Abstract
Introduction
Autophagy plays an important role in the growth and survival of hepatocellular carcinoma (HCC) cells through several target proteins or signaling pathways. Glypican-3 (GPC3) is a new reliable HCC marker, which is involved in tumor growth in HCC, primarily mediated by wnt/β-catenin signaling.
Objective
The present study aimed to identify the role of autophagy in the proliferation of HepG2 cells through GPC3/wnt/β-catenin signaling.
Results and discussion
Results demonstrated that induction of autophagy by nutrition starvation and rapamycin treatment led to the downregulation of GPC3 expression in HepG2 cells, accompanied by the decreased expression of wnt downstream target genes (β-catenin, c-myc and cyclin D1). On the other hand, inhibition of autophagy by 3-methyl adenine (3-MA) could rescue rapamycin-directed downregulation of GPC3 and wnt/β-catenin target genes and augment the proliferation of HepG2 cells. Furthermore, interference of GPC3 by siRNA suppressed wnt/β-catenin signaling and attenuated 3-MA stimulation of HepG2 cell proliferation. More interestingly, the mRNA of GPC3 remained unchanged when the protein levels of GPC3 were decreased by autophagy activation, suggesting that induction of autophagy may accelerate the degradation of GPC3.
Conclusion
These results suggest that autophagy suppresses proliferation of HepG2 cells partially by inhibition of GPC3/wnt/β-catenin signaling.
Keywords: hepatocellular carcinoma, glypican-3, autophagy, proliferation, wnt/β-catenin signaling
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common cancer and the third cause of cancer-related mortality worldwide due to high rates of recurrence and metastasis. Although several signaling pathways responsible for the pathogenesis of HCC have been put forward, the underlying molecular mechanisms still need to be elucidated. Previous studies have demonstrated that autophagy responses play an important role in hepatocellular carcinogenesis and tumor progression.1–3
Autophagy is a highly conserved process in eukaryotes, which helps in cellular degradation of biological molecules. The damaged organelles then were degraded into small molecules that are recycled for use by cells. Hence, it is recognized as an essential survival mechanism to maintain cellular homeostasis. Several studies have demonstrated that autophagy aberration occurs in diverse stress or pathological state of cells or tissues, thus contributing to various diseases, including infectious diseases, cardiovascular diseases, neurodegeneration, autoimmune diseases, and cancers.4–7 Numerous studies have confirmed that autophagy deficiency in hepatocytes may lead to HCC.8–10 Induction of autophagy by diosmetin inhibited proliferation of HCC cells and promoted its apoptosis and could be reversed by exposure to autophagy inhibitor, bafilomycin A1.11
Glypican-3 (GPC3) is a membrane-bound heparan sulfate proteoglycan, which is highly expressed in HCC tissues and recognized as a novel HCC biomarker. High expression of GPC3 was significantly associated with poor prognosis in HCC.12 Capurro et al13 demonstrated that GPC3 could promote HCC tumor growth by stimulating canonical wnt/β-catenin signaling, and silent expression of GPC3 inhibited the proliferation and induced apoptosis of HCC cells.14 According to Bao et al,8 p62 was 100% (20/20) positively expressed in GPC3-expressed HCC tissues, suggesting a significant association between autophagy and GPC3 expression. The involvement of GPC3 in autophagy-directed regulation of HCC cell proliferation is poorly understood. Hence, the present study handles the interaction of GPC3 and autophagy and the crosstalk between GPC3 and autophagy-directed regulation of HepG2 cell proliferation in detail.
Materials and methods
Chemicals and reagents
Dulbecco’s Modified Eagle’s Medium (DMEM) culture medium and fetal bovine serum (FBS) were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Lipo2000, Trizol and High Capacity cDNA Reverse transcription Kit were purchased from Thermo Fisher Scientific. Rapamycin and 3-methyl adenine (3-MA) were purchased from Sigma-Aldrich Co. (St Louis, MO, USA). FastStart Universal SYBR Green Master and protease inhibitor were purchased from Hoffman-La Roche Ltd. (Basel, Switzerland). Mouse anti-human polyclonal antibody against GPC3 was purchased from R&D Systems, Inc., (Minneapolis, MN, USA). Mouse anti-human monoclonal antibody against c-myc was purchased from Santa Cruz Biotechnology Inc., (Dallas, TX, USA). Rabbit anti-human monoclonal antibodies against LC3, Beclin-1, β-catenin and cyclin D1 were obtained from Cell Signaling Technology (Boston, MA, USA). Immobilon Western Chemiluminescent HRP substrate was purchased from EMD Millipore (Billerica, MA, USA). 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyl-2H tetrazolium bromide (MTT) cell proliferation and cytotoxicity assay kit was purchased from Beyotime (Shanghai, China).
