Abstract
Background
Staphylococcus aureus (S. aureus or SA) is a leading cause of healthcare-associated (HA-) and community-associated (CA) infection. HA-SA isolates usually cause nosocomial pneumonia, bloodstream infections, catheter-related urinary tract infections, etc. On the other hand, CA-SA isolates usually cause highly fatal diseases, such as SSTIs as well as post influenza necrotic hemorrhagic pneumonia. The differences of the infection types are partially due to the unique characteristics between HA-SA and CA-SA isolates. For example, HA-SA isolates showed strong adherence to host epithelial cells, while CA-SA isolates displayed higher virulence due to the increased activity of the important quorum-sensing system accessory gene regulator (agr). Thus, the aim of this study was to characterize the proteomic difference between HA-SA and CA-SA lineage.
Methods
In this study, the extracted peptides from those representative strains were analyzed by LC-MS/MS. The protein-protein interaction network was constructed by bioinformatics and their expressions were verified by RT-PCR and Western blot.
Results
We demonstrated that Agr system (AgrA and AgrC) and its interactive factors (PhoP, SrrB, YycG, SarX, SigB and ClpP) based on the protein–protein interaction network were expressed significantly higher in the epidemic Chinese CA-SA lineage ST398 compared to HA-SA lineage ST239 by LC-MS/MS. We further verified the increased transcription of all these genes in ST398 by RT-PCR, suggesting that the higher expression of these genes/proteins probably play role in the acute infection of CA-SA. Moreover, surface-related proteins (FnbpA, SpA, Atl, ClfA, IsaA, IsaB, LtaS, SsaA and Cna) that are repressed by the Agr system have significantly higher expression in the epidemic Chinese HA-SA clone ST239 in comparison to CA-SA lineage ST398 by LC-MS/MS. Furthermore, we confirmed the significantly increased expression of two important adhesive proteins (Atl and ClfA) in ST239 by Western blot, which may contribute to the durative infection of HA-SA.
Conclusion
The results suggest that the different proteomic profile, at least partially, contribute to the pathogenic differences between HA-SA and CA-SA.
Electronic supplementary material
The online version of this article (10.1186/s12014-017-9178-5) contains supplementary material, which is available to authorized users.
Keywords: Staphylococcus aureus, Virulent mechanism, Healthcare-associated, Community-associated, Proteomic analysis
Background
Staphylococcus aureus has been a major cause of human infections for a long time [1]. In general, healthcare-associated (HA)-SA infections primarily occur among inpatients [2, 3] and typically result in septicemia, pneumonia and device-related infections. Due to the greater resistance rates for majority of antibiotics, healthcare-associated SA (HA)-SA infections are likely to promote the emergence of methicillin-resistant SA (MRSA), therefore causing increased hospital duration and higher hospital charges than methicillin-sensitive SA (MSSA) infections [4, 5]. By contrast, community-associated (CA)-SA infections caused by both MRSA and MSSA isolates mostly occur in healthy individuals and cause severe skin and soft-tissue infections (SSTIs) and, less frequently, necrotizing pneumonia in the community [3, 6].
The MRSA challenge in China is also a serious health issue both in healthcare and community environments. Recent epidemiological surveys have demonstrated that ST239-SCCmecIII is the dominant isolate of HA-MRSA in China [7, 8], with ST5-SCCmecII ranked second [9, 10]. Comparatively, data on CA-SA in China are still limited. Studies in children suggest ST59-SCCmecIV is the predominant CA-MRSA clone [11]. In addition, there is a worldwide growing spread of the ST398 clone between animals and humans with animal contact or between humans and humans living in animal-free environments [12–14]. Human-associated ST398 is also the most prevalent CA-SA clone in adult SSTIs in China [15].
In our previous study, HA-SA ST239 isolates had improved nasal colonization ability compared with CA-SA ST398 [16]. In addition, the CA-SA ST398 clone had increased virulent ability compared with the HA-SA ST239 clone in the skin infection model [17]. Recent studies have shown the critical adaptation of the HA-MRSA ST239 clone to the highly selective nosocomial setting and its evolution to increased levels of methicillin resistance at the cost of reduced virulence, enhancing its ability to become a successful nosocomial pathogen [18]. The high methicillin resistance of HA-MRSA also attenuates the staphylococcal accessory gene regulator (agr)-regulating sensing system, leading to decreased virulence and immobility in the community, whereas the emerging CA-MRSA typically has lower production of penicillin-binding protein 2a (encoded by mecA), hence holding its full-scale virulence for successful predominance in the community environment [19].
Transcriptome studies have documented that different S. aureus strains show tremendous variations in gene transcriptional profiles causing diverse pathogenesis, despite sharing a core genome with the similarity of approximately 82% [20, 21]. Thus, it is likely that both the presence of multiple mobile genetic elements and temporal alterations in the expression of bacterial core factors contribute to the virulence in S. aureus. The ESAT-6 secretion system (Ess) of S. aureus exhibits genetic diversity and organizational variation across the species [22]. However, the higher expression of the conserved membrane protein EssB promotes the virulence and facilitates the resistance to innate host defense in the emergent CA-SA clone [17]. In addition, Protein A (SpA), as a cell-wall anchored protein, has been verified to conduce to the higher durative tissue damage in hosts infected by HA-MRSA ST239 compared with those by CA-SA ST398 [16].
However, most studies only suggest one respective prominent factor functioning in the durative damage with HA-SA ST239 infection or in the acute damage with CA-SA ST398 infection. Actually, a regulatory network including multiple factors may cause particular clinical syndromes. A key example of this is based on a previous study showing that the marked hyper-virulence of CA-SA isolates is seemingly due to differential gene expression deriving from the enhanced activity of regulators such as agr [23]. Agr up-regulated the expression of many virulence factors such as α-toxin (hla), essB, etc. [17] and repressed surface adhesins that assist the organism adhere to the host and develop biofilms [23]. These surface adhesins, including microbial surface components recognizing adhesive matrix molecules (MSCRAMMs), some non-covalently attached adhesins, and cell wall teichoic acid, mediate the adhesion to extracellular matrix and plasma proteins [24–28]. Hence, in this study, we sought to characterize the proteome of clinically significant pathogenic strains presently afflicting human beings in the nosocomial and community settings in China. We intended to identify and quantify the production of key network-based factors that enable this pathogen to promptly infect individuals and then cause diseases.
Methods
Bacterial strains and growth conditions
Bacteria were identified as staphylococci by classic microbiological methods, including Gram staining and catalase and coagulase activity in rabbit plasma. S. aureus strains were further categorized by VITEK2 automated systems (BioMérieux, France). CA-SA was defined as an isolate that was obtained either from an outpatient or from an inpatient ≤ 72 h after hospital admission and without the patient having any of the following risk factors: contact with the hospital environment in the preceding 6 months, residence in a long-term care facility in the preceding 12 months, S. aureus infection in the preceding 12 months, presence of a central vascular catheter at the time of infection. HA-SA was defined as an isolate that was obtained from an inpatient > 72 h after hospital admission, or from an outpatient or inpatient ≤ 72 h after hospital admission or having at least one of those risk factors listed above.
The HA-SA ST239 and CA-SA ST398 S. aureus isolates used for phenotypic experiments were randomly selected from the clinical isolates with typical infectious manifestations at Shanghai teaching hospitals during 2005–2014. The four HA-SA ST239 and four CA-SA ST398 S. aureus strains used for mass spectrometry were selected from the above isolates with relatively moderate phenotypes. The four HA-SA ST239 isolates were all MRSA and caused respiratory infection, while all four CA-SA ST398 isolates caused SSTIs, and only half of them were MRSA. All bacterial strains used in this study were list in Table 1. The isolates were grown in tryptic soy broth (TSB; Oxoid, Basingstoke, Hampshire, UK) at 37 °C with agitation.
Table 1.
Bacterial strains used in this study
| No. | Isolate name | CA/HA | MRSA/MSSA | MLST | Spa type | Infection type |
|---|---|---|---|---|---|---|
| Strains for mass spectrometry | ||||||
| 1 | Ji95 | HA | MRSA | ST239 | t037 | Respiratory |
| 2 | Ji99 | HA | MRSA | ST239 | t037 | Respiratory |
| 3 | 2011-1046 | HA | MRSA | ST239 | t037 | Respiratory |
| 4 | 2009-770 | HA | MRSA | ST239 | t037 | Respiratory |
| 9 | 2009-898 | CA | MSSA | ST398 | t034 | SSTIs |
| 10 | 2010-191 | CA | MRSA | ST398 | t571 | SSTIs |
| 11 | Ji92 | CA | MRSA | ST398 | t034 | SSTIs |
| 12 | 2005-109 | CA | MSSA | ST398 | t034 | SSTIs |
| Strains for RT-PCR, western-blot, adhesion, biofilm and hemolysis assays | ||||||
| 1 | Ji95 | HA | MRSA | ST239 | t037 | Respiratory |
| 2 | Ji99 | HA | MRSA | ST239 | t037 | Respiratory |
| 3 | 2011-1046 | HA | MRSA | ST239 | t037 | Respiratory |
| 4 | 2009-770 | HA | MRSA | ST239 | t037 | Respiratory |
| 5 | 2010-1030 | HA | MRSA | ST239 | t030 | SSTIs |
| 6 | 2012-169 | HA | MRSA | ST239 | t037 | SSTIs |
| 7 | 2012-32 | HA | MRSA | ST239 | t037 | Respiratory |
| 8* | 2012-97 | HA | MRSA | ST239 | t030 | Respiratory |
| 9 | 2009-898 | CA | MSSA | ST398 | t034 | SSTIs |
| 10 | 2010-191 | CA | MRSA | ST398 | t571 | SSTIs |
| 11 | Ji92 | CA | MRSA | ST398 | t034 | SSTIs |
| 12 | 2005-109 | CA | MSSA | ST398 | t034 | SSTIs |
| 13 | 2010-38 | CA | MSSA | ST398 | t034 | SSTIs |
| 14 | 2005-577 | CA | MSSA | ST398 | t571 | SSTIs |
| 15 | 2007-524 | CA | MSSA | ST398 | t034 | Respiratory |
| 16* | 2007-1089 | CA | MSSA | ST398 | t034 | SSTIs |
*No. 8 and No. 16 were not available for RT-PCR assay
Multi-locus sequence typing (MLST)
MLST was performed as previously described [29]. The PCR amplicons of seven S. aureus housekeeping genes (arcC, aroE, glpF, gmk, pta, tpi, and yqiL) were obtained from chromosomal DNA. The sequences of the PCR products were compared with the existing sequences available at the MLST website [30], and the allelic number was determined for each sequence.
S. aureus infection of epithelial cells
For the adhesion assay, S. aureus was cultured in TSB for 8 h, and the cell pellet was washed twice with F12 K medium. Human epithelial A549 cells were cultured in F12 K medium supplemented with fetal bovine serum (FBS, 10%) and l-glutamine (2 mM) in T75 flasks at 37 °C and 5% CO2. Cells were liberated from flasks using trypsin-EDTA solution (Sigma-Aldrich, St Louis, MO, USA), resuspended in culture medium and seeded at 2 × 106 cells/well in a final volume of 500 µl for 120 min at 37 °C with 5% CO2. Cells were washed 3 times in F12 K, and 108 CFU S. aureus were added and incubated with the cells for 120 min. The coverslips that were used to determine the total number of associated CFU (adherent and internalized) were dip washed three times and subsequently lysed by the addition of 500 μl of 0.1% deoxysodium cholate solution. Bacterial CFU values were enumerated by serial dilutions of epithelial cell lysates and plating onto TSA plates.
