IntOGen-mutations is a discovery tool for cancer researchers to find driver mutations in genes and pathways.
Supplementary information
The online version of this article (doi:10.1038/nmeth.2642) contains supplementary material, which is available to authorized users.
Subject terms: Data integration, Cancer genomics, Genome informatics
Abstract
The IntOGen-mutations platform (http://www.intogen.org/mutations/) summarizes somatic mutations, genes and pathways involved in tumorigenesis. It identifies and visualizes cancer drivers, analyzing 4,623 exomes from 13 cancer sites. It provides support to cancer researchers, aids the identification of drivers across tumor cohorts and helps rank mutations for better clinical decision-making.
Supplementary information
The online version of this article (doi:10.1038/nmeth.2642) contains supplementary material, which is available to authorized users.
Main
The exponential growth of data sets of somatic mutations from tumor samples1,2 demands analysis methods for a comprehensive understanding of cancer mutations, genes and pathways across tumor types. Several cancer genomics portals with data from resequenced cancer genomes exist3,4,5, but none of them systematically analyzes the data across various sequencing projects.
IntOGen-mutations is a Web platform used to identify cancer drivers across tumor types and to present the results of the systematic analysis of most currently available large data sets of tumor somatic mutations. It builds upon concepts similar to our original IntOGen platform, which focused on transcriptomic alterations and copy-number gains and losses in tumors6.
The IntOGen-mutations pipeline integrates the results of tumor genomes analyzed with different mutation-calling workflows and is scalable to hundreds of thousands of tumor genomes. It currently includes OncodriveFM7, a tool that detects genes that are significantly biased toward the accumulation of mutations with high functional impact (FM bias) without the need to estimate background mutation rate8, and OncodriveCLUST9, which picks up genes whose mutations tend to cluster in particular regions of the protein sequence with respect to synonymous mutations (CLUST bias) (Online Methods). Both tools detect signals of positive selection, which appear in genes whose mutations are selected during tumor development and are therefore likely drivers.
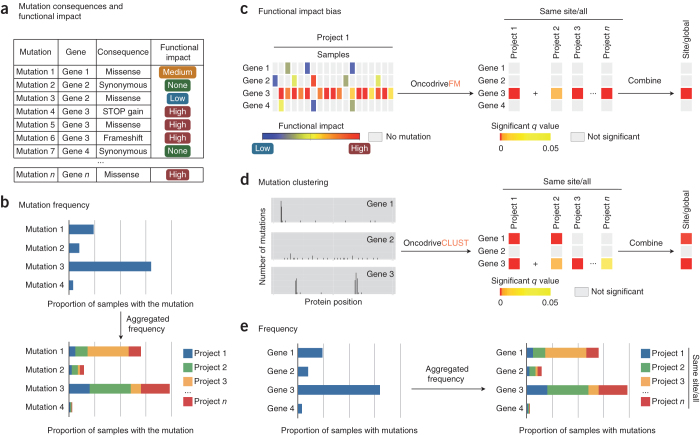
Input consists of the list of somatic mutations detected in tumor cohort resequencing projects. The pipeline first determines the consequences of these mutations using the Ensembl variant effect predictor tool10 and retrieves the functional impact scores of nonsynonymous mutations according to three well-known methods: sorting intolerant from tolerant (SIFT)11, PolyPhen2 (ref. 12) and MutationAssessor13. These scores are subsequently transformed (with transFIC14) to compensate for the differences in baseline tolerance among genes, and each mutation is classified into one of four broad groups of impact, ranging from “None” to “High,” according to its consequence type and its transFIC MutationAssessor score (Fig. 1a). The pipeline also computes each mutation's frequency of occurrence within and across projects (Fig. 1b). Mutations occurring in the same gene (or pathway) are grouped, and OncodriveFM and OncodriveCLUST identify likely drivers across the tumor samples. Genes not expressed across tumors from The Cancer Genome Atlas (TCGA) pan-cancer projects are excluded from the driver detection analysis (Online Methods). The pipeline combines the P values computed by either method for each gene into a single P value representing the FM bias or CLUST bias of the gene in tumors from one site or across all tumors (Fig. 1c,d). Finally, the pipeline computes the frequency of mutation of each gene (and pathway) within a project or cancer site (Fig. 1e).
