Detoxification of reactive carbonyl species produced by enhanced cellular sugar levels involves at least three subcellular compartments and depends on a specific glyoxalase I isoform during germination.
Abstract
Methylglyoxal (MGO) and glyoxal (GO) are toxic reactive carbonyl species generated as by-products of glycolysis. The pre-emption pathway for detoxification of these products, the glyoxalase (GLX) system, involves two consecutive reactions catalyzed by GLXI and GLXII. In Arabidopsis thaliana, the GLX system is encoded by three homologs of GLXI and three homologs of GLXII, from which several predicted GLXI and GLXII isoforms can be derived through alternative splicing. We identified the physiologically relevant splice forms using sequencing data and demonstrated that the resulting isoforms have different subcellular localizations. All three GLXI homologs are functional in vivo, as they complemented a yeast GLXI loss-of-function mutant. Efficient MGO and GO detoxification can be controlled by a switch in metal cofactor usage. MGO formation is closely connected to the flux through glycolysis and through the Calvin Benson cycle; accordingly, expression analysis indicated that GLXI is transcriptionally regulated by endogenous sugar levels. Analyses of Arabidopsis loss-of-function lines revealed that the elimination of toxic reactive carbonyl species during germination and seedling establishment depends on the activity of the cytosolic GLXI;3 isoform. The Arabidopsis GLX system involves the cytosol, chloroplasts, and mitochondria, which harbor individual components that might be used at specific developmental stages and respond differentially to cellular sugar status.
INTRODUCTION
During cellular activity, spontaneous reactions as well as side reactions of “promiscuous” enzymes can damage metabolites, yielding products that are at best useless and at worst toxic to the cell. To prevent the accumulation of such unwanted products, organisms have developed damage-control systems (Hanson et al., 2016). Reactive carbonyl species are highly reactive biological compounds that are unavoidably produced during cellular metabolism in all domains of life, from archaea and bacteria to eukaryotes (Sousa Silva et al., 2013; Semchyshyn, 2014). The physiologically most significant dicarbonyls, glyoxal (GO) and methylglyoxal (MGO), are common reactive carbonyl species that are produced under normal as well as stress or pathological conditions. GO is mainly produced by lipid peroxidation and the fragmentation of glycated proteins (Yin and Porter, 2005). MGO is generated during the oxidation of amino acids such as threonine and glycine (Thornalley, 1996; Kalapos, 2008). In addition, both reactive carbonyl species are formed by the spontaneous degradation of glucose (Thornalley et al., 1999). However, the major source of MGO appears to be the degradation of triose phosphates. A nonenzymatic deprotonation of the glycolytic key metabolites glyceraldehyde-3-phosphate (G3P) and dihydroxyacetone phosphate (DHAP) leads to the formation of an enediol phosphate intermediate, and the spontaneous elimination of the phosphate group produces MGO (Phillips and Thornalley, 1993; Chen and Thelen, 2010). This elimination reaction can also be triggered by the action of triose phosphate isomerase. During glycolysis, the enediol phosphate formed in the catalytic site of the enzyme can escape and rapidly decompose to MGO and inorganic phosphate (Richard, 1991; Phillips and Thornalley, 1993; Martins et al., 2001).
GO and MGO are electrophilic and are therefore highly reactive toward different cellular macromolecules, resulting in their irreversible modification and the formation of a variety of adducts and cross-links collectively named advanced glycation or lipoxidation end products (AGEs or ALEs; Aldini et al., 2013). The reaction of MGO with nucleic acids induces DNA strand breaks and elevated mutation frequency (Lee et al., 1995; Murata-Kamiya et al., 2000). Reactions of MGO with proteins, preferentially directed to arginine residues, lead to structural and functional changes (Sibbersen et al., 2013). In humans, AGE accumulation increases with age and is associated with several pathologies, including diabetes as well as cardiovascular and neurodegenerative diseases (Goldin et al., 2006; Thornalley, 2008; Münch et al., 2012). In plants, AGEs accumulate in leaves upon exposure to high light or elevated CO2 concentration (Qiu et al., 2008; Bechtold et al., 2009). MGO was reported to accumulate in tobacco (Nicotiana tabacum) and other plant species upon exposure to various abiotic stresses such as salinity, drought, and cold (Yadav et al., 2005).
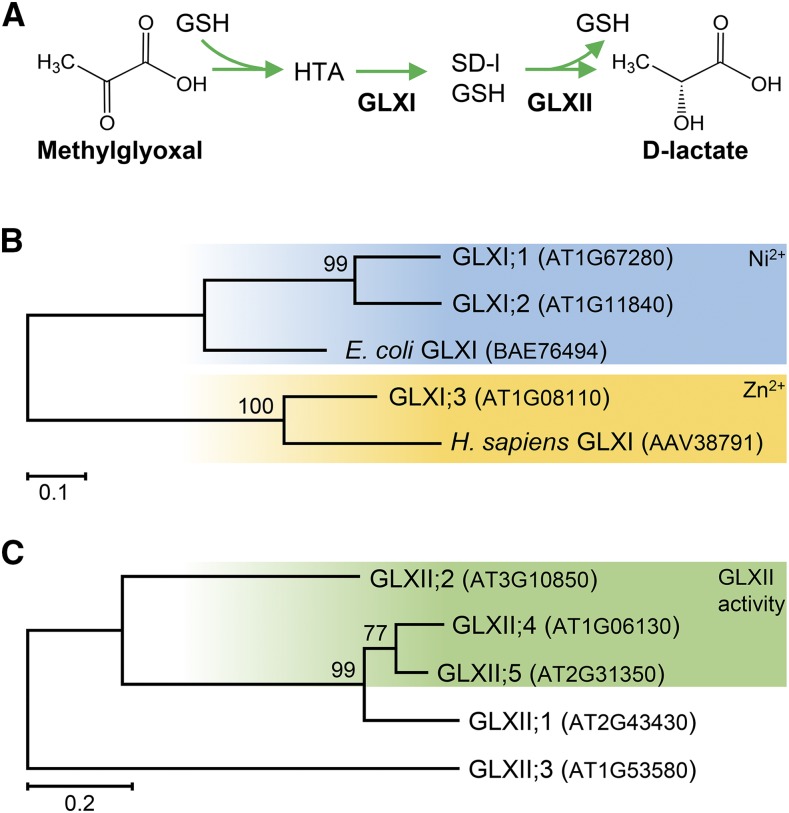
The steady state concentrations of reactive carbonyl species must be maintained at low levels to avoid their reaction with important cellular constituents. Several putative routes are described for the detoxification of MGO in different species; the conversion of MGO to d-lactate through the glyoxalase (GLX) system is assumed to be the main pathway for MGO detoxification in eukaryotic cells under normal physiological condition (Thornalley, 1990) (Figure 1A). This pathway consists of the enzymes S-d-lactoylglutathione lyase or glyoxalase I (GLXI; EC 4.4.1.5) and S-2-hydroxyacylglutathione hydrolase or glyoxalase II (GLXII; EC 3.1.2.6) acting in tandem. During the first step, MGO or GO spontaneously reacts with GSH to form a hemithioacetal, which is used as a substrate of GLXI to form S-lactoylglutathione (MGO-GSH) or S-glycolylglutathione (GO-GSH). GLXII catalyzes the conversion of S-lactoylglutathione into d-lactate or of S-glycolylglutathione into glycolate. Reduced GSH is released during this reaction. d-lactate is oxidized to pyruvate by the mitochondrial intermembrane space-localized D-lactate dehydrogenase (D-LDH), which passes the electrons to the electron transport chain via cytochrome c (CYTc) (Engqvist et al., 2009; Wienstroer et al., 2012; Welchen et al., 2016). Pyruvate finally enters the tricarboxylic acid cycle. Glycolate, derived from GO, might be metabolized by peroxisomal glycolate oxidase isoforms, which are found in all types of plant tissues.
Figure 1.
The Glyoxalase System Is the Main Pathway for MGO Detoxification in Eukaryotic Cells.
(A) MGO reacts spontaneously with GSH to form a hemithioacetal (HTA), which is the substrate of GLXI, producing S-d-lactoylglutathione (SD-l-GSH). GLXII converts SD-l-GSH into d-lactate.
(B) Simplified schematic overview of the phylogenetic relationships of all Arabidopsis GLXI proteins. GLXI;1 and GLXI;2 form a clade with the Ni2+-dependent GLXI proteins (E. coli), whereas GLXI;3 clusters together with the Zn2+-dependent GLXI proteins (Homo sapiens) (Kaur et al., 2013).
(C) The Arabidopsis genome encodes five GLXII-like proteins; however, S-d-lactoylglutathione hydrolase activity has only been reported for GLXII;2, GLXII;4, and GLXII;5. Sequences were retrieved from arabidopsis.org, and MEGA7 was used to reconstruct the maximum likelihood tree. The maximum likelihood tree is drawn to scale, with branch lengths measured in the number of substitutions per site (scale bar). Numbers at each node indicate boot strap values.
GLXI is a divalent metal ion-dependent lyase. Its homologs are categorized into two classes: Ni2+-/Co2+-dependent and Zn2+-dependent GLXI proteins. Functional GLXI proteins are composed of two GLXI domains, ensuring correct folding and activity, with potentially two metal-dependent substrate binding sites (Cameron et al., 1997; Thornalley, 2003). The two-domain GLXI isoforms remain monomeric, whereas the one-domain GLXI isoforms form homodimers (Ridderström and Mannervik, 1996; Frickel et al., 2001; Turra et al., 2015).
GLXII belongs to the β-lactamase protein family (Cricco and Vila, 1999; Crowder and Walsh, 1999) and contains a Fe3+/Zn2+ binuclear metal center that is essential for its activity (Zang et al., 2001; Wenzel et al., 2004; Marasinghe et al., 2005). Crystal structures of human and plant GLXII indicate that it is active as a dimer (Marasinghe et al., 2005). Although GLXII hydrolyzes many different glutathione thioesters, S-d-lactoylglutathione is the preferred substrate for the enzyme from most sources (Ridderström and Mannervik, 1996; Bito et al., 1997).
In plants, both GLXI and GLXII are encoded by multigene families. In Arabidopsis thaliana, 11 genes encode GLXI-like proteins (http://www.arabidopsis.org/). However, phylogenetic analysis clearly indicates that there are only three GLXI homologs encoded in the Arabidopsis genome (Kaur et al., 2013), which fall into two subclasses: (1) the Ni2+-dependent two-domain isoforms GLXI;1 and GLXI;2 and (2) the Zn2+-dependent one-domain isoform GLXI;3 (Figure 1B; Supplemental Figure 1 and Supplemental File 1). Five gene loci, GLXII;1 to GLXII;5, encode GLXII-like proteins in Arabidopsis (Figure 1C). GLXII have been extensively studied at the biochemical and structural levels (Maiti et al., 1997; Marasinghe et al., 2005). Two of these sequences do not encode functional GLXII: GLXII;1 has β-lactamase activity and responds to abiotic stress conditions (Limphong et al., 2009; Devanathan et al., 2014), while GLXII;3 (ETHE1) acts as a persulfide dioxygenase that is essential for embryo and endosperm development (Holdorf et al., 2012). In contrast, GLXII;2, GLXII;4, and GLXII;5 are active GLXII.
As MGO and GO are mainly produced from the autoxidation of glucose and conversion of triose phosphates, it is reasonable to assume that the subcellular compartments producing these metabolites possess at least a partially functional GLX system. However, definitive knowledge on the subcellular distribution of the members of this detoxification pathway is lacking. Previous studies yielded conflicting results regarding the subcellular compartmentalization of GLXI isoforms. GLXI isoforms are reported to be found in mitochondria (Salvato et al., 2014), peroxisomes (Reumann et al., 2009), chloroplasts (Tomizioli et al., 2014), the cytosol (Ito et al., 2011), and the nucleus (Kaur et al., 2017). Based on predictions, Arabidopsis GLXII;2 would localize to the cytosol, while GLXII;4 and GLXII;5 likely localize to mitochondria (Maiti et al., 1997; Zang et al., 2001; Marasinghe et al., 2005). The situation becomes more complex because several predicted GLX splice forms may be translated into isoforms with different subcellular localizations.
Despite the already established enzymatic properties of Arabidopsis GLXII isoforms, definite evidence for the existence of GLXI isoforms, their molecular properties, and their in planta relevance is fragmentary. Here, to build up a cellular model for the detoxification of MGO and GO, we analyzed the GLX system in Arabidopsis, as this species possesses extensive genetic, molecular biology, and genomic resources. We identified the physiologically predominant isoforms, providing insights into the GLX system. We show that alternative splicing of the Arabidopsis GLXI pre-mRNAs produces GLXI isoforms with different subcellular localizations. We performed a deep comparative biochemical analysis of the GLXI isoforms using MGO and GO as substrates and discovered a cofactor-dependent switch of the substrate specificities of GLXI;1 and GLXI;2. T-DNA insertion lines with defects in the GLXI homologs exhibited differential sensitivities toward reactive carbonyl species and sucrose. Our results demonstrate that the elimination of toxic reactive carbonyl species produced by enhanced cellular sugar levels is highly dependent on the cytosolic GLXI;3 isoform during germination and postgerminative seedling establishment. Moreover, we found that the transcription of the GLXI homologs is modulated by the sugar status of the plant. This hints at a need for reactive carbonyl species detoxification in conditions that induce changes in cellular sugar metabolism. In summary, our results allowed us to propose a cellular model for the detoxification of MGO and GO involving at least three subcellular compartments.
RESULTS
The Glyoxalase System of Arabidopsis Is Composed of Isoforms with Different Predicted Subcellular Localizations That Can Arise through Alternative Splicing
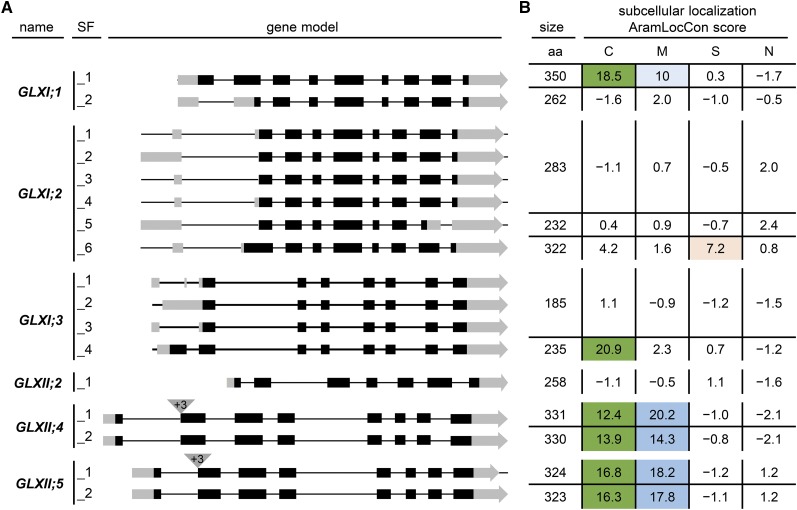
Homology models unambiguously identified three GLXI gene loci in Arabidopsis, At1g67280 (GLXI;1), At1g11840 (GLXI;2), and At1g08110 (GLXI;3) (Kaur et al., 2013) (Supplemental Figure 2). These homologous gene loci are predicted to generate several alternative splice forms corresponding to different protein isoforms, which differ at the N- or the C-terminal regions. These alterations have drastic effects on the predicted subcellular localizations of the resulting proteins (Figure 2).
Figure 2.
Arabidopsis Genes Encoding Enzymes of the GLX System.
(A) Gene models of the predicted splice forms (SF; indicated by underscore) of GLXI and GLXII. Box, exon; black, coding sequence; gray, untranslated region.
(B) Predicted subcellular localization of the proteins arising from the splice forms. aa, amino acids; C, chloroplast; M, mitochondrion; S, secretory pathway; N, nucleus. The AramLocCon score was taken from Schwacke et al. (2007) and describes a consensus subcellular localization prediction based on 20 individual prediction programs for C, M, S, or N. +3 correspond to 3 additional base pairs at the beginning of exon 2 in GLXII;4 and GLXII;5.
