Abstract
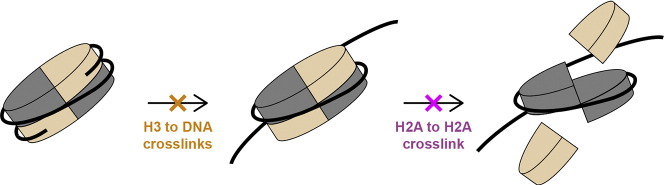
We engineered nucleosome core particles (NCPs) with two site-specific cysteine crosslinks that increase the stability of the particle. The first disulfide was introduced between the two copies of H2A via an H2A-N38C point mutation, effectively crosslinking the two H2A/H2B heterodimers together to stabilize the histone octamer against H2A/H2B dimer dissociation. The second crosslink was engineered between an R40C point mutation on the N-terminal tail of H3 and the NCP DNA ends by the introduction of a convertible nucleotide. This crosslink maintains the nucleosome DNA in a fixed translational setting relative to the histone octamer and prevents dilution-driven dissociation. The X-ray crystal structures of NCPs containing the disulfides in isolation and in combination were determined. Both disulfides stabilize the structure of the NCP without disturbing the overall structure. Nucleosomes containing these modifications will be advantageous for biochemical and structural studies as a consequence of their greater resistance to dissociation during high dilution in purification, elevated salt for crystallization and vitrification for cryogenic electron microscopy.
Abbreviations: ASP, α-satellite palindromic DNA; bp, base pair; NCP(s), nucleosome core particle(s); SELEX, systematic evolution of ligands by exponential enrichment; TFA, trifluoroacetic acid; w.t., wild type
Keywords: chromatin, DNA, histone, hexasome, X-ray crystallography
Graphical Abstract
Highlights
-
•
Crosslinked nucleosome core particles have increased stability against H2A/H2B dimer loss and DNA dissociation.
-
•
A site-specific disulfide crosslink was introduced between the two copies of H2A in the histone octamer to stabilize its quaternary structure.
-
•
Site-specific disulfide crosslinks were introduced between histone H3 and DNA within the nucleosome core particle.
-
•
Three X-ray crystal structures of crosslinked nucleosome core particles were determined at high resolution.
Introduction
The histone octamer is a highly abundant and conserved protein complex that organizes eukaryotic DNA into nucleosomes, the basal repeating unit of chromatin [1], [2]. Nucleosomes regulate access to DNA either directly via their positioning or indirectly via the recruitment of chromatin factors through their associated pattern of post-translational modifications. Structural knowledge about nucleosomes and their higher-order assembly is essential for an understanding of all genomic processes.
The histone octamer is a twofold symmetric particle comprising two copies of each of four core histones H2A, H2B, H3 and H4 [1]. The individual core histones form the stable dimer units of H2A/H2B and H3/H4 through the “handshake motif” [3]. The H3/H4 dimers further assemble into a tetramer through a symmetric four-helix bundle formed between the two copies of H3. The histone octamer is completed by the addition of two H2A/H2B dimers to the periphery of the H3/H4 tetramer via two homologous four-helix bundles between H2B and H4. H2A also makes additional interactions with the opposite H2A copy via their L1 loops and with the histone-fold core of H3/H4 via its C-terminal “docking domain.” The four-helix bundles serve to place the histone dimer pairs in a spiral path that binds 147 base pairs (bp) of DNA as a left-handed superhelix, forming the nucleosome core particle (NCP) [4]. The central 121 bp of DNA are organized by the repeating histone-fold elements α1α1 and L1L2, and the straighter 13-bp terminal portions are bound by the H3-αN histone-fold extensions. DNA is further stabilized at several positions where the histone tails pass over minor grooves or protrude through channels formed by adjacent minor grooves in the superhelix.
In vitro assembled NCPs, nucleosomes and oligonucleosomes are widely used in chromatin structural and biochemical research and depend on the availability of methodology to manufacture both recombinant histones and DNA with strong positioning properties [5], [6], [7], [8]. The histone octamer is a stable entity only at either high salt or in the presence of DNA at reduced salt [9]. Due to the differing chemical character of the four-helix bundles [4], the predominant oligomeric species at physiological salt concentrations are the H3/H4 tetramer and the H2A/H2B dimers. In vitro nucleosome assembly takes place sequentially during salt-gradient dialysis by the initial deposition of the H3/H4 tetramer onto DNA at high ionic strength to form the tetrasome, followed by the binding of two H2A/H2B dimers as the salt concentration is lowered [10], [11], [12], [13]. The lower affinity of H2A/H2B dimers can lead to the formation of a sub-stoichiometric complex lacking one H2A/H2B dimer described as a hexasome. The formation of hexasomes is perhaps exacerbated by the most commonly used DNA for reconstitution, the 601 sequence, which displays distinct asymmetry in its affinity for H2A/H2B dimers [8], [14]. This is likely a consequence of the systematic evolution of ligands by exponential enrichment procedure used to develop the 601 sequence in which salt-gradient assembly allowed the H3/H4 tetramer to dominate DNA sequence selection [15].
DNA dissociation is another problem experienced with nucleosomes when subjected to either high dilutions or high salt concentrations, and commonly occurs during purification using techniques such as size-exclusion and ion-exchange chromatography [7]. This behavior can complicate the purification of nucleosomes or nucleosomes with bound factors used for structural investigations.
Studies on the mechanics of DNA bending and its influence on nucleosome positioning are currently limited by the availability of NCP X-ray structures containing novel DNA sequences. High-resolution diffraction from NCP crystals is highly dependent on an important crystal contact formed between the DNA ends of adjacent NCPs. This contact is, in turn, sensitive to the length, stretching, translational setting and terminal phosphorylation state of the NCP DNA [4], [16], [17], [18], [19], [20], [21]. NCP structures have been limited to those based on α-satellite, 601 and mouse mammary tumor virus nucleosome A sequences of 145–147 bp, all of which adopt a single translational setting on the histone octamer. Positions for these sequences were mapped to base-pair resolution by site-directed hydroxyl radical cleavage [22], [23], [24].
We have developed two types of site-specific disulfide-stabilized nucleosomes to address the problem of nucleosome stability. The first cysteine crosslink was introduced between the two copies of H2A to stabilize the histone octamer against H2A/H2B dimer loss (Fig. 1a). We demonstrate that this crosslink stabilizes the isolated histone octamer against dimer dissociation at intermediate salt concentrations, and prevents dimer loss during nucleosome assembly using 601 DNA. A second cysteine crosslink was introduced between the DNA ends of the NCP and the histone H3 N-terminal tails (Fig. 1b). This crosslink eliminates dilution-driven dissociation and aids in the purification of intact NCP through ion exchange. Finally, we determined the crystal structures of NCPs containing both crosslinks in isolation and in combination to visualize and validate the disulfide geometry. We show that neither crosslink perturbs the canonical structure of the NCP significantly.
