ANXUR receptor-like kinases regulate two-tiered plant immunity by association with pattern recognition receptor and NLR protein complexes.
Abstract
Plants have evolved two tiers of immune receptors to detect infections: cell surface-resident pattern recognition receptors (PRRs) that sense microbial signatures and intracellular nucleotide binding domain leucine-rich repeat (NLR) proteins that recognize pathogen effectors. How PRRs and NLRs interconnect and activate the specific and overlapping plant immune responses remains elusive. A genetic screen for components controlling plant immunity identified ANXUR1 (ANX1), a malectin-like domain-containing receptor-like kinase, together with its homolog ANX2, as important negative regulators of both PRR- and NLR-mediated immunity in Arabidopsis thaliana. ANX1 constitutively associates with the bacterial flagellin receptor FLAGELLIN-SENSING2 (FLS2) and its coreceptor BRI1-ASSOCIATED RECEPTOR KINASE1 (BAK1). Perception of flagellin by FLS2 promotes ANX1 association with BAK1, thereby interfering with FLS2-BAK1 complex formation to attenuate PRR signaling. In addition, ANX1 complexes with the NLR proteins RESISTANT TO PSEUDOMONAS SYRINGAE2 (RPS2) and RESISTANCE TO P. SYRINGAE PV MACULICOLA1. ANX1 promotes RPS2 degradation and attenuates RPS2-mediated cell death. Surprisingly, a mutation that affects ANX1 function in plant immunity does not disrupt its function in controlling pollen tube growth during fertilization. Our study thus reveals a molecular link between PRR and NLR protein complexes that both associate with cell surface-resident ANX1 and uncovers uncoupled functions of ANX1 and ANX2 during plant immunity and sexual reproduction.
INTRODUCTION
Plants growing in their natural habitats are constantly exposed to various potential pathogens that can result in the detriment of growth and yield. To ward off pathogen invasion, plants have developed a two-tiered immune system in addition to preformed physical and chemical barriers (Jones and Dangl, 2006). The first branch of plant immunity, so-called pattern-triggered immunity (PTI), is triggered by microbe-associated molecular patterns (MAMPs) that are recognized by plasma membrane-localized pattern recognition receptors (PRRs) (Boller and Felix, 2009; Couto and Zipfel, 2016; Yu et al., 2017). A number of PRRs have been identified and most of them belong to receptor-like kinases (RLKs) or receptor-like proteins (Böhm et al., 2014; Couto and Zipfel, 2016). FLAGELLIN-SENSING2 (FLS2) and ELONGATION FACTOR-TU (EF-Tu) RECEPTOR (EFR), RLKs with an extracellular leucine-rich repeat domain (LRR-RLKs), recognize bacterial flagellin and EF-Tu, respectively (Gómez-Gómez and Boller, 2000; Zipfel et al., 2006). Perception of flagellin or EF-Tu triggers the rapid association of the corresponding receptors with another LRR-RLK, BRI1-ASSOCIATED RECEPTOR KINASE1 (BAK1), also known as SOMATIC EMBRYOGENESIS RECEPTOR KINASE3 (SERK3), and other SERK members (Chinchilla et al., 2007; Heese et al., 2007). The BOTRYTIS-INDUCED KINASE1 (BIK1) family receptor-like cytoplasmic kinases (RLCKs) associate with PRR complexes and are phosphorylated by BAK1 to transduce intracellular signaling (Lin et al., 2014; Lu et al., 2010; Zhang et al., 2010). In addition, the LysM-domain RLKs or receptor-like proteins recognize bacterial peptidoglycan or fungal chitin (Cao et al., 2014; Gust, 2015; Shinya et al., 2015), the lectin S-domain RLK LIPOOLIGOSACCHARIDE-SPECIFIC REDUCED ELICITATION senses bacterial lipopolysaccharide (Ranf et al., 2015), and the legume-type lectin domain-containing RLK, DOES NOT RESPOND TO NUCLEOTIDES1, is a receptor of plant endogenous eATP (Choi et al., 2014).
The second branch of plant immunity, termed effector-triggered immunity (ETI), is initiated upon recognition of pathogen effectors via intracellular immune receptors, which are often encoded by nucleotide binding domain leucine-rich repeat (NLR or NB-LRR) proteins (Jones and Dangl, 2006; Jones et al., 2016; Maekawa et al., 2011). Those pathogen effectors are translocated into the host cells and many effectors are able to suppress plant PTI or modulate host physiology to promote pathogenicity in the absence of corresponding NLRs (Dou and Zhou, 2012; Macho and Zipfel, 2015). Arabidopsis thaliana NLR proteins RPS2 and RESISTANCE TO P. SYRINGAE PV MACULICOLA1 (RPM1) initiate resistance upon recognition of Pseudomonas syringae effectors AvrRpt2 and AvrRpm1, respectively. Although they lack an apparent transmembrane domain, RPS2 and RPM1 are anchored to the plasma membrane to trigger immune responses accompanied with the hypersensitive response, a localized cell death (Axtell and Staskawicz, 2003; Gao et al., 2011). In some cases, NLR proteins directly bind to pathogen effectors; however, more often, NLR proteins sense perturbation of host proteins modified by pathogen effectors to elicit defense responses (Jones and Dangl, 2006; Jones et al., 2016; Maekawa et al., 2011). For example, AvrRpt2 cleaves RIN4 to activate the RPS2 signaling (Axtell and Staskawicz, 2003; Mackey et al., 2003), whereas AvrRpm1 induces RIN4 phosphorylation by RLCK RIPK to initiate the RPM1 signaling (Chung et al., 2011; Liu et al., 2011). Recent studies in the identification of plant immune receptors and downstream signaling events suggest a blurred boundary between PTI and ETI (Thomma et al., 2011).
To understand the mechanisms underlying plant innate immunity, we developed a series of genetic screens for components controlling immune gene transcriptional reprogramming. We have deployed the EMS-mutagenized populations of Arabidopsis transgenic plants carrying a luciferase reporter gene under the control of the FRK1 promoter (pFRK1:LUC) (Feng et al., 2015; Li et al., 2014) or WRKY46 promoter (pWRKY46:LUC). In contrast to FRK1, which is strongly induced by multiple MAMPs, WRKY46 is highly induced by Pseudomonas syringae pv tomato DC3000 (Pst) carrying avrRpm1 or avrRpt2 (Gao et al., 2013). In this study, we report that a mutation in ANXUR1 (ANX1) affects both plant PTI and ETI. ANX1 is a member of the Catharanthus roseus RLK1-like (CrRLK1L) subfamily that carries an extracellular malectin-like domain (Li et al., 2016; Lindner et al., 2012; Nissen et al., 2016). ANX1 and its closest homolog ANX2 redundantly regulate cell wall integrity during pollen tube growth whereas their closest homolog, FERONIA (FER) is involved in myriad biological processes including cell wall integrity during root hair growth, cell-cell communication during fertilization, abscisic acid signaling, and immunity (Boisson-Dernier et al., 2013, 2009; Chen et al., 2016; Duan et al., 2010; Escobar-Restrepo et al., 2007; Kessler et al., 2010; Miyazaki et al., 2009; Stegmann et al., 2017). We show here that ANX1 and ANX2 negatively regulate MAMP-induced immune responses, including mitogen-activated protein kinase (MAPK) activation, reactive oxygen species (ROS) production, and immune gene induction. ANX1 and ANX2 also negatively regulate RPM1- and RPS2-mediated ETI responses and disease resistance. ANX1 constitutively associates with FLS2 and perception of flagellin promotes ANX1 association with BAK1, which interferes with ligand-induced FLS2-BAK1 complex formation. In addition, ANX1 complexes with RPS2 and RPM1 immune receptors and appears to regulate RPS2 protein levels. Thus, ANX1 links plant PTI and ETI by association with both PRR complexes and NLR proteins. Interestingly, the anx1 mutant identified from our genetic screen with defects in both PTI and ETI displays normal pollen tube growth, suggesting uncoupled functions of ANXs during plant immunity and sexual reproduction.
RESULTS
Enhanced ETI Responses in the aggie101 Mutant
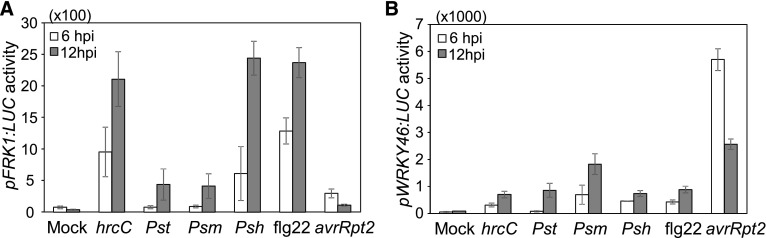
We generated transgenic plants carrying pFRK1:LUC or pWRKY46:LUC to monitor the specific elicitation of two branches of plant immune responses. Similar to the induction patterns of endogenous genes, the pFRK1:LUC activity was strongly induced by the Pst type III secretion deficient mutant hrcC, the nonadaptive bacterium P. syringae pv phaseolicola NPS3121 (Psh) and flg22, a 22-amino acid peptide derived from bacterial flagellin (Figure 1A), whereas the pWRKY46:LUC activity was preferentially induced by Pst avrRpt2 (Figure 1B). Thus, the transgenic plants carrying pFRK1:LUC or pWRKY46:LUC serve as marker lines to study immune gene transcriptional regulation in response to PTI or ETI elicitation, respectively. By screening ∼6000 mutagenized M2 pWRKY46:LUC plants upon Pst avrRpt2 infection, a series of mutants named Arabidopsis genes governing immune gene expression (aggie) with altered pWRKY46:LUC activity were identified. Here, we focus on the characterization of the aggie101 mutant, which exhibits elevated luciferase activity compared with wild-type pWRKY46:LUC transgenic plants upon Pst avrRpt2 infection (Figure 2A).
Figure 1.
Activation of Luciferase Reporters of pFRK1:LUC and pWRKY46:LUC in Transgenic Plants by Different Bacteria or flg22.
Four-week-old soil-grown plants were hand-infiltrated with water (Mock), virulent bacterium Pst or Psm, avirulent bacterium Pst avrRpt2, nonadaptive bacterium Psh at OD600 = 0.01, nonpathogenic bacterium Pst hcC at OD600 = 0.5, or 100 nM flg22. The activity of pFRK1:LUC (A) or pWRKY46:LUC (B) was measured at 6 or 12 hpi. The data are shown as means ± se (n = 12). The experiments were repeated three times with similar results.
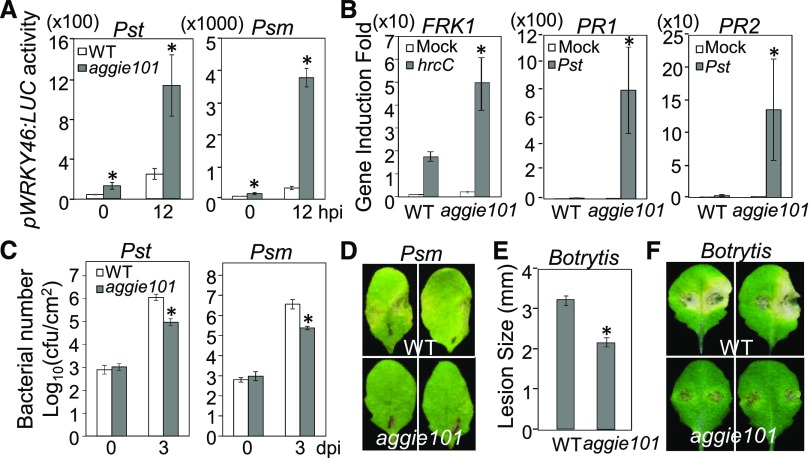
Figure 2.
