Abstract
Background:
Insulin-like growth factor-1 (IGF-1) plays an important role in the regulation of bone formation and mineralization. We aimed to perform a meta-analysis to assess the association of three IGF-1 single nucleotide polymorphisms (SNPs) rs35767, rs2288377, and rs5742612 with osteoporosis risk.
Methods:
A systematic search of PubMed, Web of Science, Embase, Medline, Scopus, CNKI, and Wanfang databases was conducted. Odds ratios (OR) and 95% confidence intervals (CI) were calculated using a fixed effects model.
Results:
Four Chinese case-control studies with a total of 2807 participants were included in this meta-analysis. The results revealed an association between rs35767 and osteoporosis risk in all study subjects (women and men) in dominant (OR 1.32, 95% CI 1.13–1.53, P < .001), recessive (OR 1.73, 95% CI 1.35–2.21, P < .001), homozygote (OR 1.89, 95% CI 1.46–2.45, P < .001), and allelic (OR 1.31, 95% CI 1.18–1.47, P < .001) models. Subgroup analysis according to gender showed that rs35767 was associated with osteoporosis risk in women under dominant (OR 1.29, 95% CI 1.08–1.54, P = .005), recessive (OR 1.59, 95% CI 1.19–2.12, P = .002), homozygote (OR 1.73, 95% CI 1.28–2.34, P < .001), and allelic (OR 1.28, 95% CI 1.12–1.47, P < .001) models. Meta-analysis did not find associations of rs2288377 and rs5742612 with osteoporosis risk. There was no evidence of between-study heterogeneity and publication bias.
Conclusion:
Our results suggest that rs35767 is associated with osteoporosis risk in Chinese, whereas there is no association of rs2288377 and rs5742612 with osteoporosis risk.
Keywords: meta-analysis, osteoporosis, polymorphism
1. Introduction
Osteoporosis is defined by the World Health Organization (WHO) as a value for bone mineral density (BMD) 2.5 standard deviations below the population average in young healthy individuals (BMD T-score of -2.5 or less).[1] It is the commonest metabolic bone disease worldwide and often remains asymptomatic and undetected until bone fracture occurs. Osteoporosis is a major public health problem and the economic burden of the disease is increasing dramatically as populations age.[2] The pathogenesis of osteoporosis is complex and multifactorial. Familial and linkage studies have suggested that genetic factors play an important role in both osteoporosis and its associated phenotypes, including BMD, bone mass, and broadband ultrasound attenuation (BUA).[3,4] Over the past 2 decades, candidate gene association studies and genome-wide association studies have identified many susceptibility genes for osteoporosis, including cytochrome P450 family 19 subfamily A member 1 (CYP19A1), low-density lipoprotein-related receptors 5 (LRP5), membrane palmitoylated protein 7 (MPP7), and Zinc finger and BTB domain-containing protein 40 (BTB40).[5] Identifying disease susceptibility genes is one of the key challenges in osteoporosis research and has attracted considerable attention from researchers, which will help to increase our understanding of the pathogenesis and pathophysiology of osteoporosis.
Insulin-like growth factor-1 (IGF-1) is the primary ligand for the cell surface tyrosine kinase signaling molecule, IGF receptor 1 (IGF1R).[6] It plays an important role in cell proliferation, differentiation, and apoptosis and is the major mediator of the effect of growth hormone (GH) on both bone growth and mineralization.[7,8] IGF-1 decreased osteoblast apoptosis and promoted osteoblastogenesis through the phosphoinositide 3-kinase (PI3K) pathway.[9] Animal studies showed that IGF-1-deficient mice developed smaller skeletons with a significant delay in mineralization at 14.5 dpc and onward.[10] In addition, conditional igf1 receptor null mice demonstrated reduced bone formation and reduced trabecular bone volume.[11] In agreement with the findings from animal studies, clinical studies showed a positive association between serum IGF-1 levels and BMD in different ethnic groups.[12–14] Low serum levels of IGF-1 were found to be associated with osteoporotic fractures.[15,16] These lines of evidence suggest an important role of IGF-1 in bone formation and mineralization. Thus, the IGF-1 gene may be a candidate gene for osteoporosis risk. Located on chromosome 12, the IGF-1 gene consists of 6 exons, including 2 leader exons, and has 2 promoters.[17] Previous case-control association studies have evaluated the association of several common single nucleotide polymorhisms (SNPs) in the IGF-1 gene with osteoporosis risk, but the evidence has not been reviewed and analyzed systematically.