Semi-quantitative biofilm assay
Semi-quantitative biofilm assays were performed as described elsewhere [31]. Subsequently, cells were fixed in Bouin fixative. The fixative was removed after 1 h, and the wells were washed with PBS. Organisms in the wells were then stained with 0.4% (wt/vol) crystal violet, and the floating stain was washed off with slowly running water. After drying, the stained biofilm was read using a MicroELISA autoreader (BioRad) at 570 nm.
Lysis of erythrocytes by culture filtrates
The supernatants were collected from bacterial cultures grown for 15 h. Hemolytic activities were determined by incubating samples with human red blood cells (2% v/v in Dulbecco’s phosphate-buffered saline, DPBS) for 1 h at 37 °C. Hemolysis was determined by measuring the optical density at 540 nm using an ELISA reader. The assay was performed in triplicate.
Label-free quantitative proteomic analysis
The overnight cultures were diluted 1:100 in 50 ml of TSB and incubated at 37 °C with shaking at 220 rpm until grown to post-exponential growth phase (8 h) (OD600 ~ 4.0). Samples were normalized according to the OD600 reading (OD600 = 2.0). The cells were collected by centrifugation at 14,000g at 4 °C for 15 min. The supernatants were removed and the pellets were washed twice with PBS and then suspended in the lysis buffer of 95 μl 20 mM Tris (pH 8.0) and exposed to 50 μg/ml lysostaphin at 37゜C for 30 min. Cells debris was removed by centrifugation at 14,000g for 30 min at 4 °C. Cell lysates were mixed with 25 μl 5X protein loading buffer and boiled for 10 min. The cell particles were removed by centrifugation at 14,000g for 5 min at 4 °C. 12% SDS-PAGE was used to pre-fractionate the proteins in the loading volume of 10 μl. The protein samples were subjected to in-gel digestion. Finally, the extracted peptides were vacuum-dried prior to LC-MS/MS analysis using a nanoflow liquid chromatography instrument (EASY-nLC 1000, Thermo Scientific) coupled to an ion trap mass spectrometer (LTQ Velos Pro. Thermo Scientific) in the data-dependent mode. The detailed LC-MS/MS settings have been described elsewhere [32]. The peptide mass peaks of the fractions obtained by LC-MS/MS were compared with the Universal Protein Resource database (UniProt) (http://www.uniprot.org/). The differentially expressed proteins were filtered by the following cutoff criteria: the P value (Mann–Whitney U test) was lower than 0.05, and the fold changes (ST239/ST398) were higher than 1.5-fold or lower than 0.67-fold. The volcano plot for significantly differentially expressed proteins was produced by Excel.
Hierarchical analysis
Hierarchical analysis of protein expression was performed using MEV (Multi Experiment View) cluster software and the Hierarchical Clustering (HCL) tool [33] selecting the distance metric of Pearson’s correlation and linkage method of average linkage clustering.
Protein–protein interaction (PPI) network construction
The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database [34] identifies the interactions of gene products, including not only the direct physical interactions of proteins but also their functional interactions. To evaluate the interactions among the differentially expressed genes, we uploaded these genes and drew a color-coded protein-protein interaction network graph. We then imported the PPI data in text format into the Cytoscape program [35] to visualize the relationships and used its network analyzer plug-in to analyze the PPI network [35].
Real-time quantitative reverse transcription-PCR (RT-PCR)
For RNA isolation, cells were collected from bacterial cultures grown to post-exponential growth phase (8 h) and then harvested and washed twice in DEPC water. Cells were disrupted by shaking with a Mini-Beadbeater (Biospec Products) at maximum speed for 30 s. Tubes were then incubated on ice for 5 min and the suspensions were centrifuged. Total RNA was isolated using an RNeasy minikit (Qiagen) as recommended in a standard protocol. Complementary DNA (cDNA) was synthesized from total RNA using the QuantiTect reverse transcription system (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Oligonucleotide primers were designed using Primer Express (Additional file 1: Table S1). The resulting cDNA and negative control samples were amplified using the QuantiTect SYBR green PCR kit (Qiagen). Reactions were performed in a MicroAmp Optical 96-well reaction plate using a 7500 Sequence Detector (Applied Biosystems, Foster City, CA, USA). Standard curves were determined for each gene using purified chromosomal DNA at concentrations of 0.005–50 ng/ml. All quantitative RT-PCR experiments were performed in duplicate, with gyrB as an internal control [36].
Construction, expression and purification of recombinant fusion protein
The isolates S0385 (GenBank accession number AM990992) and TW20 (GenBank accession number FN433596) respectively served as the reference genomes of CA-SA ST398 and HA-SA ST239 strains. The sarX gene was conserved between both of the reference genomes. Genomic DNA extracted from CA-SA ST398 isolate S0385 (GenBank accession number AM990992) served as the PCR template. The sarX gene was amplified using the forward primer 5′- CGGGATCCTTGAATACTGAGAAATTAGAAACAT-3′ and the reverse primer 5′-CGGAATTCTTAAATATTTAAAAATTGTTCTACA-3′. The respective PCR product was digested with BamHI and EcoRI. The PCR product was ligated into pET28a. The resulting plasmid were transformed into Escherichia coli Top10 strain. The correct nucleotide sequence was confirmed by sequencing. The resulting construct was transformed into Escherichia coli strain BL21 (DE3) for isopropyl-β-D-1-thiogalactopyranoside (IPTG)-induced expression according to the manufacturer’s instructions. His-tagged SarX protein was respectively affinity-purified from cleared lysates with Ni-NTA resin (Qiagen) according to manufacturer’s construction. The protein concentration was determined by the bicinchoninic acid assay (Yeasen Bio, China).
Production of rabbit antisera and purification of antibodies
Purified proteins (for SarX) or peptides (for AgrA, AgrC, Atl, ClfA, Cna, IsaA, IsaB, LtaS, YycG and FnbpA) were used as an immunogen for the production of rabbit polyclonal antisera (provided by GLbiochem China). The isolates S0385 (GenBank accession number AM990992) and TW20 (GenBank accession number FN433596) also resctively served as the peptide template of CA-SA ST398 and HA-SA ST239 strains. Synthetic peptides corresponding to amino acids of AgrA (DSKERIVYFKNKEHC), AgrC (KYKRNQEEIETYYE), ClfA (SSKEADASENSMTQ), Atl (IGEVGKYFDIPQYK), Cna (TFDDKNGKIQNGDT), IsaA (TMPGWGPTNTVDQQ), IsaB (GKDLKKENGKTKEAD), LtaS (NYTKQRQTEPNPEYY), YycG (EKELLDNFKKNITQ) and FnbpA (ELPETGGEESTNKGM) were synthesized by GLbiochem, China. All peptide fragments were respectively conserved between HA-SA ST239 and CA-SA ST398 strains. The antibody IgG were purified by the protein A affinity column Hitrap rProteinA FF (GE Healthcare) using an AKTA purifier (GE Healthcare) according to the manufacturer’s specifications. Eventually, only polyclonal antibodies for AgrA, Atl and ClfA peptides and SarX protein were successfully obtained.
Western blot analysis
For protein detection, cells were also collected from cultures of S. aureus strains grown to post-exponential growth phase (8 h). Samples were normalized according to the OD600 reading (OD600 = 2). Cells were harvested and resuspended in 50 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) and treated with lysostaphin (50 μg/ml) for 30 min at 37 °C. Samples were mixed with protein loading buffer and boiled for 10 min. Equivalent amounts of proteins were separated on 12% SDS-PAGE gels and were electrotransferred to PVDF membranes (Invitrogen). After blocking, the membranes were incubated with respective antiserum at 4 °C overnight and then were incubated with horseradish peroxidase-conjugated secondary antibody at room temperature for 1 h. To ensure consistency between blots, Sortase A (SrtA) was used as the loading control. The SrtA antibody was kindly donated by Taeok Bae, Indiana University School of Medicine-Northwest, Gary, Indiana, United States of America. All the other antibodies were generated by GLbiochem, China. Images of Western blots were acquired using a Tanon-5200 system. Densitometry analysis was performed with ImageJ software for each protein band in reference to the corresponding SrtA band. All Western blot experiments were performed in duplicate.
Statistical analysis
The Mann–Whitney U test was performed to analyze the statistical significance of difference in protein expression from mass spectrometry. Student’s t-test was used to analyze the following data resulting from RT-PCR and Western blotting. All data were analyzed using Prism (GraphPad Software, Inc., La Jolla, CA, USA), and P values less than 0.05 were deemed statistically significant. The error bars in all graphs show the standard deviation (± SD). The asterisks indicate the following: *P < 0.05, **P < 0.01, ***P < 0.001.
Results
HA-SA isolates showed increased epithelial cell adhension ability and biofilm formation compared with CA-SA isolates
The adhesion and aggregation ability of S. aureus is crucial for bacterial colonization, infection and spread in human [37]. We presumed that the differences in transmissibility of HA-SA ST239 and CA-SA ST398 may be a reflection of the expression variation in surface adhesins (e.g., MSCRAMMs) between these strain types. Here, we used human alveolar epithelial cell A549 as a model to test the adhesion ability of ST239 and ST398. We observed significantly increased adherence of HA-SA ST239 isolates compared with CA-SA ST398 isolates (P = 0.007) (Fig. 1a). Furthermore, the predominant Chinese HA-SA ST239 lineage was found to have improved biofilm formation ability compared with the CA-SA ST398 strains (Fig. 1b), suggesting that HA-SA would cause more severe intercellular bacterial aggregation. However, the interactive factors facilitating the colonization of HA-SA ST239 were unclear.
Fig. 1.