Figure 1.
Schematic representation of the analysis performed by the IntOGen-mutations analysis pipeline.
The different modules in the pipeline are executed by a workflow management system (Wok, https://bitbucket.org/bbglab/wok/). This makes IntOGen-mutations highly configurable and computationally very efficient, and it allows the addition of other methods to detect cancer drivers. The results of the pipeline are automatically loaded into a Web browser managed by the Onexus framework (Supplementary Fig. 1).
We have analyzed somatic mutations in 4,623 samples from 31 different projects covering 13 anatomical sites (mainly from the International Cancer Genome Consortium (ICGC)1 and the TCGA2) (Supplementary Tables 1,2,3). Many of the candidate driver genes are known cancer genes (annotated in the Cancer Gene Census), a status indicating that they are bona fide driver candidates; novel candidate drivers are also detected. The comparison of the results obtained with our pipeline with those reported in original publications shows a very high overlap, with some known cancer driver genes identified exclusively by IntOGen-mutations (Supplementary Note 1).
A systematic analysis of sequenced tumor genomes permits a broad view of the impact of genes in tumorigenesis across cancer types (Supplementary Fig. 2). For example, TP53, ARID1A, KRAS or PIK3CA are frequently mutated and identified as cancer drivers in most cancer sites. Other genes, such as VHL in kidney, MAPK3 and GATA3 in breast and STK11 in lung, seem to be primarily tumor-specific drivers.
IntOGen-mutations will be regularly updated with new cancer genome resequencing data. The results can be browsed through the Web (Supplementary Note 2) and with Gitools interactive heat maps15 (http://www.gitools.org/datasets/). The pipeline may be downloaded and can also be run online on our servers. It can be used to identify drivers from newly sequenced cohorts of tumor samples (Supplementary Note 3) and to interpret the mutations observed in a tumor sample for better clinical decision-making (Supplementary Note 4).
Methods
The IntOGen-mutations pipeline.
The first part of the IntOGen-mutations pipeline assesses the potential functional impact of somatic mutations detected across the cohort of tumor samples. The Ensembl variant effect predictor10 (VEP, v.70) script and precomputed cache files, downloaded from the Ensembl FTP site (ftp://ftp.ensembl.org/pub/), are used to determine the consequences of somatic mutations in annotated functional elements. The pipeline obtains SIFT11 and PolyPhen2 (ref. 12) functional impact from VEP. Precomputed MutationAssessor13 functional impacts are obtained from the MutationAssessor Web server (http://www.mutationassessor.org/) during the installation of the pipeline and are queried locally during execution. The transformation of functional impact scores to account for the baseline tolerance of genes to germline mutation (transFIC), described elsewhere14, has been reimplemented in Python as a module of the IntOGen-mutations pipeline.
The pipeline implements an expression filter to disregard genes that are not expressed across the tumor samples in the cohort. This list of expressed genes is an optional input to the pipeline, which excludes all genes outside the list from the foreground of both OncodriveFM and OncodriveCLUST (see below) while keeping their mutations in the background. In the current release of the IntOGen-mutations Web discovery tool, we have employed as a filter the list of genes expressed across any of the 12 pan-cancer data sets (ref. syn1734155).
The OncodriveFM and OncodriveCLUST approaches, also described elsewhere7,9, have been reimplemented as IntOGen-mutations pipeline modules and are available as independent programs from two Git-controlled repositories at https://bitbucket.org/bbglab/. Briefly, OncodriveFM receives as input the list of synonymous, nonsynonymous and frameshift-indel mutations and their corresponding SIFT, PolyPhen2 and MutationAssessor scores. Then it assesses whether any gene shows a trend toward the accumulation of mutations with high functional impact as compared to the background distribution of these functional impact scores in all mutations detected across the cohort of tumor samples (FM bias). For each functional impact score included in the pipeline, the method produces an empirical P value that evaluates this FM bias. These three P values are subsequently combined using Fisher's approach to produce one integrated P value for each gene. To account for possible nondependence between the three P values included in the combination, the IntOGen-mutations Web discovery tool considers as significant those with a false discovery rate (FDR) below 0.05.
OncodriveFM also computes an FM bias for pathways. Three z scores are computed in this case to assess the trend of pathways to accumulate mutations with high functional impact. The z scores are combined using Stouffer's approach, and the combined z score is transformed into an integrated P value.
