Abstract
Two key regulators of the switch to flower formation and of flower patterning in Arabidopsis are the plant-specific helix-turn-helix transcription factor LEAFY (LFY) and the MADS box transcription factor APETALA1 (AP1). The interactions between these two transcriptional regulators are complex. AP1 is both a direct target of LFY and can act in parallel with LFY. Available genetic and molecular evidence suggests that LFY and AP1 together orchestrate the switch to flower formation and early events during flower morphogenesis by altering transcriptional programs. However, very little is known about target genes regulated by both transcription factors. Here, we performed a meta-analysis of public datasets to identify genes that are likely to be regulated by both LFY and AP1. Our analyses uncovered known and novel direct LFY and AP1 targets with a role in the control of onset of flower formation. It also identified additional families of proteins and regulatory pathways that may be under transcriptional control by both transcription factors. In particular, several of these genes are linked to response to hormones, to transport and to development. Finally, we show that the gibberellin catabolism enzyme ELA1, which was recently shown to be important for the timing of the switch to flower formation, is positively feedback-regulated by AP1. Our study contributes to the elucidation of the regulatory network that leads to formation of a vital plant organ system, the flower.
Introduction
Optimal timing of the developmental phase transitions that culminate in the formation of flowers is critical for reproductive success, especially in monocarpic or annual plants. If monocarpic plants form flowers too late, they may not complete their lifecycle in the growing season (Roux et al. 2006, Anderson et al. 2011). If they form flowers too early, the accumulated resources cannot support formation of a large number of seeds, leading to poor competition in natural populations (Lunn et al. 2014). In many monocarpic plants including Arabidopsis thaliana, the switch to flower formation is biphasic (Hempel et al. 1997, 1998, Prusinkiewicz et al. 2007, Liu et al. 2013, Yamaguchi et al. 2014a). This is due to the fact that the first few new lateral primordia formed on the inflorescence after cessation of vegetative development are not competent to adopt a floral fate (Hempel et al. 1997). They instead give rise to branches subtended by cauline leaves. The degree of branching is important for optimal pollination and yield (Evers et al. 2011, Iwata et al. 2012). Primordia that form subsequent to the branch-producing prefloral inflorescence phase give rise to flowers. Once floral competence is acquired, there is no reversion to earlier primordial identities in Arabidopsis (Tooke et al. 2005, Wang et al. 2007, Melzer et al. 2008, Wagner and Meyerowitz 2011, Muller-Xing et al. 2014).
Many extrinsic and intrinsic cues direct cessation of vegetative development in Arabidopsis, which is also referred to as bolting. These cues include prolonged cold/vernalization, ambient temperature and photoperiod, age sensing, the autonomous pathway, carbohydrate signaling and hormonal cues such as gibberellin (GA) (Fornara et al. 2010, Pose et al. 2012, Song et al. 2013a, Wahl et al. 2013, Romera-Branchat et al. 2014, Verhage et al. 2014). Sensing of the stimuli occurs both in leaves and at the shoot apex. Their perception ultimately leads to accumulation of factors that promote the transition from vegetative development to formation of an inflorescence; these include the transcriptional co-regulator FLOWERING LOCUS T (FT) and the SQUAMOSA PROMOTER BINDING-LIKE (SPL) family of transcription factors (Cardon et al. 1997, Kardailsky et al. 1999, Corbesier et al. 2007, Jaeger and Wigge 2007, Schwarz et al. 2008, Wang et al. 2009, Bergonzi et al. 2013, Poethig 2013). The subsequent switch to flower formation is in large part controlled by the plant-specific transcription factor LEAFY (LFY) in Arabidopsis (Schultz and Haughn 1991, Weigel et al. 1992, Wagner et al. 1999). Recently, it was shown that the SPL transcription factor SPL9 and GA-sensitive DELLA proteins promote onset of flower formation synergistically with LFY (Yamaguchi et al. 2014a). Because GA levels increase prior to flower formation (Eriksson et al. 2006), the accumulation of DELLA proteins depends on GA catabolism. The reduction in GA levels is achieved at least in part by LFY directly inducing expression of the GA catabolism enzyme EUI-LIKE P450 A1 (ELA1) (Zhang et al. 2011, Yamaguchi et al. 2014a). The combined activity of LFY, DELLA and SPL proteins leads to strong upregulation of the MADS box transcription factor APETALA1 (AP1).
AP1 is a direct target of LFY (Parcy et al. 1998, Wagner et al. 1999). Unlike LFY, which is expressed prior to formation of the first flower in cauline leaf primordia that form during the prefloral inflorescence phase, AP1 expression is restricted to primordia that have committed to a floral fate (Mandel et al. 1992, Weigel et al. 1992). Consistent with this, photoinduction experiments revealed a significant temporal delay between LFY and AP1 upregulation (Hempel et al. 1997). The regulatory interactions that lead from LFY to AP1 induction are complex but increasingly well understood (Wagner et al. 1999, Saddic et al. 2006, Pastore et al. 2011, Yamaguchi et al. 2014a). Loss of LFY activity or AP1 activity prolongs the prefloral inflorescence phase (more cauline leaves and branches form), with more severe phenotypic defects observed in the lfy than ap1 null mutants (Weigel et al. 1992, Bowman et al. 1993). In addition, lfy ap1 double mutant plants display a very dramatic delay in the onset of flower formation, far exceeding that observed in lfy null mutants (Huala and Sussex 1992, Weigel et al. 1992). This finding has been attributed to the fact that AP1 can be induced independent of LFY, albeit later in development. The transcriptional co-activator FT plays a role in this process (Ruiz-Garcia et al. 1997, Abe et al. 2005, Wigge et al. 2005). Thus, AP1 acts not only downstream of LFY, but also in parallel with LFY. LFY and AP1 are co-expressed in young flower primordia from stage 1 onward (Mandel et al. 1992, Weigel et al. 1992). In addition to promoting onset of flower formation, LFY and AP1 also jointly regulate early events during flower morphogenesis including flower patterning (Bowman et al. 1993, Weigel and Meyerowitz 1993, Liu et al. 2009, Wu et al. 2012). However, thus far, only a handful of common LFY and AP1 target genes has been identified.
Here, we systematically identified shared LFY and AP1 targets with the aim to better understand the joint activities of the two transcription factors during early flower development. We identified several gene families and new pathways as likely directly regulated by both LFY and AP1. In addition, we show that the GA catabolism enzyme gene ELA1 is positively feedback-regulated by AP1.
Materials and methods
Identification of common target genes of LFY and AP1
Four datasets were used to identify genes bound by both LFY and AP1 (Kaufmann et al. 2010, Moyroud et al. 2011, Winter et al. 2011, Pajoro et al. 2014). The specific genotypes, developmental stages, number of peaks and number of genes identified in each of these are described in Table S1. Peak locations and assigned genes for the four datasets can be found in Tables S2 and S3.
LFY-bound genes were identified from the Moyroud et al. (2011) and Winter et al. (2011) datasets. ChIP-chip data (seedlings and inflorescences) from Winter et al. (2011) were remapped to the TAIR9/10 genome assembly by reanalyzing the CEL files (NCBI GSE28063) in CisGenome (version 2) (Ji et al. 2008) using the TAIR9 bpmap file. This resulted in 1725 seedling peaks and 1039 inflorescence peaks (False Discovery Rate (FDR) < 0.05). These were assigned to 1405 and 864 genes, respectively, using a custom Python script (Winter et al. 2011), modified to use TAIR10 annotations (as described in the TAIR10_GFF_genes.gff file, ftp://ftp.arabidopsis.org/). This script was used to assign all peaks from all LFY and AP1 datasets. The 1564 published LFY peak locations (FDR < 0.01) from Moyroud et al. (2011) were assigned to 1311 genes. The union of all LFY datasets resulted in 2460 LFY-bound genes.
AP1-bound genes were identified from the Kaufmann et al. (2010) and Pajoro et al. (2014) datasets. AP1 peak locations from Kaufmann et al. (2010) remapped to the TAIR9/10 genome assembly were downloaded from the PRI-CAT website (‘AP1_Replicate1_admin_BindingSites.csv’ and ‘AP1_Replicate2_admin_BindingSites.csv’; http://www.ab.wur.nl/pricat/quickload/A_thaliana_Jun_2009/ChIP-seq/) (Muino et al. 2011). The 3536 (FDR < 0.01) AP1 peaks from replicate 1 and the 6392 (FDR < 0.01) peaks from replicate 2 were assigned to 2411 and 4437 genes, respectively (Table S3). A gene was classified as bound by AP1 in the Kaufmann et al. (2010) dataset if it was bound by AP1 in both replicates. The 498, 956 and 1075 (FDR < 0.001) published peak locations from the 2-, 4- and 8-day induction experiments from Pajoro et al. (2014) (see Table S1) were assigned to 338, 647 and 714 genes, respectively. The union of the genes identified in all AP1 datasets resulted in 2389 AP1-bound genes.
Common LFY and AP1 targets (769 genes) were taken as the intersection of the two lists of LFY- and AP1-bound genes.
Identification of high-confidence LFY and AP1 target genes
Expression data from five datasets were used to identify high-confidence LFY and AP1 target genes (Schmid et al. 2003, 2005, Kaufmann et al. 2010, Moyroud et al. 2011, Winter et al. 2011, Pajoro et al. 2014). Genotypes, numbers of differentially expressed genes and data sources are described in detail in Table S4.
Published lists of differentially expressed genes were used for 35S:LFY-GR (Winter et al. 2011), 35S:AP1-GR (Kaufmann et al. 2010) and AP1:AP1-GR (Pajoro et al. 2014) (see Table S4). For the Kaufmann data, we filtered for the most significant probe and platform (Operon 4200, Operon 4000, or Agilent, Waltham, Massachusetts, USA) for each time point for display in Table S5. Schmid et al. (2003, 2005) raw data were analyzed using Bioconductor in R. Data were gcrma normalized (Wu et al. 2004). Non-specific filtering was performed by including only those genes that were classified as ‘Present’, using the MAS5.0 algorithm, in at least one of the arrays for a given comparison. Differentially expressed genes were identified using LIMMA (Smyth 2004). The following comparisons were tested: d7 vs d14, d14 vs d21 and d7 vs d21 (Schmid et al. 2005), and d0 vs d3, d3 vs d5, d5 vs d7 and d0 vs d7 (Schmid et al. 2003, 2005).
The 769 LFY- and AP1-bound genes were successively filtered using these expression data to identify high-confidence LFY and AP1 target genes (Tables S5 and S6).
GO term analysis
Enriched gene ontology (GO) terms were identified using ChipEnrich software (Orlando et al. 2009) (http://www.arexdb.org/software.jsp) using gene annotations downloaded from TAIR on August 28, 2014. The genes AT5G24910, AT4G23060 and AT3G55560 were manually added to the GO term ‘response to GA stimulus’. P-values were adjusted for multiple testing using the Benjamini–Hochberg method (Benjamini and Hochberg 1995) implemented in R.
Plant growth and treatment
All plants were grown on soil at 23°C in a 16 h light/8 h dark cycle. The following plant lines were previously described: lfy-1 null mutants (Weigel et al. 1992), ap1-10 (Schultz and Haughn 1993), ela1-1 (Yamaguchi et al. 2014a), ap1 cal 35S:LFY-GR (Winter et al. 2011) and ap1 cal 35S:AP1-GR (Wellmer et al. 2006). All mutants were in the Columbia background, while all inducible lines were in the Landsberg erecta background.
