Supplemental Digital Content is available in the text
Keywords: coronary artery disease, genetic risk score, major adverse cardiovascular event, single nucleotide polymorphism, statin
Abstract
Many susceptibility loci associated with coronary artery disease (CAD) have been identified using genome-wide association studies (GWAS). This study aimed to examine whether a composite of single nucleotide polymorphisms (SNPs) derived from GWAS could identify the risk of major adverse cardiovascular events (MACEs) in patients with established CAD. There were 1059 patients with CAD were included in the analysis. Of the participants, 686 were on statin treatment at the start of follow-up. A weighted genetic risk score (wGRS) was calculated as the sum of risk alleles multiplied by the hazard ratio for a particular SNP. In single variant analyses, rs579459, rs4420638, and rs2107595 were associated with an increased risk of MACE. A wGRS was further constructed to evaluate the cumulative effect of the 3 SNPs on the prognosis of CAD. The risk of MACE among patients with high and intermediate wGRS was 1.968- and 1.838-fold, respectively, higher than those with low wGRS. This effect was more evident in patients using lipid-lowering medication and with hypertension. Furthermore, the interaction analysis revealed that lipid-lowering medication and hypertension interacted with the genetic effect off wGRS on the risk of MACE in patients using lipid-lowering medication or with hypertension (Pinteraction < .001). We further analyzed the follow-up change in low-density lipoprotein cholesterol (LDL-C) level at 6 months after CAD disclosure and evaluated whether that was due to wGRS or statin use. The lowest reduction in LDL-C was observed in patients with high GRS who received statin treatment. Furthermore, LDL-C reduction of patients with intermediate wGRS was less than those with low wGRS in patients treated with statin. Taken together, a wGRS comprised of SNPs significantly predicts MACE in CAD patients receiving statin treatment and hypertension.
1. Introduction
Coronary artery disease (CAD) is one of the leading causes of death throughout the world.[1] With the social and economic transformation, improved standard of living, dietary changes, and population aging, the incidence and mortality of CAD in China are increasing persistently.[2] CAD in China has become a major threat to public health. Increasing evidence has demonstrated that secondary prevention of CAD by comprehensive risk factor modification reduces mortality and the incidence of subsequent recurrent events, and improves quality of life. It has been demonstrated that CAD is caused by genetic and environmental factors and their interactions with each other that lead to the progressive development of atherosclerotic plaques within the walls of the coronary arteries.[3] Using genome-wide association study (GWAS), more than 57 loci have been identified that contribute to risk of CAD or its major complication, myocardial infarction (MI).[4–9] Many of these loci are associated with known risks factors for CAD, such as low-density lipoprotein cholesterol (LDL-C) levels and blood pressure, which are the primary treatment targets for patients with CAD. However, the associations between these variants and clinical outcome of CAD were evaluated in comparatively few studies.
Genetic testing becomes more widely available due to the progressively reducing genotyping cost. Therefore, there is increasing interest in estimating risks of common diseases based on genetic testing.[10] Many studies have investigated the association of a genetic risk score (GRS) based on multiple loci with the risks and outcome of common diseases.[2,11–16] Furthermore, some clinical trials have been carried out to evaluate the application of genetic risk information in clinical practice,[15–17] which help refine risk estimates and prevention strategies. Therefore, the incorporation of a GRS in commonly used clinical risk scores such as the Framingham or Omnibus risk estimator will improves the risk prediction. However, it is unknown whether these genetic variants affecting risk of CAD influences clinical outcomes. To address this issue, we carried out an association study to investigate whether CAD risk associated single nucleotide polymorphisms (SNPs) identified from GWAS lead to major adverse cardiovascular events (MACEs) in CAD patients.