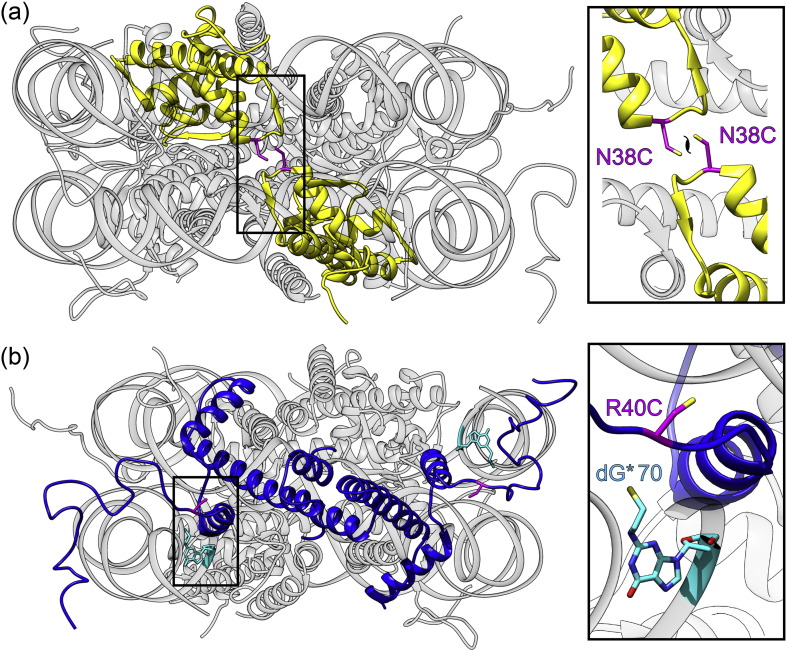
Fig. 1.
Location of site-specific crosslinks designed based on the 1.9-Å NCP structure (PDB ID 1KX5). (a) The NCP viewed up its dyad axis showing the two copies of H2A (yellow) and the modeled H2A-N38C point mutations (magenta). (b) The NCP viewed down its dyad axis showing the two copies of H3 (blue), and the modeled H3-R40C point mutations (magenta) and convertible guanosines (cyan) at position 70 on the I- and J-strand.
Results
H2A-N38C disulfide design
Disulfide crosslinks were designed using the highest-resolution X-ray structure of the NCP available (PDB ID 1KX5) [19]. Due to the inherent construction of the octamer core by four-helix bundles, an obvious candidate to prevent dimer dissociation would be to crosslink the H2A/H2B dimers directly to the H3/H4 tetramer in the vicinity of two H2B–H4 four-helix bundles. However, a simpler option exists in that the spiral arrangement of histone dimer pairs within the octamer places the two H2A/H2B dimers in close proximity. The L1 loops from the two symmetry-related chains of H2A are juxtaposed near the molecular twofold axis of the NCP (Fig. 1a). Crosslinking across this axis is convenient as it requires only a single cysteine substitution. Within the L1 loop of H2A, only asparagine 38 is at a favorable distance for introducing a cysteine crosslink (Cα–Cα distance = 5.5 Å), well within the range of naturally occurring disulfide bridges [25], [26]. Furthermore, the H2A-N38C side chains already face each other and therefore do not require major rearrangement of the peptide backbone torsion angles. The flexibility of the L1 loop can potentially accommodate any minor distortions introduced by the disulfide link.
Crosslinked H2A-N38C
The H2A-N38C substitution was introduced by site-directed mutagenesis, and the protein was initially produced via standard histone techniques and stored in lyophilized aliquots at − 80 °C [5], [7]. However, we found that during storage, cysteine-containing histones lost crosslinking efficiency, likely due to irreversible oxidation. We therefore adapted the technique of sulfitolysis used in insulin production and applied it to histone preparation [27]. In this procedure, the sulfhydryl groups are oxidized with sulfite to form S-sulfocysteine during inclusion body solubilization and are thereby protected from further oxidation during purification and storage. Complete conversion to S-sulfocysteine was verified by mass spectrometry. Cysteine sulfhydryls are later restored through reduction during histone octamer preparation. Standard histone production also relies on urea for denaturation resulting in heterogeneity introduced by carbamylation of sensitive groups, including cysteine side chains. We have therefore substituted urea with guanidine hydrochloride for all appropriate purification steps. Since guanidine hydrochloride is incompatible with ion exchange, we have replaced this step with reversed-phase HPLC using 0.3% trifluoroacetic acid (TFA) as a denaturant [28]. H2A-N38C crosslinks are only induced by oxidation after the histone octamer is assembled prior to size-exclusion chromatography, removing contaminating oligomeric species. The H2A-N38C crosslink was successfully formed as H2A subsequently migrates as a dimer on denaturing gels. As monomeric H2A comigrates with H2B in 18% acrylamide, crosslinking was quantified by HPLC and judged to be complete as free H2A was not detectable (data not shown).
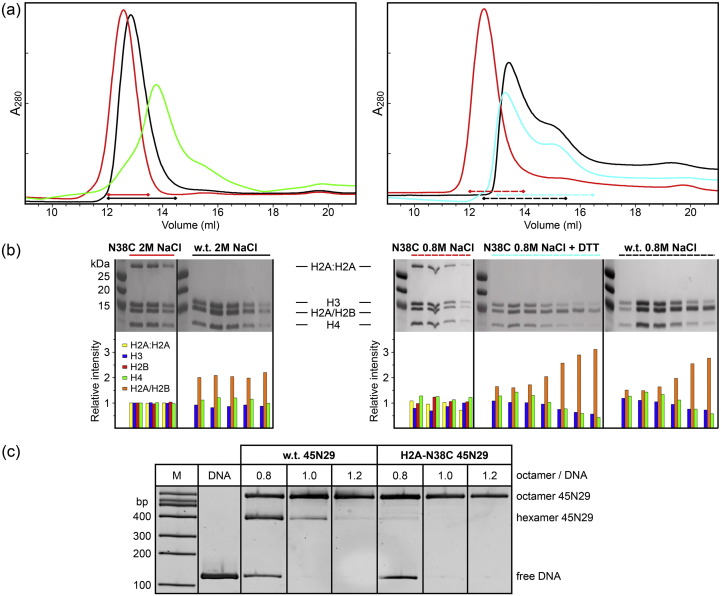
As the oligomeric state of histones is highly dependent on salt concentration and pH, we used a combination of size-exclusion chromatography and SDS PAGE to investigate the stability of H2A-N38C octamers [29]. At 2 M NaCl, pH 7.5, wild-type (w.t.) histones are predominately in an octameric form such that these conditions form the basis for octamer purification [7] (Fig. 2a). H2A-N38C octamers (oxidized) are also octameric under these conditions, but elute with a slightly higher apparent molecular weight. This difference in mobility occurs because w.t. octamers are in dynamic equilibrium and slowly dissociate during chromatography, leading to peak trailing. Consistent with an equilibrium effect, others have observed less peak trailing at 4 M NaCl or with higher concentrations of octamer [29], [30]. By preventing dimer dissociation, the H2A-N38C crosslink eliminates this effect.
Fig. 2.
Characterization of histone octamer and a nucleosome containing H2A-N38C. (a) Size exclusion on Superdex 200. The left panel shows chromatography in 2 M NaCl for oxidized H2A-N38C octamer (red) and w.t. octamer (black) at pH 7.5, and for oxidized H2A-N38C at pH 5.0 (green). The right panel shows chromatography in 0.8 M NaCl for w.t. octamer (black), oxidized H2A-N38C octamer (red) and reduced H2A-N38C octamer (cyan) at pH 7.5. (b) SDS PAGE of peak fractions from panel a. Each upper panel corresponds to a series of fractions from a peak underlined in panel a for H2A-N38C-containing or w.t. histone octamer. The bottom panels show the amount of each core histone relative to the amounts in the H2A-N38C 2 M NaCl panel as estimated by gel densitometry. The H2A-N38C disulfide of the H2A:H2A dimer is reduced on the addition of DTT (panel N38C 0.8 M NaCl versus N38C 0.8 M NaCl + DTT). Marker proteins are shown in the left lanes. (c) Native PAGE analysis of nucleosome 45N29 containing w.t. and N38C octamers. The octamer to DNA ratio used for each assembly reaction is indicated. Marker DNAs are shown in the left lane (M).
