The EIN3-PIF3 pair functions as a central integrator of multiple signals for the concerted repression of chloroplast development in soil.
Abstract
In buried seedlings, chloroplasts are arrested at the etioplast stage, but they rapidly mature upon emergence of the seedling. Etioplast-chloroplast differentiation is halted through the integration of soil-induced signals, including pressure and the absence of light, although the details on how this information converges to regulate cellular decisions remain unclear. Here, we identify an interdependent transcription module that integrates the mechanical pressure and darkness signals to control chloroplast development in Arabidopsis thaliana. Mutations of ETHYLENE-INSENSITIVE3 (EIN3), the primary transcription factor in the ethylene signaling pathway that is activated in response to mechanical pressure, cause early development of etioplasts in the dark and severe photobleaching upon light exposure. Genetic studies demonstrate that repression of etioplast differentiation by EIN3 requires PHYTOCHROME INTERACTING FACTOR3 (PIF3), a darkness-stabilized bHLH transcription factor. EIN3 and PIF3 directly interact and form an interdependent module to repress the expression of most LIGHT HARVESTING COMPLEX (LHC) genes; overexpressing even one LHC could cause premature development of etioplasts. The EIN3-PIF3 transcription module synergistically halts chloroplast development by interdependently co-occupying the promoters of LHC genes. Thus, our results define a transcriptional regulatory module and provide mechanistic insight on the concerted regulation of chloroplast development by multiple soil-induced signals.
INTRODUCTION
In nature, plant seeds are often covered by dead leaves or soil. Seedlings grown in subterranean darkness are programmed with a developmental pattern termed skotomorphogenesis, which features an elongated hypocotyl with closed and yellowish cotyledons (Wei et al., 1994; Von Arnim and Deng, 1996). In particular, the chloroplast development of dark-grown seedlings is arrested at the etioplast stage (Solymosi and Schoefs, 2010; Jarvis and López-Juez, 2013). Etioplasts and mature chloroplasts differ in the arrangements of their inner membranes and pigment molecules. The inner membranes of etioplasts are organized into a highly regular, paracrystalline structure, called the prolamellar body (PLB) (Solymosi and Schoefs, 2010; Jarvis and López-Juez, 2013). Upon reaching the soil surface, light initiates the transition of etioplasts to chloroplasts, which is characterized by the dispersal of the PLB and the formation of thylakoid membranes that contain the integral membrane proteins of the photosystems for photosynthesis (Solymosi and Schoefs, 2010; Pogson and Albrecht, 2011; Jarvis and López-Juez, 2013). The transition from etioplast to chloroplast is a key event in the deetiolation process and a point of particular vulnerability for plant survival. Upon light exposure, the chlorophyll precursor protochlorophyllide (Pchlide) in etioplasts can be transformed to molecular oxygen and generate reactive oxygen species (ROS), resulting in photooxidative damage and even photobleaching death to the seedlings (Huq et al., 2004; Moon et al., 2008; Reinbothe et al., 2010; Liu et al., 2017). Previous studies have shown that the lattice-like membranous structures of PLBs enable the activation of a unique photoprotective mechanism that involves the formation of a strong energy quencher to reduce ROS production (Green and Durnford, 1996; Schoefs and Franck, 2003; Solymosi and Schoefs, 2010). Therefore, maintaining proper PLB formation in etioplasts is crucial for seedling survival during the soil emergence process.
Light is a dominant environmental factor that activates the differentiation program of etioplasts to chloroplasts (Solymosi and Schoefs, 2010; Pogson and Albrecht, 2011; Jarvis and López-Juez, 2013). Plants have several sets of sensory photoreceptors to monitor the quantity and quality of light. Among these photoreceptors, phytochromes (phys, including five members named phyA through phyE in Arabidopsis thaliana) mediate the responses to the red and far-red regions of the light spectrum (Quail, 2002b; Rockwell et al., 2006). In etiolated seedlings, phys are synthesized as the Pr form in the cytoplasm. Upon light illumination, the Pr form is switched to the active Pfr form and is translocated into the nucleus (Quail, 2002b; Rockwell et al., 2006). Light alters the downstream gene expression by triggering the degradation of a subfamily of bHLH transcription factors, which are directly targeted by the photoactivated phys and named PHYTOCHROME INTERACTING FACTOR (PIF) (Quail, 2002a; Leivar and Quail, 2011). PIF3 plays a central role in maintaining skotomorphogenesis along with its homologs PIF1, PIF4, and PIF5 (Ni et al., 1998; Leivar et al., 2008; Leivar and Monte, 2014). Furthermore, PIF3 has been shown to repress light-induced chloroplast development (Monte et al., 2004; Stephenson et al., 2009). The cotyledons of dark-grown pif3 mutant seedlings contain higher Pchlide levels and partially predeveloped prothylakoid membranes, causing photooxidative damage in the etiolated seedlings upon initial light exposure (Monte et al., 2004; Stephenson et al., 2009; Zhong et al., 2010, 2014).
In addition to the dark environment, buried seedlings also face the challenge of mechanical pressure when pushing against the soil cover. The gaseous plant hormone ethylene is greatly induced in response to the mechanical impedance and accordingly exerts marked effects on seedling morphogenesis (Goeschl et al., 1966; Zhong et al., 2014). In Arabidopsis, ethylene is perceived by a group of five endoplasmic reticulum-localized receptors and the signal is then transduced via a multiple-step cascade to the nucleus (Ecker, 1995; Alonso and Stepanova, 2004; Ju et al., 2012; Qiao et al., 2012; Wen et al., 2012). The regulatory functions of ethylene are mediated by two plant-specific transcription factors, ETHYLENE-INSENSITIVE3 (EIN3) and ETHYLENE-INSENSITIVE3-LIKE1 (EIL1) (Chao et al., 1997), whose protein levels are tightly controlled through the 26S proteasome degradation pathway mediated by two F-box proteins, EIN3 BINDING F-BOX PROTEIN1 and 2 (EBF1/2) (Guo and Ecker, 2003; Potuschak et al., 2003; Gagne et al., 2004). During seedlings emergence from the soil, CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) channels the light fluence changes, and ethylene transduces the mechanical pressure information, cooperatively controlling the protein levels of EIN3 (Shi et al., 2016a). COP1 is an E3 ligase of EBF1/2 (Shi et al., 2016a), and ethylene independently inhibits the actions of EBF1/2 to stabilize EIN3 (An et al., 2010; Li et al., 2015; Merchante et al., 2015). As the essential transcription factor, EIN3 activates the expression of a wide range of downstream genes. Among these genes, ETHYLENE RESPONSE FACTOR (ERF1) is induced to slow hypocotyl elongation (Solano et al., 1998; Zhong et al., 2014), and HOOKLESS1 (HLS1) is activated to enhance apical hook formation (Lehman et al., 1996; Shen et al., 2016). Moreover, EIN3 inhibits Pchlide biosynthesis and directly activates the gene expression of both PROTOCHLOROPHYLLIDE OXIDOREDUCTASE A (PORA) and PORB in the cotyledon (Zhong et al., 2009, 2014). Therefore, EIN3 is also critical for maintaining proper Pchlide and PORA/PORB enzyme levels to avoid lethal photooxidative damage in deetiolation. Importantly, the photoreceptor phyB has been recently found to directly target EIN3 in a light-dependent manner and enhance the interactions between EIN3 and EBF1/2 to stimulate EIN3 degradation upon light (Shi et al., 2016b). Taken together, EIN3 emerges as a key integrator of environmental and internal signals that adaptively controls seedling growth and development to facilitate seedling soil emergence.
In this study, we focus on the regulation of the etioplast-to-chloroplast differentiation, the most important event during seedling emergence from the soil. Our results show that EIN3 and PIF3 act similarly and depend on each other to repress chloroplast development in the dark. EIN3 and PIF3 form a physically interactive transcription factor pair to directly and interdependently inhibit the transcription of most nuclear-encoded LHCA and LHCB genes. We propose that the EIN3-PIF3 module acts at an interface of light and mechanical pressure signals to regulate chloroplast development according to the environment imposed by the soil cover.
RESULTS
EIN3 Mediates a Large Part of the Light-Directed Transcriptomic Changes
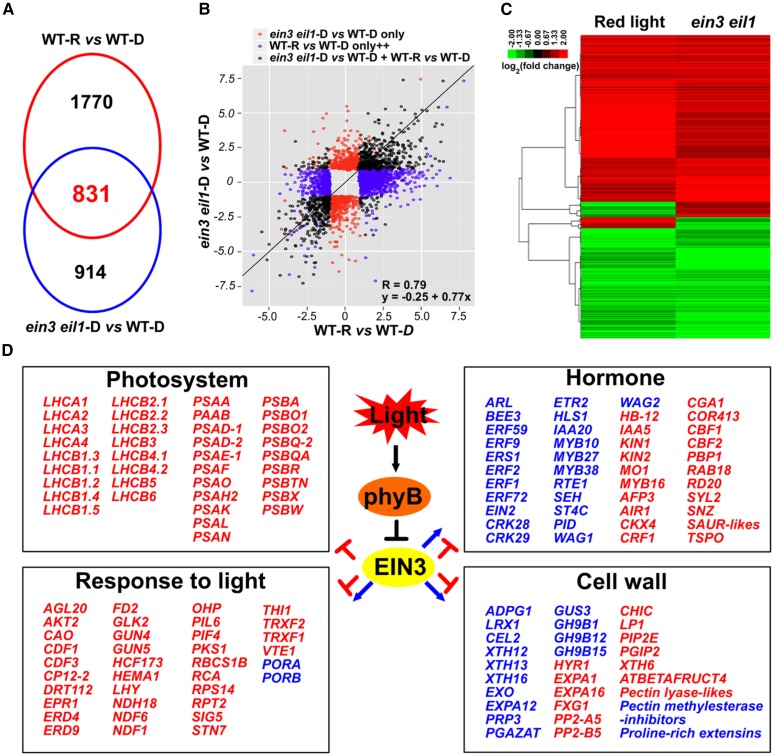
Our recent studies have found that EIN3 acts downstream of COP1 to repress seedling photomorphogenesis in the dark (Shi et al., 2016a). Upon initial light exposure, the photoactivated phyB directly interacts with EIN3 and induces its rapid degradation to promote the developmental transition of deetiolation (Shi et al., 2016b). To investigate the mechanisms by which EIN3 represses deetiolation, we examined EIN3- and red light-regulated transcriptomes via an mRNA deep-sequencing analysis. By comparing wild-type and ein3 eil1 seedlings grown side by side in the dark for 4 d, we identified 1745 genes that showed statistically significantly twofold (SSTF) changes between ein3 eil1 and the wild type (ein3 eil1-D versus WT-D), which we designated ein3 eil1-regulated genes (Figure 1A; Supplemental Data Set 1). Because light degrades EIN3 proteins to a relatively steady level within 2 h, we analyzed the transcriptomic changes in 4-d-old dark-grown wild-type seedlings upon 2 h of initial red light exposure. By comparing to seedlings maintained in the dark (WT-R versus WT-D), we found that the light exposure modulated 2601 SSTF genes, which we refer to as light-regulated genes (Figure 1A; Supplemental Data Set 1). An overlapping analysis of the ein3 eil1- and light-regulated genes revealed that 831 genes were coregulated by light and ein3 eil1 (Figure 1A; Supplemental Data Set 1). Moreover, 91% (759/831) of these genes were found to be regulated by light and ein3 eil1 in the same direction (Supplemental Figure 1A) and showed a strong positive correlation (R = 0.79; y = 0.25 + 0.77x) (Figure 1B). In addition, a cluster analysis of the coregulated genes confirmed that ein3 eil1 and light regulated these genes in highly similar patterns (Figure 1C). We therefore referred to these 759 genes that are coregulated in the same direction as downstream components of the light-phyB-EIN3 pathway. This genome-wide analysis indicates that EIN3 mediates a large portion of the transcription changes in response to light.
