Abstract
Background:
Clinically annotated specimens from cancer clinical trial participants offer an opportunity for discovery and validation of pharmacogenomic findings. The purpose of this observational study is to better understand patient/institution factors that may contribute to participation in the pharmacogenomic component of prospective cancer clinical trials.
Methods:
Patient demographic information (age, sex, self-reported race) and institutional characteristics (CALGB/CTSU site, “diversity,” and accrual) were evaluated for 8456 patients enrolled in seven CALGB phase III studies with a pharmacogenomic component. All statistical tests were two-sided.
Results:
The majority of patients (81%) consented to participate in the pharmacogenomic component. However, in a multivariable analysis, site (CALGB vs CTSU) and “institutional diversity” (percent minority cancer patients on national trials) were statistically significantly associated with participation. For both whites and nonwhites, patients from CALGB sites were more likely to participate compared with patients from CTSU sites (whites: odds ratio [OR] = 2.26, 95% confidence interval [CI] = 1.68 to 3.04, P < .001; nonwhites: OR = 1.79, 95% CI = 1.52 to 2.11, P < .001). However, as “institutional diversity” increased, the likelihood of participation in the pharmacogenomics component decreased for both white (OR = 0.94, 95% CI = 0.91 to 0.97, P < .001) and nonwhite patients (OR = 0.90, 95% CI = 0.81 to 1.00, P = .05).
Conclusions:
Most clinical trial cancer patients across geographical, racial, and practice settings are willing to participate in pharmacogenomic studies. However, to promote equitable benefit to the larger cancer community, optimization of both patient and institutional participation are needed. Institutional factors may be even more compelling than patient demographics. Prospective studies are needed to identify and address barriers/incentives to participation in pharmacogenomic research at the patient, clinician, and institutional levels.
Pharmacogenomics is the study of heritable and somatic genomic variation associated with drug response, including the prediction of toxicity and effectiveness (1–3). The field holds the promise of providing objective ways to individualize therapy, especially in the treatment of cancer patients (4,5). An efficient strategy to conduct these studies is to incorporate pharmacogenomics into prospective cancer clinical trials (6), especially large phase III studies (7). To study heritable variations in the clinical trial setting, germline DNA is extracted from blood samples of study patients who provide consent to participate in this component of the clinical trial.
The current observational study evaluates the participation of more than 8000 cancer patients in the inherited pharmacogenomics portion of seven different clinical trials from the Cancer and Leukemia Group B (CALGB) (now part of the Alliance for Clinical Trials in Oncology). Although many studies have evaluated participation in adult cancer clinical trials (6, 8–14), unique to this study is the evaluation of participation in the optional pharmacogenomics component among a group of cancer patients who have already agreed to participate in the clinical drug trial. The CALGB was among the first National Cancer Institute (NCI)–cooperative oncology groups to provide the opportunity for cancer patients to participate in optional pharmacogenomics studies, where germline DNA was collected from consenting patients to assess the relationship of heritable variations with clinical outcome (15).
In 2002, when these pharmacogenomic companion studies started, it was routine to collect tumor tissue for correlative science studies. However, it was not routine to collect germline DNA for genetic research, whether the intent was to evaluate inherited susceptibility to cancer development or to evaluate inherited pharmacogenomic variations to predict response to therapy. The pharmacogenomics studies reported in this manuscript represent one of the first times that an NCI-funded cooperative group was asking patients to contribute germline DNA to gain a better understanding of predicting response to anticancer treatments. None of these pharmacogenomic studies were utilizing pharmacogenomic results to guide therapy; all were collecting germline DNA for future pharmacogenomic research. Considering the general concerns reported in the literature at the time regarding fear of genetic research and misuse of genetic research information (16–23), our hypothesis was that participation in pharmacogenomic studies may be low because of the genetic (inherited) nature of these DNA studies. Although studies in the literature have reported on potential barriers to participation in clinical cancer trials and correlative tumor tissue studies (9,24–27), none, to our knowledge, have evaluated participation in a study of the genetics of drug response in cancer patients already participating in clinical trial research. In addition, to our knowledge, there have been no other reports evaluating participation in cancer pharmacogenomic studies in the context of multicenter clinical trials.
The primary aim of the current observational study was to evaluate the frequency with which cancer patients who had agreed to participate in a clinical cancer drug trial were participating in the optional germline pharmacogenomics component. Utilizing data already captured in the CALGB/Alliance database at the time of patient registration, we sought to identify patient and institutional factors that may contribute to participation in the pharmacogenomics component of these first seven phase III trials.
Methods
Protocol Selection
This study includes the initial seven optional pharmacogenomic studies embedded in CALGB phase III trials. The studies accrued patients from May 2002 to May 2013; all studies are now closed to accrual. All clinical trial studies, including the pharmacogenomic portion, were also available for registration by non-CALGB sites through the National Institutes of Health (NIH) Clinical Trials Support Unit (a service of the National Cancer Institute designed to facilitate access to NCI-funded clinical trials for qualified clinical sites and to support the management and conduct of those clinical trials). The trials included six different disease types: non-Hodgkin’s lymphoma (NHL) and breast, gastric, colorectal, pancreatic, and prostate cancer. The pharmacogenomic component was open to patient registration at the same time as the clinical trial in six of the seven trials. Institutional review board (IRB) approval was obtained at each site prior to patient registration to the clinical trial or optional companion studies.
Patient Selection
All patients (from CALGB and CTSU sites) registered to the seven treatment protocols were included in the analysis. Patients registered to CALGB 40101 before November 1, 2003 were excluded from the analysis because prior to this date the pharmacogenomic portion of the clinical trial had not yet been open to accrual.