The first of the three loci, At1g67280, is predicted to be spliced into two alternative mRNAs, producing GLXI;1_1 (350 amino acids) and GLXI;1_2 (262 amino acids), which differ only in the length of the N terminus of the proteins (Figure 2A). The GLXI;1_1 isoform is predicted, with high probability, to carry a chloroplastic transit peptide, whereas GLXI;1_2 does not appear to contain a subcellular localization sequence (Figure 2B, AramLocCon score; Schwacke et al., 2007). The pre-mRNA of the second locus, At1g11840, is predicted to produce six alternative splice forms, leading to three isoforms, GLXI;2_1 (derived from the splice forms GLXI;2_1 to GLXI;2_4; 283 amino acids), GLXI;2_5 (232 amino acids), and GLXI;2_6 (322 amino acids) (Figure 2A). GLXI;2_1 and GLXI;2_5 are not predicted to localize to any subcellular compartment, while GLXI;2_6 is predicted, with moderate probability, to be targeted to the secretory pathway (Figure 2B). Finally, four individual alternative splice forms are predicted from gene locus At1g08110, resulting in two isoforms, GLXI;3_1 (derived from the splice forms GLXI;3_1 to GLXI;3_3; 185 amino acids) and GLXI;3_4 (235 amino acids) (Figure 2A). The longer GLXI;3_4 isoform is predicted, with high probability, to carry a chloroplastic transit peptide (Figure 2B). In summary, the first step of the GLX system in Arabidopsis is encoded by three homologs of GLXI, producing seven putative GLXI isoforms that appear to be targeted to the cytosol, chloroplasts, and possibly the secretory pathway.
Analyses of the three gene loci encoding the second step of the GLX system, GLXII, also revealed the existence of alternative splice forms (Figure 2). The locus At3g10850 (GLXII;2) is transcribed in only one splice form, producing a protein of 258 amino acids with no predicted subcellular targeting sequence (Figures 2A and 2B). The second locus, At1g06130 (GLXII;4), is transcribed into two alternative splice forms, which differ only in the occurrence of three additional base pairs at the beginning of exon 2. These splice forms produce GLXII;4_1 (331 amino acids) and GLXII;4_2 (330 amino acids). The splice forms encoded by the third locus, At2g31350, which produce GLXII;5_1 (324 amino acids) and GLXII;5_2 (323 amino acids; Figure 2A), also differ in the occurrence of three additional base pairs at the beginning of exon 2. All four proteins are predicted to possess both a chloroplastic transit peptide and a mitochondrial targeting signal (Figure 2B). Taken together, the second step of the Arabidopsis GLX system is encoded by three homologs of GLXII, with a total of five putative GLXII isoforms that appear to be targeted to the cytosol, chloroplasts, and mitochondria.
RNA Expression Analysis Identifies the Physiologically Relevant Splice Forms in Arabidopsis
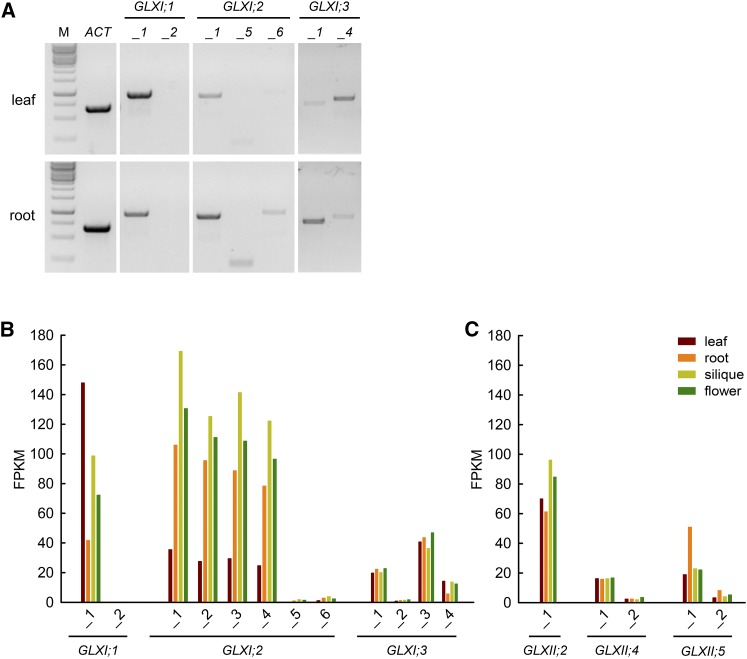
While several alternative splice forms translated into proteins with different subcellular localization are predicted for each Arabidopsis GLX homolog, it is not certain which isoforms are physiologically relevant. To assess the in vivo existence of the different isoforms, we conducted RT-PCR analysis using autotrophic (leaves) and heterotrophic (roots) organs of Arabidopsis (Figure 3A). We designed primers spanning exon-intron junctions to specifically detect the alternative splice forms leading to different isoforms. We detected the GLXI;1_1 alternative splice form in both organs analyzed, although amplification from leaf cDNA resulted in a stronger signal. Amplification products of GLXI;1_2 were never obtained (Figure 3A). Translation of GLXI;2 can result in at least three different isoforms (Figure 3A), represented by GLXI;2_1, GLXI;2_5, and GLXI;2_6. The expression of GLXI;2_1 was stronger than that of GLXI;2_5 and GLXI;2_6, which both showed very faint amplification bands (Figure 3A). In all cases, expression was higher in roots than in leaves. Finally, we obtained amplification products for both GLXI;3_1 and GLXI;3_4. Amplification of GLXI;3_1 was stronger than that of GLXI;3_4 in roots, while GLXI;3_4 amplification was stronger than that of GLXI;3_1 in leaves (Figure 3A).
Figure 3.
Transcript Abundance of GLX Homologs and Their Respective Splice Forms in Different Arabidopsis Organs.
(A) Amplification of predicted GLXI splice forms in Arabidopsis organs via RT-PCR. Specific primer pairs were used to amplify the GLXI splice forms from cDNA from leaves and roots of 3-week-old Arabidopsis plants grown on MS medium. M, molecular size standard.
(B) FPKM values of GLXI splice forms.
(C) FPKM values of GLXII splice forms. Raw data were taken from Liu et al. (2012) and mapped with high stringency to the TAIR 10 annotation (Berardini et al., 2015).
To reveal if further splice forms exist that could not be properly discriminated using exon-intron junction primers, we mapped and reanalyzed with high stringency publicly available deep-sequencing RNA-seq data derived from different plant organs (Supplemental Data Set 1) (Liu et al., 2012). In line with our RT-PCR results, the data indicated that transcription of GLXI;1 results only in the splice form GLXI;1_1 (Figure 3B), with high expression in leaves and moderate expression in roots. Transcripts of the alternative splice form GLXI;1_2 were detected with FPKM (fragments per kilobase of exon per million fragments mapped) values ranging from 0.1 to 0.3 in all tissues tested, indicating that this form is physiological irrelevant, at least in the vegetative and reproductive stages analyzed. The transcripts of GLXI;2_1 to GLXI;2_4, which translate into the same isoform, are more abundant in roots, siliques, and flowers than in leaves (Figure 3B). Notably, analysis of the RNA-seq data showed that transcripts of GLXI;2 are among the top 200 most highly expressed genes in Arabidopsis. In line with our RT-PCR analysis, the alternative splice forms GLXI;2_5 and GLXI;2_6 were mapped with FPKM values ranging from 0.1 to 3.8, suggesting a minor physiological relevance of these splice forms under the conditions tested. The splice forms GLXI;3_1 and GLXI;3_3, which produce the same isoform, are uniformly expressed in all organs tested, while GLXI;3_4 is expressed at lower levels in roots than in other organs (Figure 3B).
Using the same RNA-seq data, we further analyzed the expression of GLXII, the second component of the GLX system. The analysis indicated a moderate, uniform level of expression of GLXII;2 in all tissues tested (Figure 3C). The abundance of transcripts of the splice form GLXII;4_1 (FPKMleaf = 16) is substantially higher than that of GLXII;4_2 (FPKMleaf = 3); both splice forms are expressed uniformly in autotrophic and heterotrophic tissues. Transcript abundance of GLXII;5_1 is substantially higher than that of GLXII;5_2. Both splice forms are present with 2-fold higher FPKM values in roots compared with leaf, flower, or siliques.
In summary, in the vegetative and reproductive stages of Arabidopsis, we observed high abundances of transcripts encoding the isoforms GLXI;1_1, GLXI;2_1 (produced from the splice forms GLXI;2_1 to GLXI;2_4), GLXI;3_1 (produced from GLXI;3_1 and GLXI;3_3), and GLXI;3_4. Under the same conditions, we confirmed the occurrence of all GLXII alternative splice forms.
At1g67280, At1g11840, and At1g08110 Encode Functional GLXI Proteins with Distinct Metal Activation Profiles
Phylogenetic analysis indicated that Arabidopsis GLXI;1_1 and GLXI;2_1 cluster with other Ni2+-/Co2+-dependent GLXI homologs, while GLXI;3 clusters with Zn2+-dependent GLXI homologs (Figure 1B; Supplemental File 1) (Kaur et al., 2013). As this classification, which is based on protein sequence homologies, may not reliably indicate the physiologically relevant metal ions for catalysis, we analyzed the enzymatic activity of the GLXI isoforms using the hemithioacetals MGO-GSH and GO-GSH as substrates in the presence of various divalent metal ions (Zn2+, Mn2+, Ca2+, Mg2+, Co2+, and Ni2+).
We heterologously expressed the transcriptionally most abundant splice forms of GLXI in Escherichia coli and purified the resulting proteins to homogeneity. As alternative splicing of GLXI;3 introduces changes at the N terminus of the translated proteins, and the predicted mature GLXI;3_1 and GLXI;3_4 proteins are identical (GLXI;3_4 has a predicted chloroplastic targeting sequence of 51 amino acids; targetP v1.1; Emanuelsson et al., 2000; Figure 2), we expressed the full-length GLXI;3_1 for analysis. The isolated GLXI proteins, here named GLXI;1 (GLXI;1_1), GLXI;2 (GLXI;2_1), and GLXI;3 (GLXI;3_1 and GLXI;3_4), showed the expected molecular masses (42, 35, and 23 kD, respectively) after separation by SDS-PAGE (Supplemental Figure 3).
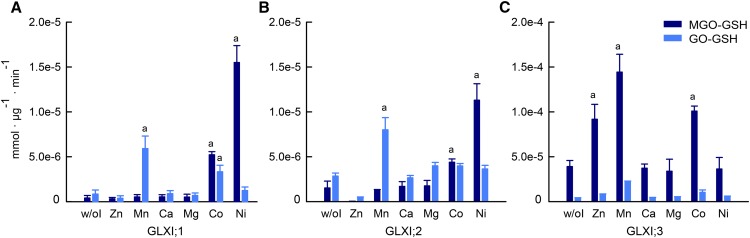
In agreement with the predicted metal cofactor preferences (Figure 1B), GLXI;1 and GLXI;2 showed the highest activities with MGO-GSH in the presence of Ni2+, followed by Co2+ (Figures 4A and 4B). Using Co2+, the activity of both isoforms dropped by around 40% compared with the fully Ni2+-activated enzymes. When using other metal cofactors, the enzymatic activities were comparable to the basal activity measured without the addition of a metal cofactor (Figures 4A and 4B). The activity of GLXI;1 and GLXI;2 was barely measurable in the presence of Zn2+, suggesting an inhibitory effect of this metal ion, which might be related to a disturbed octahedron binding geometry after binding of Zn2+ (Himo and Siegbahn, 2001; Clugston et al., 2004).
Figure 4.
Metal Activation Profile of Arabidopsis GLXI Isoforms.
Absolute activities of GLXI;1 (A), GLXI;2 (B), and GLXI;3 (C) with MGO-GSH and GO-GSH (mmol µg−1 min−1) using different bivalent metal ion cofactors. e, scientific notation (times 10 raised to the power of exponent). Values are means ± se (n = 3) of independent enzyme preparations. Testing for significant differences was performed by two-way ANOVA for GLXI;1 (substrate, P = 0.021; ion, P < 0.0001; interaction, P < 0.0001), for GLXI;2 (substrate, P = 0.20; ion, P < 0.0001; interaction, P < 0.0001), and for GLXI;3 (substrate, P = 0.016; ion, P < 0.0001; interaction, P < 0.0001) (Supplemental File 2), with subsequent Dunnett’s multiple comparisons test to w/oI (without ion) control, a = P < 0.05.
Intriguingly, when using GO-GSH as substrate, the preference of GLXI;1 and GLXI;2 for the metal ion changed. In both cases, the highest enzymatic activities were measured with Mn2+, which were 40% and 70% of the maximal activities of GLXI;1 and GLXI;2 measured with MGO-GSH, respectively (Figures 4A and 4B). In addition, GLXI;1 can also use Co2+ in the conversion of GO-GSH. With both isoforms, the use of Ni2+ in the conversion of GO-GSH yielded low relative enzymatic activities comparable to the assay without the addition of ions. The general basal enzymatic activity measured without the addition of divalent cations may be caused by trace amounts of metal ions bound to the enzyme (Mustafiz et al., 2014).
The analysis of GLXI;3 using the purified protein initially showed high activities with MGO-GSH and GO-GSH, independently of the metal cofactor used. As the Zn2+-dependent GLXI homologs are known to bind metal ions tightly (Sukdeo and Honek, 2007), it is highly likely that the metal ion was not removed during GLXI;3 isolation. Thus, to remove any remaining bound metal ions, we dialyzed GLXI;3 using EDTA. With MGO-GSH as substrate, dialyzed GLXI;3 showed maximal activity in the presence of Mn2+ and around 70% activity in the presence of Co2+ and Zn2+ (Figure 4C; Supplemental File 2). Using GO-GSH as substrate, GLXI;3 showed a preference for Mn2+; in this case, the maximal activity measured was 16% of the activity measured with MGO-GSH (Figure 4C).
The metal activation profiles obtained by this analysis indicate that GLXI;1 and GLXI;2 change their substrate specificities (MGO-GSH or GO-GSH) depending on the metal cofactor used. This metal cofactor-dependent switch in substrate specificity was not observed in the case of GLXI;3. In summary, our data suggest that the Arabidopsis GLX system is able to detoxify not only MGO but also GO and that this is dependent on the metal cofactor used for catalysis.
GLXI;1, GLXI;2, and GLXI;3 Have High Affinities for the Hemithioacetals of MGO and GO Formed in the Presence of GSH
We further investigated the kinetic properties of the GLXI isoforms using the respective optimal metal ion cofactors. We first analyzed the optimal pH range of the three Arabidopsis GLXI isoforms. All GLXI isoforms showed high activities in the broad pH range from 5.5 to 7.5. We determined that the optimal pH was 6.5 for GLXI;1 and GLXI;3 and 7.0 for GLXI;2. We analyzed the kinetic constants (Table 1) in the presence of varying concentrations of the hemithioacetals (MGO-GSH or GO-GSH), which were prepared by varying MGO or GO concentrations at fixed GSH concentration and vice versa. GLXI;1 showed high affinities toward MGO-GSH, with a Km value of 279 ± 4 µM, and toward GO-GSH, with a Km value of 359 ± 3 µM. The catalytic efficiency with MGO-GSH (kcat/Km = 32,190 s−1 M−1) was 3-fold higher than with GO-GSH (kcat/Km = 10,279 s−1 M−1). GLXI;2 also showed high affinity toward MGO-GSH, with a Km value of 330 ± 10 µmol and a catalytic efficiency of 35,430 s−1 M−1. This isoform has a comparable affinity toward GO-GSH (Km = 394 ± 5 µM) as toward MGO-GSH, but a 5.6-fold lower catalytic efficiency for GO-GSH (kcat/Km = 6286 s−1 M−1).
