Abstract
Plasma cell-free tumor DNA, or circulating tumor DNA (ctDNA), from liquid biopsy is a potential source of tumor genetic material, in the absence of tissue biopsy, for EGFR testing. Our validation study reiterates the clinical utility of ctDNA next generation sequencing (NGS) for EGFR mutation testing in non-small cell lung cancer (NSCLC). A total of 163 NSCLC cases were included in the validation, of which 132 patients had paired tissue biopsy and ctDNA. We chose to validate ctDNA using deep sequencing with custom designed bioinformatics methods that could detect somatic mutations at allele frequencies as low as 0.01%. Benchmarking allele specific real time PCR as one of the standard methods for tissue-based EGFR mutation testing, the ctDNA NGS test was validated on all the plasma derived cell-free DNA samples. We observed a high concordance (96.96%) between tissue biopsy and ctDNA for oncogenic driver mutations in Exon 19 and Exon 21 of the EGFR gene. The sensitivity, specificity, positive predictive value, negative predictive value, and diagnostic accuracy of the assay were 91.1%, 100% 100%, 95.6%, and 97%, respectively. A false negative rate of 3% was observed. A subset of mutations was also verified on droplet digital PCR. Sixteen percent EGFR mutation positivity was observed in patients where only liquid biopsy was available, thus creating options for targeted therapy. This is the first and largest study from India, demonstrating successful validation of circulating cell-free DNA as a clinically useful material for molecular testing in NSCLC.
Keywords: liquid biopsy, NSCLC, EGFR, ctDNA, NGS
Introduction
Recurrent somatic mutations in lung cancer are well known and effective targeted therapies are available. Somatic mutations in the kinase domain of EGFR leads to constitutive activation of EGFR that results in uncontrolled cell proliferation and inhibition of apoptosis which promotes tumor growth in non-small cell lung cancer (NSCLC).1 The most common activating mutations are located in Exons 18, 19, 20, and 21 which spans the TK domain of EGFR.2–4 Ever since the US Food and Drug Administration (FDA)’s approval of TKIs for first line therapy in advanced NSCLC, clinical guidelines have been recommending EGFR mutation testing in all NSCLC patients at the time of diagnosis for treatment decisions.5 The cancer incidence in India is increasing rapidly, with lung cancer being the second most common cancer after breast, with an estimated 0.1 million new cases during 2016 which is expected to increase to 0.14 million cases by 2020, accounting for 23% increase in incidence rate. Early detection of cancer and a personalized approach for treatment, with frequent assessment of therapeutic response, could improve the survival outcome in these patients, thus, reducing the mortality. With current approaches in molecular targeted therapies in NSCLC, advancements in mutation detection technologies will become a valuable addition to address the treatment planning for the increasing burden of lung cancer, and circulating tumor DNA (ctDNA) or liquid biopsy, in recent years, has been at the forefront of research and transition to a clinical setting.
Historically, tissue biopsy is the gold standard for establishing the EGFR mutation status. Based on our own experience, in a significant percentage of newly diagnosed lung cancer patients, which accounts for approximately 30%, it has not been possible to get tissue biopsy samples (K Prabhash, Tata Memorial Hospital, personal communication, November 2016). In recent years, liquid biopsy has gained importance as a potential alternative source of tumor genetic material for molecular diagnostics. The hypothesis of liquid biopsy, cell-free ctDNA, as a promising alternative to tissue biopsy for EGFR testing, has been proven and well established from the encouraging results of clinical outcomes in one of the most recent global clinical trials on NSCLC patients (Phase III LUX-Lung 3/6).6 The role of liquid biopsy is also being investigated for monitoring of treatment effect.
Allele specific PCR, Scorpion Amplified Refractory Mutation System (ARMS) PCR, droplet digital PCR (ddPCR), and next generation sequencing (NGS) are the most commonly used technologies for mutation detection in ctDNA.7–9 While the PCR-based methods can detect only specific hotspot mutations, NGS has the advantage of detecting novel mutations in addition to the hotspots. NGS has the additional advantage of being cost effective in screening for multiple genes and hotspots in a single assay. The specificity and sensitivity of the ctDNA-based mutation testing depends not only on the technology, but also to a large extent on biology of the tumor and its clinicopathological staging.
In this study, we present our findings on validation of ctDNA as a clinically useful biomarker/tumor genetic material for screening recurrent somatic mutations or hotspot mutations in EGFR (Exon 19/Exon 21) in NSCLC patients. The choice of hotspots are based on the frequency of mutations in EGFR documented in advanced NSCLC adenocarcinoma patients, wherein the presence or absence of these mutations influence treatment decisions. To the best of our knowledge, this is the largest study on validation of liquid biopsy as a predictive clinical biomarker in NSCLC patients from India.
Methods
Ethics statement
The study was approved by the Institutional Review Board (IRB) and the Ethics Committee (EC) of Tata Memorial Hospital (Mumbai, India). This study was monitored by data monitoring committee of Tata Memorial Hospital. All patients were recruited and gave written informed consent for use of their blood plasma and tissue biopsy. All clinical data and samples were received anonymously.
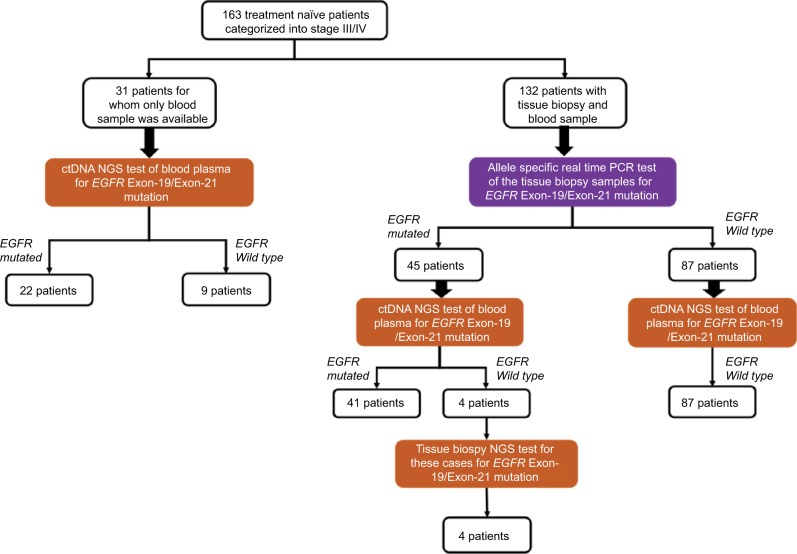
The patients were arranged in two groups. The first group consisted of 132 patients where the paired biopsy of the tissue and the liquid biopsy (blood sample) was available (Figure 1). The second group consisted of 31 patients who did not have tissue biopsy due to various technical reasons (e.g., insufficient tissue material, poor quality DNA, no DNA, low tumor yield). However, liquid biopsy (blood sample) was available for these patients which was processed for ctDNA NGS test.