Data Collection
Patient registration and data collection for the treatment trials and optional pharmacogenomic components were handled by the Alliance Statistics and Data Center using a secure server and database. Patients provided informed consent for treatment and optional companion studies; consent to each component of the trial was confirmed by each institution during the patient registration process. For patient characteristics, we utilized relevant demographic and clinical information collected at the time of patient registration to the clinical trial, including age, sex, self-reported race, and cancer diagnosis. Institutional characteristics were limited to the information retrievable from the CALGB/Alliance database and included the site through which the patient was registered to the clinical trial (CALGB or CTSU), the institutional accrual patterns to the clinical trial, and, separately, to the pharmacogenomic component. Patient participation in the optional pharmacogenomics portion of the study was confirmed by verification of consent in the CALGB database. Because patient consent was required for the optional biospecimen studies (eg, blood samples for pharmacogenomics, tumor tissue samples for correlative science) and any other companion studies (eg, health outcomes, quality of life, imaging), participation in the pharmacogenomics component of the study could be compared with participation in other companion studies.
Informed Consent Documents
All consent questions related to the optional pharmacogenomic and other laboratory tissue biomarker studies were included in a separate section of the clinical study consent document, termed “optional studies.” This section was always found at the end of the main consent document. All consents for these optional studies included checkboxes to allow patients to opt in or opt out of each correlative study (pharmacogenomic and tissue), including providing their specimens for future use (eg, you can use my sample for future genetic research, future cancer research, future noncancer research), with exact language varying slightly from study to study. All consent documents indicated that there was some risk involved in genetic research and described that samples and data were to be coded to protect individual privacy and confidentiality of information. Local IRBs at both CALGB and CTSU institutions may choose to add additional language about risk to these consents, based on local policy; however, those language changes must be in compliance with the standard template language required by NCI for cooperative group study consents, including language for optional correlative science studies. No changes can be made to the consent questions. Overall, across the seven studies, we did not observe any feature or combination of features of the consent document associated with participation to the pharmacogenomics study.
Statistical Methods
Descriptive analysis was conducted using data collected in the CALGB database. Self-reported race categories were dichotomized and collapsed into white (Caucasian) vs nonwhite (African American, Asian, and other (Native American, Pacific Islanders, Hawaiian, mixed or multiple races). Only patients with known race were included in the analyses. Only 2.5% of patients had unknown race.
To look at rate of institutional accrual to the pharmacogenomic component of the trial, we evaluated the number of patients accrued to the pharmacogenomic study at the institution per year during the time the institution was actively accruing patients to the study. A Wilcoxon rank-sum test was used to compare the distribution of number of patients accrued to either the pharmacogenomic study or the specific clinical trial per institution between CALGB and CTSU institutions. Chi-squared tests were used to compare percentages among groups (eg, pharmacogenomic participation between individual clinical trials).
We modeled the probability of consent to pharmacogenomics as a function of self-reported race (nonwhite vs white), site (CALGB vs CTSU), age (with odds ratios reported for 10-year increments), sex (male vs female), and rate of institutional accrual to the pharmacogenomic study using a generalized estimating equation (GEE) model with a logistic link (28). The GEE models perform logistic regression while accounting for the variability of participation or “clustering” among the separate clinical trials. This “cluster variable” accounts for the fact that patients are clustered within individual clinical trials. The interactions were considered within the context of the multivariable GEE model. The inference and estimation of the terms in the log-additive GEE models were carried out under an asymptotic framework, assuming that the marginal likelihood ratio statistic was asymptotically chi-square (29) with appropriate degrees of freedom, using SAS PROC GENMOD (version 9.2).
To further understand the statistical interaction between patient self-reported race (white vs nonwhite) and site (registration to the clinical trial by a CALGB institution vs the CTSU mechanism) with participation in the pharmacogenomic component of the trial, we conducted an exploratory analysis including an estimate of “institutional diversity.” To approximate racial diversity at institutions in our study we calculated a proxy, or diversity variable (minority participation fraction [MPF]), based on the percentage of nonwhite cancer patients that an institution registered to any national cooperative group trial (nonwhite cancer patients/total cancer patients [white + nonwhite] registered to a CTSU trial by that institution). This information was obtained from the NCI Regulatory Support System (RSS) database, a registry that records and archives information from all institutions that register a cancer patient through the CTSU mechanism, and includes a site identification code and patient race. The RSS database was used because of an insufficient number of patients per institution to make this estimate using the CALGB database when restricted to the 812 institutions participating in the phase III CALGB studies included in our study. Assignment of the MPF from the RSS database was limited to those institutions that registered 10 or more patients to any of the clinical trials in our study. Using the institutional identification code and applying the above criteria, 652 of the 812 (80%) institutions in our current study were assigned a MPF. The MPF (with odds ratio reported for 10% increments) was used in the final multivariable model. The relationship between MPF and participation was illustrated graphically using box plots where the MPF was categorized by 10 levels: 0% (no diversity), 1% to 9%; 10% to 19%, 20% to 29% and so on, up to 89% (the highest level of diversity [MPF] estimated).
All statistical tests were two-sided, and a P value of less than .05 was considered statistically significant.
Results
Treatment Trial Characteristics and Patient Demographics
A total of 8456 patients were analyzed. Table 1 summarizes the patient demographic and treatment trial characteristics. Registration to each clinical trial ranged from 238 to 3314 patients. Self-reported race demonstrated that the majority of patients were Caucasian (83.0%, 7019), followed by African American (11.1%, 939), Asian (2.5%, 211) and other (1.0%, 83, either Native Hawaiian or Pacific Islander, American Indian or Alaska Native, mixed or multiple race). Race was unknown for 204 patients (2.4%). Females constituted 59.3% of the total patient population, and median age for all patients was 58.3 years (55 years for females vs 63 years for males, P < .001, Wilcoxon rank-sum test).
