Natural variation in 347 Arabidopsis accessions reveals differential root plasticity responses to salinity, associated with 100 genetic loci, and roles for HKT1 and CYP79B2 in lateral root development.
Abstract
Salinity of the soil is highly detrimental to plant growth. Plants respond by a redistribution of root mass between main and lateral roots, yet the genetic machinery underlying this process is still largely unknown. Here, we describe the natural variation among 347 Arabidopsis thaliana accessions in root system architecture (RSA) and identify the traits with highest natural variation in their response to salt. Salt-induced changes in RSA were associated with 100 genetic loci using genome-wide association studies. Two candidate loci associated with lateral root development were validated and further investigated. Changes in CYP79B2 expression in salt stress positively correlated with lateral root development in accessions, and cyp79b2 cyp79b3 double mutants developed fewer and shorter lateral roots under salt stress, but not in control conditions. By contrast, high HKT1 expression in the root repressed lateral root development, which could be partially rescued by addition of potassium. The collected data and multivariate analysis of multiple RSA traits, available through the Salt_NV_Root App, capture root responses to salinity. Together, our results provide a better understanding of effective RSA remodeling responses, and the genetic components involved, for plant performance in stress conditions.
INTRODUCTION
Salt stress is a major threat in modern agriculture, affecting 20% of the cultivated area worldwide and half of the irrigated farmlands (OECD/Food and Agriculture Organization of the United Nations, 2015). Plants can adopt multiple strategies to increase their salinity tolerance, such as reduced growth rate, compartmentalization of ions, or synthesis of compatible solutes (Munns and Tester, 2008; Munns and Gilliham, 2015). One of the most robust phenotypes used for screening salinity tolerance is accumulation of sodium in shoot tissue. Exclusion of sodium from the shoot is correlated with maintenance of high rates of photosynthesis, growth, and yield (Munns and Tester, 2008). Allelic variation affecting transcription of HKT1 and CIPK13 was previously established to play a major role in sodium exclusion and, therefore, salinity tolerance (Rus et al., 2006; Munns et al., 2012; Roy et al., 2013). Another trait broadly used for identification of salt stress-related mechanisms is elongation of the main root (Wu et al., 1996; Ding and Zhu, 1997; Liu and Zhu, 1997). While main root elongation is instrumental for seedling establishment, the distribution of the root mass between main and lateral roots is also affected by salt, yet the genetic machinery underlying this process as well as its significance in salt stress tolerance are only now starting to be unraveled (Duan et al., 2013; Geng et al., 2013; Julkowska et al., 2014; Julkowska and Testerink, 2015; Kobayashi et al., 2016; Kawa et al., 2016).
The root is the first plant organ exposed to salinity stress and plays an important role in salt sensing (Galvan-Ampudia et al., 2013; Robbins et al., 2014) as well as signal transduction to the shoot tissue (Choi et al., 2014; Jiang et al., 2012). Salt stress reduces the cell cycle activity at the root meristems, resulting in growth reduction (West et al., 2004). Quiescence of lateral roots is initiated by endodermal abscisic acid (ABA) signaling, which interestingly is also involved in recovery of lateral root growth (Duan et al., 2013). In the long term, salt stress reduces main root growth more severely than lateral root elongation in the Arabidopsis thaliana accession Col-0 (Julkowska et al., 2014), causing remodeling of root system architecture (RSA) compared with control conditions. Interestingly, other Arabidopsis accessions show different RSA remodeling response to salt stress, indicating natural variation that can be exploited for candidate gene identification by means of genome-wide association studies (GWAS). Since the root system is important for efficient water uptake and ion exclusion (Faiyue et al., 2010; Ristova and Busch, 2014), changes in RSA are likely to contribute to plant performance under salt stress conditions (Julkowska and Testerink, 2015).
In this study, we explore natural variation in RSA development under control and salt stress conditions by screening 347 Arabidopsis accessions of the HapMap population (Horton et al., 2012; Li et al., 2010). Our results indicate increased phenotypic variation in main and lateral root length in salt stress conditions compared with control conditions. By performing GWAS, we identified several candidate genetic loci to be associated with the remodeling of root architecture in response to salt. Further analysis of HKT1 and CYP79B2/B3 candidate genes supports a role for them in modulation of lateral root development under salt stress conditions, increasing our understanding of root responses in contributing to plant performance under stress. To optimally exploit our complex RSA phenotyping data set beyond the analysis described in this scientific publication, we also make our data available through the Salt_NV_Root App (https://mmjulkowska.github.io/Salt_NV_RootApp/). This tool will facilitate other researchers in exploring our data set and finding new leads for further research on RSA remodeling.
RESULTS
Salt-Induced Remodeling of RSA Relies on Altered Lateral Root Development
To examine natural variation in root architecture under salt stress conditions, 347 Arabidopsis accessions of the HapMap population (Weigel and Mott, 2009; Supplemental Data Set 1) were grown on agar plates under control and two salt stress conditions (75 and 125 mM NaCl). Using EZ-Rhizo software, 17 RSA traits (Figure 1A; Supplemental Data Set 2) were extracted from the images of 8-d-old plants grown under control condition and 12-d-old plants for both salt stress conditions. Col-0 was used as the reference accession across individual experimental batches, validating the reproducibility of the experiment (Supplemental Figure 1). Using ANOVA, we identified significant effects of medium and genotype on all RSA traits except for lateral root patterning (lateral root density per branched zone) (Supplemental Data Set 2). Additionally, using two-way ANOVA, we identified significant interactions between medium and genotype in root angle traits (main root vector angle and straightness) and distribution of mass between main and lateral root (for example lateral root density per main root length) (Supplemental Data Set 2).
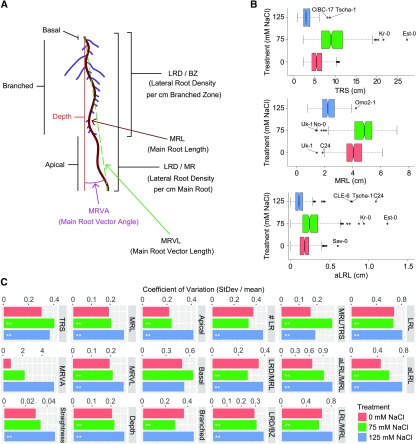
Figure 1.
Natural Variation Observed in RSA Plasticity in Response to Salt Stress.
The effect of salt stress on RSA was examined for 347 Arabidopsis accessions (Supplemental Data Set 1) on 8- and 12-d-old seedlings grown in control and salt stress conditions, respectively.
(A) Overview of RSA parameters obtained from EZ-Rhizo quantification. Additionally, lateral root length (LRL), average lateral root length (aLRL), as well as the ratios between main root (MRL), average lateral root length aLRL), and total root size (TRS) were calculated. All RSA traits are listed in Supplemental Data Set 2.
(B) Natural variation in total root size (TRS), main root length (MRL), and average lateral root length (aLRL) is presented with notched box plots. The accessions representing outliers in individual traits are indicated. Box plots of all RSA traits are presented in Supplemental Figure 2.
(C) Coefficient of variation observed in all 17 RSA traits per salt stress condition for 347 Arabidopsis accessions studied. The significant differences in variation between control and salt stress are indicated, *P value < 0.05 and **P value < 0.01, as calculated using f-tests.
Substantial natural variation was observed for all recorded RSA traits, and accessions identified as outliers differed among traits and conditions (Figure 1B; Supplemental Figure 2). The variance within the population differed between individual traits: straightness showing the smallest, and main root vector angle the largest, variance (Figure 1C). Salt stress increased the observed variance in traits related to main and lateral root length, but not in lateral root density, indicating that observed natural variation in response to salt stress is mainly due to differences between accessions in maintenance of main and lateral root growth rather than root patterning.
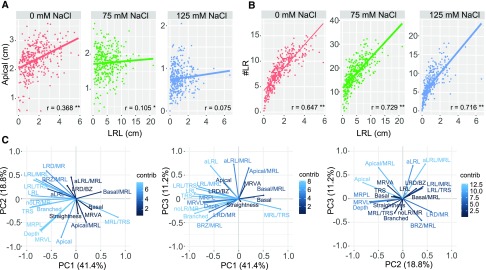
Individual components of RSA were strongly correlated under control conditions (Figures 2A and 2B; Supplemental Data Set 3). Several of these correlations, including the number of lateral roots with main root length, were significant across all conditions, indicating again that salt does not alter lateral root patterning or that the natural variation therein is limited (Supplemental Data Set 3). However, a significant correlation between the length of the apical zone and lateral root length was observed only under control conditions (Figure 2A), indicating that salt stress increased the variation in the distance from the root tip to the point at which lateral roots start to emerge and elongate, which might be related to different rates of lateral root emergence upon salt stress. The correlation between lateral root length and lateral root number was maintained in seedlings grown under salt stress conditions (Figure 2B), although a decrease in correlation strength was observed. Thus, lateral root emergence had a significant contribution to lateral root size in all conditions despite the increased variability in lateral root length (Figure 1C). The 17 RSA traits, together with the length of the individual zones relative to the main root length, were used for principal component analysis, reducing the data dimensionality to three principal components (PC1–3) (Figure 2C; Supplemental Figure 3 and Supplemental Data Set 4). Traits corresponding to lateral root development, such as number of lateral roots per main root, lateral root length, and size of the branched zone significantly contributed to PC1, explaining 41% of the variance. PC2, explaining 18.8% of the variance, corresponded to the main root-related traits, such as main root path length, vector length, and depth. The traits related to lateral root elongation, such as average lateral root length, significantly contributed to PC3, explaining 11.2% of the observed variance. Thus, the majority of the natural variation in RSA is harbored in the distribution of the root mass between main and lateral roots (PC1), maintenance of main root growth (PC2), and lateral root emergence and elongation (PC3). Summarizing the RSA results in three PCs allowed identification of the RSA traits exhibiting most natural variation and reveals phenotypic plasticity in response to salt stress, as the differences between the accessions grown under different conditions are evident even when the dimensionality of the RSA is reduced to three PCs (Supplemental Figure 3).
Figure 2.
Salt Affects the Relationship between RSA Components, with Lateral Root Development Explaining the Majority of Observed Variation.
(A) and (B) The correlation between apical zone size and total lateral root length (LRL) (A) and lateral root number (#LR) and total lateral root length (LRL) (B) for plants grown under 0, 75, and 125 mM NaCl are presented with red, green, and blue dots, respectively. The lines in corresponding color represent the linear model fitting the correlation per condition. The Pearson correlation coefficients (R) are presented in the lower right corner of each correlation graph, with **P value < 0.01 and *P value < 0.05.
(C) Principal component analysis revealed three PCs explaining 71.4% of variation. The contribution plots show how each RSA trait contributes to each PC, ranging from low contribution (dark-blue) to high contribution (light-blue). The contribution of individual traits to each PC is listed in Supplemental Data Set 4.