Cell culture
HepG2 cells were purchased from American Type Culture Collection (ATCC) (Manassas, VA, USA) and cultured in DMEM culture medium containing 10% FBS and 1% penicillin–streptomycin (10,000 U/mL penicillin and 10 mg/mL streptomycin) at 37°C in a humidified atmosphere containing 5% of CO2.
MTT assays
The viability of HepG2 cells was measured using MTT assay. Briefly, HepG2 cells were seeded into 96-well plates at a density of 2×104 cells/well and allowed to adhere and grow for 24 h. The medium was then replaced with 100 µL/well of fresh medium containing various stimuli for 24–72 h. The cells were then treated with 20 µL of 5 mg/mL MTT and incubated at 37°C for 4 h. Subsequently, the medium was removed and 150 µL DMSO was added to each well. Absorbance was measured at 490 nm after the crystals were fully dissolved.
Synthesis of siRNA targeting GPC3 and transfection
siRNA targeting GPC3 (sense: 5′-CCAGUGGUCAGUCAAAUUATT-3′ and antisense: UAAUUUGACUGACCACUGGTT) and negative control siRNA were synthesized by Shanghai Sangon Biotech (Shanghai, China). HepG2 cells were seeded into six-well plates and cultured overnight, and then transfected with siRNA at a concentration of 20 nmol/L using Lipofectamine 2000 according to the manufacturer’s protocol.
Real-time polymerase chain reaction (RT-PCR)
Cells in six-well plates were washed twice with ice-cold PBS, and total RNA was extracted using Trizol reagent according to the manufacturer’s instructions. The purity and concentration of RNA were measured using a UV spectrophotometer (Eppendorf, Hamburg, Germany). Approximately 1 µg of total RNA from each sample was used to synthesize cDNA using the High Capacity cDNA Reverse transcription Kit according to the manufacturer’s specification. Then, RT-PCR was performed by using FastStart Universal SYBR Green Master (Hoffman-La Roche). Primers used for PCR were as follows: GAPDH-F: 5′-CTGGGCTACACTGAGCACC-3′; GAPDH-R: 5′-AAGTGGTCGTTGAGGGCAATG-3′ and GPC3-F: 5′-ATTGGCAAGTTATGTGCCCAT-3′; GPC3-R: 5′-TTCGGCTGGATAAGGTTTCTTC-3′. Amplification conditions included were denaturation for 10 min at 95°C, 30 s at 95°C and 60 s at 60°C for 40 cycles. The products sizes of GAPDH and GPC3 were 130 bp and 101 bp, respectively.
Western blotting
The treated cells were washed twice with ice-cold PBS and harvested in RIPA lysis buffer containing protease inhibitor and then lysed for 30 min on ice. The lysates were centrifuged at 12,000 rpm for 15 min at 4°C, and then the supernatants were collected. Protein concentration was determined by bicinchoninic acid (BCA) analysis. Equal protein extracts were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto nitrocellulose membrane. The membranes were incubated with 5% non-fat milk for 1 h and then incubated with primary antibodies overnight at 4°C. All the antibodies were diluted at a ratio of 1:1,000 with 3% non-fat milk. After washing with Tris-buffered saline with Tween-20, membranes were incubated with HRP-conjugated secondary antibodies at room temperature for 2 h. Immunoreactive proteins were visualized using an enhanced chemiluminescent kit according to the manufacturer’s protocol.
Statistical analysis
Data were presented as mean ± standard error. Experiments were repeated at least two times. Student’s t-test was used for data analysis by using SPSS 15.0 software. P<0.05 was considered to be statistically significant.
Results
Expression of GPC3 is downregulated by autophagy
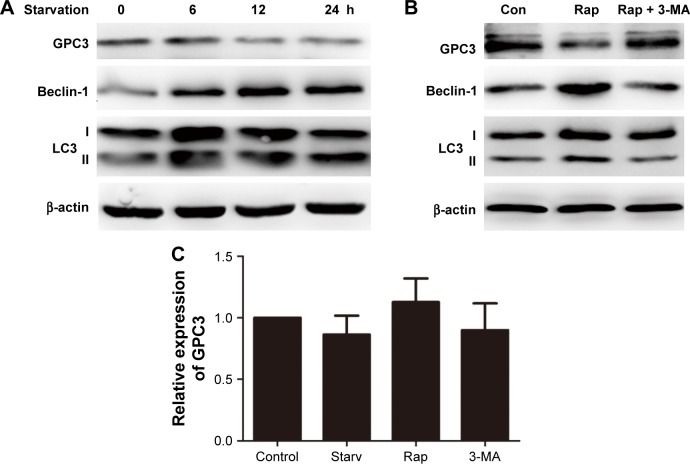
To explore the effects of autophagy on GPC3 expression in HepG2 cells, cells were cultured in serum-free medium for 6–24 h. The protein levels of GPC3 and autophagy biomarkers, LC3 and Beclin-1, were detected by Western blotting. Starvation of cells demonstrated increased autophagosomes with formation of more LC3-II from LC3-I, which acts as the critical process of autophagy activation, and the expression of Beclin-1 was significantly upregulated. Results suggest an increased expression of both LC3 and Beclin-1 under the stimulation of serum-free starvation, whereas GPC3 protein levels were decreased time-dependently (Figures 1A and S1A). To further verify the effects of autophagy on GPC3 expression in HepG2 cells, rapamycin, which is the most commonly used autophagy inducer, was applied. Similar results of starvation were observed after treatment with 100 nM rapamycin for 12 h, which was characterized by elevated LC3 and Beclin-1 and decreased GPC3 expression, whereas inhibition of autophagy by 3-MA could rescue the rapamycin-directed regulation of GPC3, LC3, and Beclin-1 (Figures 1B and S1B). To figure out whether GPC3 expression was transcriptionally inhibited by autophagy, RT-PCR was performed. Results demonstrated no significant changes in the mRNA levels of GPC3 (p>0.05) upon stimulation of starvation, rapamycin or 3-MA (Figure 1C), suggesting that autophagy reduces GPC3 protein levels probably by promoting its degradation rather than transcriptional inhibition.
Figure 1.
Autophagy reduced GPC3 protein level.
Notes: (A) Expression of GPC3 in HepG2 cells was decreased in serum Starv. (B) Expression of GPC3 induced by 100 nM rapamycin (Rap) and 5 mM 3-MA for 12 h. (C) No significant difference was detected in mRNA level of GPC3 induced by Starv, 100 nM Rap and 5 mM 3-MA for 6 h (p>0.05).
Abbreviations: Con, control; GPC3, glypican-3; Rap, rapamycin; Starv, starvation.
Autophagy negatively modulated wnt/β-catenin signaling by decreased expression of GPC3
Previous studies have shown that wnt signaling could be negatively modulated by autophagy through promoting dishevelled degradation in HEK-293T cells, MCF-1 and Hela cells.15,16 In line with those studies, Western blotting results showed that starvation or rapamycin treatment decreased the expression of several target genes (β-catenin, c-myc and cyclin D1) of wnt signaling (Figures 2A and S2A), while inhibition of autophagy by 3-MA increased the expression of these target genes. In addition, HepG2 cells transfected with siRNA-targeted GPC3 also downregulated the expres-sion of β-catenin, c-myc and cyclin D1, whereas RNAi-directed protein expression inhibition was blocked by 3-MA (Figures 2B and S2B).
Figure 2.
Autophagy suppressed wnt/β-catenin signaling.
Notes: (A) HepG2 cells were treated with serum Starv, 100 nM Rap for 12 h and 5 mM 3-MA was pretreated 1 h before adding Rap. (B) HepG2 cells transfected with 20 nM siGPC3 were incubated in the presence or absence of 3-MA.
Abbreviations: Con, control; GPC3, glypican-3; Rap, rapamycin; Starv, starvation.
Autophagy suppressed HepG2 cells’ proliferation by targeting GPC3
Previous studies have indicated that induction of autophagy can suppress the proliferation of various cells.17,18 We investigated the role of autophagy in the proliferation of HepG2 cells by MTT assay. Results revealed that starvation and rapamycin treatment of HepG2 cells showed lower cell viability compared with the untreated group (Figure 3A and Table S1). Restrained proliferation of HepG2 cells was observed in the GPC3 interfering group, which was in accordance with the previous studies,14,19 whereas inactivation of autophagy by 3-MA attenuated the proliferation repression by knockdown of GPC3 (Figure 3B and Table S2).
Figure 3.
Autophagy inhibited HepG2 cell proliferation.
Notes: (A) MTT assay was performed every 24 h, and the cells were treated with serum starvation, 100 nM rapamycin. (B) HepG2 cells transfected with 20 nM siGPC3 were incubated in the presence or absence of 3-MA.
Abbreviation: Con, control.
Discussion
Autophagy is an important mechanism that helps in maintaining energy balance by degradation of unnecessary or dysfunctional cellular organelles and proteins in all living cells. This process remains low under normal conditions, while induced by a variety of stimuli, such as infection, stress, cytokine stimulation, energy depletion, damaged organelles and accumulation of misfolded protein.20 During the last two decades, rigorous efforts have been put in force to illustrate the role of autophagy in carcinogenesis, including the development of HCC. Emerging evidence has shown a dual effect of autophagy on hepatocarginogenesis.1,2,7,10,21 On one hand, autophagy plays a protective role in the initiation of carcinogenesis by clearing abnormal proteins or mutated DNA. On the other hand, small molecular products, such as ATP and amino acids, were degraded by autophagy and contribute to the cell survival in the hypoxic–ischemia environment.
A number of studies have reported the protective role of autophagy in hepatocarcinogenesis and development. It has been revealed that p62 expression was found in HCC tissues but not in the surrounding non-tumorous hepatocytes.8 Another study has shown that reduced Beclin-1 expression was associated with the development and progression of HCC,22 and an increased susceptibility to spontaneous malignancies and accelerated HBV-related HCC was observed in Beclin-1 knockout mice.23 Deletion of either ATG5 or ATG7 gene in mice resulted in the development of hepatomegaly and hepatocellular adenoma at the age range of 6–9 months.10 Rapamycin is one of the best known autophagy inducers, which reported an anti-tumor effect in a Phase II study conducted in 25 patients with advanced HCC.24
GPC3 is a member of the glypican family that is highly expressed in HCC tissues and cell lines. Dysfunctional mutation of GPC3 was first reported by Pilia et al,25 which may lead to a rare overgrowth disease called “Simpson–Golabi–Behmel Syndrome” (SGBS). This disease manifests as an overgrowth during antenatal and postnatal period of the fetus, along with extensive abnormities of bones and viscera. Similar characteristics were observed in GPC3 knockout mice.26 A significant positive association between mRNA of HCC-related gene and GPC3 was observed in patients with liver cirrhosis,27 indicating that GPC3 plays an important role in regulation of cell growth and carcinogenesis.
Several growth signaling molecules have been reported to be involved in the tumor growth mediated by GPC3.28 Among the signaling cascades, much more attention has been paid on wnt/β-catenin signaling as it exerts a crucial role in liver development, regeneration, liver zone, as well as hepatoma carcinogenesis and progression.29 Capurro et al13 in their study demonstrated that GPC3 could stimulate the growth of HCC by increasing autocrine/paracrine canonical wnt signaling. Activated wnt signaling induces accumulation of β-catenin in cytosol which is then transported into nucleus, thus initiating the transcription of target genes, such as cyclin D1, which mediates the proliferation of HCC cells. Silent expression of GPC3 with shRNA inhibits the proliferation of human hepatoma cells.30
It has been demonstrated that autophagy in HEK-293T cells regulates wnt/β-catenin signaling.16 Autophagy induced by rapamycin or nutrient deprivation attenuated the expression of wnt reporter and target genes, while knockdown of LC3 or Beclin-1 potentiated wnt signaling. Further studies have shown that autophagy induced the ubiquitylation of Dishevelled 2 (Dvl2), which provides a signature for recognition by p62, and p62 association in turn promotes Dvl aggregation. In the present study, we have identified that autophagy negatively regulated wnt/β-catenin signaling in HepG2 cells. In addition, silent expression of GPC3 has similar effect on wnt signaling.
Conclusion
The present study has demonstrated that GPC3 was regulated by autophagy, and activation of autophagy inhibited proliferation of HepG2 cells by reduction of GPC3 protein. Abnormal level of GPC3 protein could further regulate wnt/β-catenin signaling. This study provided a novel mechanism of autophagy in tumor cell growth, and targeting GPC3 may be a potential treatment for HCC.
Highlights
To our knowledge, this is the first study revealing that induction of autophagy could reduce GPC3 protein level.
This study has first demonstrated that autophagy suppresses proliferation of HepG2 cells partially by inhibition of GPC3/wnt/β-catenin signaling.
This present study has provided a novel mechanism of hepatocarcinogenesis caused by autophagy deficiency.
Supplementary materials
(A) Autophagy induced by starvation reduced GPC3 protein level. Expression of GPC3 in HepG2 cells was decreased in serum Starv, and expression of Beclin-1 and LC3 was significantly increased. *p<0.05. (B) Autophagy reduced GPC3 protein level. Expression of GPC3 in HepG2 cells was significantly decreased under the stimulation of 100 nM Rap, while 3-MA rescued the reduction induced by Rap. *p<0.05 vs control. #p<0.05 vs Rap.
Abbreviations: Con, control; GPC3, glypican-3; Rap, rapamycin; Starv, starvation.
(A) Autophagy suppressed wnt/β-catenin signaling. Expression of β-catenin, c-myc and cyclin D1 in HepG2 cells was significantly decreased under the stimulation of serum Starv and 100 nM Rap, whereas 3-MA attenuated the inhibition of GPC3 expression induced by autophagy. *p<0.05 vs control. #p<0.05 vs Rap. (B) Autophagy suppressed wnt/β-catenin signaling. Expression of GPC3, β-catenin, c-myc and cyclin D1 was significantly decreased in HepG2 cells transfected with siGPC3, whereas RNAi-directed protein expression inhibition was blocked by 3-MA. *p<0.05 vs control. #p<0.05 vs siGPC3.
Abbreviations: Con, control; GPC3, glypican-3; Rap, rapamycin; Starv, starvation.
Table S1.
OD value of the cells induced by serum-free starvation or rapamycin
| Time | Control
|
Starvation
|
Rapamycin
|
||
|---|---|---|---|---|---|
| Mean ± SD | Mean ± SD | p-value | Mean ± SD | p-value | |
| 24 h | 0.939±0.069 | 0.843±0.068 | 0.122 | 0.740±0.115 | 0.026 |
| 48 h | 1.734±0.071 | 1.536±0.083 | 0.015 | 1.178±0.081 | 0.005 |
| 72 h | 2.222±0.105 | 1.905±0.078 | 0.001 | 1.465±0.126 | 0.000 |
Table S2.
OD value of the cells induced by siRNA transfection and 3-MA
| Time | Control
|
siGPC3
|
siGPC3 + 3-MA
|
||
|---|---|---|---|---|---|
| Mean ± SD | Mean ± SD | p-value | Mean ± SD | p-value | |
| 24 h | 0.939±0.078 | 0.755±0.065 | 0.032 | 0.819±0.072 | 0.076 |
| 48 h | 1.692±0.049 | 1.143±0.059 | 0.011 | 1.333±0.081 | 0.004 |
| 72 h | 2.034±0.099 | 1.432±0.118 | 0.000 | 1.692±0.145 | 0.000 |
Abbreviation: GPC3, glypican-3.
Acknowledgments
This study was supported by National Science Foundation of China (81570417) (Dongfang Wu) and in part by the Research and Development Project of the Science and Technology Plan of Hubei Province (2011BCB030), the Foundation for Innovative Research Team of Hubei University of Medicine (2014CXG05), and the Key Program for Precision Medicine of Taihe Hospital (2016JZ05).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Lee YJ, Jang BK. The role of autophagy in hepatocellular carcinoma. Int J Mol Sci. 2015;16(11):26629–26643. doi: 10.3390/ijms161125984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avalos Y, Canales J, Bravo-Sagua R, Criollo A, Lavandero S, Quest A. Tumor suppression and promotion by autophagy. Biomed Res Int. 2014;2014:603980. doi: 10.1155/2014/603980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen KD, Wang CC, Tsai MC, et al. Interconnections between autophagy and the coagulation cascade in hepatocellular carcinoma. Cell Death Dis. 2014;5:e1244. doi: 10.1038/cddis.2014.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agostinis P, Milito de A, Esclatine A, Simon HU. Autophagy researchers. Autophagy. 2014;10(9):1483–1486. doi: 10.4161/auto.32226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Settembre C, Ballabio A. Cell metabolism: autophagy transcribed. Nature. 2014;516(7529):40–41. doi: 10.1038/nature13939. [DOI] [PubMed] [Google Scholar]