As the H2A/H2B dimers are not directly crosslinked to the H3/H4 tetramer, it was necessary to demonstrate the salt-dependent dissociation behavior of H2A-N38C octamers while crosslinked. Histone octamer stability is progressively reduced at salt concentrations below 2 M NaCl. We selected a condition of 0.8 M NaCl, pH 7.5, as w.t. octamers were observed to dissociate into (H3/H4)2–H2A/H2B hexamers and free H2A/H2B dimers as judged by their elution behavior and quantitation of histone bands on denaturing gels (Fig. 2b). This salt concentration corresponds to the midpoint of the dimer/tetramer binding equilibrium and therefore was used to test the stability of H2A-N38C octamers. In contrast to w.t. octamers, the crosslinked H2A-N38C octamers do not dissociate in 0.8 M NaCl and elute as a single peak with the same retention time as for 2 M NaCl (Fig. 2a). Reduction of the crosslink restores the w.t. dissociation behavior and demonstrates that stabilization of the quaternary structure is dependent on an intact crosslink. Further reduction of salt concentration to 0.4 M NaCl yields w.t. octamers completely dissociated into tetramers and dimers, while H2A-N38C octamers remain intact (Fig. S1). Similar to the effect of low salt, the weakly acidic condition of pH 5.0 at 2 M NaCl also results in the dissociation of w.t. octamers into tetramers and dimers [9], [29]. However, unlike in low-salt conditions, H2A-N38C octamers dissociated into H3/H4 tetramers and H2A/H2B tetramers, which co-elute on size-exclusion chromatography (Fig. 2a).
We have previously experienced difficulty assembling nucleosomes on a DNA sequence composed of a 147-bp, 601-core sequence with 45- and 29-bp terminal extensions (45N29) [8], [31]. At a 1:1 molar ratio of octamer to DNA, hexasomes had a propensity to form, as noted by others using other 601-based constructs [14], [32] (Fig. 2c). Hexasome formation can be effectively suppressed by supplying a molar excess of either H2A/H2B dimer or octamer, but this results in the introduction of unbound histones which is undesirable for structural studies. In contrast, using H2A-N38C octamer for nucleosome assembly completely eliminated the appearance of hexasomes and resulted in the formation of nucleosomes at equal stoichiometry of octamer to DNA (Fig. 2c).
H2A-N38C NCP structure
Engineered disulfides can often fail to achieve the stability of naturally occurring disulfides and can exhibit distorted geometries or produce larger perturbations of the protein structure [25], [26]. Before H2A-N38C octamers can be used as a general tool in chromatin research, the effect of the disulfide must be quantified structurally. For this purpose, we have determined the X-ray crystal structure of an H2A-N38C disulfide containing NCP at a resolution of 2.3 Å (Table 1). For the purpose of crystallization, the H2A-N38C NCP was assembled using 147-bp α-satellite palindromic DNA (ASP), known to produce well-diffracting crystals and to aid in the comparison with the previous high resolution NCP containing all w.t. histones [19]. The H2A-N38C disulfide could be visualized in the initial difference density directly after molecular replacement, and to further exclude model bias, a composite omit map was calculated after model building and refinement (Fig. 3a). Natural disulfides can be classified according to the dihedral angles adopted by the cystine side-chains [25]. The H2A-N38C disulfide bridge belongs to the most common left-handed spiral class as it displays all negative dihedral angles (Fig. 3b). The disulfide bridge also has approximate twofold symmetry with the center of the S–S bond passing through the dyad axis of the NCP. The dihedral angle over the S–S bond (χ3) is close to − 90° and suggests an unstrained bond with good redox stability.
Table 1.
Data collection and refinement statistics for the three NCP X-ray crystal structures
| H2A-N38C | H3-DNA | H2A-N38C/H3-DNA | |
|---|---|---|---|
| Data collection | |||
| Number of crystals | 1 | 7 | 2 |
| Resolution (Å) | 47.12–2.32 | 29.54–2.80 | 29.49–2.80 |
| Resolution of last shell (Å) | 2.47–2.32 | 2.95–2.80 | 2.85–2.80 |
| Multiplicity | 3.2 | 3.3 | 3.6 |
| No. of unique hkl | 80,720 | 47,952 | 52,834 |
| Completeness % (last shell) | 86.3 (31.2) | 90.3 (68.0) | 98.4 (93.7) |
| Rmerge % (last shell) | 6.9 (21.0) | 8.4 (23.1) | 6.2 (32.8) |
| I/σ (last shell) | 10.3 (1.4) | 12.7 (2.5) | 13.9 (2.8) |
| Space group | P212121 | P212121 | P212121 |
| a (Å) | 106.75 | 106.25 | 106.56 |
| b (Å) | 182.58 | 183.13 | 183.83 |
| c (Å) | 109.64 | 109.49 | 109.49 |
| Refinement | |||
| Resolution (Å) | 47.12–2.32 | 29.5–2.80 | 29.5–2.80 |
| R-factor % (last shell) | 22.0 (38.5) | 20.0 (38.0) | 20.8 (29.2) |
| R-free % (last shell) | 25.9 (40.6) | 25.1 (42.8) | 25.4 (33.7) |
| No. of reflections work/free | 80,497 (4048) | 42,969 (4812) | 52,753 (4852) |
| No. of atoms in model | |||
| Total (Avg. B-factor (Å)) | 12,250 (75.0) | 12,218 (91.4) | 12,069 (105.0) |
| RMSD from ideality | |||
| Bond length (Å) | 0.010 | 0.011 | 0.005 |
| Bond angles (deg.) | 1.23 | 1.12 | 0.71 |
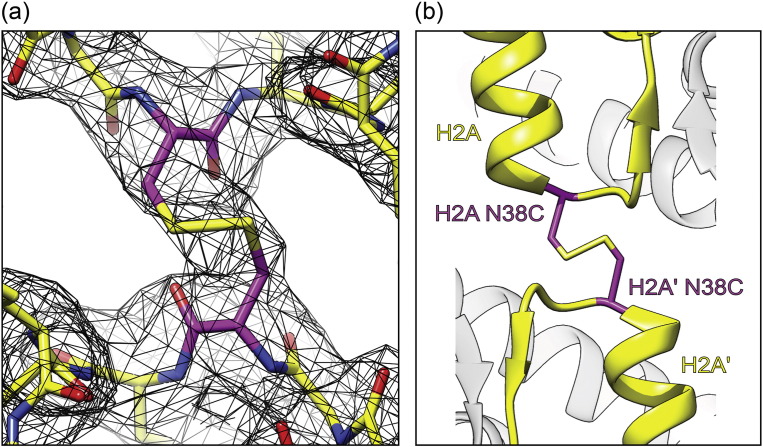
Fig. 3.
The disulfide crosslink in the H2A-N38C NCP. (a) 2FO–FC electron density for the disulfide bond (black mesh, contour 1.4 × sigma) with the NCP structure superimposed. The two N38C substitutions are highlighted (magenta). (b) Structure cartoon of the H2A-N38C crosslink and associated H2A-L1 loops (yellow).
When aligned to the ASP NCP, the H2A-N38C NCP shows excellent agreement with Cα RMSD values for each histone chain of less than 0.4 Å and C4′ RMSD values of less than 0.6 Å for both DNA chains. For comparison, these deviations are significantly less than NCP structures containing different DNA sequences [21]. The only minor difference between the two structures is located within the L1 loops. As a consequence of the disulfide bond, the H2A-N38C Cα–Cα distance is shortened from 5.45 to 5.33 Å resulting in the movement of the C38 Cα positions (0.95 Å chain C, 1.02 Å chain G) and G37 (0.67 Å chain C, 0.73 Å chain G). Importantly, these minor displacements are restricted to the buried H2A L1 loop without disturbing the canonical NCP structure.
