Abstract
Background
MicroRNAs (miRNAs) are a family of short noncoding RNAs that play important roles in posttranscriptional gene regulation. miRNAs inhibit target gene expression by blocking protein translation or by inducing mRNA degradation and therefore have the potential to modulate physiological and pathological processes.
Summary
In the kidney, miRNAs play a role in the organogenesis and in the pathogenesis of several diseases, including renal carcinoma, diabetic nephropathy, cystogenesis, and glomerulopathies. Indeed, podocytes, but also the parietal cells of the Bowman capsule are severely affected by miRNA deregulation. In addition, several miRNAs have been found involved in the development of renal fibrosis. These experimental lines of evidence found a counterpart also in patients affected by diabetic and Ig-A nephropathies, opening the possibility of their use as biomarkers. Finally, the possibility to direct target-specific miRNA to prevent the development of renal fibrosis is encouraging potential novel therapies based on miRNA mimicking or antagonism. This review reports the main studies that investigate the role of miRNAs in the kidneys, in particular highlighting the experimental models used, their potential role as biomarkers and, finally, the most recent data on the miRNA-based therapy.
Key Messages
miRNAs are crucial regulators of cell function. They are easy to detect and represent potentially good targets for novel therapies.
Keywords: miRNAs, Diabetic nephropathy, Renal cancer, Renal cystogenesis, Dicer, Drosha, Dgcr8
Introduction
MicroRNAs (miRNAs) are a family of short noncoding RNAs. They are endogenous gene regulators at posttranscriptional level, targeting complementary mRNA sequence and so promoting their degradation and/or interfering with their translation.
miRNAs biogenesis starts in the nucleus through the transcription catalyzed by polymerase II and in part by polymerase III. Pri-miRNAs are then cleaved into pre-miRNAs (about 70 nucleotides) by a multiprotein complex, called microprocessor, mainly made by RNAase III Drosha and the protein Dcrg8. Double-stranded pre-miRNAs are exported to the cytoplasm by Exportin 5 and then processed by Dicer into mature miRNA/miRNA duplexes (about 18–24 nucleotides) [1]. Finally, RNA-induced silencing complex (RISC), a multiprotein complex, incorporates only one strand of the miRNA, while the other is degraded. The RISC complex drives the base-paired match of each miRNA with its specific mRNA. From this interaction, mRNA degradation and/or block of translation occurs.
miRNAs are highly conserved among mammalians. They regulate a considerable amount of genes and are involved in several critical biological processes, including cellular proliferation, apoptosis, and differentiation. Indeed, miRNA deregulation may result in impaired cellular function and has been associated with the development of several diseases [2].
Dicer is crucial for miRNA maturation. In humans, constitutional haploinsufficiency of Dicer leads to the development of familial pleuropulmonary blastoma family tumor and dysplasia syndrome (PPB-FTDS). This association is so specific that PPB-FDS has been proposed to be renamed as Dicer syndrome [3]. PPB-FTDS patients or their family members carrying a truncating mutation develop inherited malignant tumor of pleura and lung, usually in the childhood [4]. This is associated with extrapulmonary tumors, as cystic nephroma, ovarian Sertoli-Leydig tumor, or other cystic lesions as multinodular goiter. The phenotype related to Dicer syndrome suggests a crucial role of miRNAs in regulating cell proliferation and organ development.
Indeed, several lines of evidence suggest a key role of miRNAs in organ development. This is the case for all the main target organs of the Dicer syndrome. miR-1 and miR-133 have been found fundamental for the myogenesis (miR-1) and myoblast proliferation (miR133) of both heart and muscles [5]. In addition, miR-124a and miR-9 are crucial for the development and terminal differentiation of neurons and astrocytes [6]. miRNA seems to play a role also in the development of the lung. miRNA-127 overexpression interferes with lung development [7]. Finally, in the kidney, miR-30 family is involved in the development of the pro-nephron by targeting Xlim/Lhx1, which is an important transcription factor essential for nephric duct formation and nephron differentiation [8].
The identification of miRNA in biological fluids such as blood, urine, and saliva raises the possibility that they could also be involved in cell to cell communication; namely, one cell could regulate gene expression in the neighbor cell [9]. Circulating miRNAs from nucleases are highly stable because they are encapsulated into apoptotic bodies [10], microvesicles [11], and exosomes, or complexed with the AGO proteins [12]. Valadi et al. [13] proved that miRNA and mRNA contained in the exosomes can be transferred between cells. This gene regulation mechanism seems to be used particularly by tumor cells to modulate cellular growth in the surrounding environment [14]. Exosome-induced gene regulation has also been described in tumor metastasis formation. Breast cancer cells can release exosomes containing miR-105. Once delivered in the target cells, miR-105 regulates the tight junction protein 1 (TJP1) and promotes metastasis generation [15]. However, since urinary exosomes contain some miRNAs, this kind of gene regulatory mechanism could be hypothesized also to occur between the various segments of the nephron. In addition, the presence of miRNAs in urine and in other biological fluids associated with their physiopathological specificity also opened the possibility to use them as biological markers.
