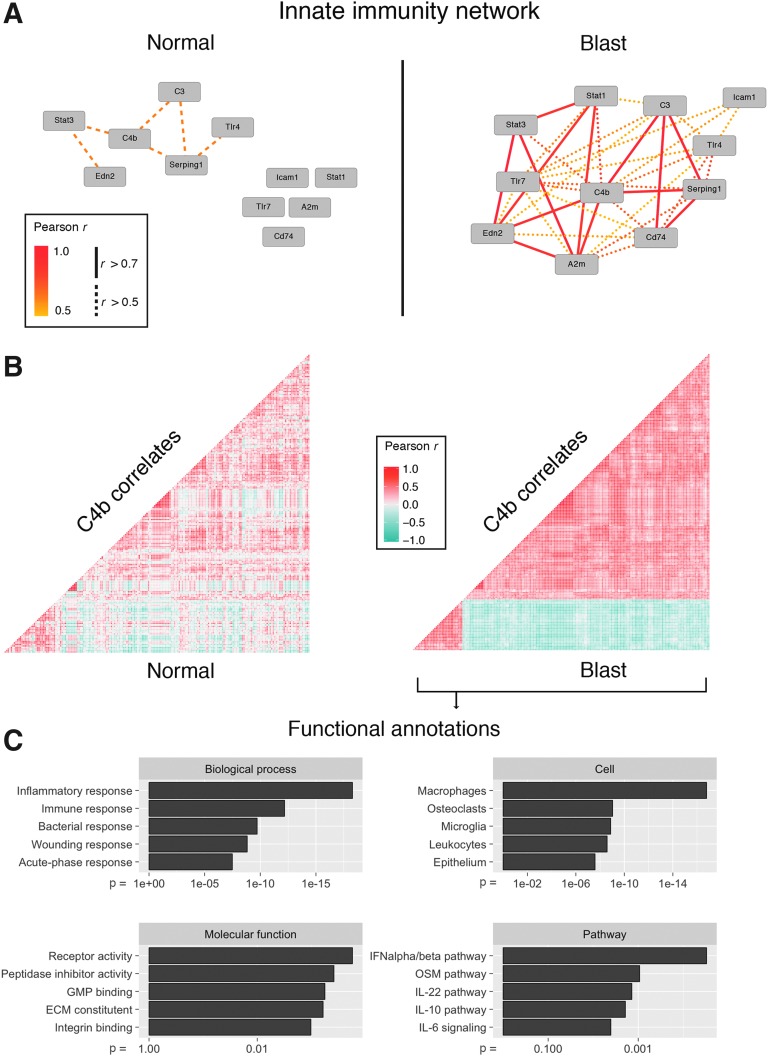
Our analysis of the effects of blast injury on gene expression defined a series of differentially expressed genes and many of these genes clustered into GO categories and KEGG pathways associated with the innate immune system. Earlier work from our group has revealed an activation of the innate immune network following ONC.20 Because ONC is a well-studied model of RGC damage, we wondered whether or not we would find activation of immune-system–related gene networks in the blast data as well. When we examined the blast injury dataset, we saw higher expression levels for many of the same genes (Table 1). Even though some of these genes did not reach significance regarding their differential expression, there was a dramatic increase in mutual correlations to genes involved in innate immunity processes (Fig. 3A). This suggested that the system is indeed activated. When we expanded this analysis for the top 200 correlates of C4b (a gene essential for the propagation of the classic complement cascade), we observed a strikingly strong mutual correlation in the blast but not in the normal condition (Fig. 3B). GO terms for these top 200 correlates of C4b revealed highly significant involvement in multiple immune-system–related biological processes and pathways (Fig. 3C), confirming the involvement of the innate immune system.
Table 1.
Genes Associated with the Innate Immune System
| Affymetrix probe | Symbol | Description | Significant at FDR <0.001 (blast vs. normal) | Mean expression [log2] | Correlation: Pearson's r | P-values (Pearson) | |||
|---|---|---|---|---|---|---|---|---|---|
| Normal | Blast | Normal | Blast | Normal | Blast | ||||
| 17343918 | C4b | Complement component 4B | No | 8.56 | 8.62 | 1 | 1 | ||
| 17346528 | C3 | Complement component 3 | No | 7.81 | 7.95 | 0.56 | 0.80 | 0.000007 | 0 |
| 17387517 | Serping1 | Serine peptidase inhibitor, clade G, member 1 | No | 9.14 | 9.07 | 0.54 | 0.76 | 0.000014 | 0 |
| 17462492 | A2m | Alpha-2-macroglobulin | No | 8.85 | 8.83 | 0.32 | 0.75 | 0.015840 | 0 |
| 17417976 | Edn2 | Endothelin 2 | No | 8.03 | 8.14 | 0.43 | 0.75 | 0.001010 | 0 |
| 17212750 | Stat1 | Signal transducer and activator of transcription 1 | No | 10.31 | 10.33 | 0.40 | 0.71 | 0.002010 | 0.000002 |
| 17350982 | Cd74 | CD74 antigen | No | 8.45 | 8.51 | 0.27 | 0.65 | 0.046220 | 0.000006 |
| 17269717 | Stat3 | Signal transducer and activator of transcription 3 | Yes | 10.92 | 11.10 | 0.54 | 0.65 | 0.000017 | 0 |
| 17546109 | Tlr7 | Toll-like receptor 7 | No | 6.36 | 6.32 | 0.09 | 0.63 | 0.379190 | 0.000013 |
| 17414836 | Tlr4 | Toll-like receptor 4 | Yes | 7.05 | 7.16 | 0.29 | 0.61 | 0.030960 | 0.000001 |
| 17515074 | Icam1 | Intercellular adhesion molecule 1 | No | 8.05 | 8.10 | 0.46 | 0.55 | 0.000370 | 0.000006 |
There is a slight increase of RNA expression and strong increase in correlation. Correlation values and their associated p-value are in relation to C4b.