In the present study, we aimed to conduct a meta-analysis of publically available data to clarify if 3 SNPs (rs35767, rs 2288377, and rs5742612) in the IGF-1 gene are associated with osteoporosis risk.
2. Materials and methods
2.1. Study identification
This meta-analysis followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) Statement. An electronic search of the literature was performed to identify case-control association studies investigating the relationship of the IGF-1 SNPs (rs35767, rs 2288377, and rs5742612) with osteoporosis risk. We searched PubMed, Web of Science, Embase, Medline, Scopus, China National Knowledge Infrastructure (CNKI), and Wanfang databases for articles published in peer-review journals. No date restrictions were placed on the search. The search strategy included using the keywords “case-control studies, polymorphism, osteoporosis, insulin-like growth factor-1, rs35767, rs2288377, rs5742612, risk.” Titles and abstracts of relevant papers identified through the search were screened by one of us and were rejected if the paper clearly did not meet the inclusion criteria. Full-text papers were then assessed for eligibility. We screened the reference lists of review articles to find relevant papers that were potentially missed by the initial search. We did not contact authors for additional data. Since we only dealt with published data in this meta-analysis, we did not obtain ethics approval from the local ethics committee.
2.2. Study inclusion/exclusion criteria
Studies were selected for review if they met the following criteria: a case-control association study, examined the association of the IGF-1 SNPs with osteoporosis risk, there were at least 2 comparison groups, and provided sufficient data for genotypic distribution in both cases and controls. We excluded studies that were published only as abstracts or conference reports. Familial-based studies were also excluded.
2.3. Data extraction and quality assessment
Data were extracted by 2 reviewers using a customized database for data extraction. The following information was collected for each study: first author, year of publication, country, number of cases and controls, gender and mean age of subjects, genotype distributions by case/control status, and genotyping method. The quality of individual studies was assessed using the Newcastle–Ottawa Scale (NOS) (http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp). Each study was judged on 8 items, categorized into 3 key areas including selection of subjects, comparability, and exposure. A score of ≤5 (out of 9) indicated a high risk of bias.[18] Disagreements were resolved by consensus.
2.4. Statistical analyses
