Abstract
Estimating the false discovery rate (FDR) among a list of tandem mass spectrum identifications is mostly done through target-decoy competition (TDC). Here we offer two new methods that can use an arbitrarily small number of additional randomly drawn decoy databases to improve TDC. Specifically, “Partial Calibration” utilizes a new meta-scoring scheme that allows us to gradually benefit from the increase in the number of identifications calibration yields and “Averaged TDC” (a-TDC) reduces the liberal bias of TDC for small FDR values and its variability throughout. Combining a-TDC with “Progressive Calibration” (PC), which attempts to find the “right” number of decoys required for calibration we see substantial impact in real datasets: when analyzing the Plasmodium falciparum data it typically yields almost the entire 17% increase in discoveries that “full calibration” yields (at FDR level 0.05) using 60 times fewer decoys. Our methods are further validated using a novel realistic simulation scheme and importantly, they apply more generally to the problem of controlling the FDR among discoveries from searching an incomplete database.
Keywords: Tandem mass spectrometry, Spectrum identification, False discovery rate, Calibration
1 Introduction
In tandem mass spectrometry analysis, the problem of inferring which peptide was responsible for generating an observed fragmentation spectrum is crucial to any subsequent analysis about the presence or quantity of peptides and proteins in the complex mixture being analyzed. Unfortunately, this spectrum identification problem is difficult to solve because, for any given spectrum, many expected fragment ions will not be observed, and the spectrum is also likely to contain a variety of additional, unexplained peaks.
The most common approach to the spectrum identification problem is peptide database search. Pioneered by SEQUEST [7], the search engine extracts from the peptide database all “candidate peptides” defined by having their mass lie within a pre-specified tolerance of the measured mass of the intact peptide (the “precursor mass”). The quality of the match between each of these candidate peptides and the observed fragmentation spectrum is then evaluated using a score function. Finally, the best-scoring peptide-spectrum match (PSM) for the given spectrum is reported, along with its score.
Sometimes the reported PSM is correct—the peptide assigned to the spectrum was present in the mass spectrometer when the spectrum was generated—and sometimes the PSM is incorrect. Ideally, we would report only the correct PSMs, but obviously we are not privy to this information: all we have is the score of the PSM, indicating its quality. Therefore, we report a thresholded list of top-scoring PSMs, together with the critical estimate of the fraction of incorrect PSMs in our reported list. This work focuses on methods for carrying out this discovery and error estimation procedure.
The problem of controlling the proportion of false discoveries has been studied extensively in the context of multiple hypotheses testing (MHT), starting with the seminal of work Benjamini and Hochberg [3]. Specifically, they introduced a simple procedure that allows us to decide which null hypotheses we reject (thus declaring them as “discoveries”) so that the FDR, which they defined as the expected value of the proportion of false discoveries (FDP), is bounded by a pre-determined level α.
The mass spectrometry community, however, relies mostly on other methods to control the FDR. The main reason is that the MHT context is predicated on associating a p-value with each tested null hypothesis, indicating how unlikely that result is assuming the hypothesis is truly a null one. Until recently, no such p-values were computed in the PSM context. Moreover, while considerable effort has of late been invested in computing such p-values [14, 1, 9, 15, 16, 12], we recently showed that there are further subtle but fundamental differences between the MHT context and the PSM one, implying that we typically cannot use FDR controlling procedures that were designed for the MHT context [?].
Instead, the most widely used FDR controlling procedure in this context is a decoy-based protocol called target-decoy competition (TDC), proposed by Elias and Gygi [5]. The target in TDC refers to the real peptide database of interest, and the decoy is a database of randomly shuffled or reversed peptides. The method consists of searching a given set of spectra against the concatenated target-decoy database and retaining the single best-scoring PSM for each spectrum. As a result of this selection, any optimal target PSM that scores less than the corresponding optimal decoy PSM is eliminated from consideration. Subsequently, at a given score threshold, the number of accepted decoy PSMs provides an estimate of the number of false discoveries, or accepted incorrect target PSMs [4, 10, 6].
The quality of an FDR controlling procedure (more precisely, of a discovery and FDR controlling procedure) can be evaluated along at least three orthogonal dimensions. First, we can gauge the procedure's power: how many (correct) target discoveries, or spectra identifications, does it report at a given FDR threshold? Second, we can analyze the accuracy of the procedure: how close is the actual FDR to the estimated one? In determining the accuracy we ask whether the method is biased or not, where liberally biased methods (those that underestimate the FDR) are particularly undesirable because they instill in the user more confidence than is due. Third, in addition to controlling bias, we also prefer methods that exhibit less variability, since exceedingly high FDP could have substantial impact on any downstream analysis.
The primary contributions of this paper are two novel procedures for improving decoy-based FDR control procedures in the context of the mass spectrum identification problem. The first procedure—partial calibration—yields improved statistical power relative to TDC; the second procedure—averaged TDC (a-TDC)—yields reduced variance. Both procedures maintain the asymptotic unbiased control of the FDR of TDC, and a-TDC mitigates much of the liberal bias of TDC observed at small FDR values [11].
Partial Calibration
The partial calibration procedure is motivated by our recently described calibration method [12]. We showed that calibrating the scores (placing the scores of all PSMs on the same scale regardless of the spectrum involved) can substantially increase the power of TDC.3 For example, we found that when calibrating the popular XCorr score [7], using the same set of spectra the number of discoveries at 1% FDR increased in the range of 12-31% [12]. However, since our calibration method relied on searching 10,000 randomly generated decoy databases (obtained by repeatedly shuffling each peptide of the target database), our procedure was computationally extremely demanding.
