Genome-wide analysis sheds light on the dependence of plastid translation on interactions between Shine-Dalgarno-like sequences in mRNA and the anti-Shine-Dalgarno sequence in 16S ribosomal RNA.
Abstract
In prokaryotic systems, the translation initiation of many, though not all, mRNAs depends on interaction between a sequence element upstream of the start codon (the Shine-Dalgarno sequence [SD]) and a complementary sequence in the 3′ end of the 16S rRNA (anti-Shine-Dalgarno sequence [aSD]). Although many chloroplast mRNAs harbor putative SDs in their 5′ untranslated regions and the aSD displays strong conservation, the functional relevance of SD-aSD interactions in plastid translation is unclear. Here, by generating transplastomic tobacco (Nicotiana tabacum) mutants with point mutations in the aSD coupled with genome-wide analysis of translation by ribosome profiling, we provide a global picture of SD-dependent translation in plastids. We observed a pronounced correlation between weakened predicted SD-aSD interactions and reduced translation efficiency. However, multiple lines of evidence suggest that the strength of the SD-aSD interaction is not the only determinant of the translational output of many plastid mRNAs. Finally, the translation efficiency of mRNAs with strong secondary structures around the start codon is more dependent on the SD-aSD interaction than weakly structured mRNAs. Thus, our data reveal the importance of the aSD in plastid translation initiation, uncover chloroplast genes whose translation is influenced by SD-aSD interactions, and provide insights into determinants of translation efficiency in plastids.
INTRODUCTION
The three genetic compartments of plant cells all contain their own protein synthesis machinery. Due to the endosymbiotic origin of plastids and mitochondria from bacteria, these organelles possess bacterial-type 70S ribosomes consisting of a 30S and a 50S subunit (Schwarz and Kössel, 1980; Yamaguchi et al., 2000; Yamaguchi and Subramanian, 2000; Tiller and Bock, 2014). Plastid rRNAs are similar to bacterial rRNAs in terms of both primary sequence and secondary and tertiary structure. Like in bacteria, the 30S subunit of the plastid ribosome contains a single rRNA species, the 16S rRNA (Schwarz and Kössel, 1980), and the 50S ribosomal subunit contains 23S and 5S rRNAs. However, in contrast to bacteria, the 50S particle of plastid ribosomes contains an additional small rRNA species, the 4.5S rRNA, that is evolutionarily derived from the 3′ part of the 23S rRNA (Edwards and Kössel, 1981; Edwards et al., 1981). The plastid 23S rRNA is further fragmented into three pieces, presumably by endoribonucleolytic cleavage at two so-called “hidden breaks” (Nishimura et al., 2010; Liu et al., 2015). The vast majority of the plastid ribosomal proteins have bacterial homologs (Tiller and Bock, 2014). In addition, there are a few plastid-specific ribosomal proteins (Tiller et al., 2012) whose functions are not well resolved. These proteins have been proposed to compensate for structural changes in the rRNAs and/or participate in translational regulation (Manuell et al., 2007; Tiller et al., 2012; Graf et al., 2017; Bieri et al., 2017).
Gene expression in plastids is mainly controlled at the posttranscriptional level, especially at the level of translation (Eberhard et al., 2002; Kahlau and Bock, 2008; Sun and Zerges, 2015). In bacteria, translation initiation represents the rate-limiting step in the control of protein synthesis, whereas regulation at later steps in the translation process is relatively rare (Duval et al., 2015). The efficiency of translation initiation largely depends on the efficiency of ribosome binding to the region upstream of the start codon (5′ untranslated region [UTR]) and on the efficiency of start codon recognition (Hall et al., 1982; McCarthy and Gualerzi, 1990; Kozak, 2005). By contrast, there is no consensus in the scientific literature as to how start codons in plastids are recognized.
The classical mechanism for translation initiation in bacteria depends on ribosome positioning within the 5′ UTR of the mRNA through hybridization of a conserved sequence motif in the 3′ end of the 16S rRNA, the so called anti-Shine-Dalgarno sequence (aSD), to a complementary sequence upstream of the start codon, the Shine-Dalgarno sequence (SD; Shine and Dalgarno, 1974; Kozak, 1999). While the binding of the ribosome to the mRNA itself is independent of the SD, the aSD-SD interaction positions the ribosome on the mRNA such that the initiator tRNAfMet(CAU) can recognize the start codon (Milón and Rodnina, 2012). Consequently, the main function of the SD is to distinguish the start codon from other AUG triplets in the mRNA, thus enabling the formation of a stable 30S initiation complex (Jin et al., 2006; Studer and Joseph, 2006). Interestingly, a substantial number of bacterial genes do not possess recognizable SDs (Chang et al., 2006; Nakagawa et al., 2010, 2017), and it has been proposed that local minima of mRNA secondary structure define start codons in these mRNAs (Scharff et al., 2011). Plastid genomes also contain both genes with and genes without SDs (Ruf and Kössel, 1988; Bonham-Smith and Bourque, 1989; Sugiura et al., 1998). The possible relevance of SDs for plastid translation has been controversially discussed in the scientific literature (Zerges, 2000; Sugiura, 2014). On the one hand, there is evidence that in plastids, SD sequences (Hirose et al., 1998; Hirose and Sugiura, 2004; Kuroda et al., 2007) and local minima of mRNA structure (Scharff et al., 2011; Zhang et al., 2012) are also important determinants of start codon recognition. In addition, bacteriophage-derived SD-containing 5′ UTRs were successfully used for high-level transgene expression in tobacco (Nicotiana tabacum) plastids (Ye et al., 2001; Oey et al., 2009). Furthermore, when compared with bacterial aSD sequences, the putative aSD sequence in the plastid 16S rRNA displays complete conservation (Supplemental Table 1). On the other hand, there are also conflicting results that have questioned the functionality and/or essentiality of SDs for translation of plastid mRNAs. For example, in some mRNAs, mutation of the SD was shown to only moderately reduce translation or even not influence it at all (Sakamoto et al., 1994; Fargo et al., 1998; Nickelsen et al., 1999). Moreover, in one case investigated in an in vitro plastid translation system, even a negative influence of the SD on protein synthesis was reported (Plader and Sugiura, 2003).
The aim of this study was to comprehensively assess the functional relevance of SDs in plastids by simultaneously investigating the translation of all mRNAs encoded in the plastid genome. To facilitate such a genome-wide approach, we targeted the aSD sequence of the plastid 16S rRNA rather than mutating the translation initiation regions of individual genes. Using stable transformation of the plastid genome, we introduced different mutations into the aSD sequence of the tobacco 16S rRNA gene and tested their effects on plant growth and development. The mutation conferring the mildest phenotype was selected for comprehensive ribosome profiling experiments to systematically identify plastid mRNAs whose translation was affected by a weakened SD-aSD interaction. Our results provide strong support for an important role of SDs in plastid translation initiation, reveal the set of plastid genes whose translation is SD dependent, and provide important insights into the relationship between mRNA secondary structure and translation efficiency.
RESULTS
Generation of Transplastomic Tobacco Lines with Mutated aSD Sequences
To assess the relevance of the SD-aSD interaction in chloroplast translation in a plastid genome-wide manner, we sought to mutate the aSD sequence in the 16S rRNA encoded by the plastid rrn16 gene. In Escherichia coli, mutated aSDs were previously introduced into ectopic copies of 16S rRNA (expressed from plasmids), leading to a second population of specialized ribosomes in the bacterial cell (Hui and de Boer, 1987), but causing lethality when expression of the mutated 16S rRNA was too strong (Jacob et al., 1987). The establishment of a parallel population of specialized ribosomes is not feasible in plastids because the highly active homologous recombination and gene conversion machineries that operate in plastids (Rogalski et al., 2006; Khakhlova and Bock, 2006) would prevent the coexistence of two rrn16 copies that differ in only a few nucleotides. We reasoned that a small genome like that of the plastid might be able to tolerate complete replacement of the wild-type 16S rRNA by a mutated copy harboring a mutation in the anti-Shine-Dalgarno sequence. It was reasonable to assume that, if the aSD plays an important role in translation initiation in the plastid, a relatively mild aSD mutation needs to be identified that is compatible with cellular viability in that it does not completely abolish translation of the many essential plastid genes (i.e., most genes involved in plastid gene expression and a few other reading frames; Drescher et al., 2000; Ahlert et al., 2003; Kode et al., 2005; Shikanai et al., 2001; Bock, 2007; Rogalski et al., 2008a, 2008b; Alkatib et al., 2012).
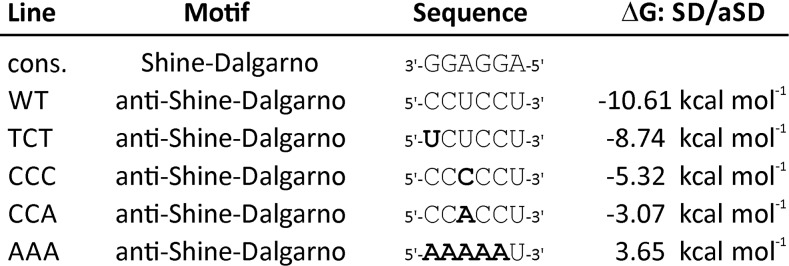
In an attempt to identify such a mutation, we tested several mutations that potentially cause reduced ability of the aSD to bind to the consensus SD. The mutations and their calculated effects on the free binding energy of the SD-aSD interaction are summarized in Figure 1. Mutation TCT affects the terminal base pair of the interaction. It changes a GC pair (with three hydrogen bonds) to a weaker GU pair (with two hydrogen bonds) and is expected to have the mildest effect, according to the moderate increase in the free binding energy of the aSD-SD interaction. Mutations CCC and CCA generate a mismatch in a central position of the SD-aSD interaction. The CCA mutation results in a purine-purine mismatch that, for steric reasons, is more destabilizing than a purine-pyrimidine mismatch, as present in the CCC mutant (Figure 1). Finally, a mutant rrn16 allele with a radically mutated aSD (AAA mutant) was constructed that should completely abolish any SD-aSD interaction (Figure 1).
Figure 1.
Sequences of Mutant Alleles of the aSD in the Plastid 16S rRNA of Tobacco Constructed in This Study and Their Calculated Effects on the Free Binding Energy of the SD-aSD Interaction in Plastid Translation.
Mutations introduced are indicated in bold letters. See text for details. cons., consensus sequence.
The different mutations were integrated into a cloned plastid DNA fragment containing the rrn16 region of the tobacco plastid genome (see Methods). An aadA cassette conferring spectinomycin resistance was inserted between the rrn16 and trnI-GAU [encoding for tRNAIle(GAU)] genes, 46 nucleotides downstream of the 3′ end of rrn16 (Figures 2A and 2B). Consequently, a 49- to 53-nucleotide-long wild-type sequence separates the mutations introduced into the aSD from the aadA cassette. This insertion site for the aadA cassette was chosen to minimize the probability of unwanted recombination events occurring between the inserted mutations and the selectable marker gene. The insertion site is upstream of a known processing site in the rRNA operon (Bisanz et al., 2003), which, however, appears to have limited functional significance in that it was reported to be removable without causing negative effects (Mühlbauer et al., 2002).
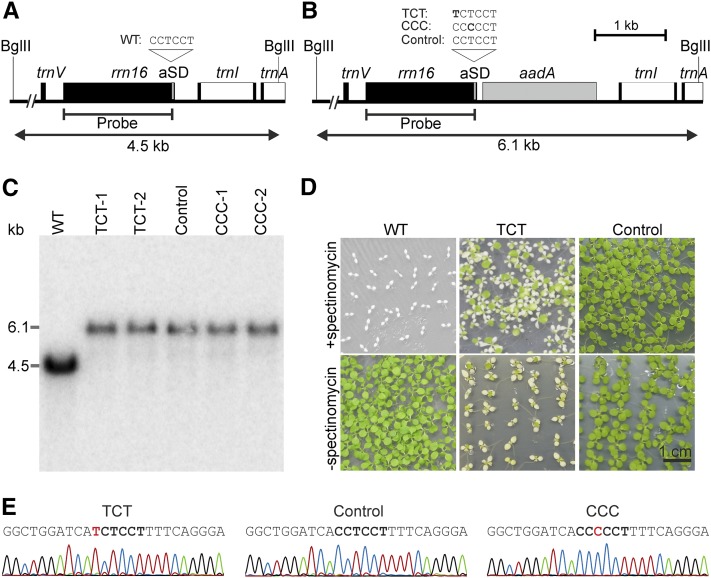
Figure 2.