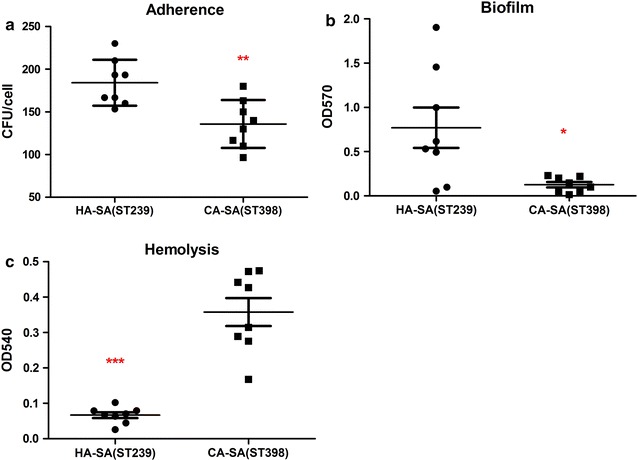
Comparison between CA-SA ST398 and HA-SA ST239 isolates in adhesion, aggregation and virulence characteristics. a Colony counts of adhesive and internalized bacteria above in A549 epithelial cells infected for 2 h. A ~ 0.4-fold increase could be seen for HA-SA ST239 isolates compared with that for CA-SA ST398 strains. b Biofilm formation. Biofilm formation abilities were calculated by the semiquantitative biofilm assay and were read using a MicroELISA autoreader (BioRad) at 570 nm. An obvious increase could be seen for HA-SA ST239 isolates compared with that for CA-SA ST398 strains. c Hemolysis (erythrocyte lysis). Hemolytic activities were determined by incubating culture filtrates with human red blood cells. A significant increase could be seen for CA-SA ST398 isolates compared with that for HA-SA ST239 strains. *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired t-test). The data is representative of three independent experiments
HA-SA isolates had decreased hemolysis ability compared with CA-SA isolates
α-toxin (Hla) can lyse red cells and is one of the most essential virulence factors in S. aureus [38]. To test the expression of Hla in the culture supernatant, the cytolytic potential was measured by analyzing the lysis of human erythrocytes. Our results showed that the hemolysis activity of CA-SA ST398 isolates strongly exceed that of HA-SA ST239 isolates (Fig. 1c), thus improving the more acute infection of CA-SA than HA-SA. Nonetheless, the interactive factors promoting the higher virulence of CA-SA ST398 remained to be determined
Proteomic analysis
SDS-PAGE analysis was performed on bacteria cells for four CA-SA ST398 isolates and four HA-SA ST239 clinical isolates at the post-exponential phase of growth (8 h). We first confirmed the comparable protein bands within four CA-SA ST398 isolates or within four HA-SA ST239 isolates (Fig. 2a). Then, Spectral counting based on LC-MS/MS proteomics was performed to investigate the difference in the protein production between four CA-SA ST398 clinical isolates and four HA-SA ST239 clinical isolates. The CA-SA ST398 group was compared with the HA-SA ST239 group to identify differentially expressed proteins. Figure 2b showed the volcano plot for the differentially expressed proteins in the ST239 and ST398 groups. These proteins with a P value by Student’s t-test lower than 0.05 and with an ST239/ST398 change higher than 1.5-fold or lower than 0.67-fold were considered as differentially expressed proteins. All the 209 differentially expressed proteins (64 up-regulated in the HA-SA ST239 group and 145 up-regulated in the CA-SA ST398 group) were listed in Tables 2 and 3 and the detailed information on differentially expressed proteins in each isolate were depicted in Additional file 1: Tables S2 and S3.
Fig. 2.
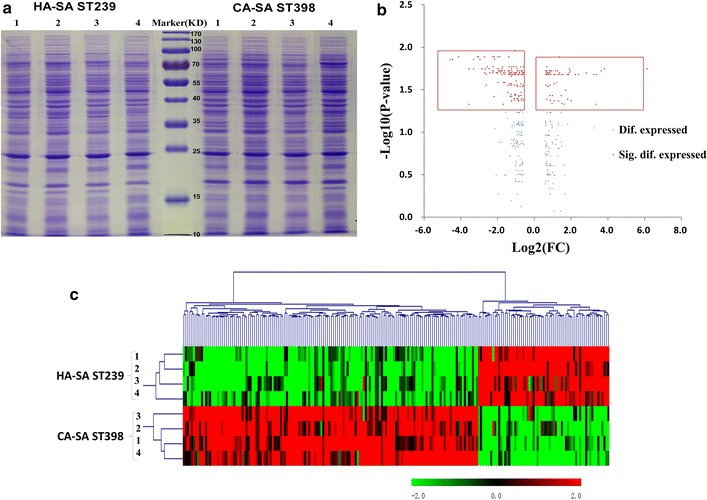
a Comparison of the total proteins between the CA-SA ST398 and HA-SA ST239 isolates by SDS-PAGE. b Volcano plot of differentially expressed proteins in the HA-SA ST239 versus CA-SA ST398 isolates. c Hierarchical cluster analysis was conducted for all the differentially expressed proteins in the HA-SA ST239 group versus the CA-SA ST398 group
Table 2.
All up-regulated proteins in the HA-SA ST239 group
| Uniprot acc no. | Prot description | FC* (239/398) | P value |
|---|---|---|---|
| Known virulence-related factors | |||
| D2NA85 | FnbpA** | 13.99 | 0.02 |
| D2N8V1 | Aerolysin/Leukocidin family protein | 7.10 | 0.02 |
| D2N8V0 | Synergohymenotropic toxin | 5.30 | 0.02 |
| D2N3G2 | SpA | 3.26 | 0.03 |
| D2NAS2 | Cna | 2.54 | 0.02 |
| D2N627 | Atl | 2.46 | 0.02 |
| D2N5J4 | ClfA | 2.11 | 0.02 |
| Staphylococcal antigen | |||
| D2N9N2 | SsaA | 4.70 | 0.02 |
| D2NAF1 | IsaA | 2.50 | 0.04 |
| D2NAM0 | IsaB | 1.73 | 0.02 |
| Other known proteins | |||
| D2N382 | Penicillin-binding protein 3 (Pbp 3) (Pspb20) | 69.88 | 0.02 |
| D2N4Y0 | Acetyl-CoA acetyltransferase | 12.51 | 0.02 |
| D2NAM6 | N-acetylmuramoyl-l-alanine amidase | 10.41 | 0.02 |
| D2N4M0 | N-acetylmuramoyl-l-alanine amidase Sle1 | 9.41 | 0.02 |
| D2N5C6 | LtaS | 4.03 | 0.04 |
| D2N9E7 | HAD-superfamily hydrolase, subfamily IIB | 3.87 | 0.02 |
| D2N751 | MsrA | 3.75 | 0.02 |
| D2NAT9 | MnmE | 3.74 | 0.02 |
| D2N835 | Universal stress protein family, putative | 3.66 | 0.02 |
| D2N6J2 | Sun | 3.51 | 0.04 |
| D2N9E2 | Morphine 6-dehydrogenase (Naloxone reductase) | 3.46 | 0.02 |
| D2NAH7 | PanD | 3.05 | 0.05 |
| D2N655 | YwbD | 2.95 | 0.02 |
| D2NAK8 | Alkaline phosphatase 3 (Alkaline phosphataseIII) (APase III) | 2.80 | 0.05 |
| D2NAM5 | Phage infection protein | 2.77 | 0.02 |
| D2N7Y0 | RuvB | 2.75 | 0.04 |
| D2N917 | Cardiolipin synthetase (Cardiolipin synthase) (CLsynthase) | 2.58 | 0.01 |
| D2N7V5 | UPF0473 protein | 2.58 | 0.02 |
| D2N410 | 5 ~-nucleotidase, lipoprotein e (P4) family | 2.47 | 0.04 |
| D2N9P4 | Inositol monophosphatase family protein | 2.40 | 0.02 |
| D2N6X6 | Glycine betaine transporter OpuD | 2.31 | 0.03 |
| D2N6I0 | PhnB | 2.24 | 0.02 |
| D2N953 | Putative transcriptional regulator superfamily | 2.10 | 0.02 |
| D2N755 | YphP | 2.10 | 0.04 |
| D2N5A8 | NagA | 2.10 | 0.03 |
| D2N7I6 | ScpB | 1.97 | 0.04 |
| D2N750 | MsrB | 1.94 | 0.05 |
| D2N657 | Phosphocarrier protein HPr | 1.92 | 0.02 |
| D2N6I4 | RpoZ | 1.84 | 0.04 |
| D2N9L8 | Butyryl-CoA dehydrogenase | 1.84 | 0.02 |
| D2N6L2 | UPF0122 protein | 1.81 | 0.04 |
| D2N7K7 | Lipoamide acyltransferase component of branched-chain alpha-keto aciddehydrogenase complex | 1.80 | 0.02 |
| D2N6J3 | RlmN | 1.80 | 0.03 |
| D2N681 | Inositol-1-monophosphatase (IMPase) (Inositol-1-phosphatase) (I-1-Pase) | 1.76 | 0.02 |
| D2N5S9 | AddA | 1.73 | 0.02 |
| D2N5P1 | HAD-superfamily subfamily IIA hydrolase | 1.69 | 0.03 |
| D2N7P4 | Superoxide dismutase | 1.68 | 0.02 |
| D2N7T3 | RsfS | 1.66 | 0.03 |
| D2N842 | Tpx | 1.64 | 0.02 |
| D2N7J2 | Oxidoreductase | 1.60 | 0.02 |
| D2N941 | UDP-N-acetylglucosamine 2-epimerase | 1.60 | 0.02 |
| D2N7L6 | NusB | 1.58 | 0.02 |
| D2N6K8 | AcpP | 1.58 | 0.02 |
| D2N4I9 | HsdM | 1.58 | 0.02 |
| D2N4U4 | RplJ | 1.55 | 0.02 |
| D2N6F6 | MurD | 1.54 | 0.02 |
| D2N5D9 | Ribonucleoside-diphosphate reductase, beta subunit | 1.54 | 0.02 |
| D2N8L2 | Aminopeptidase PepS | 1.54 | 0.03 |
| Unknown proteins | |||
| D2N9M8 | Uncharacterized protein | 3.63 | 0.02 |
| D2N5P0 | Uncharacterized protein | 2.49 | 0.04 |
| D2N4W0 | Uncharacterized protein | 2.17 | 0.02 |
| D2NA92 | Uncharacterized protein | 1.85 | 0.02 |
| D2N685 | Uncharacterized protein | 1.62 | 0.04 |
| D2N588 | Uncharacterized protein | 1.61 | 0.02 |
*The calculation of FC (239/398) in this table is dividing the average value of peptide mass of proteins in ST239 strains by that average value in ST398 strains
**Bold italics in this table is used to highlight the selected significantly differentially expressed genes for HA-SA ST239 up-regulated network construction
Table 3.