To test ELA1 induction upon dexamethasone activation of 35S:LFY-GR or 35S:AP1-GR, the synthetic steroid was dissolved in ethanol, and stored at −20°C. For mock treatment, 0.01% ethanol with 0.01% silwet L-77 was used. For steroid treatment, 0.01% ethanol with 0.01% silwet L-77 and 10 μM dexamethasone was used. Soil-grown ap1 cal 35S:LFY-GR or ap1 cal 35S:AP1-GR inflorescences were treated just after bolting by spraying.
Expression analyses
Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) was performed as previously described (Yamaguchi et al. 2009). Briefly, RNA was isolated from plants using TRIzol reagent (Invitrogen, Waltham, Massachusetts, USA) and purified using RNeasy Mini kits (Qiagen, Fitchburg, Wisconsin, USA). cDNA was synthesized using a Superscript III kit (Invitrogen).The resulting cDNA was quantified by Power SYBR Green PCR Master Mix and 7100 Real-time PCR System (Applied Biosystems, Waltham, Massachusetts, USA). Mean and SE of at least three technical replicates is shown. EIF4 was used as a loading control. Primers used were as follows: ELA1 primers: TCCGCGATGAAGTCTTTCTT and TTGTGTCCTCAAGGGCTTCT; CAL primers: CATTTCAACACCCCCATCTT and GCCGTTTGGTCTT CTTCTTG; LATE MERISTEM IDENTITY 2 (LMI2) primers: TTCAGGAATCTCGCTCCATT and CGCCACA GTAACCTCTTTCC; EIF4 primers: AAACTCAAT-GAAGTACTTGAGGGACA and TCTCAAAACCATAAG-CATAAATACCC.
In situ hybridization was performed as previously described (Wu and Wagner 2012). The ELA1 probe consisted of base pairs 112–1512 (Transcription start site (TSS) = 1). The probe was cloned into the pGEM-T Easy vector (Promega, Fitchburg, Wisconsin, USA). Antisense ELA1 probe was digested with NcoI and transcribed with the T7 polymerase using the Riboprobe Combination System (Promega) and DIG RNA labeling mix (Roche, Penzberg, Upper Bavaria, Germany).
Results
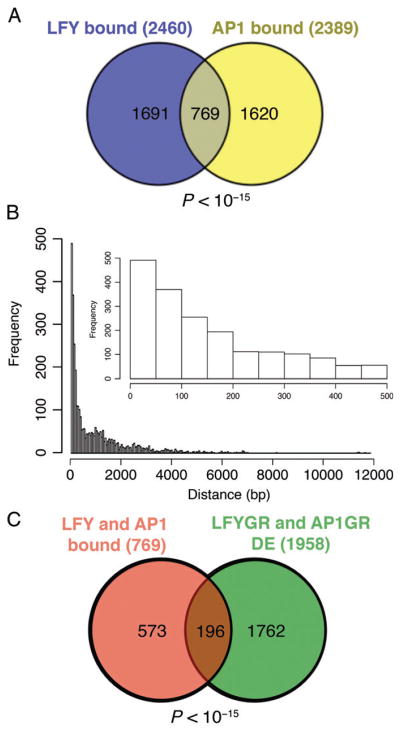
To better understand how LFY and AP1 together regulate early events in flower development, we computationally identified common direct targets of both transcription factors after reanalysis of published transcriptome and ChIP-seq or ChIP-chip datasets (Kaufmann et al. 2010, Winter et al. 2011, Moyroud et al. 2011, Pajoro et al. 2014) (Tables S1–S6, Fig. S1). We first identified genes whose regulatory regions were bound by both LFY and AP1. This analysis identified 769 genes as bound by both transcription factors, a highly significant enrichment (P-value < 10−15, χ2 test; Figs 1A and S1). In total, 31% of all LFY targets and 32% of all AP1 targets were bound by the other transcription factor (Fig. 1A). Moreover, the genomic regions occupied by LFY and AP1 were in close proximity to one another in their common target genes (Fig. 1B). More than 37% the AP1-binding sites were <200 bp away from an LFY-binding site. To discern among the 769 LFY- and AP1-bound genes those that are likely to be regulated by LFY or AP1, we next filtered for significant differential expression (FDR < 0.05) after LFY-GR or AP1-GR activation using the available public transcriptome datasets (Kaufmann et al. 2010, Winter et al. 2011, Pajoro et al. 2014) (see section Materials and methods for details). A total of 33% (249 of the 769 LFY- and AP1-bound genes) were differentially expressed upon LFY-GR activation (one developmental stage, 4 h activation; see Table S4). A total of 65% of the LFY- and AP1-bound genes (497 of the 769) were differentially expressed in at least one of the AP1-GR datasets (one developmental stage assayed, different treatment durations) (2 h to 8 days; see Table S4).
Fig. 1.
Identification of direct LFY and AP1 target genes. (A) Genes directly bound by either LFY (Moyroud et al. 2011, Winter et al. 2011) or AP1 (Kaufmann et al. 2010, Pajoro et al. 2014) or by both transcription factors. While LFY and AP1 have unique target genes, there is a significant subset of genes (769 genes; P-value < 10−15, χ2 test) whose regulatory regions are bound by both LFY and AP1. (B) Distance between AP1 peak summits and the nearest LFY-binding peak summit in the genes bound by both transcription factors. Inset: Close-up showing AP1-binding sites 500 bp or less away from LFY-binding sites. (C) Of the 769 genes bound by LFY and AP1, a significant subset (196 genes; P-value < 10−15, χ2 test) was differentially expressed (FDR < 0.01) in both LFY-GR and AP1-GR plants on the basis of public transcriptome data (Kaufmann et al. 2010, Winter et al. 2011, Pajoro et al. 2014).
Of the 769 genes bound by LFY and AP1, a total of 196 (26%) were differentially expressed after steroid activation of LFY-GR and of AP1-GR (P-value < 10−15, χ2 test; Fig. 1C, Table S5). To examine the likely functions of the 196 putative LFY and AP1 target genes, we tested for GO term enrichment among them using ChipEnrich (Orlando et al. 2009). Significantly enriched (FDR < 0.001) GO terms included ‘regulation of transcription’, ‘flower development’ and ‘stamen development’, as expected (Fig. 2). In addition, the GO terms ‘response to GA’, ‘response to auxin’, ‘response to jasmonic acid (JA)’ and ‘response to ethylene’ were enriched. ‘Auxin transport’ and ‘oligopeptide transport’ were also among the enriched GO terms, as was ‘phosphorylation’ and ‘polarity specification’. Auxin and GA have been implicated in the switch to flower formation in Arabidopsis (Yamaguchi et al. 2013, 2014a). Besides the previously described GA catabolism gene EUI-LIKE P450 A1 (ELA1) (Magome et al. 2013, Zhang et al. 2011, Yamaguchi et al. 2014a), another GA catabolism enzyme, GA2OX2 (Sun 2008), was linked to the ‘response to GA’ GO term. The genes linked to ‘response to auxin stimulus’ included regulators of the transcriptional response to auxin such as AUXIN RESPONSE FACTOR6 (ARF6) and IAA18 (Chapman and Estelle 2009), while ENHANCER OF PINOID (ENP) (Cheng et al. 2008, Furutani et al. 2014) and the ABCB1 and ABCB19 transporters (Titapiwatanakun et al. 2009, Cecchetti et al. 2015) were among the genes linked to the ‘auxin transport’ GO term. Ethylene and JA have been implicated in floral organ development and abscission (Kim 2014). Among the genes assigned to these GO terms were the transcriptional regulator JASMONATE ZIM-DOMAIN2 (JAZ2) (Chini et al. 2007) and ALLENE OXIDE CYCLASE3 (AOC3), which encodes an enzyme in the JA biosynthesis pathway (Gfeller et al. 2010). The GO term ‘regulation of polarity’ contained PINHEAD/ZWILLE (PNH) and ASYMMETRIC LEAVES1 (AS1), while TEOSINTE-LIKE1, CYCLOIDEA and PROLIFERATING CELL FACTOR1 transcription factors (TCP5, TCP10) and signaling components CORYNE (CRN) and ERECTA (ER) were linked to the ‘leaf development’ GO term.
Fig. 2.
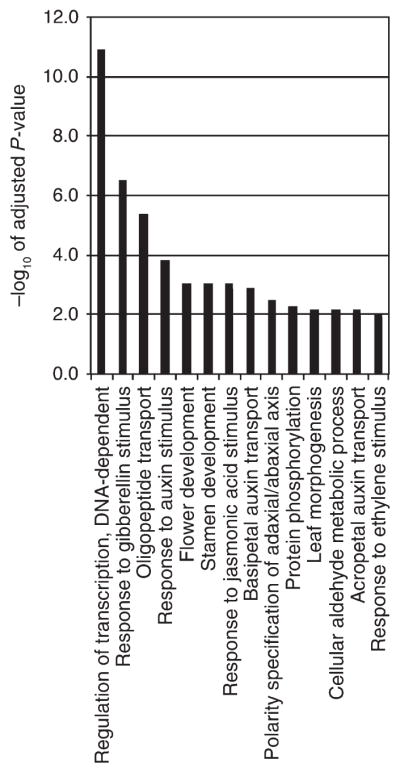
GO term enrichment in direct AP1 and LFY targets with a likely role in the meristem identity transition GO terms enriched among the 196 candidate LFY and AP1 target genes identified in Fig. 1C. For a list of these genes, see Table S5. An FDR correction was performed and FDR cutoff of less than 0.001% was implemented. −log10 P-values of all significant GO terms are shown.
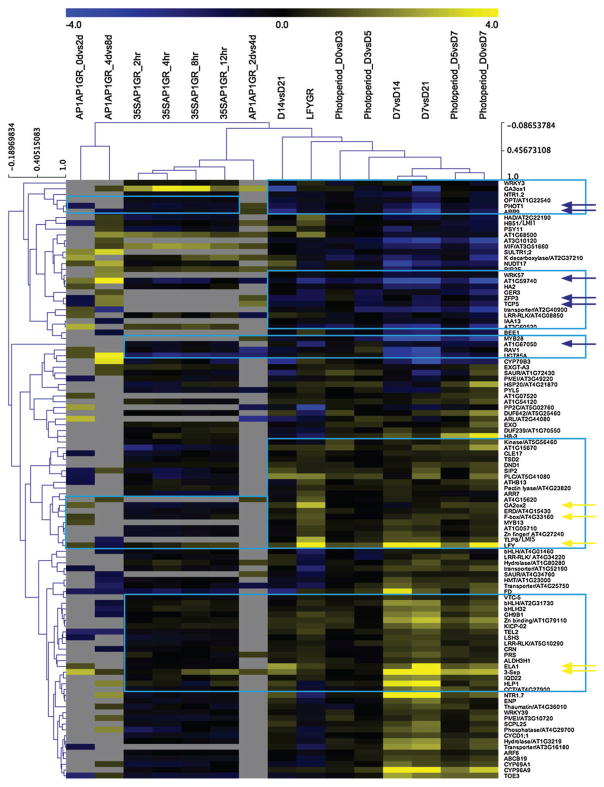
We next asked how many of the 196 candidate meristem identity/flower development genes were also significantly differentially expressed (FDR < 0.05) during the switch from vegetative to reproductive development using a publicly available shoot apex expression dataset that measured changes in gene expression between day 7, day 14 and day 21 of development (Schmid et al. 2005). Of the 196 genes, 168 genes were differentially expressed in at least one of the conditions tested (d7 vs d14, d14 vs d21 and d7 vs d21). Flower formation can also be triggered in Arabidopsis by shifting plants from non-inductive to inductive (long-day) photoperiods (Hempel et al. 1997, Schmid et al. 2003). To narrow down the list to high-confidence direct LFY and AP1 target genes, we next asked which of the 168 direct LFY and AP1 targets were also regulated by switch from non-inductive to inductive photoperiod (FDR < 0.05) using a public expression dataset (Schmid et al. 2003). Of the 168 genes, 104 genes were differentially expressed in at least one of the conditions tested (d0 vs d3, d3 vs d5, d5 vs d7 and d0 vs d7). Figure 3 shows a clustering heatmap of the expression changes observed for these 104 genes in all datasets. Two general trends are apparent. First, genes differentially expressed after steroid activation of LFY-GR co-cluster with those that change during development and during photoperiod. Second, the different datasets that probe gene expression changes upon AP1-GR activation clustered together. In addition, several groups of genes can be identified that show a coordinate regulation of gene expression (i.e. their expression increased in most conditions or it decreases in most conditions) (Fig. 3).