Figure 1.
EIN3 Mediates a Large Part of the Red Light-Induced Transcriptome Changes.
(A) Venn diagram showing the overlap of red light-regulated (WT-R versus WT-D) and ein3 eil1-regulated (ein3 eil1-D versus WT-D) genes.
(B) Correlation analysis using a scatterplot of the log2 fold change values between the red light-regulated (WT-R versus WT-D) and ein3 eil1-regulated (ein3 eil1-D versus WT-D) genes. A trend line for the shared genes (dark dots) is presented.
(C) Cluster analysis of the genes coregulated by red light and ein3 eil1. The bar represents the log2 value of the fold-change ratio.
(D) Representative genes coregulated by red light and eil3 eil1 with known functions in highly enriched GO categories. Genes that were upregulated or downregulated by light and ein3 eil1 are shown in red and blue, respectively.
To further investigate the biological function of the genes regulated by the light-phyB-EIN3 pathway, we performed a Gene Ontology (GO) enrichment analysis. The results revealed that among the light-phyB-EIN3 positively regulated genes, the most significantly enriched genes belonged to the GO categories of photosynthesis (P < 10−35), response to light stimulus (P < 10−14), and generation of precursor metabolites and energy (P < 10−11) (Figure 1D; Supplemental Figure 1B and Supplemental Data Set 2). However, the top enriched GO categories among the light-phyB-EIN3 negatively regulated genes were plant-type cell wall organization (P < 10−10), ethylene-mediated signaling pathway (P < 10−9), and two-component signal transduction system (phosphorelay) (P < 10−8) (Figure 1D; Supplemental Figure 1C and Supplemental Data Set 2). The core photosynthetic machinery is composed of two super protein complexes, i.e., photosystem I and photosystem II, and the light-harvesting complex (LHC) antenna system (Eberhard et al., 2008; Waters and Langdale, 2009). We found that most of the key genes involved in mediating the generation of photosynthesis machinery, including PSAs, PSBs, LHCA1–LHCA4, and LHCB1–LHCB6, were highly activated by the light-phyB-EIN3 pathway (Figure 1D). In contrast, the major regulators of apical hook formation (e.g., HLS1), hypocotyl growth inhibition (e.g., ERFs), and cell wall remodeling (e.g., XTHs) were largely suppressed by the light-phyB-EIN3 pathway (Figure 1D). These results suggest that the phyB-triggered EIN3 degradation plays an essential role in initiating the deetiolation process in etiolated seedlings upon light exposure.
EIN3 and PIF3 Interdependently Repress Chloroplast Development in the Dark
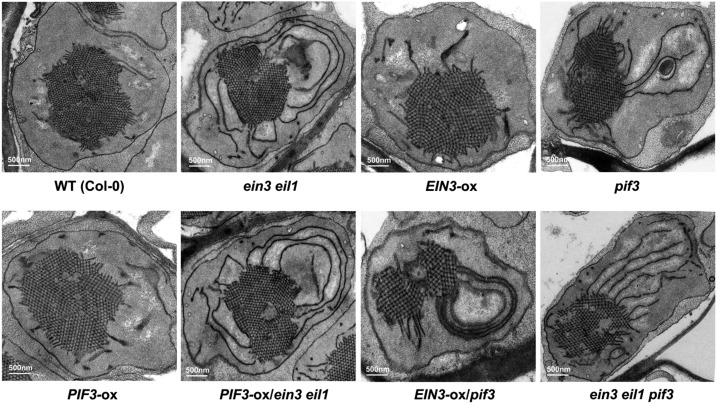
Given the highly enriched photosynthesis GO terms among the light-phyB-EIN3-regulated genes, we investigated whether EIN3 has a role in mediating chloroplast development during deetiolation. We thus examined the inner membrane structure of etioplasts in dark-grown wild-type, ein3 eil1 mutants, and EIN3-ox seedlings. Our electron microscopy examinations showed that the etioplasts in wild-type and EIN3-ox seedlings contain characteristic and highly regular PLB formations with little prothylakoid development (Figure 2). In contrast, in ein3 eil1 mutants, the PLB size of etioplasts was largely reduced, while the prothylakoid membranes were notably increased and extensive, resulting in partially developed chloroplasts in the dark (Figure 2). These ultrastructure results indicate that EIN3 is crucial for repressing chloroplast development in etiolated seedlings.
Figure 2.
EIN3 and PIF3 Interdependently Repress Etioplast Development.
Representative images of the etioplast ultrastructure in 6-d-old dark-grown wild-type, ein3 eil1, EIN3-ox, pif3, PIF3-ox, PIF3-ox/ein3 eil1, EIN3-ox/pif3, and ein3 eil1 pif3 seedlings. Bar = 500 nm.
Previous studies reported that PIF3 is a key transcription factor of the light signaling pathway that represses chloroplast development (Monte et al., 2004; Stephenson et al., 2009). In the pif3 etiolated seedlings, the etioplasts contain notably reduced PLBs and increased prothylakoid membranes (Stephenson et al., 2009), which is similar to that observed in the ein3 eil1 mutant (Figure 2). We then investigated the relationship between EIN3 and PIF3 in regulating chloroplast development. To identify the genetic relationship of EIN3 and PIF3, we constructed PIF3-ox/ein3 eil1 and EIN3-ox/pif3 by crossing. Our results showed that the etioplast ultrastructure of PIF3-ox/ein3 eil1 was similar to that of the ein3 eil1 mutant but not PIF3-ox (Figure 2), suggesting that EIN3 is required for PIF3 to repress chloroplast development. Interestingly, constitutively overexpressing EIN3 in the pif3 mutant (EIN3-ox/pif3) did not restore the early developed chloroplast phenotype of pif3 (Figure 2), indicating that PIF3 is required for the actions of EIN3. Moreover, in the ein3 eil1 pif3 triple mutant seedlings, both the PLB size and prothylakoid development were similar to those in the ein3 eil1 and pif3 mutants (Figure 2). Therefore, the genetic analysis suggests that EIN3 and PIF3 interdependently repress chloroplast development.
EIN3 and PIF3 Do Not Regulate Each Other’s Protein Stability
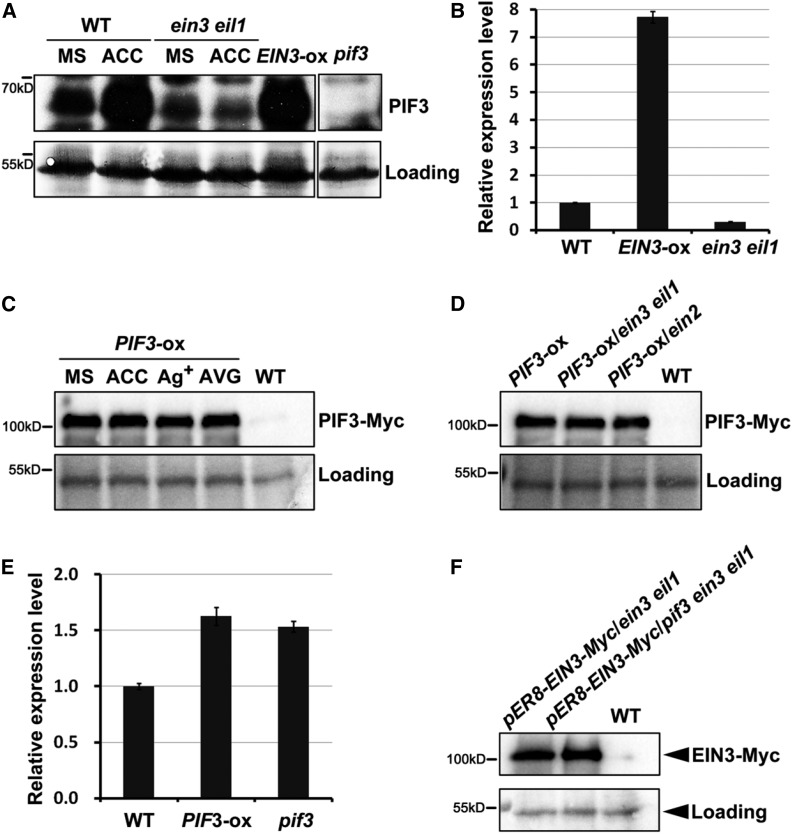
To understand the mechanism by which EIN3 and PIF3 regulate one another, we first found that the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) elevated the endogenous PIF3 protein levels in the wild type but not in the ein3 eil1 mutant dark-grown seedlings (Figure 3A). Moreover, in the EIN3-ox etiolated seedlings, the endogenous PIF3 protein levels accumulated to high levels, similar to those in the ACC-treated wild-type seedlings (Figure 3A). These results suggest that ethylene, via EIN3, upregulates the endogenous PIF3 protein levels.
Figure 3.
The Regulated Relationship between EIN3 and PIF3 at the Transcription and Protein Levels.
(A) Immunoblot results showing the endogenous PIF3 protein levels in wild-type, ein3 eil1, and EIN3-ox etiolated seedlings without (MS) or with ACC treatment. The seedlings were grown in the dark for 4 d on half-strength MS medium (MS) or half-strength MS medium supplemented with 10 μM ethylene precursor ACC. The PIF3 antibody was used to detect endogenous PIF3 proteins, and pif3 was used as a negative control for the PIF3 protein band. Ponceau S staining was used as a control for loading.
(B) RT-qPCR results showing the PIF3 gene expression levels of 4-d-old etiolated wild-type, EIN3-ox, and ein3 eil1 seedlings. Each experiment was performed at least three times with similar results and the representative results were presented. Error bars represent average value ± sd (n = 3) from technical triplicates.
(C) Immunoblot results showing the effects of ethylene on the PIF3-Myc protein levels. The PIF3-ox seedlings were grown in the dark for 4 d on half-strength MS medium (MS), half-strength MS medium supplemented with 10 μM ethylene precursor ACC, 100 μM ethylene perception inhibitor Ag+ (AgNO3), or 25 μM ethylene biosynthesis inhibitor AVG. Anti-Myc antibody was used to detect the PIF3-Myc proteins, and the wild type was used as a negative control for the PIF3-Myc protein band. Ponceau S staining was used as a control for loading.