GWAS Reveals HKT1 and CYP79B2 as Candidate Genes Explaining Natural Variation in the Response of Lateral Roots to Salt
Natural variation in 17 RSA traits and three PCs (Supplemental Data Set 5) was used as an input for GWAS using the scan_GLS algorithm (Kruijer et al., 2015). After applying correction for population structure (Kang et al., 2008) and exclusion of traits with low narrow heritability (h2 < 0.2; Supplemental Data Set 6) GWAS identified 150, 132l and 254 significantly [–log10(P value) above 5.6] associated SNPs for RSA traits in 0, 75, and 125 mM NaCl, respectively (Supplemental Data Set 7). Most SNPs were associated with the average lateral root length per main root length. The largest variation observed in the relative distribution of root mass from main root length to lateral root length corresponds with the high number of associations found for PC3 and average lateral root length measured under salt stress conditions. Identified associations were examined in detail considering their association with multiple traits, using both individual replicates and average value per accession. As the majority of the associations was mapped using the SNP set with minor allele frequency (MAF) above 0.01, we selected for the associations mapped with MAF of at least 0.01 or with the –log10(P value) score above strict Bonferroni threshold, based on the collection of single nucleotide polymorphisms (SNPs) including the rare alleles, corresponding to the score of 6.6. We further explored the associations identified with multiple RSA traits and different conditions (Supplemental Figure 4 and Supplemental Data Sets 8 to 10). Our selection yielded 100 candidate loci for RSA traits under salt stress conditions, and we subsequently compared their transcriptional changes in tissue-specific expression in response to salt (Dinneny et al., 2008), revealing 10 genes directly underlying the associated SNPs and 60 genes within the 10-kb window of the mapped SNPs to be significantly altered in the cell-type-specific expression data set in response to salt (Supplemental Data Set 11). The candidate genes that were either directly underlying the associated SNP, or located within the linkage disequilibrium of the identified SNP, were found to be significantly enriched in the genes with tissue specific alterations in response to salt stress as tested per hypergeometric test. Since natural variation in salt stress response was mostly found in traits related to the relative distribution of root mass between lateral and main roots, we selected six loci associated with average lateral root length, average lateral root length per main root length, and PC3 in salt stress (Figure 3A; Supplemental Data Set 12) for further analysis.
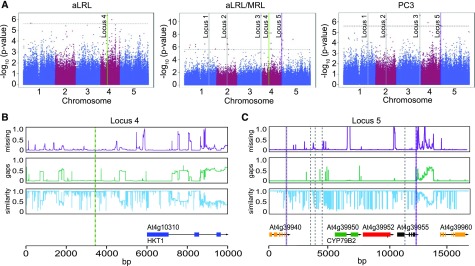
Figure 3.
GWAS Identifies Loci with High and Low Allele Diversity.
(A) GWAS was performed on average lateral root length (aLRL), ratio of average lateral and main root length (aLRL/MRL), and principle component 3 (PC3) at low salt stress conditions (75 mM NaCl) and significant associations above LOD 5.6 with MAF > 0.01 were identified. The dotted line represents the threshold of a LOD of 5.6. The six chosen loci (Supplemental Data Set 12) are marked. The validated loci containing HKT1 (locus 4) are marked with green and CYP79B2 (locus 5) with purple.
(B) and (C) Genetic variation in locus 4 (B) and 5 (C) was studied in 147 HapMap population accessions. The purple graphs represent the portion of missing data, while the green graphs represent deletions present in accessions other than Col-0. The blue graphs represent the sequence similarity compared with Col-0. The open reading frames are aligned in the lowest panel. The location of the associated SNPs from 250k SNP set is represented with highlighted dashed lines, while the SNPs mapped using the 4M SNP set are represented with gray dashed lines.
To fine-map the selected loci, the RSA phenotypes were associated with the SNPs based on the whole-genome sequencing data of the accessions (Alonso-Blanco et al., 2016), to identify additional SNPs and increase the mapping resolution. We identified between 2 and 26 additional SNPs for loci 1, 2, 5, and 6 (Supplemental Data Set 13). For the chosen loci, the regions surrounding the identified SNPs were further examined for sequence variation (Figures 3B and 3C; Supplemental Figures 5 to 8A). Genes located in the 10-kb window of the associated SNPs were examined for natural variation in their expression in root and shoot tissue under control and salt stress conditions using 48 accessions, chosen based on the SNP diversity of the candidate loci and the pronounced differences between their RSA under all conditions studied (Supplemental Figures 5 to 7, 8B, 8C, 9A, and 10; Supplemental Data Set 14). For four loci (1, 2, 3, and 6), we were not able to validate candidate genes due to lack of available mutant lines and/or limited phenotypic changes in response to salt in Col-0 background (Supplemental Figure 7E). Nevertheless, the natural variation in the expression observed among 48 accessions identified candidate background lines for future studies on these loci and can be accessed in the supplemental data (Supplemental Data Set 14).
The genomic region surrounding the 4th locus, associated with average lateral root length and the ratio of average lateral root length and main root length at 75 mM NaCl (Figures 3A and 3B; Supplemental Data Set 12), was found to contain only one gene, At4g10310, also known as Arabidopsis HIGH AFFINITY K+ TRANSPORTER1 (AtHKT1). This gene was previously identified for its role in salinity tolerance (Rus et al., 2001; Munns et al., 2012), although no role in the regulation of root system architecture was noted. The genomic region around the associated SNP contained a high number of SNPs and a number of insertions/deletions in the promoter and intron regions of HKT1 (Figure 3B). We resequenced the locus in six accessions varying in HKT1 expression (Supplemental Figures 9B and 9C; Supplemental Data Sets 15 to 17) and identified 33 cis-regulatory elements (Higo et al., 1999) that were specific for two accessions, N4 and Gr-5, showing high HKT1 expression. The location of those cis-regulatory elements was widespread between 1330 to 3530 bp upstream of the HKT1 start codon (Supplemental Data Sets 16). The coding regions of HKT1 were found to carry 11 polymorphisms compared with Col-0, of which five lead to nonsynonymous mutations and none were located in the transmembrane domains (Supplemental Data Set 17).
Locus 5 contains six SNPs, spanning from the coding region of At4g39940 up to the coding region of At4g39955 (Figure 3C; Supplemental Data Sets 12 and 13). The SNPs were associated with variation in average lateral root length in 125 mM of NaCl, average lateral root length per main root length in both salt stress conditions, and PC3 in 75 mM of NaCl. Interestingly, the expression of At4g39950, also known as CYTOCHROME P450 FAMILY 79 SUBFAMILY B2 (CYP79B2), was reported to exhibit zone-specific expression changes in response to salt (Dinneny et al., 2008) (Supplemental Data Set 11). CYP79B2 is known to convert tryptophan to indole-3-acetaldoxime (IAOx) in the biosynthesis pathway of camalexin, indole glucosinolates, and auxin (Hull et al., 2000; Mikkelsen et al., 2000; Zhao et al., 2002; Glawischnig et al., 2004; Sugawara et al., 2009), and its expression was reported at the sites of lateral root development and in the quiescent center, but no role for CYP79B2 in root development has been reported (Ljung et al., 2005). The promoter region of CYP79B2 showed extensive sequence divergence among the accessions and three SNPs were mapped to the promoter of this gene (Figure 3C), but the region was too diverse to conduct haplotype analysis. No nonsynonymous SNPs were identified in the protein coding sequence of CYP79B2.
Salt-Induced Changes in CYP79B2 Expression Correlate with Maintenance of Lateral Root Development
We further investigated the natural variation in transcriptional changes of CYP79B2, and other genes at the same locus, in response to salt stress. CYP79B2 expression significantly increased upon salt stress in both root and shoot tissue in most of the 48 accessions studied, but much variation was observed (Figure 4A; Supplemental Data Set 14). A three-way ANOVA showed that the response was the same in both tissues as no interaction effect was found, although expression in the root was significantly higher. While At4g39960 also showed significant induction of expression in salt stress in both root and shoot, the effect was more pronounced in the shoot. At4g39940 showed reduced expression in the root and induced expression in the shoot and At4g39955 showed no change upon salt stress (Supplemental Figure 9 and Supplemental Data Set 14). For At4g39952, only very recently transcripts were found, and it likely has very low expression (Liu et al., 2016). We have thus decided to focus on CYP79B2 for further analysis.
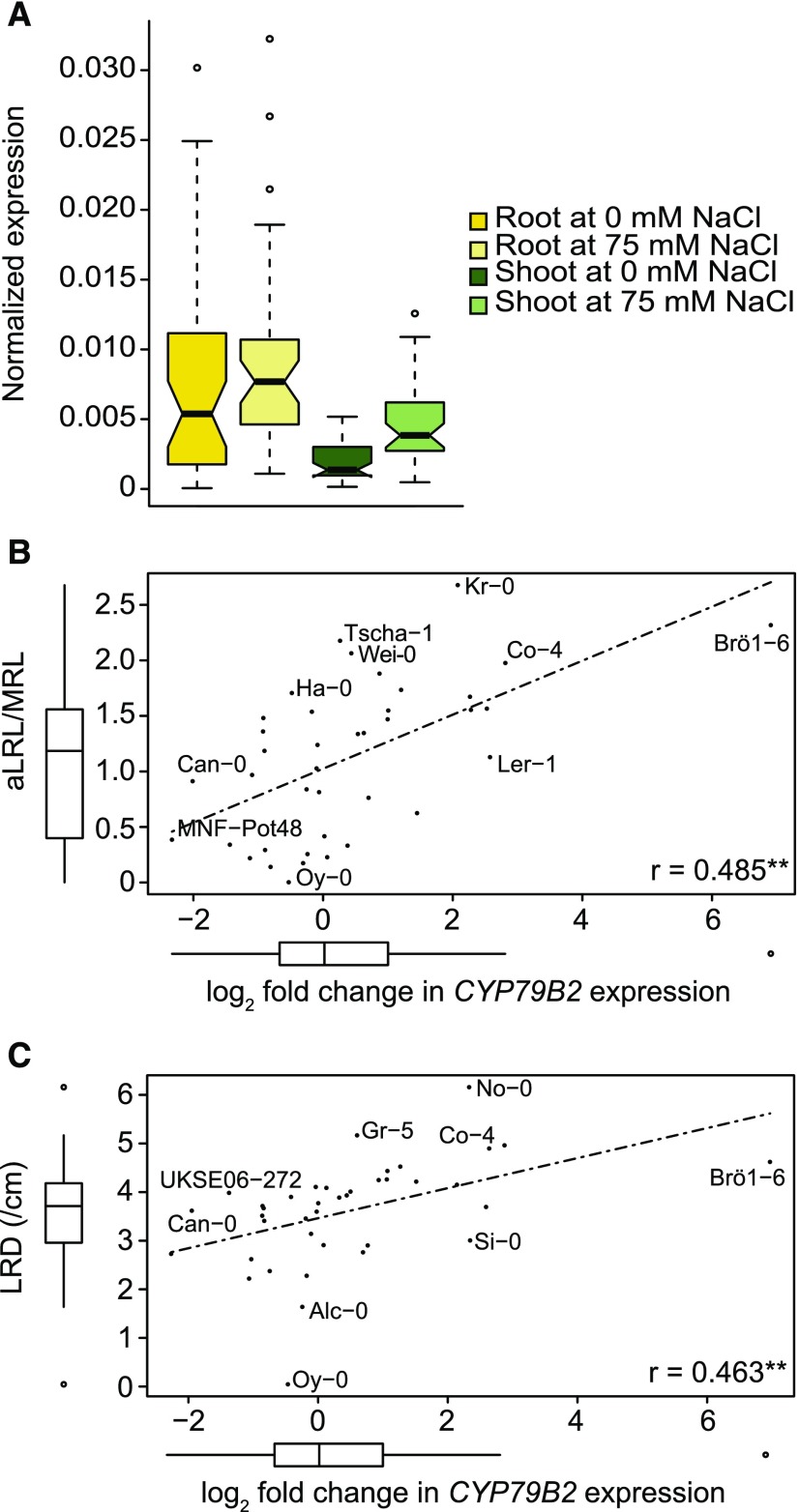
Figure 4.