The aggie101 Mutant Displays Enhanced ETI Responses and Resistance to Avirulent Bacterial Pathogens.
(A) Luciferase activity in pWRKY46:LUC (WT) and aggie101 mutant plants. Leaves from 4-week-old soil-grown plants were hand-infiltrated with water (Mock) or Pst avrRpt2 at OD600 = 0.01. Pictures were taken with an EMCCD camera at 6 hpi.
(B) Enhanced pWRKY46:LUC activity in aggie101 in response to Pst avrRpt2 or avrRpm1. Leaves from 4-week-old plants were hand-infiltrated with Pst carrying avrRpt2 (left panel) or avrRpm1 (right panel) at OD600 = 0.01, and the samples were collected at 0, 6, 12, or 24 hpi. The data are shown as means ± se from 8 to ∼12 leaves for each time point (n = 8 to ∼12).
(C) The avrRpt2-induced defense gene expression is elevated in aggie101. Four-week-old plants were hand-infiltrated with mock or Pst avrRpt2 at OD600 = 0.01, and leaf samples were collected at 6 hpi for RT-qPCR analysis. The expression of WRKY46, PR1, and PR2 was normalized to the expression of UBQ10. The data are shown as means ± sd from three biological replicates.
(D) The aggie101 mutant is more resistant to avirulent bacterial pathogens. Four-week-old plants were hand-infiltrated with Pst avrRpt2 or avrRpm1 at OD600 = 5 × 10−4 and the bacterial growth analysis was performed at 0 and 3 dpi. The data are shown as means ± sd (n = 3).
The above experiments were repeated three times with similar results. The asterisks indicate a significant difference with the wild type as determined by Student’s t test (P < 0.05).
The enhanced WRKY46 promoter activity in the aggie101 mutant was observed at various time points after infection with Pst avrRpt2 (Figure 2B). In addition, the aggie101 mutant also potentiated the pWRKY46:LUC activity in response to Pst carrying avrRpm1 (Figure 2B). Furthermore, the expression of the endogenous WRKY46 gene was elevated in the aggie101 mutant compared with wild-type plants 6 h postinoculation (hpi) of Pst avrRpt2 by RT-qPCR analysis (Figure 2C). Similarly, the induction of pathogenesis-related genes PR1 and PR2 by Pst avrRpt2 was potentiated in the aggie101 mutant (Figure 2C). The aggie101 mutant also displayed enhanced resistance to Pst carrying avrRpt2 or avrRpm1 (Figure 2D). The bacteria grew about 5- to 8-fold less in the aggie101 mutant than in wild-type plants at 3 d postinoculation (dpi). Thus, the aggie101 mutant displays enhanced ETI responses and resistance to avirulent bacterial pathogens.
Enhanced PTI Responses in the aggie101 Mutant
Notably, we consistently observed an ∼2-fold increase of pWRKY46:LUC activity in the aggie101 mutant compared with wild-type plants without infections (Figures 2B and 3A). In addition, aggie101 displayed ∼6-fold higher induction of WRKY46 promoter activity than wild-type plants in response to the virulent bacterium Pst (Figure 3A). Similarly, the induction of the WRKY46 promoter to P. syringae pv maculicola ES4326 (Psm) in the aggie101 mutant was ∼10-fold higher than that in wild-type plants (Figure 3A). The Pst hcC-mediated induction of FRK1, a PTI marker gene, was also enhanced in the aggie101 mutant as detected by RT-qPCR analysis (Figure 3B). Furthermore, the induction of PR1 and PR2 by Pst was markedly elevated in the aggie101 mutant compared with the negligible induction in the wild-type plants (Figure 3B). Consistent with these changes in gene expression, the aggie101 mutant was more resistant to virulent Pst and Psm infections (Figure 3C). The size of the bacterial population in the aggie101 mutant was about 10-fold less than that in wild-type plants at 3 dpi (Figure 3C). The disease symptom development was less pronounced in the aggie101 mutant than that in wild-type plants after Psm infection (Figure 3D). The aggie101 mutant was also more resistant to infection with the necrotrophic fungal pathogen Botrytis cinerea, as measured by lesion diameter (Figure 3E) and symptom development, compared with wild-type plants (Figure 3F). Together, these results show that the aggie101 mutant shows enhanced resistance to virulent bacterial and fungal pathogens.
Figure 3.
The aggie101 Mutant Displays Enhanced Resistance to Virulent Pathogens.
(A) Enhanced pWRKY46:LUC activity in aggie101 in response to virulent bacterial pathogens. Four-week-old pWRKY46:LUC (WT) and aggie101 plants were hand-infiltrated with virulent Pst or Psm at OD600 = 0.01, and the samples were collected at 0 and 12 hpi. The data are shown as means ± se (n = 8 to ∼12).
(B) Elevated Pst-induced defense gene expression in aggie101. Four-week-old plants were hand-infiltrated with water (Mock), Pst, or Pst hcC at OD600 = 0.01, and leaf samples were collected at 6 hpi for RT-qPCR analysis. The expression of FRK1, PR1, and PR2 was normalized to the expression of UBQ10. The data are shown as means ± sd from three biological replicates.
(C) and (D) Elevated resistance to virulent bacterial pathogens in aggie101. Four-week-old plants were hand-infiltrated with Pst or Psm at OD600 = 5 × 10−4. The samples were collected at 0 and 3 dpi for in planta bacterial multiplication assays. The data are shown as means ± sd (n = 3) (C) and representative leaves from Psm inoculated plants were detached and photographed at 3 dpi (D).
(E) and (F) The aggie101 mutant exhibits enhanced resistance to B. cinerea BO5. Four-week-old plants were drop-inoculated with B. cinerea BO5 at 105 spores/mL. The samples were collected at 3 dpi for lesion size measurement (E) and pictures (F). The data in (E) are shown as means ± se (n = 12).
The above experiments were repeated three times with similar results. The asterisks indicate a significant difference with the wild type as determined by Student’s t test (P < 0.05).
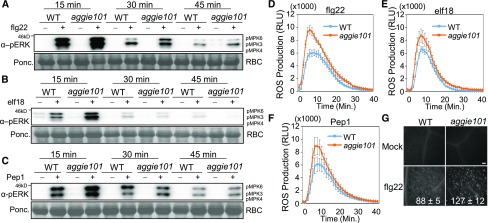
Since aggie101 displayed enhance immune gene expression and disease resistance to various pathogens, we tested whether aggie101 had elevated PTI responses triggered by different MAMPs. MAPK activation and ROS production are two early events in PTI signaling. In the wild type, the MAPKs, in particular MPK3 and MPK6, were activated by flg22, elf18, an 18-amino acid peptide of bacterial EF-Tu, and Pep1, an endogenous damage-associated molecular pattern (DAMP), at 15 min after treatment, and the induction was gradually reduced at 30 and 45 min after treatment (Figures 4A to 4C; Supplemental Figures 1A to 1C). The MAPK activation was further enhanced in the aggie101 mutant, in particular at 15 min after treatment (Figures 4A to 4C; Supplemental Figures 1A to 1C). Similarly, the aggie101 mutant exhibited an enhanced ROS burst in response to flg22, elf18, and Pep1 treatments compared with wild-type plants (Figures 4D to 4F; Supplemental Figure 1D). These results suggest that AGGIE101 likely functions upstream of MAPK activation and ROS production in PTI signaling. Callose deposition is a relatively late PTI response. The aggie101 mutant also showed more callose deposits than wild-type plants as detected by aniline blue staining at 12 h after flg22 treatment (Figure 4G). Taken together, these observations show that the aggie101 mutant has elevated responsiveness to both ETI and PTI elicitations.
Figure 4.
The aggie101 Mutant Displays Enhanced PTI Responses.
(A) to (C) Enhanced MAPK activation in aggie101 in response to MAMPs/damage-associated molecular patterns. Leaves from 4-week-old soil-grown plants were hand-infiltrated with 100 nM flg22 (A), elf18 (B), Pep1 (C), or water control, and the samples were collected at indicated time points. The MAPK activation was detected by immunoblotting with an α-pERK antibody (top), and the protein loading is shown by Ponceau S staining for Rubisco (RBC) (bottom).
(D) to (F) Enhanced accumulation of ROS in aggie101 in response to flg22 (D), elf18 (E), and Pep1 (F). ROS are presented as relative light units (RLU). Leaf discs of 4-week-old soil-grown plants were treated with 100 nM peptides. The data are shown as means ± se (n = 16).
(G) Enhanced callose deposits in aggie101 in response to flg22. Callose deposits were stained with aniline blue in 5-week-old plant leaves infiltrated with water or 1 µM flg22 for 12 h. The quantification data by Image J software are shown as means ± sd (n = 3). Bar = 0.1 mm.
The above experiments were repeated three times with similar results.
The aggie101 Mutant Harbors a Mutation in ANX1
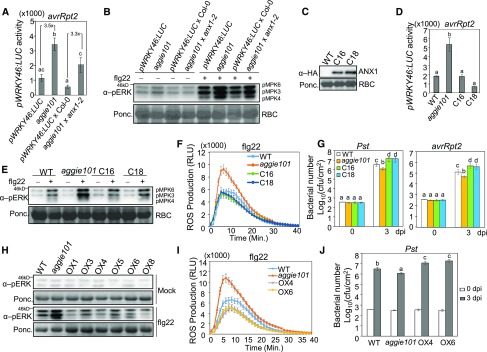
The Pst avrRpt2-induced WRKY46 promoter activity of F1 plants from a backcross of aggie101 to wild-type pWRKY46:LUC transgenic plants was similar to that of wild-type plants, indicating that the aggie101 mutation is largely recessive (Supplemental Figure 2A). We crossed the aggie101 mutant (in the Col-0 accession background) with the Ler accession and mapped aggie101 to the upper arm of chromosome 3 between markers HG18 and HG19, which are ∼49 kb apart (Supplemental Figure 2B). Next-generation sequencing of the aggie101 mutant revealed a C-to-T mutation at the 1073 bp from the predicted start codon of ANX1 (At3g04690), which results in a substitution of alanine (GCG) to valine (GTG) (ANX1A358V) at the residue 358 (Supplemental Figures 2C and 2D). ANX1 bears an extracellular malectin-like domain, which contains two malectin domains with similarity to the animal carbohydrate binding malectin proteins involved in the endoplasmic reticulum-quality control (Boisson-Dernier et al., 2011; Schallus et al., 2008). The mutation of ANX1A358V lies in the second malectin domain of ANX1. ANX1A358 is conserved in its closest homolog ANX2 (Supplemental Figure 3). Interestingly, the corresponding residue in FER is valine but not alanine (Supplemental Figure 3). To determine whether the ANX1A358V mutation is responsible for aggie101 phenotype, we crossed aggie101 with anx1-2, a T-DNA knockout mutant of ANX1 in the Col-0 background. The WRKY46 promoter activity of F1 plants of aggie101 × anx1-2 was ∼3.3-fold higher than that of the control F1 plants of pWRKY46:LUC × Col-0 upon Pst avrRpt2 infection, which is comparable with enhanced WRKY46 promoter activity in the aggie101 mutant (Figure 5A). Notably, the F1 plants of pWRKY46:LUC × Col-0 and aggie101 × anx1-2 only carry one copy of the pWRKY46:LUC transgene, thus resulting in the reduced (about half) WRKY46 promoter activity when compared with homozygous pWRKY46:LUC plants and the aggie101 mutant (Figure 5A). In addition, the aggie101 × anx1-2 F1 plants showed the enhanced flg22-induced MAPK activation compared with pWRKY46:LUC × Col-0 F1 plants (Figure 5B; Supplemental Figure 2E). These results suggest that aggie101 is allelic to anx1-2.