2. Materials and methods
2.1. Subjects
A total of 1505 CAD patients were recruited from Tongren Hospital between May 2012 and December 2014. The diagnosis of CAD was confirmed by coronary angiography. Patients with at least one stenosis ≥50% stenosis in any of the major coronary arteries (left anterior descending, left circumflex, and right coronary artery) were included in the study. The exclusion criteria included monogenic diseases, valvular heart disease, cardiomyopathy, heart failure, peripheral arterial disease, connective tissue diseases, severe liver and renal diseases, tumors, other vascular diseases, and known history of cerebrovascular attack, coronary angioplasty, and acute MI. In addition, patients were also excluded if they did not perform daily living activities independently. None of patients received lipid-lowering drugs just before the study. All cases were unrelated Han Chinese residing in Eastern China. The primary endpoint was the MACEs, including vascular death, MI, stroke, hospitalization for unstable angina, and coronary revascularization. In patients suffering multiple events, only the time of the first event was used for further analyses. The study protocol was approved by the Ethics Committee of Tongren Hospital. Written informed consent was obtained from all participants in this study before enrollment.
2.2. Genotyping
We selected 10 previously identified SNPs located in 10 chromosome regions (Chr. 6, 7, 9, 10, 11, 15, and 19) to examine their potential associations with prognosis of CAD.[5,7,18–22] All these SNP had minor allele frequency (MAF) ≥0.05 in the Chinese Han population. The list of the 10 susceptibility SNPs and the associated genes was summarized in Table S1.
Three milliliters of peripheral blood was drawn from a vein into a sterile tube containing ethylenediamine tetraacetic acid (EDTA). Genomic DNA was extracted from peripheral blood leukocytes using the Wizard genomic DNA purification kit (Promega, Madison, WI) according to the manufacturer's protocol. Genotyping was performed by the TaqMan method.
2.3. Statistical analyses
For the construction of GRS, each of the 10 SNPs was initially examined for independent association with MACEs in a Cox proportional hazards model. We then simply counted the number of risk-increasing allele for each SNP that the genotypes are coded “0” for nonrisk allele homozygotes, “1” for heterozygotes, and “2” for risk-allele homozygotes. A weighted method was used to calculate GRS for each individual as previously described.[23] The weighted GRS (wGRS) was calculated by multiplying each subject's allele score (0, 1, 2) by the hazard ratio (HR) for a particular SNP.
All study data were analyzed using SPSS 20 (SPSS, Inc., Chicago, IL). All categorical data were expressed as frequency and percentage and continuous data were presented as mean (±SD). Linear regression models were used for continuous data. Cox's proportional hazard models were used to calculate HRs and 95% confidence intervals (CIs) of SNPs and wGRS with adjustment of age, sex, body mass index (BMI), blood pressure, diabetes mellitus (DM), smoking status, statin treatment, and baseline levels of total cholesterol (TC), triglycerides (TG), high density lipoprotein cholesterol (HDL-C), LDL-C, apolipoprotein A1 (ApoA1), apolipoprotein B (ApoB), and lipoprotein(a) (Lp(a)), as appropriate. A P-value less than .05 was considered statistically significant.
3. Results
3.1. Characteristics of the study participants
Of 1505 CAD patients, 446 patients were excluded from further analyses due to incomplete clinical and follow-up data. As a result, a total of 1059 patients met all inclusion criteria and were included in the study. The demographic and clinical characteristics of 1059 patients are summarized in Table 1. Of 1059 patients, 686 cases were on statin treatment at the start of follow-up. A total of 273 events were occurred during the median follow-up period of 36 months. A total of 55 events were fatal.
Table 1.
Baseline clinical characteristics of the study patients.