All up-regulated proteins in the CA-SA ST398 group
| Uniprot acc no. | Prot_description | FC* (398/239) | P value |
|---|---|---|---|
| Known virulence-related factors | |||
| D2N8W5 | AgrC** | 23.91 | 0.01 |
| D2N8W6 | AgrA | 7.20 | 0.02 |
| D2N7I3 | SrrB | 2.02 | 0.02 |
| D2N8Z3 | SigB | 1.98 | 0.03 |
| D2N718 | Conserved virulence factor B (SAPIG1392) | 1.98 | 0.02 |
| D2N5H5 | ClpP | 1.77 | 0.02 |
| Transcriptional regulator | |||
| D2N576 | SarX | 9.91 | 0.01 |
| D2N9W6 | AraC family regulatory protein | 4.48 | 0.01 |
| D2N9P8 | Phosphosugar-binding transcriptional regulator, RpiR family | 3.99 | 0.02 |
| D2N3I9 | Transcriptional regulator, GntR family | 3.32 | 0.02 |
| D2N9Q7 | Phosphosugar-binding transcriptional regulator | 3.07 | 0.03 |
| D2N823 | PhoP | 2.36 | 0.02 |
| D2N6S6 | Glycerol uptake operon antiterminator regulatory protein | 2.41 | 0.03 |
| D2N8K6 | RecX | 2.08 | 0.04 |
| Arginine and proline metabolism | |||
| D2NAL7 | ArcA | 6.11 | 0.03 |
| D2NAL6 | ArcB | 4.18 | 0.02 |
| D2NAL4 | ArcC | 2.52 | 0.03 |
| Multidrug resistance | |||
| D2N9T7 | Multidrug resistance protein A | 4.72 | 0.02 |
| D2N552 | Multidrug resistance ABC transporter ATP-binding and permease protein | 3.84 | 0.02 |
| Two-component system | |||
| D2N9Y1 | NarH | 2.36 | 0.03 |
| D2N361 | YycG | 1.89 | 0.03 |
| Pyrimidine metabolism | |||
| D2NAJ6 | NrdD | 11.84 | 0.02 |
| D2N6H8 | PyrE | 2.36 | 0.02 |
| D2N6H3 | PyrB | 2.11 | 0.04 |
| D2N942 | Upp | 1.52 | 0.02 |
| Pyruvate metabolism | |||
| D2N3S5 | PflB | 2.37 | 0.04 |
| D2N9V0 | Mqo | 2.26 | 0.02 |
| D2N7L8 | AccC | 1.78 | 0.02 |
| Histidine metabolism | |||
| D2N9R5 | HutU | 17.44 | 0.01 |
| D2N9R4 | HutI | 3.59 | 0.02 |
| D2N9R7 | HutG | 1.82 | 0.04 |
| Glycolysis/gluconeogenesis | |||
| D2NAA0 | Fbp | 2.63 | 0.02 |
| D2N9Z9 | GpmA | 2.50 | 0.02 |
| D2N8B5 | PckA | 2.34 | 0.02 |
| Other known proteins | |||
| D2N3K5 | CapC | 21.85 | 0.01 |
| D2N3F9 | Antigen, 67 kDa | 11.84 | 0.05 |
| D2N5J2 | Probable membrane protein | 8.22 | 0.01 |
| D2NA54 | 3-oxoacyl-[acyl-carrier-protein] reductase (3-ketoacyl-acyl carrier protein reductase) | 7.79 | 0.01 |
| D2N5B9 | QueC | 6.59 | 0.01 |
| D2N3D6 | Amidohydrolase 2 | 6.51 | 0.02 |
| D2N543 | Iron dependent repressor | 6.42 | 0.02 |
| D2N8W0 | Nitroreductase family protein | 6.12 | 0.02 |
| D2N628 | Acetyl transferase | 5.96 | 0.02 |
| D2NA36 | Para-nitrobenzyl esterase | 5.36 | 0.02 |
| D2N4R9 | FolP | 4.83 | 0.02 |
| D2N3T8 | ABC transporter, substrate-binding protein | 4.82 | 0.02 |
| D2N549 | Teichoic acid biosynthesis protein X | 4.81 | 0.01 |
| D2N944 | UPF0340 protein | 4.72 | 0.02 |
| D2N4D0 | Stage 0 sporulation protein J | 4.54 | 0.01 |
| D2N5Q4 | Cytosol aminopeptidase family protein | 4.53 | 0.04 |
| D2N4P0 | Stage 0 sporulation protein YaaT | 3.98 | 0.02 |
| D2N526 | Hydrolase, alpha/beta hydrolase fold family | 3.92 | 0.02 |
| D2N723 | DapB | 3.80 | 0.02 |
| D2N3Q0 | Pts system eiibc component | 3.71 | 0.02 |
| D2N8G9 | ABC transporter EcsB | 3.67 | 0.01 |
| D2N3F6 | Aminoacylase | 3.58 | 0.02 |
| D2N585 | Lipoprotein, putative | 3.47 | 0.02 |
| D2N6B0 | SpoU rRNA Methylase family protein | 3.42 | 0.02 |
| D2N349 | NnrD | 3.39 | 0.02 |
| D2NAA6 | Glyoxalase family protein | 3.37 | 0.02 |
| D2N362 | YycH protein | 3.21 | 0.03 |
| D2N6F0 | UPF0747 protein | 3.18 | 0.02 |
| D2N3K2 | Aldehyde-alcohol dehydrogenase 2 | 3.12 | 0.02 |
| D2N9Q3 | Phosphoglycolate phosphatase | 3.04 | 0.02 |
| D2N411 | ABC transporter, permease protein | 3.01 | 0.02 |
| D2N7J1 | Asppase | 2.69 | 0.03 |
| D2N7R6 | NfeD | 2.63 | 0.01 |
| D2N4E3 | Phosphoglycerate mutase family protein | 2.59 | 0.02 |
| D2N7R2 | PhoH family protein | 2.55 | 0.02 |
| D2N5I8 | Rnr | 2.52 | 0.02 |
| D2N7Z5 | Tag | 2.48 | 0.02 |
| D2N9Z1 | Zinc-binding lipoprotein AdcA | 2.47 | 0.04 |
| D2N4S0 | FolB | 2.41 | 0.04 |
| D2N363 | YycI protein | 2.39 | 0.02 |
| D2NA39 | ABC transporter ATP-binding protein | 2.36 | 0.04 |
| D2N499 | NAD-dependent epimerase/dehydratase | 2.36 | 0.02 |
| D2N6R1 | PgsA | 2.34 | 0.02 |
| D2N5Y1 | NadK | 2.34 | 0.02 |
| D2N804 | EngB | 2.18 | 0.02 |
| D2N421 | NanE | 2.15 | 0.04 |
| D2N6C3 | MurI | 2.12 | 0.02 |
| D2N356 | DHH subfamily 1 protein | 2.11 | 0.02 |
| D2N8M1 | Map | 2.10 | 0.02 |
| D2N5H2 | UPF0042 nucleotide-binding protein | 2.10 | 0.03 |
| D2N7R9 | Ribosomal RNA small subunit methyltransferase E | 2.10 | 0.01 |
| D2N870 | Ftsk/spoiiie family protein | 2.09 | 0.02 |
| D2N9Y2 | Nitrate reductase, alpha subunit | 2.04 | 0.02 |
| D2N5V8 | FabH | 1.95 | 0.02 |
| D2NA13 | ABC transporter, ATP-binding/permease protein | 1.92 | 0.04 |
| D2N6V0 | Cardiolipin synthetase (Cardiolipin synthase) (CLsynthase) | 1.92 | 0.03 |
| D2N6W2 | Catalase | 1.90 | 0.02 |
| D2N8Q5 | Aldehyde dehydrogenase | 1.87 | 0.02 |
| D2N558 | DhaK | 1.86 | 0.04 |
| D2N9V4 | TpgX protein | 1.84 | 0.02 |
| D2N860 | Fhs | 1.82 | 0.02 |
| D2N8R8 | Acyl-coenzyme A:6-aminopenicillanic acid acyl-transferase | 1.80 | 0.04 |
| D2N5D0 | ABC transporter permease protein | 1.80 | 0.04 |
| D2N7M6 | GcvPB | 1.80 | 0.04 |
| D2N694 | Glycerophosphoryl diester phosphodiesterase | 1.79 | 0.02 |
| D2N634 | QoxB | 1.78 | 0.02 |
| D2N4Q0 | IspE | 1.78 | 0.02 |
| D2N727 | LysA | 1.77 | 0.02 |
| D2N6R4 | Rny | 1.74 | 0.02 |
| D2N9L2 | ModA | 1.71 | 0.03 |
| D2N5F5 | DegV family protein | 1.69 | 0.02 |
| D2N520 | Iron-binding protein | 1.66 | 0.04 |
| D2N9C8 | Alcohol dehydrogenase, zinc-binding domain protein | 1.64 | 0.02 |
| D2N359 | PurA | 1.64 | 0.04 |
| D2N6G7 | IleS | 1.62 | 0.02 |
| D2N9V2 | TagF domain protein | 1.61 | 0.02 |
| D2N6Y5 | Transcription antiterminator | 1.60 | 0.04 |
| D2N7P9 | ATP-dependent RNA helicase | 1.60 | 0.02 |
| D2N4Q1 | PurR | 1.58 | 0.02 |
| D2N5F8 | Ribosomal subunit interface protein | 1.57 | 0.02 |
| D2N662 | Potassium uptake protein TrkA | 1.57 | 0.02 |
| D2N9C5 | Conserved domain protein | 1.57 | 0.02 |
| D2N7J7 | Zwf | 1.56 | 0.03 |
| D2N3I0 | Acetoin(Diacetyl) reductase (Acetoin dehydrogenase) | 1.55 | 0.02 |
| D2N780 | AsnS | 1.53 | 0.02 |
| D2N8B2 | 2,5-diketo-D-gluconic acid reductase A | 1.52 | 0.02 |
| D2N5Q3 | YumB | 1.52 | 0.02 |
| D2N6S5 | MutL | 1.52 | 0.03 |
| D2N6M8 | TopA | 1.52 | 0.04 |
| D2N523 | Hydrolase, alpha/beta hydrolase fold family | 1.51 | 0.02 |
| D2N4S2 | LysS | 1.51 | 0.02 |
| Unknown proteins | |||
| D2N9S7 | Uncharacterized protein | 9.98 | 0.02 |
| D2N965 | Uncharacterized protein | 8.45 | 0.01 |
| D2N3Y5 | Uncharacterized protein | 5.51 | 0.02 |
| D2N899 | Uncharacterized protein | 4.17 | 0.02 |
| D2N9W5 | Uncharacterized protein | 3.98 | 0.01 |
| D2N4F3 | Uncharacterized protein | 3.92 | 0.03 |
| D2N692 | Uncharacterized protein | 3.24 | 0.05 |
| D2N5Y6 | Uncharacterized protein | 2.69 | 0.02 |
| D2N5H8 | Uncharacterized protein | 2.65 | 0.02 |
| D2N8E4 | Uncharacterized protein | 2.60 | 0.02 |
| D2N678 | Uncharacterized protein | 2.58 | 0.02 |
| D2N6V3 | Uncharacterized protein | 2.15 | 0.02 |
| D2N9T5 | Uncharacterized protein | 2.07 | 0.04 |
| D2N8T9 | Uncharacterized protein | 2.03 | 0.04 |
| D2N8R7 | Uncharacterized protein | 1.89 | 0.02 |
| D2N8B3 | Uncharacterized protein | 1.65 | 0.02 |
| D2N6R0 | Uncharacterized protein | 1.64 | 0.03 |
| D2N9C6 | Uncharacterized protein | 1.58 | 0.02 |
| D2N971 | Uncharacterized protein | 1.56 | 0.02 |
| D2N5K3 | Uncharacterized protein | 1.67 | 0.03 |
*The calculation of FC (398/239) in this table is dividing the average value of peptide mass of proteins in ST398 strains by that average value in ST239 strains
**Bold italics in this table is used to highlight the selected significantly differentially expressed genes for CA-SA ST398 up-regulated network construction
Hierarchical cluster analysis was further performed for all the differentially expressed proteins in the HA-SA ST239 group versus the CA-SA ST398 group since HCL method allows samples that are highly similar in quantitative profiles to be merged in an agglomerative fashion. As shown in Fig. 2c, we were able to distinguish the two different types of clinical SA by identifying those differentially expressed proteins. Although there might be some exceptions of individual isolate whose expression was not in accordance with the whole expression pattern of the certain protein, it won’t influence the clustering of the four HA-SA ST239 strains or the four CA-SA ST398 strains. The results suggested that HA-SA ST239 elicits a much different proteomic profile from CA-SA ST398. To decipher which proteins were related to the respective virulent effect between HA-SA ST239 and CA-SA ST398, string analysis was performed focusing on the network of proteins possibly involved in the pathogenesis of S. aureus.