Targeted Introduction of Point Mutations into the aSD Sequence of the Plastid 16S rRNA.
(A) Physical map of the wild-type tobacco plastid genome. Genes are represented as boxes, and relevant restriction enzyme digestion sites are indicated by thin vertical lines, with the name of the enzyme indicated above. The position of the hybridization probe and the length of the resulting RFLP fragments are also shown. The aSD is represented as a thin gray box at the 3′ end of the rrn16 gene, and its sequence is shown above the map.
(B) Map of the plastid genome region modified in the transplastomic mutants. The gray box indicates the inserted selectable marker gene aadA. The point mutations introduced into the aSD in the two mutants that could be purified to homoplasmy (TCT and CCC) are denoted in bold. Control lines contain the aadA but no mutation in the aSD.
(C) Representative RFLP analysis of wild-type tobacco and the three sets of transplastomic lines (TCT, CCC, and control). Total DNA was digested with BglII (cf. [A] and [B]) and hybridized to the radiolabeled rrn16 probe indicated in (A) and (B). All transplastomic lines are homoplasmic for the integrated selectable marker gene, as evidenced by the complete absence of the wild-type-specific band at 4.5 kb and the exclusive presence of the transgene-specific band at 6.1 kb. Moreover, they showed no phenotypic segregation upon vegetative propagation for more than 3 years, indicating homoplasmy for the introduced aSD mutation as well.
(D) Segregation assays demonstrating homoplasmy of a TCT and a control line. Seeds were germinated in the absence or presence of spectinomycin, and photographs were taken 1 month after sowing. Note that the pale seedling phenotype in the TCT line is caused by the mutation and not by sensitivity to spectinomycin, as evident from comparison of growth in medium with versus medium without spectinomycin). The CCC lines could not be included because due to their severe mutant phenotype (cf. Figure 3), they did not produce seeds.
(E) Sequencing of the 3′ end of the rrn16 gene of a TCT mutant, a control plant, and a CCC mutant to confirm the presence and homoplasmy of the point mutations in the two mutant lines. The aSD sequence is marked in bold, and the point mutations are labeled in red.
Using biolistic transformation and selection for spectinomycin resistance conferred by the chimeric aadA gene (Svab and Maliga, 1993), all aSD mutations (Figure 1) could be integrated into the tobacco plastid genome. However, homoplasmic mutants (i.e., mutants with the desired mutation in all chloroplast genomes) were obtained only for the TCT and the CCC mutations, suggesting that the CCA and AAA mutations are lethal. The CCA and AAA mutations initially remained heteroplasmic on medium with spectinomycin (balancing selection; Drescher et al., 2000), but eventually the mutations were lost due to recombination and/or gene conversion events (Khakhlova and Bock, 2006). The resulting stable homoplasmic lines contained the aadA marker but no point mutation in the 16S rRNA gene and therefore provided suitable control lines for all subsequent experiments with the TCT and CCC mutant lines. The homoplasmic state of the insertion of the aadA cassette was confirmed by restriction fragment length polymorphism (RFLP) analysis (and, for the TCT line, additionally by inheritance assays; Figures 2A to 2D), whereas the homoplasmic presence of the aSD mutations in the TCT and CCC lines and the absence of mutations from the aSD of the control lines were verified by DNA sequencing (Figure 2E).
Phenotypes of Transplastomic Lines with Mutations in the aSD Sequence
When grown under heterotrophic conditions on sucrose-containing medium, homoplasmic TCT lines were pale green, whereas homoplasmic CCC mutants displayed a much stronger phenotype. The plants grew slowly and had highly elongated, nearly white leaves. The control lines were indistinguishable from the wild type (Figure 3A). These phenotypes and the failure to isolate homoplasmic CCA and AAA lines confirm the theoretical predictions about the effects of the mutations based on the calculated hybridization affinities (free energies) of SD and aSD (Figure 1).
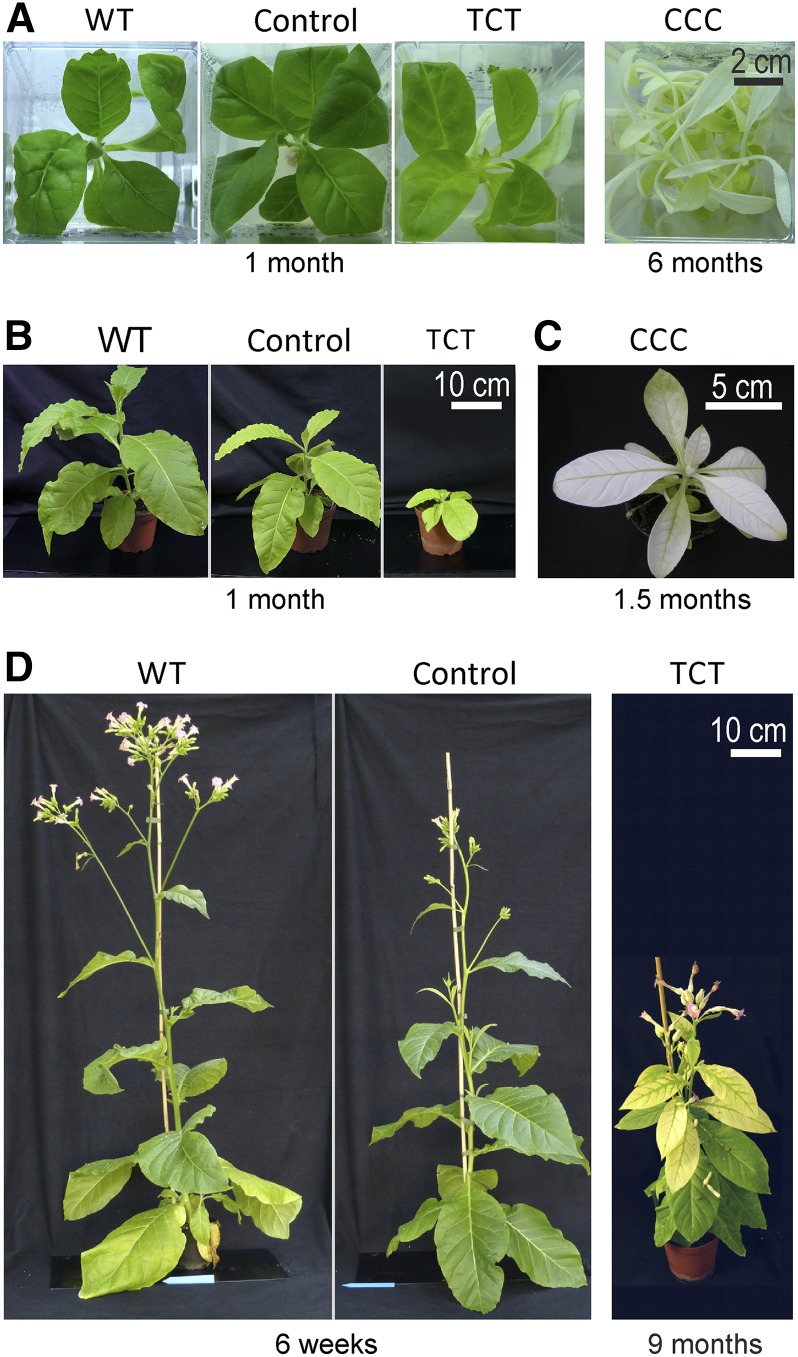
Figure 3.
Phenotypes of Transplastomic Plants Generated in This Study Compared to Wild-Type Plants.
(A) A wild-type plant, an aadA control line, and the TCT and CCC mutant lines grown on sucrose-containing medium. The plants were propagated through stem cuttings, and the different culture times required to reach the plant sizes shown here are given below the photographs.
(B) A wild-type plant in comparison to a control plant and a TCT line upon photoautotrophic growth in soil. The plants were derived from tissue culture cuttings and grown for 1 month at an average light intensity of 85 µE m−2 s−1 (as determined by several measurements during the day).
(C) A representative CCC mutant plant grown in soil for 1.5 months at 55 µE m−2 s−1.
(D) Comparison of a wild-type plant with a control plant and a TCT mutant after extended growth under photoautotrophic conditions. The plants were derived from tissue culture cuttings and grown photoautotrophically for the indicated time periods.
When grown autotrophically in soil, the phenotypic differences between the TCT and CCC mutants became even more pronounced (Figure 3), an observation that is in line with the known negative influence of impaired chloroplast translation on photosynthesis and thus photoautotrophic growth (Fleischmann et al., 2011; Ehrnthaler et al., 2014). The TCT lines developed pale green leaves and grew slowly, but ultimately flowered and produced viable seeds (Figures 3B and 3D). By contrast, the CCC mutants were extremely retarded in growth, had almost no visible pigmentation, and did not flower (Figure 3C). Under these conditions, the control line also displayed a subtle phenotype characterized by somewhat paler young leaves and a mild reduction in growth (Figures 3B and 3D). Interestingly, as the plants matured, the mutant phenotype became less severe and the plants were nearly indistinguishable from wild-type plants (Figure 3D), with the exception of a slight delay in flowering.
Homoplasmic presence of the resistance marker in the TCT mutant and the control lines was further confirmed by seed assays. When seeds were germinated on synthetic medium, the cotyledons of the TCT mutant seedlings were pale, but all seedlings grew in the presence of spectinomycin (and produced increasingly green leaves), ultimately demonstrating their homoplasmic state (Figure 2D). Due to the strong mutant phenotype, no inheritance assays could be performed with the CCC lines. However, the phenotype of the plants has remained somatically stable during more than three years of vegetative propagation in the absence of selection pressure for the transgenic plastid genome (in that the plants remained homogeneously pale without any reappearance of green sectors), indicating the absence of residual wild-type copies of the plastid genome.
Together with the impossibility to isolate homoplasmic CCA and AAA mutants, the phenotypes of the TCT and CCC lines correlate well with the predicted impairment in the SD-aSD interaction and suggest that the aSD-SD interaction plays an essential role in chloroplast translation.
16S rRNA Processing in Transplastomic Plants
The mild pale-green phenotype of the control lines (Figure 3) suggested that aadA insertion downstream of the 16S rRNA gene might affect rRNA processing. To test this possibility and to exclude a direct effect of the aSD mutations on 16S rRNA processing or stability, we analyzed rRNA processing and rRNA accumulation levels in the generated transplastomic lines. As additional controls, we included both wild-type plants and the wild-type-like E15 line harboring an aadA insertion in a different location of the plastid genome (between the psbE operon [encoding subunits of photosystem II] and the petA gene [encoding cytochrome f]; Bock et al., 1994).
RNA gel blot analysis revealed that, as suspected, the insertion of the aadA cassette downstream of the rrn16 gene caused the accumulation of additional 16S rRNA-containing transcripts, especially longer, incompletely processed transcripts that include the aadA coding region (Figures 4A to 4D). The size and relative abundance of the additional 16S rRNA transcripts were identical in the TCT mutant and the control line, pointing to insertion of the aadA cassette as the sole cause of the accumulation of these longer precursor RNA species. The very similar transcript processing and accumulation patterns of the TCT mutant and the control line also demonstrate that the severe phenotype of the TCT mutant (Figure 3) is not caused by destabilization or impaired processing of the 16S rRNA due to the introduced aSD mutation.
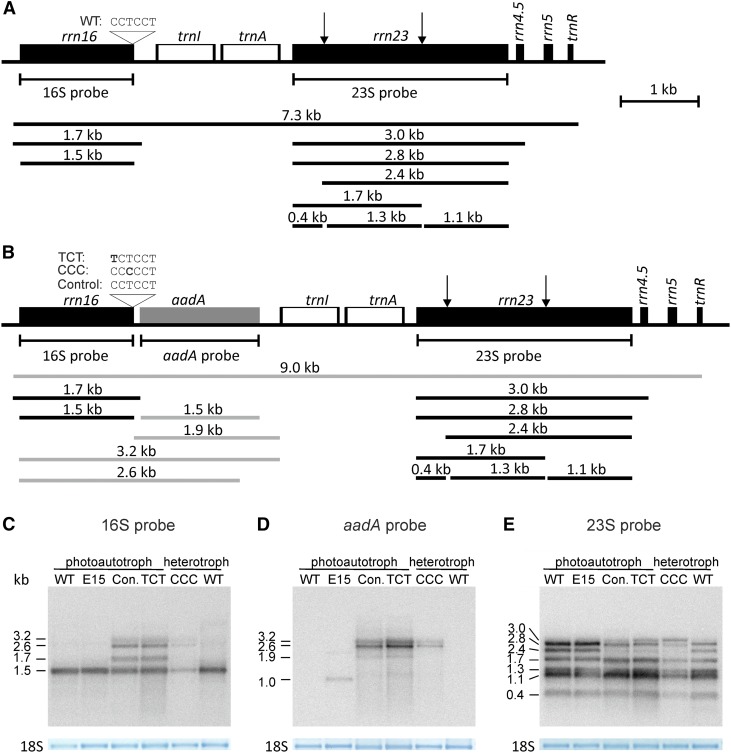
Figure 4.
rRNA Accumulation and Processing in Wild-Type Plants, aadA-Expressing Transplastomic Control Lines, and aSD Mutants.