Figure 1.
Schematic representation of patients enrolled in the study and their mutation status.
Abbreviations: ctDNA, circulating tumor DNA; NGS, next generation sequencing; PCR, polymerase chain reaction.
Patients and sample collection
A total of 163 treatment naïve patients diagnosed with advanced NSCLC, at Tata Memorial Hospital, Mumbai were enrolled in this study. These patients had both blood and tissue samples available. It was taken from the database maintained in the Medical Oncology Department at Tata Memorial Hospital.
Sample processing and DNA extraction for tissue biopsy and liquid biopsy
For liquid biopsy test 20 mL peripheral blood was collected in Streck® tubes, at the time of diagnosis, as per the IRB and the EC recommendations, an informed consent was obtained from the patients enrolled in the study. The sampling was done at the same time for tissue biopsy as well as blood for ctDNA/liquid biopsy in all the 132 patients to reduce the effect of any temporal variations, and to ensure uniformity.
Approximately 10 mL plasma was obtained for all the patients and the plasma cfDNA was isolated using standard procedure (Qiagen kit [Cat. no. 55114] Qiagen NV, Venlo, the Netherlands). The circulating nucleic acid isolation kit from Qiagen has been very robust in obtaining consistent and good quality cell-free DNA.
Similarly, in all the patients, where biopsy was available, tumor genomic DNA extracted from FFPE blocks was analyzed for EGFR mutation status by allele specific real time PCR.11
Library construction and target enrichment
Target enrichment was performed using amplicon-based method adopting custom designed amplicons for the EGFR Exon 19/Exon 21 region. The targeted region consists of recurrent hotspot mutations (including SNPs, insertions/deletions less than 25 bp) in EGFR. Input of ctDNA varied in the range of 3–50 ng for target enrichment. The amplicons were subjected to NGS library preparation using KAPA HTP library preparation kit from Illumina. The NGS libraries were sequenced at ultra-high depth of 100,000× depth using Illumina (San Diego, CA, USA) HiSeq 2500® instrument.
Sequenced data analysis
The raw fastq sequences obtained were aligned to human reference genome (GRCh37/hg19) using BWA program32,33 and processed using Picard and GATK toolkits.34,35 The low frequency variants were identified using LoFreq program. An in-house method has been developed to filter the low frequency variant to control false positive rate at ultra-high depth sequencing of the samples. We have applied a voting based method to identify the low frequency variants. In our method, we generated 70 in silico experiments of the input sample by down sampling. The in-silico instance average read depth varies from 100×–100,000×. Variant calling is performed for each in silico experiment and also on the instance with all reads. The variants predicted from each instance are aggregated. A voting score is generated for each variant. Higher voting score represents higher confidence for the variant identified in the sample. The allele frequency was calculated for all the filtered mutants. The allele frequency is defined as the ratio of mutant read count vs. total read depth at the mutant position. The filtered variants were then annotated using our in-house annotation pipeline (VariMAT). The variants present in the sample, but found in various population databases (1000G, ExAC, EVS, 1000Japanese, dbSNP, UK10K, MedVarDb [in-house database]) with ≥1% were filtered from reporting.36–41 Clinically relevant mutations were annotated using published literature, databases, and in-house proprietary databases.
Our in-house curated somatic database (OncoMD) (http://oncomd.medgenome.com/MutationViewer),42 which includes somatic variants from published literature, TCGA, and ICGC was used to identify clinically significant somatic mutations as per the AMP-ASCO-CAP guidelines.43
Technical validation of assay on reference standard samples
External reference standards kit (HD780) for cell-free tumor DNA was procured from Horizon Discovery (Cambridge, UK) technologies to derive the performance characteristics of the ctDNA NGS test. The Horizon standards included a panel of mutations: EGFR (L858R, E746-A750 del, T790M, V769-D770insASV), KRAS (G12D), NRAS (Q61K, A59T), at different allele frequencies: 5% and 1%, 0.1%, and 0% (Wild type). Tissue-based EGFR mutation status based on allele specific real time PCR30 was considered as the benchmark reference standard for all the concordance analysis and the metrics of validation between ctDNA and tissue biopsy was limited to EGFR Exon 19 and Exon 21 only. The variant class includes missense mutations and short indels.
A subset of samples which were Exon 19 (deletions) positive, were also verified with ddPCR. ddPCR™ EGFR Exon 19 Deletions Screening Kit, (Cat. no. 12002392) and Cat. no. 10031249: Wild type HEX label (Wt EGFR T790M), Cat. no. 10031246: mutant type FAM label (EGFR T790M), was purchased from Bio-Rad Laboratories Inc. (Hercules, CA, USA) and the ddPCR experiments were performed as per the manufacturer’s recommendations. All the experiments were performed on Bio-Rad QX-100 instrument. The results were analyzed using the inbuilt software Quantasoft V1.7. The ctDNA input in ddPCR varied from 0.65 ng to 26 ng, depending on the yield from plasma.
