Abstract
Purpose
Daily modulation of gene expression is critical for the circadian rhythms of many organisms. One of the modulating mechanisms is based on nocturnin, a deadenylase that degrades mRNA in a circadian fashion. The nocturnin genes are expressed broadly, but their tissue expression patterns differ between mice and the frog Xenopus laevis; this difference suggests that the extent of the regulation of nocturin gene expression varies among species. In this study, we set out to characterize the expression patterns of two zebrafish nocturnin genes; in addition, we asked whether a frog nocturnin promoter has transcriptional activity in zebrafish.
Methods
We used reverse transcription (RT)–PCR, quantitative real-time PCR (qRT-PCR), and rapid amplification of cDNA ends (RACE) analysis to determine whether the nocturnin-a and nocturnin-b genes are expressed in the eye, in situ hybridization to determine the cellular expression pattern of the nocturnin-b gene in the retina, and confocal microscopy to determine the protein expression pattern of the transgenic reporter green fluorescent protein (GFP) driven by the frog nocturnin promoter.
Results
We found that the amino acid sequences of zebrafish nocturnin-a and nocturnin-b are highly similar to those of frog, mouse, and human nocturnin homologs. Only nocturnin-b is expressed in the eye. Within the retina, nocturnin-b mRNA was expressed at higher levels in the retinal photoreceptors layer than in other cellular layers. This expression pattern echoes the restricted photoreceptor expression of nocturnin in the frog. We also found that the frog nocturnin promoter can be specifically activated in zebrafish rod photoreceptors.
Conclusions
The high level of similarities in amino acid sequences of human, mouse, frog, and zebrafish nocturnin homologs suggest these proteins maintain a conserved deadenylation function that is important for regulating retinal circadian rhythmicity. The rod-specific transcriptional activity of the frog nocturnin promoter makes it a useful tool to drive moderate and rod-specific transgenic expression in zebrafish. The results of this study lay the groundwork to study nocturnin-based circadian biology of the zebrafish retina.
Introduction
Most organisms, ranging from unicellular prokaryotic species to eukaryotic mammals, synchronize their behavioral and physiologic activities with the 24 h cycle of Earth’s rotation [1-3]. Disruption of this circadian rhythmicity can lead to sleep disorders, metabolic imbalance, and even the development of cancer [4,5]. Thus, proper regulation of circadian rhythms is critical for normal functions of organisms.
Circadian rhythmicity is regulated by complex feedback networks. In vertebrates, the circadian rhythm-regulating networks are composed of the pacemaker of the suprachiasmatic nucleus of the hypothalamus and peripheral oscillating organs [6]. Each component of the circadian system has its own internal rhythm. Interestingly, the internal rhythms of individual organs are normally not on a precise 24 h cycle [7,8]. Thus, biologic circadian rhythms need to be entrained by various environmental cues, namely, zeitgebers, to synchronize with the 24 h earth rotation cycle. Among the various zeitgebers, the light-dark cycle plays a much more prominent role than other factors such as temperature and food. In lower vertebrates, the sensation of light-dark signals is collectively performed by the retina, pineal organ, and extraretinal deep brain photoreceptors [9-11]. In contrast, in mammals, it is believed to be exclusively conducted by the retina [12-14]. The retina integrates the external signals with its internal rhythmic signals and provides the photic input to the pacemaker of the suprachiasmatic nucleus. Therefore, the retina is a pivotal component of the circadian systems [6]. The importance of the retina in the circadian systems is also manifested by the discovery of the retinal circadian clock that is independent of the suprachiasmatic nucleus [7].
As the most important sensor of the light-dark zeitgeber, the retinas themselves undergo many circadian structural changes, such as regular photoreceptor disc shedding in the evening and daily modulations of the synaptic junctions of photoreceptors [15-18]. These structural changes are based on circadian modulation of retinal metabolism, biochemistry, and gene expression profiling [19-21]. One of the many ways to modulate gene expression profiling is through the regulation of mRNA turnover. mRNA turnover can be accelerated through depolyadenylation by deadenylases, to which nocturnin belongs [22]. Interestingly, nocturnin expression displays its own rhythmicity in the photoreceptor of the frog Xenopus laevis, maximally at night [22]. Thus, the cyclic expression of nocturnin regulates the mRNA turnover of a wide spectrum of genes in the photoreceptors in a circadian fashion; some of these genes mediate the synthesis of the melatonin hormone, which can be circulated in the vascular system to modulate the rhythms of other tissues and organs in distance [23].
The function of nocturnin as a post-transcriptional regulator is not limited in the retina [24]. In mice, the nocturnin is also expressed in non-retinal tissues, such as the liver, spleen, kidney, and heart [25]. Considering that up to 10% of mammalian transcriptome is estimated to be regulated in a circadian fashion [26,27], spatial and temporal regulation of nocturnin expression, thus, is an important means for modulating the circadian rhythmicity of the entire body [24].
The retinal expression patterns and functions of nocturnin have yet to be characterized in the zebrafish. Here, we report the cloning of a zebrafish nocturnin gene and the characterization of its retinal expression patterns. In addition, we show that the promoter of the frog nocturnin gene can be specifically activated in zebrafish rod photoreceptors to drive moderate transgenic expression, suggesting certain conservation of nocturnin expression regulation between the two species in the retina. Consequently, the frog nocturnin promoter is a useful tool to transgenically express genes of interest at moderate levels in the zebrafish rod photoreceptors.