In this work, we show that the advantages calibration offers can be gradually realized starting with a relatively modest requirement of one additional decoy set and increasing according to the user's computational resources. In a nutshell, partial calibration uses the calibrating decoys to convert the raw scores into empirical p-values. However, whereas our original approach employed the commonly used method of replacing the raw score with the empirical p-value, here we keep both and use a two tiered scoring scheme. The primary score is the empirical p-value, with ties resolved by the secondary score, which is the raw score. This new primary-secondary scheme allows us to use as few as a single calibrating decoy in a meaningful way. Previously, in such a case roughly half the (null) scores would have one p-value (0) and the other half would have a different p-value (1), so very little could have been done with this data.
By allowing us to benefit from any number of calibrating decoys, partial calibration raises the question of how many calibrating decoy sets are “enough.” One option is to let our computational resources determine the number of decoys we can afford to generate for the given data. However, the downside of this strategy is that for some datasets and FDR thresholds we would spend too much effort, whereas in other cases we would still achieve sub-par results. An ideal approach would allow us to intelligently trade off between statistical power and computational expense.
Here we propose an ad hoc method, called “progressive calibration” (PC), that employs a doubling strategy to dynamically determine the number of calibrating decoys our partial calibration procedure should use. The method works by factoring in the user's computational limits, the particular dataset at hand, and the range of FDR values the user is interested in. The latter is a particularly important factor because, empirically, the law of diminishing returns, in terms of number of discoveries per number of calibrating decoys, kicks in much sooner for higher FDR levels.
Averaged TDC
The second primary procedure, a-TDC, is motivated by simulations that show that, for sets of 1000 spectra, the actual FDP among the PSMs selected by using TDC with an FDR threshold of 0.05 can readily be ±50% of that level, and this problem gets much worse for smaller-sized sets of spectra and tighter FDR levels [11]. Although calibration can somewhat reduce TDC's variability [12], even if we achieve perfect calibration we still cannot get around the inherent decoy-dependent variability of TDC.
a-TDC gets around this problem by applying TDC to the target database paired with a small number (np) of randomly drawn “competing” decoy databases and “averaging” the results. Clearly, averaging will reduce the TDC variance, but the challenge is to make sense of this averaging, especially because the list of TDC discoveries varies with each competing decoy database.
One might be tempted to define this list of “average target discoveries” as all the target PSMs that outscore the majority of their decoy competitions. That is, a target PSM is an a-TDC discovery if it is a (TDC) discovery in more than np/2 of the np concatenated target-decoy searches4. While intuitively appealing, when paired with the equally appealing averaging of the (TDC) estimated FDR, this approach quickly becomes too liberal: the FDR is underestimated.
We therefore devised a more nuanced approach which sequentially constructs its target discovery list starting from the highest target PSM score. Our method then goes through the decreasing target PSM scores, ensuring that the number of discoveries at the current score level does not deviate from the average number of (TDC) target discoveries, at the same score level, across our np independent TDC procedures. In order to meet this guarantee, a-TDC occasionally needs to filter out or reject a target PSM as it goes down the list. At that point, the PSM that is selected for rejection is the one with the smallest score among all hitherto selected target discoveries that lost the most decoy competitions (Sec. 2.4).
At this point there are several plausible ways to define the corresponding a-TDC estimate of the FDR among its list of discoveries. We settled on the ratio between the average number of decoy discoveries across the np independent TDC procedures, and the actual number of a-TDC discoveries. Importantly, this definition makes a-TDC with a single decoy (np = 1) identical to TDC.
Verification
We apply our novel procedures—partial calibration (and its adaptive variant, PC) and a-TDC—to real as well as simulated data. Most simulations of the spectrum identification problem, carried out by us as well as others, have used calibrated scores. However, because much of our work here is dedicated to the effects of partial calibration, it was crucial to develop a simulation procedure using uncalibrated scores.
The simulated data supports our claim that our procedures control the FDR on-par or better than TDC does, and both the real and simulated data show that a-TDC reduces the variability of TDC and that partial calibration can yield a sizable increase in the number of target discoveries. In addition, we observe that, for a typical FDR range of interest, PC allows us to enjoy most of the gains offered by the our original, brute-force calibration procedure, while employing significantly fewer than 10,000 decoys.
2 Methods
2.1 TDC, FDR estimation, and target discoveries
An FDR controlling procedure returns a list of discoveries together with an estimate of the FDR among the reported discoveries. In particular, TDC defines its list of T (ρ) discoveries at score level ρ as all target PSMs with score ≥ ρ that outscore their corresponding decoy competition (i.e., they remain discoveries in the search of the concatenated database). Denoting by D (ρ) the number of decoy discoveries at score level ρ in the concatenated search, TDC estimates the FDR in its target discovery list as .
Often, the user is more interested in specifying a desired FDR level τ. In this context the score threshold that corresponds to an (estimated) FDR level of τ is . We refer to T (τ), the number of target discoveries at (estimated) FDR level τ, as short for T (ρ (τ)), the number of discoveries at score level ρ(τ), and similarly for the list of actual target discoveries at FDR level τ. For computational efficiency we limited our attention to a predetermined set of FDR values (Supp. Sec. 1.1). Note that the latter relation between τ and ρ(τ) is defined for any FDR estimation method and is not specific to TDC.
2.2 Calibrating and competing decoys
Let Σ denote the set of spectra generated in the experiment. We associate with each spectrum σ ∈ Σ its optimal matching peptide in the target database, which we loosely refer to as the “target PSM,” or just the “target score,” w(σ), when we refer to the score of that PSM.
Similarly, we assume that each spectrum σ is searched against two sets of randomly drawn decoy databases that, in practice, are generated by independently shuffling each peptide in the target database. The sets are statistically identical but we refer to one, , as the “calibrating” set of decoy databases and the other, , as the “competing” set of decoys. The distinction between the two sets of decoys is based on the different roles they play. The calibrating decoys are used to calibrate the scores whereas the competing decoys are used for estimating the FDR using target decoy competition.