Interaction network construction and network analysis
We respectively evaluated 64 up-regulated genes in the HA-SA ST239 group and 145 up-regulated genes in the CA-SA ST398 group using the STRING (version 10.0) database to identify the potential interactions among these gene products. However, the network incorporated numerous isolated nodes and pair-linked nodes providing useless information. Hence, we cut out the differentially expressed genes corresponding to such types of nodes and identified 22 up-regulated proteins in HA-SA group and 95 up-regulated proteins in CA-SA group based on the potential networks (Additional file 2: Figure 1A and B). Next, a PPI network was constructed via Cytoscape software to judge the importance order of the differentially expressed genes between HA-SA ST239 and CA-SA ST398. We also combined the functional enrichment in the network and category according to the degree of knowledge of those proteins. The final HA-SA ST239 up-regulated protein network was constituted of 9 nodes and 12 edges, with one as the minimum degree of connectivity of a node and five as the maximum. For the 9 nodes in the network, the average degree of connectivity was 2.67, and the PPI enrichment P value was 7.22e−08. While the resulting CA-SA ST398 up-regulated protein network was composed 8 nodes and 11 edges, and the average degree of connectivity for the 8 nodes was 2.75, the PPI enrichment P value was 4.04e−09. Meanwhile, the KEGG enrichment analysis of the STRING database showed that there is only one significantly enriched pathway (extracellular region) in the HA-SA ST239 up-regulated network, including 9 proteins—Fibronectin-binding protein A (FnbpA), Immunoglobulin G binding SpA, Bifunctional autolysin (Atl), Clumping factor A (ClfA), Immunodominant staphylococcal antigen A (IsaA), Immunodominant staphylococcal antigen B (IsaB), Lipoteichoic acid synthase (LtaS), Staphylococcal secretory antigen A (SsaA) and Collagen adhesin (Cna)—and also only one significantly enriched pathway (two-component system) in the CA-SA ST398 up-regulated network including five proteins—Accessory gene regulator A (AgrA), Accessory gene regulator C (AgrC), Alkaline phosphatase synthesis transcriptional regulatory protein (PhoP), Staphylococcal respiratory response protein (SrrB), and Sensor histidine kinase (YycG, also named WalK) (Table 4). Based on the knowledge of proteins and their link in the enriched pathways, Staphylococcal accessory regulator X (SarX), RNA polymerase sigma factor B (SigB) and ATP-dependent Clp protease proteolytic subunit P (ClpP) were also identified as candidate key proteins in the CA-SA ST398 up-regulated network. Finally, nine proteins (FnbpA, SpA, Atl, ClfA, IsaA, IsaB, LtaS, SsaA and Cna) and eight proteins (AgrA, AgrC, PhoP, SrrB, YycG, SarX, SigB and ClpP) were, respectively, used for the model reconstruction of the HA-SA ST239 and CA-SA ST398 key virulent network alone and combined (Fig. 3).
Table 4.
KEGG-enrichment pathway and genes from String analysis
| #Pathway ID | Pathway description | Gene count | False discovery rate | Matching proteins in your network (labels) |
|---|---|---|---|---|
| Up-regulated in ST239 | ||||
| 5576 | Extracellular region | 9 | 8.08E−05 | FnbpA, SpA, Atl, ClfA, IsaA, IsaB, LtaS, SsaA, Cna |
| Up-regulated in ST398 | ||||
| 2020 | Two-component system | 5 | 3.27E−05 | AgrA, AgrC, PhoP, SrrB, YycG |
Fig. 3.
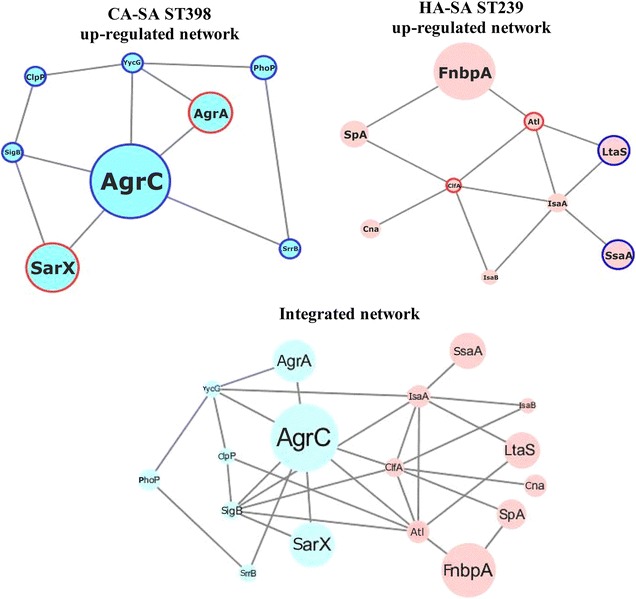
Cytoscape network reconstruction. Circle nodes represent genes. For the genes, the borders of the nodes represent the type of gene regulation determined by LC-MS/MS. The centers of the nodes indicate the gene expression changes; the diameter of the circle is proportional to the level of regulation. The blue outer circles of the nodes depict corresponding results from LC-MS/MS and qRT-PCR analyses (Fig. 4), and the red outer circles indicate corresponding results from LC-MS/MS and Western blot analysis (Fig. 5)
One node would be considered more significant than the other nodes and would be called the central node when the degree of connectivity of the node was much greater than the average. The central node of a PPI network also represents the core protein in the network. In the current study, all nodes were analyzed through a network analyzer, according to the degree of connectivity of each node which was set as ≥ the mean value to identify candidate key virulent genes. Based on the degree of connectivity of a node, the HA-SA ST239 up-regulated proteins (ClfA, Atl and IsaA) and CA-SA ST398 up-regulated proteins (AgrC, YycG and SigB) were identified as key proteins in the respective network. However, according to the results of the relatively higher level of differentially expression (the fold changes of ST239/ST398 were higher than 4.0-fold or lower than 0.25-fold), the HA-SA ST239 upregulated proteins (FnbpA, LtaS and SsaA) and CA-SA ST398 up-regulaed proteins (AgrA, AgrC and SarX) were identified as crucial candidate proteins. Nevertheless, the IsaA, AgrC, YycG, SigB, FnbpA, LtaS and SsaA could not been verified successfully in the translational level for the lack of effective antibodies. Thus, we selected these four proteins (ClfA, Atl, AgrA, SarX) for further validation of Western blotting.
Validation of the proteomic data using RT-PCR
The transcription rates of 17 candidate key genes coding for proteins with different expression between HA-SA ST239 group and CA-SA ST398 group detected by LC-MS/MS were tested by RT-PCR. This validation finally selected ten genes with significantly different transcriptional expression between the two groups. As shown in Fig. 4a, the transcriptional levels of eight genes (sarX, sigB, agrA, agrC, yycG, clpP, srrB, and phoP) were significantly greater in the CA-SA ST398 group than in the HA-SA ST239 group, consistent with the proteomic results. However, among the nine proteins (FnbpA, SpA, Atl, ClfA, IsaA, IsaB, LtaS, SsaA and Cna) which were detected up-regulated in the HA-SA ST239 group, only two genes (ltaS and ssaA) displayed higher transcriptional level in this group, indicating there could be possible differences in translational or post-translational level between ST239 and ST398. As indicated in Fig. 4b showing the fold change of the average transcriptionally expressed value of specific gene within HA-SA ST239 strains relative to the corresponding value within CA-SA ST398 strains, the greatest differentially expressed genes in the transcriptional level were sarX and agrA (respectively 7.29 and 6.53-fold in ST398 strains compared to ST239 strains).
Fig. 4.
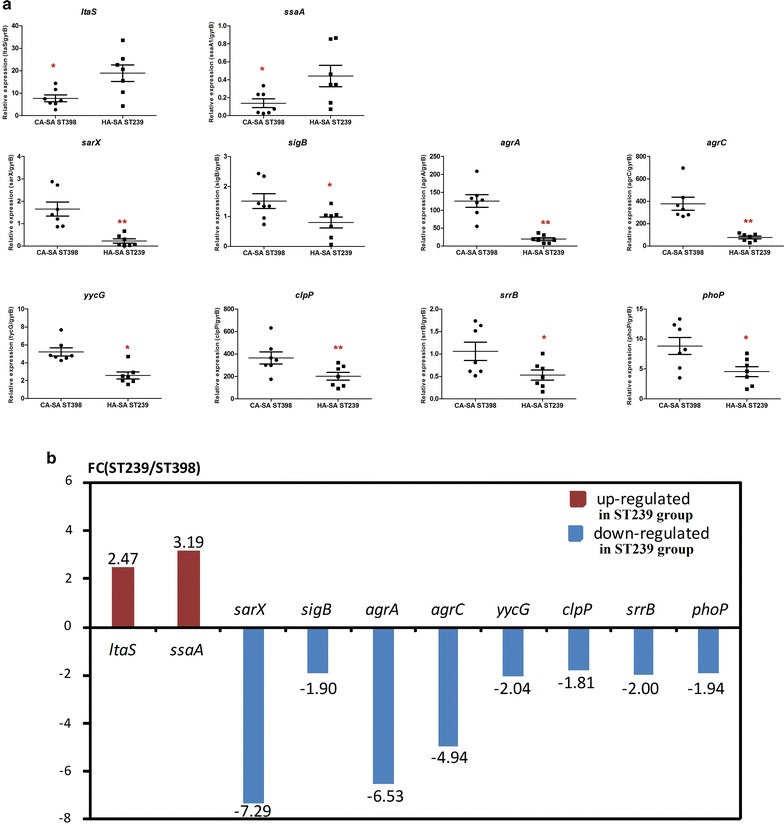
RT-PCR validation. a Transcriptional expression levels of the 10 significantly key differentially expressed genes between HA-SA and CA-SA by qRT-PCR in cultures grown to the post-exponential growth phase (8 h) from 17 candidate key differentially expressed genes. *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired t-test). The data is representative of three independent experiments. b FC (ST239/ST398) in this figure shows the fold change of the average transcriptionally expressed value of specific gene within HA-SA ST239 strains relative to that within CA-SA ST398 strains. Therefore, Down-regulated in ST239 group equivalents to Up-regulated in ST398 group
Validation of the proteomic data using western blot
To further confirm the proteomic data, we tested the expression of several proteins by Western blot. In the Western blotting analysis indicated in Fig. 5a, b, the translational expression levels of AgrA and SarX were significantly greater in the CA-SA ST398 group than in the HA-SA ST239 group (P = 0.013 and P = 0.006 respectively), nonetheless the translational expression levels of Atl and ClfA were significantly higher in the HA-SA ST239 group than in the CA-SA ST398 group (P = 0.005 and P = 0.048 respectively), which is consistent with the findings in proteomic analysis. Figure 5c showed the fold change of the average translationally expressed value of specific protein within HA-SA ST239 strains relative to the corresponding value within CA-SA ST398 strains. Among the above proteins, Atl was the most differentially expressed protein on the translational level (12.28-fold in ST239 strains compared to ST398 strains).
Fig. 5.