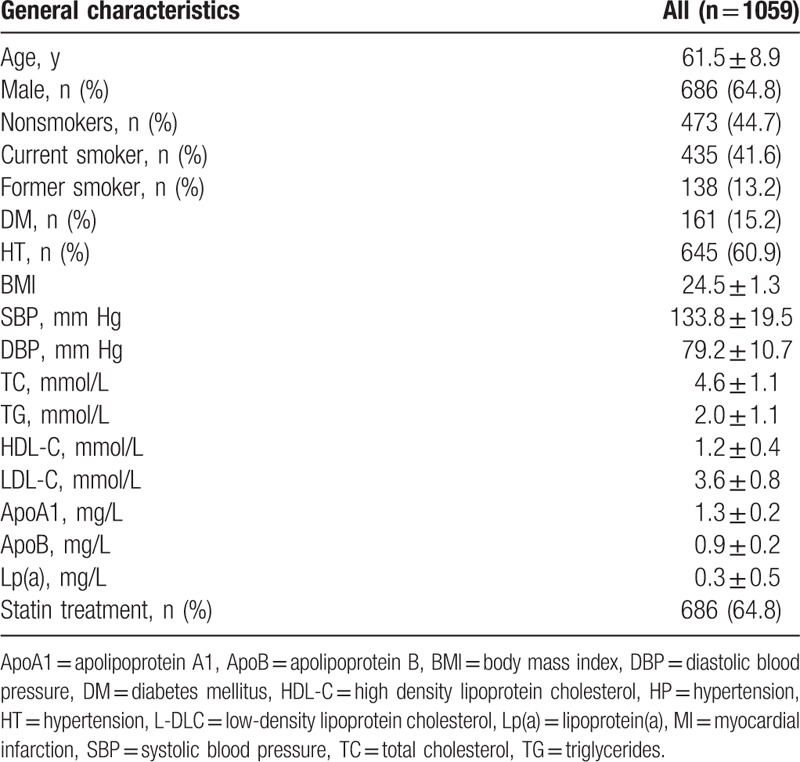
3.2. Association of CAD-related SNPs with MACE
Three SNPs (rs579459, rs4420638, and rs2107595) were associated with an increased risk of MACE. The differences remained significant even after adjusted for age, sex, BMI, blood pressure, DM, smoking status, and baseline levels of TC, TG, HDL-C, LDL-C, ApoA, ApoB, and Lp(a) (Table 2). These indicated that rs579459, rs4420638, and rs2107595 were independent prognostic factors for CAD.
Table 2.
Associations between SNPs and risk of events.
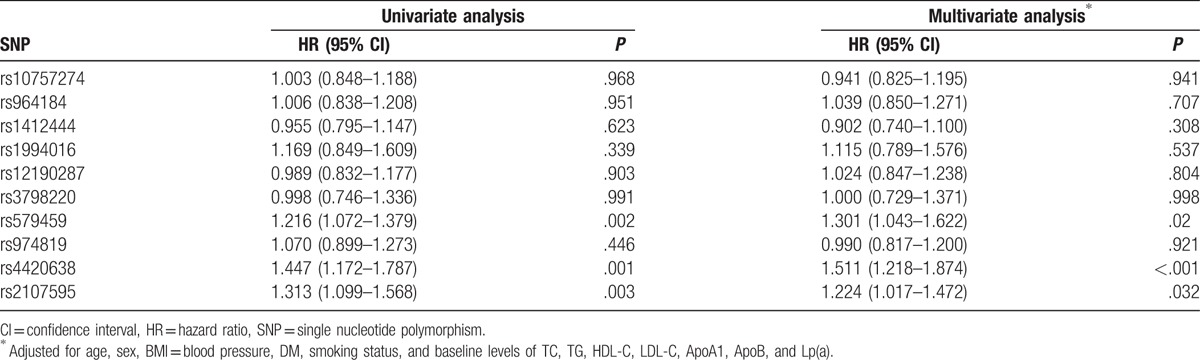
3.3. Association of wGRS with MACE
There was a positive correlation between wGRS and MACE. Higher wGRS was associated with a higher risk of MACE even after adjusted for clinical variables (Table 3). When patients were divided into low (≤1st quintile), intermediate (2nd–4th quintile), and high (>4th quintile) wGRS groups, a significant gradient of risk for MACE was observed in the study. The multivariable adjusted HR (95% CI) for incident MACE for intermediate and high wGRS were 1.817 (95% CI: 1.330–2.484, P < .001) and 1.943 (95% CI: 1.459–2.586, P < .001), respectively, as compared with low wGRS.
Table 3.
Association between GRS and risk of MACE.
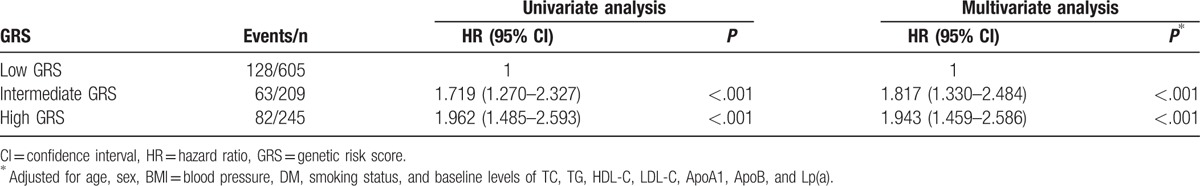
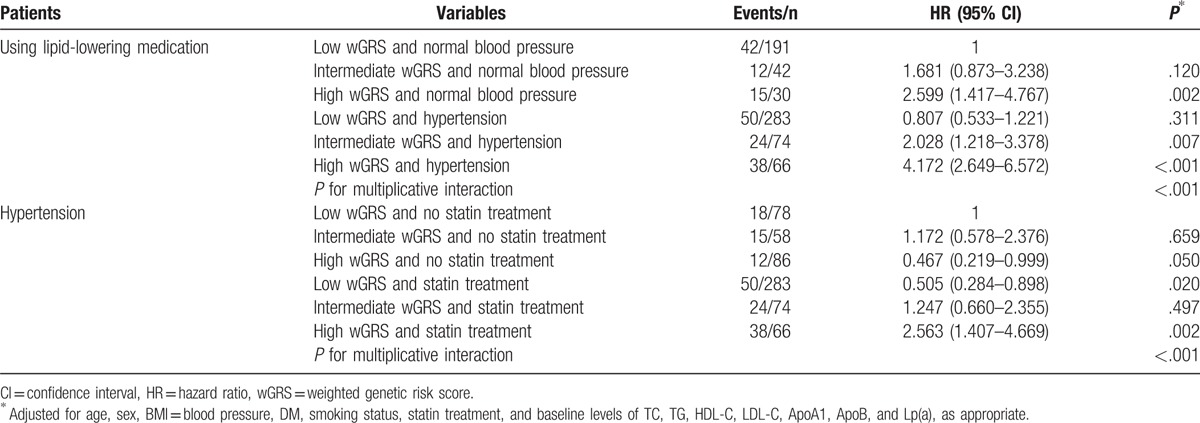
Since the treatment of dyslipidemia and hypertension can significantly reduce all-cause mortality or major cardiovascular events, we further carried out stratification analysis by lipid-lowering medication. As shown in Table 4, there was a significant interaction between wGRS and lipid-lowering medication. Among patients using lipid-lowering medication, we observed that patients with intermediate and high wGRS had over 2.138- and 4.048-fold increased risk for developing MACE (P < .001). However, no significant differences were observed in patients who did not receive lipid-lowering medication. All patients with hypertension were treated with conventional antihypertensives. There was also a significant interaction between wGRS and hypertension (Table 5). Patients with hypertension and intermediate or high wGRS had a higher risk for developing MACE compared with those with low wGRS (P < .001). No significant differences were observed in patients with normal blood pressure. Therefore, we further carried out interaction analysis in patients with hypertension or using lipid-lowering medication, and statistically significant multiplicative interactions on MACE were observed between wGRS and hypertension or lipid-lowering medication (Pinteraction < .001, Table 6). Patients with high wGRS, with or without hypertension, or using lipid-lowering medication, had a significantly elevated risk of MACE (P < .05). For patients with hypertension and low wGRS, lipid-lowering medication can significantly reduce the risk of MACE compared with no treatment (HR = 0.505, 95% CI: 0.284–0.898, P = .020). Furthermore, patients with intermediate wGRS and hypertension had higher risk of MACE compared with those with low wGRS and normal blood pressure among those using lipid-lowering medication (HR = 2.028, 95% CI: 1.218–3.378, P = .007) (Table 6).
Table 4.
Associations between GRS and risk of events stratified by lipid-lowering medication.
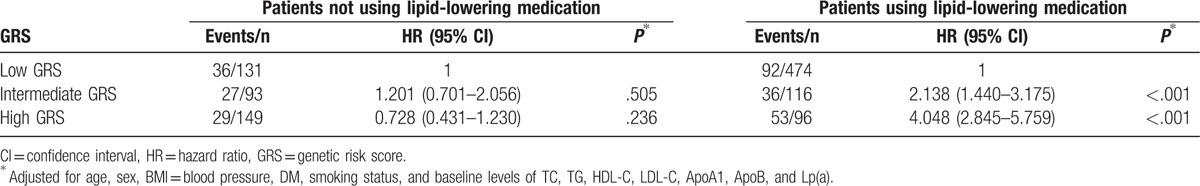
Table 5.
Associations between GRS and risk of events stratified by hypertension.
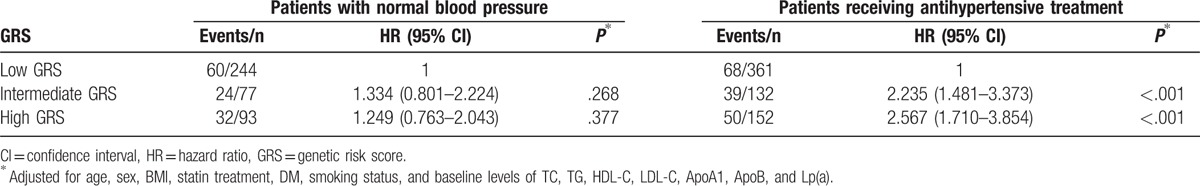
Table 6.
Interaction analysis between wGRS and hypertension or statin treatment.
3.4. Association between wGRS and LDL-C
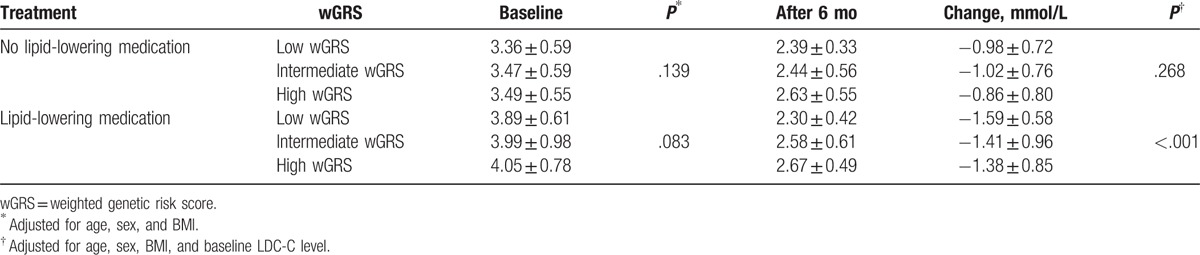
We further investigated the effect of wGRS on LDL-C reduction in patients who did or did not receive statin treatment. Patients receiving statin treatment had higher baseline LDL-C level than those not using lipid-lowering medication (P < .001). No difference in baseline LDL-C level was observed between wGRS groups. After 6 months for lipid-lowering treatment, patients with low wGRS had a 0.18 and 0.21 mmol/L LDL-C level than those with intermediate and high GRS, respectively (Table 7). The overall downward trend in LDL-C in patients with high and intermediate wGRS was significantly lower than in those with low wGRS (P < .001). For patients who did not receiving statin treatment, there was no difference in LDL-C reduction between wGRS groups (P = .268).
Table 7.
The effect of GRS on the changes in LDL-C levels after 6 months.
4. Discussion
Large-scale GWASs have identified multiple common variants that are individually related to the risk of CAD. In the present study, we found that 3 CAD-risk related SNPs (rs579459, rs4420638, and rs2107595) were also associated with the risk of MACE in patients with established CAD. Previous studies have demonstrated that both rs579459 and rs2107595 are not only associated with the risk of CAD, but also with the risk of stroke.[7,24] Furthermore, rs2107595 confers susceptibility to acute coronary syndrome in the Chinese Han population. These results indicate that rs579459 and rs2107595 increase long-term risk of MACE, especially stroke and MI, in CAD patients even receiving statin treatment. The rs4420638 influences the lipid levels and is associated with statin therapy response,[25–28] which may affect the prognosis of CAD patients treated with statin. When combined into a 3-variant risk score, higher wGRS is associated with LDL-C level and MACE, and the later only exists in patients using lipid-lowering medication. The desired LDL-C target level for patients with higher wGRS is more difficultly reached compared with those with low wGRS when patients received statin treatment. These results indicate that CAD patients with low wGRS will benefit from lipid-lowering medication, whereas patients with intermediate and high wGRS may need additional treatment to reduce the risk of MACE.
Many studies have been reported or is currently underway to develop risk prediction models to improve our ability to accurately estimate the risk and prognosis of diseases.[11–16,29] This will help identify high-risk individuals and make precision medicine for disease care and prevention. Given higher incidence and mortality of cardiovascular disease, many risk prediction models about cardiovascular disease have been developed and most models focus on traditional factors such as clinical, biochemical, and imaging parameters,[2,11,15] but few models focused on the clinical outcome of cardiovascular disease. It is important that these prognostic models were constructed based on the European population, whereas whether these predictors can be directly applied in the Chinese population? On the other hand, wGRS derived from inherited genetic variants reflect individual's lifetime risk while traditional factors only reflect risk at a single time point. Genetic variants in combination with traditional factors may improve their capacity to predict individual risk.
Traditional factors for cardiovascular risk stratification have been potentially used to tailor therapy for the individual. The findings of the present study suggest that genetic variants may also play such a role. A meta-analysis has demonstrated that use of statin in both primary and secondary prevention can remarkably reduce the relative risk of cardiovascular events.[30] Although some patients do not currently receive statin therapy according to clinical practice guidelines, they may still be at increased risk of events. Previous studies have estimated genetics and MACE for patients with established CAD in the setting of statin therapy.[31–33] In the present study, we used a multilocus wGRS compromised of CAD-risk SNPs to evaluate the score for prognostic stratification of patients with established CAD. The present analysis showed that for the patients using lipid-lowering medication, cases with low wGRS benefited from statin therapy, while others were still at high risk of events. In addition, there was no association between wGRS and MACE in patients not using lipid-lowering medication and without hypertension. The main reason was that these patients might have relatively lower risk, but also became more active and willing to lowering cholesterol by eating habits and exercise means. Similarly, the effect of wGRS on MACE was only found in patients with hypertension, not present in those with normal nor normal blood pressure. Therefore, lipid-lowering and/or hypertension treatment is not enough to reduce the risk of MACE for CAD patients with high risk and wGRS.
There are several limitations to these analyses. First, although all selected SNPs were identified from GWAS for this study, these studies that underpin our study were mostly based on Caucasian populations. It is possible that other genetic variants in these regions or novel regions may play more important roles in Chinese population. Second, there was lack of the information of dietary fat intake and physical activity of patients, which deeply influence the clinical outcomes of CAD patients. Third, the sample size is relatively small. Fourth, 446 patients were excluded from the analyses due to incomplete clinical and follow-up data and therefore selection bias may impact the findings and conclusions. Further well characterized large-scale studies are warranted to validate our findings.
5. Conclusions
In conclusion, our findings indicate that a wGRS comprised with 3 SNPs was related to the risk of MACE in CAD patients, especially those receiving statin treatment. For patients with higher wGRS who are receiving statin therapy according to clinical practice guidelines, lipid-lowering medication is not enough to reduce the risk of MACE.
Acknowledgment
We thank Dr. Haihui Sheng (National Engineering Center for Biochip at Shanghai) for data analysis and reviewing the manuscript.
Supplementary Material
Footnotes
Abbreviations: ApoA1 = apolipoprotein A1, ApoB = apolipoprotein B, BMI = body mass index, CAD = coronary artery disease, CI = confidence interval, DBP = diastolic blood pressure, DM = diabetes mellitus, GRS = genetic risk score, GWAS = genome-wide association study, HDL-C = high density lipoprotein cholesterol, HR = hazard ratio, HT = hypertension, LDLC = low-density lipoprotein cholesterol, Lp(a) = lipoprotein(a), MACE = major adverse cardiovascular event, MI = myocardial infarction, SBP = systolic blood pressure, SNP = single nucleotide polymorphism, TC = total cholesterol, TG = triglycerides, wGRS = weighted genetic risk score.
Funding: This work was supported by the Fund from Changning Committee of Science and Technology, Shanghai, China (grant no. cnkw2014z06).
CZ and PZ contributed equally to this work.
Authors’ contributions: Conception and design: CZ and LJ; provision of study materials or patients: CZ, PZ, and QS; collection and assembly of data: CZ, PZ, and QS; data analysis and interpretation: CZ, PZ, and LJ; manuscript writing: all authors; final approval of manuscript: all authors.
The authors have no conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
References
- [1].Kassebaum NJ, Bertozzi-Villa A, Coggeshall MS, et al. Global, regional, and national levels and causes of maternal mortality during 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 2014;384:980–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Antiochos P, Marques-Vidal P, McDaid A, et al. Association between parental history and genetic risk scores for coronary heart disease prediction: the population-based CoLaus study. Atherosclerosis 2016;244:59–65. [DOI] [PubMed] [Google Scholar]
- [3].Libby P, Ridker PM, Hansson GK. Progress and challenges in translating the biology of atherosclerosis. Nature 2011;473:317–25. [DOI] [PubMed] [Google Scholar]
- [4].Hu Q, Liu Q, Wang S, et al. NPR-C gene polymorphism is associated with increased susceptibility to coronary artery disease in Chinese Han population: a multicenter study. Oncotarget 2016;7:33662–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Coronary Artery Disease (C4D) Genetics Consortium. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet 2011;43:339–44. [DOI] [PubMed] [Google Scholar]
- [6].Hartiala JA, Tang WH, Wang Z, et al. Genome-wide association study and targeted metabolomics identifies sex-specific association of CPS1 with coronary artery disease. Nat Commun 2016;7:10558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Nikpay M, Goel A, Won HH, et al. A comprehensive 1,000 genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet 2015;47:1121–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kathiresan S, Voight BF, et al. Myocardial Infarction Genetics Consortium. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet 2009;41:334–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Samani NJ, Erdmann J, Hall AS, et al. Genomewide association analysis of coronary artery disease. N Engl J Med 2007;357:443–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Manolio TA. Bringing genome-wide association findings into clinical use. Nat Rev Genet 2013;14:549–58. [DOI] [PubMed] [Google Scholar]
- [11].Elosua R, Lluis-Ganella C, Subirana I, et al. cardiovascular risk factors and ischemic heart disease: is the confluence of risk factors greater than the parts? A genetic approach. Circ Cardiovasc Genet 2016;9:279–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Curtit E, Pivot X, Henriques J, et al. Assessment of the prognostic role of a 94-single nucleotide polymorphisms risk score in early breast cancer in the SIGNAL/PHARE prospective cohort: no correlation with clinico-pathological characteristics and outcomes. Breast Cancer Res 2017;19:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Harrison TM, Mahmood Z, Lau EP, et al. An Alzheimer's disease genetic risk score predicts longitudinal thinning of hippocampal complex subregions in healthy older adults. eNeuro 2016;3: e0098-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Go MJ, Lee Y, Park S, et al. Genetic-risk assessment of GWAS-derived susceptibility loci for type 2 diabetes in a 10 year follow-up of a population-based cohort study. J Hum Genet 2016;61:1009–12. [DOI] [PubMed] [Google Scholar]
- [15].Kullo IJ, Jouni H, Austin EE, et al. Incorporating a genetic risk score into coronary heart disease risk estimates: effect on low-density lipoprotein cholesterol levels (the MI-GENES Clinical Trial). Circulation 2016;133:1181–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hivert MF, Christophi CA, Franks PW, et al. Lifestyle and metformin ameliorate insulin sensitivity independently of the genetic burden of established insulin resistance variants in diabetes prevention program participants. Diabetes 2016;65:520–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Natarajan P, Young R, Stitziel NO, et al. Polygenic risk score identifies subgroup with higher burden of atherosclerosis and greater relative benefit from statin therapy in the primary prevention setting. Circulation 2017;135:2091–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lu X, Wang L, Chen S, et al. Genome-wide association study in Han Chinese identifies four new susceptibility loci for coronary artery disease. Nat Genet 2012;44:890–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Schunkert H, Konig IR, Kathiresan S, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet 2011;43:333–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Vargas-Alarcon G, Posadas-Romero C, Villarreal-Molina T, et al. Single nucleotide polymorphisms within LIPA (Lysosomal Acid Lipase A) gene are associated with susceptibility to premature coronary artery disease. a replication in the genetic of atherosclerotic disease (GEA) Mexican study. PLoS ONE 2013;8:e74703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Reilly MP, Li M, He J, et al. Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: two genome-wide association studies. Lancet 2011;377:383–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Elliott P, Chambers JC, Zhang W, et al. Genetic Loci associated with C-reactive protein levels and risk of coronary heart disease. JAMA 2009;302:37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lin X, Song K, Lim N, et al. Risk prediction of prevalent diabetes in a Swiss population using a weighted genetic score—the CoLaus study. Diabetologia 2009;52:600–8. [DOI] [PubMed] [Google Scholar]
- [24].Zhou X, Guan T, Li S, et al. The association between HDAC9 gene polymorphisms and stroke risk in the Chinese population: a meta-analysis. Sci Rep 2017;7:41538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Deshmukh HA, Colhoun HM, Johnson T, et al. Genome-wide association study of genetic determinants of LDL-c response to atorvastatin therapy: importance of Lp(a). J Lipid Res 2012;53:1000–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Varga TV, Sonestedt E, Shungin D, et al. Genetic determinants of long-term changes in blood lipid concentrations: 10-year follow-up of the GLACIER study. PLoS Genet 2014;10:e1004388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Liu Y, Zhou D, Zhang Z, et al. Effects of genetic variants on lipid parameters and dyslipidemia in a Chinese population. J Lipid Res 2011;52:354–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hu M, Lui SS, Mak VW, et al. Pharmacogenetic analysis of lipid responses to rosuvastatin in Chinese patients. Pharmacogenet Genomics 2010;20:634–7. [DOI] [PubMed] [Google Scholar]
- [29].Mega JL, Stitziel NO, Smith JG, et al. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials. Lancet 2015;385:2264–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Baigent C, Blackwell L, et al. Cholesterol Treatment Trialists’ (CTT) Collaboration. Efficacy and safety of more intensive lowering of LDL cholesterol: a meta-analysis of data from 170,000 participants in 26 randomised trials. Lancet 2010;376:1670–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Iakoubova OA, Sabatine MS, Rowland CM, et al. Polymorphism in KIF6 gene and benefit from statins after acute coronary syndromes: results from the PROVE IT-TIMI 22 study. J Am Coll Cardiol 2008;51:449–55. [DOI] [PubMed] [Google Scholar]
- [32].Chiodini BD, Franzosi MG, Barlera S, et al. Apolipoprotein E polymorphisms influence effect of pravastatin on survival after myocardial infarction in a Mediterranean population: the GISSI-Prevenzione study. Eur Heart J 2007;28:1977–83. [DOI] [PubMed] [Google Scholar]
- [33].van de Woestijne AP, van der Graaf Y, de Bakker PI, et al. LDL-c-linked SNPs are associated with LDL-c and myocardial infarction despite lipid-lowering therapy in patients with established vascular disease. Eur J Clin Invest 2014;44:184–91. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