Table 1. Kinetic Parameters of Arabidopsis GLXI Isoforms.
| Protein | Substrate | Km (µM) | kcat (−1) | kcat/Km (s−1 M−1) | pH | Preferred Cofactor |
|---|---|---|---|---|---|---|
| GLXI;1 | MGO-GSH | 279 ± 4 | 8.9 ± 0.5 | 32,190 | 5.0–7.5 | Ni2+ |
| GO-GSH | 359 ± 3 | 3.7 ± 0.1 | 10,279 | 5.5–7.0 | Mn2+ | |
| GLXI;2 | MGO-GSH | 330 ± 10 | 9.1 ± 0.6 | 35,430 | 5.5–7.5 | Ni2+ |
| GO-GSH | 394 ± 5 | 2.3 ± 0.1 | 6,286 | 5.5–7.5 | Mn2+ | |
| GLXI;3 | MGO-GSH | 476 ± 125 | 175.1 ± 29.8 | 368,294 | 5.5–7.5 | Mn2+ |
| GO-GSH | 388 ± 128 | 20.5 ± 1.1 | 52,860 | 5.5–7.5 | Mn2+ |
The kinetic constants were calculated by nonlinear regression analysis. Values are means ± se (n = 4–5) of independent enzyme preparations.
As GLXI;1 and GLXI;2 showed normal hyperbolic saturation kinetics when we used fixed concentrations of the reactive carbonyl species or GSH in the reaction medium, we determined the kinetic parameters using all of the data collected. In contrast, when we analyzed the kinetic properties of the Mn2+-dependent GLXI;3, different responses were observed when using either fixed MGO (or GO) or GSH to produce the hemithioacetal in the reaction medium. When using a fixed MGO or GO concentration and varied GSH concentrations, GLXI;3 was inhibited at high GSH/MGO ratios. The plot of the enzymatic activity versus the concentration of the substrate indicated that GLXI;3 activity was inhibited at concentrations of MGO-GSH higher than 1.0 mM and of GO-GSH higher than 1.5 mM. With MGO-GSH as substrate, we calculated a Km of 476 ± 125 µM and a catalytic efficiency of 368,294 s−1 M−1 (Table 1). When using GO-GSH, we calculated a Km of 388 ± 128 µM and a catalytic efficiency of 52,860 s−1 M−1, which is 7-fold lower than with MGO-GSH (Table 1). On the other hand, at fixed GSH and varying MGO or GO concentrations, GLXI;3 showed an “S-shaped” sigmoidal curve. We determined a Hill coefficient of n = −2.2 for MGO-GSH and n = −1.2 for GO-GSH, which indicates negative cooperativity for the binding of both substrates to the enzyme. We calculated an EC50 of 0.99 mM for MGO-GSH and of 1.23 mM for GO-GSH. These results suggest that an excess of free GSH in the reaction medium affects GLXI;3 activity. This is in line with the observed inhibition of GLXI;3 activity when using high GSH concentrations at fixed MGO or GO concentrations.
All three Arabidopsis GLXI isoforms act on MGO-GSH and GO-GSH with similar affinities and convert MGO-GSH with higher catalytic rates than GO-GSH.
Arabidopsis GLXI Homologs Functionally Complement a Yeast GLXI Loss-of-Function Mutant
As the kinetic properties of GLXI;1 and GLXI;2 differ from those of GLXI;3, we analyzed the in vivo relevance of the three GLXI isoforms in a complementation assay using a yeast (Saccharomyces cerevisiae) mutant deficient in GLOI, a Zn2+-dependent GLXI (glo1Δ; YML004c). In yeast, only one gene locus encodes GLXI; the glo1Δ knockout strain is therefore sensitive to MGO (Inoue and Kimura, 1996). We constitutively expressed GLXI;1, GLXI;2, and GLXI;3 in the glo1Δ knockout strain.
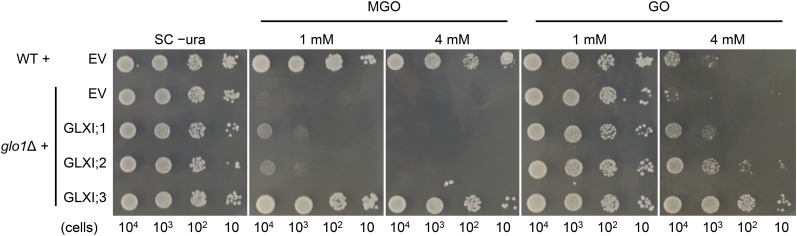
Wild-type yeast transformed with an empty vector was able to grow on 4 mM MGO, whereas glo1Δ already failed to grow on 1 mM MGO (Figure 5). Complementation of glo1Δ with Arabidopsis GLXI;1 or GLXI;2 resulted in a moderate suppression of the MGO sensitive phenotype (Figure 5). In contrast, the expression of GLXI;3 in glo1Δ resulted in a full recovery of growth on 4 mM MGO (Figure 5). As our kinetic data indicated that the conversion of MGO-GSH by GLXI;1 and GLXI;2 is Ni2+ dependent, we aimed to increase the suppression of MGO sensitivity by supplementation with 25 µM Ni2+. However, no enhanced tolerance toward MGO was observed. Notably, the use of Ni2+ with S. cerevisiae is controversial, as no specific transporter for Ni2+ has been identified. Instead, Ni2+ might be transported via a broad-specificity divalent metal transporter (Schmidt et al., 2009); furthermore, yeast lacks another common Ni2+-dependent enzyme, urease (Zhang et al., 2009).
Figure 5.
Functional Complementation of the S. cerevisiae Mutant glo1Δ.
Growth of wild-type and glo1Δ strains transformed with pDR195 as empty vector control (EV) and glo1Δ transformed with pDR195:GLXI;1, pDR195:GLXI;2, or pDR195:GLXI;3 on 1 and 4 mM MGO or GO was monitored up to 48 h at 30°C. Cell numbers are indicated at the bottom.
When we grew the wild type and glo1Δ yeast on medium containing GO, both strains were tolerant to 1 mM GO, but they showed sensitivity to this compound at a concentration of 4 mM (Figure 5). Heterologous expression of GLXI;1 and GLXI;2 resulted in moderate suppression of the GO-sensitive phenotype at 4 mM GO. Again, the expression of GLXI;3 in glo1Δ led to a strongly enhanced tolerance to 4 mM GO. We showed that the conversion of GO-GSH by all GLXI isoforms is Mn2+ dependent (Figure 4). Several Mn2+ transport systems as well as Mn2+-dependent metallo-enzymes have been described in yeast (Reddi et al., 2009). Thus, it is reasonable to assume that the endogenous Mn2+ availability influences the usage of this cofactor by the different isoforms and in turn the extent of suppression of the sensitivity toward GO of the complemented yeast strains.
In summary, growth recovery of the yeast strain glo1Δ on MGO and GO by the expression of Arabidopsis GLXI;1, GLXI;2, and GLXI;3 provides evidence that all of these isoforms are active GLXI enzymes that are able to detoxify both substrates in vivo.
The Cytosol and Chloroplasts Harbor a Full Glyoxalase System for the Detoxification of MGO and GO
Our extensive analysis of transcriptomic data revealed the occurrence of several splice forms of GLXI and GLXII pre-mRNAs, which give rise to several isoforms with diverse (and in some cases ambiguous) predicted subcellular localizations (Figure 2). With the aim of unambiguously establishing the subcellular localization of all physiologically relevant GLXI and GLXII isoforms of a model plant cell, we amplified the full-length coding sequences of all predominant splice forms from Arabidopsis leaves and roots (Figures 3B and 3C) and performed reporter gene-targeting analysis of the encoded proteins. The isolated splice forms were cloned in-frame with a C-terminal YFP tag into pNL3 or with an N-terminal YFP tag into pNL25 vectors. For isoforms with a predicted subcellular localization (AramLocCon score >3; Figure 2), the GLXI constructs were coexpressed with the corresponding organelle markers.
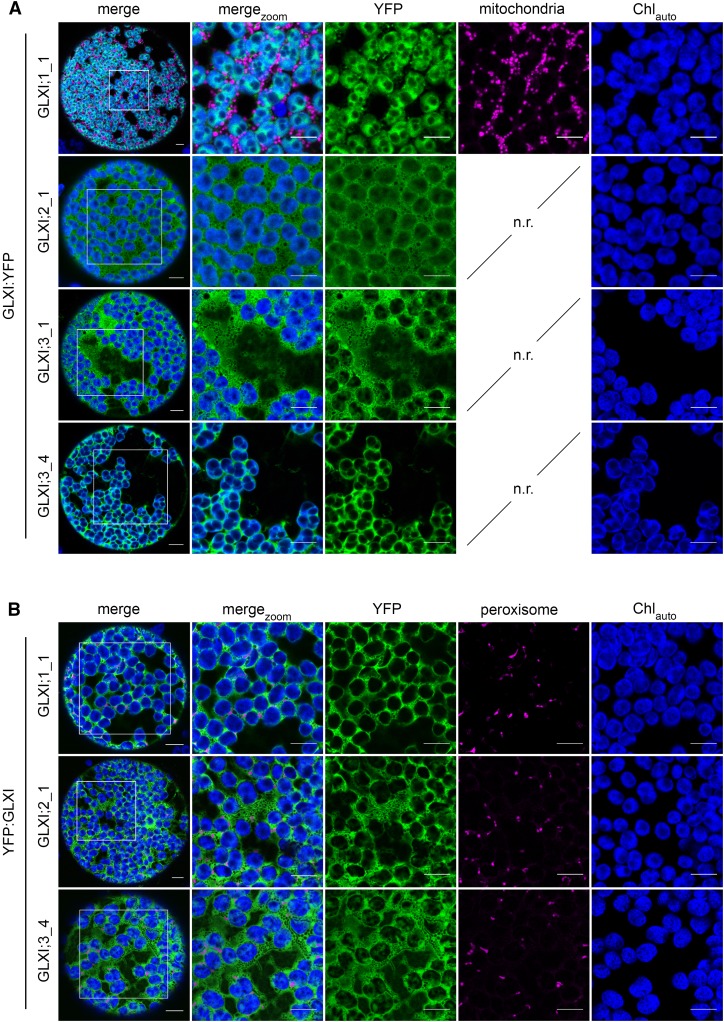
The prediction score of GLXI;1_1 indicates not only a high probability for chloroplastic localization, but also a possible mitochondrial localization. Fluorescence of the mitochondrial red fluorescent protein marker (mito:eqFP611; Forner and Binder, 2007) did not colocalize with GLXI;1_1:YFP fluorescence. In contrast, GLXI;1_1:YFP fluorescence clearly colocalized with the chlorophyll autofluorescence signal, demonstrating a chloroplastic localization (Figure 6A).
Figure 6.
Subcellular Localization of Arabidopsis GLXI Isoforms Using Tobacco Leaf Protoplasts.
(A) The coding sequence of YFP was fused in-frame with the C terminus of the full-length coding sequence of GLXI;1_1, GLXI;2_1, GLXI;3_1, and GLXI;3_4 (GLXI:YFP fusions) and expressed in tobacco leaf protoplasts. GLXI proteins with high probability of mitochondrial localization (AramLocCon Score >3) were coexpressed with the mitochondrial marker protein mito:eq611.
(B) YFP was fused in frame with the N terminus of the full coding sequence of GLXI;1_1, GLXI;2_1, and GLXI;3_4 (YFP:GLXI fusions) and coexpressed with a peroxisomal marker protein in tobacco leaf protoplasts. Images were taken after 3 d of infiltration. merge, protoplast overview; mergezoom, 50 µm detail; YFP, YFP fluorescence; mitochondria or peroxisome, fluorescence of marker proteins for the organelles; Chlauto, autofluorescence of chlorophyll; n.r., not required. Bar = 10 µm.
GLXI;2_1:YFP fluorescence signal accumulated in the cytosol, where a high number of nonfluorescent inclusions, most likely representing other organelles, could be observed (Figure 6A). Cytosolic fluorescence distribution was verified by the expression of YFP alone (Supplemental Figure 4A). We were never able to isolate a full-length transcript of GLXI;2_5, which is consistent with the extremely low FPKM values found for this splice form (close to zero) in the RNA-seq data analysis (Figure 3B); together, these findings indicate that GLXI;2_5 is expressed at extremely low levels; therefore, it is unlikely that this isoform plays a significant role in vivo. Due to the very low levels of expression of GLXI;2_6, we consider the occurrence of this splice form to be a minor event in the tissues tested (Figures 3A and 3B); nevertheless, we were able to amplify a full-length coding sequence from roots and, thus, we included GLXI;2_6 in the subcellular localization analysis. GLXI;2_6:YFP, which carries an additional 39 amino acids at the N terminus, did not localize to the cytosol, chloroplast, mitochondria, or peroxisomes, as no superimposition of the YFP signal with chlorophyll autofluorescence or the respective organelle markers was observed (Supplemental Figure 4B). In line with the high prediction score for the secretory pathway (Figure 2B), the GLXI:2_6:YFP fluorescence instead colocalized with the endoplasmic reticulum fluorescent marker protein (ER:CFP; Nelson et al., 2007; Supplemental Figure 4B).
The expression of GLXI;3_1:YFP resulted in strong accumulation of the fluorescence signal in the cytosol (Figure 6A). GLXI;3_4 has a 50-amino acid longer N terminus than GLXI;3_1 and a high prediction score for chloroplastic localization (Figure 2B). In line with this prediction, we found that GLXI;3_4:YFP fluorescence colocalized with the chlorophyll autofluorescence signal, indicating a chloroplastic localization.
As proteomic analysis revealed a GLXI in the peroxisomal fraction (Reumann et al., 2009), we also generated N-terminal YFP:GLXI fusion proteins and analyzed their possible peroxisomal localization by coexpressing them with the peroxisomal cyan fluorescent protein marker (CFP:PTS1; Linka et al., 2008; Figure 6B). The fluorescence signals from YFP:GLXI;1_1, YFP:GLXI;2_1, and YFP:GLXI;3_4 did not accumulate in distinct cytosolic bodies, nor did they colocalize with the peroxisomal marker protein. This analysis revealed a clear cytosolic distribution of the fluorescence signal when the N terminus of the GLXI isoforms was blocked (Figure 6B), indicating that none of the predominant GLXI isoforms carries a PTS1 signal.
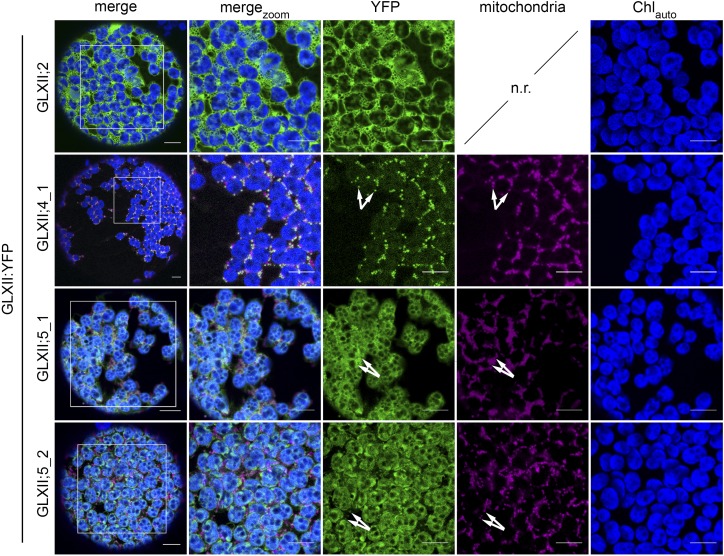
Out of the five possible splice forms representing the second component of the GLX system, we were able to amplify GLXII;2, GLXII;4_1, GLXII;5_1, and GLXII;5_2 and generate C-terminal YFP fusion constructs. In line with the lack of predicted localization to any subcellular compartment, the fluorescence signals from GLXII;2:YFP accumulated in the cytosol (Figure 7). The expression of GLXII;4_1:YFP resulted in an accumulation of fluorescence signals in subcellular structures that colocalized with the mitochondrial marker protein. Intriguingly, a fluorescence signal of lower intensity was also detected in the chloroplast, as it colocalized with chlorophyll autofluorescence (Figure 7). Similarly, GLXII;5_1:YFP and GLXII;5_2:YFP showed a clear dual targeting of the proteins to chloroplasts and mitochondria (Figure 7). To independently verify the dual localization of these proteins, we additionally generated YFP-fusions of GLXII;4_1, GLXII;5_1, and GLXII;5_2 under the control of the native Arabidopsis UBQ10 promoter. In agreement with our previous results, the analysis of fluorescence supported the dual targeting of GLXII;4_1, GLXII;5_1, and GLXII;5_2 to mitochondria and chloroplasts (Supplemental Figure 5).