FDR, false discovery rate; RNA, ribonucleic acid.
FIG. 3.
(A) Network graph displaying Pearson's r for select innate immunity genes in the normal (left) and blast injury (right) situation. Although there is little correlation in the normal condition, the innate immunity network is activated after blast. (B) Correlation matrices showing mutual Pearson's r for the top 200 correlates of the gene C4b. Each dot represents one gene. Hierarchical clustering was applied to the blast matrix and genes did not change position between conditions. There is a strong increase in the mutual correlations indicating activation of a genetic network centered around C4b in the blast condition. (C) Top five Gene Ontology (GO) terms for the C4b correlates shown in (B). The adjusted p-value is indicated on the x-axis. GO enrichment demonstrates a strong relationship of C4b correlates to the immune system. ECM, extracellular matrix,\; GMP, guanosine monophosphate synthetase; IL, interleukin; OSM, oncostatin M.
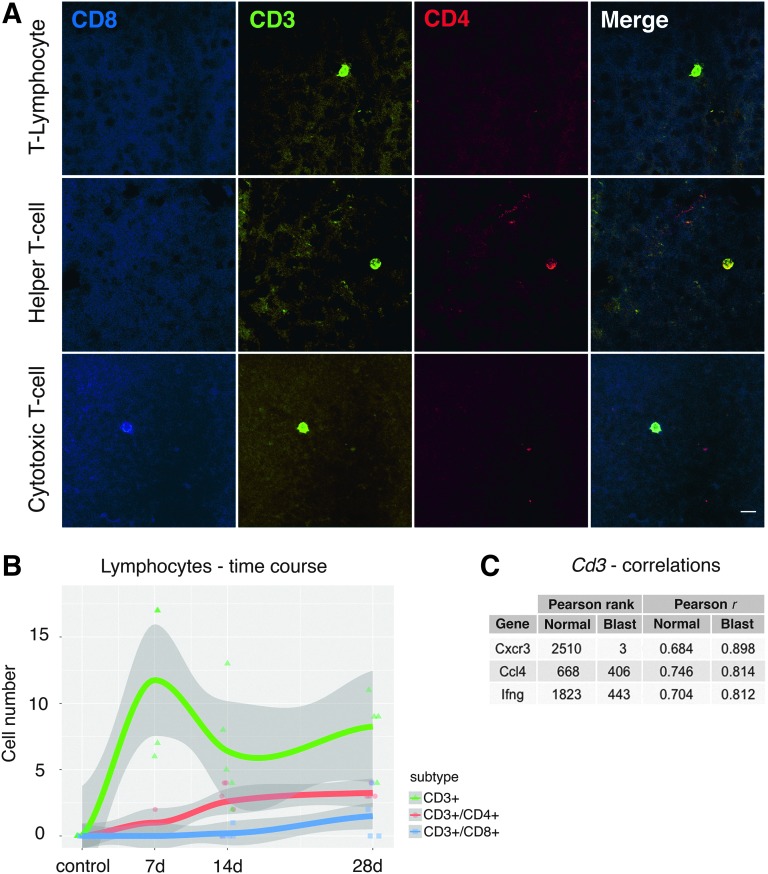
This acute activation also coincided with an increase in markers of the acquired immune system. There was a significant (adj. p < 0.001) increase in the gene expression levels of Cd3 and Cd8 (known markers of T-lymphocytes) in our microarray datasets. Others have shown infiltration of T-cells into the central nervous system (CNS) and retina under pathological conditions.26,27 To determine if this was also the case after blast, we examined the retina at 7, 14, and 28 days following blast injury. At 7 days, CD3-positive cells were observed invading the retina and most of these cells were found in the inner nuclear layer, in close proximity to the intraretinal vessels (Fig. 4A), and this increase was significant (p < 0.0001). At 14 days, there was an increase in the number of CD3/CD4 double-positive lymphocytes (T-helper cells). By 28 days after blast, many CD3-positive cells remained within the retina and in addition to the presence of CD3/CD4-positive T-helper cells, a few CD3/CD8 cytotoxic T-cells were observed (Fig. 4B). Our results are consistent with other research investigating lymphocyte invasion into the retina. For example, it was shown in a model of autoimmune uveitis that CD4+ T-cells predominate during the early phase, whereas CD8+ T-cells accumulate in later stages.28 The identification of T-cells in the injured retina supports the view that cellular immune mechanisms could be responsible for the tissue damage caused by blast injury.