Table 1.
Characteristics of Patients By Clinical Trial
| Clinical trial (DISEASE) | 40101* breast cancer | 50303 non- Hodgkin’s lymphoma | 80101 gastric cancer | 80203 metastatic colorectal cancer | 80405 metastatic colon cancer | 80303 pancreatic cancer | 90401 prostate cancer | Total |
|---|---|---|---|---|---|---|---|---|
| Characteristic | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) | No. (%) |
| Sample size | 3314 (39.2) | 423 (5.0) | 546 (6.5) | 238 (2.8) | 2283 (27.0) | 602 (7.1) | 1050 (12.4) | 8456 |
| Trial status | Closed | Open | Closed | Closed | Closed | Closed | Closed | --- |
| Race | ||||||||
| Caucasian | 2759 (83.3) | 339 (80.1) | 403 (73.8) | 207 (87.0) | 1859 (81.4) | 529 (87.9) | 923 (87.9) | 7019 (83.0) |
| African American | 362 (10.9) | 47 (11.1) | 71 (13.0) | 27 (11.3) | 273 (12.0) | 49 (8.1) | 110 (10.5) | 939 (11.1) |
| Asian | 62 (1.9) | 14 (3.3) | 45 (8.2) | 1 (0.4) | 72 (3.2) | 10 (1.7) | 7 (0.7) | 211 (2.5) |
| Other | 36 (1.1) | 7 (1.7) | 4 (0.7) | 1 (0.4) | 25 (1.1) | 5 (0.8) | 5 (0.5) | 83 (1.0) |
| Unknown | 95 (2.9) | 16 (3.8) | 23 (4.2) | 2 (0.8) | 54 (2.4) | 9 (1.5) | 5 (0.5) | 204 (2.4) |
| Minority† | ||||||||
| White | 2759 (85.7) | 339 (83.3) | 403 (77.1) | 207 (87.7) | 1859 (83.4) | 529 (89.2) | 923 (88.3) | 7019 (85.1) |
| Nonwhite | 460 (14.3) | 68 (16.7) | 120 (22.9) | 29 (12.3) | 370 (16.6) | 64 (10.8) | 122 (11.7) | 1233 (14.9) |
| Sex | ||||||||
| Male | NA | 226 (53.4) | 371 (67.9) | 140 (58.8) | 1324 (58.0) | 329 (54.7) | 1050 (100.0) | 3440 (40.7) |
| Female | 3314 (100.0) | 197 (46.6) | 175 (32.1) | 98 (41.2) | 959 (42.0) | 273 (45.3) | NA | 5016 (59.3) |
| Age, y | ||||||||
| Median | 53 | 57.5 | 58.7 | 61.3 | 59.1 | 64.2 | 69 | 58.3 |
| Range | 22.5 – 84.7 | 18.8 – 86.6 | 23.2 – 83.1 | 22 – 84.4 | 20.8 – 89.5 | 26.3 – 88.6 | 41.7 – 93.5 | 18.8–93.5 |
| Institution | ||||||||
| CTSU | 1755 (53.0) | 186 (44.0) | 384 (70.3) | 123 (51.7) | 1553 (68.0) | 282 (46.8) | 564 (53.7) | 4847 (57.3) |
| CALGB | 1559 (47.0) | 237 (56.0) | 162 (29.7) | 115 (48.3) | 730 (32.0) | 320 (53.0) | 486 (46.3) | 3609 (42.7) |
| Consent to pharmacogenomics | 2768 (83.5) | 384 (90.8) | 416 (76.2) | 219 (92.0) | 1756 (76.9) | 475 (78.9) | 864 (82.3) | 6882 (81.4) |
* Trial 40101: patients registered before November 1, 2003 are not included in the statistical analysis, as the pharmacogenomics study was not open at that time.
† Eight thousand two hundred fifty-two patients with known racial status: white = Caucasian: nonwhite = African American, Asian, and other (Pacific Islander, Hawaiian, multirace).
Patients could be registered to the clinical trial (and pharmacogenomic component) as a CALGB site or as a CTSU site (Table 1). Different patterns of accrual to the clinical trials were observed based on site (CTSU vs CALGB). Of the 8456 patients registered to the seven trials, 57.3% were registered through the CTSU mechanism (CTSU sites) and 42.7% through CALGB (CALGB sites). Of the 812 institutions participating in these trials, the majority (72.8%) registered patients through the CTSU. Compared with CTSU sites, CALGB institutions had a higher median accrual per institution to any individual clinical trial (8 vs 4 patients per study, P < .001, Wilcoxon rank-sum test), and CALGB sites were more likely than CTSU sites to participate in multiple clinical trials (of the 7 trials included) (P < .001, Chi-squared test).
Patient Consent to the Pharmacogenomic Study
Of the 8456 patients studied, 81.4% (6882) consented to participate in the pharmacogenomic component of the clinical trial (Table 1). Participation ranged from 76.2% (Trial 80101, gastric cancer) to 92.0% (Trial 80203, metastatic colorectal cancer) and was statistically different between clinical trials (P < .001, Chi-squared test).
Patient Consent to Pharmacogenomics Compared With Other Companion Studies
We compared patient participation (patient consent) to the pharmacogenomic component compared with the tumor tissue component in those trials providing both types of optional correlative science studies. Table 2 only looks at the clinical trials that had both a tumor tissue correlative science component and a pharmacogenomics component. The table looks at four categories of participation in these optional biospecimen studies: 1) patients who did not consent to contribute any biospecimen—ie, patients did not consent to participate in the pharmacogenomic component and they did not consent to the tumor tissue correlative component; 2) patients provided consent only to the pharmacogenomic component (and not the tumor tissue component); 3) patients provided consent only to the tumor tissue component (and not the pharmacogenomic component); and 4) patients provided consent to participate in both the pharmacogenomic and the tumor tissue components. Across all trials, it was more likely that a patient would participate in both the pharmacogenomic and tissue studies (n = 5200, range = 74.9%-89.5%) than either one alone. Some patients did not participate in either type of biospecimen study (n = 788, varying from 4.2%-21.8% nonparticipation depending on the trial).