Change in CYP79B2 Expression in Response to Salt Stress Correlates with Lateral Root Development during Salt Stress.
Natural variation in genes in locus 5 was studied in root and shoot tissue in 5-d-old seedlings treated with mock or 75 mM NaCl for 24 h. The box plot in (A) represents the median and extent of natural variation in CYP79B2 expression as observed for population of 48 accessions. Box plots for the other genes in the same locus can be found in Supplemental Figure 10, together with heat plots of the expression per accession, while the normalized expression of all studied genes is presented in Supplemental Data Set 14. The relative log2 fold change in expression of CYP79B2 between control and salt stress positively correlated to several root architecture traits in the accessions, including average lateral root length per main root length (aLRL/MRL) at 75 mM NaCl (B) and the lateral root density (LRD) at 75 mM NaCl (C). The Pearson correlation coefficients (R) are presented in the lower right corner of each correlation graph, with **P value < 0.01 and *P value < 0.05.
The effect of salt on CYP79B2 expression in the root showed much variation between accessions. The log2 fold change in gene expression in the root upon salt stress ranged from −2.2 in MNF-Pot-4 to 7.2 in Brö1-6 (Supplemental Data Set 14). To investigate whether the change in CYP79B2 expression upon salt stress would correspond with lateral root development, we examined the correlations between the log2 fold change in expression of CYP79B2 and associated RSA traits. The average lateral root length at 75 mM of NaCl significantly correlated with the log2 fold change in CYP79B2 expression upon salt stress (Figure 4B). In addition, the log2 fold change showed a significant positive correlation with the lateral root density in salt stress conditions (Figure 4C).
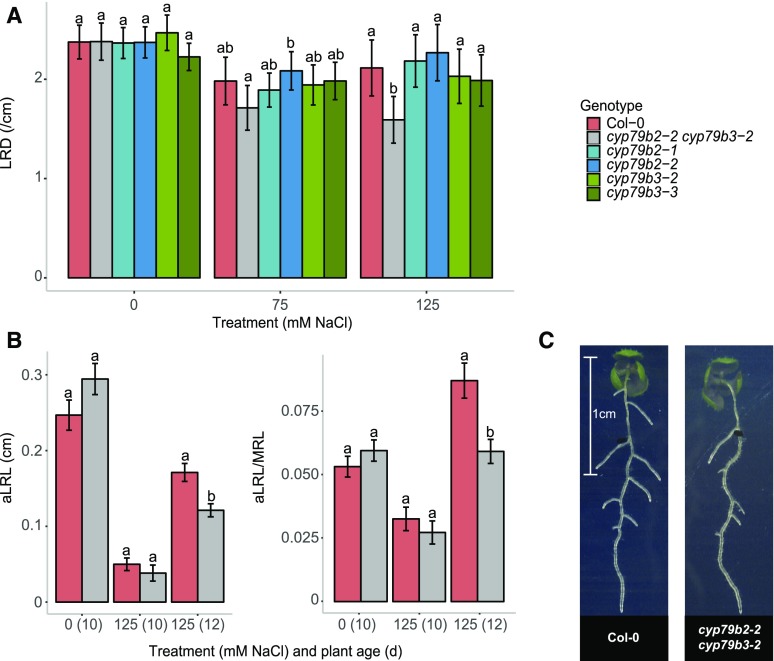
As the function of CYP79B2 is known to partially overlap with that of CYP79B3 (Hull et al., 2000; Zhao et al., 2002; Glawischnig et al., 2004), we investigated the root system architecture of T-DNA insertion lines (Supplemental Figure 11A) for both genes and the cyp79b2 cyp79b3 double mutant at different salt concentrations. The cyp79b2-1 and cyp79b2-2 lines and the double mutant cyp79b2-2 cyp79b3-2 showed a slight, but significant, longer main root independent of the conditions (Supplemental Figure 11B), but no lateral root-related differences were observed under control conditions. Interestingly, cyp79b2-2 cyp79b3-2 mutant seedlings grown on 125 mM NaCl supplemented medium developed significantly fewer lateral roots per centimeter of main root at 10 d after germination (Figure 5A). This is consistent with the observed positive correlation between fold change in CYP79B2 expression upon salt stress and lateral root density (Figure 4C). Double mutant seedlings also developed shorter lateral roots at 12 d after germination, consistent with the mapped traits in the GWAS (Figures 5B and 5C; Supplemental Figure 11B). In summary, our results show that loss of function of CYP79B2, together with the partially overlapping CYP79B3, leads to reduced development of lateral root growth under salt stress, but not in control conditions.
Figure 5.
Loss of Function of Both CYP79B2 and CYP79B3 Results in Reduced Lateral Root Development during Salt Stress.
(A) Four-day-old seedlings of Col-0, cyp79b2, cyp79b3, and cyp79b2 cyp79b3 lines were transferred to 0, 75, and 125 mM NaCl, and RSA of 10-d-old seedlings was quantified. The bar plots represent the average lateral root density (LRD) of three experiments with each 20 replicates; error bars represent pooled se. A mixed linear model was used to determine significant differences and a significant interaction between treatment and genotype was found. Letters represent significant differences (P < 0.05) following a manual-contrasts post-hoc comparison within treatment.
(B) Average lateral root length (aLRL/MRL) and average lateral root length (aLRL) for Col-0 and cyp79b2 cyp79b3 in 10-d-old seedlings treated with 0 mM NaCl and 10- and 12-d-old seedlings treated with 125 mM NaCl. Other lateral root traits are presented in Supplemental Figure 11. Bar plots represent the average trait value as observed in 20 replicates and error bars represent se. All traits were analyzed using two-way ANOVA and significant interactions were found for shown traits. Letters represent significant differences (P < 0.05) following a manual-contrasts post-hoc comparison within treatment.
(C) Pictures of representative seedlings of Col-0 and the cyp79b2 cyp79b3 double mutant at 125 mM NaCl 12-d-old seedlings.
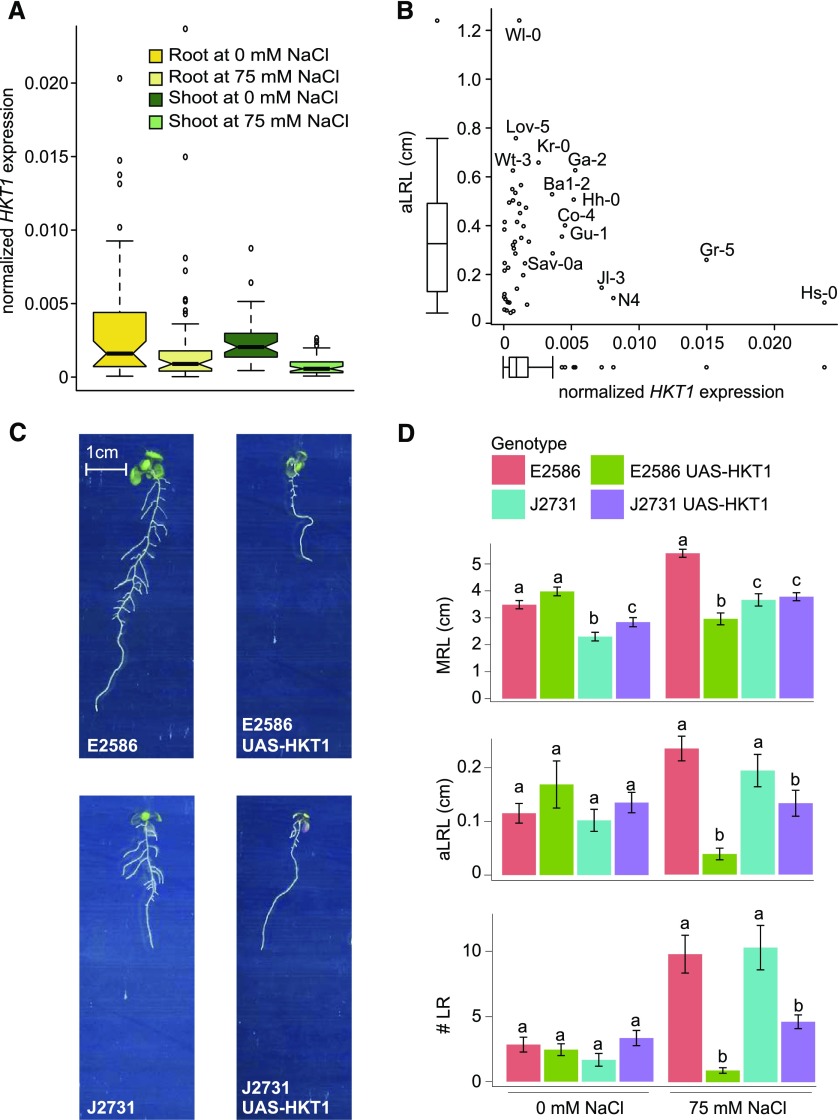
High HKT1 Expression Reduces Lateral Root Development in Saline Conditions
Extensive natural variation in HKT1 expression in the set of 48 accessions was observed in the root tissue of seedlings exposed to salt stress (Figure 6A; Supplemental Figure 9A), with Gr-5, Hs-0, N4, Ga-2, and Jl-3 showing the highest and LDV-58, CUR-3, and Tsu-0 the lowest expression. Although only few accessions exhibited high HKT1 expression, all of these tended to develop shorter lateral roots under salt stress conditions (Figure 6B). By contrast, T-DNA insertion lines with reduced HKT1 expression in the Col-0 background did not exhibit any differences in RSA development compared with wild-type Col-0 (Supplemental Figure 12), consistent with the natural variation expression data. Since ubiquitous overexpression of HKT1 was previously observed to have a detrimental effect on plant development (Møller et al., 2009), we studied the phenotypes of two available lines with enhanced HKT1 expression at the native expression site, the root pericycle (Mäser et al., 2002; Møller et al., 2009): E2586 UAS-HKT1 in Col-0 background and J2731 UAS-HKT1 in C24 background (Møller et al., 2009). No significant differences between the background lines and lines with enhanced HKT1 expression were observed under our control conditions. Interestingly though, under salt stress conditions lines with enhanced HKT1 expression developed fewer and shorter lateral roots in both Col-0 and C24 backgrounds (Figures 6C and 6D), while the high HKT1 expression caused severe reduction in main root length only in Col-0 background (E2586 UAS-HKT1) (Figures 6C and 6D). Thus, while low expression levels of HKT1, either in Arabidopsis accessions or T-DNA lines, seem to have no obvious effects on root morphology, enhanced expression can cause severe alterations in root architecture under salt stress, possibly due to toxic effects of high Na+ concentrations or changes in osmotic potential of root cells.