Figure 7.
Subcellular Localization of Arabidopsis GLXII Isoforms in Tobacco Leaf Protoplasts.
The coding sequence of YFP was fused in-frame with the C terminus of the full-length coding sequence of GLXII;2, GLXII;4_1, GLXII;5_1, and GLXII;5_2 (GLXII:YFP fusions) and expressed in tobacco leaf protoplasts. GLXII proteins with high probability of mitochondrial localization were coexpressed with the mitochondrial marker protein mito:eq611. Images were taken after 3 d of infiltration. merge, protoplast overview; mergezoom, 50 µm detail; YFP, YFP fluorescence; mitochondria, fluorescence of marker proteins for the organelles; Chlauto, autofluorescence of chlorophyll; n.r., not required. Arrows indicate colocalization of YFP and marker protein fluorescence. Bar = 10 µm.
The Expression of GLXI Is Regulated by the Cellular Sugar Status
As MGO and GO are generated by the degradation of glucose and triose phosphates, the formation of MGO and GO might be directly coupled to steady state sugar levels. Thus, we investigated the hypothesis that changes in the cellular sugar status affect GLXI gene expression.
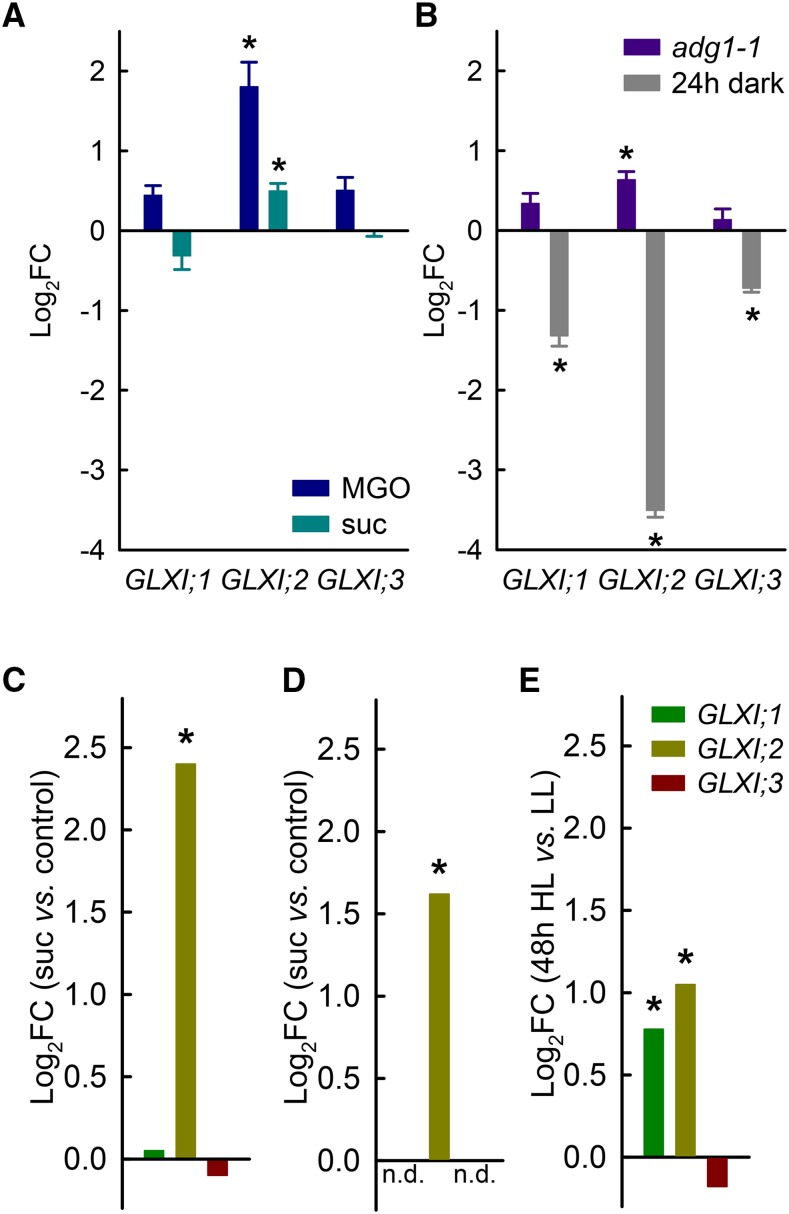
We performed transcriptional analysis of the GLXI genes via qPCR using Arabidopsis seedlings exposed to MGO or sucrose. When the seedlings were grown on 1 mM MGO, the expression of all GLXI genes was higher than in seedlings grown on Murashige and Skoog (MS; Figure 8A). GLXI;2 showed a 3.5-fold increase in expression (corresponding to a log2-fold change [log2FC] of 1.84, P < 0.05; Figure 8A). Feeding seedlings with 60 mM sucrose for 4 d induced an increase in the expression of GLXI;2, with an increase of around 1.5-fold (P < 0.05, log2FC = 0.58; Figure 8A). Given the high expression level of GLXI;2 (Figure 3B), an increase in transcript abundance of 50% corresponds to a marked increase in absolute transcript numbers, which represents a biologically significant cellular event.
Figure 8.
Transcriptional Regulation of the GLXI Isoforms by Different Stresses.
(A) Expression levels of GLXI;1, GLXI;2, and GLXI;3 relative to the untreated control (set to 0) in whole 2-week-old Arabidopsis seedlings grown in the presence of 0.5 mM MGO or 60 mM sucrose.
(B) Expression levels of GLXI;1, GLXI;2, and GLXI;3 relative to the untreated control (set to 0) in 4-week-old Arabidopsis rosettes of wild-type and ADP-glucose pyrophosphorylase loss-of-function (adg1-1) plants after prolonged darkness (24 h dark). ACTIN2 was used as the endogenous reference. Values are means ± se (n = 3, biological replicates with three to four individual plants pooled, *P < 0.05, unpaired two-tailed Student’s t test).
(C) Relative expression levels of GLXI;1, GLXI;2, and GLXI;3 after sucrose treatment of Arabidopsis seedlings (array data from Gonzali et al. [2006]).
(D) Relative expression levels of GLXI;2 in Arabidopsis leaves after treatment with 100 mM sucrose for 16 h (*P < 0.001; array data from Müller et al. [2007]).
(E) Relative expression levels of GLXI;1, GLXI;2, and GLXI;3 in Col-0 leaves after 48 h of high light (48 h HL) exposure (*P < 0.05, Bonferroni adjusted; array data from Schmitz et al. [2014]). n.d., no data; FC, fold change.
Endogenous sugar levels are affected in different loss-of-function mutants of primary carbohydrate metabolism, i.e., those with mutations affecting starch synthesis. It is well established that loss of ADP-glucose pyrophosphorylase in Arabidopsis (adg1-1) leads to high levels of sucrose, glucose, and fructose during the day, which can reach levels nearly five times higher than in the wild type (Schmitz et al., 2012; Matsoukas et al., 2013). When comparing GLXI expression levels in adg1-1 and the wild type, we again observed a significant 1.5-fold increase in GLXI;2 transcript abundance (P < 0.05, log2FC = 0.63; Figure 8B). To further analyze this point, we exposed Arabidopsis wild-type plants to prolonged darkness (24 h) to achieve a depletion of sugars and measured GLXI gene expression in these plants in comparison to that at midday in plants grown under a normal day-night cycle. We observed a significant decrease in the expression of all GLXI genes in leaves after prolonged darkness (P < 0.05; Figure 8B); in particular, GLXI;2 transcript abundance was strongly reduced (11.3-fold; log2FC = −3.5; Figure 8B). These results point to a clear link between the regulation of GLXI gene expression, especially GLXI;2, and cellular sugar status.
We further analyzed the expression of the GLXI genes in several independent microarray studies aimed at understanding sugar-controlled gene expression. Sugar feeding experiments of young Arabidopsis seedlings (Gonzali et al., 2006) revealed a statistically significant 5.3-fold induction of GXLI;2 expression versus the control (P < 0.05, log2FC = 2.40; Figure 8C). Furthermore, feeding 6-week-old Arabidopsis plants with 100 mM sucrose (Müller et al., 2007) resulted in a significant (3-fold) increase in GLXI;2 transcript levels compared with the control (log2FC = 1.62; Figure 8D). In a recent study of the acclimation response of Arabidopsis toward high light in the context of the cellular sugar status (Schmitz et al., 2014), analysis of transcription in leaves after transfer to high light (Figure 8E) showed an increase of 1.7-fold for GLXI;1 (P < 0.05; log2FC = 0.77) and 2.1-fold for GLXI;2 (P < 0.05; log2FC = 1.05). These independent studies with Arabidopsis plants at different developmental stages (Figures 8A to 8E) support our qPCR data showing that the transcription of GLXI;2, and in some cases GLXI;1, is regulated by cellular sugar status.
Elimination of Toxic Reactive Carbonyl Species Produced by Enhanced Cellular Sugar Levels Is Highly Dependent on GLXI;3
To gain deeper insights into the physiological importance of the first step of MGO and GO detoxification, we isolated T-DNA insertion lines of all GLXI homologs. Two independent insertion lines of GLXI;1 were identified via PCR. The T-DNA insertion is 113 bp upstream of the start codon in glxI;1-1 and 975 bp downstream of the start codon in glxI;1-2 (Supplemental Figure 6A). Line glxI;1-2 represents a true loss-of-function mutant, as GLXI;1_1 transcripts were not detected in these plants (Supplemental Figure 6B). For the GLXI;2 gene locus, we isolated the T-DNA insertion line glxI;2; this line contains an insertion 112 bp upstream of the start codon of GLXI;2_1 and has a GLXI;2 transcript level of 12 to 20% compared with the wild type (Supplemental Figures 6C and 6D). We isolated two independent T-DNA insertion lines of GLXI;3, glxI;3-2 and glxI;3-4, with insertions located 924 and 172 bp downstream of the start codon of GLXI;3_1, respectively. GLXI;3 transcripts were not detected in these lines (Supplemental Figures 6E and 6F). Thus, glxI;3-2 and glxI;3-4 represent mutant lines lacking GLXI;3 in both the cytosol and chloroplasts, as none of the GLXI;3_1 and GLXI;3_4 isoforms can be produced in these lines. To eliminate the Ni2+-dependent GLXI;1 and GLXI;2 activities, we generated the glxI;1-2 glxI;2 double mutant by performing genetic crosses of the single mutants.
All mutant lines showed no apparent phenotype when grown under greenhouse conditions (Supplemental Figure 7). As we observed modulation in the expression of GLXI genes in response to changes in the sugar metabolic status of the plants (Figure 8), and as enhanced cellular levels of MGO due to sugar supplementation were already observed in other systems (Padayatti et al., 2001; Miller et al., 2006), we analyzed the influence of sucrose as well as MGO and GO feeding on germination and seedling establishment in the mutant lines.
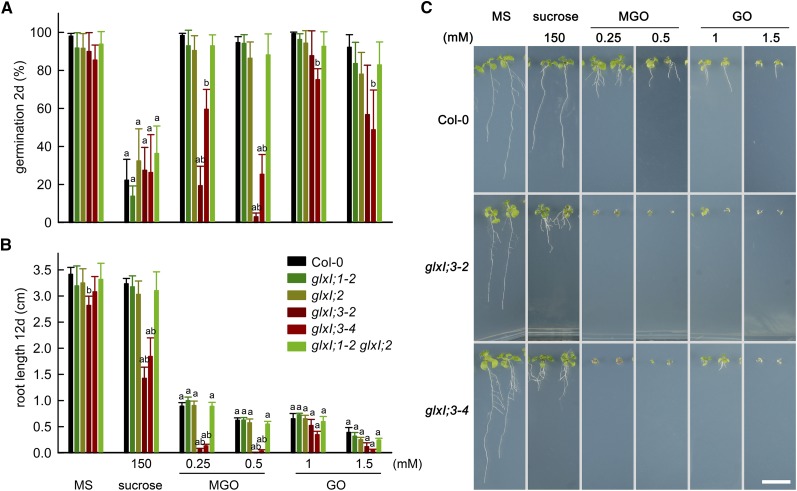
As an indicator of germination, we used the emergence of the radicle, i.e., radicle protrusion; we quantified germination at 2 d after seed imbibition (germination in %; Figure 9A). This analysis indicated that the germination capacity of glxI;3-2 and glxI;3-4 was highly reduced by the presence of MGO in the growth medium and that this response was dose dependent (Figure 9A). The presence of GO in the growth medium moderately reduced the germination capacity of glxI;3 mutants in a dose-dependent manner.
Figure 9.
Effect of Sucrose, MGO, and GO on the Germination and Seedling Establishment of Arabidopsis GLXI T-DNA Insertion Lines.
(A) Germination (in percentage of total seeds sown) at 2 d after seed imbibition. The emergence of the radicle was used as an indicator of germination. Values are means ± se (n = 4) from independent experimental trials with 50 to 100 counted seeds for each genotype. Testing for significant differences was performed by two-way ANOVA on arcsine transformed data (treatment, P < 0.0001; genotype, P < 0.0001; interaction, P = 0.0005).
(B) Root length after 12 d of germination, values are means ± se (n = 4) from independent experimental trials with 7 to 10 root measurements each. Testing for significant differences was performed by two-way ANOVA (treatment, P < 0.0001; genotype, P < 0.0001; interaction, P < 0.0001) (Supplemental File 2). Tukey’s multiple comparison test was used, where “a” indicates significant differences in treatments versus MS control and “b” indicates significant differences in genotypes versus Col-0 control.
(C) Photographs of Col-0 and glxI;3 loss of function lines after 12 d of germination in MS medium and in the presence of sucrose, MGO, and GO. Bar = 1 cm.
To quantify seedling establishment and postgerminative growth, we measured the root length in plants 12 d after germination. We found that the root lengths of glxI;3-2 and glxI;3-4 were slightly reduced (15% and 10%) compared with the wild type in MS medium. Sucrose in the growth medium did not affect postgerminative growth of Arabidopsis wild type, glxI;1, glxI;2, or the glxI;1 glxI;2 double mutant (Figure 9B; Supplemental Figure 8). However, sucrose highly affected root growth in glxI;3-2 and glxI;3-4, with reductions in root length of 45 to 55% compared with the wild type (Figure 9B). The addition of MGO to the medium led to a similar reduction in root length in the wild type, glxI;1, glxI;2, and glxI;1 glxI;2. In the case of glxI;3-2 and glxI;3-4, the presence of MGO in the medium resulted in developmental arrest during seedling establishment (Figure 9C), as also indicated by arrested root development (Figure 9B). The effects of GO on the glxI;3 mutants were also dose dependent but not as pronounced as for MGO.
Our results indicate that the Mn2+-dependent enzyme GLXI;3 is the major enzyme involved in the detoxification of MGO and GO in vivo, where its activity is specifically required for defense against sugar-derived reactive carbonyl species during seedling establishment.
DISCUSSION
The GLX system is highly conserved in nearly all domains of life and is thought to be the major pathway for the detoxification of MGO and other reactive carbonyl species such as GO (Thornalley, 1990; Ponces Freire et al., 2003). Alternative pathways for MGO detoxification have been reported in prokaryotic organisms and in human cells under pathological conditions (Supplemental Figure 9, 1–4) (Sridhara and Wu, 1969; Ray and Ray, 1982; Xu et al., 2006; Baba et al., 2009). In addition, the existence of a glutathione-independent GLX (GLXIII) that might convert MGO directly into d-lactate has been reported for plants (Supplemental Figure 9, 4) (Misra et al., 1995; Kwon et al., 2013; Ghosh et al., 2016). However, E. coli GLXIII (DJ1-d/HSP31) was recently shown to be a protein glycase, which prevents the accumulation of already formed AGEs (Mihoub et al., 2015; Richarme et al., 2015). This would make the GLX system the first level of glycation defense, followed by deglycation of proteins by GLXIII as the second level of defense.
Here, comprehensive analysis of the biochemical properties, subcellular localization, and transcriptional regulation of the GLX isoforms resulting from alternative splicing in Arabidopsis enabled us to provide a cellular model for the detoxification of MGO and GO through the GLX system. In addition, the analysis of plants lacking functional GLXI proteins allowed us to identify the physiologically relevant isoforms involved in germination and postgerminative development in Arabidopsis.
Alternative Splicing of GLXI Modifies the Subcellular Localization of the Derived Isoforms
Alternative mRNA splicing is thought to ensure transcriptome plasticity and proteome diversity in eukaryotes. A high proportion of alternative splicing events occurring at the first exon can alter the N-terminal regions of proteins, especially in Arabidopsis and rice (Oryza sativa) (Chen et al., 2007). By combining expression analysis with fluorescent reporter protein studies, we identified the subcellular localization of the predominant splice forms of the GLX system in Arabidopsis and found that alternative splicing of GLXI modifies the subcellular localization of the derived isoforms. GLXI;1 produces a unique GLXI;1 isoform that localizes to the chloroplast. Alternative splicing of GLXI;2 produces a transcriptionally predominant GLXI;2_1 isoform that localizes to the cytosol and a minor isoform (GLXI;2_6) that localizes to the endoplasmic reticulum (Figure 2; Supplemental Figure 4B). Alternative splicing of GLXI;3 renders mature proteins with identical sequences but with altered subcellular localizations: GLXI;3_1 localizes to the cytosol, while GLXI;3_4 localizes to chloroplasts (Figures 2 and 6).
GLXII;2 produces a single isoform located to the cytosol. We found that the GLXII;4 and GLXII;5 isoforms are targeted to both the mitochondria and chloroplasts (Figures 2 and 7; Supplemental Figure 5). Although the prediction scores for the subcellular localization of the two splice forms of GLXII;4 differ slightly (Figure 2B), the addition of one amino acid does not alter the localization of the isoforms in vivo (Figure 7; Supplemental Figure 5).
Alternative splicing can be regulated in a development-specific and tissue-/cell-specific manner, especially in conjunction with environmental stress (Palusa et al., 2007; Mazzucotelli et al., 2008). We speculate that the levels of the isoforms of the GLX system located in different subcellular compartments would be adjusted when plants are challenged by specific environmental pressures or metabolic imbalances. Apart from the degradation of triose phosphates, reactive carbonyl species can be produced by lipid peroxidation, fragmentation of glycated proteins, degradation of glucose, and oxidation of some amino acids. As this may occur in different cellular compartments, the existence of GLXI isoforms in organelles other than the cytosol and chloroplasts may be physiologically relevant, even if they are minor isoforms. This could be the case for GLXI;2_6, which is expressed at low levels and localizes to the endoplasmic reticulum.
The predicted minor isoform GLXI;2_5 lacks a region at the C terminus, leading to the formation of a putative noncanonical PTS1 signal (RLL; Reumann et al., 2009). However, in line with the extremely low levels of expression determined by RNA-seq, we never obtained a full-length transcript of GLXI;2_5 using leaf or root cDNA (Figure 3). Domain and 3D model predictions via Phyre2 (Kelley et al., 2015) indicated that the loss of 51 amino acids at the C terminus of GLXI;2_5 hampers the folding of at least one of the two domains that make up the protein, as only five out of eight beta-sheets can be formed. It can thus be assumed that GLXI;2_5 does not render any physiologically relevant GLXI in the conditions tested. However, we cannot rule out the possibility that the protein produced by this splice form becomes relevant under other specific conditions.
Finally, a recent study reported that a Zn2+-dependent GLXI homolog from rice (OsGLXI;8) localizes to the nucleus (Kaur et al., 2017). The Arabidopsis homolog of OsGLXI;8 is GLXI;3_4, which was also shown to be localized to the nucleus in the same study. However, in line with subcellular localization predictions that indicate with high probability that GLXI;3_4 carries a chloroplastic signal peptide (Figure 2B), our YFP fusion analysis clearly shows a chloroplastic localization of this isoform (Figure 6A). It cannot be completely ruled out that GLXI;3_4 also localizes to the nucleus under certain conditions; however, the nuclear localization of GLXI;3_4 was deduced based on experiments using chloroplast-free onion epidermis cells (Kaur et al., 2017).
The complex subcellular distribution and expression patterns of the proteins of the GLX system emphasize the importance of identifying and characterizing the predominant splice forms that occur in specific plant organs and at specific developmental stages.
Arabidopsis GLXI;3 Prefers Mn2+ as Its Cofactor and Plays a Prevalent Role in Methylglyoxal Detoxification
Based on sequence homologies, GLXI;1 and GLXI;2 are Ni2+-dependent GLXI isoforms, while GLXI;3 is a Zn2+-dependent isoforms. All three isoforms show activity with MGO-GSH as substrate. Accordingly, GLXI activities using other bivalent cations have rarely been tested, and most of the assays were confined to the use of MGO as substrate. In this study, we comparatively analyzed the properties of the GLXI isoforms using the most abundant reactive carbonyl species, MGO-GSH and GO-GSH. We found that while GLXI;3 strongly prefers Mn2+ in the conversion of MGO-GSH and GO-GSH, GLXI;1 and GLXI;2 prefer Ni2+ in the conversion of MGO-GSH and Mn2+ in the conversion of GO-GSH. Thus, GLXI;1 and GLXI;2 possess the specific feature of switching substrate specificity depending on the metal ion cofactor utilized. Variations in substrate preference depending on the metal cofactor used is a phenomenon with important implications in nature, as it may represent a way by which enzymes serve multiple functions in vivo (Lukačin et al., 2004). That the enzymes may have the capacity to switch cofactors makes it possible that the substrate spectrum of the GLXI isoforms might be even broader than previously considered.
Our kinetic studies indicated that the Arabidopsis GLXI isoforms have similar affinities toward MGO-GSH and GO-GSH, with comparable values described for GLXI isoforms of other organisms. The Km values of the Arabidopsis isoforms lie between 279 and 476 µM (Table 1), while the Km values of homologs from diverse animal sources lie between 130 and 500 µM and that of yeast is 530 µM (Thornalley, 1990; Ponces Freire et al., 2003). Nevertheless, the catalytic efficiencies for MGO-GSH and GO-GSH vary among isoforms; GLXI;1 uses MGO-GSH with a 3-fold higher catalytic efficiency than GO-GSH, while GLXI;3 uses MGO-GSH with a 7-fold higher catalytic efficiency, indicating that the isoforms exhibit different behaviors toward their substrates.
Our kinetic analysis also suggested that an excess of free GSH in the reaction medium negatively affects GLXI;3 activity. This phenomenon may occur only in vitro, but we cannot rule out that it could be relevant in vivo, given that the average cellular GSH concentration (200 nmol gFW−1; Krueger et al., 2009; Noctor et al., 2011) normally exceeds that of free MGO (0.3–3 nmol gFW−1; Chen and Thelen, 2010; Rabbani and Thornalley, 2014) by roughly 100-fold in plant tissues. Moreover, competitive inhibition of a yeast GLXI by high free GSH levels has been reported (Vander Jagt et al., 1972; Marmstal and Mannervik, 1981). This suggests that Arabidopsis GLXI;3 possesses a particular molecular property shared with the yeast homolog. Modulation of GLXI;3 activity by GSH in vivo would represent a way to regulate its enzymatic activity in response to changes in the (sub-)cellular concentrations of GSH.
The analysis performed with the heterologously expressed GLXI proteins indicated that all isoforms are active in vitro. Evidence that all isoforms are functional in vivo was obtained from the yeast complementation assay and the analysis of Arabidopsis loss-of-function mutants of the GLXI homologs. All of these studies also indicated that GLXI;3 is the physiologically relevant isoform involved in MGO detoxification. The yeast knockout strain of GLXI that expresses AtGLXI;3 showed full recovery of growth in medium containing MGO or GO (Figure 5). In addition, independent Arabidopsis loss-of-function GLXI;3 mutants were highly sensitive to the presence of MGO, GO, and sucrose during germination and postgerminative development (Figure 9; Supplemental Figure 8). This major involvement of GLXI;3 in MGO and GO detoxification is in line with the obtained kinetic data; GLXI;3 possesses very high catalytic efficiencies for the conversion of MGO-GSH and GO-GSH (Table 1). In this way, the presence of GLXI;3 guarantees the rapid removal of MGO and GO that accumulate in the cytosol and chloroplasts.
The Glyoxalase System Emerges as an Important Protective Mechanism against Reactive Carbonyl Species Derived from Enhanced Cellular Sugar Levels
Stresses that influence plant energy consumption or carbon fixation fluxes induce changes in the cellular levels of phosphorylated and soluble sugars; in turn, these are likely to enhance the formation of MGO and other reactive carbonyl species. Consequently, transcriptional regulation of the GLX system might be coupled with changes in the cellular levels of soluble sugars. In line with this idea, we found a significant transcriptional induction of GLXI;2 upon exposure of wild-type Arabidopsis seedlings to exogenously applied MGO and sucrose, as well as to increased endogenous sugar levels in the starch-free adg1-1 mutant. Moreover, gene expression of all GLXI isoforms was diminished when sugars were depleted during a prolonged dark phase (Figures 8A and 8B). All of these results are in line with the results of independent transcriptomic studies, which found that GLXI;2 is responsive to plant sugar status (Figures 8C and 8D) (Gonzali et al., 2006; Müller et al., 2007).
Furthermore, moderately high light exposure significantly increased the transcript abundance of GLXI;1 and GLXI;2 (Figure 8E) (Schmitz et al., 2014). Higher light intensities can (to some extent) increase CO2 fixation and the flux through the Calvin Benson cycle and consequently the export of triose phosphates to the cytosol; reactive carbonyl species formation is enhanced under these conditions (Takagi et al., 2014). Detoxification of MGO produced under high light would thus primarily involve the chloroplastic GLXI;1 and the cytosolic GLXI;2_1 isoforms. The involvement of these isoforms in either chloroplastic or cytosolic MGO detoxification is also supported by the transcript abundances in distinct plant organs. Chloroplastic GLXI;1 is highly expressed in green leaves and moderately expressed in heterotrophic tissues such as roots, while the cytosolic GLXI;2_1 isoform is highly expressed in heterotrophic tissues (Figures 3A and 3B). Interestingly, GLXI;3 showed no significant changes in transcript abundance, except under prolonged darkness.
GLXI expression has been reported to be responsive to different abiotic stresses. Using a bioinformatic approach, proteins showing different grades of homology to GLXI in Arabidopsis (Mustafiz et al., 2011) and soybean (Glycine max; Ghosh and Islam, 2016) were found to respond to abiotic stress conditions such as salt, cold, and drought. However, no biochemical data exist that support the GLXI enzymatic activity of these GLXI-like proteins. According to Kaur et al. (2013), only 13 of 24 soybean GLXI-like proteins identified by Ghosh and Islam and only three of 11 Arabidopsis GLXI-like proteins identified by Mustafiz et al. have significant homology with GLXI proteins with confirmed GLXI activity. In line with our results showing that GLXI;1 and GLXI;2 are transcriptionally regulated by the sugar level, Mustafiz et al. (2011) showed that the expression of GLXI;1 and GLXI;2 is moderately induced upon stress treatment, whereas GLXI;3 shows no response. As the abiotic stresses cold, drought, and salt are well known to also induce osmotic stress, it is possible that the production of soluble sugars as osmolytes under these stress conditions is responsible for the changes in expression observed for GLXI;1 and GLXI;2. In favor of this hypothesis, it was recently reported that upon the exposure of roots to osmotic stress, plants degrade starch during the day to increase sugar export to the root to regulate osmotic adjustment and growth (Thalmann et al., 2016).
In feeding experiments, it is assumed that roots would be affected first if the supplemented compound is toxic to germinating seedlings. Our feeding experiments using mutants of the different GLXI isoforms demonstrated that glxI;3 loss-of-function lines, lacking both the cytosolic and chloroplastic GLXI;3 isoforms, are severely affected in seedling establishment and root growth in response to reactive carbonyl species. Since reactive carbonyl species are highly toxic compounds, they should be rapidly eliminated after being taken up or produced by sucrose metabolism in the root. In this system, the detoxification in the roots takes place predominantly in the cytosol, indicating that the cytosolic GLXI;3_1 isoform plays a predominant role in the detoxification of MGO and GO during germination and postgerminative development. This important physiological function in heterotrophic tissues is highlighted by the observation that the glxI;3 loss-of-function lines already showed slightly reduced root growth in seedlings grown in MS control medium. The activity of the chloroplastic GLXI;3_4 and GLXI;1 would primarily be required in photosynthetic tissues, as reactive carbonyl species formation in this organelle reflects the flux through the Calvin Benson cycle.
The lack of increased sensitivity toward MGO and GO observed in the glxI;1, glxI;2 and glxI;1 glxI;2 mutants might be due to the 10 to 20% remaining transcript level in the glxI;2 insertion line and the overlapping high enzymatic activity of GLXI;3. However, based on our finding that the Ni2+-dependent GLXI;1 and GLXI;2 are able to switch substrate specificities depending on the ion cofactor and based on their conserved co-occurrence with the Mn2+-dependent GLXI;3 (Kaur et al., 2013), we hypothesize that in plants, the Ni2+-dependent GLXI isoforms may cover a different substrate spectrum from the Mn2+-dependent GLXI;3 isoforms. It is possible that during germination and seedling establishment, the GLXI;1 and GLXI;2 activities might be primarily involved in the detoxification of reactive carbonyl species other than MGO and GO.
Our results demonstrate that the cytosolic Mn2+-dependent GLXI;3 is the major isoform involved in sugar-derived reactive carbonyl species detoxification during germination and seedling establishment.
Defense against Reactive Carbonyl Species Involves at Least Three Subcellular Compartments
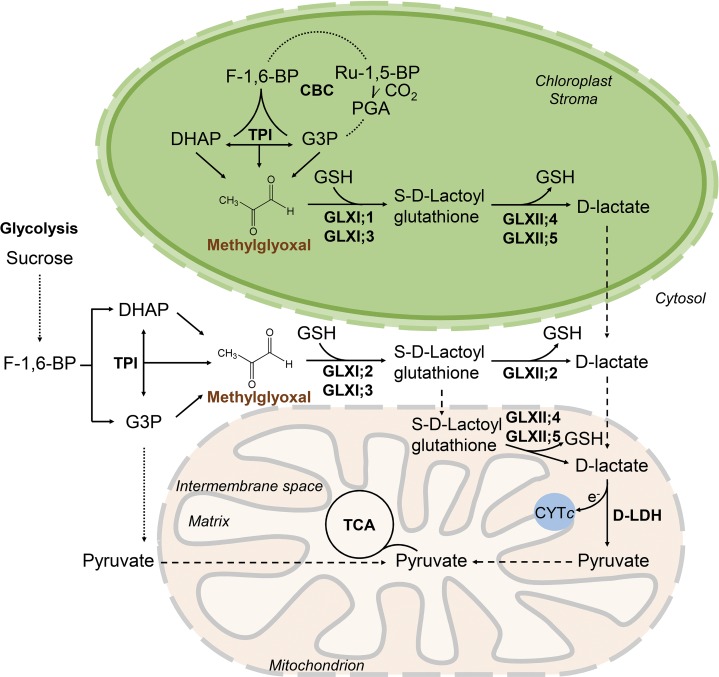
The primary source of MGO is the flux through the glycolytic pathway due to the action of triose phosphate isomerase and the occurrence of the key metabolites DHAP and G3P. However, as a unique feature of plant cells, the flux through the Calvin Benson cycle results in a second source of MGO. The steady state cellular concentrations of DHAP and G3P in Arabidopsis rosette leaves average 50 nmol gFW−1, whereas that of glucose exceeds 1 µmol gFW−1 under physiological conditions (Arrivault et al., 2009). As glycolysis and the Calvin Benson cycle are functional in the cytosol and chloroplasts, these cellular compartments require the existence of an active and direct detoxification pathway under normal physiological conditions. Based on our findings, we propose the following cellular model for the production and detoxification of MGO in Arabidopsis (Figure 10). In the cytosol, depending on the tissue, developmental stage, and cellular needs, GLXI;2 and/or GLXI;3 convert MGO and GSH into S-d-lactoylglutathione. This is further converted to d-lactate by cytosolic GLXII;2 or transported to the mitochondria, where GLXII;4 and/or GLXII;5 convert it to d-lactate. In the chloroplasts, GLXI;1 and/or GLXI;3 convert MGO and GSH into S-d-lactoylglutathione, which is further converted to d-lactate by chloroplastic GLXII;4 and/or GLXII;5. d-lactate is the substrate of mitochondrial d-lactate dehydrogenase (Welchen et al., 2016). The pyruvate produced in this reaction is fed into the tricarboxylic acid cycle, and the electrons are donated to the ETC through CYTc. Following a similar metabolic pathway, GO would be converted to glycolate, which could be transported to peroxisomes for further metabolization (Maurino and Engqvist, 2015).
Figure 10.
Proposed Production and Detoxification Pathways of MGO within a Plant Cell.
The fluxes through glycolysis and the Calvin Benson cycle (CBC) determine the rate at which MGO molecules are formed in the cytosol and chloroplast, respectively. Our results indicate that in the cytosol, GLXI;2 and/or GLXI;3 convert MGO and GSH into S-d-lactoylglutathione. This can be further converted to d-lactate by cytosolic GLXII;2 or transported to the mitochondria, where it is converted to d-lactate by GLXII;4 and/or GLXII;5. In the chloroplasts, GLXI;1 and/or GLXI;3 convert MGO and GSH into S-d-lactoylglutathione, which is further converted to d-lactate by chloroplastic GLXII;4 and/or GLXII;5. In the mitochondria, d-lactate is finally converted by d-lactate dehydrogenase (D-LDH) to pyruvate, which is fed into the tricarboxylic acid cycle (TCA). TPI, triose phosphate isomerase; PGA, 3-phosphoglycerate; F-1,6-BP, fructose-1,6-bisphosphate. Dashed arrows indicate possible transport; dotted arrows indicate summarized reactions.
This model implicates the existence of several transport processes in Arabidopsis. Both the absence of GLXI and the existence of GLXII activity in mitochondria indicate that S-d-lactoylglutathione should be (in part) transported to this organelle. Moreover, the exclusive localization of d-lactate dehydrogenase to the mitochondrial intermembrane space (Welchen et al., 2016) points not only to the necessity of a transport system for d-lactate out of the chloroplast and into the mitochondria, but also to the localization of the GLXII isoforms to the mitochondrial intermembrane space. In this scenario, the second step of the GLX system would release reduced glutathione into the intermembrane space; consequently, GLXII activity would not be the primary source of glutathione in the mitochondrial matrix, as has been speculated previously (Sciré et al., 2000).
Our results demonstrate that both the cytosol and chloroplast harbor a functional GLX system, and they suggest that the individual components play specialized roles in specific plant developmental stages and metabolic conditions induced by cellular and environmental changes. The involvement of different organelles during the detoxification of MGO implies that the corresponding enzymatic and transport activities between the cellular compartments are synchronized.
METHODS
Plant Lines and Growth Conditions
The plant lines used in this work include wild-type Arabidopsis thaliana plants, ecotype Columbia-0 (Col-0; wild type), ADP-glucose pyrophosphorylase loss-of-function mutant (adg1-1; Lin et al., 1988), and the T-DNA insertion lines glxI;1-1 (Salk-142650), glxI;1-2 (Sail-706_G03), glxI;2 (Salk-103699), glxI;3-2 (GK-239E11.02), and glxI;3-4 (Salk-131547). The T-DNA insertion lines were obtained from the Nottingham Arabidopsis Stock Centre (http://arabidopsis.info/). All T-DNA insertion lines were screened via PCR with specific genomic and T-DNA left border primers (Supplemental Table 1), and homozygous plants were isolated in all cases. Transcript abundance was measured via RT-PCR using specific primers for each isoform (Supplemental Table 1). In the case of glxI;2-1, the remaining transcript levels were quantified based on band intensity measurements relative to the wild type using Fiji (Schindelin et al., 2012). All plant lines were grown in soil at 22°C under a long-day (16 h light/8 h darkness) photoperiod using a mix of bulbs from Spectralux Plus NL 36W/840 (Radium) and Fluora L 36W/77 (Osram) with a light intensity of ∼100 µE m−2 s−1. Growth in solid sterile culture was conducted on 0.5× MS medium solidified with 0.9% plant agar (Duchefa; Murashige and Skoog, 1962) under the conditions described above. Seeds were stratified for 3 d at 4°C before being transferred to the conditions described above. Germination and root length measurements of wild-type and mutant plants were performed using sterile 0.5× MS medium supplemented with 0.25 mM MGO, 0.5 mM MGO, 1 mM GO, 1.5 mM GO, or 150 mM sucrose, with two independent batches of seeds. For stress-related transcriptional analysis, wild-type seeds were germinated on 0.5× MS medium. After 10 d, the seedlings were transferred to 0.5× MS medium containing 1 mM MGO or 60 mM sucrose and harvested after 4 d of exposure, where three to four seedlings were pooled for one biological replicate. For the transcriptional analysis, wild-type and adg1-1 plants were grown on soil for 21 d, and whole rosettes were harvested in the middle of the light phase of a 16/8-h photoperiod or after 24 h prolonged darkness, where three to four plant rosettes were pooled for one biological replicate.
RNA Isolation, Reverse Transcription, and PCR Analysis
Total RNA from wild-type or adg1-1 tissues was isolated according to Logemann et al. (1987) and controlled for quality via gel electrophoresis. After the removal of possible genomic DNA contaminations with an Ambion DNA-free DNA removal kit, 2 µg RNA was transcribed into the cDNA (cDNA) with RevertAid reverse transcriptase (Thermo Scientific) using an oligo(dT) primer according to the manufacturer’s instructions. The existence of the GLXI splice forms was analyzed by RT-PCR using the cDNA and specific primers designed to bridge the exon-intron junctions (Supplemental Table 1). cDNA levels were normalized to ACTIN2 as an endogenous control.
Transcript analysis via qPCR was performed with KAPA SYBR FAST qPCR Master Mix (Kapa Biosystems) and 2 μL of 1:10 diluted cDNA in an Applied Biosystems Step One Plus real-time PCR system according to the manufacturer’s instructions. The reference gene ACTIN2 and some primers were chosen from Czechowski et al. (2005); other primers were designed using Primer-Blast (Ye et al., 2012) (Supplemental Table 1). Differential GLXI gene expression was analyzed and expressed using the ΔΔCT method (Schmittgen and Livak, 2008) and tested for significant differences with the Student’s t test.
cDNA Isolation and Cloning into Destination Vectors
The coding sequences of the verified splice forms of the GLXI and GLXII homologs were amplified from Arabidopsis Col-0 cDNA using Phusion Polymerase (Thermo Scientific) and specific primers (Supplemental Table 1). The amplified fragments were subcloned into pCR-TOPO-Blunt (Life Technologies). The generated TOPO vectors were used as templates for further PCR-dependent fragment constructions according to Gibson et al. (2009). The primers for Gibson cloning were designed with a 20-bp overlap with the destination vector.
The vector pET16b-His was used for heterologous expression in Escherichia coli. Purified Gibson-cloning PCR fragments of GLXI;1_1 or GLXI;2_1 were ligated into the pET16b vector linearized with BamHI. As an alternative for the His-tag-based purification, the modified vector pTacStrep was used to express GLXI;3_1 under the control of the tac-promoter. The pTacStrep was linearized with XhoI to assemble the PCR-amplified GLXI;3_1 CDS fragments in-frame with the N-terminal Streptavidin-tag according to Gibson et al. (2009).
The vectors pNL3 and pNL25 (Linka et al., 2008) were used to construct the C- and N-terminal eYFP-fusions. Vectors linearized with BamHI were used in a Gibson assembly reaction with the amplified GLXI and GLXII PCR fragments using specifically designed primers (Supplemental Table 1) to yield in-frame eYFP fusions, whose expression was under the control of the constitutive CaMV 35S promoter. The vector pUBQ10_Venus was used for alternative expression of GLXII:YFP fusion proteins with dual targeting sites (Kunz et al., 2014). Full GLXII;4_1:YFP, GLXII;5_1:YFP, and GLXII;5_2:YFP constructs were amplified using pNL3-35S-GLXII:eYFP templates with specific primer pairs; these were then cloned via Gibson assembly in SacI/XhoI-linearized pUBQ10 vector.
The vector pDR195 (Rentsch et al., 1995) was used for the expression of the GLXI homologs in Saccharomyces cerevisiae. GLXI;1_1, GLXI;2_1, or GLXI;3_4 PCR fragments were ligated into BamHI-linearized pDR195 vector expressed under the control of the constitutive ScPMAI promoter (yeast plasma membrane ATPase promoter).
Heterologous Protein Expression and Purification and SDS-PAGE
For heterologous protein expression, pET16b-GLXI;1, pET16b-GLXI;2, and pTacStrep-GLXI;3 were transformed in E. coli BL21 Rosetta pLysS strain (Novagen). The bacterial cultures were grown overnight at 37°C in medium containing 34 µg/mL chloramphenicol and 50 µg/mL ampicillin up to an optical density of 0.5 at 600 nm. Protein expression was induced by the addition of 1 mM isopropyl β-d-thiogalactopyranoside for 4 h. Bacteria were harvested by centrifugation at 4000g for 15 min at 4°C and stored at −20°C until purification. His-tag-based protein purification of GLXI;1 and GLXI;2 was performed as described (Esser et al., 2014), except that DTT was kept out of all buffers used. Strep-tag-based protein purification of GLXI;3 was conducted according to the manufacturer’s instructions (IBA Life Science). The success of protein purification was controlled via Coomassie staining after SDS-PAGE (Laemmli, 1970). Protein concentration was determined using the amido black precipitation method (Schaffner and Weissmann, 1973). The activity of purified GLXI;1 and GLXI;1 decreases significantly over time. We thus performed all biochemical assays on freshly purified proteins. Only GLXI;3 was stable for at least 10 d when stored in 30% glycerol at −20°C. As GLXI;3 was dialyzed overnight, the biochemical assays were conducted the day after isolation.
Measurement of Enzymatic Activities and Determination of Kinetic Constants
GLXI activities were monitored spectrophotometrically with a Synergy HT (Biotek) in 96 well UV-Star Microplates (Greiner Bio-one). The standard assay mixture for metal activation profile was buffered with 100 mM MOPS containing 0.5 mM of metal ion, 3.5 mM MGO or GO, and 1.7 mM GSH. The conversion of MGO or GO by GLXI is dependent on the spontaneous reaction of the reactive carbonyl species with GSH, forming a hemithioacetal (HTA), which is the natural substrate of GLXI. The HTA-forming reaction was performed 30 min prior to the enzymatic assays, and a Kdiss of 3.0 mM was used to calculate the actual HTA concentration in dynamic equilibrium (Kdiss [3.0 mM] = GSHfreexMGOfree/HTA) in the mixture (Vander Jagt et al., 1972). For the determination of the kinetic constants, we varied MGO or GO at fixed GSH concentration or vice versa with the use of the determined preferred metal ion cofactor. Conversion of the HTA to S-glycolylglutathion (ε = 3.26 mM cm−1) or S-d-lactoylglutathione (ε = 3.1 mM cm−1) was monitored at 240 nm at 25°C (Marasinghe et al., 2005). The dependence of GLXI activity on pH was analyzed in a physiological pH range using MES (pH 5.0–6.0), MOPS (pH 6.5–7.5), and HEPES (pH 8–8.5). All measurements were performed in at least three independent experiments.
Complementation Studies in Yeast
Wild-type S. cerevisiae (BY4741) and the GLXI loss-of-function mutant (glo1Δ; Y00572) were obtained from Euroscarf (University Frankfurt, Germany). The vectors pDR195-GLXI;1_1, pDR195-GLXI;2_1, and pDR195-GLXI;3_4 were transformed into the yeast strains and selected for positive transformation on minimal medium according to standard protocols. For analysis of phenotype suppression, a fresh culture of positive clones and a corresponding wild type were grown in liquid minimal medium overnight at 30°C. The cultures were then diluted 100-fold in fresh minimal medium and grown to a cell density of 2·106 mL−1 calculated based on the optical density at 600 nm. After a serial dilution from 2·106 mL−1 to 2·103 mL−1, 5 μL of culture was spotted onto minimal medium containing 1 mM and 4 mM of GO or MGO. Growth was monitored after 48 h of incubation at 30°C.
Transient Expression in Tobacco Protoplasts and Confocal Laser Scanning Microscopy
The vectors pNL3-35S-GLXI;1_1:eYFP, pNL3-35S-GLXI;2_1:eYFP, pNL3-35S-GLXI;2_6:eYFP, pNL3-35S-GLXI;3_1:eYFP, pNL3-35S-GLXI;3_4:eYFP, pNL3-35S-GLXII;2:eYFP, pNL3-35S-GLXII;4_1:eYFP, pNL3-35S-GLXII;5_1:eYFP, pNL3-35S-GLXII;5_2:eYFP, pUBQ10-GLXII;4_1:eYFP, pUBQ10-GLXII;5_1:eYFP, and pUBQ10-GLXII;5_2:eYFP constructed for the expression of C-terminal fusion proteins as well as the vectors pNL25-35S-eYFP;GLXI:1_1, pNL25-35S-eYFP:GLXI;2_1, pNL25-35S-eYFP:GLXI;3_4, pNL25-35S-eYFP:GLXII;2, pNL25-35S-eYFP:GLXI;4_1, and pNL25-35S-eYFP:GLXI;5_1 constructed for the expression of N-terminal fusion proteins were transformed into Agrobacterium tumefaciens (strain GV3101 pMP90) and selected on YEB plates containing rifampicin (150 µg/mL), gentamycin (50 µg/mL), and spectinomycin (100 µg/mL) for the pNL vectors or kanamycin (50 µg/mL) for the pUBQ10 vectors.
Infiltration of Agrobacterium cultures into 5-week-old wild tobacco leaves (Nicotiana benthamiana) and protoplast isolation were performed according to Waadt and Kudla (2008). The vectors pNL3 and pNL25 carrying the GLXI or GLXII isoforms were transiently coexpressed with either the mitochondrial marker pIVD-eqFP611-pBI121 (Forner and Binder, 2007) or the peroxisomal marker pNL4-eCFP:PTS1 (Linka et al., 2008), respectively. The vector ER-cb (ER:CFP; Nelson et al., 2007) was used for the colocalization for GLXI;2_6:YFP. Fluorescence was monitored under an LSM 780 confocal laser scanning microscope (Zeiss). CFP fluorescence was excited at 458 nm and emission detected at 460 to 505 nm. YFP fluorescence was excited at 514 nm and emission detected at 517 to 560 nm. The mitochondrial marker eqFP611 was excited at 561 nm and emission was detected at 580 to 620 nm. Chlorophyll autofluorescence was detected at 640 to 720 nm.