Fig. 3.
Clustering of direct AP1 and LFY targets with a likely role in the meristem identity transition. (A) Hierarchical clustering of direct LFY- and AP1-regulated genes. Heatmap displays log2 fold changes of the 104 direct LFY and AP1 targets differentially expressed in LFY-GR, AP1-GR, in response to photoperiod induction and during the switch to flower formation (day 7, day 14 and day 21) based on public transcriptome datasets (Schmid et al. 2003, 2005, Kaufmann et al. 2010, Winter et al. 2011, Pajoro et al. 2014). See Table S4 for a detailed description of data sources. Blue boxes highlight genes coordinately regulated over multiple datasets. Arrows highlight high-confidence direct LFY- and AP1-regulated genes (see Figs 4 and 5 below). Blue: downregulation, Yellow: upregulation. Clustering was performed using TMEV freeware (http://www.tm4.org/) using default values.
Consistent with this analysis, about one third of the 104 genes (34) changed in expression coordinately (FDR < 0.05) over all four conditions, with 21 genes upregulated and 13 genes downregulated significantly (FDR < 0.05) in at least one condition during development (d7 vs d14, d14 vs d21 or d7 vs d21), during photoperiod switch (d0 vs d3, d3 vs d5, d5 vs d7 or d0 vs d7) after activation of AP1-GR, and after activation of LFY-GR (see Table S5). To narrow the list of likely coordinately LFY- and AP1-regulated direct targets down further, we asked which of the 34 genes show an expression fold change of 1.5 or greater in all conditions. This identified a total of 11 genes as AP1- and LFY-regulated targets (Fig. 3 arrows, see Table S5). Five of these were coordinately upregulated by LFY and AP1. Among the upregulated genes are the known direct LFY and AP1 targets SEPALLATA3 (SEP3) and LFY, as expected (Kaufmann et al. 2010, Winter et al. 2011, Wu et al. 2012). SEP3 acts together with LFY during flower patterning (Liu et al. 2009, Kaufmann et al. 2010, Winter et al. 2011, Wu et al. 2012). LFY autoregulates and is positively feedback-regulated by AP1 (Liljegren et al. 1999, Kaufmann et al. 2010, Winter et al. 2011). In addition, a gene encoding a cytochrome P450 enzyme CYP714A1/EUI-LIKE P450 A1 (ELA1), a GA 2 oxidase (GA2OX2) and an F-box protein were among the upregulated LFY and AP1 targets. Both GA2OX2 and ELA1 have a role in GA catabolism (Zhu et al. 2006, Zhang et al. 2011, Nomura et al. 2013). The biological function of the F-box protein we identified (At4G33160) has not yet been elucidated. The UNUSUAL FLORAL ORGANS (UFO) F-box protein acts as an LFY co-factor for the activation of the class B floral homeotic genes (Levin and Meyerowitz 1995, Lee et al. 1997, Chae et al. 2008).
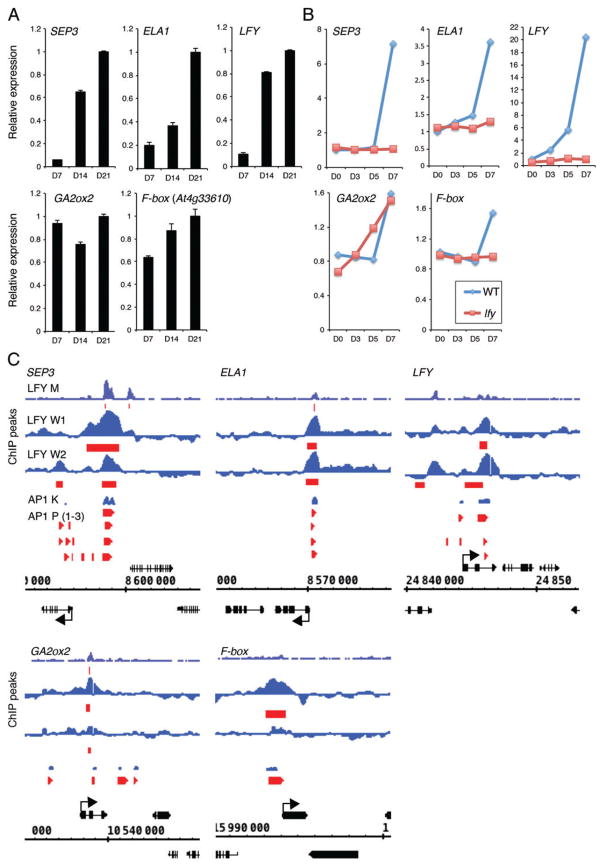
We next examined the expression of the five LFY and AP1 upregulated genes during development and photoinduction (Schmid et al. 2003, 2005) more closely. SEP3, ELA1 and LFY were strongly upregulated during the onset of reproductive development, while the F-box protein shows weaker upregulation and GA2OX2 appears to first decrease in levels (d7–d14) before it increases (d14–21) (Fig. 4A). In response to inductive photoperiod, all five genes are upregulated, with LFY and ELA1 accumulation already increasing at day 5, while all other genes did not increase until day 7 (Fig. 4B). In the lfy mutant, neither LFY activity nor AP1 RNA is present by day 7 (Fig. S2). This dataset hence provides an opportunity to assess whether upregulation by photoinduction requires these transcription factors. SEP3, ELA1 and the F-box protein were not induced by photoperiod in the lfy mutant, suggesting that LFY and AP1 are important for their upregulation in response to inductive photoperiod (Figs 4B and S2). LFY expression is slightly upregulated in the lfy mutant, as expected. The SOC1 and AGL24 transcription factors are thought to mediate photoperiod induction of LFY (Lee et al. 2008, Liu et al. 2008). GA2OX2 induction by long-day photoperiod was not at all impaired in the lfy mutant (Fig. 4B). Hence, other factors mediate photoperiod induction of GA2OX2. Finally, we examined LFY and AP1 binding to these five loci based on public ChIP-chip or ChIP-seq datasets (Kaufmann et al. 2010, Moyroud et al. 2011, Winter et al. 2011, Pajoro et al. 2014). LFY and AP1 bound to similar regions at these genes, in the 5′ intergenic region of the SEP3, ELA1 and the F-box protein loci and to the second intron of the LFY and GA2OX2 loci (Fig. 4C). In addition, sites uniquely bound by AP1 were present in LFY and GA2OX2 and a unique LFY-bound site was present at the LFY locus (Fig. 4C).
Fig. 4.
High-confidence direct LFY and AP1 upregulated targets during the switch to flower formation. (A) Expression of high-confidence direct LFY and AP1 target genes at different times of development and in different genetic backgrounds based on a public transcriptome dataset (Schmid et al. 2005). For criteria used to identify these genes, see Table S6. (B) Induction of high-confidence direct LFY and AP1 target genes by long-day photoperiod in wild-type or lfy mutant inflorescences based on a public transcriptome dataset (Schmid et al. 2003). (C) LFY and AP1 binding peaks to the regulatory regions of high-confidence direct LFY and AP1 target genes on the basis of chromatin immunoprecipitation. Screenshots were taken from the Integrated Genome Browser (IGB; http://bioviz.org/igb/) using available data for LFY [LFY M (Moyroud et al. 2011); LFY W1, W2 (Winter et al. 2011)]. Data for AP1 [AP1 K (Kaufmann et al. 2010)] were obtained through IGB (Nicol et al. 2009) using the PRI-CAT data source (http://www.ab.wur.nl/pricat/tutorial.html). AP1 K: replicate 1 data are displayed. AP1 P (1–3) (Pajoro et al. 2014): only significantly bound regions are displayed.
Among the six genes coordinately downregulated by LFY and AP1 was a gene encoding a putative oligopeptide transporter (OPT), the cytokinin-induced type-A authentic response regulator ARR9, the class II TCP transcription factor TCP5, the blue light receptor PHOTOTROPIN1 (PHOT1), a zinc finger transcription factor, ZINC FINGER PROTEIN3 (ZFP3) and a protein of unknown function (At1g67050) (Joseph et al. 2014). The OPT gene identified here has been assigned the name NFP4.3 for NITRATE TRANSPORTER PEPTIDE TRANSPORTER FAMILY, but the cargo of this transporter is unknown (Leran et al. 2014). ARR9 and TCP5 have both been linked to negative regulation of transcriptional response to cytokinin (Efroni et al. 2013, Schaller et al. 2015). PHOT1 has recently been linked to inflorescence phototropism (Kagawa et al. 2009). ZFP3 contains a single zinc finger protein and is expressed in flowers (Joseph et al. 2014). ZFP3 has recently been shown to be a negative regulator of ABA and a positive regulator of red light response (Joseph et al. 2014).
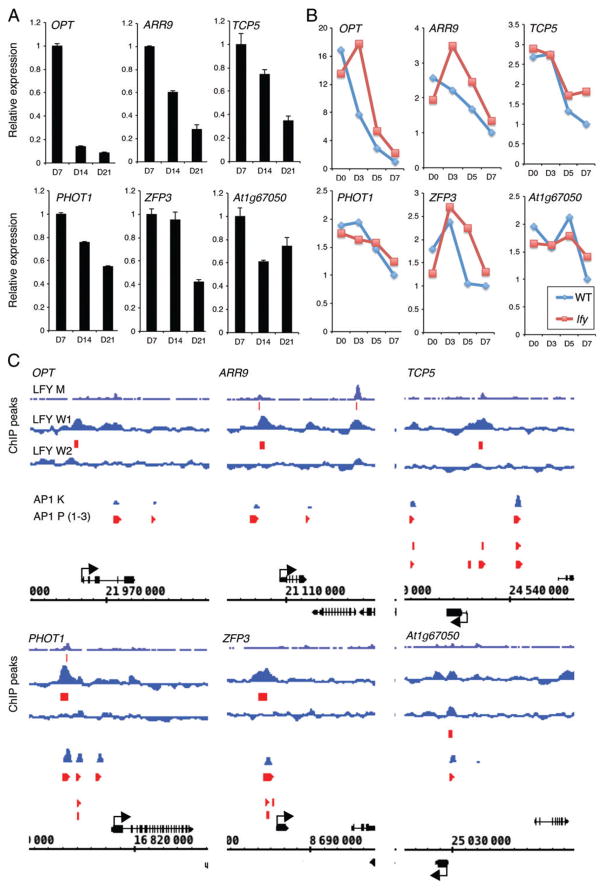
During development OPT, ARR9 and TCP5 are strongly downregulated (Fig. 5A). PHOT1 is slightly less dramatically downregulated, while repression of ZFP3 is only seen at the last time point (d21), and that of At1g67050 does not change further after d14 (Fig. 5A). In response to inductive photoperiod, OPT, ARR9, TCP5 and PHOT1 expression decreased (Fig. 5B). By contrast, ZFP3 expression first increased and then decreased, while that of At1g67050 decreased only late in the time course, between day 5 and day 7 (Fig. 5B). When we examined response to photoperiod in the lfy mutant, four genes showed elevated expression compared with the wild-type in at least one time point assayed; these were OPT, ARR9, TCP5 and ZFP3 (Fig. 5B). The remaining two genes did not require LFY and AP1 activity for response to photoperiod (Fig. 5B). Finally, we examined the LFY- and AP1-bound sites at all six loci. Common LFY- and AP1-bound regions upstream of ARR9, TCP5, PHOT1, ZFP3 and At1g67050 were identified (Fig. 5C). LFY bound to the 5′ intergenic region of OPT, while AP1 bound in the second intron and the 3′ intergenic region of this locus (Fig. 5C). Unique AP1- or LFY-bound sites were also present at four loci (ARR9, TCP5, PHOT1 and ZFP3) (Fig. 5C).