Methods
Zebrafish care
Adult AB zebrafish were raised at 28.5 °C on a 14 h:10 h light-dark cycle. Zebrafish embryos were collected and raised at 28.5 °C. All procedures for zebrafish care conformed to the University of Pittsburgh standards for care and use of animals in research (University of Pittsburgh Animal Protocol Number: 0909487B-3; Assurance Number: A3187–01). The study adhered to the ARVO Statement for Use of Animals in Research.
RT–PCR confirmation of the zebrafish nocturnin-b cDNA open reading frame
For RT–PCR analysis of nocturnin expression, total RNA molecules were either isolated at 8 AM, 12 PM, 4 PM, 8 PM, 12 AM, and 4 AM from adult fish eyes or isolated at 12 AM from 5-dpf larval eyes and 5-dpf whole larval fish. The samples were cut into small pieces and incubated with TRIzol (Thermo Fisher Scientific, Waltham, MA) for 5 min at room temperature. Then 0.2 ml of chloroform was added to every 1 ml of TRIzol tissue extraction, followed by gentle shaking for 15 s and 3 min of incubation at room temperature. The aqueous fractions of the lysates were retrieved and used for RNA precipitation with isopropyl alcohol with centrifugation. The RNA pellet was washed with 75% ethanol three times and redissolved in 100% formamide and stored at −80 °C until later usage. To make a cDNA library, the total RNA was first incubated at 65 °C for 5 min to remove their secondary structures and then reversely transcribed using an oligo dT primer and SuperScript III reverse transcriptase (Invitrogen). The expression of the zebrafish nocturnin-a and nocturnin-b genes in the eye was examined with RT–PCR reactions using the following primers (Figure 1C): noc-a F7 (5′-AGC CTT CAG AGT TCA CG ACA-3′); noc-a R7 (5′-GGC TCT GGT CAT GAT GAA CT-3′); noc-a F8 (5′-ATT TAG GAG TTC TTC ATT A-3′); noc-a R8 (5′-ACT GAA GCT GAG GTC ACA CA-3′); noc-b F4 (5′-ACA ATC TGA ATC TGG ATC AG-3′); noc-b R4 (5′-TCC CTC CAG GAC ATG TAC TT-3′); noc-b F5 (5′-GAA GAC GTC TAT AGG AAT TT-3′); noc-b R5 (5′- AAA GGT GAT CTG AAG GGT AG-3′); elf2a-F (5′-TTG AGA AGA AAA TCG GTG GTG CTG-3′); and elf2a-R (5′-GGA ACG GTG TGA TTG AGG GAA ATT C-3′). The PCR conditions were initial denaturation at 95 °C for 1 min, and 40 cycles of amplification and quantification (95 °C for 20 s, 55 °C for 20 s, and 72 °C 30 s). To confirm the sequence of the nocturnin-b open reading frame (ORF), we sequenced an RT–PCR nocturnin-b ORF DNA (cloned in a pDrive vector), which was amplified with a forward primer covering the start codon (5′-TCC GAA TTC CGT GTT CCA TGG GCA GCG GC-3′; the underlined sequence is an EcoR I site) and a reverse primer binding to its 3′ untranslated region (UTR) (5′-TCC AAG CTT CGT GAA TTC ATA CAA AAT TGC ACA G-3′; the underlined sequence is a Hind III site).
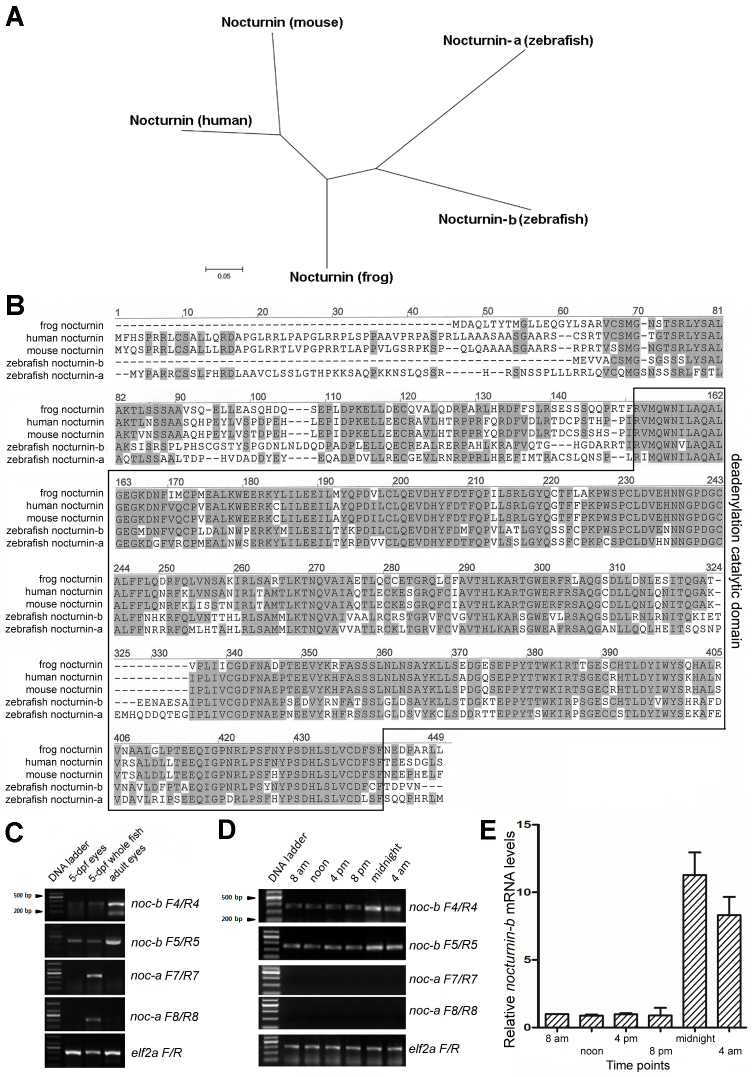
Figure 1.