FIG. 4.
(A) Micrographs of lymphocytes invading the retina. CD3 positivity identifies these cells as T-lymphocytes. Co-staining of CD3 and CD4 indicates Helper T-cells, and combined CD3/CD8 positivity identifies cytotoxic T-cells. Scale bar = 10μm. (B) Time course of lymphocyte counts in flat-mounted retinas. No lymphocytes were found in the control situation. There is an initial increase of CD3+ lymphocytes at 7 days after blast. As CD3+ cells decrease, the number of CD4+ and CD8+ cells increases over the course of a month. The gray shaded area denotes the 95% confidence interval per group. (C) Correlations of Cd3 to the cytokine-related genes Cxcr3 (chemokine receptor 3), Ccl4 (chemokine ligand 4), and Ifng (interferon gamma). A drastic increase in ranked correlation and Pearson's correlation coefficient after blast can be seen. Color image is available online at www.liebertpub.com/neu
One potential link between the activation of the innate immune system and the infiltration of lymphocytes could be through a series of soluble factors such as pro-inflammatory cytokines Cxcr3, Ccl4, and IFN-gamma, all of which are expressed in the injured retina.29–31 These cytokines and chemokines, which are released after injury by glial or endothelial cells, may play crucial roles in the recruitment of T-lymphocytes to the injured retina. Even though of these three cytokines only IFN-gamma was significantly upregulated after blast (adj. p = 2.5 e-06), this hypothesis is at least partially supported by increased correlations of all three cytokines to Cd3 in our blast database (Fig. 4C).
Ocular blast injury leads to progressive vision loss associated with loss of retinal ganglion cells
The transcriptional changes observed at 5 days following a blast injury represent a small series of molecular cascades that may result in progressive loss of visual function and the death of RGCs. Because the functional changes that eventually lead to blindness may not be apparent as early as 5 days after blast, we assayed RGC features and function at different time points.
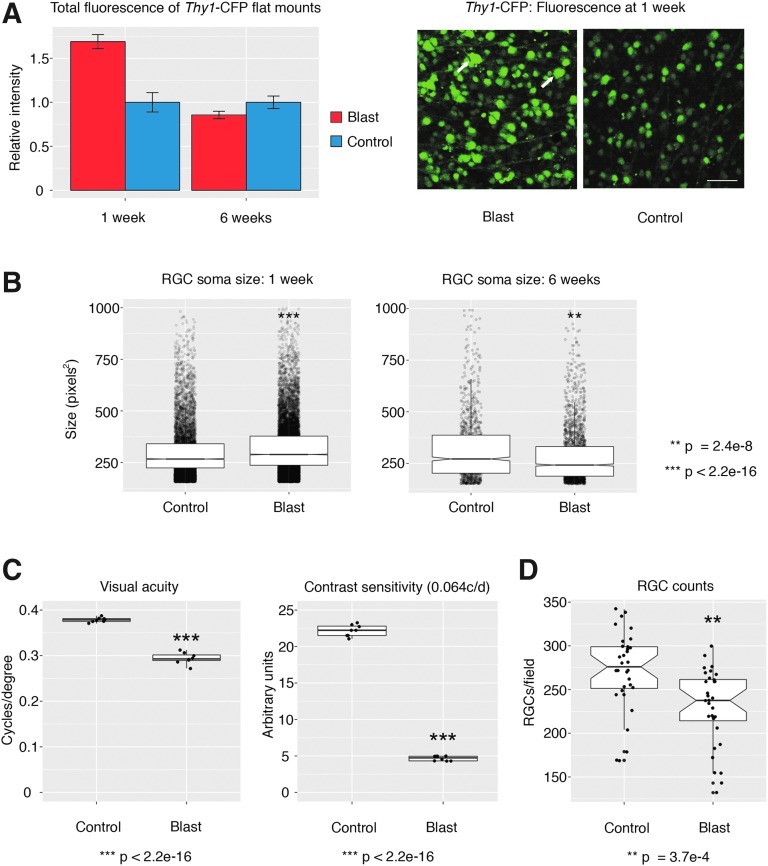
Many genes can serve as proxy for the identification of RGCs,32 and when we investigated our data for changes in these markers, we surprisingly observed higher expression of many of these after blast (Thy1, Tubb3, Pou4f2, Pou4f1, Rbpms; bold ones are significant at p < 0.001). For example, the generic RGC marker Thy1 showed a 0.2-fold log2 change (adj. p = 2.16e-6). We confirmed this upregulation by measuring the total fluorescence of flat-mounted retinas from Thy1-CFP transgenic animals at 1 week (Fig. 5A). At the same time-point, we also observed a significant increase in RGC soma size (Fig. 5B). Both total fluorescence and RGC soma size were significantly decreased at 6 weeks after blast. Functional measures at 6 weeks were also diminished in the same mice, as evidenced by a moderate drop in visual acuity and a dramatic drop in contrast sensitivity (Fig. 5C). Along with that, we observed an ∼16% loss of Thy1-CFP-positive RGCs, ultimately identifying the culprit of ongoing vision loss (Fig. 5D). Taken together, these data demonstrate the devastating effects of what appears to be a relatively modest injury. They also reinforce the importance for early treatment, as the transcriptional events that are observable as early as 5 days after blast eventually lead to blindness.