Results
Technical validation of ctDNA NGS test
As part of technical validation, we first validated our ctDNA NGS test on external reference kit (HD780) that consists of four cell-free DNA samples provided by Horizon discovery technologies. The mutation status of EGFR gene was evaluated for these reference samples using our targeted sequencing ctDNA NGS test. The sequenced data generated for these four reference standard samples were at an average depth of 100,000×. For all these four samples, technical duplicates were also generated. We obtained 100% concordance for the panel of mutations with manufacturer’s estimation of mutation allele frequencies up to 1%.
There were 132 patients for whom we had information on tissue biopsy EGFR mutation status (Exon 19 and Exon 21), which constitutes greater than 90% of all the EGFR activating mutations known in NSCLC patients.10 Benchmarking tissue biopsy as a gold standard for mutation analysis, we calculated the metrics of validation for our ctDNA NGS assay in these 132 cases.
Tissue biopsy genotyping by allele specific real time PCR
For the tissue biopsy sample set consisting of 132 patients, the tumor genomic DNA was analyzed for EGFR Exon-19 and Exon-21 mutation status in the hotspot regions using allele specific real time PCR.11 The overall clinical and demographic summary of the subjects included in this study is provided in Table 1. Of the 132 patients, 45 (34%) were found positive for EGFR mutation. Of the positive case, 36 patients were positive for EGFR Exon-19 deletions mutation and nine patients were positive for EGFR Exon-21 SNP (L858R or L861Q). The result from allele specific real time PCR was taken as gold standard result and was compared with the results obtained using ctDNA NGS test.
Table 1.
Summary of clinical and demographic details of the subjects
| Clinical/demographic details | Variables |
|---|---|
| Total number of subjects | 132 |
| Wild type for EGFR Exon-19 and Exon-21 hotspot mutation | 87 |
| Mutated for EGFR (Exon 19/Exon 21) | 45 |
| EGFR Exon-19 mutated subjects | 36 |
| EGFR Exon-21 mutated subjects | 9 |
| Clinical stage** | Stage IV |
| Gender | |
| Number of male subjects | 92 |
| Number of female subjects | 40 |
| Histology | |
| Adenocarcinoma | 113 |
| Squamous cell carcinoma | 19 |
| Smoking status | |
| Smokers | 77 |
| Non-smokers | 55 |
Note:
All patients were diagnosed with distant metastases which included bone, brain, liver, adrenal, and lung sites.
Targeted sequencing of hotspot mutations in ctDNA NGS assay
The targeted sequencing of the 163 samples was performed at an average depth of ~100,000×. More than 85% of the data passed Q30 Phred score. Of the total reads obtained, ~99% of them mapped to the reference human genome (hg19). Of the total aligned reads, 95% of them mapped to the targeted region. After performing the variant calling and filtering, the EGFR mutations overlapping the Exon-19 and Exon-21 hotspot regions were evaluated for further analysis. The result obtained is summarized in Figure 1.
Overall, out of 45 EGFR positive patients (as per tissue biopsy), ctDNA NGS test identified 41 patients as EGFR positive. Of these, we found 34 and seven patients positive for EGFR Exon-19 and Exon-21, respectively (Table 2). All 87 EGFR negative patients (as per tissue biopsy), were found to be EGFR negative using ctDNA NGS test. The overall accuracy, sensitivity, specificity, and precision of the ctDNA NGS test was 96.97%, 91.11%, 100%, and 100% respectively (Table 3). Of the total EGFR positive patients identified from ctDNA NGS test, in 47% of them the mutant allele percentage detected in ctDNA was less than 1%, with the majority of them being EGFR Exon-19 mutations (Figure 2). There were only ten patients (14.6% of total EGFR positive patients) for whom we found mutant allele presence at more than 10% in ctDNA NGS test. The least mutant allele percentage detected using ctDNA NGS test was 0.03%, whereas, the highest mutant allele percentage observed was 37.27%. Of the total EGFR Exon-19 mutated patients, 50% of them carried EGFR E746-A750del (Table 3) mutation which is one of the most frequent deletions reported in literature for NSCLC patients (COSMIC ID: COSM12980). There were four patients for whom we could not detect EGFR mutation in cfDNA NGS test. For these patients, the DNA from tumor tissue biopsy was also sequenced using the same targeted sequencing NGS protocol, and in all four patients the tissue sample was found to be positive for EGFR (Table 4).
Table 2.
Performance metrics for ctDNA NGS test (calculated based on clinical validation of 132 patient samples)
| Patient type | Patient count
|
Sensitivity (%) | Specificity (%) | Accuracy (%) | Precision (%) | |||
|---|---|---|---|---|---|---|---|---|
| True positive | False negative | True negative | False positive | |||||
| Exon-19 mutated | 34 | 2 | 96 | 0 | 94.44 | 100 | 98.48 | 100 |
| Exon-21 mutated | 7 | 2 | 123 | 0 | 77.78 | 100 | 98.48 | 100 |
| Overall mutated | 41 | 4 | 87 | 0 | 91.11 | 100 | 96.97 | 100 |
Notes: True positive: samples predicted as EGFR mutated in allele specific real time PCR and ctDNA NGS tests. False negative: samples with EGFR mutation in allele specific real time PCR but Wild type in ctDNA NGS test. True negative: samples predicted as Wild type EGFR in allele specific real time PCR and ctDNA NGS tests. False positive: samples predicted as Wild type EGFR in allele specific real time PCR test but mutated EGFR in ctDNA NGS test.
Abbreviations: ctDNA, circulating tumor DNA; NGS, next generation sequencing.
Table 3.