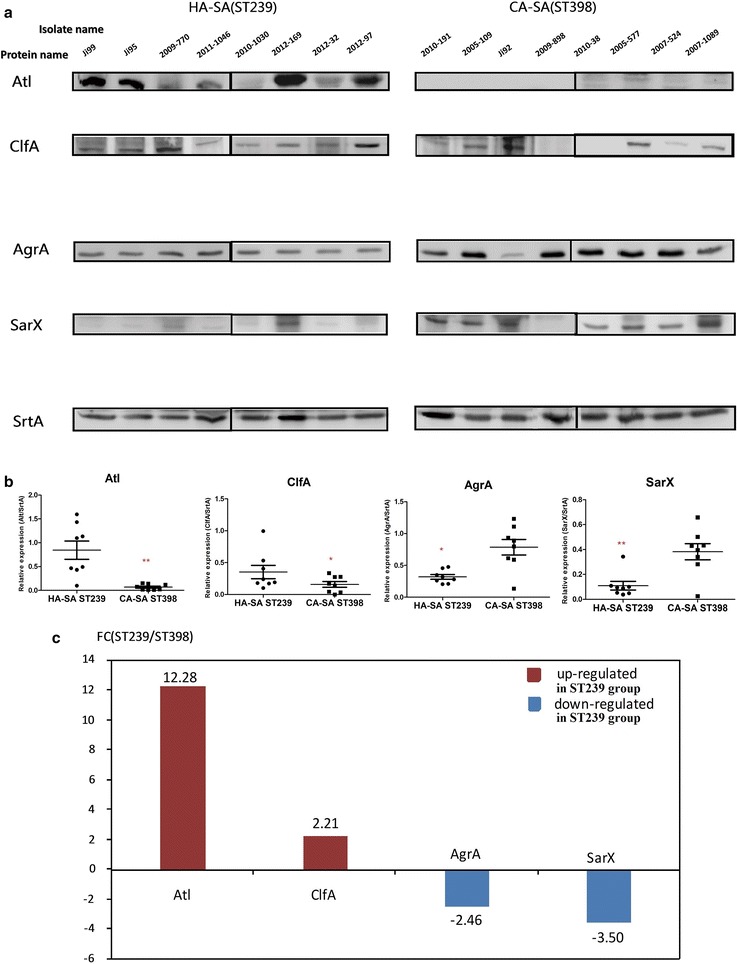
Western blot validation. a The expression levels of the four key differentially expressed proteins between HA-SA and CA-SA by densitometry of Western Blot in post-exponential phase cultures (8 h) using sortase A as control. b Densitometry plot of Western Blot. The values are normalized versus sortase A signals obtained using the same samples. *P < 0.05, **P < 0.01, ***P < 0.001 (unpaired t-test). c FC (ST239/ST398) in this figure shows the fold change of the average translationally expressed value of specific protein within HA-SA ST239 strains relative to that within CA-SA ST398 strains. Therefore, Down-regulated in ST239 group equivalents to Up-regulated in ST398 group
Discussion
The study was initiated to identify the key virulent network of HA- and CA-SA. We presented the different phenotypes between HA-SA ST239 and CA-SA ST398. HA-SA ST239 showed more adhesion ability to human alveolar epithelial cells and biofilm than CA-SA ST398 isolates, while the CA-SA ST398 exhibited stronger hemolysis ability than the HA-SA ST239. Further proteomic profile analysis showed that higher expression of surface-related proteins (FnbpA, SpA, Atl, ClfA, IsaA, IsaB, LtaS, SsaA and Cna) may involve in the durative infection in HA-SA ST239, while the higher expression of Agr system (AgrA and AgrC) and its interactive factors (PhoP, SrrB, YycG, SarX, SigB and ClpP) contribute to the higher virulence in CA-ST ST398.
It had been reported that CA-MRSA typically express low quantities of penicillin -binding protein 2a to sustain higher virulence and further predominate in the community setting [19]. Moreover, HA-MRSA strains are less virulent despite the high expression of mecA gene [19]. In our study, we set out to profile the proteome of certain clinically significant and relevant strains of SA to discover distinct sets of interactive factors contributing respectively to enhance the virulence of two SA types and cause their different clinical syndromes. Specifically, HA-SA ST239 and CA-SA ST398 representative clinical isolates were analyzed by quantitative methods at the post-exponential phase of growth (8 h), during which the toxins are initially produced and gradually accumulated in a large quantity. We conducted a high throughout and quantitative proteomic analysis to search for the virulent network of HA- and CA-SA [32].
When comparing HA-SA ST239 to CA-SA ST398 during the post-exponential phase of growth, we found that HA-SA ST239 strains significantly expressed a broad range of cell surface-associated virulence proteins (FnbpA, SpA, Atl, ClfA, IsaA, IsaB, LtaS, SsaA and Cna) at a higher level than ST398 strains. The cell surface proteins including MSCRAMMs such as FnbpA, SpA, ClfA and Cna played important roles in the adhesion, biofilm formation and eliciting of endovascular, bone and joint and prosthetic-device infections [39]. For example, FnbpA is involved in bacterial adhesion to host tissues by binding to fibronectin, and to affect biofilm formation [40, 41]. IsaA, involved in peptidoglycan hydrolysis, has been demonstrated as a virulence factor which existed both in the culture supernatant and the cell wall fraction [42]. Furthermore, the inactivation of isaA results in the up-regulation of ssaA, whose gene product has been demonstrated to own peptidoglycan hydrolase activity and is supposed to be crucial in biofilm formation [43, 44]. These surface proteins play a critical role in preventing the organism from recognition by the host immune system [45]. SpA could adhere to the Fc portion of IgG and Fab fragment of VH3-type B cell receptors, leading to bacterial immune evasion from the host [41, 46]. ClfA factor is an important staphylococcal adhesin, which could bind to dimeric host Fg complex via the carboxy-terminal component of the Fg gamma chain, thus causing staphylococcal accumulation in plasma or purified Fg [47]. As the major Fg binding protein, ClfA also promotes the adherence to immovable Fg- or fibrin-coated surfaces, facilitating its binding to both of blood clots and biomaterials [47]. FnbpA has been shown to be involved in adherence to host cells and to promote internalization by host cells [48, 49]. FnbpA has also been demonstrated to be important in in vivo infection by S. aureus [50]. The collagen (Cn)-binding protein Cna is a prototype of the MSCRAMMs that plays an important role in staphylococcal pathogenesis both as an adherence factor and as an immune evasion factor. Cna is a proven virulence factor in septic arthritis, where the strength of adhesion to collagen correlates with disease pathogenesis [51, 52]. IsaB, also as a cell-wall associated and secreted virulence factor, could trigger an immune responsive process during the life-threatening septicemia [53]. Through the analysis of microarray data, isaB production was also shown to increase in response to some certain settings, such as the existence of biofilms, neutrophil exposure, anaerobic condition and further internalization into human epithelial cells, indicating an important role in infection despite of its involvement in immune escape [54, 55]. Previous study has identified the importance of the atl gene products in the initial stages of biofilm formation [56]. Deletion of atl whether in S. aureus or S. epidermidis would cause extensive cell aggregation and following biofilm-negative phenotype [56]. This change of phenotype already indicates that Atl is a necessary factor for the successful partitioning of daughter cells after their division. LtaS participates in the synthesis of lipoteichoic acid (LTA), which is one of the major components of the cell wall in S. aureus [57]. As for the observation of increased protein levels of ClfA and Atl in ST239 isolates which was not confirmed by the transcriptional studies, two possible reasons might be responsible for the discrepancy between the transcriptional and translational level. On one hand, regulation of post-transcription, translation, transcript degradation and protein degradation could contribute as much to variation in the final protein concentration. On the other hand, there could be a significant amount of noise and error in mRNA experiment, limiting our access to get a consistent result with protein experiment [58]. Apart from the discrepancies as above, ClfA/Atl/IsaA seemingly acts as central nodes in HA-SA ST239 up-regulated network. However, only the significantly increased expression of ClfA/Atl in HA-SA ST239 group could be verified at the translational level. One would speculate that ClfA/Atl might act as a group leader to gather other team members (SpA, Cna, FnbpA, SsaA, IsaA, IsaB and LtaS) together, thus probably contributing to biofilm formation.
In addition, we found that 8 Agr-interactive network proteins (AgrA, AgrC, PhoP, SrrB, YycG, SarX, SigB and ClpP) are generally up-regulated in the CA-SA strain ST398. This finding is in accord with that the reported transcriptomic studies suggesting a hyper-vital agr in CA-SA lineage compared with those in HA-SA lineage [59]. In this study, we have strikingly shown that AgrC in the PPI network reconstruction acted as central node in the ST398 up-regulated network. The higher transcripts of both agrA and agrC and the higher translational proteins of AgrA were all confirmed in CA-SA ST398. It could be speculated that AgrAC might interact with other connected factors such as PhoP, SrrB, YycG, SarX, SigB and ClpP, however, the regulatory mechanism between them was not clear at present.
Although as the sensor of the agr locus, AgrC unexpectedly displayed a higher degree of connectivity to other factors in this bioinformatic analysis than the response regulator AgrA. One possible reason could be the strict selection rule of the significantly differentially expressed proteins for constructing the interaction networks. Some genes/proteins that were regulated by AgrA might be eliminated from the list for the network construction due the cutoff of a P value by Student’s t-test lower than 0.05 and with an ST239/ST398 change higher than 1.5-fold or lower than 0.67-fold. There were links between AgrC and five factors in the construction model (AgrA, SarX, SigB, SrrB and YycG). It was well known that AgrA/AgrC act together, however, other interactions were not so clear. As a SarA paralog, the HTH-type transcriptional regulator (SarX) was initially identified in S. aureus by Manna and Cheung (2006). They have also reported that the sarX gene in S. aureus expressed maximally and temporally at the transcriptional level in stationary phase [60]. The agr transcripts, RNAII and RNAIII, has been found to have significantly increased expression in a sarX mutant. SarX has been discovered to negatively control agr expression by binding to the agr promoter, which in turn displays strain-specific effects on regulating the biofilm formation. This finding seems to contradict the simultaneously increased expression of AgrC and SarX in CA-SA ST398 group. The transcription factor sigma B (SigB) is reported to influence the expressional production of several genes encoding stress-response proteins and virulence factors, and appears to counteract the agr system on its effect on the expressional production of virulent factors [61]. It also contradicts the simutaneously elevated expression of AgrC and SigB in CA-SA ST398 group. As for the SrrB and YycG, the AgrA/C, SrrA/B, YycG/H systems are independently well-known two-component regulatory systems. There are also no report showing the interaction between AgrC and SrrB or YycG. However, in view of the active interaction sources of STRING database include Co-occurrence, Gene Fusion, Neighborhood, Co-expression, Databases, Experiments and Textmining, it would be possible for us to fail to find the related experiments to support our results. In this model, AgrC seems to be the central nodes in CA-SA ST398 up-regulated network, indicating the contribution of Agr to the high virulence in CA-SA ST398. Unfortunately, we failed to confirm the higher expression of AgrC in CA-SA ST398 group because of the lack of AgrC antibody.
Notably, the higher amounts of agrC transcripts are present in CA-SA ST398 isolates detected by the LC MS/MS and identified by RT-PCR, albeit of the fact that agrC and agrA are both part of the same polycistronic transcript, RNAII. The transcriptional difference might come from the experimental noise or the prompt response of AgrC to autoinducing peptide (AIP, the AgrC ligand) in CA-SA ST398 lineage than other lineages. As for the discrepancy between the transcriptional and translational level of agrC/agrA expression ratio. We have checked that both of primer pairs for agrA and agrC have optimal primer efficiencies (> 90%) in their qRT-PCR setup, although the primer efficiency of agrC (~ 97%) is higher than that of agrA (~ 92%). The factor of primer efficiency might contribute little to the expression ratio discrepancy between the transcriptional and translational level. Therefore, the above reasons of post modification and significant noise could be more prone to lead to this difference.
Additionally, there could be two possible reasons for the other RNAII encoded factors AgrB and AgrD missing in the up-regulated in ST398 isolates. (1) Because of the sensitivity of the method, AgrD (~ 5KD) has low molecular weight to be detected by the LC-MS/MS method. (2) The small peptide of AIP produced by AgrD was secreted to the supernatant. (3) There could be one or several mutations in the agrB gene causing the change of amino acid in AgrB (~ 26 kDa) and the mutated AgrB protein couldn’t match to the template (S0385 and TW20).