FIG. 5.
(A) Measurements of Thy1-CFP fluorescence in retinal flat mounts at 1 and 6 weeks after blast. The micrographs on the right are representative of the hyper-fluorescence seen 1 week after blast. Arrows indicate Thy1-CFP positive cells with very large somata. Scale bar = 100 μm. Control versus Blast at 1 week, p < 0.001, n = 3 per condition. (B) Automated retinal ganglion cell (RGC) soma size measurements at 1 and 6 weeks after blast. Soma size is significantly larger at 1 week after blast but significantly smaller at 6 weeks after blast. (C) Visual acuity and contrast sensitivity 6 weeks after blast. Both measures drop significantly after compared with a control situation. (D) RGC counts in retinal Thy1-CFP flat mounts. There is a significant drop 6 weeks after mice have been subjected to blast injury. Color image is available online at www.liebertpub.com/neu
Discussion
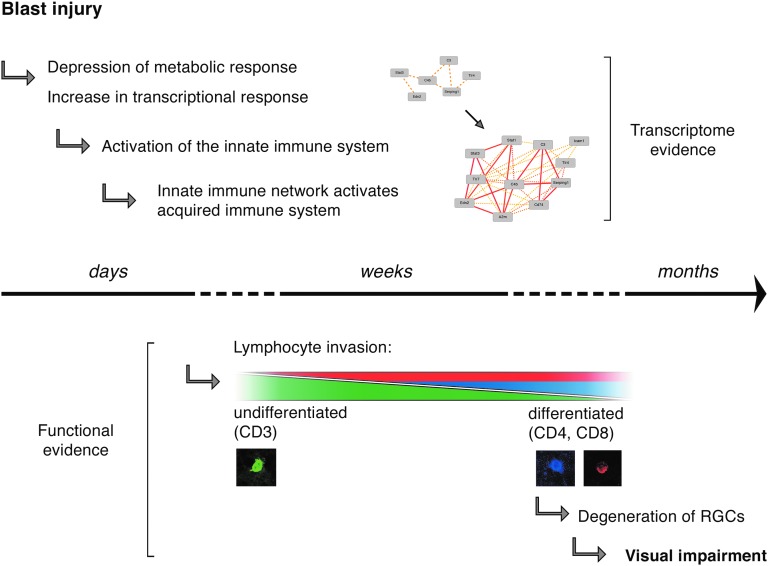
This study comprehensively characterized the in vivo effects of blast injury to the mouse retina and offers the first report on the systems genetics of ocular blast injury. A few other studies have previously investigated the molecular effects of ocular blast injury, and we note that the pressures used to inflict injury differ between models. Whereas one study reported globe rupture at pressures of 40 psi and more, this was not the case in our model.6 In preliminary experiments using our gun, we did not see globe ruptures until pressures more than 70 psi were reached. One possible explanation for this difference could be variation in build of the models or placement of the pressure transducer (see Methods). It is likely that the effective pressure reaching the back of the eye is closer to the range previously reported (<30 psi), as there was an approximate 6-mm distance between the tip of the barrel and the eye. Nevertheless, calibration to 50 psi at the tip of the barrel was necessary to avoid technical variance in the blast apparatus. This pressure is comparable to the amount of pressure sustained at the epicenter of a grenade explosion, making this model roughly equivalent to being a few steps away from a grenade or bomb explosion (as pressure decreases with the cube of distance).33 Here, we presented evidence that a single 50-psi ocular blast as measured at the tip of the airgun barrel was sufficient to lead to declining visual function. This progression was accompanied by a steady increase in the number of lymphocytes migrating into the retina (Fig. 6).
FIG. 6.
Diagram summarizing the main findings of this study. Over the course of days and weeks, the moderate transcriptional changes seen in the retina lead to activation of the innate and the acquired immune networks, which in turn results in chronic neurodegeneration and visual impairment months after injury.