Figure 5.
The AGGIE101 Encodes ANX1.
(A) aggie101 and anx1-2 are allelic for Pst avrRpt2-induced pWRKY46:LUC activation. Four-week-old pWRKY46:LUC, aggie101, and F1 plants from a cross between pWRKY46:LUC × Col-0 or aggie101 × anx1-2 (SALK_0456870) were hand-infiltrated with Pst avrRpt2 at OD600 = 0.01 for 6 h. F1 plants of pWRKY46:LUC × Col-0 were used as control for heterozygous pWRKY46:LUC transgene in F1 plants of aggie101 × anx1-2. The data are shown as means ± se (n = 12–16). The number between two bars indicates the induction fold compared with its cognate control.
(B) Enhanced MAPK activation in F1 plants of aggie101 × anx1-2 in response to flg22. Four-week-old soil-grown plants were hand-infiltrated with water or 100 nM flg22 for 15 min. MAPK activation was detected by immunoblotting with an α-pERK antibody (top), and Ponceau S stained membrane is shown for Rubisco (RBC) as controls of protein loading (bottom).
(C) The expression level of ANX1-HA proteins in two representative complementation lines (C16 and C18). ANX1-HA proteins were detected by immunoblotting with an α-HA antibody (top), and Ponceau S-stained membrane is shown for Rubisco as controls of protein loading (bottom).
(D) ANX1 restores pWRKY46:LUC activity in aggie101 to the wild-type level. Four-week-old plants were hand-infiltrated with Pst avrRpt2 at OD600 = 0.01 for 6 h. C16 and C18 are two complementation lines with 35S:ANX1-HA in the aggie101 background. The data are shown as means ± se (n = 12).
(E) Restored MAPK activation in complementation lines in response to flg22. Four-week-old soil-grown plants were hand-infiltrated with water or 100 nM flg22 for 15 min. MAPK activation was detected with an α-pERK antibody (top), and Rubisco was stained with Ponceau S for protein loading control (bottom).
(F) Restored ROS accumulation in complementation lines in response to flg22. Leaf discs of 4-week-old plants were treated with 100 nM flg22. The data are shown as means ± se (n = 16).
(G) Enhanced susceptibility to Pst and Pst avrRpt2 infections in ANX1 complementation lines. Four-week-old plants were infiltrated with bacteria at OD600 = 5 × 10−4. The samples were harvested at 0 and 3 dpi. The data are shown as means ± sd (n = 3).
(H) Reduced flg22-induced MAPK activation in ANX1 overexpression lines. Four-week-old T2 plants were infiltrated with water or 100 nM flg22 for 15 min. The MAPK activation was detected with an α-pERK antibody (top), and Rubisco was stained with Ponceau for protein loading control (bottom).
(I) Reduced flg22-induced ROS production in two homozygous T3 lines. The data are shown as means ± se (n = 16). Leaf discs of 4-week-old plants were treated with 100 nM flg22.
(J) Enhanced Pst susceptibility in two homozygous T3 lines. Four-week-old plants were infiltrated with Pst at OD600 = 5 × 10−4. The samples were harvested at 0 and 3 dpi. The data are shown as means ± sd (n = 3).
The experiments in (A) to (C) were repeated two times and others were repeated three times with similar results. The different letters indicate statistically significant difference analyzed with one-way ANOVA followed by Tukey’s test (P < 0.05).
We also silenced ANX1 in wild-type pWRKY46:LUC plants by virus-induced gene silencing (VIGS). When inoculated with Pst avrRpt2, the ANX1-silenced plants showed the enhanced WRKY46 promoter activity compared with control vector-inoculated plants (Supplemental Figure 4A). The ANX1-silenced plants also displayed the enhanced resistance to Pst avrRpt2 or Pst infections (Supplemental Figure 4B) and flg22-induced MAPK activation compared with control plants (Supplemental Figure 4C). The data suggest that ANX1 plays a negative role in AvrRpt2-mediated ETI and flg22-mediated PTI.
We further transformed the HA epitope-tagged ANX1 under the control of the CaMV 35S promoter into the aggie101 mutant. Two independent homozygous lines (C16 and C18) with moderate ANX1-HA expression were selected for further analysis (Figure 5C). The WRKY46 promoter activity of C16 and C18 plants was similar to that in wild-type pWRKY46:LUC plants after Pst avrRpt2 infection (Figure 5D). Furthermore, the flg22-induced MAPK activation and ROS production in C16 and C18 plants were lower than that of aggie101 and similar to that of the wild type (Figures 5E and 5F; Supplemental Figures 4D and 4E). We further inoculated C16 and C18 plants with Pst and Pst carrying avrRpt2. The bacterial population of Pst and Pst avrRpt2 in C16 and C18 plants was significantly higher than that of aggie101 (Figure 5G). Notably, the C16 and C18 plants were even more susceptible to Pst and Pst avrRpt2 infections than wild-type plants (Figure 5G).
Furthermore, we ectopically expressed 35S:ANX1-HA in wild-type pWRKY46:LUC plants. Multiple 35S:ANX1-HA T1 transgenic lines showed moderate ANX1-HA expression (Supplemental Figure 5A) and reduced WRKY46 promoter activity after Pst avrRpt2 infection compared with wild-type pWRKY46:LUC plants (Supplemental Figure 5B). The overexpression lines also displayed lower flg22-induced MAPK activation than the wild type (Figure 5H). Two homozygous T3 overexpression lines displayed reduced ROS production after flg22 treatment (Figure 5I) and increased bacterial growth of Pst compared with the wild type (Figure 5J). Together, our results indicate that the immunity-related phenotypes observed in aggie101 can be attributed to the anx1A358V mutation and AGGIE101 is ANX1.
ANX1 and ANX2 Negatively Regulate Plant Immunity
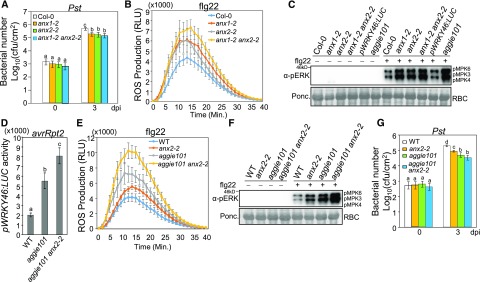
ANX1 and ANX2 function redundantly to maintain pollen tube integrity during fertilization (Boisson-Dernier et al., 2009). We examined the role of ANX1 and ANX2 in the disease resistance and PTI responses with the T-DNA insertional mutants of anx1-2 and anx2-2. The anx1-2 and anx2-2 mutants showed increased resistance to Pst infections (Figure 6A). The anx1-2 and anx2-2 mutants also showed enhanced ROS production (Figure 6B; Supplemental Figure 6A) and MAPK activation (Figure 6C; Supplemental Figure 6B) in response to flg22 compared with Col-0 plants. The anx1-2 anx2-2 double mutant had slightly, but not significantly, higher induction of flg22-induced ROS production and MAPK activation than the anx1-2 and anx2-2 single mutants (Figures 6B and 6C; Supplemental Figures 6A and 6B). The bacterial growth of Pst in anx1-2 anx2-2 was similar to that in anx1-2 and anx2-2 (Figure 6A). We also generated an aggie101 anx2-2 double mutant, which was homozygous for the pWRKY46:LUC transgene by genetic crossing (Supplemental Figures 6C and 6D). When inoculated with Pst avrRpt2, the WRKY46 promoter activity was significantly higher than that in aggie101 (Figure 6D). The flg22-induced ROS production and MAPK activation were also further enhanced in the aggie101 anx2-2 mutant compared with the aggie101 and anx2-2 single mutants (Figures 6E and 6F; Supplemental Figure 6E). The bacterial growth of Pst was slightly decreased in aggie101 anx2-2 (Figure 6G). Similarly, when we silenced ANX1 in the anx2-2 mutant by VIGS, the flg22-induced ROS production in these plants was higher than that in Col-0 silenced with ANX1 or anx2-2 mutant inoculated with a VIGS control vector (Supplemental Figure 6F). We also introduced the pWRKY46:LUC transgene into the anx2-2 mutant. The pWRKY46:LUC/anx2-2 plants showed enhanced promoter activity when inoculated with Pst or Pst carrying avrRpt2 compared with wild-type pWRKY46:LUC plants (Supplemental Figures 6G and 6H). These data indicate that both ANX1 and ANX2 negatively regulate plant immunity in a partially redundant manner.
Figure 6.
ANX1 and ANX2 Negatively Regulate Plant Immunity.
(A) The anx1-2, anx2-2, and anx1-2 anx2-2 mutants are more resistant to Pst infections. Four-week-old plants were infiltrated with Pst at OD600 = 5 × 10−4. The samples were collected at 0 and 3 dpi. The data are shown as means ± sd (n = 3). Col-0 plants were used as control for mutants.
(B) Enhanced flg22-induced ROS accumulation in anx mutants. Leaf discs of 4-week-old-plants were treated with 100 nM flg22. The data are shown as means ± se (n = 16).
(C) Enhanced flg22-induced MAPK activation in anx mutants. Four-week-old plants were infiltrated with water or 100 nM flg22 for 15 min. MAPK activation was detected with an α-pERK antibody (top), and Rubisco (RBC) was stained with Ponceau S for protein loading control (bottom).
(D) Enhanced pWRKY46:LUC activity in the aggie101 anx2-2 double mutant. Four-week-old wild-type pWRKY46:LUC, aggie101, and aggie101 anx2-2 mutants were infiltrated with Pst avrRpt2 at OD600 = 0.01 for 6 h. The data are shown as means ± se (n = 8 to ∼12).
(E) Enhanced flg22-induced ROS accumulation in aggie101 anx2-2. Leaf discs of 4-week-old wild-type pWRKY46:LUC, anx2-2, aggie101, and aggie101 anx2-2 were treated with 100 nM flg22. The data are shown as means ± se (n = 16).
(F) Enhanced flg22-induced MAPK activation in aggie101 anx2-2. Four-week-old plants were infiltrated with water or 100 nM flg22 for 15 min.