H3-DNA NCP disulfide design
We designed site-specific disulfide bridges between the histone octamer and the nucleosome DNA, with the aim of preventing both DNA dissociation and the translational repositioning of nucleosome DNA. The NCP contains several possible locations for introducing a crosslink between the histone octamer and the nucleosome DNA [19]. We focused on the N-terminal tails of H3 for crosslinking to the nucleosome DNA due to their proximity to the DNA ends (Fig. 1b). The H3 N-terminal tail protrudes through a minor groove channel formed by juxtaposed minor grooves from the DNA ends and the central gyre of the NCP superhelix. H3 H39 and R40 are in close proximity to the nucleosome DNA at ± 69 and ± 70 bp from the dyad axis. Both cysteine point mutations were separately introduced into the H3 tail by site-directed mutagenesis. A thiol group was introduced into the DNA at either position T69 or T70 on both the I- and J-strands through oligonucleotides incorporating a convertible nucleotide [33], [34], [35], [36]. 2-Fluoro-deoxyinosine (2-F-dI) was used in oligonucleotide synthesis and converted post-synthetically to contain a cystamine moiety at the purine N2 position which projects into the DNA minor groove (dG*). Restriction sites were introduced near the ends of the 147-bp α-satellite sequence used in ASP to create ligation sites for these modified oligonucleotides. The resulting NCP contains palindromic DNA capable of forming a disulfide bridge at both ends (Fig. S2). All permutations of crosslinks between histone H3-H39C and H3-R40C and DNA T69G* and T70G* were evaluated for crosslinking efficiency (data not shown), and the combination of H3-R40C and T70G* was selected for the production of H3-DNA NCP.
Crosslinked H3-DNA NCP
The major contaminants of H3-DNA NCP are expected to consist of particles lacking either one or both crosslinks. As uncrosslinked NCP dissociates at elevated ionic strength, this behavior was used to purify fully crosslinked H3-DNA NCP by anion-exchange chromatography and salt-gradient elution [5] (Fig. 4a). DNA from partially or fully dissociated NCP binds more strongly to the anion-exchange matrix than intact NCP for which the DNA charge is partially neutralized by the histones. As seen by SDS PAGE, the peak fractions corresponding to the H3-DNA NCP can be purified to homogeneity and are devoid of uncrosslinked H3 (Fig. 4b).
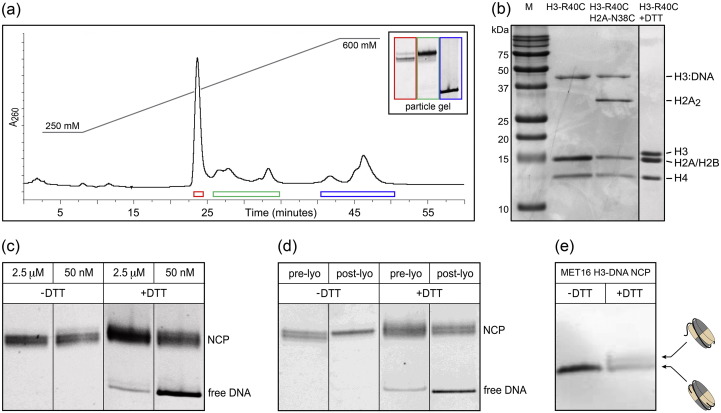
Fig. 4.
Purification and characterization of H3-DNA NCP. (a) Purification of H3-DNA NCP by anion-exchange chromatography. The linear salt-gradient (gray line) maximizes the yield of NCP containing H3-DNA disulfide crosslinks. Three elution volumes were analyzed by native PAGE (inset) and correspond to crosslinked NCP (red), partially crosslinked and uncrosslinked products (green), and mainly free DNA (blue). The ethidium bromide dye used to visualize products is relatively ineffective on the crosslinked DNA (red). (b) SDS PAGE of purified H3-DNA and H3-DNA/H2A-N38C NCPs. H3-R40C migrates as a 40-kDa species when crosslinked to NCP DNA and as free H3 in the presence of DTT. Crosslinked H2A-N38C migrates as a dimer. Marker proteins are shown in the left lane (M). (c) Effect of dilution on H3-DNA NCP. Native PAGE of crosslinked (− DTT) and reduced (+ DTT) NCP upon dilution from 2.5 μM to 50 nM NCP. (d) Effect of lyophilization on H2A-N38C/H3-DNA NCP. Native PAGE of crosslinked (− DTT) and reduced (+ DTT) NCP before (pre-lyo) and after lyophilization and resuspension in solution (post-lyo). (e) Effect on multiple DNA positions for MET16 H3-DNA NCP. Native PAGE of crosslinked (− DTT) and reduced (+ DTT) NCP containing a DNA sequence from the Saccharomycescerevisiae MET16 promoter.
Nucleosomes have been observed to partially dissociate into free DNA and histones when diluted to nanomolar concentration [37], [38]. Under physiological salt conditions, this dissociation is not strictly reversible and hence does not satisfy the conditions for a true equilibrium. Dissociated species become kinetically trapped and nucleosomes cannot spontaneously reform upon a return to their original concentration. Such conditions can therefore be used to test the stability of the H3-DNA NCP against dilution-driven dissociation. Starting with a 2.53-μM concentration of purified H3-DNA NCP, a series of dilutions was performed followed by reconcentration to permit visualization by native PAGE. As expected, the crosslinked H3-DNA NCP shows no detectable DNA dissociation even at the low nanomolar concentration (Fig. 4c). Reduction of the disulfide restores dilution-driven dissociation of NCP samples, demonstrating that prevention of DNA dissociation is critically dependent on the crosslinks between the H3 tails and the nucleosome DNA ends.
To further increase the stability of the NCP, we combined the two types of disulfide bridges to form a double crosslinked NCP (H2A-N38C/H3-DNA NCP). The dilution experiments yielded identical results for the H2A-N38C/H3-DNA NCP with respect to the stability against dilution-driven DNA dissociation (data not shown). The stability of the H2A-N38C/H3-DNA NCP was also assessed by lyophilization. Lyophilized H2A-N38C/H3-DNA NCP after resuspension showed no DNA dissociation as seen by native PAGE or compositional change as judged from SDS PAGE (not shown). By comparison, reduction of the H3-DNA crosslink prior to lyophilization resulted in the appearance of free DNA (Fig. 4d).
The H3-DNA crosslink has the potential to lock the nucleosome DNA to a single translational setting as well as suppress possible DNA stretching inside the nucleosome. When applied to the NCP crystallization, the resulting uniformity of DNA end-to-end contacts is expected to improve the diffraction quality of crystals. For this goal, we chose the DNA sequence of the well-positioned, yeast MET16 promoter nucleosome for which the dyad position was previously mapped to base-pair resolution by whole-genome chemical mapping [39]. When the H3-DNA crosslinking procedure was used for the assembly of MET16 NCP, the usual ensemble of multiple translational positions was suppressed. Reduction of the crosslinks with dithiothreitol (DTT) after assembly generated such a population (Fig. 4e). Crystals of the crosslinked MET16 NCP could be successfully grown and diffracted to 3.2-Å resolution (data not shown).