Although it is currently unknown whether this confers a regenerative or destructive effect, in many ways the pathology of ocular blast injury appears to be closely related to TBI. It is believed that in TBI, early-phase leukocyte-mediated breakdown of the blood–brain barrier eventually leads to vascular and synapse remodeling.34 The ensuing neurodegeneration manifests itself as depression or anxiety in TBI or, in the case of ocular blast, as blindness. In mice, the negative neurological outcomes seen in TBI can be mitigated through inhibition of lymphocyte-mediated signaling, whereas the decline in visual function after ocular blast injury can be reduced through immediate-early administration of non-steroidal anti-inflammatory drugs (NSAIDs) such as meloxicam (P. Michael Iuvone, unpublished data).35,36 Because this inflammatory response seems to occur in an acute and chronic phase over an extended period of time, treatment strategies have a wide therapeutic window. Early immunomodulatory treatments in the acute or subacute phase could have dramatic effects on the chronic response. Thus,it is appropriate to investigate the transcriptional changes at the transition from an acute to a chronic state, as the invasion of lymphocytes into the retina likely marks an irreversibly damaging process.
Toward that end, we monitored the retinal transcriptome 5 days after blast injury, and found that a vast number of genes was differentially expressed at that time despite none of the changes exceeding two-fold. A potential reason for this is that we analyzed whole retina, which contains >7 cell types, but our and others' results indicate that the pathological changes mostly occur in fewer cell types (RGCs and glial cells), which together make up less than 1% of cells in the retina.37 As such, most of the RNA that microarrays were normalized to is contributed by the likely unaffected photoreceptors, the most abundant cell type in the retina. Thus, even though it could be very possible that larger-fold changes exist in the affected cell populations, they are not seen in the whole retina data. Instead of focusing on a biologically relevant cutoff, we therefore strongly controlled for statistical outliers by setting a stringent FDR. Our results indicate that transcriptional changes originating from extracellular signaling pathways are dominating the ocular environment 5 days after blast, which comes at the expense of the cells' metabolic function, RNA processing, and protein production. It is not surprising that the largest number of downregulated genes after blast injury was associated with mitochondria, as dysregulated mitochondrial metabolism has long been known to play a significant role in TBI.38 Whereas actual uncoupling of adenosine triphosphate (ATP) synthesis from the respiratory chain would result in mitochondrial stress and acute cell death, other more low-grade mechanisms of mitochondrial dysfunction must be responsible for the slow neurodegeneration that manifests itself after blast injury in the retina. It has very recently been shown that mitochondrial fission is strongly increased in TBI, and that the negative effects on learning and memory could be rescued through the administration of a fission inhibitor.39 It would be interesting to investigate if similar improvements of metabolic function could be achieved in ocular blast injury.
Other changes in gene expression we observed between blasted and normal mice were seemingly related to the balance between transcription and translation. Along with a decrease in genes responsible for ribosomal or endoplasmic reticulum function, we found increased expression of many transcription factors and co-factors. This mirrors the dynamic nature of gene regulatory networks. Changes in RNA expression measured in total tissue are either due to one specific cell type adapting its transcriptional program to a stimulus, or additional new cells that became part of the whole cell population. Although it is likely that a small fraction of the changes seen are the result of lymphocyte invasion, a large part of the differentially expressed RNA will be contributed from retinal cells synthesizing regulatory molecules that prepare the cell for the changes to come.
Because ocular blast injury was previously associated with thinning of the retinal nerve fiber layer, we investigated RGC function as well as expression changes in RGC marker genes.7 Interestingly, we saw statistically significant increased expression in genes such as Thy1, which also corresponded to an increase in RGC soma size and Thy1-CFP fluorescence. This could be related to a process termed “neuronal chromatolysis,” a cellular response after axonal damage resulting in the dissolution of Nissl bodies and redistribution of cytoskeletal proteins with an apparent increase in soma size.40 As chromatolytic neurons are thought to still possess the ability to regenerate, it is interesting to speculate whether or not treatment at this time would stall neuronal apoptosis. We and others have observed that the decline in RCG number or nerve fiber layer thickness is gradual, suggesting a slow but constant underlying molecular process. It appears that this process is related to immune signaling, as our enrichment analysis identified the strongest positive change in expression in genes related to the immune system. Even when the changes in mRNA levels were not significant, increased correlation and connectivity of co-expressed genes was seen especially for immunity-related genes. This illustrates activation of genetic networks, which we have previously found to be the case in the same mouse population after ONC.20 In ONC, a fixed amount of pressure is applied to the optic nerve without interrupting the blood flow to the retina, which leads to gradual decline in RGC number.41 Therefore, both ONC and blast injury are models for RGC death, in which the immune signaling cascade appears to play a significant role. The exact molecular cascades leading to this have yet to be determined, but it is likely that cytokine signaling plays a significant role. Other studies suggest that expression of cytokines in the retina is mediated by glial cells and that an increase in cytokine signaling results in activation of retinal microglia, astrocytes, and Müller glia on site.42,43 In our analysis, we saw that C4b formed a genetic network after blast injury that was significantly enriched in the cellular GO terms “Macrophages” and “Microglia.” The parent protein of C4b is complement factor C4. It has been known now for about 20 years that microglia and astrocytes in mouse brains can synthesize complement factors44 and it also has been shown that complement genes are expressed in the retina.45 Whereas the exact origin of complement factor secretion in the retina remains unknown, our and others' results indicate a role for microglia in this process. It is interesting to note that C4b correlates were also enriched for the terms “Epithelium,” “Extracellular Matrix,” and “Integrin Binding.” This could suggest that one of the mechanisms that permit lymphocyte invasion through the otherwise tight blood–retina barrier after blast is mediated through breakdown of the blood–retina barrier by molecules secreted from microglia activated by complement factors. Similar processes have been observed in TBI as well.46,47
Ultimately, the DBA/2J mouse is known for having several immune-system–related defects compared with the C57BL/6J mouse.48,49 Among these is a condition that abrogates ocular immune privilege associated with the anterior chamber, called a dysfunctional anterior chamber associated immune deviation (ACAID).50 This syndrome was recently found to be at least in part due to a dysfunctional natural killer cell system resulting from Cd94 deficiency in DBA/2J mice.51 We examined our databases for correlations between the presence (B6 genotype) or absence (D2 genotype) of ACAID and inflammatory markers, but no significant correlation was found (data not shown). Whereas another study has detected greater influx of immune components into the anterior part of the eye in DBA/2J mice after blast injury, our data suggest no such connection for retina.43 This indicates that lymphocyte infiltration into the retina is independent of a functional or dysfunctional ACAID.