(G) Bacterial growth of Pst. Four-week-old plants were infiltrated with Pst at OD600 = 5 × 10−4. The samples were collected at 0 and 3 dpi. The data are shown as means ± sd (n = 3).
The above experiments were repeated three times with similar results. The different letters indicate statistically significant difference analyzed with one-way ANOVA followed by Tukey’s test (P < 0.05).
ANX1 and ANX2 Functions in Plant Immunity Are Developmental Stage Dependent
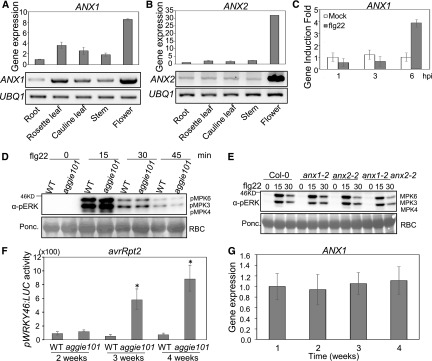
ANX1 and ANX2 were reported to be preferentially expressed in pollen (Boisson-Dernier et al., 2009). Consistent with this, we observed strong expression of ANX1 and ANX2 in flowers (Figures 7A and 7B). We were also able to detect their expression in roots, rosette leaves, cauline leaves, and stems, albeit to a lesser extent compared with their expression in flowers (Figures 7A and 7B). The ANX1 gene was also induced by flg22 treatment at 6 hpi, further supporting its role in plant immunity (Figure 7C).
Figure 7.
Developmental Stage-Dependent ANX1 Function in Plant Immunity.
(A) ANX1 transcripts in various tissues of 6-week-old soil-grown plants. The data of RT-qPCR analysis with UBQ10 as an internal control are shown on the top panel. The data of RT-PCR analysis with UBQ1 as a control are shown on the bottom panel.
(B) The expression of ANX2 transcripts in various tissues.
(C) flg22-induced ANX1 expression in Col-0. Four-week-old soil-grown Col-0 plants were hand-infiltrated with 100 nM flg22 or water control, and the samples were collected at indicated time points for RT-qPCR analysis. The data are shown as means ± sd (n = 3).
(D) flg22-induced MAPK activation in 2-week-old seedlings of the wild type and aggie101. The seedlings grown on 0.5× MS were treated with 100 nM flg22 or water control and the samples were collected at the indicated time points. The MAPK activation was detected with an α-pERK antibody (top), and Rubisco (RBC) was stained with Ponceau for protein loading control (bottom).
(E) flg22-induced MAPK activation in 2-week-old seedlings of Col-0, anx1-2, anx2-2, and anx1-2 anx2-2 double mutant.
(F) Luciferase activity in pWRKY46:LUC (WT) and aggie101 mutant plants at different stages. The 2-, 3-, or 4-week-old soil-grown plants were hand-infiltrated with Pst avrRpt2 at OD600 = 0.01, and the samples were collected at 6 hpi. The data are shown as means ± se (n = 12). The asterisks indicate a significant difference with the wild type as determined by Student’s t test (P < 0.05).
(G) The ANX1 transcripts in Col-0 plants at different developmental stages. Leaves from soil-grown plants were collected for RT-qPCR analyses using UBQ10 as a control. The data are shown as means ± sd (n = 3).
The experiments in (A) to (C) were repeated two times and others were repeated three times with similar results.
Interestingly, we observed that the enhanced immune responses in the aggie101, anx1-2, and anx2-2 mutants were much less pronounced at 2-week-old seedling stage than at 4-week-old stage. The flg22-induced MAPK activation was comparable between wild-type pWRKY46:LUC and aggie101 (Figure 7D) or between Col-0, anx1-2, anx2-2, and anx1-2 anx2-2 at multiple time points (Figure 7E) at 2-week-old seedling stage. We also tested Pst-induced WRKY46 promoter activity in wild-type and aggie101 plants at different growth stages. There was no significant difference of WRKY46 promoter activity in wild-type and aggie101 plants at 2-week-old stage (Figure 7F). At 3-week-old stage, the aggie101 mutant showed significantly higher WRKY46 promoter activity than wild-type plants after Pst infection. The difference of WRKY46 promoter activity in the wild type and the aggie101 mutant became more pronounced at the 4-week-old stage (Figure 7F). Thus, the ANX function in plant immunity appears to be developmental stage dependent. This developmental stage-dependent function was unlikely due to a change in its transcripts, since we did not observe a notable difference in ANX1 expression level from 1 to 4 weeks (Figure 7G). Notably, Nicotiana benthamiana RECEPTOR-LIKE PROTEIN REQUIRED FOR CSP22 RESPONSIVENESS, which associates with BAK1 upon bacterial cold shock protein perception, also confers age-dependent plant immunity to bacterial pathogens (Saur et al., 2016). It is possible that ANXs sense a developmentally regulated ligand to dampen PTI and ETI responses.
ANX1 Associates with Both PTI and ETI Immune Receptors
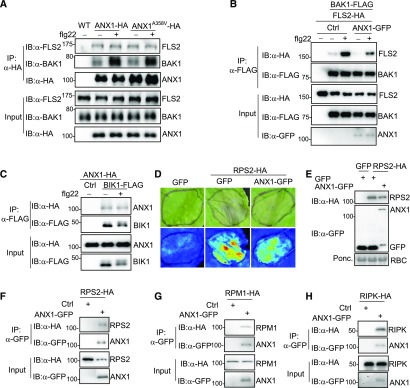
Since ANX1 is a plasma membrane-localized protein and appears to function at a very early step in PTI signaling, we tested whether ANX1 associates with the plasma membrane-localized flagellin receptor FLS2 and coreceptor BAK1. A coimmunoprecipitation (co-IP) in ANX1-HA transgenic plants with α-FLS2 or α-BAK1 antibody indicated that ANX1 associated with both endogenous FLS2 and BAK1 (Figure 8A). Interestingly, flg22 treatment markedly induced ANX1 association with BAK1 but not with FLS2 (Figure 8A). Apparently, the mutation in ANX1A358V did not affect its association with FLS2 or BAK1 (Figure 8A). FLS2, but not CERK1, a LysM domain-containing RLK, associated with ANX1 in protoplast transient assay (Supplemental Figure 7A). The cytosolic domain of ANX1 interacted with the cytosolic domain of BAK1 with a yeast two-hybrid assay (Supplemental Figure 7B). Perception of flg22 triggers rapid complex formation of FLS2 and BAK1 (Chinchilla et al., 2007; Heese et al., 2007). Coexpression of ANX1 antagonized flg22-induced FLS2-BAK1 association (Figure 8B). In addition, ANX1 also associated with BIK1, a RLCK in the FLS2-BAK1 complex (Figure 8C). Taken together, the data point to ANX1 being closely associated with the FLS2-BAK1-BIK1 complex in the resting state and flg22 treatment promoting or stabilizing the association of ANX1 with BAK1. This association would then interfere with the ligand-induced FLS2-BAK1 complex formation, thereby negatively regulating PTI signaling.
Figure 8.
ANX1 Associates with Two Tiers of Immune Receptors.
(A) ANX1 associates with FLS2/BAK1 in transgenic plants. Protein extracts from wild-type, 35S:ANX1-HA, or 35S:ANX1A358V-HA transgenic plants were immunoprecipitated with α-HA antibody (IP:α-HA) and immunoblotted with α-FLS2 (IB:α-FLS2), α-BAK1 (IB:α-BAK1), or α-HA antibody (IB:α-HA) (top three panels). The protein inputs are shown before IP (bottom three panels). Plants were treated with 100 nM flg22 for 15 min.
(B) ANX1 inhibits flg22-induced FLS2-BAK1 interaction. BAK1-FLAG and FLS2-HA were coexpressed in Arabidopsis protoplasts with or without ANX1-GFP. Protein extracts were immunoprecipitated with α-FLAG antibody (IP:α-FLAG) and immunoblotted with α-HA (IB:α-HA) or α-FLAG antibody (IB:α-FLAG) (top two panels). The protein inputs are shown with immunoblotting before immunoprecipitation (bottom three panels). The protoplasts were treated with 100 nM flg22 for 15 min.
(C) ANX1 associates with BIK1 in Arabidopsis protoplasts. ANX1-HA and BIK1-FLAG were coexpressed in protoplasts, and the protein extracts were used for immunoprecipitation and immunoblotting.
(D) ANX1 attenuates RPS2-mediated cell death in N. benthamiana. RPS2-HA without or with ANX1-GFP was expressed in N. benthamiana by Agrobacterium-mediated transient assay. GFP construct was used as a control. Cell death was visualized on the front (top panel) of leaves or under UV light with the ChemiDoc Imaging System (bottom panel) 48 h after infiltration. The infiltrated areas are labeled with black lines.
(E) The protein expression of RPS2-HA (top) and ANX1-GFP (middle) in N. benthamiana. The experiments were performed as in (D), and the samples were collected 18 h after infiltration for protein expression.
(F) and (H) ANX1 associates with RPS2, RPM1 and RIPK in N. benthamiana. ANX1-GFP was coexpressed with RPS2-HA (F), RPM1-HA (G), or RIPK-HA (H) in N. benthamiana. The samples were collected 18 h for RPS2-HA and 48 h for RPM1-HA and RIPK-HA after infiltration for co-IP assays. Note that coexpression of ANX1-GFP reduced RPS2-HA protein level (third panel in [F]).
The above co-IP experiments were repeated three times and cell death assay was repeated five times. The representative results are shown.
Ectopic expression of RPS2 in N. benthamiana induces cell death (Day et al., 2005). In line with the negative role of ANX1 in regulating RPS2-mediated disease resistance (Figure 2), we observed that coexpression of ANX1 with RPS2 attenuated RPS2-mediated cell death in N. benthamiana (Figure 8D). Interestingly, we observed a reduced RPS2 protein level when it was coexpressed with ANX1 in N. benthamiana (first panel in Figure 8E and third panel in Figure 8F), suggesting that ANX1 may affect the RPS2 protein stability. However, the ANX1A358V mutant reduced the ability to attenuate RPS2-mediated cell death in N. benthamiana (Supplemental Figure 7C). In addition, the co-IP assays indicate that ANX1 associated with RPS2 in N. benthamiana (Figure 8F) and in Arabidopsis protoplasts (Supplemental Figure 7D). We also found that ANX1 associated with RPM1 (Figure 8G) and RIPK (Figure 8H; Supplemental Figure 7E). The ANX1A358V mutant also did not affect its association with RPS2 or RIPK (Supplemental Figures 7D and 7E). In addition, the aggie101 mutant or ANX1 overexpression plants did not affect Pst avrRpm1-induced RIN4 phosphorylation (Supplemental Figure 7F). Taken together, the data indicate that ANX1 negatively regulates two-tiered plant immunity by association with both PRR and NLR immune receptor complexes.