Table 2.
Consent to contribute blood for pharmacogenomics compared with tumor tissue for biomarker studies*
| Clinical trial # | Did not consent to biospecimen No. (%) | Consent to pharmacogenomics only No. (%) | Consent to tissue only No. (%) | Consent to both pharmacogenomics and tissue No. (%) |
|---|---|---|---|---|
| 40101 | 260 (7.8) | 241 (7.3) | 286 (8.6) | 2527 (76.3) |
| 50303 | 29 (6.9) | 11 (2.4) | 10 (2.6) | 373 (88.2) |
| 80101 | 119 (21.8) | 7 (1.3) | 11 (2.0) | 409 (74.9) |
| 80203 | 10 (4.2) | 6 (2.5) | 9 (3.8) | 213 (89.5) |
| 80405 | 370 (16.2) | 78 (3.4) | 157 (6.9) | 1678 (73.5) |
* Clinical trials that did not collect blood for pharmacogenomics as well as tumor tissue for correlative science are excluded from the table (eg, 90401, 80303).
We also evaluated patient participation to the pharmaco genomic or tissue studies relative to other available optional companions (eg, quality of life, imaging). We observed no evidence to indicate that patients were agreeing to participate in other companions and not in the pharmacogenomic study. In most trials, patients were participating in the pharmacogenomic study and at least one other companion (see Supplementary Table 1, available online).
Univariate Analysis
Combining all trials, univariate logistic regression analyses demonstrated no statistical differences in participation in the pharmacogenomic study by patient age or sex. However, institutional site (CALGB vs CTSU) and self-reported patient race were associated with pharmacogenomic participation (Table 3). Patients from CALGB sites were more likely to participate compared with patients from CTSU sites (odds ratio [OR] = 2.08, 95% confidence interval [CI] = 1.64 to 2.63, P < .001). Nonwhite patients were statistically significantly less likely to participate compared with whites (OR = 0.50, 95% CI = 0.43 to 0.57, P < .001). Overall, 71.4% (880/1233) of nonwhites (African American, Asian, other) consented to the pharmacogenomic component, compared with 83.4% (5853/7019) of whites (Caucasian). Participation was statistically significantly lower among nonwhites compared with whites within both CALGB and CTSU sites (CALGB: OR = 0.43, 95% CI = 0.39 to 0.48, P < .001; CTSU: OR = 0.51, 95% CI = 0.43 to 0.61, P < .001) (Table 3).
Table 3.
Factors associated with pharmacogenomic study participation: univariate model*
| Variable | OR (95% CI) | P |
|---|---|---|
| Site: CALGB vs CTSU | 2.08 (1.64 to 2.63) | <.001 |
| Self-reported race†: nonwhite vs white | 0.50 (0.43 to 0.57) | <.001 |
| Age‡ | 1.00 (0.97 to 1.03) | .97 |
| Sex: female vs male | 0.91 (0.76 to 1.10) | .33 |
| Rate of pharmacogenomic accrual§ | 1.00 (0.96 to 1.05) | .95 |
| Self-reported race by site | ||
| CALGB: nonwhite vs white | 0.43 (0.39 to 0.48) | <.001 |
| CTSU: nonwhite vs white | 0.51 (0.43 to 0.61) | <.001 |
* Generalized estimating equation model accounts for variability of patient participation among the separate clinical trials by use of study as a “cluster variable,” because patients are clustered within individual trials. Reported P values are two-sided. CI = confidence interval; OR = odds ratio.
† Nonwhite = African American, Asian, other (Native American, Pacific Islander, mixed race); white = Caucasian; persons with unknown data for race were excluded from this analysis (n = 8252).
‡ Age was analyzed as a continuous variable, and OR is reported for 10-year increments.
§ Rate of pharmacogenomics accrual calculated as the number of patients accrued to the pharmacogenomics study at each institution per year, during the time the institution was actively accruing patients to the study and is presented in increments of 10.
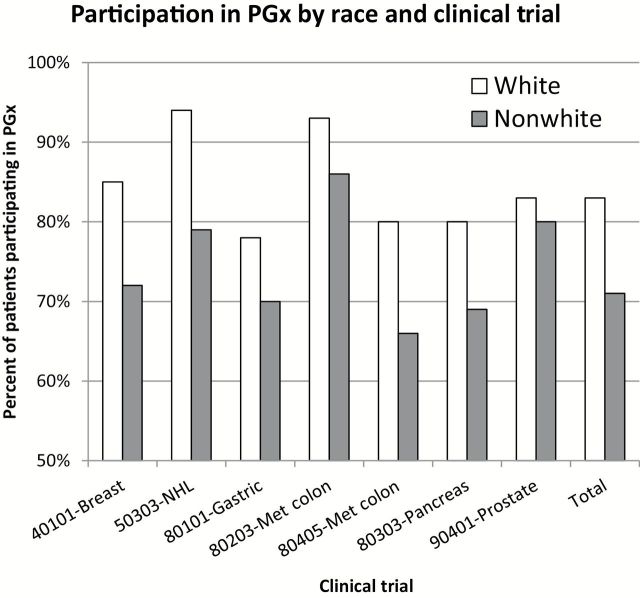
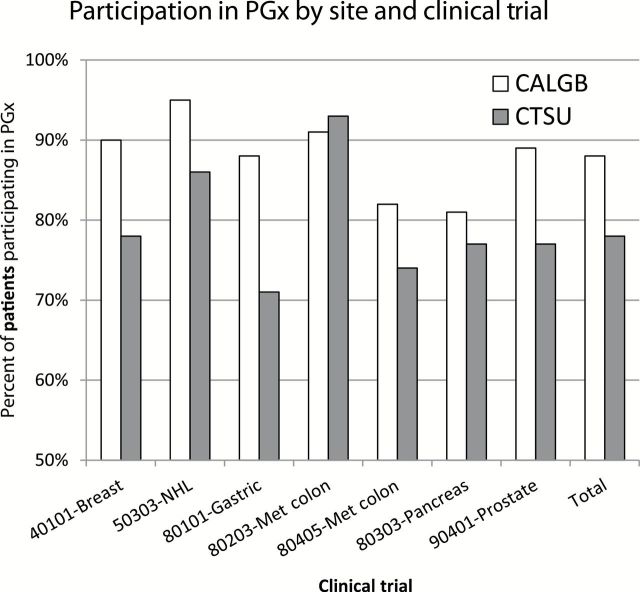
Evaluating trials separately, Figure 1 illustrates that for each individual clinical trial, participation by nonwhite patients was consistently lower compared with white patients. Figure 2 illustrates that for nearly all individual trials, patient participation was higher from CALGB sites compared with CTSU sites.