List of samples detected as positive in allele specific real time PCR test and its status in ctDNA NGS test
| Sample ID | Chromosome | hg19 position | Reference | Alt | Amino acid change | Alt allele frequency (%) |
|---|---|---|---|---|---|---|
| 31906 | chr7 | 55242479 | CAACATCTCCGAAAGCCAACAAGGA | C | 751-758del | 4.02 |
| 21321 | chr7 | 55242464 | AGGAATTAAGAGAAGC | A | E746-A750del | 21.32 |
| 23830 | chr7 | 55242464 | AGGAATTAAGAGAAGC | A | E746-A750del | 0.29 |
| 24867 | chr7 | 55242464 | AGGAATTAAGAGAAGC | A | E746-A750del | 1.14 |
| 39626 | chr7 | 55242464 | AGGAATTAAGAGAAGC | A | E746-A750del | 0.06 |
| 40278 | chr7 | 55242464 | AGGAATTAAGAGAAGC | A | E746-A750del | 0.03 |
| 40279 | chr7 | 55242464 | AGGAATTAAGAGAAGC | A | E746-A750del | 1.0 |
| 41714 | chr7 | 55242464 | AGGAATTAAGAGAAGC | A | E746-A750del | 3.37 |
| 41767 | chr7 | 55242464 | AGGAATTAAGAGAAGC | A | E746-A750del | 0.78 |
| 45203 | chr7 | 55242464 | AGGAATTAAGAGAAGC | A | E746-A750del | 0.86 |
| 46093 | chr7 | 55242464 | AGGAATTAAGAGAAGC | A | E746-A750del | 0.96 |
| 51134 | chr7 | 55242464 | AGGAATTAAGAGAAGC | A | E746-A750del | 0.51 |
| 28340 | chr7 | 55242464 | AGGAATTAAGAGAAGC | A | E746-A750del | 0.15 |
| 19779 | chr7 | 55242465 | GGAATTAAGAGAAGCA | G | E746-A750del | 3.07 |
| 23045 | chr7 | 55242465 | GGAATTAAGAGAAGCA | G | E746-A750del | 0.27 |
| 29938 | chr7 | 55242465 | GGAATTAAGAGAAGCA | G | E746-A750del | 5.08 |
| 34230 | chr7 | 55242465 | GGAATTAAGAGAAGCA | G | E746-A750del | 1.25 |
| 40089 | chr7 | 55242465 | GGAATTAAGAGAAGCA | G | E746-A750del | 0.16 |
| 42603 | chr7 | 55242465 | GGAATTAAGA | G | E746-R748del | 10.64 |
| 39100 | chr7 | 55242465 | GGAATTAAGA | G | E746-R748del | 3.57 |
| 36135 | chr7 | 55242466 | GAATTAAGAGAAGCAACA | G | E746-T751del | 0.52 |
| 35630 | chr7 | 55242466 | GAATTAAGAGAAGCAACAT | G | E746-T751del | 0.08 |
| 48807 | chr7 | 55242466 | GAATTAAGAGAAGCAACAT | G | E746-T751del | 0.08 |
| 23047 | chr7 | 55242466 | GAATTAAGAGAAGCAACATC | G | E746-T751del | 0.06 |
| 45201 | chr7 | 55242467 | AATTAAGAGAAGC | A | L747-A750del | 37.37 |
| 36137 | chr7 | 55242466 | GAATTAAGAGA | G | L747-E749del | 4.02 |
| 23508 | chr7 | 55242469 | TTAAGAGAAGCAACATCTC | T | L747-S752del | 0.54 |
| 46955 | chr7 | 55242469 | TTAAGAGAAGCAACATCTC | T | L747-S752del | 12.82 |
| 36139 | chr7 | 55242469 | TTAAGAGAAGCAACATCTC | T | L747-S752del | 0.12 |
| 35631 | chr7 | 55242466 | GAATTAAGAGAAGCAA | G | L747-T751del | 35.38 |
| 36136 | chr7 | 55242466 | GAATTAAGAGAAGCAA | G | L747-T751del | 9.9 |
| 27956 | chr7 | 55242467 | AATTAAGAGAAGCAAC | A | L747-T751del | 0.27 |
| 42982 | chr7 | 55242470 | TAAGAGAAGCA | T | R748-A-750del | 18.31 |
| 25789 | None | None | None | None | None | |
| 41354 | None | None | None | None | None | |
| 50482 | chr7 | 55242465 | AGGAATTAAGAGAAGC | A | E746-A750del | 1.0 |
| 25790 | chr7 | 55259515 | T | G | 0.31 | |
| 28574 | chr7 | 55259515 | T | G | 33.31 | |
| 28580 | chr7 | 55259515 | T | G | 11.85 | |
| 39627 | chr7 | 55259515 | T | G | 25.39 | |
| 42704 | chr7 | 55259515 | T | G | 1.42 | |
| 43563 | chr7 | 55259515 | T | G | 15.92 | |
| 33481 | None | None | None | None | None | |
| 36208 | None | None | None | None | None | |
| 34560 | chr7 | 55259525 | T | G | L861R | 1.0 |
Abbreviations: Alt, alternate allele; ctDNA, circulating tumor DNA; NGS, next generation sequencing.
Figure 2.
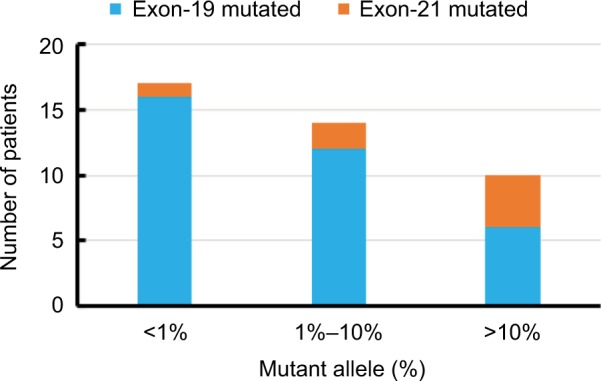
Allele frequency distribution of EGFR mutations (Exon 19 and Exon 21) from the ctDNA NGS assay.
Abbreviations: ctDNA, circulating tumor DNA; NGS, next generation sequencing.
Table 4.
False negative samples on liquid biopsy reconfirmed the mutation status on tissue biopsy by NGS
| Sample ID | Chromosome position | Reference | Alt | Amino acid change | Tissue biopsy allele frequency (%) |
|---|---|---|---|---|---|
| 25789 | 55242464 | AGGAATTAAGAGAAGC | A | p.Glu746_Ala750del | 9 |
| 33481 | 55259515 | T | G | p.Leu858Arg | 11.7 |
| 36208 | 55259515 | T | G | p.Leu858Arg | 32.9 |
| 41354 | 55242464 | AGGAATTAAGAGAAGC | A | p.Glu746_Ala750del | 4.5 |
Abbreviation: NGS, next generation sequencing.