Zebrafish nocturnin-a and nocturnin-b are similar to frog and mammalian nocturnin genes. A: An unrooted phylogenetic tree of nocturnin proteins suggests zebrafish nocturnin-b is more similar to frog nocturnin than nocturnin-a. The scale bar represents 5% estimated sequence divergence. B: An alignment of the amino acid sequences of the nocturnin homologs of human, mouse, Xenopus laevis, and zebrafish revealed high conservation: Identical amino acid residues between different nocturnin homologs are highlighted in gray. The deadenylation catalytic domain is boxed. C: An reverse transcription (RT)-PCR analysis with two primer pairs for each zebrafish nocturnin gene showed that nocturnin-b was expressed in 5-dpf and adult zebrafish eyes, as well as in 5-dpf whole larval fish, whereas nocturnin-a expression was not detectable in the eyes, although it was detectable in the 5-dpf whole larval fish. D: RT–PCR analysis of nocturnin-a and nocturnin-b expression in adult eyes at 8 AM, 12 PM, 4 PM, 8 PM, 12 AM, and 4 AM, noting prominent nocturnin-b expression but undetectable nocturnin-a expression. E: Quantitative real-time PCR (qRT-PCR) revealed the rhythmic changes in nocturnin-b expression in zebrafish adult eyes at the 8 AM, 12 PM, 4 PM, 8 PM, 12 AM, and 4 AM y-axis, fold differences between nocturnin-b and elf2a, normalizing against the level at 8 AM. Error bars (standard deviations) are based on three experiments.
RACE identification of the full-length sequence of the zebrafish nocturnin-b cDNA
The total RNA of the adult AB wild-type zebrafish eyes was extracted with an RNeasy Mini Kit (Qiagen, Hilden, Germany). Briefly, small pieces of eye tissue (30 mg) were dissociated and homogenized gently in an RLT Buffer with a MACS Dissociator (Miltenyi Biotec., Teterow, Germany). An equal volume of 70% ethanol was added to the lysates to allow selective binding of RNA to the RNeasy membranes for purification. The total RNA was eluted in RNase-free water and stored at −80 °C until later usage.
To identify the full-length sequence of the zebrafish nocturnin-b cDNA sequence, 5′ and 3′ rapid amplification of cDNA ends (RACE) analyses were performed with the total RNA using a GeneRacer Kit (Invitrogen) following the manufacturer’s protocol. For 5′ RACE, nocturnin-b-specific reverse primer 1 (5′-TCC AAG CTT CGT GAA TTC ATA CAA AAT TGC ACA G-3′) and primer 2 (5′-CTA CAT CGA GAC ATG GAG ACC ATG G-3′) were used for the first-round PCR and then nested-PCR, respectively. For 3′ RACE, nocturnin-b-specific forward primer 1 (5′-TCC GAA TTC CGT GTT CCA TGG GCA GCG GC-3′) and primer 2 (5′-CGT GGA AGA TCC GAC CCA GTG GCG AGA TT-3′) were used for the first-round PCR and the subsequent nested-PCR, respectively. Platinum High Fidelity Taq DNA Polymerase was used for all PCR reactions. The PCR conditions were initial denaturation at 94 °C for 2 min, and 35 cycles of amplification and quantification (94 °C for 30 s, 55 °C for 30 s, and 68 °C 2 min). The amplified PCR fragments of the nocturnin-b cDNA were cloned with a TOPO TA cloning kit (Invitrogen) and then sequenced (Figure 1B).
qRT-PCR
Quantitative real-time PCR (qRT-PCR) analysis of nocturnin-b at four time points during the day was performed with SYBR Green/ROX qPCR Master Mix (Fermentas, Thermo Fisher Scientific, Waltham, MA) on a Roter-Gene 6000 real-time genetic analyzer (Corbett Life Science, Qiagen, Hilden, Germany) according to the manufacturer’s instructions; elf2a was used as an internal control. The following primers were used for qRT-PCR: noc-b F4, noc-b R4, elf2a-F, and elf2a-R. Each sample was replicated three times. The qRT-PCR was performed with the following conditions: initial denaturation at 95 °C for 2 min, 40 cycles of amplification and quantification (95 °C for 5 s, 55-57 °C for 30 s), and a melting curve program 55-95 °C with 0.5 °C increment each cycle).