Figure 1.
Patient participation in pharmacogenomic (PGx) study by self-reported race. This histogram illustrates the percentage of patients participating in the pharmacogenomic component of each clinical trial (identified by clinical trial study number and disease stie), stratified by self-reported race (white: Caucasian; and non white: African American, Asian, other (American Indian, Alaska Native, or mixed race). Open bars represent whites; shaded bars represent nonwhites.
Figure 2.
Patient participation in pharmacogenomic study by site and trial. This histogram illustrates the percentage of patients participating in the pharmacogenomic (PGx) component of each clinical trial (identified by clinical trial study number and disease site), stratified by registration process (CALGB: patient was registered through a CALGB institution; CTSU: patient was registered through the Clinical Trials Support Unit [CTSU]).
Institutional Diversity
To further understand the interaction between site and race, we conducted an exploratory analysis that included a proxy for institutional racial diversity. This proxy was termed minority participation fraction (MPF) and reflected the percent minority cancer patients registered by an institution to any national cooperative cancer trial. When this proxy, MPF, was included in the multivariable regression analysis, we observed no evidence for an interaction with any of the covariates, indicating that the MPF diversity measure was an independent variable. The multivariable model presented in Table 4 is stratified by white vs nonwhite patient participation and includes the institutional MPF diversity variable. For whites and nonwhites (self-reported race), patients from CALGB sites were more likely to participate compared with patients from CTSU sites (whites: OR = 2.26, 95% CI = 1.68 to 3.04, P < .001; nonwhites: OR = 1.79, 95% CI = 1.52 to 2.11, P < .001). However, as the MPF diversity variable increased (measured in 10% increments), the likelihood of participation in the pharmacogenomics component decreased for both white (OR = 0.94, 95% CI = 0.91 to 0.97, P < .001) and nonwhite patients (OR = 0.90, 95% CI = 0.81 to 1.00, P = .05) (see Supplementary Tables 2 and 3, available online, for additional analyses by site, by individual trial).
Table 4.
Multivariable analysis stratified by race, including diversity*
| All trials | White patients (n = 6542) | Nonwhite patients (n = 1084) | ||
|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | |
| Site: CALGB vs CTSU | 2.26 (1.68 to 3.04) | <.001 | 1.79 (1.52 to 2.11) | <.001 |
| Age (10-year increase)† | 0.96 (0.92 to 0.99) | .05 | 1.03 (0.97 to 1.09) | .31 |
| Sex (female vs male) | 0.94 (0.80 to 1.10) | .44 | 0.86 (0.75 to 0.99) | .03 |
| Rate of pharmacogenomic accrual‡ | 1.01 (0.92 to1.11) | .83 | 1.04 (0.95 to 1.15) | .41 |
| Minority participation fraction (diversity variable)§ (10% increase) | 0.94 (0.91 to 0.97) | <.001 | 0.90 (0.81 to 1.00) | .05 |
* Generalized estimating equation model accounts for variability of patient participation among the separate clinical trials by use of study as a “cluster variable,” because patients are clustered within individual trials. The odds ratios presented are from the model without the interaction terms. Reported P values are two-sided. CI = confidence interval; OR = odds ratio.
† Age was analyzed as a continuous variable, and OR is reported for 10-year increments.
‡ Rate of pharmacogenomics accrual calculated as number of patients accrued to the pharmacogenomics study at each institution per year, during the time the institution was actively accruing patients to the study and is presented in increments of 10.
§ The minority participation fraction (MPF) was used as a proxy for institutional racial diversity, analyzed as a continuous variable, and OR is reported for 10% increases. The MPF reflects the proportion of nonwhite patients registered by an institution to any CTSU cooperative group study (from RSS database for Cancer Trials Support Unit). The MPF diversity variable was only assigned to sites with more than 10 patients enrolled in the seven pharmacogenomic clinical trials.
Figure 3 graphically illustrates the overall relationship between MPF and participation in the pharmacogenomic component. MPF was categorized into 10 levels (0%; 1%-9%; 10%-19%, etc., up to the highest diversity estimated [80%-89%]). Nearly 80% of institutions (507/652, 77.7%) were assigned an MPF-derived diversity score of less than 20%. High-diversity categories (50%-89% diversity) were assigned to 24 different institutions and represented 226 patients. Two groups of high-diversity institutions are observed with different participation: one group includes high-diversity institutions (60%-79%) with high median participation rates (84%-89%); the other group, (diversity categories 50%-59% and 80%-89%) had lower median participation rates (66.7% and 12.5%, respectively).