Meta-analyses were performed using Stata version 11.0. Between-study heterogeneity was assessed using the Q-test and I2 statistic, with significance set at P < .10.[19] I2 values of 25%, 50%, and 75% were considered low, medium, and high heterogeneity, respectively. Odds ratio (OR) and 95% interval confidence (CI) were calculated from the dominant, recessive, homozygote, and allelic model for each SNP having the minor allele frequency as the reference category. In case of substantial heterogeneity, random effects summary ORs were calculated using the DerSimonian method.[20] When heterogeneity was absent, the Mantel–Haenszel method was used to calculate fixed effects summary ORs.[21] The significance of the summary ORs was determined by the Z-test and a P value of less than .05 was assumed to be statistically significant. Forest plots were produced to visually assess the individual study ORs and overall ORs with corresponding 95% CIs. Summary minor allele frequency of each IGF-1 SNP in control subjects was calculated using Meta-analyst 3.13. Sensitivity analysis was performed to evaluate whether the omission of 1 study would have a disproportionate impact on the results of the meta-analyses. For the assessment of publication bias, we utilized Egger test and Begg test. Since there were fewer than 10 studies qualified for each SNP, funnel plots were not produced for evaluating publication bias.[22]
3. Results
3.1. Characteristics of published studies
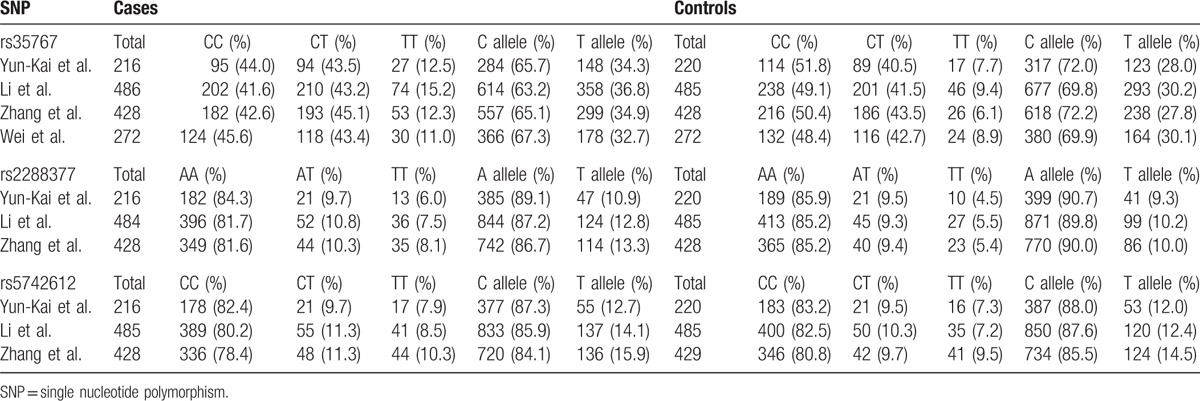
Our initial literature searches yielded 127 reports in total. Of these, 121 duplicated or nonrelevant studies were excluded after screening of the titles/abstracts. Six full-text studies were assessed for eligibility. Among these, 4 case-control studies met the inclusion criteria and were included in the final analyses.[23–26]Figure 1 summarizes the process of identifying eligible studies. All of the studies were published in English. A total of 2807 participants took part in the studies, with 1 recruiting men and women,[25] and 3 women only.[23,24,26] Four studies investigated rs35767,[23–26] 3 studies assessed rs2288377,[23–25] and 3 studies evaluated rs5742612.[23–25] The pooled minor allele frequency for rs35767, rs2288377, rs5742612 was 0.291 (95% CI: 0.275–0.308), 0.100 (95% CI: 0.088–0.113), and 0.131 (95% CI: 0.118–0.146), respectively. The main characteristics of the eligible studies are presented in Table 1. The genotype and allele frequencies for each SNP in the eligible studies are summarized in Table 2.
Figure 1.
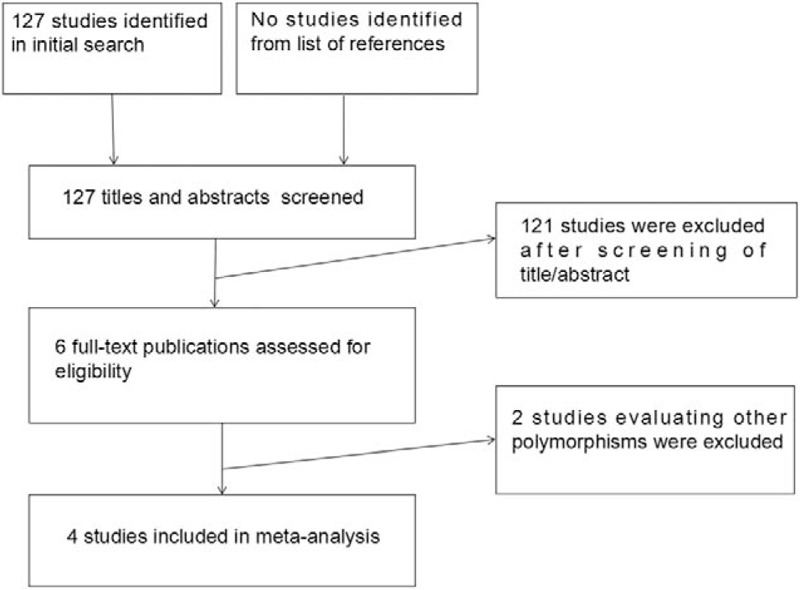
Flow diagram of the screening process of retrieved studies.
Table 1.
Summary of included studies.
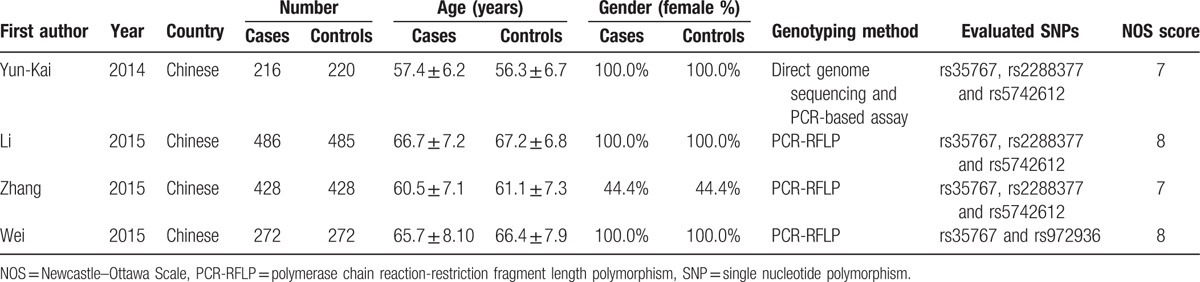
Table 2.
Genotypic and allelic distribution of each SNP.