Computational analyses of genes
We used Clustalw2 software to analyze the amino acid sequence similarities among various full-length nocturnin homologs of the following species: zebrafish (nocturnin-b, XP_697426; nocturnin-a, XP_700794); Xenopus laevis (NM001085812), mouse (NM009834), and human (NM012118). The amino acid sequence of zebrafish nocturnin-b was predicted from the sequence of its cDNA that was prepared from the adult whole eye total RNA. The unrooted phylogenetic relationships between different nocturnin proteins were analyzed with VectorNTI and the MEGA4 program using the neighbor-joining method.
Transgenesis
A 417-bp DNA fragment of the frog nocturnin promoter [28] was amplified with PCR using Xenopus genomic DNA, a forward primer (5′-CTG CTC GAG CTA GTG AAA TGT CAC TGT G-3'; Xho I site, underlined), and a reverse primer (5′-ATA GAA TTC CTG TGC TTC CCT CTC AGC AC-3′; EcoR I site, underlined). The PCR conditions were initial denaturation at 94 °C for 2 min, and 35 cycles of amplification (94 °C for 30 s, 55 °C for 30 s, and 68 °C 2 min). The Tol2-based transgenesis system was used to generate a transgenic fish line that expresses green fluorescent protein (GFP) under the control of the frog nocturnin promoter using a standard protocol as described previously [29]. A stable zebrafish transgenic line, designated Tg(nocturninfrog:GFP)pt158, was identified through PCR genotyping of F1 generation and screening of GFP expression.
Immunohistochemistry
Immunohistochemical confocal microscopic analyses were performed to examine the GFP expression patterns in the Tg(nocturninfrog:GFP)pt158 transgenic fish using protocols as described previously [30]. Anti-GFP (Sigma-Aldrich, St. Louis, MO, G1544–100UG; 1:300), zpr1 antibody (ZFIN, 1:300), and zpr3 antibody (ZFIN, 1:300) were used to visualize GFP, green/red double cones, and rods, respectively.
Western blotting
The adult fish eyes were homogenized and extracted with a lysis buffer (1X PBS [137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.3], 1% Triton X-100, and 1X protease inhibitor cocktail [Roach Diagnostics, Indianapolis, IN]) for 30 min on ice. The supernatants of the lysates were mixed with a 6X SDS loading buffer and boiled for 5 min. Protein lysates was separated with 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and analyzed with standard western blotting. The signal intensities of the GFP expression were measured with ImageJ software (National Institutes of Health, Bethesda, MA), and their relativity to α-tubulin signal intensities were determined with the GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA), with the relative intensity at 8 AM set as 1.
Nocturnin-b in situ hybridization
A zebrafish nocturnin-b cDNA fragment was amplified with RT–PCR with a forward primer (5′-TCC GAA TTC CGT GTT CCA TGG GCA GCG GC-3′; EcoR I site, underlined) and a reverse primer (5′-TCC AAG CTT ATA ACA ATC TGG CTG GAT CTT CGT-3′; Hind III site, underlined) and subsequently cloned in the pDrive vector. The resulting pDrive construct was linearized and transcribed with the T7/SP6 mMESSAGE mMACHINE Kit (Ambion, Thermo Fisher Scientific, Waltham, MA) to make the antisense and sense nocturnin-b Digoxigenin-RNA probes. After the RNA synthesis, the DNA templates were removed with DNase I digestion. The RNA probes were precipitated with ethanol and dissolved in 50% formamide and stored at −80 °C.
The adult zebrafish eyes were fixed with 4% paraformaldehyde overnight at 4 °C and cryosectioned at 300 μm thickness. The eye sections were dehydrated with a series of PBS/methanol solutions, followed by treatment with 100% acetone for 8 min at −20 °C. The samples were then rehydrated with a series of methanol/PBST (1% Triton X-100 in 1X PBS) washes and treated with 1 μg/ml proteinase K for 20 min. The proteinase K-treated sections were fixed again with 4% paraformaldehyde for 20 min at room temperature and washed with PBS. The retinal sections were then analyzed with in situ hybridization by using a previously published protocol [29]. The stained sections were then embedded in JB4 resin and cut at 4 μm thickness. The sections were stained with propidium iodide (PI; Invitrogen; 1:200) to visualize the cell nuclei. The slides were reviewed under an Axio Imager microscope (Carl Zeiss, Oberkochen, Germany).
Results and Discussion
Identification of the zebrafish nocturnin genes
To identify the zebrafish nocturnin genes, we searched the published zebrafish genome with the amino acid sequence of Xenopus nocturnin as a query [22]. The search identified two predicted zebrafish genes (XP_697426 and XP_700794) that show significant similarity to the frog nocturnin gene (Figure 1A,B). We designate the XP_700794 gene as the nocturnin-a gene and the XP_697426 gene as the zebrafish nocturnin-b gene. It is predicted that nocturnin-a encodes a polypeptide of 432 amino acids and nocturnin-b encodes a polypeptide of 371 amino acids. To determine which nocturnin gene is expressed in the eye, we next examined nocturnin-a and nocturnin-b mRNA levels in 5-dpf eyes, adult eyes, and 5-dpf whole fish with RT–PCR. We found that nocturnin-b was expressed in larval and adult eyes, whereas nocturnin-a was not, despite being expressed in the whole larval fish; these results suggest that the eye expresses nocturnin-b but not nocturnin-a, and that the two genes may promote mRNA depolyadenylation in a distinct tissue-specific fashion (Figure 1C). Furthermore, nocturnin-b expression in the eye is rhythmic, at higher levels at night than during the day (Figure 1D,E), echoing the rhythmic expression of nocturnin in the frog [22].