The aggie101 Mutation Does Not Affect ANX1 Function in Pollen Tube Growth
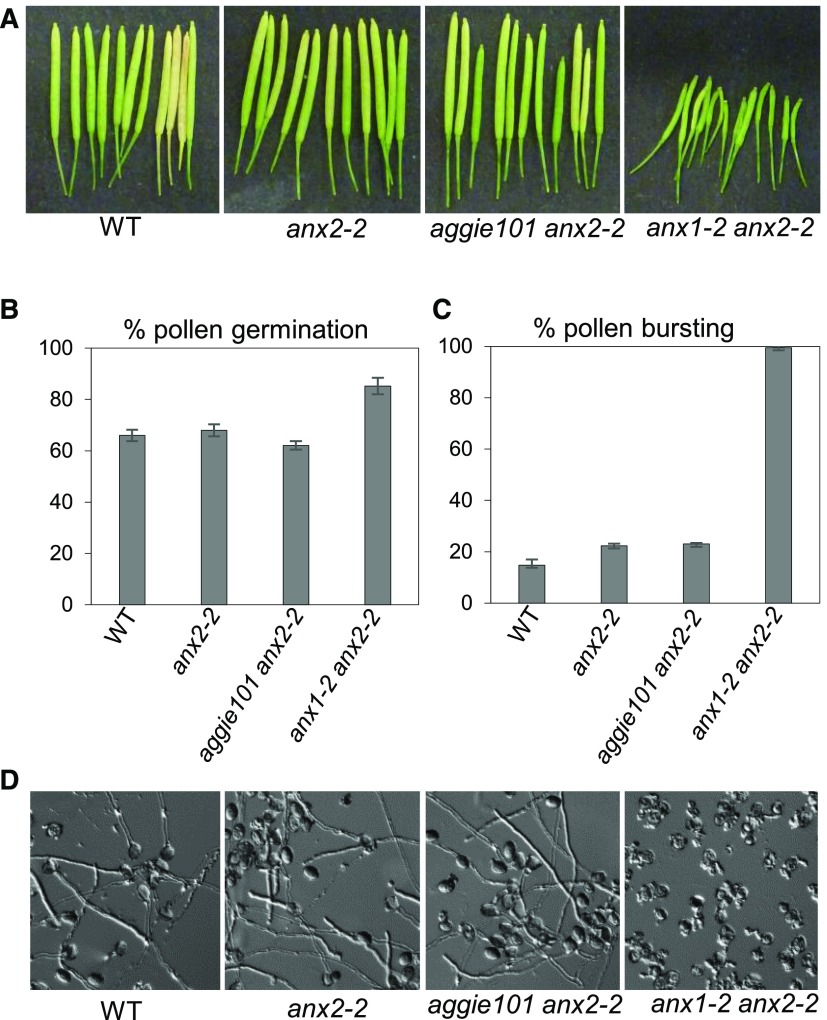
ANX1 and ANX2 function redundantly in controlling cell wall integrity during pollen tube growth with pollen of the anx1-2 anx2-2 double mutant, but not the respective single mutants, bursting precociously after germination. Consequently, anx1-2 anx2-2 plants are male sterile and produce very short siliques with few seeds (Boisson-Dernier et al., 2009; Miyazaki et al., 2009). To test if the anx1A358V mutation in the aggie101 mutant also affects pollen tube growth, we crossed aggie101 with anx2-2 to obtain the aggie101 anx2-2 double mutant. Interestingly, unlike anx1-2 anx2-2, the aggie101 anx2-2 double mutant produced wild-type-looking siliques (Figure 9A) and its pollen germinated and did not burst more than the single anx2-2 mutant or wild-type plants (Figures 9B to 9D). Apparently, the A358V mutation in ANX1 specifically affects its role in immunity but not its function in regulating pollen tube growth. Thus, the dual functions of ANX1 in immunity and pollen tube growth can be uncoupled.
Figure 9.
The aggie101 Mutation Does Not Impair ANX1 Function during Pollen Tube Growth.
(A) Siliques of the wild type, anx2-2, aggie101 anx2-2, and anx1-2 anx2-2. Note that aggie101 anx2-2 is fertile with long siliques, unlike anx1-2 anx2-2.
(B) Percentage of pollen germination for wild-type, anx2-2, aggie101 anx2-2, and anx1-2 anx2-2 plants.
(C) Percentage of pollen bursting for wild-type, anx2-2, aggie101 anx2-2, and anx1-2 anx2-2 plants.
(D) Representative images of in vitro pollen tube growth assays for wild-type, anx2-2, aggie101 anx2-2, and anx1-2 anx2-2 plants. Note that pollen grains of anx1-2 anx2-2 plants, but not the wild type, anx2-2, or aggie101 anx2-2, systematically burst, releasing their cytoplasmic content in the media, and are unable to produce intact tubes.
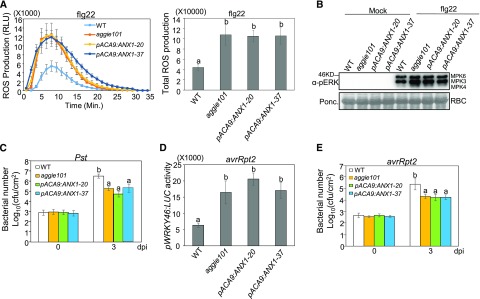
In addition, we transformed the aggie101 mutant with ANX1 under the control of the ACA9 promoter (pACA9:ANX1), which drives strong and specific gene expression in pollen (Schiøtt et al., 2004). We examined the PTI and ETI responses in two homozygous lines (pACA9:ANX1-20 and pACA9:ANX1-37). Both lines behaved similarly to the aggie101 mutant for flg22-induced ROS production (Figure 10A) and MAPK activation (Figure 10B). They were also more resistant to Pst infections as the aggie101 mutant than wild-type control plants (Figure 10C). These data indicate that expression of ANX1 in pollen, while sufficient for complementing the anx1anx2 pollen bursting phenotype (Boisson-Dernier et al., 2013), is not sufficient to complement the aggie101 defects in plant PTI responses. Furthermore, similar to the aggie101 mutant, both lines showed enhanced WRKY46 promoter activity (Figure 10D) and disease resistance (Figure 10E) to Pst avrRpt2 infections, indicating that expression of ANX1 in pollen cannot complement the aggie101 defects in plant ETI responses. Taken together, our data indicate that the dual functions of ANXs in pollen tube growth and plant immunity are largely independent.
Figure 10.
Expression of ANX1 under the Control of the Pollen-Preferential Promoter pACA9 (pACA9:ANX1) in aggie101 Did Not Restore Its Immunity-Related Defects.
(A) The pACA9:ANX1 transgenic lines did not complement aggie101 for the elevated ROS production in response to flg22 treatment. The ROS production at different time points is shown on the left and total photon count is shown on the right. Leaf discs of 4-week-old plants were treated with 100 nM flg22. The data are shown as means ± se (n = 16).
(B) The pACA9:ANX1 transgenic lines did not complement aggie101 for the elevated MAPK activation in response to flg22 treatment. Four-week-old-plants were infiltrated with water or 100 nM flg22 for 15 min. The MAPK activation was detected with an α-pERK antibody (top), and Rubisco (RBC) was stained with Ponceau for protein loading control (bottom).
(C) The pACA9:ANX1 transgenic lines did not complement aggie101 for enhanced resistance to Pst. Four-week-old plants were infiltrated with Pst at OD600 = 5 × 10−4 and the samples were collected at 0 and 3 dpi. The data are shown as means ± sd (n = 3).
(D) The pACA9:ANX1 transgenic lines did not complement aggie101 for enhanced pWRKY46:LUC activity to Pst avrRpt2. Four-week-old plants were infiltrated with Pst avrRpt2 at OD600 = 0.01, and the samples were collected at 6 hpi. The data are shown as means ± se (n = 8 to ∼12).
(E) The pACA9:ANX1 transgenic lines did not complement aggie101 for enhanced resistance to Pst avrRpt2. Four-week-old plants were infiltrated with Pst avrRpt2 at OD600 = 5 × 10−4 and the samples were collected at 0 and 3 dpi. The data are shown as means ± sd (n = 3).
The above experiments were repeated two times with similar results. The different letters indicate statistically significant difference analyzed with one-way ANOVA followed by Tukey’s test (P < 0.05).
DISCUSSION
Plasma membrane-resident malectin-like domain-containing RLKs have long been known to be key regulators in various developmental processes, including cell elongation, polarized growth, and fertilization in plants (Li et al., 2016; Nissen et al., 2016). The malectin-like domain-containing RLKs ANX1 and ANX2 play redundant roles in cell wall integrity during pollen tube growth (Boisson-Dernier et al., 2009, 2013; Miyazaki et al., 2009). In this study, we show that ANX1 and ANX2 are important regulators in plant immunity. The aggie101 mutant, which carries a mutation in the second malectin domain of ANX1, displayed enhanced defense gene activation and disease resistance in response to the P. syringae effectors AvrRpt2 and AvrRpm1 (Figure 2) and increased MAPK activation, ROS production, and immune gene induction in response to flg22, elf18, and Pep1 (Figure 4). These immunity-related phenotypes were also observed in the T-DNA insertional mutants of ANX1 and ANX2 (Figure 6). In addition, transient expression of ANX1 attenuated RPS2-mediated cell death in N. benthamiana (Figure 8D). Thus, ANX1 and ANX2 negatively regulate both PTI and ETI responses in plants. Interestingly, unlike the anx1-2 anx2-2 double mutant derived from T-DNA insertions, the aggie101 anx2-2 double mutant exhibits normal pollen germination and bursting, suggesting that the mutation in aggie101 did not affect ANX1 function during pollen tube growth (Figure 9). The data point to the uncoupled functions of ANXs in plant immunity and sexual reproduction. More importantly, ANX1 associates with two tiers of plant immune receptors: intracellular, but plasma membrane-anchored NLR proteins, RPS2 and RPM1, and cell surface-resident PRR proteins, FLS2 and its coreceptor BAK1 (Figure 8). Remarkably, flg22 treatment induced ANX1 association with BAK1 but not with FLS2 (Figure 8A), and ANX1 reduced flg22-induced FLS2-BAK1 association (Figure 8B). BAK1 is a shared coreceptor of multiple PRRs (Ma et al., 2016). It is likely that the ligand-induced ANX1-BAK1 interaction interferes with ligand-induced BAK1 dimerization with PRRs, thus negatively regulating plant PTI.