There were 31 cases for which tumor biopsy was not available. For these cases, liquid biopsy sample was available and hence ctDNA-based NGS testing was performed on the same. As an outcome of liquid biopsy alone in this subset, six patients were positive for EGFR mutation on ctDNA NGS testing (Table 5).Out of five positive cases two patients had Exon 19 deletion and the remaining three had Exon 21 L858R mutation. There was one patient who was positive for T790M mutation. The mutant allele fraction varies from 0.01% to 30.3%.
Table 5.
Mutation status of subset of patients (31) where only liquid biopsy was available for EGFR mutation assessment
| Sample ID | Chromosome 7 | Reference | Alt | Amino acid change | Liquid biopsy allele frequency (%) |
|---|---|---|---|---|---|
| 19778 | 55242464 | AGGAATTAAGAGAAGC | A | p.Glu746_Ala750del | 0.01 |
| 29939 | 55259515 | T | G | p.Leu858Arg | 0.41 |
| 34227 | 55259515 | T | G | p.Leu858Arg | 30.3 |
| 42981 | 55259515 | T | G | p.Leu858Arg | 11.53 |
| 48400 | 55242464 | AGGAATTAAGAGAAGC | A | p.Glu746_Ala750del | 0.007 |
We also verified our ctDNA NGS findings for all Exon 19 deletions and T790M mutation positive cases on plasma using ddPCR. Table 6 summarizes the comparison of mutation status across three different technologies for different patients. We found nearly 100% concordance between allele specific PCR, ctDNA NGS, and ddPCR.
Table 6.
Concordance assessment of mutation positivity in EGFR Exon 19 across different platforms
| Sample ID | Exon number | NGS (ctDNA) | qPCR (Tissue) | droplet digital PCR (ctDNA) |
|---|---|---|---|---|
| 19779 | Exon 19 | Positive | Positive | Positive |
| 21321 | Exon 19 | Positive | Positive | Positive |
| 23047 | Exon 19 | Positive | Positive | Positive |
| 23045 | Exon 19 | Positive | Positive | Positive |
| 23508 | Exon 19 | Positive | Positive | Positive |
| 23830 | Exon 19 | Positive | Positive | Positive |
| 24867 | Exon 19 | Positive | Positive | Positive |
| 29938 | Exon 19 | Positive | Positive | Positive |
| 34230 | Exon 19 | Positive | Positive | Positive |
| 39100 | Exon 19 | Positive | Positive | Positive |
| 39626 | Exon 19 | Positive | Positive | Positive |
| 40089 | Exon 19 | Positive | Positive | Positive |
| 40278 | Exon 19 | Positive | Positive | Positive |
| 40279 | Exon 19 | Positive | Positive | Positive |
| 41714 | Exon 19 | Positive | Positive | Positive |
| 41767 | Exon 19 | Positive | Positive | Positive |
| 42603 | Exon 19 | Positive | Positive | Positive |
| 45201 | Exon 19 | Positive | Positive | Positive |
| 45203 | Exon 19 | Positive | Positive | Positive |
| 46093 | Exon 19 | Positive | Positive | Positive |
| 46955 | Exon 19 | Positive | Positive | Positive |
| 50482 | Exon 19 | Positive | Positive | Positive |
| 51134 | Exon 19 | Positive | Positive | Positive |
| 27956 | Exon 19 | Positive | Positive | Positive |
| 28340 | Exon 19 | Positive | Positive | Positive |
| 31906 | Exon 19 | Positive | Positive | Positive |
| 35630 | Exon 19 | Positive | Positive | Positive |
| 35631 | Exon 19 | Positive | Positive | Positive |
| 36135 | Exon 19 | Positive | Positive | Positive |
| 36136 | Exon 19 | Positive | Positive | Positive |
| 36137 | Exon 19 | Positive | Positive | Positive |
| 36139 | Exon 19 | Positive | Positive | Positive |
| 42982 | Exon 19 | Positive | Positive | Positive |
| 48807 | Exon 19 | Positive | Positive | Positive |
Abbreviations: ctDNA, circulating tumor DNA; NGS, next generation sequencing.
We also analyzed the false negatives of ctDNA NGS on ddPCR, and noticed that one of two Exon 19 deletion cases was positive on ddPCR.
Discussion
In this study, we have validated our ctDNA NGS assay on 163 NSCLC patients from India. Our study has shown very high (96.97%) concordance between allele specific real time PCR (on tumor tissue DNA) and ctDNA NGS test performed on the plasma cell-free DNA for the EGFR mutation detection (Exon 19 and Exon 21 mutations).
For ctDNA mutation profiling, NGS was the choice of technology based on its sensitivity, specificity, robustness/reproducibility, ability to detect known and unknown mutations in several genes in one assay, and above all, its cost effectiveness/affordability in testing in Indian patient population without having a major impact on the cancer treatment management.
The precision and specificity of the ctDNA NGS assay observed was 100%. A false negative rate of 3% was observed in this study, and is one of the lowest documented in ctDNA-based EGFR testing. There were four EGFR mutation positive patients (as per tissue biopsy), where the EGFR mutation could not be detected using ctDNA NGS assay. A retrospective workup on the tumor tissue DNA using the targeted NGS assay, revealed mutation positivity, further confirming the false negative status of ctDNA for EGFR mutation. There were samples in which ctDNA NGS assay failed to detect EGFR mutations, which could be due to various factors including inherent tumor biology and extremely low mutant frequency which is beyond limit of detection for the assay.
Apart from being a non-invasive/minimally invasive test with high accuracy, the ctDNA NGS test also offers other advantages which include: a) ability to identify novel mutation, b) ability to identify low frequency mutation, c) faster turnaround time.