We next cloned and sequenced the entire cDNA of nocturnin-b to confirm the predicted amino acid sequence in the GenBank database. At the amino acid level, the two zebrafish nocturnin homologs show more than 60% similarity to their counterparts in the frog, human, and mouse (Figure 1B). In particular, the catalytic domains of the zebrafish nocturnin homologs are about 70% identical to their counterparts, suggesting a conserved catalytic function (Figure 1B, boxed area). Thus, the highly conserved nocturnin-b gene is expressed in the zebrafish eye, and the gene potentially regulates the mRNA turnover of the retinal cells in a circadian rhythm.
Expression patterns of nocturnin-b in zebrafish eyes
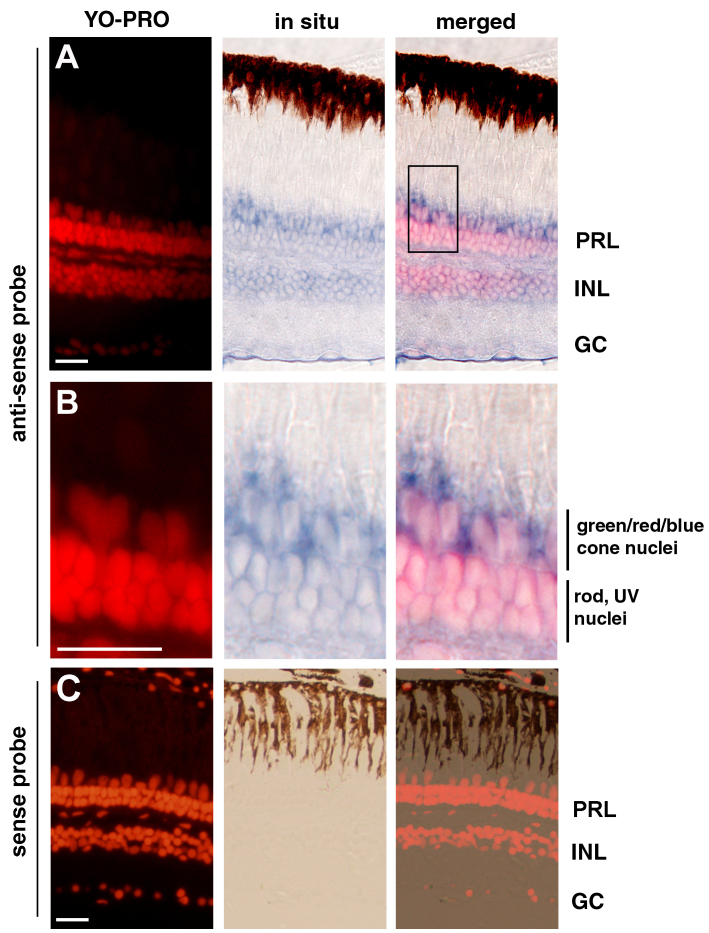
Previous studies showed that in the frog, the nocturnin gene is restrictively expressed in the retinal photoreceptors [22,28], whereas in the mouse, the gene is broadly expressed in various tissues, such as the liver, spleen, kidney, and heart [25]. This difference in expression pattern patterns prompted us to determine which cells express the nocturnin-b gene in the zebrafish retina. With in situ hybridization analysis, we found that the highest levels of the nocturnin-b gene were expressed in the photoreceptor layer (PRL), compared with the inner nuclear layer (INL) and the ganglion cell layer (GCL; Figure 2). The strongest expression of the nocturnin-b gene in zebrafish photoreceptors suggests that the nocturnin-b gene plays an important role in regulating the circadian turnover of mRNA in photoreceptors, a function that echoes the gene’s counterpart in the frog [22,28].
Figure 2.
In situ hybridization analysis showed that the nocturnin-b gene is expressed in multiple retinal cell types. A: The nocturnin-b mRNA signals, visualized with an anti-sense probe (blue), were stronger in the photoreceptor layer (PRL) than in the inner nuclear (INL) and the ganglion cell layer (GC). B: A local region of the photoreceptor layer in panel A (boxed region) is magnified to better illustrate the enrichment of the nocturnin-b mRNA signals at the inner segment regions. The cell nuclei were stained with YO-PRO (red). C: In situ hybridization with a sense probe for nocturnin-b did not stain the retina, supporting the staining specificity by the anti-sense probe in panels A and B. Scale bars, 20 μm.
Transcriptional activity of the frog nocturnin promoter in zebrafish rods
Because transcription factors interact with cis-regulatory elements to drive transcription [31], the broader expression of the nocturnin-b gene in the zebrafish retina than the gene’s counterpart in the frog retina suggests mechanistic differences in activating nocturnin transcription. For example, the cis-regulatory elements of the frog nocturnin gene may be different from those of the nocturnin-b gene, and as a result, it can be activated only in photoreceptors [28]. Could it be possible that the cis-regulatory elements of the frog nocturnin gene are also functional specifically in the zebrafish photoreceptors?