OncodriveCLUST, on the other hand, receives as input two separate lists of mutations: potentially protein-affecting mutations (nonsynonymous, stop and splice site) and silent mutations (synonymous), with their corresponding locations across the proteins' sequences. It then assesses the significance of the trend of potentially protein-affecting mutations to be clustered with respect to a background represented by the homologous trend for silent mutations.
Genes mutated in less than 1% of the samples in projects whose median of mutations per sample was below 100 were not analyzed by OncodriveFM. In projects with higher median of mutations per samples, this threshold was set to 5 samples with mutations. For OncodriveCLUST, the thresholds were 3 and 5 mutated samples, respectively. These and many other parameters of the pipeline are configurable by the user, as explained in its documentation.
In addition to third-party (and in-house) software and data, IntOGen-mutations pipeline installation requires some Python libraries. The most important of these are the numpy and scipy scientific computing libraries and the statsmodels Python statistical library.
The pipeline also relies on other external data files. During pipeline installation, all of the needed external and third-party data files are downloaded and correctly placed, and external libraries are downloaded and compiled, thereby creating a Python environment where the pipeline executes.
The analysis of the 4,623 tumor samples currently included in the IntOGen-mutations Web discovery tool takes approximately 5 h on an eight-core, 12 GB RAM computer.
Obtaining and processing somatic mutations data sets.
As mentioned in the Results section, we obtained the 31 somatic mutations data sets currently included in the IntOGen Web discovery tool from the ICGC, the TCGA and literature searches. All ICGC data sets were downloaded directly from the Data Coordination Centre (DCC) Biomart3. These data sets were already in the tab-separated format accepted by the pipeline. TCGA data sets were downloaded from the Synapse platform (syn1729383) as MAF files within the context of the PANCANCER project. These MAF files were transformed to the tab-separated format accepted by the pipeline. Finally, a manual PubMed search allowed us to identify somatic mutations data sets that had been produced by research groups outside these large initiatives. We parsed supplementary files of the papers reporting these studies to extract the lists of somatic mutations detected across tumor samples and then transformed them into the tab-separated format accepted by the pipeline.
Using IntOGen-mutations as a knowledge discovery resource (case 1).
The systematic analysis of more than 4,500 tumors across projects and tumor sites allows researchers to have a wide view of genes and pathways involved in tumorigenesis. Cancer researchers can search IntOGen-mutations to find out which genes are candidate drivers for a given tumor site or the likelihood that a given gene (or gene set) is a driver across different malignancies. Case 1 is a general use of the IntOGen-mutations Web discovery tool that is illustrated in Supplementary Note 2.
Using IntOGen-mutations to identify drivers in a cohort of tumors (case 2).
The IntOGen-mutations platform is the first tool that unites a pipeline to analyze the somatic mutations identified across a cohort of tumor samples with a Web discovery tool containing accumulated knowledge on the role of somatic mutations in tumors obtained from systematic equivalent analysis of data sets of resequenced tumor genomes. Therefore, one important use of IntOGen-mutations is to identify likely driver genes across a cohort of tumors and compare them with the list of previously detected likely drivers in the same cancer site or in general that is provided by the IntOGen-mutations Web discovery tool.
To illustrate this use case (Supplementary Note 3), we downloaded a data set of somatic mutations detected through whole-genome sequencing of 37 medulloblastoma samples16. We analyzed the 931 mutations deemed as tier 1 by the authors of the study. We submitted a data file containing the list of mutations per sample to the online version of the pipeline at http://www.intogen.org/mutations/analysis/. Upon completion of the analysis (∼5 min), we explored the results obtained on the private Web discovery tool.
In summary, the 931 mutations affected 1,290 genes, 63 of them being mutated in at least two samples. Seven genes exhibited a significant FM bias (q value <0.05), four of which were included by the authors of the original report. Of particular interest among the three FM-biased genes not cited as particularly interesting by the authors of the original report is SF3B1, which encodes a splicing factor known to drive hematopoietic malignancies17,18 and other tumors19. Exploring the results within the Web discovery tool allows quick comparison of the identified mutations and candidate driver genes with those previously found and reported in IntOGen-mutations. The pathways analysis correctly identified the Wnt signaling and focal adhesion pathways among the top-ranking FM-biased pathways.