(A) Map of the plastid rRNA operon including the different rRNA precursors and mature forms of plastid rRNAs. The binding sites of the hybridization probes and the position and sequence of the aSD are indicated. Arrows indicate the position of the “hidden break” processing sites in the 23S rRNA (Nishimura et al., 2010; Tiller et al., 2012).
(B) Map of the plastid rRNA operon in the aSD mutants and the control lines with the aadA cassette inserted downstream of rrn16. The additional RNA species including the aadA cistron are shown in gray.
(C) Accumulation and processing of the 16S rRNA in wild-type plants, the wild-type-like E15 line, the control line (Con.), and the aSD mutant plants (TCT and CCC), as determined by RNA gel blot hybridization. As a loading control, the methylene blue-stained cytosolic 18S rRNA band is shown. Note that additional 16S rRNA species accumulate in the control line and the mutant lines. The 2.6- and 3.2-kb species arise from cotranscription with the aadA cassette (cf. [B]). Wild-type, E15, control, and the TCT mutant plants were grown photoautotrophically, whereas the CCC mutant and another wild-type plant were grown heterotrophically on sucrose-containing medium.
(D) Accumulation of aadA-containing RNA species. The detected RNAs include those comprising the 16S rRNA and the aadA cassette (cf. [B] and [C]). The aadA cassette in the E15 line is inserted between petA and psbJ, and the expected transcript size is ∼1.1 kb (Bock et al., 1994).
(E) Accumulation and processing of the plastid 23S rRNA. Note that the 3-kb 23S-4.5S precursor overaccumulates in the CCC mutant line. RNA gel blot analyses were performed in two biological replicates. See text for details.
To exclude more subtle processing defects that would not be detectable by RNA gel blot analysis, we additionally mapped the 5′ and 3′ ends of the mature 16S rRNA in the wild-type and TCT lines (Supplemental Figure 1). Due to the proximity of the aSD to the 3′ end of the 16S rRNA, this was necessary to verify that 3′ end maturation of the 16S rRNA was not disturbed by the aSD mutation. However, no difference in 5′ and 3′ end formation was detected between the wild type and the TCT mutant. The sequencing of the 3′ ends also confirmed once again that the TCT mutation was present in all 16S rRNA molecules (Supplemental Figure 1). Compared with the wild type, both the control line and the TCT mutant showed slightly reduced levels of the 1.5-kb mature 16S rRNA (Figure 4C), thus suggesting a likely explanation for the mild growth phenotype of the control lines early in development (Figures 3B and 3D).
The CCC line showed strongly reduced levels of mature 16S and 23S rRNAs (Figures 4C and 4E), indicating the presence of substantially fewer plastid ribosomes. Consistent with this interpretation, the CCC plants also overaccumulate the 3-kb 23S-4.5S rRNA precursor, a processing defect that has been seen previously in various mutants affected in plastid translation (Yu et al., 2008; Tiller et al., 2012; Fristedt et al., 2014) and likely represents a secondary consequence of plastid ribosome deficiency. All transplastomic lines also showed reduced abundance of the 2.4-kb processing intermediate of the 23S rRNA (Figure 4E), but the three mature “hidden break” fragments of the 23S rRNA (Nishimura et al., 2010; Tiller et al., 2012) accumulated to wild-type levels in the TCT mutant and the control line.
In conclusion, comparison of rRNA processing and accumulation in the aadA control line and the aSD mutants suggests that the growth phenotypes of the aSD mutants and the reduced ribosome levels of the CCC lines are likely a direct consequence of inefficient translation as caused by the mutations introduced into the aSD sequence.
A Mutated aSD Sequence Causes Reduced Translation of a Subset of Plastid mRNAs
To characterize the suspected defects in plastid protein synthesis caused by the aSD mutations, we decided to comprehensively analyze translation by ribosome profiling. Ribosome profiling is a technique based on the analysis of nuclease-protected RNA footprints that are left by elongating ribosomes on their mRNA template (Ingolia et al., 2009). When mapped and quantified, footprint abundances in a given reading frame provide a valid proxy of protein synthesis (i.e., translational output; Ingolia et al., 2009). Previously, microarrays were employed to specifically map and quantify plastid transcript-derived ribosome footprints (Zoschke et al., 2013). Here, we used this approach to systematically analyze chloroplast translation in our transplastomic aSD mutants in comparison to the wild-type-like aadA-containing transplastomic line E15 (Bock et al., 1994) and a transplastomic control line with the aadA cassette inserted into the same genomic position as in the aSD mutants. Inclusion of the latter control was required to be able to distinguish between the effects of altered 16S rRNA processing (Figure 4C) and the translational defects caused by the point mutations in the aSD. We focused on the TCT mutant because secondary effects are expected to occur in the CCC mutants due to the strongly reduced accumulation of ribosomes (Figures 4C and 4E) and the poor growth of the mutant under both heterotrophic and photoautotrophic conditions (Figures 3A and 3C). To be able to exclude altered mRNA abundance as a possible cause of altered protein synthesis, the chloroplast transcript profiles of the TCT mutant and the control lines were also examined. By normalizing ribosome footprint abundances to transcript abundances, we calculated translation efficiencies, which reflect the translation activity of a given reading frame independent of the transcript level.
Chloroplast transcript levels and protein synthesis (i.e., ribosome footprints) of all lines were determined using a custom-made tiling microarray (see Methods for details). The results were highly reproducible in different biological replicates (Supplemental Figures 2A to 2F). Plastid transcript accumulation was very similar in the TCT mutant, the control line, and the wild-type-like line E15 (Supplemental Figures 2G and 2I), indicating that the aSD mutation does not appreciably affect transcription and/or mRNA stability. By contrast, plastid protein synthesis (measured by ribosome footprint abundances) in the TCT line was clearly different from that in the control and E15 lines (Supplemental Figures 2H and 2J). Given the unaltered mRNA levels, this observation points very strongly to changes in the translation efficiency of the affected plastid mRNAs.
To evaluate the changes in plastid translation activity, we first sorted the genes in the plastid genome based on the presence or absence of a predicted SD sequence (Supplemental Data Set 1). For this purpose, all genes with values <0 kcal mol−1 for the binding of the aSD to the 5′ UTR were preliminarily assumed to contain an SD. However, it should be noted that this cutoff also includes relatively weak potential interactions between aSD and 5′ UTR (Scharff et al., 2011; see Methods). We additionally considered to what extent the SD-aSD interaction would be affected by the TCT aSD mutation (Supplemental Data Set 1). According to the criteria defined above, 54 genes in the tobacco plastid genome possess a putative SD, including 30 that were predicted to suffer from weakened binding to the aSD in the TCT mutant. For 11 genes, no change was predicted and for 13 genes, a strengthened binding was predicted. Twenty-four genes have no recognizable SD in the wild type, and nine of these may acquire a weak SD in the TCT mutant (Supplemental Data Set 1).
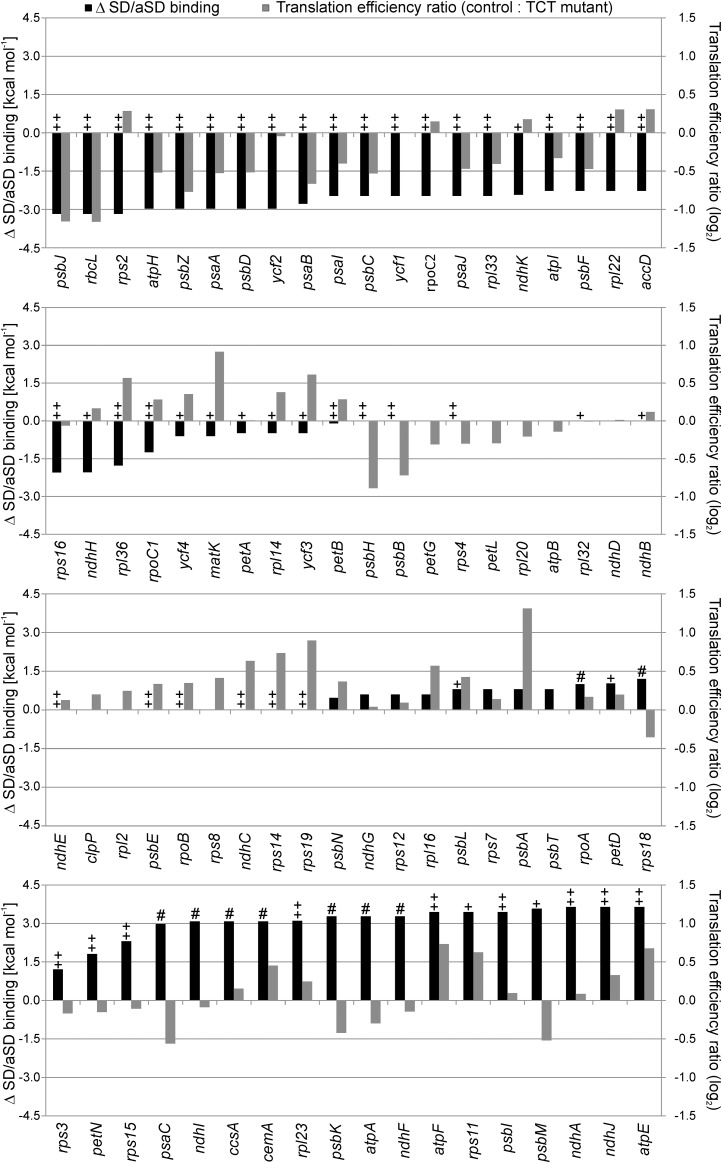
For each coding region, the relative change in median translation efficiency (ribosome footprints/transcript) between the TCT mutant and the control line was calculated and plotted together with the in silico calculated difference in the strength of SD-aSD binding (Figure 5; Supplemental Data Set 1). The genes were then sorted according to the extent of predicted impairment of the SD-aSD binding (black bars in Figure 5).
Figure 5.
Genome-Wide Comparison of the Calculated Changes in SD-aSD Binding and the Measured Changes in Translation Efficiency.
The changes in SD-aSD binding (black columns) were calculated for all plastid genes by subtracting the value for the TCT mutant (calculated by Free2Bind at 22°C; see Methods) from that calculated for the wild type. A negative value indicates a weakened binding in the TCT mutant, and a positive value a potentially strengthened SD-aSD interaction. The genes were sorted according to the difference in SD-aSD binding. The translation efficiency was calculated by normalizing the ribosome profiling data to the wild-type-like line E15 and dividing the signal intensities of the ribosome footprints by those of the transcripts for each probe. Displayed are median values for each coding region as ratio control/TCT (gray columns; log2 values). A negative value indicates reduced translation efficiency in the TCT mutant, a positive value increased translation efficiency. The ++ indicates genes with a strong putative SD (< −5 kcal mol−1) in the wild type, + genes with a relatively weak putative SD (−5 to 0 kcal mol−1), # genes that potentially gain a weak SD (−5 to 0 kcal mol−1) in the TCT mutant, and all unmarked genes lack any potential SD (>0 kcal mol−1). There were no genes that would gain a strong SD by the TCT mutation. Note that most genes with a strong SD that were predicted to display a reduction in the strength of the SD-aSD interaction in the TCT mutant showed a decline in translation efficiency (uppermost panel). By contrast, genes with unchanged or potentially improved SD-aSD binding displayed heterogeneous changes in translation efficiencies (lower three panels).