H3-DNA NCP structure
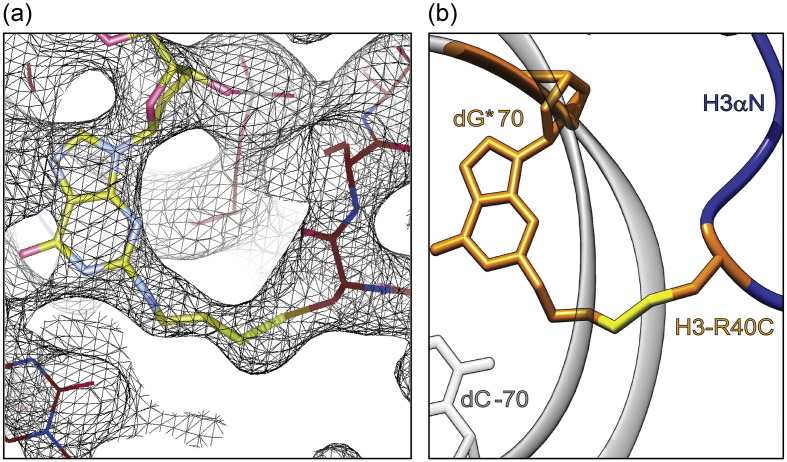
We determined the X-ray crystal structure of the H3-DNA NCP containing the disulfide between H3-R40C and dG* at position 70 to 2.8 Å (Table 1). Using the combination of limited X-ray exposure and crystal soaking in sodium nitrate as an effective free radical scavenger, the electron density of the intact disulfide could be visualized [40] (Fig. 5a). Both disulfide bridges between H3 and the NCP DNA are left-handed hooks, which occur rarely in protein structures [26] (Fig. 5b). Both disulfide χ3 angles are close to −90° indicating stable disulfide bonds. The side chain of H3-R40 in the ASP NCP structure points away from the position in the NCP DNA where the convertible nucleotide was placed [19]. Therefore, the backbone of the H3 N-terminal tail rearranges considerably to form the disulfide bond. Nevertheless, no significant distortion to the chain C-terminal to the crosslink is observed. The electron density for the H3 tail (1–38) N-terminal to the disulfide bridge is too weak to permit conformational analysis.
Fig. 5.
The disulfide crosslink in the H3-DNA NCP. (a) 2F0–FC electron density for the disulfide bond (black mesh, contour 0.7 × sigma) with the NCP structure superimposed. An H3-R40C substitution (red) bound to a convertible guanosine at DNA position 70 NCP (yellow) is highlighted. (b) Structure cartoon of the H3-DNA disulfide crosslink. The two DNA backbones (gray) show the location of the minor groove.
The structure of the H3-DNA NCP was compared to the 1.9-Å resolution ASP NCP [19]. The RMSD overall is less than 0.3 Å for both protein Cα and DNA C4’ atoms. In particular, the pseudo-continuous end-to-end-stacking of adjacent NCP in the crystal is not affected, as evidenced by the low RMSD of 0.64 Å for the terminal 7 bp of both strands.
H2A-N38C/H3-DNA NCP structure
The X-ray crystal structure of the H2A-N38C/H3-DNA NCP was also determined to 2.8 Å. The overall RMSD between the H2A-N38C/H3-DNA NCP and the 1.9-Å structure of the ASP NCP is less than 0.5 and 1 Å for the histone Cα and C4’ positions, respectively. The elevated RMSD values are also reflected in the larger B-factors of the H2A-N38C/H3-DNA NCP structure (Table 1). The H2A-N38C disulfide bridge in H2A-N38C/H3-DNA NCP aligns with the disulfide in the H2A-N38C NCP, both making a left-handed spiral on the dyad axis of the NCP.
Discussion
The dynamic properties of nucleosomes complicate their use in structural studies. In this work, we have engineered two types of site-specific disulfide crosslinks in order to stabilize the quaternary structure of the nucleosome. To improve crosslinking efficiency, we have modified histone production to include sulfitolysis and purification without urea. In fact, these techniques could be applied more generally to any cysteine-containing histone protein and are particularly important when cysteine reactivity is required, for example, in the production of semi-synthetic histones bearing chemical marks [41].
H2A-N38C octamers are a useful biochemical tool to avoid H2A/H2B dimer loss, and we demonstrate that they effectively suppress hexasome formation during nucleosome salt assembly. Since H2A/H2B dimer loss limits the overall thermodynamic stability of the nucleosome, it follows that H2A-N38C octamers should form more stable nucleosomes. In turn, the wrapping of DNA around the histone octamer is expected to sterically prevent dissociation of the crosslinked H2A/H2B dimers. Transient unwrapping of DNA from the nucleosome surface should also be more readily reversible as dimers are less likely to be lost into solution. Overall, the X-ray structure of the H2A-N38C-containing NCP is highly isomorphous with ASP NCP and therefore can be used as a direct replacement for most experiments.
A major difference between w.t. and H2A-N38C octamers occurs in the nucleosome salt-gradient assembly pathway at pH 7.5. Although the H2A-N38C/H2B dimers are only crosslinked to each other and not to the H3/H4 tetramer, H2A-N38C octamers are stable over the entire salt range encountered during nucleosome assembly, including 0.8 M NaCl where w.t. histones exist as a mixture of hexamer and free dimer. This salt concentration also corresponds roughly to the equilibrium midpoint for H2A/H2B dimer binding to tetrasomes during nucleosome assembly, in concordance with hexamers and hexasomes having the same salt-dependent interactions between H2A/H2B and H3/H4 [4], [13], [42]. In contrast, the H2A-N38C octamer likely binds DNA as a whole during salt-gradient nucleosome assembly.
The sensitivity of the dimer/tetramer interaction for both w.t. and H2A-N38C histone octamer in weakly acidic conditions indicates that the protonation state of the H4 histidine 75 imidazole group within the H2B/H4 four-helix bundle is important for assembly [29] (Fig. 2a). The disturbance of the positively charged side chain at this interface is evidently not compensated for by the H2A-N38C disulfide crosslink at the adjacent L1–L1 interface between H2A copies.
In vivo, nucleosome positions along genomic DNA depend most probably on the affinity of intact histone octamer over a length of 147 bp. The ISWI family of chromatin remodeling factors involved in DNA replication and gene repression facilitates the sampling of nucleosome positions through translocation of the octamer [31], [43]. In vitro, salt-gradient assembly of nucleosomes is biased by tetrasome formation prior to completion of the nucleosome through the addition of the H2A/H2B dimers [44]. This is reflected by the lack of conserved sequence features found over the dimer binding regions in sequences derived through SELEX, which is in stark contrast to natural nucleosomes which contain conserved sequence features over the dimer binding regions [39], [45]. Similarly, competition experiments used to measure the relative affinities of the histone octamer to different DNA sequences are established under elevated salt conditions, usually 1 M NaCl where the H2A/H2B dimers are unlikely to contribute [46], [47]. By comparison, the use of H2A-N38C octamers in these applications would yield nucleosome positions having a greater contribution from the H2A/H2B dimers and provide greater accuracy for the dependence of nucleosome positioning on DNA sequence.
Histone dimer ejection and variant exchange is an important part of the mechanism of remodeling factors such as SWI/SNF and INO80 [48]. In remodeling assays for these enzymes, H2A-N38C octamers could be used as a negative control by limiting octamer flexibility and inhibiting the exchange of histone dimers [10]. They could also possibly be used to trap intermediates of remodeling reactions. Furthermore, H2A-N38C octamers are likely locked in a left-handed ramp conformation and may contribute to an in vitro assay for the presence of speculative topological variants of nucleosomes, such as the reversome, and compositional variants, such as hemisomes [49]. The increased stability of H2A-N38C octamers may also facilitate studies involving histones from different species, histone variants and post-translationally modified histones.
