DNA modifications such as methylation serve as epigenetic markers that influence gene expression and function. The importance of similar modifications in RNA is gaining appreciation, leading to the emergence of epitranscriptomic analyses. RNA modifications appear to be a general feature of eukaryotes and are best understood in mammals, where reversible mRNA methylation is now known to regulate processes ranging from cell fate transition to stress responses (reviewed in Zhao et al., 2017). Methylation of mRNA—and other types of RNA—is also present in plants. In a recent epitranscriptomics study, David et al. (2017) cataloged over 1000 sites of cytosine methylation (m5C) in the Arabidopsis thaliana transcriptome. In mammals, N6-methyladenosine (m6A) is the most common modification of mRNA (see figure). The m6A mark is reversible: It can be placed (or “written”) by a methylation complex made of several subunits and “erased” by demethylases. So-called “readers” interpret the marks and transduce them into downstream effects via alterations in RNA splicing, maturation, translation, and degradation.
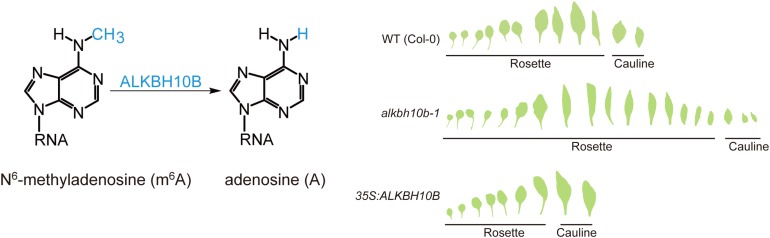
ALKBH10B demethylates m6A and regulates flowering time. N6-methyladenosine and adenosine residues in RNA (left). Number of leaves produced before flowering in the wild type compared with the albkbh10b-1 mutant and ALKBH10B overexpression line (right). (Adapted from Duan et al. [2017], Figures 2A and 4C.)
The first plant homolog of an m6A eraser was recently reported: Arabidopsis ALKBH9B, a homolog of the human m6A demethylase ALKBH5, has m6A demethylase activity and is involved in defense against viral infection (Martínez-Pérez et al., 2017). Now, Duan et al. (2017) report that another m6A demethylase functions in the regulation of flowering time.
Among five orthologs of human ALKBH5 in Arabidopsis, Duan and coworkers found that ALKBH10B had the most abundant transcripts in flowers and was expressed in most tissues tested. When ALKBH10B was expressed in Nicotiana benthamiana, the purified protein showed m6A demethylation activity that was dependent on the presence two conserved iron(II) binding residues. Similar to human ALKBH5, ALKBH10B had greater activity with a nonstructured RNA (or single-stranded RNA) substrate than with a stem-loop structured substrate in vitro.
The alkbh10b-1 mutant had higher levels of m6A-modified mRNA in both reproductive and vegetative organs, and overexpression of ALKBH10B in wild-type plants decreased m6A levels, establishing that ALKBH10B indeed functions as an RNA demethylase in vivo. The authors further showed that this activity was specific for m6A in mRNA.
Notably, the alkbh10b-1 mutant exhibited late flowering, while the overexpression line flowered early (see figure). Duan et al. found that ALKBH10B activity enhanced the transcript accumulation of the flowering promoter FT and that FT mRNA had a higher level of m6A in the mutant than in the wild type. The FT regulators SPL3 and SPL9 also had lower transcript levels and higher m6A levels in the alkbh10b-1 mutant. Furthermore, Duan et al. showed that ALKBH10B directly interacted with FT, SPL3, and SPL9 transcripts. Demethylation decreased the degradation of these transcripts, again similar to its effects on human mRNA.
Finally, Duan et al. examined global ALKBH10B-mediated demethylation, identifying 1276 sites in 1190 genes with greater m6A levels in the mutant compared with the wild type. Their work thus provides a solid foundation for understanding the roles of m6A-type methylation and demethylation in plants, which appears to share fundamental characteristics with that in mammals. In addition, it is clear that models for the regulation of flowering time must be updated to include posttranscriptional control of multiple transcripts via m6A marks.
Footnotes
Articles can be viewed without a subscription.
References
- David R., Burgess A., Parker B., Li J., Pulsford K., Sibbritt T., Preiss T., Searle I.R. (2017). Transcriptome-wide mapping of RNA 5-methylcytosine in Arabidopsis mRNAs and noncoding RNAs. Plant Cell 29: 445–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H.-C., Wei L.-H., Zhang C., Wang Y., Chen L., Lu Z., Chen P.R., He C., Jia G. (2017). ALKBH10B is an RNA N6-methyladenosine demethylase affecting Arabidopsis floral transition. Plant Cell 29: 2995–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Pérez M., Aparicio F., López-Gresa M.P., Bellés J.M., Sánchez-Navarro J.A., Pallás V. (2017). Arabidopsis m6A demethylase activity modulates viral infection of a plant virus and the m6A abundance in its genomic RNAs. Proc. Natl. Acad. Sci. USA 114: 10755–10760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B.S., Roundtree I.A., He C. (2017). Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 18: 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]