Applying IntOGen-mutations toward personalized cancer medicine (case 3).
The IntOGen-mutations pipeline can be used to rank the somatic mutations identified in the tumor of an individual patient. Researchers with a list of mutations detected in a tumor can identify mutations with functional impact, find mutations affecting cancer driver genes and identify any mutations in the patient that have been previously observed in tumors. All this information can help to suggest which genes might have driven tumorigenesis in the patient, with the final aim of informing a personalized approach to treatment.
To illustrate this case (Supplementary Note 4), we obtained the list of somatic mutations detected in one patient's metastatic colorectal cancer in a study aimed at making personalized recommendations conducive to treatment20. We ran the online version of the pipeline. As a result, we obtained a list of 42 genes affected by mutations of high or medium functional impact. We determined that six of these genes had been detected as significantly FM-biased in colorectal cancer in the IntOGen-mutations Web discovery tool. Only one of them, NRAS, had been identified as an actionable driver for therapy in the original report.
Supplementary information
Supplementary Figures 1 and 2, Supplementary Tables 1–3 and Supplementary Notes 1–4 (PDF 3044 kb)
TCGA Network Authorship List (XLSX 33 kb)
Acknowledgements
We acknowledge funding from the Spanish Ministry of Economy and Competitivity (grants SAF2009-06954 and SAF2012-36199) and the Spanish National Institute of Bioinformatics (INB). We gratefully acknowledge contributions from the TCGA Research Network and its TCGA Pan-Cancer Analysis Working Group (contributing consortium members are listed in Supplementary Note 5). The TCGA Pan-Cancer Analysis Working Group is coordinated by J.M. Stuart, C. Sander and I. Shmulevich. We are also grateful to the ICGC for the tumor genome resequencing data generated.
Author Contributions
A.G.-P. and C.P.-L. performed the analyses. C.P.-L. developed Wok and designed and coded the pipeline. A.S. helped on the development of the first version of the pipeline. J.D.-P. developed Onexus and designed and coded the Web browser. A.J.-S., D.T. and M.P.S. participated in the analysis and validation of the results. A.G.-P. and N.L.-B. drafted the manuscript and prepared the figures. N.L.-B. supervised the whole project.
Competing interests
The authors declare no competing financial interests.
References
- 1.The International Cancer Genome Consortium. Nature464, 993–998 (2010).
- 2.Weinstein, J.N. et al. Nat. Genetics 10.1038/ng.2764 (in the press).
- 3.Zhang J, et al. Database (Oxford) 2011;2011:bar026. doi: 10.1093/database/bar026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cerami E, et al. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forbes SA, et al. Nucleic Acids Res. 2010;38:D652–D657. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gundem G, et al. Nat. Methods. 2010;7:92–93. doi: 10.1038/nmeth0210-92. [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez-Perez A, Lopez-Bigas N. Nucleic Acids Res. 2012;40:e169. doi: 10.1093/nar/gks743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawrence MS, et al. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tamborero D, Gonzalez-Perez A, Lopez-Bigas N. Bioinformatics. 2013;29:2238–2244. doi: 10.1093/bioinformatics/btt395. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, et al. BMC Genomics. 2010;11:293. doi: 10.1186/1471-2164-11-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar P, Henikoff S, Ng PC. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 12.Adzhubei IA, et al. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reva B, Antipin Y, Sander C. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez-Perez A, Deu-Pons J, Lopez-Bigas N. Genome med. 2012;4:89. doi: 10.1186/gm390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perez-Llamas C, Lopez-Bigas N. PLoS ONE. 2011;6:e19541. doi: 10.1371/journal.pone.0019541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson G, et al. Nature. 2012;488:43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L, et al. N. Engl. J. Med. 2011;365:2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quesada V, et al. Nat. Genet. 2012;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 19.Ellis MJ, et al. Nature. 2012;486:353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roychowdhury S, et al. Sci. Transl. Med. 2011;3:111ra121. doi: 10.1126/scitranslmed.3003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1 and 2, Supplementary Tables 1–3 and Supplementary Notes 1–4 (PDF 3044 kb)
TCGA Network Authorship List (XLSX 33 kb)