Identification of GLXI and GLXII Splice Forms by RNA-Seq Data Analysis
An RNA-seq data set from different organs, containing 948,442,901 Illumina HiSeq 2000 single-end reads, was used in this study (Liu et al., 2012). The reads were aligned to the TAIR 10 release of the Arabidopsis transcriptome (http://www.arabidopsis.org/) using the RSEM software package specifically designed for quantifying gene and isoform abundances (Li and Dewey, 2011). To minimize incorrect read allocations during the mapping process, RSEM was run in “very-sensitive” mode (equivalent to Bowtie2 options: -D 20 -R 3 -N 0 -L 20 -i S,1,0.50), and the parameters “–no-discordant” and “–no-mixed” were passed to Bowtie2. Bowtie2 was chosen as the read mapping algorithm due to its faster computation speed than Bowtie1 and because gapped alignments are not supported (Langmead and Salzberg, 2012), further reducing the potential for misalignments around splice junctions that may negatively affect isoform transcript quantification. Gibbs sampling (Geman and Geman, 1984) (–calc-pme) was used to calculate posterior mean expression estimates within 95% confidence intervals (–calc-ci). Gibbs sampling was iterated 500 times (–gibbs-burnin 500). The RNA-seq data set used in this study was mapped separately to the TAIR 10 transcriptome and displayed as FPKM.
In Silico Analysis, Phylogenetic Tree Reconstruction, and 3D Modeling
The amino acid sequences of the GLXI and GLXII homologs were retrieved from the TAIR 10 release. Sequences were aligned with Guidance2 (Sela et al., 2015) using the MAFFT algorithm (Katoh et al., 2002). Subsequently, MEGA7 was used for molecular phylogenetic analysis (Kumar et al., 2016). We inferred the evolutionary history using maximum likelihood with the JTT substitution model (Jones et al., 1992), analyzing 1000 bootstrap replicates. The tree with the highest log likelihood was chosen for each figure.
Subcellular localization predictions of the members of the GLX system with reliable AraLocCon score (Schwacke et al., 2007) were obtained from Aramemnon (Schwacke et al., 2003). Protein structures for the GLXI predominant splice forms GLXI;1_1, GLXI;2_1, and GLXI;3_1 were predicted using Phyre2 (Kelley et al., 2015).
Accession Numbers
RNA sequencing data used in this article can be found in the NCBI’s Sequence Read Archive database under BioProject ID PRJNA168212. Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL libraries under the following accession numbers: GLXI;1 (At1g67280), GLXI;2 (At1g11840), GLXI;3 (At1g08110), DSI-1 VOC (At1g07645), GLXI-like;4 (At1g15380), GLXI-like;5 (At1g64185), GLXI-like;7 (At1g80160), GLXI-like;8 (At2g28420), GLXI-like;9 (At2g32090), GLXI-like;10 (At5g41650), GLXI-like;11 (At5g57040), HsGLXI (IAAV38791), EcGLXI (BAE76494), GLXII;1 (At2g43430), GLXII;2 (At3g10850), ETHE1/GLXII;3 (AT1G53580), GLXII;4 (At1g06130), GLXII;5 (At2g31350), and D-LDH (At5g06580).
Supplemental Data
Supplemental Figure 1. Overview of the phylogenetic relationships of Arabidopsis proteins with putative GLXI-like domains.
Supplemental Figure 2. 3D models of the Arabidopsis GLXI isoforms predicted by Phyre2.
Supplemental Figure 3. Purification of recombinant AtGLXI proteins
Supplemental Figure 4. Subcellular localization of YFP and GLXI;2_6:YFP in tobacco leaf protoplasts.
Supplemental Figure 5. Subcellular localization of GLXII;4_1:YFP, GLXII;5_1:YFP, and GLXII;5_2:YFP in tobacco leaf protoplasts.
Supplemental Figure 6. Arabidopsis GLXI T-DNA insertion lines.
Supplemental Figure 7. Arabidopsis GLXI T-DNA insertion lines grown under greenhouse conditions.
Supplemental Figure 8. Phenotypes of GLXI T-DNA mutants grown for 12 d on 0.5× MS medium and medium supplemented with sucrose, MGO, or GO.
Supplemental Figure 9. Putative enzymatic catabolic pathways for the detoxification of methylglyoxal in different organisms.
Supplemental Table 1. Overview and sequences of primer pairs used in this study.
Supplemental Data Set 1. Transcript abundances (FPKMs) of all Arabidopsis splice forms from organ-specific RNA sequencing.
Supplemental File 1. Alignments used to construct the phylogenetic trees shown in Figure 1 and Supplemental Figure 1.
Supplemental File 2. ANOVA tables.
Acknowledgments
We thank the Center for Advanced Imaging (Düsseldorf, Germany) for technical help with fluorescence microscopy, Stefan Binder (Ulm, Germany) for kindly providing the pIVD-mito:eq611 construct, Stephan Krueger (Cologne, Germany) for providing the adg1-1 mutant, and Jasmin Schlicht, Larissa Willing, and Vanessa Reichel-Deland for the help with the analysis of the T-DNA insertion lines. This work was supported by grants of the Deutsche Forschungsgemeinschaft (FOR 1186 and EXC 1028) to V.G.M.
AUTHOR CONTRIBUTIONS
V.G.M. conceived the project. J.S. and V.G.M. designed the experiments, supervised data analysis and interpretation, and wrote the manuscript. J.S., I.C.D., J.D.B., M.S., and M.H. performed the experiments and acquired the raw data. A.W.R. processed RNA-seq data.
References
- Aldini G., Vistoli G., Stefek M., Chondrogianni N., Grune T., Sereikaite J., Sadowska-Bartosz I., Bartosz G. (2013). Molecular strategies to prevent, inhibit, and degrade advanced glycoxidation and advanced lipoxidation end products. Free Radic. Res. 47 (suppl. 1): 93–137. [DOI] [PubMed] [Google Scholar]
- Arrivault S., Guenther M., Ivakov A., Feil R., Vosloh D., van Dongen J.T., Sulpice R., Stitt M. (2009). Use of reverse-phase liquid chromatography, linked to tandem mass spectrometry, to profile the Calvin cycle and other metabolic intermediates in Arabidopsis rosettes at different carbon dioxide concentrations. Plant J. 59: 826–839. [DOI] [PubMed] [Google Scholar]
- Baba S.P., Barski O.A., Ahmed Y., O’Toole T.E., Conklin D.J., Bhatnagar A., Srivastava S. (2009). Reductive metabolism of AGE precursors: a metabolic route for preventing AGE accumulation in cardiovascular tissue. Diabetes 58: 2486–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold U., Rabbani N., Mullineaux P.M., Thornalley P.J. (2009). Quantitative measurement of specific biomarkers for protein oxidation, nitration and glycation in Arabidopsis leaves. Plant J. 59: 661–671. [DOI] [PubMed] [Google Scholar]
- Berardini T.Z., Reiser L., Li D., Mezheritsky Y., Muller R., Strait E., Huala E. (2015). The Arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis 53: 474–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bito A., Haider M., Hadler I., Breitenbach M. (1997). Identification and phenotypic analysis of two glyoxalase II encoding genes from Saccharomyces cerevisiae, GLO2 and GLO4, and intracellular localization of the corresponding proteins. J. Biol. Chem. 272: 21509–21519. [DOI] [PubMed] [Google Scholar]
- Cameron A.D., Olin B., Ridderström M., Mannervik B., Jones T.A. (1997). Crystal structure of human glyoxalase I--evidence for gene duplication and 3D domain swapping. EMBO J. 16: 3386–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Thelen J.J. (2010). The plastid isoform of triose phosphate isomerase is required for the postgerminative transition from heterotrophic to autotrophic growth in Arabidopsis. Plant Cell 22: 77–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.H., Lv G., Lv C., Zeng C., Hu S. (2007). Systematic analysis of alternative first exons in plant genomes. BMC Plant Biol. 7: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clugston S.L., Yajima R., Honek J.F. (2004). Investigation of metal binding and activation of Escherichia coli glyoxalase I: kinetic, thermodynamic and mutagenesis studies. Biochem. J. 377: 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cricco J.A., Vila A.J. (1999). Class B β-lactamases: the importance of being metallic. Curr. Pharm. Des. 5: 915–927. [PubMed] [Google Scholar]
- Crowder M.W., Walsh T.R. (1999). Structure and function of metallo-β-lactamases. Recent Res. DeV. Antimicrob. Agents Chemother. 3: 105–132. [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devanathan S., Erban A., Perez-Torres R., Kopka J., Makaroff C.A. (2014). Arabidopsis thaliana glyoxalase 2-1 is required during abiotic stress but is not essential under normal plant growth. PLoS ONE 9: e95971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., Brunak S., von Heijne G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300: 1005–1016. [DOI] [PubMed] [Google Scholar]
- Engqvist M., Drincovich M.F., Flügge U.I., Maurino V.G. (2009). Two D-2-hydroxy-acid dehydrogenases in Arabidopsis thaliana with catalytic capacities to participate in the last reactions of the methylglyoxal and beta-oxidation pathways. J. Biol. Chem. 284: 25026–25037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser C., Kuhn A., Groth G., Lercher M.J., Maurino V.G. (2014). Plant and animal glycolate oxidases have a common eukaryotic ancestor and convergently duplicated to evolve long-chain 2-hydroxy acid oxidases. Mol. Biol. Evol. 31: 1089–1101. [DOI] [PubMed] [Google Scholar]
- Forner J., Binder S. (2007). The red fluorescent protein eqFP611: application in subcellular localization studies in higher plants. BMC Plant Biol. 7: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frickel E.M., Jemth P., Widersten M., Mannervik B. (2001). Yeast glyoxalase I is a monomeric enzyme with two active sites. J. Biol. Chem. 276: 1845–1849. [DOI] [PubMed] [Google Scholar]
- Geman S., Geman D. (1984). Stochastic relaxation, gibbs distributions, and the bayesian restoration of images. IEEE Trans. Pattern Anal. Mach. Intell. 6: 721–741. [DOI] [PubMed] [Google Scholar]
- Ghosh A., Islam T. (2016). Genome-wide analysis and expression profiling of glyoxalase gene families in soybean (Glycine max) indicate their development and abiotic stress specific response. BMC Plant Biol. 16: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A., Kushwaha H.R., Hasan M.R., Pareek A., Sopory S.K., Singla-Pareek S.L. (2016). Presence of unique glyoxalase III proteins in plants indicates the existence of shorter route for methylglyoxal detoxification. Sci. Rep. 6: 18358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D.G., Young L., Chuang R.-Y., Venter J.C., Hutchison C.A. III, Smith H.O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Goldin A., Beckman J.A., Schmidt A.M., Creager M.A. (2006). Advanced glycation end products: sparking the development of diabetic vascular injury. Circulation 114: 597–605. [DOI] [PubMed] [Google Scholar]
- Gonzali S., Loreti E., Solfanelli C., Novi G., Alpi A., Perata P. (2006). Identification of sugar-modulated genes and evidence for in vivo sugar sensing in Arabidopsis. J. Plant Res. 119: 115–123. [DOI] [PubMed] [Google Scholar]
- Hanson A.D., Henry C.S., Fiehn O., de Crécy-Lagard V. (2016). Metabolite damage and metabolite damage control in plants. Annu. Rev. Plant Biol. 67: 131–152. [DOI] [PubMed] [Google Scholar]
- Himo F., Siegbahn P.E.M. (2001). Catalytic mechanism of glyoxalase I: a theoretical study. J. Am. Chem. Soc. 123: 10280–10289. [DOI] [PubMed] [Google Scholar]
- Holdorf M.M., Owen H.A., Lieber S.R., Yuan L., Adams N., Dabney-Smith C., Makaroff C.A. (2012). Arabidopsis ETHE1 encodes a sulfur dioxygenase that is essential for embryo and endosperm development. Plant Physiol. 160: 226–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y., Kimura A. (1996). Identification of the structural gene for glyoxalase I from Saccharomyces cerevisiae. J. Biol. Chem. 271: 25958–25965. [PubMed] [Google Scholar]
- Ito J., Batth T.S., Petzold C.J., Redding-Johanson A.M., Mukhopadhyay A., Verboom R., Meyer E.H., Millar A.H., Heazlewood J.L. (2011). Analysis of the Arabidopsis cytosolic proteome highlights subcellular partitioning of central plant metabolism. J. Proteome Res. 10: 1571–1582. [DOI] [PubMed] [Google Scholar]
- Jones D.T., Taylor W.R., Thornton J.M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8: 275–282. [DOI] [PubMed] [Google Scholar]
- Kalapos M.P. (2008). The tandem of free radicals and methylglyoxal. Chem. Biol. Interact. 171: 251–271. [DOI] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K., Miyata T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur C., Vishnoi A., Ariyadasa T.U., Bhattacharya A., Singla-Pareek S.L., Sopory S.K. (2013). Episodes of horizontal gene-transfer and gene-fusion led to co-existence of different metal-ion specific glyoxalase I. Sci. Rep. 3: 3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur C., Tripathi A.K., Nutan K.K., Sharma S., Ghosh A., Tripathi J.K., Pareek A., Singla-Pareek S.L., Sopory S.K. (2017). A nuclear-localized rice glyoxalase I enzyme, OsGLYI-8, functions in the detoxification of methylglyoxal in the nucleus. Plant J. 89: 565–576. [DOI] [PubMed] [Google Scholar]
- Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10: 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger S., Niehl A., Lopez Martin M.C., Steinhauser D., Donath A., Hildebrandt T., Romero L.C., Hoefgen R., Gotor C., Hesse H. (2009). Analysis of cytosolic and plastidic serine acetyltransferase mutants and subcellular metabolite distributions suggests interplay of the cellular compartments for cysteine biosynthesis in Arabidopsis. Plant Cell Environ. 32: 349–367. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz H.H., Zamani-Nour S., Häusler R.E., Ludewig K., Schroeder J.I., Malinova I., Fettke J., Flügge U.I., Gierth M. (2014). Loss of cytosolic phosphoglucose isomerase affects carbohydrate metabolism in leaves and is essential for fertility of Arabidopsis. Plant Physiol. 166: 753–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon K., Choi D., Hyun J.K., Jung H.S., Baek K., Park C. (2013). Novel glyoxalases from Arabidopsis thaliana. FEBS J. 280: 3328–3339. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.T., Plump A., DeSimone C., Cerami A., Bucala R. (1995). A role for DNA mutations in diabetes-associated teratogenesis in transgenic embryos. Diabetes 44: 20–24. [DOI] [PubMed] [Google Scholar]
- Li B., Dewey C.N. (2011). RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limphong P., Nimako G., Thomas P.W., Fast W., Makaroff C.A., Crowder M.W. (2009). Arabidopsis thaliana mitochondrial glyoxalase 2-1 exhibits β-lactamase activity. Biochemistry 48: 8491–8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T.P., Caspar T., Somerville C.R., Preiss J. (1988). A starch deficient mutant of Arabidopsis thaliana with low ADPglucose pyrophosphorylase activity lacks one of the two subunits of the enzyme. Plant Physiol. 88: 1175–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linka N., Theodoulou F.L., Haslam R.P., Linka M., Napier J.A., Neuhaus H.E., Weber A.P. (2008). Peroxisomal ATP import is essential for seedling development in Arabidopsis thaliana. Plant Cell 20: 3241–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Jung C., Xu J., Wang H., Deng S., Bernad L., Arenas-Huertero C., Chua N.H. (2012). Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24: 4333–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann J., Schell J., Willmitzer L. (1987). Improved method for the isolation of RNA from plant tissues. Anal. Biochem. 163: 16–20. [DOI] [PubMed] [Google Scholar]