FER, the closest homolog of ANX1 and ANX2, regulates root growth through recognition of the secreted peptide ligand, RAPID ALKALIZATION FACTOR1 (RALF1) (Haruta et al., 2014). FER is also involved in plant resistance to biotrophic powdery mildew fungus (Kessler et al., 2010) and bacterial MAMP-, flg22-, and elf18-triggered PTI responses (Keinath et al., 2010; Stegmann et al., 2017). The fer mutant is more resistant to powdery mildew infections, suggesting a negative role of FER in the response to this biotrophic fungus (Kessler et al., 2010). FER was enriched in the detergent-resistant membrane fraction upon flg22 stimulation and the fer mutant had enhanced flg22-induced ROS production and MAPK activation (Keinath et al., 2010). A recent study indicates that FER is required for flg22-, elf18-, and chitin-triggered ROS production and positively regulates plant immunity to bacterial pathogens (Stegmann et al., 2017). Arabidopsis RALF23, a close homolog of RALF1, inhibits plant immunity via direct binding to FER, which otherwise promotes ligand-induced PRR-BAK1 complex formation (Stegmann et al., 2017). Intriguingly, both FER and ANX1 associate with FLS2 and BAK1, and in both cases, flg22 treatment increased FER and ANX1 specific association with BAK1. However, FER and ANX1 appear to behave mechanistically differently within the FLS2-BAK1 complex since FER enhances the flg22-induced FLS2-BAK1 complex formation, thereby promoting plant PTI responses, whereas ANX1 negatively regulates flg22-induced FLS2-BAK1 complex formation and plant PTI responses. It would be interesting to test in the future if FER and ANX1 compete with each other for their association with the FLS2-BAK1 complex to either enhance or dampen PTI responses and if this FER-ANX1 balance is under the control of different RALF peptides. The glycosylphosphatidylinositol-anchored protein LLG1, the chaperon of FER (Li et al., 2015), also associates with FLS2-BAK1 complex and modulates plant PTI responses (Shen et al., 2017). Finally, it is noteworthy that a RALF homolog from the fungus Fusarium oxysporum is an essential pathogenicity factor, the function of which depends on Arabidopsis FER (Masachis et al., 2016).
We have shown that ANX1A358V mutation did not affect its function in pollen tube growth (Figure 9), and expression of ANX1 under the control of a pollen-preferential promoter pACA9 in aggie101 did not restore its immunity-related defects (Figure 10). Similarly, the llg1-3 mutant, which has a defect in plant PTI responses, did not affect FER-dependent growth and development (Shen et al., 2017). The mutation of ANX1A358V in aggie101 lies in the second malectin domain of ANX1, which might be involved in ligand binding. It is possible that ANX1 perceives different ligands in regulating pollen tube growth and immunity. While RALF1 and RALF23 have been shown to bind FER ectodomain (Haruta et al., 2014; Stegmann et al., 2017), no ligand has yet been reported for ANX1. Interestingly, ANX1A358V did not affect its association with PRR and NLR complexes (Figure 8A; Supplemental Figures 7D and 7E). Similarly, LLG1G114R, the mutation in llg1-3, still normally interacted with FLS2 and EFR (Shen et al., 2017). Notably, ANX1A358 is conserved in ANX2, but the corresponding residue in FER is valine, the mutation in aggie101 (Supplemental Figure 3), which may partially explain the opposite function of FER and ANXs in PTI responses.
Besides the CrRLK1L subfamily, some LRR1 group LRR-RLKs also contain malectin-like domain followed by a short stretch of the LRR domain, which are named malectin-like/LRR-RLKs (Hok et al., 2011). Arabidopsis IMPAIRED OOMYCETE SUSCEPTIBILITY1 (IOS1), a malectin-like/LRR-RLK, is highly induced by oomycete downy mildew pathogen, Hyaloperonospora arabidopsidis, but negatively regulates resistance to H. arabidopsidis (Hok et al., 2011). IOS1 plays a positive role in Arabidopsis resistance to the bacterial pathogen P. syringae (Yeh et al., 2016). The ios1 mutants showed reduced responses to MAMPs, whereas IOS1 overexpression plants showed enhanced responses to MAMPs, including flg22 and elf18. IOS1 constitutively interacts with FLS2, EFR, and BAK1, likely promoting/stabilizing PRR-BAK1 complex formation (Yeh et al., 2016). Arabidopsis BAK1-INTERACTING RLK2 (BIR2) negatively regulates PTI responses and resistance to P. syringae (Halter et al., 2014). BIR2 constitutively interacts with BAK1 and flg22 treatment reduced BIR2-BAK1 association. Thus, BIR2 likely sequesters BAK1 away from FLS2 in the resting state (Halter et al., 2014). In contrast, we observed that flg22 treatment induced ANX1-BAK1 association (Figure 8A), thereby blocking ligand-induced FLS2-BAK1 complex formation. Our result suggests a novel regulation of the BAK1-associated PRR complexes by ANXs in the active state.
We have shown that ANX1 also negatively regulates ETI responses and complexes with plasma membrane-localized NLR protein complexes, including RPS2, RPM1, and RIPK (Figures 8F to 8 H). In line with our observations, a recent study showed that FER interacts with RIPK and RALF1 treatment promotes transphosphorylation of FER and RIPK in regulating root growth (Du et al., 2016). It remains unknown whether FER-RIPK association is involved in ETI. We further observed that coexpression of ANX1 attenuated RPS2-mediated cell death (Figure 8D) and reduced RPS2 protein level (Figures 8E and 8F). The stability of several NLR proteins, including RPS2, is regulated by SKP1-CULLIN1-F-box (SCF) complex-mediated proteasome degradation pathway (Cheng et al., 2011; Gou et al., 2012). The F-box protein CPR1 interacts with RPS2 in vivo (Cheng et al., 2011). It will be interesting to determine whether ANX1, a malectin domain-containing RLK, controls NLR RPS2 protein stability through SCFCPR1 complex. Notably, we did not observe the obvious effect of ANX1 expression on RPM1 protein level (Figure 8G), suggesting that ANX1 regulates RPM1-mediated immunity through a distinct mechanism.
Our observation that ANX1 complexes with FLS2/BAK1 and RPS2/RPM1 is in line with a previous report showing that FLS2 was found in the same complex with RPM1, RPS2, and RIN4 (Qi et al., 2011). In addition, Arabidopsis COMPROMISED RECOGNITION OF TCV1, an ATPase that associates with multiple NLR proteins and PRR FLS2, positively regulates both plant PTI and ETI (Kang et al., 2008, 2012). Thus, accumulating evidence suggests the interaction between PRR and NLR immune receptors. Our data further point to ANX1 and likely ANX2 functioning as molecular links of PRR and NLR complexes and independently regulating outputs of PTI and ETI.
METHODS
Plant Materials and Growth Condition
The Arabidopsis thaliana plants were grown on soil (Metro-Mix 366; Sungro Horticulture) or 0.5× Murashige and Skoog (MS) medium in a growth chamber under 50 to 60% relative humidity and 75 µE m−2 s−1 light with 12-h-light (23°C)/12-h-dark (22°C) cycle. Philips F40T12/DX cool white fluorescent bulbs were used for plant growth. The anx1-2 (Salk_0456870) and anx2-2 (Salk_133057) mutants in the Col-0 background were obtained from the ABRC. The anx1-2 anx2-2 double mutant was reported previously (Boisson-Dernier et al., 2009).
Generation of pWRKY46:LUC Transgenic Plants and Mutant Screens
The pWRKY46:LUC construct in a protoplast transient expression vector (Gao et al., 2013) was subcloned into the binary vector pCB302 and introduced into Arabidopsis Col-0 plants. The transgenic plants were selected for Basta resistance and analyzed with Pst avrRpt2-induced pWRKY46:LUC expression. The seeds of homozygous pWRKY46:LUC transgenic plants were mutagenized with 0.4% EMS. Approximately 6000 M2 plants were grown on soil for 4 weeks and inoculated with Pst avrRpt2 at 107 colony-forming units (cfu)/mL in 10 mM MgCl2. The inoculated leaves were collected 6 h after inoculation and placed into each well of a 96-well plate. The plate was sprayed with 0.2 mM luciferin and put in the dark for 20 min. The bioluminescence signal was read by a luminometer (Perkin-Elmer 2030 Multilabel Reader, Victor X3). Putative mutants were selected and confirmed in the M3 and M4 generations.
Map-Based Cloning and Next-Generation Sequencing
The aggie101 mutant was crossed with Ler accession, and an F2 population was used for map-based cloning. The initial mapping placed aggie101 on Chromosome 3 based on a bulked segregation analysis with a pool of 59 plants displaying aggie101 mutant phenotype with INDEL markers between Col-0 and Ler. Further analysis with 283 individual F2 plants displaying aggie101 mutant phenotype placed aggie101 mutation between markers HG18 and HG19 that are 49 kb apart. The genomic DNA of aggie101 was isolated for 125-nucleotide single-end sequencing on an Illumina HiSeq 2000 platform at Texas AgriLife Genomics and Bioinformatics Service. Forty-fold genome coverage was obtained. Illumina reads were mapped to the TAIR10 release of the Col-0 genome using CLC Genomics Workbench 6.0.1 software (http://www.clcbio.com). The subsequent quality-based variant detection was performed to identify the pattern of genome-wide single nucleotide polymorphisms. The candidate variants between HG18 and HG19 were selected, and a C-to-T mutation in the position of 1073 nucleotides of At3g04690 was identified and further confirmed with Sanger sequencing.
Genotyping of aggie101 Mutation and pWRKY46:LUC Transgene
To reveal the single nucleotide polymorphism present in aggie101, cleaved amplified polymorphic sequence primers were designated to include MaeII restriction site in aggie101. ANX1 genomic DNA from 42 bp upstream and 148 bp downstream of mutation site was amplified by PCR with forward primer (5′-GGTGGACAGGAGAGAAAGGA-3′) and reverse primer (5′-GTTTGGACCCGCAAGATTT-3′). The PCR products were incubated with 0.1 units/μL MaeII (Thermo Fisher Scientific) at 65°C for 1 h and analyzed in 4% agarose gels with wild-type PCR fragment of 191 bp and aggie101 fragments of 152 and 43 bp.
To genotype pWRKY46:LUC transgene, the specific primers from 643 bp upstream and 313 bp downstream of pWRKY46:LUC insertion site were designed (forward primer: 5′-ATCCATCGCAGCAATAACGG-3′ and reverse primer: 5′-CTCGTCAAAGCTCGGGATTG-3′). The T-DNA left border primer is 5′-CTAAGCGTCAATTTGTTTACACCAC-3′. The PCR products were analyzed in 1.5% agarose gels.
Generation of Constructs and Transgenic Plants
The construct of BIK1 in the plant expression vector (pHBT) were reported previously (Lu et al., 2010). The pACA9:ANX1 construct was reported (Boisson-Dernier et al., 2013). To generate the pHBT-35S:ANX1-HA and pHBT-35S:ANX1-GFP constructs, the ANX1 genomic DNA was amplified from the wild type with primers containing the BamHI or StuI site and cloned into the pHBT vector with an HA epitope tag at the C terminus. After confirmation with Sanger sequencing, ANX1 fragment was transferred into the pHBT vectors with a GFP epitope tag or the binary vector pCAMBIA2300 with an HA or GFP epitope tag. pCAMBIA2300-35S:ANX1 was transformed into the wild type or aggie101 with Agrobacterium tumefaciens-mediated floral dipping transformation. The transgenic plants were screened by germination on 0.5× MS medium containing 50 µg/mL kanamycin. For RIPK cloning, genomic DNA was amplified from wild-type plants with primers containing the BamHI or StuI site, cloned into the pHBT vector, and subcloned in the pCAMBIA2300 vector with an HA tag at the C terminus. For RPS2 and RPM1 cloning, cDNA was amplified from wild-type plants with primers containing the KpnI/NcoI or SmaI site, cloned into the pHBT vector, and subcloned in the pCB302 vector with an HA tag at the C terminus. The primers for cloning are listed in Supplemental Table 1.