Figure 6.
High HKT1 Expression Reduces LR Development in Salt Stress Conditions.
(A) Natural variation in the HKT1 expression was studied in root and shoot tissue in 5-d-old seedlings treated with mock (C) or 75 mM NaCl (S) conditions for 24 h. The boxplots represent the median and extent of natural variation as observed for population of 48 accessions (Supplemental Figure 11A and Supplemental Data Set 14).
(B) A trend between HKT1 expression and average lateral root length (aLRL) was observed, with accessions with high HKT1 expression developing short lateral roots.
(C) Pictures of representative 12-d-old seedlings of UAS-HKT1 lines grown with 75 mM NaCl.
(D) UAS-HKT1 lines, with enhanced HKT1 expression in the root pericycle in Col-0 (E2586) and C24 (J2731) backgrounds, were examined for salt induced changes in RSA. Four-day-old seedlings were transferred to 0 and 75 mM NaCl and the RSA of 8- and 12-d-old seedlings was quantified. The bar plots represent the average trait value as observed in 16 replicates and error bars represent se. Different letters are used to indicate the significant differences between the genotypes per condition as calculated using one-way ANOVA with Tukey’s post-hoc test with significance levels of 0.05.
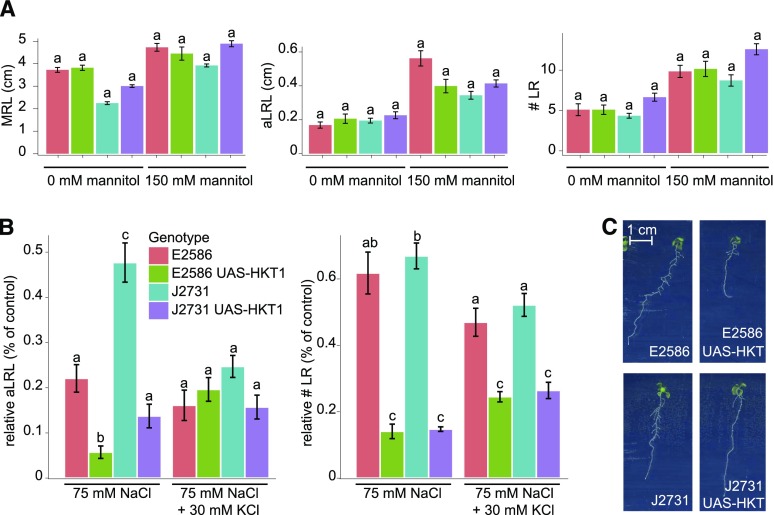
To investigate whether the severe reduction of lateral root development is caused by osmotic stress, the lines with and without enhanced HKT1 expression in both backgrounds were examined for their lateral root development when grown on media supplemented with 150 mM mannitol. No differences between the background lines and lines with enhanced HKT1 expression were observed (Figure 7A), indicating that the reduced lateral root development is due to the ionic rather than the osmotic component of salinity stress. Salt stress results in imbalance between sodium and potassium ions, and phenotypes of some salt overly sensitive mutants are known to be rescued by supplementing K+ (Zhu et al., 1998). Therefore, we examined the effect of additional K+ on the UAS-HKT1 lines. For the background lines (E2586 and J2731), extra K+ further reduced lateral root development under salt stress conditions (Figures 7B and 7C; Supplemental Figure 13). However, for the UAS-HKT1 lines, addition of potassium to the salt containing medium resulted in partial rescue of lateral root development. In conditions in which 75 mM NaCl was supplemented by 30 mM KCl, the average lateral root length of the UAS-HKT1 lines was comparable to that of the background lines (Figure 7B), while lateral root emergence was partially rescued (Supplemental Figure 13). Reduction of main root length in the Col-0 UAS-HKT1 line was not affected by additional potassium. These results indicate that potassium supplementation alleviates the effect of enhanced HKT1 expression on lateral root growth specifically in salt stress conditions, suggesting that reduced lateral root growth observed in Arabidopsis accessions and UAS-HKT1 lines with high HKT1 expression might be due to an imbalance between sodium and potassium ions.
Figure 7.
Reduction in Lateral Root Development in UAS-HKT1 Lines Is Due to the Ionic Component of Salt Stress.
(A) and (B) Lines with enhanced HKT1 in root pericycle in Col-0 (E2586) and C24 (J2731) backgrounds were studied for their main root length (MRL), average lateral root length (aLRL), and number of lateral roots (# LR) in control and osmotic stress condition (150 mM mannitol) (A) as well as media supplemented with 30 mM KCl in addition to 75 mM NaCl (B). The RSA was quantified on 10-d-old seedlings and the relative decrease in average lateral root length (aLRL) and lateral root number (# LR) was calculated relative to 0 mM NaCl (75 mM NaCl) and 30 mM KCl (75 mM NaCl + 30 mM KCl). The bar plots represent the average value as observed in 16 replicates. The error bars represent se. Different letters are used to indicate the significant differences between the genotypes per condition as calculated using one-way ANOVA (in [A]) and two-way ANOVA (in [B]) with Tukey’s post-hoc test with significance levels of 0.05.
(C) Pictures of representative 12-d-old seedlings of UAS-HKT1 lines grown on 30 mM KCl + 75 mM NaCl.
High HKT1 Expression Reduces Salt Tolerance of Col-0 during Early Development
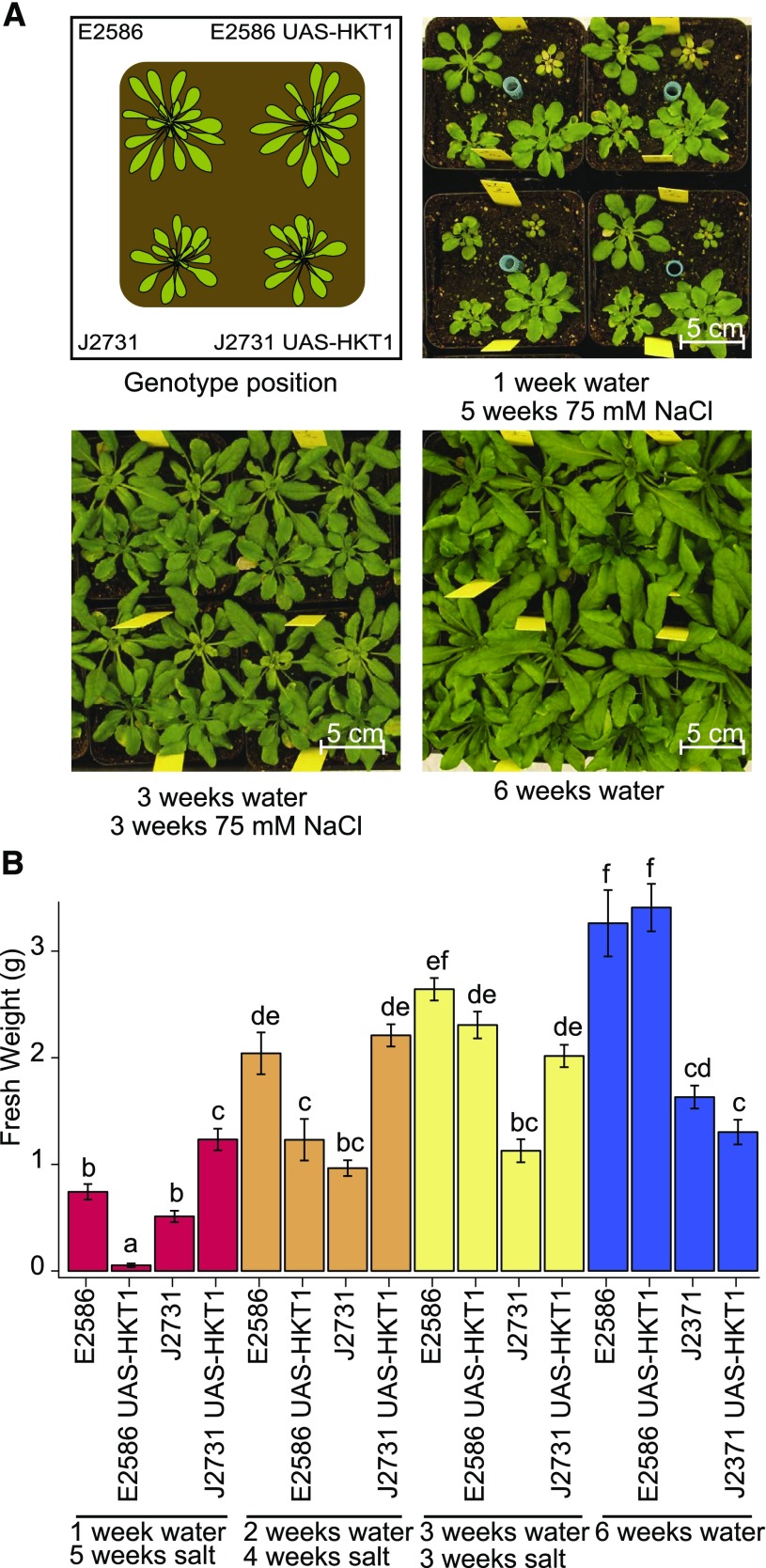
Interestingly, UAS-HKT1 lines were previously described to exhibit enhanced salt stress tolerance in hydroponic cultures (Møller et al., 2009), but the salt stress was applied at a different developmental stage than in our study. To examine the role of UAS-HKT1 expression mediated altered RSA in salt stress tolerance under more natural conditions, UAS-HKT1 and their background lines were grown in soil and were stressed at 1, 2, or 3 weeks after germination by watering with 75 mM NaCl (Figure 8). The UAS-HKT1 line in Col-0 background (E2586 UAS-HKT1) was observed to develop smaller rosettes than its background line (E2586) when plants were stressed 1 week after germination (Figure 8B). This difference between the Col-0 and UAS-HKT1 lines was less pronounced when plants were treated 2 weeks after germination or later. Consistent with a previous report (Møller et al., 2009), the UAS-HKT1 line in C24 background developed larger rosettes than the background line (J2731) independent of the timing of stress treatment. Interestingly, the rosettes of C24 background line plants grown in control conditions showed no significant difference with plants stressed for 3 or 4 weeks with salt (Figure 8B). Two-way ANOVA analysis (Supplemental Figure 14) showed a background-specific effect, where enhanced HKT1 expression is detrimental for plant development in Col-0 background but not in C24 background when salt stress is applied shortly after germination. Therefore, the correlation between high HKT1 expression and salinity tolerance is dependent on genotype by environment (GxE) effects and the developmental stage at which plants are exposed to salinity stress.