To investigate this possibility, we next examined whether a 417-bp frog nocturnin promoter, which is sufficient to express a GFP reporter gene specifically in the frog photoreceptors [28], can drive photoreceptor expression of a GFP reporter in the zebrafish by generating a stable transgenic zebrafish line Tg(nocturninfrog:GFP)pt158 (Figure 3A). In Tg(nocturninfrog:GFP) pt158, GFP was detected only in the outer nuclear layer (ONL) but not in other two retinal cellular layers. Interestingly, the GFP signals were restricted to rods: The GFP-positive cell nuclei, round and strongly TO-PRO-stained, resided in the basal half of the ONL, where rod nuclei localize (Figure 3B-E); furthermore, GFP immunostaining localized to cells that were positive for rod marker Zpr3 (Figure 3F-I). In contrast, GFP signals were not detected in the double cones and the ultraviolet (UV) cones (Figure 3E,J,M). In addition to rod-specific transcriptional activity, the frog nocturnin promoter in Tg(nocturninfrog:GFP) pt158 was more than 1,000 times weaker than the rod opsin promoter in the Tg(−3.7rho:EGFP)kj2 transgenic fish (Figure 3N) [32]. However, the GFP protein level did not show apparent rhythmic fluctuation, possibly due to the greater stability of the GFP protein (Figure 3O). Thus, the promoter of the frog nocturnin gene can be activated only in the rods in the zebrafish retina.
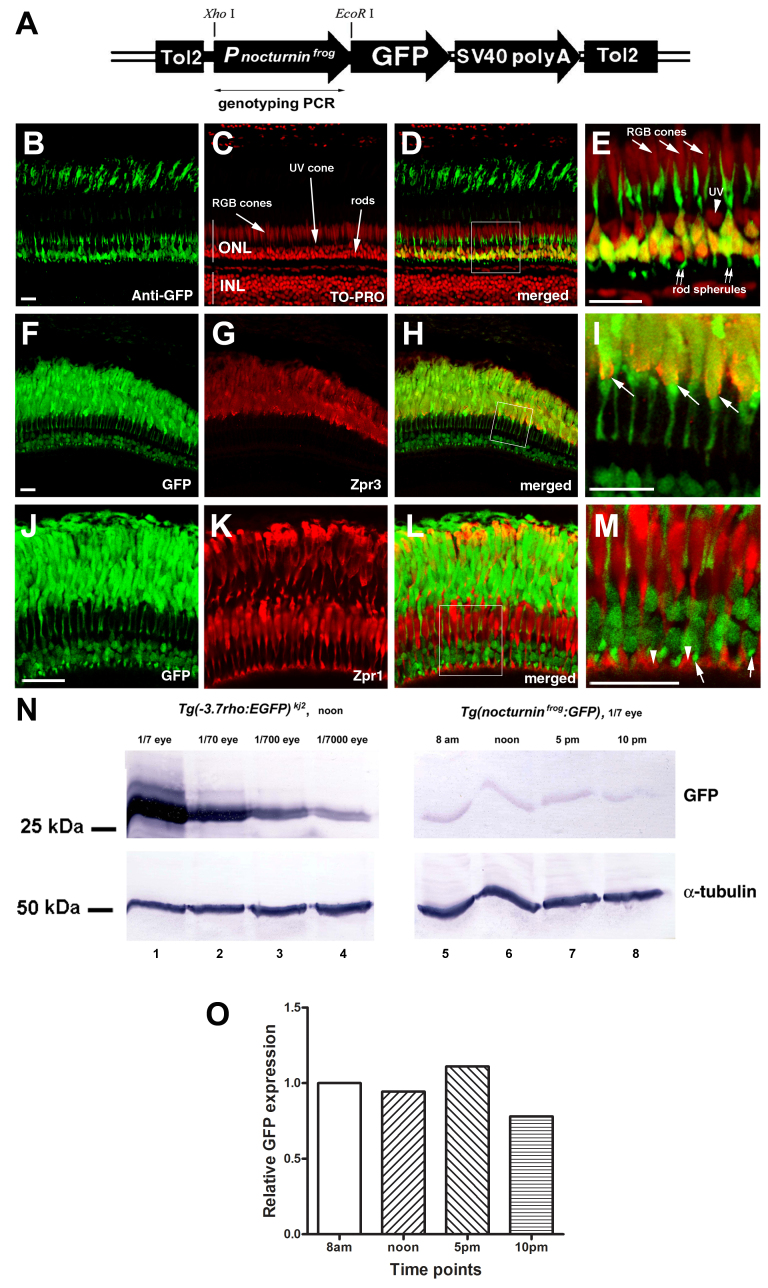
Figure 3.