The score of the optimal match to σ in is denoted by , and similarly is the score of the optimal match to σ in . For each fixed σ the distribution of (with respect to a randomly drawn decoy) is identical to that of . Importantly, since we do not assume that the score is necessarily calibrated, the said distribution can vary with the spectrum σ.
2.3 Partial calibration
By “partial calibration” we refer to a procedure that allows us to convert a raw score into a new score that is “more calibrated.” We prefer to be somewhat vague on what exactly the latter means but, intuitively, it means that the distribution of the decoy scores z (σ) is “less varied” with respect to the spectrum σ.
Our specific procedure here first uses the calibrating scores associated with the spectrum σ, , to assign to each observed competing decoy score or target score s = w (σ) a new, primary score, qσ(s). This primary score is equivalent to the p-value of s with respect to the empirical cumulative distribution function (ECDF) constructed from the calibrating scores:
The secondary score assigned to s is the score s itself.
Using our primary-secondary score we define a new linear order, ≻, on the set of all observed target and competing decoy scores as follows. Let si be a score of an optimal PSM involving the spectrum σi. Instead of using the raw scores s1 and s2 to determine the order, we now say s1 ≽ s2 if qσ1 (s1) > qσ2 (s2), or if qσ1 (s1) = qσ2 (s2) and s1 ≥ s2.
Technically, we implement the new order, ≻, by first converting all observed target scores, 𝒲 = {w (σ) : σ ∈ Σ}, and competing decoy scores, , into ranks, where the rank of 1 corresponds to the smallest observed score and the rank of |Σ| (np + 1) corresponds to the largest observed score. We then map each observed raw score s associated with the spectrum σ to the partially calibrated score
where r (s; 𝒲 ∪ Ƶp), is the rank of the raw score s in the list of |Σ| (np + 1) observed target and competing decoys scores. It is easy to see that, given the set 𝒲 ∪ Ƶp, for any observed pair of scores s1 and s2 from that set, s1 ≽ s2 if and only if ψσ (s1) ≥ ψσ (s2).
If the size of the calibrating set nb is very large then, assuming s1 ≠ s2, it is very unlikely that qσ1 (s1) = qσ2 (s2) and the new ordering will coincide with the one determined by the spectrum specific ECDFs that was used in our previously described calibration procedure [12]. At the other extreme end, when there are no calibrating decoys we revert to the ordering determined by the raw score. All other cases in some sense interpolate between these two extremes, with more weight placed on the ECDF the more refined it is.
Two points are worth noting. First, if the raw score is already calibrated then our partial calibration procedure will leave it calibrated. More precisely, recall that we defined the optimal PSM score function as calibrated if the distribution of z(σ), the score of the optimal match to σ in a randomly drawn database, 𝒟, is invariant of the spectrum σ. Assuming that the calibrating decoys are drawn at the same time as 𝒟, our new score ψ will also be calibrated. Second, the definition of ψσ (s) allows us to efficiently utilize an increasing number of calibrating decoys – a fact that we will return to when discussing progressive calibration.
2.4 Averaged TDC
The a-TDC procedure begins with repeatedly applying TDC to the target database 𝒟t paired with each of the np independently drawn (competing) decoy databases , for i = 1,…, np. Let Ti (ρ) and Di (ρ) denote the number of target, respectively, decoy discoveries at level ρ, that are reported by TDC in the ith application. Recall that Di (ρ) is used to estimate Fi (ρ), the corresponding number of false target discoveries, and note that the reported list of target discoveries typically changes with each decoy database.
Let ρi denote the decreasing target PSM scores, and let and be the average of numbers of target and decoy discoveries, respectively, at level ρi across our np TDC procedures. Our a-TDC procedure sequentially constructs its discovery list (and simultaneously its filtered target PSMs list), ensuring that its number of discoveries at level ρi, T (ρi), does not deviate from (apart from the inevitable difference due to rounding).
When a-TDC determines that it needs to filter out a target PSM, it rejects the PSM with the lowest partially calibrated score among all hitherto selected target PSMs with raw score s ≥ ρi. Note that the latter partially calibrated score is with respect to the competing decoys rather than the usual calibrating decoys, and that the rank component of the score is taken only with respect to the target scores. In other words, the rejected PSM is the one with the smallest raw score among all remaining target discoveries scoring ≥ ρi that lost the most decoy competitions. Finally, a-TDC estimates the FDR in its level ρi discovery list as (Suppl. Alg. 1).
2.5 Progressive calibration with mean cutoff criterion
Progressive calibration (PC) starts with zero calibrating decoys (raw scores) and goes through several cycles of essentially doubling the number of calibrating decoys. At the ith cycle we randomly draw 2i−1 additional calibrating decoy databases and search each of our spectra against these databases. Thus, after the ith doubling, for each spectrum we have a set of 2i − 1 calibrating decoy scores, which contains the corresponding calibrating decoy set of the previous doubling cycle. The process terminates if the cutoff criterion below was engaged, or the maximal number of calibrating decoys was reached (2047 for our simulations and 10,000 for the real data).
Adjusting the partially calibrated score to take into account the newly drawn set of decoys PSM scores, in each of PC's doubling cycles, is straightforward due to the identity
| (1) |
which in turn implies
Hence, we only need to compute the ranks, r (s; 𝒲 ∪ Ƶp) for all s observed target scores, 𝒲, and competing decoy scores, Ƶp , once and then update using (1).
We will see below (Fig. 1) that for some combinations of data and FDR levels, partial calibration can achieve near optimal results with very few calibrating decoys. In other cases, and particularly for very small FDR levels, achieving near optimal results requires many more calibrating decoys. Taking this into account, PC's stopping criterion focuses only on the mean increase in the number of discoveries for FDR levels in a range that is specified by the user (≥ 0.05 in our experiments). The exact details are in Supp. Sec. 1.2.