In general, most genes with a strong SD that were predicted to display a reduction in the strength of the SD-aSD interaction in the TCT mutant showed a decline in translation efficiency. Genes with a predicted strong SD and a predicted strongly reduced SD-aSD binding in the mutant were most severely impaired in their translation efficiency by the TCT aSD mutation (Figure 5, uppermost panel).
Polysome Loading Analyses Support Ribosome Profiling Data
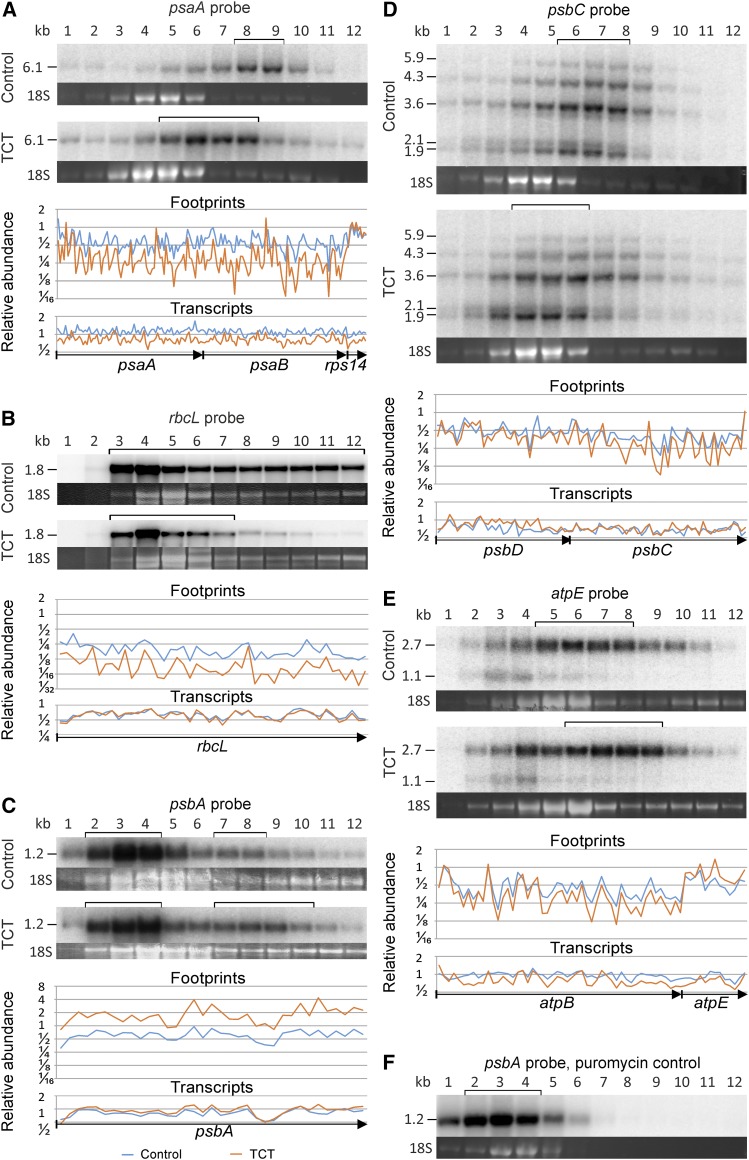
For selected genes, the observed changes in translation efficiency were verified by polysome analysis (Figure 6). To this end, polysomal complexes were purified and size-fractionated in sucrose density gradients, followed by identification of mRNA-containing fractions by RNA gel blot analysis. We first analyzed three reading frames whose translation, according to the ribosome profiling data (Figure 5), was clearly negatively affected by the aSD mutation: psaA, psbC, and rbcL (encoding a reaction center protein of photosystem I, the core antenna protein CP43 of photosystem II, and the large subunit of Rubisco, respectively). psaA-, psbC-, and rbcL-containing mRNAs showed a clear shift toward lighter gradient fractions in the TCT mutant (Figures 6A, 6B, and 6D), indicating coverage with fewer ribosomes and, hence, less efficient translation initiation. We also analyzed two transcripts whose translation appeared to be increased based on their translation efficiency, as measured by ribosome profiling (Figure 5): atpE (encoding the ε-subunit of the ATP synthase) and psbA (encoding the reaction center protein D1 of photosystem II). Analysis of polysome loading of atpE mRNA revealed migration of a portion of the transcript population into gradient fractions of higher density in the TCT mutant (Figure 6E), thus confirming more efficient translation, which is consistent with both the increased number of ribosome footprints (normalized to transcript levels) and the calculated strengthened SD-aSD interaction (Figure 5, lowermost panel). The psbA mRNA represents a somewhat special case in that (1) psbA is one of the most intensely transcribed plastid genes, and (2) psbA gene expression is strongly translationally regulated by light and redox signals (Staub and Maliga, 1993; Trebitsh and Danon, 2001; Herz et al., 2005). Only a fraction of the psbA transcript pool is actively translated, whereas the other fraction presumably serves as a buffer for rapid light and/or redox-induced translational activation. Consequently, analysis of polysome loading of the psbA mRNA revealed two pools of psbA transcripts: a large untranslated pool accumulating in the upper gradient fractions and a smaller pool of translated mRNAs that migrated more deeply into the gradient (Figures 6C and 6F). The latter was shifted into heavier fractions in the TCT mutant, confirming the more intense translation of psbA in the aSD mutant that had been detected by ribosome profiling (Figure 5).
Figure 6.
Verification of Changes in Translation by Comparative Polysome Analysis of TCT Mutant Plants and Control Plants.
The polysome analysis and the relative transcript and ribosome footprint abundances as determined by microarray analyses are shown. The brackets above the polysome blots indicate the gradient fractions in which the bulk of the transcripts accumulate. The sucrose gradient fractions are numbered from the top to the bottom of the gradient (1–12). The ethidium bromide-stained cytosolic 18S rRNA in the agarose gel prior to blotting is shown as a quality control for equal distribution of the cytosolic ribosomes in the sucrose density gradients used for analysis of the two plant lines. The transcript and ribosome footprint abundances of the TCT mutant (orange) and the control line (blue) determined by microarray analysis are normalized to the wild-type-like control line and shown as log2 values. Only data for coding regions are shown. Arrows below the plots denote the different reading frames.
(A) Analysis of psaA translation. psaA suffers from a weakened SD-aSD interaction in the aSD mutant (cf. Figure 5). psaA is, together with psaB (also affected by a weakened SD-aSD interaction) and rps14 (unaltered SD-aSD interaction), part of a tricistronic transcript.
(B) Analysis of rbcL translation. Translation of rbcL is reduced by a weakened SD-aSD interaction in the TCT mutant. rbcL represents a monocistronic transcript.
(C) Analysis of psbA translation. Both the untranslated pool of psbA mRNA (fractions 2-4) and the translated pool (fractions 7-8 and 7-10, respectively) are marked by brackets. Note the shift of psbA transcripts into fractions of higher density in the TCT mutant (see text for details), which confirms the increase in the relative abundance of footprints as detected by microarray analysis. psbA has no SD.
(D) Analysis of psbC translation. psbC is another mRNA whose translation would be affected by a weakened SD-aSD interaction in the TCT mutant. psbC is part of an operon that additionally includes psbD and psbZ (both of which are also predicted to have weakened SD-aSD interactions). The probe detects monocistronic (∼1.9 kb), dicistronic (with psbD; ∼3.6 kb), and tricistronic (with psbD and psbZ; ∼4.3 kb) psbC transcripts.
(E) Analysis of atpE translation. atpE is an example of a gene with a predicted strengthened SD-aSD interaction in the TCT mutant (Figure 5). The upper band represents dicistronic atpB/E transcripts and the lower band the monocistronic atpE message (atpB has no SD). Note the shift of a portion of the atpE transcripts into fractions of higher density in the TCT mutant corresponding to the increased relative abundance of ribosome footprints, as detected by microarray analysis.
(F) Puromycin control. Puromycin causes premature translation termination (i.e., release of the nascent peptide chain and dissociation of the two ribosomal subunits). The puromycin control reveals the gradient fractions containing untranslated mRNAs, including mRNAs present in other ribonucleoprotein complexes. The polysomes were isolated from a control line and, for direct comparison with (C), the blot was hybridized to a psbA-specific probe to demonstrate that fractions 7 to 12 contain translated psbA mRNA.
In summary, the changes in plastid translation uncovered by ribosome profiling can be readily verified by polysome loading assays, supporting the earlier notion that analysis of ribosome footprints per transcript represents a valid proxy of translation activity (Ingolia et al., 2009; Zoschke et al., 2013).
Evidence in both bacteria and chloroplasts indicates that ribosome progression can be paused by interactions between the aSD sequence and SD-like sequences within coding regions (Li et al., 2012; Zoschke et al., 2013). The genome-wide inspection of ribosome distribution patterns in the TCT mutant and the control lines revealed very similar profiles (for examples, see Figure 6 and Supplemental Figure 3), which strongly argues for an overall unaltered elongation behavior. However, it should be noted that our ribosome profiling method has a limited resolution of ∼30 nucleotides, which is not sufficient to analyze ribosome pausing in detail (and to assign the few observed local differences in ribosome occupancy to predicted changes in aSD binding to the coding region). Future analyses by deep sequencing-based ribosome profiling will be needed to address this issue more comprehensively.
Relationship between Translation Efficiency and the Strength of the SD-aSD Interaction
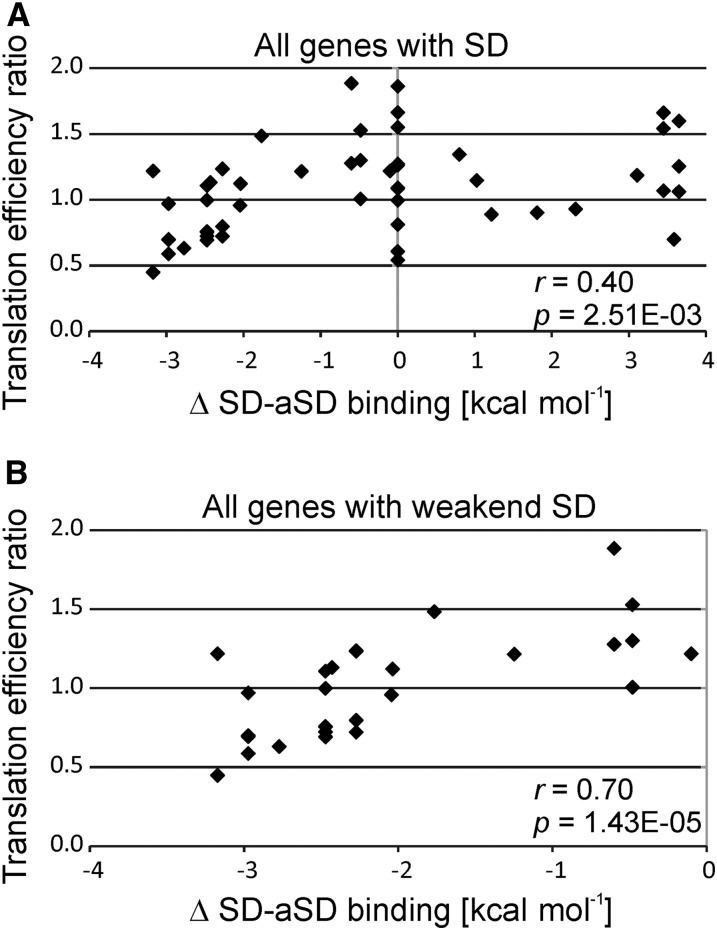
When all plastid reading frames were analyzed, a correlation between the change in SD-aSD binding and the change in the translation efficiency caused by the TCT mutation was observed (Figure 7A). The correlation was even stronger when only genes whose SD-aSD interaction is predicted to be impaired in the TCT mutant were considered (Figure 7B). However, there are also a few notable exceptions, such as the rps2 gene (encoding ribosomal protein S2), which showed improved translation in the TCT mutant despite a calculated decrease in the strength of the SD-aSD interaction (Figure 5, uppermost panel). These exceptions may point to the operation of other regulatory mechanisms in translational control that are not (only) dependent on the SD-aSD interaction and may, for example, rely on mRNA-specific translational activator and/or repressor proteins (Choquet et al., 1998; Fisk et al., 1999; McCormac and Barkan, 1999; Shen et al., 2001; Sane et al., 2005; Zoschke et al., 2012; Link et al., 2012).
Figure 7.
Correlation between Measured Translation Efficiency and Calculated Impairment of SD-aSD Binding.