In an attempt to find novel biomarkers associated with the development of diabetic nephropathy (DN), Eissa et al. [16] screened miRNA expression in the urinary exosomes from a homogenous cohort of type 2 diabetes (T2D) patients stratified according to the presence of normo-, micro-, and macroalbuminuria. In this way, miR-133b, miR-342, and miR-30a were proven to be strongly associated with the development of proteinuria in T2D patients. The predicted targets of these miRNAs were mainly involved in the pathways related to TGF-β1, MAPK, fatty acid metabolisms, and insulin signaling cascade, suggesting their primary involvement in the pathogenesis of the T2D-induced endothelial dysfunction. Indeed, the good correlation between their expression and the decline of the eGFR corroborate the hypothesis. Whether these miRNAs could serve also as prognostic factors or anticipate disease onset could not be answered.
By following a reductionist approach, the presence of miRNA-regulating genes involved in TGF-β1-dependent renal fibrosis was investigated in the urine of patients affected by Ig-A nephropathy. Compared with healthy volunteers, Ig-A-affected patients showed increased urinary level of miR-21, miR-29, and miR-93 [17]. The studied cohort of patients presented also a slight reduction in eGFR and an overt proteinuria compared to the control group, and this limited the specificity of the findings. However, this study corroborated the role of TGF-β1-mediated renal fibrosis as crucial mechanism in Ig-A nephropathy. The identification of miRNAs in many kidney-related diseases suggests they are fundamental for renal development and physiology as for the other organs [18]. To address this point, experimental models are required. So far, three different approaches have been followed to address the role of miRNAs in renal physiology:
Deregulation of miRNA genesis in selective renal cells
Identification of differentially expressed miRNAs in renal disease models by high-throughput techniques
Evaluation of the expression level of specific miRNAs predicted to be regulators of selective metabolic or pathological pathways
miRNA Deregulation Secondary to Tissue-Specific Dicer Ablation
Suppression of Dicer gene expression is the most used model to investigate the role of miRNAs. In the kidney, Dicer conditional KO models have been generated for podocytes [19], juxtaglomerular (JG) cells [20], proximal tubules [21], ureteric bud epithelium [22], and overall epithelial cells of the nephron [23] (Fig. 1).
Fig. 1.
Mechanism of miRNA deregulation secondary to cell-specific Dicer, Drosha, and Dgcr8 ablation. The main steps of miRNA biogenesis are shown on the left. The black cross indicates the site of interference promoted by Drosha, Dgcr8, or Dicer, respectively. A summary picture of the cell-specific sites used to investigate the role of miRNAs deregulation along the nephron is shown on the right.
The podocyte-specific Dicer knockout model was reported in 2008 by three research groups [19]; the deletion of Dicer was performed using the CRE-lox recombinase system, expressing the CRE under the Nphs2 promoter, a gene encoding for podocin. All the three models showed the development of progressive chronic renal failure secondary to glomerular dysfunction. Indeed, loss of podocytes due to cytoskeletal modifications and foot processes effacement with an overt proteinuria developed 2–4 weeks after birth. The defects rapidly progressed into end-stage renal diseases, and finally the mice died from severe renal failure. This experimental approach allows the identification of four miR-30 family members as fundamental for podocytes function [24].
Dicer-dependent regulation of miRNAs has been found of note also for the function of the juxta-glomerular apparatus of the nephron. This structure regulates the glomerular filtration rate according to the electrolyte composition of the lumen, mainly reducing the filtration rate when a high concentration of chloride is detected. In addition, the JG cells highly synthesize and secrete renin [20] and contribute to the regulation of the blood volume and thus systemic blood pressure [25]. Selective suppression of Dicer-floxed gene in the JG cells has been achieved by using a CRE recombinase associated with the promoter of the REN1 gene [20]. This conditional knockout (cKO) model had a loss of JG cells, followed by a consequent reduction in renal and plasma renin concentration. This determines a drop in arterial blood pressure together with severe renal development alterations such as irregular surface, low kidney weight, interstitial fibrosis, and tubular and glomerular damage. Taken together, these observations suggested that Dicer and miRNAs are essential for the maintenance of JG cells and for the proper kidney development. However, in this study, no miRNAs specific for the JG were identified [20].