In conclusion, our data reveal the genetic networks of ocular blast injury for the first time. Using a systems genetics approach, we show that the dysregulated transcriptional environment is reminiscent of the pathophysiology of TBI, with loss of metabolic function and activation of inflammatory cascades that eventually lead to decreases in visual function. Having used BXD strains for this study will potentially allow for identification of upstream modulators of this immune cascade as future work. To our knowledge, this is by far the largest microarray study on ocular blast injury, and it is our hope that the publically available data will be useful for fellow researchers who are interested in specific genes or pathways involved in the pathogenesis of blast injury.
Supplementary Material
Acknowledgments
This study was supported by the Department of Defense CDMRP Grants W81XWH1210255 and W81XWH-12-1-0436 from the U.S. Army Medical Research and Materiel Command and the Telemedicine and Advanced Technology (EEG, PMI), NEI grants R01EY178841 (EEG), R01EY004864 (PMI), P30EY06360 (Emory Vision Core, PMI), and unrestricted funds from Research to Prevent Blindness. FLS is supported by the institutional training grant T32EY007092-30 (PMI). Additional support was provided by the Emory Integrated Genomics Core (EIGC), which is subsidized by the Emory University School of Medicine as one of the Emory Integrated Core Facilities. We thank Dr. Robert Williams and Arthur Centeno (University of Tennessee Health Science Center) for maintaining the data on GeneNetwork, Dr. Xiangdi Wang and Dr. Justin Templeton for their technical assistance, as well as April Brooke Still for her help in breeding some BXD animals at Emory University.
Author Disclosure Statement
No competing financial interests exist. The study was conceived by EEG. RK collected and isolated RNA from most of the animals. FLS performed the blast procedure and all statistical and bioinformatic analyses. YL was responsible for lymphocyte immunostaining and counting. MC compiled Thy1-CFP flat mounts and helped with animal breeding. CS and PL completed the OKT experiments overseen by PMI. FLS, EEG, and YL wrote the article with input from all other authors. All authors read and approved the final manuscript.
References
- 1.Weichel E.D., and Colyer M.H. (2008). Combat ocular trauma and systemic injury. Curr. Opin. Ophthalmol. 19, 519–525 [DOI] [PubMed] [Google Scholar]
- 2.Cockerham G.C., Rice T.A., Hewes E.H., Cockerham K.P., Lemke S., Wang G., Lin R.C., Glynn-Milley C., and Zumhagen L. (2011). Closed-eye ocular injuries in the Iraq and Afghanistan wars. N. Engl. J. Med. 364, 2172–2173 [DOI] [PubMed] [Google Scholar]
- 3.Corrales G., and Curreri A. (2009). Eye trauma in boxing. Clin. Sports Med. 28, 591–607, vi. [DOI] [PubMed] [Google Scholar]
- 4.Nemet A.Y., Asalee L., Lang Y., Briscoe D., and Assia E.I. (2016). Ocular paintball injuries. Isr. Med. Assoc. J 18, 27–31 [PubMed] [Google Scholar]
- 5.Bricker-Anthony C., Hines-Beard J., and Rex T.S. (2014). Molecular changes and vision loss in a mouse model of closed-globe blast trauma. Invest. Ophthalmol. Vis. Sci. 55, 4853–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hines-Beard J., Marchetta J., Gordon S., Chaum E., Geisert E.E., and Rex T.S. (2012). A mouse model of ocular blast injury that induces closed globe anterior and posterior pole damage. Exp. Eye Res. 99, 63–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohan K., Kecova H., Hernandez-Merino E., Kardon R.H., and Harper M.M. (2013). Retinal ganglion cell damage in an experimental rodent model of blast-mediated traumatic brain injury. Invest. Ophthalmol. Vis. Sci. 54, 3440–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dougherty A.L., MacGregor A.J., Han P.P., Heltemes K.J., and Galarneau M.R. (2011). Visual dysfunction following blast-related traumatic brain injury from the battlefield. Brain Inj. 25, 8–13 [DOI] [PubMed] [Google Scholar]
- 9.Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J., Golland P., and Sabatini D.M., (2006). CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Douglas R.M., Alam N.M., Silver B.D., McGill T.J., Tschetter W.W., and Prusky G.T. (2005). Independent visual threshold measurements in the two eyes of freely moving rats and mice using a virtual-reality optokinetic system. Vis. Neurosci. 22, 677–684 [DOI] [PubMed] [Google Scholar]