Figure 3.
Estimate of institutional “diversity” and PGx participation*. This figure graphically illustrates the overall relationship between the proxy for institutional diversity (minority participation fraction [MPF]) and participation in the pharmacogenomics component. MPF calculations were based on the percentage of nonwhite cancer patients that an institution registered to any CTSU cooperative group trial (nonwhite cancer patients/total cancer patients [white + nonwhite] registered to a CTSU trial by that institution). This information was obtained from the National Cancer Institute Regulatory Support System (RSS) database. MPF derived institutional diversity was categorized into 10 levels (0%; 1%-9%; 10%-19%, etc., up to the highest diversity estimated (80%-89%). The total number of institutions associated with each MPF-derived diversity category is indicated in parentheses along the x-axis. Box plots illustrate mean (diamonds), median (line), and interquartile range (IQR) of participation for each category (bottom and top edges of the box indicate IQR betwen the 25th and 75th percentiles). The whiskers that extend from each box indicate the range of values that are outside IQR but are close enough not to be considered outliers (a distance of less than or equal to 1.5*IQR). Any points that are a distance of more than 1.5*IQR from the box are considered to be outliers and are indicated by open circles.
Discussion
This study demonstrates that pharmacogenomic studies are clearly achievable in the context of publically funded multicenter cancer clinical trials, with over 80% of patients participating across six different types of cancer. This indicates an important pathway to the myriad systematic pharmacogenomic discovery and replication studies needed to move the molecular age of cancer medicine forward (30). However, we also identified the need to optimize both patient and institution participation in these studies.
Institutional diversity (minority participation fraction) was used as a proxy for institutional racial diversity. This variable was calculated as part of an exploratory analysis to better understand the statistical interaction observed between self-reported race (white vs nonwhite) and site (CALGB vs CTSU). Within both types of registration sites, nonwhite patients had a statistically significantly lower participation compared with white patients. In a multivariable analysis, the proxy diversity variable (minority participation fraction) was an independent predictor of participation, showing no evidence of interaction with the other covariates. The observation that some institutions with “high diversity” (high MPF) had decreased participation among whites as well as nonwhites suggests that patients at institutions that serve a larger minority population may be less likely to provide the opportunity to participate in optional pharmacogenomic studies compared with those institutions that serve a lower fraction of minority patients. It also highlights that factors at the institution level, in addition to those at the patient level, may be barriers to participation in optional biospecimen studies. These are important observations, worthy of further evaluation in future prospective studies.
Guth and colleagues observed that place of care has important implications for use of a clinical genomic test in breast cancer patients. In their study, use of the Oncotype Dx test was evaluated among minority and economically disadvantaged breast cancer patients receiving care in inner city hospitals. Patients treated at municipal hospitals were statistically significantly less likely to have the Oncotype Dx test ordered compared with patients at tertiary care hospitals (31). These observations further suggest that characteristics of the individual institution, in addition to the patient, need to be carefully considered to better understand and address the causal relationships with observed disparities in care. The report of a large and growing disparity in black vs white breast cancer mortality among the largest cities in the United States (32) further underscores the need to assess factors associated with “place as well as race” (31).
Our study uniquely demonstrates that, even among individuals who have already agreed to participate in a clinical research trial, some are not participating in the genetic component of the trial. One of the most important factors for this issue may be whether or not the patient had the opportunity to participate, ie, was approached to participate in the pharmacogenomic study. As an observational study, we cannot determine if the patient was approached and declined to participate or if the patient was only asked to participate in the treatment part of the study, not the pharmacogenomic component. Some studies suggest that racial and ethnic minorities in the United States are as willing to participate in health research as non-Hispanic whites, if they are asked (9,11,33).
A complex interplay of multiple factors acting as incentives and barriers to clinical research participation exist at the patient, clinician, institution, and community levels (9,12,14,34,35). The Accrual to Clinical Trials (ACT) model offers one approach to integrate factors at each of these levels, including norms, beliefs, attitudes, awareness, and opportunity, to improving ethnic/racial minority cancer patient accrual to cancer clinical trials (34). It is likely that many of these same issues apply to participation in pharmacogenomic studies. However, issues unique to genetic or genomic studies may also exist (36–39). At the patient level this includes mistrust (stemming not only from an individual’s fear of harm while participating in the genetic study, but also from misgivings about the consequences and ultimate uses of information gathered from these studies) (23,39) and misrepresentation or misapplication of the genetic data to “reinforce racism” (22). At the clinician and institutional levels, additional factors, including awareness, perceived value, familiarity with pharmacogenomic research, and limited time, are also highly influential (6,9). Studies from our group and others indicate a wide variability of clinician knowledge, comfort with interpreting results and adoption of cancer genomic tests, especially pharmacogenomic tests (40–43). In addition, it is feasible that some institutions may have given the priority and resources to provide a robust correlative science infrastructure, while others were not able to support collection and shipment of samples. Resource-poor institutions may focus on getting patients access to the drug study and not prioritize the time and effort to include optional biospecimen studies. Time pressures, a team approach to correlative science, resources for education, and the lack of direct benefit, for either the patient or the institution, are all practical challenges for clinicians and institutions who wish to offer their patients opportunities to participate in genomic research (9). The findings that CALGB institutions recruit statistically significantly more patients to participate compared with CTSU sites and that some institutions with “higher diversity” (some of which may be resource poor because of location or other financial constraints) recruit fewer patients to participate further suggest that the expertise, resources, infrastructure, and culture of an institution may provide an environment that is conducive to enrollment on pharmacogenomic and other correlative science elements of complex clinical trials. Sufficient incentives such as institutional participation credit for companion studies may also be a factor, especially for resource-poor institutions or for institutions only registering a few patients to a trial. Indeed, during this time period, it is our understanding that CALGB institutions were eligible for greater per-patient support of companion studies than non-CALGB sites that registered patients through the CTSU.