The overall sensitivity of the ctDNA assay was 91.1%; for Exon 19 it was 94.4% and for Exon 21 it was 77.8%. The poor sensitivity of Exon 21 could be attributed to the small number of events in the study subset with Exon 21 positivity. The sensitivity of ctDNA NGS assay is a function of the average panel depth. The higher the panel coverage depth, the higher the sensitivity, thus pushing the limits of sensitivity less than 0.1 for some of the clinical samples in the study. However, with higher depth the false positivity increases, nevertheless, using our unique bioinformatics proprietary approach we have neutralized the false positivity in the assay.
Our results compare favorably in terms of higher concordance rate, sensitivity, and specificity as compared with some of the earlier studies on tumor tissue biopsy vs. ctDNA, to establish the EGFR mutation status.12–26 There is high variability in the performance metrics of plasma cell-free DNA vs. tumor tissue biopsy that could be due to various reasons including; sample type and size, the methods of sample collection, storage, isolation and amount of the ctDNA obtained, followed by variation in mutation detection methods. In spite of this variability, a recent meta-analysis on diagnostic value of plasma cfDNA in place of tissue biopsy demonstrated adequate diagnostic accuracy for plasma cell-free DNA-based EGFR testing as promising screening test for NSCLC patients.27 So far, the circulating nucleic acid isolation kit from Qiagen has been very robust in obtaining consistent and good quality cell-free DNA. Among the technologies used for mutation detection, several competing technologies are on the market, including real time PCR, ddPCR, massARRAY, and customized NGS protocols for liquid biopsy samples. One of the major advantages of NGS includes multiplexing of different genes for targeted amplification followed by sequencing. This facilitates the option to derive the mutation status of hotspot mutations as well as other novel mutations within the amplicons being sequenced. In AS-PCR and ddPCR – the technology involves detection of predetermined hotspot/driver mutations alone in each assay and not the novel mutations. Also, multiplexing in allele specific (AS)-PCR and ddPCR has limitations in terms of the diversity of fluorescent probes used for detection of different mutations in the same pool. All the metrics of ctDNA assay validation were calculated based on the mutation status and allele frequency burden estimated for clinical samples and cell-free DNA external reference standard with known mutant allele burden at 5%, 1% obtained from Horizon discovery (HD780).
NGS assay, being more exploratory as compared to known mutant target specific real time PCR assays, could detect novel/rare indels in Exon 19, hence, added value to have EGFR mutation status evaluated by NGS-based methods as and when applicable and feasible. We also observed more than one type of Exon 19 deletions in the same patient, appearing in the ctDNA at different allele frequency burden. Providing an estimate of allele frequency burden in a patient, at baseline or at any given point of time, helps in monitoring the drug response during subsequent liquid biopsy sampling events. Approximately a fraction, i.e., 20% patients who enrolled in the study, but could not get their tissue testing done, due to unavailability of tumor tissue, and only underwent the plasma ctDNA-based EGFR mutation status assessment, and there was 19.4% mutation positivity observed for Exon 19/21 in these patients. Put together, considering all the 163 patients who initially enrolled in the study, nearly 12.2% of patients could get the option of targeted therapy with EGFR TKI in this study subset, benefited by plasma ctDNA-based NGS testing, who would have been otherwise considered for standard chemotherapy.
The clinical efficacy and sensitivity of EGFR TKIs gefitinib, erlotinib, and afatinib in EGFR mutation positive (except T790M) NSCLC patients is well established.28 Recent studies have shown patients with mutation positivity on ctDNA had poor clinical outcome measured by PFS and OS as compared to those patients who were ctDNA negative and tissue DNA positive for EGFR mutation status.29,30 Such a comparison could not be assessed in our study as we had more than 97% of the patients with ctDNA positivity in concordance with tissue DNA and they are being closely followed-up. In both Caucasian and Asian populations similar high concordance between tissue EGFR status and liquid biopsy has been reported.9,12,21,27 Most of these studies on NSCLC have reported >90% concordance in EGFR mutation status between tissue biopsy and plasma cfDNA using different technologies. Kimura et al12 have shown 92.9% (39/42) concordance in EGFR mutation status between tumors and serum samples using direct sequencing and ARMS technology. In a paired tumor and plasma samples study by He et al,21 the concordance between direct sequencing and mutant-enriched PCR was 94.4% (17/18). In another study by Douillard et al, (ASSESS)9,44 the concordance for matched tissue/cytologic and plasma samples was tested with the Qiagen therascreen kit, and was highly consistent with that of Efficacy, Safety, Tolerability of Gefitinib as 1st Line in Caucasian Patients With EGFR Mutation Positive Advanced NSCLC (IFUM) study45 (concordance 95% [131 of 138], sensitivity 73% [16 of 22], specificity 99% [115 of 116], positive predictive value 94% [16 of 17], and negative predictive value 95% [115 of 121]). From our experience, and based on clinical studies published in the literature,25 the following could be some of the reasons for achieving a high concordance rate between tissue biopsy and plasma cfDNA: a) the design of the study – all the patients were recruited treatment naïve and parallel sampling of tissue and liquid biopsy was ensured to avoid any temporal variation in the mutation evolution in the plasma; b) all patients were in advanced stage of the cancer (stage IV); c) ultra-deep sequencing of the samples (median depth of 100,000×); d) in NSCLC, adenocarcinoma patients, literature11 suggests that >90% of these patients carry mutations in either Exon19 or Exon 21 of EGFR.
A few studies29,31 have shown that the yield of mutant allele burden in ctDNA at baseline was associated with aggressive disease, which could not be verified in our study due to the small number of subjects presenting high mutant allele burden. The yield of cfDNA varied across all the subjects though not very significantly. The mutant allele burden varied significantly across subjects who all presented with advanced disease. This further indicates that not all tumors with similar clinical staging would yield comparable ctDNA, and the penetrance of mutant DNA molecules from the primary or metastatic sites may vary across individuals with the same EGFR mutations and similar clinical staging at the time of diagnosis.
To summarize, until date, to the best of our knowledge, in an Indian population, the current study on 163 NSCLC patients provides the largest cohort of data demonstrating the application of liquid biopsy/ctDNA-based EGFR mutation detection in blood. Post-baseline, periodic blood sampling of these patients would enable us to assess the treatment response during the course of treatment and at the time of disease progression.