In order to recapitulate the main alterations of the human Dicer syndrome, we suppressed Dicer expression in the epithelial cell of the kidney and thyroid by generating a Dicerfloxed-Pax8CRE mouse model [23]. This mouse model develops multiple cysts in the kidneys and thyroid, resembling the goiter and the renal nephroma crucial traits of the Dicer syndrome [3]. The hallmarks of this experimental model were the progressive development of a glomerulocystic phenotype, mainly due to the enlargement of the parietal cells of the Bowman's capsule. Glomerulocystic formation leads to progressive proteinuria and renal failure. We showed that higher cellular turnover of the parietal cells of Bowman's capsule precedes the development of the cysts and that the enlargement of the Bowman's space was associated with progressive disappearance of the primary cilium. As molecular determinant of the Dicer induced cystogenesis, we identified a dysregulation of the of GSK3β/β-catenin pathways [23]. Indeed, severe downregulation of β-catenin and cytosolic retention was observed in cKO mice, and this was paralleled by alteration in GSK3β expression. In a similar experimental setting, Patel et al. [26] found a similar phenotype, and by evaluating the miRNAs expression profile they pointed out that downregulation of the members of the miR-200 family was directly linked to dysregulation of the PKD1 gene.
However, our Dicerfloxed-Pax8CRE cKo mice also developed an overt polyuria. This was associated with a reduced expression of functional markers (NKCC2 and AQP2) of TAL and CD, the two major nephron segments involved in water conservation mechanism. The progressive development of polyuria could have been associated with the progression of renal failure in this model and the reduced nephron mass. However, dysregulation of GSK3β pathway is one of the determinants of the AQP2 downregulation secondary to lithium treatment [27, 28], and so it could contribute to water reabsorption impairment independently by renal failure. Since long-term lithium treatment is associated with the development of microcysts together with polyuria and GSK3β pathway alteration, it is likely that the same mechanism is induced by Dicer-dependent miRNA deregulation. It seems that miRNA integrity pathway is fundamental also for the embryonic development of the collecting duct from the ureteric bud. Early (E11.5) selective inactivation of Dicer (promoted by HoxB7-driven CRE system) severely affects the ductal branching and leads to cyst formation [22].
miRNA Deregulation Secondary to Tissue-Specific Drosha and Dgcr8 Ablation
In addition to the ultimate maturation of miRNAs, Dicer plays other miRNA-independent functions. Indeed, Dicer acts as a DNAse promoting the genomic DNA fragmentation during apoptosis and also clearing DNA-repeated elements and other small endogenous or exogenous inhibitory RNA [29]. Thus, Dicer cKO models could not reflect exclusively miRNA deletion. To rule out potential miRNA unrelated effect, other experimental models of targeting miRNAs have been developed by suppressing the expression of either Drosha or Dgcr8, which assemble the microprocessor complex crucial in the initial steps of miRNA maturation.
Drosha is an RNase III enzyme and, together with its cofactor DiGeorge syndrome critical region 8 (Dgcr8), in the nucleus, cleaves the pri-miRNA in pre-miRNA. Dgcr8 recognizes and interacts with the hairpin structure of the pri-miRNAs and recruits Drosha. Drosha then cleaves pri-miRNAs precisely at the stem-loop structure cutting the miRNA in an about 70-nucleotide fragment.
Podocyte-specific deletion of Drosha (driven by CRE recombinase linked to Nphs2 promoter) induces a severe glomerulopathy [30]. Conditional inducible deletion of Drosha in podocytes at 2 and 3 months of age resulted in a collapsing glomerulopathy, secondary to podocyte dedifferentiation. Indeed, the podocytes lost their specific markers, namely synaptopodin, WT-1, podocin, and nephrin. This was associated with, glomerular tuft collapse, wrinkling of the basement membrane and pseudo-crescent formation, which led to proteinuria and ultimately to renal failure and death at around 6 weeks of age. Several miRNAs were found downregulated in this model and recognized as pathogenic markers of collapsing glomerulopathy. These were similar to what was found in the Dicer cKO models driven by the same promoter gene.
Models of selective inactivation of Dgcr8 are also an efficient way to interfere with miRNA expression. Selective inactivation of Dgcr8 in Pax8-expressing cells leads to broad suppression of the protein in the majority of the renal epithelial cells. Such a model resembles in the phenotype the Dicerflox-Pax8CRE mice [23, 26], developing glomerulocystic abnormalities associated with progressive renal failure and polyuria [31]. In this model, a downregulation of miR-200 family members was found associated with the phenotype as in the Dicerflox-Pax8CRE mice described by Patel et al. [26]. The studies reviewed here demonstrate that interfering with the miRNA maturation at the stage of the pri- or pre-miRNA by suppressing Drosha/Dgcr8 and Dicer, respectively, leads to similar phenotypes. These data support an equal role of the three key proteins involved in miRNA maturation in deregulating mature miRNAs and limit the causative role of miRNA-independent factors potentially associated with Dicer function.
miRNA-Based Therapy: Current Approaches
The role of miRNAs in the pathophysiology of several renal diseases may open new avenues for a miRNA-based therapy. Recently, a great interest of the scientific community has been focused on the use of chemically engineered antisense oligonucleotides targeting specific miRNAs termed mimics or antagomirs when able to reproduce or inhibit specific miRNA function, respectively.
One of the challenges to the broad use of this approach is to protect the oligonucleotides by circulating RNase- and pH-dependent degradation. Chemically modified nucleobases as phosphorothioate-containing oligonucleotides, 2′-O-methyl-(2′-O-Me) or 2′-O-methoxyethyl-oligonucleotides [32], locked nucleic acid (LNA) oligonucleotides [33], peptide nucleic acids [34], and fluorine derivatives (FANA and 2′-F) [35] could confer stability to miRNAs and facilitate their systemic delivery (Fig. 2). When these chemical modifications are at both 3′ and 5′ ends, they confer more stability from nuclease degradation, and configure a so-called “gapmer.” In this compound, a central “gap,” 8–12 base of unmodified DNA or phosphorothioate DNA, allows the formation of a DNA-RNA hybrid [36] and then recruit the RNase H for the cleavage of the DNA-RNA hybrid. For these molecules, LNA seems to offer a high degree of stability against nucleases and of binding affinity [37].
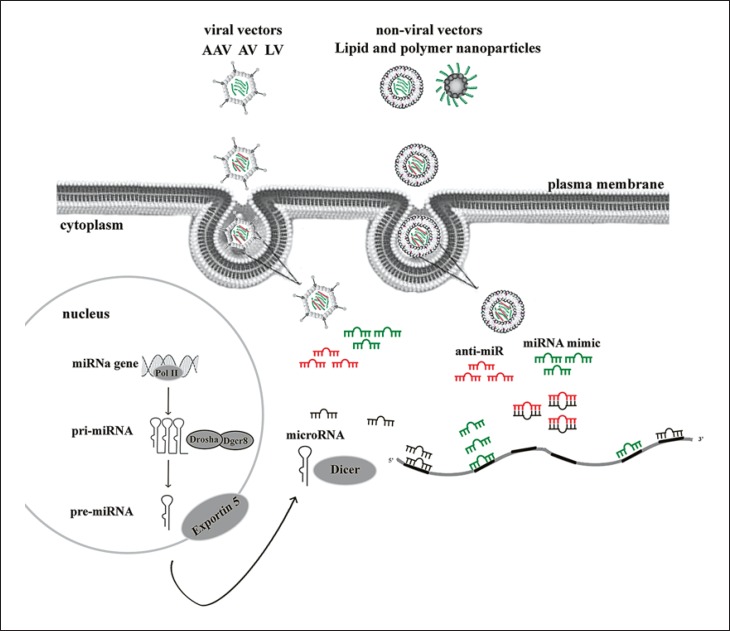
Fig. 2.
Existing strategies for miRNA delivery. miRNA mimics or antagomir developed in viral and/or nonviral vectors are internalized in the cells by endocytosis and then they release their content still stable to replace or inhibit the miRNA function. AAV, adeno-associated virus; AV, adenovirus; LV, lentivirus.
As for any drug, the crucial point in the development of a miRNA-based therapy is safety and efficacy. To guarantee this, minimizing off-target effects is mandatory. This can be a problem when injecting in vivo since the drugs have to cross several biological barriers before docking to the target cells.
However, the insertion of mimic or antagomir into the envelopes is the most used strategy. Currently, the most used carriers for miRNAs delivery in vivo can be divided into two categories, viral and nonviral [38]. The viral-mediated delivery including lentivirus, adenovirus, and adeno-associated virus (Fig. 2) has been widely used to deliver exogenous DNA and siRNA for gene therapy in the past 20 years [39]. The viral-mediated delivery approach provides a large amount and long-term persistent expression of the carried miRNA, even though the risk of random integration and off-target effects remains high [40]. Another issue is the host immune response induced by the transduced viral vectors, especially by adenovirus and adeno-associated virus [41]. Furthermore, occasional poor integration and transient expression also hamper the application of the viral delivery system [41].
To overcome the limitation of the use of viral vectors, nonviral vectors are becoming good alternatives. Chemically synthesized single-strand miRNA inhibitors or double-strand miRNA mimics are safe, cheaper, and more specific for the delivery systems and, finally, they are delivered in nonviral vehicles like lipids and nanoparticles. Lipid and nanoparticle-based delivery is the most widely and clinically advanced approach. Lipid-based delivery systems include liposomes, microemulsions, and solid lipid nanoparticles. The liposomes are carriers with an aqueous core surrounded by a phospholipid bilayer; they are biocompatible and biodegradable. The core of anionic oligonucleotides is encapsulated in cationic liposomes. After the fusion with the plasma membranes, the oligonucleotides are transferred into the cells. Cationic liposomes are highly immunogenic and can interfere with the activity of serum proteins and blood cells. Instead, nanoparticles of 50–70 nm would be a safer alternative because of their lower immunogenicity, higher biocompatibility, and a more stable profile against degradation process. These are mainly made of polyethylene glycol-coated cationic lipid bilayers, polyethyleneimine-miRNA complexes and atelocollagen-miRNA complexes (such as poly(lactic-co-glycolic acid)) [42]. Recently, nanoparticles called stable nucleic acid lipid particles [43] have been used for clinical use. The first clinically tested miRNA was miR-34 [44]. miR-34 is a well-defined miRNA tumor suppressor. It is transcriptionally induced by p53 and acts as an important effector molecule in the execution of molecular programs directed by p53 [45]. A loss of expression of miR-34 has been found in many tumor types, including lung, liver, breast, and colon carcinoma. Interestingly, the delivery of miR-34 mimics to cancer cells showed the inhibition of tumor growth and progression. The most effective delivery strategy implies the use of stable nucleic acid lipid particles [43].
Other miRNAs have been proposed as potential therapeutic modulators in cancer. Indeed, anti-miR-155 encapsulated in a poly(lactic-co-glycolic acid) nanoparticle can lead to rapid tumor regression in the brain and lungs of a miR-155 Cre-loxP tetracycline-controlled knock-in mouse model [46]; anti-miR-217 was used as tumor suppressor for the pancreatic ductal adenocarcinoma cell line [47]; finally, miR-33a-mimic and miR-145-mimic were locally and systemically delivered as tumor suppressors using polyethylenimine in a mouse model of colorectal carcinoma [48]. Other reports demonstrated the involvement of miR-122 in promoting the replication of the hepatitis C virus, resulting in an interesting therapeutic target for the treatment of this disease. It was proven that the inhibition of miR-122 using an LNA-modified oligonucleotide reduces the infection and the liver damage in the mouse model [49] and in non-human primates [50]. Currently, anti-miR-122 LNAs is used in clinical trials as a therapy for hepatitis C. Attempts of miRNA-based therapy in renal diseases were mainly to prevent the development of renal fibrosis. The most compelling evidence in this sense is represented by the use of the antagomir against miRNA-21 [51]. By weekly subcutaneous injection of the specific antagomir in a mouse model of CKD, not only the renal fibrosis decreased, but also the life span of CKD-affected mice was prolonged. This was associated with the efficacy of the antagomir in reversing both glomerular and tubular cell damage. Proper targeting in glomerular and tubular epithelial cells was assessed by mir-21 Cy-3-conjugated fluorophore injection [51]. A similar experiment was performed in a model of DN.
It is well known that one of the main features of DN is the development of renal fibrosis. Several studies highlighted that the dysregulation of the components of the miR-29 family (a, b, and c) plays an important role in the pathogenesis of renal fibrosis [52]. Chen et al. [53] studied the efficacy of miR-29b supplementation in diabetic db/db mice. They used an ultrasound-based gene delivering system to restore the miR29 level in the kidneys. With this approach, they demonstrated that miR-29b is able to block the TGF-β/Smad3 pathway, decreasing collagen matrix accumulation and reducing Sp1/NF-κB-dependent inflammation [53]. With a different miRNA delivery approach, a substantial protective role of miRNA29a in the development of diabetic glomerulopathy was proven. Indeed, supplementation of miR-29a to streptozotocin-induced diabetic mice by means of alentivirus prevented the upregulation of profibrotic factors like TGF-β1, fibronectin, and DKK1 [54]. Finally, a recent study showed that miR-29c upregulation is harmful for the renal phenotype of db/db mice, and targeting its activity by silencing its expression prevents the development of a diabetes-induced albuminuria [55]. From these results, it is clear that the miR-29 family is crucial for the glomerular damage induced in the experimental model of diabetes mellitus, and that a complex equilibrium between the relative expression levels of the single members of the miR-29 family is a cornerstone of renal fibrosis modulation.
Conclusion
Since the discovery of miRNAs, the knowledge on their function and potential usage has been exponentially increasing. The association of miRNAs with a specific state of disease makes them potential candidate biomarkers for human pathology. Even though several efforts have been made, so far, no miRNAs accomplish this role in the clinical practice. An additional ongoing and interesting application of miRNAs includes miRNA-based therapy. The interest in miRNA-based therapy currently involves mainly its application in oncology, as recently reviewed [56]. However, its potential use in rare renal disease is promising. Indeed, the pathogenesis of these diseases involves mainly a single gene defect. Posttranscriptional regulation of this deficient gene or its downstream regulators could be provided by selective appropriate miRNA mimics or antagomirs. To this aim, both the research into the role of miRNAs in specific physiological processes and the improvement of miRNA delivery means must be addressed.
Conflict of Interest Statement
The authors declare no conflict of interest.
References
- 1.Graves P, Zeng Y. Biogenesis of mammalian microRNAs: a global view. Genomics Proteomics Bioinformatics. 2012;10:239–225. doi: 10.1016/j.gpb.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chandrasekaran K, Karolina DS, Sepramaniam S, Armugam A, Wintour EM, Bertram JF, Jeyaseelan K. Role of microRNAs in kidney homeostasis and disease. Kidney Int. 2012;81:617–627. doi: 10.1038/ki.2011.448. [DOI] [PubMed] [Google Scholar]
- 3.Slade I, Bacchelli C, Davies H, Murray A, Abbaszadeh F, Hanks S, Barfoot R, Burke A, Chisholm J, Hewitt M, Jenkinson H, King D, Morland B, Pizer B, Prescott K, Saggar A, Side L, Traunecker H, Vaidya S, Ward P, Futreal PA, Vujanic G, Nicholson AG, Sebire N, Turnbull C, Priest JR, Pritchard-Jones K, Houlston R, Stiller C, Stratton MR, Douglas J, Rahman N. CER1 syndrome: clarifying the diagnosis, clinical features and management implications of a pleiotropic tumour predisposition syndrome. J Med Genet. 2011;48:273–278. doi: 10.1136/jmg.2010.083790. [DOI] [PubMed] [Google Scholar]
- 4.Dehner LP, Messinger YH, Schultz KA, Williams GM, Wikenheiser-Brokamp K, Hill DA. Pleuropulmonary blastoma: evolution of an entity as an entry into a familial tumor predisposition syndrome. Pediatr Dev Pathol. 2015;18:504–511. doi: 10.2350/15-10-1732-OA.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Izarra A, Moscoso I, Cañón S, Carreiro C, Fondevila D, Martín-Caballero J, Blanca V, Valiente I, Díez-Juan A, Bernad A. miRNA-1 and miRNA 133a are involved in early commitment of pluripotent stem cells and demonstrate antagonistic roles in the regulation of cardiac differentiation. J Tissue Eng Regen Med. 2017;11:787–799. doi: 10.1002/term.1977. [DOI] [PubMed] [Google Scholar]
- 6.Bonni A, Sun Y, Nadal-Vicens M, Bhatt A, Frank DA, Rozovsky I, Stahl N, Yancopoulos GD, Greenberg ME. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science. 1997;278:477–483. doi: 10.1126/science.278.5337.477. [DOI] [PubMed] [Google Scholar]
- 7.Bhaskaran M, Wang Y, Zhang H, Weng T, Baviskar P, Guo Y, Gou D, Liu L. MicroRNA-127 modulates fetal lung development. Physiol Genomics. 2009;37:268–278. doi: 10.1152/physiolgenomics.90268.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agrawal R, Tran U, Wessely O. The miR-30 miRNA family regulates Xenopus pronephros development and targets the transcription factor Xlim1/Lhx1. Development. 2009;136:3927–3936. doi: 10.1242/dev.037432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turchinovich A, Samatov TR, Tonevitsky AG, Burwinkel B. Circulating miRNAs: cell-cell communication function? Front Genet. 2013;4:119. doi: 10.3389/fgene.2013.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, Hristov M, Köppel T, Jahantigh MN, Lutgens E, Wang S, Olson EN, Schober A, Weber C. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal. 2009;2:ra81. doi: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- 11.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, Xiao T, Schafer J, Lee ML, Schmittgen TD, Nana-Sinkam SP, Jarjoura D, Marsh CB. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turchinovich A, Burwinkel B. Distinct AGO1 and AGO2 associated miRNA profiles in human cells and blood plasma. RNA Biol. 2012;9:1066–1075. doi: 10.4161/rna.21083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 14.Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, Yu Y, Chow A, O'Connor ST, Chin AR, Yen Y, Wang Y, Marcusson EG, Chu P, Wu J, Wu X, Li AX, Li Z, Gao H, Ren X, Boldin MP, Lin PC, Wang SE. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eissa S, Matboli M, Bekhet MM. Clinical verification of a novel urinary microRNA panel: 133b, -342 and -30 as biomarkers for diabetic nephropathy identified by bioinformatics analysis. Biomed Pharmacother. 2016;83:92–99. doi: 10.1016/j.biopha.2016.06.018. [DOI] [PubMed] [Google Scholar]
- 17.Wang G, Kwan BC, Lai FM, Chow KM, Li PK, Szeto CC. Urinary miR-21, miR-29, and miR-93: novel biomarkers of fibrosis. Am J Nephrol. 2012;36:412–418. doi: 10.1159/000343452. [DOI] [PubMed] [Google Scholar]
- 18.Lorenzen JM, Haller H, Thum T. MicroRNAs as mediators and therapeutic targets in chronic kidney disease. Rev Nephrol. 2011;7:286–294. doi: 10.1038/nrneph.2011.26. [DOI] [PubMed] [Google Scholar]
- 19.Ho J, Ng KH, Rosen S, Dostal A, Gregory RI, Kreidberg JA. Podocyte-specific loss of functional microRNAs leads to rapid glomerular and tubular injury. J Am Soc Nephrol. 2008;19:2069–2075. doi: 10.1681/ASN.2008020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sequeira-Lopez ML, Weatherford ET, Borges GR, Monteagudo MC, Pentz ES, Harfe BD, Carretero O, Sigmund CD, Gomez RA. The microRNA processing enzyme dicer maintains juxtaglomerular cells. J Am Soc Nephrol. 2010;21:460–467. doi: 10.1681/ASN.2009090964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei Q, Bhatt K, He HZ, Mi QS, Haase VH, Dong Z. Targeted deletion of Dicer from proximal tubules protects against renal ischemia-reperfusion injury. J Am Soc Nephrol. 2010;21:756–761. doi: 10.1681/ASN.2009070718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagalakshmi VK, Ren Q, Pugh MM, Valerius MT, McMahon AP, Yu J. Dicer regulates the development of nephrogenic and ureteric compartments in the mammalian kidney. Kidney Int. 2011;79:317–330. doi: 10.1038/ki.2010.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iervolino A, Trepiccione F, Petrillo F, Spagnuolo M, Scarfò M, Frezzetti D, De Vita G, De Felice M, Capasso G. Selective dicer suppression in the kidney alters GSK3β/β-catenin pathways promoting a glomerulocystic disease. PLoS One. 2015;10:e0119142. doi: 10.1371/journal.pone.0119142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi S, Yu L, Chiu C, Sun Y, Chen J, Khitrov G, Merkenschlager M, Holzman LB, Zhang W, Mundel P, Bottinger EP. Podocyte-selective deletion of dicer induces proteinuria and glomerulosclerosis. J Am Soc Nephrol. 2008;19:2159–2169. doi: 10.1681/ASN.2008030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sinning A, Radionov N, Trepiccione F, López-Cayuqueo KI, Jayat M, Baron S, Cornière N, Alexander RT, Hadchouel J, Eladari D, Hübner CA, Chambrey R. Double knockout of the Na+-driven Cl−/HCO3− exchanger and Na+/Cl− cotransporter induces hypokalemia and volume depletion. J Am Soc Nephrol. 2017;28:130–139. doi: 10.1681/ASN.2015070734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel V, Hajarnis S, Williams D, Hunter R, Huynh D, Igarashi P. MicroRNAs regulate renal tubule maturation through modulation of Pkd1. J Am Soc Nephrol. 2012;23:1941–1948. doi: 10.1681/ASN.2012030321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trepiccione F, Pisitkun T, Hoffert JD, Poulsen SB, Capasso G, Nielsen S, Knepper MA, Fenton RA, Christensen BM. Early targets of lithium in rat kidney inner medullary collecting duct include p38 and ERK1/2. Kidney Int. 2014;86:757–767. doi: 10.1038/ki.2014.107. [DOI] [PubMed] [Google Scholar]
- 28.Trepiccione F, Christensen BM. Lithium-induced nephrogenic diabetes insipidus: new clinical and experimental findings. J Nephrol. 2010;23((suppl 16)):S43–S48. [PubMed] [Google Scholar]
- 29.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhdanova O, Srivastava S, Di L, Li Z, Tchelebi L, Dworkin S, Johnstone DB, Zavadil J, Chong MM, Littman DR, Holzman LB, Barisoni L, Skolnik EY. The inducible deletion of Drosha and microRNAs in mature podocytes results in a collapsing glomerulopathy. Kidney Int. 2011;80:719–30. doi: 10.1038/ki.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartram MP, Amendola E, Benzing T, Schermer B, de Vita G, Müller RU. Mice lacking microRNAs in Pax8-expressing cells develop hypothyroidism and end-stage renal failure. BMC Mol Biol. 2016;17:11. doi: 10.1186/s12867-016-0064-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoo BH, Bochkareva E, Bochkarev A, Mou TC, Gray DM. 2′-O-methyl-modified phosphorothioate antisense oligonucleotides have reduced non-specific effects in vitro. Nucleic Acids Res. 2004;32:2008–2016. doi: 10.1093/nar/gkh516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wahlestedt C, Salmi P, Good L, Kela J, Johnsson T, Hökfelt T, Broberger C, Porreca F, Lai J, Ren K, Ossipov M, Koshkin A, Jakobsen N, Skouv J, Oerum H, Jacobsen MH, Wengel J. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc Natl Acad Sci USA. 2000;97:5633–5638. doi: 10.1073/pnas.97.10.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hyrup B, Nielsen PE. Peptide nucleic acids (PNA): synthesis, properties and potential applications. Bioorg Med Chem. 1996;4:5–23. doi: 10.1016/0968-0896(95)00171-9. [DOI] [PubMed] [Google Scholar]
- 35.Pallan PS, Greene EM, Jicman PA, Pandey RK, Manoharan M, Rozners E, Egli M. Unexpected origins of the enhanced pairing affinity of 2′-fluoro-modified RNA. Nucleic Acids Res. 2011;39:3482–3495. doi: 10.1093/nar/gkq1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watts JK, Corey DR. Silencing disease genes in the laboratory and the clinic. J Pathol. 2012;226:365–379. doi: 10.1002/path.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lennox KA, Behlke MA. Chemical modification and design of anti-miRNA oligonucleotides. Gene Ther. 2011;18:1111–1120. doi: 10.1038/gt.2011.100. [DOI] [PubMed] [Google Scholar]
- 38.Wang J, Lu Z, Wientjes MG, Au JL. Delivery of siRNA therapeutics: barriers and carriers. AAPS J. 2010;12:492–503. doi: 10.1208/s12248-010-9210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thorne B, Takeya R, Vitelli F, Swanson X. Gene therapy. Adv Biochem Eng Biotechnol. 2017 doi: 10.1007/10_2016_53. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 40.Chen Y, Zhao H, Tan Z, Zhang C, Fu X. Bottleneck limitations for microRNA-based therapeutics from bench to the bedside. Pharmazie. 2015;70:147–154. [PubMed] [Google Scholar]
- 41.Warnock JN, Daigre C, Al-Rubeai M. Introduction to viral vectors. Methods Mol Biol. 2011;737:1–25. doi: 10.1007/978-1-61779-095-9_1. [DOI] [PubMed] [Google Scholar]
- 42.Hong S, Kim KY, Wook HJ, Park SY, Lee KD, Lee DY, Lee H. Attenuation of the in vivo toxicity of biomaterials by polydopamine surface modification. Nanomedicine (Lond) 2011;6:793–801. doi: 10.2217/nnm.11.76. [DOI] [PubMed] [Google Scholar]
- 43.Misso G, Di Martino MT, De Rosa G, Farooqi AA, Lombardi A, Campani V, Zarone MR, Gullà A, Tagliaferri P, Tassone P, Caraglia M. Mir-34: a new weapon against cancer? Mol Ther Nucleic Acids. 2014;3:e194. doi: 10.1038/mtna.2014.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 45.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, MacDougald OA, Cho KR, Fearon ER. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 46.Babar IA, Cheng CJ, Booth CJ, Liang X, Weidhaas JB, Saltzman WM, Slack FJ. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc Natl Acad Sci USA. 2012;109:E1695–E1704. doi: 10.1073/pnas.1201516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao WG, Yu SN, Lu ZH, Ma YH, Gu YM, Chen J. The miR-217 microRNA functions as a potential tumor suppressor in pancreatic ductal adenocarcinoma by targeting KRAS. Carcinogenesis. 2010;31:1726–1733. doi: 10.1093/carcin/bgq160. [DOI] [PubMed] [Google Scholar]
- 48.Ibrahim AF, Weirauch U, Thomas M, Grünweller A, Hartmann RK, Aigner A. MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma. Cancer Res. 2011;71:5214–5224. doi: 10.1158/0008-5472.CAN-10-4645. [DOI] [PubMed] [Google Scholar]
- 49.Elmén J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjärn M, Hansen JB, Hansen HF, Straarup EM, McCullagh K, Kearney P, Kauppinen S. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjärn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 51.Gomez IG, Nakagawa N, Duffield JS. MicroRNAs as novel therapeutic targets to treat kidney injury and fibrosis. Am J Physiol Renal Physiol. 2016;310:F931–F944. doi: 10.1152/ajprenal.00523.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang B, Komers R, Carew R, Winbanks CE, Xu B, Herman-Edelstein M, Koh P, Thomas M, Jandeleit-Dahm K, Gregorevic P, Cooper ME, Kantharidis P. Suppression of microRNA-29 expression by TGF-β1 promotes collagen expression and renal fibrosis. J Am Soc Nephrol. 2012;23:252–265. doi: 10.1681/ASN.2011010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen HY, Zhong X, Huang XR, Meng XM, You Y, Chung AC, Lan HY. MicroRNA-29b inhibits diabetic nephropathy in db/db mice. Mol Ther. 2014;22:842–853. doi: 10.1038/mt.2013.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hsu YC, Chang PJ, Ho C, Huang YT, Shih YH, Wang CJ, Lin CL. Protective effects of miR-29a on diabetic glomerular dysfunction by modulation of DKK1/Wnt/β-catenin signaling. Sci Rep. 2016;6:30575. doi: 10.1038/srep30575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Long J, Wang Y, Wang W, Chang BH, Danesh FR. MicroRNA-29c is a signature microRNA under high glucose conditions that targets Sprouty homolog 1, and its in vivo knockdown prevents progression of diabetic nephropathy. J Biol Chem. 2011;286:11837–11848. doi: 10.1074/jbc.M110.194969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bader AG. miR-34 – a microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:120. doi: 10.3389/fgene.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]