The convertible nucleoside methodology was established to permit site-specific crosslinks between DNA-binding proteins and target DNA [50], [51], [52]. It has been successfully applied to stabilize protein–DNA complexes enabling crystallization. For example, the HIV reverse transcriptase was site-specifically crosslinked to a template DNA sequence using a convertible deoxyguanosine base [33]. We have applied this technique to stabilize the binding of DNA to the histone octamer in the NCP. The protocol for the production of crosslinkable DNA is adaptable to all nucleosome-forming DNA sequences. In addition to the convertible nucleotide, the use of type IIS restriction enzymes (e.g., BsaI) allows for the production of crosslinkable DNA without the need to alter the internal DNA sequence to accommodate a restriction site.
H3-DNA NCP exhibits complete suppression of DNA dissociation under conditions both of high salt and of nanomolar concentration. In this regard, crystallization of NCPs with bound nuclear factors may benefit from using H3-DNA crosslinking to permit exploration of a wider range of crystallization conditions including elevated salt conditions. Structural investigation of NCPs alone or in complex with factors by cryogenic electron microscopy may benefit from the increased stability of H3-DNA NCP to counteract surface tension forces and the effect of cryocooling on DNA twist during blotting and vitrification [53].
Most nucleosomes prepared using genomic DNA sequences are likely to adopt multiple translation settings, making them unsuitable for high-resolution structure determination, reminiscent of the inherent problem with mixed-sequence NCP [16]. As shown for the MET16 promoter NCP, H3-DNA crosslinking, however, provides a means to obtain a single setting and suppress DNA end-stretching. As a consequence, high-resolution crystals could be grown with evidently well-defined DNA end-to-end crystal contacts. Preparation of H3-DNA NCP with virtually any DNA sequence should be possible and would facilitate the structure solution of NCPs containing genomic sequences.
In summary, we have introduced two different site-specific crosslinks in the NCP and shown that the canonical NCP structure is undisturbed whether they are applied independently or together. All versions provide additional stability to the NCP and will enable further structural and biochemical studies on NCP and nucleosomes on their own and in combination with nuclear factors such as chromatin modifier and remodeling enzymes.
Materials and Methods
Histone preparation
Recombinant Xenopus laevis histone proteins were overexpressed in BL21 (DE3) pLysS Escherichia coli cells and purified from inclusion bodies with modifications to the established protocol [7]. Inclusion bodies for histones containing cysteine point mutants were solubilized in sulfitolysis buffer [20 mM Tris–HCl (pH 9.0), 6 M guanidine–HCl, 300 mM sodium sulfite, 60 mM sodium tetrathionate] for 24 h and 22 °C in the dark [27]. Denatured histones were purified by size-exclusion chromatography using a Sephacryl S-200 column (50 × 1000 mm; GE Healthcare) equilibrated in 20 mM sodium acetate (pH 5.2), 6 M guanidine–HCl, 200 mM NaCl, and 1 mM EDTA. Selected fractions were pooled, dialyzed extensively against water and lyophilized. Histones were redissolved in 2 M guanidine–HCl for purification by reversed-phase chromatography (TSK-Octadecyl-4PW, 21.5 × 150 mm; Tosoh Biosciences) and eluted with a 35%–65% gradient of buffer A (0.3% TFA, 5% acetonitrile) to buffer B (0.3% TFA, 90% acetonitrile). Selected fractions were pooled, dialyzed and lyophilized as 4-mg aliquots and stored at − 80 °C.
Histone octamer preparation
Histone octamer was assembled from individual histone proteins as previously described with modification [7]. Prior to Superdex 200 chromatography (16 × 600 mm; GE Healthcare), H2A-N38C histone octamer was crosslinked by dialysis into crosslinking buffer [20 mM Tris–HCl (pH 9.0), 2 M NaCl, 1 mM EDTA, 1 mM oxidized glutathione] for 24 h at 22 °C in the dark. This step was omitted for the H3-R40C and H2A-N38C/H3-R40C histone octamers, which were instead kept reduced with 1 mM DTT during size-exclusion chromatography. All octamers included the histone H3-C110A substitution so as not to interfere with disulfide formation.
For histone octamer dissociation studies, samples already purified by Superdex 200 chromatography were dialyzed into either 50 mM sodium acetate (pH 5.0) or 10 mM Tris–HCl (pH 7.5), and 0.4 M, 0.8 M or 2 M NaCl and 1 mM EDTA for 12 h. Each sample (1 mg) was injected onto a Superdex 200 column (10 × 300 mm; GE Healthcare). Fractions were analyzed by SDS PAGE (18% acrylamide). Histone band intensities were quantified by densitometry (Gel Logic, Carestream). Protein markers were Precision Plus Protein Standard (Bio-Rad).
NCP assembly and crosslinking
H2A-N38C NCP was assembled from equimolar amounts of histone octamer and 147-bp ASP as previously described, including heat shifting at 50 °C for 3 h [6], [7]. H3-DNA NCP contained modified ASP or MET16 DNA having a convertible dG* placed at position 70 from the dyad axis near the I- and J-strand 3′-ends. NCP was assembled by stepwise salt dialysis from a buffer containing 10 mM Tris–HCl (pH 7.5), 0.1 mM EDTA and 2 M KCl to a final buffer containing 10 mM KCl over 12 h at 4 °C [7]. Assembled NCP was incubated at 50 °C for 3 h during which the crosslink formation between the convertible nucleotide and the cysteine on H3 was facilitated by the addition of 4 mM sodium tetrathionate at pH 9.0. H3-DNA NCP was purified by anion-exchange chromatography (TSK-5PW-diethylaminoethyl; Tosoh Biosciences) using a salt gradient of buffer A [250 mM KCl, 20 mM Tris–HCl (pH 7.5) and 0.1 mM EDTA] to buffer B [600 mM KCl, 20 mM Tris–HCl (pH 7.5) and 0.1 mM EDTA]. H2A-N38C/H3-DNA NCP was crosslinked as for H3-DNA NCP. NCP characterization was done by mobility analyses using native PAGE (5% acrylamide). DNA markers were 2-Log DNA Ladder (New England Biolabs).
Crosslinkable DNA preparation
ASP was prepared as previously, but with an additional AvaII restriction site introduced 18 bp from the end of the nucleosome DNA [7]. Oligonucleotides containing a convertible dG nucleotide were synthesized (Microsynth, Balgach, Switzerland) with a 2-F-dI CE phosphoramidite at a selected location and with a phosphorylated 5′-end. Post-synthesis treatment with a 0.5 M cystamine solution converted the 2-F-dI to dG with a thioethyl linker at the N2 position. Subsequent treatment with 1 M 1,8-diazabicyclo[5.4.0] undec-7-ene and 32% v/v ammonia deprotected and cleaved the oligonucleotide from the solid support (The Glen Report 15.1, 2002). The modified oligonucleotide was annealed with a complementary oligonucleotide by heating to 95 °C and slow cooling over 5–6 h. The resulting double-stranded oligonucleotide had an AvaII compatible sticky end and an unphosphorylated blunt end and was purified by ion-exchange chromatography (5 ml HiTrap Q-HP; GE Healthcare) and gradient elution from 300 to 450 mM NaCl [10 mM Tris–HCl (pH 7.5), 0.1 mM EDTA]. Ligation to the digested core DNA and HinfI palindrome ligation were performed together for 16 h at 8 °C. The product was purified by ion-exchange purification as previously described [19]. The overall workflow of crosslinkable DNA production is shown in Fig. S2.