Fig. 5.
High-confidence direct LFY and AP1 downregulated targets during the switch to flower formation. (A) Expression of high-confidence direct LFY and AP1 target genes at different times of development and in different genetic backgrounds based on a public transcriptome dataset (Schmid et al. 2005). For criteria used to identify these genes, see Table S6. (B) Induction of high-confidence direct LFY and AP1 target genes by long-day photoperiod in wild-type or lfy mutant inflorescences based on a public transcriptome dataset (Schmid et al. 2003). (C) LFY and AP1 binding peaks to the regulatory regions of high-confidence direct LFY and AP1 target genes on the basis of chromatin immunoprecipitation. Screenshots were taken from the Integrated Genome Browser (IGB) using available data for LFY [LFY M (Moyroud et al. 2011); LFY W1, W2 (Winter et al. 2011)]. Data for AP1 [AP1 K (Kaufmann et al. 2010)] were obtained through IGB (Nicol et al. 2009) using the PRI-CAT data source. AP1 K: replicate 1 data are displayed. AP1 P (1–3) (Pajoro et al. 2014): only significantly bound regions are displayed.
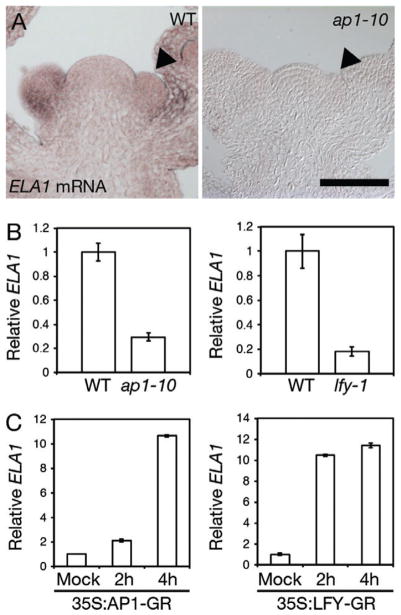
We recently reported that LFY directly activates expression of the gene encoding the GA catabolism enzyme ELA1 in inflorescences, causing a reduction in the level of bioactive GA in this tissue (Yamaguchi et al. 2014a). The reduced GA levels in turn allow a stronger upregulation of the direct LFY target AP1 and a more rapid switch to flower formation (Yamaguchi et al. 2014a). The current data suggest that AP1 also regulates ELA1 expression, perhaps as part of a positive feedback mechanism. To further investigate a role for AP1 in this pathway, we performed ELA1 in situ hybridization in wild-type and ap1 mutant inflorescences. ELA1 expression was much reduced in ap1 mutant relative to wild-type inflorescences (Fig. 6A). qRT-PCR confirmed the reduction in ELA1 expression in ap1 mutant relative to wild-type inflorescences at the time of flower formation and revealed the defect in ELA1 upregulation to be similar to that observed in lfy-1 null mutants (Fig. 6B). In addition, when we activated 35S:AP1-GR with dexamethasone, we saw an increase in ELA1 expression (Fig. 6C). The transcriptional upregulation of ELA1 upon AP1-GR activation was slower that observed after LFY-GR activation (Fig. 6C). This finding as well as prior observations that place ELA1 upstream of AP1 (Yamaguchi et al. 2014a) suggest that AP1 regulates ELA1 via positive feedback (see section Discussion for more details).
Fig. 6.
ELA1 expression is regulated by AP1. (A) In situ hybridization of ELA1 in 30-day-old wild-type and ap1-10 null mutant inflorescences. Arrows point to young flower primordia. (B) qRT-PCR-based expression of ELA1 in wild-type (WT) compared with lfy-1 null mutant or ap1-10 null mutant inflorescences shortly after the switch to flower formation (day 15). (C) qRT-PCR-based expression of ELA1 in 25-day-old ap1 cal 35S:LFY-GR or ap1 cal 35S:AP1-GR inflorescences 2 or 4 h after application of 10 μM dexamethasone relative to mock-treated inflorescences.
That ELA1 is a direct target of both LFY and AP1 further highlights the central role of this GA catabolism enzyme in the control of the onset of flower formation. We therefore hypothesized that reduced GA accumulation in the inflorescences may not only affect the accumulation of AP1 but also perhaps affect the expression of additional regulators of the switch to flower formation. To test this hypothesis, we examined whether the ela1-1 null mutant (Yamaguchi et al. 2014a) could enhance the delay in flower formation of a very strong/null ap1 mutant, ap1-10 (Schultz and Haughn 1993). It has previously been reported that neither loss of ELA1 function nor loss of AP1 function affect the cessation of vegetative development, consistent with the finding that neither gene is expressed during the transition from vegetative to prefloral inflorescence development (Mandel et al. 1992, Bowman et al. 1993, Yamaguchi et al. 2014a). Both AP1 and ELA1 promote cessation of the prefloral inflorescences phase (Mandel et al. 1992, Yamaguchi et al. 2014a) (Table 1). The duration of the prefloral, branch-forming, inflorescence phase can be estimated by counting the number of cauline leaves and secondary inflorescence branches formed. When we compared the number of cauline leaves and secondary inflorescences formed in ela1-1 ap1-10 double mutants with those in the parental lines or in the wild-type, we found that ela1-1 ap1-10 formed significantly more (P-value < 3 × 10−34, Student’s t-test) secondary inflorescences (Table 1). The number of cauline leaves was not significantly increased (P-value = 0.205, Student’s t-test). The data suggest that additional genes besides AP1 depend on low GA levels for proper regulation of expression at this important juncture in the plant life cycle.
Table 1.
Meristem identity phenotypes of ela1 mutants. Average number of cauline leaves and secondary inflorescences formed in the genotypes indicated. Mean ± SEM for one representative experiment is shown. All phenotypic experiments were performed multiple times and two-sided Student’s t-tests were performed for each experiment.
| Genotype | Cauline leaves | Secondary inflorescences |
|---|---|---|
| Wild-type (Col) | 2.8 ± 0.4 | 2.8 ± 0.4 |
| ela1-1 | 4.6 ± 0.1 | 4.6 ± 0.1 |
| ap1-10 | 3.4 ± 0.1 | 4.3 ± 0.1 |
| ap1-10 ela1-1 | 4.9 ± 0.1 | 11.3 ± 0.2*** |
P-value < 3 × 10−34.
Discussion
Here, we identify a large cohort of genes (196) as likely directly regulated by LFY and AP1 during flower development. The filtering criteria we employed for identification of these direct LFY- and AP1-regulated targets uncovered several known LFY targets including LMI1, LMI5/TLP8, RAX1, SEP3 and ELA1 (William et al. 2004, Saddic et al. 2006, Chahtane et al. 2013, Yamaguchi et al. 2014a), thus validating our approach. Our findings suggest that LFY and AP1 jointly regulate many processes in early flower development.
To identify these LFY and AP1 targets, we did not analyze gene expression changes in constitutive lfy and ap1 mutant compared with wild-type inflorescences (Schmid et al. 2005). The reason for this is that both ap1 and lfy mutant plants have dramatically different tissue composition from the wild-type (Schultz and Haughn 1991, Weigel et al. 1992, Bowman et al. 1993). These differences in tissue composition can affect gene expression more than the loss of transcription factor activity. A case in point, ap1 mutants form a large number of flower primordia relative to the wild-type and genes expressed in flower primordia, such as LFY, appear to exhibit increased expression when entire ap1 inflorescences are assayed by transcriptomic or qRT-PCR analyses. It is well known that AP1 positively feedback-regulates LFY expression (Bowman et al. 1993, Schultz and Haughn 1993, Liljegren et al. 1999). We focus here on changes in gene expression triggered by dexamethasone activation of fusion proteins of LFY or AP1 to the glucocorticoid receptor (Wagner et al. 1999, Wellmer et al. 2006, Pajoro et al. 2014) to identify LFY- and AP1-regulated genes. For a discussion of this approach, see also Yamaguchi et al. (2015). The strength of this approach is (1) that it compares gene expression in morphologically identical plants and (2) that rapid changes in gene expression can be observed (Kaufmann et al. 2010, Winter et al. 2011). Its weakness lies in the different experimental set-ups used by different investigators. To address this issue and the fact that both LFY and AP1 might be required to activate some common targets effectively (Wagner and Meyerowitz 2011), future investigations should focus on simultaneous activation of both fusion proteins in the same genetic background.
The GO term enrichment analysis suggests a prominent role for both LFY and AP1 in hormone response, in particular response to GA, auxin and JA. GA plays multiple roles in the developmental phase transitions to flower formation. In Arabidopsis, it promotes cessation of vegetative development to allow onset of the pre-floral inflorescence phase (Galvao et al. 2012, Osnato et al. 2012, Yu et al. 2012, Andres et al. 2014, Yamaguchi et al. 2014a). By contrast, GA represses the switch from the prefloral to the floral inflorescence phase (Yamaguchi et al. 2014a). Subsequent to this step, during flower development, GA levels increase again; this is required for the elaboration of several types of floral organs (Yu et al. 2004). The class C floral homeotic gene AGAMOUS may contribute to this increase in GA levels (Gomez-Mena et al. 2005). LFY and AP1 have both been directly linked to GA homeostasis (Kaufmann et al. 2010, Winter et al. 2011, Yamaguchi et al. 2014a). Among the 196 likely AP1- and LFY-regulated genes were six genes linked to GA biosynthesis (GA2OX1, GA2OX2, GA2OX4, GA20OX1, GA3OX1 and ELA1), one to GA sensing (GID1B) and one to GA response (IQD22) (Sun 2008, Zou and Sun 2011, Daviere and Achard 2013, Magome et al. 2013, Yamaguchi et al. 2014a). LFY and AP1 directly upregulate ELA1; this causes a reduction in GA levels that enables the switch to flower formation [this study, Yamaguchi et al. (2014a)]. Future studies will reveal whether AP1 and LFY also control GA homeostasis in developing flower primordia.