(A) Correlation analysis for all plastid genes with SD sequences. The relative change in the translation efficiency in the TCT mutant compared with the control line is correlated to the difference in the strength of SD-aSD binding. The values for the translation efficiency are the medians of the ribosome footprints normalized to the medians of the transcript levels for each coding region. The values for both the TCT mutant and the control line were normalized to the wild-type-like line E15, and the ratio control/TCT was used for this plot. The difference in SD-aSD binding was calculated by subtracting the value calculated for the TCT mutant (with Free2Bind; see Methods) from the value calculated for the wild-type aSD. A negative value indicates a weakened interaction and a positive value a stronger interaction in the TCT mutant. Pearson’s r and P value are given (in nEm non-superscript format for n⋅10m).
(B) Correlation analysis of plastid genes that are predicted to suffer from a weakened SD-aSD interaction in the TCT mutant.
Interestingly, the sensitivity of translation activity to a reduction in the strength of the SD-aSD interaction was particularly pronounced for plastid genes related to photosynthesis (Supplemental Figure 4). When translation in operon-derived polycistronic transcripts was investigated, the changes in translation efficiency conferred by the aSD mutation were found to be largely independent of the position of the genes in the operon (Supplemental Figure 5). This was the case in both operons comprising genes coding for subunits of the same protein complexes and operons encoding subunits of different protein complexes (Supplemental Figure 5). Taken together, these results suggest that impaired SD-aSD binding is responsible for the observed defects in plastid translation and the specific mutant phenotype of the TCT plants.
There are several mRNAs that are clearly more efficiently translated in the TCT mutant than in the wild type, for example, atpE, atpF (both encoding subunits of the ATP synthase), cemA (encoding a chloroplast envelope membrane protein), psbL (encoding the L subunit of photosystem II), and rps11 (encoding ribosomal protein S11) (Figures 5 and 6C). However, while there is a strong correlation between the predicted weakening of SD-aSD interactions and the observed changes in translation efficiency in the TCT mutant (Figure 7B), there was virtually no correlation between potentially strengthened SD-aSD interactions and the determined changes in translation efficiency (Supplemental Figure 6A; Figure 5, two bottom panels). This also holds true for genes that gained a putative SD in the TCT mutant (Supplemental Figure 6B). However, it should be noted that the SDs gained were all relatively weak (−5 to 0 kcal mol−1; Supplemental Data Set 1).
The lack of a correlation between improved (or gained) SD-aSD binding and translation efficiency (Supplemental Figure 6) and the observed correlation between weakened SD-aSD interactions and translation efficiency (Figure 7B) prompted us to investigate whether the strength of SD-aSD binding can predict the translation efficiency of a given gene. To this end, we analyzed the ribosome profiling data for the wild-type-like E15 line, the control line, the TCT mutant, and a published data set from maize (Zea mays; Chotewutmontri and Barkan, 2016). No strong correlation between the strength of the SD-aSD interaction and translation efficiency was seen (Supplemental Figure 7), suggesting that, although the SD-aSD interaction clearly influences translation of many plastid genes (Figures 5 to 7), the strength of the interaction does not directly determine the translation activity. Instead, our data suggest that the interplay with other regulatory factors, such as mRNA-specific translational regulator proteins and/or mRNA secondary structure around the start codon (see following paragraph), is likely to fine-tune and also limit the translational output for most plastid genes.
The Role of mRNA Secondary Structure in SD-Dependent Plastid Translation
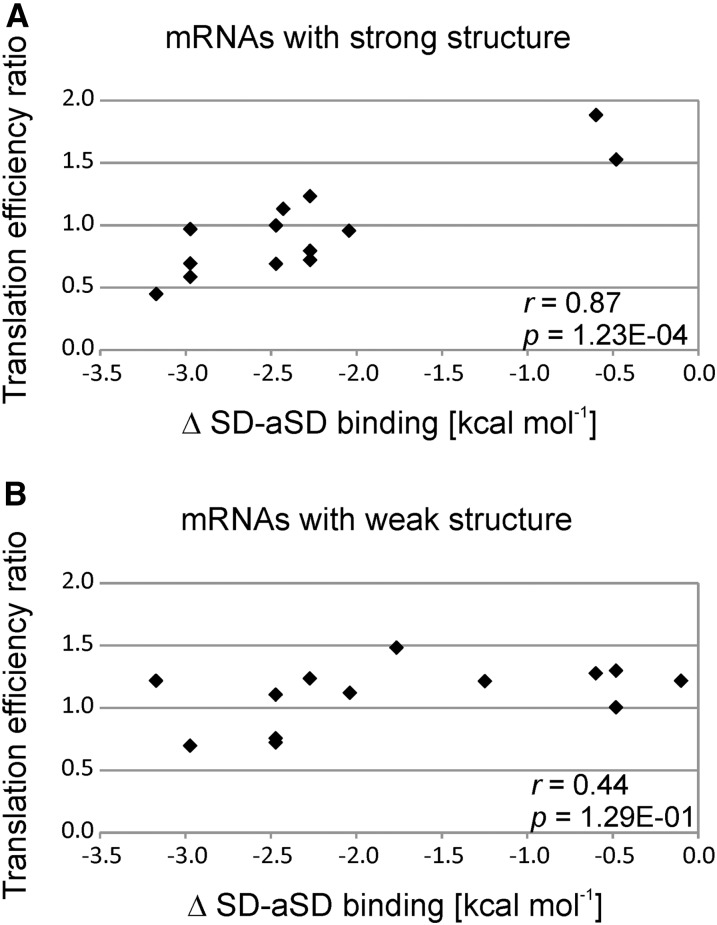
The expression of reporter genes in E. coli and in tobacco chloroplasts demonstrated that a local minimum of mRNA structure could compensate for the lack of an SD in promoting translation initiation (Scharff et al., 2011; Zhang et al., 2012). We therefore sought to test the influence of mRNA secondary structure on SD-dependent translation. To this end, we sorted the plastid genes according to the stability of the mRNA secondary structure around the start codon. Interestingly, we found that the genes with the strongest mRNA structures were impaired in their translation efficiency in the TCT mutant in strong correlation with the weakened SD-aSD binding (Figure 8A), confirming their SD-dependent translation. By contrast, the group of genes that had only weak mRNA secondary structure around the start codon showed no appreciable correlation between impaired SD-aSD binding and reduced translation efficiency in the TCT mutant (Figure 8B). This indicates that the start codons of these genes could be alternatively recognized based on the lack of mRNA secondary structure (Scharff et al., 2011); therefore, translation of these genes may not strictly depend on an SD. A good example for this group is rps2, which has both a strong SD and a local minimum of mRNA secondary structure around the start codon. Its translation efficiency was not reduced in the mutant, even though SD-aSD binding is expected to be strongly impaired (Figure 5, uppermost panel; Supplemental Figure 5 and Supplemental Data Set 1).
Figure 8.
Influence of the mRNA Structure around the Start Codon on Translation Efficiency in Relation to the Strength of the SD-aSD Interaction.
(A) Analysis of the 43% of plastid genes with the strongest mRNA structure around the start codon (i.e., the most negative minimum free energy) and weakened SD-aSD binding in the TCT mutant. Note that the change in the translation efficiency in the TCT mutant compared with the control line is highly correlated to the effect on SD-aSD binding. Pearson’s r and P value are given (in nEm non-superscript format for n⋅10m).
(B) Analysis of the 43% of genes with the weakest mRNA structure around the start codon and weakened SD-aSD binding in the TCT mutant. Note that the change in translation efficiency in the TCT mutant compared with the control line only weakly correlates with the effect on SD-aSD binding. Pearson’s r and P value are given. Genes with intermediate (uncertain) structuredness were omitted from the analysis (14% of the genes). No pronounced bias in the representation of plastid gene classes (photosynthesis-related genes versus other genes; cf. Supplemental Figure 4) was observed among strongly structured or weakly structured mRNAs.
Similar to the results observed for the strength of the SD-aSD interaction (Supplemental Figure 7), there was also no correlation between the amount of mRNA secondary structure around start codons and translation efficiency (Supplemental Figure 8). This result may indicate that many plastid genes are not translated at the maximum level that their translation initiation region would permit, but rather are additionally regulated by other factors such as mRNA-specific trans-acting regulator proteins.
DISCUSSION
The role of SD sequences in plastids is highly controversial (Zerges, 2000; Sugiura, 2014), which motivated us to systematically assess their role in translation initiation. In this study, we introduced mutations into the aSD sequence in the 16S rRNA (Figures 1 and 2) to enable a genome-wide analysis of an impaired SD-aSD interaction. Compared with the analysis of individual transcripts using conventional techniques (such as polysome-loading analysis), ribosome profiling of all plastid mRNAs does not only reveal global patterns and genome-wide changes in protein synthesis, but it also allows the detection of specific changes in translation efficiency of individual cistrons in polycistronic transcripts.
Of the four mutations tested, only the two mutations expected to have the mildest effects on the SD-aSD interaction (Figure 1) gave rise to viable homoplasmic transplastomic mutants. Of these two, the severity of the mutant phenotype was correlated with the calculated impact of the mutation on the strength of SD-aSD hybridization (Figures 1 to 3). The stronger CCC mutant, in which the central U of the aSD was mutated to C, was much more severely affected in growth than the TCT mutant (Figures 3A and 3C), with the extent of its growth impairment similar to that in the strongest plastid translation mutant currently known in tobacco, the rpl36 (gene for ribosomal protein L36) knockout (Fleischmann et al., 2011; Ehrnthaler et al., 2014). The failure to generate an even stronger point mutation by introducing a U-to-A transversion in the central position (CCA mutant) or by completely eliminating the aSD (AAA mutant; Figure 1) suggests that with the CCC mutation, the maximum tolerable reduction in translation activity in the plastid has essentially been reached. Together, the observed effects of the tested mutations in vivo and their correlation with the degree of weakening of the consensus SD-aSD interaction suggest that the aSD sequence plays an essential role in chloroplast translation.
The TCT mutant may represent one of the mildest mutations that can be introduced into the aSD because it (1) affects only the terminal base pair of the SD-aSD interaction and (2) is a pyrimidine transition (C-to-U exchange; Figure 1). Comprehensive analysis of translation activity by ribosome profiling of the TCT mutant in comparison to the control lines established a clear correlation between the predicted extent of weakening of the SD-aSD interaction and the measured reduction of translation efficiency of the affected plastid mRNAs (Figure 7B). The demonstration that the aSD is important for translation of most genes (1) that possess an SD and (2) whose SD-aSD interaction was predicted to be negatively affected in the TCT mutant (Figures 5 to 7) lends strong further support to the functional significance of the aSD for SD-containing mRNAs in plastids.
Our results are in agreement with some in vitro data obtained previously by testing mutant 5′ UTRs of specific plastid genes. For example, translation of psbC and rbcL was described as being SD-dependent in an in vitro translation system (Kuroda et al., 2007; Hirose and Sugiura, 2004). Both genes are predicted to have a weakened SD-aSD interaction in our TCT mutant and, indeed, their translation efficiency was reduced in the TCT mutant (Figure 5; Supplemental Data Set 1). Based on in vitro data, atpE translation has also been suggested to be SD dependent (Hirose and Sugiura, 2004). atpE exhibits strengthened SD-aSD binding in the TCT mutant; accordingly, its translation efficiency was found to be increased in the mutant (Figure 5; Supplemental Data Set 1).
It is important to note that not all plastid genes behave as expected based on their SD-aSD binding properties (Figure 5). Interestingly, the single gene for which, based on in vitro data, a negative influence of the SD on translation was reported (rps2; Plader and Sugiura, 2003) was also not affected by the TCT mutation (Figure 5; Supplemental Figure 5). rps2 possesses a strong SD, but in addition, its start codon resides within a local minimum of mRNA secondary structure (sixth weakest secondary structure of all plastid translation initiation regions; Supplemental Data Set 1). We propose that the latter provides an alternative route to translation initiation on the rps2 mRNA and, in this way, makes the SD-aSD interaction nonessential.
In E. coli, translation initiation rates are also strongly influenced by mRNA structure (Kudla et al., 2009; Goodman et al., 2013; Burkhardt et al., 2017), and the interplay between SD strength, mRNA secondary structure, and the action of additional translational regulator proteins makes it very difficult to predict translation efficiencies. In plastids, we found that the translation of mRNAs that are weakly structured in their translation initiation region is less dependent on the presence of an SD (Figure 8). This finding lends further support to the existence of two distinct mechanisms for start codon recognition in plastids: ribosome binding to the SD sequence followed by initiation at a start codon present at an appropriate distance downstream (Drechsel and Bock, 2011) or, alternatively, direct start codon recognition facilitated by its accessibility in an extended unstructured region of the mRNA (Scharff et al., 2011).