The MET16-promoter NCP DNA was cloned from synthetic DNA (Life Technologies). BsaI sites were introduced into the sequence 22 bp from the DNA ends. Multiple copies were generated by tandem repeat cloning [17]. Ligation of oligonucleotides with convertible nucleotides was carried out as for ASP.
NCP dilution assay
A 3-μM sample of NCP was prepared and split into 25-μl aliquots and TEK10 [10 mM Tris–HCl (pH 7.5), 1 mM EDTA, 10 mM KCl] added to each to provide a dilution series. The samples were incubated for 12 h at 4 °C and then reconcentrated to 25 μl using concentrators with 5 or 10 kDa MWCO (Amicon, Sartorius).
NCP lyophilization
A 60-μl sample of 4 μM H2A-N38C/H3-DNA NCP in TEK50 [10 mM Tris–HCl (pH 7.5), 1 mM EDTA, 50 mM KCl] and 5% trehalose was frozen in liquid nitrogen and lyophilized (Alpha 1–2 LD plus, Christ) for 16 h. For a second sample, the crosslinks were reduced by adding DTT to 20 mM and incubating for 1 h prior to freezing and lyophilization.
Crystal preparation
All NCP crystals were grown by vapor diffusion in sitting drops over the course of 1–4 weeks as previously described [19]. The wells were covered with argon gas before sealing and placed in a nitrogen atmosphere at 22 °C. Crystals were harvested and transferred stepwise into 37 mM MnCl2, 40 mM KCl, 20 mM potassium cacodylate (pH 6.0), 24% (v/v) 2-methyl-2,4-pentanediol and 2% (w/v) trehalose to improve diffraction quality and permit cryo-cooling in liquid propane at − 120 °C. The H3-DNA NCP crystals were additionally soaked in the same solution but also containing 0.5 M sodium nitrate for 4 min prior to cryo-cooling.
Crystallographic data collection
Data were measured at the Swiss Light Source synchrotron (Paul Scherrer Institute) on beamline X06DA under cryogenic conditions. A Pilates 6M-F detector was used at a X-ray wavelength of 1 Å. The H3-DNA and H2A-N38C/H3-DNA data sets were collected over multiple crystals with a total dose per crystal position of 10 MGy. Partial data sets were indexed and merged using XDS controlled by the XDShelper suite (T.J. Richmond, unpublished) [54].
Model refinement
X-ray structures were determined by molecular replacement using Phaser and the 1.9-Å resolution X-ray structure of the NCP (PDB ID 1KX5) [19], [55]. The H2A-N38C NCP was refined using CNS [56]. The H3-DNA and H2A-N38C/H3-DNA were refined using Phenix [57]. Model building was performed with Coot [58]. Structure alignments, RMSD calculations and figures were done using UCSF Chimera [59]. For alignments, only the H3/H4 chains were used in order not to bias the positions of the H2A/H2B dimer or DNA.
Accession numbers
Atomic coordinates and structure factors were deposited in the Protein Data Bank (www.pdb.org) and have accession numbers 5OMX (H2A-N38C NCP), 5ONG (H3-DNA NCP) and 5ONW (H2A-N38C/H3-DNA NCP).
Acknowledgments
Acknowledgments
We thank S. Duda for biochemical technical assistance and D. Sargent for X-ray data collection assistance. We are grateful for the support of the Swiss Light Source beamline scientists. T.D.F. was supported by the European Research Council (Advanced Grant Agreement No. 322778) awarded to T.J.R. P.D.B. was supported by the Louis-Jeantet Prize 2002 awarded to T.J.R.
Edited by Georg Schulz
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmb.2017.10.029.
Appendix A. Supplementary data
Supplementary figures
References
- 1.Kornberg R. Structure of chromatin. Annu. Rev. Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- 2.van Holde K. Springer; New York: 1989. Chromatin. [Google Scholar]
- 3.Arents G., Burlingame R., Wang B., Love W., Moudrianakis E. The nucleosomal core histone octamer at 3.1 Å resolution: a tripartite protein assembly and a left-handed superhelix. Proc. Natl. Acad. Sci. U. S. A. 1991;88:10148–10152. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luger K., Maeder A., Richmond R., Sargent D., Richmond T. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 5.Luger K., Rechsteiner T., Flaus A., Waye M., Richmond T. Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol. 1997;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- 6.Dyer P., Edayathumangalam R., White C., Bao Y., Chakravarthy S., Muthurajan U. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 7.Luger K., Rechsteiner T., Richmond T. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 8.Lowary P., Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 9.von Holt C., Brandt W., Greyling H., Lindsey G., Retief J., Rodrigues J. Isolation and characterization of histones. Methods Enzymol. 1989;170:431–523. doi: 10.1016/0076-6879(89)70061-6. [DOI] [PubMed] [Google Scholar]
- 10.Andrews A., Luger K. A coupled equilibrium approach to study nucleosome thermodynamics. Methods Enzymol. 2011;488:265–285. doi: 10.1016/B978-0-12-381268-1.00011-2. [DOI] [PubMed] [Google Scholar]
- 11.Andrews A., Luger K. Nucleosome structure(s) and stability: variations on a theme. Annu. Rev. Biophys. 2011;40:99–117. doi: 10.1146/annurev-biophys-042910-155329. [DOI] [PubMed] [Google Scholar]
- 12.Wilhelm F., Wilhelm M., Erard M., Duane M. Reconstitution of chromatin: assembly of the nucleosome. Nucleic Acids Res. 1978;5:505–521. doi: 10.1093/nar/5.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oohara I., Wada A. Spectroscopic studies on histone–DNA interactions. II. Three transitions in nucleosomes resolved by salt-titration. J. Mol. Biol. 1987;196:399–411. doi: 10.1016/0022-2836(87)90700-5. [DOI] [PubMed] [Google Scholar]
- 14.Levendosky R., Sabantsev A., Deindl S., Bowman G. The Chd1 chromatin remodeler shifts hexasomes unidirectionally. Elife. 2016;5 doi: 10.7554/eLife.21356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thastrom A., Bingham L., Widom J. Nucleosomal locations of dominant DNA sequence motifs for histone–DNA interactions and nucleosome positioning. J. Mol. Biol. 2004;338:695–709. doi: 10.1016/j.jmb.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 16.Richmond T., Finch J., Rushton B., Rhodes D., Klug A. Structure of the nucleosome core particle at 7 Å resolution. Nature. 1984;311:532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- 17.Richmond T., Searles M., Simpson R. Crystals of a nucleosome core particle containing defined sequence DNA. J. Mol. Biol. 1988;199:161–170. doi: 10.1016/0022-2836(88)90386-5. [DOI] [PubMed] [Google Scholar]
- 18.Vasudevan D., Chua E., Davey C. Crystal structures of nucleosome core particles containing the ‘601’ strong positioning sequence. J. Mol. Biol. 2010;403:1–10. doi: 10.1016/j.jmb.2010.08.039. [DOI] [PubMed] [Google Scholar]
- 19.Davey C., Sargent D., Luger K., Maeder A., Richmond T. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 20.Ong M., Richmond T., Davey C. DNA stretching and extreme kinking in the nucleosome core. J. Mol. Biol. 2007;368:1067–1074. doi: 10.1016/j.jmb.2007.02.062. [DOI] [PubMed] [Google Scholar]