A link from LFY and AP1 to auxin response and auxin transport is not surprising given the previously described roles for auxin in the switch to flower formation and in floral organ initiation (Okada et al. 1991, Bennett et al. 1995, Cheng et al. 2008, Krizek 2011, Yamaguchi et al. 2013, 2014b). In addition, recent investigations have linked LFY to regulation of polar auxin transport (Li et al. 2013, Yamaguchi et al. 2014b). Auxin and auxin response have also been implicated in the development of both male and female reproductive floral organs and of the ovules (Wu et al. 2006, Shi and Yang 2011, Reeves et al. 2012, Galbiati et al. 2013, Larsson et al. 2013, Song et al. 2013b, Cardarelli and Cecchetti 2014, Hawkins and Liu 2014). Finally, auxin promotes flower maturation, a role it executes at least in part together with JA (Nagpal et al. 2005, Tabata et al. 2010, Rubio-Somoza and Weigel 2013, Wang et al. 2013). AP1 has not as yet been directly linked to auxin response or transport. Three genes with roles in auxin transport (ENP, ABCB1 and ABCB19) and seven genes with roles in auxin response (ARF6, ARF11, IAA13, IAA17, ARGOS-LIKE, SAUR/AT1G72430 and SAUR/AT4G34760) were among the direct LFY- and AP1-regulated genes. It will be interesting to investigate whether additional auxin-dependent pathways are controlled by LFY and AP1 in young flower primordia. Such a study would be enabled by generating conditional lfy ap1 double mutants at defined stages of flower development [see Wu et al. (2012) for an example of this approach]. Most of the roles described for JA occur late in Arabidopsis flower development, although earlier roles have been described in other plant species [see Ito et al. (2007), Acosta et al. (2009) and Cai et al. (2014) for examples].
Other new families of genes and pathways could be identified among the 196 genes possibly regulated by both LFY and AP1. For example, among the genes linked to onset of flower formation and flower development were known flowering repressors and activators (TEMPRANILLO1, FLOWERING LOCUS D and NF-YB2) (Abe et al. 2005, Wigge et al. 2005, Castillejo and Pelaz 2008, Cao et al. 2014). Likewise, three related genes, ALOG family transcription factors [LIGHT-DEPENDENT SHORT HYPOCOTYLS (LSH) 1, 2 and 3] were among the direct LFY and AP1 targets. The rice LSH-like protein TAWAWA1 (TAW1) and the tomato LSH-like protein TERMINATING FLOWER (TMF) shape the inflorescence architecture in each species, a role similar to that executed by LFY and AP1 in Arabidopsis (Liu et al. 2013, Yamaguchi et al. 2014a). In agreement with this idea, the regulator of axillary meristem initiation RAX1 (Muller et al. 2006) was among the direct LFY and AP1 targets we identified.
Several of the new high-confidence direct LFY and AP1 targets we identified warrant further investigation, primarily those whose induction or repression by long-day photoperiod is altered when LFY and AP1 activity are absent. These include two upregulated genes: the F-box protein and ELA1 (see below). The F-box protein is of interest, because LFY requires the F-box UFO as a co-factor during flower patterning in Arabidopsis (Levin and Meyerowitz 1995, Lee et al. 1997, Chae et al. 2008). In other plant species, in particular in the Solanaceae (tomato and petunia), LFY activity generally depends on F-Box proteins during additional stages and in additional pathways such as the switch to flower formation (Lippman et al. 2008, Souer et al. 2008). Besides the high-confidence target gene F-box (AT4G33160), the 196 LFY/AP1 targets contained another F-box protein (AT1G78100). It is possible that UFO is not required for all LFY activity in Arabidopsis because of the presence of other F-box proteins with overlapping function.
Among the downregulated LFY and AP1 direct high-confidence targets, three showed abnormal expression upon the shift to inductive photoperiod. OPT (At1g59740) is very strongly downregulated by photoperiod and is dependent on LFY and AP1 for this response. Three other transporters of this family were among the 196 LFY/AP1 targets: AT1g22540, AT1g52190 and At3g16180. The latter two have recently been implicated in nitrate transport (Hsu and Tsay 2013). It will be of interest to determine what peptides and other molecules are differentially transported in flowers or inflorescences. A second high-confidence direct LFY and AP1 downregulated gene that displays different response to photoperiod in the absence of LFY and AP1 activity is a negative regulator of cytokinin response, ARR9 (Schaller et al. 2015). The significance of this is not understood. Recent data have suggested extensive crosstalk between cytokinin and auxin during initiation of flower primordia (Besnard et al. 2014a, 2014b, Schaller et al. 2015) and in gynoecium development (Reyes-Olalde et al. 2013, Zuniga-Mayo et al. 2014). Cytokinin may be required for AP1 function (Han et al. 2014). Finally, ZFP3 is a Zn finger nuclear factor with a role in ABA signaling (Joseph et al. 2014). Interestingly, several other genes with a link to this pathway were among the 196 direct LFY- and AP1-regulated genes, including components of the ABA receptor (PYL5, PYL6 and PP2C/At5G41080) (Cutler et al. 2010, Raghavendra et al. 2010). Whether there is a link from this stress hormone to flower initiation or flower patterning by LFY and AP1 remains to be elucidated.
Common LFY- and AP1-regulated targets can theoretically be grouped into distinct categories. A first category of shared LFY and AP1 targets are those that act downstream of both LFY and AP1 and are transcriptionally regulated by both. There is precedent for this type of interaction. For example, the gene encoding the MADS box transcription factor SEP3 is directly induced by both LFY and AP1 (Liu et al. 2009, Kaufmann et al. 2010, Winter et al. 2011). This type of regulatory interaction, where one transcription factor activates another, and together they regulate a third gene, constitutes a feed-forward loop network motif (Alon 2007). Feed-forward loops are very common in developmental switches and help to buffer against noisy inputs (Alon 2007). Because LFY acts upstream of AP1 (Weigel and Nilsson 1995, Hempel et al. 1997, Liljegren et al. 1999, Wagner et al. 1999), a second theoretical category of shared LFY and AP1 targets is comprised of genes induced by LFY that are feedback-regulated by AP1, i.e. genes that act downstream of LFY, but upstream of AP1. Known examples of genes regulated in this manner are LMI1 and LMI2. Both LMI1 and LMI2 are activated by LFY, act upstream of AP1 and are positively feedback-regulated by AP1 (William et al. 2004, Saddic et al. 2006, Kaufmann et al. 2010). Positive feedback loops are also common network motifs in developmental switches and are known to drive the network equilibrium toward the next step; in this case, commitment to floral fate. A third category of direct LFY and AP1 are those that act upstream of both LFY and AP1 and are feedback-regulated by both genes. Candidate direct LFY and AP1 targets in this category are FLOWERING LOCUS D (this study) (Kaufmann et al. 2010, Winter et al. 2011, Jaeger et al. 2013) and TFL1. TFL1 is a direct target of LFY and of AP1 (Kaufmann et al. 2010, Moyroud et al. 2011, Winter et al. 2011), but was not identified here because of the stringent peak to gene assignment criteria used. Of note, we have focused here on genes coordinately regulated by both LFY and AP1 and 67% of the 196 direct LFY- and AP1-regulated targets behave in this manner (Table S6). In some cases (33% of the 196 targets), genes are regulated in the opposite manner by the two transcription factors (Table S6). For example, the opposite regulation could be due to genes activated by LFY that are subsequently or in different tissues repressed by AP1.
We do not have conclusive evidence for the newly identified LFY and AP1 targets acting downstream of LFY and AP1 or upstream and being feedback regulated. One indicator that can be used for a preliminary classification is the temporal upregulation of the identified genes relative to that of LFY or AP1 upon photoperiod shift (Schmid et al. 2003) (see Figs 4 and 5). Of the high-confidence target genes that are dependent on LFY and AP1 for correct photoperiod response, four (ELA1, OPT, ARR9 and ZFP3) respond prior to AP1 during photoperiod shift, suggesting that these genes might be feedback-regulated by AP1. In addition, all of these genes except ELA1 may also be feedback-regulated by LFY based on these criteria. By contrast, the F-box protein-encoding gene is not induced before AP1 and hence (like SEP3) is likely a downstream target of both LFY and AP1.
In the case of the GA catabolism enzyme ELA1, prior and current data suggest that it is indeed feedback-regulated by AP1. ELA1 is an important direct target of LFY in the control of onset of flower formation (Yamaguchi et al. 2014a). Several pieces of evidence suggest that ELA1 acts upstream of AP1: in ela1 mutants AP1 upregulation is delayed, AP1 expression is more strongly induced by LFY when GA levels are reduced and induction of ELA1 precedes that of AP1 not only upon photoperiod shift but also during development (Schmid et al. 2003, Yamaguchi et al. 2014a). In this study, we saw a more rapid upregulation of ELA1 upon LFY-GR than upon AP1-GR activation. This finding suggests that AP1 may first need to upregulate a co-factor such as LFY before it can induce ELA1 expression. Thus, both LFY and AP1 are required for ELA1 upregulation with LFY acting upstream and AP1 downstream of ELA1. The positive feedback from AP1 to ELA1 likely contributes to a more robust reduction in GA levels at the time when the first flower is formed. Together with the previously described positive feedback from AP1 to transcriptional regulators that act upstream of AP1 such as LFY, LMI2 and LMI1 (Saddic et al. 2006, Kaufmann et al. 2010, Pastore et al. 2011, Winter et al. 2011), this feedback would help tip the balance toward the next step in development (Ferrell 2012), here commitment to floral fate. Our data also fit well with the prior observation that AP1 is important for the plant’s ability to respond to LFY, when LFY activity or other signals that promote formation of flowers are limiting (Liljegren et al. 1999, Wagner and Meyerowitz 2011).
Finally, we provide genetic evidence that the reduced GA level triggered by ELA1 accumulation is important for AP1 accumulation. ela1 ap1 double mutants displayed a more significant delay in the onset of flower formation than either of the parental lines. One possible explanation for this finding is that additional regulators of the switch to flower formation besides AP1 are dependent on reduced GA levels for proper expression in incipient primordia. Future studies are needed to uncover additional roles for low GA during the switch to flower formation.
Supplementary Material
Identification of LFY- and AP1-bound genes.
Expression of AP1 in lfy mutants during photoperiod induction.
Identification of LFY- and AP1-bound genes. Individual LFY and AP1 ChIP-seq and ChIP-chip datasets and sources, and the number of genes identified in each are listed below. Unless noted, published peak locations were assigned to genes using a custom Python script (Winter et al. 2011), rewritten to use TAIR10 annotations. The total number of LFY-bound genes was taken as the union of the genes identified in the three LFY ChIP datasets. See Fig. S1A for a detailed overlap analysis of the three LFY datasets. The total number of AP1-bound genes was taken as the union of the four AP1 ChIP datasets. See Fig. S1B for a detailed overlap analysis of the four AP1 ChIP datasets. The 769 LFY- and AP1-bound genes are the intersects of the total LFY-bound genes and total AP1-bound genes (see Fig. 1A).
LFY peaks and gene assignments.
AP1 peaks and gene assignments.
Summary of LFY-GR, AP1-GR, development and photoperiod transcriptome datasets used for the identification of high-confidence LFY- and AP1-bound genes.
One hundred ninety six genes bound by LFY and AP1 and differentially expressed in LFY-GR and AP1-GR.
Filtering criteria used to identify high-confidence LFY and AP1 targets. The steps below were applied to successively filter the list of 769 LFY-and AP1-bound genes (see Table S1) to obtain a list of 11 high-confidence LFY and AP1 targets. See also Table S5 for genes and expression data. See Table S4 for more information regarding the transcriptome datasets employed.
Acknowledgments
We thank Jose Muino for information about the AP1 ChIP datasets. This work was supported by NSF grant IOS 1257111 to D. W., NIH Developmental Biology Training Grant T32-HD007516 and NIH Ruth L. Kirschstein NRSA F32 Fellowship GM106690-01 to C. M. W. and Japan Society for the Promotion of Science for Research Fellowship to N. Y.