3.2. Association between the IGF-1 SNPs and osteoporosis risk
Four case-control association studies with a total of 1402 cases and 1405 controls assessed the relation of rs35767 with osteoporosis risk. The fixed-effect meta-analyses revealed that rs35767 was associated with osteoporosis risk in all study subjects (women and men) under dominant (TT + CT vs CC, OR 1.32, 95% CI 1.13–1.53, P < .001), recessive (TT vs CT + CC, OR 1.73, 95% CI 1.35–2.21, P < .001), homozygote (TT vs CC, OR 1.89, 95% CI 1.46–2.45, P < .001), and allelic (T allele vs C allele, OR 1.31, 95% CI 1.18–1.47, P < .001) models (Fig. 2 and Table 3). We further performed stratified analyses according to gender. The results showed that there was an association between rs35767 and osteoporosis risk in women under dominant (TT + CT vs CC, OR 1.29, 95% CI 1.08–1.54, P = .005), recessive (TT vs CT + CC, OR 1.59, 95% CI 1.19–2.12, P = .002), homozygote (TT vs CC, OR 1.73, 95% CI 1.28–2.34, P < .001), and allelic (T allele vs C allele, OR 1.28, 95% CI 1.12–1.47, P < .001) models (Fig. 2 and Table 3). We did not identify any between-study heterogeneity in the analyses for rs35767 (I2 = 0.0%, P > .10) (Fig. 2 and Table 3). Sensitivity analyses by excluding each study in turn ensured that no single study was solely responsible for the significance of the results.
Figure 2.
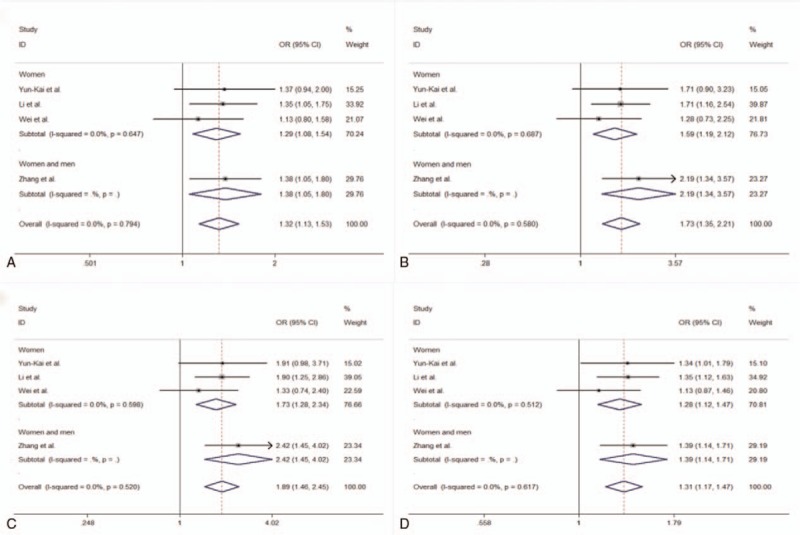
Forest plots showing meta-analysis of the association between rs35767 and osteoporosis risk. A, Meta-analysis of the association between rs35767 and osteoporosis risk under dominant model (TT + CT vs CC). B, Meta-analysis of the association between rs35767 and osteoporosis risk under recessive model (TT vs CT + CC). C, Meta-analysis of the association between rs35767 and osteoporosis risk under homozygote model (TT vs CC). D, Meta-analysis of the association between rs35767 and osteoporosis risk under allelic model (T allele vs C allele).
Table 3.
The results of the meta-analysis for the association of rs35767 with osteoporosis.