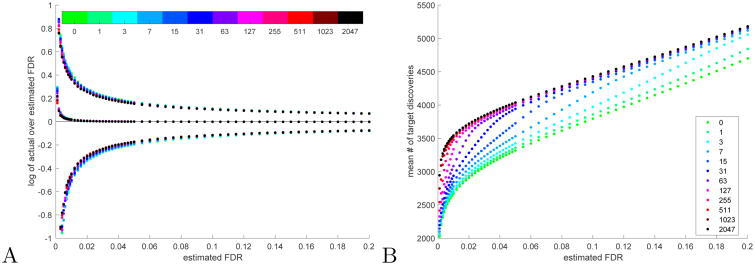
Fig. 1. Partial calibration (TDC).
A The set of middle curves, which correspond to the log of the ratio between the empirical FDR and the nominal FDR level, essentially coincide for all considered numbers of calibrating decoys (the modest liberal bias of TDC for low FDR values is discussed in Sec. 3.2). The set of lower and upper curves correspond to the log of the ratio of the 0.05 and 0.95 quantiles of the FDP to the nominal FDR level. B The mean number of (TDC) target discoveries consistently increases with the number of calibrating decoys, although the law of diminishing returns is quite evident. A-B All means and quantiles are taken with respect to 10K simulation runs using our raw score, each with 10K spectra, 50% native spectra. The number of calibrating decoys was varied from 0 to 2047 (see Methods for details).
Note that the above cutoff criterion applies regardless of whether the FDR estimation is done using TDC or a-TDC. In addition, we only engage the cutoff criterion from the third doubling cycle onward (so we use at least seven calibrating decoys).
2.6 Simulations using uncalibrated scores
One can readily simulate raw decoy scores by searching real spectra against randomly shuffled versions of real peptide databases, but it is less clear how to simulate target PSM scores while still knowing which ones are “correct” and which are “false.” Here we accomplish this by first sampling the optimal PSM scores using a variant of our previously described calibrated sampling scheme [13], where false PSMs are drawn from a null distribution and correct PSM are drawn from an alternative, beta distribution, and each spectrum has a fixed number of candidate peptides it can match. We then convert, these calibrated PSM scores to raw scores using a spectrum specific transformation modeled after a real data set, as explained next.
We begin with associating with each spectrum from the real set (here we used the yeast dataset, Supp. Sec. 1.4) an ECDF constructed from a sample of 10K optimal decoy PSM scores, which are obtained by searching the spectrum against 10K randomly shuffled versions of the target database. While we could have converted the calibrated scores to raw scores using the quantiles of this spectrum-specific ECDF, this would have limited the granularity of our score when analyzing the often encountered high scoring correct PSMs.
Therefore, to preserve the necessary granularity in our scoring function we instead relied on the observation that the distribution of the null optimal PSM scores of a specific spectrum can often be well approximated by a Gumbel EVD [16]. Specifically, we fitted a location-shifted and scaled Gumbel distribution to each of our spectrum-specific ECDFs generated by the yeast real data. We then randomly associated the fitted Gumbel distributions to our simulated spectra and used the quantiles of those fitted distributions to convert our initially sampled calibrated scores (Supp. Sec. 1.3). Using this approach our simulated raw scores inherit the uncalibrated nature of the real yeast data.
2.7 Real data analysis
We analyze three real data sets, derived from yeast, C. elegans (worm), and Plasmodium falciparum (“malaria”). For each of these three sets, we conducted 2000 independent experiments, each of which consisted of randomly drawing 10 distinct competing decoys from a pool of 1K such decoys. We then applied partial calibration to both the target and the competing decoy PSM scores, using an increasing number of calibrating decoys, which were randomly drawn from a pool of 10K such decoys. We next applied a-TDC (using the 10 drawn competing decoys) and TDC (using just the first of those competing decoys) to the increasingly calibrated data, noting the number of discoveries and virtually applying PC to the data. Further details are provided in Supp. Sec. 1.4.
3 Results
3.1 Partial calibration yields more statistical power
The effectiveness of our partial calibration scheme is demonstrated by our new raw score simulation method. For example, looking at the mean number of TDC target discoveries across 10K runs, each of which simulated a set of spectra of size 10K with 50% native spectra (these are spectra for which the correct peptide is in the target database), we find that this mean consistently increases with the number of calibrating decoys (Fig. 1B). Moreover, for FDR levels which are not very small, much of that increase can be realized with a relatively small number of calibrating decoys, e.g., at an estimated FDR level of 0.05 using no calibrating decoys (raw score) the mean number of TDC discoveries is 3317, but using 31 calibrating decoys it rises to 3726 (12% increase) and with 63 decoys it is at 3942, which is 98% of the maximal 4038 average discoveries (22% increase over the raw score) obtained when using 2047 calibrating decoys (Fig. 1B). Of course, the increase in the mean number of discoveries varies with the parameters of the problem, but our simulations show a consistent increase with the number of calibrating decoys (Supp. Fig. 1).
The accuracy of TDC, in terms of the actual FDR (as estimated by the empirical mean of the FDP across 10K samples) over the nominal threshold, seems largely unchanged by the increase in the number of calibrating decoys (Fig. 1A and Supp. Fig. 2, middle curves). At the same time, as expected, the variability of the estimate decreases slightly with the increased calibration (same figures, upper and lower set of curves).
3.2 Averaged TDC
With calibrated scores
Fig. 2A demonstrates that a-TDC reduces the variability in FDR estimation, even when the score is perfectly calibrated. This reduction is more pronounced with smaller sets of spectra and smaller FDR levels (Supp. Fig. 3). As a consequence, the variabilities in the reported number of discoveries as well as of false discoveries are also reduced (Fig. 2B, and Supp. Fig. 5 and 7). These variance reductions imply that the actual list of target discoveries should also exhibit reduced variability compared with single-decoy TDC, although we did not try to quantify this effect here.