Moreover, SigB should be co-transcribed with the anti-sigma factors (RsbV and RsbW) [62]. We have detected the RsbV and RsbW proteins by the LC-MS/MS in our original data. However, our Table 3 shows the differentially expressed proteins with a P value by Student’s t-test lower than 0.05 and with an ST239/ST398 change higher than 1.5-fold or lower than 0.67-fold. Due to our strict screening rule, although RsbV and RsbW were up-regulated in some isolates of CA-SA ST398, their expressions are not significant according to P value by Student’s t-test.
Interestingly, the highest fold-change in expression of penicillin-binding protein 3 (Pbp3) between HA-SA ST239 and CA-SA ST398 isolates may account for the inferior fitness of ST239. Compared to the four HA-MRSA ST239 isolates, only half of the four CA-SA ST398 isolates are MRSA. However, instead of Pbp2a, Pbp3 was highly expressed in HA-MRSA ST239 group. One possible illustration for the striking Pbp3 expression might be due to the polymorphism of pbp3 in HA-MRSA ST239. Chadwick et al has reported that two nucleotide polymorphisms (G88A and G2047A) in the pbp3 gene of S. aureus were highly associated with the CA-MRSA USA300 lineage [63]. The nucleotide polymorphism need to be verified by gene sequencing. Another possibility may be due to fluoroquinolone connection in HA-MRSA ST239. It is well-established that HA-MRSA strains are more resistant to various groups of antibiotics such as fluoroquinolones than CA-MRSA. Diverse fitness cost associated with high-level resistance to fluoroquinolones was demonstrated by multiple groups to contribute to the clonal dynamics of HA-MRSA [64–66]. The genetic basis of differing vitality has also been partly established [64, 67]. The connection of the fitness cost of the HA-MRSA ST239 lineage and fluoroquinolone resistance needs to be further studied.
Technically, our study was based on quantitative proteome analysis of multiple pathogenic polypeptides of four HA-SA ST239 and four CA-SA ST398 isolates simultaneously, which is a good supplement to the recently published proteomic analysis [68]. One theoretical boundedness is isolate-dependence, which might demand the recharacterization of each single strain. Nevertheless, the identical concept is effective for any phylogenic or physiologic characterization of any isolate.
Taken together, biologically, we could conclude that the higher expression of Agr system and its interactive factors (PhoP, SrrB, YycG, SarX, SigB and ClpP) based on the protein-protein interaction network in the CA-SA ST398 strains may improve the acute infection of CA-SA, while the higher expression of agr-negative regulating surface-related factors (FnbpA, SpA, Atl, ClfA, IsaA, IsaB, LtaS, SsaA and Cna) contribute to the durative infection of HA-SA.
Additional files
Additional file 1. The detailed information on differentially expressed 209 proteins in each isolate.
Additional file 2. Potential interactions based on 64 up-regulated genes in the HA-SA ST239 group and 145 up-regulated genes in the CA-SA ST398 group using the STRING (version 10.0) database.
Authors’ contributions
ML and XL conceived the study. LH, HM, QL, MH, YW and XC performed experiments and analyzed the data. ML, LH, MH and QL drafted the manuscript. LH, HM, QL, MH, YW, XC, XL and ML revised and approved the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grants 81501804, 81371875, 81671975 and 81401735), the Shanghai Committee of Science and Technology, China (Grant 15411960500), the Foundation for Innovative Research Groups of the National Natural Science Foundation of China (Grant 81421001) and “Young talent plan” sponsored by Shanghai Health and Family Planning Commission (Grants 20154Y0014 and 201440435).
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets and its additional file supporting the conclusions of this article are included within the article.
Ethics approval and consent to participate
Blood of healthy individuals for lysis of erythrocytes by bacterial culture were collected with a standard method in accordance with a protocol approved by the ethics committee of Renji Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China. All individuals consented to participate the assay and gave written informed consent for the publication of this report and any accompanying images prior to donating blood.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12014-017-9178-5) contains supplementary material, which is available to authorized users.
Lei He, Hongwei Meng, and Qian Liu have contributed equally to this article
Contributor Information
Lei He, Email: buningweishi_1985@126.com.
Hongwei Meng, Email: menghongwei21@163.com.
Qian Liu, Email: qq2005011@163.com.
Mo Hu, Email: humour@pku.edu.cn.
Yanan Wang, Email: wangyn0819@126.com.
Xiaoying Chen, Email: cxy2419@163.com.
Xiaoyun Liu, Email: xiaoyun.liu@pku.edu.cn.
Min Li, Email: ruth_limin@126.com.
References
- 1.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339(8):520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Harinstein L, Schafer J, D’Amico F. Risk factors associated with the conversion of meticillin-resistant Staphylococcus aureus colonisation to healthcare-associated infection. J Hosp Infect. 2011;79(3):194–197. doi: 10.1016/j.jhin.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 3.Naimi TS, LeDell KH, Como-Sabetti K, Borchardt SM, Boxrud DJ, Etienne J, Johnson SK, Vandenesch F, Fridkin S, O’Boyle C, et al. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA. 2003;290(22):2976–2984. doi: 10.1001/jama.290.22.2976. [DOI] [PubMed] [Google Scholar]
- 4.Xia J, Gao J, Kokudo N, Hasegawa K, Tang W. Methicillin-resistant Staphylococcus aureus antibiotic resistance and virulence. Biosci Trends. 2013;7(3):113–121. [PubMed] [Google Scholar]
- 5.Otto M. Looking toward basic science for potential drug discovery targets against community-associated MRSA. Med Res Rev. 2010;30(1):1–22. doi: 10.1002/med.20160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated meticillin-resistant Staphylococcus aureus. Lancet. 2010;375(9725):1557–1568. doi: 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H, Liu Y, Jiang X, Chen M, Wang H. Rapid change of methicillin-resistant Staphylococcus aureus clones in a Chinese tertiary care hospital over a 15-year period. Antimicrob Agents Chemother. 2010;54(5):1842–1847. doi: 10.1128/AAC.01563-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Sousa MA, Crisostomo MI, Sanches IS, Wu JS, Fuzhong J, Tomasz A, de Lencastre H. Frequent recovery of a single clonal type of multidrug-resistant Staphylococcus aureus from patients in two hospitals in Taiwan and China. J Clin Microbiol. 2003;41(1):159–163. doi: 10.1128/JCM.41.1.159-163.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao M, Wang H, Zhao Y, Mao LL, Brown M, Yu YS, O’Sullivan MV, Kong F, Xu YC. National surveillance of methicillin-resistant Staphylococcus aureus in China highlights a still-evolving epidemiology with 15 novel emerging multilocus sequence types. J Clin Microbiol. 2013;51(11):3638–3644. doi: 10.1128/JCM.01375-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li T, Song Y, Zhu Y, Du X, Li M. Current status of Staphylococcus aureus infection in a central teaching hospital in Shanghai, China. BMC Microbiol. 2013;13:153. doi: 10.1186/1471-2180-13-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geng W, Yang Y, Wu D, Huang G, Wang C, Deng L, Zheng Y, Fu Z, Li C, Shang Y, et al. Molecular characteristics of community-acquired, methicillin-resistant Staphylococcus aureus isolated from Chinese children. FEMS Immunol Med Microbiol. 2010;58(3):356–362. doi: 10.1111/j.1574-695X.2010.00648.x. [DOI] [PubMed] [Google Scholar]
- 12.Wang D, Wang Z, Yan Z, Wu J, Ali T, Li J, Lv Y, Han B. Bovine mastitis Staphylococcus aureus: antibiotic susceptibility profile, resistance genes and molecular typing of methicillin-resistant and methicillin-sensitive strains in China. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis. 2015;31:9–16. doi: 10.1016/j.meegid.2014.12.039. [DOI] [PubMed] [Google Scholar]
- 13.van Belkum A, Melles DC, Peeters JK, van Leeuwen WB, van Duijkeren E, Huijsdens XW, Spalburg E, de Neeling AJ, Verbrugh HA, Dutch Working Party on Surveillance Research of MRSA et al. Methicillin-resistant and -susceptible Staphylococcus aureus sequence type 398 in pigs and humans. Emerg Infect Dis. 2008;14(3):479–483. doi: 10.3201/eid1403.0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huijsdens XW, van Dijke BJ, Spalburg E, van Santen-Verheuvel MG, Heck ME, Pluister GN, Voss A, Wannet WJ, de Neeling AJ. Community-acquired MRSA and pig-farming. Ann Clin Microbiol Antimicrob. 2006;5:26. doi: 10.1186/1476-0711-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao C, Liu Y, Zhao M, Liu Y, Yu Y, Chen H, Sun Q, Chen H, Jiang W, Liu Y, et al. Characterization of community acquired Staphylococcus aureus associated with skin and soft tissue infection in Beijing: high prevalence of PVL + ST398. PLoS ONE. 2012;7(6):e38577. doi: 10.1371/journal.pone.0038577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong X, Qin J, Li T, Dai Y, Wang Y, Liu Q, He L, Lu H, Gao Q, Lin Y, et al. Staphylococcal protein A promotes colonization and immune evasion of the epidemic healthcare-associated MRSA ST239. Front Microbiol. 2016;7:951. doi: 10.3389/fmicb.2016.00951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Hu M, Liu Q, Qin J, Dai Y, He L, Li T, Zheng B, Zhou F, Yu K, et al. Role of the ESAT-6 secretion system in virulence of the emerging community-associated Staphylococcus aureus lineage ST398. Sci Rep. 2016;6:25163. doi: 10.1038/srep25163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baines SL, Holt KE, Schultz MB, Seemann T, Howden BO, Jensen SO, van Hal SJ, Coombs GW, Firth N, Powell DR, et al. Convergent adaptation in the dominant global hospital clone ST239 of methicillin-resistant Staphylococcus aureus. mBio. 2015;6(2):e00080. doi: 10.1128/mBio.00080-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rudkin JK, Edwards AM, Bowden MG, Brown EL, Pozzi C, Waters EM, Chan WC, Williams P, O’Gara JP, Massey RC. Methicillin resistance reduces the virulence of healthcare-associated methicillin-resistant Staphylococcus aureus by interfering with the agr quorum sensing system. J Infect Dis. 2012;205(5):798–806. doi: 10.1093/infdis/jir845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loughman JA, Fritz SA, Storch GA, Hunstad DA. Virulence gene expression in human community-acquired Staphylococcus aureus infection. J Infect Dis. 2009;199(3):294–301. doi: 10.1086/595982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M, Diep BA, Villaruz AE, Braughton KR, Jiang X, DeLeo FR, Chambers HF, Lu Y, Otto M. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci USA. 2009;106(14):5883–5888. doi: 10.1073/pnas.0900743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warne B, Harkins CP, Harris SR, Vatsiou A, Stanley-Wall N, Parkhill J, Peacock SJ, Palmer T, Holden MT. The Ess/Type VII secretion system of Staphylococcus aureus shows unexpected genetic diversity. BMC Genom. 2016;17(1):222. doi: 10.1186/s12864-016-2426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Neill E, Pozzi C, Houston P, Smyth D, Humphreys H, Robinson DA, O’Gara JP. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J Clin Microbiol. 2007;45(5):1379–1388. doi: 10.1128/JCM.02280-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menzies BE. The role of fibronectin binding proteins in the pathogenesis of Staphylococcus aureus infections. Curr Opin Infect Dis. 2003;16(3):225–229. doi: 10.1097/00001432-200306000-00007. [DOI] [PubMed] [Google Scholar]
- 25.Tung H, Guss B, Hellman U, Persson L, Rubin K, Ryden C. A bone sialoprotein-binding protein from Staphylococcus aureus: a member of the staphylococcal Sdr family. Biochem J. 2000;345(Pt 3):611–619. [PMC free article] [PubMed] [Google Scholar]
- 26.Foster TJ, Hook M. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 1998;6(12):484–488. doi: 10.1016/s0966-842x(98)01400-0. [DOI] [PubMed] [Google Scholar]
- 27.Patti JM, Allen BL, McGavin MJ, Hook M. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu Rev Microbiol. 1994;48:585–617. doi: 10.1146/annurev.mi.48.100194.003101. [DOI] [PubMed] [Google Scholar]
- 28.Clarke SR, Foster SJ. Surface adhesins of Staphylococcus aureus. Adv Microb Physiol. 2006;51:187–224. doi: 10.1016/S0065-2911(06)51004-5. [DOI] [PubMed] [Google Scholar]
- 29.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38(3):1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The Multi Locus Sequence Typing website. http://www.mlst.net. Accessed 3 Sept 2016.