- 11.Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., and Speed T.P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 [DOI] [PubMed] [Google Scholar]
- 12.Cheadle C., Vawter M.P., Freed W.J., and Becker K.G. (2003). Analysis of microarray data using Z score transformation. J. Mol. Diagn. 5, 73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wickham H. (2009). Ggplot2: Elegant Graphics for Data Analysis. Use R! 2009. Springer: New York, pps. viii, 212 [Google Scholar]
- 14.Wang J., Duncan D., Shi Z., and Zhang B. (2013). WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 41, W77–W83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King R., Lu L., Williams R.W., and Geisert E.E. (2015). Transcriptome networks in the mouse retina: an exon level BXD RI database. Mol. Vis. 21, 1235–1251 [PMC free article] [PubMed] [Google Scholar]
- 16.Gusnanto A., Calza S., and Pawitan Y. (2007). Identification of differentially expressed genes and false discovery rate in microarray studies. Curr. Opin. Lipidol. 18, 187–193 [DOI] [PubMed] [Google Scholar]
- 17.Mulligan M.K., Mozhui K., Prins P., and Williams R.W. (2017). GeneNetwork: a toolbox for systems genetics. Methods Mol. Biol. 1488, 75–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gene Ontology Consortium. (2015). Gene Ontology Consortium: going forward. Nucleic Acids Res. 43, D1049–D1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M., Sato Y., Kawashima M., Furumichi M., and Tanabe M. (2016). KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Templeton J.P., Freeman N.E., Nickerson J.M., Jablonski M.M., Rex T.S., Williams R.W., and Geisert E.E. (2013). Innate immune network in the retina activated by optic nerve crush. Invest. Ophthalmol. Vis. Sci. 54, 2599–2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmin S., Mathiesen T., Shetye J., and Biberfeld P. (1995). Intracerebral inflammatory response to experimental brain contusion. Acta Neurochir. (Wien) 132, 110–119 [DOI] [PubMed] [Google Scholar]
- 22.Langfelder P. and Horvath S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuller T.F., Ghazalpour A., Aten J.E., Drake T.A., Lusis A.J., and Horvath S. (2007). Weighted gene coexpression network analysis strategies applied to mouse weight. Mamm. Genome 18, 463–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langfelder P., Mischel P.S., and Horvath S. (2013). When is hub gene selection better than standard meta-analysis? PLoS One 8, e61505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao W., Langfelder P., Fuller T., Dong J., Li A., and Hovarth S. (2010). Weighted gene coexpression network analysis: state of the art. J. Biopharm. Stat. 20, 281–300 [DOI] [PubMed] [Google Scholar]
- 26.Charteris D.G., Champ C., Rosenthal A.R., and Lightman S.L. (1992). Behcet's disease: activated T lymphocytes in retinal perivasculitis. Br. J. Ophthalmol. 76, 499–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imagawa T., Kitagawa H., and Uehara M. (2003). Appearance of T cell subpopulations in the chicken and embryo retina. J. Vet. Med. Sci. 65, 23–28 [DOI] [PubMed] [Google Scholar]
- 28.Richardson P.R., Boulton M.E., Duvall-Young J., and McLeod D. (1996). Immunocytochemical study of retinal diode laser photocoagulation in the rat. Br. J. Ophthalmol. 80, 1092–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ha Y., Liu H., Xu Z., Yokota H., Narayanan S.P., Lemtalsi T., Smith S.B., Caldwell R.W., Caldwell R.B., and Zhang W. (2015). Endoplasmic reticulum stress-regulated CXCR3 pathway mediates inflammation and neuronal injury in acute glaucoma. Cell Death Dis. 6, e1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rutar M., Natoli R., Chia R.X., Valter K., and Provis J.M. (2015). Chemokine-mediated inflammation in the degenerating retina is coordinated by Muller cells, activated microglia, and retinal pigment epithelium. J. Neuroinflammation 12, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zinkernagel M.S., Chinnery H.R., Ong M.L., Petitjean C., Voigt V., McLenachan S., McMenamin P.G., Hill G.R., Forrester J.V., Wikstrom M.E., and Degli-Esposti M.A. (2013). Interferon gamma-dependent migration of microglial cells in the retina after systemic cytomegalovirus infection. Am. J. Pathol. 182, 875–885 [DOI] [PubMed] [Google Scholar]