Our study had areas of limitation. First, we excluded from our analysis patients with no known race. However, this only reflects 2.5% of the eligible patients registered to the clinical drug studies. Second, the diversity variable (MPF) used best available data to estimate institutional diversity, but future studies could employ a more accurate measure of minorities served. In addition, because of the need for a quantifiable calculation, the diversity variable was only calculated for those institutions registering at least 10 patients to a study, thus reflecting only 80% of the institutions in our study. Nonetheless, these findings are suggestive of important site characteristics that need to be considered in future studies. Third, the influence of financial support for correlative science could have an influence on accrual to the pharmacogenomics component. Site-specific data was not available in the public domain but could be a variable analyzed in future assessments. Despite these limitations, our findings warrant further study.
Participation in cancer pharmacogenomics and correlative studies can be influenced by a number of factors (9,34,35,44,45). The main findings of our study highlight a shift in the approach to understanding disparity, still considering patient factors but also shifting attention to factors associated with place where care is received. Our study demonstrates that race alone does not explain participation. Factors at the institutional level also need to be considered, many of which could not be measured in our observational study. These are issues that lend themselves to quantitation, intervention, and improvement, elements that are amenable to a prospective study design. The field now needs to interrogate the process of clinical trial participation at a more granular level, to prospectively identify and address causal relationships for disparate correlative science participation, and to avoid the generation of a new class of therapeutically underprivileged patients, for whom the “best” application of modern cancer care lacks evidence and guidance.
Funding
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Numbers U10CA180821 and U10CA180882 to the Alliance for Clinical Trials in Oncology and the following grants to the legacy CALGB: CA31946, CA47559, and CA41287; and PAAR grant GM61393 to the University of Chicago (Mark J. Ratain).
Supplementary Material
The study funders had no role in design of the study, the collection, analysis, or interpretation of the data, the writing of the manuscript, nor the decision to submit the manuscript for publication.
The authors thank the numerous patients, institutions, and clinicians who participated in the clinical trials and pharmacogenomic studies. The authors also thank the staff of the former CALGB Pathology Coordinating Office and Chicago Central Protocol Office for their support.
References
- 1. Paugh SW, McCorkle JR, Diouf B, Crews KR, Evans WE. Cancer pharmacogenomics. Clin Pharmacol Ther. 2011;90(3):461–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang L, McLeod HL, Weinshilboum RM. Genomics and drug response. N Engl J Med. 2011;364(12):1144–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wheeler HE, Maitland ML, Dolan ME, et al. Cancer pharmacogenomics: strategies and challenges. Nat Rev Genet. 2013;14(1):23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. O’Donnell PH, Dolan ME. Cancer pharmacoethnicity: ethnic differences in susceptibility to the effects of chemotherapy. Clin Cancer Res. 2009;15(15):4806–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Innocenti F, Cox NJ, Dolan ME. The use of genomic information to optimize cancer chemotherapy. Sem Oncology. 2011;38(2):186–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Meropol NJ, Kris MG, Winer EP. The American Society of Clinical Oncology’s Blueprint for Transforming Clinical and Translational Cancer Research. J Clin Oncol. 2012;30(7):690–691. [DOI] [PubMed] [Google Scholar]
- 7. Freedman AN, Sansbury LB, Figg WD, et al. Cancer pharmacogenomics and pharmacoepidemiology: setting a research agenda to accelerate translation. J Natl Cancer Inst. 2010;102(22):1698–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kass NE, Sugarman J, Medley AM, et al. An intervention to improve cancer patients’ understanding of early-phase clinical trials. IRB. 2009;31(3):1–10. [PMC free article] [PubMed] [Google Scholar]
- 9. Baer AR, Smith ML, Collyar D, et al. Issues surrounding biospecimen collection and use in clinical trials. J Oncol Pract. 2010;6(4):206–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Howie LJ, Peppercorn JM. The ethics of clinical trials for cancer therapy. N C Med J. 2014;75(4):270–273. [DOI] [PubMed] [Google Scholar]
- 11. Weinfurt KP, Seils DM, Lin L, et al. Research participants’ high expectations of benefit in early-phase oncology trials: are we asking the right question? J Clin Oncol. 2012;30(35):4396–4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peppercorn JM, Weeks JC, Cook EF, et al. Comparison of outcomes in cancer patients treated within and outside clinical trials: conceptual framework and structured review. Lancet. 2004;363(9405):263–270. [DOI] [PubMed] [Google Scholar]
- 13. Schnipper LE, Meropol NJ. Payment for cancer care: time for a new prescription. J Clin Oncol. 2014;32(36):4027–4028. [DOI] [PubMed] [Google Scholar]
- 14. Ford JG, Howerton MW, Lai GY, et al. Barriers to recruiting underrepresented populations to cancer clinical trials: a systematic review. Cancer. 2008;112(2):228–242. [DOI] [PubMed] [Google Scholar]
- 15. Ratain MJ, Miller AA, McLeod HL, et al. The Cancer and Leukemia Group B Pharmacology and Experimental Therapeutics Committee: a historical perspective. Clin Cancer Res. 2006;12(11 Pt 2):3612s–3616s. [DOI] [PubMed] [Google Scholar]
