Abstract
Local adaptation frequently occurs across populations as a result of migration-selection balance between divergent selective pressures and gene flow associated with life in heterogeneous landscapes. Studying the effects of selection and gene flow on the adaptation process can be achieved in systems that have recently colonized extreme environments. This study utilizes an endemic South American duck species, the speckled teal (Anas flavirostris), which has both high- and low-altitude populations. High-altitude speckled teal (A. f. oxyptera) are locally adapted to the Andean environment and mostly allopatric from low-altitude birds (A. f. flavirostris); however, there is occasional gene flow across altitudinal gradients. In this study, we used next-generation sequencing to explore genetic patterns associated with high-altitude adaptation in speckled teal populations, as well as the extent to which the balance between selection and migration have affected genetic architecture. We identified a set of loci with allele frequencies strongly correlated with altitude, including those involved in the insulin-like signaling pathway, bone morphogenesis, oxidative phosphorylation, responders to hypoxia-induced DNA damage, and feedback loops to the hypoxia-inducible factor pathway. These same outlier loci were found to have depressed gene flow estimates, as well as being highly concentrated on the Z-chromosome. Our results suggest a multifactorial response to life at high altitudes through an array of interconnected pathways that are likely under positive selection and whose genetic components seem to be providing an effective genomic barrier to interbreeding, potentially functioning as an avenue for population divergence and speciation.
Keywords: local adaptation, hypoxia, waterfowl
Introduction
Heterogeneous landscapes provide venues in which populations experiencing divergent selective pressures can differentiate into locally adapted subpopulations (Nosil et al. 2009). However, the probability for local adaptation and continued divergence depends on the strength of selection balanced by gene flow among populations; that is, migration-selection balance. Specifically, if the strength of selection is weak relative to gene flow, most local genetic variation will tend to be homogenized among populations. Alternatively, if selection is stronger than the amalgamating force of gene flow, genetic differentiation can accumulate and be maintained across specific loci experiencing strong divergent selection (Yeaman and Whitlock 2011). In essence, in the latter scenario, selection against maladapted immigrants limits the effects of gene flow and provides a means for local adaptation (Yeaman and Whitlock 2011; Feder et al. 2012). In general, studies continue to find that much of a genome is free to introgress during early stages of population divergence (Via and West 2008; Nosil et al. 2009; Feder and Nosil 2010; Via 2012). For example, specific genetic regions continue to be identified as being important in early stages of divergence and maintaining species boundaries even in the face of gene flow, including sex-linked traits (Carling and Brumfield 2008; Ellegren 2009; Storchová et al. 2010; Elgvin et al. 2011; Martin et al. 2013; Lavretsky et al. 2015), chromosomal inversions, or “supergenes” (Kirkpatrick and Barton 2006; Thompson and Jiggins 2014), and a variety of other genes associated with adaptation to extreme environments (Chapman et al. 2013; DeFaveri et al. 2013; Wit and Palumbi 2013; Soria-Carrasco et al. 2014).Thus, the probability of becoming locally adapted is influenced by this interplay between gene flow and divergent selection (Le Corre and Kremer 2012; Savolainen et al. 2013).
Species that have recently invaded extreme environments are ideal systems to study the effects of selection and gene flow on the adaptation process (Feder et al. 2012). Among these, high-altitude habitats offer an excellent opportunity to investigate the genetic basis of local adaptation (Storz et al. 2010; Cheviron and Brumfield 2012) as the characteristics of high-altitude habitats (e.g., low temperatures, ultraviolet radiation, increased water loss, and low oxygen or hypoxic conditions) are typically debilitating for low-altitude individuals that are unacclimated (Hopkins and Powell 2001). Therefore, high-altitude species represent an unparalleled system for studying adaptation in animals, not only because organisms have to overcome a clear challenge to survival and reproduction but also because the physiological mechanisms of oxygen transport are well understood (Beall 2001; Powell 2003), and selective pressures relatively well defined (Cheviron and Brumfield 2012).
In general, high-altitude species have dynamic respiratory and circulatory systems capable of responding to changes in oxygen (O2) supply and demand (Beall 2001). Adaptation to hypoxic environments has been shown to be refereed in part through the activation of hypoxia-inducible transcription factors (HIF), which start a signaling cascade of genes involved in biological processes such as angiogenesis and erythropoiesis, and assist in promoting and increasing O2 delivery to hypoxic tissues (Gorr et al. 2006; Hoogewijs et al. 2007; Rytkönen et al. 2011; Semenza 2007, 2011). Moreover, for aerobic energy production, genes involved in mitochondrial function and energy metabolism, O2 binding and delivery, and hematopoiesis are activated (Hopkins and Powell 2001; Hoogewijs et al. 2007). Ultimately, high-altitude adaptation involves coordinated changes in the expression of many genes that are involved in interacting biochemical pathways (Cork and Purugganan 2004; Cheviron and Brumfield 2012; Cheviron et al. 2012). Moreover, with different biochemical pathways, organisms can mechanistically differ in their response to hypoxic environments (Storz et al. 2010), making it important to understand how organisms adapt in different ways to these environments. At the genomic level, determining the balance between gene flow and selection is especially important when attempting to demarcate genomic regions associated with how populations have adapted to new environments. We expect established high-altitude populations to be genetically differentiated from their low-altitude counterparts at those genes/genetic regions associated with their new environment. Additionally, if gene flow is maintained between two populations, then we expect selection against maladapted genotypes to result in signatures of divergent selection at specific genes, with other parts of the genome largely undifferentiated (Feder et al. 2012; Via 2012).
In this study, we tested for genomic differentiation, and specifically for regions putatively under divergent selection between low- (<1,500 m) and high- (up to 5,000 m) altitude populations of the speckled teal (Anas flavirostris), a widespread South American waterfowl. Although high-altitude speckled teal (A. f. oxyptera) are locally adapted to the Andean environment and largely allopatric from low-altitude birds (A. f. flavirostris), speckled teal are known to disperse long-distances, which appears to result in occasional gene flow across altitudinal gradients (McCracken et al. 2009a, 2009b). Previous research suggested a degree of asymmetry in gene flow in the speckled teal, with more pronounced levels of gene flow going from low to high than high to low (McCracken et al. 2009a). Additionally, genes shown to be under positive selection such as hemoglobin (Natarajan et al. 2015) had lower levels of gene flow compared with neutral loci, attributed to countervailing selection at loci associated with high-altitude adaptation (McCracken et al. 2009a, 2009c). However, studies to date have been restricted in genomic coverage, and thus the extent and influence of gene flow in regards to local, high-altitude adaption in speckled teal remains still unexplored.
Here, we used Restriction Site-Associated DNA Sequencing (RAD-Seq) to subsample the genomes of low- and high-altitude speckled teal to examine whether highly differentiated loci are associated with possible selective mechanisms in high-altitude environments, or are consistent with neutral (allopatric) divergence. In addition, we also sequenced the α- and β-hemoglobin genes and the mitochondrial (mtDNA) control region. To identify highly differentiated loci, we calculated FST and performed genomic scans and outlier analyses using Bayesian and other methods at three marker-types with different patterns of inheritance, autosomal, Z-chromosomal sex-linked, and mitochondrial. If the two populations are diverged due to selective processes, we expected outliers to be associated with pathways related to hypoxia, and/or those involved with growth and development, metabolism, O2 transport (i.e., hemoglobin), energy production, and oxidative damage (Cheviron and Brumfield 2012; Storz and Cheviron 2016). Moreover, if there has been local adaptation in the face of continuous gene flow, then we expect a small number of genes/genetic regions to exhibit high levels of allelic differentiation between populations, whereas the rest of the genome to exhibit low levels of allelic differentiation. Conversely, if high-altitude populations have been largely allopatric during their divergence from lower altitude populations, then we expect to find a larger portion of loci across the genome to be differentiated, primarily through stochastic processes such as genetic drift, characteristic of later stages of population divergence (Feder et al. 2012).
Methods
Specimen Collection and DNA Extraction
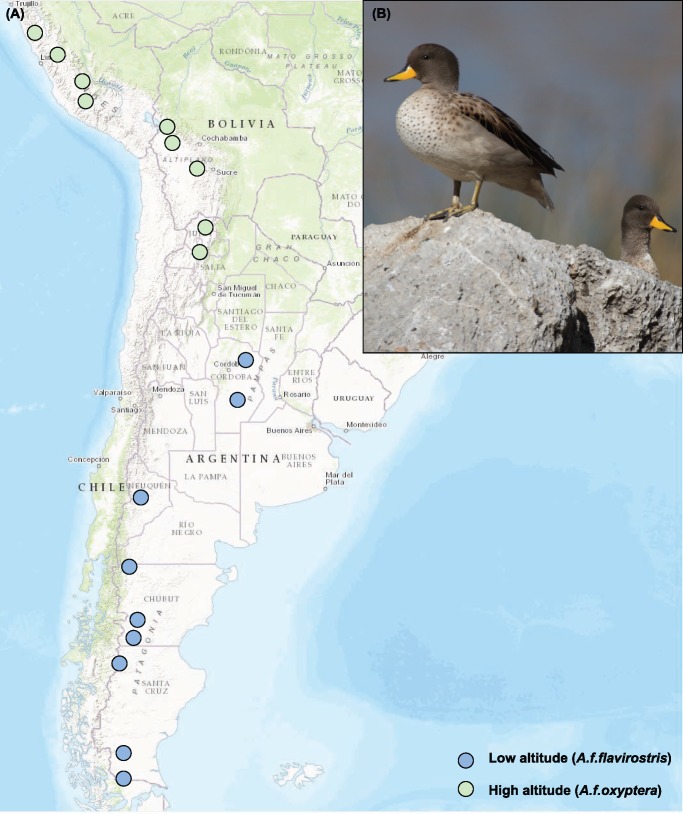
A total of 20 speckled teal were collected from low- (n = 10; elevation range 77–860 m) and high-altitude (n = 10; elevation range 3,211–4,405) sites (fig. 1 and supplementary table S1, Supplementary Material online). Animal collection was performed in adherence to Guidelines to the Use of Wild Birds in Research (Fair and Jones 2010) under permits to collect specimens from Peru (2002), Bolivia (2001), and Argentina (2001, 2003). Genomic DNA was extracted from muscle or blood using a DNeasy Tissue Kit (Qiagen, Valencia, CA) and following manufacturer’s protocols.
Fig. 1.
—Specimen collection locations of the speckled teal, Anas flavirostris (A) 10 individuals collected from high-altitude populations, A. f. oxyptera, shown in gray, and 10 individuals from low-altitude populations, A. f. flavirostris, shown in white (B) Representative photograph.
Hemoglobin and Mitochondrial Sequences
The αA-hemoglobin subunit (676 bp; HBA2), βA-hemoglobin subunit (1,578 bp; HBB), and mitochondrial sequences containing part of the control region and tRNA-Phe gene (McCracken and Wilson 2011) were obtained from GenBank, for the same 20 individuals that were used in this study (McCracken et al. 2009a), except for the high-altitude population, for which the mitochondrial sequences had not been published previously (NCBI accession numbers are provided in supplementary table S2, Supplementary Material online). For both hemoglobins, all exons and introns were sequenced. All genes were sequenced using PCR and DNA sequencing protocols as described in McCracken et al. (2009a).
RAD Sequencing and Bioinformatics
Genomic DNA was normalized to ∼2 ng/μl using a Qubit Fluorometer (Invitrogen, Grand Island, NY) and submitted to Floragenex (Eugene, OR) for RAD-Seq. In short, genomic DNA was first digested with the 8-bp SbfI restriction enzyme (CCTGCAGG) followed by barcode and adapter ligation. Individually barcoded samples were multiplexed and sequenced on the Illumina HiSeq 2000 with single-end 100-bp sequencing chemistry. Following the run, RAD sequences were demultiplexed and trimmed to yield resulting RAD sequences of 90 bp. The methods used by Floragenex are described by Miller et al. (2007), Baird et al. (2008), and Hohenlohe et al. (2010) and are summarized below.
Genotypes at each variable site were determined using Floragenex’s VCF_popgen v.4.0 pipeline to generate a customized VCF 4.1 (variant call format) database with parameters set as follows: minimum AF for genotyping = 0.075, minimum Phred score = 15, minimum depth of sequencing coverage = 10×, and allowing missing genotypes from up to 2 out of 20 individuals (10%) at each site. To filter out base calls that were not useful due to low-quality scores or insufficient coverage, genotypes at each nucleotide site were inferred using the Bayesian maximum likelihood algorithm. The genotyping algorithm incorporates the site-specific sequencing error rate, and assigns the most likely diploid genotype to each site using a likelihood ratio test and significance level of P = 0.05. Sites for which the test statistic is not significant are not assigned a genotype for that base in that individual, effectively removing data from sites for which there are too few high-quality sequencing reads. The sequencing coverage and quality scores were then summarized using the software VCFtools v.0.1.11 (Danecek et al. 2011). Custom perl scripts written by M. Campbell (University of Alaska Fairbanks) were used to retain biallelic sites only and converted to VCF file format.
Population Structure
Nucleotide diversity (π) and pairwise FST (Weir and Cockerham 1984) for each SNP between low- and high-altitude populations for RAD-Seq, hemoglobin, and the mtDNA were calculated in Arlequin v. 3.5 (Excoffier and Lischer 2010).
A haplotype network was used to visualize structure at mtDNA using TCS (Clement et al. 2000) and as implemented and visualized in the program Population Analysis with Reticulate Trees (PopART) (software available at: http://popart.otago.ac.nz). Analyses were done under default settings.
For nuclear markers, population structure was first visualized using a principal component analysis (PCA) as implemented in the software package PLINK v1.9 (Purcell et al. 2007). The PCA used in PLINK uses a two-dimensional reduction routine based on the variance-standardization relationship matrix. The top principle components are used as covariants in association analysis regressions to help correct for population stratification, whereas multidimensional scaling (MDS) coordinates help visualize genetic distances (Compagnon and Green 1994).
Next, assignment probabilities were calculated using ADMIXTURE (Alexander et al. 2009; Alexander and Lange 2011). ADMIXTURE assumes a specific number of hypothetical populations (K) and provides a maximum likelihood estimate (i.e., Q estimate) of allele frequencies for each population and admixture proportion for each individual. We analyzed values of K = 1–5 using the block-relaxation method algorithm for point estimation and terminating them when the estimates increased by <0.0001.
Identifying Outlier Loci: Hemoglobin Sequences and RAD-Seq Data
A genomic scan was performed by obtaining pairwise FST estimates calculated in Arlequin v. 3.5 (Excoffier and Lischer 2010) between low- and high-altitude populations. Instances of speciation-with-gene-flow are typically characterized by a vast majority of the genome being homogenized via gene-flow with low FST, whereas a few regions containing genes under strong divergent selection have restricted gene-flow and high FST. Therefore, empirical examples are expected to produce a characteristic L-shaped distribution of differentiation across loci in the genome, with most loci having low FST values. Whereas, populations experiencing allopatric speciation with established reproductive isolation are characterized by less gene-flow and greater divergence across much more of the genome; characterized by a distribution with less skew, more density in the center, and a more pronounced tail of higher FST values (Via 2001; Savolainen et al. 2006; Feder et al. 2012).
Next, we tested for signatures of selection using two programs, which minimized the probability of detecting false positives. First, Lositan, which implements the FDIST2 function, was used to demarcate markers putatively under positive selection (Beaumont and Nichols 1996; Antao et al. 2008): analyses were based on the Infinite Alleles Model with 50,000 simulations, a confidence interval of 0.95 and a false-discovery rate (FDR) of 0.01, as well as using the neutral mean FST and forcing mean FST options. In addition, we used a Bayesian approach as implemented in BayeScan v. 2.1 to again identify putatively selected loci, using default parameters suggested for large data sets (prods = 100). BayeScan uses a logistic regression model to separate locus-specific effects of selection from demographic differences (Foll and Gaggiotti 2008). Previous tests have shown that outliers detected by BayeScan are likely to be “better” candidate regions of the genome, because of its more conservative approach (Pérez‐Figueroa et al. 2010). Foll (2012) proposed a logarithmic scale for model choice defined as: >3 substantial (log10PO > 0.5); >10 strong (log10PO > 1.0); >32 very strong (log10PO > 1.5); and >100 decisive evidence for accepting a model (log10PO > 2.0). In our genome scans, a threshold for PO > 0.5 (substantial) was used for a marker to be considered under selection. Thus, top outlier loci were considered to be those that were identified by both methods, as previously stated. General outliers were considered to be those that were identified by both methods, including markers that were significant in Lositan analyses, and were in the top 1% of Log10(PO) values in BayeScan; these data were used for gene-flow analyses (see below).
Any markers identified as putatively being under divergent selection by both Lositan and BayeScan were next subjected to a BLAST search (database – nr, e-value < e − 10, annotation cut-off > 50) in Blast2GO (Conesa et al. 2005) using the taxonomy filter for birds (taxa: 8782, Aves) to determine gene identity. Additionally, they were categorized by chromosomal location, either as autosomal or Z-linked based on a BLAST search to the Chicken genome (Gallus gallus 5.0 reference) through NCBI, using stringent criteria (blastn, >75% identity, e-value < 10−3, max-score >40). Putative chromosomal locations of each of the autosomal categorized clusters were also identified; although we focus primarily on the Z-chromosome versus autosome comparison due to the tendency for sex chromosomes to house genes related to sexual selection, reproductive isolation, and speciation (Ellegren 2009; Ellegren et al. 2012; Lavretsky et al. 2015).
Gene ontology (GO) terms were then assigned to each of the sequences with significant BLAST hits using PANTHER (Mi et al. 2013); the GO terms associated with our outliers were subjected to term enrichment/overrepresentation analyses and visualization in AmiGO 2.0 (Carbon et al. 2009), using both the PANTHER and Go Experimental annotation data sets. In addition, different functional classifications of the gene list were performed, using the gene list analysis tool in PANTHER to determine which categories were enriched in the confirmed hit gene list relative to their representation in the reference list. For all GO term analyses, the reference list for the Chicken genome (Gallus gallus) was utilized.
Gene Flow (∂a∂i) and Coalescent Analyses
Previous multilocus work in this species suggested asymmetric gene flow from low- to high-altitude populations (McCracken et al. 2009a). We tested for and estimated rates, as well as the directionality of gene flow, with the program ∂a∂i (Gutenkunst et al. 2009, 2010), which implements a diffusion-based approach to test against specified evolutionary models with the best-fit model determined using a log likelihood-based multinomial approach. ∂a∂i analyses were done with RAD-Seq data, which were parsed into three categories that were analyzed separately, 1) All SNPs, 2) outliers-only as determined by Lositan/BayeScan analyses (see above), and 3) nonoutliers only. Given our interest in identifying differences between migration rates between outlier and nonoutlier loci, autosomal and Z-chromosome linked SNPs were analyzed together. To estimate bidirectional migration rates, the data were tested against the Isolation-with-Migration evolutionary model. Parameters in ∂a∂i were optimized prior to calculating the likelihood, which is the product of Poisson likelihoods for each parameter given an expected value from the model frequency spectrum.
In the previous study of this species, five introns were used in a multilocus coalescent analysis to estimate the effective population size parameter (θ) and bidirectional gene flow (m1 and m2) between low- and high-altitude populations (McCracken et al. 2009a). We reanalyzed these sequence data here to estimate divergence time (t) and other parameters with the IM software (Nielsen and Wakeley 2001; Hey and Nielsen 2004). Starting parameters for the IM analysis were as follows, for models incorporating and not incorporating population splitting: -q1 2 -q2 2 -qA 4 -m1 50 -m2 100 -t 2. Parameter estimates from the IM analyses were then used with MS (Hudson 2002) to create a simulated distribution of FST, conditioned on the previously estimated values of t, θ, and m, generating 1,000 randomly sampled genealogies under the Infinite Sites Model. This was performed for three different divergence estimates (-ej 0.25, 0.5, 1.0) and under three different migration rate parameters (equal, and asymmetrical in both directions). The empirical values of FST for all SNPs, outliers-only as determined by Lositan/BayeScan analyses, and nonoutliers were then compared with the distribution of simulated FST values generated by MS.
Results
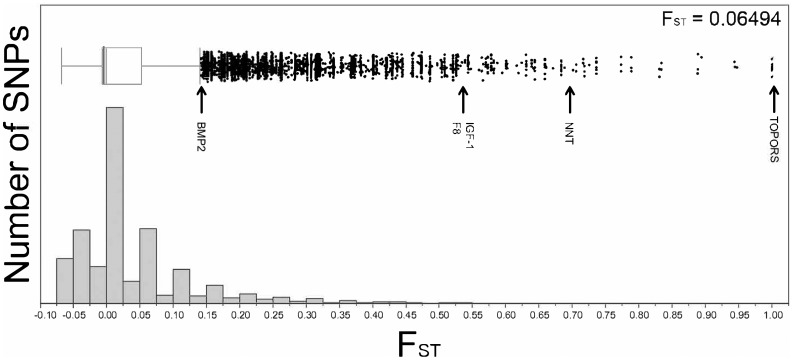
A total of 159 million RAD-Seq reads were obtained with an average depth of 38.6 (±8.0 SD) million reads per sample. After filtering, an average of 19,434 RAD clusters were recovered per sample, which represents ∼1% of a typical 1.1 gigabase avian genome (Zhang et al. 2014). Finally, a total of 47,731 polymorphic SNPs were identified. Overall, FST estimates between low- and high-altitude speckled teal populations were identical for Lositan (FST = 0.065) and Arlequin (FST = 0.065). The mtDNA with FST = 0.77 exhibited a 14-fold difference in population differentiation compared with nuclear DNA (table 1).
Table 1.
Divergence Measurements (FST) Associated with Different Subsets of Markers (mitochondrial vs. nuclear/RAD-Seq), and Their Chromosomal Location (putatively Z-linked vs. autosomal)
| Chromosomal Location | Marker Type | FST |
|---|---|---|
| Mitochondria | — | 0.770 |
| Nuclear (RAD-seq) | All | 0.065 |
| Z-linked | All | 0.123 |
| Z-linked | Outlier | 0.577 |
| Z-linked | Nonoutlier | 0.064 |
| Autosomal | All | 0.061 |
| Autosomal | Outlier | 0.442 |
| Autosomal | Nonoutlier | 0.049 |
Population Structure and Outlier Detection
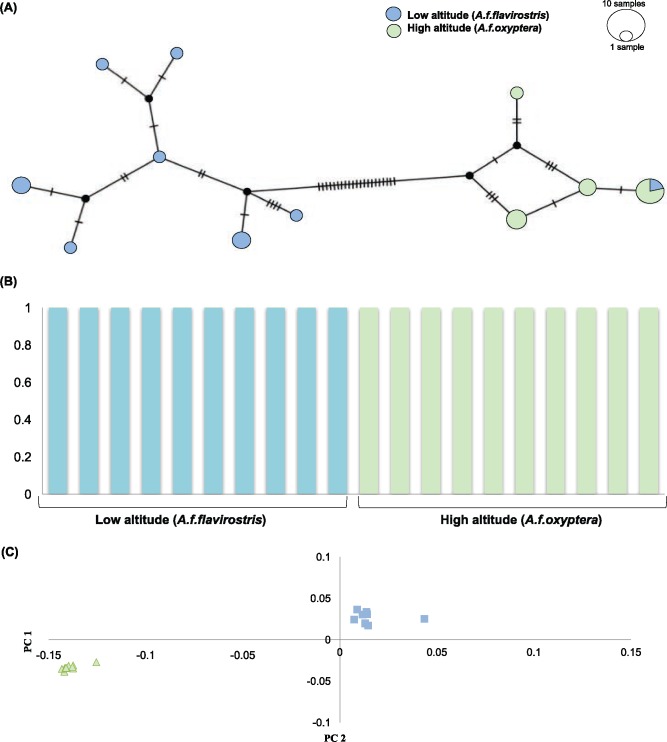
For nuclear sequences, ADMIXTURE results corresponded with PCA analyses in which low- and high-altitude individuals were highly differentiated into two clusters with 99% assignment probabilities (fig. 2B and C), despite overall low FST. The mitochondrial sequences corroborate the nuclear data showing two distinct populations, however, suggesting a much deeper level of divergence with FST = 0.77 (fig. 2A).
Fig. 2.
—Population structure between the high- and low-altitude speckled teal populations (A) mitochondrial haplotype (B) ADMIXTURE results (C) principle component analysis.
BayeScan identified ∼1% of markers (457 SNPs associated with 420 RAD clusters), as compared with Lositan’s ∼6% (3,008 SNPs, associated with 2,664 RAD clusters) as being putatively under positive selection (supplementary table S2, Supplementary Material online). Thus, our RAD-Seq data show a FST distribution with a pronounced right-tailed L-shape, with most variants showing little-to-no population differentiation and a smaller percentage of the genome comprised of highly differentiated loci (figs. 3 and 4). Loci were classified as outliers if they were identified by both methods (Lositan and BayeScan) as being significant. Ultimately, although 356 RAD-Seq loci overlapped both programs, only 34 of them returned a significant BLAST hit, and 29 of those were to a gene or transcript with an identifier. For the RAD-Seq data, the top hits with the highest prior odds were TOPORS and NNT, whereas F8, IGF-1, and BMP-2 had slightly lower prior odds (table 2). The low hit rate for our BLAST results may, in part, be because bird genomes currently have annotated 70% of the total number genes compared with the human genome; this is likely due to an enrichment of genes in GC-rich regions which are difficult to sequence, assemble, and annotate; thus they appear missing (Botero-Castro et al. 2017).
Fig. 3.
—RAD cluster distribution against measurement of population divergence (FST): markers with black arrows signify RAD-Seq markers and their respective gene identifier that were labeled as outliers for both LOSITAN and BayeScan.
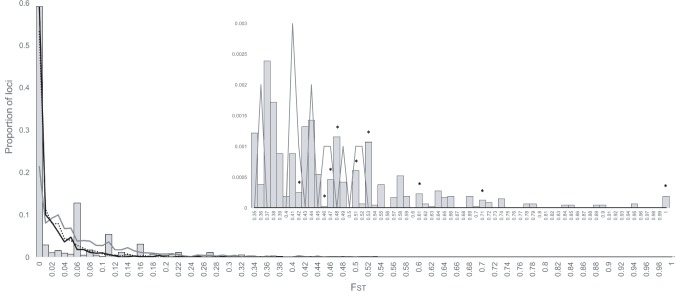
Fig. 4.
—Distribution of FST values based on MS simulations compared with empirical RAD-Seq data for divergence time of 0.5 Ma: stars = location of significant outliers, gray bars = RAD-Seq data, gray line = equal migration rate, black line = unequal migration rate (1 → 2, 10; 2 → 1, 100), dotted line = equal migration rate (1 → 2, 100; 2 → 1, 100).
Table 2.
The Gene Identification Information for RAD-Seq Markers That Were Outliers from Both LOSITAN and BayeScan Analyses, with Chromosomal Location, Gene Sequence Description, Min e-value, and Mean Similarity (blastn), as well as Gene Ontology Term Links (PANTHER)
| Gene ID | BLAST Hit Description | Min. e-Value | Mean Similarity (%) | PANTHER Family/Subfamily | Seq. Name | Chromosome |
|---|---|---|---|---|---|---|
| METRN | Anas platyrhynchos meteorin-like transcript variant mrna | 1.54E-34 | 93.33 | METEORIN (PTHR28593: SF4) | RADid_0003361_depth_116 | Autosomal |
| ARHGAP44 | Gallus gallus rho gtpase activating protein 44 transcript variant mrna | 6.54E-33 | 95.19 | RHO GTPASE-ACTIVATING PROTEIN 44 (PTHR14130: SF16) | RADid_0006028_depth_98 | Autosomal |
| TOPORSa | Anas platyrhynchos e3 ubiquitin-protein ligase topors-like partial mrna | 2.97E-37 | 100.00 | E3 UBIQUITIN-PROTEIN LIGASE TOPORS (PTHR22937: SF86) | RADid_0016436_depth_109 | Z-linked |
| ZNF469 | Anas platyrhynchos zinc finger protein 469 mrna | 1.26E-35 | 98.00 | ZINC FINGER PROTEIN 469 (PTHR21465: SF3) | RADid_0021978_depth_146 | Autosomal |
| IL20 | Anas platyrhynchos interleukin 20 mrna | 6.54E-33 | 89.50 | INTERLEUKIN-20 (PTHR10078: SF59) | RADid_0022434_depth_89 | Autosomal |
| — | Gallus gallus bac clone tam33-29c6 from chromosome complete sequence | 5.74E-21 | 89.00 | — | RADid_0027544_depth_59 | Z-linked |
| — | Opisthocomus hoazin bio-material lwl<deu | 1.04E-17 | 85.00 | — | RADid_0027947_depth_120 | Z-linked |
| CAMKK1 | Anas platyrhynchos calcium calmodulin-dependent protein kinase kinase alpha transcript variant mrna | 1.26E-35 | 98.00 | CALCIUM/CALMODULIN-DEPENDENT PROTEIN KINASE KINASE 1 (PTHR24347: SF330) | RADid_0032309_depth_138 | Autosomal |
| TRIOBP | Mus musculus trio and f-actin binding protein transcript variant mrna | 1.54E-34 | 91.78 | TRIO AND F-ACTIN-BINDING PROTEIN (PTHR17271: SF15) | RADid_0035703_depth_125 | Autosomal |
| F8b | Anas platyrhynchos coagulation factor procoagulant component mrna | 4.41E-35 | 92.67 | COAGULATION FACTOR VIII (PTHR10127: SF725) | RADid_0042196_depth_153 | Autosomal |
| SYDE2 | Anas platyrhynchos synapse defective rho homolog 2 (elegans) transcript variant mrna | 7.97E-32 | 95.00 | RHO GTPASE-ACTIVATING PROTEIN SYDE2 (PTHR23176: SF89) | RADid_0043458_depth_294 | Autosomal |
| LIPA | Anas platyrhynchos lysosomal acid lipase cholesteryl ester hydrolase-like transcript variant mrna | 6.54E-33 | 98.00 | LYSOSOMAL ACID LIPASE/CHOLESTERYL ESTER HYDROLASE (PTHR11005: SF61) | RADid_0044346_depth_103 | Autosomal |
| IGF1b | Anser anser insulin-like growth factor i (igf-i) intron 3 | 1.88E-14 | 98.00 | INSULIN-LIKE GROWTH FACTOR I (PTHR11454: SF24) | RADid_0050513_depth_248 | Autosomal |
| TRIM71 | Anas platyrhynchos e3 ubiquitin-protein ligase trim71-like mrna | 1.26E-35 | 98.00 | E3 UBIQUITIN-PROTEIN LIGASE TRIM71 (PTHR24103: SF544) | RADid_0053903_depth_95 | Autosomal |
| — | Gallus gallus bac clone ch261-26d19 from chromosome complete sequence | 1.04E-17 | 87.00 | — | RADid_0061725_depth_138 | Z-linked |
| USP24 | Anas platyrhynchos ubiquitin specific peptidase 24 mrna | 1.53E-34 | 93.71 | — | RADid_0064189_depth_48 | Autosomal |
| CLEC2L | Anas platyrhynchos c-type lectin domain family 2 member l-like transcript variant misc_rna | 1.26E-35 | 96.00 | C-TYPE LECTIN DOMAIN FAMILY 2 MEMBER L (PTHR22800: SF216) | RADid_0066143_depth_105 | Z-linked |
| ARHGEF10 | Chlorocebus sabaeus rho guanine nucleotide exchange factor 10-like transcript variant mrna | 1.75E-27 | 88.63 | RHO GUANINE NUCLEOTIDE EXCHANGE FACTOR 18 (PTHR12673: SF204) | RADid_0068177_depth_21 | Autosomal |
| VIPR1 | Ficedula albicollis vasoactive intestinal polypeptide receptor-like mrna | 9.71E-12 | 85.00 | VASOACTIVE INTESTINAL POLYPEPTIDE RECEPTOR 1 (PTHR12011: SF363) | RADid_0069283_depth_65 | Z-linked |
| MIEF1/MID51 | Anas platyrhynchos smith-magenis syndrome chromosome candidate 7-like transcript variant mrna (mitochondrial dynamic protein MID51, mitochondrial elongation factor 1 MIEF1) | 2.97E-37 | 100.00 | MITOCHONDRIAL DYNAMICS PROTEIN MID51 (PTHR16451: SF16) | RADid_0071374_depth_108 | Autosomal |
| — | Anser cygnoides clone zaas082 microsatellite sequence | 2.43E-19 | 82.00 | — | RADid_0073508_depth_100 | Z-linked |
| APEH | Ficedula albicollis vasoactive intestinal polypeptide receptor-like mrna | 5.03E-28 | 92.50 | ACYLAMINO-ACID-RELEASING ENZYME (PTHR42776: SF4) | RADid_0075723_depth_129 | Autosomal |
| TSPAN19 | Anas platyrhynchos tetraspanin 19 transcript variant mrna | 8.49E-19 | 94.07 | TETRASPANIN-19-RELATED (PTHR19282: SF274) | RADid_0075864_depth_49 | Autosomal |
| TTLL11 | Anas platyrhynchos tubulin tyrosine ligase-like member 11 transcript variant mrna | 2.97E-37 | 100.00 | TUBULIN POLYGLUTAMYLASE TTLL11 (PTHR12241: SF136) | RADid_0081068_depth_86 | Autosomal |
| DCIR | Anas platyrhynchos dendritic cell immunoreceptor complete cds dcar*null complete sequence and dendritic cell immunoactivating receptor complete cds | 4.13E-29 | 93.00 | C-TYPE LECTIN DOMAIN FAMILY 4 MEMBER A (PTHR22802: SF289) | RADid_0081629_depth_106 | Z-linked |
| MRPL54 | Anas platyrhynchos mitochondrial ribosomal protein l54 partial mrna | 1.26E-35 | 87.50 | 39S RIBOSOMAL PROTEIN L54, MITOCHONDRIAL (PTHR28595: SF2) | RADid_0084279_depth_80 | Autosomal |
| ARHGEF18 | Anas platyrhynchos rho rac guanine nucleotide exchange factor 18 transcript variant mrna | 2.97E-37 | 93.00 | RHO GUANINE NUCLEOTIDE EXCHANGE FACTOR 18 (PTHR12673: SF204) | RADid_0091284_depth_213 | Autosomal |
| MED10 | Anas platyrhynchos mediator complex subunit 10 mrna | 2.97E-37 | 100.00 | MEDIATOR OF RNA POLYMERASE II TRANSCRIPTION SUBUNIT 10 (PTHR13345: SF7) | RADid_0098634_depth_34 | Autosomal |
| BMP2b | Anas platyrhynchos bone morphogenetic protein 2-like mrna | 1.26E-35 | 86.50 | BONE MORPHOGENETIC PROTEIN 2 (PTHR11848: SF210) | RADid_0105984_depth_195 | Autosomal |
| DCIR | Anas platyrhynchos dendritic cell immunoreceptor complete cds dcar*null complete sequence and dendritic cell immunoactivating receptor complete cds | 2.78E-31 | 85.70 | C-TYPE LECTIN DOMAIN FAMILY 4 MEMBER A (PTHR22802: SF289) | RADid_0107007_depth_106 | Autosomal |
| NNTa | Chaetura pelagica nicotinamide nucleotide transhydrogenase mrna | 3.39E-30 | 90.83 | NICOTINAMIDE NUCLEOTIDE TRANSHYDROGENASE (PTHR10160: SF25) | RADid_0107938_depth_49 | Z-linked |
| — | Gallus gallus bac clone ch261-78c3 from chromosome complete sequence | 2.28E-13 | 91.00 | — | RADid_0115589_depth_45 | Z-linked |
| TLE2 | Aquila chrysaetos canadensis transducin-like enhancer of split 2 transcript variant mrna | 1.75E-27 | 91.48 | AMINO-TERMINAL ENHANCER OF SPLIT-RELATED (PTHR10814: SF33) | RADid_0118842_depth_161 | Autosomal |
| DCIR | Anas platyrhynchos dendritic cell immunoreceptor complete cds dcar*null complete sequence and dendritic cell immunoactivating receptor complete cds | 2.60E-25 | 87.50 | C-TYPE LECTIN DOMAIN FAMILY 4 MEMBER A (PTHR22802: SF289) | RADid_0124734_depth_48 | Z-linked |
Candidates of top interest.
Candidates of medium interest.
Outlier detection within the hemoglobin sequences was performed by calculating pairwise FST for each SNP via a locus-by-locus AMOVA. This identified four SNPs on the α-chain and four SNPs on the β-chain subunit of the major hemoglobin. All four of these variants have previously been identified, and for several on the β chain a putatively beneficial role in hemoglobin function has been identified via experimental tests revealing increased O2-binding affinity in the high-altitude population: α77 (pos 379, FST = 0.95), β116 (pos 346, FST = 1.00), β133 (pos 397, FST = 1.00), and β13 (pos 37, FST = 0.42) (Natarajan et al. 2015).
GO Terms of Outliers
Gene ontology terms were found for 27 of the 28 genes (table 2 and supplementary table S3a, Supplementary Material online); the gene list analysis summarizes the represented GO term groups in the form of those associated with pathway, protein, cellular component, biological process, and molecular function. Pathway, molecular function, and biological process yielded the most illuminating categories, whereas protein and cellular component were very general (supplementary table S3b, Supplementary Material online). The categories with the highest numbers in the “pathway” class were Gonadotropin-releasing hormone receptor pathway (2), followed by CCRK signaling, Wnt signaling pathway, Insulin/IGF pathway-protein kinase B signaling cascade, Insulin/IGF pathway-mitogen activated protein kinase kinase/MAP kinase cascade, Androgen/estrogene/progesterone biosynthesis, TGF-beta signaling pathway, and blood coagulation (1 each). In the “biological process” and “molecular function” classes, metabolic process (12), and catalytic activity (12) were their highest represented categories, respectively. However, the results of AmiGO statistical representation yielded no significantly overrepresented categories, likely due to the small size of the gene list.
Chromosomal Location
In general, 29 out of the 35 chromosomes were represented in our RAD-Seq data. The only chromosomes that were missing some cluster representation were Chromosome 16 and 30–33, which are some of the smallest microchromosomes. Approximately 4.8% of the total clusters “mapped” to still unplaced scaffolds within the Chicken genome.
Of the 19,434 RAD clusters, 889 clusters were identified as putatively Z-linked (i.e., having a significant BLAST hit to the Chicken Z-chromosome); therefore, the rest of the 18,549 RAD-Seq clusters were classified as putatively autosomal, although there is no way of knowing for sure given the short length of the RAD clusters (i.e., 90 bp). Compared with the autosomal clusters, the Z-linked clusters had higher FST by a factor of two; Z-linked FST = 0.123 (1,797 SNPs from 889 seqs) and autosomal FST = 0.061 (45,944 SNPs from 18,549 seqs). This Z:Autosomal ratio of 2.01 is higher than expected (expected ≤ 1.33) under a neutral model of evolution (Whitlock and McCauley 1999).
The chromosomal locations of the outliers were also identified: of the total outliers (356), 1.67% were mapped to the autosomes (310/18,549), compared with 5.17% outliers on the Z-chromosome (46/889), approximately a 3-fold difference. The Chicken Z-chromosome is 82,363,669 bp representing ∼7.68% of the total Chicken genome (1,072,544,763 bp, Gallus gallus, NCBI Annotation Release 103). Thus, given what we know about the size of the Chicken genome, out of the 356 the overlapping outliers, we classified 49 as belonging on the Z-chromosome (12.9%), which is a significantly larger portion of outliers than expected by chance (X2 = 5.5102, P value = 0.019). The two top hits in the outlier analyses were located on the Z-chromosome (NNT, TOPORS).
Gene Flow Estimates
Although low- and high-altitude populations are differentiated, the ∂a∂i results support extensive gene flow between populations under an Isolation-With-Migration model, specifically asymmetric gene flow from low to high altitude (table 3), corroborating the previous coalescent study (McCracken et al. 2009a). These asymmetric gene flow estimates correspond with the same directionality obtained from mtDNA in which 3% (n = 2) of high-altitude individuals were recovered with low-altitude haplotypes, whereas no low-altitude individuals were recovered with high-altitude haplotypes (fig. 2A). It is important to note that these introgressed mitochondrial haplotype were collected from the far northwest Argentina, which is close to where the low- and high-altitude subspecies distributions are adjacent.
Table 3.
Gene Flow Estimates for Both Nonoutlier and Outlier Loci with Isolation-with-Migration (IM) Model in ∂a∂i Showing Two Independent Runs between Low- and High-Altitude Populations
|
Ne |
Migration Rate |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Theta | Likelihood | S | Time | Low | High | High–> Low | Low–> High | ||
| Nonoutliers | IM—run 1 | 9598.944863 | −6375.446714 | 0.07592961 | 4.99593192 | 1.24293272 | 0.46102421 | 0.60743074 | 5.5158448 |
| IM—run 2 | 9598.944863 | −6375.446714 | 0.06175858 | 1.68199377 | 1.10955537 | 0.37120589 | 0.44283411 | 9.99918778 | |
| IM average | 9598.944863 | — | 0.068844095 | 3.338962845 | 1.176244045 | 0.41611505 | 0.525132425 | 7.75751629 | |
| Outliers | IM—run 1 | 87.12585977 | −996.6523572 | 0.80475161 | 9.99245969 | 0.90984298 | 0.17140578 | 0.24171386 | 2.34512902 |
| IM—run 2 | 87.12585977 | −996.6523572 | 0.124112 | 9.99098621 | 2.9884493 | 0.23662533 | 0.22651885 | 1.58623989 | |
| IM average | 87.12585977 | — | 0.464431805 | 9.99172295 | 1.94914614 | 0.204015555 | 0.234116355 | 1.965684455 | |
Note.—Averaged migration rates between the marker types for the two populations are shown in bold text. Theta, Waterson’s θ or nucleotide diversity; Ne, effective population size; migration rate, individual migrants per generation.
The analyses also suggest a substantial reduction in gene flow in outlier loci compared with nonoutlier loci, showing a 2-fold decrease from high- to low-altitude estimates (0.525 → 0.234 migrants per generation), and a 4-fold decrease from low- to high-altitude (7.76→1.97 migrants per generation; table 2). These outlier regions may represent those portions of the genome that are under selective constraint due to their being associated with adaptations to O2 deprivation and/or cold temperatures at high-altitude. Finally, the simulated distribution of FST in the MS analysis (fig. 4 and supplementary fig. S1, Supplementary Material online) indicates that we observed significantly more outlier loci, and a more right-shifted distribution of FST, than expected under a neutral model of evolution, in which all loci evolve per drift-mutation equilibrium with migration, but without influence of migration-selection balance.
Discussion
Our data reveal that despite low levels of genomic divergence (FST = 0.065) and high gene flow, the low- and high-altitude populations of speckled teal are genetically distinct. Evidence of this can be seen in the population structure associated with both mitochondrial and nuclear markers (table 1 and fig. 2A–C). In the nuclear DNA, this pattern is due, in part, to relatively few regions (i.e., regions with outliers) associated with genes known to be involved in responses to ROS production/oxidative damage (TOPORS, NNT), response to hypoxia through the HIF pathway (TOPORS, IGF, BMP), and response to hypoxia through heme factors/blood (F8, HBA2, HBB, BMP-2). These genes were found to involve autosomal (IGF, BMP, HBA2, HBB, F8), Z-linked (NNT, TOPORS) and mitochondrial elements (NNT = nuclear encoded, mitochondrial embedded) (fig. 3 and table 2). As predicted, these outliers also showed depressed gene flow between the populations, compared with the rest of the genome, suggesting an important role for migration-selection balance in the evolution of these loci in these two populations (table 3). Lastly, our results also point at a dominant role for genes located on the Z-chromosome (fig. 3 and table 2).
Migration-Selection Balance and High-Altitude Adaptation in Speckled Teal
The interplay between selection and gene flow is important for population divergence in the presence of gene flow (Feder et al. 2012). Under such a scenario, the magnitude of selection dictates whether populations continue to diverge or evolve as a single group. Here, we identified a relatively small proportion of markers to be under positive/divergent selection with restricted levels of gene flow between low- and high-altitude speckled teal populations.
Under neutrality, where genetic drift acts on markers with different effective population sizes (Ne) (Whitlock and McCauley 1999), there are different expectations for FST estimates associated with certain marker types (i.e., mitochondrial vs. nuclear, sex chromosome vs. autosome). The expectations associated with Ne are useful in illuminating whether the evolution of these different markers types has been shaped through different selective pressures on specific chromosome types (Charlesworth 2009). Previous work on waterfowl support that ducks fit the ¼ rule for mtDNA and ¾ rule for Z-linked markers (Lavretsky et al. 2015, 2016). Specifically, under a scenario of equal reproductive success between males and females, the mitochondria will have ¼ the effective population size of the nuclear genome, resulting in a 4-fold difference in FST between mtDNA and nuDNA (Caballero 1995; Whitlock and McCauley 1999; Dean et al. 2015). There is a similar expectation for Z chromosome/autosomal comparisons. Under equal reproductive success between males and females, the Z chromosome will have ¾ the effective population size of autosomes, resulting in an expected Z:Autosomal FST ratio of ≤1.33 (Caballero 1995; Whitlock and McCauley 1999; Dean et al. 2015).
However, our results show significant deviations from predictions under neutrality, with mtDNA: nuDNA resulting in a 14-fold difference, and a 2-fold difference for Z:Autosomal (table 1), suggesting a substantial role for evolutionary forces such as selection, drift, or gene flow. There are several factors that may partly explain these patterns. First, the observed difference is most extreme for the mtDNA, which could be because of the faster sorting rate of mtDNA allowing it to accumulate fixed differences faster than nuDNA (Zink and Barrowclough 2008), or because many waterfowl including speckled teal have very large population sizes (Frankham 2007; Wilson et al. 2011), although this explanation is less likely because the high-altitude population is not small. Second, the extreme divergence in the mitochondria, despite similar estimates of gene flow, may suggest that this pattern has resulted from selective processes, since the mitochondrial genome is frequently a source for selective sweeps and strong purifying selection on genetic variation associated with adaptations to hypoxic environments (Xu et al. 2007; Scott et al. 2011; Yu et al. 2011; Li et al. 2013b; Zhang et al. 2013). This could then contribute to mito-nuclear incompatibilities hampering interbreeding between the two populations (Burton and Barreto 2012), with introgression further limited by the selective pressure exerted by hypoxia (Wolff et al. 2014). However, recent evidence suggests this is not the case, as there was no discernable evidence for positive selection on the mitochondrial genome of speckled teal, and an overall lack of nonsynonymous variation associated with any protein-coding regions (Graham AM and McCracken KG, unpublished data). Lastly, it is perhaps most likely that male-biased dispersal and female philopatry common in ducks underlies the substantial structuring of mtDNA in the speckled teal (Avise et al. 1992; Peters et al. 2007, 2012; Zink and Barrowclough 2008; Flint et al. 2009).
The Z:Autosome ratio also is larger (2.01) than predicted under neutrality (1.33), although not as large as the mtDNA:nuDNA ratio, suggesting a role for selective pressures for the Z chromosome. For example, significant gene flow, predominantly from low- to high altitude, was detected by ∂a∂i analyses regardless of marker set; however, there was a substantial decrease in gene flow across outlier loci as compared with nonoutliers (table 3). Similarly restricted gene flow was detected in Hb sequences compared with other nuclear loci in speckled teal and other Andean waterfowl (McCracken et al. 2009a). We also found that more significant outliers were on the Z chromosome than expected, and such loci have been implicated in population divergence and speciation in many bird species (Saether et al. 2007; Carling and Brumfield 2009; Pryke 2010; Storchová et al. 2010; Elgvin et al. 2011; Ellegren et al. 2012; Stölting et al. 2013; Lavretsky et al. 2015). A higher density of Z-linked genes in these types of analysis is thought to be the result of a higher rate of evolution relative to autosomes (Mank et al. 2007; Ellegren 2009), as well as through its role in sexual selection, and reproductive isolation (Ritchie 2007). However, it could also be because of a general reduction in recombination rates across the Z chromosome, which can cause blocks of linkage disequilibrium, serving to further offset the disrupting force of gene flow (Saether et al. 2007; Qvarnström and Bailey 2009).
Given that the two speckled teal populations are phenotypically distinct in both plumage and body size, it seems likely that the outlier analyses have pinpointed genes underlying mechanisms involved in speciation through adaptive divergence. The two genes with the highest divergence were Z-linked, and potentially have a direct link to adaptations involving high-altitude functions (see next section). Two possibilities are that 1) hypoxia and the high-altitude environment are driving divergence directly in these genes or closely linked genes, or 2) that reproductive isolation is driving phenotypic divergence and these outliers are effectively hitchhiking on that selective pressure on the sex chromosomes. However, with our current data, we are not able to clarify further which of the two possibilities is responsible, thus warranting future research. Nonetheless, in the case of the speckled teal, we see a significant role for nonneutral processes involving Z-linked and autosomal markers—a result consistent with recent findings in other birds (Dean et al. 2015; Dhami et al. 2016) as well as other duck species (Lavretsky et al. 2015, 2016).
Overall, our results suggest selection is acting to prevent admixture at loci of adaptive importance, likely through selection against alleles originated from maladapted migrants (i.e., low-altitude individuals immigrating to the highlands or vice versa). This appears to be occurring despite these speckled teal populations being relatively divergent, estimated between 0.5 and 1.0 Ma or older, with gene flow likely playing a recurrent role throughout their recent history. Thus, at the genomic level, these populations do not seem to be suffering from an erosion of locally adaptive alleles through swamping. Ultimately, this matches predictions of divergent selection generating extrinsic reproductive isolation (Yeaman and Whitlock 2011; Feder et al. 2012), due to poor reproductive success of low-altitude migrants at high altitude compared with high-altitude residents with high-altitude genotypes. Extra-limital examples of low-altitude waterfowl species (e.g., A. discors, A. bahamensis) occasionally are observed at high altitude; however, low egg hatchability due to embryonic susceptibility to hypoxia appears to present a serious selective pressure potentially limiting reproductive success, which ultimately may influence a species’ ability to colonize the high-altitude environment (Visschedijk et al. 1980; Leon-Velarde et al. 1984; Monge and Leon-Velarde 1991; Carey et al. 1994). In addition, the asymmetrical gene flow found in this study corroborates prior work on speckled teal, 1) which showed that there were higher levels of gene flow from low to high altitude than the reverse (McCracken et al. 2009a), 2) and that other genes under positive selection (i.e., hemoglobin) had much lower levels of gene flow compared with neutral loci. In the latter case, this was attributed to countervailing selection at loci that have now been shown experimentally to be associated with mechanisms related to high-altitude adaptation (McCracken et al. 2009a, 2009c). Both the asymmetry of gene flow and the low hatchability of low-altitude genotypes at high-altitude suggest that crucial genes for survival at high-altitudes are thus sequestered in high-altitude locations, whereas the rest of the genome is seemingly free to admix. In addition, this asymmetry could be enhanced by the fact that the low altitude population is considered more migratory than their high-altitude counterparts (McCracken et al. 2009a). Therefore, our results support strong roles for both selection and gene flow in shaping the genomic architecture of speckled teal populations in response to genetic adaptations to the high-altitude Andean environment.
High-Altitude Adaptation and Z-Linked Genes: TOPORS and NNT
It is well known that exposure to low O2 leads to a cascade of metabolic and physiological changes. At high altitudes, oxygen utilization has crucial consequences for cellular function, especially through the production of reactive oxygen species (ROS) typically produced through inefficiencies during mitochondrial respiration (Turrens 2003; Palmer and Clegg 2014). The imbalance of ROS and antioxidant capacity is driven by hypoxic stress and increases at high altitudes; therefore, mechanisms to protect against oxidative damage or reduce ROS production would be advantageous (Storz et al. 2010). Our data present two gene regions with functions associated with a response to oxidative damage, TOPORS and NNT.
The region that we identified as having the highest allelic divergence between the two populations was TOPORS (FST = 1.0), which is an E3 ubiquitin-protein ligase known to regulate p53 (Rajendra et al. 2004; Weger et al. 2005) and DNA Topoisomerase I (Haluska 1999; Weger et al. 2005). Both of these genes/pathways are utilized as part of an intracellular nonimmune surveillance mechanism that controls cellular response to a variety of stress conditions, including DNA damage and hypoxia, among others, leading to cell growth arrest and apoptosis (Levine 1997; Sermeus and Michiels 2011; Bertozzi et al. 2014). Although TOPORS has not previously been identified as candidate gene involved in adaptation to high-altitude environments, p53 has been implicated in response to low O2, including, hypoxic microenvironments created by tumors (Royds et al. 1998), underground burrows of rodents (Ashur-Fabian 2004; Quina et al. 2015), and even high-altitude environments (Zhao et al. 2013). In addition to roles previously stated, TOPORS has also been implicated in playing a key role in regulating primary cilia-dependent development and function in the retina (Chakarova et al. 2011), potentially in response to increased levels of ultraviolet radiation exposure, as well as oxidative stress, at higher altitudes (Paul and Gwynn-Jones 2003). Therefore, the role of TOPORS in hypoxia and DNA damage presents a potentially pleiotropic response to both low O2 and DNA repair from oxidative and ultraviolet radiation damage.
The other gene of interest is the nuclear-encoded, but mitochondrially embedded NAD (P) transhydrogenase (NNT), which is essential for oxidative phosphorylation (OXPHOS). Under normal conditions, NNT uses energy from the mitochondrial proton gradient to produce high concentrations of NADPH; the resulting NADPH is used for biosynthesis, as well as in reactions inside the mitochondria required to remove reactive oxygen species (Figueira et al. 2013). To date, this is the first time NNT has been identified as an outlier/gene of interest in relation to high-altitude adaptation, or low-O2 environments. Previously, mutations in NNT have been shown to increase the amount of oxidative damage due to its inability to regulate ROS within the mitochondria (Freeman et al. 2006; Huang et al. 2006). Although, other mechanisms involved in DNA damage have been implicated in other high-altitude species (Subramani et al. 2015; Yang et al. 2015; Qiao et al. 2016; Zhang et al. 2016), it has been in response to ultraviolet radiation.
Autosomal Genes Associated with High-Altitude Adaptation: IGF, BMP, HBA2, and HBB
One of the biggest selective pressures for species invading high-altitude environments is the low levels of O2 in the atmosphere causing hypoxia (Storz et al. 2010). Hypoxia triggers a conservation of energy through a global reduction in protein expression, as well as a switch to anaerobic metabolism via a family of transcription factors involved in the HIF pathway (Semenza 2007). This pathway is widely considered the “master regulator” of oxygen sensing, and because of its central role in mediating a system-wide response to low O2, is frequently found to be of importance in high-altitude species (Bigham et al. 2009, 2013; Beall et al. 2010; Hanaoka et al. 2012; Li et al. 2014; Wang et al. 2014).
Although our study did not identify members of the HIF pathway specifically, we did find that the insulin-like signaling pathway (IGF-1, IGFBP-5, MAPK) and skeletal morphogenesis (BMP2) are likely under positive selection between these two populations. Not only have both the IGF and BMP pathways been shown to interact with each other (Nakae et al. 2001; Guntur and Rosen 2013) but they are also involved in the system-wide response to hypoxia mediated via the HIF pathway (Feldser et al. 1999; Fukuda et al. 2002; Wang et al. 2007). Both IGF signaling (Li et al. 2013a; Welch et al. 2014; Berg et al. 2015) and bone morphogenesis (Qu et al. 2013; Yang et al. 2016) have been implicated in the acclimation to various environmental changes, including adaptations to low O2. Additionally, both the IGF and BMP pathways are known to interact with the p53 circuit (Harris and Levine 2005), suggesting the potential for multiple outliers in interconnected pathways having an effect on high-altitude adaptation in the speckled teal.
Hemoglobin has frequently been of interest in relationship to adaptive responses to low O2 (Weber 2007; Storz and Moriyama 2008; Storz et al. 2009; Beall et al. 2010; Grispo et al. 2012; Projecto-Garcia et al. 2013; Revsbech et al. 2013; Cheviron et al. 2014; Tufts et al. 2015), and has been shown to be under selection in a variety of organisms, including Andean ducks (McCracken et al. 2009a, 2009b, 2009c, 2010). Our study was able to identify eight significant SNP variants between low- and high-altitude populations in the HBA2 and HBB subunits; of those genotypic variants identified, two have been experimentally characterized (βA116-Ser and βA133-Met) as causing an increase in the HbA isoform’s O2-binding affinity (McCracken et al. 2009a, 2009c) in high-altitude speckled teal (Natarajan et al. 2015).
In addition to selective pressures on the hemoglobin subunits, another candidate for adaptation to high altitude is the blood coagulation factor VIII or F8, as found in our outlier analyses. This gene encodes for a large plasma glycoprotein, most notably responsible for hemophilia in humans (Gouw et al. 2012). However, it has not been identified as a candidate in other high-altitude organism studies, even though it is well established that plasma concentration of F8, and other clotting factors, are elevated in humans encountering hypoxic situations (O’Brodovich et al. 1984; Chohan 2014).
Using RAD-Seq in Outlier Analyses of the Speckled Teal
We acknowledge that our data set represents a small percentage (∼1%) of the speckled teal genome, and that there are likely many other candidate gene regions for high-altitude adaptation for this species (Lowry et al. 2017a), including genes involved in the HIF pathway, as well as mitochondrial elements (Scheinfeldt and Tishkoff 2010; Storz et al. 2010; Beall et al. 2012). Still, the incomplete nature of RAD-Seq data does not necessarily detract from the results gathered for several reasons, including the limitations associated with sequencing in nonmodel organisms, and those with large or complex genomes, as well as the fact that it is technically impossible to identify all loci subjected to selection regardless of methodology (McKinney et al. 2017). Thus, RAD-Seq still remains a powerful and efficient approach for studying selection in natural populations (Andrews et al. 2016; Catchen et al. 2017), as long as the potential limitations of such data are acknowledged, including concerns with physical linkage (Lowry et al. 2017b). It is important to note that the patterns we observed in autosomal loci (in addition to some of the putative outlier genes) might be the result of genetic hitchhiking, or genetic draft, arising from linkage to genes or regions on the same chromosome that are the “actual” targets of selection. However, in birds, linkage disequilibrium (LD) decays quickly even over short distances, reflecting high recombination rates (Wong et al. 2004; Backström et al. 2006; Balakrishnan and Edwards 2009; Stapley et al. 2010); therefore, this possibility seems less likely.
Ultimately, we are careful to suggest that these outliers are not singularly causative, but instead represent just a sampling of the genes possibly under selection in high-altitude/hypoxic environments. In addition, the outlier loci identified in this study are largely not without precedent, and have been previously characterized as outliers based on prior whole-genome analyses or functional assessments in other organisms. The other loci found as candidates (those without asterisks; table 2) possibly represent those with closer potential linkage to loci not captured in the current data set. Therefore, the patterns seen in this study are likely representative of the genome as a whole, and represent a springboard for future projects in this species looking to elucidate the genetic architecture associated with selection pressures resulting from O2 deprivation.
Conclusions
Our analyses suggest that adaptations to high-altitude environments are resulting in genomic divergence, despite longstanding and recurrent gene flow between populations of speckled teal. This has created a genome-wide signature in which patterns of migration-selection balance are prevalent across various portions of the genome. We have identified a set of loci putatively under selection with differences in allele frequencies strongly correlated with high- and low-altitude habitats—most notably those involved in the insulin-like signaling pathway, bone morphogenesis, metabolic processes through the mitochondria (oxidative phosphorylation), responders to hypoxia-induced DNA damage, and feedback loops to the HIF pathway. Although gene flow was found on all linkage groups (mitochondrial, autosomal, and Z chromosome), many outlier loci in the nuclear genome were found to have depressed gene flow estimates, compared with other loci. We also discussed Z-linked loci and their role in the population differentiation of incipient diverging species; our data suggest that Z-linked loci may be simultaneously under selection due to their mechanistic role in high-altitude adaptation as well as phenotypic divergence.
Together the data identify a proportion of the genome, known to be linked to previously studied examples of high-altitude adaption, that are likely under positive directional selection in the high-altitude speckled teal population. Overall, our results suggest a multifactorial response to life at high altitudes through an array of interconnected pathways, that are not only under positive selection but whose genetic components seem to be providing at least a partial genomic barrier to gene flow and continued interbreeding, functioning as an avenue for population divergence and speciation, even if the speciation process has stalled short of completion (Peters et al. 2012). Ultimately, this study illustrates another example of how populations are able to invade novel, and sometimes adapt to extreme habitats, and provides the most comprehensive genomic study of this Andean species.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Author contributions
A.M.G. and K.G.M. designed the study; V.M.F., A.J.G., and K.G.M. provided funding; K.G.M., and R.E.W. performed the research/generated the data; A.M.G. and P.L. analyzed the data and wrote the manuscript. All authors commented on the manuscript.
Supplementary Material
Acknowledgments
We thank the many people and provincial and federal governments in Argentina, Bolivia, and Peru who assisted us with fieldwork for many years (019-2002-INRENA-DGFFS-DCB). Funding was provided by Alaska EPSCoR (NSF EPS-0092040, EPS-0346770), the National Science Foundation (DEB-0444748 and IOS-0949439), Frank M. Chapman Fund at the American Museum of Natural History, and the James A. Kushlan Endowment for Waterbird Biology and Conservation from the University of Miami. This work was supported in part by the high-performance computing and data storage resources operated by the Research Computing Systems Group at the University of Alaska Fairbanks, Geophysical Institute.
Literature Cited
- Alexander DH, Lange K.. 2011. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinformatics 12:246.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19(9):1655–1664.http://dx.doi.org/10.1101/gr.094052.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews KR, Good JM, Miller MR, Luikart G, Hohenlohe PA.. 2016. Harnessing the power of RADseq for ecological and evolutionary genomics. Nat Rev Genet. 17(2):81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antao T, Lopes A, Lopes RJ, Beja-Pereira A, Luikart G.. 2008. LOSITAN: a workbench to detect molecular adaptation based on a Fst-outlier method. BMC Bioinformatics 9:323.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashur-Fabian O. 2004. Evolution of p53 in hypoxia-stressed Spalax mimics human tumor mutation. Proc Natl Acad Sci U S A. 101(33):12236–12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise JC, Alisauskas RT, Nelson WS, Ankney CD.. 1992. Matriarchal population genetic structure in an avian species with female natal philopatry. Evolution 46(4):1084–1096.http://dx.doi.org/10.1111/j.1558-5646.1992.tb00621.x [DOI] [PubMed] [Google Scholar]
- Backström N, Qvarnström A, Gustafsson L, Ellegren H.. 2006. Levels of linkage disequilibrium in a wild bird population. Biol Lett. 2(3):435–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NA, et al. 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One 3(10):e3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan CN, Edwards SV.. 2009. Nucleotide variation, linkage disequilibrium and founder-facilitated speciation in wild populations of the zebra finch (Taeniopygia guttata). Genetics 181(2):645–660.http://dx.doi.org/10.1534/genetics.108.094250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall CM. 2001. Adaptations to altitude: a current assessment. Annu Rev Anthropol. 30(1):423–456.http://dx.doi.org/10.1146/annurev.anthro.30.1.423 [Google Scholar]
- Beall CM, et al. 2010. Natural selection on EPAS1 (HIF2α) associated with low hemoglobin concentration in Tibetan highlanders. Proc Natl Acad Sci U S A. 107:11459–11464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall CM, Jablonski NG, Steegmann A.. 2012. Human adaptation to climate: temperature, ultraviolet radiation, and altitude. Hum Biol. 163–224. [Google Scholar]
- Beaumont MA, Nichols RA.. 1996. Evaluating loci for use in the genetic analysis of population structure. Proc R Soc Lond B Biol Sci. 263(1377):1619–1626. [Google Scholar]
- Berg PR, et al. 2015. Adaptation to low salinity promotes genomic divergence in Atlantic cod (Gadus morhua L.). Genome Biol Evol. 7(6):1644–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertozzi D, et al. 2014. The natural inhibitor of DNA topoisomerase I, camptothecin, modulates HIF-1α activity by changing miR expression patterns in human cancer cells. Mol Cancer Ther. 13(1):239–248. [DOI] [PubMed] [Google Scholar]
- Bigham AW, et al. 2009. Identifying positive selection candidate loci for high-altitude adaptation in Andean populations. Hum Genomics 4(2):79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigham AW, et al. 2013. Andean and Tibetan patterns of adaptation to high altitude. Am J Hum Biol. 25(2):190–197.http://dx.doi.org/10.1002/ajhb.22358 [DOI] [PubMed] [Google Scholar]
- Botero-Castro F, Figuet E, Tilak M-K, Nabholz B, Galtier N.. 2017. Avian genomes revisited: hidden genes uncovered and the rates vs. traits paradox in birds. Mol Biol Evol. 34(12):3123–3131. [DOI] [PubMed] [Google Scholar]
- Burton RS, Barreto FS.. 2012. A disproportionate role for mtDNA in Dobzhansky–Muller incompatibilities? Mol Ecol. 21(20):4942–4957. [DOI] [PubMed] [Google Scholar]
- Caballero A. 1995. On the effective size of populations with separate sexes, with particular reference to sex-linked genes. Genetics 139(2):1007–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon S, et al. 2009. AmiGO: online access to ontology and annotation data. Bioinformatics 25(2):288–289.http://dx.doi.org/10.1093/bioinformatics/btn615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey C, Dunin-Borkowski O, Leon-Velarde F, Espinoza D, Monge C.. 1994. Gas exchange and blood gases of Puna teal (Anas versicolor puna) embryos in the Peruvian Andes. J Comp Physiol B 163(8):649–656. [Google Scholar]
- Carling MD, Brumfield RT.. 2008. Haldane’s rule in an avian system: using cline theory and divergence population genetics to test for differential introgression of mitochondrial, autosomal, and sex-linked loci across the Passerina bunting hybrid zone. Evolution 62(10):2600–2615. [DOI] [PubMed] [Google Scholar]
- Carling MD, Brumfield RT.. 2009. Speciation in Passerina buntings: introgression patterns of sex‐linked loci identify a candidate gene region for reproductive isolation. Mol Ecol. 18(5):834–847. [DOI] [PubMed] [Google Scholar]
- Catchen JM, et al. 2017. Unbroken: RADseq remains a powerful tool for understanding the genetics of adaptation in natural populations. Mol Ecol Resour. 17(3):362–365.http://dx.doi.org/10.1111/1755-0998.12669 [DOI] [PubMed] [Google Scholar]
- Chakarova CF, et al. 2011. TOPORS, implicated in retinal degeneration, is a cilia-centrosomal protein. Hum Mol Genet. 20(5):975–987.http://dx.doi.org/10.1093/hmg/ddq543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MA, Hiscock SJ, Filatov DA.. 2013. Genomic divergence during speciation driven by adaptation to altitude. Mol Biol Evol. 30(12):2553–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. 2009. Effective population size and patterns of molecular evolution and variation. Nat Rev Genet. 10(3):195–205.http://dx.doi.org/10.1038/nrg2526 [DOI] [PubMed] [Google Scholar]
- Cheviron Z, Brumfield R.. 2012. Genomic insights into adaptation to high-altitude environments. Heredity 108(4):354–361.http://dx.doi.org/10.1038/hdy.2011.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheviron ZA, Bachman GC, Connaty AD, McClelland GB, Storz JF.. 2012. Regulatory changes contribute to the adaptive enhancement of thermogenic capacity in high-altitude deer mice. Proc Natl Acad Sci U S A. 109(22):8635–8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheviron ZA, et al. 2014. Integrating evolutionary and functional tests of adaptive hypotheses: a case study of altitudinal differentiation in hemoglobin function in an Andean sparrow, Zonotrichia capensis. Mol Biol Evol. 31(11):2948–2962.http://dx.doi.org/10.1093/molbev/msu234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chohan I. 2014. Blood coagulation changes at high altitude. Defence Sci J. 34:361–379.http://dx.doi.org/10.14429/dsj.34.6083 [Google Scholar]
- Clement M, Posada D, Crandall KA.. 2000. TCS: a computer program to estimate gene genealogies. Mol Ecol. 9(10):1657–1659.http://dx.doi.org/10.1046/j.1365-294x.2000.01020.x [DOI] [PubMed] [Google Scholar]
- Compagnon R, Green C.. 1994. PLINK user’s manual. Ecole Polytechnique Federale de Lausanna.
- Conesa A, et al. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21(18):3674–3676.http://dx.doi.org/10.1093/bioinformatics/bti610 [DOI] [PubMed] [Google Scholar]
- Cork JM, Purugganan MD.. 2004. The evolution of molecular genetic pathways and networks. Bioessays 26(5):479–484.http://dx.doi.org/10.1002/bies.20026 [DOI] [PubMed] [Google Scholar]
- Danecek P, et al. 2011. The variant call format and VCFtools. Bioinformatics 27(15):2156–2158.http://dx.doi.org/10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean R, Harrison PW, Wright AE, Zimmer F, Mank JE.. 2015. Positive selection underlies Faster-Z evolution of gene expression in birds. Mol Biol Evol. 32(10):2646–2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFaveri J, Jonsson PR, Merilä J.. 2013. Heterogeneous genomic differentiation in marine threespine sticklebacks: adaptation along an environmental gradient. Evolution 67(9):2530–2546. [DOI] [PubMed] [Google Scholar]
- Dhami KK, Joseph L, Roshier DA, Peters JL.. 2016. Recent speciation and elevated Z‐chromosome differentiation between sexually monochromatic and dichromatic species of Australian teals. J Avian Biol. 47(1):92–102. [Google Scholar]
- Elgvin TO, et al. 2011. Hybrid speciation in sparrows II: a role for sex chromosomes? Mol Ecol. 20(18):3823–3837. [DOI] [PubMed] [Google Scholar]
- Ellegren H. 2009. Genomic evidence for a large-Z effect. Proc R Soc Lond B Biol Sci. 276(1655):361–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H, et al. 2012. The genomic landscape of species divergence in Ficedula flycatchers. Nature 491(7426):756–760. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HE.. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 10(3):564–567.http://dx.doi.org/10.1111/j.1755-0998.2010.02847.x [DOI] [PubMed] [Google Scholar]
- Fair JM, Jones J.. 2010. Guidelines to the use of wild birds in research. Ornithological Council.
- Feder JL, Egan SP, Nosil P.. 2012. The genomics of speciation-with-gene-flow. Trends Genet. 28(7):342–350.http://dx.doi.org/10.1016/j.tig.2012.03.009 [DOI] [PubMed] [Google Scholar]
- Feder JL, Nosil P.. 2010. The efficacy of divergence hitchhiking in generating genomic islands during ecological speciation. Evolution 64(6):1729–1747.http://dx.doi.org/10.1111/j.1558-5646.2009.00943.x [DOI] [PubMed] [Google Scholar]
- Feldser D, et al. 1999. Reciprocal positive regulation of hypoxia-inducible factor 1α and insulin-like growth factor 2. Cancer Res. 59(16):3915–3918. [PubMed] [Google Scholar]
- Figueira TR, et al. 2013. Mitochondria as a source of reactive oxygen and nitrogen species: from molecular mechanisms to human health. Antioxid Redox Signal. 18:2029–2074.http://dx.doi.org/10.1089/ars.2012.4729 [DOI] [PubMed] [Google Scholar]
- Flint PL, et al. 2009. Breeding-season sympatry facilitates genetic exchange among allopatric wintering populations of northern pintails in Japan and California. Condor 111(4):591–598.http://dx.doi.org/10.1525/cond.2009.090100 [Google Scholar]
- Foll M. 2012. BayeScan v2. 1 user manual. Ecology 20:1450–1462. [Google Scholar]
- Foll M, Gaggiotti O.. 2008. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180(2):977–993.http://dx.doi.org/10.1534/genetics.108.092221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham R. 2007. Efective population size/adult population size ratios in wildlife: a review. Genet Res. 89(5–6):491–503.http://dx.doi.org/10.1017/S0016672308009695 [DOI] [PubMed] [Google Scholar]
- Freeman H, Shimomura K, Cox R, Ashcroft F.. 2006. Nicotinamide nucleotide transhydrogenase: a link between insulin secretion, glucose metabolism and oxidative stress. Biochem Soc Trans. 34(Pt 5):806–810. [DOI] [PubMed] [Google Scholar]
- Fukuda R, et al. 2002. Insulin-like growth factor 1 induces hypoxia-inducible factor 1-mediated vascular endothelial growth factor expression, which is dependent on MAP kinase and phosphatidylinositol 3-kinase signaling in colon cancer cells. J Biol Chem. 277(41):38205–38211. [DOI] [PubMed] [Google Scholar]
- Gorr TA, Gassmann M, Wappner P.. 2006. Sensing and responding to hypoxia via HIF in model invertebrates. J Insect Physiol. 52(4):349–364.http://dx.doi.org/10.1016/j.jinsphys.2006.01.002 [DOI] [PubMed] [Google Scholar]
- Gouw SC, et al. 2012. F8 gene mutation type and inhibitor development in patients with severe hemophilia A: systematic review and meta-analysis. Blood 119(12):2922–2934. [DOI] [PubMed] [Google Scholar]
- Grispo MT, et al. 2012. Gene duplication and the evolution of hemoglobin isoform differentiation in birds. J Biol Chem. 287(45):37647–37658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guntur AR, Rosen CJ.. 2013. IGF-1 regulation of key signaling pathways in bone. BoneKEy Rep. 2:437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD.. 2009. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 5(10):e1000695.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD.. 2010. Diffusion approximations for demographic inference. DaDi Available from Nature Precedings http://hdl.handle.net/10101/npre.2010.4594.1 (2010).
- Haluska P. 1999. Interaction between human topoisomerase I and a novel RING finger/arginine-serine protein. Nucleic Acids Res. 27(12):2538–2544.http://dx.doi.org/10.1093/nar/27.12.2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka M, et al. 2012. Genetic variants in EPAS1 contribute to adaptation to high-altitude hypoxia in Sherpas. PLoS One 7(12):e50566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SL, Levine AJ.. 2005. The p53 pathway: positive and negative feedback loops. Oncogene 24(17):2899–2908.http://dx.doi.org/10.1038/sj.onc.1208615 [DOI] [PubMed] [Google Scholar]
- Hey J, Nielsen R.. 2004. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics 167(2):747–760.http://dx.doi.org/10.1534/genetics.103.024182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenlohe PA, et al. 2010. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet. 6(2):e1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogewijs D, et al. 2007. From critters to cancers: bridging comparative and clinical research on oxygen sensing, HIF signaling, and adaptations towards hypoxia. Integr Comp Biol. 47(4):552–577.http://dx.doi.org/10.1093/icb/icm072 [DOI] [PubMed] [Google Scholar]
- Hopkins SR, Powell FL.. 2001. Common themes of adaptation to hypoxia In: Roach RC, Wagner PD, Hackett PH, editors. Hypoxia. Advances in Experimental Medicine and Biology, vol 502 Boston, MA: Springer. [DOI] [PubMed] [Google Scholar]
- Huang T-T, et al. 2006. Genetic modifiers of the phenotype of mice deficient in mitochondrial superoxide dismutase. Hum Mol Genet. 15(7):1187–1194.http://dx.doi.org/10.1093/hmg/ddl034 [DOI] [PubMed] [Google Scholar]
- Hudson RR. 2002. Generating samples under a Wright–Fisher neutral model of genetic variation. Bioinformatics 18(2):337–338.http://dx.doi.org/10.1093/bioinformatics/18.2.337 [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M, Barton N.. 2006. Chromosome inversions, local adaptation and speciation. Genetics 173(1):419–434.http://dx.doi.org/10.1534/genetics.105.047985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavretsky P, et al. 2015. Speciation genomics and a role for the Z chromosome in the early stages of divergence between Mexican ducks and mallards. Mol Ecol. 24(21):5364–5378. [DOI] [PubMed] [Google Scholar]
- Lavretsky P, et al. 2016. Becoming pure: identifying generational classes of admixed individuals within lesser and greater scaup populations. Mol Ecol. 25(3):661–674. [DOI] [PubMed] [Google Scholar]
- Le Corre V, Kremer A.. 2012. The genetic differentiation at quantitative trait loci under local adaptation. Mol Ecol. 21(7):1548–1566.http://dx.doi.org/10.1111/j.1365-294X.2012.05479.x [DOI] [PubMed] [Google Scholar]
- Leon-Velarde F, Whittembury J, Carey C, Monge C.. 1984. Permeability of eggshells of native chickens in the Peruvian Andes In: Respiration and metabolism of embryonic vertebrates. The Netherlands: Springer; p. 245–257. [Google Scholar]
- Levine AJ. 1997. p53, the cellular gatekeeper for growth and division. Cell 88(3):323–331.http://dx.doi.org/10.1016/S0092-8674(00)81871-1 [DOI] [PubMed] [Google Scholar]
- Li M, et al. 2013a. Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat Genet. 45(12):1431–1438. [DOI] [PubMed] [Google Scholar]
- Li Y, et al. 2013b. High altitude adaptation of the schizothoracine fishes (Cyprinidae) revealed by the mitochondrial genome analyses. Gene 517:169–178. [DOI] [PubMed] [Google Scholar]
- Li Y, et al. 2014. Population variation revealed high-altitude adaptation of Tibetan mastiffs. Mol Biol Evol. 31(5):1200–1205.http://dx.doi.org/10.1093/molbev/msu070 [DOI] [PubMed] [Google Scholar]
- Lowry DB, et al. 2017a. Breaking RAD: an evaluation of the utility of restriction site‐associated DNA sequencing for genome scans of adaptation. Mol Ecol Resour. 17:142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry DB, et al. 2017b. Responsible RAD: striving for best practices in population genomic studies of adaptation. Mol Ecol Resour; 17:366–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE, Axelsson E, Ellegren H.. 2007. Fast-X on the Z: rapid evolution of sex-linked genes in birds. Genome Res. 17(5):618–624.http://dx.doi.org/10.1101/gr.6031907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SH, et al. 2013. Genome-wide evidence for speciation with gene flow in Heliconius butterflies. Genome Res. 23(11):1817–1828.http://dx.doi.org/10.1101/gr.159426.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken KG, Barger CP, Sorenson MD.. 2010. Phylogenetic and structural analysis of the HbA (alpha(A)/beta(A)) and HbD (alpha(D)/beta(A)) hemoglobin genes in two high-altitude waterfowl from the Himalayas and the Andes: bar-headed goose (Anser indicus) and Andean goose (Chloephaga melanoptera). Mol Phylogenet Evol. 56(2):649–658.http://dx.doi.org/10.1016/j.ympev.2010.04.034 [DOI] [PubMed] [Google Scholar]
- McCracken KG, et al. 2009a. Signatures of high-altitude adaptation in the major hemoglobin of five species of Andean dabbling ducks. Am Nat. 174(5):631–650. [DOI] [PubMed] [Google Scholar]
- McCracken KG, et al. 2009b. Parallel evolution in the major haemoglobin genes of eight species of Andean waterfowl. Mol Ecol. 18(19):3992–4005. [DOI] [PubMed] [Google Scholar]
- McCracken KG, et al. 2009c. Gene flow in the face of countervailing selection: adaptation to high-altitude hypoxia in the beta a hemoglobin subunit of yellow-billed pintails in the Andes. Mol Biol Evol. 26(4):815–827. [DOI] [PubMed] [Google Scholar]
- McCracken KG, Wilson RE. 2011. Gene flow and hybridization between numerically imbalanced populations of two duck species in the Falkland Islands. PLoS One, 6(8):e23173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney GJ, Larson WA, Seeb LW, Seeb JE.. 2017. RADseq provides unprecedented insights into molecular ecology and evolutionary genetics: comment on Breaking RAD by Lowry et al.(2016). Mol Ecol Resour. 17(3):356–361.http://dx.doi.org/10.1111/1755-0998.12649 [DOI] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Casagrande JT, Thomas PD.. 2013. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 8(8):1551–1566.http://dx.doi.org/10.1038/nprot.2013.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MR, Dunham JP, Amores A, Cresko WA, Johnson EA.. 2007. Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Res. 17(2):240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monge C, Leon-Velarde F.. 1991. Physiological adaptation to high altitude: oxygen transport in mammals and birds. Physiol Rev. 71(4):1135–1172. [DOI] [PubMed] [Google Scholar]
- Nakae J, Kido Y, Accili D.. 2001. Distinct and overlapping functions of insulin and IGF-I receptors. Endocr Rev. 22(6):818–835.http://dx.doi.org/10.1210/edrv.22.6.0452 [DOI] [PubMed] [Google Scholar]
- Natarajan C, et al. 2015. Convergent evolution of hemoglobin function in high-altitude Andean waterfowl involves limited parallelism at the molecular sequence level. PLoS Genet. 11(12):e1005681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Wakeley J.. 2001. Distinguishing migration from isolation: a Markov chain Monte Carlo approach. Genetics 158(2):885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P, Funk DJ, Ortiz-Barrientos D.. 2009. Divergent selection and heterogeneous genomic divergence. Mol Ecol. 18(3):375–402.http://dx.doi.org/10.1111/j.1365-294X.2008.03946.x [DOI] [PubMed] [Google Scholar]
- O’Brodovich HM, Andrew M, Gray G, Coates G.. 1984. Hypoxia alters blood coagulation during acute decompression in humans. J Appl Physiol. 56:666–670. [DOI] [PubMed] [Google Scholar]
- Palmer BF, Clegg DJ.. 2014. Oxygen sensing and metabolic homeostasis. Mol Cell Endocrinol. 397(1–2):51–58. [DOI] [PubMed] [Google Scholar]
- Paul ND, Gwynn-Jones D.. 2003. Ecological roles of solar UV radiation: towards an integrated approach. Trends Ecol Evol. 18(1):48–55.http://dx.doi.org/10.1016/S0169-5347(02)00014-9 [Google Scholar]
- Pérez‐Figueroa A, García‐Pereira M, Saura M, Rolán‐Alvarez E, Caballero A.. 2010. Comparing three different methods to detect selective loci using dominant markers. J Evol Biol. 23(10):2267–2276. [DOI] [PubMed] [Google Scholar]
- Peters JL, Bolender KA, Pearce JM.. 2012. Behavioural vs. molecular sources of conflict between nuclear and mitochondrial DNA: the role of male‐biased dispersal in a Holarctic sea duck. Mol Ecol. 21(14):3562–3575. [DOI] [PubMed] [Google Scholar]
- Peters JL, Omland KE, Johnson K.. 2007. Population structure and mitochondrial polyphyly in North American Gadwalls (Anas strepera). Auk 124:444–462.http://dx.doi.org/10.1642/0004-8038(2007)124[444:PSAMPI]2.0.CO;2 [Google Scholar]
- Powell FL. 2003. Functional genomics and the comparative physiology of hypoxia. Annu Rev Physiol. 65:203–230.http://dx.doi.org/10.1146/annurev.physiol.65.092101.142711 [DOI] [PubMed] [Google Scholar]
- Projecto-Garcia J, et al. 2013. Repeated elevational transitions in hemoglobin function during the evolution of Andean hummingbirds. Proc Natl Acad Sci U S A. 110(51):20669–20674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryke SR. 2010. Sex chromosome linkage of mate preference and color signal maintains assortative mating between interbreeding finch morphs. Evolution 64(5):1301–1310. [DOI] [PubMed] [Google Scholar]
- Purcell S, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81(3):559–575.http://dx.doi.org/10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Q, et al. 2016. Transcriptome sequencing of Crucihimalaya himalaica (Brassicaceae) reveals how Arabidopsis close relative adapt to the Qinghai-Tibet Plateau. Sci Rep. 6(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, et al. 2013. Ground tit genome reveals avian adaptation to living at high altitudes in the Tibetan plateau. Nat Commun. 4. [DOI] [PubMed] [Google Scholar]
- Quina A, et al. 2015. p53 gene discriminates two ecologically divergent sister species of pine voles. Heredity 115(5):444–451.http://dx.doi.org/10.1038/hdy.2015.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qvarnström A, Bailey RI.. 2009. Speciation through evolution of sex-linked genes. Heredity 102(1):4–15. [DOI] [PubMed] [Google Scholar]
- Rajendra R, et al. 2004. Topors functions as an E3 ubiquitin ligase with specific E2 enzymes and ubiquitinates p53. J Biol Chem. 279(35):36440–36444. [DOI] [PubMed] [Google Scholar]
- Revsbech IG, et al. 2013. Hemoglobin function and allosteric regulation in semi-fossorial rodents (family Sciuridae) with different altitudinal ranges. J Exp Biol. 216(Pt 22):4264–4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie MG. 2007. Sexual selection and speciation. Annu Rev Ecol Evol Syst. 38(1):79–102.http://dx.doi.org/10.1146/annurev.ecolsys.38.091206.095733 [Google Scholar]
- Royds J, Dower S, Qwarnstrom E, Lewis C.. 1998. Response of tumour cells to hypoxia: role of p53 and NFkB. Mol Pathol. 51(2):55..http://dx.doi.org/10.1136/mp.51.2.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rytkönen KT, Williams TA, Renshaw GM, Primmer CR, Nikinmaa M.. 2011. Molecular evolution of the metazoan PHD–HIF oxygen-sensing system. Mol Biol Evol. 28(6):1913–1926. [DOI] [PubMed] [Google Scholar]
- Saether SA, et al. 2007. Sex chromosome-linked species recognition and evolution of reproductive isolation in flycatchers. Science 318(5847):95–97.http://dx.doi.org/10.1126/science.1141506 [DOI] [PubMed] [Google Scholar]
- Savolainen O, Lascoux M, Merilä J.. 2013. Ecological genomics of local adaptation. Nat Rev Genet. 14(11):807–820. [DOI] [PubMed] [Google Scholar]
- Savolainen V, et al. 2006. Sympatric speciation in palms on an oceanic island. Nature 441(7090):210–213.http://dx.doi.org/10.1038/nature04566 [DOI] [PubMed] [Google Scholar]
- Scheinfeldt LB, Tishkoff SA.. 2010. Living the high life: high-altitude adaptation. Genome Biol. 11(9):133..http://dx.doi.org/10.1186/gb-2010-11-9-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott GR, et al. 2011. Molecular evolution of cytochrome c oxidase underlies high-altitude adaptation in the bar-headed goose. Mol Biol Evol. 28(1):351–363.http://dx.doi.org/10.1093/molbev/msq205 [DOI] [PubMed] [Google Scholar]
- Semenza GL. 2007. Hypoxia-inducible factor 1 (HIF-1) pathway. Sci Signal. 2007(407):cm8.. [DOI] [PubMed] [Google Scholar]
- Semenza GL. 2011. Oxygen sensing, homeostasis, and disease. N Engl J Med. 365(6):537–547. [DOI] [PubMed] [Google Scholar]
- Sermeus A, Michiels C.. 2011. Reciprocal influence of the p53 and the hypoxic pathways. Cell Death Dis. 2:e164.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria-Carrasco V, et al. 2014. Stick insect genomes reveal natural selection’s role in parallel speciation. Science 344(6185):738–742. [DOI] [PubMed] [Google Scholar]
- Stapley J, Birkhead TR, Burke T, Slate J.. 2010. Pronounced inter-and intrachromosomal variation in linkage disequilibrium across the zebra finch genome. Genome Res. 20(4):496–502.http://dx.doi.org/10.1101/gr.102095.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stölting KN, et al. 2013. Genomic scan for single nucleotide polymorphisms reveals patterns of divergence and gene flow between ecologically divergent species. Mol Ecol. 22(3):842–855. [DOI] [PubMed] [Google Scholar]
- Storchová R, Reif J, Nachman MW.. 2010. Female heterogamety and speciation: reduced introgression of the Z chromosome between two species of nightingales. Evolution 64(2):456–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Cheviron ZA.. 2016. Functional genomic insights into regulatory mechanisms of high-altitude adaptation In: Hypoxia. Springer; p. 113–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Moriyama H.. 2008. Mechanisms of hemoglobin adaptation to high altitude hypoxia. High Alt Med Biol. 9(2):148–157.http://dx.doi.org/10.1089/ham.2007.1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, et al. 2009. Evolutionary and functional insights into the mechanism underlying high-altitude adaptation of deer mouse hemoglobin. Proc Natl Acad Sci U S A. 106(34):14450–14455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Scott GR, Cheviron ZA.. 2010. Phenotypic plasticity and genetic adaptation to high-altitude hypoxia in vertebrates. J Exp Biol. 213(Pt 24):4125–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani PA, Hameed B, Michael RD.. 2015. Effect of UV-B radiation on the antibody response of fish–Implication on high altitude fish culture. J Photochem Photobiol B Biol. 143:1–4.http://dx.doi.org/10.1016/j.jphotobiol.2014.12.021 [DOI] [PubMed] [Google Scholar]
- Thompson M, Jiggins C.. 2014. Supergenes and their role in evolution. Heredity 113(1):1–8.http://dx.doi.org/10.1038/hdy.2014.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufts DM, et al. 2015. Epistasis constrains mutational pathways of hemoglobin adaptation in high-altitude pikas. Mol Biol Evol. 32(2):287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrens JF. 2003. Mitochondrial formation of reactive oxygen species. J Physiol. 552(Pt 2):335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via S. 2001. Sympatric speciation in animals: the ugly duckling grows up. Trends Ecol Evol. 16(7):381–390.http://dx.doi.org/10.1016/S0169-5347(01)02188-7 [DOI] [PubMed] [Google Scholar]
- Via S. 2012. Divergence hitchhiking and the spread of genomic isolation during ecological speciation-with-gene-flow. Philos Trans R Soc B Biol Sci. 367(1587):451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via S, West J.. 2008. The genetic mosaic suggests a new role for hitchhiking in ecological speciation. Mol Ecol. 17(19):4334–4345.http://dx.doi.org/10.1111/j.1365-294X.2008.03921.x [DOI] [PubMed] [Google Scholar]
- Visschedijk A, Ar A, Rahn H, Piiper J.. 1980. The independent effects of atmospheric pressure and oxygen partial pressure on gas exchange of the chicken embryo. Respir Physiol. 39(1):33–44.http://dx.doi.org/10.1016/0034-5687(80)90012-2 [DOI] [PubMed] [Google Scholar]
- Wang G-D, et al. 2014. Genetic convergence in the adaptation of dogs and humans to the high-altitude environment of the Tibetan plateau. Genome Biol Evol. 6(8):2122–2128.http://dx.doi.org/10.1093/gbe/evu162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wan C, Gilbert SR, Clemens TL.. 2007. Oxygen sensing and osteogenesis. Ann N Y Acad Sci. 1117:1–11.http://dx.doi.org/10.1196/annals.1402.049 [DOI] [PubMed] [Google Scholar]
- Weber RE. 2007. High-altitude adaptations in vertebrate hemoglobins. Respir Physiol Neurobiol. 158(2–3):132–142. [DOI] [PubMed] [Google Scholar]
- Weger S, Hammer E, Heilbronn R.. 2005. Topors acts as a SUMO-1 E3 ligase for p53 in vitro and in vivo. FEBS Lett. 579(22):5007–5012. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC.. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38(6):1358–1370. [DOI] [PubMed] [Google Scholar]
- Welch AJ, et al. 2014. Polar bears exhibit genome-wide signatures of bioenergetic adaptation to life in the arctic environment. Genome Biol Evol. 6(2):433–450.http://dx.doi.org/10.1093/gbe/evu025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock MC, McCauley DE.. 1999. Indirect measures of gene flow and migration: FST≠ 1/(4Nm+ 1). Heredity 82(2):117–125. [DOI] [PubMed] [Google Scholar]
- Wilson RE, et al. 2011. Speciation, subspecies divergence, and paraphyly in the cinnamon teal and blue-winged teal. Condor 113(4):747–761.http://dx.doi.org/10.1525/cond.2011.110042 [Google Scholar]
- Wit P, Palumbi SR.. 2013. Transcriptome‐wide polymorphisms of red abalone (Haliotis rufescens) reveal patterns of gene flow and local adaptation. Mol Ecol. 22(11):2884–2897. [DOI] [PubMed] [Google Scholar]
- Wolff JN, Ladoukakis ED, Enríquez JA, Dowling DK.. 2014. Mitonuclear interactions: evolutionary consequences over multiple biological scales. Philos Trans R Soc B Biol Sci. 369(1646):20130443.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong GK-S, et al. 2004. A genetic variation map for chicken with 2.8 million single-nucleotide polymorphisms. Nature 432:717–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, et al. 2007. High altitude adaptation and phylogenetic analysis of Tibetan horse based on the mitochondrial genome. J Genet Genomics 34(8):720–729.http://dx.doi.org/10.1016/S1673-8527(07)60081-2 [DOI] [PubMed] [Google Scholar]
- Yang J, et al. 2016. Whole-genome sequencing of native sheep provides insights into rapid adaptations to extreme environments. Mol Biol Evol. 33(10):2576–2592.http://dx.doi.org/10.1093/molbev/msw129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, et al. 2015. Comparative transcriptomic analysis revealed adaptation mechanism of Phrynocephalus erythrurus, the highest altitude Lizard living in the Qinghai-Tibet Plateau. BMC Evol Biol. 15:101.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeaman S, Whitlock MC.. 2011. The genetic architecture of adaptation under migration–selection balance. Evolution 65(7):1897–1911.http://dx.doi.org/10.1111/j.1558-5646.2011.01269.x [DOI] [PubMed] [Google Scholar]
- Yu L, Wang X, Ting N, Zhang Y.. 2011. Mitogenomic analysis of Chinese snub-nosed monkeys: evidence of positive selection in NADH dehydrogenase genes in high-altitude adaptation. Mitochondrion 11(3):497–503.http://dx.doi.org/10.1016/j.mito.2011.01.004 [DOI] [PubMed] [Google Scholar]
- Zhang G, et al. 2014. Comparative genomic data of the Avian Phylogenomics Project. GigaScience 3(1):26.http://dx.doi.org/10.1186/2047-217X-3-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, et al. 2016. Genome resequencing identifies unique adaptations of Tibetan chickens to hypoxia and high-dose ultraviolet radiation in high-altitude environments. Genome Biol Evol. 8(3):765–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z-Y, Chen B, Zhao D-J, Kang L.. 2013. Functional modulation of mitochondrial cytochrome c oxidase underlies adaptation to high-altitude hypoxia in a Tibetan migratory locust. Proc R Soc B Biol Sci. 280(1756):20122758.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, et al. 2013. Codon 104 variation of p53 gene provides adaptive apoptotic responses to extreme environments in mammals of the Tibet plateau. Proc Natl Acad Sci U S A. 110(51):20639–20644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink RM, Barrowclough GF.. 2008. Mitochondrial DNA under siege in avian phylogeography. Mol Ecol. 17(9):2107–2121.http://dx.doi.org/10.1111/j.1365-294X.2008.03737.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.