Figure 8.
The Effect of Enhanced HKT1 Expression on Salinity Tolerance Is Developmental Stage and Genetic Background Dependent.
(A) E2586, E2586 UAS-HKT1, J2731, and J2731 UAS-HKT1 lines were germinated in soil under short-day conditions and watered from above with 75 mM NaCl 1, 2, or 3 weeks after germination. The pictures represent 6-week-old plants and the scheme in the upper left panel shows the distribution of the genotypes in soil pots.
(B) Fresh weight of the rosette was determined 6 weeks after germination. The bars represent average fresh weight observed over 15 biological replicates and error bars represent se. Different letters are used to indicate groups that are significantly different from each other as determined with two-way ANOVA using pairwise post-hoc Tukey’s test with significance levels of 0.05.
DISCUSSION
Salt stress not only reduces the development and growth of roots (West et al., 2004; Liu and Zhu, 1997; Kobayashi et al., 2016; Shelden et al., 2013; Tu et al., 2014), but also causes reprogramming and redistribution of the root mass between main and lateral roots (Julkowska et al., 2014). By studying natural variation for root architecture in Arabidopsis, we observed that the natural variation in RSA responses to salt stress is exhibited mainly through the emergence and elongation, rather than patterning, of lateral roots. The variance observed in traits related to root mass distribution between main and lateral root increased with salt stress exposure (Figure 1D), while the correlations between main root length and number of lateral roots remained unaltered by salt stress (Supplemental Data Set 3). Those trends are in agreement with the data on dynamic changes in RSA performed on a smaller number of Arabidopsis accessions (Julkowska et al., 2014) and with the reduction of lateral root development under salt stress conditions observed in an earlier study (Kawa et al., 2016). In this study, we describe the traits with the most pronounced phenotypic plasticity in response to salt stress to be related to main root length, number of lateral roots, size of the apical zone, and average lateral root length, as they make most significant contributions to principal components describing natural variation observed (Supplemental Data Set 4) and are subjected to GxE interactions (Supplemental Data Set 2). Collecting multitrait phenotypes and performing multivariate analysis, as presented in this work, will improve our understanding of how plant development affects performance under stress conditions. The data presented in this study are available through the Salt_NV_Root App for the user to explore in more detail, compare individual accessions, and perform cluster analysis on the RSA traits of interest. This tool can thus provide additional insight for further exploration of natural variation in complex RSA traits and their relationship to plant performance.
While many GWAS studies published to date focus on single traits, such as ion accumulation, main root growth, or compatible solute accumulation (Strauch et al., 2015; Baxter et al., 2010; Lachowiec et al., 2015; Slovak et al., 2014; Verslues et al., 2014), there is an increasing focus on multitrait response phenotypes, including RSA response to stress (Rosas et al., 2013; Kawa et al., 2016). An advantage of performing GWAS on multitrait phenotypes, also apparent in our study, is that additional confidence can be gained when the same candidate loci are mapped using different traits and/or different stress conditions or when the multitrait phenotypes are reduced to principle components that map to the overlapping loci (Figure 3A). Interestingly, 100 candidate loci identified by GWAS in this study contain genes involved in ethylene and ABA signaling, such as EIN2 and SnRK2.7 (Supplemental Data Set 11), indicating natural variation in hormonal control pathways that were earlier described to play a role in RSA responses to salt stress (Duan et al., 2013; Geng et al., 2013). The significant overlap between the identified candidate genes and salt stress-induced changes in gene expression (Dinneny et al., 2008; Supplemental Data Set 11) implies that the allelic variation might accumulate in the regions important for transcriptional regulation of genes involved in root remodeling in response to salt. Also, a meta-analysis on genetic architecture of abiotic and biotic stress responses, which included a selection of our salt stress response data, implied genetic correlations with other abiotic stress responses (Thoen et al., 2017).
Our genetic and physiological data confirm two identified candidate genes, CYP79B2 and HKT1, to be involved in lateral root development under salinity stress, which suggests that genes identified in our GWAS are indeed involved in reshaping RSA under salt stress conditions. Significant correlations were found between salt-induced expression of CYP79B2 and lateral root development under salinity stress (Figure 4). Further analysis of CYP79B2 and CYP79B3, which are known to have partially overlapping functions, showed that these genes are required for the maintenance of lateral root growth under salt stress conditions, but not in control conditions (Figure 5). As both CYP79B2 and CYP79B3 convert tryptophan to IAOx, these data suggest the involvement of the IAOx pathway in shifting investments from main to lateral roots during salt stress. Both CYP79B2 and CYP79B3 were previously observed to be expressed at the site of newly developing lateral roots (Ljung et al., 2005). The IAOx pathway is Brassica specific and can produce three types of active compounds: camalexin, indole glucosinolates, and indole-3-acetic acid (Hull et al., 2000; Zhao et al., 2002; Mikkelsen et al., 2000; Glawischnig et al., 2004; Sugawara et al., 2009). Both camalexin and indole glucosinolates act as defense compounds induced in both shoot and root upon pathogen attack (Glawischnig, 2007; Halkier and Gershenzon, 2006; Brown et al., 2003; Lemarié et al., 2015), but no clear link to root development has been found. The IAOx pathway has been shown to contribute to auxin production under heat stress conditions (Zhao et al., 2002). NITRILASE1 (NIT1), proposed to catalyze the last step of the IAOx pathway leading to auxin biosynthesis, was previously described to be involved in maintenance of lateral root development (Lehmann et al., 2017), and the expression of NIT1 and NIT2 was observed to be upregulated in response to salt stress (Bao and Li, 2002). We propose that the natural variation in promoter region of CYP79B2 results in altered expression induction by salt stress and that the IAOx pathway is necessary for maintenance of lateral root development under salt stress, most likely through production of auxin, although a role for other IAOx pathway generated compounds cannot be excluded.
The association found in the promoter region of HKT1 provides a link between ion sequestration and lateral root development. Our results suggest that some accessions with high HKT1 expression develop shorter lateral roots under salt stress conditions (Figure 6B). Using transgenic lines with enhanced expression at the native root expression site (UAS-HKT1 lines), we confirmed that high HKT1 expression indeed reduces lateral root formation under salt stress in Col-0 and C24 background (Figures 6C and 6D). Potassium uptake was previously described to repress lateral root formation under control and drought conditions, as kup268 and gork mutants showed enhanced root development (Osakabe et al., 2013). Our results suggest that the effect of potassium is stress dependent, as additional potassium enhanced lateral root development only in lines with enhanced HKT1 expression, but not in the background lines (Figures 7B and 7C). Although sodium accumulation in the root stele is important for ion exclusion from shoot tissue (Møller et al., 2009; Kotula et al., 2015; Munns et al., 2012), in the young seedlings the high stelar sodium accumulation could potentially result in ABA-dependent lateral root quiescence (Duan et al., 2013) or even damage to the lateral root primordia. The exact mechanisms underlying the reduced lateral root development in lines overexpressing HKT1 remain to be verified in future studies.
The salinity tolerance of the C24 UAS-HKT1 line was reported before to be enhanced when 3-week-old plants were exposed to salinity (Møller et al., 2009), and high HKT1 expression was previously found to cause sodium exclusion from leaf tissue and therefore higher salt stress tolerance (Rus et al., 2006; Baxter et al., 2010; Munns et al., 2012). Previously, natural accessions that exhibited reduced lateral root emergence were associated with increased sensitivity to salt stress, while those with many, but shorter, later roots showed better maintenance of Na+/K+ ratio under salt stress (Julkowska et al., 2014). Here, we found that enhanced HKT1 expression reduced RSA development when 4-d-old seedlings were exposed to salt on agar plates and while also negatively affecting salinity tolerance in the Col-0 background when these plants were exposed to salt in soil early in their development (Figure 8), which might be ascribed to differences in sodium storage capacity of 4-d-old roots versus 3-week-old roots. On the other hand, enhanced stelar expression of HKT1 in the C24 line was associated with increased salinity tolerance in soil independent of the developmental stage at the application of salt stress (Figure 8), in agreement with previously published hydroponics data (Møller et al., 2009). These results imply that the benefit of tissue-specific sodium compartmentalization depends on maintenance of the ion balance between sodium and potassium as well as genetic background and the developmental stage at which plants are exposed to salinity.
The allelic variation responsible for high HKT1 expression was widespread across the promoter region, with 33 predicted cis-regulatory elements shared between accessions with high HKT1 expression (Supplemental Figure 9 and Supplemental Data Set 16). The 33 predicted cis-regulatory elements did not overlap with either the minimal promoter of HKT1 (Mäser et al., 2002) or ABI4 binding sites (Shkolnik-Inbar et al., 2013). Identification of a promoter region responsible for enhanced HKT1 expression will require identification of transcription factors regulating HKT1 expression and their binding sites, rather than examining extensive and widespread allelic variation. Although high HKT1 expression was earlier associated with increased salt stress tolerance (Møller et al., 2009; Munns et al., 2012), the plants used in those studies were much further along in their development at the time of salt stress exposure. Our results suggest that this strategy is not advantageous when plants are exposed to salinity early in their development (Figure 8), in accordance with several coastal accessions carrying a low-expression HKT1 allele (Baxter et al., 2010). The complex relationship between high HKT1 expression and plant tolerance to salinity shown in this study provides a rationale for high variation of the HKT1 promoter region in natural accessions of Arabidopsis as well as other plant species (Munns et al., 2012; Negrão et al., 2013) and fine-tuning of HKT1 expression levels depending on local soil conditions and timing of exposure to salt stress. Correlations between RSA and plant survival (Katori et al., 2010) and sodium exclusion (Baxter et al., 2010) are very weak (Supplemental Figure 15), which might be partially due to different experimental setups used. Additionally, the relationship between RSA and salt tolerance depends on the developmental and the genetic context, as we have shown in this study using UAS-HKT1 lines. Further investigation of the other candidate genes identified in this study and examining their contribution to RSA development and salt stress tolerance in a uniform genetic background will provide more insight in how salt stress affects root morphology and how those changes would increase salt stress tolerance of crops.
METHODS
Plant Material and Growth Conditions
The Arabidopsis thaliana HapMap collection (Weigel and Mott, 2009) was obtained from the ABRC (www.abrc.osu.edu). The 360 accessions were propagated under long-day conditions (21°C, 70% humidity, 16/8-h light/dark cycle, 125 μmol/m2/s, Sylvania Britegrow F58W 1084 lamps), with 8 weeks vernalization (between 4 and 8°C, 70% humidity, 16/8-h light/dark cycle) starting at the 3rd week after germination to ensure flowering of all the accessions. Accessions that failed to germinate or flower were excluded from the screen, resulting in 347 accessions in total (Supplemental Data Set 1). Seeds used for the experiments were between 2 months and 1 year old.