(D) Immunoblot results showing the PIF3-Myc protein levels in the PIF3-ox (35S:PIF3-Myc/Col-0), PIF3-ox/ein3 eil1, and PIF3-ox/ein2 etiolated seedlings. The seedlings were grown in the dark for 4 d on half-strength MS medium. Anti-Myc antibody was used to detect the PIF3-Myc proteins and the wild type was used as a negative control for the PIF3-Myc protein band. Ponceau S staining was used as a control for loading.
(E) RT-qPCR results showing the EIN3 gene expression levels in 4-d-old etiolated wild-type (Col-0), PIF3-ox, and pif3 seedlings. Each experiment was performed at least three times with similar results and the representative results were presented. Error bars represent average value ± sd (n = 3) from technical triplicates.
(F) Immunoblot results show the EIN3-Myc protein levels in pER8-EIN3-Myc/ein3 eil1 and pER8-EIN3-Myc/pif3 ein3 eil1 etiolated seedlings. The seedlings were grown in the dark for 4 d on half-strength MS medium supplemented with 10 µM β-estradiol inducer. Anti-Myc antibody was used to detect the PIF3-Myc proteins and the wild type was used as a negative control for the PIF3-Myc protein band. Ponceau S staining was used as a control for loading.
Then, we examined the transcriptional and posttranscriptional regulation of PIF3 by EIN3. Ethylene has been reported to activate PIF3 transcription in previous studies (Zhong et al., 2012, 2014), which was consistent with our RT-qPCR results that EIN3 markedly promotes the gene expression of PIF3 (Figure 3B). To examine the posttranscriptional regulation of PIF3, we grew the PIF3-ox transgenic lines constitutively expressing PIF3 (35S:PIF3-Myc) on Murashige and Skoog (MS) medium or MS medium supplemented with the ethylene precursor ACC, the ethylene pathway inhibitor Ag+ (AgNO3), and the ethylene synthesis inhibitor aminoethoxyvinylglycine (AVG). The immunoblotting results showed that neither ACC nor Ag+/AVG affected the protein stability of PIF3 (Figure 3C). EIN2 is a central positive regulator of ethylene signaling pathway upstream of EIN3 (Alonso et al., 1999). We further introduced PIF3-ox into the ein3 eil1 or ein2 mutants by crossing and found that the PIF3-Myc protein levels were similar across the wild-type, ein3 eil1, and ein2 backgrounds (Figure 3D). Collectively, these results indicate that ethylene elevates PIF3 protein levels in an EIN3-dependent manner at the transcriptional level but not the posttranscriptional level. On the other hand, the RT-qPCR results showed that compared with wild type, the gene expression levels of EIN3 were not notably altered in either PIF3-ox or pif3 (Figure 3E). Moreover, we used the EIN3 complementary transgenic lines in the ein3 eil1 and pif3 ein3 eil1 backgrounds and found that the EIN3 protein levels were not affected regardless of the existence of PIF3 (Figure 3F). These data indicate that PIF3 does not regulate the gene expression or protein stability of EIN3.
EIN3 and PIF3 Proteins Interact Directly with Each Other
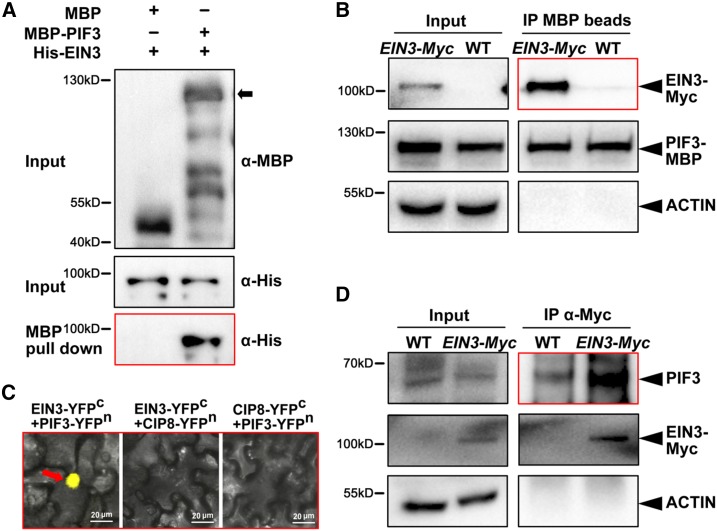
We then investigate whether EIN3 interacts with PIF3 and each regulates the protein activity of the other. Our pull-down experiment results showed that the recombinant purified MBP-PIF3 protein, but not the MBP protein, interacts with the His-EIN3 protein, demonstrating the existence of a direct interaction between the EIN3 and PIF3 proteins in vitro (Figure 4A). In the semi-in vivo pull-down assay, we used two different tagged EIN3 transgenic lines 35S:EIN3-Myc/ein3 eil1 and 35S:EIN3-GFP/ein3 eil1. The recombinant purified MBP-PIF3 proteins were incubated in cell extracts and pulled down by amylose beads. Our results showed that both EIN3-Myc and EIN3-GFP proteins were pulled down by MBP-PIF3 (Figure 4B; Supplemental Figure 2A), indicating that EIN3 specifically interacts with PIF3. To verify the interaction in vivo, we employed a transient bimolecular fluorescence complementation (BiFC) assay. We fused EIN3 with the C-terminal (EIN3-YFPc) and PIF3 with the N-terminal (PIF3-YFPn) of YFP. By coexpressing the split YFP construct pairs in Nicotiana benthamiana leaves, we found that EIN3-YFPc and PIF3-YFPn reconstituted the YFP signals in the nucleus (Figure 4C). We used a nucleus-localized protein COP1-INTERACTING PROTEIN8 (CIP8) as the negative control (Torii et al., 1999). CIP8 is a valid negative control, as CIP8-fused split YFP proteins were capable of interacting with its known interacting protein COP1 in the nucleus of N. benthamiana leaf cells (Supplemental Figure 2B). CIP8 did not reconstitute the YFP signals with either EIN3-YFPc or PIF3-YFPn, indicating the specific interactions between EIN3 and PIF3 in planta (Figure 4C). Furthermore, we performed in vivo coimmunoprecipitation assays in 4-d-old etiolated Arabidopsis seedlings. Since EIN3 activates PIF3 gene expression to elevate endogenous PIF3 protein abundance (Figures 3A and 3B), we used the inducible transgenic plants pER8-EIN3-Myc/ein3 eil1 grown on half-strength MS medium supplemented with 1 µM β-estradiol to adjust PIF3 protein to levels comparable to that in the wild type. The results showed that endogenous PIF3 could be strongly coimmunoprecipitated by EIN3-Myc (Figure 4D). Collectively, our in vitro and in vivo data demonstrate that EIN3 and PIF3 directly interact with each other in the nucleus.
Figure 4.
EIN3 and PIF3 Proteins Directly Interact with Each Other.
(A) The pull-down assay demonstrates the physical interaction between EIN3 and PIF3 in vitro. Purified MBP-PIF3 or MBP proteins were used to pull down His-EIN3 proteins using amylose beads. Anti-MBP and anti-His antibodies were used for the immunoblotting.
(B) Semi-in vivo pull-down assay indicates the interaction between EIN3 and PIF3. Purified PIF3-MBP was incubated in extracts from 4-d-old EIN3–Myc/ein3 eil1 or wild-type etiolated seedlings. Amylose beads were used for the precipitation. Anti-MBP, anti-Myc, and anti-ACTIN antibodies were used for the immunoblotting. ACTIN was used as a negative control for loading.
(C) BiFC assay shows the EIN3-PIF3 interaction in the nucleus of N. benthamiana leaf cells. Full-length PIF3 and EIN3 proteins were fused to the split N-terminal (YFPn) or C-terminal (YFPc) fragments of YFP, respectively. Nuclear-localized CIP8-YFPn and CIP8-YFPc fused proteins were used as negative controls. Bar = 20 µm.
(D) Coimmunoprecipitation assay demonstrates the interaction between EIN3 and PIF3 in vivo. Inducible pER8-EIN3-Myc/ein3 eil1 and wild-type plants were grown on the half-strength MS medium supplemented with 1 µM β-estradiol inducer in the dark for 4 d. Under this condition, pER8-EIN3-Myc/ein3 eil1 etiolated seedlings induced the suitable EIN3-Myc proteins to elevate endogenous PIF3 protein levels comparable with that in the wild type. Anti-Myc antibody was used for precipitation. Anti-PIF3, anti-Myc, and anti-ACTIN antibodies were used for the immunoblotting. ACTIN was used as a negative control for loading.
EIN3 and PIF3 Synergistically Suppress the Expression of LHC Genes
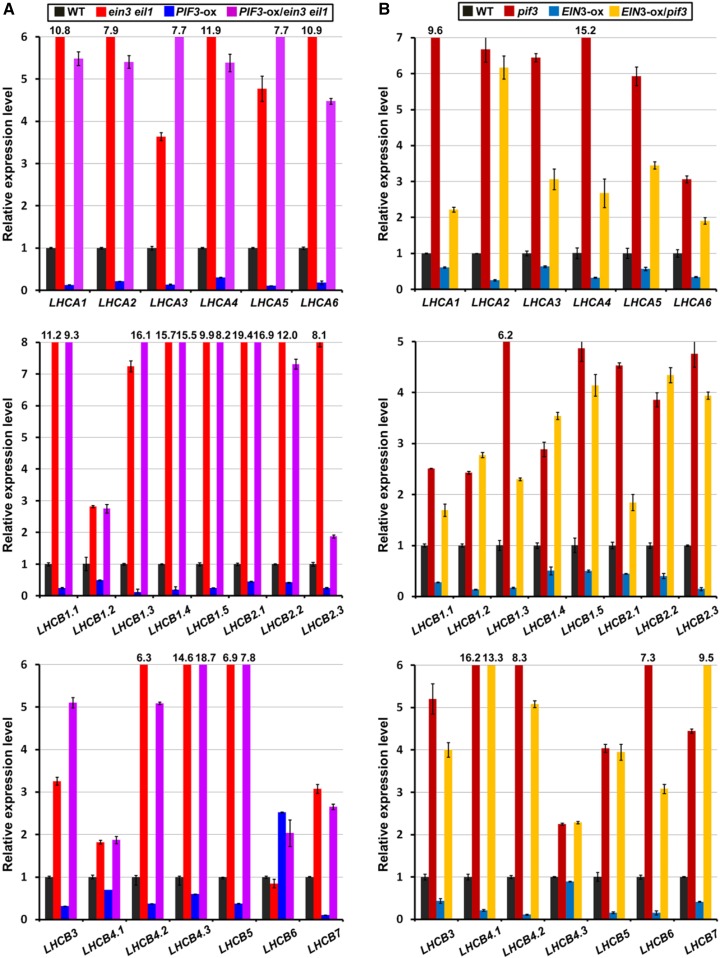
LHCs, including LHCI and LHCII, are pigment binding proteins that reside in the thylakoid membrane. These proteins capture light energy and transfer the energy to photosynthetic reaction centers for conversion into chemical energy (Eberhard et al., 2008; Waters and Langdale, 2009). In higher plants, there are 14 different types of LHC proteins (Lhca1– Lhca6 and Lhcb1– Lhcb8). Under standard conditions, LHCI is composed of Lhca1–Lhca4, and LHCII is organized in minor (Lhcb4–Lhcb6) and major (Lhcb1–Lhcb3) antenna systems (Jansson, 1994; Jansson et al., 1997). As LHCs are essential for photosynthesis initiation, we investigated whether EIN3 and PIF3 regulate the expression of LHCA and LHCB genes. We performed RT-qPCR to examine the LHCA and LHCB gene expression levels in a series of homozygous seedlings, including ein3 eil1, PIF3-ox, PIF3-ox/ein3 eil1, pif3, EIN3-ox, and EIN3-ox/pif3. Our results revealed that almost all LHCA1–LHCA6 and LHCB1–LHCB7 genes were dramatically elevated in the ein3 eil1 and pif3 mutants (Figure 5), while in the PIF3-ox and EIN3-ox seedlings, the expression of these genes was largely reduced (Figure 5). In the PIF3-ox/ein3 eil1 seedlings, the LHCA and LHCB gene expression levels were markedly increased, which was similar to that observed in the ein3 eil1 mutants, indicating that EIN3/EIL1 are required for PIF3 to repress target gene expression (Figure 5A). On the other hand, in the EIN3-ox/pif3 seedlings, the expression of most LHCA and LHCB genes was greatly increased to levels comparable to those of pif3, suggesting that the EIN3 repression of LHC gene transcription requires the coexistence of PIF3 (Figure 5B). The gene expression regulation patterns in these single and double mutants demonstrate the synergistic interrelationships between EIN3 and PIF3, which is consistent with their regulation of chloroplast development.