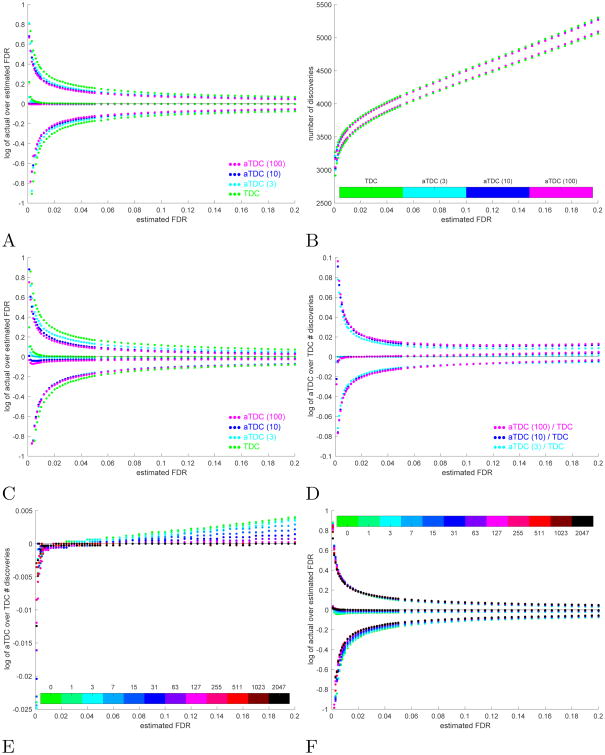
Fig. 2. Averaged TDC (a-TDC). A-D.
Comparing the FDR controlling procedure of a-TDC with 1 (TDC), 3, 10, and 100 competing decoys. E-F a-TDC with 10 competing decoys. A Plotted are the log of the ratios of the mean (empirical FDR, middle curves) as well as the 0.05 and 0.95 quantiles (upper and lower curves) of the FDP in the target discovery lists of each of the four procedures, to the nominal FDR level. Scores are calibrated. B The 0.05 and 0.95 quantiles of the number of target discoveries. Scores are calibrated. C Same as panel A, except the simulations were done using the raw (uncalibrated) score. D Shown are the logarithm of the median (middle curves), 0.05 and 0.95 quantiles of the number of true target discoveries reported by a-TDC (with 3, 10, and 100 decoys) over the corresponding number reported by TDC at the same FDR threshold. Scores are uncalibrated. E The log of the median of the ratio of the number of true a-TDC to TDC discoveries show that the power advantage of a-TDC over TDC diminishes with the increase in the number of calibrating decoys. F Coincidentally, a-TDC becomes less conservative: the middle set of curves show that the log of the empirical FDR (mean of FDP) over the nominal level increases toward 0 for small FDR levels. (The 0.05 and 0.95 quantiles are also provided.) A-F All quantiles are taken with respect to 10K simulations, each with 10K spectra, 50% native spectra.
Interestingly, a-TDC also typically mitigates much of the previously noted liberal bias of TDC [11], as can be seen in the set of middle curves of Fig. 2A and Supp. Fig. 3, which compare the empirical FDR (average of the FDP with respect to 10K independently drawn sets) with the selected FDR threshold (nominal level) using TDC as well as a-TDC with 3, 10 and 100 decoys.
With raw scores
As expected, when using a raw, uncalibrated score, a-TDC reduces TDC's variability even slightly more effectively (Fig. 2C, and Supp. Fig. 4, 6, and 8). Unexpectedly however, a-TDC also becomes slightly conservative as the number of competing decoys increases (Fig. 2C, and Supp. Fig. 4). In spite of this trend, with the exception of very small estimated FDR levels, where TDC is clearly liberally biased, a-TDC is typically making at least as many true discoveries as does TDC. Moreover, there are cases in which the number of true discoveries increases with the number of competing decoys that a-TDC utilizes, and in particular, in those cases it is typically making more true discoveries than TDC does (Fig. 2D and Supp. Fig. 9).
The a-TDC procedure yields more true discoveries than TDC when using an uncalibrated score because a-TDC benefits from the same effect that partial calibration does: by having a better way to order the PSMs. While, strictly speaking, a-TDC is not reordering the PSMs as partial calibration does, a-TDC selects the target PSMs for filtering based on the partially calibrated score with respect to the competing decoys; hence, a-TDC engages in implicit calibration.
a-TDC benefits from partial calibration
We next investigated the benefits of combining our two procedures, a-TDC and partial calibration. We find that, similar to TDC, a-TDC can gain a significant boost in statistical power, as can be seen by the increase in the number of target discoveries in Supp. Fig. 10. As expected from our analyses of a-TDC's performance, a-TDC's power advantage over TDC diminishes with the increase in the number of calibrating decoys (Fig. 2E, Supp. Fig. 11). Indeed, when the score is perfectly calibrated a-TDC should not have more power than TDC does; regardless, for all degrees of calibration, a-TDC does exhibit reduced variability (e.g., Supp. Fig. 12). At the same time, a-TDC become less conservative with the increase in the number of calibrating decoys (Fig. 2F, Supp. Fig. 13).
3.3 Progressive calibration dynamically decides how many decoys are sufficient
Progressive calibration can be quite effective in significantly reducing the number of calibrating decoys that we use while achieving near-optimal power. In Fig. 3A-B) we repeatedly simulated identifying 10K spectra (50% native) and used PC coupled with TDC to control the FDR. The experiment was repeated 10K times, and the average number of calibrating decoys determined by our PC procedure was only 117. Still, in terms of power little was lost: comparing the ratio of the number of (TDC) target discoveries our PC procedure made to the number of (TDC) target discoveries attained by the maximally considered 2047 calibrating decoys we find a median of 99.3% and 0.95 quantile of 98% for all nominal FDR levels ≥ 0.05. In other words, while using about 20 times fewer calibrating decoys, PC delivered 98% of the target discoveries at any estimated FDR level ≥ 0.05 in 95% of our 10K experiments. For results using additional spectrum set sizes and proportions of native spectra see Supp. Fig. 14.
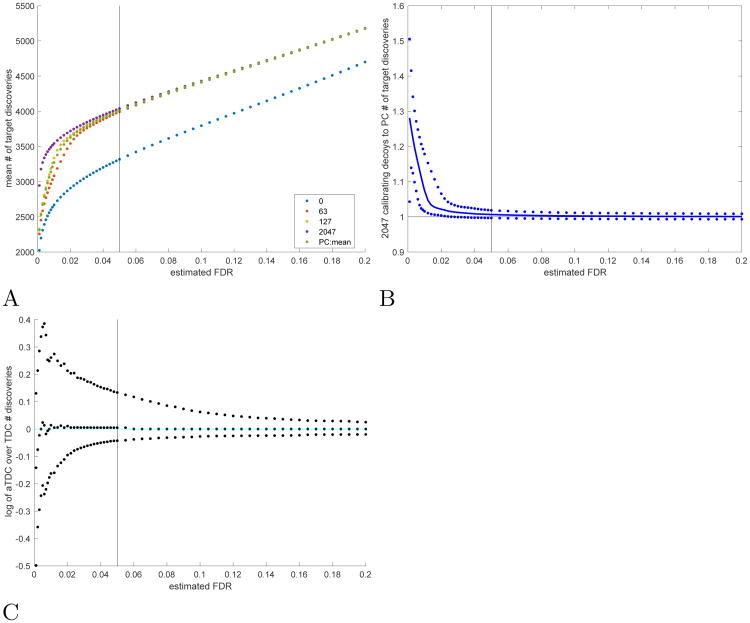
Fig. 3. Progressive calibration (PC).
A The mean number of TDC discoveries using: 0 (raw score), 63, 127, 2047 calibrating decoys as well as the number determined by PC (63 and 127 are the number of decoys in the two cycles that bound the mean number of decoys used by PC in this experiment: 117). B The 0.05, 0.5, and 0.95 quantiles of the ratio of the number of TDC discoveries when using the maximal number of 2047 calibrating decoys to the number of discoveries found by PC. C The same quantiles of the ratio of a-TDC (10 competing decoys) to TDC correct discoveries (both with PC). A-C The vertical bars are located at 0.05, the minimal FDR level of interest for PC in this setup. All means and quantiles are taken with respect to 10K simulations using raw scores, each with 10K spectra in A-B, and 500 spectra in C, 50% native spectra in all.
Our cutoff criterion is not infallible. Indeed, our simulations show that when the set of spectra or the number of correct PSMs is small, then progressive calibration might fail to achieve the near-optimal results TDC can achieve with “full” calibration. For example, for n = 500 and a native spectrum proportion of 10% (Supp. Fig. 15), the median increase (across 10K experiments) in the number of TDC target discoveries made when using 2047 calibrating decoys compared with using PC at FDR level 0.05 is 14%. The corresponding 0.95 quantile is 60% additional discoveries; that is, in 5% of the experiments the increase in discoveries at FDR 0.05 is 60% or higher. One should, however, keep in mind that the mean number of additional discoveries the more intensive calibration effort yields here at FDR 0.05 is about 5. Similarly, with 500 spectra and 50% native spectra, we see in 5% of the experiments an increase higher than 16% when using TDC with 2047 decoys (the median increase is only 1.5%).
It is not surprising that combining a-TDC with PC still offers reduced variability in FDR estimation compared to combining TDC with PC (Supp. Fig. 19). However, with its reduced variability a-TDC can also help us here in better identifying those cases where increased calibration can yield a non-negligible number of additional discoveries. For example, in the same experiment described above with 500 spectra, of which 50% are native, we find that in 95% of the runs the increase in a-TDC discoveries using 2047 decoys over using PC at a nominal FDR level 0.05 is no more than 6.8% (Supp. Fig. 18), compared with 16% when using TDC. This experiment demonstrates that a-TDC is less likely to prematurely terminate the doubling cycle, and it translates here to observing in 5% of the experiments a 14% or more increase in the number of correct a-TDC target discoveries compared with TDC at the same FDR level of 0.05 (Supp. Fig. 3C, Supp. Fig. 20). Of course, this increase in correct discoveries does not come for free: when using TDC to control the FDR, PC used an average of 123 calibrating decoys, whereas with a-TDC that number was 134.
3.4 Analysis of real data
Thus far, we have described analyses that were carried out with simulated data sets. We also carried out similar analyses using the three real data sets described in Supp. Sec. 1.4. Of course, when analyzing real data we do not know which of the discoveries are false, so instead we compare the reported number of discoveries. As outlined below, the results qualitatively agree with our simulations findings.
Consistent with the analysis of our simulations using raw scores, we see that the number of both TDC and a-TDC target discoveries increases with the number of calibrating decoys (Supp. Fig. 21). Specifically, using the malaria data and an estimated FDR level of 0.05, the mean number of TDC target discoveries gradually increases from 2434 when using no calibrating decoys to 2845 when using 10K calibrating decoys (17%, Fig. 4A). We observed a similar increase in the average number of discoveries at the same FDR level when using a-TDC: 2433 to 2844 when increasing from 0 to 10K calibrating decoys. Also consistent with our simulations, we see that regardless of how well calibrated are our scores, a-TDC reduces the variability in the number of discoveries (Fig. 4B, Supp. Fig. 22).
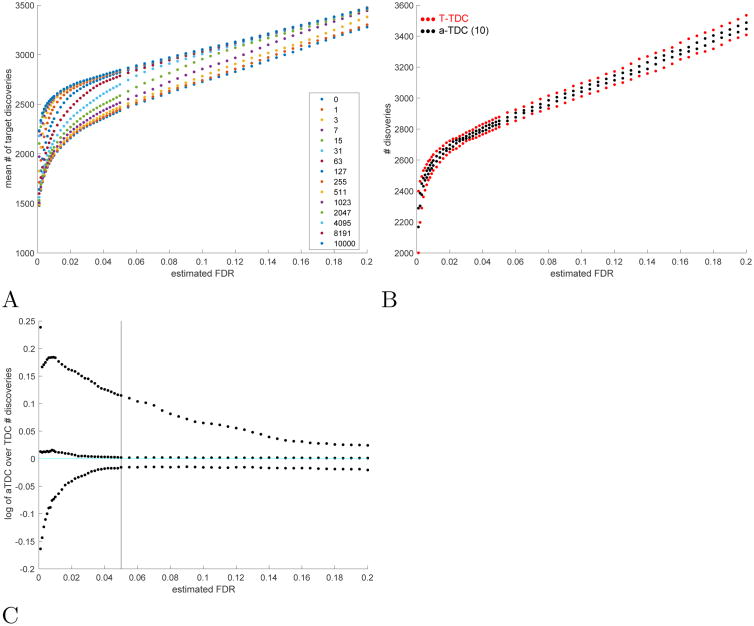
Fig. 4. Malaria data.
A Partial calibration: the mean number of target discoveries in the malaria dataset increases with the number of calibrating decoys. B a-TDC is less variable than TDC: the 0.05 and 0.95 quantiles of the number of a-TDC / TDC discoveries are compared (scores are calibrated using all 10K calibrating decoys). C Using PC a-TDC can have more power than TDC: plotted are the log of the ratios of the 0.05, 0.5 and 0.95 quantiles of the number of a-TDC (10 competing decoys) discoveries over the corresponding numbers for TDC discoveries. A-C All quantiles are taken with respect to 2000 randomly drawn sets of competing decoys, as described in Supp. Sec. 1.4.
We also observe in the real data that PC, especially combined with a-TDC, yields near optimal power with a significantly smaller number of calibrating decoys (Supp. Fig. 23). Specifically, the average number of calibrating decoys used by PC with a-TDC (10 competing decoys) are: yeast 262 (278 with TDC), worm 235 (165), and malaria 165 (141). The corresponding power, expressed in terms of the median of the percentage of the number of discoveries made when using all 10K calibrating decoys, is: yeast 99.2% (99.3% for TDC), worm 98.8% (96.9%), malaria 99.3% (98.9%). This shows that typically PC yields almost maximal power (with respect to calibration) using a much smaller number of calibrating decoys.
Again we find that combining PC with a-TDC can reduce the number of premature stops in PC's doubling process. For example, in 100 of our 2K runs (5%) using the malaria data set, the number of TDC discoveries at FDR level 0.05 was at least 13% higher when using all 10K calibrating decoys than when TDC used PC. Applying a-TDC, the corresponding increase was only 2.8% (Supp. Fig. 23) and this translated to a 12.2% increase in the number of a-TDC discoveries in 5% of our 2K runs (Supp. Fig. 4C).
Finally, even using a-TDC, PC can sometime terminate its decoy doubling procedure earlier than we would like. For example, applying a-TDC to the worm data set using all 10K calibrating decoys, we found that in 100 of our 2K runs (5%) there are at least 7.2% more discoveries than when using the number of decoys determined by PC (Supp. Fig. 23).
4 Discussion
We offer two novel methods that rely on additional decoy databases to improve on TDC, the most commonly used FDR controlling procedure for tandem mass spectrum identification. The partial calibration procedure increases the power of TDC by using a primary-secondary score that implicitly interpolates between our original calibration procedure based on the spectrum-specific ECDF and the raw score. This primary-secondary score's flexibility allows us to gradually enjoy the increase in power that our original calibration procedure offers while investing significantly fewer computational resources: the procedure works even with a single calibrating decoy.
As we noted before [12], we are not the first to point out the value of calibration [10]. However, our approach is different because it does not assume a specific parametric family [15, 16] or require the introduction of a new score function [14, 9]. These previous approaches are less general, and in some cases they might partially fail [12], In contrast, our approach is generally applicable, albeit at a computational cost.
Our new a-TDC procedure helps reduce the decoy-dependent variability of TDC, both in terms of the composition of the reported list of discoveries, as well as in the associated FDR estimation. The impact of a-TDC is particularly noticeable for smaller datasets, and those are also the ones where the additional computational load of a-TDC is less prohibitive.
Interestingly, Barber and Candés recently proved that a slightly modified version of TDC, where one replaces TDC's estimated FDR of with , does not suffer from the liberal bias that TDC exhibits for small FDR levels [2]. Our experiments above show that a-TDC is also able to mitigate much of the liberal bias of TDC and suggest that a-TDC does not result in the loss of power that is associated with the Barber and Candés correction.
An alternative to a-TDC to reduce variability would be to use multiple decoys in a concatenated search. In such an approach, instead of using, say, 10 decoy databases, each the same size as the target database, one can use a single decoy database that is 10 times larger than the target database. A simple adjustment to the estimated FDR makes this approach feasible; however, it has the obvious downside that the larger the decoy set is, the more target discoveries are lost. In comparison, the number of target discoveries a-TDC filters out is the average number that is filtered out by each of the individual TDC procedures (each using equal-sized sets of decoys and targets).