Seeds were surface sterilized in a desiccator of 1.6 liters volume using 20 mL household bleach and 600 μL 40% HCl for 3 h and were put in the laminar flow for 1.5 h to remove toxic vapors. The seeds were stratified in 0.1% agar at 4°C in the dark for 72 h and sown on square Petri dishes (12 × 12 cm) containing 50 mL of control growth medium consisting of 0.5× Murashige and Skoog medium, 0.5% sucrose, 0.1% MES monohydrate, and 1% Daishin agar, pH 5.8 (KOH), dried for 1 h in a laminar flow. Plates were placed vertically at a 70° angle under long-day conditions (21°C, 70% humidity, 16/8-h light/dark cycle). Four-day-old seedlings were transferred to square Petri dishes containing basic medium supplemented with 0, 75, or 125 mM NaCl. Each plate contained four seedlings of two genotypes (two seedlings per genotype). Plates were placed in the growth chamber following a random design. The plates were scanned with Epson perfection V700 scanner at 200 dpi every other day until the 8th day after transfer. The 8-d-old seedlings grown in 0 mM NaCl were used for phenotyping of RSA in control conditions while for both salt stress conditions (75 and 125 mM NaCl) phenotypes of 12-d-old seedlings were scored. The pictures were analyzed with EZ-Rhizo software (Armengaud et al., 2009). The entire population of 347 accessions was screened over six individual experiments with Col-0 as internal reference (Supplemental Figure 1). The RSA phenotypes of individual accessions were calculated from four biological replicates, except for Nd-1, Tommegap, and Tottarp, where the RSA phenotype in control conditions was measured only for two biological replicates.
Analysis of Natural Variation in RSA Phenotypes
The collected data on RSA phenotypes were cleared of outliers by removing the accessions of which the sd within the genotype was larger than the sd within the HapMap population, and RSA phenotyping was repeated for these accessions in the next experimental batch. The natural variation in the studied population was further explored using average values per accessions as an input for notched boxplots and calculating coefficient of variance per condition studied. The correlations among different RSA traits at different conditions are presented in Supplemental Data Set 3. Seventeen RSA parameters and three additional parameters consisting of the length of apical, branched, and basal zone represented as the portion of main root length (apical/MRL, branched/MRL, and basal/MRL respectively) were reduced to three PC by performing PCA (Supplemental Figures 2C and 3). The raw data were first normalized per trait by applying Z-score normalization in individual phenotypes. The principal components were calculated using Mat Lab. The importance of individual RSA traits for each PC is presented in Supplemental Data Set 4. Individual PCs and the raw phenotypic data were used as input for GWAS (Supplemental Data Set 5).
The entire data set and the multivariate analysis were integrated into Shiny App based “Salt_NV_Root App,” available at https://mmjulkowska.github.io/Salt_NV_RootApp/.
GWAS on RSA Phenotypes
The RSA phenotypes were linked to published genomic data on accessions from a 250k SNP chip with average SNP density of one SNP in 500 bp (Horton et al., 2012). The narrow sense heritability of individual traits at different conditions was determined (Supplemental Data Set 6). Apart from one trait (excluded from further analysis), the estimated heritability for all traits was above 0.2. We performed GWAS using the entire SNP data set as well as excluding the SNPs with minor allele frequency of 0.01, 0.05, and 0.10 to avoid misleading associations. The associations between each SNP and individual RSA phenotypes were tested using a scan_GLS program (Kruijer et al., 2015), based on EMMA-X (Kang et al., 2008). The method implements Gao and Bonferroni corrections for multiple testing to minimize the false discovery rates. Both methods were applied on phenotypic values per accession with α of 0.01 and 0.05 and MAF of 0.00, 0.01, 0.05, and 0.10. We used 5.6 as the threshold value, as there were around 20,000 SNPs with MAF > 0.01 and the threshold was therefore −log10(0.05/20000) = 5.6. The overview of numbers of associated loci identified with individual traits with LOD > 5.6, including all rare alleles is presented in Supplemental Data Set 7.
The list of putative loci was selected using metrics considering the association when the individual values of each replica were mapped and when average values per genotype were used for GWAS. Furthermore, we selected for the associations mapped using only the SNP subset with MAF > 0.01, as the majority of the associations were identified with this SNP set, while excluding the rare alleles, represented by less than three accessions, that could skew the associations found. For the loci that were mapped with a SNP set including rare alleles, we used the more stringent Bonferroni threshold for further selection of associations, based on all SNPs used (including the rare SNPs), determined as log10(0.05/200 000) = 6.6 (Supplemental Data Sets 8 to 10). We selected the SNPs that were mapped with both average and individual replica trait values and –log10(P value) above the Bonferroni threshold or mapped with the SNP subset of MAF > 0.01 (Supplemental Figure 4). Our selection yielded 49 associations with traits measured under control conditions and 154 associations specific to RSA traits measured under salt stress conditions (Supplemental Figure 4 and Supplemental Data Sets 8 to 10). The candidates from all conditions studied were compared and the associations overlapping between individual traits and conditions were identified.
The enrichment analysis of the 100 candidate loci identified under salt stress conditions was performed with the hypergeometric test, using the phyper() function in R. We used 27,655 protein-coding genes as the “population size” parameter, with 1632 genes identified by Dinneny et al. (2008) as responsive to salt stress. For testing the genes directly underlying the associated SNP, we used 10 genes that we identified to be altered in their expression, among 100 possible genes. For testing the genes in linkage disequilibrium with the identified SNP, we calculated the average number of genes per 10 kb upstream and downstream from identified SNP (27,655 genes per 119,146,348 bp resulting in 0.0002321095 gene per bp, and 4.64219 genes per 20 to 10kb upstream and 10 kb downstream from the identified SNP). Subsequently, we used the average gene density to determine the number of possible candidate genes for 100 identified loci, ending up with 464,219 possible candidate genes. Therefore, the hypergeometric test for the neighboring candidate genes was run with 70 genes that we identified to be altered in their expression among 464,219 possible genes.
The associations with average lateral root length and the ratio between average lateral root length were studied in greater detail (Supplemental Data Set 12). To further fine-map the chosen loci, the RSA phenotypes were linked to the genotypic databased on whole-genome sequencing data of the different accessions (Alonso-Blanco et al., 2016) and covered 4,000,000 SNPs. For the accessions that were not sequenced, genome information was imputed based on the 250k SNP chip data (Horton et al., 2012). Those imputations are known to have negligible effect on the GWAS outcomes (Cao et al., 2011). The GWAS was performed on the average trait value per genotype and the SNPs corresponding to the six loci that we identified (Supplemental Data Set 12) are presented in Supplemental Data Set 13.
For the selected associations (Supplemental Data Set 12), sequence information of 147 accessions belonging to the HapMap population was downloaded from the 1001 genomes project website (1001genomes.org) and aligned with ClustalO as described by Julkowska et al. (2016).
Expression Analysis of Accessions
Forty-eight Arabidopsis accessions were selected based on different haplotypes of selected candidate loci as determined from SNP data and the pronounced differences between their RSA under all conditions studied (Supplemental Data Set 14). The accessions were used for studying the natural variation in the expression of candidate genes (Supplemental Data Set 14). The 4-d-old seedlings of 48 different accessions were transferred to plates supplemented with 0 or 75 mM NaCl. After 24 h, the seedlings were harvested, snap-frozen in liquid nitrogen, and divided into root and shoot fraction. RNA extraction was performed with TRI reagents (Sigma-Aldrich) with additional chloroform cleaning step, followed by TURBO DNase treatment (Ambion). The RNA was checked for the integrity on 2% agarose gel. The cDNA was synthesized from 1 µg total RNA using reverse transcriptase (Fermentas). The cDNA was diluted to ∼10 ng/µL. The diluted cDNA was used in a specific target amplification reaction and subjected to PCR on a Biomark genetic analysis system on a 96 × 96 Dynamic array, according to the manufacturer’s instructions (Fluidigm). The primer sequence for qPCR was designed focusing on the 3ʹ end and targeting the conserved sites with no/little natural variation in the sequence as found out in 1001 sequence browser. The sequences of the primers used can be found in Supplemental Data Set 18. For the loci 1, 2, 3, 4, and 6, the transcript levels of At1G07920, At1G13320, At3G04120, At5G12240, and At5G46630 were used for normalization of expression. Expression of the genes in the locus 5 was measured in a separate experiment, using At1G07920, At1G13320, At5G12240, and At5G46630 as reference transcripts for normalization, without At3G04120.
Samples of low quality were removed and Ct values ≥ 30 were set to 30 to reduce the background noise. The expression levels of target genes were calculated by ΔCt = 2−Ct value target/2−Ct-value reference. To normalize the expression for all five/four reference genes used, geometrical mean was calculated from delta-Ct-normalized expression for each reference gene and the average expression per accession, tissue, and conditions were calculated for each putative candidate gene. For regression analysis between traits and expression, highly influential observations were not considered based on Cook’s distance (if >0.5).
T-DNA Insertion Lines Genotyping and Phenotyping
The lines were ordered from the European Arabidopsis Stock Centre (nasc.org.uk), except for the cyp79b2-2 cyp79b3-2 line (Sugawara et al., 2009), which was kindly provided by Hiroyushi Kasahara (RIKEN Center for Sustainable Resource Science, Yokohama, Japan). The full list of T-DNA insertion lines used is to be found in Supplemental Data Set 19. The T-DNA insertion lines were genotyped by extracting DNA from leaf material ground in liquid nitrogen using 10% Chelex (Bio-Rad) in MiliQ followed by 15 min incubation at 95°C and 15 min centrifugation at maximum speed in the standard tabletop centrifuge. The supernatant was used as an input for the PCR reaction. The primers used for T-DNA insertion lines identification are listed in Supplemental Data Set 19. The RSA phenotypes of HKT1 T-DNA insertion lines were studied as described above for the phenotyping of Arabidopsis accessions (n = 16). A one-way ANOVA was used to analyze these data, as the control plants and salt treatment plants were not the same age. A Tukey post-hoc test was used to analyze differences between genotypes within treatments. The RSA phenotypes of CYP79B2 and CYP79B3 T-DNA insertion lines were studied as described above, but no sucrose was added in the medium (n = 20) and RSA traits were quantified with Smartroot (Lobet et al., 2011). For analyses of the single and double mutant lines on day 10 (Figure 5A; Supplemental Figure 11B), data of three experiments were pooled to increase statistical power. To correct for this, experimental batch was included as random factor in a mixed linear model. For the analysis of double mutant lines on day 10 and day 12, data of one representative experiment are shown (Figure 6B; Supplemental Figure 12C). Data were analyzed using a three-way ANOVA. For both, if a significant interaction was found, a Bonferroni-corrected post-hoc within treatment (and day) analysis was used to analyze differences between genotypes.