- 31.Vuong C, Gerke C, Somerville GA, Fischer ER, Otto M. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J Infect Dis. 2003;188(5):706–718. doi: 10.1086/377239. [DOI] [PubMed] [Google Scholar]
- 32.Hu M, Liu Y, Yu K, Liu X. Decreasing the amount of trypsin in in-gel digestion leads to diminished chemical noise and improved protein identifications. J Proteomics. 2014;109C:16–25. doi: 10.1016/j.jprot.2014.06.017. [DOI] [PubMed] [Google Scholar]
- 33.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34(2):374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 34.The STRING database V9.1. http://string-db.org/. Accessed 5 Oct 2016.
- 35.Cytoscape software V3.4.0. http://www.cytoscape.org/. Accessed 30 Jan 2017.
- 36.Li M, Du X, Villaruz AE, Diep BA, Wang D, Song Y, Tian Y, Hu J, Yu F, Lu Y, et al. MRSA epidemic linked to a quickly spreading colonization and virulence determinant. Nat Med. 2012;18(5):816–819. doi: 10.1038/nm.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edwards AM, Potts JR, Josefsson E, Massey RC. Staphylococcus aureus host cell invasion and virulence in sepsis is facilitated by the multiple repeats within FnBPA. PLoS Pathog. 2010;6(6):e1000964. doi: 10.1371/journal.ppat.1000964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhakdi S, Tranum-Jensen J. Alpha-toxin of Staphylococcus aureus. Microbiol Rev. 1991;55(4):733–751. doi: 10.1128/mr.55.4.733-751.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin YC, Peterson ML. New insights into the prevention of staphylococcal infections and toxic shock syndrome. Expert Rev Clin Pharmacol. 2010;3(6):753–767. doi: 10.1586/ecp.10.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Neill E, Pozzi C, Houston P, Humphreys H, Robinson DA, Loughman A, Foster TJ, O’Gara JP. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J Bacteriol. 2008;190(11):3835–3850. doi: 10.1128/JB.00167-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Foster TJ, Geoghegan JA, Ganesh VK, Hook M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol. 2014;12(1):49–62. doi: 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakata N, Terakubo S, Mukai T. Subcellular location of the soluble lytic transglycosylase homologue in Staphylococcus aureus. Curr Microbiol. 2005;50(1):47–51. doi: 10.1007/s00284-004-4381-9. [DOI] [PubMed] [Google Scholar]
- 43.Liu Y, Manna AC, Pan CH, Kriksunov IA, Thiel DJ, Cheung AL, Zhang G. Structural and function analyses of the global regulatory protein SarA from Staphylococcus aureus. Proc Natl Acad Sci USA. 2006;103(7):2392–2397. doi: 10.1073/pnas.0510439103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stapleton MR, Horsburgh MJ, Hayhurst EJ, Wright L, Jonsson IM, Tarkowski A, Kokai-Kun JF, Mond JJ, Foster SJ. Characterization of IsaA and SceD, two putative lytic transglycosylases of Staphylococcus aureus. J Bacteriol. 2007;189(20):7316–7325. doi: 10.1128/JB.00734-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Foster TJ. Immune evasion by staphylococci. Nat Rev Microbiol. 2005;3(12):948–958. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- 46.Kim HK, Cheng AG, Kim HY, Missiakas DM, Schneewind O. Nontoxigenic protein A vaccine for methicillin-resistant Staphylococcus aureus infections in mice. J Exp Med. 2010;207(9):1863–1870. doi: 10.1084/jem.20092514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McDevitt D, Francois P, Vaudaux P, Foster TJ. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol Microbiol. 1994;11(2):237–248. doi: 10.1111/j.1365-2958.1994.tb00304.x. [DOI] [PubMed] [Google Scholar]
- 48.Garzoni C, Kelley WL. Staphylococcus aureus: new evidence for intracellular persistence. Trends Microbiol. 2009;17(2):59–65. doi: 10.1016/j.tim.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 49.Hauck CR, Ohlsen K. Sticky connections: extracellular matrix protein recognition and integrin-mediated cellular invasion by Staphylococcus aureus. Curr Opin Microbiol. 2006;9(1):5–11. doi: 10.1016/j.mib.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 50.Kerdudou S, Laschke MW, Sinha B, Preissner KT, Menger MD, Herrmann M. Fibronectin binding proteins contribute to the adherence of Staphylococcus aureus to intact endothelium in vivo. Thromb Haemost. 2006;96(2):183–189. [PubMed] [Google Scholar]
- 51.Nilsson IM, Patti JM, Bremell T, Hook M, Tarkowski A. Vaccination with a recombinant fragment of collagen adhesin provides protection against Staphylococcus aureus-mediated septic death. J Clin Investig. 1998;101(12):2640–2649. doi: 10.1172/JCI1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Patti JM, Bremell T, Krajewska-Pietrasik D, Abdelnour A, Tarkowski A, Ryden C, Hook M. The Staphylococcus aureus collagen adhesin is a virulence determinant in experimental septic arthritis. Infect Immun. 1994;62(1):152–161. doi: 10.1128/iai.62.1.152-161.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mackey-Lawrence NM, Potter DE, Cerca N, Jefferson KK. Staphylococcus aureus immunodominant surface antigen B is a cell-surface associated nucleic acid binding protein. BMC Microbiol. 2009;9:61. doi: 10.1186/1471-2180-9-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garzoni C, Francois P, Huyghe A, Couzinet S, Tapparel C, Charbonnier Y, Renzoni A, Lucchini S, Lew DP, Vaudaux P, et al. A global view of Staphylococcus aureus whole genome expression upon internalization in human epithelial cells. BMC Genom. 2007;8:171. doi: 10.1186/1471-2164-8-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fuchs S, Pane-Farre J, Kohler C, Hecker M, Engelmann S. Anaerobic gene expression in Staphylococcus aureus. J Bacteriol. 2007;189(11):4275–4289. doi: 10.1128/JB.00081-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Biswas R, Voggu L, Simon UK, Hentschel P, Thumm G, Gotz F. Activity of the major staphylococcal autolysin Atl. FEMS Microbiol Lett. 2006;259(2):260–268. doi: 10.1111/j.1574-6968.2006.00281.x. [DOI] [PubMed] [Google Scholar]
- 57.Yan J, Liu Y, Gao Y, Dong J, Mu C, Li D, Yang G. RNAIII suppresses the expression of LtaS via acting as an antisense RNA in Staphylococcus aureus. J Basic Microbiol. 2015;55(2):255–261. doi: 10.1002/jobm.201400313. [DOI] [PubMed] [Google Scholar]
- 58.Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics. 2001;17(6):509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 59.Jones MB, Montgomery CP, Boyle-Vavra S, Shatzkes K, Maybank R, Frank BC, Peterson SN, Daum RS. Genomic and transcriptomic differences in community acquired methicillin resistant Staphylococcus aureus USA300 and USA400 strains. BMC Genom. 2014;15:1145. doi: 10.1186/1471-2164-15-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Manna AC, Cheung AL. Expression of SarX, a negative regulator of agr and exoprotein synthesis, is activated by MgrA in Staphylococcus aureus. J Bacteriol. 2006;188(12):4288–4299. doi: 10.1128/JB.00297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Senn MM, Giachino P, Homerova D, Steinhuber A, Strassner J, Kormanec J, Fluckiger U, Berger-Bachi B, Bischoff M. Molecular analysis and organization of the sigmaB operon in Staphylococcus aureus. J Bacteriol. 2005;187(23):8006–8019. doi: 10.1128/JB.187.23.8006-8019.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Schaik W, Tempelaars MH, Zwietering MH, de Vos WM, Abee T. Analysis of the role of RsbV, RsbW, and RsbY in regulating {sigma}B activity in Bacillus cereus. J Bacteriol. 2005;187(16):5846–5851. doi: 10.1128/JB.187.16.5846-5851.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chadwick SG, Prasad A, Smith WL, Mordechai E, Adelson ME, Gygax SE. Detection of epidemic USA300 community-associated methicillin-resistant Staphylococcus aureus strains by use of a single allele-specific PCR assay targeting a novel polymorphism of Staphylococcus aureus pbp3. J Clin Microbiol. 2013;51(8):2541–2550. doi: 10.1128/JCM.00417-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Horvath A, Dobay O, Kardos S, Ghidan A, Toth A, Paszti J, Ungvari E, Horvath P, Nagy K, Zissman S, et al. Varying fitness cost associated with resistance to fluoroquinolones governs clonal dynamic of methicillin-resistant Staphylococcus aureus. Eur J Clin Microbiol Infect Dis. 2012;31(8):2029–2036. doi: 10.1007/s10096-011-1536-z. [DOI] [PubMed] [Google Scholar]
- 65.Knight GM, Budd EL, Whitney L, Thornley A, Al-Ghusein H, Planche T, Lindsay JA. Shift in dominant hospital-associated methicillin-resistant Staphylococcus aureus (HA-MRSA) clones over time. J Antimicrob Chemother. 2012;67(10):2514–2522. doi: 10.1093/jac/dks245. [DOI] [PubMed] [Google Scholar]
- 66.Holden MT, Hsu LY, Kurt K, Weinert LA, Mather AE, Harris SR, Strommenger B, Layer F, Witte W, de Lencastre H, et al. A genomic portrait of the emergence, evolution, and global spread of a methicillin-resistant Staphylococcus aureus pandemic. Genome Res. 2013;23(4):653–664. doi: 10.1101/gr.147710.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fuzi M. Dissimilar fitness associated with resistance to fluoroquinolones influences clonal dynamics of various multiresistant bacteria. Front Microbiol. 2016;7:1017. doi: 10.3389/fmicb.2016.01017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dreisbach A, Hempel K, Buist G, Hecker M, Becher D, van Dijl JM. Profiling the surfacome of Staphylococcus aureus. Proteomics. 2010;10(17):3082–3096. doi: 10.1002/pmic.201000062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. The detailed information on differentially expressed 209 proteins in each isolate.
Additional file 2. Potential interactions based on 64 up-regulated genes in the HA-SA ST239 group and 145 up-regulated genes in the CA-SA ST398 group using the STRING (version 10.0) database.
Data Availability Statement
The datasets and its additional file supporting the conclusions of this article are included within the article.