Note that there is some overlap in the goals of partial calibration and a-TDC: a-TDC can increase the number of discoveries in some cases, and calibration can also reduce variability. However, a-TDC will further reduce the variability even if the score is perfectly calibrated. In light of this observation, it would be particularly interesting to look into the balancing act of allocating extra decoys to a-TDC vs. partial calibration. An altogether different direction for future research on a-TDC is the theoretical asymptotic analysis of its performance as the number of competing decoys increases (and its potential connection with [8]).
We further introduce progressive calibration (PC), a method that attempts to find from the data what is the “right” amount of partial calibration we need to invest in. Based on a simple test of the increase in the number of target discoveries in each of its decoy-doubling cycles, PC can typically yield near-optimal power with significant computational savings. The current stopping criterion employed by PC is ad hoc and could benefit from a deeper analysis in the future including, for example, considering a criterion based on the change in the discovery lists themselves rather than just the number of discoveries.
We analyzed all our methods using a novel simulation procedure that allows us to sample datasets that are realistically modeled after real ones. In particular, our samples capture the uncalibrated nature of commonly used scores like XCorr. Our findings in simulated data are echoed in the analysis of three real data sets, showing that our methods can positively impact real biological analysis.
We note that here we looked at improving TDC using additional, randomly shuffled decoys. it would be interesting to compare the resulting enhanced performance with adjusting the mix-max competing FDR controlling method [11] to allow it to utilize multiple decoys as well.
As noted, TDC is the standard procedure for controlling the FDR, although it is typically carried out using reversed rather than shuffled databases. We see no inherent difference between shuffling the peptides and reversing them, and moreover, while not exactly considering the shuffling procedure, Elias and Gygi noted that [5], “Despite their differences, the four decoy databases considered here—protein reversal, peptide pseudo-reveral, random and Markov chain— yielded similar estimations of total correct identifications, and produced similar numbers of correct identifications.” More generally, the theoretical question of the applicability of TDC, which was raised in [8], has no particularly satisfying answer at this point. We currently view this as a modeling question: you cannot prove your model is suitable; rather, at best you can argue that it is. Regardless, the methods presented here improve on TDC whenever it is applicable.
Finally, we stress that these methods apply more generally than the spectrum identification problem. Indeed, as we recently argued, using TDC in this context is a special case of the problem of controlling the FDR among discoveries from searching an incomplete database [?]. In particular, our methods are relevant to controlling the FDR in peptide and protein identification, as well as in problems that arise in metagenomics sequence homology search and forensics.
Supplementary Material
Footnotes
More rigorously we say that the scoring function of an optimal PSM is “calibrated” if the distribution of the score of an optimal PSM in a randomly drawn decoy database is invariant of the spectrum itself.
For reference, Supp. Tab. 1 provides a summary of our notations.
References
- 1.Alves G, Ogurtsov AY, Yu YK. RAId aPS: MS/MS analysis with multiple scoring functions and spectrum-specific statistics. PLoS ONE. 2010;5(11):e15438. doi: 10.1371/journal.pone.0015438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barber RF, Candes EJ. Controlling the false discovery rate via knockoffs. The Annals of Statistics. 2015;43(5):2055–2085. [Google Scholar]
- 3.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995;57:289–300. [Google Scholar]
- 4.Cerqueira FR, Graber A, Schwikowski B, Baumgartner C. Mude: A new approach for optimizing sensitivity in the target-decoy search strategy for large-scale peptide/protein identification. Journal of Proteome Research. 2010;9(5):2265–2277. doi: 10.1021/pr901023v. [DOI] [PubMed] [Google Scholar]
- 5.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nature Methods. 2007;4(3):207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 6.Elias JE, Gygi SP. Target-decoy search strategy for mass spectrometry-based proteomics. Methods in Molecular Biology. 2010;604:55–71. doi: 10.1007/978-1-60761-444-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eng JK, McCormack AL, Yates JR., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. Journal of the American Society for Mass Spectrometry. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 8.Gupta N, Bandeira N, Keich U, Pevzner P. Target-decoy approach and false discovery rate: When things may go wrong. Journal of the American Society for Mass Spectrometry. 2011;22(7):1111–1120. doi: 10.1007/s13361-011-0139-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howbert JJ, Noble WS. Computing exact p-values for a cross-correlation shotgun proteomics score function. Molecular and Cellular Proteomics. 2014;13(9):2467–2479. doi: 10.1074/mcp.O113.036327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeong K, Kim S, Bandeira N. False discovery rates in spectral identification. BMC Bioinformatics. 2012;13(Suppl. 16):S2. doi: 10.1186/1471-2105-13-S16-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keich U, Kertesz-Farkas A, Noble WS. Improved false discovery rate estimation procedure for shotgun proteomics. Journal of Proteome Research. 2015;14(8):3148–3161. doi: 10.1021/acs.jproteome.5b00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keich U, Noble WS. On the importance of well calibrated scores for identifying shotgun proteomics spectra. Journal of Proteome Research. 2015;14(2):1147–1160. doi: 10.1021/pr5010983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kertesz-Farkas A, Keich U, Noble WS. Tandem mass spectrum identification via cascaded search. Journal of Proteome Research. 2015;14(8):3027–3038. doi: 10.1021/pr501173s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S, Gupta N, Pevzner PA. Spectral probabilities and generating functions of tandem mass spectra: a strike against decoy databases. Journal of Proteome Research. 2008;7:3354–3363. doi: 10.1021/pr8001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klammer AA, Park CY, Noble WS. Statistical calibration of the sequest XCorr function. Journal of Proteome Research. 2009;8(4):2106–2113. doi: 10.1021/pr8011107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spirin V, Shpunt A, Seebacher J, Gentzel M, Shevchenko A, Gygi S, Sunyaev S. Assigning spectrum-specific p-values to protein identifications by mass spectrometry. Bioinformatics. 2011;27(8):1128–1134. doi: 10.1093/bioinformatics/btr089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.