The expression levels of gene of interest in T-DNA insertion lines were studied by qPCR analysis. RNA was extracted from whole seedlings grown on agar plates for 12 d in control conditions with TRI reagents (Sigma Aldrich) with additional chloroform cleaning step, followed by TURBO DNase treatment (Ambion). The RNA was examined for the integrity on 2% agarose gel. The cDNA was synthesized from 1 µg total RNA using reverse transcriptase (Fermentas), diluted to ∼10 ng/μL and used for qPCR using the Eva-Green kit (Solis Biodyne) and an Applied Biosystems sds7500 machine with three biological replicates and two technical replicates. The expression was normalized using AT1G13320 transcript levels. The expression levels of putative candidate genes were calculated by ΔCt = 2−(Ct value target)/2−(Ct-value reference). The sequences of the primers used are listed in Supplemental Data Set S18.
Sequencing the HKT1 Locus and Parsimony Calculations
The sequencing of HKT1 was performed on seven accessions. The accessions were chosen based on their RSA phenotype and relative expression of HTK1. The primers used are listed in Supplemental Data Set 15. The sequences of individual accessions were aligned using MegAlign software and aligned to each other in MegAlign ClustalW algorithm (Supplemental Figure 16). The sequences encoding exons are highlighted in yellow, introns in purple, and untranslated regions in red. The identified polymorphisms were investigated by examining the patterns in the promoter region using the web-based database for Plant Cis-acting Regulatory DNA Elements (PLACE) (Higo et al., 1999), by examining the presence/absence of cis-regulatory element in the sequence flanking the polymorphism. Translating exon sequences into the protein sequences in JalView was performed to examine the nonsynonymous changes in amino acid sequence. The polymorphisms located in intron region were examined whether they interfere with the splicing acceptor, donor, or branching site. The polymorphisms found outside of those areas were assumed to have no major effect. The polymorphisms identified in promoter and exon regions were encoded with P-numbers and in exons with E-numbers for individual locus separately and are listed in Supplemental Data Sets 16 and 17.
The evolutionary history was inferred for each locus separately using the Maximum Parsimony method. The most parsimonious tree with length is shown. The MP tree was obtained using the Subtree-Pruning-Regrafting algorithm with search level 0 in which the initial trees were obtained by the random addition of sequences (10 replicates). The MP trees are drawn to scale, with branch lengths calculated using the average pathway method and are in the units of the number of changes over the whole sequence. The analysis involved seven nucleotide sequences from different Arabidopsis accessions. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA5 (Tamura et al., 2011).
Salinity Tolerance Assessment in Transpiring Conditions (Soil)
Seeds of UAS-HKT1 and the background lines to be tested were stratified for 48 h at 4°C and sown in soil in pots. Seeds of four different lines were put in one pot (four plants per pot) and were germinated under short-day conditions (21°C, 70% humidity, 11/13-h light/dark cycle). After 1, 2, or 3 weeks, the seedlings were treated with 75 mM NaCl applied from above every second day for 6 weeks. After 7 weeks of growth, the fresh weight of the rosette was measured. The statistical analysis was performed for one- and two-way ANOVA with Scheffe’s post-hoc test for significance.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the accession numbers listed in Supplemental Data Set 1.
Supplemental Data
Supplemental Figure 1. Experimental setup and internal reference accession data for screening natural variation in RSA responses to salt stress.
Supplemental Figure 2. Natural variation in all RSA phenotypes studied.
Supplemental Figure 3. Principle components used for GWAS.
Supplemental Figure 4. Selection of putative candidate loci from GWAS associations.
Supplemental Figure 5. Natural variation in Locus 1 associated with ratio of average lateral root length to main root length and principal component 3 at 75 mM NaCl.
Supplemental Figure 6. Natural variation in locus 2 associated with ratio of average lateral root length to main root length and principal component 3 at 75 mM NaCl.
Supplemental Figure 7. Natural variation in locus 3 associated with ratio of average lateral root length to main root length and principal component 3 at 75 mM NaCl.
Supplemental Figure 8. Natural variation in locus 6 on chromosome 5 associated with average lateral root length, ratio of average lateral root length to main root length, and principal component 3 at 125 mM NaCl.
Supplemental Figure 9. Natural variation in HKT1 expression in root and shoot tissue in control and salt stress conditions.
Supplemental Figure 10. Natural variation in CYP79B2 and surrounding genes in root and shoot tissue in control and salt stress conditions.
Supplemental Figure 11. RSA phenotypes of knockout mutants of CYP79B2 and CYP79B3.
Supplemental Figure 12. Reduced expression of HKT1 does not result in changes in RSA.
Supplemental Figure 13. RSA phenotype of UAS-HKT1 lines is partially rescued by addition of K+ at 75 mM NaCl.
Supplemental Figure 14. High HKT1 expression results in reduced salinity tolerance during early exposure to salt stress and is dependent on the background.
Supplemental Figure 15. Correlation between salt induced changes in RSA and salt tolerance.
Supplemental Figure 16. The alignment of HKT1 promoter and coding region sequence in seven sequenced accessions.
Supplemental Data Set 1. List of all Arabidopsis accessions screened for RSA in control and salt stress conditions.
Supplemental Data Set 2. Overview of 17 root system architecture traits measured.
Supplemental Data Set 3. Correlations between RSA traits in different growth conditions.
Supplemental Data Set 4. The scores of individual RSA traits for three principal components used for GWAS.
Supplemental Data Set 5. The average of RSA traits for individual accessions of Arabidopsis in three different conditions.
Supplemental Data Set 6. Heritability for individual RSA traits
Supplemental Data Set 7. Overview of number of significant associations identified.
Supplemental Data Set 8. List of significant associations with RSA traits at 0 mM NaCl.
Supplemental Data Set 9. List of significant associations with RSA traits at 75 mM NaCl.
Supplemental Data Set 10. List of significant associations with RSA traits at 125 mM NaCl.
Supplemental Data Set 11. List of the candidate genes identified with RSA traits at 75 or 125 mM NaCl with significant alterations in the cell-type-specific expression in response to salt.
Supplemental Data Set 12. Overview of the candidate genes selected for examination in Figure 4.
Supplemental Data Set 13. Fine-mapping of the six selected loci (Supplemental Data Set 12) by the use of a 10M SNP library.
Supplemental Data Set 14. The relative expression of accessions studied for natural variation in candidate gene expression.
Supplemental Data Set 15. List of primers used for sequencing the loci of putative candidate genes.
Supplemental Data Set 16. The list of polymorphisms found in the promoter region of At4g10310 and predicted changes in cis-regulatory elements as analyzed with PLACE.
Supplemental Data Set 17. The list of polymorphisms found in the exon regions of At4g10310.
Supplemental Data Set 18. List of primers used for expression study of candidate genes
Supplemental Data Set 19. Overview of t-DNA insertion lines studied including the primers used for genotyping T-DNA insertion lines.
Acknowledgments
We thank Willem Kruijer from Wageningen University for help with GWAS and Dorota Kawa and Jessica Meyer from University of Amsterdam for their technical support. We thank Hiroyushi Kasahara (RIKEN Center for Sustainable Resource Science, Yokohama, Japan) for the provided materials. This work was supported by Netherlands Organisation for Scientific Research ALW Graduate Program Grant 831.15.004 and by STW Learning from Nature Project 10987.
AUTHOR CONTRIBUTIONS
C.T. conceived the project and was responsible for project administration. C.T. and G.-J.d.B. acquired the funding for this project. J.J.B.K., M.M.J., H.H., A.K., and C.T. developed methodology. M.M.J., I.T.K., S.M., and R.F. collected the data. J.J.B.K., A.K., and M.T. provided resources for GWAS and reverse genetics. M.M.J., I.T.K., and S.M. performed validation of candidate genes. M.M.J., I.T.K., and C.T. performed data curation. M.M.J. and C.T. wrote the original manuscript, which was later reviewed and edited by all authors. M.M.J. developed the Salt_NV_RootApp. C.T., M.T., G.-J.d.B., and M.A.H. supervised the project.
Footnotes
Articles can be viewed without a subscription.
References
- Alonso-Blanco C., et al. ; 1001 Genomes Consortium (2016). 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166: 481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengaud P., Zambaux K., Hills A., Sulpice R., Pattison R.J., Blatt M.R., Amtmann A. (2009). EZ-Rhizo: integrated software for the fast and accurate measurement of root system architecture. Plant J. 57: 945–956. [DOI] [PubMed] [Google Scholar]
- Bao F., Li J.Y. (2002). Evidence that the auxin signaling pathway interacts with plant stress response. Acta Bot. Sin. 44: 532–536. [Google Scholar]
- Baxter I., Brazelton J.N., Yu D., Huang Y.S., Lahner B., Yakubova E., Li Y., Bergelson J., Borevitz J.O., Nordborg M., Vitek O., Salt D.E. (2010). A coastal cline in sodium accumulation in Arabidopsis thaliana is driven by natural variation of the sodium transporter AtHKT1;1. PLoS Genet. 6: e1001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P.D., Tokuhisa J.G., Reichelt M., Gershenzon J. (2003). Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 62: 471–481. [DOI] [PubMed] [Google Scholar]
- Cao J., et al. (2011). Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 43: 956–963. [DOI] [PubMed] [Google Scholar]
- Choi W.G., Toyota M., Kim S.H., Hilleary R., Gilroy S. (2014). Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc. Natl. Acad. Sci. USA 111: 6497–6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Zhu J.K. (1997). Reduced Na+ uptake in the NaCl-hypersensitive sos1 mutant of Arabidopsis thaliana. Plant Physiol. 113: 795–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny J.R., Long T.A., Wang J.Y., Jung J.W., Mace D., Pointer S., Barron C., Brady S.M., Schiefelbein J., Benfey P.N. (2008). Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320: 942–945. [DOI] [PubMed] [Google Scholar]
- Duan L., Dietrich D., Ng C.H., Chan P.M.Y., Bhalerao R., Bennett M.J., Dinneny J.R. (2013). Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell 25: 324–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faiyue B., Vijayalakshmi C., Nawaz S., Nagato Y., Taketa S., Ichii M., Al-Azzawi M.J., Flowers T.J. (2010). Studies on sodium bypass flow in lateral rootless mutants lrt1 and lrt2, and crown rootless mutant crl1 of rice (Oryza sativa L.). Plant Cell Environ. 33: 687–701. [DOI] [PubMed] [Google Scholar]
- Galvan-Ampudia C.S., Julkowska M.M., Darwish E., Gandullo J., Korver R.A., Brunoud G., Haring M.A., Munnik T., Vernoux T., Testerink C. (2013). Halotropism is a response of plant roots to avoid a saline environment. Curr. Biol. 23: 2044–2050. [DOI] [PubMed] [Google Scholar]
- Geng Y., Wu R., Wee C.W., Xie F., Wei X., Chan P.M.Y., Tham C., Duan L., Dinneny J.R. (2013). A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell 25: 2132–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glawischnig E. (2007). Camalexin. Phytochemistry 68: 401–406. [DOI] [PubMed] [Google Scholar]
- Glawischnig E., Hansen B.G., Olsen C.E., Halkier B.A. (2004). Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 8245–8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkier B.A., Gershenzon J. (2006). Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57: 303–333. [DOI] [PubMed] [Google Scholar]
- Higo K., Ugawa Y., Iwamoto M., Korenaga T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27: 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton M.W., et al. (2012). Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat. Genet. 44: 212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull A.K., Vij R., Celenza J.L. (2000). Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc. Natl. Acad. Sci. USA 97: 2379–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C., Belfield E.J., Mithani A., Visscher A., Ragoussis J., Mott R.C., Smith J.A., Harberd N.P. (2012). ROS-mediated vascular homeostatic control of root-to-shoot soil Na delivery in Arabidopsis. EMBO J. 31: 4359–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julkowska M.M., Testerink C. (2015). Tuning plant signaling and growth to survive salt. Trends Plant Sci. 20: 586–594. [DOI] [PubMed] [Google Scholar]
- Julkowska M.M., Hoefsloot H.C.J., Mol S., Feron R., de Boer G.J., Haring M.A., Testerink C. (2014). Capturing Arabidopsis root architecture dynamics with ROOT-FIT reveals diversity in responses to salinity. Plant Physiol. 166: 1387–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julkowska M.M., Klei K., Fokkens L., Haring M.A., Schranz M.E., Testerink C. (2016). Natural variation in rosette size under salt stress conditions corresponds to developmental differences between Arabidopsis accessions and allelic variation in the LRR-KISS gene. J. Exp. Bot. 67: 2127–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.M., Zaitlen N.A., Wade C.M., Kirby A., Heckerman D., Daly M.J., Eskin E. (2008). Efficient control of population structure in model organism association mapping. Genetics 178: 1709–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katori T., Ikeda A., Iuchi S., Kobayashi M., Shinozaki K., Maehashi K., Sakata Y., Tanaka S., Taji T. (2010). Dissecting the genetic control of natural variation in salt tolerance of Arabidopsis thaliana accessions. J. Exp. Bot. 61: 1125–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawa D., Julkowska M.M., Sommerfeld H.M., Ter Horst A., Haring M.A., Testerink C. (2016). Phosphate-dependent root system architecture responses to salt stress. Plant Physiol. 172: 690–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y., Sadhukhan A., Tazib T., Nakano Y., Kusunoki K., Kamara M., Chaffai R., Iuchi S., Sahoo L., Kobayashi M., Hoekenga O.A., Koyama H. (2016). Joint genetic and network analyses identify loci associated with root growth under NaCl stress in Arabidopsis thaliana. Plant Cell Environ. 39: 918–934. [DOI] [PubMed] [Google Scholar]
- Kotula L., Clode P.L., Striker G.G., Pedersen O., Läuchli A., Shabala S., Colmer T.D. (2015). Oxygen deficiency and salinity affect cell-specific ion concentrations in adventitious roots of barley (Hordeum vulgare). New Phytol. 208: 1114–1125. [DOI] [PubMed] [Google Scholar]
- Kruijer W., Boer M.P., Malosetti M., Flood P.J., Engel B., Kooke R., Keurentjes J.J.B., van Eeuwijk F.A. (2015). Marker-based estimation of heritability in immortal populations. Genetics 199: 379–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachowiec J., Shen X., Queitsch C., Carlborg Ö. (2015). A genome-wide association analysis reveals epistatic cancellation of additive genetic variance for root length in Arabidopsis thaliana. PLoS Genet. 11: e1005541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann T., Janowitz T., Sánchez-Parra B., Alonso M.P., Trompetter I., Piotrowski M., Pollmann S. (2017). Arabidopsis NITRILASE 1 contributes to the regulation of root growth and development through modulation of auxin biosynthesis in seedlings. Front. Plant Sci. 8: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemarié S., Robert-Seilaniantz A., Lariagon C., Lemoine J., Marnet N., Levrel A., Jubault M., Manzanares-Dauleux M.J., Gravot A. (2015). Camalexin contributes to the partial resistance of Arabidopsis thaliana to the biotrophic soilborne protist Plasmodiophora brassicae. Front. Plant Sci. 6: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Huang Y., Bergelson J., Nordborg M., Borevitz J.O. (2010). Association mapping of local climate-sensitive quantitative trait loci in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 21199–21204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Zhu J.K. (1997). An Arabidopsis mutant that requires increased calcium for potassium nutrition and salt tolerance. Proc. Natl. Acad. Sci. USA 94: 14960–14964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.M., Zhao J.Y., Lu P.P., Chen M., Guo C.H., Xu Z.S., Ma Y.Z. (2016). The E-subgroup pentatricopeptide repeat protein family in Arabidopsis thaliana and confirmation of the responsiveness PPR96 to abiotic stresses. Front. Plant Sci. 7: 1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung K., Hull A.K., Celenza J., Yamada M., Estelle M., Normanly J., Sandberg G. (2005). Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 17: 1090–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobet G., Pagès L., Draye X. (2011). A novel image-analysis toolbox enabling quantitative analysis of root system architecture. Plant Physiol. 157: 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäser P., et al. (2002). Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett. 531: 157–161. [DOI] [PubMed] [Google Scholar]
- Mikkelsen M.D., Hansen C.H., Wittstock U., Halkier B.A. (2000). Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J. Biol. Chem. 275: 33712–33717. [DOI] [PubMed] [Google Scholar]
- Møller I.S., Gilliham M., Jha D., Mayo G.M., Roy S.J., Coates J.C., Haseloff J., Tester M. (2009). Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell 21: 2163–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R., Gilliham M. (2015). Salinity tolerance of crops - what is the cost? New Phytol. 208: 668–673. [DOI] [PubMed] [Google Scholar]
- Munns R., Tester M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59: 651–681. [DOI] [PubMed] [Google Scholar]
- Munns R., James R.A., Xu B., Athman A., Conn S.J., Jordans C., Byrt C.S., Hare R.A., Tyerman S.D., Tester M., Plett D., Gilliham M. (2012). Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat. Biotechnol. 30: 360–364. [DOI] [PubMed] [Google Scholar]
- Negrão S., Almadanim M.C., Pires I.S., Abreu I.A., Maroco J., Courtois B., Gregorio G.B., McNally K.L., Oliveira M.M. (2013). New allelic variants found in key rice salt-tolerance genes: an association study. Plant Biotechnol. J. 11: 87–100. [DOI] [PubMed] [Google Scholar]
- OECD/Food and Agriculture Organization of the United Nations (2015). OECD-FAO Agricultural Outlook 2015. (Paris: OECD Publishing; ), 10.1787/agr_outlook-2015-en. [DOI] [Google Scholar]
- Osakabe Y., et al. (2013). Osmotic stress responses and plant growth controlled by potassium transporters in Arabidopsis. Plant Cell 25: 609–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristova D., Busch W. (2014). Natural variation of root traits: from development to nutrient uptake. Plant Physiol. 166: 518–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins N.E. II, Trontin C., Duan L., Dinneny J.R. (2014). Beyond the barrier: communication in the root through the endodermis. Plant Physiol. 166: 551–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas U., Cibrian-Jaramillo A., Ristova D., Banta J.A., Gifford M.L., Fan A.H., Zhou R.W., Kim G.J., Krouk G., Birnbaum K.D., Purugganan M.D., Coruzzi G.M. (2013). Integration of responses within and across Arabidopsis natural accessions uncovers loci controlling root systems architecture. Proc. Natl. Acad. Sci. USA 110: 15133–15138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S.J., Huang W., Wang X.J., Evrard A., Schmöckel S.M., Zafar Z.U., Tester M. (2013). A novel protein kinase involved in Na(+) exclusion revealed from positional cloning. Plant Cell Environ. 36: 553–568. [DOI] [PubMed] [Google Scholar]
- Rus A., Baxter I., Muthukumar B., Gustin J., Lahner B., Yakubova E., Salt D.E. (2006). Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genet. 2: e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rus A., Yokoi S., Sharkhuu A., Reddy M., Lee B.H., Matsumoto T.K., Koiwa H., Zhu J.K., Bressan R.A., Hasegawa P.M. (2001). AtHKT1 is a salt tolerance determinant that controls Na(+) entry into plant roots. Proc. Natl. Acad. Sci. USA 98: 14150–14155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelden M.C., Roessner U., Sharp R.E., Tester M., Bacic A. (2013). Genetic variation in the root growth response of barley genotypes to salinity stress. Funct. Plant Biol. 40: 516. [DOI] [PubMed] [Google Scholar]
- Shkolnik-Inbar D., Adler G., Bar-Zvi D. (2013). ABI4 downregulates expression of the sodium transporter HKT1;1 in Arabidopsis roots and affects salt tolerance. Plant J. 73: 993–1005. [DOI] [PubMed] [Google Scholar]
- Slovak R., Göschl C., Su X., Shimotani K., Shiina T., Busch W. (2014). A scalable open-source pipeline for large-scale root phenotyping of Arabidopsis. Plant Cell 26: 2390–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch R.C., Svedin E., Dilkes B., Chapple C., Li X. (2015). Discovery of a novel amino acid racemase through exploration of natural variation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 112: 11726–11731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara S., Hishiyama S., Jikumaru Y., Hanada A., Nishimura T., Koshiba T., Zhao Y., Kamiya Y., Kasahara H. (2009). Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 5430–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoen M.P.M., et al. (2017). Genetic architecture of plant stress resistance: multi-trait genome-wide association mapping. New Phytol. 213: 1346–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y., Jiang A., Gan L., Hossain M., Zhang J., Peng B., Xiong Y., Song Z., Cai D., Xu W., Zhang J., He Y. (2014). Genome duplication improves rice root resistance to salt stress. Rice (N. Y.) 7: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues P.E., Lasky J.R., Juenger T.E., Liu T.W., Kumar M.N. (2014). Genome-wide association mapping combined with reverse genetics identifies new effectors of low water potential-induced proline accumulation in Arabidopsis. Plant Physiol. 164: 144–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D., Mott R. (2009). The 1001 genomes project for Arabidopsis thaliana. Genome Biol. 10: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West G., Inzé D., Beemster G.T.S. (2004). Cell cycle modulation in the response of the primary root of Arabidopsis to salt stress. Plant Physiol. 135: 1050–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.J., Ding L., Zhu J.K. (1996). SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8: 617–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Hull A.K., Gupta N.R., Goss K.A., Alonso J., Ecker J.R., Normanly J., Chory J., Celenza J.L. (2002). Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 16: 3100–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.-K., Liu J., Xiong L. (1998). Genetic analysis of salt tolerance in arabidopsis. Evidence for a critical role of potassium nutrition. Plant Cell 10: 1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]