- 32.Struebing F.L., Lee R.K., Williams R.W., and Geisert E.E. (2016). Genetic Networks in Mouse Retinal Ganglion Cells. Front. Genet. 7, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glasstone S., and Dolan P. (1977). The effects of nuclear weapons. United States Department of Defense [Google Scholar]
- 34.Schwarzmaier S.M., Zimmermann R., McGarry N.B., Trabold R., Kim S.W., and Plesnila N. (2013). In vivo temporal and spatial profile of leukocyte adhesion and migration after experimental traumatic brain injury in mice. J. Neuroinflammation 10, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wood R.L., and Rutterford N.A. (2006). Demographic and cognitive predictors of long-term psychosocial outcome following traumatic brain injury. J. Int. Neuropsychol. Soc. 12, 350–358 [DOI] [PubMed] [Google Scholar]
- 36.Zhao S., Yu Z., Liu Y., Bai Y., Jiang Y., van Leyen K., Yang Y.G., Lok J.M., Whalen M.J., Lo E.H., and Wang X. (2016). CD47 deficiency improves neurological outcomes of traumatic brain injury in mice. Neurosci. Lett. 643, 125–130 [DOI] [PubMed] [Google Scholar]
- 37.Jeon C.J., Strettoi E., and Masland R.H. (1998). The major cell populations of the mouse retina. J. Neurosci. 18, 8936–8946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vink R., Head V.A., Rogers P.J., McIntosh T.K., and Faden A.I. (1990). Mitochondrial metabolism following traumatic brain injury in rats. J. Neurotrauma 7, 21–27 [DOI] [PubMed] [Google Scholar]
- 39.Fischer T.D., Hylin M.J., Zhao J., Moore A.N., Waxham M.N., and Dash P.K. (2016). Altered mitochondrial dynamics and TBI pathophysiology. Front. Syst. Neurosci. 10, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen D.H. (1978). Qualitative and quantitative study of synaptic displacement in chromatolyzed spinal motoneurons of the cat. J. Comp. Neurol. 177, 635–664 [DOI] [PubMed] [Google Scholar]
- 41.Templeton J.P., and Geisert E.E. (2012). A practical approach to optic nerve crush in the mouse. Mol. Vis. 18, 2147–2152 [PMC free article] [PubMed] [Google Scholar]
- 42.Stahl T., C. Mohr C., Kacza J., Reimers C., Pannicke T., Sauder C., Reichenbach A., and Seeger J. (2003). Characterization of the acute immune response in the retina of Borna disease virus infected Lewis rats. J. Neuroimmunol. 137, 67–78 [DOI] [PubMed] [Google Scholar]
- 43.Bricker-Anthony C., Hines-Beard J., D'Surney L., and Rex T.S. (2014). Exacerbation of blast-induced ocular trauma by an immune response. J. Neuroinflammation 11, 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haga S., Aizawa T., Ishii T., and Ikeda K. (1996). Complement gene expression in mouse microglia and astrocytes in culture: comparisons with mouse peritoneal macrophages. Neurosci. Lett. 216, 191–194 [DOI] [PubMed] [Google Scholar]
- 45.Luo C., Chen M., and Xu H. (2011). Complement gene expression and regulation in mouse retina and retinal pigment epithelium/choroid. Mol. Vis. 17, 1588–1597 [PMC free article] [PubMed] [Google Scholar]
- 46.Baskaya M.K., Rao A.M., Dogan A., Donaldson D., and Dempsey R.J. (1997). The biphasic opening of the blood-brain barrier in the cortex and hippocampus after traumatic brain injury in rats. Neurosci. Lett. 226, 33–36 [DOI] [PubMed] [Google Scholar]
- 47.Balu R. (2014). Inflammation and immune system activation after traumatic brain injury. Curr. Neurol. Neurosci. Rep. 14, 484. [DOI] [PubMed] [Google Scholar]
- 48.Casanova T., Van de Paar E., Desmecht D., and Garigliany M.M. (2015). Hyporeactivity of alveolar macrophages and higher respiratory cell permissivity characterize DBA/2J mice infected by influenza A virus. J. Interferon Cytokine Res. 35, 808–820 [DOI] [PubMed] [Google Scholar]
- 49.Miyairi I., Tatireddigari V.R., Mahdi O.S., Rose L.A., Belland R.J., Lu L., Williams R.W., and Byrne G.I. (2007). The p47 GTPases Iigp2 and Irgb10 regulate innate immunity and inflammation to murine Chlamydia psittaci infection. J. Immunol. 179, 1814–1824 [DOI] [PubMed] [Google Scholar]
- 50.Streilein J.W., and Niederkorn J.Y. (1981). Induction of anterior chamber-associated immune deviation requires an intact, functional spleen. J. Exp. Med. 153, 1058–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chattopadhyay S., O'Rourke J., and Cone R.E. (2008). Implication for the CD94/NKG2A-Qa-1 system in the generation and function of ocular-induced splenic CD8+ regulatory T cells. Int. Immunol. 20, 509–516 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.