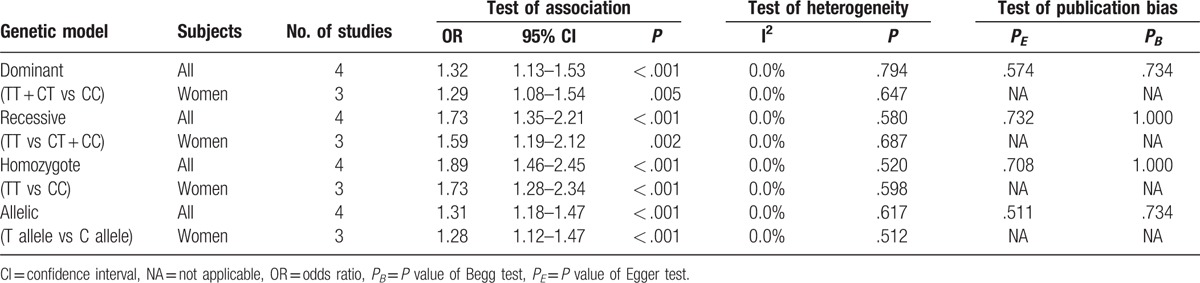
Three studies including 1128 cases and 1133 controls to date assessed the relationship of rs2288377 with osteoporosis risk. Pooling data from these showed that rs2288377 was associated with osteoporosis risk in all study subjects (women and men) under dominant (TT + AT vs AA, OR 1.26, 95% CI 1.01–1.58, P = .042), recessive (TT vs AT + AA, OR 1.44, 95% CI 1.02–2.03, P = .037), homozygote (TT vs AA, OR 1.46, 95% CI 1.04–2.06, P = .031), and allelic (T allele vs A allele, OR 1.31, 95% CI 1.08–1.57, P = .005) models (Table 4). However, in subgroup analysis according to gender, the results did not suggest an association between rs2288377 and osteoporosis risk in women under dominant (TT + AT vs AA, OR 1.23, 95% CI 0.93–1.64, P = .151), recessive (TT vs AT + AA, OR 1.36, 95% CI 0.87–2.11, P = .173), homozygote (TT vs AA, OR 1.38, 95% CI 0.89–2.15, P = .154), and allelic (T allele vs A allele, OR 1.26, 95% CI 1.00–1.60, P = .054) models (Table 4). The single study containing women and men subjects did not find an association between rs2288377 and osteoporosis risk (not shown).[25] Sensitivity analyses demonstrated that after the exclusion of the study by Li et al or the study by Zhang et al, the association between rs2288377 and osteoporosis risk in all study subjects was not statistically significant (P > .05), suggesting that the results of meta-analysis in all study subjects were not stable. There was no between-study heterogeneity among the studies (I2 = 0.0%, P > .10) (Table 4).
Table 4.
The results of the meta-analysis for the association of rs2288377 with osteoporosis.
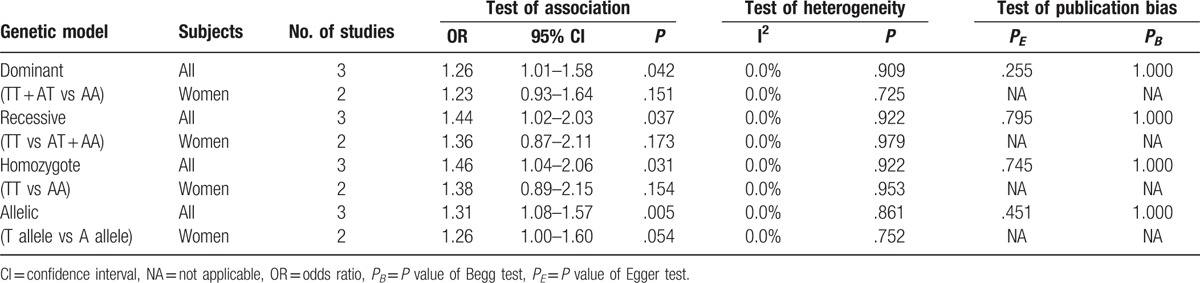
For rs5742612, 3 studies with 1129 cases and 1134 controls were included in the meta-analysis. The pooled effect estimates from all studies did not suggest an association between this SNP and osteoporosis risk in all study subjects (women and men) under dominant (TT + CT vs CC, OR 1.13, 95% CI 0.92–1.40, P = .241), recessive (TT vs CT + CC, OR 1.13, 95% CI 0.84–1.51, P = .435), homozygote (TT vs CC, OR 1.14, 95% CI 0.85–1.54, P = .383), and allelic (T allele vs C allele, OR 1.13, 95% CI 0.95–1.34, P = .164) models (Table 5). In stratified analysis by gender, rs5742612 was not associated with osteoporosis risk in women under any of the genetic models (Table 5). There was no evidence of significant between-study heterogeneity (I2 = 0.0%, P > .10). Sensitivity analysis for the association between rs5742612 and osteoporosis risk did not change our results.
Table 5.
The results of the meta-analysis for the association of rs5742612 with osteoporosis.