Frog nocturnin promoter drives rod-specific transcription in the zebrafish retina. A: A schematic illustrates the configuration of the transgene cassette to express green fluorescent protein (GFP) in the zebrafish under the control of the frog nocturnin promoter, which covers 398 bp upstream of and 21 bp downstream of the translation start site of the nocturnin gene. This construct, thus, drives the expression of a fusion protein between GFP and the first seven amino acids of frog nocturnin. SV40 polyA stands for the 3′- untranslated region (UTR) of the SV40 gene and its polyadenylation signals. The transgene is flanked by two Tol2 transposon elements (Tol2). The diagram is not drawn to scale. A double-headed arrow indicates the region that was amplified when the fish was PCR genotyped. B–E: In Tg(nocturninfrog:GFP) pt158, GFP immunostaining highlighted rods. GFP was enriched in the rod cell nuclei in the basal half of the outer nuclear layer (green; arrows). Double arrows indicate the GFP signals in the rod spherules. In contrast, no GFP expression was detected in the UV cone nuclei (arrowhead) and the RGB cones (red, green, and blue cones). The cell nuclei were stained with TO-PRO (red). Panel E is a magnification of the boxed region in panel D. F-I: In Tg(nocturninfrog:GFP) pt158, GFP localized to cells positive for the rod marker Zpr3 at the outer segments (arrows). Panel I is a magnification of the boxed region in panel H. J–M: In Tg(nocturninfrog:GFP) pt158, GFP signals did not colocalize with double cone marker Zpr1. Panel M is a magnification of the boxed region in panel L. Arrowheads indicate the double cone pedicle synaptic junctions, and arrows indicate the rod spherules. The GFP signals at the outer segments were stronger when GFP was visualized directly with its own florescence (panels F and J) than indirectly with immunostaining (panel B). This difference is believed to be due to the strong autofluorescence of the visual pigments at the outer segments: When GFP expression was as weak as in Tg(nocturninfrog:GFP) pt158, to directly visualize GFP expression with its own florescence under confocal microscopy, we needed to use a much stronger laser power, which made the autofluorescence of the visual pigments more prominent. Thus, the GFP expression patterns in Tg(nocturninfrog:GFP) pt158 could be more reliably determined by its presence in the synaptic junctions, cell body, and inner segments. N: Western blotting showed that even after a 1,000-fold dilution with wild-type eye extracts, the GFP level in the Tg(−3.7rho:EGFP)kj2 transgenic fish eye was still significantly higher than that in Tg(nocturninfrog:GFP) pt158 (compare lane 6 with lane 4). α-tubulin blotting served as a loading control. O: A histogram of the intensity ratio between Tg(nocturninfrog:GFP) pt158 GFP and α-tubulin signals (N) normalized against the ratio at 8 AM, shows no apparent rhythmic fluctuation in GFP levels among the four time points. Scale bars, 20 μm.
The frog nocturnin promoter contains an E-box-like nocturnin element, which functions as a cyclic AMP response element to recruit the phosphorylated CREB transcription factor for its activation [28]. The rod-specific expression of GFP in Tg(nocturninfrog:GFP) pt158 suggests that the frog nocturnin promoter may carry sufficient cis-regulatory elements for its activation in the zebrafish rod photoreceptors but not necessarily for other zebrafish retinal cell types.
The characteristics of GFP expression in Tg(nocturninfrog:GFP) pt158 make the frog nocturnin promoter desirable for moderate transgenic expression in the zebrafish rod photoreceptors. The zebrafish rod opsin promoters have been frequently used to express transgenes in rods thanks to the promoters’ superb rod photoreceptor specificity [32,33]. However, zebrafish rod opsin promoters can be problematic because their strong transcriptional activities may cause unwanted overexpression phenotypes in certain situations. Thus, the frog nocturnin promoter becomes particularly useful when low-level transgenic expression is desired in zebrafish rod photoreceptors.
Acknowledgments
This study was supported by a NIH core grant (5P30EY008098), NIH R01EY025638 (to XW), NIH R21EY023665 (to XW), a Research to Prevent Blindness (RPB) Wasserman Merit Award (to XW), an unrestricted RPB grant (to the Ophthalmology Department of the University of Pittsburgh), and a grant from Natural Science Foundation of Guangdong Province (2015A030313440 to XY). Dr. Xiaojun Yang (yangx@stu.edu.cn) and Dr.Xiangyun Wei (weix@upmc.edu) are co-corresponding authors for this paper.