Figure 5.
EIN3 and PIF3 Depend on One Another to Repress LHC Gene Expression.
(A) RT-qPCR results show the relative expression levels of LHCA and LHCB genes in wild-type, ein3 eil1, PIF3-ox, and PIF3-ox/ein3 eil1 4-d-old etiolated seedlings. Each experiment was performed at least three times with similar results and the representative results were presented. Error bars represent average value ± sd (n = 3) from technical triplicates.
(B) RT-qPCR results show the relative expression levels of LHCA and LHCB genes in wild-type, pif3, EIN3-ox, and EIN3-ox/pif3 4-d-old etiolated seedlings. Each experiment was performed at least three times with similar results and the representative results were presented. Error bars represent average value ± sd (n = 3) from technical triplicates.
EIN3 and PIF3 Bind the Promoters of LHC Genes in an Interdependent Manner
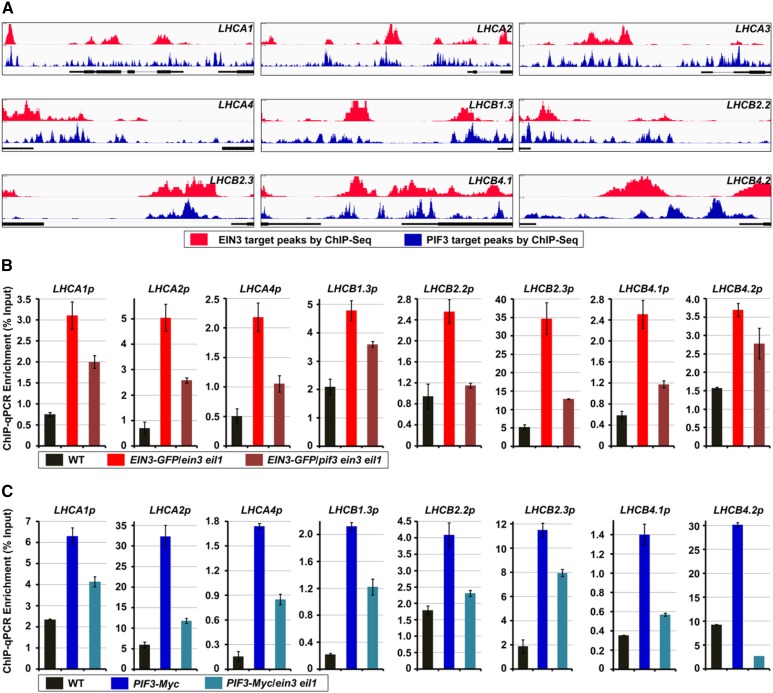
To obtain a detailed understanding of the transcription regulatory functions of EIN3 and PIF3, we used high-throughput chromatin immunoprecipitation-sequencing (ChIP-seq) to comprehensively analyze the genomic occupancy of EIN3 and PIF3 (Chang et al., 2013; Zhang et al., 2013). The global genomic analysis revealed that EIN3 and PIF3 are both associated with the promoters of LHCA and LHCB genes (Figure 6A). We further found that the binding peaks of EIN3 and PIF3 around the LHCA and LHCB promoter regions were largely coincident with each other, implying that EIN3 and PIF3 might coassociate with the same chromosomal regions to synergistically regulate the target gene expression (Figure 6A).
Figure 6.
PIF3 and EIN3 Independently Bind to the Promoters of LHC Genes.
(A) Visualization of EIN3 (in red) and PIF3 (in blue) ChIP-seq data of the genomic regions surrounding the LHCA and LHCB representative genes.
(B) ChIP-qPCR assay shows the relative enrichment of representative LHC gene promoter fragments bound by EIN3-GFP in the ein3 eil1 and pif3 ein3 eil1 backgrounds. The wild-type, EIN3-GFP/ein3 eil1, and EIN3-GFP/pif3 ein3 eil1 4-d-old etiolated seedlings were extracted, and an anti-GFP antibody was used for the precipitation. The wild type was used as the normalized control. Each experiment was performed at least three times with similar results and the representative results were presented. Error bars represent average value ± sd (n = 3) from technical triplicates.
(C) ChIP-qPCR assay shows the relative enrichment of representative LHC gene promoter fragments bound by PIF3-Myc in wild-type and ein3 eil1 backgrounds. The wild-type, PIF3-Myc, and PIF3-Myc/ein3 eil1 4-d-old etiolated seedlings were extracted, and an anti-Myc antibody was used for the precipitation. The wild type was used as the normalized control. Each experiment was performed at least three times with similar results and the representative results were presented. Error bars represent average value ± sd (n = 3) from technical triplicates.
To address this possibility, we performed ChIP-qPCR assays to examine the relationship between EIN3 and PIF3 in occupying the target LHCA and LHCB gene promoter regions. We first used the EIN3-GFP transgenic plants in the ein3 eil1 and pif3 ein3 eil1 backgrounds to quantify the binding of target genes by EIN3. Compared with the wild-type seedlings, the LHCA and LHCB promoter fragments were highly enriched in the EIN3-GFP/ein3eil1 seedlings (Figure 6B), indicating that the EIN3 proteins are associated with these genomic regions in vivo. However, in the pif3 ein3 eil1 background, the binding of the target LHCA and LHCB gene promoter fragments by EIN3 was largely compromised (Figure 6B; Supplemental Figure 3), suggesting that PIF3 is required for the associations between the target LHCA and LHCB gene promoters and EIN3. On the other hand, the ChIP-qPCR results using PIF3-Myc in the wild-type and ein3 eil1 backgrounds showed that the PIF3 proteins precipitate much more target DNA fragments of the LHCA and LHCB promoters in the wild type than in the ein3 eil1 backgrounds (Figure 6C; Supplemental Figure 3), demonstrating that the binding of target genes by PIF3 requires the presence of EIN3. Combining these data, we propose that EIN3 and PIF3 interdependently bind the target LHCA and LHCB gene promoters to synergistically regulate transcription.
LHC Proteins Initiate Etioplast Differentiation without Affecting the Pchlide Level or POR Expression in the Dark
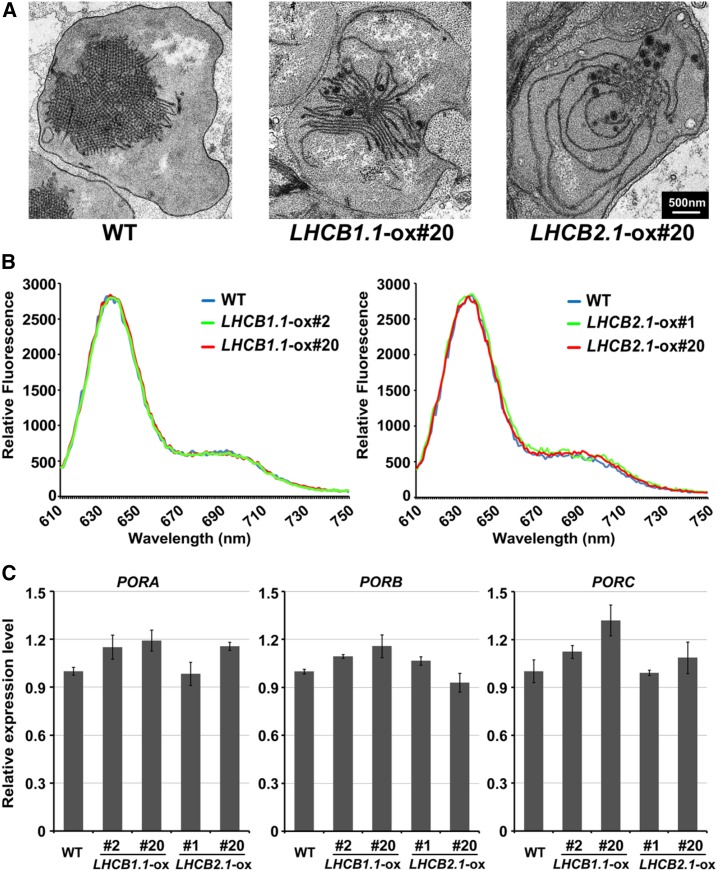
During the etioplast-to-chloroplast differentiation, thylakoid membranes are formed alone with the photosynthetic complexes, which requires the coordinated import of pigment binding proteins, such as Lhca/Lhcb proteins, and the biosynthesis of mature pigments (Pogson and Albrecht, 2011; Jarvis and López-Juez, 2013). Given the synergistic relationship between EIN3 and PIF3 in repressing the etioplast-to-chloroplast development and LHC gene expression, we speculated whether the LHC proteins regulate chloroplast development. We constructed constitutively expressed LHCB1.1 and LHCB2.1 driven by the 35S promoters and examined the ultrastructure of the etioplasts. We found that in the LHCB1.1-ox and LHCB2.1-ox etiolated seedlings, the crystal structures of PLB were diminished, but the prothylakoid membranes were largely increased and extended (Figure 7A), a differentiation phenotype similar to that observed in ein3 eil1 and pif3 (Figures 2 and 7A). Previous studies have shown that EIN3 and PIF proteins suppress Pchlide accumulation and POR gene transcription (Huq et al., 2004; Shen et al., 2008; Shin et al., 2009; Stephenson et al., 2009; Zhong et al., 2009). Interestingly, the Pchlide levels in LHCB1.1-ox and LHCB2.1-ox were similar to those in the wild type in the dark (Figure 7B). Moreover, the overexpression of LHCB1.1 and LHCB2.1 did not affect the PORA, PORB, or PORC transcripts (Figure 7C). Taken together, these results suggest that Lhcb1.1 and Lhcb2.1 repress the etioplast-to-chloroplast development in the dark, but do not regulate Pchlide accumulation or POR gene expression.
Figure 7.
Overexpressing LHC Genes Induces Etioplast Differentiation but Does Not Alter Pchlide Accumulation or POR Gene Expression in the Dark.
(A) Representative images of the etioplast ultrastructure in 6-d-old dark-grown wild-type, LHCB1.1-ox #20-1, and LHCB2.1-ox #20-4 seedlings. Bar = 500 nm.
(B) Fluorescence of Pchlide in 5-d-old etiolated wild-type, LHCB1.1-ox, and LHCB2.1-ox seedlings. Two independent transgenic lines were used.
(C) RT-qPCR results showing the relative expression levels of PORA, PORB, and PORC genes in the wild-type, LHCB1.1-ox, and LHCB2.1-ox 5-d-old etiolated seedlings. Two independent transgenic lines were used. Each experiment was performed at least three times with similar results and the representative results are presented. Error bars represent average value ± sd (n = 3) from technical triplicates.
Overexpressing LHC Genes in the Dark Leads Photooxidative Damage to the Seedlings during Deetiolation
When etiolated seedlings emerge from darkness into initial light exposure, they undergo one of the most dramatic and vulnerable developmental transitions in plants, i.e., deetiolation or greening. It has been reported that PIF3 acts downstream of EIN3 to repress Pchlide accumulation (Zhong et al., 2014). In this study, we revealed that EIN3 and PIF3 cooperatively maintain PLB formation (Figure 2). Since both Pchlide and PLB formation are critical for preventing photooxidative damage in greening, we wanted to know the relationship of EIN3 and PIF3 in regulating seedling deetiolation. We examined the greening rates of the wild type, PIF3-ox, ein3 eil1, and PIF3-ox/ein3 eil1 in a time-course experiment, in which the seedlings were grown in the dark for 3 to 6 d before light exposure (Supplemental Figure 4). We found that with 3-d dark incubation, all the seedlings turned green normally. With 4-d dark incubation, the greening rate of ein3 eil1 declined to 30%, and 90% of PIF3-ox/ein3 eil1 seedlings turned green that was slightly less than the wild type and PIF3-ox. When grown in the dark for 5 d, all of the PIF3-ox seedlings and most of the wild-type seedlings turned green normally, but ein3 eil1 were totally photobleached and dead. PIF3-ox/ein3 eil1 could largely rescue the greening defects of ein3 eil1, with the greening rate of 80%. When the dark incubation period was prolonged to 6 d, over 90% of PIF3-ox seedlings still turned green. By contrast, the greening rate of PIF3-ox/ein3 eil1 declined to around 20% (Supplemental Figure 4). Therefore, although PIF3 rescued the overaccumulated Pchlide of ein3 eil1, the abnormal etioplast structure of PIF3-ox/ein3 eil1 under prolonged dark incubation could lead to severe greening defects.
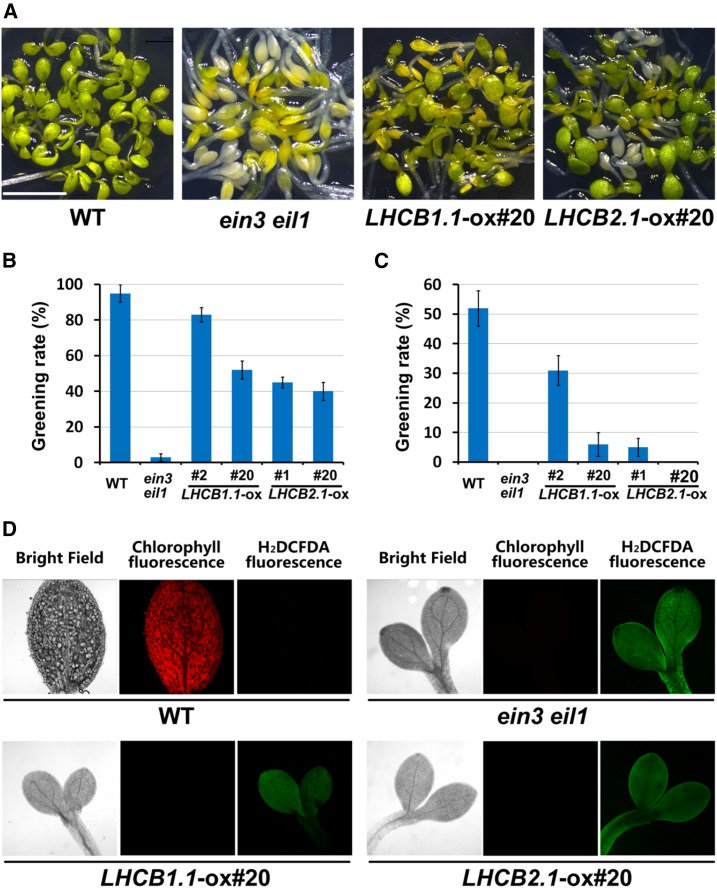
Given the prematured PLBs in the LHCB1.1-ox and LHCB2.1-ox etiolated cotyledons, we then investigated whether Lhcb1.1 and Lhcb2.1 regulate the greening process during the dark-to-light transition. We grew wild-type, ein3 eil1, LHCB1.1-ox, and LHCB2.1-ox seedlings side by side in the dark for 5 d and then exposed them to light illumination for an additional 2 d. We found that more than 90% of the wild-type seedlings turned green normally, while almost all ein3 eil1 seedlings became bleached and died (Figures 8A and 8B). Of the LHCB1.1-ox #20, LHCB2.1-ox #1, and LHCB2.1-ox #20 seedlings, more than half exhibited obvious greening defects and became bleached or yellowish (Figures 8A and 8B). LHCB1.1-ox #2 showed milder greening defects, and 80% of these seedlings turned green (Figures 8A and 8B). Furthermore, when we prolonged the dark-grown period up to 6 d, the greening rates of the wild type, LHCB1.1-ox, and LHCB2.1-ox dramatically decreased. Only half of the wild-type seedlings turned green, and <30% of the LHCB1.1-ox #2 seedlings turned green (Figure 8C; Supplemental Figure 5). Almost all LHCB1.1-ox #20, LHCB2.1-ox #1, and LHCB2.1-ox #20 seedlings became bleached similarly to ein3 eil1 (Figure 8C; Supplemental Figure 5). We further examined the ROS levels as indicated by H2DCFDA when the 5-d-old seedlings were transferred from dark to light. Compared with the wild type, the seedlings of ein3 eil1, LHCB1.1-ox, and LHCB2.1-ox, which suffered photobleached cotyledons, displayed much higher levels of ROS and lower levels of chlorophyll (Figure 8D). Therefore, these results indicate that the repression of LHC gene expression in the dark is critical for inhibiting ROS production and ensuring proper greening of etiolated seedlings in the dark-to-light transition.
Figure 8.
Overexpressing LHC Genes Causes Seedling Lethal Photooxidation upon Light Exposure.
(A) to (C) Representative images of cotyledons (A) and greening rates ([B] and [C]) of wild-type, ein3 eil1, LHCB1.1-ox, and LHCB2.1-ox seedlings. The seedlings were grown in the dark for 5 ([A] and [B]) or 6 (C) d and then transferred to white light exposure for additional 2 d. Error bars represent average value ± se (n = 3) from three independent biological replicates.
(D) Representative fluorescence images of ROS (stained by H2DCFDA) and chlorophyll in wild-type, ein3 eil1, LHCB1.1-ox, and LHCB2.1-ox seedlings. The seedlings were grown in the dark for 5 d and then transferred to white light for additional 2 d.
DISCUSSION
The EIN3/PIF3-LHC Model Is a Key Mechanism Underlying the Adaptive Regulation of Chloroplast Development in Seedling Emergence
After seed germination in soil, the chloroplast development in buried seedlings is arrested at the etioplast stage. Upon reaching the surface, seedlings confront at least two major environmental changes, light and the mechanical pressure of soil, and undergo the most dramatic transition of etioplast to chloroplast. However, the mechanism by which seedlings regulate chloroplast development under buried and out-of-soil conditions remains unclear.
In this study, we reveal the molecular mechanism of the heterotypic transcription factors EIN3 and PIF3 in interdependently regulating chloroplast development during seedling emergence from soil. EIN3 and PIF3 directly interact with each other and occupy the promoters of LHC genes to repress their transcription in a concerted manner. Therefore, when seedlings grow in soil, the mechanical pressure-induced EIN3 and darkness-stabilized PIF3 proteins accumulate in the nucleus and form a collective unit to suppress etioplast maturation by intensively inhibiting LHC gene expression. Upon emerging from the soil, the mechanical pressure is removed, and light activates photoreceptor phyB to induce the rapid degradation of both EIN3 and PIF3, so that the repression of LHC gene expression is relieved in a timely manner to initiate the etioplast-chloroplast transition (Figure 9). It should be noted that the experiments were performed on the medium in darkness under lab conditions. Although EIN3 proteins are stabilized in the dark and the mechanical pressure confronted by roots stimulates ethylene production (Kays et al., 1974; Shi et al., 2016a, 2016b), the protein levels of EIN3 dynamically change during seedling emerging from the soil. Therefore, investigations of chloroplast development with real soil covering under natural conditions are worthwhile in the future.
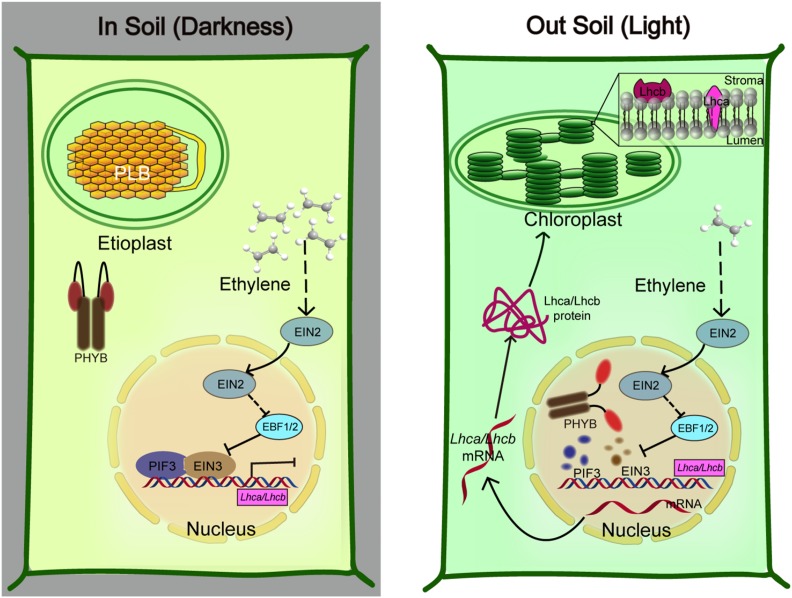
Figure 9.
A Working Model of the PIF3-EIN3 Module Integrated Regulation of Chloroplast Development in Soil (Darkness) and out of Soil (Light).
Left: Proposed model of the signaling pathway in which the EIN3-PIF3 module inhibits the transcription of LHC genes for PLB maintenance. The germinating seedlings under soil confront the darkness and mechanical pressure simultaneously. In the dark, phyB is in the inactive form in the cytoplasm. Darkness-stabilized PIF3 and mechanical pressure-elevated EIN3 proteins accumulate in the nucleus. EIN3 and PIF3 directly interact with each other and interdependently bind the promoters of LHC genes, strongly repressing LHC gene expression. Right: Proposed model of the rapid establishment of chloroplast maturation when etiolated seedlings emerge from soil. The removal of mechanical stress largely decreases ethylene production and EIN3 accumulation, while light-activated phyB promotes both EIN3 and PIF3 protein degradations. Thus, the repression by EIN3-PIF3 on the LHC genes is rapidly relieved to timely initiate the PLB-to-thylakoid membrane differentiation, achieving chloroplast maturation for photosynthesis.
EIN3/EIL1 Transcriptionally Control Etioplast Differentiation along with PIF3
The chloroplast is essential for plant photosynthesis and is associated with metabolic pathways. Its development must be coordinated with plant growth to ensure the rapid initiation of photosynthesis without causing photooxidative damage upon seedling emergence. Among the several thousand chloroplast proteins, more than 95% are encoded by the nuclear genome (Pogson and Albrecht, 2011; Jarvis and López-Juez, 2013). Thus, the tightly controlled gene expression of nuclear-encoded proteins is critical for proper chloroplast development. Previous studies have reported that both environmental and hormonal factors regulate chloroplast development. PIF1 and PIF3 act similarly and additively to repress chlorophyll synthesis and chloroplast development in the dark (Huq et al., 2004; Monte et al., 2004; Shen et al., 2008; Stephenson et al., 2009). Light degrades PIFs to upregulate lots of nuclear genes encoding chloroplast proteins (Shen et al., 2008; Leivar et al., 2009; Stephenson et al., 2009). Plant hormone gibberellin-suppressed DELLA proteins regulate chlorophyll and carotenoid biosynthesis to repress etioplast-chloroplast transition partially by sequestering PIFs (Cheminant et al., 2011), while cytokinin promotes the etioplast-chloroplast transition via the B-type ARR transcription factors (Cortleven et al., 2016). In this study, we reveal that EIN3/EIL1 represent a class of key transcription factors that repress chloroplast differentiation, indicating the crucial functions of ethylene in chloroplast development. By combining extensive genetic, biochemical, and molecular results, we show that EIN3 and PIF3 form an interdependent unit and serves as an integrator of ethylene and light signaling pathways to cooperatively regulate chloroplast development at the transcriptional level.
During chloroplast development, maintaining the prolamellar body (PLB) can be considered an important milestone in which the formation of thylakoid membranes distinguishes the etioplast and chloroplast (Solymosi and Schoefs, 2010; Pogson and Albrecht, 2011; Jarvis and López-Juez, 2013). Light is the dominant environmental factor that drives the differentiation of PLBs into thylakoid membranes. The maintenance of PLBs is critical for etiolated seedlings to survive the initial dark-to-light transition (Green and Durnford, 1996; Schoefs and Franck, 2003; Solymosi and Schoefs, 2010). Proteomic studies of isolated PLBs have shown that the major membrane protein of the PLBs is Pchlide oxidoreductase (POR), the rate-limiting enzyme catalyzing phototoxic Pchlide into mature chlorophyll (Blomqvist et al., 2008; Solymosi and Schoefs, 2010). An ultrastructure analysis of mutants impaired in Pchlide and carotenoid suggests that Pchlide and carotenoid are important for PLB formation (Park et al., 2002; Solymosi and Schoefs, 2010). Here, we find that most of the LHC genes are dramatically suppressed by the EIN3-PIF3 module in the dark. Constitutively overexpressing LHC genes in etiolated seedlings causes partially developed thylakoid membranes. Thus, the strict repression of LHC gene expression maintains the PLB, thereby avoiding premature thylakoid membrane formation in the dark and sequentially photooxidation upon light. Future studies of how LHC proteins initiate the PLB-to-thylakoid membrane transition will greatly advance our understanding of chloroplast development.
The EIN3-PIF3 Module Specifically Represses Chloroplast Development in Soil by Integrating Multiple Soil-Caused Signals
Plants have evolved the capacity to respond to a wide range of environmental signals and integrate them with internal hormone signals to ensure coherent cellular responses. How multiple signals are integrated into cellular decisions is a central question in plant biology. The coordinated actions of light and hormones have been extensively studied at multiple levels (e.g., transcription activation, protein stability) (de Lucas et al., 2008; Feng et al., 2008; Bai et al., 2012; Oh et al., 2012; Zhong et al., 2012; Shi et al., 2016a, 2016b). Notably, recent findings shed light on the central modules of transcription factors in cell elongation. The BZR1, ARF6, and PIF4 proteins interact with each other and synergistically regulate the expression of common genes to converge the brassinosteroid, auxin, and light-signaling pathways (Bai et al., 2012; Oh et al., 2012, 2014). In animal tissue- and organ-specific development, transcription factors commonly interact with each other and form collective heterotypic transcription factor units, thereby combining multiple information signals to cis-regulatory elements (De Val et al., 2008; Junion et al., 2012; Luna-Zurita et al., 2016). For example, the TBX5-NKX2-5-GATA4 interdependent transcription module coordinately determines the mouse cardiac differentiation (Luna-Zurita et al., 2016).
In this study, we propose that chloroplast development is regulated by light and ethylene via the EIN3-PIF3 transcription module, which integrates these regulatory pathways. Buried seedlings simultaneously confront two major environmental signals, darkness and mechanical pressure. The EIN3-PIF3 interdependent module provides a molecular mechanism, through which etioplast-chloroplast differentiation is halted when the darkness and mechanical pressure signals coexist. After emerging from the soil, the seedlings undergo diurnal light-dark changes or could be mechanically pressured by surrounding objects. The EIN3-PIF3 module will be impaired and allow continued chloroplast biogenesis to achieve photoautotrophic capacity. Therefore, this EIN3-PIF3 interdependent module ensures the appropriate etioplast-chloroplast development to adapt to the complicated environmental changes that occur during seedling emergence. Taking these studies together, the interdependent transcription factor module might be a common mechanism adopted by plants and animals to converge external and internal signals in regulating specific developmental processes. Future efforts to identify and elucidate these modules will greatly improve our understanding of the coherent cell activity controlled by various signals.
METHODS
Plant Materials and Growth Conditions
All of the wild-type Arabidopsis thaliana seeds used in this study were Columbia-0 ecotype. The ein3 eil1, ein2, pif3 ein3 eil1, PIF3-ox, PIF3-ox/ein3 eil1, pif3, EIN3-ox, EIN3-ox/pif3, pER8-EIN3-Myc/ein3 eil1, 35S:EIN3-Myc/ein3 eil1, and 35S:EIN3-GFP/ein3 eil1 seeds have been previously used (Zhong et al., 2009, 2010, 2012; Shi et al., 2016a, 2016b). We generated multiple mutants by crossing, and the homozygous lines were confirmed by genotyping. All seeds were surface sterilized using 75% ethanol containing 0.1% Triton X-100 for 15 min and then washed five times with sterile water. The sterilized seeds were plated on a half-strength MS medium (2.2 g/L MS salts, 5 g/L sucrose, and 8 g/L agar, pH 5.7) unless specified otherwise. Plants were grown under a long-day photoperiod (16 h light/8 h dark; Philips cool white fluorescence tube).
Transmission Electron Microscopy
The plant samples were fixed in the fixation buffer I (5% glutaraldehyde and 0.1 M phosphate, pH 7.4) for 4 h at room temperature and then were fixed in the fixation buffer II (2% osmium tetroxide and 0.1 M phosphate, pH 7.4) at 4°C overnight. After one wash in phosphate buffer and two washes in distilled water two times, the samples were stained for 1 h in 1% (m/v) uranyl acetate. After another wash in distilled water, the samples were dehydrated through a graded alcohol series (30%, 50%, 70%, and 85%) and then embedded in Spurr’s resin (Sigma-Aldrich). The ultrathin sections were collected in copper grids with a single slot after cutting with an ultra-microtome (UC7; Leica). The sections were stained with uranyl acetate and lead citrate, observed, and photographed under an electron microscope (Tecnai G2 20 TWIN; FEI) at 120 kV. Three individual cotyledons were used for each genotype and at least 10 cells per cotyledon were observed. Representative etioplast results of each genotype are shown.
RNA Extraction, RT-qPCR, and Transcriptome Analysis
The seedlings were harvested in liquid nitrogen under a dim-green safe light in the dark room and then were ground to powder. The Spectrum Plant Total RNA kit (Sigma-Aldrich) was used to extract the total RNA. Spectrophotometry and a gel electrophoretic analysis were performed to detect the RNA quality. Two micrograms of the RNA was employed to synthesize cDNA using the ReverTra Ace qPCR RT Master Mix (Toyobo). RT-qPCR was performed on the ABI Fast 7500 real-time system using SYBR Green Mix (Takara). A mix of all tested genotypes in one assay was used for the diluted temples to determine the PCR efficiency of each primer set (Supplemental Table 1). The expression values were adjusted according to the actual PCR efficiencies, and the final expression values were normalized to two reference genes PP2A and SAND (Remans et al., 2014). All RT-qPCR experiments were biologically repeated at least three times using independent pools of samples, and representative results are shown.
For the transcriptome analysis, mRNA was extracted using the same procedure as that employed for the detection of gene expression. Information regarding the library preparation and high-throughput sequencing can be found at the website of the Yale Center for Genome Analysis (http://ycga.yale.edu/index.aspx). A bioinformatic analysis was performed as previously described (Shi et al., 2013). GO enrichment analysis was performed using the agriGO online tool (http://bioinfo.cau.edu.cn/agriGO/index.php) (Du et al., 2010).