Abbreviations
- AP1
APETALA1
- ARF
AUXIN RESPONSE FACTOR
- ARR9
cytokinin-induced type-A authentic response regulator 9
- ELA1
EUI-LIKE P450 A1
- ENP
ENHANCER OF PINOID
- FT
FLOWERING LOCUS T
- GA
gibberellin
- GO
gene ontology
- JA
jasmonic acid
- LFY
LEAFY
- LMI
LATE MERISTEM IDENTITY 1
- LSH
LIGHT-DEPENDENT SHORT HYPOCOTYLS
- OPT
oligopeptide transporter
- PHOT
PHOTOTROPIN1
- qRT-PCR
quantitative reverse transcriptase polymerase chain reaction
- SEP3
SEPALLATA3
- SPL
SQUAMOSA PROMOTER BINDING-LIKE
- UFO
UNUSUAL FLORAL ORGAN
- ZFP3
ZINC FINGER PROTEIN3
Footnotes
Author contributions
C. M. W., N. Y. and D. W. conceived of this study; C. M. W. performed the computational analyses; N. Y. performed the phenotypic and qRT-PCR analyses; M.-F. W. performed the in situ hybridization and D. W. wrote the manuscript with input from C. M. W. and N. Y.
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309:1052–1056. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- Acosta IF, Laparra H, Romero SP, Schmelz E, Hamberg M, Mottinger JP, Moreno MA, Dellaporta SL. tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science. 2009;323:262–265. doi: 10.1126/science.1164645. [DOI] [PubMed] [Google Scholar]
- Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- Anderson JT, Willis JH, Mitchell-Olds T. Evolutionary genetics of plant adaptation. Trends Genet. 2011;27:258–266. doi: 10.1016/j.tig.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres F, Porri A, Torti S, Mateos J, Romera-Branchat M, Garcia-Martinez JL, Fornara F, Gregis V, Kater MM, Coupland G. SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition. Proc Natl Acad Sci USA. 2014;111:E2760–E2769. doi: 10.1073/pnas.1409567111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate – a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- Bennett SRM, Alvarez J, Bossinger G, Smyth DR. Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J. 1995;8:505–520. [Google Scholar]
- Bergonzi S, Albani MC, Loren V, van Themaat E, Nordstrom KJ, Wang R, Schneeberger K, Moerland PD, Coupland G. Mechanisms of age-dependent response to winter temperature in perennial flowering of Arabis alpina. Science. 2013;340:1094–1097. doi: 10.1126/science.1234116. [DOI] [PubMed] [Google Scholar]
- Besnard F, Refahi Y, Morin V, Marteaux B, Brunoud G, Chambrier P, Rozier F, Mirabet V, Legrand J, Laine S, Thevenon E, Farcot E, Cellier C, Das P, Bishopp A, Dumas R, Parcy F, Helariutta Y, Boudaoud A, Godin C, Traas J, Guedon Y, Vernoux T. Cytokinin signalling inhibitory fields provide robustness to phyllotaxis. Nature. 2014a;505:417–421. doi: 10.1038/nature12791. [DOI] [PubMed] [Google Scholar]
- Besnard F, Rozier F, Vernoux T. The AHP6 cytokinin signaling inhibitor mediates an auxin-cytokinin crosstalk that regulates the timing of organ initiation at the shoot apical meristem. Plant Signal Behav. 2014b;9:e28788. doi: 10.4161/psb.28788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR. Control of flower development in Arabidopsis thaliana by Apetala1 and interacting genes. Development. 1993;119:721–743. [Google Scholar]
- Cai Q, Yuan Z, Chen M, Yin C, Luo Z, Zhao X, Liang W, Hu J, Zhang D. Jasmonic acid regulates spikelet development in rice. Nat Commun. 2014;5:3476. doi: 10.1038/ncomms4476. [DOI] [PubMed] [Google Scholar]
- Cao S, Kumimoto RW, Gnesutta N, Calogero AM, Mantovani R, Holt BF., 3rd A distal CCAAT/NUCLEAR FACTOR Y complex promotes chromatin looping at the FLOWERING LOCUS T promoter and regulates the timing of flowering in Arabidopsis. Plant Cell. 2014;26:1009–1017. doi: 10.1105/tpc.113.120352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardarelli M, Cecchetti V. Auxin polar transport in stamen formation and development: how many actors? Front Plant Sci. 2014;5:333. doi: 10.3389/fpls.2014.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon GH, Hohmann S, Nettesheim K, Saedler H, Huijser P. Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: a novel gene involved in the floral transition. Plant J. 1997;12:367–377. doi: 10.1046/j.1365-313x.1997.12020367.x. [DOI] [PubMed] [Google Scholar]
- Castillejo C, Pelaz S. The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr Biol. 2008;18:1338–1343. doi: 10.1016/j.cub.2008.07.075. [DOI] [PubMed] [Google Scholar]
- Cecchetti V, Brunetti P, Napoli N, Fattorini L, Altamura MM, Costantino P, Cardarelli M. ABCB1 and ABCB19 auxin transporters have synergistic effects on early and late Arabidopsis anther development. J Integr Plant Biol. 2015 doi: 10.1111/jipb.12332. [DOI] [PubMed] [Google Scholar]
- Chae E, Tan QK, Hill TA, Irish VF. An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development. Development. 2008;135:1235–1245. doi: 10.1242/dev.015842. [DOI] [PubMed] [Google Scholar]
- Chahtane H, Vachon G, Le Masson M, Thevenon E, Perigon S, Mihajlovic N, Kalinina A, Michard R, Moyroud E, Monniaux M, Sayou C, Grbic V, Parcy F, Tichtinsky G. A variant of LEAFY reveals its capacity to stimulate meristem development by inducing RAX1. Plant J. 2013;74:678–689. doi: 10.1111/tpj.12156. [DOI] [PubMed] [Google Scholar]
- Chapman EJ, Estelle M. Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet. 2009;43:265–285. doi: 10.1146/annurev-genet-102108-134148. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Qin G, Dai X, Zhao Y. NPY genes and AGC kinases define two key steps in auxin-mediated organogenesis in Arabidopsis. Proc Natl Acad Sci USA. 2008;105:21017–21022. doi: 10.1073/pnas.0809761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernandez G, Adie B, Chico JM, Lorenzo O, Garcia-Casado G, Lopez-Vidriero I, Lozano FM, Ponce MR, Micol JL, Solano R. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, Coupland G. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- Daviere JM, Achard P. Gibberellin signaling in plants. Development. 2013;140:1147–1151. doi: 10.1242/dev.087650. [DOI] [PubMed] [Google Scholar]
- Efroni I, Han SK, Kim HJ, Wu MF, Sang Y, Steiner E, Hong JC, Eshed Y, Wagner D. Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev Cell. 2013;24:438–445. doi: 10.1016/j.devcel.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S, Bohlenius H, Moritz T, Nilsson O. GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell. 2006;18:2172–2181. doi: 10.1105/tpc.106.042317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers JB, van der Krol AR, Vos J, Struik PC. Understanding shoot branching by modelling form and function. Trends Plant Sci. 2011;16:464–467. doi: 10.1016/j.tplants.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Ferrell JE., Jr Bistability, bifurcations, and Waddington’s epigenetic landscape. Curr Biol. 2012;22:R458–R466. doi: 10.1016/j.cub.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F, de Montaigu A, Coupland G. SnapShot: control of flowering in Arabidopsis. Cell. 2010;141:550550e1–2. doi: 10.1016/j.cell.2010.04.024. [DOI] [PubMed] [Google Scholar]
- Furutani M, Nakano Y, Tasaka M. MAB4-induced auxin sink generates local auxin gradients in Arabidopsis organ formation. Proc Natl Acad Sci USA. 2014;111:1198–1203. doi: 10.1073/pnas.1316109111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbiati F, Sinha Roy D, Simonini S, Cucinotta M, Ceccato L, Cuesta C, Simaskova M, Benkova E, Kamiuchi Y, Aida M, Weijers D, Simon R, Masiero S, Colombo L. An integrative model of the control of ovule primordia formation. Plant J. 2013;76:446–455. doi: 10.1111/tpj.12309. [DOI] [PubMed] [Google Scholar]
- Galvao VC, Horrer D, Kuttner F, Schmid M. Spatial control of flowering by DELLA proteins in Arabidopsis thaliana. Development. 2012;139:4072–4082. doi: 10.1242/dev.080879. [DOI] [PubMed] [Google Scholar]
- Gfeller A, Dubugnon L, Liechti R, Farmer EE. Jasmonate biochemical pathway. Sci Signal. 2010;3:cm3. doi: 10.1126/scisignal.3109cm3. [DOI] [PubMed] [Google Scholar]
- Gomez-Mena C, de Folter S, Costa MM, Angenent GC, Sablowski R. Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development. 2005;132:429–438. doi: 10.1242/dev.01600. [DOI] [PubMed] [Google Scholar]
- Han Y, Zhang C, Yang H, Jiao Y. Cytokinin pathway mediates APETALA1 function in the establishment of determinate floral meristems in Arabidopsis. Proc Natl Acad Sci USA. 2014;111:6840–6845. doi: 10.1073/pnas.1318532111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins C, Liu Z. A model for an early role of auxin in Arabidopsis gynoecium morphogenesis. Front Plant Sci. 2014;5:327. doi: 10.3389/fpls.2014.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel FD, Weigel D, Mandel MA, Ditta G, Zambryski PC, Feldman LJ, Yanofsky MF. Floral determination and expression of floral regulatory genes in Arabidopsis. Development. 1997;124:3845–3853. doi: 10.1242/dev.124.19.3845. [DOI] [PubMed] [Google Scholar]
- Hempel FD, Zambryski PC, Feldman LJ. Photoinduction of flower identity in vegetatively biased primordia. Plant Cell. 1998;10:1663–1676. doi: 10.1105/tpc.10.10.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PK, Tsay YF. Two phloem nitrate transporters, NRT1.11 and NRT1.12, are important for redistributing xylem-borne nitrate to enhance plant growth. Plant Physiol. 2013;163:844–856. doi: 10.1104/pp.113.226563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huala E, Sussex IM. LEAFY interacts with floral homeotic genes to regulate Arabidopsis floral development. Plant Cell. 1992;4:901–913. doi: 10.1105/tpc.4.8.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Ng KH, Lim TS, Yu H, Meyerowitz EM. The homeotic protein AGAMOUS controls late stamen development by regulating a jasmonate biosynthetic gene in Arabidopsis. Plant Cell. 2007;19:3516–3529. doi: 10.1105/tpc.107.055467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata T, Nagasaki O, Ishii HS, Ushimaru A. Inflorescence architecture affects pollinator behaviour and mating success in Spiranthes sinensis (Orchidaceae) New Phytol. 2012;193:196–203. doi: 10.1111/j.1469-8137.2011.03892.x. [DOI] [PubMed] [Google Scholar]
- Jaeger KE, Wigge PA. FT protein acts as a long-range signal in Arabidopsis. Curr Biol. 2007;17:1050–1054. doi: 10.1016/j.cub.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Jaeger KE, Pullen N, Lamzin S, Morris RJ, Wigge PA. Interlocking feedback loops govern the dynamic behavior of the floral transition in Arabidopsis. Plant Cell. 2013;25:820–833. doi: 10.1105/tpc.113.109355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph MP, Papdi C, Kozma-Bognar L, Nagy I, Lopez-Carbonell M, Rigo G, Koncz C, Szabados L. The Arabidopsis ZINC FINGER PROTEIN3 interferes with abscisic acid and light signaling in seed germination and plant development. Plant Physiol. 2014;165:1203–1220. doi: 10.1104/pp.113.234294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa T, Kimura M, Wada M. Blue light-induced phototropism of inflorescence stems and petioles is mediated by phototropin family members phot1 and phot2. Plant Cell Physiol. 2009;50:1774–1785. doi: 10.1093/pcp/pcp119. [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. Activation tagging of the floral inducer FT. Science. 1999;286:1962–1965. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Wellmer F, Muino JM, Ferrier T, Wuest SE, Kumar V, Serrano-Mislata A, Madueno F, Krajewski P, Meyerowitz EM, Angenent GC, Riechmann JL. Orchestration of floral initiation by APETALA1. Science. 2010;328:85–89. doi: 10.1126/science.1185244. [DOI] [PubMed] [Google Scholar]