In conclusion, this study successfully demonstrates the clinical validation of ctDNA-based molecular profiling by NGS methods. All the pre-analytical and analytical variables that may affect the results of ctDNA and its comparison with tissue typing have been carefully addressed and sampling events have been synchronized for all the patients to avoid such variations. The decision to use EGFR Exon 19 and Exon 21, was to investigate the role of major oncogenic drivers involved in lung carcinogenesis with a potential role in personalized therapy planning in NSCLC patients. The co-existing nature of different mutant subgroups reveals the inherent genetic complexity, providing alternative options to guide treatment decisions. Our performance metrics are on par with the most sensitive methods documented in literature. A non-zero false negativity has been a limitation in ctDNA-based molecular profiling. Further advancements in ctDNA isolation and mutation detection technologies may overcome the barrier of false negativity. Until date, ctDNA remains a marker for prognosis and clinical follow-up. Its sensitivity and robustness in detecting cancer specific mutations in blood has not been well established in diverse clinical presentations of early stage cancers. Considering its dynamic variability across different stages of cancer, it will be a long time before we can use ctDNA as a standard clinical biomarker for early diagnosis or early cancer screening or as a cancer predisposition tool.
Acknowledgments
This work was internally funded. The authors would like to thank the patients for being part of this clinical research study.
We would also like to thank Dr Vijay Patil and Dr Shripad D Banavali, Tata Memorial Hospital, for the technical discussions and suggestions. We thank Dr Bharti Mittal, MedGenome for the ddPCR experiments.
Footnotes
Author contributions
VHV and RV conceptualized and designed the study. VHV and TR acquired the NGS data and analyzed the data. AC performed the real time PCR experiments, RG designed the bioinformatics algorithm for detecting somatic mutations in liquid biopsy. RG and NM established the bioinformatics pipeline for liquid biopsy. VHV and TR designed the ddPCR experiments; VN, AJ, and AD contributed toward subject recruitment. VHV drafted the manuscript. KP and RLV provided critical inputs in revising the manuscript. All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7(3):169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 2.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 3.Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 4.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101(36):13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.National Comprehensive Cancer Network (NCCN) Drugs & Biologics Compendium. [Accessed January 13, 2009]. Available from: www.nccn.org/professionals/drug_compendium/content/contents.asp.
- 6.Wu YL, Sequist LV, Hu CP, et al. EGFR mutation detection in circulating cell-free DNA of lung adenocarcinoma patients: analysis of LUX-Lung 3 and 6. Br J Cancer. 2017;116(2):175–185. doi: 10.1038/bjc.2016.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janku F, Angenendt P, Tsimberidou AM, et al. Actionable mutations in plasma cell-free DNA in patients with advanced cancers referred for experimental targeted therapies. Oncotarget. 2015;6(14):12809–12821. doi: 10.18632/oncotarget.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yung TK, Chan KC, Mok TS, Tong J, To KF, Lo YM. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res. 2009;15(6):2076–2084. doi: 10.1158/1078-0432.CCR-08-2622. [DOI] [PubMed] [Google Scholar]
- 9.Douillard JY, Ostoros G, Cobo M, et al. Gefitinib treatment in EGFR mutated caucasian NSCLC: circulating-free tumor DNA as a surrogate for determination of EGFR status. J Thorac Oncol. 2014;9(9):1345–1353. doi: 10.1097/JTO.0000000000000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitsudomi T, Yatabe Y. Epidermal growth factor receptor in relation to tumor development: EGFR gene and cancer. FEBS J. 2010;277(2):301–308. doi: 10.1111/j.1742-4658.2009.07448.x. [DOI] [PubMed] [Google Scholar]
- 11.Choughule A, Noronha V, Joshi A, et al. Epidermal growth factor receptor mutation subtypes and geographical distribution among Indian non-small cell lung cancer patients. Indian J Cancer. 2013;50(2):107–111. doi: 10.4103/0019-509X.117023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimura H, Suminoe M, Kasahara K, et al. Evaluation of epidermal growth factor receptor mutation status in serum DNA as a predictor of response to gefitinib (IRESSA) Br J Cancer. 2007;97(6):778–784. doi: 10.1038/sj.bjc.6603949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moran T, Paz-Ares L, Isla D, et al. High correspondence between EGFR mutations in tissue and in circulating DNA from non-small-cell lung cancer (NSCLC) patients (p) with poor performance status (PS) J Clin Oncol. 2007;25(18 Suppl):7505. [Google Scholar]
- 14.Bai H, Mao L, Wang HS, et al. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non-small-cell lung cancer. J Clin Oncol. 2009;27(16):2653–2659. doi: 10.1200/JCO.2008.17.3930. [DOI] [PubMed] [Google Scholar]
- 15.Esposito A, Bardelli A, Criscitiello C, et al. Monitoring tumor-derived cell-free DNA in patients with solid tumors: Clinical perspectives and research opportunities. Cancer Treat Rev. 2014;40(5):648–655. doi: 10.1016/j.ctrv.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 16.Murtaza M, Dawson SJ, Tsui DW, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497(7447):108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 17.Wang S, Han X, Hu X, et al. Clinical significance of pretreatment plasma biomarkers in advanced non-small cell lung cancer patients. Clin Chim Acta. 2014;430:63–70. doi: 10.1016/j.cca.2013.12.026. [DOI] [PubMed] [Google Scholar]