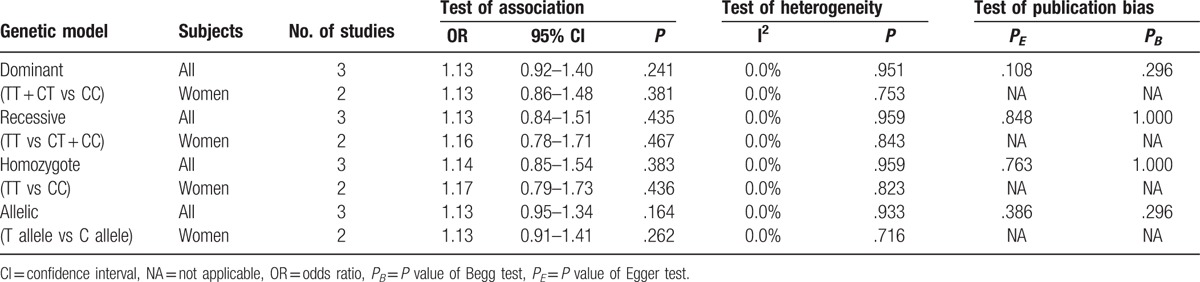
3.3. Publication bias
Begg test and Egger test did not indicate any evidence of publication bias (Tables 3–5). As there were less than 10 studies for each IGF-1 polymorphism, we did not produce funnel plots to assess publication bias.[22]
4. Discussion
IGF-1 is a 70 amino acid single chain polypeptide synthesized by many tissues, particularly by the liver in response to GH stimulation. It is one of the most important growth factors for the development and growth of the skeleton and maintenance of bone mass. In vitro studies demonstrated that IGF-1 treatment effectively inhibited apoptosis and increased proliferation and differentiation of primary osteoblasts.[27,28] In animal models of osteoporosis, treatment with IGF-1 enhanced the recruitment of osteoblastic cells, increased trabecular bone formation, and prevented trabecular bone loss.[29] Compared to control littermate mice at 8 weeks of age, IGF-1-deficient mice had significantly reduced femoral areal and volumetric BMD.[30] Consistent with the animal data, clinical studies found that serum levels of IGF-1 were remarkably downregulated in osteoporosis patients and associated with increased risk of fractures.[16,31] In addition, bone marrow IGF-1 concentrations were 40% lower in individuals with osteoporosis than control subjects.[32] Furthermore, recombinant IGF-1 treatment increased osteoblastic function and stimulated both bone resorption and formation in healthy older women.[33] Given the pivotal role of IGF-1 in bone health, the IGF-1 gene has become a candidate to study in osteoporosis.
rs35767, rs2288377, and rs5742612 are 3 common SNPs in the IGF-1 gene. In the past 5 years, several case-control association studies have investigated their relationship with osteoporosis risk. However, overall evidence is mixed, and no systematic reviews or meta-analyses are available. In the present study, we conducted a meta-analysis of 4 case-control studies with a total of 2807 participants to evaluate the association of these SNPs with osteoporosis risk. The results showed that rs35767 was associated with the risk of osteoporosis in the Chinese population, whereas current evidence was not sufficient to support an association of rs2288377 and rs5742612 with osteoporosis risk. To the best of our knowledge, this is the first meta-analysis on the topic.
It was noteworthy that although meta-analysis found an association between rs2288377 and osteoporosis risk in all subjects (women and men), in subgroup analysis by gender, the results showed no association between this SNP and osteoporosis risk in women, and the single study containing women and men found rs2288377 was not associated with osteoporosis under dominant, recessive, and homozygote models.[25] In addition, sensitivity analysis demonstrated that the association between rs2288377 and osteoporosis risk was not statistically significant after the exclusion of the study by Li et al or the study by Zhang et al.[24,25] Subgroup analysis and sensitivity analysis suggested that the results of rs2288377 were not stable, and future studies were needed to derive a more definitive conclusion. Our meta-analysis did not identify any between-study heterogeneity among the eligible studies (P > .10, I2 = 0.0%), and there was no evidence of publication bias. Based on strict selection of studies and careful evaluation, the results of our meta-analysis could be reliable.
BMD has been utilized as the phenotype of choice for defining heritable markers for osteoporosis. Previous genetic studies have shown that regulation of BMD is determined by the effects of genetic variations.[4] Among the included studies, the study by Yun-kai demonstrated that carriers of the rs35767 T allele had lower values of BMD at L1-L4 vertebrae, femoral neck, total hip, and trochanter than those carrying the C allele.[23] Similar findings were obtained from the study by Zhang et al, which showed that BMD levels at L1-L4 vertebrae, femoral neck, total hip, and trochanter were significantly decreased in the CT + TT genotypes of rs35767 in comparison with the CC genotypes in osteoporosis patients.[25] It was of interest to evaluate the association between rs35767 and BMD. However, there was discrepancy between the 2 studies in the selection of cases and controls for evaluating BMD. Yun-kai et al divided subjects according to allele status,[23] whereas the study by Zhang et al utilized genotypes.[25] Due to discrepancy in study design and limited data availability (n < 3), we did not perform subgroup analysis for BMD. No included studies assessed the relation of rs35767, rs2288377, and rs5742612 with osteoporotic fractures.