- 21.Frouws T., Duda S., Richmond T. X-ray structure of the MMTV-A nucleosome core. Proc. Natl. Acad. Sci. U. S. A. 2016;113:1214–1219. doi: 10.1073/pnas.1524607113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flaus A., Luger K., Tan S., Richmond T. Mapping nucleosome position at single base-pair resolution by using site-directed hydroxyl radicals. Proc. Natl. Acad. Sci. U. S. A. 1996;93:1370–1375. doi: 10.1073/pnas.93.4.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorigo B., Schalch T., Bystricky K., Richmond T. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 24.Flaus A., Richmond T. Positioning and stability of nucleosomes on MMTV 3′LTR sequences. J. Mol. Biol. 1998;275:427–441. doi: 10.1006/jmbi.1997.1464. [DOI] [PubMed] [Google Scholar]
- 25.Richardson J. The anatomy and taxonomy of protein structure. Adv. Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt B., Ho L., Hogg P. Allosteric disulfide bonds. Biochemistry. 2006;45:7429–7433. doi: 10.1021/bi0603064. [DOI] [PubMed] [Google Scholar]
- 27.Nilsson J., Jonasson P., Sameulsson E., Stahl S., Uhlén M. Integrated production of human insulin and its C-peptide. J. Biotechnol. 1996;48:241–250. doi: 10.1016/0168-1656(96)01514-3. [DOI] [PubMed] [Google Scholar]
- 28.Gurley L., Spall W., Valdez J., Jackson P., Meyne J., Ray F. HPLC of Biological Macromolecules. Marcel Dekker, Inc.; New York: 1990. HPLC of Histones; pp. 529–570. [Google Scholar]
- 29.Eickbush T., Moudrianakis E. The histone core complex: an octamer assembled by two sets of protein–protein interactions. Biochemistry. 1978;17:4955–4964. doi: 10.1021/bi00616a016. [DOI] [PubMed] [Google Scholar]
- 30.Ruiz-Carrillo A., Jorcano J. An octamer of core histones in solution: central role of the H3–H4 tetramer in the self-assembly. Biochemistry. 1979;18:760–768. doi: 10.1021/bi00572a004. [DOI] [PubMed] [Google Scholar]
- 31.Yamada K., Frowus T., Angst B., Fitzgerald D., DeLuca C., Schimmele K. Structure and mechanism of the chromatin remodelling factor ISW1a. Nature. 2011;472:448–453. doi: 10.1038/nature09947. [DOI] [PubMed] [Google Scholar]
- 32.Hall M., Shungdrovsky A., Bai L., Fulbright R., Lis J., Wang M. High-resolution dynamic mapping of histone–DNA interactions in a nucleosome. Nat. Struct. Mol. Biol. 2009;16:124–129. doi: 10.1038/nsmb.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang H., Chopra R., Verdine G., Harrison S. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 34.Banerjee A., Yang W., Karplus M., Verdine G. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature. 2005;434:612–618. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- 35.Malecka K., Ho W., Marmorstein R. Crystal structure of a p53 core tetramer bound to DNA. Oncogene. 2009;28:325–333. doi: 10.1038/onc.2008.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Alten D., Erlanson D., Verdine G., Joshua-Tor L. A structural snapshot of base-pair opening in DNA. Proc. Natl. Acad. Sci. U. S. A. 1999;96:11809–11814. doi: 10.1073/pnas.96.21.11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ausio J., Seger D., Eisenberg H. Nucleosome core particle stability and conformational change. J. Mol. Biol. 1984;176:77–104. doi: 10.1016/0022-2836(84)90383-8. [DOI] [PubMed] [Google Scholar]
- 38.Thastrom A., Gottesfeld J., Luger K., Widom J. Histone–DNA binding free energy cannot be measured in dilution-driven dissociation experiments. Biochemistry. 2004;43:736–741. doi: 10.1021/bi0302043. [DOI] [PubMed] [Google Scholar]
- 39.Brogaard K., Xi L., Wang J., Widom J. A map of nucleosome positions in yeast at base-pair resolution. Nature. 2012;486:496–501. doi: 10.1038/nature11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De la Mora E., Carmichael I., Garman E.F. Effective scavenging at cryotemperatures: further increasing the dose tolerance of protein crystals. J. Synchrotron Radiat. 2011;18:346–357. doi: 10.1107/S0909049511007163. [DOI] [PubMed] [Google Scholar]
- 41.Chiang K., Jensen M., McGinty R., Muir T. A semisynthetic strategy to generate phosphorylated and acetylated histone H2B. Chembiochem. 2009;10:2182–2187. doi: 10.1002/cbic.200900238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burton D., Butler M., Hyde J., Phillips D., Skidmore C., Walker I. The interaction of core histones with DNA: equilibrium binding studies. Nucleic Acids Res. 1978;5:3643–3663. doi: 10.1093/nar/5.10.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fei J., Torigoe S.E., Brown C.R., Khuong M.T., Kassavetis G.A., Boeger H. The prenucleosome, a stable conformational isomer of the nucleosome. Genes Dev. 2015;29:2563–2575. doi: 10.1101/gad.272633.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rhodes D., Laskey R. Assembly of nucleosomes and chromatin in vitro. Methods Enzymol. 1989;170:575–585. doi: 10.1016/0076-6879(89)70065-3. [DOI] [PubMed] [Google Scholar]
- 45.Satchwell S., Drew H., Travers A. Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol. 1986;191:659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 46.Shrader T., Crothers D. Artificial nucleosome positioning sequences. Proc. Natl. Acad. Sci. U. S. A. 1989;86:7418–7422. doi: 10.1073/pnas.86.19.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shrader T., Crothers D. Effects of DNA sequence and histone–histone interactions on nucleosome placement. J. Mol. Biol. 1990;216:69–84. doi: 10.1016/S0022-2836(05)80061-0. [DOI] [PubMed] [Google Scholar]
- 48.Cairns B. Chromatin remodeling: insights and intrigue from single-molecule studies. Nat. Struct. Mol. Biol. 2007;14:989–996. doi: 10.1038/nsmb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lavelle C., Recouvreux P., Wong H., Bancaud A., Viovy J., Prunell A. Right-handed nucleosome: myth or reality? Cell. 2009;139:1217–1218. doi: 10.1016/j.cell.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 50.MacMillan A., Verdine G. Synthesis of functionally tethered oligodeoxynucleotides by the convertible nucleoside approach. J. Organomet. Chem. 1990;55:5931–5933. [Google Scholar]
- 51.Ferentz A., Verdine G. Disulfide-crosslinked oligonucleotides. J. Am. Chem. Soc. 1991;113:4000–4002. [Google Scholar]
- 52.MacMillan A., Verdine G. Engineering tethered DNA molecules by the convertible nucleoside approach. Tetrahedron. 1991;47:2603–2616. [Google Scholar]
- 53.Chua E., Vogirala V., Inian O., Wong A., Nordenskiöld L., Plitzko J. 3.9 Å structure of the nucleosome core particle determined by phase-plate cryo-EM. Nucleic Acids Res. 2016;44:8013–8019. doi: 10.1093/nar/gkw708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kabsch W. XDS. Acta Cryst D Biol. Cryst. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCoy A., Grosse-Kunstleve R., Adams P., Winn M., Storoni L., Read R. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brunger A., Adams P., Clore G., DeLano W., Gros P., Grosse-Kunstleve R. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Cryst. D Biol. Cryst. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 57.Adams P., Afonine P., Bunkoczi G., Chen V., Davis I., Echols N. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. D Biol. Cryst. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emsley P., Lohkamp B., Scott W., Cowtan K. Features and development of coot. Acta Cryst. D Biol. Cryst. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Petterson E., Goddard T.D., Huang C., Couch G., Greenblatt D., Meng E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures