Abstract
Objectives
During recent decades, the number of invasive fungal infections among immunosuppressed patients has increased significantly, whereas the number of effective systemic antifungal drugs remains low and unsatisfactory. The aim of this study was to characterize a novel antifungal compound, CW-8/haemofungin, which we previously identified in a screen for compounds affecting fungal cell wall integrity.
Methods
The in vitro characteristics of haemofungin were investigated by MIC evaluation against a panel of pathogenic and non-pathogenic fungi, bacteria and mammalian cells in culture. Haemofungin mode-of-action studies were performed by screening an Aspergillus nidulans overexpression genomic library for resistance-conferring plasmids and biochemical validation of the target. In vivo efficacy was tested in the Galleria mellonella and Drosophila melanogaster insect models of infection.
Results
We demonstrate that haemofungin causes swelling and lysis of growing fungal cells. It inhibits the growth of pathogenic Aspergillus, Candida, Fusarium and Rhizopus isolates at micromolar concentrations, while only weakly affecting the growth of mammalian cell lines. Genetic and biochemical analyses in A. nidulans and Aspergillus fumigatus indicate that haemofungin primarily inhibits ferrochelatase (HemH), the last enzyme in the haem biosynthetic pathway. Haemofungin was non-toxic and significantly reduced mortality rates of G. mellonella and D. melanogaster infected with A. fumigatus and Rhizopus oryzae, respectively.
Conclusions
Further development and in vivo validation of haemofungin is warranted.
Introduction
The incidence of life-threatening invasive fungal infections has risen significantly during the past 30 years.1,2 Most are caused by species of Cryptococcus, Candida and Aspergillus.3 It is estimated that invasive aspergillosis and candidiasis affect between 10% and 25% of all leukaemic and bone marrow transplant patients, with an alarmingly high mortality rate of ~50%.4,5 However, despite the growing needs, treatments for invasive fungal infections remain unsatisfactory, with existing classes of antifungals showing toxicity, narrow specificity, increasing resistance or limited formulation.6 Therefore, there is a pressing need to develop novel antifungals that inhibit fungus-specific targets such as the fungal cell wall.
We previously screened a diverse chemical library of 35000 drug-like molecules (ChemDiv, San Diego, CA, USA) to identify inhibitors of Aspergillus fumigatus growth.7 The resulting antifungal compounds were next tested for cell wall-damaging activity using the alcA-PKC mutant. Of these, one group, the CANBEFs, was described previously.7 In this report, we describe the detailed analysis of another compound identified in this screen, CW-8, which we named here haemofungin. It demonstrated cell wall-damaging properties, promising in vitro antifungal activity against a panel of pathogenic fungi and in vivo efficacy in insect models of fungal infection. Its mode of action, inhibition of haem biosynthesis, was elucidated.
Materials and methods
Strains and media
The strains used in this study are detailed in Table S1 (available as Supplementary data at JAC Online). Conidia were harvested in 0.2% (v/v) Tween 80, resuspended in double-distilled water (DDW) and counted with a haemocytometer. Moulds were grown in rich YAG medium containing 0.5% (w/v) yeast extract, 1% (w/v) glucose, 10 mM MgCl2, supplemented with 0.1% (v/v) trace elements solution, and 0.2% (v/v) vitamin mix or in defined minimal medium (MM) containing 70 mM NaNO3, 1% (w/v) glucose, 12 mM potassium phosphate pH 6.8, 4 mM MgSO4, 7 mM KCl and trace elements. For MM containing glycerol, glucose was replaced with 0.2% (v/v) glycerol. Complete medium (CM) was prepared by adding 0.1% yeast extract, 0.2% peptone and 0.1% tryptone (all w/v) to MM. Yeasts were grown in YPD rich medium composed of 1% (w/v) yeast extract, 2% (w/v) peptone and 2% glucose (w/v). Bacteria were grown in LB broth composed of 1% tryptone, 0.5% yeast extract and 1% NaCl (all w/v).
Pan-fungal and bacterial screen
The fungal strains listed in Table S1 were tested for susceptibility according to CLSI standard M27-A3 or M38-A2 protocols, respectively.8,9 Identification of cell wall-active antifungal compounds using the Aspergillus nidulans alcA-PKC mutant was performed as previously described.7
Cell culture
Hit compounds were assessed for toxicity to mammalian cells using the human cancer cell line A549 (ATCC CLL 185), derived from a human lung carcinoma, and mouse embryo fibroblast cell line NIH-3T3 (ATCC CRL-1658) as previously described.7 Cell viability was measured by the XTT assay kit (Biological Industries, Beit Haemek, Israel).
Microscopy and staining
The effect of the hit compounds on fungal ultrastructure was assessed by light and fluorescence microscopy after cell wall and vital staining as previously described.7 Colocalization studies were carried out by incubation of 8 h germinated conidia adherent on glass coverslips with 1 μM MitoTracker Green FM (Life Technologies) for 1 h and haemofungin (2 μM) for a further 15 min, both at room temperature. After two DDW washes, imaging was carried out on a Leica TCS SPF5 confocal microscope (excitation 488 nm, unmixing 500–650 nm and deconvolution).
Synergy chequerboard assay
Chequerboard tests were performed in standard 96-well plates (Costar; Corning, Corning, NY, USA) according to CLSI M38-A2 microdilution methodology.8
Screening an A. nidulans overexpression genomic library for resistance-conferring plasmids
A library of A. nidulans transformants containing a genomic library cloned into the multicopy non-integrating vector pRG3-AMA1 of A. nidulans10,11 was screened for resistant strains. We have used this method to successfully identify the cellular target of two antifungal drugs.12,13 Transformation was undertaken by protoplasting as previously described.14 Screening was performed in the presence of 4 μM haemofungin as described previously.7 Isolation of the resistance-conferring plasmids and identification of the resistance-conferring gene in the genomic DNA insert were undertaken as previously described.11
Northern blot analysis
Freshly harvested A. fumigatus conidia were grown for 20 h at 37°C with shaking in liquid medium (MM or CM). Haemofungin (5 μM) was added during the last 1 h. For northern blot analysis, RNA was isolated with TRI Reagent (Sigma) and peqGOLD Phase Trap (peqlab) reaction tubes; 10 μg of total RNA was separated in formaldehyde-containing agarose gels, blotted onto Hybond-N+ membranes (Amersham Biosciences) and hybridized with digoxigenin-labelled probes. Hybridization probes were amplified by PCR with primers listed in Table S2.
Measurement of protoporphyrin IX (PPIX) levels
Freshly harvested A. fumigatus conidia were grown for 20 h at 37°C with shaking in YAG liquid medium. Haemofungin (2 μM) was added during the last 2 h. Samples were flash-frozen in liquid N2, lyophilized overnight, ground to powder and dissolved in 1:1 methanol/DMSO. Chromatographic separation was carried out on an Agilent 1100 series HPLC fitted with a Phenomenex Gemini C18 110A (150×4.60 mm) 5 μm column. Next, 100 μL of sample in 1:1 methanol/DMSO was injected onto the column. The compound was eluted with a 20 min linear gradient (0–20 min) of 80% (0.1% TFA in H2O), 20% acetonitrile to 100% acetonitrile followed by a 5 min isocratic elution (20–25 min) with 100% acetonitrile and finally a return to the initial conditions (25–32 min). The flow rate was 1.0 mL/min and the porphyrins were detected with a diode array detector at 398 nm.
In vivo antifungal activity: Galleria mellonella model
Groups of 10 caterpillars of the greater wax moth G. mellonella in the final instar larval stage, weighing 250–330 mg, were employed in all assays. Larvae were infected by injecting 10 μL of saline containing 1×106 conidia of A. fumigatus strains Af293 or CEA10 into the haemocoel through the last proleg with a 50 μL Hamilton syringe. At 2 h post-infection, larvae were injected with 10 μL of saline containing haemofungin. Larval survival was assessed daily for up to 10 days post-treatment.
In vivo antifungal activity: Drosophila melanogaster model
Toll trans-heterozygotes (i.e. Tl−/− flies) were generated by crossing flies carrying a thermosensitive allele of Toll (Tlr632) with flies carrying a null allele of Toll (Tll-RXA). The assay was performed as previously described.7 Toll flies infected with Rhizopus oryzae were fed with fly food containing 14.4 mg/mL haemofungin for 7 days. Survival was assessed until day 8 after infection.
Results
Screening for cell wall-destabilizing antifungal compounds
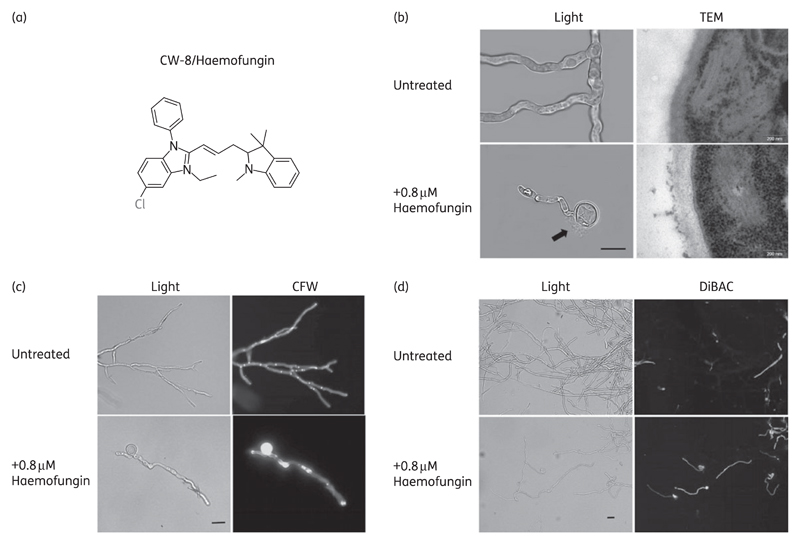
In a previously described screen for compounds with antifungal activity, we identified 16 ‘hit’ compounds that completely inhibited fungal germination and growth at a concentration of 25 μM.7 To identify potential cell wall-specific compounds, these ‘hits’ were further characterized in the A. nidulans alcA-PKC mutant. We have previously shown that this PKC-inducible strain exhibits hypersensitivity to cell wall-damaging compounds under repressing (MM-glucose) conditions, but not under inducing (MM-glycerol) conditions.15,16 One of the 16 compounds {CW-8/haemofungin; 5-chloro-3-ethyl-1-phenyl-2-[3-(1,3,3-trimethyl-2,3-dihydro-1H-indol-2-ylidene)prop-1-en-1-yl]-1H-1,3-benzodiazol-3-ium} (Figure 1a) displayed 4- and 8-fold hypersensitivity in MICs and minimum effective concentrations (MECs) under repressive conditions compared with inducing conditions for the A. nidulans alcA-PKC mutant (Table S3), supporting the conclusion that it affects the cell wall. Caspofungin, a known wall-perturbing drug, was used as a positive control and voriconazole as a negative control (no difference in MICs and MECs under repressing or inducing conditions). Haemofungin was therefore chosen for further analysis.
Figure 1.
Haemofungin causes morphological changes characteristic of damage to the cell wall of A. fumigatus. (a) Molecular structure of haemofungin, a benzimidazole derivative. (b–d) Freshly harvested spores of A. fumigatus strain Af293 were incubated for 24 h in the presence of 0.8 μM haemofungin and analysed microscopically by light, fluorescence or TEM microscopy, CFW cell wall staining or DiBAC staining of dead cells. Cell swelling and lysis [arrow, (b), light microscopy], abnormal cell wall morphology (TEM) and increased CFW staining of wall polysaccharides reveal haemofungin-induced wall damage. DiBAC staining indicates partial cell death in discrete areas of the hyphae. Black bar = 10 μm.
Haemofungin is active against most pathogenic fungi
Haemofungin was tested on a wide range of pathogenic fungal strains, mammalian cell lines and bacteria in culture (Table 1). Haemofungin was active against Candida spp. (3.13 μM<MIC<12.5 μM), A. fumigatus and Aspergillus niger (3.13 μM<MIC<6.25 μM) and Fusarium spp. (3.13 μM<MIC<12.5 μM) as well as most Rhizopus spp. (3.13 μM<MIC<12.5 μM). Haemofungin only markedly inhibited the proliferation of mammalian cells in culture at 25 μM (80% inhibition) and inhibited two of the four bacterial species tested, indicating that these compounds are not entirely fungus specific. The fungicidal activity of haemofungin was tested against A. fumigatus Af293, Candida albicans CBS 562 and Saccharomyces cerevisiae BY4741, respectively, exhibiting MFCs 2- to 4-fold higher than their MICs for these organisms.
Table 1.
Activity spectrum of haemofungin against fungi, bacteria and mammalian cell lines
| MIC (μM) | MEC (μM) | |
|---|---|---|
| C. albicans ATCC 2901 | 3.13 | 1.56 |
| C. albicans ATCC 90028 | 3.13 | 1.56 |
| C. albicans ATCC 18804/CBS 562 | 3.13 | 1.56 |
| Candida rugosa 3929 | 6.25 | 3.13 |
| Candida parapsilosis ATCC 22019 | 3.13 | 0.20 |
| Candida tropicalis ATCC 20336 | 12.50 | 1.56 |
| Candida glabrata 59343 | 6.25 | 0.78 |
| Candida krusei ATCC 6258 | 3.13 | 1.56 |
| R. oryzae 3465 | >12.50 | 3.13 |
| Rhizopus microspores 3484 | 3.13 | 0.78 |
| Rhizopus arrhizus 176 | 12.50 | 6.25 |
| Fusarium oxysporum 600711 | 12.50 | 0.20 |
| Fusarium solani 603251 | 3.13 | 0.10 |
| F. solani 600679 | 6.25 | 0.78 |
| A. fumigatus ATCC 13073 | 6.25 | 0.20 |
| A. fumigatus Af293 | 3.13 | 0.40 |
| Aspergillus flavus strain #1 | >12.50 | 0.20 |
| A. flavus strain #2 | >12.50 | 0.78 |
| A. niger strain #1 | 3.13 | 1.56 |
| A. niger strain #2 | 3.13 | 0.78 |
| A. niger strain #3 | 6.25 | 1.56 |
| S. cerevisiae A2 | 6.25 | — |
| S. cerevisiae BY4741 | 6.25 | — |
| Escherichia coli | >25 | — |
| Staphylococcus epidermidis | 0.78 | — |
| Staphylococcus aureus | >25 | — |
| Bacillus cereus | 3.13 | — |
| A549 | 25 (80%) | — |
| NIH-3T3 | 25 (80%) | — |
MIC= the lowest drug concentration to completely arrest germination and growth; MEC= the lowest drug concentration to cause visibly aberrant growth or a significant reduction in growth.
Haemofungin at a concentration of up to 25 μM did not induce sheep red blood cell haemolysis even after 24 h of incubation, indicating it does not function as a membrane-disrupting agent (data not shown). Haemofungin was highly active in various media, including MM, rich YPD or YAG, and defined cell-culture medium RPMI 1640 or DMEM, with or without 10% serum (data not shown).
Haemofungin causes morphological changes characteristic of damage to the cell wall of A. fumigatus
Further characterization of the effects of haemofungin on fungal structure was carried out on A. fumigatus strain Af293. The effect of haemofungin on cell wall polysaccharide deposition, ultrastructure and viability was determined by transmission electron microscopy (TEM), calcofluor white (CFW) staining and DiBAC staining of dead cells, respectively. After 24 h of growth in the presence of subinhibitory concentrations of haemofungin, the Af293 strain displayed abnormalities in cell wall ultrastructure, characterized by swelling and lysis of cell bodies and abnormal thickening and fragmentation of the outer cell wall (Figure 1b). Similar swelling was seen in S. cerevisiae yeast cells in the presence of MIC levels of haemofungin (data not shown). CFW chitin staining demonstrated that haemofungin increased the deposition of chitin in the cell wall of the swollen cell bodies and along the hyphae (Figure 1c). DiBAC vital staining revealed numerous dead (fluorescing) hyphal segments after haemofungin treatment, in contrast to the untreated control (Figure 1d).
Haemofungin and caspofungin interact synergistically
We hypothesized that the mode of action of haemofungin differs from that of existing antifungals and that consequently they might display beneficial synergy when combined. Therefore, haemofungin was tested for interaction with the antifungal drugs caspofungin (an inhibitor of glucan synthase), voriconazole (an inhibitor of ergosterol biosynthesis) and amphotericin B (a membrane-disrupting compound) and with the protein kinase C inhibitor staurosporine (which blocks the cell wall integrity pathway) by means of a chequerboard modification of the guidelines presented in CLSI document M38-A.17 Whereas there was no interaction between haemofungin combined with voriconazole, amphotericin B or staurosporine, there was a synergistic interaction between haemofungin and the cell wall inhibitor caspofungin (FICI = 0.27; Table S4). These results suggest that both haemofungin and caspofungin damage the cell wall, leading to synergy or alternatively that drug entry is enhanced when they are combined.
Overexpression of AN7752 in A. nidulans confers resistance to haemofungin
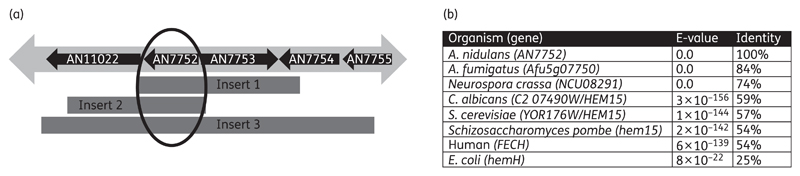
We screened an A. nidulans high-copy vector overexpression library plated under the selection of haemofungin at 4 μM (2× MIC). Eleven resistant colonies (MIC=8 μM) were identified and the multicopy library vector isolated from all of them. Retransformation of 3 of these 11 plasmids into haemofungin-susceptible A. nidulans resulted in resistance (MIC of haemofungin=8 μM compared with MIC=2 μM for a control A. nidulans strain transformed with the empty library vector). The insert in all three plasmids was sequenced and contained several ORFs; only one of them, AN7752, was shared by all three inserts (Figure 2a), identifying it as a likely candidate for being the gene responsible for resistance to haemofungin. Furthermore, transposon mapping revealed that disruption of AN7752 resulted in loss of resistance to haemofungin. AN7752 is an uncharacterized ORF in A. nidulans with many known orthologues in other fungi (Figure 2b). The S. cerevisiae orthologue is YOR176W/HEM15, a confirmed ferrochelatase, a mitochondrial enzyme catalysing the eighth and final step in the haem biosynthetic pathway.
Figure 2.
Mapping and identification of the A. nidulans AN7752 gene encoding HemH/ferrochelatase that confers haemofungin resistance upon overexpression. (a) Schematic representation of the inserts in each of the three resistance-conferring high-copy plasmids. Only the gene AN7752 is shared by all three plasmids. (b) Amino acid similarity between AN7752 and homologues of select fungal, human and Escherichia coli ferrochelatase proteins according to pBLAST.
Addition of haemin attenuates haemofungin-induced growth inhibition
We reasoned that if ferrochelatase is the primary target of haemofungin in fungal cells, addition of haemin, a downstream product of haemin biosynthesis, would block haemofungin-induced inhibition. In contrast, addition of PPIX, the ferrochelatase substrate, or aminolaevulinic acid (ALA), produced in the first step of haemin biosynthesis would not. We therefore tested the effect of adding increasing concentrations of haemin, PPIX or ALA to broth microdilutions of haemofungin in a 96-well plate with 5000 A. nidulans conidia per well (Table S5). Specifically, the addition of haemin rescued the growth of A. nidulans in the presence of haemofungin. Addition of 25 μg/mL haemin increased the haemofungin MIC 8-fold from 4 to 32 μM, whereas addition of PPIX or ALA did not. These results strengthen the hypothesis that haemofungin is an inhibitor of ferrochelatase activity. Similar results were seen with A. fumigatus (Af293), S. cerevisiae (BY4741) and C. albicans (CBS 562), suggesting that haemofungin works in a similar way in these fungi (data not shown). Further analyses described below were undertaken on A. fumigatus, because, unlike A. nidulans, it is a significant human fungal pathogen.
Haemofungin increases expression of key haem biosynthesis genes
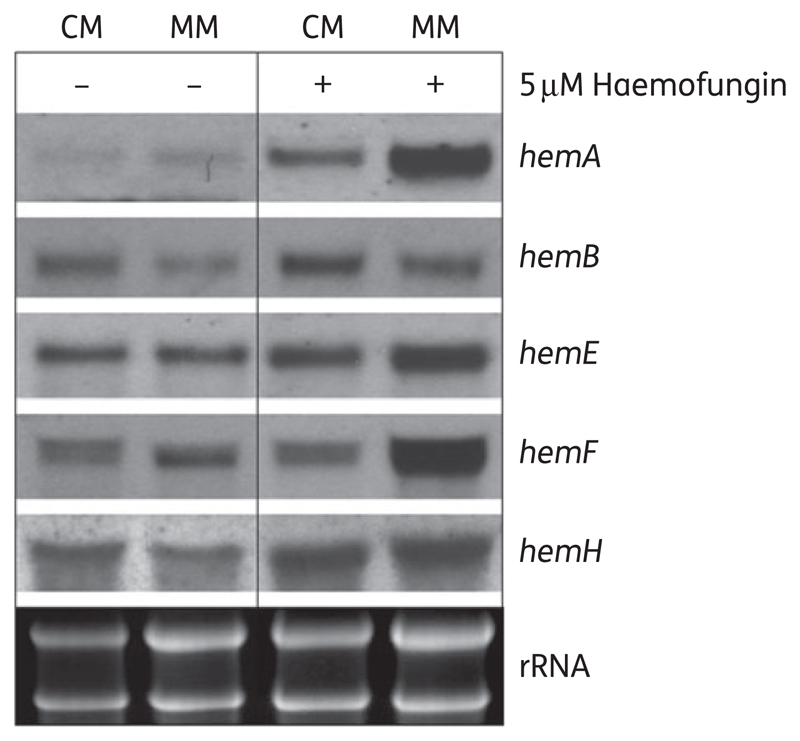
Fungal haem biosynthesis is a tightly coordinated process in which the end product haemin represses the expression of key genes in the pathway.18 We predicted that haemofungin treatment, by decreasing endogenous haem levels, would activate the transcription of genes in the fungal haem biosynthetic pathway. To test this, we grew A. fumigatus conidia overnight at 37°C in liquid culture (CM or MM) and then added 5 μM haemofungin for another 1 h of incubation. Total RNA was prepared and analysed by northern blotting for expression of hemA, hemB, hemE, hemF and hemH (Table S2). The results revealed increased levels of hemA, hemE and hemH mRNA in the presence of haemofungin in both CM and MM. hemF mRNA was only increased in the presence of haemofungin in MM. The most affected gene, hemA, encodes ALA synthase, the first committed and rate-limiting enzyme in haem biosynthesis.18 Together, these findings imply that a reduction in haem levels caused by haemofungin leads to the up-regulation of key haem biosynthetic genes (Figure 3).
Figure 3.
Haemofungin induces the expression of key haem biosynthetic genes in A. fumigatus. A. fumigatus overnight culture grown in liquid medium (either CM or MM) was treated with 5 μM haemofungin for 1 h. Total RNA was extracted and analysed by northern blotting with probes specific for the haem biosynthetic genes hemA, hemB, hemE, hemF and hemH. The expression of all genes except hemB was increased in the presence of haemofungin. Bottom panel: total RNA loading control.
Haemofungin causes accumulation of the precursor PPIX in A. fumigatus
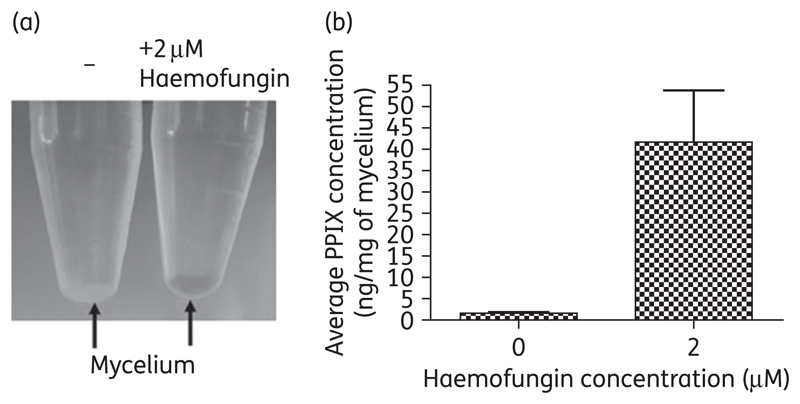
We anticipated that if haemofungin inhibits HemH/ferrochelatase activity, its presence would result in the accumulation of porphyrin precursors and in particular its substrate PPIX. To test this, we grew A. fumigatus conidia overnight at 37°C in liquid YAG and then added 2 μM haemofungin for another 2 h of incubation. The haemofungin-treated mycelium became gradually reddish orange, suggesting that it was accumulating porphyrins (Figure 4a). HPLC analysis of mycelial extracts showed significant (P = 0.03) accumulation of PPIX in the presence of haemofungin (Figure 4b). Despite repeated attempts, using either HPLC analysis or commercial haemin assay kits, we were unable to reliably measure the levels of fungal haemin. Therefore, we could not demonstrate a parallel reduction in haemin levels after haemofungin treatment.
Figure 4.
Treatment with haemofungin causes accumulation of PPIX in A. fumigatus. A. fumigatus overnight culture grown in YAG liquid medium was treated with 2 μM haemofungin for 2 h. The mycelium was (a) spun down and directly viewed for accumulation of porphyrins and (b) lyophilized and analysed for PPIX content by HPLC (C18) column separation.
Haemofungin localizes in the mitochondrial membranes of A. fumigatus
While analysing the effects of haemofungin on fungal cells by means of fluorescence microscopy, we discovered that haemofungin fluoresces brightly in discrete subcellular regions under blue light (488 nm). We suspected that the fluorescing areas coincided with mitochondrial localization. To test this, we analysed by confocal fluorescence microscopy A. fumigatus hyphae incubated in the presence of both haemofungin and the mitochondria-specific dye MitoTracker Green. We found that haemofungin and MitoTracker fluorescence colocalize (Figure S1). Interestingly, HemH, the putative target of haemofungin, is a transmembrane protein localized in the mitochondrial membrane.18 These results suggest that haemofungin specifically accumulates in the immediate vicinity of its target, HemH, possibly enhancing its efficacy.
Haemofungin reduces mortality in G. mellonella larvae infected with A. fumigatus and in D. melanogaster Tl flies infected with R. oryzae
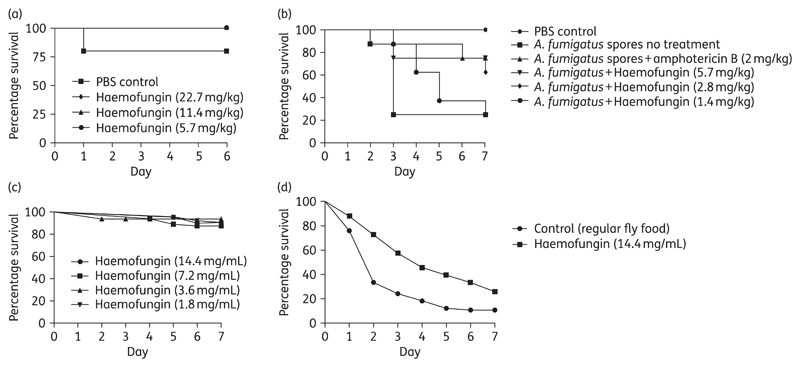
Toxicity of haemofungin injected into Galleria larvae indicated that it was non-toxic up to a concentration of 22.7 mg/kg (Figure 5a). To test the efficacy of haemofungin in treating invasive aspergillosis in Galleria larvae, groups of 10 larvae were infected with 5×106 A. fumigatus conidia per larva and, treated with 1.4, 2.8, 5.7, 11.4 or 22.7 mg/kg haemofungin or 2 mg/kg amphotericin B (positive control), or PBS for the untreated group. Treatment with amphotericin B at 2 mg/kg increased larval survival (P=0.03). The two highest doses of haemofungin (11.4 and 22.7 mg/kg) caused increased larval mortality (data not shown). Encouragingly, the efficacy of 5.7 mg/kg haemofungin was similar to that of the gold standard antifungal amphotericin B at 2 mg/kg (P=0.03). Haemofungin demonstrated a clear dose-dependent increase in efficacy as the dose was increased from 1.4 to 2.8 to 5.7 mg/kg (equivalent to 2.5, 5 and 10 μM, respectively) (Figure 5b).
Figure 5.
Toxicity and efficacy analysis of haemofungin. For analysis of toxicity, G. mellonella larvae (a) were injected once with up to 22.7 mg/kg haemofungin and Drosophila Tl−/− flies (c) were fed with fly food containing up to 14.4 mg/mL haemofungin for 7 days. For analysis of survival, G. mellonella larvae (b) and Drosophila Tl−/− flies (d) were infected with A. fumigatus (Af293) or R. oryzae conidia and treated with up to 5.7 mg/kg and 14.4 mg/mL haemofungin, respectively. Treatment with amphotericin B was used as a positive control in G. mellonella.
Haemofungin was not toxic to Toll flies fed food containing up to 14.4 mg/mL of the compound (Figure 5c). Haemofungin was effective in treating Toll flies infected with R. oryzae. Flies were infected with Rhizopus conidia and fed with fly food containing 14.4 mg/mL haemofungin or regular fly food for 7 days. Results showed a significant difference in survival (P=0.0002) between flies infected with 1×106 conidia/mL of Rhizopus and fed with haemofungin-containing fly food (14.4 mg/mL) versus flies infected with 1×106 conidia/mL of Rhizopus and fed with regular fly food (Figure 5d).
Discussion
In this report, we characterized the activity and mode of action of haemofungin, a novel antifungal compound we identified in a screen of 35000 synthetic drug-like molecules in the pathogenic mould A. fumigatus.7 Haemofungin has not been previously described in the scientific literature as having antifungal activity.
In vitro, haemofungin had potent inhibitory activity. It damaged the morphology of the fungal cell wall, causing cell body swelling and lysis and completely inhibited growth of most pathogenic yeast and moulds at low concentrations (3.1–12.5 μM). Importantly, it was non-toxic and highly active in vivo in two insect models of fungal infection.
To determine the mode of action of haemofungin, we used an overexpression screen in A. nidulans.12,13 High-copy number expression of AN7752, homologous to the essential S. cerevisiae protein HemH/ferrochelatase, confers 4-fold increased haemofungin resistance in A. nidulans. This protein catalyses the last step of haem biosynthesis, the insertion of ferrous iron (Fe2+) into PPIX and is located in the inner mitochondrial membrane with its active site facing the matrix space.18
Several more lines of evidence further supported the conclusion that haemofungin directly inhibits Hem15p/ferrochelatase activity: (i) addition of haem, the end product of the reaction, blocked the antifungal activity of haemofungin in A. nidulans, A. fumigatus, C. albicans and S. cerevisiae; (ii) the level of PPIX, the substrate of ferrochelatase, increased ~20-fold in haemofungin-treated A. fumigatus hyphae; (iii) the mRNA levels of the key A. fumigatus genes catalysing haem biosynthesis, hemA, hemE and hemH (the A. fumigatus homologue of S. cerevisiae HEM15), were increased in the presence of haemofungin, indicating that the cells were reacting to a shortage in available haemin by up-regulating the components of its biosynthetic pathway; and (iv) haemofungin specifically localized to the A. fumigatus mitochondrial membrane where HemH/ferrochelatase is found.
The ability to synthesize haemin is essential for cell survival. Haemin is a cofactor in the respiratory cytochromes a–c, the cytochrome P450 enzymes involved in sterol biosynthesis and in ligninases, peroxidases and catalases.18 In yeast, seven of the eight haem biosynthetic pathway genes (except HEM14) are essential. In A. niger, hemH is essential but growth can be partially restored by the addition of haemin. Interestingly, the hemH ferrochelatase null A. niger mutant is remarkably similar in phenotype to haemofungin-treated A. fumigatus, displaying both swollen cell bodies and PPIX accumulation.19
Previous attempts to develop antifungals targeting haemin biosynthesis have yielded the plant-derived alkaloid sampangine with broad, non-selective antiproliferative and antifungal activities, apparently by hyperactivating HEM4 and inhibiting haem biosynthesis.20 In contrast, haemofungin exerted its antifungal activity by inhibiting HemH/ferrochelatase, a target that to date has only been inhibited by non-specific chemicals such as 3,5-diethoxycarbonyl-1,4-dihydrocollidine.21 Cell death following inhibition of haem biosynthesis is primarily attributed the accumulation of porphyrin intermediates that are toxic.20 The cell wall damage, swelling and lysis that we observed after treatment with haemofungin may be a secondary effect due to porphyrin poisoning.
HemH/ferrochelatase is well conserved between fungi and humans with ~54% identity at the amino acid level, suggesting it is not an ideal antifungal target. However, this is not an insurmountable obstacle to further development of more selective inhibitors: fungal ERG11, the target of the antifungal azoles, shares ~40% identity with human lanosterol 14α-demethylase, but nevertheless, fungus-specific azoles have been developed. Higher specific for haemofungin-like compounds could be achieved by generating scaffold derivatives and identifying lead compounds with greater selectivity for fungal ferrochelatase. In its present form, haemofungin may prove useful as a research tool for inducing rapid changes in haemin flux in living cells. It should also be tested in cancer cells that synthesize haemin at higher levels due to their increased metabolic requirements.
In summary, we have identified and characterized a novel broad-spectrum antifungal compound, haemofungin, which interferes with the process of haem biosynthesis. Further characterization in mammalian models of infection is needed to determine its potential.
Supplementary data
Tables S1 to S5 and Figure S1 are available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/).
Funding
This study was supported by the Binational Science Foundation (BSF) 2011322 grant (to N. O. and D. P. K.). A.-M. D. and H. H. were supported by the Austrian Science Fund (FWF) grant P25978. A.-M. D. is an associate student of the HOROS doctoral programme.
Footnotes
Transparency declarations
None to declare.
References
- 1.Ben-Ami R, Lewis RE, Kontoyiannis DP. Invasive mould infections in the setting of hematopoietic cell transplantation: current trends and new challenges. Curr Opin Infect Dis. 2009;22:376–84. doi: 10.1097/QCO.0b013e32832db9f3. [DOI] [PubMed] [Google Scholar]
- 2.Yapar N. Epidemiology and risk factors for invasive candidiasis. Ther Clin Risk Manag. 2014;10:95–105. doi: 10.2147/TCRM.S40160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown GD, Denning DW, Gow NA, et al. Hidden killers: human fungal infections. Sci Transl Med. 2012;4:165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 4.Denning DW. Early diagnosis of invasive aspergillosis. Lancet. 2000;355:423–4. doi: 10.1016/S0140-6736(00)82003-6. [DOI] [PubMed] [Google Scholar]
- 5.Groll AH, Shah PM, Mentzel C, et al. Trends in the postmortem epidemiology of invasive fungal infections at a university hospital. J Infect. 1996;33:23–32. doi: 10.1016/s0163-4453(96)92700-0. [DOI] [PubMed] [Google Scholar]
- 6.Paiva JA, Pereira JM. New antifungal antibiotics. Curr Opin Infect Dis. 2013;26:168–74. doi: 10.1097/QCO.0b013e32835ebcb7. [DOI] [PubMed] [Google Scholar]
- 7.Mircus G, Albert N, Ben-Yaakov D, et al. Identification and characterization of a novel family of selective antifungal compounds (CANBEFs) that interfere with fungal protein synthesis. Antimicrob Agents Chemother. 2015;59:5631–40. doi: 10.1128/AAC.00850-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi—Second Edition: Approved Standard M38-A2. CLSI, Wayne; PA, USA: 2008. [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts—Third Edition: Approved Standard M27-A3. CLSI, Wayne; PA, USA: 2008. [Google Scholar]
- 10.Osherov N, Mathew J, May GS. Polarity-defective mutants of Aspergillus nidulans. Fungal Genet Biol. 2000;31:181–8. doi: 10.1006/fgbi.2000.1236. [DOI] [PubMed] [Google Scholar]
- 11.Osherov N, May G. Conidial germination in Aspergillus nidulans requires RAS signaling and protein synthesis. Genetics. 2000;155:647–56. doi: 10.1093/genetics/155.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu W, May GS, Lionakis MS, et al. Extra copies of the Aspergillus fumigatus squalene epoxidase gene confer resistance to terbinafine: genetic approach to studying gene dose-dependent resistance to antifungals in A. fumigatus. Antimicrob Agents Chemother. 2004;48:2490–6. doi: 10.1128/AAC.48.7.2490-2496.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osherov N, Kontoyiannis DP, Romans A, et al. Resistance to itraconazole in Aspergillus nidulans and Aspergillus fumigatus is conferred by extra copies of the A. nidulans P-450 14α-demethylase gene, pdmA. J Antimicrob Chemother. 2001;48:75–81. doi: 10.1093/jac/48.1.75. [DOI] [PubMed] [Google Scholar]
- 14.Osmani SA, May GS, Morris NR. Regulation of the mRNA levels of nimA, a gene required for the G2-M transition in Aspergillus nidulans. J Cell Biol Funding. 1987;104:1475–504. doi: 10.1083/jcb.104.6.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ronen R, Sharon H, Levdansky E, et al. The Aspergillus nidulans pkcA gene is involved in polarized growth, morphogenesis and maintenance of cell wall integrity. Curr Genet. 2007;51:321–9. doi: 10.1007/s00294-007-0129-y. [DOI] [PubMed] [Google Scholar]
- 16.Mircus G, Hagag S, Levdansky E, et al. Identification of novel cell wall destabilizing antifungal compounds using a conditional Aspergillus nidulans protein kinase C mutant. J Antimicrob Chemother. 2009;64:755–63. doi: 10.1093/jac/dkp270. [DOI] [PubMed] [Google Scholar]
- 17.Espinel-Ingroff A, Fothergill A, Ghannoum M, et al. Quality control and reference guidelines for CLSI broth microdilution susceptibility method (M38-A document) for amphotericin B, itraconazole, posaconazole, and voriconazole. J Clin Microbiol. 2005;43:5243–6. doi: 10.1128/JCM.43.10.5243-5246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franken AC, Lokman BC, Ram AF, et al. Heme biosynthesis and its regulation: towards understanding and improvement of heme biosynthesis in filamentous fungi. Appl Microbiol Biotechnol. 2011;91:447–60. doi: 10.1007/s00253-011-3391-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franken AC, Werner ER, Haas H, et al. The role of coproporphyrinogen III oxidase and ferrochelatase genes in heme biosynthesis and regulation in Aspergillus niger. Appl Microbiol Biotechnol. 2013;97:9773–85. doi: 10.1007/s00253-013-5274-2. [DOI] [PubMed] [Google Scholar]
- 20.Huang Z, Chen K, Xu T, et al. Sampangine inhibits heme biosynthesis in both yeast and human. Euk Cell. 2011;10:1536–44. doi: 10.1128/EC.05170-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brady AM, Lock EA. Inhibition of ferrochelatase and accumulation of porphyrins in mouse hepatocyte cultures exposed to porphyrinogenic chemicals. Arch Toxicol. 1992;66:175–81. doi: 10.1007/BF01974011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.