This meta-analysis has potential limitations. First, all of the included studies were performed in the Chinese population. We did not find relevant studies from other regions. The association between these IGF-1 SNPs and osteoporosis risk needed to be evaluated in other ethnic groups, including Koreans, Japanese, and Europeans. Second, although we included all published case-control association studies on the topic through extensive literature search, the sample size of eligible studies in this meta-analysis was still relatively small. In addition, among the 4 eligible studies, 3 were performed on postmenopausal female subjects. Future research containing both male and female subjects with a larger sample size is required to validate our findings. Third, we only evaluated the relation of SNPs with osteoporosis risk. Other variants in the IGF-1 gene including the promoter CA-repeat polymorphism were not included in this meta-analysis.
In summary, we found that the IGF-1 SNP rs35767 was associated with osteoporosis risk in the Chinese population, whereas there was no association of rs2288377 and rs5742612 with osteoporosis risk.
Footnotes
Abbreviations: BMD = bone mineral density, BTB40 = BTB domain-containing protein 40, BUA = broadband ultrasound attenuation, CI = confidence interval, CNKI = China national knowledge infrastructure, CYP19A1 = cytochrome P450 family 19 subfamily A member 1, GH = growth hormone, IGF-1 = insulin-like growth factor-1, IGF1R = insulin-like growth factor-1 receptor, LRP5 = low-density lipoprotein-related receptors 5, MPP7 = membrane palmitoylated protein 7, NOS = Newcastle–Ottawa scale, OR = odds ratio, PI3K = phosphoinositide 3-kinase, PRISMA = preferred reporting items for systematic reviews and meta-analyses, SNP = single nucleotide polymorphism, WHO = World Health Organization.
The authors have no conflicts of interest to disclose.
References
- [1].Alejandro P, Constantinescu F. A review of osteoporosis in the older adult. Clin Geriatr Med 2017;33:27–40. [DOI] [PubMed] [Google Scholar]
- [2].Drake MT, Clarke BL, Lewiecki EM. The pathophysiology and treatment of osteoporosis. Clin Ther 2015;37:1837–50. [DOI] [PubMed] [Google Scholar]
- [3].Xie W, Ji L, Zhao T, et al. Identification of transcriptional factors and key genes in primary osteoporosis by DNA microarray. Med Sci Monit 2015;21:1333–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mafi Golchin M, Heidari L, Ghaderian SM, et al. Osteoporosis: a silent disease with complex genetic contribution. J Genet Genomics 2016;43:49–61. [DOI] [PubMed] [Google Scholar]
- [5].Sabik OL, Farber CR. Using GWAS to identify novel therapeutic targets for osteoporosis. Transl Res 2017;181:15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bikle DD, Tahimic C, Chang W, et al. Role of IGF-I signaling in muscle bone interactions. Bone 2015;80:79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Pass C, MacRae VE, Ahmed SF, et al. Inflammatory cytokines and the GH/IGF-I axis: novel actions on bone growth. Cell Biochem Funct 2009;27:119–27. [DOI] [PubMed] [Google Scholar]
- [8].Li W, Yang SY, Hu ZF, et al. Growth factors enhance endothelial progenitor cell proliferation under high-glucose conditions. Med Sci Monit 2009;15:BR357–363. [PubMed] [Google Scholar]
- [9].Cao JJ, Kurimoto P, Boudignon B, et al. Aging impairs IGF-I receptor activation and induces skeletal resistance to IGF-I. J Bone Miner Res 2007;22:1271–9. [DOI] [PubMed] [Google Scholar]
- [10].Sheng MH, Lau KH, Baylink DJ. Role of osteocyte-derived insulin-like growth factor I in developmental growth, modeling, remodeling, and regeneration of the bone. J Bone Metab 2014;21:41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhang M, Xuan S, Bouxsein ML, et al. Osteoblast-specific knockout of the insulin-like growth factor (IGF) receptor gene reveals an essential role of IGF signaling in bone matrix mineralization. J Biol Chem 2002;277:44005–12. [DOI] [PubMed] [Google Scholar]
- [12].Langlois JA, Rosen CJ, Visser M, et al. Association between insulin-like growth factor I and bone mineral density in older women and men: the Framingham Heart Study. J Clin Endocrinol Metab 1998;83:4257–62. [DOI] [PubMed] [Google Scholar]