- Kim J. Four shades of detachment: regulation of floral organ abscission. Plant Signal Behav. 2014;9:e976154. doi: 10.4161/15592324.2014.976154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA. Auxin regulation of Arabidopsis flower development involves members of the AINTEGUMENTA-LIKE/PLETHORA (AIL/PLT) family. J Exp Bot. 2011;62:3311–3319. doi: 10.1093/jxb/err127. [DOI] [PubMed] [Google Scholar]
- Larsson E, Franks RG, Sundberg E. Auxin and the Arabidopsis thaliana gynoecium. J Exp Bot. 2013;64:2619–2627. doi: 10.1093/jxb/ert099. [DOI] [PubMed] [Google Scholar]
- Lee I, Wolfe DS, Nilsson O, Weigel D. A LEAFY co-regulator encoded by UNUSUAL FLORAL ORGANS. Curr Biol. 1997;7:95–104. doi: 10.1016/s0960-9822(06)00053-4. [DOI] [PubMed] [Google Scholar]
- Lee J, Oh M, Park H, Lee I. SOC1 translocated to the nucleus by interaction with AGL24 directly regulates leafy. Plant J. 2008;55:832–843. doi: 10.1111/j.1365-313X.2008.03552.x. [DOI] [PubMed] [Google Scholar]
- Leran S, Varala K, Boyer JC, Chiurazzi M, Crawford N, Daniel-Vedele F, David L, Dickstein R, Fernandez E, Forde B, Gassmann W, Geiger D, Gojon A, Gong JM, Halkier BA, Harris JM, Hedrich R, Limami AM, Rentsch D, Seo M, Tsay YF, Zhang M, Coruzzi G, Lacombe B. A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci. 2014;19:5–9. doi: 10.1016/j.tplants.2013.08.008. [DOI] [PubMed] [Google Scholar]
- Levin JZ, Meyerowitz EM. UFO: an Arabidopsis gene involved in both floral meristem and floral organ development. Plant Cell. 1995;7:529–548. doi: 10.1105/tpc.7.5.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhou Y, Liu X, Yu P, Cohen JD, Meyerowitz EM. LEAFY controls auxin response pathways in floral primordium formation. Sci Signal. 2013;6:ra23. doi: 10.1126/scisignal.2003937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren SJ, Gustafson-Brown C, Pinyopich A, Ditta GS, Yanofsky MF. Interactions among APETALA1, LEAFY, and TERMINAL FLOWER1 specify meristem fate. Plant Cell. 1999;11:1007–1018. doi: 10.1105/tpc.11.6.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman ZB, Cohen O, Alvarez JP, Abu-Abied M, Pekker I, Paran I, Eshed Y, Zamir D. The making of a compound inflorescence in tomato and related nightshades. PLoS Biol. 2008;6:e288. doi: 10.1371/journal.pbio.0060288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Chen H, Er HL, Soo HM, Kumar PP, Han JH, Liou YC, Yu H. Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development. 2008;135:1481–1491. doi: 10.1242/dev.020255. [DOI] [PubMed] [Google Scholar]
- Liu C, Xi W, Shen L, Tan C, Yu H. Regulation of floral patterning by flowering time genes. Dev Cell. 2009;16:711–722. doi: 10.1016/j.devcel.2009.03.011. [DOI] [PubMed] [Google Scholar]
- Liu C, Teo ZW, Bi Y, Song S, Xi W, Yang X, Yin Z, Yu H. A conserved genetic pathway determines inflorescence architecture in Arabidopsis and rice. Dev Cell. 2013;24:612–622. doi: 10.1016/j.devcel.2013.02.013. [DOI] [PubMed] [Google Scholar]
- Lunn JE, Delorge I, Figueroa CM, Van Dijck P, Stitt M. Trehalose metabolism in plants. Plant J. 2014;79:544–567. doi: 10.1111/tpj.12509. [DOI] [PubMed] [Google Scholar]
- Magome H, Nomura T, Hanada A, Takeda-Kamiya N, Ohnishi T, Shinma Y, Katsumata T, Kawaide H, Kamiya Y, Yamaguchi S. CYP714B1 and CYP714B2 encode gibberellin 13-oxidases that reduce gibberellin activity in rice. Proc Natl Acad Sci USA. 2013;110:1947–1952. doi: 10.1073/pnas.1215788110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature. 1992;360:273–277. doi: 10.1038/360273a0. [DOI] [PubMed] [Google Scholar]
- Melzer S, Lens F, Gennen J, Vanneste S, Rohde A, Beeckman T. Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat Genet. 2008;40:1489–1492. doi: 10.1038/ng.253. [DOI] [PubMed] [Google Scholar]
- Moyroud E, Minguet EG, Ott F, Yant L, Pose D, Monniaux M, Blanchet S, Bastien O, Thevenon E, Weigel D, Schmid M, Parcy F. Prediction of regulatory interactions from genome sequences using a biophysical model for the Arabidopsis LEAFY transcription factor. Plant Cell. 2011;23:1293–1306. doi: 10.1105/tpc.111.083329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muino JM, Hoogstraat M, van Ham RC, van Dijk AD. PRI-CAT: a web-tool for the analysis, storage and visualization of plant ChIP-seq experiments. Nucleic Acids Res. 2011;39:W524–W527. doi: 10.1093/nar/gkr373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller D, Schmitz G, Theres K. Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell. 2006;18:586–597. doi: 10.1105/tpc.105.038745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Xing R, Clarenz O, Pokorny L, Goodrich J, Schubert D. Polycomb-group proteins and FLOWERING LOCUS T maintain commitment to flowering in Arabidopsis thaliana. Plant Cell. 2014;26:2457–2471. doi: 10.1105/tpc.114.123323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P, Ellis CM, Weber H, Ploense SE, Barkawi LS, Guilfoyle TJ, Hagen G, Alonso JM, Cohen JD, Farmer EE, Ecker JR, Reed JW. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development. 2005;132:4107–4118. doi: 10.1242/dev.01955. [DOI] [PubMed] [Google Scholar]
- Nicol JW, Helt GA, Blanchard SG, Jr, Raja A, Loraine AE. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25:2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Magome H, Hanada A, Takeda-Kamiya N, Mander LN, Kamiya Y, Yamaguchi S. Functional analysis of Arabidopsis CYP714A1 and CYP714A2 reveals that they are distinct gibberellin modification enzymes. Plant Cell Physiol. 2013;54:1837–1851. doi: 10.1093/pcp/pct125. [DOI] [PubMed] [Google Scholar]
- Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y. Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell. 1991;3:677–684. doi: 10.1105/tpc.3.7.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando DA, Brady SM, Koch JD, Dinneny JR, Benfey PN. Manipulating large-scale Arabidopsis microarray expression data: identifying dominant expression patterns and biological process enrichment. Methods Mol Biol. 2009;553:57–77. doi: 10.1007/978-1-60327-563-7_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osnato M, Castillejo C, Matias-Hernandez L, Pelaz S. TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nature Commun. 2012;3:808. doi: 10.1038/ncomms1810. [DOI] [PubMed] [Google Scholar]
- Pajoro A, Madrigal P, Muino JM, Matus JT, Jin J, Mecchia MA, Debernardi JM, Palatnik JF, Balazadeh S, Arif M, O’Maoileidigh DS, Wellmer F, Krajewski P, Riechmann JL, Angenent GC, Kaufmann K. Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol. 2014;15:R41. doi: 10.1186/gb-2014-15-3-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F, Nilsson O, Busch MA, Lee I, Weigel D. A genetic framework for floral patterning. Nature. 1998;395:561–566. doi: 10.1038/26903. [DOI] [PubMed] [Google Scholar]
- Pastore JJ, Limpuangthip A, Yamaguchi N, Wu MF, Sang Y, Han SK, Malaspina L, Chavdaroff N, Yamaguchi A, Wagner D. LATE MERISTEM IDENTITY2 acts together with LEAFY to activate APETALA1. Development. 2011;138:3189–3198. doi: 10.1242/dev.063073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig RS. Vegetative phase change and shoot maturation in plants. Curr Top Dev Biol. 2013;105:125–152. doi: 10.1016/B978-0-12-396968-2.00005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pose D, Yant L, Schmid M. The end of innocence: flowering networks explode in complexity. Curr Opin Plant Biol. 2012;15:45–50. doi: 10.1016/j.pbi.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Prusinkiewicz P, Erasmus Y, Lane B, Harder LD, Coen E. Evolution and development of inflorescence architectures. Science. 2007;316:1452–1456. doi: 10.1126/science.1140429. [DOI] [PubMed] [Google Scholar]
- Raghavendra AS, Gonugunta VK, Christmann A, Grill E. ABA perception and signalling. Trends Plant Sci. 2010;15:395–401. doi: 10.1016/j.tplants.2010.04.006. [DOI] [PubMed] [Google Scholar]
- Reeves PH, Ellis CM, Ploense SE, Wu MF, Yadav V, Tholl D, Chetelat A, Haupt I, Kennerley BJ, Hodgens C, Farmer EE, Nagpal P, Reed JW. A regulatory network for coordinated flower maturation. PLoS Genet. 2012;8:e1002506. doi: 10.1371/journal.pgen.1002506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Olalde JI, Zuniga-Mayo VM, Chavez Montes RA, Marsch-Martinez N, de Folter S. Inside the gynoecium: at the carpel margin. Trends Plant Sci. 2013;18:644–655. doi: 10.1016/j.tplants.2013.08.002. [DOI] [PubMed] [Google Scholar]
- Romera-Branchat M, Andres F, Coupland G. Flowering responses to seasonal cues: what’s new? Curr Opin Plant Biol. 2014;21C:120–127. doi: 10.1016/j.pbi.2014.07.006. [DOI] [PubMed] [Google Scholar]
- Roux F, Touzet P, Cuguen J, Le Corre V. How to be early flowering: an evolutionary perspective. Trends Plant Sci. 2006;11:375–381. doi: 10.1016/j.tplants.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Rubio-Somoza I, Weigel D. Coordination of flower maturation by a regulatory circuit of three microRNAs. PLoS Genet. 2013;9:e1003374. doi: 10.1371/journal.pgen.1003374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Garcia L, Madueno F, Wilkinson M, Haughn G, Salinas J, Martinez-Zapater JM. Different roles of flowering-time genes in the activation of floral initiation genes in Arabidopsis. Plant Cell. 1997;9:1921–1934. doi: 10.1105/tpc.9.11.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saddic LA, Huvermann B, Bezhani S, Su Y, Winter CM, Kwon CS, Collum RP, Wagner D. The LEAFY target LMI1 is a meristem identity regulator and acts together with LEAFY to regulate expression of CAULIFLOWER. Development. 2006;133:1673–1682. doi: 10.1242/dev.02331. [DOI] [PubMed] [Google Scholar]
- Schaller GE, Bishopp A, Kieber JJ. The Yin–Yang of hormones: cytokinin and auxin interactions in plant development. Plant Cell. 2015;27:44–63. doi: 10.1105/tpc.114.133595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU. Dissection of floral induction pathways using global expression analysis. Development. 2003;130:6001–6012. doi: 10.1242/dev.00842. [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Schultz EA, Haughn GW. Leafy, a homeotic gene that regulates inflorescence development in Arabidopsis. Plant Cell. 1991;3:771–781. doi: 10.1105/tpc.3.8.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz EA, Haughn GW. Genetic analysis of the floral induction process (FLIP) in Arabidopsis. Development. 1993;119:745–765. [Google Scholar]
- Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol Biol. 2008;67:183–195. doi: 10.1007/s11103-008-9310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi DQ, Yang WC. Ovule development in Arabidopsis: progress and challenge. Curr Opin Plant Biol. 2011;14:74–80. doi: 10.1016/j.pbi.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Song J, Irwin J, Dean C. Remembering the prolonged cold of winter. Curr Biol. 2013a;23:R807–R811. doi: 10.1016/j.cub.2013.07.027. [DOI] [PubMed] [Google Scholar]
- Song S, Qi T, Huang H, Xie D. Regulation of stamen development by coordinated actions of jasmonate, auxin, and gibberellin in Arabidopsis. Mol Plant. 2013b;6:1065–1073. doi: 10.1093/mp/sst054. [DOI] [PubMed] [Google Scholar]
- Souer E, Rebocho AB, Bliek M, Kusters E, de Bruin RA, Koes R. Patterning of inflorescences and flowers by the F-Box protein DOUBLE TOP and the LEAFY homolog ABERRANT LEAF AND FLOWER of petunia. Plant Cell. 2008;20:2033–2048. doi: 10.1105/tpc.108.060871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP. Gibberellin metabolism, perception and signaling pathways in Arabidopsis. Arabidopsis Book. 2008;6:e0103. doi: 10.1199/tab.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata R, Ikezaki M, Fujibe T, Aida M, Tian CE, Ueno Y, Yamamoto KT, Machida Y, Nakamura K, Ishiguro S. Arabidopsis auxin response factor6 and 8 regulate jasmonic acid biosynthesis and floral organ development via repression of class 1 KNOX genes. Plant Cell Physiol. 2010;51:164–175. doi: 10.1093/pcp/pcp176. [DOI] [PubMed] [Google Scholar]
- Titapiwatanakun B, Blakeslee JJ, Bandyopadhyay A, Yang H, Mravec J, Sauer M, Cheng Y, Adamec J, Nagashima A, Geisler M, Sakai T, Friml J, Peer WA, Murphy AS. ABCB19/PGP19 stabilises PIN1 in membrane microdomains in Arabidopsis. Plant J. 2009;57:27–44. doi: 10.1111/j.1365-313X.2008.03668.x. [DOI] [PubMed] [Google Scholar]
- Tooke F, Ordidge M, Chiurugwi T, Battey N. Mechanisms and function of flower and inflorescence reversion. J Exp Bot. 2005;56:2587–2599. doi: 10.1093/jxb/eri254. [DOI] [PubMed] [Google Scholar]
- Verhage L, Angenent GC, Immink RG. Research on floral timing by ambient temperature comes into blossom. Trends Plant Sci. 2014;19:583–591. doi: 10.1016/j.tplants.2014.03.009. [DOI] [PubMed] [Google Scholar]
- Wagner D, Meyerowitz E. Switching on flowers: transient LEAFY induction reveals novel aspects of flower development in Arabidopsis. Front Plant Sci. 2011;2:60. doi: 10.3389/fpls.2011.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D, Sablowski RW, Meyerowitz EM. Transcriptional activation of APETALA1 by LEAFY. Science. 1999;285:582–584. doi: 10.1126/science.285.5427.582. [DOI] [PubMed] [Google Scholar]
- Wahl V, Ponnu J, Schlereth A, Arrivault S, Langenecker T, Franke A, Feil R, Lunn JE, Stitt M, Schmid M. Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science. 2013;339:704–707. doi: 10.1126/science.1230406. [DOI] [PubMed] [Google Scholar]
- Wang Q, Sajja U, Rosloski S, Humphrey T, Kim MC, Bomblies K, Weigel D, Grbic V. HUA2 caused natural variation in shoot morphology of A. thaliana. Curr Biol. 2007;17:1513–1519. doi: 10.1016/j.cub.2007.07.059. [DOI] [PubMed] [Google Scholar]
- Wang JW, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138:738–749. doi: 10.1016/j.cell.2009.06.014. [DOI] [PubMed] [Google Scholar]
- Wang J, Yan DW, Yuan TT, Gao X, Lu YT. A gain-of-function mutation in IAA8 alters Arabidopsis floral organ development by change of jasmonic acid level. Plant Mol Biol. 2013;82:71–83. doi: 10.1007/s11103-013-0039-y. [DOI] [PubMed] [Google Scholar]
- Weigel D, Meyerowitz EM. Activation of floral homeotic genes in Arabidopsis. Science. 1993;261:1723–1726. doi: 10.1126/science.261.5129.1723. [DOI] [PubMed] [Google Scholar]
- Weigel D, Nilsson O. A developmental switch sufficient for flower initiation in diverse plants. Nature. 1995;377:495–500. doi: 10.1038/377495a0. [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. LEAFY controls floral meristem identity in Arabidopsis. Cell. 1992;69:843–859. doi: 10.1016/0092-8674(92)90295-n. [DOI] [PubMed] [Google Scholar]
- Wellmer F, Alves-Ferreira M, Dubois A, Riechmann JL, Meyerowitz EM. Genome-wide analysis of gene expression during early Arabidopsis flower development. PLoS Genet. 2006;2:e117. doi: 10.1371/journal.pgen.0020117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309:1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- William DA, Su Y, Smith MR, Lu M, Baldwin DA, Wagner D. Genomic identification of direct target genes of LEAFY. Proc Natl Acad Sci USA. 2004;101:1775–1780. doi: 10.1073/pnas.0307842100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter CM, Austin RS, Blanvillain-Baufume S, Reback MA, Monniaux M, Wu MF, Sang Y, Yamaguchi A, Yamaguchi N, Parker JE, Parcy F, Jensen ST, Li H, Wagner D. LEAFY target genes reveal floral regulatory logic, cis motifs, and a link to biotic stimulus response. Dev Cell. 2011;20:430–443. doi: 10.1016/j.devcel.2011.03.019. [DOI] [PubMed] [Google Scholar]
- Wu MF, Wagner D. RNA in situ hybridization in Arabidopsis. Methods Mol Biol. 2012;883:75–86. doi: 10.1007/978-1-61779-839-9_5. [DOI] [PubMed] [Google Scholar]
- Wu Z, Irizarry R, Gentleman R, Martinez Murillo F, Spencer F. A Model Based Background Adjustment for Oligonucleotide Expression Arrays. Johns Hopkins University, Department of Biostatistics; 2004. Working Papers, Working Paper 1. [Google Scholar]
- Wu MF, Tian Q, Reed JW. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development. 2006;133:4211–4218. doi: 10.1242/dev.02602. [DOI] [PubMed] [Google Scholar]
- Wu MF, Sang Y, Bezhani S, Yamaguchi N, Han SK, Li Z, Su Y, Slewinski TL, Wagner D. SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc Natl Acad Sci USA. 2012;109:3576–3581. doi: 10.1073/pnas.1113409109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A, Wu MF, Yang L, Wu G, Poethig RS, Wagner D. The microRNA-regulated SBP-box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev Cell. 2009;17:268–278. doi: 10.1016/j.devcel.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Wu MF, Winter CM, Berns MC, Nole-Wilson S, Yamaguchi A, Coupland G, Krizek BA, Wagner D. A molecular framework for auxin-mediated initiation of flower primordia. Dev Cell. 2013;24:271–282. doi: 10.1016/j.devcel.2012.12.017. [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, Winter CM, Wu MF, Kanno Y, Yamaguchi A, Seo M, Wagner D. Gibberellin acts positively then negatively to control onset of flower formation in Arabidopsis. Science. 2014a;344:638–641. doi: 10.1126/science.1250498. [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, Wu MF, Winter C, Wagner D. LEAFY together with polar auxin transport coordinates Arabidopsis flower development. Plants. 2014b;3:251–265. doi: 10.3390/plants3020251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Winter CM, Wellmer F, Wagner D. Identification of direct targets of plant transcription factors using the GR fusion technique. Methods Mol Biol. 2015;1284:123–138. doi: 10.1007/978-1-4939-2444-8_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Ito T, Zhao Y, Peng J, Kumar P, Meyerowitz EM. Floral homeotic genes are targets of gibberellin signaling in flower development. Proc Natl Acad Sci USA. 2004;101:7827–7832. doi: 10.1073/pnas.0402377101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Galvao VC, Zhang YC, Horrer D, Zhang TQ, Hao YH, Feng YQ, Wang S, Schmid M, Wang JW. Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell. 2012;24:3320–3332. doi: 10.1105/tpc.112.101014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang B, Yan D, Dong W, Yang W, Li Q, Zeng L, Wang J, Wang L, Hicks LM, He Z. Two Arabidopsis cytochrome P450 monooxygenases, CYP714A1 and CYP714A2, function redundantly in plant development through gibberellin deactivation. Plant J. 2011;67:342–353. doi: 10.1111/j.1365-313X.2011.04596.x. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Nomura T, Xu Y, Zhang Y, Peng Y, Mao B, Hanada A, Zhou H, Wang R, Li P, Zhu X, Mander LN, Kamiya Y, Yamaguchi S, He Z. ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell. 2006;18:442–456. doi: 10.1105/tpc.105.038455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X, Sun TP. IQD, a negative regulator of GA response, plays a role in the regulatory network among the GA, calcium and auxin pathways. 22nd International Conference on Arabidopsis Research; Madison, Wisconsin. 2011. [Google Scholar]
- Zuniga-Mayo VM, Reyes-Olalde JI, Marsch-Martinez N, de Folter S. Cytokinin treatments affect the apical-basal patterning of the Arabidopsis gynoecium and resemble the effects of polar auxin transport inhibition. Front Plant Sci. 2014;5:191. doi: 10.3389/fpls.2014.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identification of LFY- and AP1-bound genes.
Expression of AP1 in lfy mutants during photoperiod induction.
Identification of LFY- and AP1-bound genes. Individual LFY and AP1 ChIP-seq and ChIP-chip datasets and sources, and the number of genes identified in each are listed below. Unless noted, published peak locations were assigned to genes using a custom Python script (Winter et al. 2011), rewritten to use TAIR10 annotations. The total number of LFY-bound genes was taken as the union of the genes identified in the three LFY ChIP datasets. See Fig. S1A for a detailed overlap analysis of the three LFY datasets. The total number of AP1-bound genes was taken as the union of the four AP1 ChIP datasets. See Fig. S1B for a detailed overlap analysis of the four AP1 ChIP datasets. The 769 LFY- and AP1-bound genes are the intersects of the total LFY-bound genes and total AP1-bound genes (see Fig. 1A).
LFY peaks and gene assignments.
AP1 peaks and gene assignments.
Summary of LFY-GR, AP1-GR, development and photoperiod transcriptome datasets used for the identification of high-confidence LFY- and AP1-bound genes.
One hundred ninety six genes bound by LFY and AP1 and differentially expressed in LFY-GR and AP1-GR.
Filtering criteria used to identify high-confidence LFY and AP1 targets. The steps below were applied to successively filter the list of 769 LFY-and AP1-bound genes (see Table S1) to obtain a list of 11 high-confidence LFY and AP1 targets. See also Table S5 for genes and expression data. See Table S4 for more information regarding the transcriptome datasets employed.