Previous work in both bacteria and plastids has revealed the importance of appropriate spacing between the SD and the translation initiation codon (Chen et al., 1994; Hirose and Sugiura, 2004). For example, using an in vitro translation system, it was shown that the SD-like sequence of the plastid rps12 mRNA (encoding ribosomal protein S12), which is too far upstream of the start codon (−42 nucleotides) to act as a functional SD, has no influence on translation activity. Our in vivo results described here are in good agreement with the in vitro findings in that rps12 translation is not reduced in the TCT mutant, likely because the rps12 start codon is recognized through a local minimum of mRNA secondary structure (Figure 5; Supplemental Data Set 1).
Our data also show that the strength of SD-aSD binding alone cannot predict the translation efficiency of plastid genes (Supplemental Figure 7). This is also the case in E. coli (Li et al., 2014; Burkhardt et al., 2017). It appears likely that the translation of most genes is regulated by additional transcript-specific mechanisms to fine-tune protein synthesis and adjust it to changing cellular demands. Therefore, plastid translation may only rarely be active to the maximum level that is theoretically possible based on the properties of the translation initiation region. This assumption is supported by the extraordinarily high levels of foreign protein accumulation attained in chloroplasts by transgene expression from foreign expression elements (e.g., phage-derived 5′ UTRs), which likely remain unregulated in plastids (Ye et al., 2001; Zhou et al., 2008; Oey et al., 2009).
In both prokaryotes and eukaryotes, translation efficiency and transcript stability are correlated (Sachs, 1993; Sunohara et al., 2004; Deana and Belasco, 2005; Presnyak et al., 2015; Barahimipour et al., 2015; Neymotin et al., 2016) in that untranslated (or inefficiently translated) mRNAs are often prone to rapid degradation. By contrast, in chloroplasts, transcript accumulation is often not strongly influenced by altered translation efficiency. Our comparative ribosome profiling experiments revealed many plastid transcripts whose translation was altered (i.e., either increased or decreased; Figure 5; Supplemental Figure 9). However, altered translation efficiency (i.e., ribosomes per transcript) did not result in appreciable changes in mRNA accumulation levels (Supplemental Figure 2). This suggests that, unlike in most other systems, mRNA turnover in plastids is largely independent of the translational status, and ribosomes do not exert a strong protective role. Hence, in the absence of ribosomes, plastid transcripts must be largely protected from degradation by other factors, such as terminal secondary structures and/or protective RNA binding proteins (Stern and Gruissem, 1987; Suay et al., 2005; Pfalz et al., 2009; Kupsch et al., 2012; reviewed in Barkan and Small, 2014).
In conclusion, our work highlights both similarities and differences in the translation processes in plastids and bacteria. Both plastids and bacteria possess genes with and genes without SDs (Chang et al., 2006; Nakagawa et al., 2010; Scharff et al., 2011). In both systems, SD-aSD binding and local minima of mRNA structure determine start codon recognition and, in this way, the efficiency of translation initiation. Differences between bacterial translation and plastid translation lie in the smaller effects of RNA structure on SD-dependent translation in chloroplasts and in the trans-acting factors that are employed to regulate translation. While translational control in bacteria is exerted by regulatory proteins, riboswitches, or small RNAs (Duval et al., 2015), plastid translation is mainly fine-tuned by nucleus-encoded gene-specific RNA binding proteins, many of which were established during the coevolution of the host and endosymbiont (Barkan, 2011; Greiner and Bock, 2013; Barkan and Small, 2014).
METHODS
Plant Material and Growth Conditions
Nicotiana tabacum cv Petit Havana plants for the plastid transformation experiments were grown under aseptic conditions on Murashige and Skoog medium (Murashige and Skoog, 1962) supplemented with 30 g/L sucrose at a light intensity of 50 µE m−2 s−1 (bulb type: Philips MASTER, white, TL-D SUPER80 58W/840 DIM 1SL/25). Regenerated transplastomic plants were grown on the same medium and under identical conditions, with the exception of homoplasmic CCC lines, which were grown at 25 µE m−2 s−1. For the analysis of translation, plants were grown in soil at 75 µE m−2 s−1 and 70% relative humidity in long-day conditions (16 h light at 22°C/8 h dark at 18°C). Leaf tissue was harvested from plants of a similar developmental stage (four true leaves) 30 min after the start of the light cycle and immediately snap-frozen in liquid nitrogen. Samples were stored at −80°C until use. For each analysis, leaf tissue from two individual plants of identical genotype was pooled and represented one biological replicate.
Construction of Plastid Transformation Vectors
The plastid transformation vector for introduction of the TCT mutation (Figure 1) into the rrn16 gene of the tobacco chloroplast genome was constructed as follows: A portion of the rrn16 gene (corresponding to nucleotide positions 103,208–104,298 and 138,322–139,422 in the inverted repeat region of the tobacco plastid genome; NC_001879.2) was amplified as the flanking region for homologous recombination with the primers Prrn16for (adding a HindIII restriction site) and Prrn16-mut1ASDrev (introducing the point mutation into the aSD sequence and a SalI restriction site 46 nucleotides downstream of the 3′ end of the rrn16 gene; for primer sequences, see Supplemental Table 2). The second flanking region for homologous recombination includes part of the trnI-GAU gene (corresponding to nucleotide positions 104,299–105,286 and 137,344–138,331, respectively). It was amplified with the primers PtrnIfor (introducing the same SalI restriction site 46 nucleotides downstream of rrn16 described above and an additional KpnI restriction site downstream of the SalI site) and PtrnIrev (Supplemental Table 2). The two flanking regions were combined by PCR using a 30-nucleotide overlapping sequence and the primers Prrn16for and PtrnIrev. The resulting amplification product was inserted into plasmid vector pUC18, from which the EcoRI site had been removed by digestion with EcoRI, followed by conversion to blunt ends by a fill-in reaction using the Klenow fragment of Escherichia coli DNA polymerase I. The blunted linear vector was then digested with HindIII and ligated to the PCR product digested with the same enzyme. The resulting plasmid clone contains the SalI and KpnI restriction sites in the spacer between rrn16 and trnI-GAU. Between these restriction sites, an aadA cassette (comprising the aadA coding region from E. coli, the psbA promoter and 5′ UTR, and the rbcL 3′ UTR from Chlamydomonas reinhardtii) was inserted, which had been cut out from plasmid pLS1 (Fleischmann et al., 2011) with the same enzyme pair, thus generating the final plastid transformation vector (TCT vector). The TCT vector was used as the starting point to construct the vectors for introduction of the other aSD mutations. The CCC mutation (Figure 1) was inserted by amplification of part of the rrn16 gene with primers Prrn16-EcoRIfor and Prrn16-mut2ASDrev (Supplemental Table 2). The latter primer includes the mutated aSD. The PCR product was digested with EcoRI and SalI and inserted into the TCT vector cut with the same enzymes. The vectors carrying the CCA mutation (CCA vector) and the A stretch (AAA vector) replacing the aSD sequence (Figure 1) were generated by the same approach, but using the reverse primers Prrn16-mut3ASDrev and Prrn16-delta2ASDrev, respectively. The sequences of all vectors were confirmed by complete resequencing.
Plastid Transformation and Selection of Transplastomic Lines
Leaves from aseptically grown N. tabacum cv Petit Havana plants were bombarded with plasmid DNA-coated 0.6-µm gold particles using a helium-driven biolistic gun (PDS1000He; Bio-Rad). Primary transformants were selected on plant regeneration medium (RMOP medium; Svab and Maliga, 1993) containing 500 µg/mL spectinomycin. Spontaneous spectinomycin-resistant mutants were identified based on their sensitivity to streptomycin (500 µg/mL; Bock, 2001) and discarded. All transplastomic lines were tested for the presence of the inserted point mutations by amplifying and sequencing the corresponding region of the 16S rRNA gene with primers Prrn16for and PaadA136 (for primer sequences, see Supplemental Table 2). These tests were repeated after each selection round because the point mutation and the selectable marker gene are separated by a 49- to 53-bp wild-type sequence and therefore can be uncoupled by gene conversion (Khakhlova and Bock, 2006). Transplastomic lines containing only the aadA cassette inserted between rrn16 and trnI-GAU but no aSD mutation were used as controls. As an additional wild-type-like control, a transplastomic line with an aadA cassette inserted between petA and psbJ was included (Bock et al., 1994).
Isolation and Analysis of Nucleic Acids
For RFLP analyses, total plant DNA was extracted using a cetyltrimethylammonium bromide-based method (Doyle and Doyle, 1990), digested with the restriction enzyme BglII, separated by electrophoresis in 1% agarose gels, and blotted onto Hybond N nylon membranes (GE Healthcare). For RNA gel blot analyses, total RNA was isolated from fresh leaves using the peqGOLD TriFast reagent (Peqlab), electrophoretically separated in denaturing formaldehyde-containing 1.2% agarose gels, and blotted onto Hybond N nylon membranes (GE Healthcare). For detection of nucleic acids on membranes, [α-32P]dCTP-labeled probes were produced by random priming (Multiprime DNA labeling kit; GE Healthcare) using amplified and gel-purified PCR products as template (for primer sequences, see Supplemental Table 2). Hybridizations were performed at 65°C using standard protocols. Polysome analyses were performed as described previously (Ehrnthaler et al., 2014) using sucrose gradients for the separation of polysomal complexes. The ends of the 16S rRNA were determined by circularizing the RNA molecules with T4 RNA ligase. cDNA covering the joined ends was produced by reverse transcription using primer P16S_endmapper_F and Superscript III reverse transcriptase (Thermo Fisher). Following digestion of the RNA with RNase H, the cDNA was amplified with primers P16S_Endmapper_F2 and P16S_Endmapper_R2 (Supplemental Table 2). The resulting PCR product was inserted into a standard cloning vector (PCR 2.1-TOPO; Thermo Fisher) and transformed into E. coli, and plasmids isolated from individual clones were sequenced.
Ribosome Profiling
Ribosome profiling was essentially performed according to published protocols (Zoschke et al., 2013). Briefly, 400 mg snap-frozen tobacco leaves from two individual plants were ground with a mortar and pestle, and the frozen powder was lysed with 4 mL ribosome extraction buffer (Zoschke et al., 2013). A 0.5-mL aliquot of the lysate was used to isolate RNA for transcript profiling. The rest of the lysate (∼4 mL) was treated with nuclease to produce monosomes that harbor ribosome footprints as described previously (Zoschke et al., 2013). Subsequently, the lysate was layered onto a 1-mL sucrose cushion and centrifuged for 1.5 h at 55,000g at 4°C in a 55 Ti rotor (Beckman) to pellet the monosomes. Next, ribosome footprints were extracted and purified as described (Zoschke et al., 2013). Ribosome footprints from the TCT mutant and control plants were labeled with the fluorescent dye Cy3 and ribosome footprints from E15 plants were labeled with the fluorescent dye Cy5. The labeled samples were competitively hybridized onto custom microarrays, as described (Zoschke et al., 2013). Transcripts from the different genotypes were labeled and hybridized in the same manner. Fluorescence intensities on hybridized microarrays were determined as described (Zoschke et al., 2013). Our custom tiling microarrays comprise 50mer oligonucleotide probes that overlap by 20 nucleotides and cover each chloroplast reading frame (probe sequences and positions are given in Supplemental Data Set 2). Two biological replicates were analyzed for each plant line, including four technical replicates (i.e., four probe replicates on the microarray).
Ribosome profiling data were analyzed as described (Zoschke et al., 2013). The annotation of two coding regions was corrected following comparison of their sequences to those of other plants: accD (now position 59,849 to 61,336 of the tobacco plastid genome, NC_001879.2; compared with Caroca et al., 2013) and psbI (8402 to 8512). Translation efficiencies were calculated for each probe by dividing the signal of the ribosome footprints by the signal of the total mRNA. The data for each probe were normalized by dividing the value for each probe from the wild-type-like E15 line (Bock et al., 1994) by the corresponding value from the TCT mutant. The data for the control line (with the aadA inserted in the same location as in the TCT mutant but the wild-type aSD sequence) were treated the same way. For each coding region, the median of all probes covering it was calculated. The change in translation efficiency was calculated by dividing the rate E15/control by the rate E15/TCT.
Calculation of aSD-SD Hybridization Strengths
The strength of SD-aSD binding was analyzed by in silico hybridizing the aSD 5′-CCUCCU-3′ to the region from −22 to −2 of each 5′ UTR at 22°C using Free2bind (Starmer et al., 2006). Differences in SD-aSD binding were calculated by subtracting the value for the mutant from that for the wild type. A relaxed threshold of <0 kcal mol−1 was used to define SDs. The minimum free energy of the mRNA structure around the start codon was calculated using a 50-nucleotide running window passing over a region from 100 nucleotides upstream to 100 nucleotides downstream of the start codon using RNAfold (Gruber et al., 2008).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under accession number NC_001879.2 (tobacco chloroplast genome).
Supplemental Data
Supplemental Figure 1. Determination of the ends of the mature 16S rRNA in the wild type and the TCT mutant.
Supplemental Figure 2. Analysis of plastid transcript accumulation and protein synthesis.
Supplemental Figure 3. Distribution of ribosome footprint abundance in coding regions.
Supplemental Figure 4. Correlation between impairment of the SD-aSD interaction and altered translation efficiency of plastid genes sorted according to their function.
Supplemental Figure 5. Impaired translation of individual genes residing in operons in the TCT mutant.
Supplemental Figure 6. Lack of correlation between translation efficiency and improved or gained SD-aSD binding.
Supplemental Figure 7. Lack of a general correlation between SD strength and translation efficiency.
Supplemental Figure 8. Absence of a correlation between the lack of mRNA structure around the start codon and translation efficiency.
Supplemental Figure 9. Plot of the measured changes in translation efficiency of all plastid genes in the TCT mutant compared with control plants.
Supplemental Table 1. Conservation of anti-Shine-Dalgarno sequences from bacteria to plastids.
Supplemental Table 2. List of oligonucleotides used as PCR primers in this study.
Supplemental Data Set 1. Translation efficiency, strength of mRNA structure around the start codon, strength of the SD-aSD binding, and differences in SD-aSD binding between the mutants and the wild type.
Supplemental Data Set 2. Data from array-based ribosome profiling experiments and control hybridizations with total RNA.
Acknowledgments
We thank Michael Tillich for help with microarray design, Ines Gerlach (both from the Max Planck Institute of Molecular Plant Physiology) for excellent technical assistance, and Maxim Ivanov (Copenhagen Plant Science Centre, University of Copenhagen) for providing scripts. This work was supported by grants from the Deutsche Forschungsgemeinschaft to R.B. (FOR 2092, BO 1482/17-1, and SFB-TR 175) and R.Z. (ZO 302/4-1 and SFB-TR 175) and by the Max Planck Society.
AUTHOR CONTRIBUTIONS
L.B.S., M.E., L.C., M.J., C.H., and R.Z. performed research. L.B.S., M.E., L.C., M.J., J.G., S.R., R.Z., and R.B. designed research and analyzed data. L.B.S., R.Z., and R.B. wrote the article with input from all other authors.
References
- Ahlert D., Ruf S., Bock R. (2003). Plastid protein synthesis is required for plant development in tobacco. Proc. Natl. Acad. Sci. USA 100: 15730–15735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkatib S., Scharff L.B., Rogalski M., Fleischmann T.T., Matthes A., Seeger S., Schöttler M.A., Ruf S., Bock R. (2012). The contributions of wobbling and superwobbling to the reading of the genetic code. PLoS Genet. 8: e1003076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barahimipour R., Strenkert D., Neupert J., Schroda M., Merchant S.S., Bock R. (2015). Dissecting the contributions of GC content and codon usage to gene expression in the model alga Chlamydomonas reinhardtii. Plant J. 84: 704–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (2011). Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 155: 1520–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A., Small I. (2014). Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65: 415–442. [DOI] [PubMed] [Google Scholar]
- Bieri P., Leibundgut M., Saurer M., Boehringer D., Ban N. (2017). The complete structure of the chloroplast 70S ribosome in complex with translation factor pY. EMBO J. 36: 475–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisanz C., Bégot L., Carol P., Perez P., Bligny M., Pesey H., Gallois J.-L., Lerbs-Mache S., Mache R. (2003). The Arabidopsis nuclear DAL gene encodes a chloroplast protein which is required for the maturation of the plastid ribosomal RNAs and is essential for chloroplast differentiation. Plant Mol. Biol. 51: 651–663. [DOI] [PubMed] [Google Scholar]
- Bock R. (2001). Transgenic plastids in basic research and plant biotechnology. J. Mol. Biol. 312: 425–438. [DOI] [PubMed] [Google Scholar]
- Bock R. (2007). Structure, function, and inheritance of plastid genomes. Top. Curr. Genet. 19: 29–63. [Google Scholar]
- Bock R., Kössel H., Maliga P. (1994). Introduction of a heterologous editing site into the tobacco plastid genome: the lack of RNA editing leads to a mutant phenotype. EMBO J. 13: 4623–4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonham-Smith P.C., Bourque D.P. (1989). Translation of chloroplast-encoded mRNA: potential initiation and termination signals. Nucleic Acids Res. 17: 2057–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt D.H., Rouskin S., Zhang Y., Li G.-W., Weissman J.S., Gross C.A. (2017). Operon mRNAs are organized into ORF-centric structures that predict translation efficiency. eLife 6: e22037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caroca R., Howell K.A., Hasse C., Ruf S., Bock R. (2013). Design of chimeric expression elements that confer high-level gene activity in chromoplasts. Plant J. 73: 368–379. [DOI] [PubMed] [Google Scholar]
- Chang B., Halgamuge S., Tang S.L. (2006). Analysis of SD sequences in completed microbial genomes: non-SD-led genes are as common as SD-led genes. Gene 373: 90–99. [DOI] [PubMed] [Google Scholar]
- Chen H., Bjerknes M., Kumar R., Jay E. (1994). Determination of the optimal aligned spacing between the Shine-Dalgarno sequence and the translation initiation codon of Escherichia coli mRNAs. Nucleic Acids Res. 22: 4953–4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet Y., Stern D.B., Wostrikoff K., Kuras R., Girard-Bascou J., Wollman F.-A. (1998). Translation of cytochrome f is autoregulated through the 5′ untranslated region of petA mRNA in Chlamydomonas chloroplasts. Proc. Natl. Acad. Sci. USA 95: 4380–4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotewutmontri P., Barkan A. (2016). Dynamics of chloroplast translation during chloroplast differentiation in maize. PLoS Genet. 12: e1006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deana A., Belasco J.G. (2005). Lost in translation: the influence of ribosomes on bacterial mRNA decay. Genes Dev. 19: 2526–2533. [DOI] [PubMed] [Google Scholar]
- Doyle J.J., Doyle J.L. (1990). Isolation of plant DNA from fresh tissue. Focus 12: 13–15. [Google Scholar]
- Drechsel O., Bock R. (2011). Selection of Shine-Dalgarno sequences in plastids. Nucleic Acids Res. 39: 1427–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drescher A., Ruf S., Calsa T. Jr., Carrer H., Bock R. (2000). The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J. 22: 97–104. [DOI] [PubMed] [Google Scholar]
- Duval M., Simonetti A., Caldelari I., Marzi S. (2015). Multiple ways to regulate translation initiation in bacteria: Mechanisms, regulatory circuits, dynamics. Biochimie 114: 18–29. [DOI] [PubMed] [Google Scholar]
- Eberhard S., Drapier D., Wollman F.-A. (2002). Searching limiting steps in the expression of chloroplast-encoded proteins: relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J. 31: 149–160. [DOI] [PubMed] [Google Scholar]
- Edwards K., Kössel H. (1981). The rRNA operon from Zea mays chloroplasts: nucleotide sequence of 23S rDNA and its homology with E. coli 23S rDNA. Nucleic Acids Res. 9: 2853–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K., Bedbrook J., Dyer T., Kössel H. (1981). 4.5S rRNA from Zea mays chloroplasts shows structural homology with the 3′ end of prokaryotic 23S rRNA. Biochem. Int. 2: 533–538. [Google Scholar]
- Ehrnthaler M., Scharff L.B., Fleischmann T.T., Hasse C., Ruf S., Bock R. (2014). Synthetic lethality in the tobacco plastid ribosome and its rescue at elevated growth temperatures. Plant Cell 26: 765–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fargo D.C., Zhang M., Gillham N.W., Boynton J.E. (1998). Shine-Dalgarno-like sequences are not required for translation of chloroplast mRNAs in Chlamydomonas reinhardtii chloroplasts or in Escherichia coli. Mol. Gen. Genet. 257: 271–282. [DOI] [PubMed] [Google Scholar]
- Fisk D.G., Walker M.B., Barkan A. (1999). Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 18: 2621–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann T.T., Scharff L.B., Alkatib S., Hasdorf S., Schöttler M.A., Bock R. (2011). Nonessential plastid-encoded ribosomal proteins in tobacco: a developmental role for plastid translation and implications for reductive genome evolution. Plant Cell 23: 3137–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristedt R., Scharff L.B., Clarke C.A., Wang Q., Lin C., Merchant S.S., Bock R. (2014). RBF1, a plant homolog of the bacterial ribosome-binding factor RbfA, acts in processing of the chloroplast 16S ribosomal RNA. Plant Physiol. 164: 201–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman D.B., Church G.M., Kosuri S. (2013). Causes and effects of N-terminal codon bias in bacterial genes. Science 342: 475–479. [DOI] [PubMed] [Google Scholar]
- Graf M., Arenz S., Huter P., Dönhöfer A., Novácek J., Wilson D.N. (2017). Cryo-EM structure of the spinach chloroplast ribosome reveals the location of plastid-specific ribosomal proteins and extensions. Nucleic Acids Res. 45: 2887–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner S., Bock R. (2013). Tuning a ménage à trois: co-evolution and co-adaptation of nuclear and organellar genomes in plants. BioEssays 35: 354–365. [DOI] [PubMed] [Google Scholar]
- Gruber A.R., Lorenz R., Bernhart S.H., Neuböck R., Hofacker I.L. (2008). The Vienna RNA websuite. Nucleic Acids Res. 36: W70–W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M.N., Gabay J., Débarbouillé M., Schwartz M. (1982). A role for mRNA secondary structure in the control of translation initiation. Nature 295: 616–618. [DOI] [PubMed] [Google Scholar]
- Herz S., Füssl M., Steiger S., Koop H.-U. (2005). Development of novel types of plastid transformation vectors and evaluation of factors controlling expression. Transgenic Res. 14: 969–982. [DOI] [PubMed] [Google Scholar]
- Hirose T., Sugiura M. (2004). Functional Shine-Dalgarno-like sequences for translational initiation of chloroplast mRNAs. Plant Cell Physiol. 45: 114–117. [DOI] [PubMed] [Google Scholar]
- Hirose T., Kusumegi T., Sugiura M. (1998). Translation of tobacco chloroplast rps14 mRNA depends on a Shine-Dalgarno-like sequence in the 5′-untranslated region but not on internal RNA editing in the coding region. FEBS Lett. 430: 257–260. [DOI] [PubMed] [Google Scholar]
- Hui A., de Boer H.A. (1987). Specialized ribosome system: preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proc. Natl. Acad. Sci. USA 84: 4762–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N.T., Ghaemmaghami S., Newman J.R.S., Weissman J.S. (2009). Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob W.F., Santer M., Dahlberg A.E. (1987). A single base change in the Shine-Dalgarno region of 16S rRNA of Escherichia coli affects translation of many proteins. Proc. Natl. Acad. Sci. USA 84: 4757–4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Zhao Q., Gonzalez de Valdivia E.I., Ardell D.H., Stenström M., Isaksson L.A. (2006). Influences on gene expression in vivo by a Shine-Dalgarno sequence. Mol. Microbiol. 60: 480–492. [DOI] [PubMed] [Google Scholar]
- Kahlau S., Bock R. (2008). Plastid transcriptomics and translatomics of tomato fruit development and chloroplast-to-chromoplast differentiation: chromoplast gene expression largely serves the production of a single protein. Plant Cell 20: 856–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakhlova O., Bock R. (2006). Elimination of deleterious mutations in plastid genomes by gene conversion. Plant J. 46: 85–94. [DOI] [PubMed] [Google Scholar]
- Kode V., Mudd E.A., Iamtham S., Day A. (2005). The tobacco plastid accD gene is essential and is required for leaf development. Plant J. 44: 237–244. [DOI] [PubMed] [Google Scholar]
- Kozak M. (1999). Initiation of translation in prokaryotes and eukaryotes. Gene 234: 187–208. [DOI] [PubMed] [Google Scholar]
- Kozak M. (2005). Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361: 13–37. [DOI] [PubMed] [Google Scholar]
- Kudla G., Murray A.W., Tollervey D., Plotkin J.B. (2009). Coding-sequence determinants of gene expression in Escherichia coli. Science 324: 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupsch C., Ruwe H., Gusewski S., Tillich M., Small I., Schmitz-Linneweber C. (2012). Arabidopsis chloroplast RNA binding proteins CP31A and CP29A associate with large transcript pools and confer cold stress tolerance by influencing multiple chloroplast RNA processing steps. Plant Cell 24: 4266–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H., Suzuki H., Kusumegi T., Hirose T., Yukawa Y., Sugiura M. (2007). Translation of psbC mRNAs starts from the downstream GUG, not the upstream AUG, and requires the extended Shine-Dalgarno sequence in tobacco chloroplasts. Plant Cell Physiol. 48: 1374–1378. [DOI] [PubMed] [Google Scholar]
- Li G.-W., Oh E., Weissman J.S. (2012). The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature 484: 538–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G.-W., Burkhardt D., Gross C., Weissman J.S. (2014). Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157: 624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link S., Engelmann K., Meierhoff K., Westhoff P. (2012). The atypical short-chain dehydrogenases HCF173 and HCF244 are jointly involved in translational initiation of the psbA mRNA of Arabidopsis. Plant Physiol. 160: 2202–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., et al. (2015). The conserved endoribonuclease YbeY is required for chloroplast ribosomal RNA processing in Arabidopsis. Plant Physiol. 168: 205–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuell A.L., Quispe J., Mayfield S.P. (2007). Structure of the chloroplast ribosome: novel domains for translation regulation. PLoS Biol. 5: e209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy J.E.G., Gualerzi C. (1990). Translational control of prokaryotic gene expression. Trends Genet. 6: 78–85. [DOI] [PubMed] [Google Scholar]
- McCormac D.J., Barkan A. (1999). A nuclear gene in maize required for the translation of the chloroplast atpB/E mRNA. Plant Cell 11: 1709–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milón P., Rodnina M.V. (2012). Kinetic control of translation initiation in bacteria. Crit. Rev. Biochem. Mol. Biol. 47: 334–348. [DOI] [PubMed] [Google Scholar]
- Mühlbauer S.K., Lössl A., Tzekova L., Zou Z., Koop H.-U. (2002). Functional analysis of plastid DNA replication origins in tobacco by targeted inactivation. Plant J. 32: 175–184. [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue culture. Physiol. Plant. 15: 473–497. [Google Scholar]
- Nakagawa S., Niimura Y., Miura K., Gojobori T. (2010). Dynamic evolution of translation initiation mechanisms in prokaryotes. Proc. Natl. Acad. Sci. USA 107: 6382–6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S., Niimura Y., Gojobori T. (2017). Comparative genomic analysis of translation initiation mechanisms for genes lacking the Shine-Dalgarno sequence in prokaryotes. Nucleic Acids Res. 45: 3922–3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neymotin B., Ettore V., Gresham D. (2016). Multiple transcript properties related to translational affect mRNA degradation rates in Saccharomyces cerevisiae. G3 (Bethesda) 6: 3475–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickelsen J., Fleischmann M., Boudreau E., Rahire M., Rochaix J.-D. (1999). Identification of cis-acting RNA leader elements required for chloroplast psbD gene expression in Chlamydomonas. Plant Cell 11: 957–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K., Ashida H., Ogawa T., Yokota A. (2010). A DEAD box protein is required for formation of a hidden break in Arabidopsis chloroplast 23S rRNA. Plant J. 63: 766–777. [DOI] [PubMed] [Google Scholar]
- Oey M., Lohse M., Kreikemeyer B., Bock R. (2009). Exhaustion of the chloroplast protein synthesis capacity by massive expression of a highly stable protein antibiotic. Plant J. 57: 436–445. [DOI] [PubMed] [Google Scholar]
- Pfalz J., Bayraktar O.A., Prikryl J., Barkan A. (2009). Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 28: 2042–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plader W., Sugiura M. (2003). The Shine-Dalgarno-like sequence is a negative regulatory element for translation of tobacco chloroplast rps2 mRNA: an additional mechanism for translational control in chloroplasts. Plant J. 34: 377–382. [DOI] [PubMed] [Google Scholar]
- Presnyak V., Alhusaini N., Chen Y.-H., Martin S., Morris N., Kline N., Olson S., Weinberg D., Baker K.E., Graveley B.R., Coller J. (2015). Codon optimality is a major determinant of mRNA stability. Cell 160: 1111–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski M., Ruf S., Bock R. (2006). Tobacco plastid ribosomal protein S18 is essential for cell survival. Nucleic Acids Res. 34: 4537–4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski M., Karcher D., Bock R. (2008a). Superwobbling facilitates translation with reduced tRNA sets. Nat. Struct. Mol. Biol. 15: 192–198. [DOI] [PubMed] [Google Scholar]
- Rogalski M., Schöttler M.A., Thiele W., Schulze W.X., Bock R. (2008b). Rpl33, a nonessential plastid-encoded ribosomal protein in tobacco, is required under cold stress conditions. Plant Cell 20: 2221–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf M., Kössel H. (1988). Occurrence and spacing of ribosome recognition sites in mRNAs of chloroplasts from higher plants. FEBS Lett. 240: 41–44. [Google Scholar]
- Sachs A.B. (1993). Messenger RNA degradation in eukaryotes. Cell 74: 413–421. [DOI] [PubMed] [Google Scholar]
- Sakamoto W., Chen X., Kindle K.L., Stern D.B. (1994). Function of the Chlamydomonas reinhardtii petd 5′ untranslated region in regulating the accumulation of subunit IV of the cytochrome b6/f complex. Plant J. 6: 503–512. [DOI] [PubMed] [Google Scholar]
- Sane A.P., Stein B., Westhoff P. (2005). The nuclear gene HCF107 encodes a membrane-associated R-TPR (RNA tetratricopeptide repeat)-containing protein involved in expression of the plastidial psbH gene in Arabidopsis. Plant J. 42: 720–730. [DOI] [PubMed] [Google Scholar]
- Scharff L.B., Childs L., Walther D., Bock R. (2011). Local absence of secondary structure permits translation of mRNAs that lack ribosome-binding sites. PLoS Genet. 7: e1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz Z., Kössel H. (1980). The primary structure of 16S rDNA from Zea mays chloroplast is homologous to E. coli 16S rRNA. Nature 283: 739–742. [Google Scholar]
- Shen Y., Danon A., Christopher D.A. (2001). RNA binding-proteins interact specifically with the Arabidopsis chloroplast psbA mRNA 5′ untranslated region in a redox-dependent manner. Plant Cell Physiol. 42: 1071–1078. [DOI] [PubMed] [Google Scholar]
- Shikanai T., Shimizu K., Ueda K., Nishimura Y., Kuroiwa T., Hashimoto T. (2001). The chloroplast clpP gene, encoding a proteolytic subunit of ATP-dependent protease, is indispensable for chloroplast development in tobacco. Plant Cell Physiol. 42: 264–273. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. (1974). The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. USA 71: 1342–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starmer J., Stomp A., Vouk M., Bitzer D. (2006). Predicting Shine-Dalgarno sequence locations exposes genome annotation errors. PLOS Comput. Biol. 2: e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub J.M., Maliga P. (1993). Accumulation of D1 polypeptide in tobacco plastids is regulated via the untranslated region of the psbA mRNA. EMBO J. 12: 601–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D.B., Gruissem W. (1987). Control of plastid gene expression: 3′ inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell 51: 1145–1157. [DOI] [PubMed] [Google Scholar]
- Studer S.M., Joseph S. (2006). Unfolding of mRNA secondary structure by the bacterial translation initiation complex. Mol. Cell 22: 105–115. [DOI] [PubMed] [Google Scholar]
- Suay L., Salvador M.L., Abesha E., Klein U. (2005). Specific roles of 5′ RNA secondary structures in stabilizing transcripts in chloroplasts. Nucleic Acids Res. 33: 4754–4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura M. (2014). Plastid mRNA translation. Methods Mol. Biol. 1132: 73–91. [DOI] [PubMed] [Google Scholar]
- Sugiura M., Hirose T., Sugita M. (1998). Evolution and mechanism of translation in chloroplasts. Annu. Rev. Genet. 32: 437–459. [DOI] [PubMed] [Google Scholar]
- Sun Y., Zerges W. (2015). Translational regulation in chloroplasts for development and homeostasis. Biochim. Biophys. Acta 1847: 809–820. [DOI] [PubMed] [Google Scholar]
- Sunohara T., Jojima K., Tagami H., Inada T., Aiba H. (2004). Ribosome stalling during translation elongation induces cleavage of mRNA being translated in Escherichia coli. J. Biol. Chem. 279: 15368–15375. [DOI] [PubMed] [Google Scholar]
- Svab Z., Maliga P. (1993). High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl. Acad. Sci. USA 90: 913–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller N., Bock R. (2014). The translational apparatus of plastids and its role in plant development. Mol. Plant 7: 1105–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller N., Weingartner M., Thiele W., Maximova E., Schöttler M.A., Bock R. (2012). The plastid-specific ribosomal proteins of Arabidopsis thaliana can be divided into non-essential proteins and genuine ribosomal proteins. Plant J. 69: 302–316. [DOI] [PubMed] [Google Scholar]
- Trebitsh T., Danon A. (2001). Translation of chloroplast psbA mRNA is regulated by signals initiated by both photosystems II and I. Proc. Natl. Acad. Sci. USA 98: 12289–12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K., Subramanian A.R. (2000). The plastid ribosomal proteins. Identification of all the proteins in the 50 S subunit of an organelle ribosome (chloroplast). J. Biol. Chem. 275: 28466–28482. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K., von Knoblauch K., Subramanian A.R. (2000). The plastid ribosomal proteins. Identification of all the proteins in the 30 S subunit of an organelle ribosome (chloroplast). J. Biol. Chem. 275: 28455–28465. [DOI] [PubMed] [Google Scholar]
- Ye G.-N., Hajdukiewicz P.T.J., Broyles D., Rodriguez D., Xu C.W., Nehra N., Staub J.M. (2001). Plastid-expressed 5-enolpyruvylshikimate-3-phosphate synthase genes provide high level glyphosate tolerance in tobacco. Plant J. 25: 261–270. [DOI] [PubMed] [Google Scholar]
- Yu F., Liu X., Alsheikh M., Park S., Rodermel S. (2008). Mutations in SUPPRESSOR OF VARIEGATION1, a factor required for normal chloroplast translation, suppress var2-mediated leaf variegation in Arabidopsis. Plant Cell 20: 1786–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerges W. (2000). Translation in chloroplasts. Biochimie 82: 583–601. [DOI] [PubMed] [Google Scholar]
- Zhang J., Ruf S., Hasse C., Childs L., Scharff L.B., Bock R. (2012). Identification of cis-elements conferring high levels of gene expression in non-green plastids. Plant J. 72: 115–128. [DOI] [PubMed] [Google Scholar]
- Zhou F., Badillo-Corona J.A., Karcher D., Gonzalez-Rabade N., Piepenburg K., Borchers A.-M.I., Maloney A.P., Kavanagh T.A., Gray J.C., Bock R. (2008). High-level expression of human immunodeficiency virus antigens from the tobacco and tomato plastid genomes. Plant Biotechnol. J. 6: 897–913. [DOI] [PubMed] [Google Scholar]
- Zoschke R., Kroeger T., Belcher S., Schöttler M.A., Barkan A., Schmitz-Linneweber C. (2012). The pentatricopeptide repeat-SMR protein ATP4 promotes translation of the chloroplast atpB/E mRNA. Plant J. 72: 547–558. [DOI] [PubMed] [Google Scholar]
- Zoschke R., Watkins K.P., Barkan A. (2013). A rapid ribosome profiling method elucidates chloroplast ribosome behavior in vivo. Plant Cell 25: 2265–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]