- 16. Offit K, Groeger E, Turner S, et al. The “duty to warn” a patient’s family members about hereditary disease risks. JAMA. 2004;292(12):1469–1473. [DOI] [PubMed] [Google Scholar]
- 17. Beskow LM, Botkin JR, Daly M, et al. Ethical issues in identifying and recruiting participants for familial genetic research. Am J Med Genet. 2004;130a(4):424–431. [DOI] [PubMed] [Google Scholar]
- 18. Juengst ET. FACE facts: why human genetics will always provoke bioethics. J Law Med Ethics. 2004;32(2):267–275,191. [DOI] [PubMed] [Google Scholar]
- 19. Hsieh A. A nation’s genes for a cure to cancer: evolving ethical, social and legal issues regarding population genetic databases. Columbia J Law Soc Probl. 2004;37(3):359–411. [PubMed] [Google Scholar]
- 20. Clayton EW. Ethical, legal, and social implications of genomic medicine. N Engl J Med. 2003;349(6):562–569. [DOI] [PubMed] [Google Scholar]
- 21. Henderson G, Garrett J, Bussey-Jones J, et al. Great expectations: views of genetic research participants regarding current and future genetic studies. Genet Med. 2008;10(3):193–200. [DOI] [PubMed] [Google Scholar]
- 22. Bussey-Jones J, Henderson G, Garrett J, et al. Asking the right questions: views on genetic variation research among black and white research participants. J Gen Intern Med. 2009;24(3):299–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bussey-Jones J, Garrett J, Henderson G, et al. The role of race and trust in tissue/blood donation for genetic research. Genet Med. 2010;12(2):116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Helft PR, Daugherty CK. Are we taking without giving in return? The ethics of research-related biopsies and the benefits of clinical trial participation. J Clin Oncol. 2006;24(30):4793–4795. [DOI] [PubMed] [Google Scholar]
- 25. Peppercorn J, Shapira I, Collyar D, et al. Ethics of mandatory research biopsy for correlative end points within clinical trials in oncology. J Clin Oncol. 2010;28(15):2635–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peppercorn J. Toward Improved Understanding of the Ethical and Clinical Issues Surrounding Mandatory Research Biopsies. J Clin Oncol. 2013;31(1):1–2. [DOI] [PubMed] [Google Scholar]
- 27. Zhang T, Schneider A, Hamilton E, et al. Prevalence and impact of correlative science in breast cancer phase II trials. Breast Cancer Res Treat. 2013;139(3):845–850. [DOI] [PubMed] [Google Scholar]
- 28. Liang K-Y, Zeger S. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73(1):13–22. [Google Scholar]
- 29. Zeger SL, Liang KY. Longitudinal data analysis for discrete and continuous outcomes. Biometrics. 1986;42(1):121–130. [PubMed] [Google Scholar]
- 30. McLeod HL. Cancer pharmacogenomics: early promise, but concerted effort needed. Science. 2013;339(6127):1563–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guth AA, Fineberg S, Fei K, et al. Utilization of Oncotype DX in an Inner City Population: Race or Place? Int J Breast Cancer. 2013;2013:653805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hunt BR, Whitmans S, Hurlbert MS. Increasing Black:White disparities in breast cancer mortality in the 50 largest cities in the United States. Cancer Epid. 2014;38(2):118–23. [DOI] [PubMed] [Google Scholar]
- 33. Wendler D, Kington R, Madans J, et al. Are racial and ethnic minorities less willing to participate in health research? PLoS Med. 2006;3(2):e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Paskett ED, Reeves KW, McLaughlin JM, et al. Recruitment of minority and underserved populations in the United States: the Centers for Population Health and Health Disparities experience. Contemp Clinl Trials. 2008;29(6):847–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Warnecke RB, Oh A, Breen N, et al. Approaching health disparities from a population perspective: the National Institutes of Health Centers for Population Health and Health Disparities. Am J Public Health. 2008;98(9):1608–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Carpenter WR, Godley PA, Clark JA, et al. Racial differences in trust and regular source of patient care and the implications for prostate cancer screening use. Cancer. 2009;115(21):5048–5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Corbie-Smith G, Thomas SB, St George DM. Distrust, race, and research. Arch Intern Med. 2002;162(21):2458–2463. [DOI] [PubMed] [Google Scholar]
- 38. Corbie-Smith G, Williams IC, Blumenthal C, et al. Relationships and communication in minority participation in research: multidimensional and multidirectional. J Natl Med Assoc. 2007;99(5):489–498. [PMC free article] [PubMed] [Google Scholar]
- 39. Murphy EJ, Wickramaratne P, Weissman MM. Racial and ethnic differences in willingness to participate in psychiatric genetic research. Psychiatr Genet. 2009;19(4):186–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dodson C. Knowledge and attitudes concerning pharmacogenomics among healthcare professionals. Per Med. 2011;8(4):421–428. [DOI] [PubMed] [Google Scholar]
- 41. Stanek EJ, Sanders CL, Taber KA, et al. Adoption of pharmacogenomic testing by US physicians: results of a nationwide survey. Clin Pharmacol Ther. 2012;91(3):450–458. [DOI] [PubMed] [Google Scholar]
- 42. Dressler LG, Deal AM, Patel J, et al. Cancer pharmacogenomics, adoption by oncologists and patient benefit. Per Med. 2014;11(2):143–153. [DOI] [PubMed] [Google Scholar]
- 43. Gray SW, Hicks-Courant K, Cronin A, et al. Physicians’ attitudes about multiplex tumor genomic testing. J Clin Oncol. 2014;32(13):1317–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stark N, Paskett E, Bell R, et al. Increasing participation of minorities in cancer clinical trials: summary of the “Moving Beyond the Barriers” Conference in North Carolina. JNatl Med Assoc. 2002;94(1):31–39. [PMC free article] [PubMed] [Google Scholar]
- 45. Swanson GP, Carpenter WR, Thompson IM, et al. Urologists’ attitudes regarding cancer clinical research. Urology. 2007;70(1):19–24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.