Immunoblotting
For the chemical treatment, the seeds were plated on half-strength MS medium supplemented with 10 µM ACC, 100 μΜ AgNO3, or 25 μΜ AVG as previously described (An et al., 2010; Shen et al., 2016). The pER8:EIN3-Myc/ein3 eil1 and pER8:EIN3-Myc/pif3 ein3 eil1 seeds were plated on half-strength MS medium containing 10 μΜ β-estradiol to induce EIN3-Myc expression. For the immunoblotting, the 4-d-old etiolated seedlings were harvested in liquid nitrogen and ground to powder. The powder was homogenized in a protein extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 0.1% Tween 20, 1 mM PMSF, and one Roche complete EDTA-free Protease Inhibitor Cocktail Tablet/50 mL). The sample harvest and protein extraction were performed under a dim-green safe light in the dark room. The extracts were boiled in SDS loading buffer, followed by centrifugation at 14,000g for 10 min twice. The proteins were separated on 8% or 10% SDS-PAGE gels. The anti-Myc (Sigma-Aldrich; M4439, 1:5000 dilution), anti-GFP (Clontech; MMS-118P, 1:3000 dilution), and anti-PIF3 (1:1000 dilution) antibodies were used for the immunoblotting analysis. To generate PIF3 antibody, we made the PIF3 antigen as previously reported (Al-Sady et al., 2006) and the antibody was produced by ABclonal (China). The protein bands were then visualized using the standard enhanced chemiluminescence method.
BiFC
Full-length coding sequences of PIF3, CIP8, or COP1 were fused in-frame with the N terminus of YFP (PIF3-YFPn, CIP8-YFPn, or COP1-YFPn). The full-length coding sequence of EIN3, CIP8, or COP1 was fused in-frame with the C terminus of YFP (EIN3-YFPc, CIP8-YFPn, or COP1-YFPn). The indicated plasmid pairs were infiltrated into Nicotiana benthamiana leaves by the Agrobacterium tumefaciens strain as described previously (Feng et al., 2008). The YFP fluorescence signals were observed and imaged under a Carl Zeiss confocal laser scanning microscope (LSM510 Meta). YFP fluorescence was excited by a 514-nm laser and detected between 517 and 589 nm.
Pull-Down Assay
One microgram of MBP-PIF3, His-EIN3, or empty MBP recombinant purified proteins was added to the binding buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 1 mM EDTA) as previously indicated (Shi et al., 2016a). The solutions were incubated at 4°C with gentle rotation for 2 h. The MBP fusion proteins were precipitated using 20 μL amylose resins (NEB) and then centrifuged at 2000g for 1 min. The resins were washed four times with the washing buffer (50 mM Tris-HCl, pH 7.5, 0.5% Triton X-100, 10% glycerol, and 1 mM EDTA). Anti-His (Cell Signaling; 2366S, 1:2000 dilution) and anti-MBP (NEB; E8032S, 1:1000 dilution) antibodies were used for the immunoblotting detection.
Semi-in Vivo Pull-Down Assay
The semi-in vivo pull-down assay was performed as previously described (Shi et al., 2016a). Briefly, 1 μg of the purified recombinant PIF3-MBP proteins was added to 500 µg total soluble plant protein solutions and incubated at 4°C with gentle rotation for 1 h. Then, 20 μL amylose resins were added to the solution and incubated at 4°C with gentle rotation for 2 h. The plant total protein extraction and pull-down were performed under a dim-green safe light in the dark room. After the pull-down, the beads were washed three times with the washing buffer (50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 10% glycerol, 0.1% Tween 20, and 1 mM EDTA) and then subjected to immunoblotting detection. For the immunoblotting, the anti-MBP (NEB; E8032S, 1:1000 dilution), anti-GFP (Clontech; MMS-118P, 1:3000 dilution), and anti-ACTIN (Sigma-Aldrich; A0480, 1:8000 dilution) antibodies were used.
Coimmunoprecipitation Assay
The pER8:EIN3-Myc/ein3 eil1 and wild-type seeds were plated on half-strength MS medium containing 1 μΜ β-estradiol for 4 d in the dark. The seedlings were harvested in liquid nitrogen and ground to powder. Immunoprecipitation was performed as previously described (Shi et al., 2015). The sample harvest, protein extraction, and immunoprecipitation were performed under a dim-green safe light in the dark room. Anti-Myc (Sigma-Aldrich) and Protein G Sepharose (Sigma-Aldrich) were used for immunoprecipitation. For the immunoblotting, the anti-Myc (Sigma-Aldrich; M4439, 1:5000 dilution), anti-PIF3 (1:1000 dilution), and anti-ACTIN (Sigma-Aldrich; A0480, 1:8000 dilution) antibodies were used.
ChIP Assays and ChIP-Seq Data Analysis
The EIN3-GFP/ein3 eil1, EIN3-GFP/pif3 ein3 eil1, PIF3-ox, PIF3-ox/ein3 eil1, and wild type were grown in the dark for 4 d. The seedlings were harvested and cross-linked in a 1% formaldehyde solution for 30 min in a vacuum. The cross-linking, protein extraction, and immunoprecipitation were performed under a dim-green safe light in the dark room. The following procedures were performed as previously described (Shi et al., 2013). Anti-GFP antibody (Clontech; MMS-118P, 1:3000 dilution), protein G beads (GE), and anti-c-Myc Affinity Gel (Sigma-Aldrich) were used for the immunoprecipitation.
The ChIP-seq data were originally from previous studies (Chang et al., 2013; Zhang et al., 2013), and the peaks were visualized using the Integrative Genomics Viewer (http://software.broadinstitute.org/software/igv/).
Seedling Dark-to-Light Greening Rate
To determine the seedling greening rate, more than 100 seeds per sample were plated. The seedlings were grown in darkness for the indicated number of days and then transferred to continuous white light (100 µmol/m2s) for additional 2 d. The greening rates were calculated as previously described (Zhong et al., 2009). Briefly, the greening rate was calculated through dividing the number of normal greening seedlings (with intact green cotyledons) by the total germinated seedlings. Three independent biological repeats were performed and the error bar was calculated from the biological triplicates.
Protochlorophyllide Measurement
The seedlings were grown in darkness for the indicated number of days and then 20 dark-grown etiolated seedlings were collected under a dim-green safe light in the dark room. The seedlings were soaked in 1 mL of the extraction buffer (90% acetone containing 0.1% NH3) (Zhong et al., 2014). The extraction was performed in the dark at room temperature for 24 h. The supernatant was transferred to a new tube after centrifuging, and the fluorescence emission spectra were measured. The excitation wavelength was 443 nm, and the emission spectra were recorded from 610 to 740 nm with 1 nm bandwidth. Three independent biological repeats were performed, and the representative results were shown.
Histochemical ROS Staining
More than 80 seedlings were grown in darkness for the indicated number of days and then transferred to continuous white light (100 µmol/m2s) for additional 2 d. The seedlings were merged in the Tris buffer (10 mM Tris-HCl, pH 7.2, and 100 μM H2DCFDA) in the dark for 5 min and washed five times with the Tris buffer (Zhong et al., 2009). Fluorescence microscopic images were observed under a Carl Zeiss confocal laser scanning microscope (LSM510 Meta) with an excitation wavelength of 488 nm and emission wavelength of 530 nm. Chlorophyll autofluorescence was excited at 660-nm wavelength.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL data libraries under the following accession numbers: EIN3 (AT3G20770), EIL1 (AT2G27050), PIF3 (AT1G09530), PORA (AT5G54190), PORB (AT4G27440), PORC (AT1g03630), EIN2 (AT5G03280), COP1 (AT2G32950), CIP8 (AT5G64920), EBF1 (AT2G25490), EBF2 (AT5G25350), LHCA1 (AT3G54890), LHCA2 (AT3G61470), LHCA3 (AT1G61520), LHCA4 (AT3G47470), LHCA5 (AT1G45474), LHCA6 (AT1G19150), LHCB1.1 (AT1G29920), LHCB1.2 (AT1G29910), LHCB1.3 (AT1G29930), LHCB1.4 (AT2G34430), LHCB1.5 (AT2G34420), LHCB2.1 (AT2G05100), LHCB2.2 (AT2G05070), LHCB2.3 (AT3G27690), LHCB3 (AT5G54270), LHCB4.1 (AT5G01530), LHCB4.2 (AT3G08940), LHCB4.3 (AT2G40100), LHCB5 (AT4G10340), LHCB6 (AT1G15820), LHCB7 (AT1G76570), PP2A (AT1g13320), and SAND (AT2G28390).
Supplemental Data
Supplemental Figure 1. GO analysis of light and ein3 eil1 coregulated genes.
Supplemental Figure 2. EIN3 and PIF3 proteins directly interact with each other.
Supplemental Figure 3. EIN3-GFP and PIF3-Myc protein levels are similar in different backgrounds.
Supplemental Figure 4. The relationships between PIF3 and EIN3 in the greening of etiolated seedlings.
Supplemental Figure 5. LHCB1.1-ox and LHCB2.1-ox display severe photobleaching and death during the initial dark-to-light transition.
Supplemental Table 1. RT-qPCR primer sequences and efficiencies.
Supplemental Data Set 1. Red light and ein3 eil1-regulated genes identified in the mRNA sequencing analysis.
Supplemental Data Set 2. GO analysis of the genes coregulated by red light and ein3 eil1.
Acknowledgments
We thank Shoucheng Liu and Mohan Lv from Peking University for valuable help with the bioinformatic analysis. We also thank Ying-Chun Hu for professional technical assistance with the electron microscopy sample preparation and image analysis at the Core Facilities of College of Life Sciences, Peking University. This work was supported by grants from the National Key Research and Development Program of China (2016YFA0502900) and the National Science Foundation of China (31770304 and 31770208). S.Z. and H.S. were supported by an open project from the State Key Laboratory of Protein and Plant Gene Research. X.L. was supported by a China Postdoctoral Science Foundation Grant (2016M600857) and the Outstanding Postdoctoral Fellowship of Peking-Tsinghua Center for Life Sciences.
AUTHOR CONTRIBUTIONS
H.S. and S.Z. designed the research. H.S., X.L., R.L., and Y.L. performed the experiments. H.S., S.Z., and X.L. analyzed the data. H.S., S.Z., and X.L. wrote the article.
References
- Alonso J.M., Stepanova A.N. (2004). The ethylene signaling pathway. Science 306: 1513–1515. [DOI] [PubMed] [Google Scholar]
- Alonso J.M., Hirayama T., Roman G., Nourizadeh S., Ecker J.R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152. [DOI] [PubMed] [Google Scholar]
- Al-Sady B., Ni W., Kircher S., Schäfer E., Quail P.H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23: 439–446. [DOI] [PubMed] [Google Scholar]
- An F., et al. (2010). Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22: 2384–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M.Y., Shang J.X., Oh E., Fan M., Bai Y., Zentella R., Sun T.P., Wang Z.Y. (2012). Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14: 810–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomqvist L.A., Ryberg M., Sundqvist C. (2008). Proteomic analysis of highly purified prolamellar bodies reveals their significance in chloroplast development. Photosynth. Res. 96: 37–50. [DOI] [PubMed] [Google Scholar]
- Chang K.N., et al. (2013). Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. eLife 2: e00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Q., Rothenberg M., Solano R., Roman G., Terzaghi W., Ecker J.R. (1997). Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144. [DOI] [PubMed] [Google Scholar]
- Cheminant S., Wild M., Bouvier F., Pelletier S., Renou J.P., Erhardt M., Hayes S., Terry M.J., Genschik P., Achard P. (2011). DELLAs regulate chlorophyll and carotenoid biosynthesis to prevent photooxidative damage during seedling deetiolation in Arabidopsis. Plant Cell 23: 1849–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortleven A., Marg I., Yamburenko M.V., Schlicke H., Hill K., Grimm B., Schaller G.E., Schmülling T. (2016). Cytokinin regulates the etioplast-chloroplast transition through the two-component signaling system and activation of chloroplast-related genes. Plant Physiol. 172: 464–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484. [DOI] [PubMed] [Google Scholar]
- De Val S., et al. (2008). Combinatorial regulation of endothelial gene expression by ets and forkhead transcription factors. Cell 135: 1053–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Zhou X., Ling Y., Zhang Z., Su Z. (2010). agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38: W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard S., Finazzi G., Wollman F.A. (2008). The dynamics of photosynthesis. Annu. Rev. Genet. 42: 463–515. [DOI] [PubMed] [Google Scholar]
- Ecker J.R. (1995). The ethylene signal transduction pathway in plants. Science 268: 667–675. [DOI] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne J.M., Smalle J., Gingerich D.J., Walker J.M., Yoo S.D., Yanagisawa S., Vierstra R.D. (2004). Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc. Natl. Acad. Sci. USA 101: 6803–6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeschl J.D., Rappaport L., Pratt H.K. (1966). Ethylene as a factor regulating the growth of pea epicotyls subjected to physical stress. Plant Physiol. 41: 877–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green B.R., Durnford D.G. (1996). The chlorophyll-carotenoid proteins of oxygenic photosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 685–714. [DOI] [PubMed] [Google Scholar]
- Guo H., Ecker J.R. (2003). Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677. [DOI] [PubMed] [Google Scholar]
- Huq E., Al-Sady B., Hudson M., Kim C., Apel K., Quail P.H. (2004). Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941. [DOI] [PubMed] [Google Scholar]
- Jansson S. (1994). The light-harvesting chlorophyll a/b-binding proteins. Biochim. Biophys. Acta 1184: 1–19. [DOI] [PubMed] [Google Scholar]
- Jansson S., Stefansson H., Nystrom U., Gustafsson P., Albertsson P.A. (1997). Antenna protein composition of PS I and PS II in thylakoid sub-domains. Biochim. Biophysi. Acta 1320: 297–309. [Google Scholar]
- Jarvis P., López-Juez E. (2013). Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14: 787–802. [DOI] [PubMed] [Google Scholar]
- Ju C., et al. (2012). CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 19486–19491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junion G., Spivakov M., Girardot C., Braun M., Gustafson E.H., Birney E., Furlong E.E. (2012). A transcription factor collective defines cardiac cell fate and reflects lineage history. Cell 148: 473–486. [DOI] [PubMed] [Google Scholar]
- Kays S.J., Nicklow C.W., Simons D.H. (1974). Ethylene in relation to response of roots to physical impedance. Plant Soil 40: 565–571. [Google Scholar]
- Lehman A., Black R., Ecker J.R. (1996). HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell 85: 183–194. [DOI] [PubMed] [Google Scholar]
- Leivar P., Quail P.H. (2011). PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16: 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E. (2014). PIFs: systems integrators in plant development. Plant Cell 26: 56–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Tepperman J.M., Monte E., Calderon R.H., Liu T.L., Quail P.H. (2009). Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Oka Y., Liu T., Carle C., Castillon A., Huq E., Quail P.H. (2008). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18: 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Ma M., Feng Y., Li H., Wang Y., Ma Y., Li M., An F., Guo H. (2015). EIN2-directed translational regulation of ethylene signaling in Arabidopsis. Cell 163: 670–683. [DOI] [PubMed] [Google Scholar]
- Liu X., Li Y., Zhong S. (2017). Interplay between light and plant hormones in the control of Arabidopsis seedling chlorophyll biosynthesis. Front. Plant Sci. 8: 1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna-Zurita L., et al. (2016). Complex interdependence regulates heterotypic transcription factor distribution and coordinates cardiogenesis. Cell 164: 999–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchante C., Brumos J., Yun J., Hu Q., Spencer K.R., Enríquez P., Binder B.M., Heber S., Stepanova A.N., Alonso J.M. (2015). Gene-specific translation regulation mediated by the hormone-signaling molecule EIN2. Cell 163: 684–697. [DOI] [PubMed] [Google Scholar]
- Monte E., Tepperman J.M., Al-Sady B., Kaczorowski K.A., Alonso J.M., Ecker J.R., Li X., Zhang Y., Quail P.H. (2004). The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc. Natl. Acad. Sci. USA 101: 16091–16098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J., Zhu L., Shen H., Huq E. (2008). PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 9433–9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M., Tepperman J.M., Quail P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667. [DOI] [PubMed] [Google Scholar]
- Oh E., Zhu J.Y., Wang Z.Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14: 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Zhu J.Y., Bai M.Y., Arenhart R.A., Sun Y., Wang Z.Y. (2014). Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 3: e03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H., Kreunen S.S., Cuttriss A.J., DellaPenna D., Pogson B.J. (2002). Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. Plant Cell 14: 321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson B.J., Albrecht V. (2011). Genetic dissection of chloroplast biogenesis and development: an overview. Plant Physiol. 155: 1545–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak T., Lechner E., Parmentier Y., Yanagisawa S., Grava S., Koncz C., Genschik P. (2003). EIN3-dependent regulation of plant ethylene hormone signaling by two arabidopsis F box proteins: EBF1 and EBF2. Cell 115: 679–689. [DOI] [PubMed] [Google Scholar]
- Qiao H., Shen Z., Huang S.S., Schmitz R.J., Urich M.A., Briggs S.P., Ecker J.R. (2012). Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science 338: 390–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail P.H. (2002a). Photosensory perception and signalling in plant cells: new paradigms? Curr. Opin. Cell Biol. 14: 180–188. [DOI] [PubMed] [Google Scholar]
- Quail P.H. (2002b). Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 3: 85–93. [DOI] [PubMed] [Google Scholar]
- Reinbothe C., El Bakkouri M., Buhr F., Muraki N., Nomata J., Kurisu G., Fujita Y., Reinbothe S. (2010). Chlorophyll biosynthesis: spotlight on protochlorophyllide reduction. Trends Plant Sci. 15: 614–624. [DOI] [PubMed] [Google Scholar]
- Remans T., Keunen E., Bex G.J., Smeets K., Vangronsveld J., Cuypers A. (2014). Reliable gene expression analysis by reverse transcription-quantitative PCR: reporting and minimizing the uncertainty in data accuracy. Plant Cell 26: 3829–3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell N.C., Su Y.S., Lagarias J.C. (2006). Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57: 837–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoefs B., Franck F. (2003). Protochlorophyllide reduction: mechanisms and evolutions. Photochem. Photobiol. 78: 543–557. [DOI] [PubMed] [Google Scholar]
- Shen H., Zhu L., Castillon A., Majee M., Downie B., Huq E. (2008). Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20: 1586–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Li Y., Pan Y., Zhong S. (2016). Activation of HLS1 by mechanical stress via ethylene-stabilized EIN3 is crucial for seedling soil emergence. Front. Plant Sci. 7: 1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Liu R., Xue C., Shen X., Wei N., Deng X.W., Zhong S. (2016a). Seedlings transduce the depth and mechanical pressure of covering soil using COP1 and ethylene to regulate EBF1/EBF2 for soil emergence. Curr. Biol. 26: 139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Shen X., Liu R., Xue C., Wei N., Deng X.W., Zhong S. (2016b). The red light receptor phytochrome B directly enhances substrate-E3 ligase interactions to attenuate ethylene responses. Dev. Cell 39: 597–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Wang X., Mo X., Tang C., Zhong S., Deng X.W. (2015). Arabidopsis DET1 degrades HFR1 but stabilizes PIF1 to precisely regulate seed germination. Proc. Natl. Acad. Sci. USA 112: 3817–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Zhong S., Mo X., Liu N., Nezames C.D., Deng X.W. (2013). HFR1 sequesters PIF1 to govern the transcriptional network underlying light-initiated seed germination in Arabidopsis. Plant Cell 25: 3770–3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Kim K., Kang H., Zulfugarov I.S., Bae G., Lee C.H., Lee D., Choi G. (2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 106: 7660–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R., Stepanova A., Chao Q., Ecker J.R. (1998). Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12: 3703–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solymosi K., Schoefs B. (2010). Etioplast and etio-chloroplast formation under natural conditions: the dark side of chlorophyll biosynthesis in angiosperms. Photosynth. Res. 105: 143–166. [DOI] [PubMed] [Google Scholar]
- Stephenson P.G., Fankhauser C., Terry M.J. (2009). PIF3 is a repressor of chloroplast development. Proc. Natl. Acad. Sci. USA 106: 7654–7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii K.U., Stoop-Myer C.D., Okamoto H., Coleman J.E., Matsui M., Deng X.W. (1999). The RING finger motif of photomorphogenic repressor COP1 specifically interacts with the RING-H2 motif of a novel Arabidopsis protein. J. Biol. Chem. 274: 27674–27681. [DOI] [PubMed] [Google Scholar]
- Von Arnim A., Deng X.W. (1996). Light control of seedling development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 215–243. [DOI] [PubMed] [Google Scholar]
- Waters M.T., Langdale J.A. (2009). The making of a chloroplast. EMBO J. 28: 2861–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N., Kwok S.F., von Arnim A.G., Lee A., McNellis T.W., Piekos B., Deng X.W. (1994). Arabidopsis COP8, COP10, and COP11 genes are involved in repression of photomorphogenic development in darkness. Plant Cell 6: 629–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X., Zhang C., Ji Y., Zhao Q., He W., An F., Jiang L., Guo H. (2012). Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res. 22: 1613–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Mayba O., Pfeiffer A., Shi H., Tepperman J.M., Speed T.P., Quail P.H. (2013). A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 9: e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Shi H., Xi Y., Guo H. (2010). Ethylene is crucial for cotyledon greening and seedling survival during de-etiolation. Plant Signal. Behav. 5: 739–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Shi H., Xue C., Wei N., Guo H., Deng X.W. (2014). Ethylene-orchestrated circuitry coordinates a seedling’s response to soil cover and etiolated growth. Proc. Natl. Acad. Sci. USA 111: 3913–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Zhao M., Shi T., Shi H., An F., Zhao Q., Guo H. (2009). EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc. Natl. Acad. Sci. USA 106: 21431–21436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Shi H., Xue C., Wang L., Xi Y., Li J., Quail P.H., Deng X.W., Guo H. (2012). A molecular framework of light-controlled phytohormone action in Arabidopsis. Curr. Biol. 22: 1530–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]