Pathogen Infection Assays
Pseudomonas syringae pv tomato DC3000 (Pst), P. syringae pv maculicola ES4326 (Psm), and Pst carrying avrRpt2 or avrRpm1 were grown overnight at 28°C in King’s B medium with appropriate antibiotics. Bacteria were collected, washed, and diluted to the desired density with water. For infection assays, the leaves from 4-week-old plants were hand-infiltrated with different bacteria at a concentration of 5 × 104 cfu/mL using a needleless syringe. Bacterial counting was performed from six leaves of different plants as three replicates at 0 and 3 dpi. Two leaf discs were ground in 100 μL water and serial dilutions were plated on tryptic soy agar medium with appropriate antibiotics. Bacterial colony forming units (cfu) were counted 2 d after incubation at 28°C. Each data point is shown as triplicates. The disease symptom was recorded from the representative infected leaves at the indicated time points. Culture of Botrytis cinerea strain BO5-10 and its infection on Arabidopsis were performed as described previously (Lin et al., 2014).
RT-PCR and RT-qPCR Analyses
Total RNA was isolated from seedlings grown on 0.5× MS plates or leaves of soil-grown plants with TRIzol reagent (Invitrogen). RNA was reverse transcribed to synthesize first-strand cDNA with M-MuLV reverse transcriptase and oligo(dT) primer following RNase-free DNase I (New England Biolabs) treatment. RT-PCR analyses were performed using Taq DNA polymerase. Fragments of target genes were amplified using the primers listed in Supplemental Table 1. UBQ1 was used as an internal control. RT-qPCR analyses were performed using iTaq SYBR green Supermix (Bio-Rad) supplemented with ROX in an ABI GeneAmp PCR System 9700. The expression of tested genes was normalized to the expression of UBQ10.
Co-IP Assay
The HA, FLAG, or GFP epitope-tagged pair of plasmids was expressed in 1 mL of Arabidopsis Col-0 protoplasts (2 × 105/mL) for 8 h and then applied with water control or 100 nM flg22 for 15 min. The FLAG tagged proteins were immunoprecipitated with 5 μL of α-FLAG agarose beads (Sigma-Aldrich) in 250 μL co-IP buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.5, 5 mM EDTA, 1% Triton X-100, 2 mM Na3VO4, 2 mM NaF, 1 mM DTT, and 1:200 complete protease inhibitor cocktail from Sigma-Aldrich). A small aliquot of samples (20 µL) in co-IP buffer was used for input control before adding α-FLAG agarose beads. The co-IP samples were incubated for 3 h at 4°C. The beads were collected and washed three times with washing buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.5, 5 mM EDTA, and 0.1% Triton) and once with 50 mM Tris-HCl, pH 7.5. The samples were analyzed by immunoblot with an appropriate antibody.
The binary vectors were transformed into Agrobacterium strain GV3101. Agrobacterium-mediated transient expression in Nicotiana benthamiana was performed as described with some modifications (Meng et al., 2015). Briefly, Agrobacterium VG3101 (OD600 = 1) carrying different vectors was syringe-infiltrated into 5-week-old N. benthamiana leaves. N. benthamiana leaves were collected at 36 hpi for co-IP. The co-IP and immunoblot were performed as above-described for protoplasts. The co-IP for seedlings of transgenic plants was performed as described (Lin et al., 2014). Briefly, Arabidopsis seedlings grown on 0.5× MS were ground in liquid nitrogen. The extract was added co-IP buffer, kept on ice for 15 min, and centrifuged at 14,500g for 10 min at 4°C three times to get rid of debris. The supernatants were incubated with α-HA antibody for 2 h at 4°C and then protein G-agarose beads (Roche) with gentle shaking for another 3 h at 4°C. The washing and immunoblot were performed as described above for protoplasts.
Cell Death Assay in N. benthamiana
Agrobacterium-mediated cell death assay in N. benthamiana was performed as described previously (de Oliveira et al., 2016). Briefly, Agrobacterium GV3101 harboring the desired plasmids was suspended in solution containing 10 mM MgCl2, 10 mM MES, and 200 µM acetosyringone to an OD600 = 0.75. The culture was kept in the dark for 3 h at room temperature. Then, a 1:1 (v/v) mixture of agrobacterial culture with different constructs was hand-infiltrated into 4- to 5-week-old N. benthamiana leaves. N. benthamiana leaves were harvested at the indicated time for detecting protein expression by immunoblot with α-HA or α-FLAG antibody. The cell death was monitored over 48 h. Detached leaves were exposed under UV to visualize phenolic compound using Molecular Imager Gel Doc XR+ (Bio-Rad) and photographed at 48 hpi. At least three individual leaves were included for each combination in each repeat.
MAPK Assay
Leaves of 4-week-old soil-grown plants or 2-week-old seedlings were inoculated with water control, 100 nM flg22, 100 nM elf18, or 100 nM Pep1 for the indicated times. Samples were grounded in 40 μL of extraction buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.5, 5 mM EDTA, 1% Triton X-100, 2 mM Na3VO4, 2 mM NaF, 1 mM DTT, and 1:200 complete protease inhibitor cocktail from Sigma-Aldrich). Supernatant was collected after 12,000 rpm centrifugation for 5 min at 4°C and protein samples with 1× SDS buffer were loaded on 10% SDS-PAGE gel to detect pMPK3, pMPK6, and pMPK4 by immunoblot with α-pERK1/2 antibody (Cell Signaling; no. 9101).
ROS Assay
At least four leaves from 5-week-old plants were excised into 36 leaf discs of 0.25 cm2, following an overnight incubation in a 96-well plate with 200 μL of water to eliminate the wounding effect. Water was replaced by 100 μL of reaction solution containing 50 μM of luminol and 10 μg/mL of horseradish peroxidase (Sigma-Aldrich) supplemented with 100 nM flg22, elf18, or Pep1. The measurement was conducted immediately after adding the solution with a luminometer (Perkin-Elmer 2030 Multilabel Reader, Victor X3) with a 1-min interval reading time over a period of 30 min. The measured value for ROS production from 36 leaf discs per treatment was indicated as means of relative light units.
Callose Deposition
Callose deposition was assayed as described (Lu et al., 2011), with modifications. Briefly, two to three leaves of 5-week-old wild-type and aggie101 mutant plants were infiltrated with 1 µM of flg22 or water, and leaves were detached 12 h after infiltration. The detached leaves were merged in alcoholic lactophenol (1 volume of phenol:glycerol:lactic acid:water [1:1:1:1] and 2 volumes of ethanol) overnight. Samples were sequentially rinsed with 95%, 50% ethanol, and water and then cleared leaves were stained with 0.01% aniline blue in 0.15 M phosphate buffer (pH 9.5), and the deposition of callose was observed with a fluorescence microscope equipped with UV filter. The number of deposits was counted using ImageJ 1.49v software (http://imagej.nih.gov/ij/).
Statistical Analysis
The statistical analysis was performed with R Studio software (RStudio) one-way ANOVA followed by Tukey test or Student’s t tests for significant differences. Different samples and biological repeats were obtained from different plants.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ANX1 (AT3G04690), ANX2 (AT5G28680), FLS2 (AT5G46330), BAK1 (AT4G33430), BIK1 (AT2G39660), RPS2 (AT4G26090), RPM1 (AT3G07040), RIPK (AT2G05940), RIN4 (AT2G04410), WRKY46 (AT2G46400), and FRK1 (AT2G19190).
Supplemental Data
Supplemental Figure 1. The aggie101 mutant displays enhanced PTI responses.
Supplemental Figure 2. Map-based cloning of aggie101 and scheme of ANX1.
Supplemental Figure 3. The A358 residue of ANX1 is conserved in ANX2.
Supplemental Figure 4. Analysis of ANX1 VIGS plants and complementation lines.
Supplemental Figure 5. Overexpression of ANX1 suppresses defense responses.
Supplemental Figure 6. ANX1 and ANX2 function in plant immunity and identification of aggie101 anx2-2 mutant.
Supplemental Figure 7. ANX1 associates with PRRs and NLRs.
Supplemental Table 1. Primers used in this study.
Supplemental File 1. ANOVA tables.
Acknowledgments
We thank the ABRC for the Arabidopsis T-DNA insertional lines, Antje Heese for α-FLS2 antibody, Jenny Russinova for α-BAK1 antibody, Gitta Coaker for α-RIN4 antibody, and members of the laboratories of L.S. and P.H. for the comments of the experiments. The work was supported by National Science Foundation (IOS-1252539) and the National Institutes of Health (NIH; R01GM092893) to P.H., by the NIH (1R01GM097247) and the Robert A. Welch Foundation (A-1795) to L.S., and by the Deutsche Forschungsgemeinschaft (BO 4470/1-1) to A.B.-D. Z.H. and G.X. were partially supported by China Scholarship Council, and H.M. was partially supported by the Institute for Basic Science, Republic of Korea (IBS-R013-G2).
AUTHOR CONTRIBUTIONS
H.M., A.B.-D., L.S., and P.H. designed the research. H.M., B.F., Z.H., A.B.-D, C.M.F., X.M., Y.H., J.Z., G.X., and T.W. performed research and analyzed data. H.M., L.S., and P.H. wrote the article with input from the other authors.