- 18.Jing CW, Wang Z, Cao HX, Ma R, Wu JZ. High resolution melting analysis for epidermal growth factor receptor mutations in formalin-fixed paraffin-embedded tissue and plasma free DNA from non-small cell lung cancer patients. Asian Pac J Cancer Prev. 2013;14(11):6619–6623. doi: 10.7314/apjcp.2013.14.11.6619. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, Liu D, Li S, et al. Comparison of EGFR signaling pathway somatic DNA mutations derived from peripheral blood and corresponding tumor tissue of patients with advanced non-small-cell lung cancer using liquidchip technology. J Mol Diagn. 2013;15(6):819–826. doi: 10.1016/j.jmoldx.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 20.Kim HR, Lee SY, Hyun DS, et al. Detection of EGFR mutations in circulating free DNA by PNA-mediated PCR clamping. J Exp Clin Cancer Res. 2013;32(1):50. doi: 10.1186/1756-9966-32-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He C, Liu M, Zhou C, Zhang J, Ouyang M, Zhong N, Xu J. Detection of epidermal growth factor receptor mutations in plasma by mutant-enriched PCR assay for prediction of the response to gefitinib in patients with non-small-cell lung cancer. Int J Cancer. 2009;125(10):2393–2399. doi: 10.1002/ijc.24653. [DOI] [PubMed] [Google Scholar]
- 22.Kuang Y, Rogers A, Yeap BY, et al. Noninvasive detection of EGFR T790M in gefitinib or erlotinib resistant non-small cell lung cancer. Clin Cancer Res. 2009;15(8):2630–2636. doi: 10.1158/1078-0432.CCR-08-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brevet M, Johnson ML, Azzoli CG, Ladanyi M. Detection of EGFR mutations in plasma DNA from lung cancer patients by mass spectrometry genotyping is predictive of tumor EGFR status and response to EGFR inhibitors. Lung Cancer. 2011;73(1):96–102. doi: 10.1016/j.lungcan.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goto K, Ichinose Y, Ohe Y, et al. Epidermal growth factor receptor mutation status in circulating free DNA in serum: from IPASS, a phase III study of gefitinib or carboplatin/paclitaxel in non-small cell lung cancer. J Thorac Oncol. 2012;7(1):115–121. doi: 10.1097/JTO.0b013e3182307f98. [DOI] [PubMed] [Google Scholar]
- 25.Liu X, Lu Y, Zhu G, et al. The diagnostic accuracy of pleural effusion and plasma samples versus tumour tissue for detection of EGFR mutation in patients with advanced non-small cell lung cancer: comparison of methodologies. J Clin Pathol. 2013;66(12):1065–1069. doi: 10.1136/jclinpath-2013-201728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao X, Han RB, Zhao J, et al. Comparison of epidermal growth factor receptor mutation statuses in tissue and plasma in stage I-IV non-small cell lung cancer patients. Respiration. 2013;85(2):119–125. doi: 10.1159/000338790. [DOI] [PubMed] [Google Scholar]
- 27.Luo J, Shen L, Zheng D. Diagnostic value of circulating free DNA for the detection of EGFR mutation status in NSCLC: a systematic review and meta-analysis. Sci Rep. 2014;4:6269. doi: 10.1038/srep06269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yap TA, Popat S. Toward precision medicine with next-generation EGFR inhibitors in non-small-cell lung cancer. Pharmgenomics Pers Med. 2014;7:285–295. doi: 10.2147/PGPM.S55339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mok T, Wu YL, Lee JS, et al. Detection and dynamic changes of EGFR mutations from circulating tumor DNA as a predictor of survival outcomes in NSCLC patients treated with first-line intercalated erlotinib and chemotherapy. Clin Cancer Res. 2015;21(14):3196–3203. doi: 10.1158/1078-0432.CCR-14-2594. [DOI] [PubMed] [Google Scholar]
- 30.Karachaliou N, Mayo-de las CC, Queralt C, et al. Association of EGFR L858R mutation in circulating free DNA with survival in the EURTAC trial. JAMA Oncol. 2015;1(2):149–157. doi: 10.1001/jamaoncol.2014.257. [DOI] [PubMed] [Google Scholar]
- 31.Lee YJ, Yoon KA, Han JY, et al. Circulating cell-free DNA in plasma of never smokers with advanced lung adenocarcinoma receiving gefitinib or standard chemotherapy as first-line therapy. Clin Cancer Res. 2011;17(15):5179–5187. doi: 10.1158/1078-0432.CCR-11-0400. [DOI] [PubMed] [Google Scholar]
- 32.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer LR, Zweig AS, Hinrichs AS, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013;41(Database issue):D64–69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.The 1000 Genomes Project Consortium. Auton A, Brooks LD. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. esp.gs.washington.edu [homepage on the Internet] NHLBI Grand Opportunity Exome Sequencing Project (ESP) [Accessed October 11, 2017]. Available from: https://esp.gs.washington.edu/drupal.
- 39.Nagasaki M, Yasuda J, Katsuoka F, et al. Rare variant discovery by deep whole-genome sequencing of 1,070 Japanese individuals. Nat Commun. 2015;6:8018. doi: 10.1038/ncomms9018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moayyeri A, Hammond CJ, Hart DJ, Spector TD. The UK Adult Twin Registry (TwinsUK Resource) Twin Res Hum Genet. 2013;16(1):144–149. doi: 10.1017/thg.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. ncbi.nlm.nih.gov [homepage on the Internet] dbSNP short genetic variations. [Accessed October 11, 2017]. Available from: http://www.ncbi.nlm.nih.gov/SNP/
- 42.Bueno R, Stawiski EW, Goldstein LD, et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat Genet. 2016;48(4):407–416. doi: 10.1038/ng.3520. [DOI] [PubMed] [Google Scholar]
- 43.Li MM, Datto M, Duncavage EJ, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19(1):4–23. doi: 10.1016/j.jmoldx.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reck M, Hagiwara K, Han B, et al. ctDNA Determination of EGFR Mutation Status in European and Japanese Patients with Advanced NSCLC: The ASSESS Study. J Thorac Oncol. 2016;11(10):1682–1689. doi: 10.1016/j.jtho.2016.05.036. [DOI] [PubMed] [Google Scholar]
- 45.Douillard JY, Ostoros G, Cobo M, et al. First-line gefitinib in Caucasian EGFR mutation-positive NSCLC patients: a phase-IV, open-label, single-arm study. Br J Cancer. 2014;110(1):55–62. doi: 10.1038/bjc.2013.721. [DOI] [PMC free article] [PubMed] [Google Scholar]