- 6.Amaravadi RK. Cancer. Autophagy in tumor immunity. Science. 2011;334(6062):1501–1502. doi: 10.1126/science.1216428. [DOI] [PubMed] [Google Scholar]
- 7.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bao L, Chandra PK, Moroz K, et al. Impaired autophagy response in human hepatocellular carcinoma. Exp Mol Pathol. 2014;96(2):149–154. doi: 10.1016/j.yexmp.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takamura A, Komatsu M, Hara T, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25(8):795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dash S, Chava S, Chandra PK, Aydin Y, Balart LA, Wu T. Autophagy in hepatocellular carcinomas: from pathophysiology to therapeutic response. Hepat Med. 2016;8:9–20. doi: 10.2147/HMER.S63700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Ren H, Liu B, Zhang Q, Li M, Zhu R. Diosmetin inhibits cell proliferation and induces apoptosis by regulating autophagy via the mammalian target of rapamycin pathway in hepatocellular carcinoma HepG2 cells. Oncol Lett. 2016;12(6):4385–4392. doi: 10.3892/ol.2016.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L, Pan L, Yao M, Cai Y, Dong Z, Yao D. Expression of oncofetal antigen glypican-3 associates significantly with poor prognosis in HBV-related hepatocellular carcinoma. Oncotarget. 2016;7(27):42150–42158. doi: 10.18632/oncotarget.9892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Capurro MI, Xiang YY, Lobe C, Filmus J. Glypican-3 promotes the growth of hepatocellular carcinoma by stimulating canonical Wnt signaling. Cancer Res. 2005;65(14):6245–6254. doi: 10.1158/0008-5472.CAN-04-4244. [DOI] [PubMed] [Google Scholar]
- 14.Miao HL, Pan ZJ, Lei CJ, et al. Knockdown of GPC3 inhibits the proliferation of Huh7 hepatocellular carcinoma cells through down-regulation of YAP. J Cell Biochem. 2013;114(3):625–631. doi: 10.1002/jcb.24404. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Wang F, Han L, et al. GABARAPL1 negatively regulates Wnt/beta-catenin signaling by mediating Dvl2 degradation through the autophagy pathway. Cell Physiol Biochem. 2011;27(5):503–512. doi: 10.1159/000329952. [DOI] [PubMed] [Google Scholar]
- 16.Gao C, Cao W, Bao L, et al. Autophagy negatively regulates Wnt signalling by promoting dishevelled degradation. Nat Cell Biol. 2010;12(8):781–790. doi: 10.1038/ncb2082. [DOI] [PubMed] [Google Scholar]
- 17.Ahn JS, Ann EJ, Kim MY, et al. Autophagy negatively regulates tumor cell proliferation through phosphorylation dependent degradation of the Notch1 intracellular domain. Oncotarget. 2016;7(48):79047–79063. doi: 10.18632/oncotarget.12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li S, Chen JW, Xie X, et al. Autophagy inhibitor regulates apoptosis and proliferation of synovial fibroblasts through the inhibition of PI3K/AKT pathway in collagen-induced arthritis rat model. Am J Transl Res. 2017;9(5):2065–2076. [PMC free article] [PubMed] [Google Scholar]
- 19.Ruan J, Liu F, Chen X, et al. Inhibition of glypican-3 expression via RNA interference influences the growth and invasive ability of the MHCC97-H human hepatocellular carcinoma cell line. Int J Mol Med. 2011;28(4):497–503. doi: 10.3892/ijmm.2011.704. [DOI] [PubMed] [Google Scholar]
- 20.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duffy A, Le J, Sausville E, Emadi A. Autophagy modulation: a target for cancer treatment development. Cancer Chemother Pharmacol. 2015;75(3):439–447. doi: 10.1007/s00280-014-2637-z. [DOI] [PubMed] [Google Scholar]
- 22.Osman NA, Abd El-Rehim DM, Kamal IM. Defective Beclin-1 and elevated hypoxia-inducible factor (HIF)-1alpha expression are closely linked to tumorigenesis, differentiation, and progression of hepatocellular carcinoma. Tumour Biol. 2015;36(6):4293–4299. doi: 10.1007/s13277-015-3068-0. [DOI] [PubMed] [Google Scholar]
- 23.Qu X, Yu J, Bhagat G, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112(12):1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Decaens T, Luciani A, Itti E, et al. Phase II study of sirolimus in treatment-naive patients with advanced hepatocellular carcinoma. Dig Liver Dis. 2012;44(7):610–616. doi: 10.1016/j.dld.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 25.Pilia G, Hughes-Benzie RM, MacKenzie A, et al. Mutations in GPC3, a glypican gene, cause the Simpson-Golabi-Behmel overgrowth syndrome. Nat Genet. 1996;12(3):241–247. doi: 10.1038/ng0396-241. [DOI] [PubMed] [Google Scholar]
- 26.Cano-Gauci DF, Song HH, Yang H, et al. Glypican-3-deficient mice exhibit developmental overgrowth and some of the abnormalities typical of Simpson-Golabi-Behmel syndrome. J Cell Biol. 1999;146(1):255–264. doi: 10.1083/jcb.146.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X, Wang SK, Zhang K, et al. Expression of glypican 3 enriches hepatocellular carcinoma development-related genes and associates with carcinogenesis in cirrhotic livers. Carcinogenesis. 2015;36(2):232–242. doi: 10.1093/carcin/bgu245. [DOI] [PubMed] [Google Scholar]
- 28.Akutsu N, Yamamoto H, Sasaki S, et al. Association of glypican-3 expression with growth signaling molecules in hepatocellular carcinoma. World J Gastroenterol. 2010;16(28):3521–3528. doi: 10.3748/wjg.v16.i28.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wands JR, Kim M. WNT/beta-catenin signaling and hepatocellular carcinoma. Hepatology. 2014;60(2):452–454. doi: 10.1002/hep.27081. [DOI] [PubMed] [Google Scholar]
- 30.Liu S, Li Y, Chen W, et al. Silencing glypican-3 expression induces apoptosis in human hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2012;419(4):656–661. doi: 10.1016/j.bbrc.2012.02.069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Autophagy induced by starvation reduced GPC3 protein level. Expression of GPC3 in HepG2 cells was decreased in serum Starv, and expression of Beclin-1 and LC3 was significantly increased. *p<0.05. (B) Autophagy reduced GPC3 protein level. Expression of GPC3 in HepG2 cells was significantly decreased under the stimulation of 100 nM Rap, while 3-MA rescued the reduction induced by Rap. *p<0.05 vs control. #p<0.05 vs Rap.
Abbreviations: Con, control; GPC3, glypican-3; Rap, rapamycin; Starv, starvation.
(A) Autophagy suppressed wnt/β-catenin signaling. Expression of β-catenin, c-myc and cyclin D1 in HepG2 cells was significantly decreased under the stimulation of serum Starv and 100 nM Rap, whereas 3-MA attenuated the inhibition of GPC3 expression induced by autophagy. *p<0.05 vs control. #p<0.05 vs Rap. (B) Autophagy suppressed wnt/β-catenin signaling. Expression of GPC3, β-catenin, c-myc and cyclin D1 was significantly decreased in HepG2 cells transfected with siGPC3, whereas RNAi-directed protein expression inhibition was blocked by 3-MA. *p<0.05 vs control. #p<0.05 vs siGPC3.
Abbreviations: Con, control; GPC3, glypican-3; Rap, rapamycin; Starv, starvation.
Table S1.
OD value of the cells induced by serum-free starvation or rapamycin
| Time | Control
|
Starvation
|
Rapamycin
|
||
|---|---|---|---|---|---|
| Mean ± SD | Mean ± SD | p-value | Mean ± SD | p-value | |
| 24 h | 0.939±0.069 | 0.843±0.068 | 0.122 | 0.740±0.115 | 0.026 |
| 48 h | 1.734±0.071 | 1.536±0.083 | 0.015 | 1.178±0.081 | 0.005 |
| 72 h | 2.222±0.105 | 1.905±0.078 | 0.001 | 1.465±0.126 | 0.000 |
Table S2.
OD value of the cells induced by siRNA transfection and 3-MA
| Time | Control
|
siGPC3
|
siGPC3 + 3-MA
|
||
|---|---|---|---|---|---|
| Mean ± SD | Mean ± SD | p-value | Mean ± SD | p-value | |
| 24 h | 0.939±0.078 | 0.755±0.065 | 0.032 | 0.819±0.072 | 0.076 |
| 48 h | 1.692±0.049 | 1.143±0.059 | 0.011 | 1.333±0.081 | 0.004 |
| 72 h | 2.034±0.099 | 1.432±0.118 | 0.000 | 1.692±0.145 | 0.000 |
Abbreviation: GPC3, glypican-3.