References
- Axtell M.J., Staskawicz B.J. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112: 369–377. [DOI] [PubMed] [Google Scholar]
- Böhm H., Albert I., Fan L., Reinhard A., Nürnberger T. (2014). Immune receptor complexes at the plant cell surface. Curr. Opin. Plant Biol. 20: 47–54. [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A., Kessler S.A., Grossniklaus U. (2011). The walls have ears: the role of plant CrRLK1Ls in sensing and transducing extracellular signals. J. Exp. Bot. 62: 1581–1591. [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A., Lituiev D.S., Nestorova A., Franck C.M., Thirugnanarajah S., Grossniklaus U. (2013). ANXUR receptor-like kinases coordinate cell wall integrity with growth at the pollen tube tip via NADPH oxidases. PLoS Biol. 11: e1001719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier A., Roy S., Kritsas K., Grobei M.A., Jaciubek M., Schroeder J.I., Grossniklaus U. (2009). Disruption of the pollen-expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development 136: 3279–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406. [DOI] [PubMed] [Google Scholar]
- Cao Y., Liang Y., Tanaka K., Nguyen C.T., Jedrzejczak R.P., Joachimiak A., Stacey G. (2014). The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 3: 03766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., et al. (2016). FERONIA interacts with ABI2-type phosphatases to facilitate signaling cross-talk between abscisic acid and RALF peptide in Arabidopsis. Proc. Natl. Acad. Sci. USA 113: E5519–E5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y.T., Li Y., Huang S., Huang Y., Dong X., Zhang Y., Li X. (2011). Stability of plant immune-receptor resistance proteins is controlled by SKP1-CULLIN1-F-box (SCF)-mediated protein degradation. Proc. Natl. Acad. Sci. USA 108: 14694–14699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J.D.G., Felix G., Boller T. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500. [DOI] [PubMed] [Google Scholar]
- Choi J., Tanaka K., Cao Y., Qi Y., Qiu J., Liang Y., Lee S.Y., Stacey G. (2014). Identification of a plant receptor for extracellular ATP. Science 343: 290–294. [DOI] [PubMed] [Google Scholar]
- Chung E.H., da Cunha L., Wu A.J., Gao Z., Cherkis K., Afzal A.J., Mackey D., Dangl J.L. (2011). Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe 9: 125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto D., Zipfel C. (2016). Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16: 537–552. [DOI] [PubMed] [Google Scholar]
- Day B., Dahlbeck D., Huang J., Chisholm S.T., Li D., Staskawicz B.J. (2005). Molecular basis for the RIN4 negative regulation of RPS2 disease resistance. Plant Cell 17: 1292–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira M.V.V., et al. (2016). Specific control of Arabidopsis BAK1/SERK4-regulated cell death by protein glycosylation. Nat. Plants 2: 15218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou D., Zhou J.M. (2012). Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe 12: 484–495. [DOI] [PubMed] [Google Scholar]
- Du C., et al. (2016). Receptor kinase complex transmits RALF peptide signal to inhibit root growth in Arabidopsis. Proc. Natl. Acad. Sci. USA 113: E8326–E8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Q., Kita D., Li C., Cheung A.Y., Wu H.M. (2010). FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc. Natl. Acad. Sci. USA 107: 17821–17826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Restrepo J.M., Huck N., Kessler S., Gagliardini V., Gheyselinck J., Yang W.C., Grossniklaus U. (2007). The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science 317: 656–660. [DOI] [PubMed] [Google Scholar]
- Feng B., Liu C., de Oliveira M.V., Intorne A.C., Li B., Babilonia K., de Souza Filho G.A., Shan L., He P. (2015). Protein poly(ADP-ribosyl)ation regulates Arabidopsis immune gene expression and defense responses. PLoS Genet. 11: e1004936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Chen X., Lin W., Chen S., Lu D., Niu Y., Li L., Cheng C., McCormack M., Sheen J., Shan L., He P. (2013). Bifurcation of Arabidopsis NLR immune signaling via Ca2+-dependent protein kinases. PLoS Pathog. 9: e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., Chung E.H., Eitas T.K., Dangl J.L. (2011). Plant intracellular innate immune receptor Resistance to Pseudomonas syringae pv. maculicola 1 (RPM1) is activated at, and functions on, the plasma membrane. Proc. Natl. Acad. Sci. USA 108: 7619–7624. Erratum. Proc. Natl. Acad. Sci. USA 108: 8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L., Boller T. (2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5: 1003–1011. [DOI] [PubMed] [Google Scholar]
- Gou M., Shi Z., Zhu Y., Bao Z., Wang G., Hua J. (2012). The F-box protein CPR1/CPR30 negatively regulates R protein SNC1 accumulation. Plant J. 69: 411–420. [DOI] [PubMed] [Google Scholar]
- Gust A.A. (2015). Peptidoglycan perception in plants. PLoS Pathog. 11: e1005275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halter T., et al. (2014). The leucine-rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr. Biol. 24: 134–143. [DOI] [PubMed] [Google Scholar]
- Haruta M., Sabat G., Stecker K., Minkoff B.B., Sussman M.R. (2014). A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343: 408–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese A., Hann D.R., Gimenez-Ibanez S., Jones A.M.E., He K., Li J., Schroeder J.I., Peck S.C., Rathjen J.P. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 104: 12217–12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hok S., Danchin E.G., Allasia V., Panabières F., Attard A., Keller H. (2011). An Arabidopsis (malectin-like) leucine-rich repeat receptor-like kinase contributes to downy mildew disease. Plant Cell Environ. 34: 1944–1957. [DOI] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Jones J.D., Vance R.E., Dangl J.L. (2016). Intracellular innate immune surveillance devices in plants and animals. Science 354: aaf6395. [DOI] [PubMed] [Google Scholar]
- Kang H.G., et al. (2012). CRT1 is a nuclear-(t)ranslocated MORC endonuclease that participates in multiple levels of plant immunity. Nat. Commun. 3: 1297. [DOI] [PubMed] [Google Scholar]
- Kang H.G., Kuhl J.C., Kachroo P., Klessig D.F. (2008). CRT1, an Arabidopsis ATPase that interacts with diverse resistance proteins and modulates disease resistance to turnip crinkle virus. Cell Host Microbe 3: 48–57. [DOI] [PubMed] [Google Scholar]
- Keinath N.F., Kierszniowska S., Lorek J., Bourdais G., Kessler S.A., Shimosato-Asano H., Grossniklaus U., Schulze W.X., Robatzek S., Panstruga R. (2010). PAMP (pathogen-associated molecular pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J. Biol. Chem. 285: 39140–39149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler S.A., Shimosato-Asano H., Keinath N.F., Wuest S.E., Ingram G., Panstruga R., Grossniklaus U. (2010). Conserved molecular components for pollen tube reception and fungal invasion. Science 330: 968–971. [DOI] [PubMed] [Google Scholar]
- Li C., Wu H.M., Cheung A.Y. (2016). FERONIA and her pals: Functions and mechanisms. Plant Physiol. 171: 2379–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., et al. (2015). Glycosylphosphatidylinositol-anchored proteins as chaperones and co-receptors for FERONIA receptor kinase signaling in Arabidopsis. eLife 4: 06587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., et al. (2014). Modulation of RNA polymerase II phosphorylation downstream of pathogen perception orchestrates plant immunity. Cell Host Microbe 16: 748–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Li B., Lu D., Chen S., Zhu N., He P., Shan L. (2014). Tyrosine phosphorylation of protein kinase complex BAK1/BIK1 mediates Arabidopsis innate immunity. Proc. Natl. Acad. Sci. USA 111: 3632–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner H., Müller L.M., Boisson-Dernier A., Grossniklaus U. (2012). CrRLK1L receptor-like kinases: not just another brick in the wall. Curr. Opin. Plant Biol. 15: 659–669. [DOI] [PubMed] [Google Scholar]
- Liu J., Elmore J.M., Lin Z.J., Coaker G. (2011). A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 9: 137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Lin W., Gao X., Wu S., Cheng C., Avila J., Heese A., Devarenne T.P., He P., Shan L. (2011). Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Wu S., Gao X., Zhang Y., Shan L., He P. (2010). A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 107: 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Xu G., He P., Shan L. (2016). SERKing coreceptors for receptors. Trends Plant Sci. 21: 1017–1033. [DOI] [PubMed] [Google Scholar]
- Macho A.P., Zipfel C. (2015). Targeting of plant pattern recognition receptor-triggered immunity by bacterial type-III secretion system effectors. Curr. Opin. Microbiol. 23: 14–22. [DOI] [PubMed] [Google Scholar]
- Mackey D., Belkhadir Y., Alonso J.M., Ecker J.R., Dangl J.L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112: 379–389. [DOI] [PubMed] [Google Scholar]
- Maekawa T., Kufer T.A., Schulze-Lefert P. (2011). NLR functions in plant and animal immune systems: so far and yet so close. Nat. Immunol. 12: 817–826. [DOI] [PubMed] [Google Scholar]
- Masachis S., Segorbe D., Turrà D., Leon-Ruiz M., Fürst U., El Ghalid M., Leonard G., López-Berges M.S., Richards T.A., Felix G., Di Pietro A. (2016). A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nat. Microbiol. 1: 16043. [DOI] [PubMed] [Google Scholar]
- Meng X., Chen X., Mang H., Liu C., Yu X., Gao X., Torii K.U., He P., Shan L. (2015). Differential function of Arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr. Biol. 25: 2361–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki S., Murata T., Sakurai-Ozato N., Kubo M., Demura T., Fukuda H., Hasebe M. (2009). ANXUR1 and 2, sister genes to FERONIA/SIRENE, are male factors for coordinated fertilization. Curr. Biol. 19: 1327–1331. [DOI] [PubMed] [Google Scholar]
- Nissen K.S., Willats W.G., Malinovsky F.G. (2016). Understanding CrRLK1L function: Cell walls and growth control. Trends Plant Sci. 21: 516–527. [DOI] [PubMed] [Google Scholar]
- Qi Y., Tsuda K., Glazebrook J., Katagiri F. (2011). Physical association of pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) immune receptors in Arabidopsis. Mol. Plant Pathol. 12: 702–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranf S., Gisch N., Schäffer M., Illig T., Westphal L., Knirel Y.A., Sánchez-Carballo P.M., Zähringer U., Hückelhoven R., Lee J., Scheel D. (2015). A lectin S-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana. Nat. Immunol. 16: 426–433. [DOI] [PubMed] [Google Scholar]
- Saur I.M., Kadota Y., Sklenar J., Holton N.J., Smakowska E., Belkhadir Y., Zipfel C., Rathjen J.P. (2016). NbCSPR underlies age-dependent immune responses to bacterial cold shock protein in Nicotiana benthamiana. Proc. Natl. Acad. Sci. USA 113: 3389–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schallus T., Jaeckh C., Fehér K., Palma A.S., Liu Y., Simpson J.C., Mackeen M., Stier G., Gibson T.J., Feizi T., Pieler T., Muhle-Goll C. (2008). Malectin: a novel carbohydrate-binding protein of the endoplasmic reticulum and a candidate player in the early steps of protein N-glycosylation. Mol. Biol. Cell 19: 3404–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiøtt M., Romanowsky S.M., Baekgaard L., Jakobsen M.K., Palmgren M.G., Harper J.F. (2004). A plant plasma membrane Ca2+ pump is required for normal pollen tube growth and fertilization. Proc. Natl. Acad. Sci. USA 101: 9502–9507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q., Bourdais G., Pan H., Robatzek S., Tang D. (2017). Arabidopsis glycosylphosphatidylinositol-anchored protein LLG1 associates with and modulates FLS2 to regulate innate immunity. Proc. Natl. Acad. Sci. USA 114: 5749–5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinya T., Nakagawa T., Kaku H., Shibuya N. (2015). Chitin-mediated plant-fungal interactions: catching, hiding and handshaking. Curr. Opin. Plant Biol. 26: 64–71. [DOI] [PubMed] [Google Scholar]
- Stegmann M., Monaghan J., Smakowska-Luzan E., Rovenich H., Lehner A., Holton N., Belkhadir Y., Zipfel C. (2017). The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355: 287–289. [DOI] [PubMed] [Google Scholar]
- Thomma B.P.H.J., Nürnberger T., Joosten M.H.A.J. (2011). Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23: 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh Y.H., Panzeri D., Kadota Y., Huang Y.C., Huang P.Y., Tao C.N., Roux M., Chien H.C., Chin T.C., Chu P.W., Zipfel C., Zimmerli L. (2016). The Arabidopsis malectin-like/LRR-RLK IOS1 is critical for BAK1-dependent and BAK1-independent pattern-triggered immunity. Plant Cell 28: 1701–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Feng B., He P., Shan L. (2017). From chaos to harmony: Responses and signaling upon microbial pattern recognition. Annu. Rev. Phytopathol. 55: 109–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., et al. (2010). Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301. [DOI] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J.D., Boller T., Felix G. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760. [DOI] [PubMed] [Google Scholar]