- Lukačin R., Matern U., Specker S., Vogt T. (2004). Cations modulate the substrate specificity of bifunctional class I O-methyltransferase from Ammi majus. FEBS Lett. 577: 367–370. [DOI] [PubMed] [Google Scholar]
- Maiti M.K., Krishnasamy S., Owen H.A., Makaroff C.A. (1997). Molecular characterization of glyoxalase II from Arabidopsis thaliana. Plant Mol. Biol. 35: 471–481. [DOI] [PubMed] [Google Scholar]
- Marasinghe G.P.K., Sander I.M., Bennett B., Periyannan G., Yang K.W., Makaroff C.A., Crowder M.W. (2005). Structural studies on a mitochondrial glyoxalase II. J. Biol. Chem. 280: 40668–40675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmstal E., Mannervik B. (1981). Evaluation of the two-substrate pathway of glyoxalase I from yeast by use of carbonic anhydrase and rapid-kinetic studies. FEBS Lett. 13: 301–304. [Google Scholar]
- Martins A.M.T.B.S., Cordeiro C.A.A., Ponces Freire A.M.J. (2001). In situ analysis of methylglyoxal metabolism in Saccharomyces cerevisiae. FEBS Lett. 499: 41–44. [DOI] [PubMed] [Google Scholar]
- Matsoukas I.G., Massiah A.J., Thomas B. (2013). Starch metabolism and antiflorigenic signals modulate the juvenile-to-adult phase transition in Arabidopsis. Plant Cell Environ. 36: 1802–1811. [DOI] [PubMed] [Google Scholar]
- Maurino V.G., Engqvist M.K. (2015). 2-Hydroxy acids in plant metabolism. The Arabidopsis Book 13: e0182, doi/10.1199/tab.0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzucotelli E., Mastrangelo A.M., Crosatti C., Guerra D., Stanca A.M., Cattivelli L. (2008). Abiotic stress response in plants: When post-transcriptional and post-translational regulations control transcription. Plant Sci. 174: 420–431. [Google Scholar]
- Mihoub M., Abdallah J., Gontero B., Dairou J., Richarme G. (2015). The DJ-1 superfamily member Hsp31 repairs proteins from glycation by methylglyoxal and glyoxal. Biochem. Biophys. Res. Commun. 463: 1305–1310. [DOI] [PubMed] [Google Scholar]
- Miller A.G., Smith D.G., Bhat M., Nagaraj R.H. (2006). Glyoxalase I is critical for human retinal capillary pericyte survival under hyperglycemic conditions. J. Biol. Chem. 281: 11864–11871. [DOI] [PubMed] [Google Scholar]
- Misra K., Banerjee A.B., Ray S., Ray M. (1995). Glyoxalase III from Escherichia coli: a single novel enzyme for the conversion of methylglyoxal into D-lactate without reduced glutathione. Biochem. J. 305: 999–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R., Morant M., Jarmer H., Nilsson L., Nielsen T.H. (2007). Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol. 143: 156–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch G., Westcott B., Menini T., Gugliucci A. (2012). Advanced glycation endproducts and their pathogenic roles in neurological disorders. Amino Acids 42: 1221–1236. [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15: 473–497. [Google Scholar]
- Murata-Kamiya N., Kamiya H., Kaji H., Kasai H. (2000). Methylglyoxal induces G:C to C:G and G:C to T:A transversions in the supF gene on a shuttle vector plasmid replicated in mammalian cells. Mutat. Res. 468: 173–182. [DOI] [PubMed] [Google Scholar]
- Mustafiz A., Singh A.K., Pareek A., Sopory S.K., Singla-Pareek S.L. (2011). Genome-wide analysis of rice and Arabidopsis identifies two glyoxalase genes that are highly expressed in abiotic stresses. Funct. Integr. Genomics 11: 293–305. [DOI] [PubMed] [Google Scholar]
- Mustafiz A., Ghosh A., Tripathi A.K., Kaur C., Ganguly A.K., Bhavesh N.S., Tripathi J.K., Pareek A., Sopory S.K., Singla-Pareek S.L. (2014). A unique Ni2+-dependent and methylglyoxal-inducible rice glyoxalase I possesses a single active site and functions in abiotic stress response. Plant J. 78: 951–963. [DOI] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136. [DOI] [PubMed] [Google Scholar]
- Noctor G., Queval G., Mhamdi A., Chaouch S., Foyer C.H. (2011). Glutathione. Arabidopsis Book 9: e0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padayatti P.S., Jiang C., Glomb M.A., Uchida K., Nagaraj R.H. (2001). High concentrations of glucose induce synthesis of argpyrimidine in retinal endothelial cells. Curr. Eye Res. 23: 106–115. [DOI] [PubMed] [Google Scholar]
- Palusa S.G., Ali G.S., Reddy A.S. (2007). Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. Plant J. 49: 1091–1107. [DOI] [PubMed] [Google Scholar]
- Phillips S.A., Thornalley P.J. (1993). The formation of methylglyoxal from triose phosphates. Investigation using a specific assay for methylglyoxal. Eur. J. Biochem. 212: 101–105. [DOI] [PubMed] [Google Scholar]
- Ponces Freire A., Ferreira A., Gomes R., Cordeiro C. (2003). Anti-glycation defences in yeast. Biochem. Soc. Trans. 31: 1409–1412. [DOI] [PubMed] [Google Scholar]
- Qiu Q.S., Huber J.L., Booker F.L., Jain V., Leakey A.D.B., Fiscus E.L., Yau P.M., Ort D.R., Huber S.C. (2008). Increased protein carbonylation in leaves of Arabidopsis and soybean in response to elevated [CO2]. Photosynth. Res. 97: 155–166. [DOI] [PubMed] [Google Scholar]
- Rabbani N., Thornalley P.J. (2014). Measurement of methylglyoxal by stable isotopic dilution analysis LC-MS/MS with corroborative prediction in physiological samples. Nat. Protoc. 9: 1969–1979. [DOI] [PubMed] [Google Scholar]
- Ray S., Ray M. (1982). Purification and characterization of NAD and NADP-linked alpha-ketoaldehyde dehydrogenases involved in catalyzing the oxidation of methylglyoxal to pyruvate. J. Biol. Chem. 257: 10566–10570. [PubMed] [Google Scholar]
- Reddi A.R., Jensen L.T., Culotta V.C. (2009). Manganese homeostasis in Saccharomyces cerevisiae. Chem. Rev. 109: 4722–4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentsch D., Laloi M., Rouhara I., Schmelzer E., Delrot S., Frommer W.B. (1995). NTR1 encodes a high affinity oligopeptide transporter in Arabidopsis. FEBS Lett. 370: 264–268. [DOI] [PubMed] [Google Scholar]
- Reumann S., Quan S., Aung K., Yang P., Manandhar-Shrestha K., Holbrook D., Linka N., Switzenberg R., Wilkerson C.G., Weber A.P., Olsen L.J., Hu J. (2009). In-depth proteome analysis of Arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes. Plant Physiol. 150: 125–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard J.P. (1991). Kinetic parameters for the elimination reaction catalyzed by triosephosphate isomerase and an estimation of the reaction’s physiological significance. Biochemistry 30: 4581–4585. [DOI] [PubMed] [Google Scholar]
- Richarme G., Mihoub M., Dairou J., Bui L.C., Leger T., Lamouri A. (2015). Parkinsonism-associated protein DJ-1/Park7 is a major protein deglycase that repairs methylglyoxal- and glyoxal-glycated cysteine, arginine, and lysine residues. J. Biol. Chem. 290: 1885–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridderström M., Mannervik B. (1996). Optimized heterologous expression of the human zinc enzyme glyoxalase I. Biochem. J. 314: 463–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvato F., Havelund J.F., Chen M., Rao R.S.P., Rogowska-Wrzesinska A., Jensen O.N., Gang D.R., Thelen J.J., Møller I.M. (2014). The potato tuber mitochondrial proteome. Plant Physiol. 164: 637–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner W., Weissmann C. (1973). A rapid, sensitive, and specific method for the determination of protein in dilute solution. Anal. Biochem. 56: 502–514. [DOI] [PubMed] [Google Scholar]
- Schindelin J., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K., Wolfe D.M., Stiller B., Pearce D.A. (2009). Cd2+, Mn2+, Ni2+ and Se2+ toxicity to Saccharomyces cerevisiae lacking YPK9p the orthologue of human ATP13A2. Biochem. Biophys. Res. Commun. 383: 198–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- Schmitz J., Schöttler M.A., Krueger S., Geimer S., Schneider A., Kleine T., Leister D., Bell K., Flügge U.-I., Häusler R.E. (2012). Defects in leaf carbohydrate metabolism compromise acclimation to high light and lead to a high chlorophyll fluorescence phenotype in Arabidopsis thaliana. BMC Plant Biol. 12: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz J., Heinrichs L., Scossa F., Fernie A.R., Oelze M.L., Dietz K.J., Rothbart M., Grimm B., Flügge U.I., Häusler R.E. (2014). The essential role of sugar metabolism in the acclimation response of Arabidopsis thaliana to high light intensities. J. Exp. Bot. 65: 1619–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R., Fischer K., Ketelsen B., Krupinska K., Krause K. (2007). Comparative survey of plastid and mitochondrial targeting properties of transcription factors in Arabidopsis and rice. Mol. Genet. Genomics 277: 631–646. [DOI] [PubMed] [Google Scholar]
- Schwacke R., Schneider A., van der Graaff E., Fischer K., Catoni E., Desimone M., Frommer W.B., Flügge U.I., Kunze R. (2003). ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 131: 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciré A., Tanfani F., Saccucci F., Bertoli E., Principato G. (2000). Specific interaction of cytosolic and mitochondrial glyoxalase II with acidic phospholipids in form of liposomes results in the inhibition of the cytosolic enzyme only. Proteins 41: 33–39. [PubMed] [Google Scholar]
- Sela I., Ashkenazy H., Katoh K., Pupko T. (2015). GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 43: W7–W14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semchyshyn H.M. (2014). Reactive carbonyl species in vivo: generation and dual biological effects. Sci. World J. 2014: 417842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibbersen C., Palmfeldt J., Hansen J., Gregersen N., Jørgensen K.A., Johannsen M. (2013). Development of a chemical probe for identifying protein targets of α-oxoaldehydes. Chem. Commun. (Camb.) 49: 4012–4014. [DOI] [PubMed] [Google Scholar]
- Sousa Silva M., Gomes R.A., Ferreira A.E., Ponces Freire A., Cordeiro C. (2013). The glyoxalase pathway: the first hundred years... and beyond. Biochem. J. 453: 1–15. [DOI] [PubMed] [Google Scholar]
- Sridhara S., Wu T.T. (1969). Purification and properties of lactaldehyde dehydrogenase from Escherichia coli. J. Biol. Chem. 244: 5233–5238. [PubMed] [Google Scholar]
- Sukdeo N., Honek J.F. (2007). Pseudomonas aeruginosa contains multiple glyoxalase I-encoding genes from both metal activation classes. Biochim. Biophys. Acta 1774: 756–763. [DOI] [PubMed] [Google Scholar]
- Takagi D., Inoue H., Odawara M., Shimakawa G., Miyake C. (2014). The Calvin cycle inevitably produces sugar-derived reactive carbonyl methylglyoxal during photosynthesis: a potential cause of plant diabetes. Plant Cell Physiol. 55: 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalmann M.R., Pazmino D., Seung D., Horrer D., Nigro A., Meier T., Kölling K., Pfeifhofer H.W., Zeeman S.C., Santelia D. (2016). Regulation of leaf starch degradation by abscisic acid is important for osmotic stress tolerance in plants. Plant Cell 28: 1860–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornalley P.J. (1990). The glyoxalase system: new developments towards functional characterization of a metabolic pathway fundamental to biological life. Biochem. J. 269: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornalley P.J. (1996). Pharmacology of methylglyoxal: formation, modification of proteins and nucleic acids, and enzymatic detoxification--a role in pathogenesis and antiproliferative chemotherapy. Gen. Pharmacol. 27: 565–573. [DOI] [PubMed] [Google Scholar]
- Thornalley P.J. (2003). Glyoxalase I--structure, function and a critical role in the enzymatic defence against glycation. Biochem. Soc. Trans. 31: 1343–1348. [DOI] [PubMed] [Google Scholar]
- Thornalley P.J. (2008). Protein and nucleotide damage by glyoxal and methylglyoxal in physiological systems--role in ageing and disease. Drug Metabol. Drug Interact. 23: 125–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornalley P.J., Langborg A., Minhas H.S. (1999). Formation of glyoxal, methylglyoxal and 3-deoxyglucosone in the glycation of proteins by glucose. Biochem. J. 344: 109–116. [PMC free article] [PubMed] [Google Scholar]
- Tomizioli M., et al. (2014). Deciphering thylakoid sub-compartments using a mass spectrometry-based approach. Mol. Cell. Proteomics 13: 2147–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turra G.L., Agostini R.B., Fauguel C.M., Presello D.A., Andreo C.S., González J.M., Campos-Bermudez V.A. (2015). Structure of the novel monomeric glyoxalase I from Zea mays. Acta Crystallogr. D Biol. Crystallogr. 71: 2009–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Jagt D.L., Han L.P.B., Lehman C.H. (1972). Kinetic evaluation of substrate specificity in the glyoxalase-I-catalyzed disproportionation of -ketoaldehydes. Biochemistry 11: 3735–3740. [DOI] [PubMed] [Google Scholar]
- Waadt R., Kudla J. (2008). In plant visualization of protein interactions using bimolecular fluorescence complementation (BiFC). Cold Spring Harb. Protoc. 3: 1–8. [DOI] [PubMed] [Google Scholar]
- Welchen E., Schmitz J., Fuchs P., García L., Wagner S., Wienstroer J., Schertl P., Braun H.-P., Schwarzländer M., Gonzalez D.H., Maurino V.G. (2016). D-Lactate dehydrogenase links methylglyoxal degradation and electron transport through cytochrome C. Plant Physiol. 172: 901–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel N.F., Carenbauer A.L., Pfiester M.P., Schilling O., Meyer-Klaucke W., Makaroff C.A., Crowder M.W. (2004). The binding of iron and zinc to glyoxalase II occurs exclusively as di-metal centers and is unique within the metallo-β-lactamase family. J. Biol. Inorg. Chem. 9: 429–438. [DOI] [PubMed] [Google Scholar]
- Wienstroer J., Engqvist M.K., Kunz H.H., Flügge U.I., Maurino V.G. (2012). D-Lactate dehydrogenase as a marker gene allows positive selection of transgenic plants. FEBS Lett. 586: 36–40. [DOI] [PubMed] [Google Scholar]
- Xu D., Liu X., Guo C., Zhao J. (2006). Methylglyoxal detoxification by an aldo-keto reductase in the cyanobacterium Synechococcus sp. PCC 7002. Microbiology 152: 2013–2021. [DOI] [PubMed] [Google Scholar]
- Yadav S.K., Singla-Pareek S.L., Ray M., Reddy M.K., Sopory S.K. (2005). Methylglyoxal levels in plants under salinity stress are dependent on glyoxalase I and glutathione. Biochem. Biophys. Res. Commun. 337: 61–67. [DOI] [PubMed] [Google Scholar]
- Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L. (2012). Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Porter N.A. (2005). New insights regarding the autoxidation of polyunsaturated fatty acids. Antioxid. Redox Signal. 7: 170–184. [DOI] [PubMed] [Google Scholar]
- Zang T.M., Hollman D.A., Crawford P.A., Crowder M.W., Makaroff C.A. (2001). Arabidopsis glyoxalase II contains a zinc/iron binuclear metal center that is essential for substrate binding and catalysis. J. Biol. Chem. 276: 4788–4795. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Rodionov D.A., Gelfand M.S., Gladyshev V.N. (2009). Comparative genomic analyses of nickel, cobalt and vitamin B12 utilization. BMC Genomics 10: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]