- [13].Rhee EJ, Oh KW, Lee WY, et al. Age, body mass index, current smoking history, and serum insulin-like growth factor-I levels associated with bone mineral density in middle-aged Korean men. J Bone Miner Metab 2004;22:392–8. [DOI] [PubMed] [Google Scholar]
- [14].Zhao HY, Liu JM, Ning G, et al. Relationships between insulin-like growth factor-I (IGF-1) and OPG, RANKL, bone mineral density in healthy Chinese women. Osteoporos Int 2008;19:221–6. [DOI] [PubMed] [Google Scholar]
- [15].Yamaguchi T, Kanatani M, Yamauchi M, et al. Serum levels of insulin-like growth factor (IGF); IGF-binding proteins-3, -4, and -5; and their relationships to bone mineral density and the risk of vertebral fractures in postmenopausal women. Calcif Tissue Int 2006;78:18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ohlsson C, Mellström D, Carlzon D, et al. Older men with low serum IGF-1 have an increased risk of incident fractures: the MrOS Sweden study. J Bone Miner Res 2011;26:865–72. [DOI] [PubMed] [Google Scholar]
- [17].Wong HL, Koh WP, Probst-Hensch NM, et al. Insulin-like growth factor-1 promoter polymorphisms and colorectal cancer: a functional genomics approach. Gut 2008;57:1090–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wells GA, Shea B, O’Connell D, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. Available at: http://www.ohri.ca/programs/clinical_epidemiology/oxford.htm. (Date of access: July 21, 2017). [Google Scholar]
- [19].Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med 2002;21:1539–58. [DOI] [PubMed] [Google Scholar]
- [20].DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials 1986;7:177–88. [DOI] [PubMed] [Google Scholar]
- [21].Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 1959;22:719–48. [PubMed] [Google Scholar]
- [22].Higgins JP, Altman DG, Gøtzsche PC, et al. The Cochrane Collaboration's tool for assessing risk of bias in randomised trials. BMJ 2011;343:d5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yun-Kai L, Hui W, Xin-Wei Z, et al. The polymorphism of Insulin-like growth factor-I (IGF-I) is related to osteoporosis and bone mineral density in postmenopausal population. Pak J Med Sci 2014;30:131–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Li F, Xing WH, Yang XJ, et al. Influence of polymorphisms in insulin-like growth factor-1 on the risk of osteoporosis in a Chinese postmenopausal female population. Int J Clin Exp Pathol 2015;8:5727–32. [PMC free article] [PubMed] [Google Scholar]
- [25].Zhang W, Zhang LC, Chen H, et al. Association between polymorphisms in insulin-like growth factor-1 and risk of osteoporosis. Genet Mol Res 2015;14:7655–60. [DOI] [PubMed] [Google Scholar]
- [26].Wei YK, Ma HL, Guo YZ, et al. Association of the IGF-1 rs35767 and rs972936 polymorphisms with the risk of osteoporosis in a Chinese postmenopausal female population. Genet Mol Res 2015;14:14325–30. [DOI] [PubMed] [Google Scholar]
- [27].Hill PA, Tumber A, Meikle MC. Multiple extracellular signals promote osteoblast survival and apoptosis. Endocrinology 1997;138:3849–58. [DOI] [PubMed] [Google Scholar]
- [28].Tumber A, Meikle MC, Hill PA. Autocrine signals promote osteoblast survival in culture. J Endocrinol 2000;167:383–90. [DOI] [PubMed] [Google Scholar]
- [29].Machwate M, Zerath E, Holy X, et al. Insulin-like growth factor-I increases trabecular bone formation and osteoblastic cell proliferation in unloaded rats. Endocrinology 1994;134:1031–8. [DOI] [PubMed] [Google Scholar]
- [30].Mohan S, Richman C, Guo R, et al. Insulin-like growth factor regulates peak bone mineral density in mice by both growth hormone-dependent and -independent mechanisms. Endocrinology 2003;144:929–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kurland ES, Rosen CJ, Cosman F, et al. Insulin-like growth factor-I in men with idiopathic osteoporosis. J Clin Endocrinol Metab 1997;82:2799–805. [DOI] [PubMed] [Google Scholar]
- [32].Xian L, Wu X, Pang L, et al. Matrix IGF-1 maintains bone mass by activation of mTOR in mesenchymal stem cells. Nat Med 2012;18:1095–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ghiron LJ, Thompson JL, Holloway L, et al. Effects of recombinant insulin-like growth factor-I and growth hormone on bone turnover in elderly women. J Bone Miner Res 1995;10:1844–52. [DOI] [PubMed] [Google Scholar]