References
- 1.Pittendrigh CS. Temporal organization: reflections of a Darwinian clock-watcher. Annu Rev Physiol. 1993;55:16–54. doi: 10.1146/annurev.ph.55.030193.000313. [DOI] [PubMed] [Google Scholar]
- 2.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–41. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 3.Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–41. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer. 2003;3:350–61. doi: 10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- 5.Hastings MH, Reddy AB, Maywood ES. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4:649–61. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- 6.Ralph MR, Foster RG, Davis FC, Menaker M. Transplanted suprachiasmatic nucleus determines circadian period. Science. 1990;247:975–8. doi: 10.1126/science.2305266. [DOI] [PubMed] [Google Scholar]
- 7.Tosini G, Menaker M. Circadian rhythms in cultured mammalian retina. Science. 1996;272:419–21. doi: 10.1126/science.272.5260.419. [DOI] [PubMed] [Google Scholar]
- 8.Barbot W, Wasowicz M, Dupressoir A, Versaux-Botteri C, Heidmann T. A murine gene with circadian expression revealed by transposon insertion: self-sustained rhythmicity in the liver and the photoreceptors. Biochim Biophys Acta. 2002;1576:81–91. doi: 10.1016/s0167-4781(02)00296-8. [DOI] [PubMed] [Google Scholar]
- 9.Underwood H. Circadian organization in nonmammalian vertebrates. In Handbook of Behavioral Neurobiology: Circadian Clocks (Kluwer Academic/Plenum publishers), 2001; pp. 111–140. [Google Scholar]
- 10.Deguchi T. A circadian oscillator in cultured cells of chicken pineal gland. Nature. 1979;282:94–6. doi: 10.1038/282094a0. [DOI] [PubMed] [Google Scholar]
- 11.Besharse JC, Iuvone PM. Circadian clock in Xenopus eye controlling retinal serotonin N-acetyltransferase. Nature. 1983;305:133–5. doi: 10.1038/305133a0. [DOI] [PubMed] [Google Scholar]
- 12.Meijer JH. Photic entrainment in mammals. In Handbook of Behavioral Neurobiology: Circadian Clocks (Kluwer Academic/Plenum publishers), 2001; pp. 183–222. [Google Scholar]
- 13.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–42. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamazaki S, Goto M, Menaker M. No evidence for extraocular photoreceptors in the circadian system of the Syrian hamster. J Biol Rhythms. 1999;14:197–201. doi: 10.1177/074873099129000605. [DOI] [PubMed] [Google Scholar]
- 15.Cooper NG, McLaughlin BJ. Structural correlates of physiological activity in chick photoreceptor synaptic terminals: effects of light and dark stimulation. J Ultrastruct Res. 1982;79:58–73. doi: 10.1016/s0022-5320(82)90052-1. [DOI] [PubMed] [Google Scholar]
- 16.Adly MA, Spiwoks-Becker I, Vollrath L. Ultrastructural changes of photoreceptor synaptic ribbons in relation to time of day and illumination. Invest Ophthalmol Vis Sci. 1999;40:2165–72. [PubMed] [Google Scholar]
- 17.Anderson FE, Green CB. Symphony of rhythms in the Xenopus laevis retina. Microsc Res Tech. 2000;50:360–72. doi: 10.1002/1097-0029(20000901)50:5<360::AID-JEMT5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 18.LaVail MM. Rod outer segment disc shedding in relation to cyclic lighting. Exp Eye Res. 1976;23:277–80. doi: 10.1016/0014-4835(76)90209-8. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi JS. Circadian rhythms: from gene expression to behavior. Curr Opin Neurobiol. 1991;1:556–61. doi: 10.1016/s0959-4388(05)80028-5. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi JS. Circadian-clock regulation of gene expression. Curr Opin Genet Dev. 1993;3:301–9. doi: 10.1016/0959-437x(93)90038-q. [DOI] [PubMed] [Google Scholar]
- 21.Pierce ME, Sheshberadaran H, Zhang Z, Fox LE, Applebury ML, Takahashi JS. Circadian regulation of iodopsin gene expression in embryonic photoreceptors in retinal cell culture. Neuron. 1993;10:579–84. doi: 10.1016/0896-6273(93)90161-j. [DOI] [PubMed] [Google Scholar]
- 22.Green CB, Besharse JC. Identification of a novel vertebrate circadian clock-regulated gene encoding the protein nocturnin. Proc Natl Acad Sci USA. 1996;93:14884–8. doi: 10.1073/pnas.93.25.14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayasaka N, LaRue SI, Green CB. In vivo disruption of Xenopus CLOCK in the retinal photoreceptor cells abolishes circadian melatonin rhythmicity without affecting its production levels. J Neurosci. 2002;22:1600–7. doi: 10.1523/JNEUROSCI.22-05-01600.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stubblefield JJ, Terrien J, Green CB. Nocturnin: at the crossroads of clocks and metabolism. Trends Endocrinol Metab. 2012;23:326–33. doi: 10.1016/j.tem.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Osterbur DL, Megaw PL, Tosini G, Fukuhara C, Green CB, Besharse JC. Rhythmic expression of Nocturnin mRNA in multiple tissues of the mouse. BMC Dev Biol. 2001;1:9. doi: 10.1186/1471-213X-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–20. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 27.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 28.Liu X, Green CB. A novel promoter element, photoreceptor conserved element II, directs photoreceptor-specific expression of nocturnin in Xenopus laevis. J Biol Chem. 2001;276:15146–54. doi: 10.1074/jbc.M009970200. [DOI] [PubMed] [Google Scholar]
- 29.Zou J, Beermann F, Wang J, Kawakami K, Wei X. The Fugu tyrp1 promoter directs specific GFP expression in zebrafish: tools to study the RPE and the neural crest-derived melanophores. Pigment Cell Res. 2006;19:615–27. doi: 10.1111/j.1600-0749.2006.00349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei X, Zou J, Takechi M, Kawamura S, Li L. Nok plays an essential role in maintaining the integrity of the outer nuclear layer in the zebrafish retina. Exp Eye Res. 2006;83:31–44. doi: 10.1016/j.exer.2005.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–26. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 32.Hamaoka T, Takechi M, Chinen A, Nishiwaki Y, Kawamura S. Visualization of rod photoreceptor development using GFP-transgenic zebrafish. Genesis. 2002;34:215–20. doi: 10.1002/gene.10155. [DOI] [PubMed] [Google Scholar]
- 33.Fadool JM. Development of a rod photoreceptor mosaic revealed in transgenic zebrafish. Dev Biol. 2003;258:277–90. doi: 10.1016/s0012-1606(03)00125-8. [DOI] [PubMed] [Google Scholar]